Abstract
The nuclear factor I (NFI) family of transcription factors play an important role in normal development of multiple organs. Three NFI family members are highly expressed in the brain, and deletions or sequence variants in two of these, NFIA and NFIX, have been associated with intellectual disability (ID) and brain malformations. NFIB, however, has not previously been implicated in human disease. Here, we present a cohort of 18 individuals with mild ID and behavioral issues who are haploinsufficient for NFIB. Ten individuals harbored overlapping microdeletions of the chromosomal 9p23-p22.2 region, ranging in size from 225 kb to 4.3 Mb. Five additional subjects had point sequence variations creating a premature termination codon, and three subjects harbored single-nucleotide variations resulting in an inactive protein as determined using an in vitro reporter assay. All individuals presented with additional variable neurodevelopmental phenotypes, including muscular hypotonia, motor and speech delay, attention deficit disorder, autism spectrum disorder, and behavioral abnormalities. While structural brain anomalies, including dysgenesis of corpus callosum, were variable, individuals most frequently presented with macrocephaly. To determine whether macrocephaly could be a functional consequence of NFIB disruption, we analyzed a cortex-specific Nfib conditional knockout mouse model, which is postnatally viable. Utilizing magnetic resonance imaging and histology, we demonstrate that Nfib conditional knockout mice have enlargement of the cerebral cortex but preservation of overall brain structure and interhemispheric connectivity. Based on our findings, we propose that haploinsufficiency of NFIB causes ID with macrocephaly.
Keywords: intellectual disability, haploinsufficiency, chromosome 9p22.3, chromosome 9p23, NFIB, nuclear factor I, macrocephaly, megalencephaly, developmental delay, agenesis of the corpus callosum
Introduction
The nuclear factor one (NFI) site-specific DNA-binding proteins represent a family of transcription factors important for the development of multiple organ systems including the brain.1, 2, 3, 4, 5, 6, 7, 8, 9 Recent reports have highlighted a role for two NFI family members, NFIA (MIM: 600727) and NFIX (MIM: 164005), in individuals with intellectual disability (ID). Deletions of chromosome 1p32p31 (chromosome 1p32p31 deletion syndrome [MIM: 613735]) as well as deletions or sequence variants of NFIA lead to a phenotype with developmental delay, macrocephaly, ID, dysgenesis of the corpus callosum, ventriculomegaly or congenital hydrocephalus, and craniofacial dysmorphisms.10, 11, 12, 13, 14, 15, 16, 17, 18, 19 Haploinsufficiency of NFIX causes Sotos syndrome 2 or Malan syndrome (MIM: 614753), which is characterized by developmental delay, macrocephaly, ID, postnatal overgrowth, and mild craniofacial anomalies.20, 21, 22, 23, 24, 25, 26 In addition, specific sequence variants affecting the 3′ region of NFIX cause Marshall-Smith syndrome (MSS) (MIM: 602535), a more severe phenotype with severe intellectual disability, progressive dysostosis, respiratory difficulties, and a characteristic facial dysmorphism, and is presumably a result of a dominant-negative mechanism.24, 27, 28, 29
During normal brain development in mice, the expression patterns of Nfia and Nfix overlap with that of another family member, Nfib.30, 31, 32 Knockout mice for any one of the three Nfi genes exhibit comparable brain defects, including dysgenesis of the corpus callosum and enlarged ventricles, which implies a common, but not redundant, function in brain development.2, 5, 9, 33, 34 Considering the similar brain phenotypes of the Nfia, Nfib, and Nfix knockout mice, NFIB (MIM: 600728) is another strong candidate gene for intellectual disability and/or brain abnormalities in humans. To date, only a single 1.6 Mb deletion encompassing NFIB and five other genes has been reported in a subject with developmental delay and agenesis of the corpus callosum.9, 35 Here, we present a detailed clinical characterization of eight individuals with sequence variants in NFIB and ten individuals carrying overlapping microdeletions in the chromosomal region 9p23-p22.2, with the smallest region of overlap solely containing NFIB. Five individuals harbored nonsense mutations in NFIB, while three other individuals had de novo missense mutations within the DNA binding domain that cause a loss of function as demonstrated using a luciferase reporter assay. Furthermore, using a novel Nfib conditional knockout mouse model, we demonstrated that loss of Nfib results in an enlarged cortex, providing a direct link between a haploinsufficiency of NFIB in individuals and macrocephaly.
Subjects and Methods
Study Cohort
All 18 affected individuals were recruited independently through GeneMatcher or a network of collaborating clinicians and geneticists.36 They presented for genetic evaluation due to developmental delay or intellectual disability.37 Subjects’ ages ranged from 3 to 33 years (median 8 years) at the time of assessment. Except for a pair of siblings and two mother-child pairs, all of the cases were sporadic and there were no instances of parental consanguinity. A summary of the clinical features of affected individuals is given in Tables 1 and 2. Facial features of eight individuals are shown in Figure 1, showing mild dysmorphic facial features. Detailed clinical information is provided in the Supplemental Note.
Table 1.
Clinical Features in Individuals with NFIB Haploinsufficiency (Nucleotide Variations)
| Subject | P1 | P2 | P3 | P4 | P5 | P6a | P6b | P7 |
|---|---|---|---|---|---|---|---|---|
| NFIB variant | p.Arg37∗ | p.Arg89∗ | p.Lys114Thr | p.Lys126Glu | p.Leu132Pro | p.Asn254∗ | p.Asn254∗ | p.Ile355Serfs∗48 |
| Inheritance | de novo | father unavailable | father unavailable | de novo | de novo | de novo | maternally inherited | de novo |
| Sex | M | M | M | M | M | F | M | F |
| Age at last examination | 16 y | 7 y | 8 y | 32 y | 7 y | 33 y | 7 y | 3 y |
| Prenatal Growth | ||||||||
| Birth weight (g) (SD) | 2,750 (−1.30 SD; 10th) | 4,000 (+0.89 SD; 81th) | 2,495 (−1.70 SD; 4th) | 3,080 (−0.77 SD; 22th) | 3,280 (−0.45 SD; 33th) | 2,960 (−0.89 SD; 19th) | 3,900 (+0.69 SD; 75th) | 3,020 (−0.77 SD; 22th) |
| Body length at birth (cm) (SD) | ND | ND | ND | 50 (−0.06 SD; 48th) | 52 (+0.70 SD; 76th) | ND | ND | 48 (−0.81 SD; 21th) |
| OFC (cm) (centile) | ND | ND | ND | 39.5 (+2.2 SD; 99th) | 33 (−1.27 SD; 10th) | ND | ND | 37 (+1.22 SD; 89th) |
| Postnatal Growth | ||||||||
| Height (cm) (SD) | 173 (+0.20 SD; 58th) | 124.9 (+0.58 SD; 72th) | 137 (+2.80 SD; > 99th) | 192.5 (+2.25 SD; 99th) | 127 (+0.93 SD; 83th) | ND | ND | 95.8 (+0.28 SD; 61th) |
| Weight (kg) | 56 (−0.42 SD; 34th) | 23 (−0.02 SD; 49th) | 43.6 (+4.17 SD; >99th) | 65 (−0.49 SD; 31th) | 22.4 (−0.25 SD; 40th) | ND | ND | 14.3 (+0.20 SD; 58th) |
| OFC (cm) (SD) | 60.5 (+3.76 SD; >99th) | 57.2 (+3.94 SD; >99th) | 58 (+5.17 SD; >99th) | 63 (+5.52 SD; >99th) | 55 (+2.17 SD; 99th) | ND | ND | 55 (+4.72 SD; >99th) |
| Neurodevelopmental Characteristics | ||||||||
| Muscular hypotonia | ND | Y | N | Y | Y | N | Y | Y |
| Motor delay | Y | N | Y | Y | Y | N | Y | Y |
| Speech delay | Y | Y | Y | Y | Y | Y | Y | Y |
| Intellectual disability | borderline | mild-moderate | mild-moderate | mild-moderate | borderline, developmental coordination disorder | mild | mild | mild |
| Attention deficit | N | Y | N | N | Y | NA | NA | N |
| Behavioral anomalies | short temper at younger age, problems with falling asleep | impulsivity | aggression, hyperactivity, impulsivity, autism | anxiety | autism, anxiety | NA | NA | N |
| Seizures | N | N | N | N | N | N | N | N |
| Neurological deficits | N | N | N | N | headaches | ND | ND | N |
| Brain MRI Findings | ||||||||
| Corpus callosum anomaly | agenesis | mild thinning | N | N | partial hypoplasia | ND | ND | N |
| Ventriculomegaly | ND | N | N | N | N | ND | ND | Y |
| Other | ND | mildly infolded perisylvian regions likely due to thin white matter; mild cerebellar tonsillar ectopia | arachnoid cyst | cyst of septum pellucidum; diffuse cerebral atrophy; dorsal meningocele | ND | ND | ND | two small nodules of gray matter heterotopia along the frontal horns of the lateral ventricles; subtle irregularity and thickening along the posterior perisylvian cortex, right greater than left, which raises the possibility of polymicrogyria |
| Internal Malformations | ||||||||
| Genitourinary system | N | N | N | right cryptorchidism | N | N | N | N |
| Gastrointestinal system | N | N | N | N | N | N | N | N |
| Heart | N | N | N | N | N | N | N | innocent murmur |
| Other malformations | N | N | N | N | N | N | ND | |
Abbreviations: Y, trait present; N, trait absent; ND, not determined; NA, not applicable; ID, intellectual disability; SD, standard deviation; gw, weeks of gestation.
Table 2.
Clinical Features in Indifviduals with NFIB Haploinsufficiency (Deletions)
| Subject | P8a | P8b | P9 | P10a | P10b | P11 | P12 | P13 | P14 | P15 | Summary All Subjects |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Aberration | 9p23p22.3 (14098659_14324147)x1 | 9p23p22.3 (14098659_14324147)x1 | 9p22.3p23 (14102175_14386038)x1 | 9p23p22.3 (13974415_14286259)x1 | 9p23p22.3 (13974415_14286259)x1 | 9p23p22.3 (13106806_14639971)x1 | 9p23p22.3 (13034407_14653394)x1 | 9p23p22.2 (14178768_16619009)x1 | 9p23p22.2 (13563537_18491752)x1 | 9p23p22.2 (13739630_18023839)x1 | |
| Deletion size | 225 kb | 225 kb | 284 kb | 312 kb | 312 kb | 1.5 Mb | 1.6 Mb | 2.4 Mb | 4.9 Mb | 4.3 Mb | |
| Inheritance | familial | familial | de novo | de novo | maternally inherited | de novo | unknown | de novo | de novo | de novo | |
| Sex | F | F | M | F | F | M | M | F | M | F | |
| Age at last examination | 6 y | 8 y | 7 y | 20 y | 3 y 5 m | 8 y 4 m | 21 y | 7 y 10 m | 10 y | 17 y | |
| Prenatal Growth >2 SDS | |||||||||||
| Birth weight (g) (SD) | ND | 3,470 (−0.1 SD; 54th) | 3,500 (−0.10 SD; 46th) | 4,400 (+2.13 SD; 98th) | 3,400 (−0.05 SD;48th) | 2,750 (−1.30 SD; 10th) | ND | 4,030 (+1.32 SD; 91th) | 3,200 (−0.58 SD; 28th) | 3,370 (−0.11 SD; 46th) | 1/16 |
| Body length at birth (cm) (SD) | ND | ND | 50 (−0.06 SD; 48th) | 58 (+3.1 SD; > 99th) | ND | 49 (−0.44 SD; 33th) | ND | 56 (+2.38 SD; 99th) | 49 (−0.44 SD; 33th) | ND | 2/7 |
| OFC (cm) (centile) | ND | ND | 37 (+0.68 SD; 75th) | 39 (+1.92 SD; 97th) | 37 (+1.22SD; 89th) | 34 (−0.83 SD; 20th) | ND | 37 (+1.22 SD; 89th) | ND | ND | 1/8 |
| Postnatal Growth >2 SDS | |||||||||||
| Height (cm) (SD) | 122.6 (+1.46 SD; 93th) | 131 (+0.56 SD; 71th) | 131 (+1.70 SD; 96th) | 165 (+0.27 SD; 61th) | 92 (−0.68 SD; 25th) | 131.5 (+0.62 SD; 73th) | 177.8 (−0.17 SD; 57th) | 137 (+2.68 SD; >99th) | 144 (+0.80 SD; 79th) | (75 centile) | 3/16 |
| Weight (kg) | 26.2 (+1.32 SD; 91th) | 28.9 (+0.49 SD; 69th) | 22 (−0.41 SD; 34th) | 68.5 (+0.68 SD; 75th) | 18.9 (+2.36 SD; 99th) | 25.5 (−0.04 SD; 48th) | ND | 43.9 (+3.82 SD; >99th) | 41 (+0.99 SD; 84th) | (50–75 centile) | 3/15 |
| OFC (cm) (SD) | 55.6 (+3.35 SD; >99th) | 55.2 (+2.71 SD; >99th) | 55 (+2.17 SD; 99th) | 61 (+6.01; >99th) | 55.5 (+4.60 SD; >99th) | 52.5 (+0.15 SD; 56th) | 61.5 (+4.46 SD; >99th) | 57 (+4.39 SD; >99th) | 54.5 (+1.19 SD; 88th) | (50–75 centile) | 13/16 |
| Neurodevelopmental Characteristics | |||||||||||
| Muscular hypotonia | N | N | Y | Y | Y | N | N | Y | Y | Y | 11/17 |
| Motor delay | ND | ND | Y | Y | Y | N | Y | N | Y | N | 11/16 |
| Speech delay | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 18/18 |
| Intellectual disability | learning disability (not formally tested) | borderline mild ID | learning disability | mild | mild | mild | learning disability | mild | mild | learning disability | 18/18 |
| Attention deficit | Y | Y | Y | Y | NA | Y | Y | Y | Y | Y | 11/15 |
| Behavioral anomalies | reactive attachment disorder, passive, pleasing | autistic features, social difficulties | N | short temper | NA | anxiety, hetero-agressivity | ASD, impulse control problems, trouble gauging emotions | Y | ASD and psychotic episodes | Y | 13/15 |
| Seizures | N | N | N | N | N | N | N | N | N | N | 0/18 |
| Neurological deficits | N | N | N | ND | ND | N | insensitivity to pain, temperature dysregulation | N | N | N | 2/14 |
| Brain MRI Findings | 9/11 | ||||||||||
| Corpus callosum anomaly | ND | N | N | ND | ND | ND | complete agenesis | complete agenesis | N | ND | 5/11 |
| Ventriculomegaly | ND | N | N | ND | ND | ND | Y | N | N | ND | 2/11 |
| Other | ND | slight asymmetric hemispheres | NA | ND | ND | ND | moderately decreased white matter volume; bilateral probst bundles | mildely decreased white matter volume | NA | ND | 7/11 |
| Internal Malformations | 7/17 | ||||||||||
| Genitourinary system | N | N | N | N | N | ND | N | N | cryptorchidism | N | 2/17 |
| Gastrointestinal system | N | N | N | N | N | ND | N | N | N | N | 0/17 |
| Heart | small VSD | narrow pulmonary artery, normalized at 2 y | N | N | PDA | ND | N | N | N | bicuspid aortic valve | 5/17 |
| Other malformations | N | N | N | N | N | NA | N | N | N | N | 0/17 |
Abbreviations: Y, trait present; N, trait absent; ND, not determined; NA, not applicable; ID, intellectual disability; SD, standard deviation; gw, weeks of gestation.
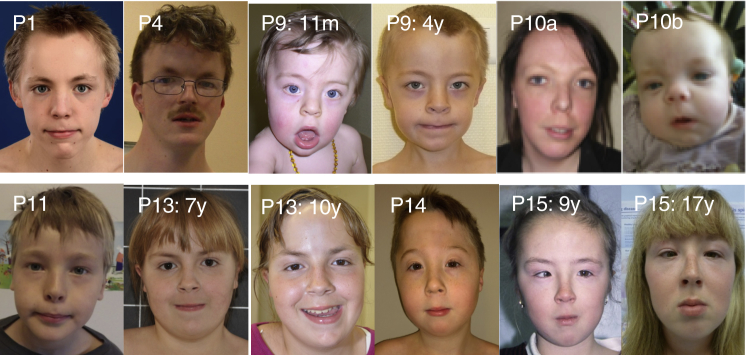
Figure 1.
Clinical Features of Individuals with NFIB Haploinsufficiency
Craniofacial phenotype of individuals P1, P4, P9, P10a, P10b, P11, P13, P14, and P15. The pictures show individual P9 at the age of 11 months and at the age of 4. Individual P13 is represented at the ages of 7 and 10 years, while P15 is represented at the ages of 9 and 17 years. Note the long face with the high forehead, sparse eyebrows, down-slanting palpebral fissures and mild blepharophimosis, a narrow nasal bridge, anteverted nares, and a long and smooth philtrum.
All genetic studies were done on genomic DNA extracted from blood samples. Array CGH/microarray-based molecular karyotyping was generally done on a clinical basis with informed consent given by the patient, parents, or a legal guardian according to the regulations of the respective countries. Whole-exome sequencing (WES) was performed in part of the cohort on a clinical basis with appropriate consent according to national regulations and in the other individuals on a research basis. All procedures that were done in a research environment had been approved by the responsible institutional committee on human experimentation. Moreover, signed consent for scientific evaluation and publication of genetic and clinical data (including photograph) was given by each participating individual or their legal guardians.
Molecular Karyotyping and Method of Confirmation
Molecular karyotyping was performed in subjects 8a, 9, and 13 using an Affymetrix genome-wide human SNP 6.0 array and in subject 8b using an Affymetrix genome-wide human SNP CytoScan HD array (Affymetrix, part of Thermo Fisher Scientific). Molecular karyotyping for subject 10a was carried out using a CytoSure ISCA v2 8x60k array and in subject 15 using a CytoSure ISCA v2 180K array (Oxford Gene Technology). In subject 11 molecular karyotyping was performed using the Agilent genome-wide SNP 60K array (Agilent Technologies). Molecular karyotyping in subject 12 was performed by the Illumina Infinium II HumanHap610 BeadChip (Illumina) and in subject 14 using the BlueGnome CytoChip (v1.1) BAC array (BlueGnome).
Experimental procedures were performed according to the manufacturer’s instructions. Image data of the Affymetrix array were analyzed with the Affymetrix Genotyping console 3.0.1 and 4.1 and the Chromosome Analysis Suite v1.2 and v3.0. Image data of the OGT array were analyzed with the CytoSure Interpret software and of the BlueGnome array with the BlueGnome BlueFuse analyzing-software. For the Illumina array, analysis and CNV identification were done using the PennCNV software.
Results were interpreted using external and internal data resources. External resources include the Database of Genomic Variants (DGV),38 DECIPHER,39 ECARUCA,40 ClinVar,41 and OMIM.
The deletions in individuals 8a, 8b, 10a, 10b, and 13 were confirmed by multiplex-ligation dependent probe amplification (MLPA) using self-designed probes in exons 2, 4, and 11 of NFIB transcript ENST00000380953.5 (GenBank: NM_001190737.1) and in exon 1 of NFIB transcript ENST0000390934.8 (alternative first exon; GenBank: NM_001190738.1) (Figure S1). In subject 9 validation was performed by FISH analyses with a mix of locus-specific probes. The amplification of the sequence overlapping the deletion breakpoints in the subject was achieved by long-range PCR. In individuals 11 and 12 validation was performed by qPCR.
Sequencing and Filtering of Variants
Whole-exome capture and sequencing were performed in subjects 1, 2, 3, and 4 (singletons), 6a/6b (child-parent pair), and 5 and 7 (child-parent trios) using the SeqCap EZ MedExome (Roche NimbleGen), Agilent SureSelect Human All Exon V4/V5, Agilent Clinical Research Exome Kit (Agilent Technologies), or Nextera Rapid Capture Exome (Illumina). Libraries were sequenced on an Illumina HiSeq (2000, 2500, or 4000) according to the manufacture’s recommendation for 75 bp, 100 bp, or 125 bp paired-end reads, respectively. Sequence reads were aligned to the human reference genome (hg19) using different versions of the Burrows-Wheeler Alignment software (BWA v.0.5.8-v.0.6.2 to BWA-Mem v.0.7.5-8). The Genome Analysis toolkit software package (GATK v.1.6.9 to v.2.6-4) or SAMtools v.0.1.18.10 were used for base quality score recalibration, indel realignment, and variant discovery (both single-nucleotide variants and indels). Common variants (defined as variants with >1% frequency in dbSNP, 1000 Genomes Browser, NHLBI Exome Sequencing Project Exome Variant Server, and/or the Exome Aggregation Consortium ExAC Browser) were excluded. De novo, homozygous, compound heterozygous, heterozygous, or X-linked variants (on the basis of inheritance pattern based on the family structure and reported phenotype) present in exons or at exon/intron boundaries (±6 nt in the intron) were examined. In silico analysis of the sequence variants was performed using the open access software PredictSNP2,42 Mutation Taster,43 PROVEAN/SIFT,44 and PolyPhen-2.45 Sequence alterations are reported according to the Human Genome Variation Society (HGVS) nomenclature guidelines. All relevant variants identified by NGS were validated by conventional Sanger sequencing in forward and reverse direction (Figure S1).
Generation and Validation of Mutant NFIB Expression Constructs for Luciferase Assays
The NFIB missense single-nucleotide variations identified in individuals with ID were cloned into the pCAG-Nfib-IRES-GFP plasmid containing a HA-tagged mouse Nfib coding sequence (CCDS38790.1).46 The mouse NFIB protein is 99% identical to the 420 aa human protein encoded by isoform 3 (GenBank: NM_005596.3/CCDS6474.1), with only two amino acid substitutions that were not located near any of the mutated sites. Primers recognizing the plasmid backbone (Table S1) were used in combination with reciprocal primers containing the missense variant to generate two fragments that were amplified using the Phusion High-Fidelity DNA Polymerase (New England Biolabs) and pCAG-Nfib-IRES-GFP as a template. The two fragments were annealed and then amplified using plasmid-specific primers to generate a full-length Nfib insert containing the missense mutation. Mutant Nfib inserts were digested with NotI (New England Biolabs), ligated into the NotI-digested pCAG-Nfib-IRES-GFP backbone, and verified by Sanger sequencing. Protein size and expression were validated by western blots of transfected cells.
Dual-Luciferase Reporter Assays
Neuro-2A47 or U25148 cells were seeded into a 96- or 48-well plate 24 hr prior to transfection at a density of 30%–50% and maintained in DMEM (Sigma-Aldrich) supplemented with 10% v/v fetal bovine serum (SAFC Biosciences, part of Sigma-Aldrich). The pGFAPp-Luc firefly luciferase reporter construct49 was co-transfected with either the empty pCAG-IRES-GFP or the wild-type or mutant pCAG-Nfib-IRES-GFP construct into seeded cells using FuGENE 6 transfection reagent (Promega). The pNL1.1.TK NanoLuc luciferase vector (Promega) was co-transfected with all transfections as an internal control to normalize for transfection efficiency. Luciferase activity was assayed 48 hr after transfection using the Nano-Glo Dual-Luciferase Reporter Assay system (Promega) and the POLARstar OPTIMA plate reader (BMG Labtech, part of Thermo Fisher Scientific). Statistical significance was determined using the Student’s t test.
Mouse Brain Collection and Analyses
All mice were housed and handled in accordance with the National Health and Medical Research Council’s Australian Code of Practice for the Care and Use of Animals for Scientific Purposes and with approval from the University of Queensland Animal Ethics Committee. The FLR-flanked neomycin cassette was removed in the Nfib conditional strain Nfibtm2Rmg to generate Nfibtm2.1Rmg.50 Nfibtm2.1Rmg;Gt(ROSA)26Sortm14(CAG-tdTomato)Hze;Emx1-iCre51, 52 mice, henceforth referred to as Nfibflxdn;tdTom;Emx1iCre, were housed at the Queensland Brain Institute animal facility on a 12 hr dark/light cycle with water and food provided ad libitum. Nfibflxdn;tdTom;Emx1iCre animals were bred to generate progeny that were all homozygous for both Nfibflxdn and tdTom and either negative (wild-type; WT) or positive (conditional knockout; cKO) for Emx1iCre. All animals were genotyped by the Australian Equine Genetics Research Centre (AEGRC; University of Queensland, Brisbane, QLD, Australia).
Animals anesthetized with an intraperitoneal injection of sodium pentobarbitone (Lethabarb; Virbac; 185 mg/kg body weight) were transcardially perfused with saline, followed by 4% w/v paraformaldehyde solution, as previously described.53 After 5–7 days of post-fixation at 4°C–7°C, the brains were dissected from the skull and measured using calipers. Magnetic resonance images were acquired at the Centre for Advance Imaging at the University of Queensland using a 16.4 Tesla vertical bore, small animal MRI system (Bruker Biospin; ParaVision v5.0). Acquisition, brain measurements, and tractography are described in detail in the Supplemental Methods. In short, brain orientation was standardized between mice, and the lengths and cross-sectional areas of specific brain structures were measured in either the mid-sagittal plane or the coronal plane at Bregma level −2.18 mm. All analyses were normalized to total brain height (dorso-ventral size) or length (rostro-caudal size) to account for size variations between individual mice. Probabilistic tractography was performed to detect cortical connectivity and topographic organization of the corpus callosum.54, 55 The structural MR image for each mouse brain was divided into 20 segments via the multi-atlas segmentation tool provided by Advanced Normalization Tools (v 2.2.0, stnava). Nine hand parcellated mouse brain atlases from the Magnetic Resonance Microimaging Neurological Atlas formed the basis of the multi-atlas segmentation.56 The segmented images were analyzed in MATLAB (v R2015b, MathWorks) to determine the number of voxels within the bounds of each segment and calculate their total volume for each mouse brain. For detailed information, see Supplemental Methods.
For histological analyses, dissected brains were embedded in 3%–3.5% Difco Noble agar (Becton, Dickinson and Company) in distilled water and sectioned coronally at 50 μm on a vibratome (Leica). The sections were then mounted onto SuperfrostPlus slides (Menzel-Gläser) and dried at room temperature until fully adherent. Sections were incubated in 4′,6-diamidino-2-phenylindole (DAPI) (1:1,000; Thermo Fisher Scientific) in 0.2% Triton X-100 in PBS for 5 min. The sections were then washed for 3 × 20 min with PBS and coverslipped using ProLong Gold anti-fade reagent (Thermo Fisher Scientific). Fluorescence imaging was performed with a Metafer VSlide Scanner fitted with a Zeiss Axio Imager Z2 (Metasystems). Images were pseudocolored, cropped, sized, and enhanced for contrast and brightness with Imaris 8.2.1 software (Bitplane), ImageJ (NIH), and Illustrator (Adobe Systems). All relevant measurements were made using ImageJ.
For animals of postnatal day (P) 25 or older, only females were included to exclude sex-based size differences. For each stage, 3–8 cKO and WT littermates were measured. Significance was determined using a one-way ANOVA.
Results
Molecular Karyotyping Identifies Disruption of NFIB as the Common Consequence of 9p23-p22.2 Microdeletions
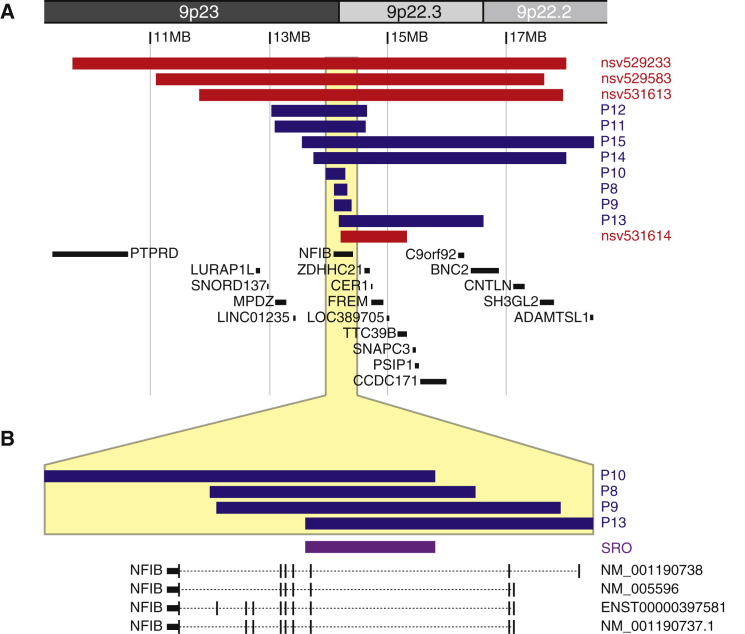
We identified overlapping deletions in the 9p23-p22.2 chromosomal region in ten individuals with mild ID and other behavioral features (Table 2 and Supplemental Note).37 These included two siblings (P8a and 8b), a mother and daughter (P10a and 10b), and six other unrelated individuals (P9, P11–P15) with deletions ranging in size from 225 kb to 4.9 Mb (Figure 2A). Individual 12 was previously reported without a full clinical description.9, 35
Figure 2.
The Smallest Region of Overlap in 9p23-p22 Deletions within NFIB
(A and B) Graphical depiction of the deletions overlapping with NFIB on chromosome 9p23-p22 detected in individuals P8–P15 (blue). In red, four additional cases from the ClinVar database are depicted. The smallest region of overlap, depicted in purple in (B), contains only exon 3 of NFIB. Adapted from UCSC Genome Browser hg19.
The deletions in individuals 10a, 10b, and 13 delineate the smallest region of overlap, comprising a genomic segment of 107 kb (chr9: 14,178,768–14,286,259; hg19) that encompasses only NFIB (Figure 2B). The breakpoints of the deletions are non-recurrent. Three individuals (P8a, 8b, 9) presented with intragenic deletions affecting only NFIB. By MLPA analysis, using self-designed probes, we confirmed that the affected siblings 8a and 8b harbored an intragenic deletion affecting exons 1–10 of the NFIB transcript ENST00000380953.5 (Figure S1). Parental samples were not available to determine whether or not this deletion was inherited. The de novo 284 kb deletion in individual 9 likewise encompassed exons 1 to 10 of NFIB, but the deletion breakpoints differed. In individuals 10a and 10b, the results of MLPA analysis indicated that the proximal deletion breakpoint was located within intron 2, thus mapping the deletion to exons 3 to 11 of NFIB. Individual 10a inherited the deletion, which originated de novo in her affected mother, 10b. In four other simplex case subjects, the deletion was confirmed to be de novo, while parental samples were not available for one individual (P12).
Exome Sequencing Identifies De Novo Missense and Nonsense Variants in NFIB
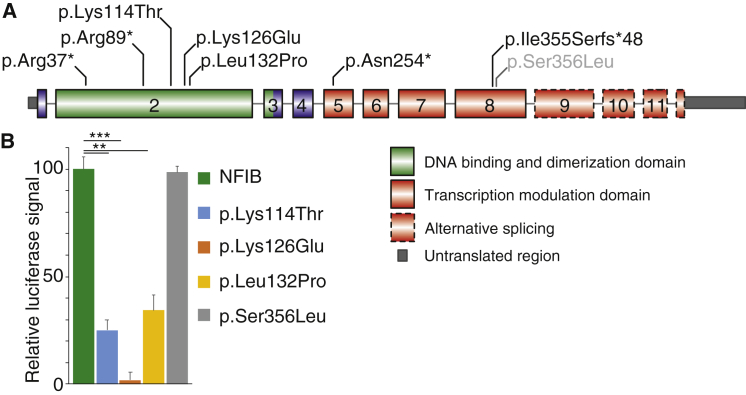
In eight individuals from seven families with ID, who had previous molecular karyotyping with normal results, WES was performed and revealed sequence variants in NFIB (GenBank: NM_001190737.1; Figure 3A). These included four variants predicting premature termination codons: c.109C>T (p.Arg37∗) (P1), c.265C>T (p.Arg89∗) (P2), c.758_759dupTG (p.Asn254∗) (P6a and b), and c.1063_1076del (p.Ile355Serfs∗48) (P7). The other three observed variants were missense changes predicted to affect highly conserved amino acid residues within the DNA-binding and dimerization domain of NFIB: c.341A>C (p.Lys114Thr) (P3), c.376A>G (p.Lys126Glu) (P4), and c.395T>C (p.Leu132Pro) (P5). None of these variants were present in public databases (dbSNP, 1000 Genomes Browser, NHLBI Exome Sequencing Project Exome Variant Server, the Exome Aggregation Consortium ExAC Browser). Parental DNA studies confirmed that variants were de novo in individuals 1, 4, 5, and 7 as well as in individual 6a who transmitted the variant to her similarly affected son, individual 6b. Parental samples were not available for individuals 2 and 3.
Figure 3.
Sequence Varations Identified in NFIB
(A) Schematic representation of the identified sequence variations in NFIB in individuals P1–7 (black) and in the fetal case (gray), based on GenBank: NM_001190737.1 (ENST00000380953.5).
(B) The missense variants found in individuals 3, 4, and 5 were cloned into an NFIB overexpression construct. Using the GFAP-promoter reporter construct in a dual-luciferase assay in U251 and Neuro-2A cells, their ability to induce promoter activity was tested. The data represent one of these experiments with the relative luciferase signal increase to the vector control normalized to wild-type NFIB. All three missense variants—p.Lys114Thr, p.Lys126Glu, and p.Leu132Pro—resulted in significantly less promoter activity, while another missense variant (p.Ser356Leu) did not alter activity. The data depicted are representative of three independent experiments, each consisting of 4–6 technical replicates. Error bars represent standard error and the significance was determined by one-way ANOVA. ∗∗∗p < 0.0005, ∗∗p < 0.005.
Additionally, targeted exome sequencing detected the NFIB variant c.1067C>T (p.Ser356Leu) in a 29-week fetus with hypoplasia of the corpus callosum and pulmonary sequestration.57 This variant was also present in the mother, who had no physical abnormalities and normal intellect (Supplemental Note).
To predict the impact of these variants on NFIB function, we performed in silico analyses. Based on PredictSNP2, the variants were classified as damaging (Table S2).42 For the four variants that introduce premature stop codons, a truncated protein is predicted, which removes (part of) the transactivation domain. In addition, these variants might contribute to mRNA clearing via nonsense-mediated RNA decay, as shown previously for NFIX.29 Notably, a C-terminally truncated protein with strongly reduced transcriptional ability was previously reported in humans.58 The three observed missense variants were consistently predicted to affect protein function by all employed prediction programs.
Functional Analyses Reveal Loss of Function of De Novo Missense Variants in NFIB
To further investigate the functional impact of the missense variants p.Lys114Thr, p.Lys126Glu, p.Leu132Pro, and p.Ser356Leu, these sequence variants were cloned into an NFIB expression construct.46, 59 We then analyzed their ability to activate the human GFAP promoter,49 a well-established transcriptional target of NFIB, to determine whether mutant NFIB displays a disrupted function.5, 7, 9, 60 In a luciferase reporter assay in which luciferase is driven from the GFAP promoter, wild-type NFIB increased luciferase activation, as previously published (Figure 3B).9 When the three mutated proteins were tested in this assay, we observed that all three resulted in a significant reduction in luciferase activity compared to the wild-type protein. In contrast, the missense variant p.Ser356Leu displayed luciferase activity similar to the wild-type protein. Based on these results, we confirmed that the p.Lys114Thr, p.Lys126Glu, and p.Leu132Pro missense variants confer loss of function, while the p.Ser356Leu variant showed no functional impact in this assay, consistent with the assumption that this maternally transmitted variant is likely non-pathogenic.
Nfib Deletion Results in Increased Cortical Size
In addition to mild ID, macrocephaly was observed in all individuals with NFIB sequence variants (P1–P7) and in individuals with haploinsufficiency due to deletions encompassing just NFIB (P8a–P10b; Table 2). In Nfib knockout mouse embryos, the perturbed development of the dorsal telencephalon has been shown to be associated with increased generation of progenitor cells.5, 7, 8, 9, 61 This increase in progenitors may directly impact the overall size of the cortex and therefore head size. However, this has not been confirmed previously, because Nfib deletion in mice is perinatally lethal due to defects in lung maturation.7
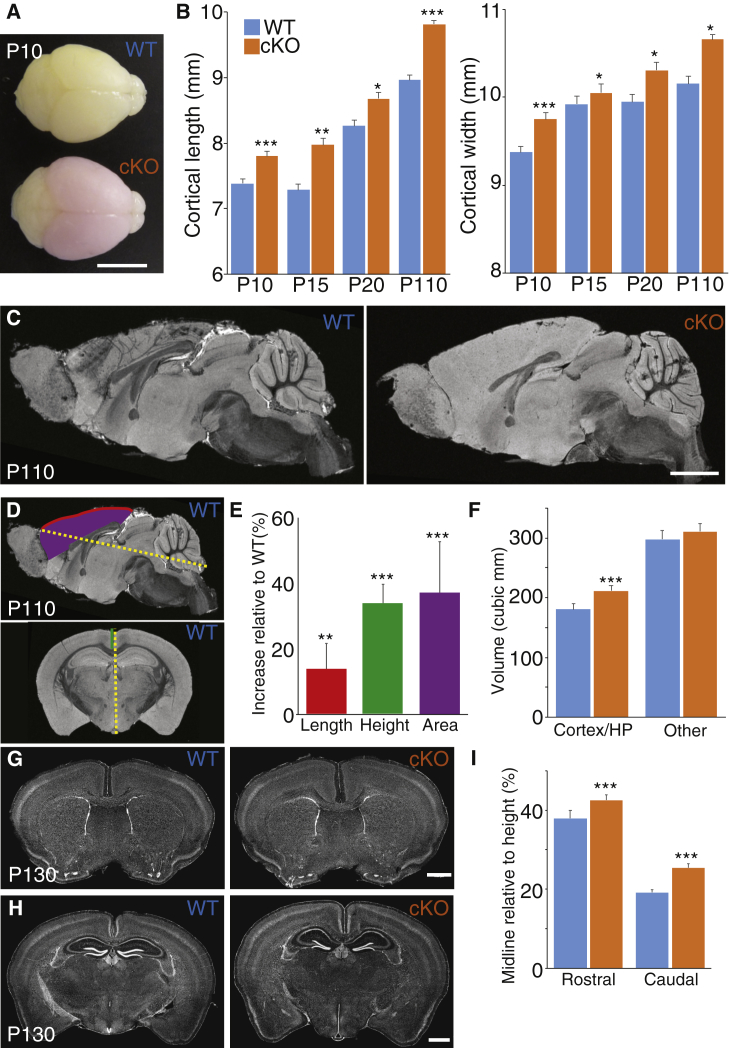
To determine whether disruption of NFIB expression increases cortical size, we generated a dorsal telencephalon-specific Nfib conditional knockout model, Nfibflxdn;tdTom;Emx1iCre, including a red fluorescent marker protein to indicate regions of Cre-recombination and Nfib deletion (Figure 4A).50, 51, 52 Analyses of the size of the cortex of these mice at postnatal day 10, 15, 25, and 110 demonstrate that conditional knockout of Nfib in Cre-positive mice (cKO) resulted in a significant increase in cortical length and width (medio-lateral size) compared to their Cre-negative littermates (WT) at all ages (Figure 4B; one-way ANOVA).
Figure 4.
Cortical-Specific Knockout of Nfib Results in an Enlargement of the Cortex
(A) Representative brains of homozygous Nfib conditional littermates with Cre (conditional knockout; cKO) and without Cre (wild-type; WT) at postnatal day (P) 10. The cKO brain expresses tdTomato red fluorescent protein as a reporter for Cre-mediated Nfib knockout. Scale bar is 5 mm.
(B) Based on measurements of dissected brains from cKO and WT littermates, the cortical length and width were significantly increased in cKO compared to WT animals at P10, P15, P25, and P110. Error bars represent standard error and the significance was determined by a one-way ANOVA. P10, n = 8 for both groups; P15, n = 5 and 6; P25, n = 3 and 5; P110, n = 8 and 4 for WT and cKO, respectively. ∗∗∗p < 0.0005, ∗∗p < 0.005, ∗p < 0.05.
(C) Fast low-angle shot (FLASH) MRI images of the same P110 animals measured in (B) revealed no major changes of the overall brain structure of cKO brains compared to WT littermates. Scale bar is 2 mm.
(D and E) The overall length (red line) and surface area (purple) of the cortex normalized to the overall brain length (yellow) was increased. Similarly, the height of the cortex at the midline (green line in coronal section), as measured from the corpus callosum to the caudal tip of the brain and normalized to the total height (yellow line in coronal section), was also increased. Significance was determined by Welch’s corrected t test. n = 7 and 8. ∗∗∗p < 0.0005, ∗∗p < 0.005.
(F) Based on MRI, the volumes of the cortex and hippocampus (HP) increased more than 16%, while the volume of the other brain structures only slightly increased. Significance was determined by Welch’s corrected t test. n = 7 and 8. ∗∗∗p < 0.0005.
(G and H) Histological analyses of independent brains at P130 confirmed no major structural changes, including in cortical lamination. Scale bar is 1 mm.
(I) Similar to the measurements in MRI images, the cortical height at the midline was increased. Significance was determined by Welch’s corrected t test. P10, n = 4 and 6. ∗∗∗p < 0.0005. All error bars represent standard deviation.
To further determine whether Nfib deletion altered the overall structure of the cortex, fast low-angle shot (FLASH; Figure S2) and diffusion-weighted spin-echo magnetic resonance imaging (dMRI; Figures 4C and 4D)54, 55 as well as histological analyses (Figures 4G and 4H) were performed. No major structural changes were observed in the brain. Specific measurements on brain structures in MRI images and histological sections did reveal that the corpus callosum was slightly shorter and thinner in Nfib cKO animals (Figures 4C, 4G, and 4H), although these findings were not statistically significant.
The cortex was expanded in length (rostro-caudal dimension; Figure 4E) and height, when measuring from the corpus callosum to the caudal tip (Figures 4E and 4I). However, the overall radial thickness of the cortex and individual cortical layers were not altered compared to wild-type littermates. This suggests that the increased cortical size is mainly due to a lateral expansion, which is in line with the increased number of radial glia observed in Nfib knockout mice during early cortical development.7, 34, 46
Based on MRI, the overall brain volume of the cKO animals was almost 10% larger than that of their Cre-negative littermates. After segmentation of the brain in different regions,56 the cortical and hippocampal volume contributed 70% of the total increase in volume (Figure 4F), while the increase of all other brain structures, including the cerebellum and thalamus, was not significant. Hence, these data suggest that the increased head circumference and macrocephaly identified in individuals with NFIB haploinsufficiency may correspond to megalencephaly secondary to lateral expansion of the cortex during fetal development.
Discussion
Here, we report overlapping heterozygous microdeletions in the chromosomal region 9p23-p22.2 and sequence variants affecting NFIB in individuals with a neurodevelopmental phenotype characterized by ID, macrocephaly, and other features. The pathogenic significance is supported by the confirmation of de novo occurrence in 9 out of 11 simplex case subjects, where parental samples were available. In three familial case subjects, the NFIB deletion or sequence variant segregated with the phenotype. Individual 10b inherited the deletion from her similarly affected mother, 10a, in which the deletion had occurred de novo. Similarly, individual 6b inherited the sequence variant from his affected mother. The affected siblings (subjects 8a and 8b) also likely inherited the deletion from an affected parent, but clinical details on the biological parents were insufficient and DNA samples were unavailable.
The entire region involved in the various 9p23-p22.2 deletions presented here encompasses 16 genes (chr9:13,106,806–18,491,752; Figure 2A), of which haploinsufficiency is predicted for five (NFIB, ZDHHC21, PSIP1, BNC2, SH3GL2).62 However, since the smallest region of overlap contained only exon 3 of NFIB and because microdeletions in individuals 8a, 8b, and 9 are intragenic deletions of NFIB (Figure 2B), NFIB is the only gene shared between the case subjects. The pLI score (probability of LoF intolerance) for NFIB is 0.99, indicating a very high likelihood for haploinsufficiency of this gene.63
The role of NFIB haploinsufficiency as the causative event is further supported by the identification of seven sequence variants of NFIB in six simplex and one familial case subject with similar phenotypes. Four of the observed variants create premature termination codons in exons 2, 5, and 8, respectively, thus plausibly suggesting loss of function of the mutant allele. The three missense sequence variations, p.Lys114Thr, p.Lys126Glu, and p.Leu132Pro, were predicted as also likely damaging mutations in silico (Table S2), and we were able to confirm the impaired function of mutant proteins in vitro by demonstrating a severe reduction of transcriptional activity compared to wild-type NFIB. In contrast, the missense variant p.Ser356Leu, detected by WES in a fetus with brain anomalies but also in the unaffected mother and predicted damaging in silico, did not demonstrate a loss of function in our in vitro test assay. These findings can be regarded as a confirmation of the validity of this assay to detect functionally defective NFIB proteins with respect to DNA binding and transcription activation.
Clinical Presentation
All 18 individuals with haploinsufficiency of NFIB presented with mild intellectual disability or learning disability and speech delay. Motor delay (12/16) and muscular hypotonia (11/17) were also commonly reported (Tables 1 and 2). NFIB deletion and sequence variant carriers also showed attention deficit disorder (11/14) and variable behavioral anomalies including autistic behavior, anxiety, psychotic episodes, and aggression. Furthermore, 13/16 individuals presented with macrocephaly (>97th centile; Figures S3A and S3B).
Variable structural brain anomalies were reported based on brain imaging (9/11), including dysgenesis of the corpus callosum as the most common, albeit not consistent, finding (5/11) (Tables 1 and 2 and Figure S4). As brain imaging was not available in some cases, and since high-quality imaging and specific analysis may be required to properly identify corresponding anomalies in humans, the full spectrum of structural brain anomalies in the affected individuals remains to be analyzed in detail.
Although striking craniofacial anomalies were not part of the phenotype, affected individuals did share some minor facial dysmorphic features including a long face with high forehead, sparse eyebrows, down-slanting palpebral fissures and blepharophimosis, a narrow nasal bridge, anteverted nares, a long and smooth philtrum, and small ears (Figure 1). No major malformations in other organ systems were recorded, although five individuals had minor heart defects and two male individuals had cryptorchidism. No lung defects have been reported in any of the individuals with pathogenic NFIB changes, contrasting with the frequency of lung maturation defects in both heterozygous and homozygous knockout mouse embryos.7, 64 Whether this reflects perinatal lethality, partial retention of protein function in sequence variants, or developmental differences between species remains unclear.
Despite the varying size of the deletions encompassing NFIB, the severity of cognitive impairment was similar in all individuals with 9p23-p22.2 microdeletions and was comparable to that seen in point mutation cases. However, individuals with larger deletions displayed a slightly different clinical presentation. For instance, facial anomalies were more pronounced in these individuals (Figure 1). It is therefore possible that other dosage-sensitive genes in the larger-sized deletions might be contributing to the expanded phenotype of these individuals (Figure 2A). Similarly, these other genes could mitigate the macrocephaly, as the three individuals without macrocephaly had large deletions involving multiple genes.
Developmental Origin of Defects in NFIB Deficiency
Much of our understanding about the function of NFIB is based on analyses of embryos from Nfib knockout mouse models. These studies have revealed the importance of NFIB for normal cortical development. Nfib knockout embryos display multiple defects, including agenesis of the corpus callosum, enlarged ventricles, and hippocampal anomalies.5, 7, 9, 46, 61 On a cellular level, cortical progenitor cells remain self-renewing for longer in Nfib knockout mice, and neurogenesis and gliogenesis are delayed.8, 34 However, it has not previously been possible to study the postnatal structural and functional consequences of this developmental brain phenotype as these animals are not viable due to lung failure.7, 64
In this study, we use a conditional knockout mouse model with Nfib deletion localized to the dorsal telencephalon. As a consequence, homozygous conditional knockout animals remain viable postnatally. These animals displayed an isolated increase in cortical size without other structural brain defects. Overall, cortical lamination and interhemispheric wiring is similar to those of wild-type animals, which is consistent with observations in complete null embryos of the constitutive knockout model.8 Although further analysis is required, the increased radial glial population due to Nfib deletion in mice appears to be the likely cause of the observed increase in cortical size.34, 46 Developmentally, this increase results in ventricular enlargement, especially near the midline, where NFIB expression is higher than in lateral regions of the cortex.7, 34
In contrast to the previous constitutive Nfib knockout model, which displays fully penetrant complete agenesis of the corpus callosum and aberrant fiber tracts,5, 7, 9 homozygous conditional knockout mice have a corpus callosum. We recently identified that complete agenesis in Nfib knockout mice is caused by a defect in midline glial development that disrupts interhemispheric midline remodeling.65 In heterozygous embryos of the constitutive Nfib knockout model, complete callosal agenesis has not been observed, although sporadically embryos displayed milder forms of callosal dysgenesis.7 The corpus callosum in homozygous conditional knockout mice is slightly shorter and thinner (Figures 4C and 4E) but exhibits no interhemispheric wiring defects. In these conditional knockout mice, Nfib was not deleted in the midline glia, therefore permitting midline fusion and the formation of the corpus callosum.9, 35
Overlapping Function of NFIA, NFIB, and NFIX
In mice, Nfia, Nfib, and Nfix display a very similar expression pattern during early brain development, although their expression becomes more distinct at later ages.30, 32, 66 Each of the individual Nfi knockout mice display comparable cortical defects, particularly for Nfia and Nfib knockout embryos.1, 2, 5, 6, 7, 8, 9, 46, 61 In this context, all three family members function non-redundantly and additively.34, 67 Hence, the number of Nfi alleles corresponds with the severity of the observed cortical phenotype, such that Nfib homozygous knockout and Nfia;Nfib double heterozygous knockout mice display similar severity of phenotypes.34, 67 This overlap in biological function may explain the similarities between the individuals with NFIB haploinsufficiency described here and those with NFIA or NFIX haploinsufficiency (Figure S3C).
The NFI proteins share a highly homologous N-terminal DNA-binding and dimerization domain, mainly encoded by exon 2,3 and disease-causing mutations may occur in homologous amino acids between family members. Indeed, missense variants, corresponding to the p.Lys114Thr and p.Lys126Glu observed in subjects 3 and 4, has been recently reported in an individual with Malan syndrome in the respective codon of NFIX p.Lys113Glu, p.Lys125Glu, and p.Lys125Gln.21, 68 The region surrounding these lysines could be important for NFI function, as we observed a p.Leu132Pro missense change in NFIB in subject 5, while p.Arg115Trp, p.Arg116Gly, p.Arg116Gln, p.Arg116Prf, p.Arg121Pro, and p.Arg128Gln have been reported in NFIX.68, 69, 70, 71
Association of NFIB with Intellectual Disability and Other Neurological Disease
More cases with mild intellectual disability and variable behavioral anomalies may potentially be linked to NFIB loss or altered expression. In the ClinVar database,72 four entries with deletions overlapping NFIB (dbVar: nsv529583, nsv531613, nsv531614, nsv529233) are listed (Figure 2A). The clinical information provided for these individuals was limited but indicated developmental delay in 3/4 case subjects and unspecified abnormalities of the central nervous system in the fourth (dbVar: nsv529233). In line with our findings, all four deletions listed in this database were classified as pathogenic. There are further genomic alterations of NFIB reported in individuals with autism spectrum disorder (ASD): a paternally inherited intronic loss of 8,341 bp in intron 2, a sequence variant affecting the 3′ splice region of exon 4, and a balanced cytogenetic abnormality affecting NFIB.73, 74, 75
In addition, genome-wide association studies have implicated single-nucleotide polymorphisms within or near NFIB with behavioral phenotypes. For instance, intronic SNP rs4741351 has been associated with decreased learning attainment and rs1322987 with delayed story telling.76, 77 Furthermore, SNPs in NFIB have also been associated with bipolar disorder and schizophrenia.78, 79, 80, 81 Interestingly, an ASD-associated intronic SNP in Engrailed 2 removes an NFIB-binding site and results in reduced Engrailed 2 expression,82 suggesting that this gene is potentially an important downstream target of NFIB. Taken together, NFIB may be more broadly implicated in ID and behavioral phenotypes than in the cohort presented in this paper.
NFIB Haploinsufficiency Causes a Syndrome with Macrocephaly and Intellectual Disability
We report 18 individuals with ID in which we identified alterations of NFIB, including partial and whole gene deletions with a variable number of neighboring genes, as well as seven pathogenic sequence variants. Based on these findings, we propose that haploinsufficiency of NFIB is the common underlying pathogenic mechanism, thus introducing NFIB mutations as causative for ID. The 18 affected individuals shared a similar phenotype of mild ID, muscular hypotonia, speech delay, attention deficit disorder, and variable behavioral anomalies. Head circumference was above the mean in 16/16, and 13/16 individuals had absolute macrocephaly. Other structural brain anomalies including corpus callosum dysgenesis may be present. Although congenital malformations and facial anomalies that allow clinical recognition of the disease are not part of the presentation, all individuals share some minor dysmorphic features. We propose that NFIB haploinsufficiency causes a macrocephaly-intellectual disability syndrome overlapping with NFIA and NFIX haploinsufficiency phenotypes.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We would like to thank the families for their collaboration and contribution to this project. We thank Prof. William D. Richardson and Dr. Nicoletta Kessaris, Wolfson Institute for Biomedical Research, University College London, for providing the Emx1iCre mice for this project. We would like to thank Dr. Nyoman Kurniawan of the Centre for Advanced Imaging, the University of Queensland, for his technical expertise and assistance in conducting the ex vivo mouse brain MRI. In additional, we would also like to thank the members of the IRC5 consortium for their support.
This work was supported by grants from the National Health and Medical Research Council Australia (GNT1100443 to L.J.R.), the French Ministry of Health (PHRC national 2008/2008-A00515-50), Regional Council of Burgundy/Dijon University hospital (PARI 2012), The Genesis Foundation for Children, the US National Institutes of Health under NINDS grants (1R01NS092772 and 234567890 to W.B.D.; 1R01NS058721 to W.B.D. and E.H.S.; and K08NS092898 to G.M.M.), and Jordan’s Guardian Angels (G.M.M.). J.W.C.L. was supported by an International Postgraduate Research Scholarship and UQ Centennial Scholarship. R.M.G. was supported by NYSTEM grants (C026714, C026429, and C030133). R.J.D. was supported by Brain Injured Children’s Aftercare Recovery Endeavours (BICARE) Fellowship. L.J.R. was supported by an NHMRC Principal Research Fellowship (GNT1005751). M.Z. was supported by a grant from the German Ministry of Education and Research (BMBF) (GeNeRARe 01GM1519A). We acknowledge the Linkage Infrastructure, Equipment and Facilities (LIEF) grant (LE100100074) awarded to the Queensland Brain Institute for the Slide Scanner and the facilities of the National Imaging Facility (NIF) at the Centre for Advanced Imaging, University of Queensland, used in the animal experiments. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding sources.
Published: November 1, 2018
Footnotes
Supplemental Data include a Supplemental Note on case reports, four figures, two tables, Supplemental Methods, and Acknowledgments, and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.10.006.
Contributor Information
Jens Bunt, Email: j.bunt@uq.edu.au.
Martin Zenker, Email: martin.zenker@med.ovgu.de.
Accession Numbers
The ClinVar accession numbers for the NFIB sequence variants and NFIB deletions reported in this paper are as follows: c.109C>T (p.Arg37∗), SCV000803743.1; c.265C>T (p.Arg89∗), SCV000803744.1; c.341A>C (p.Lys114Thr), SCV000803745.1; c.376A>G (p.Lys126Glu), SCV000803746.1; c.395T>C (p.Leu132Pro), SCV000803747.1; c.758_759dupTG (p.Asn254∗), SCV000803748.1; c.1063_1076del (p.Ile355Serfs∗48), SCV000803749.1; c.1067C>T (p.Ser356Leu), SCV000803750.1; arr[hg19] 9p23p22.3(14098659_14324147)x1, SCV000809030; arr[hg19] 9p23p22.3(14102175_14386038)x1, SCV000809031; arr[hg19] 9p23p22.3(13974415_14286259)x1, SCV000809032; arr[hg19] 9p23p22.3(13106806_14639971)x1, SCV000809033; arr[hg19] 9p23p22.3(13034407_14653394)x1, SCV000809034; arr[hg19] 9p23p22.2(14178768_16619009)x1, SCV000809035; arr[hg19] 9p23p22.2(13563537_18491752)x1, SCV000809036; arr[hg19] 9p23p22.2(13739630_18023839)x1, SCV000809037.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
Database of Genomic Variants (DGV), http://dgv.tcag.ca/dgv/app/home
DECIPHER, https://decipher.sanger.ac.uk/
Ensembl Genome Browser, http://www.ensembl.org/index.html
ExAC Browser, http://exac.broadinstitute.org/
Head circumference calculator, https://simulconsult.com/resources/measurement.html?type=head
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PredictSNP2, https://loschmidt.chemi.muni.cz/predictsnp2/
PROVEAN, http://provean.jcvi.org
Supplemental Data
References
- 1.Campbell C.E., Piper M., Plachez C., Yeh Y.T., Baizer J.S., Osinski J.M., Litwack E.D., Richards L.J., Gronostajski R.M. The transcription factor Nfix is essential for normal brain development. BMC Dev. Biol. 2008;8:52. doi: 10.1186/1471-213X-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.das Neves L., Duchala C.S., Tolentino-Silva F., Haxhiu M.A., Colmenares C., Macklin W.B., Campbell C.E., Butz K.G., Gronostajski R.M. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc. Natl. Acad. Sci. USA. 1999;96:11946–11951. doi: 10.1073/pnas.96.21.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gronostajski R.M. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 4.Mason S., Piper M., Gronostajski R.M., Richards L.J. Nuclear factor one transcription factors in CNS development. Mol. Neurobiol. 2009;39:10–23. doi: 10.1007/s12035-008-8048-6. [DOI] [PubMed] [Google Scholar]
- 5.Piper M., Moldrich R.X., Lindwall C., Little E., Barry G., Mason S., Sunn N., Kurniawan N.D., Gronostajski R.M., Richards L.J. Multiple non-cell-autonomous defects underlie neocortical callosal dysgenesis in Nfib-deficient mice. Neural Dev. 2009;4:43. doi: 10.1186/1749-8104-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shu T., Butz K.G., Plachez C., Gronostajski R.M., Richards L.J. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J. Neurosci. 2003;23:203–212. doi: 10.1523/JNEUROSCI.23-01-00203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steele-Perkins G., Plachez C., Butz K.G., Yang G., Bachurski C.J., Kinsman S.L., Litwack E.D., Richards L.J., Gronostajski R.M. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol. Cell. Biol. 2005;25:685–698. doi: 10.1128/MCB.25.2.685-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betancourt J., Katzman S., Chen B. Nuclear factor one B regulates neural stem cell differentiation and axonal projection of corticofugal neurons. J. Comp. Neurol. 2014;522:6–35. doi: 10.1002/cne.23373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gobius I., Morcom L., Suárez R., Bunt J., Bukshpun P., Reardon W., Dobyns W.B., Rubenstein J.L., Barkovich A.J., Sherr E.H., Richards L.J. Astroglial-mediated remodeling of the interhemispheric midline is required for the formation of the corpus callosum. Cell Rep. 2016;17:735–747. doi: 10.1016/j.celrep.2016.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C.P., Su Y.N., Chen Y.Y., Chern S.R., Liu Y.P., Wu P.C., Lee C.C., Chen Y.T., Wang W. Chromosome 1p32-p31 deletion syndrome: prenatal diagnosis by array comparative genomic hybridization using uncultured amniocytes and association with NFIA haploinsufficiency, ventriculomegaly, corpus callosum hypogenesis, abnormal external genitalia, and intrauterine growth restriction. Taiwan. J. Obstet. Gynecol. 2011;50:345–352. doi: 10.1016/j.tjog.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Ji J., Salamon N., Quintero-Rivera F. Microdeletion of 1p32-p31 involving NFIA in a patient with hypoplastic corpus callosum, ventriculomegaly, seizures and urinary tract defects. Eur. J. Med. Genet. 2014;57:267–268. doi: 10.1016/j.ejmg.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Koehler U., Holinski-Feder E., Ertl-Wagner B., Kunz J., von Moers A., von Voss H., Schell-Apacik C. A novel 1p31.3p32.2 deletion involving the NFIA gene detected by array CGH in a patient with macrocephaly and hypoplasia of the corpus callosum. Eur. J. Pediatr. 2010;169:463–468. doi: 10.1007/s00431-009-1057-2. [DOI] [PubMed] [Google Scholar]
- 13.Lu W., Quintero-Rivera F., Fan Y., Alkuraya F.S., Donovan D.J., Xi Q., Turbe-Doan A., Li Q.G., Campbell C.G., Shanske A.L. NFIA haploinsufficiency is associated with a CNS malformation syndrome and urinary tract defects. PLoS Genet. 2007;3:e80. doi: 10.1371/journal.pgen.0030080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Negishi Y., Miya F., Hattori A., Mizuno K., Hori I., Ando N., Okamoto N., Kato M., Tsunoda T., Yamasaki M. Truncating mutation in NFIA causes brain malformation and urinary tract defects. Hum Genome Var. 2015;2:15007. doi: 10.1038/hgv.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyboe D., Kreiborg S., Kirchhoff M., Hove H.B. Familial craniosynostosis associated with a microdeletion involving the NFIA gene. Clin. Dysmorphol. 2015;24:109–112. doi: 10.1097/MCD.0000000000000079. [DOI] [PubMed] [Google Scholar]
- 16.Rao A., O’Donnell S., Bain N., Meldrum C., Shorter D., Goel H. An intragenic deletion of the NFIA gene in a patient with a hypoplastic corpus callosum, craniofacial abnormalities and urinary tract defects. Eur. J. Med. Genet. 2014;57:65–70. doi: 10.1016/j.ejmg.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Zhao W.W. Intragenic deletion of RBFOX1 associated with neurodevelopmental/neuropsychiatric disorders and possibly other clinical presentations. Mol. Cytogenet. 2013;6:26. doi: 10.1186/1755-8166-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikhail F.M., Lose E.J., Robin N.H., Descartes M.D., Rutledge K.D., Rutledge S.L., Korf B.R., Carroll A.J. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am. J. Med. Genet. A. 2011;155A:2386–2396. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- 19.Revah-Politi A., Ganapathi M., Bier L., Cho M.T., Goldstein D.B., Hemati P., Iglesias A., Juusola J., Pappas J., Petrovski S. Loss-of-function variants in NFIA provide further support that NFIA is a critical gene in 1p32-p31 deletion syndrome: A four patient series. Am. J. Med. Genet. A. 2017;173:3158–3164. doi: 10.1002/ajmg.a.38460. [DOI] [PubMed] [Google Scholar]
- 20.Dong H.Y., Zeng H., Hu Y.Q., Xie L., Wang J., Wang X.Y., Yang Y.F., Tan Z.P. 19p13.2 Microdeletion including NFIX associated with overgrowth and intellectual disability suggestive of Malan syndrome. Mol. Cytogenet. 2016;9:71. doi: 10.1186/s13039-016-0282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurrieri F., Cavaliere M.L., Wischmeijer A., Mammì C., Neri G., Pisanti M.A., Rodella G., Laganà C., Priolo M. NFIX mutations affecting the DNA-binding domain cause a peculiar overgrowth syndrome (Malan syndrome): a new patients series. Eur. J. Med. Genet. 2015;58:488–491. doi: 10.1016/j.ejmg.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Jezela-Stanek A., Kucharczyk M., Falana K., Jurkiewicz D., Mlynek M., Wicher D., Rydzanicz M., Kugaudo M., Cieslikowska A., Ciara E. Malan syndrome (Sotos syndrome 2) in two patients with 19p13.2 deletion encompassing NFIX gene and novel NFIX sequence variant. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2016;160:161–167. doi: 10.5507/bp.2016.006. [DOI] [PubMed] [Google Scholar]
- 23.Klaassens M., Morrogh D., Rosser E.M., Jaffer F., Vreeburg M., Bok L.A., Segboer T., van Belzen M., Quinlivan R.M., Kumar A. Malan syndrome: Sotos-like overgrowth with de novo NFIX sequence variants and deletions in six new patients and a review of the literature. Eur. J. Hum. Genet. 2015;23:610–615. doi: 10.1038/ejhg.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malan V., Rajan D., Thomas S., Shaw A.C., Louis Dit Picard H., Layet V., Till M., van Haeringen A., Mortier G., Nampoothiri S. Distinct effects of allelic NFIX mutations on nonsense-mediated mRNA decay engender either a Sotos-like or a Marshall-Smith syndrome. Am. J. Hum. Genet. 2010;87:189–198. doi: 10.1016/j.ajhg.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oshima T., Hara H., Takeda N., Hasumi E., Kuroda Y., Taniguchi G., Inuzuka R., Nawata K., Morita H., Komuro I. A novel mutation of NFIX causes Sotos-like syndrome (Malan syndrome) complicated with thoracic aortic aneurysm and dissection. Hum Genome Var. 2017;4:17022. doi: 10.1038/hgv.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimojima K., Okamoto N., Tamasaki A., Sangu N., Shimada S., Yamamoto T. An association of 19p13.2 microdeletions with Malan syndrome and Chiari malformation. Am. J. Med. Genet. A. 2015;167A:724–730. doi: 10.1002/ajmg.a.36959. [DOI] [PubMed] [Google Scholar]
- 27.Aggarwal A., Nguyen J., Rivera-Davila M., Rodriguez-Buritica D. Marshall-Smith syndrome: Novel pathogenic variant and previously unreported associations with precocious puberty and aortic root dilatation. Eur. J. Med. Genet. 2017;60:391–394. doi: 10.1016/j.ejmg.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Martinez F., Marín-Reina P., Sanchis-Calvo A., Perez-Aytés A., Oltra S., Roselló M., Mayo S., Monfort S., Pantoja J., Orellana C. Novel mutations of NFIX gene causing Marshall-Smith syndrome or Sotos-like syndrome: one gene, two phenotypes. Pediatr. Res. 2015;78:533–539. doi: 10.1038/pr.2015.135. [DOI] [PubMed] [Google Scholar]
- 29.Schanze D., Neubauer D., Cormier-Daire V., Delrue M.A., Dieux-Coeslier A., Hasegawa T., Holmberg E.E., Koenig R., Krueger G., Schanze I. Deletions in the 3′ part of the NFIX gene including a recurrent Alu-mediated deletion of exon 6 and 7 account for previously unexplained cases of Marshall-Smith syndrome. Hum. Mutat. 2014;35:1092–1100. doi: 10.1002/humu.22603. [DOI] [PubMed] [Google Scholar]
- 30.Chaudhry A.Z., Lyons G.E., Gronostajski R.M. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev. Dyn. 1997;208:313–325. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 31.Bunt J., Lim J.W., Zhao L., Mason S., Richards L.J. PAX6 does not regulate Nfia and Nfib expression during neocortical development. Sci. Rep. 2015;5:10668. doi: 10.1038/srep10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plachez C., Lindwall C., Sunn N., Piper M., Moldrich R.X., Campbell C.E., Osinski J.M., Gronostajski R.M., Richards L.J. Nuclear factor I gene expression in the developing forebrain. J. Comp. Neurol. 2008;508:385–401. doi: 10.1002/cne.21645. [DOI] [PubMed] [Google Scholar]
- 33.Driller K., Pagenstecher A., Uhl M., Omran H., Berlis A., Gründer A., Sippel A.E. Nuclear factor I X deficiency causes brain malformation and severe skeletal defects. Mol. Cell. Biol. 2007;27:3855–3867. doi: 10.1128/MCB.02293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bunt J., Osinki J., Lim J.W.C., Vidovic D., Ye Y., Zalucki O., O’Connor T., Harris L., Gronostajski R., Richards L.J. Combined allelic dosage of Nfia and Nfib regulates cortical development. BNA. 2017 doi: 10.1177/2398212817739433. Published online November 22, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sajan S.A., Fernandez L., Nieh S.E., Rider E., Bukshpun P., Wakahiro M., Christian S.L., Rivière J.B., Sullivan C.T., Sudi J. Both rare and de novo copy number variants are prevalent in agenesis of the corpus callosum but not in cerebellar hypoplasia or polymicrogyria. PLoS Genet. 2013;9:e1003823. doi: 10.1371/journal.pgen.1003823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X., Snijders A., Segraves R., Zhang X., Niebuhr A., Albertson D., Yang H., Gray J., Niebuhr E., Bolund L., Pinkel D. High-resolution mapping of genotype-phenotype relationships in cri du chat syndrome using array comparative genomic hybridization. Am. J. Hum. Genet. 2005;76:312–326. doi: 10.1086/427762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacDonald J.R., Ziman R., Yuen R.K., Feuk L., Scherer S.W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vulto-van Silfhout A.T., van Ravenswaaij C.M., Hehir-Kwa J.Y., Verwiel E.T., Dirks R., van Vooren S., Schinzel A., de Vries B.B., de Leeuw N. An update on ECARUCA, the European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations. Eur. J. Med. Genet. 2013;56:471–474. doi: 10.1016/j.ejmg.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 41.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bendl J., Musil M., Štourač J., Zendulka J., Damborský J., Brezovský J. PredictSNP2: a unified platform for accurately evaluating SNP effects by exploiting the different characteristics of variants in distinct genomic regions. PLoS Comput. Biol. 2016;12:e1004962. doi: 10.1371/journal.pcbi.1004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 44.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 45.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piper M., Barry G., Harvey T.J., McLeay R., Smith A.G., Harris L., Mason S., Stringer B.W., Day B.W., Wray N.R. NFIB-mediated repression of the epigenetic factor Ezh2 regulates cortical development. J. Neurosci. 2014;34:2921–2930. doi: 10.1523/JNEUROSCI.2319-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olmsted J.B., Carlson K., Klebe R., Ruddle F., Rosenbaum J. Isolation of microtubule protein from cultured mouse neuroblastoma cells. Proc. Natl. Acad. Sci. USA. 1970;65:129–136. doi: 10.1073/pnas.65.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westermark B. The deficient density-dependent growth control of human malignant glioma cells and virus-transformed glia-like cells in culture. Int. J. Cancer. 1973;12:438–451. doi: 10.1002/ijc.2910120215. [DOI] [PubMed] [Google Scholar]
- 49.Zhou B.Y., Liu Y., Kim Bo., Xiao Y., He J.J. Astrocyte activation and dysfunction and neuron death by HIV-1 Tat expression in astrocytes. Mol. Cell. Neurosci. 2004;27:296–305. doi: 10.1016/j.mcn.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 50.Chang C.Y., Pasolli H.A., Giannopoulou E.G., Guasch G., Gronostajski R.M., Elemento O., Fuchs E. NFIB is a governor of epithelial-melanocyte stem cell behaviour in a shared niche. Nature. 2013;495:98–102. doi: 10.1038/nature11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu Y.C., Osinski J., Campbell C.E., Litwack E.D., Wang D., Liu S., Bachurski C.J., Gronostajski R.M. Mesenchymal nuclear factor I B regulates cell proliferation and epithelial differentiation during lung maturation. Dev. Biol. 2011;354:242–252. doi: 10.1016/j.ydbio.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kessaris N., Fogarty M., Iannarelli P., Grist M., Wegner M., Richardson W.D. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat. Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piper M., Harris L., Barry G., Heng Y.H., Plachez C., Gronostajski R.M., Richards L.J. Nuclear factor one X regulates the development of multiple cellular populations in the postnatal cerebellum. J. Comp. Neurol. 2011;519:3532–3548. doi: 10.1002/cne.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim J.W., Donahoo A.L., Bunt J., Edwards T.J., Fenlon L.R., Liu Y., Zhou J., Moldrich R.X., Piper M., Gobius I. EMX1 regulates NRP1-mediated wiring of the mouse anterior cingulate cortex. Development. 2015;142:3746–3757. doi: 10.1242/dev.119909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moldrich R.X., Pannek K., Hoch R., Rubenstein J.L., Kurniawan N.D., Richards L.J. Comparative mouse brain tractography of diffusion magnetic resonance imaging. Neuroimage. 2010;51:1027–1036. doi: 10.1016/j.neuroimage.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma Y., Hof P.R., Grant S.C., Blackband S.J., Bennett R., Slatest L., McGuigan M.D., Benveniste H. A three-dimensional digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Neuroscience. 2005;135:1203–1215. doi: 10.1016/j.neuroscience.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 57.Alby C., Malan V., Boutaud L., Marangoni M.A., Bessières B., Bonniere M., Ichkou A., Elkhartoufi N., Bahi-Buisson N., Sonigo P. Clinical, genetic and neuropathological findings in a series of 138 fetuses with a corpus callosum malformation. Birth Defects Res. A Clin. Mol. Teratol. 2016;106:36–46. doi: 10.1002/bdra.23472. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y., Bernard H.U., Apt D. NFI-B3, a novel transcriptional repressor of the nuclear factor I family, is generated by alternative RNA processing. J. Biol. Chem. 1997;272:10739–10745. doi: 10.1074/jbc.272.16.10739. [DOI] [PubMed] [Google Scholar]
- 59.Stringer B.W., Bunt J., Day B.W., Barry G., Jamieson P.R., Ensbey K.S., Bruce Z.C., Goasdoué K., Vidal H., Charmsaz S. Nuclear factor one B (NFIB) encodes a subtype-specific tumour suppressor in glioblastoma. Oncotarget. 2016;7:29306–29320. doi: 10.18632/oncotarget.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brun M., Coles J.E., Monckton E.A., Glubrecht D.D., Bisgrove D., Godbout R. Nuclear factor I regulates brain fatty acid-binding protein and glial fibrillary acidic protein gene expression in malignant glioma cell lines. J. Mol. Biol. 2009;391:282–300. doi: 10.1016/j.jmb.2009.06.041. [DOI] [PubMed] [Google Scholar]
- 61.Barry G., Piper M., Lindwall C., Moldrich R., Mason S., Little E., Sarkar A., Tole S., Gronostajski R.M., Richards L.J. Specific glial populations regulate hippocampal morphogenesis. J. Neurosci. 2008;28:12328–12340. doi: 10.1523/JNEUROSCI.4000-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gründer A., Ebel T.T., Mallo M., Schwarzkopf G., Shimizu T., Sippel A.E., Schrewe H. Nuclear factor I-B (Nfib) deficient mice have severe lung hypoplasia. Mech. Dev. 2002;112:69–77. doi: 10.1016/s0925-4773(01)00640-2. [DOI] [PubMed] [Google Scholar]
- 65.Gobius I., Suárez R., Morcom L., Paolino A., Edwards T.J., Kozulin P., Richards L.J. Astroglial-mediated remodeling of the interhemispheric midline during telencephalic development is exclusive to eutherian mammals. Neural Dev. 2017;12:9. doi: 10.1186/s13064-017-0086-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen K.S., Harris L., Lim J.W.C., Harvey T.J., Piper M., Gronostajski R.M., Richards L.J., Bunt J. Differential neuronal and glial expression of nuclear factor I proteins in the cerebral cortex of adult mice. J. Comp. Neurol. 2017;525:2465–2483. doi: 10.1002/cne.24206. [DOI] [PubMed] [Google Scholar]
- 67.Harris L., Zalucki O., Gobius I., McDonald H., Osinki J., Harvey T.J., Essebier A., Vidovic D., Gladwyn-Ng I., Burne T.H. Transcriptional regulation of intermediate progenitor cell generation during hippocampal development. Development. 2016;143:4620–4630. doi: 10.1242/dev.140681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Priolo M., Schanze D., Tatton-Brown K., Mulder P.A., Tenorio J., Kooblall K., Acero I.H., Alkuraya F.S., Arias P., Bernardini L. Further delineation of Malan syndrome. Hum. Mutat. 2018;39:1226–1237. doi: 10.1002/humu.23563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoneda Y., Saitsu H., Touyama M., Makita Y., Miyamoto A., Hamada K., Kurotaki N., Tomita H., Nishiyama K., Tsurusaki Y. Missense mutations in the DNA-binding/dimerization domain of NFIX cause Sotos-like features. J. Hum. Genet. 2012;57:207–211. doi: 10.1038/jhg.2012.7. [DOI] [PubMed] [Google Scholar]
- 70.Tatton-Brown K., Loveday C., Yost S., Clarke M., Ramsay E., Zachariou A., Elliott A., Wylie H., Ardissone A., Rittinger O., Childhood Overgrowth Collaboration Mutations in epigenetic regulation genes are a major cause of overgrowth with intellectual disability. Am. J. Hum. Genet. 2017;100:725–736. doi: 10.1016/j.ajhg.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Tippin Davis B., Baxter R.M., Zeng W., Mroske C., Parra M.C. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 72.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prasad A., Merico D., Thiruvahindrapuram B., Wei J., Lionel A.C., Sato D., Rickaby J., Lu C., Szatmari P., Roberts W. A discovery resource of rare copy number variations in individuals with autism spectrum disorder. G3 (Bethesda) 2012;2:1665–1685. doi: 10.1534/g3.112.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iossifov I., Ronemus M., Levy D., Wang Z., Hakker I., Rosenbaum J., Yamrom B., Lee Y.H., Narzisi G., Leotta A. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Redin C., Brand H., Collins R.L., Kammin T., Mitchell E., Hodge J.C., Hanscom C., Pillalamarri V., Seabra C.M., Abbott M.A. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat. Genet. 2017;49:36–45. doi: 10.1038/ng.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okbay A., Beauchamp J.P., Fontana M.A., Lee J.J., Pers T.H., Rietveld C.A., Turley P., Chen G.B., Emilsson V., Meddens S.F., LifeLines Cohort Study Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cirulli E.T., Kasperaviciūte D., Attix D.K., Need A.C., Ge D., Gibson G., Goldstein D.B. Common genetic variation and performance on standardized cognitive tests. Eur. J. Hum. Genet. 2010;18:815–820. doi: 10.1038/ejhg.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Le-Niculescu H., Patel S.D., Bhat M., Kuczenski R., Faraone S.V., Tsuang M.T., McMahon F.J., Schork N.J., Nurnberger J.I., Jr., Niculescu A.B., 3rd Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2009;150B:155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- 79.Willour V.L., Seifuddin F., Mahon P.B., Jancic D., Pirooznia M., Steele J., Schweizer B., Goes F.S., Mondimore F.M., Mackinnon D.F., Bipolar Genome Study Consortium A genome-wide association study of attempted suicide. Mol. Psychiatry. 2012;17:433–444. doi: 10.1038/mp.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Segurado R., Detera-Wadleigh S.D., Levinson D.F., Lewis C.M., Gill M., Nurnberger J.I., Jr., Craddock N., DePaulo J.R., Baron M., Gershon E.S. Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: Bipolar disorder. Am. J. Hum. Genet. 2003;73:49–62. doi: 10.1086/376547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gupta C.N., Chen J., Liu J., Damaraju E., Wright C., Perrone-Bizzozero N.I., Pearlson G., Luo L., Michael A.M., Turner J.A., Calhoun V.D. Genetic markers of white matter integrity in schizophrenia revealed by parallel ICA. Front. Hum. Neurosci. 2015;9:100. doi: 10.3389/fnhum.2015.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi J., Ababon M.R., Matteson P.G., Millonig J.H. Cut-like homeobox 1 and nuclear factor I/B mediate ENGRAILED2 autism spectrum disorder-associated haplotype function. Hum. Mol. Genet. 2012;21:1566–1580. doi: 10.1093/hmg/ddr594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.