Abstract
Because of poor differentiation among the members of genus Comamonas using phenotypic methods, human infections caused by C. kerstersii are sporadically reported in the literature. Here, we represent the first complete genome sequence of C. kerstersii 8943, which caused peritonitis in a patient with continuous ambulatory peritoneal dialysis (CAPD). The complete genome with no gaps was obtained using third-generation Pacific Biosciences (PacBio) RSII sequencing system with single-molecule real-time (SMRT) analysis. Protein-coding genes, rRNAs and tRNAs were predicted. Functional annotations of the genome using different databases revealed several genes related to pathogenicity including antibiotic resistance genes and prophages. Our work demonstrates that whole genome sequencing can enhance the resolution of clinical investigations and our data can be used as a reference genome during the rapid diagnosis of C. kerstersii infections in the future.
Subject terms: DNA sequencing, Prokaryote, Genome, Bacterial genomics
Background & Summary
Comamonas kerstersii, first described in 2003, has been recognized as non-pathogenic1. Human infections caused by C. kerstersii were unusual and have been reported only recently. Currently, only five reports of C. kerstersii-related infections are available. In 2013, Almuzara et al. reported four cases of C. kerstersii-induced intra-abdominal infection, which represented the first report of human C. kerstersii infections2. Shortly after the first report, cases of C. kerstersii related bacteraemia and abdominal infections were documented3,4. Recently, C. kerstersii were reported to be involved in psoas abscess, pelvic peritonitis and acute perforated appendicitis5,6. The infections caused by C. kerstersii may be underestimated in previous literatures as the phenotypic methods used for the identification of bacteria cannot provide effective measurement for distinguishing between C. kerstersi and other Comamonas species2.
A strain (J29) of C. kerstersii was isolated from the dialysis effluent of a patient with continuous ambulatory peritoneal dialysis (CAPD)-peritonitis using sheep blood agar plates. No growth was detected when cultivated anaerobically, indicating that it is a strict aerobe. This strain was initially identified using VITEK 2 system (bioMerieux, France) applying GN ID card. However, the result showed an unidentified organism. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) (Bruker Daltonics, Germany) identified this strain as C. kerstersii with a log(score) value of 2.258. To further confirm the bacterial identity, we applied whole genome sequencing, which is a useful tool that enables precise detection of fastidious organisms. Initially, we used Illumina HiSeq platform and ABySS software. However, we only got a draft genome with 148 contigs (ASM129444v1). Because of the importance of and our interest in the study of this strain, we intended to get its complete genome sequence. In the present study, this strain (J29), also termed as 8943, was sequenced using the third-generation Pacific Biosciences (PacBio) RSII sequencing system and a single-molecule real-time (SMRT) analysis. Finally, we obtained the complete genome sequence of this strain. Through RNAmmer, we got the 16S rRNA sequence of C. kerstersii 8943 and phylogenetic analysis clearly indicated that this strain shared 100% similarity with C. kerstersii LMG 3475T (AJ430347) with regard to 16S rRNA sequence. Our study represented the first complete genome sequence of C. kerstersii. Our data reported here will provide the genome reference for diagnosing the presence of C. kerstersii in infectious diseases in future, and will be used in the comparative genomic analysis within Comamonas genus to elucidate the pathogeny of this bacterium.
Methods
DNA extraction
C. kerstersii 8943 was cultivated in tryptic soy broth, which contains 17.0 g/L tryptone (pancreatic digest of casein), 3.0 g/L soytone (peptic digest of soybean), 2.5 g/L glucose, 5.0 g/L sodium chloride and 2.5 g/L dipotassium phosphate. After shaking at 37 °C for 24 h, bacterial cells were harvested by centrifugation at 5,000 rpm for 10 min. Genomic DNA was extracted using QIAamp DNA Mini Kit (Qiagen, Germany) according to manufacturer’s instructions. The quality and integrity of genomic DNA was assessed using 1% agarose gel electrophoresis and densitometry compared to the appropriate size standards. Meanwhile, DNA yield and purity were measured using NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, USA) and Qubit®2.0 fluorometer (Thermo Fisher Scientific, USA).
Whole genome sequencing
Qualified genomic DNA was sheared using a Covaris® g-TUBE® shearing device (Covaris, USA) (>10 kb insert sizes). After shearing, the approximate sizes of the DNA were determined using Agilent® 2100 Bioanalyzer (Agilent Technologies, USA). The fragmented DNA was then purified using 0.45×AMPure® PB beads (Pacific Biosciences, USA). The ends of the fragmented DNA were repaired using the PacBio Template Prep Kit (Pacific Biosciences, USA) before ligating to the hairpins (SMRTbell™ templates). The resulting SMRTbell library was quantitated via Qubit. Before sequencing, sequencing primers were annealed to both ends of the SMRTbell template and DNA sequencing polymerases were bound to the templates to form the template-polymerase complex. Single-molecule real-time (SMRT®) sequencing was performed using a Pacific Biosciences RSII sequencer (PacBio, Menlo Park, CA) according to the manufacturer’s instructuions (MagBead Standard Seq v2 loading, 1×180 min movie) using P4-C2 chemistry.
Genome assembling and annotation
Hierarchical Genome Assembly Process (HGAP) pipeline was used to a generate high quality de novo assembly of the genome with default parameters7. Shorter reads were aligned against the longest reads to correct random errors and generate the pre-assembled reads that were both long and of high accuracy. The quality of the assembled genome was assessed using CheckM v1.0.98. The circulation of the assembled genome was verified by aligning the complete genome with the draft genome of C. kerstersii 8943 (ASM129444v1). Open reading frames (ORFs) were predicted using Glimmer v3.02. rRNAs and tRNAs were predicted using RNAmmer9 and tRNAscan-SE10, respectively. The phylogenetic tree was constructed according to the neighbor-joining method using Molecular Evolutionary Genetics Analysis (MEGA) 7.0 software11. Antibiotic resistance genes were annotated using BLAST-2.7.1+ program12 against the Antibiotic Resistance Gene-ANNOTation (ARG-ANNOT) database13 with an e-value cut-off of 1e-5 and identity cut-off of 90%. Prophage regions were identified by PHAge Search Tool Enhanced Release (PHASTER)14. The circular map of the genome was generated using DNAPlotter software15 and multiple genome alignment was performed using BLAST Ring Image Generator (BRIG)16. The genomic average nucleotide identity (ANI) was calculated using Orthologous Average Nucleotide Identity Tool (OAT)17.
Code availability
Most of the custom codes used in the generation or processing of our data are stated in the Methods section. Detailed information including versions of software and database are provided in Table 1.
Table 1. Versions of software and database used in this study.
| Software/Database | Application | Version/Date |
|---|---|---|
| HGAP | Genome assembly | SMART analysis 2.2.1 |
| CheckM | Genome quality assessment | 1.0.9 |
| RNAmmer | rRNA prediction | 1.2 |
| tRNAscan-SE | tRNA prediction | 1.3 |
| BLAST | Sequence comparison | 2.7.1+ |
| ARG-ANNOT | Antibiotic resistance genes prediction | May 2018 |
| PHASTER | Prophages prediction | May 2018 |
| DNAPlotter | Genome map | Artemis 17.0.1 |
| BRIG | Genome alignment | 0.95 |
| MEGA | Phylogenetic analysis | 7.0.26 |
| OAT | Average nucleotide identity calculation | Jul 2018 |
Data Records
Whole genome sequence of C. kerstersii 8943 has been deposited in GenBank (Data Citation 1). All of the reads for C. kerstersii 8943 genome have been deposited in the NCBI Sequence Read Archive (Data Citation 2).
Technical Validation
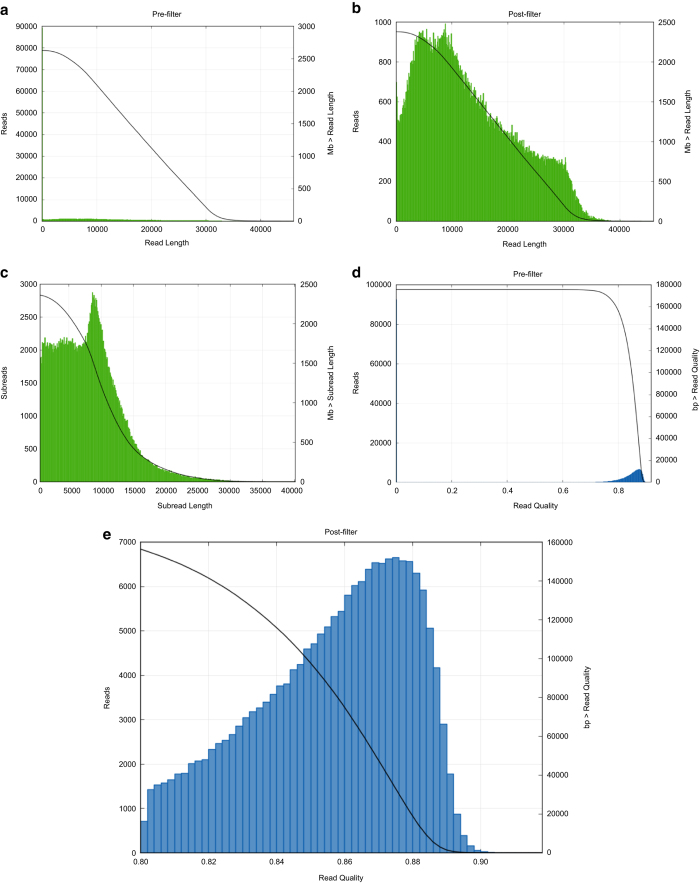
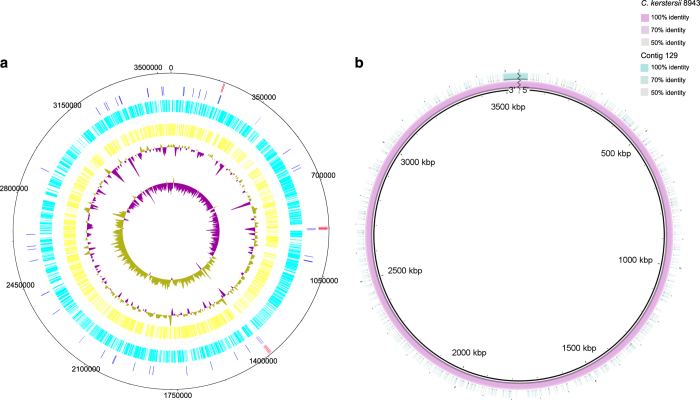
To maintain the quality of the assembly, we applied pre-assembly, de novo assembly and assembly polishing steps. Raw reads generated through sequencing were filtered to obtain clean reads, resulting in a total of 300,584 clean reads with an average size of 13,001 bp (Fig. 1). Further, we applied a sub-read filtering step by removing the adapter from the raw reads to obtain clean sub-reads that have a mean length of 7,968 bp and an N50 of 10,227 bp (Fig. 1). The de novo assembly generated a chromosome of 3,547,915 bp with a GC content of 59.6%. The genome was predicted to contain 3,155 protein-coding genes, 16 rRNAs and 101 tRNAs (Fig. 2a). The assessment of genome quality showed that the genome exhibited 97.52 % completeness, 0.85 % contamination and 0 % strain heterogeneity, indicating that the assembled genome was of high quality. The alignment of the complete genome with the draft genome of C. kerstersii 8943 (ASM129444v1) showed that both the start and the end of the complete genomic sequences were mapping to the same contig (contig 129) of ASM129444v1, indicating that the genome was circular (Fig. 2b). 16S rRNAs were used to construct a phylogenetic tree with other Comamonas species and the result reconfirmed the phylogenetic position of this strain as C. kerstersii (Fig. 3).
Figure 1. Quality control of the sequencing data.
(a) Read length distribution before filtering. (b) Read length distribution after filtering. (c) Subread length distribution after filtering. (d) Read quality distribution before filtering. (e) Read quality distribution after filtering.
Figure 2. Circular map of C. kerstersii 8943 chromosome.
(a) Genomic features appearing from inside to outsider are as follows: GC skew, GC plot, forward CDS (yellow), reverse CDS (cyan), tRNAs (blue) and rRNAs (red). (b) Assembled chromosome of C. kerstersii 8943 (pink), contig 129 of ASM129444v1 (green) and divider line of the chromosome (black zigzag line).
Figure 3. Phylogenetic tree depicting the relationship of C. kerstersii 8943 with other Comamonas species based on 16S rRNA sequences.

The GenBank accession numbers for 16S rRNA of the type strains used in analysis are indicated in parentheses.
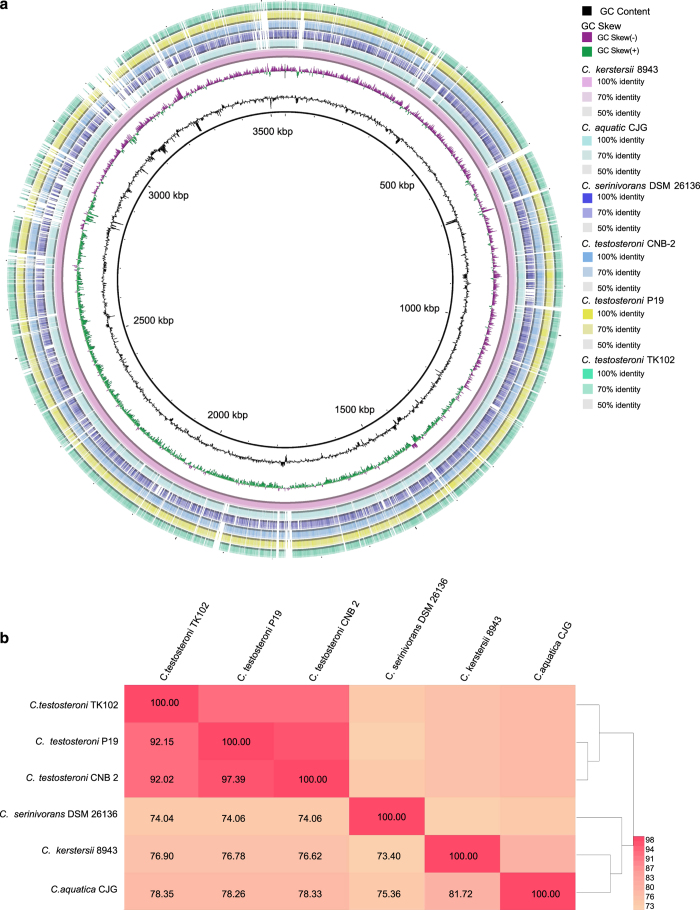
All the protein coding genes could be functionally annotated. A beta-lactamase encoding gene, blaOXA-1, was predicted in the genome. In accordance, C. kerstersii 8943 was tested to be resistant to ampicillin, a beta-lactam antibiotic. Besides C. kerstersii 8943 reported in the present study, there are five other completed genomes of Comamonas species which have been deposited in GenBank. Comparative genomic analysis was conducted with all the six genomes and the results supported the different isolation sources of C. kerstersii 8943 (isolated from human) and other Comamonas species (isolated from environment) (Fig. 4a). The ANI values among the six genomes indicated that there were great differences between C. kerstersii 8943 and other Comamonas species (Fig. 4b). The genome size of C. kerstersii 8943 was the smallest among the six Comamonas species and this was consistent with previous reports, which have shown that symbiotic bacteria usually harbour smaller genomes compared with free-living bacteria18,19. As a pathogen, C. kerstersii 8943 was found to have more intact prophage regions than other environmentally derived Comamonas species (Table 2). Moreover, 10 antibiotic resistance genes with high amino acid identity (>90%) were found in the genome of C. kerstersii 8943, whereas no antibiotic resistance genes were annotated in the other five Comamonas species (Table 2). These genes, including tetA, strB, sul1, blaOXA-1, strA, sul2, catB3 and floR, enable the survival of C. kerstersii 8943 in a clinical environment.
Figure 4. Comparative genomic analysis of C. kerstersii 8943 and other Comamonas species.
(a) BLAST atlas of the genomes. (b) ANI values between species.
Table 2. Feature differences between C. kerstersii 8943 and other Comamonas species.
| Species | Isolation source | Genome size (bp) | Intact prophage regions | Antibiotic resistance genes |
|---|---|---|---|---|
| C. kerstersii 8943 | dialysis effluent of a patient | 3,547,915 | 2 | 10 |
| C. aquatic CJG | water | 3,764,434 | 0 | 0 |
| C. serinivorans DSM 26136 | compost | 4,522,913 | 1 | 0 |
| C. testosteroni CNB-2 | activated sludge | 5,373,644 | 1 | 0 |
| C. testosteroni P19 | waste water | 5,632,239 | 1 | 0 |
| C. testosteroni TK102 | soil | 6,062,703 | 1 | 0 |
In summary, we applied the above-mentioned software and parameters in the quality control of this dataset. Therefore, theresulting data should be error-free. In addition, the annotation analysis performed using this dataset was in accordance with experimental results. Furthermore, comparative genomic studies using this dataset indicate its high level of accuracy and practicability.
Additional information
How to cite this article: Jiang, X. et al. Complete genome sequencing of Comamonas kerstersii 8943, a causative agent for peritonitis. Sci. Data. 5:180222 doi: 10.1038/sdata.2018.222 (2018).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Nos. 81741098 and 41406140); the National Key Research and Development Program of China (No. 2016YFD0501105); and the Zhejiang Provincial Key Research and Development Program (No. 2015C03032). We would like to thank Dr. Hong Cheng for the assistance in genome annotation.
Footnotes
The authors declare no competing interests.
Data Citations
- 2017. NCBI Assembly. GCA_002056725.1
- 2018. NCBI Sequence Read Archive. SRP150107
References
- Wauters G., De Baere T., Willems A., Falsen E. & Vaneechoutte M. Description of Comamonas aquatica comb. nov. and Comamonas kerstersii sp. nov. for two subgroups of Comamonas terrigena and emended description of Comamonas terrigena. Int J Syst Evol Microbiol 53, 859–862 (2003). [DOI] [PubMed] [Google Scholar]
- Almuzara M. N. et al. Intra-abdominal infections due to Comamonas kerstersii. J Clin Microbiol 51, 1998–2000 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opota O. et al. Bacteremia caused by Comamonas kerstersii in a patient with diverticulosis. J Clin Microbiol 52, 1009–1012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas J. S., Fitchett J. & O'Hara G. Comamonas kerstersii and the perforated appendix. J Clin Microbiol 52, 3134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almuzara M. et al. Unusual presentations of Comamonas kerstersii infection. New Microbes New Infect 19, 91–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. H., Ma H. X., Dong Z. Y. & Shen M. H. Comamonas kerstersii bacteremia in a patient with acute perforated appendicitis: A rare case report. Medicine 97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L. et al. Complete genome sequencing of the luminescent bacterium, Vibrio qinghaiensis sp. Q67 using PacBio technology. Sci Data 5, 170205 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P. & Tyson G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K. et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35, 3100–3108 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M. & Eddy S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25, 955 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G. & Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount D. W. Using the basic local alignment search tool (BLAST). CSH Protoc 2007, pdb. top17 (2007). [DOI] [PubMed] [Google Scholar]
- Gupta S. K. et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58, 212–220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D. et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44, W16–W21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T., Thomson N., Bleasby A., Berriman M. & Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25, 119–120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan N. F., Petty N. K., Zakour N. L. B. & Beatson S. A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Kim Y. O., Park S.-C. & Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66, 1100–1103 (2016). [DOI] [PubMed] [Google Scholar]
- Konstantinidis K. T. & Tiedje J. M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. U. S. A. 101, 3160–3165 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X. W. et al. Comparative genomic study of three species within the genus Ornithinibacillus, reflecting the adaption to different habitats. Gene 578, 25–31 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2017. NCBI Assembly. GCA_002056725.1
- 2018. NCBI Sequence Read Archive. SRP150107