Abstract
Current guideline recommendations for pharmacogenetic testing for clopidogrel by the American Heart Association/American College of Cardiology (AHA/ACC) contradict the Clinical Pharmacogenetics Implementation Consortium and the US FDA. The AHA/ACC recommends against routine pharmacogenetic testing for clopidogrel because no randomized controlled trials have demonstrated that testing improves patients’ outcomes. However the AHA/ACC and the National Comprehensive Cancer Network (NCCN) recommend other pharmacogenetic tests in the absence of randomized controlled trials evidence. Using clopidogrel as a case example, we compared the evidence for other pharmacogenetic tests recommended by the AHA/ACC and NCCN. In patients that received percutaneous coronary intervention, the evidence supporting pharmacogenetic testing for clopidogrel is stronger than other pharmacogenetic tests recommended by the AHA/ACC and NCCN.
Keywords: : American Heart Association/American College of Cardiology, clopidogrel, epidermal growth factor receptor inhibitors, guidelines, long QT syndrome, National Comprehensive Cancer Network, pharmacogenetics, recommendations, thiopurines
Cardiovascular clinical practice in the USA heavily relies on clinical practice guidelines published by the American Heart Association/American College of Cardiology (AHA/ACC) [1]. Reliance on those guidelines is complicated when it comes to pharmacogenetics. Currently, the AHA/ACC Guideline recommendations for pharmacogenetics contradict the pharmacogenetic recommendations from two other major organizations: the US FDA and the Clinical Pharmacogenetics Implementation Consortium (CPIC) [2–5]. The 2016 AHA/ACC Guideline focused update on duration of dual antiplatelet therapy in patients with coronary artery disease states the following: “to date, no randomized controlled trial (RCT) has demonstrated that routine platelet function testing or genetic testing to guide P2Y12 inhibitor therapy improves outcome; thus, the routine use of platelet function and genetic testing is not recommended [3]”. Clopidogrel is an antiplatelet P2Y12 inhibitor that is primarily metabolized by cytochrome P450 2C19 (gene: CYP2C19). The CPIC does not recommend whether or not to order pharmacogenetic tests, but if the patient's CYP2C19 genotype is known, they recommend alternative antiplatelet therapy (e.g., ticagrelor or prasugrel) in patients that are poor or intermediate CYP2C19 metabolizers. The US FDA states in a ‘black box’ warning that tests are available to identify patients who are CYP2C19 poor metabolizers, and to consider the use of another platelet P2Y12 inhibitor in patients identified as CYP2C19 poor metabolizers [5]. A ‘black box’ warning is the strictest warning provided by the US FDA to call attention to serious or life-threatening risks. To our knowledge, such contradicting recommendations regarding pharmacogenetics is unique to cardiology, and thus clinicians taking care of the millions of Americans with cardiovascular disease are left without clear guidance on the use of pharmacogenetics for their patients.
It is puzzling that the AHA/ACC recommends against routine genetic testing to guide clopidogrel therapy while recommending other genetic tests that also lack prospective RCT data showing that genetic testing improves outcomes; for example, in patients with hypertrophic cardiomyopathy [6], nonischemic cardiomyopathy [7] and arrhythmia syndromes [7], where the genetic test results are used for similar purposes, such as risk stratification, therapy decisions (e.g., medications and implantable cardiac defibrillator [ICD]) and to inform the family members of the affected patient. The particular case example for which we will evaluate and compare the evidence with the evidence for CYP2C19 genotype-guided clopidogrel therapy is genetic testing in long QT syndrome (LQTS).
The LQTS is a disorder in which the QT interval detected on a patient's electrocardiogram is abnormally long, which can lead to arrhythmias causing syncope and even death. The LQTS can be acquired (caused by certain medications or comorbid conditions) or inherited (caused by genetic mutations). The three most common types of inherited LQTS are LQTS1, LQTS2 and LQTS3, which are distinguished by mutations in the following genes, respectively: KCNQ1, KCNQ2/HERG and SCNA5 [8]. The 2017 AHA/ACC/Heart Rhythm Society (HRS) Guideline for Management of Patients With Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death states: “Genotyping can have therapeutic implications for some arrhythmogenic phenotypes such as long QT syndrome and Fabry's disease, where a monogenic pathogenic mutation has been clearly identified: the risk to mutation positive individuals has been extensively studied and effective therapy relevant to the mutation can be instituted.”
Multiple modes of therapy are used to treat the different genetic forms of LQTS, such as lifestyle interventions (to avoid particular triggers), an ICD, and medications. As we are using genetic testing in LQTS as the case example for comparison with genotype-guided clopidogrel therapy, we only focus on the first-line pharmacologic therapy for different genetic types of LQTS. β-blockers are recommended as first line in all patients with LQTS, but the 2017 AHA/ACC/HRS Guideline for the Evaluation and Management of Patients With Syncope states: “The response to β-blockers depends on the genotype and not all β-blockers are the same. Patients with LQTS1 appear to respond better than patients with LQTS2 and LQTS3 [9].” Obviously the clinical scenarios for LQTS and clopidogrel indications are very different, but LQTS was selected as the cardiovascular case example because the guidelines specifically mention differences in drug response based on patient genotype.
It is also puzzling that the AHA/ACC recommends against pharmacogenetics due to the lack of RCT data, but analogous organizations, such as the National Comprehensive Cancer Network (NCCN), do not. In the absence of RCT data, the NCCN recommends genotype-guided thiopurine and epidermal growth factor receptor (EGFR) inhibitor therapy in patients with acute lymphoblastic leukemia (ALL) and colon cancer, respectively [10,11]. The level of evidence required for the use of pharmacogenetics in clinical practice has been widely debated across therapeutic areas [12–20]. The purpose of this perspective paper is to stimulate discussion on the question: does cardiology hold pharmacogenetics to an inconsistent standard? To address this question, we compared the levels of evidence for the following four genotype-guided pharmacotherapies: clopidogrel for patients that received percutaneous coronary intervention [3], β-blockers for patients with different types of genetic LQTS [9], thiopurines for patients with ALL [11] and EGFR inhibitors for patients with colon cancer [10]. Instead of comparing genotype-guided pharmacotherapy with other factors that are readily available in the electronic medical record (e.g., patient age, renal or hepatic function and drug–drug interactions) [19], we purposefully compared the evidence for pharmacogenetic tests with other pharmacogenetic tests so that any arguments for the potential practical barriers to pharmacogenetic testing would be moot (e.g., high cost, lack of reimbursement, test unavailability, long turn-around time for test results).
Evidence for CYP2C19 genotype-guided clopidogrel for patients that received percutaneous coronary intervention
Several literature reviews on clopidogrel pharmacogenetics have been published [21–23], and the Pharmacogenomics Knowledgebase (PharmGKB®) has curated the available literature [24]. Therefore, an in-depth review of the literature on clopidogrel pharmacogenetics is beyond the scope of this paper. Other genes have been implicated in clopidogrel response (e.g., CES1, ABCB1) [24], but we only focus on CYP2C19 because only CYP2C19 is the focus of the US FDA and CPIC recommendations. Many in vitro, preclinical and clinical studies with surrogate end points support the effects of CYP2C19 genetic variation on clopidogrel response [4], but clinical practice guidelines are based on the highest levels of evidence. Thus, we prioritized studies with the following characteristics, indicating the highest quality of clinical evidence: randomized, controlled, prospective, multicenter, large sample size, clinical outcomes and meta-analyses.
Starting with the gold standard RCTs, one RCT was published prior to the AHA/ACC's recommendation against routine CYP2C19 genotype-guided clopidogrel therapy, and one RCT was published after. The RCT that was published prior to the AHA/ACC's recommendation was a prospective RCT of 600 patients with coronary artery disease undergoing percutaneous coronary intervention (PCI) at a single center in China [25]. Patients were randomized to conventional antiplatelet therapy or CYP2C19 genotype-guided. The primary outcome was a composite of major adverse cardiac or cerebrovascular events, including death from any cause, myocardial infarction, stroke, and ischemia-driven target vessel revascularization, for 180 days. The incidence of the primary outcome was 9.0% in the conventional treatment group, and 2.7% in the CYP2C19 genotype-guided group (p = 0.001). Bleeding events tended to be lower in the CYP2C19 genotype-guided group than in the conventional treatment group (1.3 vs 3.7%; p = 0.073). Thus, despite a prospective RCT demonstrating significantly improved outcomes (and similar bleeding rates) with CYP2C19 genotype-guided clopidogrel therapy, the AHA/ACC still recommended against routine pharmacogenetic testing for clopidogrel.
Another clopidogrel pharmacogenetic RCT has been recently published: the PHARMCLO trial [26]. PHARMCLO was a prospective, multicenter, single-blind RCT conducted in Italy between 2013 and 2015, including 888 patients who were hospitalized due to ST-segment elevation or non-ST-segment elevation acute coronary syndromes. Patients were randomized to CYP2C19 and ABCB1 genotype-guided antiplatelet therapy or the standard of care. The primary outcome was a composite of cardiovascular death and the first occurrence of nonfatal myocardial infarction, nonfatal stroke and Bleeding Academic Research Consortium (BARC) categories three to five defined major bleeding within 12 months of randomization. The trial was stopped prematurely, at only 25% of prespecified enrollment, because of the lack of in vitro diagnosis certification of the genotyping instrument. However, despite only enrolling a fraction of the anticipated sample size, the trial demonstrated a statistically significant benefit for genotype-guided antiplatelet therapy. The primary outcome occurred in 15.9% of patients in the genotype-guided arm and in 25.9%, in the standard of care arm (HR = 0.58; 95% CI: 0.43– 0.78; p < 0.001). Notably, the PHARMCLO trial was published after the AHA/ACC published its recommendation against routine pharmacogenetic testing for clopidogrel, and the AHA/ACC has yet to publish an updated statement including the data from PHARMCLO. Two other RCTs on clopidogrel pharmacogenetics are currently ongoing [27,28], but the trials are not expected to be completed for three more years.
Many large, non-RCT studies of CYP2C19 genotype-guided clopidogrel therapy with clinical outcomes have been performed, including prospective, nonrandomized trials, retrospective analyses of RCTs, and clinical outcomes evaluations of real-world implementations. Starting with the prospective, albeit nonrandomized, trials evaluating CYP2C19 genotype-guided clopidogrel therapy, the data from two trials have been released. The first was the GIANT trial [29]. The results of the GIANT trial have not been published, but the results were presented at 2013 Transcatheter Cardiovascular Therapeutics meeting [29]. The GIANT trial included n = 1499 patients from 53 centers in Europe. In patients that received a stent within 24 h of an acute myocardial infarction, their physicians were given the patients’ CYP2C19 genotype results within 48 h. The decision to adjust the patient's clopidogrel therapy based on CYP2C19 genotype was up to the physician. The rate of the primary end point (death, myocardial infarction or stent thrombosis) at 1 year was similar between patients that were extensive or ultra-rapid CYP2C19 metabolizers treated with standard dosing of clopidogrel (75 mg daily; n = 1118; rate of primary end point = 3%) and patients that were poor or intermediate CYP2C19 metabolizers that had their antiplatelet therapy adjusted by their physician (n = 272; rate of primary end point = 3%; p < 0.0001 for noninferiority). When adjusting clopidogrel therapy in the poor or intermediate CYP2C19 metabolizers, the physicians decided to treat with either prasugrel 10 mg daily or increased the dose of clopidogrel to 150 mg daily. The rate of the primary end point at 1 year was 5-times higher (15%) in the patients, which were poor or intermediate CYP2C19 metabolizers that did not have their antiplatelet therapy adjusted by their physician (n = 55; p = 0.04 vs poor/intermediate CYP2C19 metabolizers in which therapy was adjusted).
The second prospective, nonrandomized trial was a study that included 628 patients with coronary artery disease that underwent successful PCI at a single hospital in China [30]. Patients were consecutively divided into two groups: the routine therapy group and the individualized therapy group. The routine therapy group received conventional dual antiplatelet therapy of aspirin plus 75 mg clopidogrel daily. The individualized therapy group underwent CYP2C19 genotyping, and extensive CYP2C19 metabolizers received clopidogrel 75 mg daily (n = 133), intermediate CYP2C19 metabolizers received clopidogrel 150 mg daily (n = 139) and poor CYP2C19 metabolizers received ticagrelor 90 mg twice daily (n = 37). After adjustment for several clinical risk factors, the rates of major adverse cardiovascular events (death, myocardial infarction and target vessel revascularization) were significantly higher in the routine therapy group compared with the individualized therapy group at 1 month (odds ratio [OR]: 4.92 [1.48–16.29]; p = 0.009), 6 months (OR: 2.48 [1.04–5.92]; p = 0.042) and 12 months (OR: 2.36 [1.10–5.03]; p = 0.027). Therefore, two prospective, nonrandomized trials support CYP2C19 genotype-guided clopidogrel therapy. Importantly, bleeding rates were not significantly higher in the CYP2C19 genotype-guided therapy groups in either trial.
Several retrospective analyses of prospective clopidogrel RCTs have been performed, which led to several meta-analyses of those studies. The results of the meta-analyses were mixed, which prompted Osnabrugge et al. to publish ‘a systematic review and critical assessment’ of the meta-analyses; a somewhat meta-meta-analysis [31]. Osnabrugge et al. evaluated 11 meta-analyses published through August 2013. They found that the results of the meta-analyses were discordant, which was due to variability in the handling of conference abstracts and more recent primary studies, between-study heterogeneity and publication bias. Therefore, Osnabrugge et al. concluded that “personalized antiplatelet management based on genotyping is not supported by the currently available evidence”.
Many argue that the patient population and outcome are the major reasons for the discordant results in the meta-analyses [32,33]. Specifically, CYP2C19 genotype-guided clopidogrel therapy is mostly (or only) important in patients undergoing PCI [34], and thus stent thrombosis should be included in the primary end point. Indeed, Sorich et al. stratified their meta-analyses by PCI [35], and they found that CYP2C19 genotype was only statistically significant in patients that received PCI [35]. PCI was not one of the characteristics that was a focus of the meta-meta-analysis by Osnabrugge et al. [31]; therefore, it is unknown whether the results from Osnabrugge et al. would have differed if they had only included patients that underwent PCI. Moreover, since the publication of the meta-meta-analysis by Osnabrugge et al. [31], two additional meta-analyses have been published [35,36]. Both of those meta-analyses found statistically significant results, showing significantly improved clinical outcomes in patients treated with clopidogrel based on CYP2C19 genotypes. Whether an updated meta-meta-analysis including those two additional meta-analyses would yield different results is unknown.
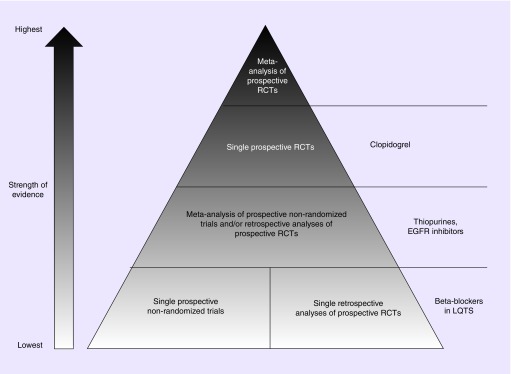
In summary, two prospective RCTs [25,26], two prospective nonrandomized trials [29,30] and two recent retrospective meta-analyses of prospective clopidogrel RCTs support CYP2C19 genotype-guided clopidogrel therapy in patients that underwent PCI. Based on those findings, it is our opinion and the opinion of others [32,33] that the weight of the literature supports CYP2C19 genotype-guided clopidogrel therapy in patients that received PCI. Indeed, even before the PHARMCLO trial was published, institutions began implementing genotype-guided clopidogrel therapy, despite the negative recommendation by the AHA/ACC [37,38], and they also found significantly improved clinical outcomes for their patients [39,40]. We have summarized the highest levels of evidence and recommendations for CYP2C19 genotype-guided clopidogrel therapy in patients that received PCI in Figure 1 and Table 1.
Figure 1. . Comparison of the highest levels of clinical evidence for the four selected pharmacogenetic case examples.
Clopidogrel for patients that received percutaneous coronary intervention; β-blockers for patients with inherited LQTS; thiopurines for patients with acute lymphoblastic leukemia; and epidermal growth factor inhibitors for patients with colon cancer.
LQTS: Long QT syndrome; RCT: Randomized controlled trial.
Table 1. . Comparison of clinical recommendations for the four pharmacogenetic case examples in this article.
| Pharmacogenetic case examples | US FDA | CPIC | Clinical Practice Guidelines |
|---|---|---|---|
| CYP2C19-clopidogrel | √ | √ | X |
| LQTS genotype-β-blockers | √ | ||
| TPMT-thiopurines | √ | √ | √ |
| KRAS-EGFR inhibitors | √ | √ | |
√ = Acknowledges difference in drug response by genotype; X = recommends against the routine use of genetic testing to guide therapy; [blank] = no mention of difference in drug response by genotype or recommendations for genetic testing.
CPIC: Clinical Pharmacogenetics Implementation Consortium; LQTS:L Long QT syndrome.
Evidence for differential β-blocker response among patients with different genetic LQTS types
The primary literature cited by the AHA/ACC/HRS to support differential β-blocker response among patients with different genetic LQTS types includes 14 studies [9]; none of which are prospective RCTs demonstrating that genetic testing for LQTS improves patient outcomes. The data for the four largest studies, (n >1000; largest n = 1648), are all different analyses of the same patient sample (the International LQTS Registry) [41–44]. The largest study that did not come from the International LQTS Registry was an observational study of Italian families (n = 674) [45], but the study did not assess differences in β-blocker effectiveness according to LQTS genotypes. The remaining studies all have n <500, and even two of the studies supporting the AHA/ACC's recommendation only had n = 10 and n = 20. The recommendation by the AHA/ACC that patients with LQTS1 appear to respond better to β-blockers than patients with LQTS2 and LQTS3 is based on only two studies: an observational study of n = 335 total Italian patients [46] and an observational study of n = 1530 patients from the International LQTS Registry [44]. To our knowledge, a true meta-analysis of differential β-blocker response among patients with different genetic LQTS types has not been published. We found only one meta-analysis evaluating the effectiveness of β-blockers in treating the different types of LQTS genotypes [47]. The meta-analysis only included two studies when evaluating β-blocker effectiveness in the different types of LQTS, but those two studies were both derived from the same International LQTS Registry. Thus, we do not agree that the study is a true meta-analysis because the data for the two different studies came from the same patient sample. We have summarized the levels of evidence and recommendations for differential β-blocker response among patients with different genetic LQTS types in Figure 1 and Table 1.
Our goal was to compare the levels of evidence for two different genetic tests in AHA/ACC Guidelines to avoid any discussion of the potential practical barriers that may have influenced the guideline recommendations. In other words, the same practical barriers that would be expected for clopidogrel genetic testing would be the same as for those expected for genetic testing for patients with LQTS. However, we believe that these potential practical barriers are worth noting when comparing these two specific examples, since the practical barriers may actually favor genetic testing for clopidogrel over genetic testing to guide therapy for LQTS. The prevalence of inherited LQTS is estimated to be 1:2000, which is approximately 15-times lower than the number of patients that receive PCI in the USA [48]. Thus, genetic testing to guide clopidogrel therapy could have a wider public health impact than genetic testing for LQTS. Moreover, the yield of a definitive test result for LQTS is lower than for CYP2C19 genotyping for clopidogrel. The three most common types of LQTS (LQTS1, LQTS2 and LQTS3) are only detected in 75% of patients [8]. Thus, for one out of every four patients with LQTS, genetic testing does not yield a useful result. Whereas for clopidogrel, real-world clinical implementation data revealed that 99.6% of CYP2C19 genetic tests yielded definitive results (i.e., poor, intermediate or other CYP2C19 metabolizer categories) [49]. In real-world clinical practice, as many as 29% of patients are poor or intermediate CYP2C19 metabolizers, in which an adjustment from standard clopidogrel therapy is recommended by CPIC [37]. And finally, the distinction of LQTS1, LQTS2 and LQTS3 requires sequencing of three genes, whereas CYP2C19 genotype-guided clopidogrel therapy only requires genotyping a few, well-established loci. In comparison with genotyping, sequencing is typically more expensive, not as widely available and takes longer to complete. Another practical benefit of genotyping CYP2C19 versus sequencing for LQTS is that CYP2C19 genotype can potentially be informative for other aspects of the patient's care. CYP2C19 metabolizes many other drugs in addition to clopidogrel, including several antidepressants and the antifungal voriconazole [50–52]. Notably, genetic testing for LQTS has benefits beyond just guiding pharmacologic therapies, as the type of genetic LQTS also informs decisions regarding lifestyle changes or ICD placement to help prevent future events. In summary, we do not believe that differences in potential practical barriers to pharmacogenetic testing account for the discordance in recommendations for clopidogrel versus LQTS, and in fact, the practical barriers favor genotype testing for clopidogrel.
Evidence for genotype-guided therapies in cancer guidelines
It could be argued that the reason for the discrepancies in recommendations among the AHA/ACC, US FDA and CPIC are the differences in the purposes of the organizations. As a regulatory agency, the US FDA may have a lower bar for creating warnings concerning patient safety. The CPIC may have a lower bar for using genotype-guided drug therapy since it is “an international consortium of individual volunteers and a small dedicated staff who are interested in facilitating use of pharmacogenetic tests for patient care [53]”. Therefore, a fairer comparison may be to compare the AHA/ACC clinical practice guidelines with clinical practice guidelines from an analogous organization, such as the NCCN. Therefore, we summarized the levels of evidence and recommendations for two other pharmacogenetic examples in clinical practice guidelines for cancer: thiopurines for the treatment of patients with ALL and EGFR inhibitors for the treatment of patients with colon cancer.
Genotype-guided thiopurine therapy in patients with ALL
The thiopurines are a class of medications used for a variety of indications, including immunosuppression and malignancies (leukemias and lymphomas). Three thiopurines are currently available: azathioprine, mercaptopurine (6-MP) and thioguanine. 6-MP is predominately used in lymphoid malignancies, and thus we will focus on 6-MP as the case example since we are focusing on the clinical practice guidelines for ALL. The most consistent, dose-related toxicity of 6-MP is myelosuppression, which can manifest as anemia, leukopenia and/or thrombocytopenia, and it can be life-threatening. One of the metabolic pathways for the inactivation of 6-MP involves thiopurine S-methyltransferase (gene: TPMT) [11]. Approximately 0.3% of Caucasian– and African–Americans carry two nonfunctional TPMT alleles, leading to little or no detectable enzyme activity. Approximately 10% of patients carry one nonfunctional allele, leading to low or intermediate TPMT activity [11]. Thus, the treatment of patients carrying one or two nonfunctional TPMT alleles results in higher levels of active metabolites of 6-MP, increasing their risk for myelosuppression. The NCCN recommends the following in the 2017 Guidelines for ALL [11]: “Determination of patient TPMT genotype using genomic DNA is recommended to optimize 6-MP dosing, especially in patients who experience myelosuppression at standard doses.” The NCCN cites two references to support that recommendation: the 2011 and 2013 CPIC Guidelines for TPMT genotype-guided thiopurine dosing [54,55]. The CPIC extrapolated data from clinical studies of the individual thiopurines to all of the thiopurines (because they are all metabolized by TPMT), and they extrapolated data from clinical studies of only a few diseases to all of the indications for which thiopurines are used. The CPIC guidelines for TPMT genotype-guided thiopurine dosing [54,55] and the PharmGKB® [24] summarize the available literature. Thus, like our literature review for clopidogrel, an in-depth review of the literature for TPMT genotype-guided thiopurine dosing is beyond the scope of this paper, but we have summarized the highest levels of clinical evidence.
Most of the primary literature on TPMT and thiopurines consists of small observational studies (n <100) and retrospective case-control studies. The strongest evidence for the association between TPMT genotype and thiopurine toxicity comes from meta-analyses of these studies. The largest meta-analysis to date of genotyped patients (n = 1309) included three prospective cohort and six cross-sectional studies in patients with irritable bowel disease, and they concluded that TPMT polymorphisms were significantly associated with bone marrow toxicity (OR: 5.93, 95% CI: 2.96–11.88; p < 0.00001) [56]. In one of the largest observational prospective studies to date (n = 207), Ansari et al. similarly concluded that heterozygous TPMT genotype strongly predicted myelotoxicity in irritable bowel disease patients given azathioprine without dose adjustment over 6 months (26% homozygous vs 0.5% wild-type TPMT, p < 0.001) [57]. A single, prospective RCT was conducted to assess the clinical utility of prospective TPMT genotyping prior to thiopurine therapy [58], and Newman et al. did not find that TPMT genotyping reduced the incidence of adverse drug reactions. Therefore, despite the fact that the only prospective RCT conducted did not demonstrate that TPMT genotyping improves patient outcomes, the NCCN still recommends TPMT genotype-guided thiopurine therapy in ALL, relative to the weight of all other non-RCT data. We summarized the highest level of evidence and guideline recommendations for TPMT genotype-guided thiopurine therapy in ALL in Figure 1 and Table 1.
Genotype-guided EGFR inhibitor therapy in patients with colon cancer
Another example where clinical practice guidelines for cancer recommend genotype-guided therapy in the absence of prospective RCT data is for EGFR inhibitors (cetuximab and panitumumab) in patients with colon cancer. The 2017 NCCN Guidelines for colon cancer state: “All patients with metastatic colorectal cancer should have tumor tissue genotyped for RAS (KRAS and NRAS) and BRAF mutations. Patients with any known KRAS mutation (exon 2 or nonexon 2) or NRAS mutation should not be treated with either cetuximab or panitumumab.” The KRAS plays a role as a cellular signal transducer in response to the stimulation of cell surface receptors, including EGFR. Approximately 30–50% of colorectal tumors harbor KRAS mutations [59]. The NCCN cited three studies to support their recommendation for KRAS genotype-guided EGFR inhibitor therapy: studies by Lièvre et al. [60], Amado et al. [61] and Douillard et al. [62]. The study by Lièvre et al. was a prospective, observational study of n = 89 patients with metastatic colorectal cancer at six centers in France [60]. The study was a follow-up to their prior study, with similar results, that only included n = 30 patients [63]. They confirmed that patients with mutations in KRAS did not benefit as much from therapy with cetuximab as patients without mutations in KRAS (median progression-free survival = 10.1 vs 31.4 weeks; p = 0.0001). The study by Amado et al. was a retrospective analysis of a Phase III RCT comparing panitumumab monotherapy with best supportive care in patients with chemotherapy-refractory metastatic colorectal cancer (n = 427) [61]. They found that panitumumab was significantly more effective in patients with tumors without KRAS mutations compared with patients with tumors with KRAS mutations (hazard ratio [HR] = 0.45; 95% CI: 0.34–0.59 vs HR = 0.99; 95% CI: 0.73–1.36; p < 0.0001). And finally, the study by Douillard et al. was a retrospective analysis of n = 512 patients in the PRIME trial [62]; a randomized Phase III study of panitumumab plus oxaliplatin, fluorouracil and leucovorin (FOLFOX4) as compared with FOLFOX4 alone in patients with previously untreated metastatic colorectal cancer. They found that the nonexon 2 KRAS mutations were significantly associated with a lack of response to panitumumab in addition to FOLFOX4. We found several meta-analyses supporting the efficacy of EGFR inhibitors in patients with colorectal tumors without KRAS mutations [64–67], including a Cochrane review [68]. We summarized the highest level of evidence and guideline recommendations for KRAS mutation-guided EGFR inhibitor therapy in patients with colorectal cancer in Figure 1 and Table 1.
Discussion
Many barriers have slowed the widespread implementation of pharmacogenetic testing into clinical practice. Many of the practical barriers have been overcome in recent years [37,38], but inconsistencies among clinical recommendations from various organizations is still one of those major barriers. In this perspective paper, our goal was to stimulate conversation to address the question: does cardiology hold pharmacogenetics to an inconsistent standard? We used CYP2C19 genotype-guided clopidogrel therapy in patients that received PCI as a case example, and we compared the highest levels of evidence supporting CYP2C19 genotype-guided clopidogrel therapy with a different genetic test recommended by the AHA/ACC and two pharmacogenetic tests recommended by the NCCN. In these selected case examples, it is our opinion that the level of evidence supporting CYP2C19 genotype-guided clopidogrel therapy in patients that received PCI is at least as strong as the other genetic tests recommended by the AHA/ACC and NCCN. Many others agree with our opinion, as several institutions have implemented pharmacogenetic testing for clopidogrel despite the negative recommendation by the AHA/ACC [37,38].
With the current evidence in the literature, the reason why the AHA/ACC would recommend against routine genetic testing for clopidogrel but state that genetic testing in LQTS has therapeutic implications is unclear. The potential consequences of using clopidogrel in patients with CYP2C19 loss-of-function genotypes can be as serious as using β-blockers in LQTS patients with the least β-blocker responsive LQTS genotypes (e.g., death). Perhaps the AHA/ACC does not believe that a prospective RCT evaluating genetic testing in LQTS would ever be feasible. Inherited LQTS is relatively rare (estimated prevalence 1:2000), and thus it may be more challenging, if not impossible, to recruit a sufficient sample size for an RCT. Therefore, lower levels of evidence are sufficient for the AHA/ACC/HRS to make recommendations for genetic testing in LQTS. Another possibility may be that the AHA/ACC draws connections to similar RCTs that failed to find significant effects. For example, platelet function tests exist to measure clopidogrel effectiveness. However, none of the prospective RCTs evaluating platelet function test-guided antiplatelet therapy showed that testing improved patient outcomes [69]. Another possibility is that the AHA/ACC extrapolated the findings from the COGENT trial [70] to CYP2C19 genotype-guided clopidogrel therapy. The COGENT trial was a prospective RCT of patients with coronary artery disease, treated with clopidogrel alone versus clopidogrel plus omeprazole, a CYP2C19 inhibitor [70]. The COGENT trial did not show a significant difference in clinical outcomes between patients treated clopidogrel with or without a CYP2C19 inhibitor [70], which could possibly be extrapolated to patients treated with clopidogrel and different CYP2C19 genotypes. However the number of patients that received PCI in the COGENT trial was not reported (the indication for which CYP2C19 genotype-guided therapy is most consistent), and the dose of omeprazole used in the trial was only 20 mg daily. In order to have a similar reduction in the activation of clopidogrel that occurs in patients that are CYP2C19 intermediate or poor metabolizers, the dose of omeprazole in the COGENT trial would have had to have been 80 mg [5,71]. Moreover, the COGENT trial was stopped early due to the abrupt cessation of funding from the study sponsor [72]. Therefore, we do not believe that the results of the COGENT trial can be extrapolated to the effects of CYP2C19 genetic polymorphisms on clopidogrel efficacy.
In addition to differential β-blocker response among patients with different genetic LQTS types, the AHA/ACC also recommends genetic testing for patients with hypertrophic cardiomyopathy and nonischemic cardiomyopathy [6,7] in the absence of RCT data demonstrating that genetic testing improves patient outcomes. Perhaps the AHA/ACC views CYP2C19 genetic testing differently than those other genetic tests because CYP2C19 genetic polymorphisms are not directly related to the disease, but only to the response to a drug. Indeed, in the field of pharmacogenetics, we often differentiate pharmacokinetic genetic variants from pharmacodynamic genetic variants [73]. In this case, the genetic test for clopidogrel is purely a pharmacokinetic variant. However, in many cases and perhaps more in the future, pharmacodynamic variants are directly related not only to the response to a drug but also to the underlying disease process, and thus they could be viewed similarly as disease-associated genetic variants [73]. The reasons that the AHA/ACC recommends genetic testing for those diseases are to aid in diagnosis, risk stratification, therapy decisions and to inform the patients’ family members. Despite being a purely pharmacokinetic variant, we believe that CYP2C19 genetic testing can aid with exactly those reasons as well; it is just a matter of perspective. For example, the diagnosis could be: a CYP2C19 poor metabolizer; the risk stratification could be: highest risk of death, myocardial infarction or stent thrombosis after PCI when treated with clopidogrel; and the therapy decision could be: change therapy from clopidogrel to ticagrelor. Family members of CYP2C19 poor metabolizers may also want to know their risk of being a CYP2C19 poor or intermediate metabolizer as well. As previously mentioned, CYP2C19 metabolizes several other drugs [50–52], and thus family members may also want to be informed of their risks for being a poor CYP2C19 metabolizer. The therapy decisions that are made based on CYP2C19 genetic test results may actually be more benign than therapy decisions that are made based on genetic testing for LQTS or other cardiac diseases. For CYP2C19, the therapy decision would simply be changing from one oral P2Y12 inhibitor to another oral P2Y12 inhibitor. Whereas for LQTS, the genetic test results not only affect the decision of whether or not to treat with a β-blocker, but also whether or not to implant an ICD and implement certain lifestyle changes [9].
It is also unclear why the level of evidence required by the AHA/ACC for pharmacogenetic tests, a prospective RCT, is higher than the levels of evidence used by an analogous organization, such as the NCCN. ‘Genetic exceptionalism’ is a well-known phenomenon in clinical practice, in which genetic information is treated differently than nongenetic clinical information [74,75]. In particular, genetic tests are held to a higher standard than nongenetic tests. It does not seem that the AHA/ACC falls victim to genetic exceptionalism, since the AHA/ACC recommends genetic testing for several cardiovascular indications: in patients with hypertrophic cardiomyopathy [6], nonischemic cardiomyopathy [7], and arrhythmia syndromes [7]. However perhaps the AHA/ACC falls victim to a specific sub-type of genetic exceptionalism, in other words, pharmacogenetic exceptionalism, in which the AHA/ACC requires prospective RCTs for pharmacogenetic tests, but not other types of genetic tests. Some speculate that the difference in evidence standards between cancer and cardiology for pharmacogenetics has to do with the availability of intermediate phenotypes for monitoring drug effectiveness [76]. Unlike most cancer pharmacotherapies, cardiovascular pharmacotherapies have intermediate phenotypes or surrogate measures for drug effectiveness that can be quickly and easily measured (e.g., international normalized ratio [INR] for warfarin). However the potential advantage of genetic testing over measuring intermediate phenotypes is that genetic testing can aid in prediction and prevention. This is in contrast to administering a drug and waiting for the patient's response, and potentially placing the patient at risk for an adverse event. Indeed, some warfarin pharmacogenetic RCTs have shown that using a patient's genetics can help the patient get to a therapeutic INR significantly faster [77] and reduce the risk of an INR >4 [78].
Another possible explanation for the different levels of evidence required by the AHA/ACC for pharmacogenetic tests than the NCCN is the increased fear of cancer and chemotherapy over heart disease. Despite heart disease being the number one cause of death in both men and women, cancer is feared more than heart disease [79]. The increased fear of cancer over heart disease has been associated with differences in patients’ attitudes and behaviors and even the prioritization of research funding [79]. Some speculate that the increased amount of research funding for cancer over cardiology has led to more rapid advances in cancer than other fields [76]. Given the well-known adverse effects of chemotherapy (nausea, vomiting and alopecia), patients may also fear the treatments associated with cancer more than the treatments associated with heart disease.
In light of the AHA/ACC recommending other genetic tests over pharmacogenetic tests in the absence of prospective RCTs, one question that needs to be considered is whether the requirement of a prospective RCT for every pharmacogenetic test is even feasible. Requiring a prospective RCT for every pharmacogenetic test could be analogous to requiring a prospective RCT for every drug–drug interaction. Many pharmacogenetic tests are based on variability in cytochrome P450 enzyme function (determined from in vitro and pharmacokinetic studies), and the mechanism of many drug–drug interactions is similarly based on variability in cytochrome P450 enzyme function (also determined from in vitro and pharmacokinetic studies). Like drug–drug interactions, the number of potential pharmacogenetic interactions would require hundreds of prospective RCTs, and the cost would be exorbitant. Currently, there are already ten cardiovascular medications with pharmacogenetic information included in their prescribing information by the US FDA, which would already require ten new prospective RCTs. Clinicians and researchers are beginning to realize the limitations of the prospective RCT requirement. “We cannot answer every question from an RCT…The clinical trials enterprise has gone awry…It is become unnecessarily expensive, cumbersome and arcane…It is inarguable that the system has become so costly and onerous that most of the important questions go unasked,”(quote from Rob Califf, cardiologist, clinical trialist and former the US FDA commissioner) [80]. In the absence of RCT data, when it comes to managing drug–drug interactions in patient care, we err on the side of patient safety and rely on lower levels of evidence. As more and more pharmacogenetic associations are discovered, we may need to similarly err on the side of patient safety and rely on lower levels of evidence for the clinical implementation of pharmacogenetics as well.
The major limitation of this perspective paper is that it was not a quantitative analysis or systematic review. As this is a perspective paper on clinical practice guidelines, we only summarized the highest levels of clinical evidence available for only four selected case examples to demonstrate our perspective; albeit, we cited the literature supporting our statements throughout this paper. The US FDA has incorporated pharmacogenetics into the prescribing information for several cardiovascular drugs, but since the AHA/ACC specifically recommends against the routine use of pharmacogenetic testing for P2Y12 inhibitor therapy, this paper only used clopidogrel as the case example. Some may argue that this perspective is irrelevant because the AHA/ACC recommends other P2Y12 inhibitors (ticagrelor or prasugrel) over clopidogrel for patients with acute coronary syndromes undergoing PCI [81]. However, the trials that compared the other P2Y12 inhibitors with clopidogrel did not use CYP2C19 genotype-guided clopidogrel therapy, and thus it is unknown whether the relative benefit from clopidogrel was diminished by the presence of intermediate and poor CYP2C19 metabolizers in the clopidogrel treatment arm [82]. Moreover, despite the AHA/ACC's recommendation to use other P2Y12 inhibitors over clopidogrel, clopidogrel is still the most widely used P2Y12 inhibitor [83]. Both clopidogrel and prasugrel have generic status, but the cash price for generic prasugrel is still approximately 5-times higher than generic clopidogrel (∼$300 vs 60 per month) [84]. Cost–effectiveness analyses have shown that despite adding on a genetic test, CYP2C19 genotype-guided clopidogrel therapy is still advantageous compared with using the other P2Y12 inhibitors [85]. Another limitation is that we generalized the term clinical ‘recommendation’, as there can be different recommendations regarding ordering a pharmacogenetic test versus using genetic test results that are already available for the patient. The CPIC states the following: “CPIC Guidelines are designed to help clinicians understand how available genetic test results should be used to optimize drug therapy rather than whether tests should be ordered.” And despite the US FDA including CYP2C19 as a black box warning for clopidogrel, the wording of the warning regarding testing does not give straightforward guidance to clinicians, simply stating ‘tests are available’; not whether or not a genetic test should be ordered. The AHA/ACC states “the routine use of platelet function and genetic testing is not recommended,” which also does not distinguish between ordering a genetic test versus using genetic test results that are already available.
In conclusion, it is our opinion that the AHA/ACC holds CYP2C19 genotype-guided clopidogrel therapy to an inconsistent standard, given that the AHA/ACC and NCCN recommend other genetic and pharmacogenetic tests in the absence of prospective RCT data. We do not expect the AHA/ACC to have recommendations consistent with other types of organizations, such as the US FDA or CPIC. However, we do call for consistency among recommendations from within the AHA/ACC, in which they recommend other genetic tests in the absence of RCTs [6,7].
Future perspective
We believe that in 5–10 years we will routinely use a patient's genetic profile to optimize their pharmacotherapy, analogously to how we currently use the myriad of other individual patient factors (e.g., patients’ age, kidney function, liver function, drug–drug interactions) [19]. Like those other factors, we will prioritize patient safety, and implement pharmacogenetics into clinical practice with lower levels of evidence than the gold standard, prospective RCT. We believe that the field will evolve to that new norm due to a combination of the following rapidly evolving factors: the rising cost of RCTs, the declining cost of genotyping, the advancements in health informatics, the number of significant gene–drug interactions being discovered, advancements in the availability of observational ‘big data’ and the technologies and methodologies in order to analyze that data, and the pharmacogenetic testing programs that are already underway at many healthcare systems [37,38].
Executive summary.
Current guideline recommendations by the American Heart Association/American College of Cardiology (AHA/ACC) for pharmacogenetics directly contradict the US FDA and Clinical Pharmacogenetics Implementation Consortium.
The AHA/ACC currently recommends against routine pharmacogenetic testing because no randomized controlled trials (RCTs) have demonstrated that testing improves patients’ outcomes.
However, the AHA/ACC and the National Comprehensive Cancer Network (NCCN) give recommendations for other genetic and pharmacogenetic tests in the absence of prospective RCT data.
Therefore, the purpose of this perspective paper was to compare the levels of evidence for four examples of genetic and pharmacogenetic recommendations by the AHA/ACC and the NCCN.
Body
The highest levels of clinical evidence supporting the following four case examples were summarized and compared: clopidogrel for patients that received percutaneous coronary intervention (not routinely recommended by the AHA/ACC); β-blockers for patients with different types of genetic long QT syndromes (recommended by the AHA/ACC); thiopurines for patients with acute lymphoblastic leukemia (recommended by the NCCN) and EGFR inhibitors for patients with colon cancer (recommended by the NCCN).
The clinical evidence supporting CYP2C19 genotype-guided clopidogrel therapy was stronger than the evidence supporting other genetic and pharmacogenetic tests that are recommended by the AHA/ACC and NCCN.
Conclusion
It is our opinion that the AHA/ACC holds CYP2C19 genotype-guided clopidogrel therapy to an inconsistent standard, given that the AHA/ACC and NCCN recommend other genetic and pharmacogenetic tests in the absence of prospective RCTs.
Footnotes
Financial & competing interests disclosure
JA Luzum was supported by the NIH Loan Repayment Program through the National Heart, Lung, and Blood Institute (L30 HL110279-02) and a New Investigator Award from the American Association of Colleges of Pharmacy. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Smaha LA American Heart Association. The American Heart Association get with the guidelines program. Am. Heart J. 2004;148(5 Suppl.):S46–S48. doi: 10.1016/j.ahj.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Society for Cardiovascular Angiography and Interventions, Society of Thoracic Surgeons, Writing Committee Members et al. ACCF/AHA Clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association. Circulation. 2010;122(5):537–557. doi: 10.1161/CIR.0b013e3181ee08ed. [DOI] [PubMed] [Google Scholar]
- 3.Levine GN, Bates ER, Bittl JA, et al. 2016 ACC/AHA Guideline focused update on duration of dual antiplatelet therapy in patients with coronary artery disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2016;68(10):1082–1115. doi: 10.1016/j.jacc.2016.03.513. [DOI] [PubMed] [Google Scholar]
- 4.Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther. 2013;94(3):317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.US FDA: Clopidogrel Prescribing Information. 2017. www.accessdata.fda.gov/drugsatfda_docs/label/2017/020839s068lbl.pdf
- 6.Gersh BJ, Maron BJ, Bonow RO, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2011;124(24):2761–2796. doi: 10.1161/CIR.0b013e318223e230. [DOI] [PubMed] [Google Scholar]
- 7.Al-Khatib SM, Stevenson WG, Ackerman MJ, et al. 2017 AHA/ACC/HRS Guideline for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Circulation. 2017;1(17):S1547–S1527. doi: 10.1161/CIR.0000000000000549. [DOI] [PubMed] [Google Scholar]
- 8.Tester DJ, Ackerman MJ. Genetics of long QT syndrome. Methodist Debakey Cardiovasc. J. 2014;10(1):29–33. doi: 10.14797/mdcj-10-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen WK, Sheldon RS, Benditt DG, et al. 2017 ACC/AHA/HRS Guideline for the Evaluation and Management of Patients With Syncope: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Circulation. 2017;136(5):e60–e122. doi: 10.1161/CIR.0000000000000499. [DOI] [PubMed] [Google Scholar]
- 10.National Comprehensive Cancer Network (NCCN): Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Colon Cancer. 2017. https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf
- 11.National Comprehensive Cancer Network (NCCN): Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Acute Lymphoblastic Leukemia. 2017. https://www.nccn.org/professionals/physician_gls/pdf/all.pdf
- 12.Gillis NK, Innocenti F. Evidence required to demonstrate clinical utility of pharmacogenetic testing: the debate continues. Clin. Pharmacol. Ther. 2014;96(6):655–657. doi: 10.1038/clpt.2014.185. [DOI] [PubMed] [Google Scholar]
- 13.Ratain MJ, Johnson JA. Meaningful use of pharmacogenetics. Clin. Pharmacol. Ther. 2014;96(6):650–652. doi: 10.1038/clpt.2014.188. [DOI] [PubMed] [Google Scholar]
- 14.Janssens AC, Deverka PA. Useless until proven effective: the clinical utility of preemptive pharmacogenetic testing. Clin. Pharmacol. Ther. 2014;96(6):652–654. doi: 10.1038/clpt.2014.186. [DOI] [PubMed] [Google Scholar]
- 15.Luzum JA, Lanfear DE. Pharmacogenetic risk scores for perindopril clinical and cost effectiveness in stable coronary artery disease: when are we ready to implement? J. Am. Heart Assoc. 2016;4(3):e003440. doi: 10.1161/JAHA.116.003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altman RB. Pharmacogenomics: “noninferiority” is sufficient for initial implementation. Clin. Pharmacol. Ther. 2011;89(3):348–350. doi: 10.1038/clpt.2010.310. [DOI] [PubMed] [Google Scholar]
- 17.Biltaji E, Kumar SS, Enioutina EY, Sherwin CMT. Can ad hoc analyses of clinical trials help personalize treatment decisions? Br. J. Clin. Pharmacol. 2017;83(11):2337–2338. doi: 10.1111/bcp.13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nissen SE. Pharmacogenomics and clopidogrel: irrational exuberance? JAMA. 2011;306(24):2727–2728. doi: 10.1001/jama.2011.1865. [DOI] [PubMed] [Google Scholar]
- 19.Bottorff MB, Bright DR, Kisor DF. Commentary: should pharmacogenomic evidence be considered in clinical decision making? Focus on select cardiovascular drugs. Pharmacotherapy. 2017;37(9):1005–1013. doi: 10.1002/phar.1979. [DOI] [PubMed] [Google Scholar]
- 20.Drozda K, Pacanowski MA. Clinical trial designs to support clinical utility of pharmacogenomic testing. Pharmacotherapy. 2017;37(9):1000–1004. doi: 10.1002/phar.1971. [DOI] [PubMed] [Google Scholar]
- 21.Zeb I, Krim N, Bella J. Role of CYP2C19 genotype testing in clinical use of clopidogrel: is it really useful? Expert Rev. Cardiovasc. Ther. 2018;16(5):369–377. doi: 10.1080/14779072.2018.1459186. [DOI] [PubMed] [Google Scholar]
- 22.Brown SA, Pereira N. Pharmacogenomic impact of CYP2C19 variation on clopidogrel therapy in precision cardiovascular medicine. J. Pers. Med. 2018;8(1) doi: 10.3390/jpm8010008. pii:E8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cavallari LH. Personalizing antiplatelet prescribing using genetics for patients undergoing percutaneous coronary intervention. Expert Rev. Cardiovasc. Ther. 2017;15(8):581–589. doi: 10.1080/14779072.2017.1355236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hewett M, Oliver DE, Rubin DL, et al. PharmGKB: the pharmacogenetics knowledge base. Nucleic Acids Res. 2002;30(1):163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie X, Ma YT, Yang YN, et al. Personalized antiplatelet therapy according to CYP2C19 genotype after percutaneous coronary intervention: a randomized control trial. Int. J. Cardiol. 2013;168(4):3736–3740. doi: 10.1016/j.ijcard.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Notarangelo FM, Maglietta G, Bevilacqua P, et al. Pharmacogenomic approach to selecting antiplatelet therapy in acute coronary syndromes: PHARMCLO trial. J. Am. Coll. Cardiol. 2018;71(17):1869–1877. doi: 10.1016/j.jacc.2018.02.029. [DOI] [PubMed] [Google Scholar]
- 27. Clinicaltrials.gov: Tailoredantiplatelet therapy following PCI (TAILOR-PCI) 2015. https://clinicaltrials.gov/ct2/show/NCT01742117 NCT01742117.
- 28. Clinicaltrials.gov. Cost–effectiveness of genotype guided treatment with antiplatelet drugs in STEMI patients: optimization of treatment (POPular genetics) 2017. https://clinicaltrials.gov/ct2/show/NCT01761786 NCT01761786.
- 29.Chevalier B, Montalescot G, Hulot JS, Belle L, Cayla G GIANT study investigators. CYP2C19 genetic profiling for thienopyridine management after primary percutaneous coronary intervention: results of the GIANT study NCT01134380. Transcatheter Cardiovascular Therapeutics Meeting. 2013 https://www.tctmd.com/slide/giant-prospective-registry-study-cyp2c19-genetic-profiling-thienopyridine-management-after [Google Scholar]
- 30.Shen DL, Wang B, Bai J, et al. Clinical value of CYP2C19 genetic testing for guiding the antiplatelet therapy in a Chinese population. J. Cardiovasc. Pharmacol. 2016;67(3):232–236. doi: 10.1097/FJC.0000000000000337. [DOI] [PubMed] [Google Scholar]
- 31.Osnabrugge RL, Head SJ, Zijlstra F, et al. A systematic review and critical assessment of 11 discordant meta-analyses on reduced-function CYP2C19 genotype and risk of adverse clinical outcomes in clopidogrel users. Genet. Med. 2015;17(1):3–11. doi: 10.1038/gim.2014.76. [DOI] [PubMed] [Google Scholar]
- 32.Shuldiner AR, Vesely MR, Fisch A. CYP2C19 genotype and cardiovascular events. JAMA. 2012;307(14):1482. doi: 10.1001/jama.2012.443. author reply 1484–1485. [DOI] [PubMed] [Google Scholar]
- 33.Mega JL, Topol EJ, Sabatine MS. CYP2C19 genotype and cardiovascular events. JAMA. 2012;307(14):1482–1483. doi: 10.1001/jama.2012.444. author reply 1484–1485. [DOI] [PubMed] [Google Scholar]
- 34.Mega JL, Simon T, Collet JP, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304(16):1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorich MJ, Rowland A, McKinnon RA, Wiese MD. CYP2C19 genotype has a greater effect on adverse cardiovascular outcomes following percutaneous coronary intervention and in Asian populations treated with clopidogrel: a meta-analysis. Circ. Cardiovasc. Genet. 2014;7(6):895–902. doi: 10.1161/CIRCGENETICS.114.000669. [DOI] [PubMed] [Google Scholar]
- 36.Mao L, Jian C, Changzhi L, et al. Cytochrome CYP2C19 polymorphism and risk of adverse clinical events in clopidogrel-treated patients: a meta-analysis based on 23,035 subjects. Arch. Cardiovasc. Dis. 2013;106(10):517–527. doi: 10.1016/j.acvd.2013.06.055. [DOI] [PubMed] [Google Scholar]
- 37.Luzum JA, Pakyz RE, Elsey AR, et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: outcomes and metrics of pharmacogenetic implementations across diverse healthcare systems. Clin. Pharmacol. Ther. 2017;102(3):502–510. doi: 10.1002/cpt.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Empey PE, Stevenson JM, Tuteja S, et al. Multisite investigation of strategies for the implementation of CYP2C19 genotype-guided antiplatelet therapy. Clin. Pharmacol. Ther. 2017 doi: 10.1002/cpt.1006. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cavallari LH, Lee CR, Beitelshees AL, et al. Multisite investigation of outcomes with implementation of CYP2C19 genotype-guided antiplatelet therapy after percutaneous coronary intervention. JACC Cardiovasc. Interv. 2018;11(2):181–191. doi: 10.1016/j.jcin.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee CR, Sriramoju VB, Cervantes A, et al. Clinical outcomes and sustainability of using CYP2C19 genotype-guided antiplatelet therapy after percutaneous coronary intervention. Circ. Genom. Precis Med. 2018;11(4):e002069. doi: 10.1161/CIRCGEN.117.002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Locati EH, Zareba W, Moss AJ, et al. Age- and sex-related differences in clinical manifestations in patients with congenital long-QT syndrome: findings from the International LQTS Registry. Circulation. 1998;97(22):2237–2244. doi: 10.1161/01.cir.97.22.2237. [DOI] [PubMed] [Google Scholar]
- 42.Jons C, Moss AJ, Goldenberg I, et al. Risk of fatal arrhythmic events in long QT syndrome patients after syncope. J. Am. Coll. Cardiol. 2010;55(8):783–788. doi: 10.1016/j.jacc.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 43.Liu JF, Jons C, Moss AJ, et al. Risk factors for recurrent syncope and subsequent fatal or near-fatal events in children and adolescents with long QT syndrome. J. Am. Coll. Cardiol. 2011;57(8):941–950. doi: 10.1016/j.jacc.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abu-Zeitone A, Peterson DR, Polonsky B, McNitt S, Moss AJ. Efficacy of different β-blockers in the treatment of long QT syndrome. J. Am. Coll. Cardiol. 2014;64(13):1352–1358. doi: 10.1016/j.jacc.2014.05.068. [DOI] [PubMed] [Google Scholar]
- 45.Priori SG, Schwartz PJ, Napolitano C, et al. Risk stratification in the long-QT syndrome. N. Engl. J. Med. 2003;348(19):1866–1874. doi: 10.1056/NEJMoa022147. [DOI] [PubMed] [Google Scholar]
- 46.Priori SG, Napolitano C, Schwartz PJ, et al. Association of long QT syndrome loci and cardiac events among patients treated with β-blockers. JAMA. 2004;292(11):1341–1344. doi: 10.1001/jama.292.11.1341. [DOI] [PubMed] [Google Scholar]
- 47.Ahn J, Kim HJ, Choi JI, et al. Effectiveness of β-blockers depending on the genotype of congenital long-QT syndrome: a meta-analysis. PLoS ONE. 2017;12(10):e0185680. doi: 10.1371/journal.pone.0185680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim LK, Feldman DN, Swaminathan RV, et al. Rate of percutaneous coronary intervention for the management of acute coronary syndromes and stable coronary artery disease in the United States (2007 to 2011) Am. J. Cardiol. 2014;114(7):1003–1010. doi: 10.1016/j.amjcard.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 49.The Pharmacogenomics Knowledgebase (PharmGKB®): implementation resources for pharmacogenomics. 2017. https://www.pharmgkb.org/page/pgxImplementationResources
- 50.Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 2015;98(2):127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharmacol. Ther. 2016 doi: 10.1002/cpt.597. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moriyama B, Obeng AO, Barbarino J, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP2C19 and voriconazole therapy. Clin. Pharmacol. Ther. 2016 doi: 10.1002/cpt.583. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clinical Pharmacogenetics Implementation Consortium (CPIC) 2018. https://cpicpgx.org/
- 54.Relling MV, Gardner EE, Sandborn WJ, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin. Pharmacol. Ther. 2011;89(3):387–391. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Relling MV, Gardner EE, Sandborn WJ, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin. Pharmacol. Ther. 2013;93(4):324–325. doi: 10.1038/clpt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong XW, Zheng Q, Zhu MM, Tong JL, Ran ZH. Thiopurine S-methyltransferase polymorphisms and thiopurine toxicity in treatment of inflammatory bowel disease. World J. Gastroenterol. 2010;16(25):3187–3195. doi: 10.3748/wjg.v16.i25.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ansari A, Arenas M, Greenfield SM, et al. Prospective evaluation of the pharmacogenetics of azathioprine in the treatment of inflammatory bowel disease. Aliment. Pharmacol. Ther. 2008;28(8):973–983. doi: 10.1111/j.1365-2036.2008.03788.x. [DOI] [PubMed] [Google Scholar]
- 58.Newman WG, Payne K, Tricker K, et al. A pragmatic randomized controlled trial of thiopurine methyltransferase genotyping prior to azathioprine treatment: the TARGET study. Pharmacogenomics. 2011;12(6):815–826. doi: 10.2217/pgs.11.32. [DOI] [PubMed] [Google Scholar]
- 59.Kafatos G, Niepel D, Lowe K, et al. RAS mutation prevalence among patients with metastatic colorectal cancer: a meta-analysis of real-world data. Biomark Med. 2017 doi: 10.2217/bmm-2016-0358. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lievre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J. Clin. Oncol. 2008;26(3):374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 61.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26(10):1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 62.Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N. Engl. J. Med. 2013;369(11):1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 63.Lievre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 64.van Helden EJ, Menke-van der Houven van Oordt CW, Heymans MW, Ket JCF, van den Oord R, Verheul HMW. Optimal use of anti-EGFR monoclonal antibodies for patients with advanced colorectal cancer: a meta-analysis. Cancer Metastasis Rev. 2017;36(2):395–406. doi: 10.1007/s10555-017-9668-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwak MS, Cha JM, Yoon JY, et al. Prognostic value of KRAS codon 13 gene mutation for overall survival in colorectal cancer: direct and indirect comparison meta-analysis. Medicine. 2017;96(35):e7882. doi: 10.1097/MD.0000000000007882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ridouane Y, Lopes G, Ku G, Masud H, Haaland B. Targeted first-line therapies for advanced colorectal cancer: a Bayesian meta-analysis. Oncotarget. 2017;8(39):66458–66466. doi: 10.18632/oncotarget.20185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Therkildsen C, Bergmann TK, Henrichsen-Schnack T, Ladelund S, Nilbert M. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: a systematic review and meta-analysis. Acta Oncol. 2014;53(7):852–864. doi: 10.3109/0284186X.2014.895036. [DOI] [PubMed] [Google Scholar]
- 68.Chan DLH, Segelov E, Wong RS, et al. Epidermal growth factor receptor (EGFR) inhibitors for metastatic colorectal cancer. Cochrane Database Syst. Rev. 2017;6:CD007047. doi: 10.1002/14651858.CD007047.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montalescot G, Range G, Silvain J, et al. High on-treatment platelet reactivity as a risk factor for secondary prevention after coronary stent revascularization: a landmark analysis of the ARCTIC study. Circulation. 2014;129(21):2136–2143. doi: 10.1161/CIRCULATIONAHA.113.007524. [DOI] [PubMed] [Google Scholar]
- 70.Bhatt DL, Cryer BL, Contant CF, et al. Clopidogrel with or without omeprazole in coronary artery disease. N. Engl. J. Med. 2010;363(20):1909–1917. doi: 10.1056/NEJMoa1007964. [DOI] [PubMed] [Google Scholar]
- 71.Mega JL, Close SL, Wiviott SD, et al. Cytochrome P-450 polymorphisms and response to clopidogrel. N. Engl. J. Med. 2009;360(4):354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 72.Wood S. Heartwire from Medscape: COGENT 1 trial scrapped, sponsor declares bankruptcy. 2009. https://www.medscape.com/viewarticle/587213
- 73.Pasternak AL, Ward KM, Luzum JA, Ellingrod VL, Hertz DL. Germline genetic variants with implications for disease risk and therapeutic outcomes. Physiol. Genomics. 2017;49(10):567–581. doi: 10.1152/physiolgenomics.00035.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Relling MV, Altman RB, Goetz MP, Evans WE. Clinical implementation of pharmacogenomics: overcoming genetic exceptionalism. Lancet Oncol. 2010;11(6):507–509. doi: 10.1016/S1470-2045(10)70097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Green MJ, Botkin JR. “Genetic exceptionalism” in medicine: clarifying the differences between genetic and nongenetic tests. Ann. Intern. Med. 2003;138(7):571–575. doi: 10.7326/0003-4819-138-7-200304010-00013. [DOI] [PubMed] [Google Scholar]
- 76.Kranzler HR, Smith RV, Schnoll R, Moustafa A, Greenstreet-Akman E. Precision medicine and pharmacogenetics: what does oncology have that addiction medicine does not? Addiction. 2017;112(12):2086–2094. doi: 10.1111/add.13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pirmohamed M, Burnside G, Eriksson N, et al. A randomized trial of genotype-guided dosing of warfarin. N. Engl. J. Med. 2013;369(24):2294–2303. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 78.Gage BF, Bass AR, Lin H, et al. Effect of genotype-guided warfarin dosing on clinical events and anticoagulation control among patients undergoing hip or knee arthroplasty: the GIFT randomized clinical trial. JAMA. 2017;318(12):1115–1124. doi: 10.1001/jama.2017.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vrinten C, Wardle J. Is cancer a good way to die? A population-based survey among middle-aged and older adults in the United Kingdom. Eur. J. Cancer. 2016;56:172–178. doi: 10.1016/j.ejca.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schattner E. Dr. Robert Califf shares ideas about real-world evidence and health data. Forbes. 2017 https://www.forbes.com/sites/elaineschattner/2017/06/28/dr-robert-califf-shares-ideas-about-real-world-evidence-and-health-data/#26df20d14eb3 [Google Scholar]
- 81.O'Gara PT, Kushner FG, Ascheim DD, et al. 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2013;127(4):e362–e425. doi: 10.1161/CIR.0b013e3182742cf6. [DOI] [PubMed] [Google Scholar]
- 82.Wallentin L, Becker RC, Budaj A, et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N. Engl. J. Med. 2009;361(11):1045–1057. doi: 10.1056/NEJMoa0904327. [DOI] [PubMed] [Google Scholar]
- 83.Fan W, Plent S, Prats J, Deliargyris EN. Trends in P2Y12 inhibitor use in patients referred for invasive evaluation of coronary artery disease in contemporary US practice. Am. J. Cardiol. 2016;117(9):1439–1443. doi: 10.1016/j.amjcard.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 84.Estimated cash prices of generic clopidogrel and generic prasugrel. 2018. www.goodrx.com
- 85.Plumpton CO, Roberts D, Pirmohamed M, Hughes DA. A systematic review of economic evaluations of pharmacogenetic testing for prevention of adverse drug reactions. Pharmacoeconomics. 2016;34(8):771–793. doi: 10.1007/s40273-016-0397-9. [DOI] [PubMed] [Google Scholar]