Abstract
Background
One of the most important processes in a machine learning-based natural language processing is to represent words. The one-hot representation that has been commonly used has a large size of vector and assumes that the features that make up the vector are independent of each other. On the other hand, it is known that word embedding has a great effect in estimating the similarity between words because it expresses the meaning of the word well. In this study, we try to clarify the correlation between various terms in the biomedical texts based on the excellent ability of estimating similarity between words shown by word embedding. Therefore, we used word embedding to find new biomarkers and microorganisms related to a specific diseases.
Methods
In this study, we try to analyze the correlation between diseases-markers and diseases-microorganisms. First, we need to construct a corpus that seems to be related to them. To do this, we extract the titles and abstracts from the biomedical texts on the PubMed site. Second, we express diseases, markers, and microorganisms’ terms in word embedding using Canonical Correlation Analysis (CCA). CCA is a statistical based methodology that has a very good performance on vector dimension reduction. Finally, we tried to estimate the relationship between diseases-markers pairs and diseases-microorganisms pairs by measuring their similarity.
Results
In the experiment, we tried to confirm the correlation derived through word embedding using Google Scholar search results. Of the top 20 highly correlated disease-marker pairs, about 85% of the pairs have actually undergone a lot of research as a result of Google Scholars search. Conversely, for 85% of the 20 pairs with the lowest correlation, we could not actually find any other study to determine the relationship between the disease and the marker. This trend was similar for disease-microbe pairs.
Conclusions
The correlation between diseases and markers and diseases and microorganisms calculated through word embedding reflects actual research trends. If the word-embedding correlation is high, but there are not many published actual studies, additional research can be proposed for the pair.
Keywords: Word embedding, Canonical Correlation Analysis (CCA), Lexical similarity, t-distributed stochastic neighbor embedding (t-SNE), Bio-marker, Microorganisms
Background
A biomarker, or biological marker, generally refers to a measurable indicator of some biological state or condition. Biomarkers are often measured and evaluated to examine normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention [1]. A microorganism or microbe is a microscopic organism, which may be single-celled or multicellular [2]. Microorganisms are divided into prokaryotes, eukaryotes, and viruses. These microorganisms and biomarkers are known to have a strong relationship with human health and disease. One of the most accurate methods for identifying biomarkers or microorganisms affecting disease is clinical detection [3, 4]. This clinical approach has the drawback of being accurate but costing too much. Therefore, in this paper, we want to extract the related information from previously published texts, not the information that the patient holds directly, to understand the relationship between biomarkers, microorganisms and diseases [5]. First, diseases, markers and microorganisms are represented using word embedding. If we calculate the similarity between these expressions, the relationship between actual markers and microorganisms and diseases can be grasped to some extent. In this study, the word embedding used to understand the relationship between words shows a remarkable performance improvement in the field of natural language processing such as syntax parsing or sentiment analysis [6].
In this paper, we extracted documents containing biomarkers, microorganisms, and disease terms from PubMed [7]. The corpus was constructed by extracting only the title and summary part. With this corpus, we want to understand the relationship between diseases-biomarkers and diseases-microorganisms. Canonical Correlation Analysis (CCA) [8] is used as a method of representing a word. The result of embedding using the CCA is first mapped in two dimensions using t-distributed stochastic neighbor embedding (t-SNE) [9] and visualized in a two-dimensional space. We also estimate the correlation between diseases-markers, and diseases-microorganisms by calculating the cosine similarity of two-dimensionally reduced vectors. In order to verify the results of this study, we use the Google Scholar to check how active the research is actually in the top 20 pairs with high similarity. In other words, we tried to show the validity of the correlation by linking the estimated correlation with the activation level of actual research.
Bio-NLP
Natural language processing (NLP) is a field of computer science, artificial intelligence and computational linguistics concerned with the interactions between computers and human (natural) languages, and, in particular, concerned with programming computers to fruitfully process large natural language corpora [10]. Most commonly known applications include text analytics, Q & A, and machine translation [11, 12]. Biomedical text mining (also known as Bio-NLP) refers to text mining applied to texts and literature of the biomedical and molecular biology domain [13]. This field is based on natural language processing and bioinformatics. Recently, biomedical text has been rapidly growing, and research on Bio-NLP has attracted much attention. Bio-NLP can be used to identify the relationship between diseases-biomarkers and diseases-microorganisms in biomedical text. Using this information, it is possible to extract and use prior information about microorganisms or biomarkers that have affected the patient’s disease [14] implemented a bio-text mining system based on natural language processing that automatically extracts biomedical interaction information from biomedical text.
Bio-NLP related works
There are many ways to find biomarkers and microorganisms that are deeply related to human health and disease. There are four main ways to identify biomarkers. First, it uses the genome, or second, it uses protein information. Third, metabolites use metabolism to detect hidden mutations. Finally, there is a method of using lipid omics, a large-scale study of cellular lipid pathways and networks in biological systems. Since the whole sequence of the human genome has been analyzed, genome-based techniques have improved diagnostic techniques for cancer or disease [15]. Genome-based methods are used to evaluate gene and protein expression profiles in cancer cells [16, 17]. The use of protein information in the discovery of new biomarkers has been a popular method. Because protein information can be used to characterize proteins associated with cancer that have been modified or not [18, 19]. In addition, biomedical markers are discovered using bioinformatics [20].
Microorganisms, like biomarkers, can be found in many ways [21] uses a combination of DNA electrochemical sensors and PCR-amplification strategies to detect microorganisms [22] also describes various physical methods for detecting microorganisms. In addition to these methods, biomarkers and microorganisms can be discovered through machine learning, data mining, unsupervised learning, and word embedding [23–25].
Word embedding, which can measure similarities between words by representing words as vectors, has recently contributed to improving the performance of machine learning models used for natural language processing, in addition to biomarkers and microorganisms discovery. For example, the performance of NER (Named Entity Recognition) has been improved by using the results of word embedding as a features of conditional random field (CRF) [26–28]. This method is useful for many tasks of natural language processing such as machine translation and speech recognition [29, 30] also demonstrated effectiveness in the bio-NLP domain using Word2Vec and GloVe among the Word Embedding models in the biomedical domain.
Word-embeddings
Word embedding is a technique of learning the vector representation of every word in a given corpus. Previous studies of word embedding have expressed words in one-hot forms. In the one-hot form, when there is a dictionary of the vocabulary size of n, the size of the vector becomes very large because each word has the same size as the size of the dictionary. In the vector, only the position of the corresponding word is represented by 1, and the rest is represented by 0 [31]. The one-hot vector assumes that the feature elements of each vector are completely independent of each other. However, the one-hot method has two problems. First, the size of the vector is very large because each word has the same size of vector as the size of the vocabulary. Second, because there is no form of similarity between the word representations, we cannot understand what the words are related to. Word-embedding is a method of vectorizing the meaning of a word itself in a k-dimensional space to compensate for the drawbacks of this one-hot form. If the words are represented by word embedding, the similarity between these words can be measured. In addition, it can be deeper inferred by performing vector operations with vectorized semantics. Word-embedding also makes the operations simpler because words can be represented as low-dimensional vectors. Figure 1 shows the One-hot vectors and a word embedding vector.
Fig. 1.
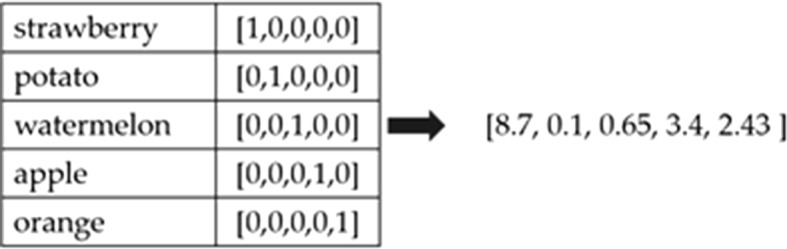
One-hot vectors and a word embedding vector
CCA (Canonical Correlation Analysis)
CCA is a technique known by Hotelling [32], which analyzes the correlation of variables. CCA is a statistical method used to investigate the relationship between two words, and simultaneously analyzes the correlations between the variables in the set and the variables in the other set. That is, the correlation of the variable group (X, Y) is grasped and a k-dimensional projection vector for maximizing the correlation is searched [33] showed that CCA can be a useful tool for investigating the relationship between two words.
In this paper, we use the CCA model to identify the relationship between specific diseases and biomarkers or microorganisms among the several models of word embedding. The CCA model is the best reflecting the global characteristics of the whole corpus among the various models. In [34], the performance of CCA is reported to be higher than that of Word2Vec’s Skip-gram in NER. Also, in [35], three representative models of word embedding (Word2Vec, CCA, GloVe) showed excellent category classification ability in biomedical domain. In this study, we constructed a corpus with title and abstract parts of the PubMed biomedical papers and showed good performance in classification of various categories such as disease names, symptoms, and biomarkers. Here, words embedded by the CCA [36] model are extracted more smoothly than by Word2Vec [37] and GloVe [38] models. For this reason, we use the CCA model for word embedding.
Methods
In this paper, we analyze the title and abstract of biomedical domain papers in the PubMed site to analyze the relationship between diseases-markers and diseases-microorganisms. The corpus for the analysis of the relationship was divided into a marker corpus and a microbial corpus. Of course, there are some documents that are included in both corpus.
Biomedical data
Figure 2 shows a paper in nxml format stored at the PubMed site. Although PubMed site also contains papers in PDF format, only papers in nxml format were used in this study because of convenience of use. In the paper file of Fig. 2, only the title part and the abstract part are extracted and a corpus is constructed. At this time, the title or abstract should include a marker or microorganism terms to be collected into the corpus.
Fig. 2.
A paper stored in the PubMed site
Tables 1 and 2 show a list of diseases and biomarkers, and diseases/symptoms/organs and microorganisms, that we want to correlate, respectively. Markers in Table 1 are markers that are known to be associated with the ovarian cancer [38], and diseases are high-frequency diseases in the corpus. Table 2 lists the most frequently occurring terms in corpus.
Table 1.
A list of diseases and biomarkers used in this research
| Diseases | Hepatitis, conjunctivitis, tuberculosis, hypertension, stomatitis, pneumothorax, glaucoma, meningitis, diabetes mellitus, cystitis, leukemia, adenocarcinoma, cancer, gastritis, tumor, asthma, dementia, pneumonia |
| Biomarkers | apoa-i, apoa-iii, CA125, CA15-3, CA19-9, CEA, Cortisol, CRP, CYFRA21-1, EGFR, FSH, HE4, IL-6, IL-8, MIF, MMP-7, Myoglobin, OPN, Prolactin, Tenascin-C, TTR |
Table 2.
A list of diseases/symptoms/organs and microorganisms
| Disease/symptom and organ | Liver, necrosis, meningitis, colitis, malaria, abdominal, diarrhea, foodborne, kidney, endocarditis, aspergillosis, fever, stomach, spleen, colorectal, bowel, candidiasis, crohn, lung, pneumonia |
| Microorganism | Archaea, aeruginosa, listeria, mycobacteria, actinobacteria, burkholderia, pathogen, vibrio, salmonella, cerevisiae, cyanobacteria, enterobacteria, typhimurium, lactobacillus, campylobacter, klebsiella, pneumoniae, proteobacteria, mrsa |
Process of disease analysis
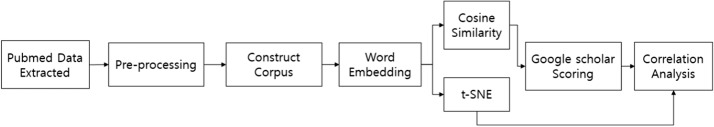
The disease analysis process in this paper consists of four steps in total shown in Fig. 3. First, we construct corpus by extracting useful data from documents in PubMed site. Second, applying word embedding to the generated corpus transforms vocabularies into appropriate vector representations. Third, cosine similarity is applied to vectors representing diseases, markers and microorganisms, and the similarity between them is calculated. Finally, we analyze the relationship between biomarkers and diseases, diseases and microorganisms, and calculate the scores via Google Scholars to verify the validity of these results. Based on this score in the future, we will be able to present new markers and microorganisms related to disease based on the difference between the similarity score and the Google Scholars score.
Fig. 3.
Process of disease analysis
Corpus preprocessing
Generally, biomarkers or disease names can be used in many forms. In particular, biomarkers often contain two or more words to represent a single marker. In this study, a corpus was formed by substituting a single word for a marker of plural words. Table 3 shows the full names and acronyms of the markers, where the full name is converted to an abbreviation and entered the embedding process. In the future, one expression that represents one marker or disease must be set in advance and all the various expressions should be replaced with this one expression.
Table 3.
Full names and their abbreviation for biomarkers
| Biomarker full name | Biomarker abbreviation |
|---|---|
| Cancer antigen 125 | CA125 |
| Cancer antigon 19-9 | CA19-9 |
| Epidermal frowth factor receptor | EGFR |
| Apolipoprotein A1 | Apoa-i |
| Apolipoprotein C3 | Apoc-iii |
| C-reactive protein | CRP |
| Follicle stimulating Hormone | FSH |
| Cancer antigon 15-3 | CA15-3 |
| interleukin-6 | IL-6 |
| interleukin-8 | IL-8 |
| Carcinoembryonic antigen | CEA |
| Osteopontin | OPN |
| Human epididymis protein 4 | HE4 |
| Matrix metalloproteinase-7 | MMP-7 |
Word-embedding and cosine similarity calculation
In this paper, the relationship between disease and biomarker, and disease and microorganism is understood by using word embedding model. In this study, CCA model is used among several word embedding models. To analyze the relationship between two words, we first generate a vector representation of a word using CCA, and then calculate the cosine similarity between these vectors. Figure 4 shows the actual embedding result of words calculated using CCA. In the experiment of this paper, we first embed a 100-dimensional vector. Figure 4 shows that the first number of vectors is much larger than the other numbers. In other words, this value makes it difficult to calculate the exact similarity. Therefore, in the present study, these vectors are transformed into two dimensions using t-SNE, and the similarity is calculated based on the results. In addition, visual analysis can be made possible by visualizing the converted result in two dimensions.
Fig. 4.
Examples of word embedding
Cosine similarity is a measure of similarity between two non-zero vectors of an inner product space that measures the cosine of the angle between them [39]. The cosine similarity is calculated by dividing the inner product of two vectors by the product of the sizes of two vectors, given the vectors A and B. The calculated similarity has a value between − 1 and 1 and is calculated in the following manner.
Result analysis
Two-dimensional mapped vocabularies using t-SNE can be represented by a point in two-dimensional space. In this study, visualized results are used when analyzing the relationship between terms. At the same time, when the calculation result of the cosine similarity is obtained, 20 pairs having the highest similarity and 20 pairs having the lowest similarity are extracted. In this study, we examined the results of searches on the actual Google Scholar to verify the usefulness of these similarities. First, after extracting the top 20 documents from Google Scholar search results, we calculated how frequently the terms appear in the titles and abstracts of these documents. The correlation between the calculation results and the cosine similarity was investigated to verify the usefulness of the proposed method.
Results
Biomaker analysis
This section describes the results of analyzing the relationship between biomarkers and diseases. The analysis was conducted according to the procedure described in the previous section. Figure 5 shows the mapping of biomarkers and diseases to a point in 2D space. Here, the blue letter indicates the biomarker, and the red letter indicates the disease. As shown in Fig. 5, biomarkers and diseases are relatively linearly discriminated. In addition, stomatitis and crp are located very close to each other, but ca125 and tuberculosis are located very far apart.
Fig. 5.
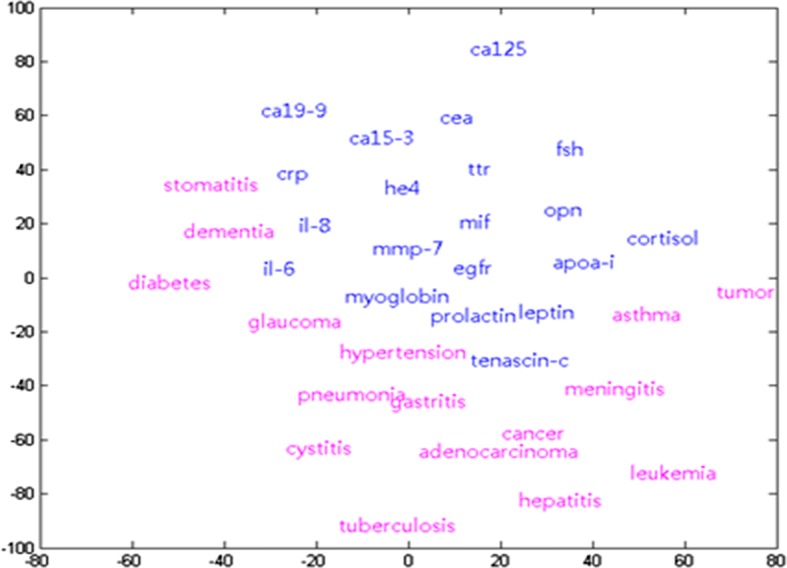
Mapping results of biomarkers and diseases
Table 4 shows the highest five similarity and the lowest five similarity among the results of calculating the similarity between two-dimensionally mapped vectors. Table 4 shows that cancer has the highest similarity to the HE4 biomarker. In other words, among the biomarkers in the literature, HE4 has a higher correlation with cancer than other markers. On the other hand, since glaucoma has the lowest similarity to the Apoc-iii marker, glaucoma and Apoc-iii have little relation to each other.
Table 4.
The highest and lowest cosine similarities between biomarkers and diseases
| Best | Worst | ||||
|---|---|---|---|---|---|
| Disease | Biomarker | Similarity | Disease | Biomarker | Similarity |
| Cancer | HE4 | 0.9993 | Glaucoma | Apoc-iii | − 0.9998 |
| Adenocarcinoma | HE4 | 0.9992 | Leukemia | CA19-9 | − 0.9997 |
| Dementia | Cortisol | 0.9991 | Stomatitis | Myoglobin | − 0.9988 |
| Stomatitis | CRP | 0.9988 | Conjunctivitis | FSH | − 0.9986 |
| Dementia | CRP | 0.9982 | Pneumothorax | FSH | − 0.9980 |
In order to test whether the results in Table 4 actually indicate a correlation between disease and markers, this study uses the results of Google Scholar search. Table 5 summarizes the results of the top 20 search results by searching the pair extracted from Table 4 with the Google Scholar. Table 5 summarizes the results of Google Scholar search for the top 5 similarity pairs.
Table 5.
Reults of Googole Scholar for the pairs of the highest similarities
| Disease | Bioamarker | Title_disease | Title_marker | Abs_dis | Abst_marker | Title_both | Abst_both |
|---|---|---|---|---|---|---|---|
| Cancer | HE4 | 15 | 20 | 73 | 144 | 15 | 19 |
| Adenocarcinoma | HE4 | 9 | 18 | 44 | 159 | 9 | 10 |
| Dementia | Cortisol | 18 | 17 | 56 | 79 | 16 | 17 |
| Stomatitis | CRP | 16 | 5 | 35 | 37 | 1 | 6 |
| Dementia | CRP | 17 | 7 | 96 | 55 | 6 | 14 |
Here, Title_disease and Title_marker refer to the number of papers in which the disease and markers appear in the title, respectively. Abs_dis and Abst_marker also show the number of papers in which the disease and markers appear in the abstract, respectively. Finally, Title_both and Abst_both mean the number of articles with both disease and markers in the title and abstract. Take Cancer-HE4 as an example. Cancer appears in the titles of 15 papers out of the top 20 papers, and HE4 appears 20 times in the titles of the 20 papers. In addition, cancer appears 73 times in the abstract of the top 20 papers and HE4 appears 144 times in the abstract of the papers. However, both of them appear at the same time in the title of 15 papers and summary of 19 papers. Of these five pairs, stomatitis-CRP pairs are much less common than the other pairs. This case may be regarded as an error of this methodology, but the closely related stomatitis and CRP may not be the subject of full-scale research. In the latter case, it would be possible to suggest CRP as a new marker related to stomatitis through this study.
The results in Table 5 show the roughness of the two keywords in the title and abstract, but it is difficult to make elaborate comparison of numbers. Therefore, in this study, the degree of co-occurrence is quantified as one number, making the comparison easier. The following formula is a formula that expresses the degree to which two keywords in the title co-occurrence. Abs_dis and Abs_marker can be used to express the degree of co-occurrence in the abstract.
If Table 5 is reconstructed in this way, it is as shown in Table 6. Table 7 shows the result of applying the same experiment to five pairs with the lowest similarity.
Table 6.
A new table which convert the Table 5 with score
| Disease | Biomarker | Similarity | Words | Score_title | Score_abstract |
|---|---|---|---|---|---|
| Cancer | HE4 | 0.99934 | 1 | 17.32 | 102.53 |
| Adenocarcinoma | HE4 | 0.99927 | 1 | 12.73 | 83.64 |
| Dementia | Cortisol | 0.99915 | 2 | 17.49 | 66.51 |
| Stomatitis | CRP | 0.99889 | 6 | 8.94 | 35.99 |
| Dementia | CRP | 0.99827 | 1 | 10.91 | 72.66n |
| Stomatitis | Leptin | 0.99751 | 2 | 10.95 | 87.46 |
| Pneumonia | Myoglobin | 0.99562 | 5 | 10.82 | 34.64 |
| Hypertension | Leptin | 0.99493 | 1 | 15.87 | 77.63 |
| Hypertension | Prolactin | 0.99259 | 1 | 13.49 | 74.46 |
| Hypertension | IL-6 | 0.99230 | 1 | 15.43 | 54.12 |
| Gastritis | CRP | 0.99203 | 2 | 9.49 | 79.37 |
| Stomatitis | Cortisol | 0.99183 | 8 | 10.95 | 27.66 |
| Tumor | CA125 | 0.99157 | 1 | 16.43 | 112.98 |
| Tumor | CEA | 0.98999 | 0 | 12.73 | 159.05 |
| Stomatitis | Prolactin | 0.98292 | 1 | 11.22 | 37.34 |
| Hypertension | Myoglobin | 0.97564 | 8 | 10.25 | 36.06 |
| Meningitis | CRP | 0.97791 | 1 | 14.87 | 110.89 |
| Stomatitis | IL-6 | 0.97592 | 4 | 13.08 | 70.70 |
| Asthma | EGFR | 0.97468 | 2 | 12.49 | 59.90 |
| Tumor | EGFR | 0.97095 | 0 | 12.33 | 150.40 |
| Average | 0.98956 | 2.4 | 12.88 | 76.69 | |
Table 7.
The score table of pairs with the lowest similarity
| Disease | Biomarker | Similarity | Words | Score_title | Score_abstract |
|---|---|---|---|---|---|
| Glaucoma | Apoc-iii | − 0.99998 | X | 0.00 | 0.00 |
| Leukemia | CA19-9 | − 0.99997 | X | 4.47 | 0.00 |
| Stomatitis | Myoglobin | − 0.99884 | X | 4.47 | 12.96 |
| Conjunctivitis | FSH | − 0.99868 | X | 1.41 | 0.00 |
| Pneumothorax | FSH | − 0.99809 | 14 | 0.00 | 6.48 |
| Hyperlipidemia | CEA | − 0.99768 | 7 | 4.36 | 19.80 |
| Pneumonia | CA125 | − 0.99736 | 2 | 7.35 | 57.16 |
| Miliaria | CYFRA21-1 | − 0.99709 | X | 0.00 | 0.00 |
| Glaucoma | CA125 | − 0.99546 | 62 | 1.41 | 5.66 |
| Glaucoma | CYFRA21-1 | − 0.99508 | X | 1.73 | 9.38 |
| Hyperlipidemia | CYFRA21-1 | − 0.99353 | 21 | 0.00 | 16.97 |
| Cataract | CEA | − 0.99336 | X | 5.29 | 6.00 |
| Hepatitis | CA125 | − 0.99277 | 5 | 7.48 | 62.26 |
| Cystitis | CA125 | − 0.98972 | 19 | 3.00 | 12.00 |
| Glaucoma | CA125 | − 0.98945 | 7 | 5.48 | 35.78 |
| Hyperlipidemia | APOA-I | − 0.98930 | X | 2.24 | 8.60 |
| Cataract | MIF | − 0.98917 | X | 1.73 | 5.29 |
| Cataract | CYFRA21-1 | − 0.98915 | X | 0.00 | 7.62 |
| Tinnitus | CA125 | − 0.97812 | 5 | 8.49 | 57.50 |
| Hyperlipidemia | CA19-9 | − 0.97871 | 7 | 6.00 | 29.93 |
| Average | − 0.99307 | – | 3.24 | 17.66 | |
Here, ‘Words’ refers to the minimum distance between two words when two words appear together in the abstract. For example, ‘cancer’ and ‘HE4’ of Table 6, which shows 1 in the ‘Words’ column, appear in succession more than once in 20 abstracts. And between ‘stomatitis’ and ‘CRP’, more than 7 words take place. The larger the value, the less the two words appear together. In Table 7, it is seen that the Words value is X, which means that the two words do not appear together in the abstracts.
Microorganism analysis
Microbiological analysis identifies the relationship between microbial terms and disease/symptom/organ. Figure 6 shows the distribution of these terms in a two-dimensional space. Here, the red letter indicates the microorganism term and the blue letter indicates the disease/symptom/organ term. Again, microbial vocabulary and the rest of the vocabulary show clear boundaries.
Fig. 6.
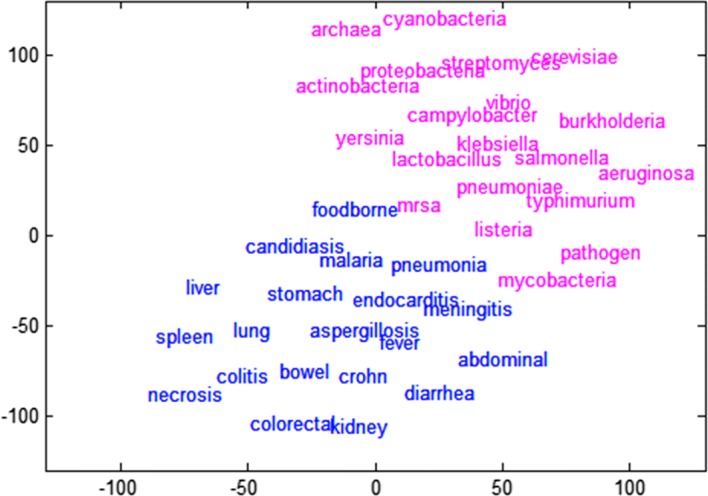
Mapping results of biomarkers and diseases
Table 8 shows the 5 pairs with the highest cosine similarity and the lowest 5 pairs. Foodborne and campylobacter showed the highest similarity and pneumonia and mycobacteria showed the lowest similarity.
Table 8.
The highest and lowest cosine similarities between microorganism and diseases/symptoms/organs
| Best | Worst | ||||
|---|---|---|---|---|---|
| Disease | Microorganism | Similarity | Disease | Microorganism | Similarity |
| Foodborne | Campylobacter | 0.9998 | Pneumonia | Mycobacteria | − 0.9997 |
| Pneumonia | Pneumoniae | 0.9994 | Kidney | Cerevisiae | − 0.9997 |
| Endocarditis | Mrsa | 0.9983 | Foodborne | Klebsiella | − 0.9993 |
| Foodborne | Mrsa | 0.9933 | Bowel | Listeria | − 0.9992 |
| Abdominal | Streptomyces | 0.9923 | Spleen | Campylobacter | − 0.9992 |
Table 9 summarizes the Google Scholars search results for the top 5 similarity pairs. Here, the foodborne-mrsa pair and the abdominal-streptomvces pair show a relatively low Title_both value and Abst_both value as compared to the other pairs. In this case, it would be advisable to conduct a clinical study on the corresponding mrsa (methicillin-resistant Staphylococcus aureus) from the foodborne standpoint.
Table 9.
Reults of Googole Scholar for the pairs of the highest similarities
| Disease | Microorganism | Title_disease | Title_micro | Abs_dis | Abst_micro | Title_both | Abst_both |
|---|---|---|---|---|---|---|---|
| Foodborne | Campylobacter | 15 | 17 | 28 | 59 | 13 | 14 |
| Pneumonia | Pneumoniae | 20 | 17 | 67 | 66 | 17 | 19 |
| Endocarditis | Mrsa | 14 | 17 | 75 | 85 | 14 | 14 |
| Foodborne | Mrsa | 8 | 15 | 21 | 87 | 3 | 9 |
| Abdominal | Streptomyces | 7 | 19 | 17 | 50 | 7 | 7 |
Tables 10 and 11 show the scores for the pair with the highest similarity and the pair with the lowest similarity listed in Table 8. Here too, the score was calculated for 20 pairs of upper and lower sides as in the marker experiment.
Table 10.
Scores of the most similar pairs
| Disease | Microorganism | Similarity | Words | Score_title | Score_abstract |
|---|---|---|---|---|---|
| Foodborne | Campylobacter | 0.9998 | 0 | 15.97 | 40.64 |
| Pneumonia | Pneumoniae | 0.9994 | 0 | 18.44 | 66.50 |
| Endocarditis | Mrsa | 0.9983 | 0 | 15.43 | 79.84 |
| Foodborne | Mrsa | 0.9933 | 1 | 10.95 | 42.74 |
| Abdominal | Streptomyces | 0.9923 | 4 | 11.53 | 29.15 |
| Average | 0.9966 | 1 | 14.46 | 51.77 | |
Table 11.
Scores of the least similar pairs
| Disease | Microorganism | Similarity | Words | Score_title | Score_abstract |
|---|---|---|---|---|---|
| Pneumonia | Mycobacteria | − 0.9997 | 7 | 8.66 | 44.18 |
| Kidney | Cerevisiae | − 0.9997 | 5 | 10.82 | 18.57 |
| Foodborne | Klebsiella | − 0.9993 | 1 | 8.25 | 22.05 |
| Bowel | Listeria | − 0.9992 | 8 | 10.00 | 22.72 |
| Spleen | Campylobacter | − 0.9992 | 4 | 8.94 | 35.20 |
| Average | − 0.9994 | 5 | 9.33 | 28.54 | |
Discussion
Tables 6 and 7 show the scores for the top 20 and the bottom 20 pairs of similarity criteria resulted from the experiments about biomarker-diseases pairs. In the Google Scholar search, about 85% of the top twenty pairs showed high scores, but in the bottom twenty pairs, only 15% showed high scores. We give a ‘high’ score to the pairs if the number of ‘Words’ column in Tables 6 and 7 do not exceeds 5 and a ‘low’ score if it is exceeds 5. Only three rows in Table 6 did not receive a high score, and only three rows in Table 7 received a high score. We can confirm clearly these trends from Tables 6 and 7. Therefore, we also consider similarity based on word embedding to have a significant correlation with existing research.
We are able to see results similar to those above in microorganism analysis. Here too, we have confirmed that much research has been done on 85% of the top 20 pairs and 5% of bottom pairs. However, in the case of microorganisms, the difference of average scores between the upper similarity pairs and the lower similarity pairs was smaller than that of the biomarker pairs. The reason is that the collected corpus is biased towards the field of molecular biology related to genes or proteins, and therefore the research results related to microorganisms are not sufficient in the corpus.
Conclusions
In this paper, we tried to analyze the correlation between biomarkers and microorganisms and specific diseases and symptoms. For the correlation analysis, we constructed a large corpus and constructed the word embedding for each word in the corpus. CCA was used for word embedding, and cosine similarity was used for correlation analysis. In order to verify the validity of the correlation values extracted from this study, we used the results of Google Scholar. Experimental results show that 85% of highly correlated pairs were searched with high frequency in Google Scholar. On the other hand, only 15% of the low-correlated pairs have been actively studies.
In the future, we will try to analyze the correlation by applying more various word embedding methods. The CCA reflects the global characteristics the best, but it does not reflect the local characteristics. Therefore, a methodology to overcome this is needed. In this study, we analyzed all the vocabulary words by word embedding. In the future, however, we will study how to use the deep learning to learn the correlation itself.
Declarations
Authors’ contributions HJS took parts in word embedding design and implementation. BHY prepared for the whole experiment, including data and programming. YSY helped BHY in programming and data preparation. CYP advised BHY in programming. JDK expertized in biomedical IT convergence research. He analyzed input and output data. YSK is a corresponding author. YSK designed this research and directed this research team. All authors read and approved the final manuscript.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2015R1A2A2A01007333), and by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2017M3C4A7068188).
Competing interests
The authors declare that they have no competing interests.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and future Planning (2015R1A2A2A01007333), and by Hallym University Research Fund, 2017 (HRF-201704-013).
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
About this supplement
This article has been published as part of BioMedical Engineering OnLine Volume 17 Supplement 2, 2018: Proceedings of the International Conference on Biomedical Engineering Innovation (ICBEI) 2016. The full contents of the supplement are available online at https://biomedical-engineering-online.biomedcentral.com/articles/supplements/volume-17-supplement-2.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- NLP
natural language processing
- Bio-NLP
biomedical natural language processing
- CCA
Canonical Correlation Analysis
- t-SNE
t-distributed stochastic neighbor embedding
- CRF
conditional random field
- NER
Named Entity Recognition
- GloVe
global vector
Contributor Information
Hye-Jeong Song, Email: hjsong@hallym.ac.kr.
Byeong-Hun Yoon, Email: yqudgns1222@gmail.com.
Young-Shin Youn, Email: pour657@gmail.com.
Chan-Young Park, Email: cypark@hallym.ac.kr.
Jong-Dae Kim, Email: kimjd@hallym.ac.kr.
Yu-Seop Kim, Email: yskim01@hallym.ac.kr.
References
- 1.Biomarker—Wikipedia. https://en.wikipedia.org/wiki/Biomarker. Accessed 11 Apr 2018.
- 2.Microorganism—Wikipedia. https://en.wikipedia.org/wiki/Microorganism. Accessed 11 Apr 2018.
- 3.Feng QQ, Mujun Y, Nancy BK. Molecular biomarkers for cancer detection in blood and bodily fluids. Crit Rev Clin Lab Sci. 2006;43:497–560. doi: 10.1080/10408360600922632. [DOI] [PubMed] [Google Scholar]
- 4.Srinivas PR, Verma M, Zhao Y, Srivastava S. Proteomics for cancer biomarker discovery. Clin Chem. 2002;48:1160–1169. [PubMed] [Google Scholar]
- 5.Nam KM, Song HJ, Kim JD, Park CY, Kim YS. Detection of alternative ovarian cancer biomarker via word embedding. Int J Softw Eng Appl. 2016;10:1–12. [Google Scholar]
- 6.Li S, Jiang Y. Semi-supervised sentiment classification using ranked opinion words. Int J Database Theory Appl. 2013;6:51–62. doi: 10.14257/ijdta.2013.6.6.05. [DOI] [Google Scholar]
- 7.PubMed—NCBI. https://www.ncbi.nlm.nih.gov/pubmed/. Accessed 11 Apr 2018.
- 8.Stratos K, Collins M, Hsu D. Model-based word embeddings from decompositions of count matrices. Proceedings of the 53rd annual meeting of the association for computational linguistics and the 7th International joint conference on natural language processing. association for computational linguistics; 2015. p. 1282–91.
- 9.Maaten L, Geoffrey H. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 10.Natural language processing—Wikipedia. https://en.wikipedia.org/wiki/Natural_language_processing. Accessed 11 Apr 2018.
- 11.Maas AL, Daly RE, Pham PT, Huang D, Ng AY, Potts C. Learning word vectors for sentiment analysis. Proceedings of the 49th annual meeting of the association for computational linguistics: human language technologies-volume 1. Association for Computational Linguistics; 2011. p. 142–50.
- 12.Tang D, Wei F, Yang N, Zhou M, Liu T, Qin B. Learning sentiment-specific word embedding for twitter sentiment classification. Association for Computational Linguistics; 2014. p. 1555–65.
- 13.Biomedical text mining—Wikipedia. https://en.wikipedia.org/wiki/Biomedical_text_mining. Accessed 11 Apr 2018.
- 14.Park KM, Hwang KB. A bio-text mining system based on natural language processing. J KIISE. 2011;17:205–213. [Google Scholar]
- 15.Safaei A, Rezaei TM, Sobhi S, Akbari ME. Breast cancer biomarker discovery: proteomics and genomics approaches. Iran J Cancer Prev. 2013;6:45–53. [Google Scholar]
- 16.Reyzer ML, Caprioli R. MALDI mass spectrometry for direct tissue analysis: a new tool for biomarker discovery. J Proteome Res. 2005;4:1138–1142. doi: 10.1021/pr050095+. [DOI] [PubMed] [Google Scholar]
- 17.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 18.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 19.Mann M, Hendrickson RC, Pandey A. Analysis of proteins and proteomes by mass spectrometry. Annu Rev Biochem. 2001;70:437–743. doi: 10.1146/annurev.biochem.70.1.437. [DOI] [PubMed] [Google Scholar]
- 20.Verma S. Bioinformatics Approaches to Biomarker Discovery. 2007.
- 21.Pedrero M, Campuzano S, Pingarrón JM. Electrochemical genosensors based on PCR strategies for microorganisms detection and quantification. Anal Methods. 2011;3:780–789. doi: 10.1039/c0ay00755b. [DOI] [Google Scholar]
- 22.Nelson WH. Physical methods for microorganisms detection. Boca Raton: CRC Press; 1991. [Google Scholar]
- 23.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Zhang A, Han Y, Wang P, Sun H, Song G, et al. Urine metabolomics analysis for biomarker discovery and detection of jaundice syndrome in patients with liver disease. Mol Cell Proteomics. 2012;11:370–380. doi: 10.1074/mcp.M111.016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beger RD, Sun J, Schnackenberg LK. Metabolomics approaches for discovering biomarkers of drug-induced hepatotoxicity and nephrotoxicity. Toxicol Appl Pharmacol. 2010;243:154–166. doi: 10.1016/j.taap.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 26.Song Y, Kim EJ, Lee GG, Yi BK. POSBIOTM-NER in the shared task of BioNLP/NLPBA 2004. Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications. Association for Computational Linguistics; 2004. p. 100–3.
- 27.Turian J, Ratinov L, Bengio Y. Word representations: a simple and general method for semi-supervised learning. Proceedings of the 48th annual meeting of the association for computational linguistics. Association for Computational Linguistics; 2010. p. 384–94.
- 28.Seok MR, Song HJ, Park CY, Kim JD, Kim YS. Named entity recognition using word embedding as a feature. Int J Softw Eng Appl. 2016;10:93–104. [Google Scholar]
- 29.Qiu L, Caop Y, Nie Z, Yu Y, Rui, Y, Learning word representation considering proximity and ambiguity. proceedings of the twenty-eighth aaai conference on artificial intelligence; 2014. p. 1572–8.
- 30.Muneeb TH, Sahu SK, Anand A. Evaluating distributed word representations for capturing semantics of biomedical concepts. In: Proceedings of ACL-IJCNLP. 2015. p. 158.
- 31.Collobert R, Weston J, Bottou L, Karlen M, Kavukcuoglu K, Kuksa P. Natural language processing (almost) from scratch. J Mach Learn Res. 2011;12:2493–2537. [Google Scholar]
- 32.Hotelling H. Relations between two sets of variates. Biometrika. 1936;28:321–377. doi: 10.1093/biomet/28.3-4.321. [DOI] [Google Scholar]
- 33.Jang MK, Kim YS, Park CY, Song HJ, Kim JD. Integration of menopausal information into the multiple biomarker diagnosis for early diagnosis of ovarian cancer. Int J Biosci Biotechnol. 2013;5:215–222. [Google Scholar]
- 34.Seok MR, Song HJ, Park CY, Kim JD, Kin YS. Comparison of NER performance using word embeddings. In: The 4th international conference on artificial intelligence and application. 2015. p. 754–88.
- 35.Youn YS, Nam KM, Song HJ, Kim JD, Park CY, Kim YS. Classification performance of bio-marker and disease word using word representation models. Int J Biosci Biotechnol. 2016;8:295–302. [Google Scholar]
- 36.Weenink D. Canonical correlation analysis. In: Proceedings of the Institute of Phonetic Sciences of the University of Amsterdam. 2003. p. 81–99.
- 37.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. advances in neural information processing systems. 2013. p. 3111–9.
- 38.Pennington J, Socher R, Manning C. Glove: Global vectors for word representation. Proceedings of the Empiricial Methods in Natural Language Processing; 2014. p. 1532–43.
- 39.Cosine similarity—Wikipedia. https://en.wikipedia.org/wiki/Cosine_similarity. Accessed 11 Apr 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.