Abstract
Pharmacogenomics is a tool for practitioners to provide precision pharmacotherapy using genomics. All providers are likely to encounter genomic data in practice with the expectation that they are able to successfully apply it to patient care. Pharmacogenomics tests for genetic variations in genes that are responsible for drug metabolism, transport, and targets of drug action. Variations can increase the risk for drug toxicity or poor efficacy. Pharmacogenomics can, therefore, be used to help select the best medication or aid in dosing. Nephrologists routinely treat cardiovascular disease and manage patients after kidney transplantation, two situations for which there are several high-evidence clinical recommendations for commonly used anticoagulants, antiplatelets, statins, and transplant medications. Successful use of pharmacogenomics in practice requires that providers are familiar with how to access and use pharmacogenomics resources. Similarly, clinical decision making related to whether to use existing data, whether to order testing, and if data should be used in practice is needed to deliver precision medicine. Pharmacogenomics is applicable to virtually every medical specialty, and nephrologists are well positioned to be implementation leaders.
Keywords: Anticoagulants, Cardiovascular Diseases, Clinical Decision-making, drug metabolism, drug transporter, Drug-related Side Effects and Adverse Reactions, Genetic Variation, Genomics, Humans, Hydroxymethylglutaryl-CoA Reductase Inhibitors, kidney transplantation, Nephrologists, nephrology, Patient Care, Pharmacogenetics, Pharmacogenomics, pharmacokinetics, Precision Medicine
Introduction
Few medical interventions are as accessible to the clinician as pharmacotherapy. However, interpatient variability in drug pharmacokinetics (absorption, metabolism, distribution, and elimination) and pharmacodynamics (concentration-effect relationships) challenges drug selection and dosing (1). Precision medicine, driven by advances in genomics technology, promises a means to mitigate these unpredictable medication responses. Nephrologists have been champions of the use of biomarkers to tailor medication dosing for decades with their use of measurements, like creatinine clearance, to estimate kidney function (2). Combined with other measures, such as weight, age, and population-based nomograms, most nephrologists are already using precision medicine routinely in daily practice.
Despite efforts to incorporate tailored dosing, an estimated 2.2 million adverse drug reactions occur in the United States annually, and medication efficacy rates vary considerably (3). Difficulty in predicting medication response has led to the paradigm of frequent dose titration and iteration among medications. Collectively, these place a significant burden on the patient, the provider, and the health care system. One potential solution is the use of individual genomic data to guide prescribing, which is termed pharmacogenomics (4).
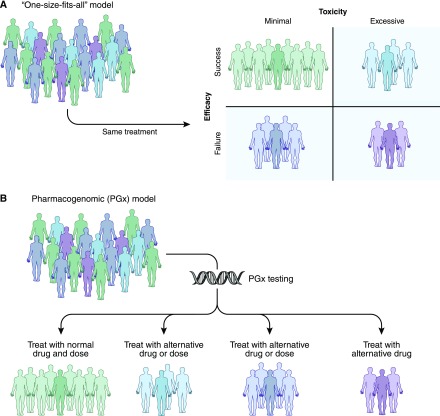
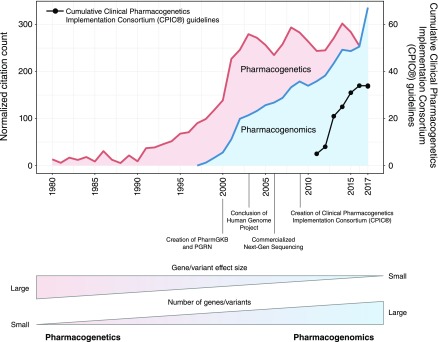
Rather than a one size fits all dosing, pharmacogenomics may enable a priori identification of which patients are likely to experience therapeutic failure or toxicity, leading to individualized pharmacotherapy (Figure 1). The past two decades have yielded a rapid influx of genomic data, with over 20,000 new pharmacogenomics citations in PubMed, in excess of 3500 gene-drug variant associations reported, and nearly 200 medications with pharmacogenomics information in their Food and Drug Administration (FDA)–approved drug product labeling (5,6). Common genetic variations predict activity of drug-metabolizing enzymes, drug affinity for treatment targets, and risk for immune reaction to medications among others (7,8). Furthermore, a number of medical centers have implemented clinical pharmacogenomics services and are providing new solutions to complement biometric-based dosing and clinician judgement to deliver more precision prescribing (9). As this area has grown, pharmacogenomics has transitioned from single gene/variant and drug response associations (originally coined as “pharmacogenetics”) to a broader analysis of multiple genetic variants from many genes and environmental factors to personalize medication therapy (10). Figure 2 describes the growth and transition of pharmacogenomics over the past 38 years.
Figure 1.
Individualized therapy through pharmacogenomics may predict patients who should receive a different dose or alternative medication. (A) Shows how a conventional one size fits all dosing model may lead to patients with therapeutic failure, drug toxicity, or both (purple shaded). (B) In contrast, shows that a pharmacogenomics-guided dosing model may allow prediction of more appropriate dose or drug alternatives.
Figure 2.
The rise of pharmacogenomics is evident from the growth in genomic technology and the use of multiple genes and variants to understand drug response. Numbers of citations indexed in PubMed that contain the words “pharmacogenetics” (red) and “pharmacogenomics” (blue), excluding review papers, and normalized to the average number of total yearly citations in PubMed. The black line shows the cumulative number of published guidelines by the Clinical Pharmacogenetics Implementation Consortium (CPIC). These show the perspective change from studying the effect of a single gene variant with a large effect size on drug response (pharmacogenetics) to the effect of many genetic variants on many drugs (pharmacogenomics). This transition from the “etic” to “omic” terminology that began in the late 1990s signaled the beginning of precision medicine with pharmacogenomics. PGRN, Pharmacogenomics Research Network; PharmGKB, Pharmacogenomics Knowledge Base.
In this review, we provide an overview of the clinical use of pharmacogenomics, focusing on specific medications highly relevant to the nephrologist. The scientific basis for pharmacogenomics, decision-making processes for using pharmacogenomics in practice, and clinician-friendly pharmacogenomics resources will be presented. Finally, we will discuss the current state of pharmacogenomics research to highlight emerging concepts.
Science of Pharmacogenomics as a Driver of Medication Response Variability
Consider the following patient who illustrates the potential of pharmacogenomics. J.H., a 60-year-old black man with a history of ESKD, received a kidney transplant last month. He was started on 0.2 mg/kg per day tacrolimus, and over the past 2 weeks in the hospital, he has been titrated up to 0.6 mg/kg per day; however, his most recent tacrolimus trough level is subtherapeutic at 2 ng/ml. Today, he has severe flank pain and is showing signs of severe acute rejection. Ultimately, J.H. suffers from allograft loss and is reinitiated on hemodialysis. The monthly quality improvement meeting concludes that his acute rejection may have been averted with more aggressive immunosuppression and possibly, a higher tacrolimus starting dose but that the titration was in line with best practice. Nothing that was known of J.H. indicated that he would need an increased dose of tacrolimus to obtain therapeutic concentrations, but you wonder if there is a method for predicting patients, like J.H., a priori.
Humans carry 23 pairs of chromosomes inherited from the maternal and paternal lineage to make up the genome. With the exception of sex chromosomes, we, therefore, have two copies of every gene. An allele refers to a single or multiple nucleotides on a single chromosome, which may be notated as the actual nucleotide (i.e., ATCG) or as an abstracted representation of the nucleotide(s) (e.g., star alleles). If at a given genomic position, two alleles are the same, then the person would be homozygous at that particular position, and if the alleles are different, they are heterozygous. Like many patients of African ancestry, J.H. carried a single-nucleotide polymorphism (SNP), a single-nucleotide base change, on both copies at marker rs776746 on his seventh chromosome. At this position, about 90% of whites have a cytosine on both chromosomes (two “C” alleles, the “C/C” genotype, termed CYP3A5 *3/*3), whereas nearly 50% of individuals with African ancestry carry thiamine on both chromosomes (two “T” alleles, “T/T” genotype, termed CYP3A5 *1/*1) (11). From this star alleles nomenclature, we can predict a clinically useful phenotype, which results from interactions between the genotype and the environment. For example, a person with CYP3A5 *3/*3 does not express the CYP3A5 protein and is thus called a “poor metabolizer,” whereas a person with CYP3A5 *1/*1 alleles does express the CYP3A5 protein and is called a “normal metabolizer.” In the case of both (e.g., CYP3A5 *1/*3), the person has an intermediate phenotype and is called an “intermediate metabolizer” (12). For reference, a list of commonly used pharmacogenomics alleles in this paper and their corresponding genetic variations are included in Table 1.
Table 1.
Selected gene alleles, their causative variations, and associated phenotypes
| Gene and Allele | Causative Variation(s) | Phenotype |
|---|---|---|
| CYP2C9 | ||
| *2 | rs1799853 (T) | Decreased function |
| *3 | rs1057910 (C) | Decreased function |
| CYP4F2 | ||
| *3 | rs2108622 (T) | Decreased function |
| CYP3A5 | ||
| *3 | rs776746 (C) | Decreased function |
| CYP2C19 | ||
| *2 | rs4244285 (A) | Decreased function |
| *3 | rs4986893 (A) | Decreased function |
| *17 | rs12248560 (T) | Increased function |
| TPMT | ||
| *2 | rs1800462 (G) | Decreased function |
| *3A | rs1800460 (T); rs1142345 (C) | Decreased function |
| *3B | rs1800460 (T) | Decreased function |
| *3C | rs1142345 (C) | Decreased function |
| *4 | rs1800584 (T) | Decreased function |
| VKORC1 | ||
| −1639G>A | rs9923231 (T) | Increased sensitivity to warfarin |
| SLCO1B1 | ||
| *5 | rs4149056 (C) | Decreased function |
| HLA-B | ||
| *58:01 | N/A | Increased SCAR risk |
TPMT, thiopurine methyltransferase; N/A, not applicable; SCAR, severe cutaneous adverse reaction.
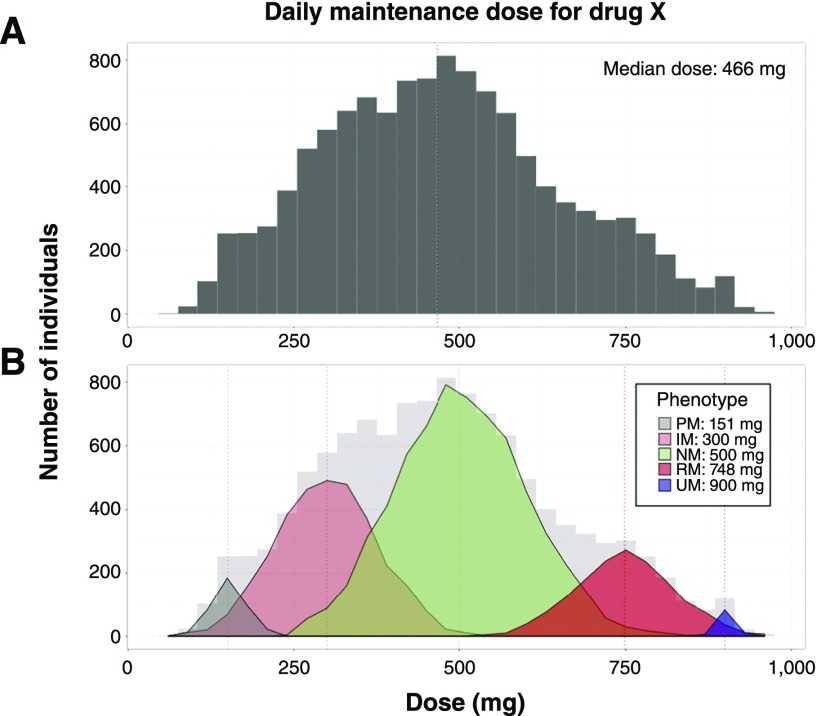
CYP3A5 is a member of the cytochrome p-450 enzyme system. It catalyzes the oxidation of tacrolimus to inactive metabolites (12). Patients who express CYP3A5 tend to require higher than usual tacrolimus doses to reach therapeutic concentrations (13). If J.H.’s CYP3A5 status was known at the initiation of therapy, then a more informed decision about tacrolimus dosing could be made. Knowing basic genetic information requires that clinicians recognize individual variability early in clinical care. This point is further illustrated in Figure 3, showing how unrecognized subpopulations may be unmasked by pharmacogenomics. In this situation, the theoretical median dose requirements may vary widely between these subpopulations and the ungenotyped aggregate.
Figure 3.
Simulated dosing data for drug X after patient titration to effect shows distinct subpopulations based on genetics. A shows the distribution of total daily dose among the aggregate population, suggesting a mostly normal distribution of doses. B shows population substructure on the basis of genetic variants in the gene responsible for metabolism of drug X for poor metabolizers (PMs), intermediate metabolizers (IMs), normal metabolizers (NMs), rapid metabolizers (RMs), and ultrapid metabolizers (UMs). Patients with decreased metabolism of drug X (PMs and IMs) have a lower effective dose, whereas patients with increased metabolism (RMs and UMs) require higher doses. This shows the utility of pharmacogenomics-based dosing in addition to clinical factors to identify subpopulations.
Pharmacogenomics Resources
Easily accessible web resources are available to the clinician to help interpret pharmacogenomics information. These include data aggregation sites, evidence-based clinical guidelines, and regulatory data (14). Pharmacogenomics information may be found in the FDA-approved prescribing information, although these data may appear in different sections (e.g., dosing information, clinical pharmacology, etc.) (6). The Pharmacogenomics Knowledge Base (www.pharmgkb.org/) is a comprehensive annotated pharmacogenomics resource that includes clinical guidelines, FDA labeling, and pharmacogenomics-related pathways (5). Organizations that compile pharmacogenomics evidence to develop clinical guidelines include the Clinical Pharmacogenetics Implementation Consortium (CPIC; www.cpicpgx.org) and the Dutch Pharmacogenetics Working Group (15). The CPIC was created to overcome implementation barriers by developing standardized clinical pharmacogenomics guidelines. Over 20 guidelines have been published since 2012, and they are publicly available to aid clinicians in translating genetic laboratory test results into actionable prescribing decisions (16).
Clinical Pharmacogenomics for the Nephrologist
In the following sections, drug-gene pairs with clinical guidelines and a high level of evidence in conditions commonly treated by the nephrologist are presented. The focus is on cardiovascular disease and transplantation versus an exhaustive list of drugs and genes. Readers are encouraged to investigate the primary literature described herein as a means to further learn about pharmacogenomics in relevant therapeutic areas. A summary of the gene-drug pairs and clinical guidelines discussed is provided in Table 2.
Table 2.
Summary of the gene-drug pairs and clinical guidelines relevant to nephrology
| Drug | Gene | Clinical Guidance Summary | Ref. |
|---|---|---|---|
| Warfarin | CYP2C9 | Use lower dose if a poor or intermediate metabolizer (e.g., *2/*2, *1/*2) | 23 |
| Warfarin | CYP4F2 | Use lower dose if decreased activity (*3) | 23 |
| Warfarin | VKORC1 | Use lower dose if increased sensitivity (−1639G>A) | 23 |
| Clopidogrel | CYP2C19 | Use alternative antiplatelet agent if poor or intermediate metabolizer (e.g., *2/*2, *1/*2); monitor for bleeding if ultrarapid metabolizer (*1/*17, *17/*17) | 35 |
| Simvastatin | SLCO1B1 | Use lower dose or alternative agent in patients with decreased transporter activity (*5, *15, *17) | 37 |
| Azathioprine | TPMT | Patients with decreased TPMT function have higher risk for toxicity | 39 |
| Tacrolimus | CYP3A5 | Carriers of at least one functional (*1) allele may require higher doses | 13 |
| Voriconazole | CYP2C19 | Use an alternative agent in CYP2C19 rapid/ultrarapid metabolizer (*1/*17, *17/*17); use alternative agent or lower dose in CYP2C19 poor metabolizer (*2/*2, *3/*3) | 40 |
| Allopurinol | HLA-B | User an alternative uric acid–lowering agent in patients who carry at least one *58:01 allele | 53 |
TPMT, thiopurine methyltransferase.
Cardiovascular Disease
Cardiovascular disease is a leading cause of death for patients suffering from CKD. Hallmarks of cardiovascular disease secondary to CKD are cardiac remodeling, atherosclerosis, and arteriosclerosis (17). Pharmacogenomics of cardiovascular disease is an active area of research and clinical implementation, with evidence-based guidelines for antiplatelets, anticoagulation, and hyperlipidemia (16,18,19).
Antiplatelet Agents and Anticoagulants
Warfarin.
Warfarin is a vitamin K antagonist that inhibits coagulation by inhibiting the formation of coagulation factors II, VII, IX, and X and proteins C and S (20). It is a narrow therapeutic index drug with high interpatient variability and a delayed time to action (i.e., dose changes are not reflected in laboratory values for approximately 72 hours) (21). Frequent monitoring of the international normalized ratio (INR) over days to weeks is needed to determine the right dose. Patients with impaired kidney function are further known to require lower dosages of warfarin, have worse control of anticoagulation, and are at a higher risk for major hemorrhage (22).
Warfarin pharmacokinetics and pharmacodynamics are affected by multiple genotypes. Genetic variations affecting CYP2C9 and CYP4F2 metabolism and VKORC1 sensitivity are known to predict the dose needed to attain optimal anticoagulation (a therapeutic INR) but are not without controversy (23). In 2013, divergent clinical trial results significantly diminished enthusiasm for routine warfarin pharmacogenomics in all patients. Although the European Pharmacogenetics of Anticoagulation Therapy Study showed that use of a pharmacogenomics algorithm increased time in therapeutic range versus fixed dosing at 12 weeks in a predominantly white population, the Clarification of Optimal Anticoagulation through Genetics Trial showed no improvement of a pharmacogenomics algorithm over a clinical algorithm at 4 weeks in a more diverse American population (24,25). Most recently, however, the multicenter, randomized Genetics Informatics Trial (GIFT) showed that genotype-guided warfarin dosing improved clinical outcomes versus clinically guided dosing. The rate of a composite of major bleeding, INR of four or greater, venous thromboembolism, or death was reduced from 14.7% to 10.8% in elderly patients undergoing elective hip or knee arthroplasty (26). Collectively, these trials show the importance of generalizability of results; measuring hard clinical outcomes versus surrogates, like INR; ethnic diversity in clinical trials; and the genotype coverage of pharmacogenomics testing. Future work will no doubt study cost-effectiveness and the effect of broader genotyping. In fact, in a recent prospective observational trial, de Oliveira Almeida et al. (27) found that other genetic variants (in APOE, ABCB1, and UGT1A1) were also associated with warfarin dose.
Current evidence-based CPIC guidelines for warfarin dosing include CYP2C9, VKORC1, and CYP4F2, and they are specific to patient self-identified ancestry. In non-African ancestry patients, the highest evidence is available for patients who carry at least one reduced function CYP2C9 allele (e.g., *2, *3), which predicts decreased hepatic clearance and lower dose requirements. Patients carrying a VKORC1–1639G>A allele are expected to have higher sensitivity to warfarin, thus requiring a lower dose. Individuals who have both of these variations require much lower doses of warfarin. Carriers of CYP4F2 *3 allele may also require a 5%–10% increase in dose. In patients with African ancestry, because nearly one half of individuals may carry CYP2C9 *5, 6, *8, *11, or rs12777823 variants, genotype-guided warfarin dosing is only recommended if testing covers these variants (23).
The FDA-approved product labeling contains recommendations for initial dosing with a convenient table on the basis of CYP2C9 and VKORC1 (28). Finally, Gage et al. (29), who led the GIFT, also maintain a web-based application (www.warfarindosing.org), which incorporates additional clinical and genetic data to provide tailored warfarin dosing in an easy-to-use interface. Neither tool currently incorporates kidney function in these recommendations.
Clopidogrel.
Antiplatelet medications (prasugrel, ticagrelor, and clopidogrel) are indicated for patients who receive coronary artery stenting (30). They may also be used after kidney artery stenting, although the evidence for this is less robust (31). These drugs carry differing risks for bleeding, treatment failure, and cost, and their use is challenged by the lack of a well validated biomarker of treatment response. The most commonly prescribed drug, clopidogrel, is a prodrug that requires metabolic activation by CYP2C19 among other enzymes. Patients with decreased metabolic activity at CYP2C19 have decreased generation of the active metabolite and decreased platelet inhibition (32,33). Conversely, patients with increased CYP2C19 activity (rapid and ultrarapid metabolizers) may have increased generation of the active metabolite for clopidogrel and thus, a theoretically higher platelet inhibition and increased risk for bleeding. The National Institutes of Health–funded Implementing Genomics in Practice network’s multicenter observational trial investigated patient outcomes with pharmacogenomics-guided antiplatelet therapy after percutaneous coronary intervention and stenting. Patients carrying at least one nonfunctional allele at CYP2C19 who were treated with clopidogrel versus alternative therapy were at higher risk for major adverse cardiovascular events (hazard ratio, 2.26; 95% CI: 1.18 to 4.32; P=0.01) (34). This suggests that pharmacogenomics testing for CYP2C19 may provide a significant clinical benefit in real world clinical use. The CPIC guideline for clopidogrel therapy recommends that patients with at least one decreased function allele (*2, *3, etc.) receive an alternative agent due to risk for decreased response. Additionally, the guideline recommends that patients with increased metabolism (*1/*17 and *17/*17) be monitored for increased bleeding risk, although it does not recommend different dosing (35). Implementation of routine CYP2C19 testing in cardiac catheterization laboratories is feasible and has been a popular first pharmacogenomics implementation at several health systems (9).
Hyperlipidemia
Hepatic hydroxymethyl glutaryl–CoA reductase inhibitors (e.g., simvastatin, atorvastatin, and “statins”) are commonly used as cholesterol-lowering agents, but they are known for rare but significant myotoxicity that can progress to rhabdomyolysis. Although the risk for toxicity is low for most statin medications, high-dose (80 mg) simvastatin may carry a slightly higher risk than other statins (36). Additionally, this risk has been associated with a pharmacogenomics marker, which could allow clinicians to either prescribe a lower dose or use an alternative agent when it is detected (36). The polymorphic transporter gene SLCO1B1 is responsible for uptake of simvastatin from the blood to hepatocytes, where it is metabolized. SLCO1B1 function is critical for simvastatin transport from blood to the liver, and when function is diminished, simvastatin blood concentrations are higher (increased systemic exposure) (36). Patients who carry at least one reduced function allele in SLCO1B1 (*5, *15, or *17) should receive an alternative agent or a reduced dose of simvastatin. Providers should also consider routine creatinine kinase monitoring in this situation (37). Although other statins carry a risk for myotoxicity, SLCO1B1 plays little to no role in prediction because of drug lipophilicity and the predominant route of elimination (kidney elimination versus being a substrate for hepatic SLCO1B1) (37).
Transplantation
Kidney transplant is the treatment of choice for stage 5 CKD, and it is another area where pharmacogenomics can augment current precision medicine practices (38). Post-transplant medications have a narrow therapeutic index with interpatient variability that can be partially explained by genetic determinants. Evidence-based guidelines exist for several medications found in the post-transplant regimen, including azathioprine, tacrolimus, and voriconazole (13,39,40).
Azathioprine
Azathioprine is an antimetabolite used post-transplantation for long-term maintenance immunosuppressive therapy, and it is highlighted as a strategy to decrease costs for patients with kidney transplant (38). Azathioprine is a prodrug converted to mercaptopurine that undergoes methylation to inactive metabolites through polymorphic thiopurine methyltransferase (TPMT) (39). TPMT activity is influenced by genotype, and dosing recommendations are available to mitigate potential toxicities resulting from predictable pharmacokinetic changes (41,42).
Dose adjustments for initial dosing in patients with less or nonfunctional TPMT enzyme activity aim to reduce the risk of severe myelosuppression. Evidence-based guidelines recommend that patients who are TPMT heterozygous (one of the following alleles: *2, *3A, *3B, *3C, and *4) receive lower initial dosing of any thiopurine medication (azathioprine, mercaptopurine, or thioguanine). Patients with the homozygous variant genotype (two of the following alleles: *2, *3A, *3B, *3C, and *4) are at a substantial risk for severe, potentially life-threatening myelosuppression due to the accumulation of active metabolites (43). For patients with two nonfunctional TPMT alleles, guidelines recommend alternative therapy or an extreme dose decrease of azathioprine (39).
Tacrolimus
Tacrolimus is a calcineurin inhibitor that remains at the cornerstone of long-term immunosuppressant therapy post-transplantation. Clinical use is characterized by routine therapeutic drug monitoring due to its narrow therapeutic window and wide interpatient variability (38). Tacrolimus undergoes oxidative metabolism to inactive metabolites by CYP3A4/3A5 enzymes and can be affected by genetic variations in CYP3A5 (44). Most whites (80%–85%) do not express CYP3A5 and may fall within standard, label-recommended dosing, whereas patients who express CYP3A5, prevalent in the black population as exemplified in the previous patient, may require higher doses (11,45).
A modified initial tacrolimus dosing on the basis of CYP3A5 metabolizer status is suggested if the genotype is known. In patients who express one or two functional copies of CYP3A5 (e.g., *1 combinations; normal or intermediate metabolizers), guidelines recommend a starting dose 1.5–2.0 times higher than the typical starting dose, not to exceed 0.3 mg/kg per day. The goal for consideration of CYP3A5 metabolizer status in addition to other clinical factors is to reach therapeutic concentrations more quickly (13).
Voriconazole
Voriconazole is a triazole antifungal agent that is indicated in patients with kidney transplants and invasive fungal infection (46). Similar to aforementioned transplant agents, voriconazole has wide interpatient variability and a narrow therapeutic index, necessitating therapeutic drug monitoring (47). Metabolism of voriconazole occurs predominantly through CYP2C19 (40).
Evidence-based guidelines recommend alternative therapy or altered dosing for certain genotypes in efforts to avoid treatment failure and reduce the risk of adverse effects. In patients with CYP2C19 ultrarapid or rapid metabolizer status (*17/*17 or *1/*17, respectively), guidelines recommend an alternative agent due to unlikely achievement of target concentrations. Patients with CYP2C19 poor metabolizer status (two alleles of either *2 or *3) have a higher risk of adverse effects due to diminished metabolism, and an alternative agent not dependent on the CYP2C19 pathway is recommended (39).
Hyperuricemia
Elevated uric acid (hyperuricemia) is a frequent comorbidity in patients with CKD, and it is a contributor to the disease progression (48,49). Allopurinol is commonly prescribed to lower uric acid levels, and it is believed to be one of the leading causes of drug-related severe cutaneous adverse reactions (SCARs), including toxic epidermal necrolysis and Steven Johnson Syndrome (50). Risk for medication-induced SCAR has been associated with certain variants of the HLA-B gene from the MHC locus (51). Patients who carry at least one HLA-B*58:01 allele are at higher risk for SCAR from allopurinol (52). This allele was first discovered in East Asian populations and has since been detected and associated with SCAR from allopurinol in European populations (52,53). The CPIC guideline for allopurinol recommends against the use of allopurinol in patients who carry at least one HLA-B*58:01 allele (53,54), but it does not provide a recommendation on whether to test patients preemptively (48,49).
Growing Evidence for Diabetes Treatments
Diabetic nephropathy is a leading cause for CKD and ESKD (55). As such, nephrologists treat many patients with comorbid type 2 diabetes mellitus who are managed on metformin. Although there are no clinical guidelines for the use of pharmacogenomics to tailor therapy with metformin, evidence has been growing that supports the use of the SNP rs11212617 in an intergenic (nongene) region of the genome called the chromosome 11 open reading frame 65 region. At this SNP, the presence of at least one “A” allele is associated with decreased response to metformin (56). In the future, this variation or others affecting pharmacokinetics (e.g., transporters) may be useful for predicting which patients will require altered doses of metformin for adequate hemoglobin A1c control.
Clinical Decision Making and the Use of Pharmacogenomics in Practice
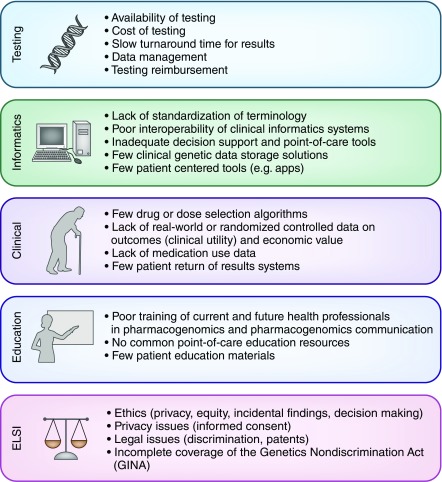
Several barriers prevent more widespread pharmacogenomics clinical implementation in everyday practice (Figure 4). Providers need to understand pharmacogenomics concepts for successful precision medicine clinical decision making, specifically whether they can apply the pharmacogenomics data within their current practice model, how the data should be integrated with other clinical parameters, and if a referral to a specialist should be made (e.g., pharmacist, medical geneticist, or genetic counselor) (57). The addition of these data also creates a growing need for provider education. Strategies to train practicing providers and health care students to use pharmacogenomics in practice have been reviewed elsewhere (58) and range from clinical decision support at the point of care to education courses that allow learners to undergo personal genomic testing as a means of learning with one’s own data (59).
Figure 4.
Pharmacogenomics implementation is limited by challenges in testing, informatics, clinical constraints, lack of education, and ELSI. ELSI, ethical, legal, and social implications.
Providers frequently face challenges with availability of testing and the origin of data, which may be from a new provider-initiated order, existing data from a previous pharmacogenomics result report, or patient-provided results from direct to consumer testing (e.g., 23andMe, Mountainview, CA). It is necessary to ensure that testing was performed using an FDA-approved test or that it was done in a Clinical Laboratory Improvement Amendments environment with appropriate clinical validation (60). Germline (i.e., genome from birth) genetic testing results generally do not change; thus, it may be cost saving to reuse test results. The testing technology used and testing coverage may vary (61). Although genome sequencing costs are plummeting, most clinical pharmacogenomics testing is accomplished using genotyping arrays targeting specific variants. It is prudent to evaluate what genes and variants were covered by the testing platform. For example, some commercial tests for CYP2C19 only test for *2 and *3, although at least eight additional low-prevalence star alleles are associated with decreased function and *17 is associated with increased activity (62).
The decision to order testing relies on the expected clinical utility of the data, availability and turnaround time of testing, and timeline for pharmacotherapy initiation. This can also be influenced by the potential broad application of data returned from a test, which may support future prescribing decisions (63). Deciding whether to test should also incorporate patient ancestry, specifically in patients in whom race/ethnicity can inform the probability of carrying one or more pharmacogenomic markers. For example, J.H. in the patient case was of African ancestry, which suggests higher probability that he carried at least one CYP3A5*1 allele (11). This is also evident for HLA-B*15:02, which predicts SCAR associated with carbamazepine and is found more often in those with Han Chinese ancestry (64). Regarding timing, warfarin therapy usually should not be delayed pending genetic data, but rather, the standard clinical algorithms should be initiated. If existing data are available or a genetic test with a rapid turnaround (<24 hours) is available, data may be actionable early enough to guide prescribing. However, for medications where genetic data can predict significant toxicity (e.g., HLA-B), it may be prudent to wait for test results (65). Table 3 provides a practical summary of the decision-making process for whether to use pharmacogenomics.
Table 3.
Clinical decision-making process for integrating pharmacogenomics in practice
| Factor | Questions |
|---|---|
| Patient or population | Is the variant likely relevant in the patient or population? |
| How common is the variant? | |
| Quality of evidence | What is the strength of the evidence for the use of data? |
| Are clinical guidelines or FDA recommendations available? | |
| Testing | Is testing available? |
| Is the coverage appropriate? | |
| What is the turnaround time of results? | |
| Data availability | Does pharmacogenomics data already exist? |
| Is the data quality sufficient to use? | |
| Drug factors | How important is the gene/variant for the pharmacokinetics or pharmacodynamics of the drug? |
| Does the drug have a narrow therapeutic index? | |
| Is the drug a prodrug or active? | |
| Will the variant decrease efficacy and/or increase toxicity? | |
| Clinical factors | Are there other factors relevant to the decision, like timing of drug start or previous use of the medication? |
| How do comorbid clinical conditions affect expected phenotypes? | |
| Are there drug-drug interactions that affect expected phenotypes? |
FDA, Food and Drug Administration.
Determining value of getting data before prescribing versus starting the medication immediately is tantamount to the argument of reactive versus preemptive pharmacogenomics testing. Reactive testing is the practice of ordering a pharmacogenomics test only when needed, which ensures that the testing is indicated (and increases the likelihood of payer reimbursement) at the expense of having to wait for results to be returned. Conversely, preemptive pharmacogenomics testing means that the worry of turnaround time is nonexistent, because the data are available before prescribing (66). Broad testing of many genes (i.e., a pharmacogenomics panel) using a preemptive testing strategy is commonly advocated as the final step to make routine use of pharmacogenomics in practice cost effective (67). However, inadequate resources to support frontline providers for pharmacogenomics decision making and challenges in achieving payer reimbursement for preemptive testing limit these strategies (68).
Unique challenges and opportunities to integrating pharmacogenomics into the care of patients with kidney disease also exist. CKD is known to alter pharmacodynamic and pharmacokinetic relationships of several medications, particular those that rely on kidney elimination. In general, this scales with CKD stage and can be especially challenging in patients receiving dialysis (69). Nephrologists must also consider the systemic changes in patients with CKD, such as the changes in hepatic drug metabolism and other nonkidney clearance pathways that occur in patients with CKD (69). Phenocoversion is when there is a genotype-phenotype mismatch (e.g., a normal metabolizer having a phenotype that looks like a poor metabolizer). The classic example is a drug interaction over-riding or masking the genotype-predicted phenotype. However, CKD could also be an extrinsic factor that may affect final drug response phenotypes. Future nephrology research should evaluate how to manage patients with CKD in this clinical scenario.
Despite the barriers to clinical pharmacogenomics, several large academic medical centers and increasingly, community providers are launching pharmacogenomics implementation programs (9,63). It is likely that the future will bring a greater expansion of precision medicine and enhanced data sharing between providers and patients. This will also add new opportunities to improve patient pharmacotherapy outcomes and additional risk that will need to be managed at all levels of clinical care and research (70,71).
Conclusions
Nephrologists care for patients with significant comorbidities and are challenged by wide interpatient variability in medication responses. They are ideally positioned to champion integration of pharmacogenomics to achieve precision medicine in the many disease areas affected by kidney disease. As pharmacogenomics knowledge expands, nephrologists will need to have familiarity with the state of the pharmacogenomics science, available pharmacogenomics resources and guidelines, contemporary application of pharmacogenomics data for specific drugs, and clinical decision-making approaches to using pharmacogenomics data.
Disclosures
None.
Acknowledgments
This work is supported by National Institutes of Health grant 5TL1TR001858-02 (to S.M.A.), the American Foundation for Pharmaceutical Education (S.M.A.), and an anonymous donor (P.E.E.).
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1.Aymanns C, Keller F, Maus S, Hartmann B, Czock D: Review on pharmacokinetics and pharmacodynamics and the aging kidney. Clin J Am Soc Nephrol 5: 314–327, 2010 [DOI] [PubMed] [Google Scholar]
- 2.Cockcroft DW, Gault MH: Prediction of creatinine clearance from serum creatinine. Nephron 16: 31–41, 1976 [DOI] [PubMed] [Google Scholar]
- 3.Simoncelli T: Paving the Way for Personalized Medicine: FDA’s Role in a New Era of Medical Product Development, Rockville, MD, US Food and Drug Administration, 2013 [Google Scholar]
- 4.Empey PE: Pharmacogenomics to achieve precision medicine. Am J Health Syst Pharm 73: 1906–1907, 2016 [DOI] [PubMed] [Google Scholar]
- 5.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE: Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92: 414–417, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. FDA: Table of Pharmacogenomic Biomarkers in Drug Labeling. 2018. Available at: https://www.fda.gov/Drugs/ScienceResearch/ucm572698.htm. Accessed February 15, 2018.
- 7.Ahmed S, Zhou Z, Zhou J, Chen SQ: Pharmacogenomics of drug metabolizing enzymes and transporters: Relevance to precision medicine. Genomics Proteomics Bioinformatics 14: 298–313, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavlos R, Mallal S, Phillips E: HLA and pharmacogenetics of drug hypersensitivity. Pharmacogenomics 13: 1285–1306, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Empey PE, Stevenson JM, Tuteja S, Weitzel KW, Angiolillo DJ, Beitelshees AL, Coons JC, Duarte JD, Franchi F, Jeng LJB, Johnson JA, Kreutz RP, Limdi NA, Maloney KA, Owusu Obeng A, Peterson JF, Petry N, Pratt VM, Rollini F, Scott SA, Skaar TC, Vesely MR, Stouffer GA, Wilke RA, Cavallari LH, Lee CR; IGNITE Network: Multisite investigation of strategies for the implementation of CYP2C19 genotype-guided antiplatelet therapy [published online ahead of print December 26, 2017]. Clin Pharmacol Ther doi: 10.1002/cpt.1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roden DM, Altman RB, Benowitz NL, Flockhart DA, Giacomini KM, Johnson JA, Krauss RM, McLeod HL, Ratain MJ, Relling MV, Ring HZ, Shuldiner AR, Weinshilboum RM, Weiss ST; Pharmacogenetics Research Network: Pharmacogenomics: Challenges and opportunities. Ann Intern Med 145: 749–757, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobson PA, Oetting WS, Brearley AM, Leduc R, Guan W, Schladt D, Matas AJ, Lamba V, Julian BA, Mannon RB, Israni A; DeKAF Investigators: Novel polymorphisms associated with tacrolimus trough concentrations: Results from a multicenter kidney transplant consortium. Transplantation 91: 300–308, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamba J, Hebert JM, Schuetz EG, Klein TE, Altman RB: PharmGKB summary: Very important pharmacogene information for CYP3A5. Pharmacogenet Genomics 22: 555–558, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birdwell KA, Decker B, Barbarino JM, Peterson JF, Stein CM, Sadee W, Wang D, Vinks AA, He Y, Swen JJ, Leeder JS, van Schaik R, Thummel KE, Klein TE, Caudle KE, MacPhee IA: Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin Pharmacol Ther 98: 19–24, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G, Zhang Y, Ling Y, Jia J: Web resources for pharmacogenomics. Genomics Proteomics Bioinformatics 13: 51–54, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swen JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, Mulder H, Rongen GA, van Schaik RH, Schalekamp T, Touw DJ, van der Weide J, Wilffert B, Deneer VH, Guchelaar HJ: Pharmacogenetics: From bench to byte--an update of guidelines. Clin Pharmacol Ther 89: 662–673, 2011 [DOI] [PubMed] [Google Scholar]
- 16.Relling MV, Klein TE: CPIC: Clinical Pharmacogenetics Implementation Consortium of the pharmacogenomics research network. Clin Pharmacol Ther 89: 464–467, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarnak MJ: Cardiovascular complications in chronic kidney disease. Am J Kidney Dis 41[Suppl]: 11–17, 2003 [DOI] [PubMed] [Google Scholar]
- 18.Giudicessi JR, Kullo IJ, Ackerman MJ: Precision cardiovascular medicine: State of genetic testing. Mayo Clin Proc 92: 642–662, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson JA, Cavallari LH: Pharmacogenetics and cardiovascular disease--implications for personalized medicine. Pharmacol Rev 65: 987–1009, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fareed J, Thethi I, Hoppensteadt D: Old versus new oral anticoagulants: Focus on pharmacology. Annu Rev Pharmacol Toxicol 52: 79–99, 2012 [DOI] [PubMed] [Google Scholar]
- 21.Kuruvilla M, Gurk-Turner C: A review of warfarin dosing and monitoring. Proc Bayl Univ Med Cent 14: 305–306, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Limdi NA, Beasley TM, Baird MF, Goldstein JA, McGwin G, Arnett DK, Acton RT, Allon M: Kidney function influences warfarin responsiveness and hemorrhagic complications. J Am Soc Nephrol 20: 912–921, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson JA, Caudle KE, Gong L, Whirl-Carrillo M, Stein CM, Scott SA, Lee MT, Gage BF, Kimmel SE, Perera MA, Anderson JL, Pirmohamed M, Klein TE, Limdi NA, Cavallari LH, Wadelius M: Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing: 2017 Update. Clin Pharmacol Ther 102: 397–404, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pirmohamed M, Burnside G, Eriksson N, Jorgensen AL, Toh CH, Nicholson T, Kesteven P, Christersson C, Wahlström B, Stafberg C, Zhang JE, Leathart JB, Kohnke H, Maitland-van der Zee AH, Williamson PR, Daly AK, Avery P, Kamali F, Wadelius M; EU-PACT Group: A randomized trial of genotype-guided dosing of warfarin. N Engl J Med 369: 2294–2303, 2013 [DOI] [PubMed] [Google Scholar]
- 25.Kimmel SE, French B, Kasner SE, Johnson JA, Anderson JL, Gage BF, Rosenberg YD, Eby CS, Madigan RA, McBane RB, Abdel-Rahman SZ, Stevens SM, Yale S, Mohler ER 3rd, Fang MC, Shah V, Horenstein RB, Limdi NA, Muldowney JA 3rd, Gujral J, Delafontaine P, Desnick RJ, Ortel TL, Billett HH, Pendleton RC, Geller NL, Halperin JL, Goldhaber SZ, Caldwell MD, Califf RM, Ellenberg JH; COAG Investigators: A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med 369: 2283–2293, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gage BF, Bass AR, Lin H, Woller SC, Stevens SM, Al-Hammadi N, Li J, Rodríguez T Jr, Miller JP, McMillin GA, Pendleton RC, Jaffer AK, King CR, Whipple BD, Porche-Sorbet R, Napoli L, Merritt K, Thompson AM, Hyun G, Anderson JL, Hollomon W, Barrack RL, Nunley RM, Moskowitz G, Dávila-Román V, Eby CS: Effect of genotype-guided warfarin dosing on clinical events and anticoagulation control among patients undergoing hip or knee arthroplasty: The GIFT randomized clinical trial. JAMA 318: 1115–1124, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Oliveira Almeida VC, Ribeiro DD, Gomes KB, Godard AL: Polymorphisms of CYP2C9, VKORC1, MDR1, APOE and UGT1A1 genes and the therapeutic warfarin dose in Brazilian patients with thrombosis: A prospective cohort study. Mol Diagn Ther 18: 675–683, 2014 [DOI] [PubMed] [Google Scholar]
- 28. Amneal Pharmaceuticals LLC: WARFARIN SODIUM—Warfarin Tablet, [Rev. 8/2017], 2017. Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=558b7a0d-5490-4c1b-802e-3ab3f1efe760. Accessed February 15, 2018.
- 29.Gage BF, Eby C, Johnson JA, Deych E, Rieder MJ, Ridker PM, Milligan PE, Grice G, Lenzini P, Rettie AE, Aquilante CL, Grosso L, Marsh S, Langaee T, Farnett LE, Voora D, Veenstra DL, Glynn RJ, Barrett A, McLeod HL: Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther 84: 326–331, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine GN, Bates ER, Bittl JA, Brindis RG, Fihn SD, Fleisher LA, Granger CB, Lange RA, Mack MJ, Mauri L, Mehran R, Mukherjee D, Newby LK, O'Gara PT, Sabatine MS, Smith PK, Smith SC Jr: 2016 ACC/AHA guideline focused update on duration of dual antiplatelet therapy in patients with coronary artery disease: A report of the American College of Cardiology/American Heart Association Task Force on clinical practice guidelines. J Am Coll Cardiol 68: 1082–1115, 2016 [DOI] [PubMed] [Google Scholar]
- 31.Mousa AY, Broce M, Campbell J, Nanjundappa A, Stone PA, Abu-Halimah S, Srivastava M, Bates MC, Aburahma AF: Clopidogrel use before renal artery angioplasty with/without stent placement resulted in tertiary procedure risk reduction. J Vasc Surg 56: 416–423, 2012 [DOI] [PubMed] [Google Scholar]
- 32.Scott SA, Sangkuhl K, Gardner EE, Stein CM, Hulot JS, Johnson JA, Roden DM, Klein TE, Shuldiner AR; Clinical Pharmacogenetics Implementation Consortium: Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther 90: 328–332, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim KA, Park PW, Hong SJ, Park JY: The effect of CYP2C19 polymorphism on the pharmacokinetics and pharmacodynamics of clopidogrel: A possible mechanism for clopidogrel resistance. Clin Pharmacol Ther 84: 236–242, 2008 [DOI] [PubMed] [Google Scholar]
- 34.Cavallari LH, Lee CR, Beitelshees AL, Cooper-DeHoff RM, Duarte JD, Voora D, Kimmel SE, McDonough CW, Gong Y, Dave CV, Pratt VM, Alestock TD, Anderson RD, Alsip J, Ardati AK, Brott BC, Brown L, Chumnumwat S, Clare-Salzler MJ, Coons JC, Denny JC, Dillon C, Elsey AR, Hamadeh IS, Harada S, Hillegass WB, Hines L, Horenstein RB, Howell LA, Jeng LJB, Kelemen MD, Lee YM, Magvanjav O, Montasser M, Nelson DR, Nutescu EA, Nwaba DC, Pakyz RE, Palmer K, Peterson JF, Pollin TI, Quinn AH, Robinson SW, Schub J, Skaar TC, Smith DM, Sriramoju VB, Starostik P, Stys TP, Stevenson JM, Varunok N, Vesely MR, Wake DT, Weck KE, Weitzel KW, Wilke RA, Willig J, Zhao RY, Kreutz RP, Stouffer GA, Empey PE, Limdi NA, Shuldiner AR, Winterstein AG, Johnson JA, Network I: Multisite Investigation of Outcomes With Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. JACC Cardiovasc Interv 11: 181–191, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, Klein TE, Sabatine MS, Johnson JA, Shuldiner AR; Clinical Pharmacogenetics Implementation Consortium: Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 Update. Clin Pharmacol Ther 94: 317–323, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Armitage J, Bowman L, Wallendszus K, Bulbulia R, Rahimi K, Haynes R, Parish S, Peto R, Collins R; Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine (SEARCH) Collaborative Group: Intensive lowering of LDL cholesterol with 80 mg versus 20 mg simvastatin daily in 12,064 survivors of myocardial infarction: A double-blind randomised trial. Lancet 376: 1658–1669, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramsey LB, Johnson SG, Caudle KE, Haidar CE, Voora D, Wilke RA, Maxwell WD, McLeod HL, Krauss RM, Roden DM, Feng Q, Cooper-DeHoff RM, Gong L, Klein TE, Wadelius M, Niemi M: The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 Update. Clin Pharmacol Ther 96: 423–428, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kidney Disease: Improving Global Outcomes Transplant Work Group: KDIGO clinical practice guideline for the care of kidney transplant recipients. Am J Transplant 9[Suppl 3]: S1–S155, 2009 [DOI] [PubMed] [Google Scholar]
- 39.Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui CH, Yee SW, Stein CM, Carrillo M, Evans WE, Klein TE; Clinical Pharmacogenetics Implementation Consortium: Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther 89: 387–391, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moriyama B, Obeng AO, Barbarino J, Penzak SR, Henning SA, Scott SA, Agúndez J, Wingard JR, McLeod HL, Klein TE, Cross SJ, Caudle KE, Walsh TJ: Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C19 and voriconazole therapy [published online ahead of print December 16, 2016]. Clin Pharmacol Ther doi: 10.1002/cpt.583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Relling MV, Hancock ML, Rivera GK, Sandlund JT, Ribeiro RC, Krynetski EY, Pui CH, Evans WE: Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst 91: 2001–2008, 1999 [DOI] [PubMed] [Google Scholar]
- 42.Black AJ, McLeod HL, Capell HA, Powrie RH, Matowe LK, Pritchard SC, Collie-Duguid ES, Reid DM: Thiopurine methyltransferase genotype predicts therapy-limiting severe toxicity from azathioprine. Ann Intern Med 129: 716–718, 1998 [DOI] [PubMed] [Google Scholar]
- 43.Budhiraja P, Popovtzer M: Azathioprine-related myelosuppression in a patient homozygous for TPMT*3A. Nat Rev Nephrol 7: 478–484, 2011 [DOI] [PubMed] [Google Scholar]
- 44.Renders L, Frisman M, Ufer M, Mosyagin I, Haenisch S, Ott U, Caliebe A, Dechant M, Braun F, Kunzendorf U, Cascorbi I: CYP3A5 genotype markedly influences the pharmacokinetics of tacrolimus and sirolimus in kidney transplant recipients. Clin Pharmacol Ther 81: 228–234, 2007 [DOI] [PubMed] [Google Scholar]
- 45.Beermann KJ, Ellis MJ, Sudan DL, Harris MT: Tacrolimus dose requirements in African-American and Caucasian kidney transplant recipients on mycophenolate and prednisone. Clin Transplant 28: 762–767, 2014 [DOI] [PubMed] [Google Scholar]
- 46.Veroux M, Corona D, Gagliano M, Sorbello M, Macarone M, Cutuli M, Giuffrida G, Morello G, Paratore A, Veroux P: Voriconazole in the treatment of invasive aspergillosis in kidney transplant recipients. Transplant Proc 39: 1838–1840, 2007 [DOI] [PubMed] [Google Scholar]
- 47.Guinea J, Escribano P, Marcos-Zambrano LJ, Peláez T, Kestler M, Muñoz P, Vena A, López-Fabal F, Bouza E: Therapeutic drug monitoring of voriconazole helps to decrease the percentage of patients with off-target trough serum levels. Med Mycol 54: 353–360, 2016 [DOI] [PubMed] [Google Scholar]
- 48.Kang DH, Nakagawa T, Feng L, Watanabe S, Han L, Mazzali M, Truong L, Harris R, Johnson RJ: A role for uric acid in the progression of renal disease. J Am Soc Nephrol 13: 2888–2897, 2002 [DOI] [PubMed] [Google Scholar]
- 49.Ramirez MEG, Bargman JM: Treatment of asymptomatic hyperuricemia in chronic kidney disease: A new target in an old enemy - A review. J Adv Res 8: 551–554, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Halevy S, Ghislain PD, Mockenhaupt M, Fagot JP, Bouwes Bavinck JN, Sidoroff A, Naldi L, Dunant A, Viboud C, Roujeau JC; EuroSCAR Study Group: Allopurinol is the most common cause of Stevens-Johnson syndrome and toxic epidermal necrolysis in Europe and Israel. J Am Acad Dermatol 58: 25–32, 2008 [DOI] [PubMed] [Google Scholar]
- 51.Chung WH, Wang CW, Dao RL: Severe cutaneous adverse drug reactions. J Dermatol 43: 758–766, 2016 [DOI] [PubMed] [Google Scholar]
- 52.Hung SI, Chung WH, Liou LB, Chu CC, Lin M, Huang HP, Lin YL, Lan JL, Yang LC, Hong HS, Chen MJ, Lai PC, Wu MS, Chu CY, Wang KH, Chen CH, Fann CS, Wu JY, Chen YT: HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc Natl Acad Sci U S A 102: 4134–4139, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hershfield MS, Callaghan JT, Tassaneeyakul W, Mushiroda T, Thorn CF, Klein TE, Lee MT: Clinical Pharmacogenetics Implementation Consortium guidelines for human leukocyte antigen-B genotype and allopurinol dosing. Clin Pharmacol Ther 93: 153–158, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saito Y, Stamp LK, Caudle KE, Hershfield MS, McDonagh EM, Callaghan JT, Tassaneeyakul W, Mushiroda T, Kamatani N, Goldspiel BR, Phillips EJ, Klein TE, Lee MT; Clinical Pharmacogenetics Implementation Consortium: Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for human leukocyte antigen B (HLA-B) genotype and allopurinol dosing: 2015 Update. Clin Pharmacol Ther 99: 36–37, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gross JL, de Azevedo MJ, Silveiro SP, Canani LH, Caramori ML, Zelmanovitz T: Diabetic nephropathy: Diagnosis, prevention, and treatment. Diabetes Care 28: 164–176, 2005 [DOI] [PubMed] [Google Scholar]
- 56.Zhou K, Bellenguez C, Spencer CC, Bennett AJ, Coleman RL, Tavendale R, Hawley SA, Donnelly LA, Schofield C, Groves CJ, Burch L, Carr F, Strange A, Freeman C, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin A, Craddock N, Deloukas P, Dronov S, Duncanson A, Edkins S, Gray E, Hunt S, Jankowski J, Langford C, Markus HS, Mathew CG, Plomin R, Rautanen A, Sawcer SJ, Samani NJ, Trembath R, Viswanathan AC, Wood NW, Harries LW, Hattersley AT, Doney AS, Colhoun H, Morris AD, Sutherland C, Hardie DG, Peltonen L, McCarthy MI, Holman RR, Palmer CN, Donnelly P, Pearson ER; GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group; Wellcome Trust Case Control Consortium 2; MAGIC investigators: Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet 43: 117–120, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pletcher BA, Toriello HV, Noblin SJ, Seaver LH, Driscoll DA, Bennett RL, Gross SJ: Indications for genetic referral: A guide for healthcare providers. Genet Med 9: 385–389, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weitzel KW, Aquilante CL, Johnson S, Kisor DF, Empey PE: Educational strategies to enable expansion of pharmacogenomics-based care. Am J Health Syst Pharm 73: 1986–1998, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams SM, Anderson KB, Coons JC, Smith RB, Meyer SM, Parker LS, Empey PE: Advancing pharmacogenomics education in the core PharmD curriculum through student personal genomic testing. Am J Pharm Educ 80: 3, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen B, Gagnon M, Shahangian S, Anderson NL, Howerton DA, Boone JD; Centers for Disease Control and Prevention (CDC): Good laboratory practices for molecular genetic testing for heritable diseases and conditions. MMWR Recomm Rep 58: 1–37, 2009 [PubMed] [Google Scholar]
- 61.Katsanis SH, Katsanis N: Molecular genetic testing and the future of clinical genomics. Nat Rev Genet 14: 415–426, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ned Mmsc Phd RM: Genetic testing for CYP450 polymorphisms to predict response to clopidogrel: Current evidence and test availability. Application: Pharmacogenomics. PLoS Curr 2: pii: RRN1180, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dunnenberger HM, Biszewski M, Bell GC, Sereika A, May H, Johnson SG, Hulick PJ, Khandekar J: Implementation of a multidisciplinary pharmacogenomics clinic in a community health system. Am J Health Syst Pharm 73: 1956–1966, 2016 [DOI] [PubMed] [Google Scholar]
- 64.Ferrell PB Jr, McLeod HL: Carbamazepine, HLA-B*1502 and risk of Stevens-Johnson syndrome and toxic epidermal necrolysis: US FDA recommendations. Pharmacogenomics 9: 1543–1546, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin MA, Klein TE, Dong BJ, Pirmohamed M, Haas DW, Kroetz DL; Clinical Pharmacogenetics Implementation Consortium: Clinical pharmacogenetics implementation consortium guidelines for HLA-B genotype and abacavir dosing. Clin Pharmacol Ther 91: 734–738, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dunnenberger HM, Crews KR, Hoffman JM, Caudle KE, Broeckel U, Howard SC, Hunkler RJ, Klein TE, Evans WE, Relling MV: Preemptive clinical pharmacogenetics implementation: Current programs in five US medical centers. Annu Rev Pharmacol Toxicol 55: 89–106, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verbelen M, Weale ME, Lewis CM: Cost-effectiveness of pharmacogenetic-guided treatment: Are we there yet? Pharmacogenomics J 17: 395–402, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weitzel KW, Cavallari LH, Lesko LJ: Preemptive panel-based pharmacogenetic testing: The time is now. Pharm Res 34: 1551–1555, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nolin TD: A synopsis of clinical pharmacokinetic alterations in advanced CKD. Semin Dial 28: 325–329, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kulynych J, Greely HT: Clinical genomics, big data, and electronic medical records: Reconciling patient rights with research when privacy and science collide. J Law Biosci 4: 94–132, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McGuire AL, Basford M, Dressler LG, Fullerton SM, Koenig BA, Li R, McCarty CA, Ramos E, Smith ME, Somkin CP, Waudby C, Wolf WA, Clayton EW: Ethical and practical challenges of sharing data from genome-wide association studies: The eMERGE Consortium experience. Genome Res 21: 1001–1007, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]