Abstract
Background
CYP/CYP450 2C19 (CYP2C19) is a highly polymorphic enzyme and exhibits individual differences in metabolic activity. The purpose of this research was mainly to explore the catalytic activities of 30 CYP2C19 variants on the substrate voriconazole in vitro, including 24 novel CYP2C19 variants (2C19.2E-.2H, .2J, .3C, .29-.33, L16F, 35FS, R124Q, R125G, T130M, N231T, M255T, R261W, N277K, S303N, I327T, N403I, and A430V) found in Chinese Han population for the first time.
Methods
These CYP2C19 variants were expressed in Spodoptera frugiperda (Sf) 21 insect cells using the baculovirus-mediated expression system. The substrate voriconazole was incubated with the abovementioned proteins at 37°C for 30 minutes in an appropriate designed system. Then through detecting its major metabolite voriconazole N-oxide by ultra-performance liquid chromatography tandem mass spectrometry, available data were obtained to explain the influence of CYP2C19 polymorphisms on voriconazole.
Results
From the results, when compared to CYP2C19.1, most variants exhibited either reduced Vmax and/or increased Km value, indicating that the intrinsic clearance (Vmax/Km) values of most variants were significantly altered. The catalytic activities of 20 novel variants exhibited decreases in different degrees compared to CYP2C19.1, with relative clearance values ranging from 1.11% to 83.78%. However, L16F exhibited the increased catalytic activity for 135.68%. In addition, the kinetic parameters of four variants (2C19.2H, .3, 35FS, and R124Q) could not be detected, due to the defective gene.
Conclusion
This is the first study to report the effects of CYP2C19 polymorphisms on vori-conazole metabolism in vitro, and we hope these data could lay the foundation for the early clinical research and individualized treatment.
Keywords: polymorphisms, CYP2C19, voriconazole, voriconazole N-oxide, drug metabolism
Introduction
The CYP/CYP450 2C19 (CYP2C19) plays an important role in the metabolism of a large portion of therapeutic drugs, including multiple types of drugs, such as anticonvulsant drug mephenytoin, proton pump inhibitor omeprazole, the antiplatelet drug clopidogrel, antidepressant drug escitalopram, antiviral drug nelfinavir, and the antifungal drug voriconazole.1 Similar with the other human CYP2C members, CYP2C19 exhibits high genetic polymorphisms, which play a key role for the biological function and result in different pharmacogenetic phenotypes termed as poor, intermediate, extensive, and ultra-rapid metabolizers.2 It is well known that high drug exposure and undesirable adverse effects will occur even if poor metabolizers (PMs) are administered at normal doses of certain drugs.3–5 To date, an increasing number of CYP2C19 genes have been discovered and reported on the Human CYP Allele Nomenclature Committee home page (http://www.cypalleles.ki.se/cyp2c19.htm), and the number is growing. Recently, 30 CYP2C19 alleles had been detected in 2,127 unrelated healthy Chinese Han, which included 24 novel alleles that had not been reported and 6 old alleles that had been discovered previously.6 Among the 24 novel alleles, the Human CYP Allele Nomenclature Committee had named 11 alleles as new alleles (CYP2C19*2E-*2H, CYP2C19*2J, CYP2C19*3C, and CYP2C19*29-*33).6
Voriconazole, as the first line available second-generation antifungal triazole drug, has a broad-spectrum antifungal activity against clinically significant pathogens, including invasive and pulmonary aspergillosis, invasive fluconazole-resistant candidiasis, and infections caused by emerging pathogens, like Scedosporium and Fusarium spp.7 Voriconazole has advantages when given in both intravenous and oral dosage forms, and shows good absorption and tolerability.8 Disadvantages of voriconazole are unpredictable, nonlinear pharmacokinetics and extensive individual differences in drug concentrations. One of the side effects of voriconazole is that it is associated with high serum concentrations, and therefore therapeutic drug monitoring is essential when voriconazole is prescribed.9,10
In vitro studies have shown that both CYP2C19 and CYP3A4 are involved in the metabolism of voriconazole,11 and CYP2C19 has a great contribution toward the significant changes in the pharmacokinetics.12 Indeed, the pharmacoki-netics of voriconazole are largely influenced by CYP2C19 genotypes in vivo.13 Several studies have demonstrated the relationship between voriconazole serum concentration and genotype, not only with the allele CYP2C19*2,3,14 but more related to CYP2C19*3.15,16
Until now, whether these abovementioned 30 CYP2C19 variants alter the enzymatic activity toward voriconazole has not been assessed. This paper mainly engaged in researching the catalytic activities of 6 variants that have been reported previously and 24 novel CYP2C19 variants (2C19.2E-.2H, .2J, .3C, .29-.33, L16F, 35FS, R124Q, R125G, T130M, N231T, M255T, R261W, N277K, S303N, I327T, N403I, A430V) found in the Chinese Han population on the metabolism of voriconazole in vitro. We expect this work can greatly extend the understanding of functional consequences of CYP2C19 genetic polymorphisms and may prove the value of CYP2C19 genotype-based therapy with voriconazole.
Materials and methods
Chemicals and materials
Voriconazole, voriconazole N-Oxide, and fluconazole were purchased from Melone Biotechnology Co., Ltd. (Beijing, China). The UPLC® BEH C18 column (2.1 mm × 50 mm, 1.7 µm) used for liquid chromatography was supplied by the Waters (Millipore, Bedford, MA, USA). NADPH was provided by Promega (Madison, WI, USA). P450 Cytochrome b5 and recombinant human CYP2C19 enzymes (the wild-type CYP2C19.1 and 30 CYP2C19 variants) effectively generated in insect cell Sf21 were gifted by Beijing Hospital, National Health and Family Planning Commission of Republic of China.17 Methanol and acetonitrile were of liquid chromatography grade and were obtained from Merck Chemicals Co., Ltd. (Darmstadt, Germany). Formic acid and other chemicals were all of analytical grade and were prepared from Sigma-Aldrich Co. (St. Louis, MO, USA) and Chemical Industries (Beijing, China), respectively. Ultrapure water used for the experiment was supplied from Milli-Q water purification system (Millipore, Bedford, MA, USA).
Incubation conditions
The total volume of the incubation mixture was 200 µL, which included 5 pmol of the wild-type CYP2C19.1 or other CYP2C19 recombinant variants, 5 pmol of purified cytochrome b5, 10 mM of potassium phosphate buffer (PH 7.4), and 3.5 µL voriconazole (the final concentration ranged from 2 to 100 µM). After incubating at 37°C for 5 minutes, 1 mM NADPH regenerating system was added to start the reaction before the mixture was continued to be incubated for 30 minutes. Finally, reactions in the tubes were terminated by cooling to –80°C. Then 400 µL of acetonitrile was added quickly and followed by the addition of 30 µL of internal standard (IS) fluconazole. Initially, IS at a concentration of 5 µg/mL was dissolved in acetonitrile solution. After vortexing for 2 minutes, the samples were immediately centrifuged at 13,000 rpm for 10 minutes at 4°C environment. The supernatant of each tube was 1:1 diluted with water, and 2 µL was subjected to the ultra-performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS) system for analysis. Incubations were carried out in triplicate and data were showed as mean ± SD.
Chromatographic and mass spectrometric conditions
The liquid chromatographic separation was conducted on an UPLC® BEH C18 column (2.1 mm ×50 mm, 1.7 µm) at 40°C. The gradient elution solvents consisted of acetonitrile (A) and 0.1% formic acid in water (B). The gradient program was conducted in a linear state as follows: 0–0.5 minutes (25%–95% A), 0.5–1.2 minutes (95%–95% A), 1.2–1.4 minutes (95%–25% A), and 1.4–3.0 minutes (25%–25% A). The total run time was 3.0 minutes with the flow rate of 0.40 mL/min.
Analysis was carried out by ultra-performance liquid chromatography tandem mass spectrometry (UPLC-MS/ MS). Mass spectrometric detection was performed on a XEVO TQD triple quadrupole mass spectrometer equipped with an electrospray ionization interface in positive ionization mode. The multiple reaction monitoring modes of transitions as quantitative analysis were m/z 307.1→238.1, m/z 350.1→281.0, and m/z 366.1→224.1 for fluconazole, voriconazole, and voriconazole N-oxide, respectively. Both intra-day and inter-day precision was below 15%, and accuracy was within ±15%; this bioanalytical method was proved to be precise and accurate. Under abovementioned appropriate conditions, the retention times of fluconazole, voriconazole, and N-oxide voriconazole were 0.96, 1.39, and 1.22 minutes, respectively. The Masslynx 4.1 software (Waters Corp., Milford, MA, USA) was used for controlling the instrument and acquiring the data.
Statistical analysis
GraphPad version 5 (Prism version 5; GraphPad Software Inc., San Diego, CA, USA) was used to calculate the kinetic parameters (Km and Vmax) by nonlinear regression curve fitting. Statistical analysis was conducted by SPSS software package (version 17.0; SPSS Inc., Chicago, IL, USA), and P<0.05 was considered statistically significant. One-way ANOVA was employed for comparison between groups. Dunnett’s test was performed to evaluate the differences in catalytic activity between CYP2C19.1 and other variants.
Results
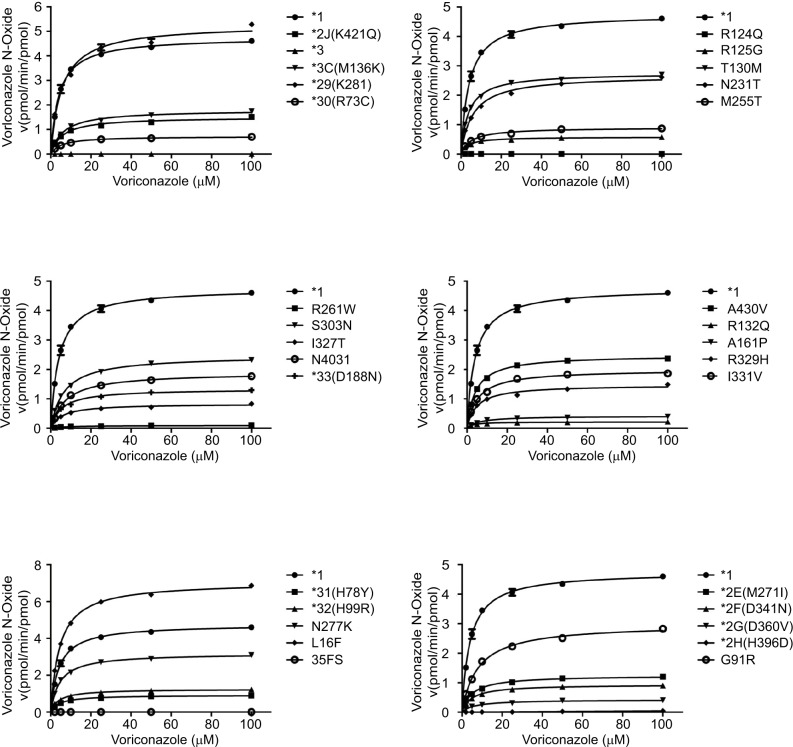
For the study purpose, we specifically analyzed the available data of the wild-type CYP2C19.1 and 30 variants for the in vitro functional analysis. The Michaelis Menten plots are shown in Figure 1, and the corresponding kinetic parameters are summarized in Table 1. For four variants (CYP2C19.2H, .3, 35FS, and R124Q), the metabolite concentrations of voriconazole were below the detection limit that the kinetic parameters could not be determined, which exhibited their null functions in this study. Among the tested alleles, 25 allelic variants exhibited significantly reduced enzymatic ability compared to the wild-type CYP2C19.1 (count as 100%) toward voriconazole. In detail, 22 variants (CYP2C19.2E, .2F, .2G, .2J, .3C, .30, .31, .32, .33, R125G, N231T, M255T, R261W, S303N, I327T, N403I, A430V, I331V, .23, .6, .2C, and .18) exhibited significantly reduced intrinsic clearance (Vmax/Km) which varied from 1.11% to 49.29%; three variants (CYP2C19.29, T130M, and N277K) exhibited mildly decreased Vmax/Km values (60.90%–83.78%). Specially, L16F exhibited significantly increased relative clearance value (135.68%). R261W variant exhibited extremely reduced Vmax value (0.11) and significantly increased Km value (about twofold when compared to CYP2C19.1). Thus, the results showed that the Vmax/Km value decreased to 1.11%, as well as that the Vmax/Km value of CYP2C19.2G, .6, and .2C decreased to 5.61%–6.89%. On the contrary, L16F exhibited greatly increased Vmax value (7.15) when compared to that of CYP2C19.1 but showed similar Km value; hence, the Vmax/Km value increased to 135.68%.
Figure 1.
Michaelis–Menten curves of the enzymatic activities of the wild-type CYP2C19.1 and 30 variants on voriconazole metabolism (data are presented as mean ± SD of three separate experiments).
Table 1.
Kinetic parameters for N-oxidation activity of CYP2C19.1 and other 30 CYP2C19 variants on the metabolism of voriconazole
| Allele | Protein | Effect | Frequencies (%)6 | Vmax (pmol/ min/pmol of P450) | Km (μM) | Intrinsic clearance (Vmax/Km) | Relative clearance (% of wild type) |
|---|---|---|---|---|---|---|---|
|
| |||||||
| CYP2C19*1 | CYP2C19.1 | 4.85±0.08 | 4.31±0.38 | 1.13±0.12 | 100.00 | ||
| CYP2C19*2E | CYP2C19.2E | Splicing defect; M271I; I331V | 0.02 | 1.26±0.01* | 5.24±0.41 | 0.24±0.02* | 21.38 |
| CYP2C19*2F | CYP2C19.2F | Splicing defect; I331V; D341N | 0.05 | 0.99±0.06* | 5.89±1.48* | 0.17±0.03* | 15.25 |
| CYP2C19*2G | CYP2C19.2G | Splicing defect; I331V; D360V | 0.05 | 0.43±0.01* | 6.84±0.15* | 0.06±0.00* | 5.61 |
| CYP2C19*2H | CYP2C19.2H | Splicing defect; I331V; H396D | 0.02 | ND | ND | ND | ND |
| CYP2C19*2J | CYP2C19.2J | Splicing defect; I331V; K421Q | 0.02 | 1.59±0.02* | 6.80±0.38* | 0.23±0.02* | 20.78 |
| CYP2C19*3 | CYP2C19.3 | W212X; I331V | 5.34 | ND | ND | ND | ND |
| CYP2C19*3C | CYP2C19.3C | M136K,W212X; I331V | 0.07 | 1.84±0.03* | 6.62±0.54* | 0.28±0.02* | 24.63 |
| CYP2C19*29 | CYP2C19.29 | K28I; I331V | 0.02 | 5.37±0.10* | 5.68±0.10* | 0.95±0.03* | 83.78 |
| CYP2C19*30 | CYP2C19.30 | R73C | 0.02 | 0.73±0.00* | 5.32±0.38 | 0.14±0.01* | 12.17 |
| CYP2C19*31 | CYP2C19.31 | H78Y; I331V | 0.02 | 0.93±0.01* | 4.51±0.19 | 0.21±0.01* | 18.36 |
| CYP2C19*32 | CYP2C19.32 | H99R; I331V | 0.02 | 1.26±0.02* | 4.22±0.38 | 0.30±0.02* | 26.57 |
| CYP2C19*33 | CYP2C19.33 | D188N; I331V | 0.02 | 1.36±0.03* | 5.70±0.74* | 0.24±0.03* | 21.25 |
| L16F | L16F | 0.02 | 7.15±0.10* | 4.67±0.18 | 1.53±0.04* | 135.68 | |
| 35FS | 35FS | 0.02 | ND | ND | ND | ND | |
| R124Q | R124Q | 0.02 | ND | ND | ND | ND | |
| R125G | R125G | 0.02 | 0.62±0.01* | 4.18±0.63 | 0.15±0.03* | 13.36 | |
| T130 M | T130 M | 0.05 | 2.76±0.03* | 3.73±0.07 | 0.74±0.01* | 65.65 | |
| N231 T | N231 T | 0.02 | 2.78±0.05* | 6.62±0.25* | 0.42±0.02* | 37.19 | |
| M255 T | M255 T | 0.02 | 0.92±0.02* | 5.48±0.03 | 0.17±0.00* | 14.92 | |
| R261W | R261W | 0.02 | 0.11±0.00* | 8.50±1.22* | 0.01±0.00* | 1.11 | |
| N277 K | N277 K | 0.07 | 3.26±0.06* | 4.74±0.15 | 0.69±0.02* | 60.90 | |
| S303 N | S303 N | 0.02 | 2.56±0.03* | 7.38±0.54* | 0.35±0.02* | 30.80 | |
| I327T | I327T | 0.02 | 0.83±0.04* | 5.56±0.88 | 0.15±0.02* | 13.43 | |
| N403I | N403I | 0.05 | 1.91±0.04* | 7.58±0.54* | 0.25±0.01* | 22.34 | |
| A430V | A430V | 0.02 | 2.54±0.08* | 4.57±0.60 | 0.56±0.06* | 49.29 | |
| CYP2C19*1B | CYP2C19.1B | I331V | 90.16 | 1.98±0.01* | 5.19±0.28 | 0.38±0.02* | 33.84 |
| CYP2C19*23 | CYP2C19.23 | G91R; I331V | 0.05 | 3.08±0.07* | 8.70±0.39* | 0.35±0.01* | 31.40 |
| CYP2C19*6 | CYP2C19.6 | R132Q; I331V | 0.09 | 0.22±0.00* | 2.90±0.40* | 0.08±0.01* | 6.89 |
| CYP2C19*2C | CYP2C19.2C | A161P, splicing defect, I331V | 0.05 | 0.41±0.01* | 5.76±0.23* | 0.07±0.00* | 6.37 |
| CYP2C19*18 | CYP2C19.18 | R329H; I331V | 0.02 | 1.54±0.02* | 5.54±0.21 | 0.28±0.01* | 24.59 |
Notes: Data are presented as mean ± SD of three independent experiments.
P<0.05 compared to the wild type.
Abbreviation: ND, not determined.
Discussion
Increasing evidences suggested that CYP2C19 is highly polymorphic among different individuals and races, which could lead to interindividual and interethnic variability in drug metabolism. Among the variants of CYP2C19, two principal alleles CYP2C19*2 (681G>A) and CYP2C19*3 (636G>A) are the most common and have been reported as PM phenotype, so both of them were extensively studied in different populations.18,19 Genetic polymorphism of CYP2C19 may cause phenotypic variability, affect drug metabolism and efficacy, and even lead to adverse drug reactions. Among the individuals who are homozygous for null variants, the “PM” phenotype for drugs clearing will possibly result in a high risk of adverse drug reactions. On the contrary, the “ultra-rapid metabolizer” phenotype is considered to be a cause of poor therapeutic efficacy when taken with standard therapeutic dose of drugs.20–22
To date, many researchers have conducted genetic studies on the polymorphism of CYP2C19 in Chinese Han populations; however, the vast majority of these studies have been involved on two common defective alleles, CYP2C19*2 and *3,23–25 which accounted for >90% of PMs in Oriental populations.26–28 This is the first study to assess the enzymatic activities of 30 CYP2C19 variants, including 24 novel variants,6 which exhibited high expression in Sf21 cells toward voriconazole in vitro. Although the frequencies of most of these newly discovered mutations are very low (usually <0.1%),6 it is imperative to study their functional characteristics as there are more than 1.4 billion people living in the mainland China.
In this study, the enzymatic activity of CYP2C19.1 was defined as the control group, with CYP2C19.3 and 35FS used as quality controls for the functional analysis in order to ensure the accuracy and reliability of this study. According to the previous experiments, CYP2C19.3 did not exhibit any protein activity when investigated in vitro, and thus no metabolic activity was observed on voriconazole. This phenomenon resulted from a truncated protein created by a premature stop codon with a single nucleotide exchange of G>A at position 636.29 Similarly, because of the insertion of five nucleotides CCTAC at the 101 position of the cDNA, which results in a frameshift in protein translation, the activity of 35FS variant could not be detected by immunoblotting.6
For evaluating the influence of 30 CYP2C19 allelic variants on the metabolism of voriconazole, we investigated these variants in detail. As previously reported, nonsynonymous mutations in CYP2C9 gene may affect the ability of protein expression in mammalian cell lines.30–32 In the present study, a similar phenomenon was observed, where T130M variant with nonsynonymous point mutation exhibited significantly decreased metabolic activities (65.65%) for voriconazole when compared to CYP2C19.1. For this variant, the reason for the reduction in enzymatic activity may be related to the low protein expression level.17
Among the rested variants, most of the variants exhibited obviously decreased Vmax/Km values, but the protein expression level was not the main reason, because in R261W variant the value decreased to 1.11% in spite of showing protein expression similar to the CYP2C19.1 variant.17 For CYP2C19*6 allele, a single base pair mutation (G395A) in exon 3 results in a change in the coding region for Arg132Gln, and the activity of this variant toward two CYP2C19 substrates (mephenytoin and tolbutamide) could be negligible.33 The 6.89% decrease in our results is consistent with the results of these previous experiments. Furthermore, CYP2C19*2H has an amino acid change from His to Asp at position 396 in exon 8, which is induced by one nucleotide substitution (C>G) at position 1,186 in the complementary DNA (cDNA).6 It exhibited absent metabolic activity, indicating that the amino acid in this site has vital importance. However, compared with the wild type, K28I exhibited lower protein expression in Sf21 cells while the metabolic capacity toward voriconazole was 83.78%.17 These results suggested that if the same amount of proteins was used in the in vitro analysis, K28I may show moderate or even high clearance values. In addition, the catalytic activity of many variants (such as R124Q and R125G) was significantly reduced, while the protein expression levels were similar to or even higher than those of wild-type alleles. In the previous literature, it had been reported that when R124Q and R125H proteins in CYP2C9 were expressed in COS-7 or 293FT cells, the enzymatic activities of CYP2C9 could be significantly reduced.30,34 In fact, CYP2C19 and CYP2C9 differ by only 43 residues (constituted of 490 amino acids).35 Therefore, mutations at the same site of CYP2C19 and CYP2C9 may have similar functional effects on the enzyme. L16F, in which amino acid Leu16 is replaced with Phe in exon 1, shows lower protein expression level when compared to the wild-type protein and exhibited similar Km value but higher Vmax value, which in turn led to increased Vmax/Km value of voriconazole (135.68%) when compared to wild-type CYP2C19.1. This result was consistent with the previous experiment on the probe drug omeprazole.36
CYP2C19*23 involves a G271C point mutation where Gly91 is replaced with Arg. However, Gly91 is located within a region that is important for surface channel access to heme, and this substitution introduces a large charged amino acid that changes the hydrophobicity.37 Thus, the Gly91Arg substitution could cause a significant conformational change at the substrate recognition site, and hence voriconazole showed much lower Vmax/Km value (31.4%) than CYP2C19.1.
For these variants, we believe that the nonsynonymous mutation in CYP2C9 genes might change the stability and/ or the folding efficiency of protein, which in turn influence the protein function. In the course of drug metabolism, the protein conformation is important and influences the specific affinity between substrate and enzyme, and different substrates has different metabolic results by the same enzyme. This phenomenon could be found when compared the results of this study with newly studies on the effects of CYP2C19 variants toward to the metabolism of nebivolol and methadone in vitro.17,38
In conclusion, this study researched the enzymatic activity of 30 CYP2C19 variants on the metabolism of voriconazole in vitro, especially the activity of 24 new variants. This study, which is the first to determine the effect of abovementioned variants on the metabolism of voriconazole, provides a new visual perspective for CYP2C19 genetic polymorphism and its effect on the substrate drug voriconazole. Moreover, this study could provide the fundamental information for the rational use of voriconazole in a clinical setting. In addition, it is also significant to promote other studies on drug metabolic abilities associated with these novel CYP2C19 genetic variants in future, develop targeted therapy, and achieve personalized medicine.
Acknowledgments
The authors thank the members of the Beijing Institute of Geriatrics of the Ministry of Health for their advice and assistance. This study was supported by grants from the Wenzhou City Science and Technology Bureau (Grant No Y20170687) and Medical and Health Science and Technology Program of Zhejiang Province in China (Grant No 2017KY461).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Myrand SP, Sekiguchi K, Man MZ, et al. Pharmacokinetics/genotype associations for major cytochrome P450 enzymes in native and first- and third-generation Japanese populations: comparison with Korean, Chinese, and Caucasian populations. Clin Pharmacol Ther. 2008;84(3):347–361. doi: 10.1038/sj.clpt.6100482. [DOI] [PubMed] [Google Scholar]
- 2.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Suan D, O’Connor K, Booth DR, Liddle C, Stewart GJ. Voricon-azole toxicity related to polymorphisms in CYP2C19. Intern Med J. 2011;41(4):364–365. doi: 10.1111/j.1445-5994.2011.02454.x. [DOI] [PubMed] [Google Scholar]
- 4.Xu RA, Lin GY, Hu LF, et al. Successful management of voriconazole-associated hyponatremia with therapeutic drug monitoring. Antimicrob Agents Chemother. 2013;57(5):2422–2423. doi: 10.1128/AAC.00146-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto N, Murakami H, Nishina T, et al. The effect of CYP2C19 polymorphism on the safety, tolerability, and pharmacokinetics of tivantinib (ARQ 197): results from a phase I trial in advanced solid tumors. Ann Oncol. 2013;24(6):1653–1659. doi: 10.1093/annonc/mdt014. [DOI] [PubMed] [Google Scholar]
- 6.Hu LM, Dai DP, Hu GX, et al. Genetic polymorphisms and novel allelic variants of CYP2C19 in the Chinese Han population. Pharmacogenom-ics. 2012;13(14):1571–1581. doi: 10.2217/pgs.12.141. [DOI] [PubMed] [Google Scholar]
- 7.Ghannoum MA, Kuhn DM. Voriconazole – better chances for patients with invasive mycoses. Eur J Med Res. 2002;7(5):242–256. [PubMed] [Google Scholar]
- 8.Donnelly JP, de Pauw BE. Voriconazole – a new therapeutic agent with an extended spectrum of antifungal activity. Clin Microbiol Infect. 2004;10(Suppl 1):107–117. doi: 10.1111/j.1470-9465.2004.00838.x. [DOI] [PubMed] [Google Scholar]
- 9.Malani AN, Kerr LE, Kauffman CA. Voriconazole: How to Use This Antifungal Agent and What to Expect. Semin Respir Crit Care Med. 2015;36(5):786–795. doi: 10.1055/s-0035-1562903. [DOI] [PubMed] [Google Scholar]
- 10.Bressán IG, Mendez ML, Gimenez MI. Validation of a Reversed-Phase Ultra-High-Performance Liquid Chromatographic Method With Photo-diode Array Detection for the Determination of Voriconazole in Human Serum and Its Application to Therapeutic Drug Monitoring. Ther Drug Monit. 2018;40(2):276–283. doi: 10.1097/FTD.0000000000000491. [DOI] [PubMed] [Google Scholar]
- 11.Murayama N, Imai N, Nakane T, Shimizu M, Yamazaki H. Roles of CYP3A4 and CYP2C19 in methyl hydroxylated and N-oxidized metabolite formation from voriconazole, a new anti-fungal agent, in human liver microsomes. Biochem Pharmacol. 2007;73(12):2020–2026. doi: 10.1016/j.bcp.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Hyland R, Jones BC, Smith DA. Identification of the cytochrome P450 enzymes involved in the N-oxidation of voriconazole. Drug Metab Dispos. 2003;31(5):540–547. doi: 10.1124/dmd.31.5.540. [DOI] [PubMed] [Google Scholar]
- 13.Weiss J, Ten Hoevel MM, Burhenne J, et al. CYP2C19 genotype is a major factor contributing to the highly variable pharmacokinetics of voriconazole. J Clin Pharmacol. 2009;49(2):196–204. doi: 10.1177/0091270008327537. [DOI] [PubMed] [Google Scholar]
- 14.Berge M, Guillemain R, Trégouet DA, et al. Effect of cytochrome P450 2C19 genotype on voriconazole exposure in cystic fibrosis lung transplant patients. Eur J Clin Pharmacol. 2011;67(3):253–260. doi: 10.1007/s00228-010-0914-2. [DOI] [PubMed] [Google Scholar]
- 15.Scholz I, Oberwittler H, Riedel KD, et al. Pharmacokinetics, metabolism and bioavailability of the triazole antifungal agent voriconazole in relation to CYP2C19 genotype. Br J Clin Pharmacol. 2009;68(6):906–915. doi: 10.1111/j.1365-2125.2009.03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S, Kim BH, Nam WS, et al. Effect of CYP2C19 polymorphism on the pharmacokinetics of voriconazole after single and multiple doses in healthy volunteers. J Clin Pharmacol. 2012;52(2):195–203. doi: 10.1177/0091270010395510. [DOI] [PubMed] [Google Scholar]
- 17.Zhou XY, Hu XX, Li MF, et al. Functional characterization of CYP2C19 variants in nebivolol 4-hydroxlation in vitro. Drug Test Anal. 2018;10(5):807–813. doi: 10.1002/dta.2334. [DOI] [PubMed] [Google Scholar]
- 18.Hiratsuka M. In vitro assessment of the allelic variants of cytochrome P450. Drug Metab Pharmacokinet. 2012;27(1):68–84. doi: 10.2133/dmpk.dmpk-11-rv-090. [DOI] [PubMed] [Google Scholar]
- 19.Chiba K, Shimizu K, Kato M, et al. Prediction of inter-individual variability in the pharmacokinetics of CYP2C19 substrates in humans. Drug Metab Pharmacokinet. 2014;29(5):379–386. doi: 10.2133/dmpk.dmpk-13-rg-137. [DOI] [PubMed] [Google Scholar]
- 20.Desta Z, Zhao X, Shin JG, Flockhart DA. Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin Pharmacokinet. 2002;41(12):913–958. doi: 10.2165/00003088-200241120-00002. [DOI] [PubMed] [Google Scholar]
- 21.Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte – an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662–673. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 22.Favela-Mendoza AF, Martinez-Cortes G, Hernandez-Zaragoza M, et al. Genetic variability of CYP2C19 in a Mexican population: contribution to the knowledge of the inheritance pattern of CYP2C19*17 to develop the ultrarapid metabolizer phenotype. J Genet. 2015;94(1):3–7. doi: 10.1007/s12041-015-0477-1. [DOI] [PubMed] [Google Scholar]
- 23.Hou X, Shi J, Sun H. Gene polymorphism of cytochrome P450 2C19*2 and clopidogrel resistance reflected by platelet function assays: a meta-analysis. Eur J Clin Pharmacol. 2014;70(9):1041–1047. doi: 10.1007/s00228-014-1714-x. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Liu N, Li W, Shao H, Zhi H, Li J. Relationship of CYP2C19*2 and CYP2C19*3 gene polymorphism with clopidogrel response variability and recurrent cardiovascular events in Chinese patients undergoing percutaneous coronary intervention. Pharmacology. 2013;91(3–4):165–172. doi: 10.1159/000346736. [DOI] [PubMed] [Google Scholar]
- 25.Gan CQ, Wang XY, Cao YD, Ye WX, Liu H, Sun YY. Association of CYP2C19*3 gene polymorphism with breast cancer in Chinese women. Genet Mol Res. 2011;10(4):3514–3519. doi: 10.4238/2011.December.5.3. [DOI] [PubMed] [Google Scholar]
- 26.Chen L, Qin S, Xie J, et al. Genetic polymorphism analysis of CYP2C19 in Chinese Han populations from different geographic areas of mainland China. Pharmacogenomics. 2008;9(6):691–702. doi: 10.2217/14622416.9.6.691. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Barceló M, Chow LY, Kum Chiu HF, et al. Frequencies of defective CYP2C19 alleles in a Hong Kong Chinese population: detection of the rare allele CYP2C19*4. Clin Chem. 1999;45(12):2273–2274. [PubMed] [Google Scholar]
- 28.Shu Y, Zhou HH. Individual and ethnic differences in CYP2C19 activity in Chinese populations. Acta Pharmacol Sin. 2000;21(3):193–199. [PubMed] [Google Scholar]
- 29.de Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46(4):594–598. [PubMed] [Google Scholar]
- 30.Dai DP, Xu RA, Hu LM, et al. CYP2C9 polymorphism analysis in Han Chinese populations: building the largest allele frequency database. Pharmacogenomics J. 2014;14(1):85–92. doi: 10.1038/tpj.2013.2. [DOI] [PubMed] [Google Scholar]
- 31.Niinuma Y, Saito T, Takahashi M, et al. Functional characterization of 32 CYP2C9 allelic variants. Pharmacogenomics J. 2014;14(2):107–114. doi: 10.1038/tpj.2013.22. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi M, Saito T, Ito M, et al. Functional characterization of 21 CYP2C19 allelic variants for clopidogrel 2-oxidation. Pharmacogenom-ics J. 2015;15(1):26–32. doi: 10.1038/tpj.2014.30. [DOI] [PubMed] [Google Scholar]
- 33.Ibeanu GC, Goldstein JA, Meyer U, et al. Identification of new human CYP2C19 alleles (CYP2C19*6 and CYP2C19*2B) in a Caucasian poor metabolizer of mephenytoin. J Pharmacol Exp Ther. 1998;286(3):1490–1495. [PubMed] [Google Scholar]
- 34.Lee MY, Borgiani P, Johansson I, et al. High warfarin sensitivity in carriers of CYP2C9*35 is determined by the impaired interaction with P450 oxidoreductase. Pharmacogenomics J. 2014;14(4):343–349. doi: 10.1038/tpj.2013.41. [DOI] [PubMed] [Google Scholar]
- 35.Reynald RL, Sansen S, Stout CD, Johnson EF. Structural characterization of human cytochrome P450 2C19: active site differences between P450s 2C8, 2C9, and 2C19. J Biol Chem. 2012;287(53):44581–44591. doi: 10.1074/jbc.M112.424895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai DP, Hu LM, Geng PW, et al. In vitro functional analysis of 24 novel CYP2C19 variants recently found in the Chinese Han population. Xenobiotica. 2015;45(11):1030–1035. doi: 10.3109/00498254.2015.1028512. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Q, Yu XM, Lin HB, et al. Genetic polymorphism, linkage disequilibrium, haplotype structure and novel allele analysis of CYP2C19 and CYP2D6 in Han Chinese. Pharmacogenomics J. 2009;9(6):380–394. doi: 10.1038/tpj.2009.31. [DOI] [PubMed] [Google Scholar]
- 38.Lan T, Yuan LJ, Hu XX, et al. Effects of CYP2C19 variants on methadone metabolism in vitro. Drug Test Anal. 2017;9(4):634–639. doi: 10.1002/dta.1997. [DOI] [PubMed] [Google Scholar]