Abstract
Primary defects in motile cilia result in dysfunction of the apparatus responsible for generating fluid flows. Defects in these mechanisms underlie disorders characterized by poor mucus clearance, resulting in susceptibility to chronic recurrent respiratory infections, often associated with infertility; laterality defects occur in about 50% of such individuals. Here we report biallelic variants in LRRC56 (known as oda8 in Chlamydomonas) identified in three unrelated families. The phenotype comprises laterality defects and chronic pulmonary infections. High-speed video microscopy of cultured epithelial cells from an affected individual showed severely dyskinetic cilia but no obvious ultra-structural abnormalities on routine transmission electron microscopy (TEM). Further investigation revealed that LRRC56 interacts with the intraflagellar transport (IFT) protein IFT88. The link with IFT was interrogated in Trypanosoma brucei. In this protist, LRRC56 is recruited to the cilium during axoneme construction, where it co-localizes with IFT trains and is required for the addition of dynein arms to the distal end of the flagellum. In T. brucei carrying LRRC56-null mutations, or a variant resulting in the p.Leu259Pro substitution corresponding to the p.Leu140Pro variant seen in one of the affected families, we observed abnormal ciliary beat patterns and an absence of outer dynein arms restricted to the distal portion of the axoneme. Together, our findings confirm that deleterious variants in LRRC56 result in a human disease and suggest that this protein has a likely role in dynein transport during cilia assembly that is evolutionarily important for cilia motility.
Keywords: ciliopathies, left-right asymmetry, cilia, flagella, dynein arms, trypanosome, intraflagellar transport, leucine-rich repeat protein
Introduction
Cilia are highly conserved eukaryotic organelles that are classified into motile and non-motile forms. Motile cilia and flagella, a hallmark of eukaryotes, display remarkable structural and molecular conservation.1 Most motile cilia exhibit a 9+2 configuration—a pair of single microtubules surrounded by nine peripheral doublets. Connected to each doublet of peripheral microtubules is an inner and outer dynein arm, consisting of multiple dynein chains that provide ATPase-mediated energy for movement.2 Dynein arms are preassembled in the cytosol and are transported to an assembly site at the distal end of the growing axoneme. This requires intraflagellar transport (IFT),3, 4, 5 an evolutionary conserved bidirectional transport system that delivers axoneme building blocks6, 7 such as tubulin to the flagellar tip.8
Defective motile cilia underlie the pathophysiology of individuals with impaired mucociliary clearance, which increases susceptibility to respiratory complications, including sinusitis, bronchitis, pneumonia, and otitis media.2 Chronic infections frequently lead to progressive pulmonary damage and bronchiectasis. Spermatozoal dysmotility in affected men causes infertility.2 These clinical features characterize the hallmark disease of motile cilia, primary ciliary dyskinesia (MIM: PS244400); guidelines for the diagnosis of PCD have recently been published.9 Approximately half of all individuals with PCD display laterality defects varying from partial to complete situs inversus, a consequence of dysfunctional embryonic nodal cilia.2 At least 36 genes currently account for the heterogeneous genetic disorder PCD, which displays mainly autosomal-recessive inheritance and is characterized by cilia dyskinesis and structural defects observed by TEM of a nasal biopsy. However, this is not always the case. Variants in CCNO (MIM: 607752) result in a congenital mucociliary clearance disorder with reduced generation of motile cilia (CILD29 [MIM: 615872]), which is not associated with laterality defects.10 Mutations in DNAH11 (MIM: 603339) result in CILD7 (MIM: 611884) due to an abnormally rapid ciliary beat frequency without a discernible structural defect.11 These data highlight the molecular complexity underlying the formation and function of cilia.2
Here, we report biallelic variants in LRRC56 that result in a disease entity within the group of mucociliary clearance disorders, possibly distinct from PCD. It is associated with bronchiectasis and laterality defects. In humans, defective ciliary function was detected only by HSVA of cultured material. We show that human LRRC56 interacts with the IFT subunit, IFT88. Functional studies using Trypanosoma brucei reveal that LRRC56 is recruited during axoneme construction in an IFT-dependent manner and is required for the addition of dynein arms to the distal segment of the flagellum. Our findings add LRRC56 to an expanding list of genes whose disruption results in an atypical ciliary phenotype and reveal a mechanism whereby disruption of LRRC56 leads to defective IFT-dependent targeting of dynein to cilia and loss of ciliary motility.
Material and Methods
Subject Evaluation
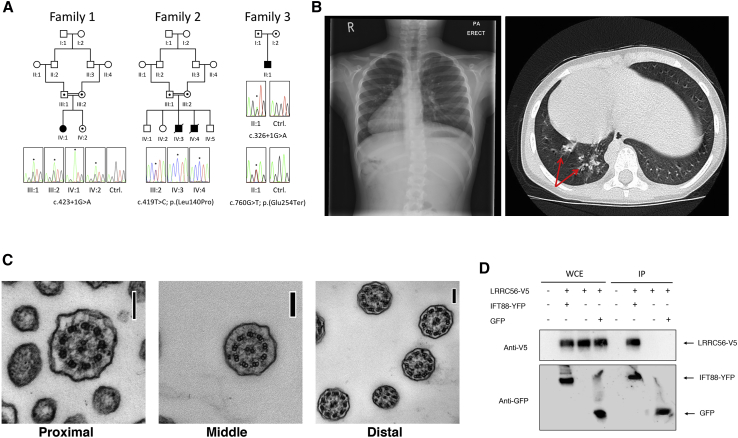
Two unrelated families were independently ascertained with features suggestive of a ciliopathy (Figure 1A). Family 1 consisted of a single female whose parents are first cousins of UK Pakistani ethnicity. She presented with chronic chest infections; nasal biopsy was obtained and respiratory epithelial cultures prepared. These were investigated by transmission electron microscopy (TEM) and high-speed video microscopy. Family 2 consisted of two affected individuals, the offspring of first-cousin consanguineous parents from Kuwait, ascertained during pregnancy to have lethal congenital heart disease. Both pregnancies were terminated and post mortem pathological investigations were performed. The families provided signed informed consent to participate in studies approved by the Leeds East Research Ethics Committee (07/H1306/113; family 1) and the Review Ethics Board of the Children’s Hospital of Eastern Ontario (11/04E; family 2). Review of clinical samples previously investigated by targeted next generation sequencing subsequently identified a further case subject with biallelic variants (family 3).
Figure 1.
LRRC56 Mutations Cause Chronic Infective Lung Disease and Laterality Defects
(A) The homozygous splice-site mutation (c.423+1G>A, GenBank: NM_198075.3) disrupts an invariant splice site in family 1 individual IV:1. The unaffected sibling IV:2 is heterozygous for the mutation. The homozygous missense mutation (c.419T>C, GenBank: NM_198075.3) was identified in the two affected siblings IV:3 and IV:4 from family 2. Consistent with autosomal-recessive inheritance, the mutations described were detected in a heterozygous state in the unaffected parents (data not shown). The third individual (family 3 II:1) was a compound heterozygote for the variants c.760G>T and c.326+1G>A (GenBank: NM_198075.3).
(B) The family 1 proband (IV:1) had dextrocardia, documented by chest X-ray (left). High-resolution axial computed tomography of the thorax in the same individual demonstrates mild bronchiectasis (red arrows) with adjacent inflammatory consolidation in the right lower lobe. Dextrocardia is also visible (CT scan; right).
(C) Cross section through the axoneme from cultured respiratory cells from family 1 individual IV:1. The position of the section is indicated, bar = 100 nm. Normal axonemal structure is visible, with intact dynein arms.
(D) Recombinant human LRRC56 and intraflagellar transport protein IFT88 interact in vitro. HEK293 cells were co-transfected with plasmids encoding human LRRC56 and IFT88 tagged with V5 and YFP, respectively (1.5 μg of each). After 48 hr, immunoprecipitation (IP) was performed with transfected and untransfected cells using Cell-TRAP magnetic beads bound to an anti-GFP antibody fragment. Protein from input whole-cell extracts (WCE, left) and immunoprecipitated proteins (IP, right) were blotted using anti-V5 or anti-GFP. The IFT88-GFP fusion is a fairly large protein and in our conditions, slight differences in migration are frequently observed. This does not impact on the underlying hypothesis being tested. A β-actin control is also shown.
Genetic Analysis
In view of the consanguinity in families 1 and 2, genetic analysis was performed under an autosomal-recessive model. Whole-exome sequencing of genomic DNA was performed at the University of Leeds and the Children’s Hospital of Eastern Ontario. Target enrichment was performed, following manufacturer’s protocols, using SureSelect hybridization capture reagents with v5 and v4 Human All Exon probes for family 1 and 2, respectively (Agilent Technologies). Enriched library preparations were sequenced on either HiSeq 2000/2500 platforms (Illumina) using paired end 100-bp reads. Bioinformatic data processing was performed as previously described, with all variants being reported against human reference genome build hg19.12, 13 Genomic regions corresponding to runs of autozygosity were identified from pipeline-produced variant call format (VCF) files using the tool AgileMultiIdeogram (see Web Resources). These were subsequently used to filter Alamut Batch-annotated variant reports. Additional filtering criteria included the exclusion of common variants (those with a minor allele frequency ≥ 1%) represented in the NHLBI Exome Variant Server (EVS) or an in-house cohort of >1,500 control exomes and the exclusion of genes with biallelic functional variants reported to the Exome Aggregation Consortium (ExAC). Pathogenic variants and segregation in the families were confirmed using Sanger sequencing with an ABI3130xl. Primer sequences and thermocycling conditions are available upon request.
RNA Splicing Assay
Total RNA was extracted from peripheral blood of the affected proband in family 1 using the QIAamp RNA blood mini kit (QIAGEN). A gene-specific primer spanning the boundary of LRRC56 exons 10 and 11 (GenBank: NM_198075.3) (dCTTGGCCAGCACCATGGGTGAG) was used to perform first-strand cDNA synthesis with a SuperScript II RT kit (Life Technologies). Two PCR amplicons were generated from the resulting cDNA using an exon 6 forward primer (dCAACCTGGACCAACTGAAGC) combined with either an exon 8 (dCCTCCAGGGTGAGCATGG) or exon 10 (dCCAGGTCCTCAGAAAGCAGG) reverse primer. PCR products were used to create Illumina-compatible sequencing libraries with NEBNext Ultra reagents (New England Biolabs) and sequenced on an Illumina MiSeq using a paired-end 150 bp read length configuration.
Co-IP Experiments
Human LRRC56 was amplified from an untagged image clone (SC123392, Origene Technologies), inserted into the pDONR201 Gateway cloning vector (Invitrogen) and subsequently pDEST40 destination vector according to the manufacturer’s instructions. The human IFT88-eYFP construct was provided by Professor Colin Johnson.14 Both plasmids were sequenced and maxi-prepped prior to transfection. HEK293 cells were co-transfected with 1.5 μg of each plasmid using Lipofectamine 2000 (Invitrogen). Cells were lysed 48 hr post transfection, using NP40 cell lysis buffer, containing 1× protease and phosphatase inhibitors, and protein extracted as per standard protocols. Immunoprecipitation (IP) of eYFP was performed with 1,000 μg of both transfected and untransfected cell extracts using the GFP-Trap_M kit (Chromotek) according to the manufacturer’s instructions. Whole-cell extracts (WCE) and immunoprecipitates (IP) were blotted using mouse anti-V5 (Invitrogen, 1/2,000) or mouse anti-GFP that detects YFP (Invitrogen, 1/1,000). A secondary goat anti-mouse HRP (Dako) was used at 1/10,000 and detected using the SuperSignal West Femto kit (Thermo Fisher Scientific).
Ciliary Function Measurements
In IV:1, family 1, ciliary beat frequency (CBF) was measured using a digital high-speed video imaging system, as described previously.15, 16 Ciliary beat pattern was investigated in three different planes: a sideways profile, beating directly toward the observer, and from directly above. Dyskinesia was defined as an abnormal beat pattern that included reduced beat amplitude, stiff beat pattern, flickering motion, or a twitching motion. Ciliary beat pattern is associated with specific ultrastructural defects in primary ciliary dyskinesia. Dyskinesia index was calculated as the percentage of dyskinetic cilia within the sample (number of dyskinetic readings/total number of readings for sample × 100). All measurements were taken with the solution temperature between 36.5°C and 37.5°C and the pH between 7.35 and 7.45. Normal ranges for CBF and percentage dyskinetic cells are 8–17 Hz and 4%–49%, respectively, as previously reported.17
Air-Liquid Interface Cell Culture
The modified method we used has previously been described.18 Briefly, nasal brush biopsy samples were grown on collagen (0.1%, Vitrogen)-coated tissue culture trays (12 well) in Bronchial Epithelial Growth Media (BEGM, Lonza) for 2–7 days. The confluent unciliated basal cells were expanded into collagen-coated 80 cm2 flasks and the BEGM was replaced every 2–3 days. The basal cells were then seeded at approximately 1–3 × 105 cells per cm2 on a collagen-coated 12-mm diameter transwell clear insert (Costar, Corning) under BEGM for 2 days. After confluency, the basal cell monolayer was fed on the basolateral side only with ALI-media (50% BEGM and 50% Hi-glucose minimal essential medium containing 100 nM retinoic acid). The media was exchanged every 2–3 days and the apical surface liquid was removed by gentle washing with phosphate-buffered saline when required. When cilia were observed on the cultures, they were physically removed from the transwell insert by gentle scraping with a spatula and washing with 1 mL of HEPES (20 mM)-buffered medium 199 containing penicillin (50 μg/mL), streptomycin (50 μg/mL), and Fungizone (1 μg/mL). The recovered ciliated epithelium was then dissociated by gentle pipetting and 100 μL of the cell suspension was examined inside a chamber slide and ciliary beat frequency and pattern assessed as described above.15, 16 The remaining 900 μL was fixed in 4% glutaraldehyde for transmission electron microscopy (TEM) analysis of cilia and ciliated epithelium.
TEM of Human Samples
TEM was performed as previously described,19 using a Jeol 1200 instrument. For TEM, the ciliated cultures were fixed with glutaraldehyde (4% w/v) in Sorensen’s phosphate buffered (pH 7.4). After post-fixation in osmium tetroxide (1% w/v), samples were dehydrated through graded ethanol series and immersed in hexamethyldisilazane. Sections were cut at 90 nm, with cross-sections categorized for height in the cilium using histological parameters. The bottom (2 μm) of the axoneme cross-sections were located because they were associated with microvilli. The middle (2 μm) cross sections were identified by wide cross-sections with the outer dynein (ODA) away from the axoneme membrane (with no microvilli present). The top 1 μm of the cilia was represented by cross-sections which have a slightly smaller diameter compared to the middle sections. In addition, the axonemal membrane was tightly wrapped to the microtubules and the ODA. The tips (0.6 μm) appear as thin cross sections and have no dynein arms.
Trypanosome Cell Culture
All cloned cell lines used for this work were derivatives of T. brucei strain Lister 427 and were cultured in SDM79 medium supplemented with hemin and 10% fetal calf serum.20 Cell lines IFT88RNAi (targeting an essential protein for anterograde IFT),21 IFT140RNAi (essential protein for retrograde IFT),22 DNAI1RNAi (component of the dynein arm),23 and ODA7RNAi (cytoplasmic assembly machinery of the dynein arm)24 have previously been described. They all express double-stranded RNA under the control of tetracycline-inducible T7 promoters.25, 26
Expression of Endogenous LRRC56 Fused to YFP
Endogenous tagging of the T. brucei Tb427.10.15260 was carried out by direct integration into LRRC56 using the p2675TbLRRC56 plasmid. This vector is derived from the p2675 plasmid and contains a 1,495 bp fragment of the trypanosome LRRC56 sequence (1–1,495) downstream of YFP.27 Transfection was achieved by nucleofection of T. brucei cells using program X-014 of the AMAXA Nucleofector apparatus (Lonza) as previously described,28 with 10 μg plasmid linearized with AfeI in the target LRRC56 sequence, for homologous recombination with the target allele. The mutant allele was obtained following T776/C base substitution (Genecust Europe) in the p2675TbLRRC56 to substitute leucine 259 for proline. The resulting p2675TbLRRC56L259P plasmid was linearized with NheI prior to nucleofection. As a result, the expression of the YFP fluorescent fusion protein is under the control of the endogenous 3′ untranslated region of the LRRC56 resulting in a similar expression level as the wild-type allele since most gene expression regulation is determined by 3′ UTR sequences.29 Transgenic cell lines were obtained following puromycin selection and cloning by serial dilution.
LRRC56 Deletion or Replacement
Sequential LRRC56 replacement in T. brucei was used to obtain lrrc56−/− cells. Sequences containing either the puromycin (PURO) or the blasticidin (BLA) drug resistance gene flanked by 300 bp long upstream and downstream regions of the LRRC56 were synthesized and cloned in a pUC57 plasmid (Genecust Europe). Amplicons were generated by PCR with primers P1: dTTTGAAGGTGCTGTGTGAGGG and P2 dAGGTAGAGGGAGGCGTTGAG (Eurogentec) which anneal to the sequences 300 bp upstream of the LRRC56 start codon and downstream of LRRC56 stop codon, respectively. Prior to nucleofection, PCR fragments containing either the blasticidin (BLA) resistance gene or the G418 neomycin (NEO) resistance gene were purified using Nucleospin gel and PCR clean-up kit (Macherey Nagel). Single-allele knockout cells were obtained following nucleofection with the BLA amplicon containing LRRC56 flanking sequences, blasticidin selection, and cloning. LRRC56 allele deletion was confirmed using PCR with 5′ UTR LRRC56-specific primer P3 dTCACCATCACGCCCTTTTGT and BLA-specific primer P4 dCTGGCGACGCTGTAGTCTT. Replacement of the remaining LRRC56 allele was performed with either the YFP::LRRC56 or the mutated YFP::LRRC56L259P in this single knockout cell line using linearized p2675TbLRRC56 or p2675TbLRRC56L259P plasmid and puromycin selection as described above. Double LRRC56 knockout was achieved following nucleofection of the cell line with a single allele knockout carrying the mutated YFP::LRRC56L259P with NEO amplicon containing LRRC56 flanking sequences and validation obtained following PCR analysis of G418-resistant cells with LRRC56-specific primers P5 dCCGTAGCATCATCCGAGACC and P6 dACTATTTGCGTCAGGTGGCA. Primers amplifying a 1,511-bp region of the unrelated aquaporin 1 gene served as positive controls. For whole-genome sequencing, genomic DNA was extracted using the QIAGEN DNeasy kit. Short insert Illumina sequencing libraries were constructed and sequenced at the Beijing Genomics Institute, generating approximately 6.6 million 100-bp paired end reads. Reads were aligned to the T. brucei TREU927 genome (TriTrypDB release 35)30 using bowtie2 in very-sensitive-local alignment mode, with an 88.6% alignment rate.30, 31 Alignment files were sorted, merged, and indexed using SAMtools.32 Aligned reads were visualized and analyzed, including counting reads per CDS using the Artemis pathogen sequence browser.33
Indirect Immunofluorescence Assay (IFA)
Cultured trypanosomes were washed twice in SDM79 medium without serum and spread directly on poly-L-lysine-coated slides (Thermoscientific, Menzel-Gläser) before fixation. For methanol fixation, cells were air-dried and fixed in methanol at −20°C for 5 min followed by a rehydration step in PBS for 15 min. For paraformaldehyde-methanol fixation, cells were left for 10 min to settle on poly-L-lysine-coated slides. Adhered cells were washed briefly in PBS before being incubated for 5 min at room temperature with a 4% PFA solution in PBS at pH 7 and fixed with methanol at −20°C for an additional 5 min followed by a rehydration step in PBS for 15 min. For immunodetection, slides were incubated for 1 hr at 37°C with the appropriate dilution of the first antibody in 0.1% BSA in PBS; mAb25 recognizes the axonemal protein TbSAXO1;34 a monoclonal antibody against the IFT-B protein IFT172,22 and a mouse polyclonal antiserum against DNAI1.24 YFP::LRRC56 was observed upon fixation by immunofluorescence using a 1:500 dilution of a rabbit anti-GFP antibody that detects YFP (Life Technologies). After several 5 min washes in PBS, species and subclass-specific secondary antibodies coupled to the appropriate fluorochrome (Alexa 488, Cy3 or Cy5, Jackson ImmunoResearch) were diluted 1/400 in PBS containing 0.1% BSA and were applied for 1 hr at 37°C. After washing in PBS as indicated above, cells were stained with a 1 μg/mL solution of the DNA-dye DAPI (Roche) and mounted with Slowfade antifade reagent (Invitrogen). Slides were either stored at −20°C or immediately observed with a DMI4000 microscope (Leica) with a 100× objective (NA 1.4) using a Hamamatsu ORCA-03G camera with an EL6000 (Leica) as light source. Image acquisition was performed using Micro-manager software and images were analyzed using ImageJ (National Institutes of Health)
Motility Analyses
Volumes of 250 μL at 5 × 106 trypanosomes/mL in warm medium were distributed in 24-well plates. Samples were analyzed under 10× objective of a DM IL LED microscope (Leica) coupled to a DFC3000G camera (Leica). Movies were recorded (200 frames, 50 ms of exposure) using LASX Leica software, converted to .avi files, and analyzed with the medeaLAB CASA Tracking v.5.5 software (medea AV GmbH).35
TEM Analysis of Trypanosome Samples
Cells were fixed with 2.5% glutaraldehyde directly in the suspension culture for 10 min and then treated with 4% paraformaldehyde, 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 1 hr. Cells were post-fixed for 30 min (in the dark) in 1% osmium tetroxide (OsO4), in 0.1 M cacodylate buffer (pH 7.2), washed and incubated in 2% uranyl acetate for 1 hr at room temperature. Samples were washed, dehydrated in a series of acetone solutions of ascending concentrations, and embedded in Polybed 812 resin. Cytoskeletons were extracted by treating cells with Nonidet 1% with protease inhibitor cocktail (Sigma, P8340) diluted in PBS for 20 min, and washed in PHEM 0.1 M (pH 7.2) for 10 min. Fixation was performed in 2.5% glutaraldehyde, 4% paraformaldehyde, and 0.5% tannic acid in 0.1 M cacodylate buffer (pH 7.2). Cells were post-fixed, incubated in uranyl acetate, dehydrated, and infiltrated as cited above. Ultrathin sections were stained with uranyl acetate and lead citrate.
Results
Clinical Characterization of Families
Individual IV:1 (family 1) was born at term by normal vaginal delivery, and there was no family history of note. After 36 hr, she developed tachypnoea and was nursed for 48 hr in 40% humidified oxygen environment. She was discharged home on day 5 of life. At age 3 months, she developed rhinorrheoa and experienced several episodes of documented respiratory tract infection, the first at age 5 months. She subsequently developed a chronic cough and recurrent middle ear infections from age 8 months. At age 18 months, she developed pneumonia. Chest X-ray and CT scan revealed bronchiectasis and dextrocardia (Figure 1B). Nasal nitric oxide levels measured under conditions of tidal breathing averaged 6 nL/min. The clinical presentation and investigations suggested a diagnosis of PCD as outlined in the European Respiratory Society guidelines.9 However, TEM analysis of nasal biopsy samples along different segments of the cilia (tip, middle, and base) showed apparently intact dynein arms (Figure 1C). Direct high-speed videomicroscopy analysis of biopsy material revealed a ciliary beat frequency of 10.87 Hz (10.4–11.34 Hz) and particulate clearance was observed (Videos S1 and S2). Although cell culture at an air-liquid interface produced a healthy ciliated epithelium, the ciliary beat pattern was in fact dyskinetic (19% twitching, 33% stiff, 48% static), with a beat frequency of 5.39 Hz (95% CI 4.29–6.49 Hz, normal range > 11 Hz36) (Videos S3 and S4 and control Video S5). TEM revealed no obvious structural defect along the shaft of the cilia. The findings were not compatible with the ERS guidelines on the diagnosis of PCD.9 However, the combination of chronic lung infections, middle ear infection, and cardiac laterality defect suggested an underlying disorder of motile cilia.
Cilia can be seen clearly moving rapidly, which also moved the edge and overlying particles are cleared. This indicated normal ciliated activity. Recorded at 30 fps under 1000x magnification.
Cilia can be seen clearly moving particles on the left hand side of the edge. This indicates that the cilia show normal function. Recorded at 500 fps under 1000x magnification.
Here the cilia seem uncoordinated and no particulate clearance can be seen. This is indicated abnormal cilia movement. Recorded at 30 fps under x1000 magnification.
The cilia can be seen clearly and their movement showed uncoordinated, slow and stiff cilia. This indicated abnormal cilia movement. Recorded at 500fps under x1000 magnification.
Family 2 was from Kuwait and included two affected fetuses. Both pregnancies were terminated because the fetuses were found to have lethal congenital heart disease. Autopsy revealed similar findings in both individuals, which included situs inversus of the thoracic and abdominal organs, with a complex congenital heart malformation characterized by double outlet right ventricle (data not shown). Fetus 1 showed atrial situs inversus. There was a persistent left-sided superior vena cava that, along with the inferior vena cava, drained to the left-sided (morphologically right) atrium. There was also a right-sided superior vena cava that drained to the right-sided (morphologically left) atrium. There was a large dominant right-sided ventricle showing right morphology; no definite, even rudimentary, left ventricle was identified. The main outflow tract of the large dominant ventricle was to the aorta. There was pulmonic hypoplasia. The aortic arch was right sided. Fetus two showed almost the exact same cardiac phenotype except that a hypoplastic left ventricle was identified, probably because there was a high muscular ventricular septal defect (VSD) through which it could decompress. Thus, atrial situs was inversus but ventricular situs was solitus, with atrioventricular discordance. The hypoplastic ventricle had no outflow other than the VSD. In addition, fetus two did not have a right-sided superior vena cava.
The third case consisted of a single individual clinically investigated by NGS. Individual II:1 (family 3) is 27 years of age, the product of a normal pregnancy born to unrelated UK parents. Only brief clinical details are available. He was found to have situs inversus soon after birth and suffered neonatal respiratory disease. During the first year of life, he developed a chronic cough and recurrent lower respiratory tract infections (LRTI), and he also suffered recurrent middle ear disease. These symptoms have persisted into adult life. The combination of laterality defects and recurrent LRTI and middle ear disease led to further investigation, including nasal ciliary biopsies. The first biopsy at age 11 months reported normal ciliary structure, although at that time (1992) no motility studies were performed. Subsequent investigation at age 26 revealed a normal NO estimation (384 ppb), and a further ciliary biopsy was obtained at this age. Investigation revealed normal ciliary beat pattern with good particulate clearance. CBF was in the normal range at 13.4 Hz. EM revealed normal dynein arms and microtubules with no evidence of ciliary disorientation. Cilia length was normal at 5.6 microns. The clinical report concluded that this was not consistent with a diagnosis of PCD. However, the ciliary phenotype was very similar to that seen in family 1. This individual was subsequently investigated by targeted next generation sequencing.
Genetic Analyses Revealed Mutations in LRRC56
Autozygosity mapping identified 46 and 33 regions of homozygosity in each proband from family 1 and 2, respectively (Table S1). In neither family did we identify disease-causing variants in genes with known association to PCD and mucociliary clearance disorder (Table S2). Assessment of variant pathogenicity was evaluated according to ACMG best practice guidelines.37
Variant filtering, using the autozygous intervals, and public/in-house databases (to eliminate variants with minor allele frequency > 1%) identified a single homozygous variant in each family, located in the same gene, LRRC56 (GenBank: NM_198075.3: family 1 c.423+1G>A; family 2 c.419T>C [p.Leu140Pro]). Neither variant is recorded in ExAC. A search of an ethnically matched control exome cohort revealed a single heterozygous carrier of the c.423+1G>A mutation among 1,541 normal subjects. Sanger sequencing confirmed the variants as well as segregation in both families. c.423+1G>A is predicted to abrogate the intron 7 donor site; a high-throughput sequencing analysis of RT-PCR products was designed to quantify the proportion of correctly or incorrectly spliced transcripts as determined by the presence of the exon 8 “GA” nucleotides and consequently confirmed that the LRRC56 variant is aberrantly spliced and not predicted to encode a functional protein (Figure S1). The missense variant c.419T>C (p.Leu140Pro) affects a highly conserved residue (Figure S2), predicted to be deleterious by SIFT (0),38 PolyPhen (1.0),39 and CADD (24.7) scores.40
NGS investigation using a targeted reagent consisting of 32 genes revealed that individual II:1, family 3 was heterozygous for c.760G>T (p.Glu254Ter) and the splicing variant c.326+1G>A. c.760G>T is found at a frequency of 5/140,890 in the GnomAD database; c.326+1G>A is not recorded on GnomAD.
LRRC56 Protein Is Conserved in Eukaryotes with Motile Cilia Constructed by Intraflagellar Transport (IFT)
Human LRRC56 encodes a 542 amino acid protein determined from transcriptomic studies and immunohistochemistry to be expressed in many organs, mostly in testis and pituitary gland,41 although significant number of tags were also detected in other organs including lungs and respiratory epithelial cells (The Human Protein Atlas).41 Interestingly, data from single-cell RNA sequencing indicated that LRRC56 transcription in lungs is restricted to ciliated cells.42 The protein contains five leucine-rich repeat domains conserved across species whose motile cilia are assembled by intraflagellar transport (IFT).43 LRRC56 is the human ortholog of the Chlamydomonas reinhardtii gene oda8, which is thought to play a role in the maturation and/or transport of outer dynein arm (ODA) complexes during flagellum assembly and has a biochemical distribution similar to IFT proteins.43 ODA8 is proposed to function together with two other proteins termed ODA5 and ODA10 to form an accessory complex involved in assembly and transport of ODA during axoneme assembly.43 However, evidence for a physical interaction is lacking, and the exact role of ODA8 remains to be defined. To evaluate a possible association of LRRC56 with the multi-subunit IFT machinery, HEK293 cells were co-transfected with plasmids encoding human LRRC56 tagged with V5 and the reference IFT protein IFT88 fused to eYFP.14 Immunoprecipitation using an anti-GFP antibody co-precipitated both IFT88-eYFP and LRRC56-V5, supporting an association of LRRC56 with the IFT machinery (Figure 1D).
The functional role of LRRC56 was further investigated in T. brucei, an organism that possesses a 9+2 axoneme and is amenable to genetic manipulation.24, 44 The T. brucei LRRC56 ortholog (Tb427.10.15260) comprises 751 amino acids and shares 42% sequence identity in the conserved leucine-rich repeat (LRR) region (Figure S2).
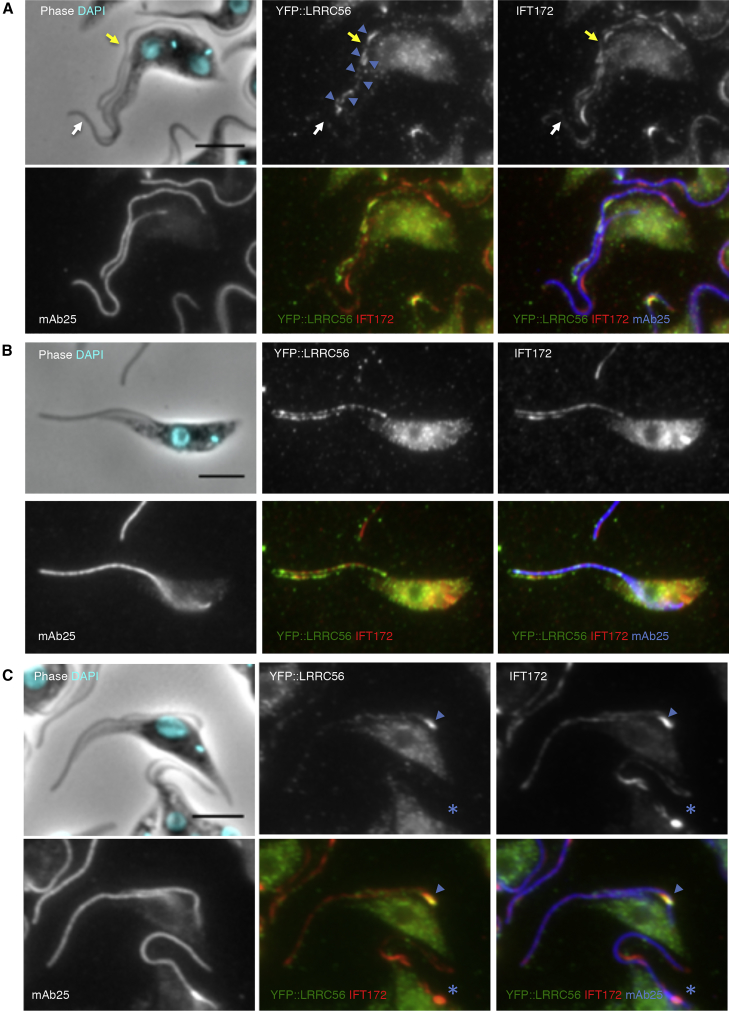
Since T. brucei maintains its mature flagellum during formation of the new one, it is possible to monitor both maintenance and assembly in the same cell.45 YFP-tagged LRRC56 (YFP::LRCC56) was expressed in cells upon endogenous tagging and the protein was detected in the distal portion of the new flagellum, whereas the old flagellum lacked the protein (Figure 2A). Following division, cells with one flagellum displayed two different profiles: either a strong signal toward the distal end of the flagellum (Figure 2B) or no signal (not shown). The first scenario reflects daughter cells that inherited a new flagellum, while the latter reflects those daughter cells that inherited an old flagellum. This observation is consistent with LRRC56 being recruited during flagellar assembly before its removal during flagellum maturation, after cell division. In cells with a single flagellum, the LRRC56 protein is present on two distinct parallel tracks (green signal on merged image) only in the distal flagellar portion of the axoneme (in blue) and colocalize partially with IFT proteins also found on these two tracks but all along the flagellum (red signal on merged image). IFT proteins (red) are clearly detected where no green signal is present.46 Co-localization with the IFT protein IFT172, but not with the axoneme marker mAb25, suggests that LRRC56 associates with IFT and not dynein arms. Many, but not all, IFT trains contained LRRC56 (Figure 2), suggesting that it is a cargo rather than a core component. The LRRC56 signal is lost upon detergent extraction of the cytoskeleton, which strips the membrane and IFT particles but not the dynein arms (data not shown), further supporting association with IFT.
Figure 2.
LRRC56 Associates with IFT Trains and Not with the Axoneme
(A) An YFP::LRRC56-expressing cell that assembles its new flagellum (yellow arrow) shows staining (anti-GFP, green on merged images) in the new flagellum (blue arrowheads) that co-localizes with the anti-IFT172 (red on merged images) but not with the anti-axoneme marker (mAb25, blue on merged images). No YFP staining is visible in the old flagellum (white arrow) whereas IFT172-positive trains are clearly present. IFT trains are predominantly found on microtubules doublets 4 and 7,22 so the visual aspect of two separate tracks around the axoneme. DNA is stained with DAPI (cyan) showing the presence of two kinetoplasts (mitochondrial DNA) and two nuclei typical of cells at late stage of their cycle.45
(B) Cytokinesis results in two daughter cells each containing a unique flagellum.45 The one inheriting the new flagellum remains positive for YFP::LRRC56 that still shows association with IFT trains (same staining as in A).
(C) The same immunofluorescence assay was performed on IFT140RNAi cells expressing YFP::LRRC56 after 24 hr in RNAi conditions. DNA staining shows that the top cell is mitotic and assembles a new flagellum. The IFT172 staining reveals the presence of a stalled IFT train that contains a considerable quantity of YFP::LRRC56 (arrowhead). The bottom cell is at a further stage of its cell cycle, yet its new flagellum is much shorter, indicating that RNAi knockdown occurred at a very early phase of construction (star). In these conditions, IFT172 staining shows that the very short flagellum contains a large amount of accumulated IFT material, yet no signal is visible for YFP::LRRC56 (star).
To further investigate the link with IFT, the distribution of YFP::LRRC56 was studied in trypanosome mutant cell lines in which tetracycline-inducible RNAi knockdown of an IFT-B (IFT88RNAi) or an IFT-A (IFT140RNAi) member results in either defective anterograde or retrograde transport, respectively.21, 22 In IFT88RNAi cells, the flagellar YFP::LRRC56 signal disappeared when anterograde transport was disrupted and the protein was found predominantly in the cytoplasm (Figures S3A and S3B). In the retrograde mutant IFT140RNAi, trains travel into the new flagellum but fail to be recycled to the base.22 The YFP::LRRC56 distribution pattern turned out to be more complex (Figures S3C and S3D) and required more detailed investigation. Cultures are not synchronized and RNAi knockdown can impact cells at different stages of flagellum construction. If this happened when flagella started to grow, IFT proteins accumulated in very short flagella, but LRRC56 was rarely detected (Figure 2C). However, when IFT arrest took place at later stages of elongation, LRRC56 was frequently found in high concentration, always associated with stalled IFT trains (Figure 2C). The presence of such an amount of IFT material in a spatially constrained area is never seen in control cell lines. Formal demonstration of stalled IFT material in the IFT140RNAi cell line has been published using live cell analysis.47 These results reflect the association of LRRC56 as cargo of IFT trains, with a progressive increase during flagellar construction. After 2 days of tetracycline-inducible RNAi knockdown, the LRRC56 is no longer detected in the flagellum (Figures S3C and S3D).
LRRC56 localization was unchanged in DNAI1RNAi and ODA7/DNAAF1RNAi mutants that are defective in their dynein arm constitution or cytoplasmic preassembly, respectively (Figure S4). This shows that LRRC56 does not require the presence of dynein arms to be associated with flagella. Together, these findings reveal that LRRC56 likely performs an IFT-associated function in motile cilia.
Absence or Mutation of T. brucei LRRC56 Is Responsible for Motility Defects Explained by Absence of Outer Dynein Arms in the Distal Segment of the Axoneme
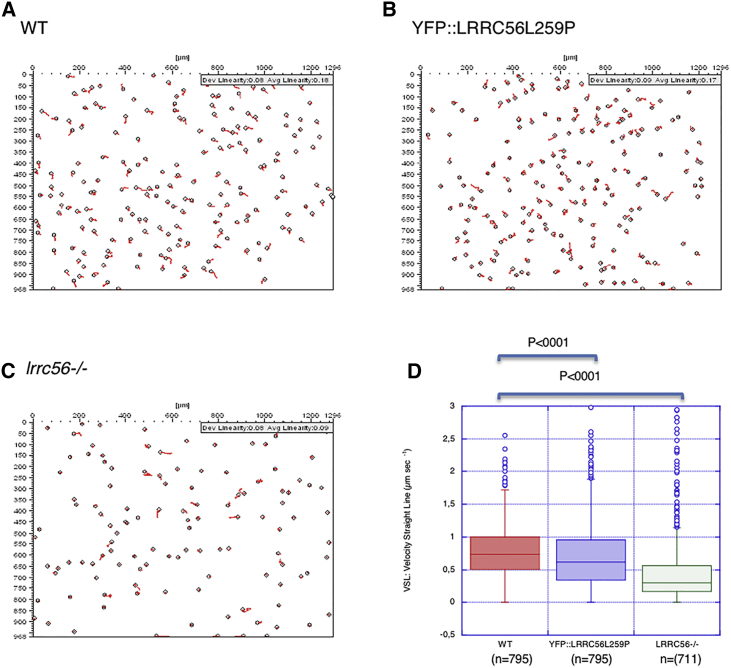
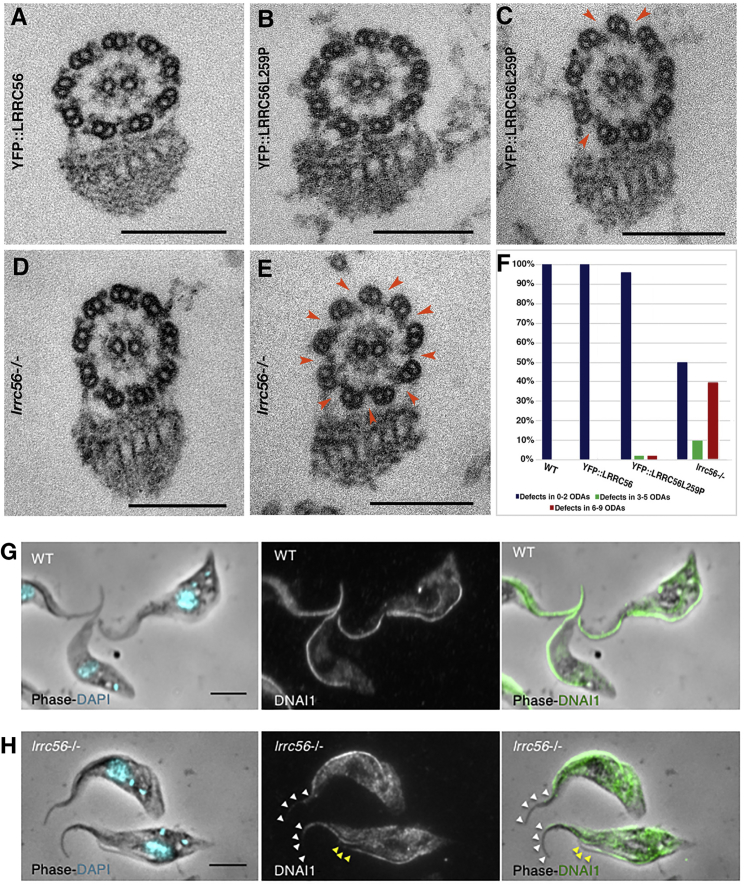
We next chose to assess any impact caused by the absence or mutation of LRRC56. An LRCC56-null mutant was generated by double gene knockout (Figure S5A). Given the small size of the T. brucei genome (35 Mb), whole-genome sequencing was performed and unambiguously demonstrated that LRRC56 had been replaced by the drug-resistance cassettes (Figure S5B). Not a single LRRC56 read was detected whereas hundreds were found in control cells. Visual examination showed a significant reduction in flagellar beating and cell swimming, with ∼10% of cells struggling to complete cell division and remaining attached by their posterior extremities (Figure S5C). These phenotypes indicate slow cytokinesis and increased generation times which, in trypanosomes, is typical of motility defects (Figure S5D).23, 48, 49 Microscopy revealed that lrrc56−/− cell motility is characterized by an erratic swimming pattern with altered propagation of the tip-to-base wave, increased frequency of base-to-tip waves, and frequent tumbling typical of outer dynein arm mutants (Figure 3).23, 50 Examination of the axoneme structure by TEM using cytoskeletons extracted with detergent, a routine procedure to analyze the fine structure of the axoneme51 revealed that more than one third of sections analyzed from lrrc56−/− cells showed absence of 6 to 9 ODAs (Figure 4). However, information on the positioning of ODAs along the length of the flagellum is not available from this assay. Hence a second analysis was performed using whole cell samples directly fixed in suspension.52 This gives lower-quality images for the fine structural details of the axoneme, but the position of the flagellum along the cell body can be determined as described.53 Briefly, sections close to the proximal part of the flagellum are found at the posterior end where the cell body is larger and those close to the distal end are toward the anterior portion of the cell body that is much thinner. The distal tip of the flagellum is not attached and so sections there are easy to identify. A total of 33 sections were obtained in these conditions. When they were in the proximal region, only 20% of the images indicated a defect in outer dynein arms. By contrast, this proportion increased to 75% and 88% when sections were in the distal region or at the tip of the flagellum, respectively. This was found in the distal portions of both old and new flagella. This suggests a role for LRRC56 in the assembly of dynein arms in the distal portion of the axoneme. Immunostaining with an anti-DNAI1 (dynein arm intermediate chain 1 also known as IC78) antibody,23, 24 which recognizes an essential structural component of the ODA, confirmed that only the proximal axonemal region stained positively in lrrc56−/− cells (Figure 4H). Control cells stained from the base to the tip of all axonemes (Figure 4G), confirming that LRRC56 is essential for distal assembly of ODA. Compared to control cells (Figure 4G), LRRC56-null mutant cells also appear to have shorter cilia (Figure 4H). Indeed, a small but significant flagellum length size reduction of about 3.8 μm as determined by IFA using mAb25 antibody was observed (data not shown). Although we don’t have clear explanation for this size reduction, this phenotype is unlikely to impinge upon our conclusion about distal loss of ODA, since a mutant defective for IFT kinesin motors assemble short flagella that still harbor unaltered ODA all along the flagellum.46
Figure 3.
Absence of LRRC56 or Expression of the p.Leu259Pro Variant Reduces Flagellum Beating and Cell Motility
(A–C) Tracking analysis35 showing the movement of individual trypanosomes in wild-type control (A), in cells expressing only YFP::LRRC56L259P (B) and in lrrc56−/− cells (C). Sustained motility is observed only in control conditions.
(D) Quantification of the straight-line movement confirms the visual impression that motility was reduced in a statistically significant manner in YFP::LRRC56L259P cells and almost abolished in lrrc56−/−. Total number of cells, mean, and standard deviation are indicated on the figure. Statistical analysis was performed using t test.
Figure 4.
LRRC56 Is Required for Assembly of ODA in the Distal Portion of the Axoneme
(A–E) Sections are shown through the flagella of detergent-extracted cytoskeletons from various cell lines. Stripping the flagellum membrane and matrix facilitates the analysis of structures.23, 51, 60 Sections through control YFP::LRRC56-expressing cells (A) possess all nine ODA, whereas a mixture of sections with normal profiles or with several missing ODA (orange arrowheads) is encountered in YFP::LRRC56L259P-expressing cells (B and C) or lrrc56−/− cells (D and E).
(F) Sections were grouped in three categories: defects in two ODA or less (blue), in five ODA or less (green), and in six ODA or more (red). The total counted number of sections is 50 for each sample. Full details are given in Table S3.
(G and H) IFA with the anti-DNAI1 antibody stains the whole axoneme of wild-type cells (G) as expected.24 However, the staining was limited to the proximal portion in lrrc56−/− cells in both growing (missing portion shown by yellow arrowheads) and mature (white arrowheads) flagella (H).
We next evaluated in trypanosomes the impact of the p.Leu259Pro substitution which corresponds to the p.Leu140Pro observed in family 2. One LRRC56 allele was deleted and the other allele was replaced endogenously with a p.Leu259Pro modification in YFP-tagged LRRC56 (YFP::LRRC56L259P cells). The YFP::LRRC56L259P protein localizes normally to the distal portion of the new flagellum, showing that this residue is not required for proper expression and localization of LRRC56 (Figure S6). However, cells showed reduced motility (Figure 3B), albeit not to the same extent as the double knockout (Figures 3C and 3D). Ultra-structural analysis of YFP::LRRC56L259P cells by TEM revealed a reduction in the number of ODAs, but that was less frequent compared to lrrc56−/− flagella (Figures 4B, 4C, and 4F). We conclude that the protein carrying the substitution can associate with IFT trains, but functions less efficiently than its normal counterpart in supporting dynein arm assembly.
Discussion
Whole-exome sequencing in two unrelated consanguineous families enabled us to identify homozygous variants in LRRC56. The common clinical phenotype in the two families was cardiac laterality defects, while in family 1 the affected individual also presented with recurrent pulmonary infections, and on investigation was found to have bronchiectasis. This combination of clinical features was suggestive of PCD, but a uniformly dyskinetic beat pattern was not observed. Investigation of cultured nasal epithelial cells from the affected individual in family 1 revealed a dyskinetic ciliary beat pattern, but no structural ciliary defects. We have observed this phenomenon before in our analysis of samples sent for the investigation of PCD (R.A.H., unpublished data).
In family 1, individual IV:1 with a homozygous splicing variant in LRRC56, we confirmed by further analysis that the mutated transcript is aberrantly spliced and is not predicted to encode a functional protein. In family 2, a homozygous missense variant was identified. Functional investigation demonstrated a role for LRRC56 in the assembly of ODA in the protist T. brucei. The affected individual in family 3 displayed a combination of laterality defects, recurrent LRTI, and middle ear disease. The respiratory cellular phenotype observed on his nasal ciliary biopsies recalled that seen in individual IV:1, family 1. This individual was found to be a compound heterozygote for variants predicted to result in absence of functional LRRC56.
We then modeled the effect of a null variant and the homozygous missense variant in this organism. We demonstrate that absence of LRRC56 or the presence of the homozygous missense variant seen in family 2 both result in motility defects, caused by loss of ODAs in the distal segment of the axoneme. This observation is consistent with our interrogation of the role of LRRC56 in this organism.
Previous studies have shown that LRRC56 is found only in species with motile cilia that rely on IFT for axonemal assembly.43 LRRC56 shows considerable species variability; for example, the human protein is only 61% identical with its mouse counterpart, compared with 86% for DNAI1. We have also noted that LRRC56 is more divergent between close species of trypanosomes than IFT proteins or dynein arm components. The reported biochemical distribution of LRRC56 is similar to that of IFT subunits, with about 50% of the protein associated with the membrane and matrix fraction.43 Our pull-down experiments carried out in human cells expressing tagged LRRC56 and IFT88 revealed that LRRC56 interacts with the IFT machinery. In T. brucei, LRRC56 is recruited to the flagellar compartment at advanced stages of construction and co-localizes with many (but not all) IFT trains. This flagellar localization is dependent on IFT, as confirmed by analysis of cell lines defective in either anterograde or retrograde IFT. These results support a model in which LRRC56 associates with IFT trains and may function as a cargo adaptor to transport dynein arms toward the distal end of cilia and flagella.
The absence of LRRC56 has an impact on ciliary motility, as observed here in cultured cells from one affected individual and in T. brucei. This is consistent with the oda8 (LRRC56 homolog)-null mutant in Chlamydomonas, although the impact on the presence of dynein arms was remarkably variable. ODAs are absent throughout the length of the flagellum in Chlamydomonas54 whereas they are missing at only the distal part of the T. brucei flagellum. In both cases, this loss of ODA interferes with proper initiation of flagellum beating, resulting in reduced motility and dyskinetic flagellar beating. In samples from the only person that was available for analysis, sections were analyzed in different segments of the cilia (proximal, intermediate, distal), but dynein arms were not visibly affected.
Although the central core of the protein containing the leucine-rich repeat domains is well conserved, other segments are variable across species, with large and often unique N- and C-terminal extensions. This may translate into a common central function, such as association of LRRC56 with the IFT machinery for dynein arm transport, but could also result in considerable structural variation, potentially supporting species-specific interaction partners. Indeed, the composition of dynein arms (number and type of heavy, intermediate, and light dynein chains) varies between species, as demonstrated by both biochemical and genetic analysis.48 The outer dynein arm composition differs between the Chlamydomonas ODAs that contain three heavy dynein chains chains (α, β, and γ) and the human and trypanosome ODA, which each contain only two heavy chains (α and β).48
In Chlamydomonas, early flagellum growth is very rapid (up to 10 μm per hour)55 and might necessitate a greater contribution from ODA8 to ODA transport compared to the slower growth rate observed in trypanosomes56 and respiratory epithelial cells.57 This could explain the absence of ODA throughout the axoneme compared to only the distal part in trypanosomes. We cannot exclude the possibility that discrete structural defects of dynein arms occur in the individual whose cilia were analyzed. For example, DNAH11 nonsense mutations are associated with subtle dynein arm modifications that can be detected only by advanced TEM imaging and tomography.11 In this regard, approximately 30% of PCD-affected individuals showing ciliary dysfunction have no or very subtle ciliary ultrastructure abnormalities when investigated with standard transmission electron microscopy58
Overall, our findings underline the evolutionary complexity of outer dynein arms assembly and trafficking. We have shown that bi-allelic mutations in LRRC56 are responsible for laterality defect in three unrelated families. We provide evidence of a disorder of mucociliary clearance with accompanying cardiac laterality defects caused by mutations in a gene encoding a protein required for ODA transport, rather than composition or assembly. Our work expands the list of ciliary genes involved in human disorders59 while also providing insight into the role of LRRC56 and cilia biology in human development.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We wish to thank the families reported here for their willingness to participate in these research efforts. We thank the Ultrastructural BioImaging Plateforme for providing access to the TEM equipment. We thank Bruno Louis and Jean-François Papon, Institut National de la Santé et de la Recherche Médicale, U955, Créteil, France, for help in video-microscopy. Research at the Institut Pasteur is funded by an ANR grant (ANR-14-CE35-0009-01), by a French Government Investissement d’Avenir programme, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (ANR-10-LABX-62-IBEID), and by La Fondation pour la Recherche Médicale (Equipe FRM DEQ20150734356). This work was supported, in part, by the Care4Rare Canada Consortium funded by Genome Canada, the Canadian Institutes of Health Research (CIHR), the Ontario Genomics Institute, Ontario Research Fund, Genome Quebec, and Children’s Hospital of Eastern Ontario Foundation and the Canadian Rare Diseases: Models and Mechanisms Network funded by CIHR and Genome Canada. The authors wish to acknowledge the contribution of the high-throughput sequencing platform of the McGill University and Génome Québec Innovation Centre, Montréal, Canada. Research at the University of Leeds was supported by grants MR/M009084/1 and MR/L01629X/1 awarded by the UK Medical Research Council.
Published: November 1, 2018
Footnotes
Supplemental Data include six figures, five videos, and three tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.10.003.
Contributor Information
Philippe Bastin, Email: philippe.bastin@pasteur.fr.
Eamonn G. Sheridan, Email: e.sheridan@leeds.ac.uk.
Web Resources
AgileMultiIdeogram, http://dna.leeds.ac.uk/agile/AgileMultiIdeogram/
Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/
CASA Tracking V.5.5, https://safe.nrao.edu/wiki/bin/view/Software/CASA/WebHome
Clustal Omega, http://www.ebi.ac.uk/Tools/msa/clustalo/
ExAC Browser, http://exac.broadinstitute.org/
ImageJ, https://imagej.nih.gov/ij/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Protein BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins
SAMtools, http://www.htslib.org/
The Human Protein Atlas (LRRC56), https://www.proteinatlas.org/ENSG00000161328-LRRC56/tissue
TriTrypDB: The Kinetoplastid Genomics Resource, http://tritrypdb.org/tritrypdb/
Supplemental Data
The shared region on chromosome 11 that contains LRRC56 is highlighted in bold. Positions reported for the human genome build hg19.
References
- 1.Ralston K.S., Lerner A.G., Diener D.R., Hill K.L. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot. Cell. 2006;5:696–711. doi: 10.1128/EC.5.4.696-711.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchison H.M., Valente E.M. Motile and non-motile cilia in human pathology: from function to phenotypes. J. Pathol. 2017;241:294–309. doi: 10.1002/path.4843. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed N.T., Gao C., Lucker B.F., Cole D.G., Mitchell D.R. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J. Cell Biol. 2008;183:313–322. doi: 10.1083/jcb.200802025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou Y., Qin H., Follit J.A., Pazour G.J., Rosenbaum J.L., Witman G.B. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin H., Diener D.R., Geimer S., Cole D.G., Rosenbaum J.L. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishikawa H., Marshall W.F. Intraflagellar transport and ciliary dynamics. Cold Spring Harb. Perspect. Biol. 2017;9:9. doi: 10.1101/cshperspect.a021998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taschner M., Lorentzen E. The intraflagellar transport machinery. Cold Spring Harb. Perspect. Biol. 2016;8:8. doi: 10.1101/cshperspect.a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craft J.M., Harris J.A., Hyman S., Kner P., Lechtreck K.F. Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J. Cell Biol. 2015;208:223–237. doi: 10.1083/jcb.201409036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas J.S., Barbato A., Collins S.A., Goutaki M., Behan L., Caudri D., Dell S., Eber E., Escudier E., Hirst R.A. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur. Respir. J. 2017;49:1601090. doi: 10.1183/13993003.01090-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallmeier J., Al-Mutairi D.A., Chen C.-T., Loges N.T., Pennekamp P., Menchen T., Ma L., Shamseldin H.E., Olbrich H., Dougherty G.W. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat. Genet. 2014;46:646–651. doi: 10.1038/ng.2961. [DOI] [PubMed] [Google Scholar]
- 11.Dougherty G.W., Loges N.T., Klinkenbusch J.A., Olbrich H., Pennekamp P., Menchen T., Raidt J., Wallmeier J., Werner C., Westermann C. DNAH11 localization in the proximal region of respiratory cilia defines distinct outer dynein arm complexes. Am. J. Respir. Cell Mol. Biol. 2016;55:213–224. doi: 10.1165/rcmb.2015-0353OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaulieu C.L., Majewski J., Schwartzentruber J., Samuels M.E., Fernandez B.A., Bernier F.P., Brudno M., Knoppers B., Marcadier J., Dyment D., FORGE Canada Consortium FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am. J. Hum. Genet. 2014;94:809–817. doi: 10.1016/j.ajhg.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson C.M., Crinnion L.A., Murphy H., Newbould M., Harrison S.M., Lascelles C., Antanaviciute A., Carr I.M., Sheridan E., Bonthron D.T., Smith A. Deficiency of the myogenic factor MyoD causes a perinatally lethal fetal akinesia. J. Med. Genet. 2016;53:264–269. doi: 10.1136/jmedgenet-2015-103620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartill V.L., van de Hoek G., Patel M.P., Little R., Watson C.M., Berry I.R., Shoemark A., Abdelmottaleb D., Parkes E., Bacchelli C. DNAAF1 links heart laterality with the AAA+ ATPase RUVBL1 and ciliary intraflagellar transport. Hum. Mol. Genet. 2018;27:529–545. doi: 10.1093/hmg/ddx422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chilvers M.A., Rutman A., O’Callaghan C. Ciliary beat pattern is associated with specific ultrastructural defects in primary ciliary dyskinesia. J. Allergy Clin. Immunol. 2003;112:518–524. doi: 10.1016/S0091-6749(03)01799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirst R.A., Rutman A., Williams G., O’Callaghan C. Ciliated air-liquid cultures as an aid to diagnostic testing of primary ciliary dyskinesia. Chest. 2010;138:1441–1447. doi: 10.1378/chest.10-0175. [DOI] [PubMed] [Google Scholar]
- 17.Chilvers M.A., O’Callaghan C. Analysis of ciliary beat pattern and beat frequency using digital high speed imaging: comparison with the photomultiplier and photodiode methods. Thorax. 2000;55:314–317. doi: 10.1136/thorax.55.4.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray T.E., Guzman K., Davis C.W., Abdullah L.H., Nettesheim P. Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1996;14:104–112. doi: 10.1165/ajrcmb.14.1.8534481. [DOI] [PubMed] [Google Scholar]
- 19.Hirst R.A., Yesilkaya H., Clitheroe E., Rutman A., Dufty N., Mitchell T.J., O’Callaghan C., Andrew P.W. Sensitivities of human monocytes and epithelial cells to pneumolysin are different. Infect. Immun. 2002;70:1017–1022. doi: 10.1128/IAI.70.2.1017-1022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brun R., Schönenberger Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 21.Kohl L., Robinson D., Bastin P. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 2003;22:5336–5346. doi: 10.1093/emboj/cdg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Absalon S., Blisnick T., Kohl L., Toutirais G., Doré G., Julkowska D., Tavenet A., Bastin P. Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes. Mol. Biol. Cell. 2008;19:929–944. doi: 10.1091/mbc.E07-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Branche C., Kohl L., Toutirais G., Buisson J., Cosson J., Bastin P. Conserved and specific functions of axoneme components in trypanosome motility. J. Cell Sci. 2006;119:3443–3455. doi: 10.1242/jcs.03078. [DOI] [PubMed] [Google Scholar]
- 24.Duquesnoy P., Escudier E., Vincensini L., Freshour J., Bridoux A.-M., Coste A., Deschildre A., de Blic J., Legendre M., Montantin G. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2009;85:890–896. doi: 10.1016/j.ajhg.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z., Morris J.C., Drew M.E., Englund P.T. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- 26.Wirtz E., Leal S., Ochatt C., Cross G.A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 27.Kelly S., Reed J., Kramer S., Ellis L., Webb H., Sunter J., Salje J., Marinsek N., Gull K., Wickstead B., Carrington M. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol. Biochem. Parasitol. 2007;154:103–109. doi: 10.1016/j.molbiopara.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burkard G., Fragoso C.M., Roditi I. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;153:220–223. doi: 10.1016/j.molbiopara.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Clayton C.E. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 31.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 34.Dacheux D., Landrein N., Thonnus M., Gilbert G., Sahin A., Wodrich H., Robinson D.R., Bonhivers M. A MAP6-related protein is present in protozoa and is involved in flagellum motility. PLoS ONE. 2012;7:e31344. doi: 10.1371/journal.pone.0031344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brasseur A., Rotureau B., Vermeersch M., Blisnick T., Salmon D., Bastin P., Pays E., Vanhamme L., Pérez-Morga D. Trypanosoma brucei FKBP12 differentially controls motility and cytokinesis in procyclic and bloodstream forms. Eukaryot. Cell. 2013;12:168–181. doi: 10.1128/EC.00077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stannard W.A., Chilvers M.A., Rutman A.R., Williams C.D., O’Callaghan C. Diagnostic testing of patients suspected of primary ciliary dyskinesia. Am. J. Respir. Crit. Care Med. 2010;181:307–314. doi: 10.1164/rccm.200903-0459OC. [DOI] [PubMed] [Google Scholar]
- 37.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 39.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 42.Treutlein B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., Quake S.R. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desai P.B., Freshour J.R., Mitchell D.R. Chlamydomonas axonemal dynein assembly locus ODA8 encodes a conserved flagellar protein needed for cytoplasmic maturation of outer dynein arm complexes. Cytoskeleton (Hoboken) 2015;72:16–28. doi: 10.1002/cm.21206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincensini L., Blisnick T., Bastin P. 1001 model organisms to study cilia and flagella. Biol. Cell. 2011;103:109–130. doi: 10.1042/BC20100104. [DOI] [PubMed] [Google Scholar]
- 45.Sherwin T., Gull K. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1989;323:573–588. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- 46.Bertiaux E., Mallet A., Fort C., Blisnick T., Bonnefoy S., Jung J., Lemos M., Marco S., Vaughan S., Trepout S. Bidirectional intraflagellar transport is restricted to only two microtubule doublets in the trypanosome flagellum. J. Cell Biol. 2018 doi: 10.1083/jcb.201805030. Published online October 1, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fort C., Bonnefoy S., Kohl L., Bastin P. Intraflagellar transport is required for the maintenance of the trypanosome flagellum composition but not its length. J. Cell Sci. 2016;129:3026–3041. doi: 10.1242/jcs.188227. [DOI] [PubMed] [Google Scholar]
- 48.Ralston K.S., Kabututu Z.P., Melehani J.H., Oberholzer M., Hill K.L. The Trypanosoma brucei flagellum: moving parasites in new directions. Annu. Rev. Microbiol. 2009;63:335–362. doi: 10.1146/annurev.micro.091208.073353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Broadhead R., Dawe H.R., Farr H., Griffiths S., Hart S.R., Portman N., Shaw M.K., Ginger M.L., Gaskell S.J., McKean P.G., Gull K. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 2006;440:224–227. doi: 10.1038/nature04541. [DOI] [PubMed] [Google Scholar]
- 50.Baron D.M., Kabututu Z.P., Hill K.L. Stuck in reverse: loss of LC1 in Trypanosoma brucei disrupts outer dynein arms and leads to reverse flagellar beat and backward movement. J. Cell Sci. 2007;120:1513–1520. doi: 10.1242/jcs.004846. [DOI] [PubMed] [Google Scholar]
- 51.Hughes L.C., Ralston K.S., Hill K.L., Zhou Z.H. Three-dimensional structure of the Trypanosome flagellum suggests that the paraflagellar rod functions as a biomechanical spring. PLoS ONE. 2012;7:e25700. doi: 10.1371/journal.pone.0025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lacomble S., Vaughan S., Gadelha C., Morphew M.K., Shaw M.K., McIntosh J.R., Gull K. Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by electron microscope tomography. J. Cell Sci. 2009;122:1081–1090. doi: 10.1242/jcs.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dawe H.R., Farr H., Portman N., Shaw M.K., Gull K. The Parkin co-regulated gene product, PACRG, is an evolutionarily conserved axonemal protein that functions in outer-doublet microtubule morphogenesis. J. Cell Sci. 2005;118:5421–5430. doi: 10.1242/jcs.02659. [DOI] [PubMed] [Google Scholar]
- 54.Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chylamydomonas reinhardtii. J. Cell Biol. 1988;107:2253–2258. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lefebvre P.A., Rosenbaum J.L. Regulation of the synthesis and assembly of ciliary and flagellar proteins during regeneration. Annu. Rev. Cell Biol. 1986;2:517–546. doi: 10.1146/annurev.cb.02.110186.002505. [DOI] [PubMed] [Google Scholar]
- 56.Bastin P., MacRae T.H., Francis S.B., Matthews K.R., Gull K. Flagellar morphogenesis: protein targeting and assembly in the paraflagellar rod of trypanosomes. Mol. Cell. Biol. 1999;19:8191–8200. doi: 10.1128/mcb.19.12.8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirst R.A., Jackson C.L., Coles J.L., Williams G., Rutman A., Goggin P.M., Adam E.C., Page A., Evans H.J., Lackie P.M. Culture of primary ciliary dyskinesia epithelial cells at air-liquid interface can alter ciliary phenotype but remains a robust and informative diagnostic aid. PLoS ONE. 2014;9:e89675. doi: 10.1371/journal.pone.0089675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shapiro A.J., Leigh M.W. Value of transmission electron microscopy for primary ciliary dyskinesia diagnosis in the era of molecular medicine: Genetic defects with normal and non-diagnostic ciliary ultrastructure. Ultrastruct. Pathol. 2017;41:373–385. doi: 10.1080/01913123.2017.1362088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reiter J.F., Leroux M.R. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 2017;18:533–547. doi: 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishikawa T. Axoneme structure from motile cilia. Cold Spring Harb. Perspect. Biol. 2017;9:9. doi: 10.1101/cshperspect.a028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cilia can be seen clearly moving rapidly, which also moved the edge and overlying particles are cleared. This indicated normal ciliated activity. Recorded at 30 fps under 1000x magnification.
Cilia can be seen clearly moving particles on the left hand side of the edge. This indicates that the cilia show normal function. Recorded at 500 fps under 1000x magnification.
Here the cilia seem uncoordinated and no particulate clearance can be seen. This is indicated abnormal cilia movement. Recorded at 30 fps under x1000 magnification.
The cilia can be seen clearly and their movement showed uncoordinated, slow and stiff cilia. This indicated abnormal cilia movement. Recorded at 500fps under x1000 magnification.
The shared region on chromosome 11 that contains LRRC56 is highlighted in bold. Positions reported for the human genome build hg19.