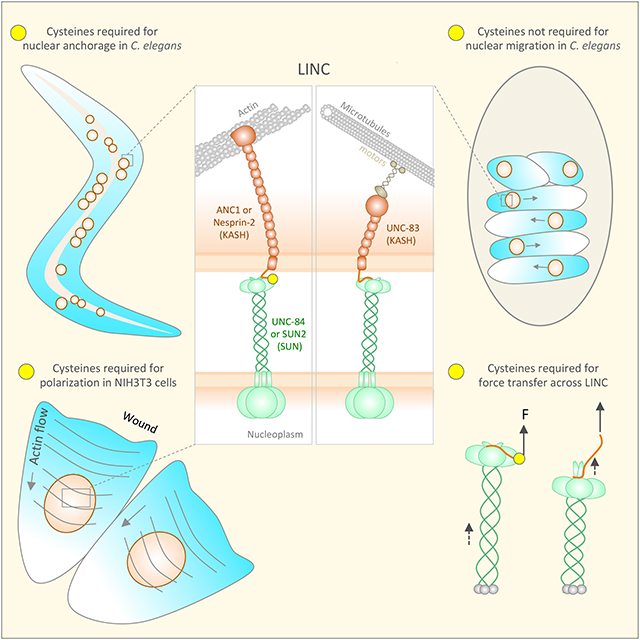
Summary
Many nuclear positioning events involve linker of nucleoskeleton and cytoskeleton (LINC) complexes, which transmit forces generated by the cytoskeleton across the nuclear envelope. LINC complexes are formed by trans-luminal interactions between inner nuclear membrane SUN proteins and outer nuclear membrane KASH proteins, but how these interactions are regulated is poorly understood. We combine in vivo C. elegans genetics, in vitro wounded fibroblast polarization, and in silico molecular dynamic simulations to elucidate mechanisms of LINC complexes. The extension of the KASH domain by a single alanine residue or the mutation of the conserved tyrosine at −7 completely blocked the nuclear migration function of C. elegans UNC-83. Analogous mutations at −7 of mouse nesprin-2 disrupted rearward nuclear movements in NIH3T3 cells, but did not disrupt ANC-1 in nuclear anchorage. Furthermore, conserved cysteines predicted to form a disulfide bond between SUN and KASH proteins are important for the function of certain LINC complexes and might promote a developmental switch between nuclear migration and nuclear anchorage. Mutations of conserved cysteines in SUN or KASH disrupted ANC-1 dependent nuclear anchorage in C. elegans and Nesprin-2G dependent nuclear movements in polarizing fibroblasts. However, the SUN cysteine mutation did not disrupt nuclear migration. Moreover, molecular dynamic simulations showed that a disulfide bond is necessary for the maximal transmission of cytoskeleton-generated forces by LINC complexes in silico. Thus, we have demonstrated functions for SUN-KASH binding interfaces, including a predicted intermolecular disulfide bond, as mechanistic determinants of nuclear positioning and may represent targets for regulation.
Keywords: Nuclear envelope, nuclear positioning, LINC complex
eTOC blurb
Cain et al. test the function of mutant SUN and KASH proteins in C. elegans nuclear positioning, NIH3T3 fibroblast polarization, and simulations of LINC complexes under mechanical strain to gain mechanistic insights into how SUN-KASH interactions might be regulated to transfer forces from the cytoskeleton to the nucleus.
Graphical Abstract
Introduction
The precise intracellular positioning of the nucleus is essential for many cellular and developmental processes including fertilization, cell migration, cell polarization, gametogenesis, neuronal development, and muscle development. Defects in nuclear positioning result in developmental disorders in tissues, including the central nervous system, reproductive organs, and skeletal muscle [1–4]. Most nuclei are positioned by the cytoskeleton, which is physically coupled to the nuclear envelope via a conserved nuclear envelope-spanning molecular bridge known as the linker of nucleoskeleton and cytoskeleton (LINC) complex [5].
The LINC complex is formed by a trans-luminal direct interaction between Sad1/UNC-84 (SUN) proteins in the inner nuclear membrane and Klarsicht/ANC-1/Syne homology (KASH) proteins spanning the outer nuclear membrane (Fig 1) [6–9]. SUN proteins contain a conserved KASH protein-binding luminal C-terminal SUN domain [10] and divergent N termini that interact with lamins and chromatin within the nucleoplasm [8, 11, 12]. KASH proteins contain a conserved C-terminal KASH domain, comprising of a transmembrane domain followed by the ~10–32 residue luminal KASH peptide [13]. The divergent N termini of KASH proteins extend into the cytoplasm and engage the cytoskeleton [5, 14, 15]. Thus, LINC complexes enable force transmission across the nuclear envelope [5, 14].
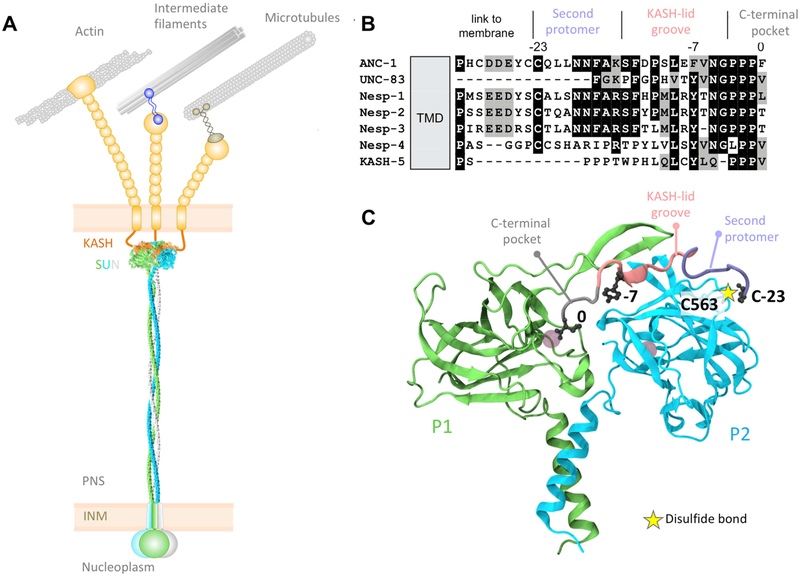
Figure 1. SUN and KASH proteins form a bridge across the nuclear envelope.
(A) SUN trimers (gray, blue, and green) cross the inner nuclear membrane (INM) and span most of the peri-nuclear space (PNS). SUN proteins interact with the luminal domains of KASH proteins (orange and tan), extending from the outer nuclear membrane (ONM). Cytoplasmic domains of KASH proteins interact with a variety of cytoskeletal components. (B) Sequence alignment of C. elegans and human KASH domains. (C) A close up of the interaction between a KASH peptide (Nesprin-2; gray, pink, and purple) and two SUN2 protomers (green and blue). The gray circles in the SUN protomers represent cations. Based on Protein Data Bank accession number 4DXS [16].
Crystal structures of mammalian SUN2 in complex with KASH peptides from Nesprin-1 or −2 provide insights into the molecular mechanisms of LINC complex-dependent mechanotransmission and nuclear positioning [16, 17]. The structures predict that SUN2 functions as a homo-trimer where each protomer consists of a non-conventional coiled-coil region followed by a β-sandwich core with a □20 residue β-hairpin extension, known as the KASH lid. Three independent KASH peptides interact with a single SUN2 homo-trimer [16, 17]. Our recent studies show that the luminal domain of SUN2 does homo-trimerize in living cells [18, 19].
Each KASH peptide forms three binding interfaces with a SUN trimer (Fig 1C) [16, 20]. First, the last four residues of KASH proteins (0 to −3, gray in Fig 1C), consisting of the PPPX motif, fit into a binding pocket formed primarily by a single SUN protomer (P1, green in Fig 1C). Second, positions −4 to −14 of KASH proteins (pink in Fig 1C) fit into a groove between two SUN protomers (P1 and P2), with the conserved hydrophobic residues at −7 and −9 facing into the cleft between P1 and P2. Third, residues −15 to −23 (purple in Fig 1C) bind along the surface of P2. At position −23 of the KASH peptide, a conserved cysteine is oriented to form a disulfide bond (yellow star in Fig 1C) with a conserved cysteine in SUN2 (C563 in human SUN2, C577 in mouse SUN2, and C953 in C. elegans UNC-84) [20]. Furthermore, an intermolecular disulfide bond was detected between transiently expressed SUN and KASH domains in HeLa cells [16, 20]. Although this covalent bridge was determined by in vitro pull-downs to be dispensable for the SUN2-KASH interaction, it was hypothesized to be important for LINC complexes to resist the considerable cytoskeleton-generated forces necessary for nuclear positioning [16, 20]. However, the physiological relevance of each of the three conserved SUN-KASH interaction domains and the conserved disulfide bond during intracellular nuclear movement and positioning remains to be tested experimentally. Here, we examine the effect of disrupting SUN-KASH interactions in two established experimental as-says for intracellular nuclear positioning: nuclear migration and anchorage in developing Caenorhabditis elegans hypodermal cells [21, 22] and rearward nuclear positioning during centrosome orientation in migrating fibroblasts [23–25]. Furthermore, we used molecular dynamics simulations of SUN2-KASH complexes under tension to model how disrupting SUN-KASH interactions might influence the ability of LINC complexes to withstand the mechanical forces required for nuclear positioning.
Nuclear positioning in C. elegans consists of two related processes—nuclear migration to a specific location and then anchoring in place. Nuclear migration in hyp7 precursors requires the SUN protein UNC-84 and the KASH protein UNC-83, which recruits microtubule motors to the surface of nuclei [10, 26, 27]. After the hyp7 syncytium is formed, UNC-84 and the KASH protein ANC-1 anchor nuclei to the actin cytoskeleton [10, 13]. In mammals, KASH proteins Nesprin-4 and KASH5 play analogous roles recruiting microtubule motors to the surface of nuclei [28, 29]. In other mammalian cell types, giant isoforms of Nesprin-1 and −2 tether mammalian nuclei to actin networks [30–32]. However, It remains unknown how UNC-84-UNC-83 complexes that move nuclei eventually yield to UNC-84-ANC-1 complexes to mediate nuclear anchorage. Interest ingly, the KASH domain in UNC-83 is much shorter than in ANC-1; the UNC-83 peri-nuclear luminal domain is only 17 residues and lacks the cysteine at −23 (Fig 1B) [6]. We hypothesized that an intermolecular disulfide bond formed between conserved cysteines at −23 of KASH peptides and at 563 of human SUN2 (residue 577 of mouse SUN2) or 953 of C. elegans UNC-84 plays a central role in LINC complex function. In order to elucidate potential molecular mechanisms for LINC complex regulation, we addressed the following: How important is the interaction between conserved residues in SUN and the PPPX motif of KASH? What happens when the interaction between the aromatic residue at the −7 position of KASH and the cleft between P1 and P2 is disrupted? And, what is the physiological relevance of the disulfide bond between SUN and KASH domains?
Results
Extension of the C terminus of the UNC-83 KASH domain disrupts nuclear migration in vivo.
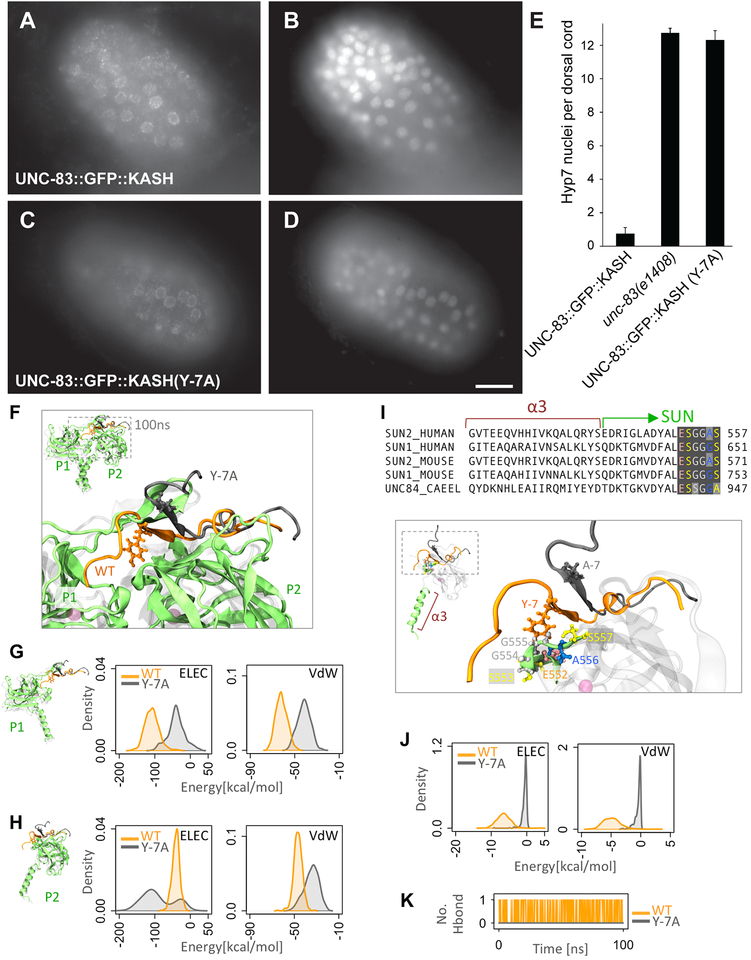
The crystal structures of mammalian SUN-KASH complexes show the C-terminus of KASH pointed into the SUN protomer P1, where it is coordinated by conserved hydroxyl groups [16, 17] (Fig 1C). Furthermore, the addition of a single alanine to the end of KASH peptides was shown to abolish in vitro pull-down interactions between SUN2 and KASH [16], while the extension of KASH peptides by 13 residues blocks their function in binding assays and acts as a dominant negative construct in tissue culture cells [33]. Therefore, we predicted that the extension of the KASH domain of UNC-83 by one residue would disrupt the interaction between UNC-84 and UNC-83 and disrupt nuclear migration in vivo. We used UNC-83::GFP::KASH, which has normal function for nuclear migration [34], as a starting point (Fig 2A,D) and added a single alanine onto the C terminus of UNC-83 to express UNC-83::GFP::KASH+A at endogenous levels. UNC-83::GFP::KASH+A failed to localize to the nuclear envelope, similar to when the entire KASH domain of UNC-83 was deleted [6]. As a control, we measured the relative fluorescence in the GFP channel in a standardized region of interest. UNC-83::GFP::KASH+A had 48.7±7.6 (mean ± standard deviation) arbitrary units of fluorescence per embryo compared to 27.1±6.5 arbitrary units in wild-type embryos (t-test; p < 0.0001), suggesting that the UNC-83::GFP::KASH+A construct was expressed, but failed to localize to the nuclear envelope. The resulting animal had a severe nuclear migration defect (Fig 2C–D). Thus, the KASH+A mutation blocks SUN-KASH function in vivo.
Figure 2. Extension of the KASH domain of UNC-83 blocks nuclear migration.
(A-C) GFP expression in comma-stage embryos with the version of UNC-83 indicated. Anterior is left, dorsal up. (A) UNC-83::GFP::KASH. (B) Wild-type UNC-83. (C) UNC-83::GFP::KASH+A. (D) Quantification of nuclear migration defects by counting average number of hyp7 nuclei in the dorsal cords of L1 animals. Error bars are 95% CI; scale bar is 10 μm.
Mutation of the tyrosine at −7 of KASH disrupts SUN-KASH function in vivo
Residues −5 to −14 of KASH proteins (pink in Fig 1) fit into a cleft between two SUN protomers. The conserved hydrophobic aromatic residue at −7 faces directly into the cleft in the crystal structure and a KASH(Y-7A) mutation blocked SUN2-KASH interactions in vitro [16]. Overexpressed mutant UNC-83(Y-7A) in a transgenic animal localized the nuclear envelope, but did not rescue unc-83(null) nuclear migration defects [6]. To rule out the effects of overexpression and test the role of Y-7 more directly, we used CRISPR/Cas9 genome editing to introduce the Y-7A mutation into the unc-83 locus. The resulting unc-83(Y-7A) mutant animal completely blocked nuclear migration in hyp7 cells (Fig 3A–E). However, UNC-83(Y-7A) localized normally to the nuclear envelope of hyp7 cells (Fig 3C–D), suggesting that unlike UNC-83(KASH+A), the UNC-83(Y-7A) mutant protein can partially interact with the SUN domain of UNC-84 for localization, but that the interaction fails to withstand forces across the nuclear envelope required to move nuclei in these cells.
Figure 3. Mutation of tyrosine −7 of KASH disrupts SUN-KASH interactions.
(A,C) Anti-GFP immunolocalization and (B,D) DAPI staining in pre-bean embryos around the time of hyp7 nuclear migration. (E) Quantification of nuclear migration defects as in Fig 2. Error bars are 95% CI; scale bar is 10 μm. (F-K) Modeling the effect of Y-7A on SUNKASH interactions. (F) Superimposed structures of wild type (orange) and Y-7A (gray) after 100ns of molecular dynamics simulation time. Only two of three SUN2 protomers are shown (P1 and P2). (G-H) Electrostatic (ELEC) and Van der Waals (VdW) energies between KASH (residues 0 to −17) and (G) the P1 or (H) P2 SUN2 protomer for WT (orange) and Y-7A (gray) (shown as densities over 100ns simulation time). (I) Sequence and structure of α3 and first few residues of the SUN domain showing that the major interacting partners of Y-7 on SUN P2 (E552-S557) are highly conserved. (J) ELEC and VdW energies between residue −7 and residues E552-S557 on SUN2 P2 for WT (orange) and Y-7A (gray) (shown as densities over 100ns simulation time). (K) Number of H-bonds between residue −7 and residues E552-S557 on SUN2 P2 for WT (orange) and Y-7A (gray).
We used molecular dynamics simulations to examine the extent to which the KASH(Y-7A) mutation might compromise SUN-KASH interactions. We performed simulations of the human SUN2 trimer bound to wild-type Nesprin-2 or Nesprin-2(Y-7A) KASH peptides. Within a 100 ns of simulation time, the KASH(Y-7A) peptides partially detached from the SUN2 trimer (Fig 3F). We calculated the non-bonded interaction energies between the KASH(Y-7A) peptide and P1 or P2 over the 100 ns simulation and compared it with wild type (Fig 3G–H). Because the KASH domain of UNC-83 only contains residues 0 to −17, we included the analogous 18 residues of the Nesprin-2 KASH peptide in our energy calculations. The Van der Waals (VdW) and electrostatic (ELEC) interactions between the 17-residues of KASH(Y-7A) and P1 were significantly reduced compared with wild type (Fig 3G). Because the KASH(Y-7A) peptide was partially released from P1, it attempted to form new electrostatic contacts with P2 during the simulation time before it partially detached from P2 as well (hence the two peaks in the density of ELEC energies of KASH(Y-7A) with P2 in Fig 3H). Note that the KASH(Y-7A) peptide did not fully detach from P1 or P2 during our simulation (Fig 3G–H), which supports our in vivo data. We further examined SUN-KASH crystal structures to identify the main interacting partners of SUN2 for the tyrosine at −7 of KASH. The side chain of tyro-sine −7 reaches into the trimer and interacts with a highly conserved motif of SUN2 (E552-S557) in P2 (Fig 3I). An alanine at position −7 is not able to reach this motif. To verify this, we computed the non-bonded interaction energies between the residue at position −7 and the E552-S557 motif in P2. This interaction was completely lost with the KASH(Y-7A) mutation (Fig 3J). Additionally, a hydrogen bond between tyrosine −7 of KASH and the E552-S557 motif was lost in simulations with the Y-7A mutation (Fig 3K). Thus, these simulations support in vivo data that the tyrosine at −7 of KASH is necessary to transfer forces across the nuclear envelope.
The cysteine at −23 of the ANC-1 KASH domain is required for nuclear anchorage
Residues −15 to −23 of KASH peptides (purple in Fig 1) traverse along the surface of P2 before they disengage with SUN proteins and extend toward the outer nuclear membrane. This interaction domain, has been proposed to stabilize LINC complexes and allow for maximal force to be transferred across the nuclear envelope [36]. We hypothesized that mutating the conserved cysteine at −23 of the KASH domain of ANC-1 would disrupt nuclear anchorage in C. elegans.
Previous assays for nuclear anchorage defects were qualitative and tended to overestimate anchorage defects [10, 37]. To quantify nuclear anchorage, we expressed nuclear GFP in adult syncytial hypodermal cells. In wild-type young adults, an average percentage of 2.4 ± 1.3 (mean ± 95% CI) GFP-positive nuclei per lateral side of an animal were clustered (Fig 4A,H). In contrast an anc-1(e1873) null animal had an average of 57.3 ± 6.1 percent (p-value < 0.0001 in a t-test) clustered nuclei per lateral side (Fig 4B,H). Also, ANC-1 nuclei appear smaller and misshapen. Surprisingly, this assay showed that unc-84(n369) null animals have a much less severe nuclear anchorage defect with only 26.5 ± 5.3 percent clustered nuclei per lateral side (p-value < 0.0001 in a t-test compared to wild type or anc-1(e1873)) (Fig 4C,H). This suggests that ANC-1 functions to anchor nuclei through at least two pathways—one through UNC-84 and a second, KASH-independent pathway that will require future investigation.
Figure 4. Mutating conserved cysteines in UNC-84 or ANC-1 disrupts nuclear anchorage.
(A-F) Lateral views of L4 or young adult animals expressing GFP in hypodermal nuclei in the following genotypes: (A) wild type; (B) anc-1(e1873); (C) unc-84(n369); (D) anc-1(CC-23–24AA); (E) unc-84::gfp; (F) unc-84(C953A), and (G) anc-1(F-7A). (H) Quantification of nuclear anchorage defects assayed by counting average number of hypodermal nuclei in clusters of two or more nuclei on one lateral side of an animal. Error bars are 95% CI; *** indicates a p-value<0.0001 in a t-test when compared to wild type; all sample sizes are ≥ 20 animals. Scale bar is 25 μm.
To test the contribution of the cysteine at −23 of the ANC-1 KASH domain to nuclear anchorage, we edited the conserved cysteine at −23 of the ANC-1 KASH domain and an unconserved cysteine at −24 (Fig 1B) to alanines to make anc-1(CC-23,−24AA). The double cysteine mutant had a significant nuclear anchorage defect of 11.3 ± 6.0 percent clustered nuclei (p<0.0001 in a t-test compared to wild type) (Fig 4D, H). Thus, the cysteines near −23 of ANC-1 are necessary for normal anchoring of nuclei.
The cysteine at 953 of the SUN domain of UNC-84 is required for nuclear anchorage, but dispensable for nuclear migration
Although ANC-1 has a full-length KASH domain with the conserved cysteine at −23, UNC-83 lacks the third interaction domain and connects from residue −18 directly to the trans-membrane domain [6]. We therefore hypothesized that an intermolecular disulfide bridge between SUN and KASH would be dispensable for nuclear migration, but needed for nuclear anchorage. Genome editing was used to make unc-84::gfp(C953A). We previously reported that the CRISPR/Cas9-edited strain unc-84::gfp is functional (Figs 5B-C and 3E,H) [38]. In two independent lines, UNC-84::GFP(C953A) localized in a pattern indistinguishable from otherwise wild-type UNC-84::GFP (Fig 5A,C), recruited UNC-83 to the nuclear envelope (Fig 5B,D), and displayed nearly normal levels of nuclear migration (Fig 5E). In contrast, unc-84::gfp(C953A) mutant animals had significant nuclear anchorage defects with 16.2 ± 4.0 percent and 17.2 ± 2.6 percent clustered nuclei per lateral side of an adult (p<0.0001 compared to UNC-84::GFP for both t-tests) (Fig 4F–H). We conclude that the conserved cysteine at 953 of UNC-84 is dispensable for nuclear migration, but required for normal nuclear anchorage.
Figure 5. The conserved cysteine in UNC-84 is not required for nuclear migration.
(A, C) Anti-GFP and (B, D) anti-UNC-83 immunolocalization in coma-stage embryos. Anterior is left, dorsal is up. Scale bar is 10μm. (E) Quantification of nuclear migration defects as in Fig 2. Error bars are 95% CI; scale bar is 10 μm.
Cysteines 577 of mouse SUN2 and −23 of Nesprin-2 are required for rearward nuclear positioning in migrating NIH3T3 fibroblasts
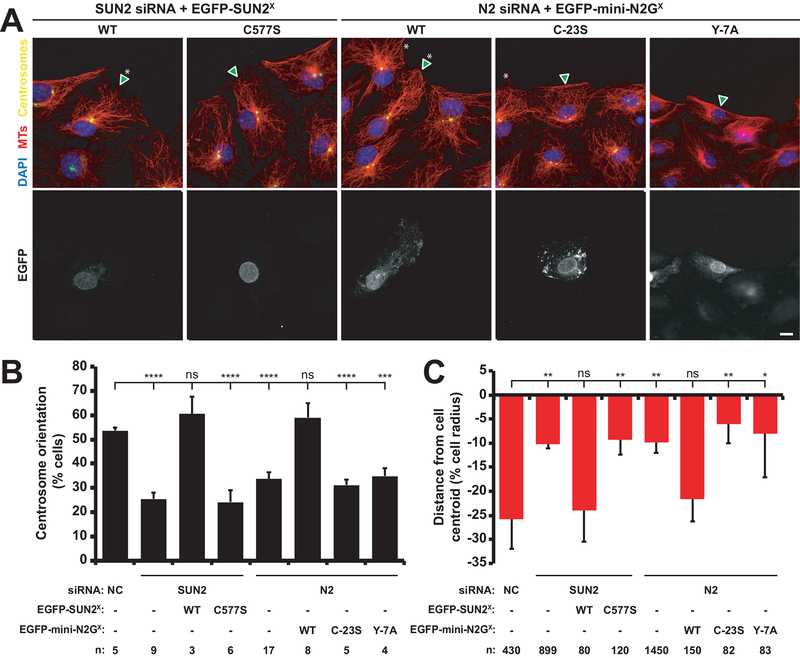
Our results in C. elegans are consistent with a role for a covalent link between UNC-84 and ANC-1 in nuclear anchoring. We next tested whether a similar role could be observed during mammalian nuclear positioning. Actin-dependent rearward nuclear positioning in wounded monolayers of fibroblasts polarizing for migration is an established assay for LINC complex-dependent nuclear positioning [23, 39, 40]. Following serum-starvation, monolayers of NIH3T3 fibroblasts are wounded and stimulated with lysophosphatidic acid (LPA), which induces the movement of nuclei away from the wound edge [24]. This process is mediated by the retrograde flow of perinuclear actin cables, which are coupled to the nuclear envelope via linear arrays of LINC complexes [23, 41]. As previously shown, knockdown of either SUN2 or Nesprin-2 blocked rear-ward nuclear movements and centrosome orientation (Fig 6) [23]. The SUN2 and Nesprin-2 siRNA defects were efficiently rescued by transient expression of EGFPSUN2WT or EGFP-mini-Nesprin-2GWT, respectively (Fig 6) [23].
Figure 6. SUN2 C577 and Nesprin-2G C-23 are required for rearward nuclear positioning during centrosome orientation.
(A) Representative epifluorescence images of centrosome orientation in NIH3T3 fibroblasts treated with siRNA and expressing the indicated cDNA constructs (arrowhead and bottom row of images). Asterisk: oriented centrosome. Scale bar: 10 μm. (B) Centrosome orientation in the cells described in A. (C) Average nuclear positions from the cells described in B. The cell center is defined as “0”. Positive values are toward the leading edge; negative values, away. Two-tailed t-tests were used to calculate p values (*p < 0.05, **p < 0.01; ***p < 0.001; ****p < 0.0001). The error bars are 95% confidence intervals.
The siRNA rescue assays were used to test the importance of the predicted disulfide bond between SUN2 and Nesprin-2G. Mutating either conserved cysteine, in EGFP-SUN2C577S or EGFP-mini-Nesprin-2GC−23S, blocked the rescue of centrosome orientation (Fig 6B) and the rearward nuclear positioning defects (Fig 6C) caused by the depletion of SUN2 or Nesprin-2G, respectively. These results suggest that the ability of SUN2 to form an intermolecular disulfide bond with Nesprin-2G is critical for actin-mediated rearward nuclear positioning in migrating fibroblasts.
The role of the conserved aromatic residues at −7 of KASH domains in the presence of the disulfide bond
We next tested the extent to which conserved aromatic residues at −7 of KASH domains (Fig 1B) were required in the context of a longer KASH domain predicted to form disulfide bonds with their SUN partners. Mutation of the phenylalanine at −7 in the ANC-1 KASH domain did not cause a nuclear anchorage defect in adult C. elegans hypodermal cells; an average of 4.3 ± 1.2 (mean ± 95% CI) percent of hypodermal nuclei per lateral side of an animal were clustered (Fig 4G,H), which is not significantly different from the wild type number of 2.4 ± 1.3 percent (p-value = 0.058 in a t-test compared to wild type). Thus, the presence of an intermolecular disulfide bond could at least partially compensate for a mutation in the KASH −7 aromatic residue in the UNC-84-ANC-1 interaction. In contrast, expression of an EGFP-mini-nesprin-2G construct harboring a mutation of the tyrosine at −7 in the KASH domain failed to rescue reward nuclear positioning in nesprin-2-depleted NIH3T3 fibroblasts during centrosome orientation (Fig 6). These results suggest that the conserved aromatic residues at the −7 position of KASH domains may be necessary for the transmission of forces generated by the actin cyto-skeleton to move nuclei whereas they are dispensable for resisting the forces experienced by nuclei anchored to the actin cytoskeleton in moving C. elegans animals. In agreement with this, our molecular dynamics results showed that despite the detachment of resides 0 to −17 in the KASH(Y-7A) mutation, the remaining residues (−18 to −23) remained intact due to the presence of the intermolecular disulfide bond (Fig 3).
Modeling force transfer across LINC complexes by molecular dynamics
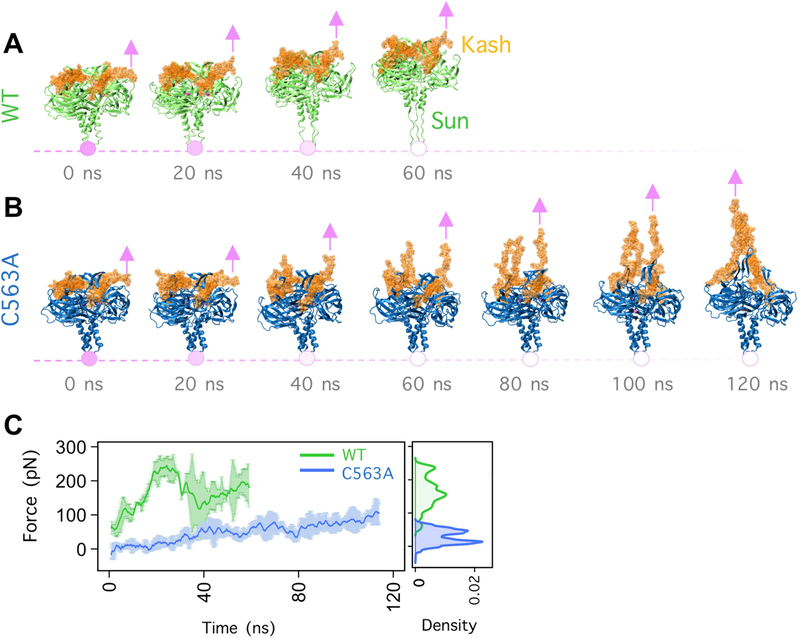
Our working model predicts that mechanical forces generated in the cytoskeleton are transferred across the nuclear envelope through LINC complexes and that disulfide bonds between SUN and KASH domains are important for the transfer of forces. We therefore modeled how SUN-KASH interactions would be affected with and without the disulfide bond. We used molecular dynamics to simulate constant velocity pulling on the amino-terminal residue of the KASH peptide of human Nesprin-2 attached to SUN2 at a constant velocity of 0.05 m/s in wild type (Fig 7A) and the SUN2 (C563A) mutation (Fig 7B). Each simulation was run three times. In a previous study we applied tension on KASH domains by exerting a constant force on each KASH peptide [36]. As opposed to constant force simulations, in this study constant-velocity pulling was used and the changes in the magnitude of forces that the complex was able to withstand were extracted. In the SUN2 (C563A) mutation model, the KASH peptide begins to detach from the SUN protomer under virtually zero forces and both KASH peptides and the KASH lids of SUN domains are stretched to accommodate the pulling (Fig 7B–C). In contrast, the wild-type KASH peptide remains closely associated with the SUN trimer throughout the simulation, withstanding significantly higher forces, up to 250 pN (N=29,700, p-value < 0.0001 obtained from a two-sample Kolmogorov-Smirnov test) (Figure 7C). Instead, the pulling forces are transferred past the SUN-KASH interaction and SUN domain and are relieved by stretching the coiled-coil domains (formed by α3) of SUN2 (Fig 7A,C,D).
Figure 7. Modeling force transfer across LINC complexes.
Snapshots of the trajectory of (A) WT and (B) C563A, SUN2-KASH2 complex under constant velocity pulling. Pink arrows represent the location and direction of the applied constant velocity. Pink circles represent the fixed atoms. (C) Forces on each KASH peptide required to displace the SUN-KASH complex at a constant velocity for WT (green) and C563A (blue) averaged over three molecular dynamics simulation runs. Error bars are the range. On the right, a density plot shows the forces on each KASH peptide at each time point throughout the three simulations.
Discussion
Interactions between conserved SUN and KASH domains form the core of the nuclear envelope-spanning LINC complex [3–5, 14, 42]. Here, we performed structure/functional analyses of the three interaction interfaces between SUN and KASH proteins. The C-terminal 4 residues of KASH proteins (0 to −4) interact with a binding pocket in SUN protomer P1, residues −4 to −14 interact with a cleft that is formed between the “KASH lid” of P1 and the core of the adjacent P2 protomer, and residues −15 to −23 interact with the surface of P2 [16]. We found that mutating any of the three interfaces disrupted the formation of LINC complexes and/or reduced the transfer of forces generated in the cytoplasm to the nucleoskeleton. Our results uncovered potential regulatory mechanisms for the formation and function of LINC complexes.
Mutational analysis of the two interfaces at the C terminus of KASH domains (consisting of KASH residues 0 to −14) showed they were necessary in vivo to move C. elegans hypodermal nuclei. These results are supported by the previous findings that the C-terminal 18 residues of the Nesprin-2 KASH domain are sufficient for binding SUN2 in vitro [16] and that the extension of the C terminus of KASH prevents its ability to interact with SUN2 in vitro [16, 33].
We also tested the extent to which the conserved aromatic residue at position −7 of KASH proteins functions in LINC assembly in our various experimental models and found significant differences. In C. elegans hyp7 precursors, UNC-83(Y-7A) expressed at endogenous levels localized to the nuclear envelope, but nuclear migration failed (Fig 3A–E). These results suggest UNC-83(Y-7A) can weakly interact with UNC-84 at a level sufficient for localization to the nuclear envelope, but the interaction is unable to sustain the forces transferred across the LINC complex during nuclear migration. Our molecular dynamics simulations support this model. Without the tyrosine at position −7, KASH was unable to reach into the cleft between two SUN protomers, resulting in the loss of several important interactions between the KASH tyrosine and a conserved motif on SUN (E552-S557 of SUN2; Fig 3). Likewise, the −7 tyrosine in the mouse nesprin-2G KASH domain was required for rearward nuclear migration and centrosome orientation in NIH3T3 cells (Fig 6). In contrast, hypodermal syncytial nuclei were anchored normally in anc-1(F-7A) mutant animals. Perhaps the cysteine at −23 of KASH is sufficient to overcome the ANC-1(F-7A) mutation for nuclear anchorage, but both the −23 cysteine and the −7 tyrosine are required in NIH3T3 cell rearward nuclear movements. Together, these findings suggest that the forces transmitted by LINC complexes required to move nuclei in polarizing fibroblasts may be greater than those required to anchor C. elegans hypodermal nuclei.
The third KASH-SUN interaction interface (−15 to −23) may solidify KASH-SUN complexes by forming an intermolecular disulfide bond between highly conserved cysteine residues in certain SUN and KASH pairs. Such a disulfide bond was observed between transiently expressed SUN and KASH proteins in HeLa cells [16]. We propose a model where the regulation of this third interface might enable a SUN protein homo-trimer to switch between different KASH proteins. In C. elegans, the SUN protein UNC-84 would first interact with the KASH protein UNC-83, which lacks the conserved cysteine, to move hypodermal nuclei. Then, UNC-84 would primarily interact with ANC-1 to anchor hypodermal nuclei. Since nuclear anchorage occurs over longer timescales, it might be advantageous to stabilize the UNC-84-ANC-1 interaction with a disulfide bond. The conserved cysteines in SUN2 and nesprin-2 were also necessary for actin-dependent rearward nuclear positioning during centrosome orientation in NIH3T3 fibro-blasts. In this manner, the rearward nuclear positioning in fibroblasts may be analogous to nuclear anchorage in the C. elegans hypodermis, as they both processes use giant KASH proteins to tether nuclei to actin networks.
In further support of our proposed model for the role of an intermolecular disulfide bond in LINC complex function, molecular dynamic simulations suggested that the disulfide bond allows for maximal force transduction across the nuclear envelope. In the absence of a disulfide bond, constant pulling rapidly stretched KASH domains and the KASH lid of SUN domains. Only ~50 pN of force was transferred across the interface without the disulfide bond. In contrast, the presence of SUN-KASH intermolecular disulfide bonds transferred the forces on the KASH peptides directly to the coiled-coil trimerization domain in the luminal parts of SUN proteins while maintaining the stability of the SUN-KASH interface in our simulations. In this case, forces could be transferred across the LINC complex and ultimately to the nucleoskeleton and/or chromatin. These findings suggest that higher forces and/or a more stable interaction may be required to anchor nuclei to actin networks more permanently, versus the interactions to microtubules during C. elegans nuclear migration that are only required for brief times. It is likely that other nuclear movements or other roles of LINC complexes in human development and disease will have a similar mode of regulation.
If the cell regulates the formation and reduction of the predicted SUN-KASH intermolecular disulfide bond, an interesting question is how it might do so. There are three protein disulfide isomerases in C. elegans. However, these three genes are understudied and their expression patterns could be developmentally regulated. A second possibility is that pulling across a disulfide bond could expose it to reduction [43]. Third, it has been proposed that the AAA+ ATPase Torsin could regulate the formation of LINC complexes [39, 44–46]. Future experiments could distinguish between these and other possible regulatory mechanisms.
The KASH domains in C. elegans UNC-83 and mammalian KASH5 are well conserved, but significantly shorter than those in other KASH proteins. Nevertheless, both bind canonical SUN proteins UNC-84 in C. elegans, or SUN1 and SUN2 in mammals [6, 7, 14, 28]. UNC-83 and KASH5 are transiently required during development, while ANC-1 and the other Nesprins tend to play longer-term roles. For example, UNC-83 is required for a few specific nuclear migration events that take ~10–30 minutes during C. elegans development [26, 27]. After migration, nuclei remain anchored in place for days by way of the interaction of ANC-1 with the actin cytoskeleton [13]. Similarly, KASH5 only functions a short time during meiotic prophase I [28], while other Nesprins function just before and after this narrow window [4]. These findings suggest that evolution has maintained the disulfide bond in C. elegans and mammalian LINC complexes that function long term and/or require maximal force transduction, but selected against the disulfide bond in LINC complexes that function transiently. The disulfide bond and third interaction domain may have been lost in KASH5 and UNC-83 after the divergence of nematodes, insects and vertebrates, as both canonical KASH proteins in Drosophila, Klarsicht and MSP-300, have the conserved cysteine [5].
In summary, we have demonstrated here that all three interaction interfaces between conserved SUN and KASH proteins contribute to LINC complex function in nuclear positioning. Our findings from C. elegans development, mammalian cultured fibro-blasts, and molecular dynamic simulations, have led to a more complete mechanistic understanding of SUN-KASH interactions, which form the core of LINC complexes. Since LINC complexes are central to a multitude of cell and developmental functions and have been implicated in multiple pathologies [4, 5, 14, 47, 48], our mechanistic findings are likely to be important for understanding a variety of cell and developmental processes.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daniel A. Starr (dastarr@ucdavis.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Caenorhabditis elegans were cultured on nematode growth medium plates spotted with OP50 bacteria and maintained at 15°C or room temperature (approximately 22°C). N2 was used as the wild-type control strain [49]. Some strains were provided by the Caenorhabditis Genetics Center, funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). Only healthy animals that were not used in previous procedures and were naïve to testing were used. See the Key Resources Table for strain list.
Low-passage NIH3T3 fibroblasts (authenticated by ATCC and of unknown sex) were cultured in DMEM with 10% bovine calf serum (Thermo Fischer). For polarization assays, cells were serum starved for 2 days, wounded and stimulated with 10 μM LPA (Avanti Polar Lipids, Inc.) [24, 50]. Cells were grown at 37°C in 5% CO2.
METHOD DETAILS
C. elegans CRISPR/Cas9 editing
Knock-in strains were constructed using CRISPR/Cas9-mediated genome editing [35, 51–53]. Some Cas9 targeting sequences were cloned into pDD162 (peft-3::Cas9 + Empty sgRNA, a gift from Bob Goldstein, Addgene plasmid # 47549) [35], using Q5 site-directed mutagenesis (New England Biolabs). For others, guideRNAs were synthesized and pre-complexed with purified Cas9 protein. Targeting sequences for each strain are listed in Table S1.
unc-83::gfp::kash(+A) was generated using unc-19(ed3) rescue [34, 35]. An ala-nine codon was inserted before the stop into pSL718 [34] using Q5 site-directed muta-genesis to create the homologous repair template pSL724, which was used with Cas9-sgRNA construct pSL715 and injection markers as reported [35] to make UD478: unc-19(ed3)I, yc28[unc-83::gfp::kash+A, LoxP::unc-19(+)::LoxP]V.
The unc-84::gfp(C953A) mutation was introduced into unc-84::gfp (UD476) by CRISPR co-conversion with single stranded oligodeoxyribonucleotide repair templates containing unc-84(C953A) and dpy-10(cn64) [52, 54], using Cas9-sgRNA constructs pSL771 and AP568–2 (gift from Geraldine Seydoux, Addgene plasmid #70047) [54]. Two independent integrated lines were isolated and analyzed UD497 and UC498:yc35[unc-84(C953A)::gfp + LoxP]X.
The anc-1(yc48), anc-1(yc56), and unc-83(yc50) mutations were made by directly injecting RNA/Cas9 complexes into C. elegans gonads [52, 53, 55]. To make anc-1(yc48), 17.5 μM of pre-complexed Alt-R CRISPR anc-1 crRNA #1 (synthesized by Integrated DNA technologies, see Table S1), tracrRNA (Integrated DNA Technologies), and Cas9 purified protein (QB3 Berkeley) were injected with 6 μM ssDNA repair templates (Integrated DNA Technologies, see Table S1). anc-1(yc48) mutates cysteine residues at −23 and −24 of the KASH domain to alanines to make ANC-1(CC-23–24AA) in strain UD558. To make unc-83(yc50) and anc-1(yc56), 4 μg crRNA against unc-83 or crRNA #2 against anc-1, 1.25 μg crRNA against dpy-10, 10 μg tracrRNA (Dharmacon), 25 μg of Cas9 protein, 1.1 μg ssDNA repair template (See Table S1), and 0.15 μg ssDNA repair for dpy-10 in a total of 10 μL was injected into young adult gonads [53]. unc-83(yc50) mutates the conserved tyrosine at residue 967 of UNC-83 to make UNC-83(Y-7A) in strain UD568. anc-1(yc56) mutates the conserved phenylalanine at position −7 from the C terminus of ANC-1 to make ANC-1(F-7A).
A fluorescent marker to identify hypodermal nuclei in the adult animal was constructed by insertion of a 1 kb fragment upstream of the col-19 gene [56] into pPD96.04 (gift from Andrew Fire, Addgene plasmid #1502) to create pcol-19::gfp::lacZ (pSL779). N2 animals were injected with pSL779 at 40 ng/μl, pBluescript SK+ at 50ng/μl, and pCFJ90 at 2 ng/μl to make strain UD522: ycEx249[pcol-19::gfp::lacZ, pmyo-2::mCherry]. Transgenic animals were identified by red fluorescent pharynx in larval stages. Green hypodermal nuclei were observed in transgenic animals beginning at the L4 molt. The marker was introduced into other strains by crossing to UD522 transgenic males.
Nuclear migration and anchorage assays in C. elegans
Defects in hyp7 nuclear migration were quantified by counting the number of hyp7 nuclei found abnormally, using Nomarski optics, in the dorsal cords of L1 or L2 hermaphrodites. We scored all the hyp7 nuclei abnormally in the dorsal cord between the second bulb of the pharynx and the anus [11, 27]. Hyp7 nuclear clustering was assayed in adult animals expressing pcol-19::gfp::lacZ. Young adult hermaphrodites were mounted on 2% agarose pads with 10 mM tetramisole in M9 buffer. Nuclei were counted as clustered if within 10% nuclear diameter proximity to another nucleus in the same focal plane along the longitudinal axis of the worm, as determined by DIC microscopy. Contacts between nuclei on the perpendicular axis were not counted, as the marker could not distinguish seam cell nuclei in proximity to hyp7 nuclei from clusters of hyp7 nuclei. Only nuclei situated between the pharynx and the anus were counted, as nuclei near the mouth and at the very end of the tail were observed to cluster in wild type animals. Only one lateral side of each animal was scored.
For both the nuclear migration and anchorage assays, C. elegans animals of the appropriate age were randomly selected at the dissecting microscope for scoring using the compound microscope and all scorable animals on the slide were counted. Animals were only scored if the entire animal was easily viewable. Although the genotypes were not blinded, animals from multiple plates were included in the data sets. In one case, the unc-84::gfp(C953A) mutant, two independently generated lines were assayed.
NIH3T3 fibroblast manipulations
siRNA experiments were performed using 50nM siRNA duplexes (Shanghai GenePharma), which were transfected using Lipofectamine RNAiMAX (Thermo Fisher). The noncoding, Nesprin-2G, and SUN2 siRNA sequences were previously validated and listed in Table S1 [23]. EGFP-SUN2C577S was made by in vitro mutagenesis of EGFP-SUN2WT [23]. EGFP-mini-Nesprin-2GC−23S was similarly generated from EGFP-mini-Nesprin-2WT [23]. The primers used to generate these constructs are in Table S1. Purified plasmid DNA was microinjected into NIH 3T3 cell nuclei at concentrations between 5 and 30 μg/ml [39]. Injections were performed on a TS100 microscope (Nikon) equipped with a chrome-free infinity Apochromat long working distance apodized dark low 20Å~/NA 0.4 objective (3-mm working distance) and a Narishige NT-88 ß [27, 57].
Immunofluorescence
For C. elegans experiments: Anti-UNC-84 mouse monoclonal antibody L72/6 [57] was used at 1:100 dilution. Mouse monoclonal anti-UNC-83 antibody 1209D7 [27] was used undiluted. Rabbit polyclonal anti-GFP antibody NB600–308 (Novus Biologicals) was used at 1:500 dilution. AlexaFluor secondary antibodies 488 donkey anti-rabbit, 555 goat anti-mouse, and 594 donkey anti-mouse (Thermo Fisher) were used at 1:500 dilution. Images were captured with a HCX Plan Apochromat 63, 1.40 NA objective on a compound microscope (DM6000) with LAS X software. Images were uniformly enhanced using the background subtraction and brightness/contrast commands in ImageJ.
For experiments with NIH3T3 cells: NIH3T3 fibroblasts were cultured in DMEM with 10% bovine calf serum (Thermo Fischer Scientific) on number 1.5 coverslips. Fibroblasts were serum starved for 2 d, wounded, and stimulated with 10 μM LPA for 2 hrs before being fixed in in −20° methanol for 10 m in. The fixed cells were then stained as follows. Anti-EGFP mouse monoclonal antibody JL-8 (Clontech) was used at a 1:200 dilution, anti-percentrin rabbit polyclonal antibody PRB-432C (Covance) was used at 1:200 dilution. Anti-tyrosinated α-tubulin rat monoclonal antibody YL1/2 (supernatant collected from hybridomas purchased from the European Collection of Animal Cell Cultures, Salisbury, UK) was used at 1:50 dilution. Goat anti-mouse, -rabbit, and -rat secondary antibodies conjugated to AlexaFluor488, AlexaFluor555, AlexaFluor594, AlexaFluor650 (Thermo Fisher) were used at 1:200 dilution. Stained cells were mounted on slides using Fluoromount (Thermo Fisher).
Images were obtained with a Nikon Eclipse Ni-E microscope driven by NIS-Elements software using a Nikon oil immersion 40Å~/1.30 NA Plan Fluor Eco-Glass oil immersion objective lens (0.20-mm working distance), a Lumencor SOLA solid state white-light excitation subsystem, and a Photometrics CoolSNAP ES2 12-bit 20-MHz digital monochrome charge-coupled device camera. A custom DAPI filter for the SOLA light source was used, which consisted of an ET 395/25Å~ excitation filter and an ET 460/50-m emission filter purchased from Chroma Technology Corp (Bellows Falls, VT). EGFP (C-FL EGFP hard coat high signal to noise Zero Shift; Nikon), Texas Red (C-FL Texas Red hard coat high signal to noise Zero Shift; Nikon), and Cy5 (C-FL CY5 hard coat high signal to noise Zero Shift; Nikon) filter sets were also used.
To quantify centrosome orientation, a fibroblast is divided by drawing a “v” that begins and ends at the left- and right-most wound-edge, respectively. The middle of the “v” is defined by the center of the nucleus. The cell is scored as having an oriented centrosome if the centrosome resides within the “v” between the nucleus and the wound-edge. Since the “v” occupies □1/3 of the cell, □33% of wound-edge cells will have an oriented centrosome. Experiments were repeated at least 3 times.
To quantify nuclear positioning, fluorescence images of the stained cells described above were pseudocolored, combined and aligned such that the wound-edge was parallel to the x-axis using ImageJ. Custom-made MATLAB software (MathWorks, Natick, MA) was then used to calculate the cell centroid and equivalent radius of the wound-edge cells, which were outlined by hand. Segmentation was used to identify the centroid of the nucleus and therefore the position of the organelle. A vector representing the distance from the nuclear centroid to the cell centroid was drawn and resolved into x and y coordinates (parallel and perpendicular to the leading edge, respectively). Measurements were normalized to cell size to allow for comparison between cells. Only the y-coordinate was used in plots as the x-coordinate (position of the nuclear centroid or centrosome along the x-axis) did not change significantly. The difference between the cell centroid and the nucleus centroid or centrosome was then divided by the radius to determine the percentage of the cell radius the nucleus traveled.
Regarding the wounded fibroblast monolayer assay for rearward nuclear positioning during centrosome orientation, at least three independent biological replicates were performed and analyzed for each experiment. To compensate for the lack of blinding, five images of at least eighty cells were collected from three different wounds and all of the scorable wound edge cells were analyzed for the non-injected control experiments. For the injected experiments, all of the scorable EGFP-positive wound edge cells were analyzed. Cells were deemed unscorable if they were not entirely visualized within the field of view of an image.
Molecular Dynamics
All molecular dynamics simulations were performed with NAMD using the CHARMM27 force field [58] and visualized using VMD software [59]. The crystal structure used in these simulations was obtained from the protein data bank (PDB ID: 4DXS). Mutations were introduced using the VMD mutator plugin. All modeled structures were then solvated in water and an ion concentration of 150 mM of KCl and 50 μM of CaCl2 was added to each system. Next, periodic boundary conditions were applied in all three directions and the systems were minimized for 5000 steps and equilibrated for ~3ns at 1 atm and 310 K using Langevin’s piston and Hoover’s method [58], For constant velocity pulling experiments, we attached a dummy atom to the position of the center of mass of the amino-terminal residue of the KASH peptide (C-23) via a virtual spring and fixed the C-terminal residue of SUN2. We then measured the forces between the dummy atom and C-23 using NAMD [58] as the dummy atom was moved at a constant velocity of 0.05 m/s.
QUANTIFICATION AND STATISTICAL ANALYSIS
The C. elegans nuclear migration and anchorage data in Figures 2–6 are displayed as means with 95% CI as error bars; t-tests were performed on indicated comparisons and the sample sizes were at least 20. For Figure 6 pertaining to the NIH3T3 cells, sample sizes are indicated within the figure. Molecular dynamic simulations were performed in triplicate, plots were prepared using R software. The mean and range of data are shown in Figure 7C. A two-sample Kolmogorov-Smirnov test was performed to determine whether the observed difference in average forces obtained from molecular dynamics simulations is significantly lower in C563A versus WT. This test was conducted using the ks.test function in R stats package.
Supplementary Material
Highlights.
Mutating conserved residues at SUN-KASH interfaces disrupt LINC function.
Cysteines in LINC are required to anchor, but not to move C. elegans nuclei.
Cysteines in LINC are needed for nuclear movement in wound-edge fibroblasts.
Simulations show disulfide bridges maximize forces transferred across LINC.
Acknowledgements
We thank Thomas Schwartz, Ulrike Kutay, and members of the Starr, Mofrad, and Luxton laboratories for helpful discussions. This work was supported by the National Institutes of Health (R01GM073874 to D.A.S., R21NS095109 and R42DA037622 to G.W.G.L., and AR007612 to N.J.H.), the National Science Foundation (CAREER award CBET-0955291 to M.R.K.M.), the Natural Sciences and Engineering Research Council of Canada (to Z.J.), the Dystonia Medical Research Foundation (G.W.G.L.), and the American Cancer Society Illinois Division (PF-13–094-01-CGC to N.E.C.).
Abbreviations
- SUN
Sad1 UNC-84
- KASH
Klarsicht, ANC-1, and Syne homology
- LINC
linker of the nucleoskeleton and cytoskeleton
- P1
SUN protomer 1
- P2
SUN protomer 2
- VdW
Van der Waals
- ELEC
electrostatic
- LPA
lysophosphatidic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
N.E.C. is currently employed as a scientific editor at Cell Reports, which is a part of Cell Press.
References
- 1.Bone CR, and Starr DA (2016). Nuclear migration events throughout development. J. Cell. Sci 129, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gundersen GG, and Worman HJ (2013). Nuclear Positioning. Cell 152, 1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burke B, and Roux KJ (2009). Nuclei take a position: managing nuclear location. Developmental Cell 17, 587–597. [DOI] [PubMed] [Google Scholar]
- 4.Ungricht R, and Kutay U (2017). Mechanisms and functions of nuclear envelope remodelling. Nature Reviews Molecular Cell Biology 18, 229–245. [DOI] [PubMed] [Google Scholar]
- 5.Starr DA, and Fridolfsson HN (2010). Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol 26, 421–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGee MD, Rillo R, Anderson AS, and Starr DA (2006). UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol. Biol. Cell 17, 1790–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, and Hodzic D (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol 172, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, and Shackleton S (2006). SUN1 interacts with nuclear lam-in A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Molecular and cellular biology 26, 3738–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, and Karakesisoglou I (2005). The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell. Sci 118, 3419–3430. [DOI] [PubMed] [Google Scholar]
- 10.Malone CJ, Fixsen WD, Horvitz HR, and Han M (1999). UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development 126, 3171–3181. [DOI] [PubMed] [Google Scholar]
- 11.Bone CR, Tapley EC, Gorjanacz M, and Starr DA (2014). The Caenorhabditis elegans SUN protein UNC-84 interacts with lamin to transfer forces from the cytoplasm to the nucleoskeleton during nuclear migration. Mol. Biol. Cell 25, 2853–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothballer A, and Kutay U (2013). The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma 122, 415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Starr DA, and Han M (2002). Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science 298, 406–409. [DOI] [PubMed] [Google Scholar]
- 14.Luxton GWG, and Starr DA (2014). KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Current opinion in cell biology 28, 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janota CS, Calero-Cuenca FJ, Costa J, and Gomes ER (2017). Snap-Shot: Nucleo-cytoskeletal Interactions. Cell 169, 970–970. e1. [DOI] [PubMed] [Google Scholar]
- 16.Sosa BA, Rothballer A, Kutay U, and Schwartz TU (2012). LINC Complexes Form by Binding of Three KASH Peptides to Domain Interfaces of Trimeric SUN Proteins. Cell 149, 1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Shi Z, Jiao S, Chen C, Wang H, Liu G, Wang Q, Zhao Y, Greene MI, and Zhou Z (2012). Structural insights into SUN-KASH complexes across the nuclear envelope. Cell Res 22, 1440–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hennen J, Hur K-H, Saunders CA, Luxton GWG, and Mueller JD (2017). Quantitative Brightness Analysis of Protein Oligomerization in the Nuclear Envelope. Biophysical Journal 113, 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hennen J, Saunders CA, Mueller JD, and Luxton GWG (2018). Fluorescence fluctuation spectroscopy reveals differential SUN protein oligomerization in living cells. Mol. Biol. Cell 29, 1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sosa BA, Kutay U, and Schwartz TU (2013). Structural insights into LINC complexes. Curr. Opin. Struct. Biol 23, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou K, and Hanna-Rose W (2010). Movers and shakers or anchored: Caenorhabditis elegans nuclei achieve it with KASH/SUN. Developmental Dynamics 239, 1352–1364. [DOI] [PubMed] [Google Scholar]
- 22.Starr DA, and Han M (2005). A genetic approach to study the role of nuclear envelope components in nuclear positioning. Novartis Found. Symp 264, 208–19; discussion 219–230. [PubMed] [Google Scholar]
- 23.Luxton GWG, Gomes ER, Folker ES, Vintinner E, and Gundersen GG (2010). Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 329, 956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomes ER, Jani S, and Gundersen GG (2005). Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell 121, 451–463. [DOI] [PubMed] [Google Scholar]
- 25.Chang W, Antoku S, and Gundersen GG (2016). Wound-Healing Assays to Study Mechanisms of Nuclear Movement in Fibroblasts and Myoblasts In Electron Microscopy Methods in Molecular Biology (New York, NY: Springer New York; ), pp. 255–267. [DOI] [PubMed] [Google Scholar]
- 26.Fridolfsson HN, and Starr DA (2010). Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J. Cell Biol 191, 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, and Han M (2001). unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development 128, 5039–5050. [DOI] [PubMed] [Google Scholar]
- 28.Horn HF, Kim DI, Wright GD, Wong ESM, Stewart CL, Burke B, and Roux KJ (2013). A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J. Cell Biol 202, 1023–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, and Burke B (2009). Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. U.S.A 106, 2194–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhen Y-Y, Libotte T, Munck M, Noegel AA, and Korenbaum E (2002). NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell. Sci 115, 3207–3222. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Q, Skepper JN, Yang F, Davies JD, Hegyi L, Roberts RG, Weissberg PL, Ellis JA, and Shanahan CM (2001). Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J. Cell. Sci 114, 4485–4498. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Xu R, Zhu B, Yang X, Ding X, Duan S, Xu T, Zhuang Y, and Han M (2007). Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development 134, 901–908. [DOI] [PubMed] [Google Scholar]
- 33.Stewart-Hutchinson PJ, Hale CM, Wirtz D, and Hodzic D (2008). Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp. Cell Res 314, 1892–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bone CR, Chang Y-T, Cain NE, Murphy SP, and Starr DA (2016). Nuclei migrate through constricted spaces using microtubule motors and actin networks in C. elegans hypodermal cells. Development 143, 4193–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickinson DJ, Ward JD, Reiner DJ, and Goldstein B (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nature Publishing Group 10, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jahed Z, Shams H, and Mofrad MRK (2015). A Disulfide Bond Is Required for the Transmission of Forces through SUN-KASH Complexes. Biophysical Journal 109, 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Alessandro M, Hnia K, Gache V, Koch C, Gavriilidis C, Rodriguez D, Nicot A-S, Romero NB, Schwab Y, Gomes E, et al. (2015). Amphiphysin 2 Orchestrates Nucleus Positioning and Shape by Linking the Nuclear Envelope to the Actin and Microtubule Cytoskeleton. Developmental Cell 35, 186–198. [DOI] [PubMed] [Google Scholar]
- 38.Lawrence KS, Tapley EC, Cruz VE, Li Q, Aung K, Hart KC, Schwartz TU, Starr DA, and Engebrecht J (2016). LINC complexes promote homologous recombination in part through inhibition of nonhomologous end joining. J. Cell Biol 215, 801–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saunders CA, Harris NJ, Willey PT, Woolums BM, Wang Y, McQuown AJ, Schoenhofen A, Worman HJ, Dauer WT, Gundersen GG, et al. (2017). TorsinA controls TAN line assembly and the retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement. J. Cell Biol 216, 657–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Folker ES, Östlund C, Luxton GWG, Worman HJ, and Gundersen GG (2011). Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc. Natl. Acad. Sci. U.S.A 108, 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luxton GWG, Gomes ER, Folker ES, Worman HJ, and Gundersen GG (2011). TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus 2, 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Razafsky D, and Hodzic D (2009). Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J. Cell Biol 186, 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiita AP, Ainavarapu SRK, Huang HH, and Fernandez JM (2006). Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. PNAS 103, 7222–7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Starr DA, and Rose LS (2017). TorsinA regulates the LINC to moving nuclei. J. Cell Biol 216, 543–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerace L (2004). TorsinA and torsion dystonia: Unraveling the architecture of the nuclear envelope. Proc. Natl. Acad. Sci. U.S.A 101, 8839–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sosa BA, Demircioglu FE, Chen JZ, Ingram J, Ploegh HL, and Schwartz TU (2014). How lamina-associated polypeptide 1 (LAP1) activates Torsin. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calvi A, and Burke B (2015). LINC Complexes and Their Role in Human Disease (Chichester, UK: John Wiley & Sons, Ltd; ). [Google Scholar]
- 48.Cartwright S, and Karakesisoglou I (2014). Nesprins in health and disease. Semin. Cell Dev. Biol 29, 169–179. [DOI] [PubMed] [Google Scholar]
- 49.Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palazzo AF, Joseph HL, Chen YJ, Dujardin DL, Alberts AS, Pfister KK, Vallee RB, and Gundersen GG (2001). Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr. Biol 11, 1536–1541. [DOI] [PubMed] [Google Scholar]
- 51.Dickinson DJ, Pani AM, Heppert JK, Higgins CD, and Goldstein B (2015). Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics 200, 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arribere JA, Bell RT, Fu BXH, Artiles KL, Hartman PS, and Fire AZ (2014). Efficient Marker-Free Recovery of Custom Genetic Modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paix A, Schmidt H, and Seydoux G (2016). Cas9-assisted recombineering in C. elegans: genome editing using in vivo assembly of linear DNAs. Nucleic Acids Res 44, e128–e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paix A, Folkmann A, Rasoloson D, and Seydoux G (2015). High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics 201, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richardson CD, Ray GJ, DeWitt MA, Curie GL, and Corn JE (2016). Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nature Biotechnology 34, 339–344. [DOI] [PubMed] [Google Scholar]
- 56.Liu Z, Kirch S, and Ambros V (1995). The Caenorhabditis elegans hetero-chronic gene pathway controls stage-specific transcription of collagen genes. Development 121, 2471–2478. [DOI] [PubMed] [Google Scholar]
- 57.Cain NE, Tapley EC, McDonald KL, Cain BM, and Starr DA (2014). The SUN protein UNC-84 is required only in force-bearing cells to maintain nuclear envelope architecture. J. Cell Biol 206, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, and Schulten K (2005). Scalable molecular dynamics with NAMD. Journal of Computational Chemistry 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Humphrey W, Dalke A, and Schulten K (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics 14, 33–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.