Abstract
Most population isolates examined to date were founded from a single ancestral population. Consequently, there is limited knowledge about the demographic history of admixed population isolates. Here we investigate genomic diversity of recently admixed population isolates from Costa Rica and Colombia and compare their diversity to a benchmark population isolate, the Finnish. These Latin American isolates originated during the 16th century from admixture between a few hundred European males and Amerindian females, with a limited contribution from African founders. We examine whole-genome sequence data from 449 individuals, ascertained as families to build mutigenerational pedigrees, with a mean sequencing depth of coverage of approximately 36×. We find that Latin American isolates have increased genetic diversity relative to the Finnish. However, there is an increase in the amount of identity by descent (IBD) segments in the Latin American isolates relative to the Finnish. The increase in IBD segments is likely a consequence of a very recent and severe population bottleneck during the founding of the admixed population isolates. Furthermore, the proportion of the genome that falls within a long run of homozygosity (ROH) in Costa Rican and Colombian individuals is significantly greater than that in the Finnish, suggesting more recent consanguinity in the Latin American isolates relative to that seen in the Finnish. Lastly, we find that recent consanguinity increased the number of deleterious variants found in the homozygous state, which is relevant if deleterious variants are recessive. Our study suggests that there is no single genetic signature of a population isolate.
Keywords: population isolate, population genetics, identity by descent, linkage disequilibrium, pedigree, deleterious mutations
Introduction
The use of population isolates to map Mendelian and complex diseases has been a key feature of medical genomics. In addition to experiencing the bottleneck involved with the migration out of Africa, some populations underwent subsequent bottlenecks and remained in relative seclusion afterward. These populations formed present-day isolates.1 The genomes of population isolates are thought to exhibit several hallmark features of genetic variation. Due to bottlenecks associated with their founding, it is thought that isolates should carry lower levels of genetic diversity and lower haplotype diversity than closely related non-isolated populations. Drift experienced by isolates is magnified by small population size, which generates more linkage disequilibrium (LD) than in non-isolated populations. In addition to increased LD, individuals from isolated populations tend to share more regions of the genome identical by descent (IBD) due to small population size. Further, due to the isolation after founding and recent mating practices, isolates may have larger regions of the genome found in runs of homozygosity (ROHs) due to recent inbreeding. Lastly, bottlenecks and inbreeding should impact patterns of deleterious variation.2, 3, 4 Consequently, one would predict that individuals from isolates will have fewer segregating sites, and the remaining deleterious variants will be segregating at a higher frequency.5 Indeed, genomic studies over the last decade have documented several of these signatures.6, 7, 8 However, it is known that not all isolates share the same demographic history. Therefore, it is essential that we understand how the factors shaping genetic variation in a population are influenced by the unique demographic history of the population.
One archetypal human population isolate that has been extensively studied is the Finnish.7, 9, 10, 11 Finland was populated through two separate major migrations. Briefly, a small number of founders, relative isolation, serial bottlenecks, and recent expansion in Finland has allowed drift to play a large role in shaping the gene pool of this population.11 The aforementioned demographic history of Finland has led to an increase in the prevalence of rare heritable Mendelian diseases, which has made this population particularly fruitful for identifying disease-associated variants.10, 12 Most of the studies in Finland employed LD mapping in affected families and well-curated genealogical records to identify causal and candidate variants.10 More recently, it has been possible to apply population-based linkage analyses to identify disease-associated variants as an alternative to genome-wide association studies (GWASs)13 due to the availability of whole-genome sequence data in conjunction with extensive electronic health records.
A number of studies have shown that power to detect causal variants can be improved by studying population isolates other than the Finnish.8, 14, 15, 16 For example, the Greenlandic Inuit experienced an extreme bottleneck which caused a depletion of rare variants and segregating sites in their genome.16 The remaining segregating variants are maintained at higher allele frequencies and a larger proportion of these SNPs are deleterious when compared to non-isolated populations. Another study of South Asian populations showed similar results. Specifically, South Asian populations have experienced more severe founder effects than the Finnish,15 thus creating an excess of rare alleles associated with recessive disease. A study of European population isolates compared the isolates with the closest non-isolated population from similar geographic regions8 and found that the total number of segregating sites was depleted across all isolates relative to the comparison non-isolate. Of the sites that were segregating in isolates, between ∼30,000 and 122,000 sites existed at an appreciable frequency (minor allele frequency [MAF] > 5.6%), while remaining rare (MAF < 1.4%) in all the non-isolate population samples.8 The authors surmised that these common and low-frequency variants could be useful in GWASs for novel associations, as they included SNPs that had been previously associated with cardio-metabolic traits.8, 17
While there have been many studies of genetic variation in population isolates, the studies described above have focused on populations where the founders all came from the same ancestral population. The founders of Latin American population isolates have come from distinct continental populations. We sampled individuals from mountainous regions of Costa Rica and Colombia where geographic barriers resulted in populations remaining isolated since their founding in the 16th and 17th centuries, until the mid-20th century.18 Both groups share a similar demographic history, having originated primarily from admixture between a few hundred European males and Amerindian females, with a limited contribution from African founders. After the founding event, both populations experienced a subsequent bottleneck and then a recent expansion, within the last 300 years, wherein the expansion increased the population size more than 1,000-fold since the initial founding event.18 The effect that admixture has had on overall patterns of genetic variation in isolates remains elusive, and it remains unclear whether these populations share the typical genomic signatures seen in population isolates. While the small founding population size could reduce diversity, because the Costa Rican and Colombian isolates were founded from multiple diverse populations, they could potentially have increased in diversity relative to other population isolates. Lastly, the impact of admixture on deleterious variation also remains unclear.
To better understand patterns of genetic variation in admixed isolated populations, we compared the Colombian and Costa Rican population isolates to a benchmark isolate, the Finnish, as well as other 1000 Genomes Project populations.19 We observe that relative to the Finnish, Latin American isolates have increased genetic diversity but an excess of IBD segments. Moreover, we detect an increase in the proportion of an individual’s genome that falls within a long ROH in Latin American isolates relative to all other sampled populations and an enrichment of deleterious variation within these long ROHs. Demographic simulations and analysis of extended pedigrees indicate that the enrichment of long ROHs is primarily a consequence of recent inbreeding in Latin American isolates. Next, we examine the relationship between the proportion of European, Native American, and African ancestry and the amount of the genome within an ROH, as well as the relationship to an individual’s pedigree inbreeding coefficient. Further, we examine demography across both recent and ancient timescales in these isolates. Our work sheds light on how the distinct demographic histories of population isolates affect both genetic diversity and the distribution of deleterious variation across the genome.
Subjects and Methods
Pedigree Data for Costa Rican and Colombian Individuals
Our study included 10 Costa Rican (CR) and 12 Colombian (CO) multi-generational pedigrees ascertained to include individuals affected by bipolar disorder 1. The sampled families are clumped geographically to some degree, and it is worth noting that the Central Valley of Costa Rica and Antioquia are population isolates but each population contains several million people. In Costa Rica there is only one psychiatric hospital, and the Antioquia Department of Colombia has few hospitals, so most case subjects were originally identified in the largest hospital in a city of more than 3 million people. More extensive details about the curation of pedigree data and clinical assessments of diagnosis can be found in Fears et al.20
Identifying Unrelated Individuals
We defined unrelated individuals as those who are at most third-degree relatives. We chose this threshold of relatedness because the families from CR and CO are known to be cryptically related. We used KING21 to identify 30 unrelated individuals from CR and CO. 24 of the 30 unrelated individuals in CO are founders in the pedigree and 15 of the 30 unrelated individuals in CR are founders, and each family sampled is represented by at least one individual, but some families had as many as seven individuals. The algorithm implemented in KING estimates familial relationships by modeling the genetic distance between a pair of individuals as a function of allele frequency and kinship coefficient, assuming that SNPs are in Hardy-Weinberg equilibrium.
Further, we also used PC-AiR22 and PC-Relate23 to estimate relatedness as these two methods are robust to population structure, cryptic relatedness, and admixture. We found that 28 of the 30 CO unrelated individuals and 26 of the 30 CR unrelated individuals were contained in the list of unrelated individuals from PC-AiR.22 Complete overlap was not expected because we retained third-degree relatives when using KING to allow for cryptic relatedness of families sampled from Costa Rica and Colombia due to their demographic history.
Lastly, we used KING to identify 30 unrelated individuals from the following 1000 Genomes Project19 populations: Yoruba (YRI), CEPH-European (CEU), Finnish (FIN), Colombian (CLM), Peruvian (PEL), Puerto Rican (PUR), and Mexican from Los Angeles (MXL). We used these 30 unrelated individuals per population for all analyses unless otherwise stated (Figure S1).
Genotype Data Processing
We generated a joint variant call file (VCF) containing single-nucleotide polymorphisms (SNPs) from two separate datasets. The first dataset contained 210 whole-genome sequences sampled from the aforementioned 1000 Genomes Project populations.19 The second dataset contained 449 whole-genome sequences from Costa Rican and Colombian individuals. Variants in the second dataset were called following the GATK best practices pipeline24 with the HaplotypeCaller of GATK. All multi-allelic SNVs and variants that failed Variant Quality Score Recalibration were removed. Genotypes with genotype quality score ≤ 20 were set to missing. Further quality control on variants was performed using a logistic regression model that was trained to predict the probability of each variant having good or poor sequencing quality. Individuals with poor sequencing quality and possible sample mix-ups were removed, and all sequenced individuals had high genotype concordance rate between whole-genome sequences and genotypes from microarray data. All sequenced individuals had consistency between the reported sex and sex determined from X chromosome as well as between empirical estimates of kinship and theoretical estimates. More information on sequencing and quality control procedures is discussed in Sul et al.25
We used the following protocol to merge these two datasets. First, we used guidelines from the 1000 Genomes Project strict mask to filter the Costa Rican and Colombian VCFs as well as the 1000 Genomes Project VCFs. Then, we used GATK to remove sites from both sets of VCFs that were not bi-allelic SNPs or monomorphic. Next, we merged the 1000 Genomes Project VCFs with the Costa Rican and Colombian VCFs into a single joint-VCF for each chromosome. We used only autosomes for our analyses. Lastly, we filtered the merged joint-VCF to only contain sites that were present in at least 90% of individuals. There were a total of 57,597,196 SNPs and 1,891,453,144 monomorphic sites in the final dataset. We ensured that the merged datasets were comparable by examining the number of derived putatively neutral alleles across the 30 unrelated individuals in all sampled populations and we found few differences between populations, which is consistent with theory5 (Figure S2).
Calculating Genetic Diversity
We computed two measures of genetic diversity from sites called across all 30 unrelated individuals from each population: pi (π) and Watterson’s theta (θw). The average number of pairwise differences per site (π) was calculated across the genome as:
where n is the total number of chromosomes sampled, p is the frequency of a given allele, and L is the length in base pairs of the sampled region. θw was computed by counting the number of segregating sites and dividing by Watterson’s constant, or the n-1 harmonic number.26
Site Frequency Spectrum (SFS)
Site frequency spectra were generated using the 30 unrelated individuals from each population. SNPs with missing data were removed from these analyses. There was a total of 16 SNPs out of the 57,597,196 SNPs that were removed due to missing data.
Linkage Disequilibrium Decay
We calculated LD between pairs of SNPs for all unrelated individuals. First, we applied a filter to remove SNPs that were not at a frequency of at least 10% across all populations. Next, pairwise r2 values were calculated using VCFTools.27 SNP pairs were then binned according to physical distance (bp) between each other and r2 was averaged within each bin.
Identifying Identity by Descent Segments
To detect regions of the genome that have shared IBD segments between pairs of individuals, we first removed singleton SNPs in each population since singletons are not informative about IBD. Then, we called IBD segments using IBDSeq.28 IBDSeq is a likelihood-based method that is designed to detect IBD segments in unphased sequence data. We chose to use IBDSeq because other methods that require computational phasing could be biased when applied to Latin American population isolates, as they do not have a publicly available reference population to aid in phasing. We compared IBDSeq to two well-known methods, Beagle29 and GERMLINE,30 to determine whether it was feasible to use IBDSeq on an admixed population (Figure S3). Data for Beagle and GERMLINE were phased beforehand with SHAPEIT31 (see Web Resources) using the 1000 Genomes as the reference panel. Beagle produced the shortest IBD segments while GERMLINE produced the longest IBD segments. IBDSeq produced segments with a length distribution similar to what we observed in Beagle, though the average segment length was slightly larger, which we expected given that IBDSeq was created to call longer segments that would have previously been broken up when using Beagle for phasing. We used the default parameters for IBDSeq.
Next, we filtered the pooled IBD segments to remove artifacts. First, we calculated the physical distance spanned by each IBD segment. Then, we totaled the number of SNPs that fell within each segment. We observed an appreciable number of IBD segments that were extremely long but sparsely covered by SNPs (Figure S4). IBD segments were removed if the proportion of the IBD segment covered by SNPs was not within one standard deviation (0.0043) of the mean proportion covered (0.0221) across all IBD segments (Figure S4). Strong deviations from the mean could indicate that the IBD segment spans a region of the genome with low mappability where we are only calling the SNPs at the outer ends of the segment. Therefore, the true segment length might be much shorter than what is being calculated by IBDSeq. Lastly, we converted from physical distance to genetic distance using the deCODE genetic map.32 A file that contains all the IBD segments (unfiltered) alongside code used to filter can be found on GitHub (see Web Resources).
Enrichment Analyses of IBD Segments
To determine whether certain populations contain more IBD segments than others, we followed the IBD score procedure outlined by Nakatsuka and collegues.15 A population’s IBD score was calculated by computing the total length of all IBD segments between 3 and 20 cM. The score difference is the difference between the query population’s IBD score and the Finnish IBD score. The score ratio is the ratio of each population’s IBD score relative to the Finnish IBD score. The significance of enrichment relative to the Finnish was evaluated using a permutation test for each population, where IBD segment length was held fixed and labels of the two populations were permuted. We recalculated the score on a total of 10,000 permutations to generate a null-distribution of scores for each isolate. The code can be found on GitHub (see Web Resources).
Estimating Effective Population Size
We used the output files from IBDSeq to estimate the recent effective population size through time from the 30 unrelated individuals from each sampled population. We estimated effective population size by using the default settings in IBDNe.33 We set the minimal IBD segment length equal to 2 cM since that is the suggested setting when using sequence data. We assumed a generation time of 30 years.
Identifying Runs of Homozygosity
Runs of homozygosity were identified for each individual using VCFTools, which implements the procedure from Auton et al.34 Next, we examined the number of callable sites that lie within each ROH. We found that there was a bi-modal distribution of coverage for ROHs, where some ROHs appeared to contain almost no callable sites, while others had much higher coverage. We only kept ROHs that were at least 2 Mb in length, which we called long runs of homozygosity, and were at least 60% covered by callable sites (Figure S5). A file that contains the final ROHs can be found on GitHub (see Web Resources).
Calculating Inbreeding Coefficients
SNP-based inbreeding coefficients were calculated using VCFTools.27 VCFTools calculates the inbreeding coefficient F per individual using the equation F = , where O is the observed number of homozygotes, E is the expected number of homozygotes (given population allele frequency), and N is the total number of genotyped loci.
Pedigree-based inbreeding coefficients were computed using the R package kinship2.35
Demographic Simulations
In order to investigate how aspects of the population history affect current day genetic diversity in Latin American isolated populations, we simulated genetic variation data using the forward simulation software SLiM 2.1.36 We simulated a sequence length of 10 Mb under uniform recombination rate of 1 × 10−8 crossing-over events per chromosome per base position per generation and under a mutation rate of 1.5 × 10−8 mutations per chromosome per base position per generation. Every simulation contained intergenic, intronic, and exonic regions, but only nonsynonymous new mutations experienced natural selection in accordance with the distribution of selection coefficients estimated in Kim et al.37 Within coding sequences, we set nonsynonymous and synonymous mutations to occur at a ratio of 2.31:1.37, 38 The chromosomal structure of each simulation was randomly generated, following the specification in the SLiM manual (7.3), which is modeled after the distribution of intron and exon lengths in Deutsch and Long.39
We assumed an effective population size in the ancestral African population of 10,000 individuals, and a reduction in size to 2,000 individuals, starting 50,000 years ago (assuming 30 years per generation), reflecting the colonization of the European, Asian, and American continents. The population then recovers to a size of 10,000 individuals 5,000 years ago. The colonization bottleneck is assumed to occur 500 years ago by an admixture event with a European population (70% admixture proportion) and is followed by an immediate reduction in population size to 1,000 individuals. The recent expansion in population size is modeled by an increase in population size to 10,000 individuals 200 years ago. We simulated data with recent inbreeding and without recent inbreeding. In the former case, inbreeding started at the time of the European colonization 500 years ago and continues until the present. Inbreeding is implemented with the “mateChoice” function in SLiM. Here, 50% of the time, mating occurs randomly. However, in the remaining case subjects, mating occurs between close relatives with a relatedness coefficient bigger than 0.25. This produces levels of consanguinity similar to those seen empirically as measured by F (see Results). We also tested whether such high observed values of F can be explained by random mating during an extreme bottleneck with a bottleneck to 100 individuals, and a bottleneck to 64 individuals, during colonization 500 to 200 years ago. To increase the speed of the simulations, we reduced mutation rate by a factor of 5, and verified the results of the simulations with theoretical predictions of the relationship between F and population size over time.40 Finally, we sampled a total of 60 random individuals and calculated summary statistics on the sample data. The simulation script can be found on GitHub (see Web Resources).
Annotation of Variants
The ancestral allele was determined using the 6-primate EPO alignment (see Web Resources) and we restricted to only those sites called with the highest confidence. After filtering, 54,049,081 SNPs remained. Subsequently, exonic SNPs were annotated using the SeattleSeq Annotation website (see Web Resources). A total of 693,301 SNPs were annotated as either nonsynonymous or synonymous. We further classified these sites as either putatively neutral or deleterious using Genomic Evolutionary Rate Profiling (GERP) scores.41 GERP scores are generated using a multiple-sequence alignment of the hg19 reference to 33 other mammalian species. When calculating the rejected substitutions (RS) score, which we will refer to as the GERP score, the hg19 reference genome is removed to eliminate confounding due to deleterious derived alleles. A GERP score less than 2 was considered as putatively neutral and a GERP score greater than 4 was considered as putatively deleterious for the 404,302 classified SNPs.
Counting Deleterious Variants
We used three different statistics to count the number of deleterious mutations per individual. First, we tabulated the number of deleterious variants (the number of heterozygous plus the number homozygous derived genotypes). Second, we counted the total number of derived deleterious alleles (the number of heterozygous genotypes plus twice the number of homozygous derived genotypes). Third, we computed the total number of derived deleterious homozygous genotypes. A table that contains the counts of all deleterious and neutral variants can be found on GitHub (see Web Resources).
Testing for an Enrichment of Deleterious Variation in ROHs
We were interested in whether there is an enrichment of nonsynonymous mutations in ROHs over non-ROH regions for the three different ways of counting deleterious variants outlined above. To account for differences in neutral variation, we standardized by synonymous variation, which is assumed to be neutral. Then, we calculated the ratio of nonsynonymous over synonymous variation in ROH regions divided by the ratio of nonsynonymous over synonymous variation outside of ROHs. We computed significance using a permutation test, where the position of each SNP and its annotation as synonymous versus nonsynonymous was fixed and the positions of the vector of ROH annotations were randomly placed throughout the genome. Thus, the frequency distribution of synonymous and nonsynonymous SNPs, as well as the total amount of ROH and non-ROH annotations, is kept constant when compared to the unpermuted data. We recalculated the ratio for a total of 10,000 permutations to form a null-distribution of ratios and then computed significance.
Calculating Ancestry Proportions
We estimated genome-wide ancestry proportions in members of the CR and CO pedigrees using LAMP.42 We generated ancestry estimates for all 838 pedigree members with SNP array genotype data (detailed information on the SNP array data can be found in Pagani et al.43). The ancestral reference populations were the CEU (n = 112) and YRI (n = 113) from HapMap,44, 45 as well as 52 Native American samples from Central or South America. The Native American samples are the Chibchan-speaking subset of those used in Reich et al.,46 selected to originate from geographical regions relevant to CR/CO and to have virtually no European or African admixture (European and African ancestry < 0.00025). The allele frequencies were calculated for each reference population and were used as input files for LAMP alongside the following configuration parameters: offset = 0.2, recombrate = 1e−8, generations = 20, alpha = 0.24,0.72,0.04, ldcutoff = 0.1. Then, we computed global ancestry estimates from the LAMP output file.
Ancestry proportions in Table 1 for 1000 Genomes and Latin American populations were estimated using ADMIXTURE.47 The analysis for the 1000 Genomes populations used 665,105 LD-pruned SNPs, an unsupervised learning model, and the number of source populations was set to K = 3 (Table 1). The analysis for the Latin American isolates used a supervised learning model with K = 3 source populations, composed of the European, African, and Native American populations mentioned above and 57,180 LD-pruned SNPs.
Table 1.
Ancestry Proportions for Each Sampled Population
| Population | Native American | African | European |
|---|---|---|---|
| YRI | 0.00 | 100.00 | 0.00 |
| CEU | 0.00 | 0.11 | 99.89 |
| FIN | 0.78 | 0.16 | 99.06 |
| PEL | 88.24 | 2.20 | 9.56 |
| CLM | 27.95 | 9.10 | 62.95 |
| CO | 20.43 | 6.64 | 72.93 |
| CR | 27.3 | 2.20 | 70.50 |
| MXL | 44.48 | 5.73 | 49.79 |
| PUR | 14.25 | 18.59 | 67.16 |
This table summarizes the average global ancestry percentages for each of the sampled populations found using ADMIXTURE.47 Admixture proportions in CO and CR were estimated using supervised model with reference populations. Admixture proportions in other populations were inferred using an unsupervised model (see Subjects and Methods). Population abbreviations are as in Figure 1.
Accounting for Relatedness
We tested for correlations among several quantities computed for each individual in the Latin American population isolates. Because some of these individuals are closely related, the data points in our linear regression are no longer independent. We used the R-package GenABEL48 to incorporate kinship when performing statistical tests for our correlations. We used the polygenic_hglm() function where the formula input was the equation for our linear model of interest and the kinship.matrix input was a kinship matrix computed from our pedigree computed using kinship2.35 Our input took the following form: kinship.matrix (FPED ∼Length of genome in ROH, kin = kinshipMatrix, data = df). We also computed p values from a genetic relatedness matrix (GRM) created using PC-AiR22 and PC-Relate;23 both sets of p values can be found in Table S1.
Results
Genetic Variation in Population Isolates
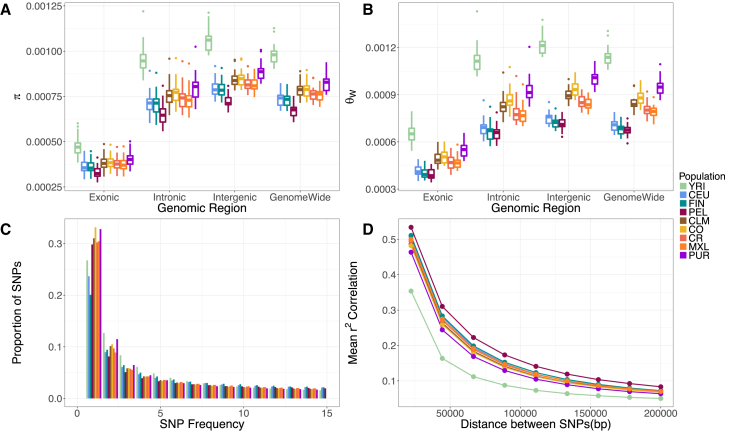
We first compared levels of genetic diversity in a sample of 30 unrelated individuals across the 1000 Genomes populations19 and the CO and CR isolates. We split the genome into several different genomic regions and in each region summarized genetic variation using both the average number of pairwise differences (π) and Watterson’s theta (θw) (Figures 1A and 1B). Overall, we found differences in diversity across the functional categories of sequence studied in all populations, with coding regions exhibiting the lowest diversity and intergenic regions the highest. These patterns are consistent with the role of purifying selection affecting coding diversity.37 However, if we look genome-wide or focus on intronic regions, we see intermediate levels of diversity (Tables S2 and S3). We suspect that these categories are more strongly influenced by linked selection.49, 50, 51
Figure 1.
Patterns of Genetic Variation in the Colombian and Costa Rican Populations Compared to the 1000 Genomes Populations
(A) Diversity measured using the average pairwise differences between sequences, π.
(B) Diversity measured using the number of segregating sites, Watterson’s theta (θW).
(C) The site frequency spectrum (SFS) for each population (truncated at a SNP frequency of 15; full SFS Figure S6).
(D) Average LD (r2) between pairs of SNPs.
All statistics were calculated using 30 unrelated individuals per population (see Subjects and Methods). Boxplots in (A) and (B) show the distribution over 22 autosomes. Abbreviations: YRI; Yoruba 1000 Genomes; CEU, Ceph-European 1000 Genomes; FIN, Finnish 1000 Genomes; PEL, Peruvian 1000 Genomes; CLM, Colombian 1000 Genomes; CO, Colombia; CR, Costa Rica; MXL, Mexican from Los Angeles 1000 Genomes; and PUR, Puerto Rican 1000 Genomes.
As we are interested in the role of demography in shaping genetic diversity, we focused on comparisons of intergenic levels of diversity as those are most likely to be neutrally evolving (Figures 1A and 1B). Overall, the YRI had the highest level of diversity (π ≈ 0.0010; θw ≈ 0.0012) (Tables S2 and S3). The European populations (CEU and FIN) had lower levels of diversity. The CEU and FIN had similar levels of π (approximately 0.0004), despite the FIN being considered an isolated population. However, the FIN had reduced numbers of SNPs as reflected by lower mean values of θw (CEU ≈ 0.00075 and FIN ≈ 0.00072). The CO and CR had levels of diversity comparable to that of several other Latin American populations in the 1000 Genomes Project (CLM and MXL). We found no clear pattern of the population isolates (FIN, CO, CR) having lower diversity than their most similar non-isolated population. Instead, diversity levels tended to be higher across all the sampled Latin American populations (CLM, CO, CR, MXL, and PUR) when compared to the European populations. One exception to this pattern is the PEL population, who had the lowest neutral levels of diversity (π ≈ 0.0007; θw ≈ 0.0007).
Next, we examined the proportional site frequency spectrum (SFS; Figures 1C and S6). Latin American populations had the highest proportion of singletons, as seen previously.52 The CO and CR had similar proportions of singletons when compared to other 1000 Genomes Project Latin American populations. Conversely, the FIN had the lowest proportion of singletons in comparison to all sampled populations. The depletion of singletons relative to common variation supports the presence of a stronger founder effect during the FIN population history.11
We also examined patterns of linkage disequilibrium (LD), since LD is affected by population size and recent bottlenecks.53, 54 Figure 1D shows the mean decay of r2 with physical distance over 2 Mb intervals across the genome in each population. We found that the YRI had the lowest levels of LD for each bin of physical distance, and the PEL formed the upper bound of the LD decay curves. The remaining Latin American populations (PUR, MXL, CLM, CO, CR) clustered together, close to the YRI, while the CEU and FIN are shifted toward higher values, like those seen in the PEL.
The FIN were previously shown to have more extensive haplotype blocks in their genome in comparison to the Latin American isolates.6 In line with these findings, we observed faster LD decay in the Latin American isolates relative to the FIN. When considering pairs of SNPs 150 kb or more apart, rates of LD decay become quite similar across all the sampled populations. Analogous to other diversity statistics, LD in the CO and CR closely resembled those of non-isolated Latin American populations. Once again, we found there is no clear pattern of having lower diversity or more LD that holds across all the population isolates (FIN, CO, CR) when compared to their most similar non-isolated population.
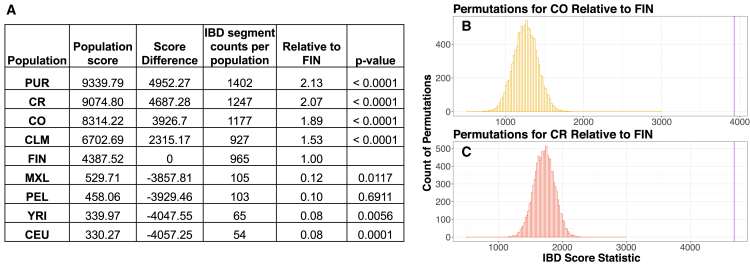
Latin American Isolates Carry More IBD Segments than the Finnish
Next, we used IBD sharing between pairs of individuals to gain insight about more recent demographic events within populations (Figure 2). We quantified the amount of IBD within each population by computing an IBD score. Each population’s IBD score was calculated by totaling the length of IBD segments between 3 cM and 20 cM. We expressed IBD scores for each population as the ratio of the IBD score for a given population relative to the IBD score in the FIN (Figure 2A). We also tabulated the total count of IBD segments for each population. The CEU showed the lowest number of both called IBD segments and the lowest IBD score relative to the FIN (p = 0.0001). Latin American populations formed the upper bounds of both total IBD segments called and IBD enrichment scores (Figure 2A). The PUR had the largest number of IBD segments (1,402) and had a 2.1-fold increase in IBD score relative to the FIN (p < 1 × 10−4). The CO and CR isolates had a 1.8-fold and 2-fold increase in their IBD scores relative to the FIN (p < 1 × 10−4), as well as carrying more IBD segments than the FIN (Figures 2B and 2C). However, there were some Latin American populations that exhibited depletions in both IBD segments and IBD scores relative to the FIN. The MXL and PEL have the lowest number of IBD segments for the Latin American populations. Previous work has shown that a larger effective population size in admixed populations likely drove the depletion of IBD segments in these two Latin American populations.55
Figure 2.
Latin American Population Isolates (CR and CO) Have Significantly More Identity by Descent (IBD) Segments Relative to the Finnish (FIN)
IBDSeq was used to generate IBD segments for the 30 unrelated individuals in each population.
(A) Population score was calculated by summing all IBD segments between 3 cM and 20 cM for each population. Score difference is the population score minus the FIN population score. IBD enrichment for each population score is reported as relative to the FIN (i.e., FIN score is 1.0).
(B and C) Histogram of 10,000 permutation tests of Colombia (p < 1.0 × 10−04) and Costa Rica (p < 1.0 × 10−04) population scores versus Finnish score. The observed score for each population is demarcated by the purple line.
Population abbreviations are as in Figure 1.
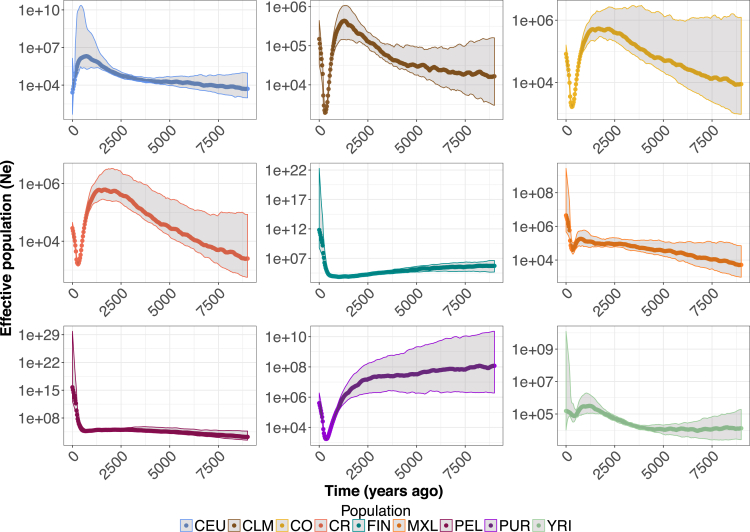
Inferring the Demographic History of Latin American Isolates
We next leveraged the patterns of IBD described above to estimate the effective population size (Ne) through time using IBDNe33 on the 30 unrelated individuals from each population (Figure 3). The use of only 30 unrelated individuals caused limitations for accurate estimation of Ne (see Discussion), but the general population size trajectory is likely to be robust to the number of individuals used. First, we found that recent demography differs vastly between the European populations (FIN and CEU). In general, CEU experienced population expansions over much of their demographic history. It was only in the most recent generations that they experienced a decrease in Ne. The FIN, on the other hand, have experienced a long population decline since their founding, approximately 4,000 years ago, followed by a recent population expansion.
Figure 3.
Recent Effective Population Size Differs across Populations
IBDNe33 (see Subjects and Methods) was used to infer effective population size (Ne) over the last 9,000 years for each population. Shaded regions denote 95% confidence intervals. Note the FIN shows a long slow decline followed by recent growth. The CO and CR show sharp bottlenecks approximately 500 years ago followed by recent growth. While the overall trends in the population size trajectories appear to be robust to the use of smaller sample sizes in IBDNe, current estimates of Ne are likely inaccurate. Population abbreviations are as in Figure 1.
When analyzing the Latin American isolates, we detected a recent bottleneck, approximately 500 years ago (Figure 3). This bottleneck could correspond to the recorded bottleneck that followed the founding of these populations, and it appears to be much shorter and more severe than the bottleneck seen in the FIN. The strength and duration of bottlenecks varied across each of the Latin American populations. For example, we observed a more severe bottleneck in the CR, CO, CLM, and PUR than in PEL or MXL. However, we detected a subsequent period of growth across all populations following the bottleneck. The rate of growth differed across each population, and the PEL appeared to be growing at a much more rapid rate than any of the other Latin American populations.
Exploring Recent Consanguinity
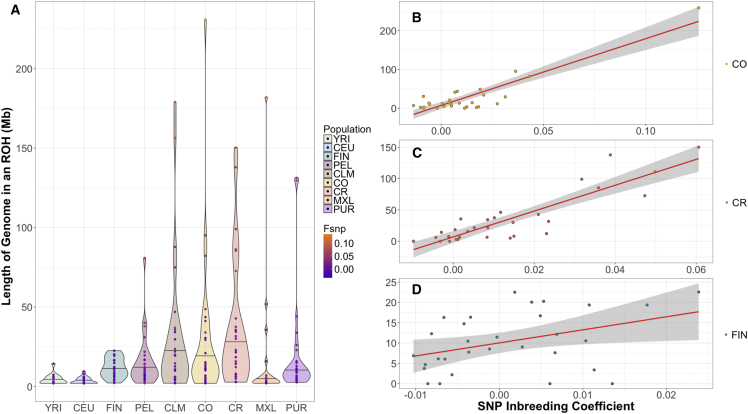
Isolated populations may have experienced recent consanguinity. To test for this, we began by examining SNP-based inbreeding coefficients (FSNP) (Figure S7). YRI individuals had the lowest median inbreeding coefficients (−0.0001) and the CO and CR isolates had the highest median inbreeding coefficients (0.0087 and 0.0086, respectively). Further, the CO and CR also had the highest maximum FSNP values in the entire sample of unrelated individuals from any population (Figure S7). Median levels of FSNP in the CEU (−0.0004) suggested that they are more homozygous than the FIN (−0.0007), which may be a result of how 1000 Genomes samples were selected. The PEL had the largest variance in FSNP across any of the sampled populations.
Next, we examined patterns of long runs (>2 Mb, see Subjects and Methods) of homozygosity, since ROHs have been linked to recent consanguinity.56, 57, 58, 59, 60 The YRI and CEU had the lowest amount of their genome contained within an ROH (Figure 4A). The FIN had higher median (median = 11 Mb and SD = 6.3 Mb) amounts of their genome within an ROH in comparison to the CEU (median = 2.4 Mb and SD = 2.1 Mb). Latin American isolates had the highest median amount of the genome contained within an ROH. Specifically, the CR had the highest median at 21.7 Mb (SD = 40.9 Mb). Further, the Latin American isolates also had the greatest variance in the amount of the genome contained within an ROH. For example, one of the CO individuals had approximately 230 Mb of their genome contained in long ROHs.
Figure 4.
Length of the Genome in a Run of Homozygosity (ROH) Varies across Populations and Correlates with SNP Inbreeding Coefficient
The length of the genome in an ROH was calculated for each unrelated individual (n = 30 per population) by summing the physical distance (Mb) of each ROH > 2 Mb.
(A) The length of the genome in an ROH varies by population. The black line within the violin marks the median. FSNP for each individual was overlaid within the ROH violin plot. A blue hue indicates the lowest FSNP and orange indicates the highest FSNP.
(B) Length of the genome in an ROH is strongly correlated with FSNP in Colombians (R2 = 0.8060, p = 1.1 × 10−11).
(C) Length of the genome in an ROH is strongly correlated with FSNP in Costa Ricans (R2 = 0.7740, p = 9.5 × 10−11).
(D) Length of the genome in an ROH is positively correlated with FSNP in Finnish (R2 = 0.1288, p = 0.03).
Population abbreviations are as in Figure 1.
As expected, we found that the amount of the genome contained in a long ROH strongly correlated with an individual’s FSNP (CO: R2 = 0.8060, p = 1.1 × 10−11; CR: R2 = 0.7740, p = 9.5 × 10−11; FIN: R2 = 0.1288, p = 0.03) (Figures 4B–4D). Indeed, individuals with higher values of FSNP tended to have more of their genome within an ROH. Further, the individual with the highest FSNP (0.133) also had the largest amount of their genome in long ROH (230 Mb).
The total number of ROH segments per individual followed a similar pattern as the total amount of genome within an ROH (Figure S8). For example, in populations with low values of FSNP, ROH segments were not frequent. One YRI individual and three CEU individuals carried an ROH > 4 Mb, whereas more than 50% of CO and CR individuals carried an ROH > 4 Mb. Additionally, the longest ROHs identified (>20 Mb) occurred only in Latin American populations, who have the largest values of FSNP (Figure S8).
Importantly, the FIN individuals had significantly fewer ROH segments than the CO and CR, and most individuals had an FSNP close to 0; while the Latin American isolates had the most ROH in comparison to any other sampled population, as well as the largest values of FSNP (Figure 4).
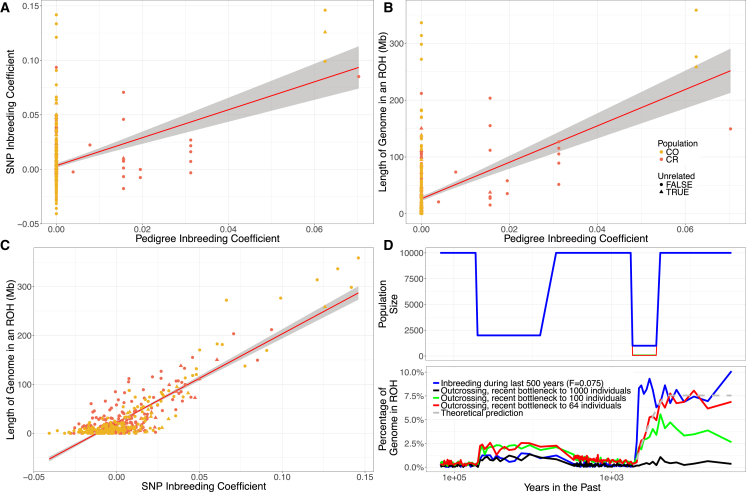
Determining the Mechanisms that Generate Runs of Homozygosity
In principle, ROHs can be generated either by recent consanguinity over the last few generations or by older historical processes, such as bottlenecks.56, 58, 60, 61, 62, 63 Based on both historical data18 and inference from IBDNe analyses, Latin American population isolates show evidence of recent population bottlenecks. Therefore, we used two complementary strategies to test whether recent consanguinity or bottlenecks drove the observed increase in ROHs in the Latin American isolates. First, we used the extensive pedigree data for 449 sequenced individuals to calculate a pedigree inbreeding coefficient (FPED). Most individuals had a FPED of 0 (Figure 5). However, there were several individuals with values of FPED as high as 0.07 in CR and 0.06 in CO. We observed a significant correlation between FSNP and FPED (R2 = 0.1520 and p < 2 × 10−16), even after accounting for the non-independence of individuals based on their kinship (Figure 5A; see Subjects and Methods). These correlations suggest that the recent consanguinity captured within the last few generations in the pedigree was a relevant factor to increase ROHs in the CO and CR populations. However, once we remove the four most influential individuals, the correlation between FSNP and FPED is no longer significant. These four individuals also account for approximately 7.5% of individuals with FPED > 0, so the reduction in sample size could also explain some component of the reduction in signal. FSNP was a substantially better predictor of the amount of an individual’s genome that falls within an ROH (R2 = 0.7540 and p < 2 × 10−16) than FPED (R2 = 0.2180 and p < 2 × 10−16) (Figures 5B and 5C), likely due to the fact that FSNP captured distant background relatedness within the population as well as the realized level of consanguinity, rather than the expected value.64 Further, because the pedigrees were ascertained and analyzed separately, connections between pedigrees were not accounted for in FPED but were likely captured by FSNP.
Figure 5.
Recent Consanguinity Creates ROHs in Costa Rica and Colombia
Triangles represent the individuals that were sampled in the unrelated dataset (n = 30).
(A) FSNP is correlated with the pedigree inbreeding coefficient (FPED; R2 = 0.1520, p < 2 × 10−16) in the full data.
(B) The length of the genome in an ROH is correlated with FPED (R2 = 0.2180, p < 2 × 10−16).
(C) The length of the genome in an ROH is correlated with FSNP (R2 = 0.7540, p < 2 × 10−16).
(D) Forward simulations show that recent consanguinity during the last 500 years was important for the generation of ROHs in the Latin American isolates. Top panel shows the changes in population size used in the simulations. Bottom panel shows how the percent of the simulated genome within an ROH changes over time for different demographic scenarios.
Population abbreviations are as in Figure 1.
As a second approach to determine the mechanism driving the increase in ROHs in the CO and CR populations, we conducted forward in time demographic simulations. We simulated a 10 Mb region under a demographic model that reflected changes in effective population size during the human expansion across the European, Asian, and American continents, as well as the more recent bottleneck during the Spanish colonization about 500 years ago (Figure 5D; see Subjects and Methods). Consanguineous nonrandom mating in the population was modeled to begin 500 years ago, leading to a mean value of FROH of about 0.075. This level of inbreeding matches the level of inbreeding in some of the CO and CR individuals, based on calculations using pedigree data.
Next, we investigated how severe the bottleneck caused by the Spanish colonization would have needed to be to generate such high levels of ROHs, when assuming random mating instead of consanguineous mating. We found that a recent population bottleneck to 1,000 individuals, as suggested by historical data for the Central Valley population of CR,71 is not capable of generating the large amounts of the genome within an ROH (>2 Mb) that we observed for some of the individuals (Figure 5D). We tested several more scenarios with severe bottlenecks where population size decreased to 100 and 64 individuals. A bottleneck to 100 individuals led to an FROH of only 0.003, which is considerably less than that estimated from the empirical data (Figure 5D). When we estimated FROH from simulation with 64 individuals, we observed the predicted value of 0.075 immediately following the bottleneck (i.e., 7.5% of the genome are in an ROH) and the value did not noticeably drop during the last 200 years even with the subsequent expansion of population size (Figure 5D). This matches theoretical predictions where the inbreeding coefficient, F, is related to the inbreeding effective population size (Ne) and number of generations40 (t) according to the formula F = 1 − (1 − 1/(2Ne))t.
Thus, bottlenecks or population structure would need to reduce inbreeding effective population size to approximately 60 individuals for multiple generations to generate ROHs that are comparable to the empirical data. However, we believe this reduction the effective population size, to 64 individuals, is rather unlikely because such a low effective population size is not predicted by our estimates of Ne during the recent bottleneck (Ne > 1,000; see Figure 3), nor by the recent genetic estimates of Ne in the Americas predicted by Browning and colleagues.65 Further, historical data suggest that the lowest census population size for just Native Americans was 300 individuals in CO18 and 1,400 in CR71, which is considerably more than 64 individuals, and does not include the unknown number of European and African American individuals. Since we observe considerable amounts of ROHs even in the larger CR population, we conclude that recent consanguineous nonrandom mating was paramount for generating the long ROH that we observed in the Latin American isolates.
Global Ancestry
We looked at the relationship between intergenic π and proportion of ancestry per population (Figure S9). We saw that populations with the largest proportions of European and Native American ancestry tended to have lower diversity, and as we expected, populations with higher African ancestry had higher diversity (Figure S9).
Since the Latin American isolates originated from an admixture event between Native Americans, Africans, and Europeans, we tested for a correlation between different inbreeding metrics and the proportion of European, African, and Native American ancestry (Figure S10). We used the entire sequenced Costa Rican and Colombian dataset (n = 449) for the local ancestry analyses and accounted for relatedness of individuals in all the following reported p values (see Subjects and Methods). First, we examined the correlation between FPED and global ancestry. We found that European ancestry was positively correlated with FPED (R2 = 0.0204; p value = 0.0052) while Native American ancestry was negatively correlated with FPED (R2 = 0.0126; p value = 0.0245). African ancestry was also negatively correlated with FPED (R2 = 0.0085; p value = 0.0496).
Next, we examined the correlation between FSNP and global ancestry. Similar to what we observed with FPED, European ancestry was positively correlated with FSNP (R2 = 0.1120; p = 4.76 × 10−12), Native American ancestry was negatively correlated with FSNP (R2 = 0.0705; p = 2.79 × 10−07), and African ancestry was negatively correlated with FSNP (R2 = 0.0545; p = 3.49 × 10−08). We expected that the correlation between FSNP and global ancestry would be stronger than FPED and global ancestry, since FSNP captures the realized inbreeding coefficient rather than the expected inbreeding coefficient.
Lastly, we examined whether ancestry was correlated with the amount of the genome within an ROH (Figure S10). The correlation between ancestry and amount of the genome within an ROH followed the same trend as the correlation between ancestry, FPED, and FSNP. Native American ancestry and African ancestry were negatively correlated with the amount of the genome within a long ROH (R2 = 0.1193; p = 9.04 × 10−12 and R2 = 0.0467; p = 2.50 × 10−07, respectively). European ancestry was positively correlated with the amount of an individual’s genome within an ROH (R2 = 0.1500; p = 1.02 × 10−15).
Recent Consanguinity Is Correlated with an Increase of Deleterious Variation
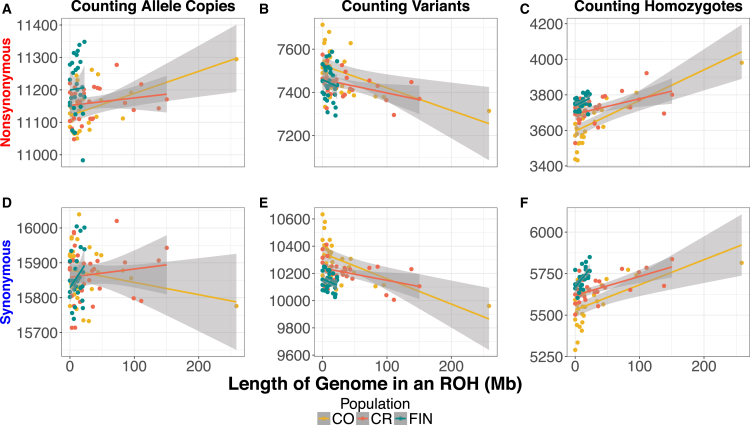
It is well known that demography impacts patterns of deleterious variation in populations.2, 5, 52, 56, 66, 67, 68, 69, 70 Thus, we compared patterns of putatively deleterious variation in the CO and CR to those in the FIN. Variants were classified as putatively deleterious or putatively neutral using GERP scores (see Subjects and Methods). Recall that we consider three ways of counting deleterious variants in the genome of an individual: first, counting the number of heterozygous genotypes plus twice the number of homozygous derived genotypes (i.e., the total number of derived deleterious alleles); second, counting the number of heterozygous and homozygous derived genotypes (counting variants); and third, counting only the number of homozygote derived genotypes (counting homozygotes). The first quantity is most relevant if deleterious alleles are additive, while the third is most relevant if they are recessive. First, we looked at absolute counts of derived deleterious variation across isolates (Figure S11). Then, we used linear regression to test whether there was a relationship between the amount of an individuals’ genome in an ROH and the number of nonsynonymous sites in the genome for each counting method (Figure 6).
Figure 6.
The Relationship between ROHs and Coding Variation in the Colombian, Costa Rican, and Finnish Samples
The count of nonsynonymous and synonymous mutations per individual as a function of the length of the genome in an ROH in the Colombian (CO), Costa Rican (CR), and Finnish (FIN) populations.
(A) Number of nonsynonymous alleles per individual.
(B) Number of nonsynonymous variants per individual.
(C) Number of homozygous nonsynonymous genotypes per individual.
(D) The number of synonymous alleles per individual.
(E) The number of synonymous variants per individual.
(F) The number of homozygous synonymous genotypes per individual.
Population abbreviations are as in Figure 1.
The FIN carried approximately 1% more derived deleterious nonsynonymous alleles per individual than CO and CR (p = 0.0007; p = 0.0013, Wilcoxon rank-sum test). However, there was no significant difference in the number of putatively neutral synonymous derived alleles per individual. These results suggest that the difference seen for putatively deleterious variants is not driven by data artifacts (Figure S11), and the FIN indeed have a slightly higher additive genetic load than the CO or CR. Turning to the number of variants per individual, FIN individuals carried significantly more deleterious nonsynonymous variants than the CR but not the CO (p = 0.0110). However, CO and CR did not differ significantly in the number of deleterious variants carried per individual (Figure S11). When we examined neutral synonymous variants, CO had significantly more variants than either FIN or CR (p = 8.56 × 10−06; p = 0.0054, respectively). Finally, when counting the number of homozygous derived genotypes, we found that the FIN carried 3.3% more deleterious variants in the homozygous state per individual than CO but not the CR (p = 0.0003) (Figure S11). Additionally, the FIN carried significantly more neutral homozygous genotypes per individual than either population (CO p = 1.01 × 10−05; CR p = 6.96 × 10−05). The increased deleterious and neutral variation in homozygous form is an expected consequence of the long-term bottleneck that the FIN experienced during their founding.
We next tested whether the amount of the genome in an individual contained within an ROH was correlated with the number of nonsynonymous mutations carried by the individual. Counting nonsynonymous (NS) or synonymous (SYN) allele copies did not show any correlation with the amount of an individual’s genome that falls within an ROH for the CR or FIN (Figures 6A, 6D, and S12–S15). However, in the CO, as the amount of the genome within an ROH increased, individuals tended to carry more NS alleles, though this correlation was strongly driven by a single individual, who also had the highest FSNP and FPED (R2 = 0.2393; p = 0.0036; Figure S12), and when this individual was removed the correlation no longer remained significant. Importantly, the number of SYN alleles per individual was not correlated with the amount of the genome in an ROH (p = 0.2261).
When counting variants per individual, we observed a significant negative correlation with the amount of an individuals’ genome that falls within an ROH in the Latin American isolates (Figures 6B, 6E, and S12–S14). The negative correlation is a result of heterozygous sites being lost when an ROH is formed due to inbreeding. Conversely, when counting homozygous genotypes per individual, we observed a significant positive correlation with the amount of an individual’s genome that falls within an ROH in both the Latin American isolates and FIN (Figures 6C, 6F, and S12–S15). Homozygous genotypes were the only statistic that correlated significantly with the amount of the genome in an ROH across all isolated populations for both SYN and NS sites. We observed a stronger correlation between the number of NS homozygous genotypes and the amount of an individual’s genome within an ROH in the Latin American isolates (R2 = 0.5000 [CO] and R2 = 0.2165 [CR]; p = 7.546−06 [CO] and p = 0.0059 [CR]) compared to the FIN (R2 = 0.1130 and p = 0.0389) (Figures S12–S15). This pattern exists because the majority of CO and CR individuals carried a larger proportion of their genome within an ROH, while the FIN individuals do not harbor many ROHs.
We next asked whether there was an enrichment or depletion of NS variants relative to SYN variants within versus outside of an ROH using a permutation test on the three different counting approaches (see Subjects and Methods). When variants or allele copies were counted, none of the populations produced significant results (Table 2). When homozygous genotypes were counted, ROHs in the MXL and CR were enriched for homozygous NS genotypes relative to SYN homozygous genotypes (p = 0.0052 and p = 0.0169) (Table 2). Additionally, if we pooled the CR and CO populations, we also observed a significant enrichment of homozygous NS genotypes within an ROH compared to non-ROH regions of the genome (p = 0.0011).
Table 2.
Enrichment of Nonsynonymous Homozygous Derived Genotypes within ROHs
| Population | Allele Copies Odds Ratio | Allele Copies p Value | Variants Odds Ratio | Variants p Value | Homozygotes Odds Ratio | Homozygotes p Value |
|---|---|---|---|---|---|---|
| YRI | 1.059 | 0.664 | 1.048 | 0.762 | 1.129 | 0.417 |
| CEU | 1.203 | 0.105 | 1.208 | 0.138 | 1.252 | 0.082 |
| FIN | 0.937 | 0.324 | 0.92 | 0.265 | 1.003 | 0.957 |
| PEL | 0.986 | 0.797 | 0.972 | 0.638 | 1.038 | 0.54 |
| CLM | 0.99 | 0.755 | 0.964 | 0.337 | 1.066 | 0.097 |
| CO | 1.008 | 0.828 | 0.985 | 0.714 | 1.074 | 0.097 |
| CR | 1.015 | 0.607 | 0.991 | 0.806 | 1.085 | 0.0169∗ |
| CO & CR | 1.025 | 0.283 | 1.002 | 0.806 | 1.088 | 0.0011∗ |
| MXL | 1.112 | 0.052 | 1.089 | 0.169 | 1.19 | 0.005∗ |
| PUR | 0.981 | 0.635 | 0.965 | 0.411 | 1.047 | 0.301 |
This table summarizes the results of our enrichment analyses for each population sampled as well as a combined super-population of Colombians and Costa Ricans (CO & CR). Odds ratios were calculated as the ratio of nonsynonymous variants relative to synonymous variants within versus outside of an ROH for each counting method. Asterisk (∗) used to indicate significant p values after permutation test was conducted (see Subjects and Methods). Population abbreviations are as in Figure 1.
We tested whether FSNP was correlated with the amount of deleterious variation per individual. We used only isolates for these regressions because we are particularly interested in how recent consanguinity affected deleterious variation in the genome. We observed the exact same pattern with FSNP as with ROHs (Figure S16). Briefly, counting NS or SYN allele copies did not show any correlation with FSNP for the CR or FIN, but there was a significant correlation with NS allele copies in CO which was driven by a single outlier individual (Figures S16–S19). The correlation with NS allele copies and FSNP in CO did not remain once the outlier individual was removed. Counting NS and SYN variants per individual produced a significant negative correlation with FSNP in the Latin American isolates (Figures S14 and S17–S19). Counting the number of NS and SYN homozygous genotypes per individual was positively correlated with FSNP in the both Latin American isolates and FIN (Figures S16–S19). Again, counting homozygotes was the only method with significant results across all isolated populations for both SYN and NS variants. The ability to recapitulate the pattern we observed in ROHs using FSNP was reassuring and adds further support to the strong relationship between recent consanguinity and ROHs.
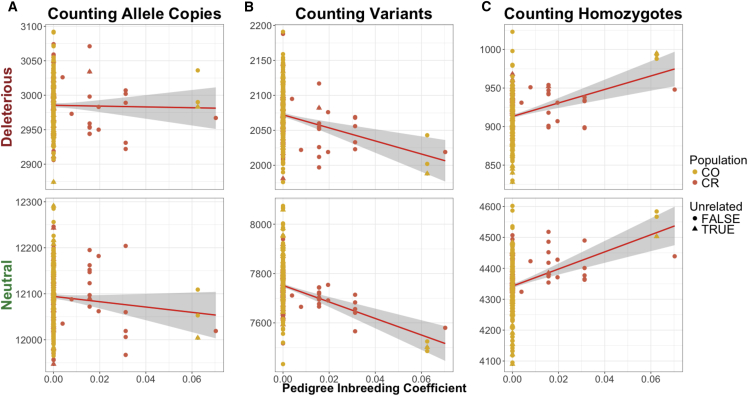
Lastly, because we had multi-generational pedigrees for the Latin American isolates, we examined the correlation between putatively deleterious variation and recent consanguinity as measured by FPED. All the following reported p values account for kinship (see Subjects and Methods). When we pooled the CO and CR individuals together, we did not observe any relationship between counting derived deleterious allele copies and FPED after correcting for kinship (Figure 7A). Moreover, we observed a negative correlation between FPED and the number of deleterious variants per individual (R2 = 0.0375, p = 6.02 × 10−06). The number of neutral variants per individual was also negatively correlated with FPED (p = 2.26 × 10−10) (Figure 7B). Finally, we observed a positive correlation between FPED and derived deleterious homozygotes (R2 = 0.0575, p = 1.0 × 10−06) as well as between FPED and the number of neutral derived homozygotes per individual (p = 1.03 × 10−08) (Figure 7C). These results suggest that recent consanguinity during the last few generations has increased the number of derived deleterious homozygous genotypes in these two populations.
Figure 7.
Pedigree Inbreeding Coefficient (FPED) Is Correlated with Deleterious Variation
Triangles represent the individuals that were sampled in the unrelated dataset (n = 30). Variants were predicted as either putatively deleterious (nonsynonymous) SNPs or putatively neutral (synonymous) SNPs using GERP.41 Correlation between FPED and the number of mutations per individual in Colombians and Costa Ricans.
(A) Number of derived alleles per individual.
(B) Number of variants per individual.
(C) Number of homozygous derived genotypes per individual. The first row depicts the correlation between deleterious sites using each counting method and FPED for sequenced individuals from Latin American isolates. The second row depicts the correlation between neutral sites using each counting method and FPED in the same individuals.
Population abbreviations are as in Figure 1.
Discussion
Here we present a comprehensive study of genetic diversity, demographic history, identity-by-descent, runs of homozygosity, and deleterious mutations in multiple admixed isolated populations. We show that admixture sufficiently increases genetic diversity of the Colombian and Costa Rican isolates such that each isolate has diversity levels comparable to a non-isolated population. However, we still observe characteristics in the Latin American isolates that are hallmarks of an archetypal isolate, such as: an excess of IBD segments, cryptic relatedness within the population, and an enrichment of long ROHs. Further, we demonstrate that long ROHs contain an enrichment of deleterious variants carried in the homozygous state, which has potential implications for fitness and disease risk.
Taken together, our results support historical data which state that a recent admixture event, within the last 500 years, founded the Colombian and Costa Rican population isolates. After founding, a bottleneck corresponding to the Spanish settlement occurred and each population has increased in size until the present day.18, 71 We see evidence of these processes in the inference of demography from IBD patterns. Importantly, the bottleneck experienced in the Latin American isolates was not as prolonged as that experienced by the Finnish. Further, the Finnish bottleneck occurred thousands of years ago. The difference in bottleneck time scales likely accounts for some portion of the higher genetic diversity observed in Latin American population isolates in comparison to the Finnish. In other words, the bottlenecks captured by IBDNe in the Latin Americans are too recent to markedly impact levels of heterozygosity. Further, the admixture process experienced by the Latin American isolates could increase levels of genetic diversity,52 especially because some individuals have appreciable levels of African ancestry.
We see little difference in patterns of genetic variation in the 1000 Genomes Colombian samples (CLM) and the isolated Colombian sample (CO) studied in this project. The CLM have similar levels of diversity and LD relative to the isolated CO. There is a modest increase in IBD segments and ROHs in the isolated CO relative to CLM. The Latin American isolates occupy areas that were considered as being geographically isolated at the time of sampling (the Central Valley of Costa Rica and the department of Antioquia in Colombia18) while the 1000 Genomes sample was taken from Medellín, which is included within the Antioquia region.72, 73, 74, 75 Thus, these results are a bit surprising as the CO samples studied in this project were from a more remote area and the individuals sampled in the 1000 Genomes Project were from a more cosmopolitan area. This finding likely indicates that the more ancient histories (prior to several hundred years ago) were likely more similar between these populations and have a greater influence on the patterns of genetic variation studied here.
Our results beg the question, what constitutes a population isolate? For example, is it a requirement that population isolates have low genetic diversity relative to the source population? Under this definition, the Latin American population isolates would not qualify as population isolates. The bottleneck in the Costa Ricans and Colombians seems to have had little effect on their genetic diversity, as their diversity levels are comparable to non-isolated Latin American populations. The Finnish, on the other hand, experienced a long-term bottleneck that has resulted in a depletion of segregating sites, and of the remaining segregating sites, there is an enrichment of deleterious variants relative to non-isolated populations,7, 11 and would clearly qualify as an isolate. However, if one measures isolation based on IBD, we see that there is an enrichment of IBD segments in the Latin American isolates relative to the Finnish. Further, looking at ROHs, Latin American individuals from population isolates have a larger burden of ROH than Finnish, thus increasing the chances of identifying more shared genomic regions in the Latin American isolates than the Finnish. By this metric, the Latin American population isolates would qualify as population isolates. Thus, both the Costa Rican and Colombian populations and the Finnish are isolates but in different ways. For example, the Costa Ricans and Colombians are historical isolates, meaning these populations are not currently isolated but they exhibit many traits of an isolate, whereas the Finnish are contemporary isolates, meaning the population is still isolated. Our work suggests that isolated populations have distinct demographic histories that impact genetic variation in different ways.
We find that Latin American isolates have the largest ROH burden in comparison to any other sampled population, which corroborates results from a recent review where authors state that populations with small Ne and recent consanguinity will harbor the largest amount of ROHs.62 Because previous research has shown a strong correlation between recent inbreeding, quantified by both FSNP and FPED, and long runs of homozygosity, we were particularly interested in the mechanism behind the generation of long ROHs.56, 57, 58, 59, 60, 61, 76, 77, 78 We used simulations to test which demographic scenarios could produce long ROHs (Figure 5). These simulations and availability of extended pedigree data were crucial, because the FSNP metric can also be influenced by a recent bottleneck. If small population size or admixture was responsible for generating the ROHs, these processes would not be reflected in FPED. Thus, we would not expect to find a correlation between FPED and the amount of the genome in ROHs. The observed correlation between FPED and the amount of the genome in ROHs suggests that recent consanguinity (as measured by FPED) is related to the extent of long ROHs in the genome. Further, our simulations show that neither admixture nor a recent population bottleneck, unless unrealistically severe (see Results), could generate the high levels of long ROHs that are observed in some individuals. It was only when we incorporated non-random mating into the simulation that levels of ROHs comparable to what we observed in our data were produced.
Our results demonstrate that the Latin American population isolates have experienced more recent consanguinity than other population isolates, like the Finnish. Further, in Finland it has previously been shown that the frequency of consanguinity, due to first-cousin marriages, is quite low and the best predictors of these unions were socio-economic class and ethnicity, rather than geographic barriers or population density.79 On the other hand, for the two Latin American isolates, consanguinity could be a consequence of increased geographic barriers preventing movement of individuals over more dispersed areas. It is also important to point out that it is unclear the extent to which ascertaining individuals from large pedigrees may impact the number of ROHs in our sample. Thus, the finding of an increase in ROHs may not be generalizable to Colombian and Costa Rican populations as a whole. However, we observed a similar pattern of increased ROHs in the CLM, which suggests that the pedigree ascertainment of the CO and CR may not be generating the increase in ROHs.
We also tested how recent consanguinity affects deleterious variation in the genome. When counting homozygous derived deleterious genotypes, we found a positive correlation between the number of nonsynonymous homozygous genotypes and the amount of an individual’s genome within an ROH (Figure 6). Further, we observed an enrichment for nonsynonymous homozygous derived genotypes relative to synonymous homozygous derived genotypes within ROHs versus the rest of the genome (Table 2). This enrichment could be a result of nonsynonymous mutations generally segregating at lower frequency and typically being carried as a single copy in an individual. When an ROH is formed, the chromosome that was carrying the mutation is copied, thus allowing the mutation to increase the number of homozygotes within the ROH.56, 61 Since long ROHs are a product of recent consanguinity and these populations have experienced recent consanguinity, we see a corresponding increase in the burden of deleterious variants in the genomes of Costa Rican and Colombian isolates. Because we are more likely to see deleterious variants in the homozygous form in areas of the genome that fall within an ROH, our work is particularly relevant for alleles associated with recessive diseases. Lastly, we provide a mechanism for how recent consanguinity can reduce fitness in natural populations.80, 81, 82 Specifically, if gene knockouts and deleterious mutations tend to be recessive,83, 84, 85, 86, 87, 105 as suggested by several studies, then recent consanguinity will increase the number of homozygous derived deleterious variants carried by an individual in a long ROH, thus leading to an overall reduction of fitness in the sampled population.3
Utilizing estimated ancestry proportions from across the genome, we tested for a correlation between an individual’s ancestry and the amount of their genome that falls within an ROH, complementing the work of Szpiech et al.88 We found a positive correlation between the proportion of European ancestry and the amount of an individual’s genome within a run of homozygosity. These results are consistent with the Latin American isolates originating from a small number of European founders, which would decrease genetic diversity and increase homozygosity for those areas of the genome containing European haplotypes. We observed a negative correlation between Native American ancestry and the amount of the genome contained within an ROH (Figure S10). This finding appears to be at odds with previous research61, 62 but largely agrees with conclusions drawn by Moreno and colleagues.89 Thus, we believe that some of this difference may be due to distinct sampling strategies of the Native American source population in our study compared to previous work. The reference Native American population we used was composed of Chibchan-speaking individuals from Reich et al.46 Chibchan-speaking populations inherited their Native American ancestry from admixture between Southern and Northern American lineages.46 Because our reference Native American population is admixed and Native American populations tend to be small, it is likely that drift has affected different alleles in source populations89 that formed the current Chibchan-speaking populations. The Chibchan-speaking populations may have more diversity and fewer fixed homozygous sites than previously sampled Native American populations, which could explain the negative correlation we observed between ancestry and ROHs.
While we found evidence of recent bottlenecks and expansions within Latin American isolates using IBDNe33 (Figure 3), our demographic inferences have some limitations. For example, the most current estimates of Ne are unrealistically large or small. The inaccurate estimates of Ne may be due to low sample size, since we only used 30 individuals and it has been suggested that IBDNe works best for larger sample sizes (>200 individuals).33 Indeed, the wide 95% confidence intervals around the most recent time points in the FIN suggests much uncertainty regarding the recent effective size of the last five generations and this estimate should not be taken literally. However, a recent study by the creators of IBDNe examined ancestry-specific effective population sizes through time by applying IBDNe to different ancestry segments.65 Importantly, in that study, the overall genome-wide trajectories of Ne largely mirror those seen for the individual ancestry components.65 Thus, we believe that it is appropriate to apply IBDNe to admixed populations. Further, we believe that the demographic patterns that we were able to detect in the Latin American populations (PUR, CO, CR, CLM, and MXL) are robust, as these patterns were recapitulated using a different larger dataset in the same paper.65
In our study, the populations with the highest IBD scores were admixed (PUR, CO, CR, and CLM). Furthermore, because IBD segments may contain useful information for identifying regions of the genome that contain disease-associated mutations, especially within individuals with the highest amounts of consanguinity, it may be useful to deconvolute ancestry for each segment when identifying disease-associated mutations because disease prevalence may differ in each parental population. One population that may be of particular interest is the PUR, who demonstrated the largest enrichment of IBD segments while still exhibiting some of the highest levels of diversity. The PUR also stood out in several recent studies. Browning and colleagues found that the PUR had smaller founder sizes than other Latin American populations,65 while Belbin and colleagues used IBD segment mapping in Puerto Ricans sampled from BioMe biobank to identify a gene, COL27A1, that is involved in a common collagen disorder.90
Population isolates have frequently been used for studying Mendelian15, 91, 92, 93, 94, 95 and complex diseases.14, 17, 96, 97, 98, 99, 100 Our work shows that the genetic diversity and genomic background of population isolates varies immensely. Therefore, it is imperative that we understand the unique genetic diversity and demography belonging to each population isolate. When attempting to identify an isolate, one could use a composite test with a number of features of interest: enrichment of IBD and/or ROHs relative to an archetypal isolate, increase in shared IBD segments, enrichment of deleterious variation at intermediate allele frequencies, or small bottleneck effective population size. For example, if we knew beforehand that there was a history of consanguineous unions within the study population, then we would expect an enrichment of ROHs in the composite test. Researchers could shape their study design to target the enrichment of ROHs as a tool for disease mapping. This method has previously been used to identify human knockouts, discover novel loci associated with disease, and understand gene function.90, 100, 101, 102, 103 Further, ROHs could be particularly helpful to better understand disease architecture104 since ROHs may harbor more recessive mutations that do not have full penetrance. Thus, our work highlights the importance of understanding the demographic history of isolated populations, as differences in demographic history will greatly impact their patterns of genetic variation.
Consortia
Members of the Costa Rica/Colombia Consortium for Genetic Investigation of Bipolar Endophenotypes: Lori Altshuler, Carmen Araya, Xinia Araya, George Bartzokis, Carrie E. Bearden, Gabriel Bedoya, Julio Bejarano, Rita M. Cantor, Gabriel Castrillón, Giovanni Coppola, Javier Escobar, Scott C. Fears, Nelson B. Freimer, Juliana Gomez-Makhinson, Alden Y. Huang, Sun-Goo Hwang, Barbara Kremeyer, Maria C. Lopez, Carlos Lopez-Jaramillo, Gabriel Macaya, Julio Molina, Gabriel Montoya, Patricia Montoya, Loes M. Olde Loohuis, Jorge Ospina-Duque, YoungJun Park, Vasily Ramensky, Margarita Ramirez, Victor I. Reus, Neil Risch, Andrés Ruiz-Linares, Chiara Sabatti, Susan K. Service, Mitzi Spesny, Jae Hoon Sul, Terri M. Teshiba, and Zhongyang Zhang.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
The authors would like to acknowledge Charleston Chiang, Jesse Garcia, Malika Kumar, and Sonya McKeown for contributing their time and thoughtful discussion. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under grant numbers DGE-1144087 and DGE-1650604 awarded to J.A.M., as well as partial support from NIH grants K01ES028064 awarded to J.H.S., R01MH095454, P30NS062691, and R01MH075007 awarded to N.F., and R35GM119856 awarded to K.E.L.
Published: October 25, 2018
Footnotes
Supplemental Data include 19 figures and 3 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.09.013.
Contributor Information
Kirk E. Lohmueller, Email: klohmueller@ucla.edu.
Costa Rica/Colombia Consortium for Genetic Investigation of Bipolar Endophenotypes:
Scott C. Fears, Susan K. Service, Barbara Kremeyer, Carmen Araya Lic, Xinia Araya Lic, Julio Bejarano, Margarita Ramirez Lic, Gabriel Castrillón, Maria C. Lopez, Gabriel Montoya, Patricia Montoya, Terri M. Teshiba, Lori Altshuler, George Bartzokis, Javier Escobar, Jorge Ospina-Duque, Neil Risch, Andrés Ruiz-Linares, Rita M. Cantor, Carlos Lopez-Jaramillo, Gabriel Macaya, Julio Molina, Victor I. Reus, Chiara Sabatti, Nelson B. Freimer, Carrie E. Bearden, Jae Hoon Sul, Alden Y. Huang, Vasily Ramensky, Sun-Goo Hwang, YoungJun Park, Zhongyang Zhang, Loes M. Olde Loohuis, Mitzi Spesny, Juliana Gomez-Makhinson, Gabriel Bedoya, and Giovanni Coppola
Web Resources
6-primate EPO alignment, ftp://ftp.ensembl.org/pub/release-75/fasta/ancestral_alleles/
ADMIXTURE: http://www.genetics.ucla.edu/software/admixture/download.html
GATK, https://software.broadinstitute.org/gatk/download/archive
IBDNe, http://faculty.washington.edu/browning/ibdne.html#download
KING (version 2.1), http://people.virginia.edu/∼wc9c/KING/history.htm
Latin American Isolates data, https://github.com/jaam92/LatinAmericanIsolates
ROH simulation script, https://github.com/LohmuellerLab/ROH_Latin_American_Isolates
SeattleSeq Annotation 138, http://snp.gs.washington.edu/SeattleSeqAnnotation138/
SHAPEIT, http://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html#download
Supplemental Data
References
- 1.Peltonen L., Palotie A., Lange K. Use of population isolates for mapping complex traits. Nat. Rev. Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 2.Lohmueller K.E., Indap A.R., Schmidt S., Boyko A.R., Hernandez R.D., Hubisz M.J., Sninsky J.J., White T.J., Sunyaev S.R., Nielsen R. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlesworth D., Willis J.H. The genetics of inbreeding depression. Nat. Rev. Genet. 2009;10:783–796. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- 4.Lohmueller K.E. The impact of population demography and selection on the genetic architecture of complex traits. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004379. e1004379.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simons Y.B., Turchin M.C., Pritchard J.K., Sella G. The deleterious mutation load is insensitive to recent population history. Nat. Genet. 2014;46:220–224. doi: 10.1038/ng.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Service S., DeYoung J., Karayiorgou M., Roos J.L., Pretorious H., Bedoya G., Ospina J., Ruiz-Linares A., Macedo A., Palha J.A. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat. Genet. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- 7.Lim E.T., Würtz P., Havulinna A.S., Palta P., Tukiainen T., Rehnström K., Esko T., Mägi R., Inouye M., Lappalainen T., Sequencing Initiative Suomi (SISu) Project Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue Y., Mezzavilla M., Haber M., McCarthy S., Chen Y., Narasimhan V., Gilly A., Ayub Q., Colonna V., Southam L. Enrichment of low-frequency functional variants revealed by whole-genome sequencing of multiple isolated European populations. Nat. Commun. 2017;8:15927. doi: 10.1038/ncomms15927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kittles R.A., Perola M., Peltonen L., Bergen A.W., Aragon R.A., Virkkunen M., Linnoila M., Goldman D., Long J.C. Dual origins of Finns revealed by Y chromosome haplotype variation. Am. J. Hum. Genet. 1998;62:1171–1179. doi: 10.1086/301831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peltonen L., Jalanko A., Varilo T. Molecular genetics of the Finnish disease heritage. Hum. Mol. Genet. 1999;8:1913–1923. doi: 10.1093/hmg/8.10.1913. [DOI] [PubMed] [Google Scholar]
- 11.Wang S.R., Agarwala V., Flannick J., Chiang C.W.K., Altshuler D., Hirschhorn J.N., GoT2D Consortium Simulation of Finnish population history, guided by empirical genetic data, to assess power of rare-variant tests in Finland. Am. J. Hum. Genet. 2014;94:710–720. doi: 10.1016/j.ajhg.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Chapelle A., Wright F.A. Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc. Natl. Acad. Sci. USA. 1998;95:12416–12423. doi: 10.1073/pnas.95.21.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin A.R., Karczewski K.J., Kerminen S., Kurki M.I., Sarin A.-P., Artomov M., Eriksson J.G., Esko T., Genovese G., Havulinna A.S. Haplotype sharing provides insights into fine-scale population history and disease in Finland. Am. J. Hum. Genet. 2018;102:760–775. doi: 10.1016/j.ajhg.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panoutsopoulou K., Hatzikotoulas K., Xifara D.K., Colonna V., Farmaki A.-E., Ritchie G.R.S., Southam L., Gilly A., Tachmazidou I., Fatumo S. Genetic characterization of Greek population isolates reveals strong genetic drift at missense and trait-associated variants. Nat. Commun. 2014;5:5345. doi: 10.1038/ncomms6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakatsuka N., Moorjani P., Rai N., Sarkar B., Tandon A., Patterson N., Bhavani G.S., Girisha K.M., Mustak M.S., Srinivasan S. The promise of discovering population-specific disease-associated genes in South Asia. Nat. Genet. 2017;49:1403–1407. doi: 10.1038/ng.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedersen C.T., Lohmueller K.E., Grarup N., Bjerregaard P., Hansen T., Siegismund H.R., Moltke I., Albrechtsen A. The effect of an extreme and prolonged population bottleneck on patterns of deleterious variation: insights from the Greenlandic Inuit. Genetics. 2017;205:787–801. doi: 10.1534/genetics.116.193821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tachmazidou I., Dedoussis G., Southam L., Farmaki A.-E., Ritchie G.R., Xifara D.K., Matchan A., Hatzikotoulas K., Rayner N.W., Chen Y., UK10K consortium A rare functional cardioprotective APOC3 variant has risen in frequency in distinct population isolates. Nat. Commun. 2013;4:2872. doi: 10.1038/ncomms3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvajal-Carmona L.G., Ophoff R., Service S., Hartiala J., Molina J., Leon P., Ospina J., Bedoya G., Freimer N., Ruiz-Linares A. Genetic demography of Antioquia (Colombia) and the central valley of Costa Rica. Hum. Genet. 2003;112:534–541. doi: 10.1007/s00439-002-0899-8. [DOI] [PubMed] [Google Scholar]
- 19.1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fears S.C., Service S.K., Kremeyer B., Araya C., Araya X., Bejarano J., Ramirez M., Castrillón G., Gomez-Franco J., Lopez M.C. Multisystem component phenotypes of bipolar disorder for genetic investigations of extended pedigrees. JAMA Psychiatry. 2014;71:375–387. doi: 10.1001/jamapsychiatry.2013.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conomos M.P., Miller M.B., Thornton T.A. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conomos M.P., Reiner A.P., Weir B.S., Thornton T.A. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sul J.H., Service S.K., Huang A.Y., Ramensky V., Hwang S.-G., Teshiba T.M., Park Y., Ori A.P.S., Zhang Z., Mullins N. Contribution of common and rare variants to bipolar disorder susceptibility in extended pedigrees from population isolates. bioRxiv. 2018 doi: 10.1038/s41398-020-0758-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watterson G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 27.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Browning B.L., Browning S.R. Detecting identity by descent and estimating genotype error rates in sequence data. Am. J. Hum. Genet. 2013;93:840–851. doi: 10.1016/j.ajhg.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browning B.L., Browning S.R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics. 2013;194:459–471. doi: 10.1534/genetics.113.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gusev A., Lowe J.K., Stoffel M., Daly M.J., Altshuler D., Breslow J.L., Friedman J.M., Pe’er I. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 2009;19:318–326. doi: 10.1101/gr.081398.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaneau O., Marchini J., Zagury J.-F. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 32.Kong, Thorleifsson G., Gudbjartsson D.F., Masson G., Sigurdsson A., Jonasdottir A., Walters G.B., Jonasdottir A., Gylfason A., Kristinsson K.T. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 33.Browning S.R., Browning B.L. Accurate non-parametric estimation of recent effective population size from segments of identity by descent. Am. J. Hum. Genet. 2015;97:404–418. doi: 10.1016/j.ajhg.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auton A., Bryc K., Boyko A.R., Lohmueller K.E., Novembre J., Reynolds A., Indap A., Wright M.H., Degenhardt J.D., Gutenkunst R.N. Global distribution of genomic diversity underscores rich complex history of continental human populations. Genome Res. 2009;19:795–803. doi: 10.1101/gr.088898.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinnwell J.P., Therneau T.M., Schaid D.J. The kinship2 R package for pedigree data. Hum. Hered. 2014;78:91–93. doi: 10.1159/000363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haller B.C., Messer P.W. SLiM 2: flexible, interactive forward genetic simulations. Mol. Biol. Evol. 2017;34:230–240. doi: 10.1093/molbev/msw211. [DOI] [PubMed] [Google Scholar]
- 37.Kim B.Y., Huber C.D., Lohmueller K.E. Inference of the distribution of selection coefficients for new nonsynonymous mutations using large samples. Genetics. 2017;206:345–361. doi: 10.1534/genetics.116.197145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huber C.D., Kim B.Y., Marsden C.D., Lohmueller K.E. Determining the factors driving selective effects of new nonsynonymous mutations. Proc. Natl. Acad. Sci. USA. 2017;114:4465–4470. doi: 10.1073/pnas.1619508114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long M., Deutsch M. Association of intron phases with conservation at splice site sequences and evolution of spliceosomal introns. Mol. Biol. Evol. 1999;16:1528–1534. doi: 10.1093/oxfordjournals.molbev.a026065. [DOI] [PubMed] [Google Scholar]
- 40.Kempthorne O. John Wiley And Sons, Inc.; New York: 1957. An Introduction to Genetic Statistics. [Google Scholar]
- 41.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., NISC Comparative Sequencing Program Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sankararaman S., Sridhar S., Kimmel G., Halperin E. Estimating local ancestry in admixed populations. Am. J. Hum. Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pagani L., St Clair P.A., Teshiba T.M., Service S.K., Fears S.C., Araya C., Araya X., Bejarano J., Ramirez M., Castrillón G. Genetic contributions to circadian activity rhythm and sleep pattern phenotypes in pedigrees segregating for severe bipolar disorder. Proc. Natl. Acad. Sci. USA. 2016;113:E754–E761. doi: 10.1073/pnas.1513525113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reich D., Patterson N., Campbell D., Tandon A., Mazieres S., Ray N., Parra M.V., Rojas W., Duque C., Mesa N. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aulchenko Y.S., Ripke S., Isaacs A., van Duijn C.M. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 49.Lohmueller K.E., Albrechtsen A., Li Y., Kim S.Y., Korneliussen T., Vinckenbosch N., Tian G., Huerta-Sanchez E., Feder A.F., Grarup N. Natural selection affects multiple aspects of genetic variation at putatively neutral sites across the human genome. PLoS Genet. 2011;7:e1002326. doi: 10.1371/journal.pgen.1002326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai J.J., Macpherson J.M., Sella G., Petrov D.A. Pervasive hitchhiking at coding and regulatory sites in humans. PLoS Genet. 2009;5:e1000336. doi: 10.1371/journal.pgen.1000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hernandez R.D., Kelley J.L., Elyashiv E., Melton S.C., Auton A., McVean G., Sella G., Przeworski M., 1000 Genomes Project Classic selective sweeps were rare in recent human evolution. Science. 2011;331:920–924. doi: 10.1126/science.1198878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kidd J.M., Gravel S., Byrnes J., Moreno-Estrada A., Musharoff S., Bryc K., Degenhardt J.D., Brisbin A., Sheth V., Chen R. Population genetic inference from personal genome data: impact of ancestry and admixture on human genomic variation. Am. J. Hum. Genet. 2012;91:660–671. doi: 10.1016/j.ajhg.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stumpf M.P., Goldstein D.B. Demography, recombination hotspot intensity, and the block structure of linkage disequilibrium. Curr. Biol. 2003;13:1–8. doi: 10.1016/s0960-9822(02)01404-5. [DOI] [PubMed] [Google Scholar]
- 54.Pritchard J.K., Przeworski M. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gravel S., Zakharia F., Moreno-Estrada A., Byrnes J.K., Muzzio M., Rodriguez-Flores J.L., Kenny E.E., Gignoux C.R., Maples B.K., Guiblet W., 1000 Genomes Project Reconstructing Native American migrations from whole-genome and whole-exome data. PLoS Genet. 2013;9:e1004023. doi: 10.1371/journal.pgen.1004023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pemberton T.J., Szpiech Z.A. Relationship between deleterious variation, genomic autozygosity, and disease risk: insights from The 1000 Genomes Project. Am. J. Hum. Genet. 2018;102:658–675. doi: 10.1016/j.ajhg.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang J.T.L., Goldberg A., Edge M.D., Behar D.M., Rosenberg N.A. Consanguinity rates predict long runs of homozygosity in Jewish populations. Hum. Hered. 2016;82:87–102. doi: 10.1159/000478897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szpiech Z.A., Xu J., Pemberton T.J., Peng W., Zöllner S., Rosenberg N.A., Li J.Z. Long runs of homozygosity are enriched for deleterious variation. Am. J. Hum. Genet. 2013;93:90–102. doi: 10.1016/j.ajhg.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McQuillan R., Leutenegger A.-L., Abdel-Rahman R., Franklin C.S., Pericic M., Barac-Lauc L., Smolej-Narancic N., Janicijevic B., Polasek O., Tenesa A. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kirin M., McQuillan R., Franklin C.S., Campbell H., McKeigue P.M., Wilson J.F. Genomic runs of homozygosity record population history and consanguinity. PLoS ONE. 2010;5:e13996. doi: 10.1371/journal.pone.0013996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pemberton T.J., Absher D., Feldman M.W., Myers R.M., Rosenberg N.A., Li J.Z. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 2012;91:275–292. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ceballos F.C., Joshi P.K., Clark D.W., Ramsay M., Wilson J.F. Runs of homozygosity: windows into population history and trait architecture. Nat. Rev. Genet. 2018;19:220–234. doi: 10.1038/nrg.2017.109. [DOI] [PubMed] [Google Scholar]
- 63.Lemes R.B., Nunes K., Carnavalli J.E.P., Kimura L., Mingroni-Netto R.C., Meyer D., Otto P.A. Inbreeding estimates in human populations: applying new approaches to an admixed Brazilian isolate. PLoS ONE. 2018;13:e0196360. doi: 10.1371/journal.pone.0196360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kardos M., Taylor H.R., Ellegren H., Luikart G., Allendorf F.W. Genomics advances the study of inbreeding depression in the wild. Evol. Appl. 2016;9:1205–1218. doi: 10.1111/eva.12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Browning S.R., Browning B.L., Daviglus M.L., Durazo-Arvizu R.A., Schneiderman N., Kaplan R.C., Laurie C.C. Ancestry-specific recent effective population size in the Americas. PLoS Genet. 2018;14:e1007385. doi: 10.1371/journal.pgen.1007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kimura M., Maruyama T., Crow J.F. The mutation load in small populations. Genetics. 1963;48:1303–1312. doi: 10.1093/genetics/48.10.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- 68.Hodgkinson A., Casals F., Idaghdour Y., Grenier J.-C., Hernandez R.D., Awadalla P. Selective constraint, background selection, and mutation accumulation variability within and between human populations. BMC Genomics. 2013;14:495. doi: 10.1186/1471-2164-14-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peischl S., Dupanloup I., Kirkpatrick M., Excoffier L. On the accumulation of deleterious mutations during range expansions. Mol. Ecol. 2013;22:5972–5982. doi: 10.1111/mec.12524. [DOI] [PubMed] [Google Scholar]
- 70.Fu W., Gittelman R.M., Bamshad M.J., Akey J.M. Characteristics of neutral and deleterious protein-coding variation among individuals and populations. Am. J. Hum. Genet. 2014;95:421–436. doi: 10.1016/j.ajhg.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Escamilla M.A., Spesny M., Reus V.I., Gallegos A., Meza L., Molina J., Sandkuijl L.A., Fournier E., Leon P.E., Smith L.B., Freimer N.B. Use of linkage disequilibrium approaches to map genes for bipolar disorder in the Costa Rican population. Am. J. Med. Genet. 1996;67:244–253. doi: 10.1002/(SICI)1096-8628(19960531)67:3<244::AID-AJMG2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 72.Wang S., Ray N., Rojas W., Parra M.V., Bedoya G., Gallo C., Poletti G., Mazzotti G., Hill K., Hurtado A.M. Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet. 2008;4:e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bedoya G., Montoya P., García J., Soto I., Bourgeois S., Carvajal L., Labuda D., Alvarez V., Ospina J., Hedrick P.W., Ruiz-Linares A. Admixture dynamics in Hispanics: a shift in the nuclear genetic ancestry of a South American population isolate. Proc. Natl. Acad. Sci. USA. 2006;103:7234–7239. doi: 10.1073/pnas.0508716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Safford F., Palacios M. Oxford University Press; USA: 2002. Colombia: Fragmented Land, Divided Society. [Google Scholar]
- 75.Carvajal-Carmona L.G., Soto I.D., Pineda N., Ortíz-Barrientos D., Duque C., Ospina-Duque J., McCarthy M., Montoya P., Alvarez V.M., Bedoya G., Ruiz-Linares A. Strong Amerind/white sex bias and a possible Sephardic contribution among the founders of a population in northwest Colombia. Am. J. Hum. Genet. 2000;67:1287–1295. doi: 10.1016/s0002-9297(07)62956-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scott E.M., Halees A., Itan Y., Spencer E.G., He Y., Azab M.A., Gabriel S.B., Belkadi A., Boisson B., Abel L., Greater Middle East Variome Consortium Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat. Genet. 2016;48:1071–1076. doi: 10.1038/ng.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Di Gaetano C., Fiorito G., Ortu M.F., Rosa F., Guarrera S., Pardini B., Cusi D., Frau F., Barlassina C., Troffa C. Sardinians genetic background explained by runs of homozygosity and genomic regions under positive selection. PLoS ONE. 2014;9:e91237. doi: 10.1371/journal.pone.0091237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li L.-H., Ho S.-F., Chen C.-H., Wei C.-Y., Wong W.-C., Li L.-Y., Hung S.-I., Chung W.-H., Pan W.-H., Lee M.-T.M. Long contiguous stretches of homozygosity in the human genome. Hum. Mutat. 2006;27:1115–1121. doi: 10.1002/humu.20399. [DOI] [PubMed] [Google Scholar]
- 79.Jorde L.B., Pitkänen K.J. Inbreeding in Finland. Am. J. Phys. Anthropol. 1991;84:127–139. doi: 10.1002/ajpa.1330840203. [DOI] [PubMed] [Google Scholar]
- 80.Wright S. University of Chicago Press; 1984. Evolution and the Genetics of Populations, Volume 3: Experimental Results and Evolutionary Deductions. [Google Scholar]
- 81.Charlesworth B., Charlesworth D. The genetic basis of inbreeding depression. Genet. Res. 1999;74:329–340. doi: 10.1017/s0016672399004152. [DOI] [PubMed] [Google Scholar]
- 82.Wang J., Hill W.G., Charlesworth D., Charlesworth B. Dynamics of inbreeding depression due to deleterious mutations in small populations: mutation parameters and inbreeding rate. Genet. Res. 1999;74:165–178. doi: 10.1017/s0016672399003900. [DOI] [PubMed] [Google Scholar]
- 83.Balick D.J., Do R., Cassa C.A., Reich D., Sunyaev S.R. Dominance of deleterious alleles controls the response to a population bottleneck. PLoS Genet. 2015;11:e1005436. doi: 10.1371/journal.pgen.1005436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mukai T., Chigusa S.I., Mettler L.E., Crow J.F. Mutation rate and dominance of genes affecting viability in Drosophila melanogaster. Genetics. 1972;72:335–355. doi: 10.1093/genetics/72.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simmons M.J., Crow J.F. Mutations affecting fitness in Drosophila populations. Annu. Rev. Genet. 1977;11:49–78. doi: 10.1146/annurev.ge.11.120177.000405. [DOI] [PubMed] [Google Scholar]
- 86.Phadnis N., Fry J.D. Widespread correlations between dominance and homozygous effects of mutations: implications for theories of dominance. Genetics. 2005;171:385–392. doi: 10.1534/genetics.104.039016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Agrawal A.F., Whitlock M.C. Inferences about the distribution of dominance drawn from yeast gene knockout data. Genetics. 2011;187:553–566. doi: 10.1534/genetics.110.124560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Szpiech Z.A., Mak A.C., White M.J., Hu D., Eng C., Burchard E.G., Hernandez R.D. Ancestry-dependent enrichment of deleterious homozygotes in runs of homozygosity. bioRxiv. 2018 doi: 10.1016/j.ajhg.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moreno-Estrada A., Gignoux C.R., Fernández-López J.C., Zakharia F., Sikora M., Contreras A.V., Acuña-Alonzo V., Sandoval K., Eng C., Romero-Hidalgo S. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science. 2014;344:1280–1285. doi: 10.1126/science.1251688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Belbin G.M., Odgis J., Sorokin E.P., Yee M.-C., Kohli S., Glicksberg B.S., Gignoux C.R., Wojcik G.L., Van Vleck T., Jeff J.M. Genetic identification of a common collagen disease in puerto ricans via identity-by-descent mapping in a health system. eLife. 2017;6:e25060. doi: 10.7554/eLife.25060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Myerowitz R., Costigan F.C. The major defect in Ashkenazi Jews with Tay-Sachs disease is an insertion in the gene for the alpha-chain of beta-hexosaminidase. J. Biol. Chem. 1988;263:18587–18589. [PubMed] [Google Scholar]
- 92.Hästbacka J., de la Chapelle A., Mahtani M.M., Clines G., Reeve-Daly M.P., Daly M., Hamilton B.A., Kusumi K., Trivedi B., Weaver A. The diastrophic dysplasia gene encodes a novel sulfate transporter: positional cloning by fine-structure linkage disequilibrium mapping. Cell. 1994;78:1073–1087. doi: 10.1016/0092-8674(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 93.Ruiz-Perez V.L., Ide S.E., Strom T.M., Lorenz B., Wilson D., Woods K., King L., Francomano C., Freisinger P., Spranger S. Mutations in a new gene in Ellis-van Creveld syndrome and Weyers acrodental dysostosis. Nat. Genet. 2000;24:283–286. doi: 10.1038/73508. [DOI] [PubMed] [Google Scholar]
- 94.Verhoeven K., Villanova M., Rossi A., Malandrini A., De Jonghe P., Timmerman V. Localization of the gene for the intermediate form of Charcot-Marie-Tooth to chromosome 10q24.1-q25.1. Am. J. Hum. Genet. 2001;69:889–894. doi: 10.1086/323742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Valente E.M., Bentivoglio A.R., Dixon P.H., Ferraris A., Ialongo T., Frontali M., Albanese A., Wood N.W. Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35-p36. Am. J. Hum. Genet. 2001;68:895–900. doi: 10.1086/319522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McInnes L.A., Service S.K., Reus V.I., Barnes G., Charlat O., Jawahar S., Lewitzky S., Yang Q., Duong Q., Spesny M. Fine-scale mapping of a locus for severe bipolar mood disorder on chromosome 18p11.3 in the Costa Rican population. Proc. Natl. Acad. Sci. USA. 2001;98:11485–11490. doi: 10.1073/pnas.191519098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ober C., Tan Z., Sun Y., Possick J.D., Pan L., Nicolae R., Radford S., Parry R.R., Heinzmann A., Deichmann K.A. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N. Engl. J. Med. 2008;358:1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stacey S.N., Sulem P., Jonasdottir A., Masson G., Gudmundsson J., Gudbjartsson D.F., Magnusson O.T., Gudjonsson S.A., Sigurgeirsson B., Thorisdottir K., Swedish Low-risk Colorectal Cancer Study Group A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat. Genet. 2011;43:1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gudmundsson J., Sulem P., Gudbjartsson D.F., Masson G., Agnarsson B.A., Benediktsdottir K.R., Sigurdsson A., Magnusson O.T., Gudjonsson S.A., Magnusdottir D.N. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat. Genet. 2012;44:1326–1329. doi: 10.1038/ng.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Saleheen D., Natarajan P., Armean I.M., Zhao W., Rasheed A., Khetarpal S.A., Won H.-H., Karczewski K.J., O’Donnell-Luria A.H., Samocha K.E. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544:235–239. doi: 10.1038/nature22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Botstein D., Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat. Genet. 2003;33(Suppl):228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 102.Lencz T., Lambert C., DeRosse P., Burdick K.E., Morgan T.V., Kane J.M., Kucherlapati R., Malhotra A.K. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc. Natl. Acad. Sci. USA. 2007;104:19942–19947. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mezzavilla M., Vozzi D., Badii R., Alkowari M.K., Abdulhadi K., Girotto G., Gasparini P. Increased rate of deleterious variants in long runs of homozygosity of an inbred population from Qatar. Hum. Hered. 2015;79:14–19. doi: 10.1159/000371387. [DOI] [PubMed] [Google Scholar]
- 104.Ku C.S., Naidoo N., Teo S.M., Pawitan Y. Regions of homozygosity and their impact on complex diseases and traits. Hum. Genet. 2011;129:1–15. doi: 10.1007/s00439-010-0920-6. [DOI] [PubMed] [Google Scholar]
- 105.Huber C.D., Durvasula A., Hancock A.M., Lohmueller K.E. Gene expression drives the evolution of dominance. Nat. Commun. 2018;9:2750. doi: 10.1038/s41467-018-05281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.