Abstract
Testicular microlithiasis (TM) is one of the symptoms of testicular dysgenesis syndrome (TDS). TM is particularly interesting as an informative marker of testicular germ cell tumors (TGCTs). KIT ligand gene (KITLG), BCL2 antagonist/killer 1 (BAK1), and sprouty RTK signaling antagonist 4 (SPRY4) genes are associated with a high risk of TGCTs, whereas bone morphogenetic protein 7 gene (BMP7), transforming growth factor beta receptor 3 gene (TGFBR3), and homeobox D cluster genes (HOXD) are related to TDS. Using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis, we investigated allele and genotype frequencies for KITLG (rs995030, rs1508595), SPRY4 (rs4624820, rs6897876), BAK1 (rs210138), BMP7 (rs388286), TGFBR3 (rs12082710), and HOXD (rs17198432) in 142 TGCT patients, 137 TM patients, and 153 fertile men (control group). We found significant differences in the KITLG GG_rs995030 genotype in TM (P = 0.01) and TGCT patients (P = 0.0005) compared with the control. We also revealed strong associations between KITLG_rs1508595 and TM (G allele, P = 0.003; GG genotype, P = 0.01) and between KITLG_rs1508595 and TGCTs (G allele, P = 0.0001; GG genotype, P = 0.0007). Moreover, there was a significant difference in BMP7_rs388286 between the TGCT group and the control (T allele, P = 0.00004; TT genotype, P = 0.00006) and between the TM group and the control (T allele, P = 0.04). HOXD also demonstrated a strong association with TGCTs (rs17198432 A allele, P = 0.0001; AA genotype, P = 0.001). Furthermore, significant differences were found between the TGCT group and the control in the BAK1_rs210138 G allele (P = 0.03) and the GG genotype (P = 0.01). KITLG and BMP7 genes, associated with the development of TGCTs, may also be related to TM. In summary, the KITLG GG_rs995030, GG_rs1508595, BMP7 TT_rs388286, HOXD AA_rs17198432, and BAK1 GG_rs210138 genotypes were associated with a high risk of TGCT development.
Keywords: associations, genotype, high risk, testicular dysgenesis syndrome, testicular microlithiasi
INTRODUCTION
Problems with male reproductive health are extremely common. They can appear in newborns (cryptorchidism, hypospadias), the young (hypogonadism, delayed puberty), and adults (subfertility/infertility, poor sperm quality, testicular germ cell tumors, and testicular atrophy). It has been estimated that one in six young men from European countries suffers from at least one of these disorders.1
Testicular microlithiasis (TM) is detected during scrotal ultrasound examination in 5% of men.2 However, its true prevalence remains unknown because it is often asymptomatic. It is considered that deregulated phagocytic activity of sertoli cells is one of the key events in TM pathogenesis. In this condition, apoptotic testicular germinal epithelium is not destroyed by phagocytosis in the seminiferous tubules but undergoes calcification and then forms microcalcinates. It has been hypothesized that this is the mechanism behind the development of testicular dysgenesis syndrome (TDS).3 Testicular germ cell tumors (TGCTs), hypospadias, cryptorchidism, hypogonadism, TM, and impaired spermatogenesis could be clinical features of TDS presenting in a combination of congenital disorders and appearing during fetal testis development. These clinical signs are thought to be connected to androgen deficiency in the male embryo.4 It has also now been clarified that clinical risk factors of testicular germ cell tumors, such as cryptorchidism, TM, and low sperm count and quality, are results of the simultaneous action of adverse genetic and environmental factors. To reveal a correlation between these clinical conditions, long-term observation with detailed anamnesis from birth to adulthood for each patient is required, but this is difficult to perform.
Genome-wide association studies (GWASs) have enabled the identification of a number of genes (KIT ligand gene [KITLG], sprouty RTK signaling antagonist 4 gene [SPRY4], BCL2 antagonist/killer 1 gene [BAK1], bone morphogenetic protein 7 gene [BMP7], transforming growth factor beta receptor 3 gene [TGFBR3], and homeobox D cluster genes [HOXD]) involved in embryonic development of the testis and human spermatogenesis. These genes could be responsible for the clinical appearance of TDS and reproductive disorders associated with TDS and TGCT development.5
To evaluate common genetic factors related to the development of both TGCTs and TM, we investigated KITLG, SPRY4, BAK1, BMP7, TGFBR3, and HOXD and performed a comparative analysis of allelic variants and genotypes of patients with TGCTs/TM and those of fertile men.
PATIENTS AND METHODS
The examined cohort featured 432 men, consisting of 142 patients with TGCTs, 137 patients with TM, and 153 fertile men (control group). The TM patient group consisted of young men, aged 16 to 26 years, who had undergone a medical examination before military service. Individuals with cryptorchidism, hydrocele, and varicocele were excluded from the study. The TGCT group included 142 patients (seminomas, n = 53; nonseminomas, n = 74; and mixed tumors, n = 15) recruited from male patients, aged 25–45 years, undergoing postsurgical treatment at the chemotherapy department of N. N. Blokhin Russian Cancer Research Centre (Moscow, Russia). Control group consisted of fertile men with no testicular tumors or other andrology diseases.
The study was conducted in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. The Institutional Ethics Committee of Russian Medical Academy of Postgraduate Education approved all studies. Study participants gave informed consent to the presented study.
Testicular microlithiasis was diagnosed by scrotal ultrasound examination using Voluson 730 PRO V (General Electric Company, Vienna, Austria) and Mindray DC-7 (Shenzhen Mindray Bio-Medical Electronics, Shenzhen, China) equipment with a high-frequency linear array transducer (7–12 MHz). The diagnostic criterion for TM was the detection of more than three inclusions (with a diameter of 1–3 mm) without acoustic shadowing in testicular parenchyma.6
Peripheral blood samples from patients and fertile men (semen donors) were used for DNA analysis. Genomic DNA was extracted from peripheral blood leukocytes using a standard method, as described previously.7
In polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis, PCR amplification was performed in 25-μl reaction mixtures (Syntol, Moscow, Russia), containing 1–2 μl of DNA, 2.0 mmol l−1 MgCl2, 1.5 mmol l−1 of each dNTP, 2 pmol l−1 of each primer, 2.5 μl of PCR buffer, and 1 unit of HotStarTaq DNA polymerase. PCR conditions, primer sequences, temperatures, and the lengths of PCR products are listed in Table 1. Subsequently, PCR products were treated with a restriction enzyme (Syntol) at 37°C for 2–5 h (Supplementary Table 1 (492.4KB, tif) and 2 (194.8KB, tif) ).
Table 1.
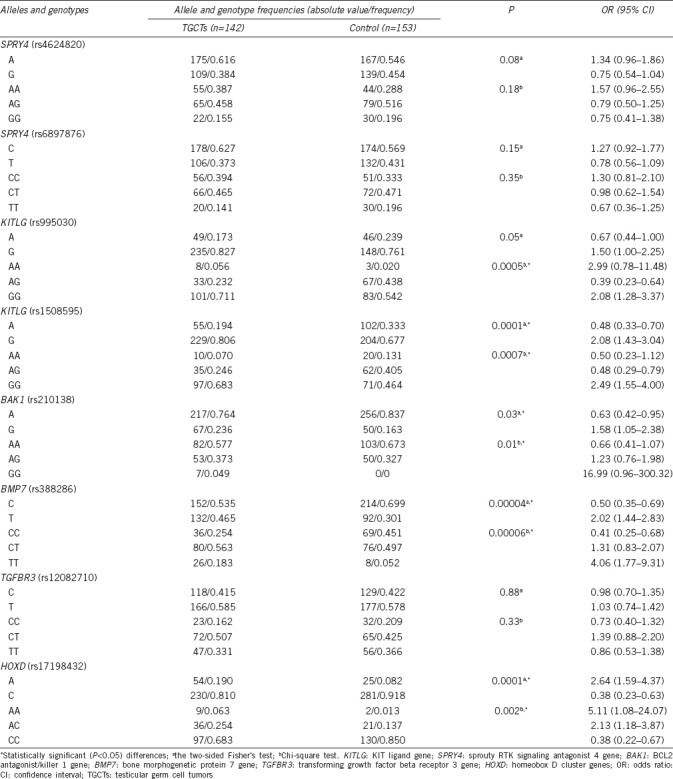
Allele and genotype frequencies of KIT ligand gene (rs995030, rs1508595), sprouty RTK signaling antagonist 4 gene (rs4624820, rs6897876), BCL2 antagonist/killer 1 gene (rs210138), bone morphogenetic protein 7 gene (rs388286), transforming growth factor beta receptor 3 gene (rs12082710), and homeobox D cluster genes (rs17198432) in testicular germ cell tumor patients
Polymerase chain reaction conditions for genotyping of KIT ligand gene (rs995030, rs1508595), sprouty RTK signaling antagonist 4 gene (rs4624820, rs6897876), BCL2 antagonist/killer 1 gene (rs210138), bone morphogenetic protein 7 gene (rs388286), transforming growth factor beta receptor 3 gene (rs12082710), and homeobox D cluster genes (rs17198432)
Polymerase chain reaction-restriction fragment length polymorphism and enzymes with length of restriction fragments
Restriction products were separated by vertical electrophoresis in 8% polyacrylamide gel (Acros Organic, Morris Plains, NJ, USA; Figure 1). To visualize the PCR products, the gel was stained with 1.1 mmol l−1 AgNO3 (Merck, Darmstadt, Germany) for 10–15 min, and then with 0.75 mol l−1 NaOH, 0.5 mol l−1 HCHO, and 2.3 mmol l−1 NaBH4 (Merck) for 10–15 min.
Figure 1.
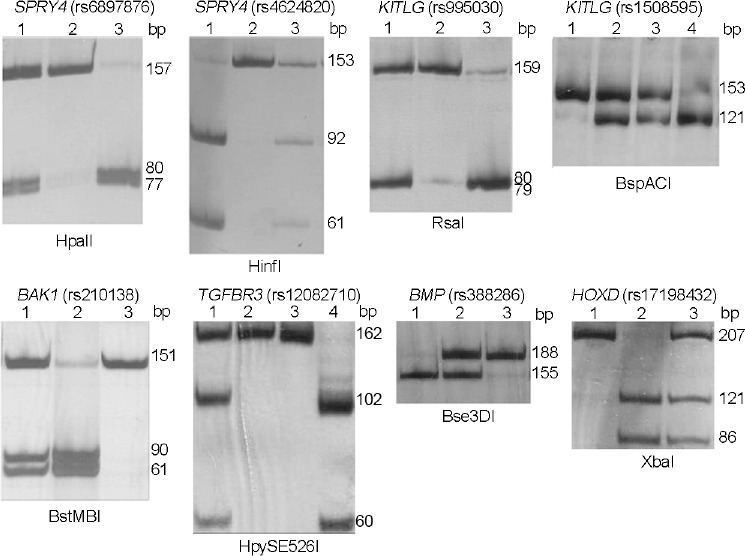
PCR-RFLP analysis of allelic variants of SPRY4 (rs4624820, rs6897876), KITLG (rs995030, rs1508595), BAK1 (rs210138), TGFBR3 (rs12082710), BMP7 (rs388286), and HOXD (rs17198432). Genotypes: SPRY4 (rs6897876): 1 – CT, 2 – TT, 3 – CC; SPRY4 (rs4624820): 1 – GG, 2 – AA, 3 – AG; KITLG (rs995030): 1 – AG, 2 – GG, 3 – AA; KITLG (rs1508595): 1 – AA, 2 – AG, 3 – AG, 4 – GG; BAK1 (rs210138): 1 – AG, 2 – GG, 3 – AA; TGFBR3 (rs12082710): 1 – CT, 2, 3 – CC, 4 – TT; BMP7 (rs388286): 1 – TT, 2 – CT, 3 – CC; and HOXD (rs17198432): 1 – AA, 2 – CC, 3 – AC. SPRY4: sprouty RTK signaling antagonist 4 gene; KITLG: KIT ligand gene; BAK1: BCL2 antagonist/killer 1 gene; TGFBR3: transforming growth factor beta receptor 3 gene; BMP7: bone morphogenetic protein 7 gene; HOXD: homeobox D cluster genes; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism.
Statistical analysis of the data was performed using GraphPad Prism version 6.0 software (GraphPad Software, San Diego, CA, USA). Associations were evaluated by comparison of allele and genotype frequencies among the TGCT, TM, and control groups. A test for Hardy–Weinberg equilibrium was performed using Chi-square criteria α = 0.05 and degree of freedom (df) = 1. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were estimated. Associations of disorders with allelic variants (genotypes) were determined using the two-sided Fisher's test, the Chi-square test with Yates's correction for continuity (df = 2, mode of inheritance), or the Mann–Whitney U test, when comparing the allele frequency and genotype distribution for each polymorphism in the TGCT and TM groups versus the control. The false discovery rate (FDR) was controlled using the Benjamini–Hochberg method (FDR test). A comparative analysis of genotype frequencies was performed using a two-sided Fisher's test. Statistical significance was assumed for P < 0.05 in all tests.
RESULTS
Using PCR-RFLP analysis, we developed a detection method (Figure 1) and investigated the KITLG (rs995030, rs1508595), SPRY4 (rs4624820, rs6897876), BAK1 (rs210138), BMP7 (rs388286), TGFBR3 (rs12082710), and HOXD (rs17198432) loci in 142 patients with TGCTs, 137 patients with TM, and 153 fertile men. The alleles and genotypes revealed in the test and the control groups are presented in Table 1 and 2.
Table 2.
Allele and genotype frequencies of KIT ligand gene (rs995030, rs1508595), sprouty RTK signaling antagonist 4 gene (rs4624820, rs6897876), BCL2 antagonist/killer 1 gene (rs210138), bone morphogenetic protein 7 gene (rs388286), transforming growth factor beta receptor 3 gene (rs12082710), and homeobox D cluster genes (rs17198432) in testicular microlithiasis patients
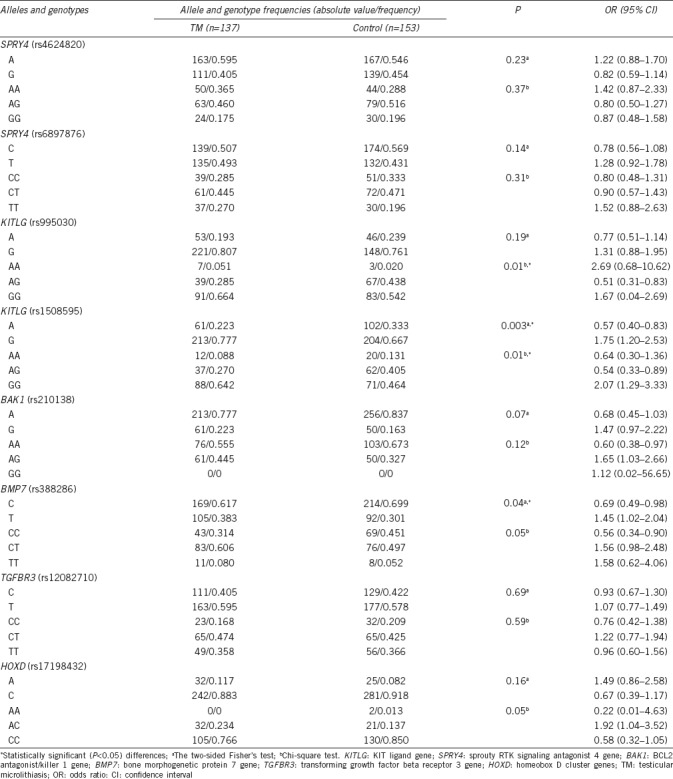
We found significant differences in the prevalence of the KITLG genotype GG rs995030 in patients with TM (P = 0.01) and TGCTs (P = 0.0005) compared with the level in the control group. Strong associations of TM with KITLG (rs1508595) were detected for both the G allele (P = 0.003) and the GG genotype (P = 0.01) and also for TGCTs with the G allele (P = 0.0001) and the GG genotype (P = 0.0007). For BMP7 (rs388286), significant differences from the control group were found for both the TGCT group (T allele, P = 0.00004; TT genotype, P = 0.00006) and the TM group (T allele, P = 0.04).
For HOXD (rs17198432), significant associations were demonstrated only with TGCTs (A allele, P = 0.0001; AA genotype, P = 0.001). For BAK1 (rs210138), significant differences were also found for the G allele (P = 0.03) and the GG genotype (P = 0.01) in the TGCT group compared with the control. However, there were no remarkable differences in TGFBR3 (rs12082710) and SPRY4 (rs4624820, rs6897876) allele and genotype frequencies between the TGCT/TM groups and the control.
KITLG and BMP7 genotypes were remarkably different from those in the control for both TGCT and TM groups (Table 1 and 2, respectively). The P value in the TM group was higher than that in the TGCT group but was still significant.
Thus, we have determined that some genotypes, i.e., KITLG, BMP7, HOXD, and BAK1, correlate with a high risk of TGCTs development (Table 1). According to GWASs, KITLG, BAK, and SPRY4 genes are associated with TGCTs, and BMP7, TGFBR3, and HOXD genes are associated with the development of TDS.5,8 The results of our study indicate that KITLG and BMP7 are also associated with TM. Evidently, some of these genes are common in TGCTs, TM, and TDS (Figure 2).
Figure 2.
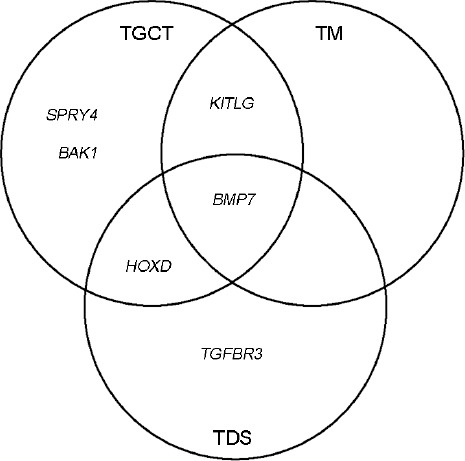
A Venn diagram showing genes (KITLG, BAK, SPRY4, BMP7, TGFBR3, and HOXD) involved in the development of testicular germ cell tumors (TGCTs), testicular microlithiasis (TM), and testicular dysgenesis syndrome (TDS). SPRY4: sprouty RTK signaling antagonist 4 gene; KITLG: KIT ligand gene; BAK1: BCL2 antagonist/killer 1 gene; TGFBR3: transforming growth factor beta receptor 3 gene; BMP7: bone morphogenetic protein 7 gene; HOXD: homeobox D cluster genes.
DISCUSSION
Recently, several GWASs have revealed a number of genetic variations with important roles in TGCT development, including variations in KITLG, SPRY4, BAK1, Doublesex- and mab-3-related transcription factor 1 gene (DMRT1), telomerase reverse transcriptase gene (TERT), activating transcription factor 7 interacting protein gene (ATF7IP), hematopoietic prostaglandin D synthase gene (HPGDS), mitotic arrest deficient 1 like 1 gene (MAD1L1), ring finger and WD repeat domain 3 gene (RFWD3), testis expressed 14, intercellular bridge forming factor gene (TEX14), and protein phosphatase, Mg2+/Mn2+ dependent 1E gene (PPM1E).8,9,10,11,12 Some TGCT-associated genes are functionally related to the early stages of human gametogenesis (KITLG, BMP7, deleted in azoospermia-like gene [DAZL], PR/SET domain 14 gene [PRDM14]) or embryonic cell differentiation and gonad development (DMRT1, HPGDS). There are also genes that control the processes of DNA repair (minichromosome maintenance complex component 3 associated protein gene [MCM3AP], RAD51 paralog C gene [RAD51C], ring finger and WD repeat domain 3 gene [RFWD3]), chromosome segregation, microtubule assembly (mitotic arrest deficient 1 like 1 gene [MAD1L1], testis expressed 14, intercellular bridge forming factor gene [TEX14]), and tumorigenesis (KITLG, BAK1, PRDM14).
The strongest association with TGCTs was identified for the KITLG gene, which is located on 12q22 and encodes a KIT ligand. The signaling pathway KIT/KITLG (Kit receptor signaling pathway) is critical for the migration and survival of primordial germ cells (PGCs), and its deregulation leads to the appearance of ectopically migrated PGCs. The abnormal development of postmigratory PGCs (gonocytes) can become a source of intratubular germ cell neoplasia and eventually TGCTs.13 The role of the KIT/KITLG signaling pathway in the development of both familial and sporadic TGCTs was confirmed by an association with SPRY, BAK1, and PDE11A genes, which are functionally related to this pathway.11 In our investigation, we demonstrated significant differences between the TGCT and control groups in the alleles and genotypesof KITLG, BAK1, BMP7, and HOXD. This confirms the importance of these genes for TGCT development. Ferlin et al.14 previously demonstrated the association of the KITLG gene with TGCT development, particularly testicular seminoma. However, to the best of our knowledge, this study is the first to present an association between TGCTs and BMP7 and HOXD. According to the GWAS data, BMP7 (rs388286) and HOXD (rs17198432) are genetic factors unrelated to TGCTs but related to TDS development.5
Interestingly, the HOXD locus (rs17198432 alleles and genotypes) is associated with TGCT development. HOXD genes are located in a cluster on chromosome 2 (locus 2q31.1) in the region between the ATP synthase membrane subunit C locus 3 gene (ATP5G3) and Lunapark, ER junction formation actor gene (KIAA1715) (https://www.genecards.org; https://genome.ucsc.edu). The ATP5G3 gene encodes a subunit of mitochondrial ATP synthase, which catalyzes ATP synthesis using an electrochemical proton gradient across the inner membrane during oxidative phosphorylation.15 The KIAA1715 gene plays a specific role in the formation and interactions of the endoplasmic reticulum and also presumably participates in the development of limbs and the central nervous system in mammals. The role of these genes in tumorigenesis and TDS development is unclear. However, there is a homeotic HOXD gene cluster located distally in the telomere direction. This gene cluster, as well as other homologues of homeobox A cluster genes (HOXA; 7p14), homeobox B cluster genes (HOXB; 17q21), and homeobox C cluster genes (HOXC;12q13), encodes a highly conserved family of transcription factors, which play an important role in the morphogenesis of multicellular organisms.16 Deletions of HOXD genes or their 5’-regions lead to severe abnormalities of limbs and gonads; point mutations in the HOXD13 gene result in synpolydactyly and brachydactyly.16 The homeotic HOXD gene cluster could also be related to reproductive problems and germ cell tumors; however, the exact mechanism behind this remains unknown.
TM, together with testicular atrophy, cryptorchidism, and male subfertility/infertility, is considered to be a clinical condition associated with a high risk of developing TGCTs.17 It was proposed that TM, as well as TGCTs, commonly results from TDS.3 Several reports have described cases of the simultaneous presence of TM and TGCTs in patients and have noted the increased prevalence of TM in men with familial TGCTs.18,19,20
The detection of common genetic factors associated with the pathogenesis of these disorders may indicate a connection between them and probable common mechanisms behind their development. Among the genes reported to be associated with TDS (according to GWASs), namely, BMP7, TGFBR3, HOXD, and KITLG, only KITLG and BMP7 demonstrated a significant association with TM in the present study. Thus, we have confirmed the importance of the KIT/KITLG signaling pathway, not only for TGCTs but also for TM development.
Another gene associated with both TM and TGCTs is BMP7. This gene encodes a growth factor related to ligands of the bone morphogenetic protein (BMP) family, which is involved in the TGF-β signaling pathway and participates in tissue formation during embryonic development.21 By binding with TGF-β receptors, ligands of the BMP family activate transcription factors of the SMAD family, which regulate the expression of specific target genes. Reproductive functions of BMP include the modulation of testosterone biosynthesis, embryonic cell maturation, sperm quality, and intactness of reproductive tissues.22 The BMP/TGFβ signaling pathway is responsible for the maintenance of pluripotency and self-renewal of embryonic stem cells, and for embryonic development of PGCs, which are precursors of immature germ cells. Moreover, since it is a growth factor, BMP can promote tumorigenesis in adulthood by maintaining cell proliferation.23
Therefore, the KITLG and BMP7 ligands play important roles in PGC development and differentiation in human embryos, but their deregulation could lead to the activation of cell proliferation and consequently to TGCTs in adulthood. Our data suggest that there is an etiological relationship between TGCTs and TM via TDS, based on the common genetic factors behind their emergence. It was hypothesized that TM and TGCTs could be events secondary to a single common disorder called tubular degeneration, which could explain the correlation between TGCTs and TM.24 In accordance with this hypothesis, TM is not a risk factor associated with a predisposition to testicular cancer development in itself, but rather a marker of tubular degeneration. Presumably, TM could be a marker only in those men who already have cryptorchidism, low sperm quality, or other risk factors for TGCTs.25
Another explanation of our results is that some patients with TM will develop TGCTs in the future. However, it is not possible to predict the risk of tumor formation because of the young age of the patients in our set. In addition, specific environmental factors also influence the process of tumor formation (tumorigenesis). Moreover, as those patients make up a relatively small proportion of the TM patients in our study, we observed a lower P value than that for the TGCT group (Table 1 and 2).
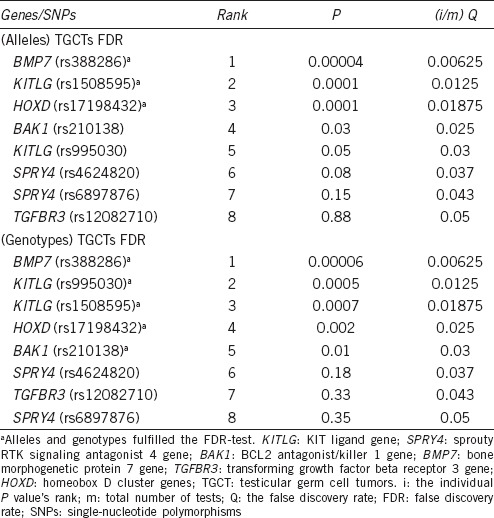
To obtain more robust P values and prevent false-positive results, the FDR test was used, which makes it possible to reject some controversial results. The FDR test in the TGCT group confirmed the data for BMP7 (rs388286), KITLG (rs1508595), HOXD (rs17198432), and KITLG (rs995030) alleles and genotypes, but none of the evaluated polymorphisms in the TM group fulfilled the requirements of the FDR test (Table 3). However, the low significance of the results could be explained by the sample set or by an indirect effect of these genes in the development of TDS. Therefore, these allelic and genotypic variants should not be overlooked as potential genetic risk factors associated with TDS.
Table 3.
Benjamini–Hochberg procedure (false discovery rate test) for investigated alleles and genotypes
In conclusion, the present study demonstrated that variants of certain genes related to the KIT/KITLG and BMP/TGFβ signaling pathways and important for the development of TGCTs are more common in TM patients than in fertile men. However, the lack of strong associations with other TDS-associated genes indicates that TM may not be unequivocally associated with TDS symptoms.
AUTHOR CONTRIBUTIONS
ISD performed DNA isolation and molecular analyses, interpreted results, and drafted the manuscript. EVI and AAT performed clinical examination of TGCT patients and collected biological samples from tumor patients. OYL, EAV, and ABO performed clinical examination of TM patients and collected biological samples from TM patients. DNG diagnosed of TM by ultrasound examination. OBL supervised the study. VVR and DSM performed statistical analysis and interpreted results. VBC overviewed results and finalized the manuscript. MVN designed the study, interpreted the results, and supervised the study. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declared no competing interests.
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Skakkebaek NE, Rajpert-De Meyts E, Buck Louis GM, Toppari J, Andersson AM, et al. Male reproductive disorders and fertility trends: influences of environment and genetic susceptibility. Physiol Rev. 2016;96:55–97. doi: 10.1152/physrev.00017.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterson AC, Bauman JM, Light DE, McMann LP, Costabile RA. The prevalence of testicular microcalcospherites in an asymptomatic population of men 18 to 35 years old. J Urol. 2001;166:2061–4. [PubMed] [Google Scholar]
- 3.Skakkebaek NE, Rajpert-De Meyts E, Main KM. Testicular dysgenesis syndrome: an increasingly common developmental disorder with environmental aspects. Hum Reprod. 2001;16:972–8. doi: 10.1093/humrep/16.5.972. [DOI] [PubMed] [Google Scholar]
- 4.Sharpe RM, Skakkebaek NE. Testicular dysgenesis syndrome: mechanistic insights and potential new downstream effects. Fertil Steril. 2008;89(Suppl 2):e33–8. doi: 10.1016/j.fertnstert.2007.12.026. [DOI] [PubMed] [Google Scholar]
- 5.Dalgaard MD, Weinhold N, Edsgärd D, Silver JD, Pers TH, et al. A genome-wide association study of men with symptoms of testicular dysgenesis syndrome and its network biology interpretation. J Med Genet. 2012;49:58–65. doi: 10.1136/jmedgenet-2011-100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shanmugasundaram R, Singh JC, Kekre NS. Testicular microlithiasis: is there an agreed protocol? Indian J Urol. 2007;23:234–9. doi: 10.4103/0970-1591.33442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989. pp. 931–57. [Google Scholar]
- 8.Chung CC, Kanetsky PA, Wang Z, Hildebrandt MA, Koster R, et al. Meta-analysis identifies four new loci associated with testicular germ cell tumor. Nat Genet. 2013;45:680–5. doi: 10.1038/ng.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanetsky PA, Mitra N, Vardhanabhuti S, Li M, Vaughn DJ, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41:811–5. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–7. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rapley EA, Turnbull C, Al Olama AA, Dermitzakis ET, Linger R, et al. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41:807–8. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Litchfield K, Shipley J, Turnbull C. Common variants identified in genome-wide association studies of testicular germ cell tumour: an update, biological insights and clinical application. Andrology. 2015;3:34–46. doi: 10.1111/andr.304. [DOI] [PubMed] [Google Scholar]
- 13.Rijlaarsdam MA, Looijenga LH. An oncofetal and developmental perspective on testicular germ cell cancer. Semin Cancer Biol. 2014;29:59–74. doi: 10.1016/j.semcancer.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Ferlin A, Pengo M, Pizzol D, Carraro U, Frigo AC, et al. Variants in KITLG predispose to testicular germ cell cancer independently from spermatogenic function. Endocr Relat Cancer. 2012;19:101–8. doi: 10.1530/ERC-11-0340. [DOI] [PubMed] [Google Scholar]
- 15.Yan WL, Lerner TJ, Haines JL, Gusella JF. Sequence analysis and mapping of a novel human mitochondrial ATP synthase subunit 9 cDNA (ATP5G3) Genomics. 1994;24:375–7. doi: 10.1006/geno.1994.1631. [DOI] [PubMed] [Google Scholar]
- 16.Quinonez SC, Innis JW. Human HOX gene disorders. Mol Genet Metab. 2014;111:4–15. doi: 10.1016/j.ymgme.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Elzinga-Tinke JE, Sirre ME, Looijenga LH, van Casteren N, Wildhagen MF, et al. The predictive value of testicular ultrasound abnormalities for carcinoma in situ of the testis in men at risk for testicular cancer. Int J Androl. 2010;33:597–603. doi: 10.1111/j.1365-2605.2009.00997.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoei-Hansen CE, Sommer P, Rajpert-De Meyts E, Skakkebaek NE. A rare diagnosis: testicular dysgenesis with carcinoma in situ detected in a patient with ultrasonic microlithiasis. Asian J Androl. 2005;7:445–7. doi: 10.1111/j.1745-7262.2005.00020.x. [DOI] [PubMed] [Google Scholar]
- 19.Coffey J, Huddart RA, Elliott F, Sohaib SA, Parker E, et al. Testicular microlithiasis as a familial risk factor for testicular germ cell tumour. Br J Cancer. 2007;97:1701–6. doi: 10.1038/sj.bjc.6604060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korde LA, Premkumar A, Mueller C, Rosenberg P, Soho C, et al. Increased prevalence of testicular microlithiasis in men with familial testicular cancer and their relatives. Br J Cancer. 2008;99:1748–53. doi: 10.1038/sj.bjc.6604704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams D, Karolak M, Robertson E, Oxburgh L. Control of kidney, eye and limb expression of Bmp7 by an enhancer element highly conserved between species. Dev Biol. 2007;311:679–90. doi: 10.1016/j.ydbio.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossa A, Munger S, Capel B. Bmp7 regulates germ cell proliferation in mouse fetal gonads. Sex Dev. 2007;1:127–37. doi: 10.1159/000100034. [DOI] [PubMed] [Google Scholar]
- 23.Ciller IM, Palanisamy SK, Ciller UA, McFarlane JR. Postnatal expression of bone morphogenetic proteins and their receptors in the mouse testis. Physiol Res. 2016;65:673–82. doi: 10.33549/physiolres.933193. [DOI] [PubMed] [Google Scholar]
- 24.Richenberg J, Belfield J, Ramchandani P, Rocher L, Freeman S, et al. Testicular microlithiasis imaging and follow-up: guidelines of the ESUR scrotal imaging subcommittee. Eur Radiol. 2015;25:323–30. doi: 10.1007/s00330-014-3437-x. [DOI] [PubMed] [Google Scholar]
- 25.Winter TC, Kim B, Lowrance WT, Middleton WD. Testicular microlithiasis: what should you recommend? AJR Am J Roentgenol. 2016;206:1164–9. doi: 10.2214/AJR.15.15226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Polymerase chain reaction conditions for genotyping of KIT ligand gene (rs995030, rs1508595), sprouty RTK signaling antagonist 4 gene (rs4624820, rs6897876), BCL2 antagonist/killer 1 gene (rs210138), bone morphogenetic protein 7 gene (rs388286), transforming growth factor beta receptor 3 gene (rs12082710), and homeobox D cluster genes (rs17198432)
Polymerase chain reaction-restriction fragment length polymorphism and enzymes with length of restriction fragments


