Abstract
Background
Investigating the roles of lncRNA prostate cancer-associated transcript 6 (PCAT6) in modulating the growth and aggressiveness of non-small-cell lung carcinoma (NSCLC) cell.
Method
The levels of PCAT6 in NSCLC tissues and cell lines were determined by quantitative real-time PCR assay. MTT as well as colony formation assays were applied to explore the effect of PCAT6 on the growth of NSCLC cell in vitro. Wound healing and Transwell assays were utilized to analyze the impact of PCAT6 on the migration and invasion of NSCLC cell. Bioinformatics analysis and luciferase reporter assay were used to prove that miR-330-5p was the target of PCAT6. Colony formation, wound healing, and Transwell invasion assays were applied to demonstrate that PCAT6 promoted NSCLC cell growth, migration, and invasion through binding miR-330-5p. Finally, xenograft model was used to explore the role of PCAT6 in the tumor growth of NSCLC cell in vivo.
Results
PCAT6 was highly overexpressed in NSCLC tissues and cells compared with normal tissues and non-tumorigenic bronchial epithelial cell line, BEAS-2B. Downregulation of PCAT6 markedly reduced the proliferation, migration, and invasion of NSCLC cell. Moreover, down-expression of PCAT6 significantly increased the level of miR-330-5p in NSCLC cell. Further functional experiments indicated that down-expression of miR-330-5p reversed the inhibitory effect of PCAT6 on NSCLC cell growth, migration, and invasion.
Conclusion
Our results reveal that lncRNA PCAT6 facilitates the proliferation, migration, and invasion of NSCLC cell via competitively binding to miR-330-5p.
Keywords: PCAT6, MiR-330-5p, NSCLC, growth, migration, invasion
Introduction
Non-small-cell lung carcinoma (NSCLC) is the most common type of lung cancer and remains the leading cause of cancer-induced death worldwide.1,2 Despite advances in the treatment for patients with NSCLC, a certain amount of patients with NSCLC experience distant metastasis. Hence, it is needed to investigate the precise mechanisms behind NSCLC metastasis.3 Recently, intensive studies identified a large number of markers of NSCLC metastasis. Previous evidences have demonstrated that lncRNAs serve as regulators in the tumorigenesis and pregres-sion of NSCLC.4
Previous investigations demonstrate that lncRNAs are aberrantly expressed in multiple tumor types.5 lncRNAs regulate the expression of target genes at either transcriptional, posttranscriptional, or epigenetic levels.6 Recent studies identified that a great deal of lncRNAs serve as endogenous RNA (ceRNA) and thereby prevent miRNAs from binding to their target genes.7 Substantial evidences demonstrate that lncRNAs have vital functions in tumor processes, including cell proliferation, epithelial–mesenchymal transition (EMT), invasion and metastasis.8
LncRNA prostate cancer-associated transcript 6 (PCAT6) enhances cellular proliferation and colony formation of prostate cancer cells in an androgen-independent way.9 In lung cancer, PCAT6 is overregulated in cancer tissues compared with the matched normal tissues, and positively correlated with the metastasis of patients with lung adenocarcinoma.10,11 In addition, PCAT6 is negatively associated with the overall survival of patients with lung cancer. Nevertheless, the precise role of PCAT6 in the growth, migration, and invasion of NSCLC cell remain not yet explored.
In this study, we revealed that PCAT6 was highly overexpressed in NSCLC. Down-expression of PCAT6 suppressed the malignancy phenotypes of NSCLC cell, including growth, migration, and invasion. Moreover, we demonstrated that downregulation of PCAT6 markedly increased the level of miR-330-5p in NSCLC cell. Bioinformatics analysis and luciferase reporter assay proved that miR-330-5p bound to PCAT6. Altogether, these findings suggested that down-expression of PCAT6 inhibited the growth, migration, and invasion of NSCLC cell via regulating miR-330-5p.
Materials and methods
NSCLC cell lines and tissues
Four NSCLC cell lines (H1650, HCC827, A549, and H1975), the non-tumorigenic bronchial epithelial cell line (BEAS-2B) and 293 T cell were purchased from GuangZhou Jennio Biotech Co., Ltd (GuangZhou, GuangDong, China). NSCLC cells were cultured in RPMI-1640 containing 10% fetal bovine serum (FBS) (Wisent, Quebec, Canada), 100 µg/mL streptomycin and 100 µg/mL penicillin. A total of 293 T cells were cultured in DMEM (Wisent). BEAS-2B cells were cultured in serum-free LHC-8 medium (Invitrogen, Carlsbad, CA, USA). All cells were cultured in an incubator with 95% air and 5% CO2 at 37°C. Twenty-six pairs of NSCLC (Female: 10, Male: 16) and corresponding adjacent normal tissues were obtained from patients who received surgery for NSCLC at People’s Hospital of Jingjiang. None of the patients received chemotherapy or radiotherapy prior to operation. The NSCLC and non-cancerous tissues were confirmed by two pathologists. This research was approved by the Institutional Research Committee of the People’s Hospital of Jingjiang. The written informed consent for participation in the study was obtained from all patients before participation in this study.
Microarray analysis
LncRNAS in The Cancer Genome Atlas (TCGA) databases (https://tcga-data.nci.nih.gov/tcga/) were analyzed by using The UCSC Cancer Genomics Browser (https://genome-cancer.soe.ucsc.edu/proj/site/hgHeatmap/). The TCGA lung cancer (LUNG) RNAseq (IlluminaHiSeq) dataset was used to compare the dysregulation of lncRNAs in lung cancer and normal tissues. Heatmap mode was used to display the expression data, which are sorted using sample type (lung tumor or normal).
Cell transfection
The siRNA oligonucleotides were synthesized by Shanghai GenePharma Co. Ltd. (Shanghai, China), and the siRNA sequence of PCAT6 was as follow: siRNA negative control (si-NC) (5′-GCGACCAA CGCCTTGA TTG-3′) and si-PCAT6 (5′-GGTGTCTCCATCCTCATTC-3′). The si-NC was used as negative control. Cells were cultured in six-well plates overnight and then transiently transfected with PCAT6-siRNA using Lipofectamine 2000 kit (Invitrogen, Grand Island, NY, USA). In miRNAs transfection, the cells were transfected with miR-330-5p (100 nM) and negative control miRNA (miR-NC) for 24 hours using Oligofectamine (Invitrogen). For miRNA inhibitor transfection, cells were transfected with negative control miRNA inhibitor or miR-330-5p inhibitor using Lipofectamine 2000 kit (Invitrogen). The specific siRNA against PCAT6 (si-PCAT6) was designed and synthesized by GenePharma Co. Ltd. A negative control siRNA (si-Con) was synchronously synthesized. For the generation of stable cell lines, A549 cells were transfected si-PCAT6 and the transfected cells were selected with 1 µg/mL puromycin. After ~4 weeks, puromycin-resistant A549 cells were established and collected for in vivo experiments.
Quantitative real-time PCR (qRT-PCR) assay
Total RNA was extracted from NSCLC tissues or cells using TRIzol reagent (Thermo, San Jose, CA, USA). The expression of PCAT6 was quantified using a SYBR Premix EX Taq™ II kit (TaKaRa, Dalian, China) on ABI Prism 7500 (Applied Biosystems, Foster City, CA, USA). miRNA was extracted using a PureLink™ miRNA isolation kit (Thermo, Carlsbad, CA, USA), and the level of miRNA was measured using a TaqMan microRNA assay kit (Thermo). The primer sequences were as follow: GAPDH, 5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′ (reverse); PCAT6, 5′-CAGGAACCCCCTCCTTACTC-3′ (forward) and 5′-CTAGGGATGTGTCCGAAGGA-3′ (reverse).
Bioinformatics analysis
The binding site between miR-330-5p and PCAT6 was analyzed using online tool starBase (http://starbase.sysu. edu.cn/index.php).
Luciferase reporter assay
The pGL3-PCAT6 wild type (PCAT6-wt) plasmid was constructed by insertion of PCAT6 cDNA fragment which contained miR-330-5p binding sites into pGL3 luciferase reporter vector (Promega, Madison, WI, USA). The mutant type of pGL3-PCAT6 (PCAT6-mut) was constructed by insertion of PCAT6 cDNA fragment which contained mutated binding sites into pGL3 luciferase reporter vector. A total of 293 T cells were cotransfected with PCAT6-wt or PCAT6-mut and miR-330-5p using the Lipofectamine 2000 (Invitrogen). After 48 hours, the luciferase activity was detected using luciferase reporter assay system (Promega).
MTT assay
Cell proliferation was detected using the cell proliferation reagent kit (MTT, Promega) according to the manufacturer’s instructions.12
Colony formation assay
Cells (1,000 cells/well) were cultured into six-well plates and incubated for 4 weeks. Then, cell colonies were fixed using 1% paraformaldehyde and were stained with 0.1% crystal violet. The cell colonies were counted in five independent fields.13
Migration assay
Cells were seeded into six-well plates for 24 hours. Next, a wound in each group was scratched using a 100 µL pipette tip. The wounds were photographed at 0 hour or 48 hours. The distance between each group was calculated using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The rate of wound healing was calculated. The percentage of wound-healing rate = (0 hour width of scratch − 48 hours width of scratch)/0 hour width of scratch×100%.
Invasion assay
1×105 cells were seeded into the upper chamber of Transwell that was pre-coated with Matrigel (Sigma, Beijing, China). Complete medium containing 20% FBS was placed in the lower chamber of Transwell. After 24 hours, the invaded cells were stained with 0.1% crystal violet and counted.14
Xenograft model
5×106 si-PCAT6 or si-Con-transfected cells were suspended in 100 µL PBS, and subcutaneously inoculated into nude mice (n=6 for each group). After 35 days, all mice were sacrificed and tumors from each group were weighted. Experimental protocol was approved by the Ethics Committee of The People’s Hospital of Jingjiang in accordance with Institutional Guidelines and the Guide for the Care and Use of Laboratory Animals (NIH publication no 85–23, revised 1996).
Statistical analysis
Data are presented as Mean ± SD and evaluated using Graph-Pad Prism software. Statistical analysis was conducted with 2-tailed Student’s t-test or one-way ANOVA followed by post hoc Dunnett’s test. Pearson’s correlation analysis was carried out to determine the relationship between PCAT6 and miR-330-5p. P<0.05 was considered statistically significant.
Results
lncRNA PCAT6 is overexpressed in NSCLC
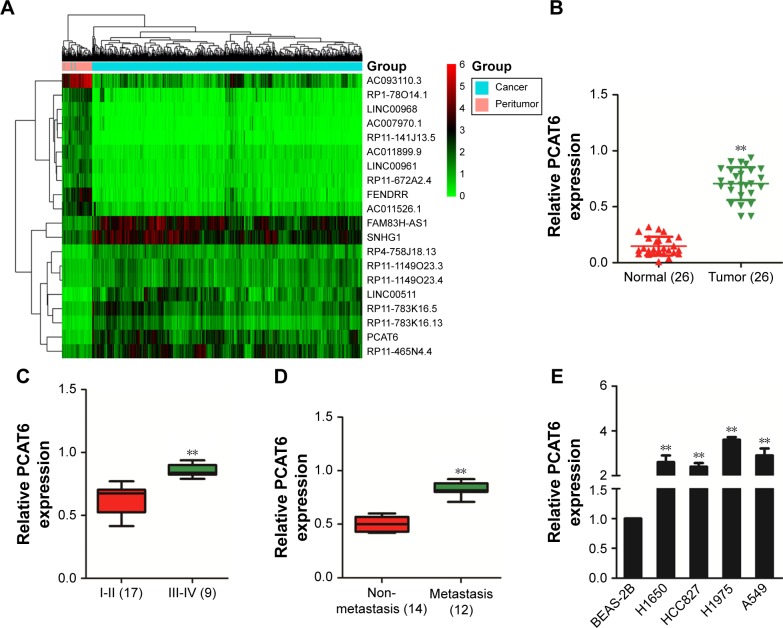
To identify the expression pattern of lncRNA in NSCLC, microarray analysis (TCGA) was applied to explore the differential expressed lncRNAs in NSCLC tissues (tumor) and the adjacent tissues (peritumor).15 As shown in Figure 1A, the levels of lncRNA PCAT6, FAM83H Antisense RNA 1 (FAM83H-AS1), and small nucleolar RNA host gene 1 (SNHG1) were higher in NSCLC (tumor) than that in the paired peritumor tissues. Previous investigations indicate that overexpression of FAM83H-AS1 is associated with the poor survival of patients with lung cancer. Down-regulation of FAM83H-AS1 impairs the proliferation and invasion of lung cancer cell via regulating the tyrosine-protein kinase Met/epidermal growth factor receptor (MET/EGFR) signaling pathway.16 In addition, SNHG1 promotes the progression of NSCLC by upregulating Metadherin via sponging miR-145 -5p.17 Nevertheless, the precise roles of PCAT6 in growth, migration, and invasion of NSCLC are yet to be well explored. Next, we determined the level of PCAT6 in 26 pairs of NSCLC tissues and matched non-cancerous tissues via qRT-PCR assay. As shown in Figure 1B, the level of PCAT6 was higher in cancer tissues compared with the adjacent normal tissues (P<0.01). In addition, we compared the level of PCAT6 between patients with different stages and found that PCAT6 was higher in stage III-IV (III and IV) than that in stage I-II (I and II) NSCLC (Figure 1C). Meanwhile, the expression of PCAT6 was significantly higher in patients with metastasis than patients with non-metastasis (Figure 1D). Finally, the level of PCAT6 in NSCLC cell lines (H1650, HCC827, H1975, and A549) and non-tumorigenic bronchial epithelial cell line (BEAS-2B) was detected. As shown in Figure 1E, PCAT6 was remarkably overexpressed in NSCLC cells compared with BEAS-2B cell. Altogether, these results indicated that lncRNA PCAT6 was up-regulated in NSCLC.
Figure 1.
PCAT6 is overexpressed in NSCLC.
Notes: (A) Heatmap: the dysexpression pattern of lncRNAs in NSCLC tissues compared with non-cancer tissues. (B) The level of PCAT6 was measured in NSCLC tissues and adjacent normal tissues using qRT-PCR. **P<0.01 compared with normal, Student’s t-test. (C) PCAT6 was overexpressed in advanced NSCLC tissues. **P<0.01 compared with stage I–II, Student’s t-test. (D) PCAT6 was overexpressed in patients with metastasis compared with patients with non-metastasis. **P<0.01 compared with non-metastasis, Student’s t-test. (E) The level of PCAT6 in four NSCLC cell lines A549, H1650, HCC827, and H1975, and BEAS-2B cell was detected by qRT-PCR. **P<0.01 compared with BEAS-2B, Student’s t-test.
Abbreviations: NSCLC, non-small-cell lung carcinoma; PCAT6, prostate cancer-associated transcript 6; qRT-PCR, quantitative real-time polymerase chain reaction.
Down-expression of PCAT6 inhibits the growth of NSCLCcells
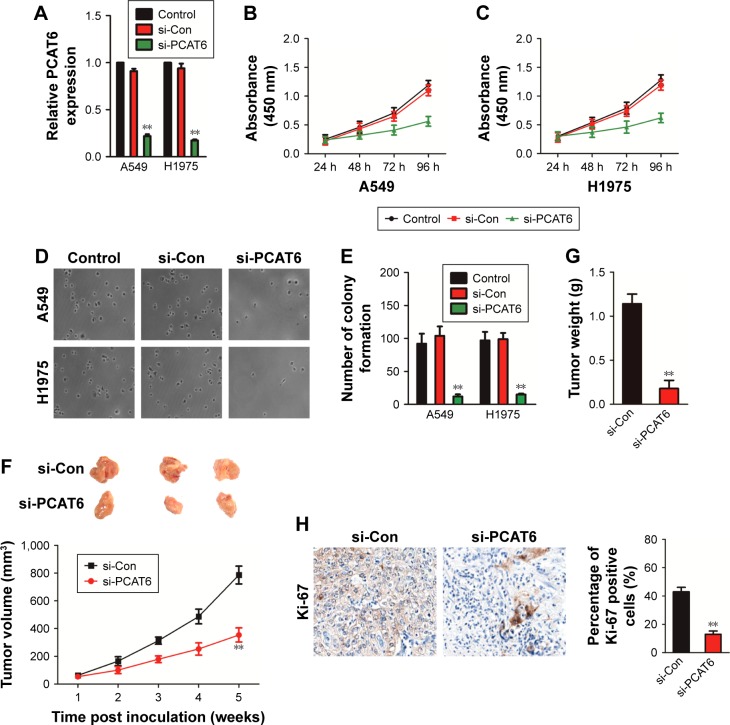
To determine the functions of PCAT6 in NSCLC, siRNA PCAT6 (si-PCAT6) was transfected into A549 and H1975 cells, and the expression of PCAT6 was demonstrated using qRT-PCR assay (Figure 2A). Then, the impact of PCAT6 on the growth of A549 and H1975 cell was assessed by MTT assay. As shown in Figure 2B and C, the cell proliferation was significantly inhibited by down-expression of PCAT6. Consistently, down-expression of PCAT6 remarkably inhibited the colony formation of both A549 and H1975 cells (Figure 2D and E). To further explore the impact of PCAT6 in NSCLC cell growth in vivo, A549/si-Con and A549/si-PCAT6 cells were subcutaneously inoculated into nude mice. The growth of A549 cells in nude mice were significantly inhibited by down-expression of PCAT6 (Figure 2F). Meanwhile, the average weight of tumor, which was formed by A549/si-Con-transfected cells was significantly greater than the tumor, which was derived from A549/si-PCAT6-transfected cells (Figure 2G). Finally, tumor tissues were subjected to immunohistochemical staining using Ki-67 antibody. As expected, the level of Ki-67 was lower in the tumor that was formed by PCAT6 down-expressing A549 cells compared with the si-Con-transfected A549 cells (Figure 2H). Collectively, lncRNA PCAT6 inhibited NSCLC cell growth in vitro and in vivo.
Figure 2.
Knockdown of PCAT6 inhibits NSCLC cell growth.
Notes: (A) si-PCAT6 or si-Con was transfected into A549 and H1975 cells. The levels of PCAT6 were determined by qRT-PCR assay. **P<0.01 compared with control, Student’s t-test. (B, C) The cell proliferation of A549 and H1975 cells that was transfected with si-PCAT6 was determined by MTT assay, one-way ANOVA. (D, E) The colony formation analysis of si-Con or si-PCAT6 transfected A549 and H1975 cells. **P<0.01 compared with si-Con, Student’s t-test. (F) Knockdown of PCAT6 significantly inhibits A549 cell growth in xenograft model, one-way ANOVA. **P<0.01 compared with si-Con. (G) Tumor weight, **P<0.05 compared with si-Con, Student’s t-test. (H) The levels of Ki-67 in tumor that was formed by si-PCAT6 or si-Con-transfected A549 cells were detected by immunohistochemical staining. **P<0.01 compared with si-Con, Student’s t-test.
Abbreviations: NSCLC, non-small-cell lung carcinoma; PCAT6, prostate cancer-associated transcript 6; qRT-PCR, quantitative real-time polymerase chain reaction.
Knockdown of PCAT6 inhibits the migration and invasion of NSCLCcell
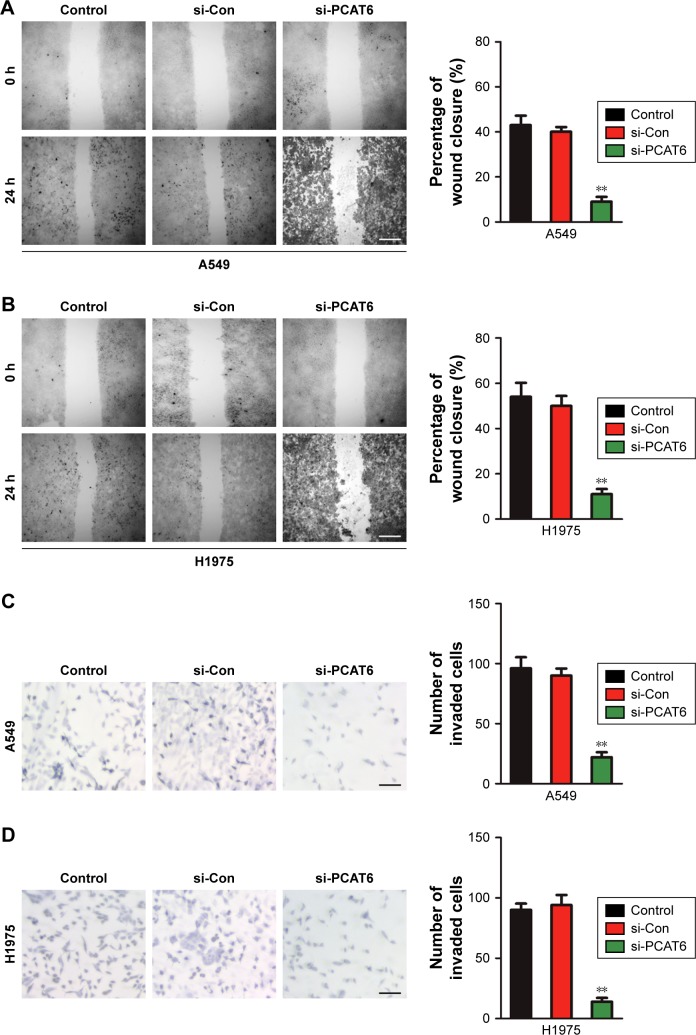
To explore the functional roles of PCAT6 in the migration and invasion of NSCLC cell, we performed the wound healing and Transwell invasion assays. As shown in Figure 3A and B, downregulation of PCAT6 markedly suppressed the migration of A549 and H1975 cells. Similarly, the invasion capacity of A549 and H1975 cells was inhibited by PCAT6 knockdown (Figure 3C and D). Altogether, these results indicated that down-expression of PCAT6 decreased the migration and invasion abilities of NSCLC cell.
Figure 3.
Down-expression of PCAT6 inhibits the migration and invasion of NSCLC cell.
Notes: (A, B) Knockdown of PCAT6 inhibited the migration of A549 and H1975 cells. (C, D) Knockdown of PCAT6 suppressed the invasion of NSCLC cell. **P<0.01 compared with control, Student’s t-test.
Abbreviations: NSCLC, non-small-cell lung carcinoma; PCAT6, prostate cancer-associated transcript 6.
PCAT6 is a target of miR-330-5p
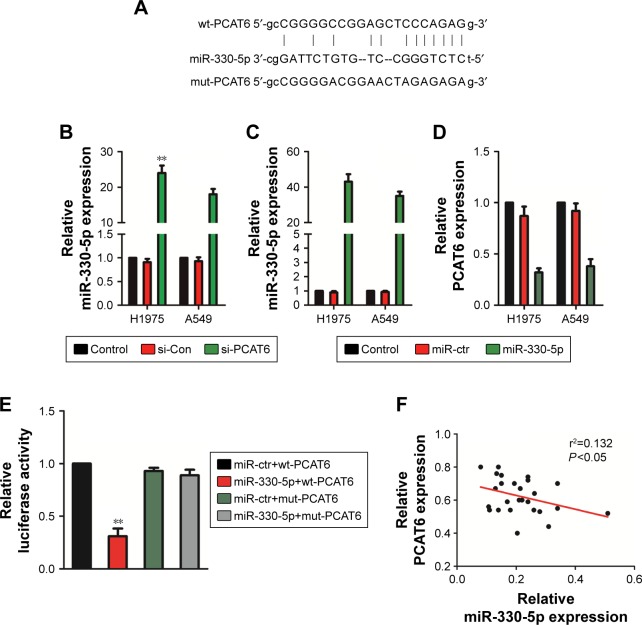
Previous investigations have demonstrated that lncR-NAs serve as competing endogenous RNAs (ceRNAs) and sponge miRNAs, regulating the expression of target mRNA. To find out the specific miRNA that was regulated by PCAT6, we performed bioinformatics analysis and luciferase reporter assay. First, the results of bioinformatics analysis starBase (http://starbase.sysu.edu.cn/index.php) suggested that miR-330-5p, which contained the putative binding sites, was the target of PCAT6 (Figure 4A). Next, we assessed the level of miR-330-5p in NSCLC cell that was transfected with si-PCAT6, and we found that the level of miR-330-5p was markedly increased in PCAT6 down-expressing A549 and H1975 cells (Figure 4B). Meanwhile, A549 and H1975 cells were transfected with miR-330-5p (Figure 4C) and we found that upregulation of miR-330-5p decreased the level of PCAT6 (Figure 4D). In order to further prove PCAT6 was the target of miR-330-5p, the luciferase reporter assay was applied. The fragment of PCAT6 containing miR-330-5p binding sites (wt-PCAT6) or mutant fragment (mut-PCAT6) was inserted into plasmid. Then, 293 T cell was cotransfected with plasmid and miR-330-5p or miR-ctr. As shown in Figure 4E, the luciferase activity in wt-PCAT6-transfected cell was significantly decreased by miR-330-5p. However, the inhibitory effect of miR-330-5p was abolished in cell that was transfected with mut-PCAT6. Finally, there was a negative relationship between PCAT6 and miR-330-5p in NSCLC tissues (Figure 4F). Hence, these results suggested that lncRNA PCAT6 directly bound to miR-330-5p.
Figure 4.
PCAT6 is the target of miR-330-5p.
Notes: (A) Bioinformatics analysis using online tool showed the putative binding sites between miR-330-5p and PCAT6. (B) The level of miR-330-5p was detected using qRT-PCR assay in A549 and H1975 cells. **P<0.01 compared with control, Student’s t-test. (C) The levels of miR-330-5p in miR-330-5p overexpressing A549 and H1975 cells were determined by qRT-PCR. **P<0.01 compared with control, Student’s t-test. (D) The expressions of PCAT6 in miR-330-5p overexpressing A549 and H1975 cells were determined by qRT-PCR. **P<0.01 compared with control, Student’s t-test. (E) wt-PCAT6 or mut-PCAT6 was transfected into 293 T cell combination with miR-330–5p. The luciferase activity was measured. **P<0.01 compared with miR-ctr+wt-PCAT6, Student’s t-test. (F) The association between PCAT6 and miR-330-5p in NSCLC tissues was analyzed using qRT-PCR assay.
Abbreviations: NSCLC, non-small-cell lung carcinoma; PCAT6, prostate cancer-associated transcript 6; qRT-PCR, quantitative real-time polymerase chain reaction.
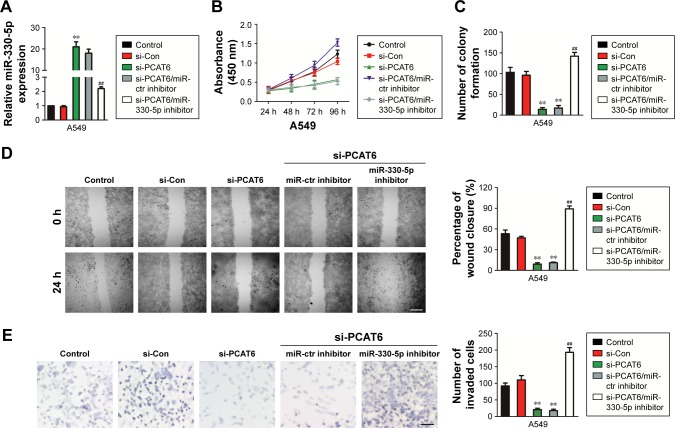
Downregulation of miR-330-5p reverses the suppressive effects of si-PCAT6
Although previous findings have proved that PCAT6 is the target of miR-330-5p, the impact of miR-330-5p on the PCAT6-mediated growth, migration, and invasion of NSCLC cell needed deeper investigation. As shown in Figure 5A, miR-330-5p inhibitor transfection significantly decreased the level of miR-330-5p in A549 cell that was transfected with si-PCAT6. In addition, knockdown of miR-330-5p in A549 cell that was pre-transfected with si-PCAT6 markedly neutralized the suppressive effects of si-PCAT6 on the proliferation and colony formation of A549 cell (Figure 5B and C). In addition, miR-330-5p inhibitor markedly increased the migration and invasion of A549 cell that was suppressed by si-PCAT6 (Figure 5D and E). Altogether, these data suggested that downregulation of PCAT6 inhibited NSCLC cell growth, migration, and invasion through regulating miR-330-5p.
Figure 5.
Regulation of NSCLC cell growth, migration, and invasion by PCAT6 requires miR-330-5p.
Notes: (A) The level of miR-330-5p was measured using qRT-PCR in A549 cell that was transfected with si-PCAT6 alone, or si-PCAT6 combination with miR-330-5p inhibitor. (B) Cell proliferation assay was conducted in A549 cell, one-way ANOVA. (C) Cell colony formation assay was conducted in A549 cell. (D, E) The migration and invasion of A549 cell that was transfected with si-PCAT6 alone, or si-PCAT6 combination with miR-330–5 p inhibitor was determined using wound healing and Transwell assays. **P<0.01 compared with control, ##P<0.01 compared with si-PCAT6, Student’s t-test.
Abbreviations: NSCLC, non-small-cell lung carcinoma; PCAT6, prostate cancer-associated transcript 6; qRT-PCR, quantitative real-time polymerase chain reaction.
Discussion
Recently, substantial studies have suggested that plenty of lncRNAs are dysregulated in a wide range of cancer types.18,19 For example, Linc01433 is overexpressed in NSCLC tissues compared with normal lung tissues. Overexpression of linc01433 in lung cancer cell increases growth, metastasis, and EMT.20 Meanwhile, the expression of ASAP1-IT1 is overregulated in NSCLC cell lines and tissues, which can promote the growth and metastasis of NSCLC cell via regulating the phosphatase and tensin homolog/protein kinase B signal pathway.21 Furthermore, PCAT6 is markedly upexpressed in lung cancer and is closely correlated with the metastasis of lung cancer.11 Although previous study reveals that down-expression of PCAT6 significantly suppresses the proliferation of lung cancer cell and it could serve as a potential biomarker in NSCLC, the precise mechanism by which PCAT6 regulates NSCLC cell growth, migration, and invasion remains not yet investigated.10 In the current study, we revealed that lncRNA PCAT6 was overexpressed in NSCLC and associated with the growth, migration, and invasion of NSCLC cell, which suggested that PCAT6 played crucial roles in the progression of NSCLC.
Consistent with the previous studies, we found that the expression of PCAT6 was markedly increased in NSCLC tissues compared with adjacent non-cancerous tissues.11 To investigate the function of PCAT6 in NSCLC, A549 and H1975 cells were transfected with the si-PCAT6. Next, downregulated PCAT6 inhibited the growth of A549 and H1975 cells as demonstrated by both MTT and colony formation assays. Having identified the crucial role of PCAT6 in the proliferation of NSCLC cell in vitro, we investigated the impact of PCAT6 on the growth of NSCLC cell in vivo. We found that downregulation of PCAT6 significantly inhibited the tumor growth of NSCLC cell in the xenograft model. In addition, wound healing and Transwell invasion assays were applied to explore the role of PCAT6 in the migration and invasion of NSCLC cell. The results demonstrated that the migration and invasive capacities of NSCLC cells were significantly decreased by the down-expression of PCAT6.
Emerging evidences demonstrated that lncRNA acts as miRNA sponge and competitively bind to miRNA.22 To explore whether PCAT6 serves as a miRNA sponge, the online bioinformatics analysis tool, starBase was selected.23 The results revealed that there were binding sites between miR-330-5p and PCAT6, and the level of miR-330-5p was negatively associated with PCAT6 in NSCLC tissues. Moreover, the level of miR-330-5p was higher in PCAT6 down-expressing A549 and H1975 cell. Luciferase reporter assay further demonstrated that the luciferase activity was decreased by miR-330-5p in wt-PCAT6-transfected 293 T cell. Therefore, these findings proved that PCAT6 bound to miR-330-5p.
A growing number of studies reveal that miRNAs serve as oncogenes or anti-oncogenes in many cancers.24 For example, miR-330-5p is downexpressed in glioblastoma cell lines and upregulation of miR-330-5p has an inhibitory effect on the growth, invasion, and migration of glioblastoma cell.25 Furthermore, miR-330-5p is markedly lower in colorectal cancer tissues than that in corresponding non-tumorous tissues and downregulation of miR-330-5p affects the development of colorectal cancer through regulating the expression of integ-rin alpha-5.26 In order to reveal the precise roles of PCAT-6/miR-330-5p in the growth, migration and invasion of NSCLC cell, A549 cell was cotransfected with si-PCAT6 and miR-330-5p inhibitor. The rescue experiments demonstrated that miR-330-5p inhibitor reserved the inhibitory effects of si-PCAT6 on NSCLC cell, which revealed that down-expression of PCAT6 inhibited the proliferation, migration, and invasion of NSCLC cell by regulating miR-330-5p.
We demonstrated that PCAT6 was overexpressed in NSCLC cells and clinical tissues. Downregulation of PCAT6 inhibited the growth, migration, and invasion of NSCLC cell. Furthermore, down-expression of PCAT6 significantly suppressed the tumor growth of NSCLC cell in vivo. In addition, miR-330-5p was the direct target of PCAT6 and PCAT6 inhibited the proliferation, migration, and invasion of NSCLC cell through regulating miR-330-5p. Our study first revealed the possible link between PCAT6 and miR-330-5p in NSCLC.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Reinmuth N. New data: new options for front-line therapy in NSCLC? ESMO Open. 2018;3(3):e000369. doi: 10.1136/esmoopen-2018-000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xing Z, Wang HY, Su WY, Wy S, et al. Wnt3 knockdown sensitizes human non-small cell type lung cancer (NSCLC) cells to cisplatin via regulating the cell proliferation and apoptosis. Eur Rev Med Pharmacol Sci. 2018;22(5):1323–1332. doi: 10.26355/eurrev_201803_14474. [DOI] [PubMed] [Google Scholar]
- 3.Gong W, Cao Y, Wang Y, et al. Upregulation of LncRNA FEZF-AS1 is associated with advanced clinical stages and family history of cancer in patients with NSCLC. Pathol Res Pract. 2018;214(6):857–861. doi: 10.1016/j.prp.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Shen QM, Wang HY, Xu S. LncRNA GHET1 predicts a poor prognosis of the patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22(8):2328–2333. doi: 10.26355/eurrev_201804_14823. [DOI] [PubMed] [Google Scholar]
- 5.Esmatabadi MJD, Motamedrad M, Sadeghizadeh M. Down-regulation of lncRNA, GAS5 decreases chemotherapeutic effect of dendrosomal curcumin (DNC) in breast cancer cells. Phytomedicine. 2018;42:56–65. doi: 10.1016/j.phymed.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 6.Xiao B, Zhang W, Chen L, et al. Analysis of the miRNA-mRNA-lncRNA network in human estrogen receptor-positive and estrogen receptor-negative breast cancer based on TCGA data. Gene. 2018;658:28–35. doi: 10.1016/j.gene.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Tuo Z, Zhang J, Xue W. LncRNA TP73-AS1 predicts the prognosis of bladder cancer patients and functions as a suppressor for bladder cancer by EMT pathway. Biochem Biophys Res Commun. 2018;499(4):875–881. doi: 10.1016/j.bbrc.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Zheng P, Li H, Xu P, et al. High lncRNA HULC expression is associated with poor prognosis and promotes tumor progression by regulating epithelial-mesenchymal transition in prostate cancer. Arch Med Sci. 2018;14(3):679–686. doi: 10.5114/aoms.2017.69147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.du Z, Fei T, Verhaak RG, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20(7):908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan L, Zhang L, Fan K, et al. Knockdown of Long Noncoding RNA PCAT6 Inhibits Proliferation and Invasion in Lung Cancer Cells. Oncol Res. 2016;24(3):161–170. doi: 10.3727/096504016X14618564639178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan L, Zhang L, Fan K, Wang JJ. Diagnostic significance of circulating long noncoding RNA PCAT6 in patients with non-small cell lung cancer. Onco Targets Ther. 2017;10:5695–5702. doi: 10.2147/OTT.S149314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng SM, Chang YC, Liu CY, et al. YM155 down-regulates survivin and XIAP, modulates autophagy and induces autophagy-dependent DNA damage in breast cancer cells. Br J Pharmacol. 2015;172(1):214–234. doi: 10.1111/bph.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amaral CL, Freitas LB, Tamura RE, et al. S6Ks isoforms contribute to viability, migration, docetaxel resistance and tumor formation of prostate cancer cells. BMC Cancer. 2016;16:602. doi: 10.1186/s12885-016-2629-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He W, Zhang H, Wang Y, et al. CTHRC1 induces non-small cell lung cancer (NSCLC) invasion through upregulating MMP-7/MMP-9. BMC Cancer. 2018;18(1):400. doi: 10.1186/s12885-018-4317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu HS, Somvanshi S, Patel E, et al. Pan-Cancer Analysis of lncRNA Regulation Supports Their Targeting of Cancer Genes in Each Tumor Context. Cell Rep. 2018;23312(1):297–312. doi: 10.1016/j.celrep.2018.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Feng S, Su W, et al. Overexpression of FAM83H-AS1 indicates poor patient survival and knockdown impairs cell proliferation and invasion via MET/EGFR signaling in lung cancer. Sci Rep. 2017;7:42819. doi: 10.1038/srep42819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Q, Shan S, Li Y, Zhu D, Jin W, Ren T. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. Faseb J. 2018;32(7):3957–3967. doi: 10.1096/fj.201701237RR. [DOI] [PubMed] [Google Scholar]
- 18.Wang ZD, Shen LP, Chang C, et al. Long noncoding RNA lnc-RI is a new regulator of mitosis via targeting miRNA-210-3p to release PLK1 mRNA activity. Sci Rep. 2016;6:25385. doi: 10.1038/srep25385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Wu X, Gu L, et al. Long non-coding RNA ATB promotes malignancy of esophageal squamous cell carcinoma by regulating miR-200b/Kindlin-2 axis. Cell Death Dis. 2017;8(6):e2888. doi: 10.1038/cddis.2017.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian B, Wang X, Mao C, et al. Long non-coding RNA linc01433 promotes migration and invasion in non-small cell lung cancer. Thorac Cancer. 2018;9(5):589–597. doi: 10.1111/1759-7714.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Shi SB, Zhu Y, Qian TT, Wang HL. Long non-coding RNA ASAP1-IT1 promotes cell proliferation, invasion and metastasis through the PTEN/AKT signaling axis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22(1):142–149. doi: 10.26355/eurrev_201801_14111. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Cao L, Wu J, Wang Q. Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. Int J Oncol. 2018;52(1):77–88. doi: 10.3892/ijo.2017.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo J, Feng Z, Huang Z, Wang H, Lu W. MicroRNA-217 functions as a tumour suppressor gene and correlates with cell resistance to cisplatin in lung cancer. Mol Cells. 2014;37(9):664–671. doi: 10.14348/molcells.2014.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng L, Ma J, Ji H, Liu Y, Hu W. miR-330-5p suppresses glioblastoma cell proliferation and invasiveness through targeting ITGA5. Biosci Rep. 2017;37(3):1–9. doi: 10.1042/BSR20170019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Xu W, Jiang H, Zhang F, Gao J, Hou J. MicroRNA-330 inhibited cell proliferation and enhanced chemosensitivity to 5-fluorouracil in colorectal cancer by directly targeting thymidylate synthase. Oncol Lett. 2017;13(5):3387–3394. doi: 10.3892/ol.2017.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]