Abstract
The relative contribution of intrinsic genetic factors and extrinsic environmental ones to cancer aetiology and natural history is a lengthy and debated issue. Gene–environment interactions (G x E) arise when the combined presence of both a germline genetic variant and a known environmental factor modulates the risk of disease more than either one alone. A panel of experts discussed our current understanding of cancer aetiology, known examples of G × E interactions in cancer, and the expanded concept of G × E interactions to include somatic cancer mutations and iatrogenic environmental factors such as anti-cancer treatment. Specific genetic polymorphisms and genetic mutations increase susceptibility to certain carcinogens and may be targeted in the near future for prevention and treatment of cancer patients with novel molecularly based therapies. There was general consensus that a better understanding of the complexity and numerosity of G × E interactions, supported by adequate technological, epidemiological, modelling and statistical resources, will further promote our understanding of cancer and lead to novel preventive and therapeutic approaches.
Facts
Gene–environment interaction indicates that combination of a genetic and an environmental factor modulates the risk of cancer more than either one alone.
Lifetime risk of cancers of many different types was shown to strongly correlate with the total number of divisions of the normal self-renewing stem cells.
Specific genetic polymorphisms and mutations increase susceptibility to certain carcinogens.
p53 is a prototype genetic factor for cancer, it can be somatically or inheritably mutated, as well as subjected to polymorphism.
All carriers of inherited BAP1 mutations have developed one or more cancers during their lifetime. Exposure to asbestos and UV-light further increases their risk of developing mesothelioma, melanoma and skin cancers: GXE interaction.
Questions
Why environmental carcinogens cause cancer only in a fraction of exposed individuals?
Alterations in TP53 and BAP1 genes both predispose to cancer, but with a different spectrum. What are the underlining molecular determinants of this specificity?
Will single cell analysis resolve the difficulties in diagnosis and study associated to tumour heterogeneity?
Can we design prophylactic or therapeutic anti-cancer approaches based on genetic of polymorphisms and mutations?
The origin of cancer: studies clash over ‘intrinsic’ versus ‘extrinsic’ risk
Hypotheses on the origin of cancer are integral part of the history of medicine. The first three evidences supporting the presence of extrinsic chemical risk factors date back to the XVIII century: John Hill’s record on tobacco snuff and nasal cancer; Percivall Pott’s association between exposure to soot and scrotal cancer; Samuel von Sömmerring’s observation of a relationship between lip carcinoma and clay-pipe smoking [1]. Recognition of the existence of intrinsic genetic risk factors is more recent, and originated from analyses of familial clustering of cancer cases and is experimentally supported by numerous efforts to generate murine models of hereditary cancers [2]. Epidemiological studies have demonstrated that environmental carcinogens cause cancer only in a fraction of exposed individuals; the biological reasons are often unknown.
Taking advantage of the extremely large amount of data generated in the last 20 years, bioinformatics represents a powerful tool to investigate both the interactions between genes and environment in determining cancer risk and the origin of cancer itself. In the last few years, a great interest has been raised on this topic by a series of articles. Tomasetti & Vogelstein’s in 2015 reported that lifetime risk of cancers of many different types is strongly correlated with the total number of divisions of the normal self-renewing stem cells maintaining tissues’ homoeostasis [3], thus supporting a prevalent ‘bad-luck’ intrinsic origin of cancer. Wu et al. instead proposed that the overwhelming majority of cancers develop following exogenous damage, with only ~ 10–30% of the risk attributable to intrinsic factors, thus supporting the ‘toxic insults’ theory [4]. These studies provide correlations, without entering into causal molecular mechanisms, but the message of these papers could not be in more strident contradiction, with the effect of igniting a public debate on this very hot topic.
Unique occurrences of cancer epidemics in remote regions of the world have provided definitive proof for the gene X environment hypothesis. In particular, studies of an epidemic of mesothelioma in Cappadocia, Turkey, where over 50% of the population exposed to carcinogenic erionite fibers dies of mesothelioma, the most devastating cancer epidemic ever recorded in medicine, revealed that susceptibility to mesothelioma was transmitted in a Mendelian fashion and that the cause of the epidemic was gene–environment interaction [5–7].
Expanding the concept of gene–environment interaction (G × E)
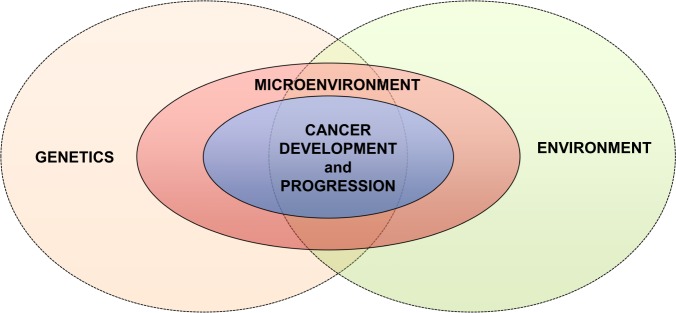
In the complex picture of cancer pathogenesis and progression, it is now clear that both external environmental and genetic factors affect the so-called microenvironment, i.e. the local cellular milieu where cancer develops, which has a critical role in determining cell fate (Fig. 1).
Fig. 1.
Gene–environment and cancer pathogenesis. Both genetics and environment interact to affect the microenvironment from which ultimately causes cancer to develop in the first place and then progress
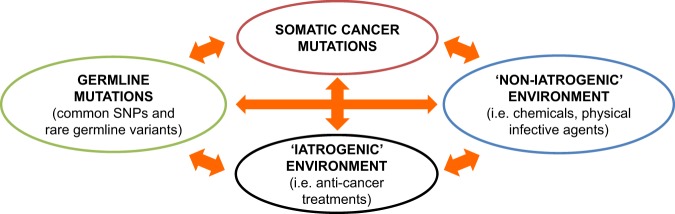
Gene–environment interactions arise when the combined presence of both a genetic and an environmental factor modulates the risk of cancer more than either one alone. The traditional definition of G × E interaction most often implies ‘intrinsic’ genetic mutations (i.e. polymorphisms and rare germline mutations) and ‘non-iatrogenic’ environmental factors. Moreover, in cancer patients two additional levels of complexity arise: ‘acquired’ genetic mutations and ‘acquired iatrogenic’ environmental factors (e.g. drugs, radiotherapy) contribute to the cancer phenotype (Figs 2 and 3). Understanding the mutual relationships between all these factors is crucial both from a scientific and from a clinical perspective.
Fig. 2.
The complexity of gene–environment interactions. Including somatic cancer mutations and anti-cancer treatments further increases the layers of gene–environment interactions, providing us with the opportunity to increase our understand the of complexity of the cancer phenomenon
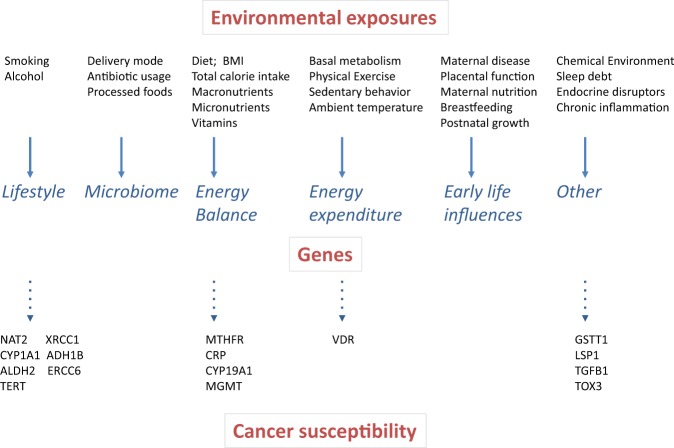
Fig. 3.
Environmental pressure and cancer susceptibility. A number of extrinsic factors associated to lifestyle, early-life influence or pre-existing chronic conditions exerts pressure on cancer insurgence. The genetic background, including somatic mutations, germline mutations and single nucleotide polymorphism contributes to the outcome of the impact of these environmental factors on the organism. Some extrinsic factors can also promote alteration of the genetic information producing mutagenic events that trigger cancer
The International Weinman Conferences, held at the University of Hawaii Cancer Center, Honolulu, HI, in 2017 (January 26, 27 and November 30–December 1) were conceived to stimulate discussions among experts from different fields of cancer research on complex inter-relations between genetics, external environment and microenvironment relevant to cancer aetiology—thus trying to find common ground in the lengthy conflict between ‘bad-luck’ and ‘toxic insults’, and to cancer progression and therapy. Here, we describe exemplificative topics on G × E interactions in both solid and haematologic malignancies presented by the authors, and the conclusions they reached following the discussions.
Genome-wide association studies, environment and molecular biology
Despite the unclear contribution of each single factor, it is commonly accepted that the risk of developing cancer is determined by a complex interplay of both genetic and environmental factors [8–10]. There are many well-established environmental and lifestyle risk factors for various cancers, including smoking, alcohol, ionizing and solar radiations, exposure to carcinogenic fibres present in the environment, such as asbestos and erionite, as well as occupational exposures to these same fibres, infectious agents, obesity, and physical inactivity, which together are thought to account for up to 60% of cancer deaths in the United States [11]. However, only a small fraction of individuals exposed to these risk factors develop cancer, suggesting the co-existence of intrinsic genetic susceptibility.
Recent genome-wide association studies (GWAS) and next-generation sequencing studies have identified many common and rare germline genetic variations that contribute to increased cancer risks. Many of these cancer susceptibility loci are located in or near genes involved in DNA repair, or epigenetic regulators of gene transcription; however several examples exist of variants in genes involved in carcinogen metabolism. A paradigmatic example is the interaction between N-acetyltransferase 2 (NAT2) gene polymorphisms and smoking in elevating bladder cancer risk. NAT2 is a phase II detoxification enzyme that catalyses the metabolic inactivation of aromatic amines and heterocyclic amines, constituents of cigarette smoking and other environmental exposure. NAT2 polymorphisms allow classification of the human population into rapid, intermediate and slow ‘acetylator’ phenotypes. Numerous population studies and a meta-analysis [12] confirmed that NAT2 slow acetylators, resulting in slow detoxification of tobacco carcinogens, have a 1.4-fold increased risk of bladder cancer, and identified a significant synergistic interaction between NAT2 genotype and smoking: individuals with the NAT2 slow acetylating genotype who are also heavy smokers exhibited the highest risk of developing bladder cancer [12]. Another eminent example is represented by the interaction between aldehyde dehydrogenase (ALDH2) gene variants, alcohol metabolism and oesophageal cancer [13].
Based on these and other data, there is a clear need for so-called ‘gene–environment-wide association studies’ which take into account environmental factors together with genetic variations to explain complex traits and phenomena [14]. Moreover, while in selected cases such as those represented by NAT2 and ALDH2, it is possible to infer the mechanistic link between the genetic variants and the consequent molecular alterations, a further step is needed to improve our understanding of G × E interactions from a molecular perspective, as shown by recent efforts in this direction [15].
Taking advantage of unique epidemiological resource
The study of G × E interaction requires the capability of discerning as much as possible the contribution of each of the two elements to the observed phenotype. Compared to small families and close relatives, which often share environment and lifestyle besides genes, such interactions can be more clearly elucidated studying distant relatives and high-risk pedigrees.
A unique resource that combines extensive genealogy with cancer and exposure data, the Utah Population Data Base (UPDB), provides the opportunity to better understand the contribution of genes, of the environment, and their combined effects to cancer. The UPDB represents the genealogy of the Utah Mormon pioneers and their descendants. The original Utah genealogy included 1.6 million individuals linked in genealogies 6–7 generations deep [16], and has been expanded from the 1970s with relationship data from Utah Vital Statistics records. State-wide SEER cancer data since 1966 for over 300,000 individuals have been linked to this database. The UPDB today represents 7 million Utah residents, of which 3 million have 3–16 generations of data. The power of the UPDB for G × E interaction studies lies in the simultaneous record of exposure data (such as tobacco use or radon exposure, both available in the UPDB), data for cancer incidence, and data on genetic relationships.
Health-related predisposition genes can be identified, and G × E interactions can be elucidated in high-risk pedigrees using these powerful genealogical resources. Analysis of UPDB high-risk pedigrees identified BRCA1, BRCA2, and CDKN2A as cancer predisposition genes [17–19], and studies of environmental exposures in individuals sharing genetic predisposition variants have clarified G × E interactions in cancer [20–22].
Similar efforts in creating large databases including both environmental and genetics data are steadily increasing in number, as they provide invaluable opportunities to study the complexity of cancer. Among the most recent examples, are the Iceland cohort [23], the Swedish Family-Cancer Database [24], and the US Veterans Genealogy Project [25].
The case of malignant mesothelioma and BAP1
Malignant mesothelioma (MM) is often used as the “standard” example of a cancer caused by G × E interaction. Epidemiologically, MM is an aggressive polyclonal cancer [26] associated with occupational and/or environmental exposure to tumorigenic mineral fibres, such as asbestos, erionite, or other non-regulated asbestos-like fibres [27–30]. However, “only” ~ 5% of asbestos workers exposed to asbestos continuously for over 10 years developed MM, suggesting that monogenic or polygenic genetic predisposition also contributes to MM risk [27]. Therefore, inherited genetic variants in genes encoding for proteins involved in the DNA damage response and/or in the inflammatory response may likely modulate the risk of asbestos-induced MM. Once they reach the mesothelium, asbestos fibres cause mesothelial cell death and chronic inflammation, associated with the release of damage-associated molecular patterns (DAMPs) molecules and cytokines [27]. Among these DAMPs, the active and passive release of different isoforms of high-mobility group box 1 (HMGB1) protein [31] sustains the chronic inflammatory response, with secretion of TNF-α and reactive oxygen species (ROS) [32, 33] (Fig. 4). Moreover, asbestos fibres can also directly induce ROS production because of the iron they contain, which behaves as a catalyst for free-radical generation [34, 35]. These molecules in turn activate NF-κB, leading to the survival of mesothelial cells that have accumulated genetic damage [32]. Accordingly, anti-inflammatory drugs and HMGB1 inhibitors impair MM growth in vitro and in experimental animal models and could represent prophylactic and therapeutic approaches [36–38].
Fig. 4.
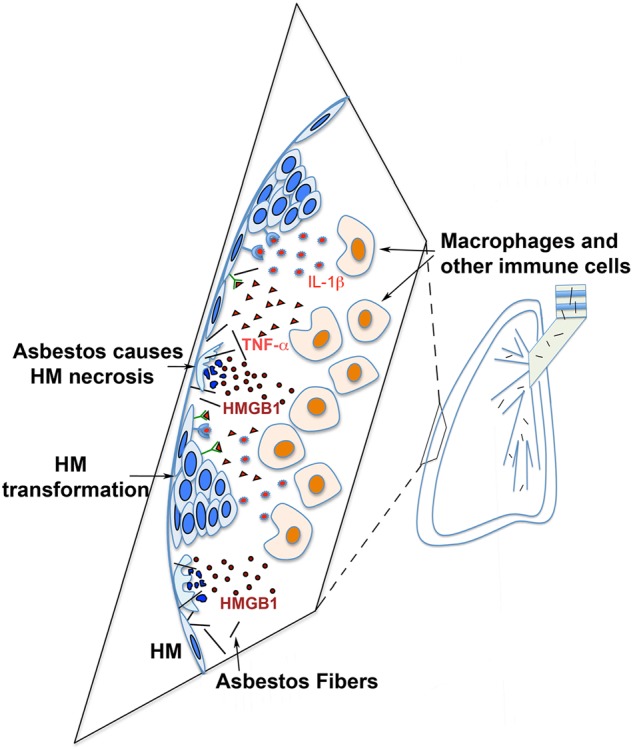
Asbestos pathogenesis leads to mesothelioma. Asbestos fibers travel to pleura and cause human mesothelial cells (HM) to die of necrosis, leading to the release of HMGB1 into the extracellular space. HMGB1 induces macrophages and other inflammatory cells to accumulate and triggers the inflammatory response and leads to the secretion of cytokines such as TNF-α and IL-1β, which stimulate survival signalling pathways that increase survival of asbestos-damaged HM. This allows key genetic alterations to accumulate within HM that sustain asbestos-induced DNA damage that over time may lead to HM transformation and mesothelioma development
BRCA1-associated protein 1, BAP1 is the most frequently mutated gene in MM, as about 2/3 of sporadic cases—i.e., MM not developing in carriers of germline BAP1 mutations—carry somatic BAP1 mutations [39, 40]. Besides single nucleotide variations and other intragenic alterations, frequently MM have large copy number variations and micro-deletions reminiscent of chromothripsis. Most common are minute deletions (ranging between 100 bp and 3000 bp) in chromosome band 3p21 in genes encoding for epigenetic modifiers such as BAP1 and others, e.g. SETD2, SCAP, SMARCC1, PBRM1 [39]. Because BAP1 mutations are very rare in lung cancer, the presence or absence of wild-type BAP1, easily detected as presence or absence of nuclear BAP1 immunostaining helps pathologists differentiate MM from lung cancer [41]. A better characterization of the most common molecular modifications in MM should pave the way to novel and more specific therapies [42].
In this respect, germline BAP1 mutations significantly increase the risk of MM, even among individuals not exposed to asbestos [43, 44]. Intriguingly, patients with MM who also carry germline BAP1 mutations have prolonged survival [45].
How to reconcile the environmental and the genetic factor in MM pathogenesis? The incidence of germline BAP1 mutations is low among sporadic MM patients, suggesting that other yet unknown genetic mutations might contribute to MM risk [46]. Based on the experimental evidence in mice, germline BAP1 mutations significantly increase the risk of asbestos-induced MM after exposure to low levels of asbestos, levels that rarely cause MM in wild-type mice [47, 48]. Germline BAP1 mutations are not only associated with MM, but also with a wider recently identified cancer predisposition syndrome which includes cutaneous and uveal melanoma, squamous and basal cell carcinoma (all associated to sunlight exposure) as well as to clear cell renal cell carcinoma (ccRCC) a malignancy not yet associated with a specific carcinogen. Moreover, although less frequent, carriers of germline BAP1 mutations develop also several other malignancies, such as breast cancer, cholangiocarcinoma, sarcomas, brain tumors, etc., suggesting that BAP1 mutations increase the overall cancer risk although those associated to environmental carcinogens predominate [49].
The case of clear cell renal cell carcinoma and BAP1
BAP1 is commonly mutated at the somatic level in ccRCC, and is one of two genes that underlie the first molecular genetic classification of ccRCC [50]. Mutations in BAP1 (found in ~ 15% of ccRCC) tend to anti-correlate with mutations in PBRM1 (found in ~ 50% of ccRCC). Both BAP1 and PBRM1 are two-hit tumour suppressor genes and they reside on chromosome 3p. Chromosome 3p harbours the VHL tumour suppressor gene, which is mutated (or silenced) in the majority of ccRCC. VHL mutation is an initiating event and this mutation is followed by loss of 3p, which eliminates the second copy of VHL as well as one copy of BAP1 and PBRM1 [51]. In mice, inactivation of VHL is insufficient for tumour development, but RCC develops if VHL and either BAP1 or PBRM1 are targeted [52]. In humans, PBRM1-deficient tumours tend to be of low grade and BAP1-deficient tumours are typically of high grade (and associated with poor survival), and this is reproduced in mice [52]. These findings provide insight into the mechanism of renal cancer development and pave the way for tailored treatment approaches.
Besides BAP1 [53] renal cancers can be associated with germline mutations in a variety of genes, such as VHL, FH and genes encoding for the different subunits of the succinate dehydrogenase [54]. It is also notable, that some families with BAP1 germline mutations appear to be almost exclusively affected by renal cancer [53, 55]. However, compared to other cancer types, the interactions between genetic and environmental factors in the pathogenesis of renal cancer is less understood. Recent studies have proposed G × E interactions in the aetiology of renal cancer between specific loci and consumption of cooked meat, which translates into higher intake of dietary mutagenic polycyclic aromatic hydrocarbons/heterocyclic amines [56]. Understanding the interplay between these environmental factors and germline mutations predisposing to renal cancer is an expanding area of research [57].
p53 family members in cancer predisposition and development: between macro- and micro-environment
The most well studied tumour suppressor in cancer development is TP53, which encodes the tumour suppressor protein p53 [58–64]. Mutations in the p53 effector pathway significantly influence the relative incidence of tumour development and progression [65–67]. Indeed, p53 is the most frequently mutated protein among all human cancers, being mutated in over 50% of cancers [68, 69]. Notably, TP53 mutations and polymorphisms are often associated with the emergence of oncogenic “gain–of–function” activities by the protein product, which further contribute to cancer development. For example, the expression of mutant forms of p53 (missense mutations) affects cellular motility and invasion through several pathways, often involving the other members of the p53 family [70–75]. These evidences are derived from both somatic mutations and rare inherited mutations occurring in the Li-Fraumeni cancer syndrome [66].
Moreover, recent bioinformatics analyses of whole genome data showed that frequent germinal isoforms of the TP53 gene have a profound effect on the relative incidence of many cancers.
Also distinct single nucleotide polymorphisms (SNPs) of TP53 are strongly associated with cancer development, making the entire p53-pathway very relevant to cancer development [76]. TP53 mutations are a potent ‘intrinsic’ driver of cancer development. However, TP53 variants have also emerged in G × E interactions studies. For example, TP53 polymorphisms appear to modulate human papillomavirus (HPV)-induced cancer risk, through differential targeting of the p53 variant proteins by the E6 viral oncoprotein [77].
The p53 family, which includes p53, p63 and p73, exerts a number of critical biological functions, such as regulating cellular processes critical for development, homeostasis and more importantly tumour suppression. Some of these functions are shared by all members of the p53 family, others are exclusive functions of specific members [78–80]. The canonical pathway for activation of p53 family members follows the DNA damage insults. However, a revised model for p53 family suggests that several types of insults, such as hypoxia, nutrient fluctuation, oxygen radicals, etc., lead to activation of these proteins and regulation of a wide range of biological processes [81]. This makes the p53 family particularly relevant in interaction with the environment. For example, G × E interactions have been proposed between TP73 polymorphisms and smoking and obesity in modifying lung cancer risk [82].
The molecular connections between specific genetic variants and functional consequences are still unclear. We know that, similarly to the best-characterised member p53, both p73 and p63 are transcribed as two distinct isoforms TA-isoforms and ΔN-isoforms, containing, or not, the N-terminal transactivation domain [81]. Both p63 and p73 are involved in female infertility maternal reproduction [83] and in cancer development and progression. For example, ΔNp63 might control cancer development through regulation of Sonic Hedgehog signalling [84] and regulation of the metabolism via Hexokinase II [85]. Conversely, in vivo experiments have shown a tumour suppressor role for TAp73 and an oncogenic role for ΔNp73, whose deletion is associated with neurological phenotype [86, 87]. Moreover, recently, the transcription factor TAp73 has been demonstrated to oppose hypoxia-induced factor (HIF)-1 activity promoting its polyubiquitination and consequent proteasomal degradation, and thus affecting tumour angiogenesis [81, 88].
It is still unclear whether additional genetic polymorphisms and/or environmental factors might modulate the ratio of transcribed mRNA and/or transduced protein of the different isoforms of the p53 family. A deeper understanding of these processes will be beneficial for preventing negative outcome in the interaction between genes and environment and for improving pharmaceutical intervention for cancer and toxicity-associated disease.
The p53 family also regulates the interaction between cells and their microenvironment through regulation of cellular metabolism, promoting oxidative metabolism, glutaminolysis and serine biosynthesis, thus promoting GSH production to buffer oxidative stress [89–95]. The key function that the p53 family exerts in control of redox status represents a pivotal contribution to interaction with environmental stressors and this can be expanded beyond the tumour protective role of this family of genes [96]. The influence of p73 and p53 on serine biosynthesis and one-carbon metabolism not only represents a barrier against tumour transformation, but could also influence the response of cancer cells to anti-metabolic drugs that target enzymatic activities of these pathways [97–99].
An additional layer of complexity in the interaction of p53 family with the microenvironment is added by the recent discovery of an influence of p53 on the mevalonate pathway. The mevalonate pathway has been implicated in multiple aspects of tumour biology, including growth, proliferation, recurrence and metastasis [100]. Mutant p53 can upregulate expression of mevalonate pathway genes, possibly through interaction with SREBP-2, the key transcriptional factor of many sterol biosynthesis genes, and this determines a susceptibility of the mevalonic pathway to the p53 status in the cancer cells [74]. One of the important outputs of the mevalonate pathway is RhoA geranylgeranylation which can lead to stabilized mutant p53 [101]. Interestingly, statins also destabilize mutant p53, although the mechanism appears to involve counteracting DNAJA1 interaction with mutant p53 [102]. Statins target the activity of the HMG-CoA reductase, the rate-limiting enzyme in the mevalonate pathway. These compounds, widely used for treatment of hypercholesterolemia, have been reported to be protective for certain cancers [103, 104]. Pre-clinical studies indicate that targeting mevalonic pathway, alone or in combination, may offer designed therapeutic options for patients bearing p53 mutant tumours.
A full characterisation of the interaction between genetic landscape and response to treatment targeting metabolic pathways would be desirable to improve therapeutic efficacy in cancer patients.
Regulation of IP3 receptors through oncogenes and tumor suppressors
IP3 receptors (IP3Rs) are IP3-activated Ca2+ release channels located on intracellular Ca2+ stores mainly at the endoplasmic reticulum (ER), and play critical roles in the regulation of the Ca2+ release from the store [105–107]. Recently IP3Rs have been found to have pivotal roles in the regulation of cell death and survival by controlling Ca2+ transfer from ER to mitochondria. There are three isoforms in human and rodents, IP3R1, IP3R2 and IP3R3, with different tissue distributions and different IP3 binding affinity [108–113]. IP3Rs are huge tetrameric channels with each monomer composed of about 2700 amino acids. IP3Rs bind several proteins that influence IP3Rs activities. IP3Rs are divided into five domains: The N-terminal suppressor domain, the IP3 binding domain, the regulatory/coupling domain, the channel domain with six transmembrane regions, and the C-terminal gatekeeper domain IRBIT pseudo ligand of IP3 binds to the IP3-binding core. The C-terminal gatekeeper domain is targeted by various regulatory proteins including Bcl-2, Bcl-XL, Mcl-1, cytochrome c, K-Ras4B, Akt, Huntingtin, BRCA1, GIT, protein and 4.1 N and 80K-H [114–123].
The tumor suppressors PTEN and BAP1 regulate IP3R stability and intracellular Ca2+ dynamics, and increase apoptosis sensitivity of cancer cells [124, 125]. X-ray crystallographic and mutagenesis analyses, together with functional analyses, have revealed a leaflet structure in the large regulatory cytosolic domain that is critical in transmitting the IP3-dependent global conformational change to the channel domain [126].
IP3Rs at ER-mitochondria contact sites
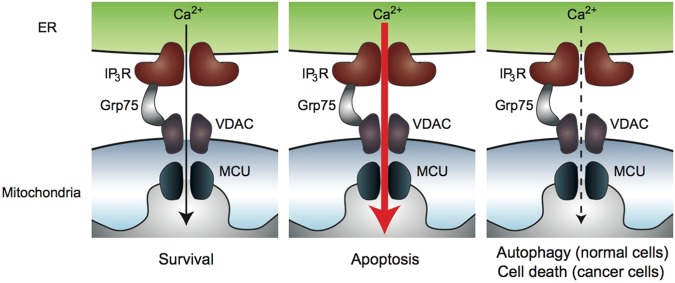
The localization of IP3Rs in specialized microdomain of ER called mitochondria-associated ER membranes (MAMs) is critical in the regulation of cell death and survival. Mitochondrial Ca2+ uptake systems needs close proximity to high Ca2+ microdomains of IP3R-mediated Ca2+ release sites [127–129]. MAMs are the physical contact site between ER and mitochondria where Ca2+ transfer and exchange of phospholipids takes place [128, 130–133]. IP3Rs form complexes with voltage-dependent anion channel 1 (VDAC1) located in the mitochondrial outer membranes through a chaperon protein Grp75 [134, 135]. Ca2+ released from IP3Rs flows into mitochondria through VDAC and mitochondrial Ca2+ uniporter (MCU) located in the mitochondrial inner membranes [136, 137]. Ca2+ transfer from ER to mitochondria through MAMs regulate cell death and survival in cancer cells (Fig. 5). IP3R-mediated Ca2+ release plays pivotal roles in apoptosis pathway [138–141]. Excessive Ca2+ influx into mitochondria causes opening of mitochondrial permeability transition pore that results in inhibition of oxidative phosphorylation, production of reactive oxygen species (ROS), and loss of mitochondrial membrane integrity [142]. This leads to the release of pro-apoptotic proteins such as cytochrome c from mitochondria into cytosol. Cytochrome c triggers the sequential activation of caspases, resulting in apoptotic cell death. Cytochrome c also interacts with the IP3R and increases its activity, causing positive feedback amplification of apoptotic signalling [117, 143]. Suppression of the IP3R activity by Bcl-2 prevents excessive Ca2+ transfer into mitochondria and suppresses apoptosis. Disruption of the interaction between IP3R and Bcl-2 has therapeutic potentials against cancer [144, 145].
Fig. 5.
Ca2+ transfer from ER to mitochondria at the MAM sites. Ca2+ released from ER through IP3Rs flows into mitochondria matrix through VDAC in the outer mitochondrial membranes and MCU in the inner mitochondrial membranes. Optimal levels of Ca2+ transfer to mitochondria is necessary for mitochondrial metabolism and energy production (left). Excessive Ca2+ flux into mitochondria results release of pro-apoptotic proteins into cytosol, and results in apoptosis (middle). Suppression of basal mitochondrial Ca2+ uptake induces reduction in ATP production and autophagy in normal cells. Autophagy is not enough for survival of cancer cells (right) [155]
IRBIT, a new sensor for the environment interaction: novel opportunities in the tumorigenesis and apoptosis
IRBIT was discovered as a molecule that binds to the IP3-binding pocket of IP3R [146, 147]. In contrast, an IRBIT homologue Long-IRBIT does not interacts with IP3R1 because its long N-terminal appendage inhibits the interaction [148, 149]. IRBIT is a multifunctional protein that regulates diverse proteins including Na+-HCO3- co-transporter, CFTR, Na+-H+ exchanger [150], CaM kinase IIα [151], and PIP kinases [152]. The N-terminal intrinsically disordered protein region of IRBIT plays critical roles in its specific target recognition. Therefore, IRBIT works as a nexus of cell signaling. In addition, a member of Bcl-2 family proteins, a group of proteins that control the apoptosis, has been revealed to associate with IRBIT [153]. Bcl2l10, one of the anti-apoptotic proteins belonging to the Bcl-2 family, binds to a part of the IP3-binding domain distinct from the IP3 binding pocket. Bcl2l10, anti-apoptotic factor, and IRBIT interact and exert an additive inhibition of IP3R. The interplay between IRBIT and Bcl2l10 is involved in regulation of apoptosis. These proteins associate in a complex in MAMs for Ca2+ transfer between ER and mitochondria. When apoptotic stress is added, IRBIT is dephosphorylated and released off with Bcl2l10 from the IP3Rs thus the enhancement of Ca2+ transfer from ER to mitochondria occurs and promotes apoptosis. In addition IRBIT was reported to suppress ribonucleotide reductase in higher eukaryotes [154] working as a tumor suppressor. These data strongly suggest that IRBIT as a new regulator of cell death [155]
Mechanisms: how genetic alterations induce susceptibility to specific carcinogens
Specific genes appear to have more prominent roles in different tissue and they influence susceptibility to specific carcinogens, ensuing in a specific spectrum of different cancer types that characterize the different cancer syndromes. Accordingly, some genetic mutations cause certain tumour types, some instead are associated with a wider spectrum of cancers. For example, the Lynch syndrome, the most common among cancer syndromes, can be caused by mutations of several genes, MLH1, MSH2, MSH6, PMS2, and EPCAM, and it is characterized by a prevalence of colorectal carcinomas (lifetime risk varies between 40–80% depending on the underlying condition), although other tumour types may develop. In this cancer syndrome, genetic mutations and chronic inflammation work in concert in causing malignancy, although the exact mechanisms remain to be elucidated. BRCA1 and BRCA2 mutations lead mostly to an excess of breast and ovarian carcinomas (lifetime risk is ~ 50%) while other tumour types are also observed, but less frequently. TP53 mutations lead to the Li-Fraumeni cancer syndrome, characterized by an excess of almost any cancer type, particularly breast cancers (in women), leukaemias, sarcomas, and adrenocortical carcinomas. BAP1 mutations have also a wide spectrum of tumour types, as they give rise to the BAP1 cancer syndrome characterized by an excess of mesothelioma, eye and skin melanomas, all types of cutaneous carcinomas, renal cell carcinomas and sarcomas, while other cancer types are rare. To date, all known members of families affected by the BAP1 cancer syndrome who inherited BAP1 mutations have developed one or more cancers in their lifetime [46].
Studies of the BAP1 cancer syndrome have elucidated how reduced BAP1 levels increase susceptibility to environmental carcinogens. BAP1 is a deubiquitylase that regulates DNA repair by homologous recombination [156, 157]. It was found that BAP1 has a dual nuclear and cytoplasmic activity. In the cytoplasm BAP1 deubiquitylates and stabilizes the IP3R3 channels, the channels that control the release of Ca2+ from the endoplasmic reticulum to the mitochondria, resulting in increased intra-mitochondrial Ca2+ levels and consequent apoptosis. Exposure of normal cells derived from donors carrying inherited heterozygous germline BAP1 mutations to asbestos, UV light, and ionizing radiation-induced increased DNA damage in BAP1+/− cells, compared to normal cells; the damage could not be properly repaired because of the reduced levels of nuclear BAP1 and impaired DNA repair. Reduction (+/−) or absence (−/−) of BAP1 in the cytoplasm lead to a significant reduction of IP3R3 receptor channels due to the unopposed activity of the ubiquitin ligase and consequent degradation of the IP3R3 receptor by the proteasome. Thus, although these cells accumulate mutations, they cannot execute apoptosis due to reduced Ca2+ levels in the mitochondria. This resulted in significantly increased cellular transformation of BAP1+/− cells compared to control wild-type cells following exposure to these environmental carcinogens, an effect that was reversed upon transfection of wild-type BAP1 into mutant cells [125].
Malignant growth is facilitated by the additional discovery that “normal” primary cells from individuals carrying germline BAP1 mutations derive energy through aerobic glycolysis and thus display a set of metabolic alterations known as “Warburg effect” which to date has been considered a hallmark of cancer cells. Thus, the Warburg effect does not always occur as an adaptive process that follows carcinogenesis, but may also predate by many years, and probably facilitate, carcinogenesis. Therefore, cells from BAP1 mutation carriers do not need to go through a selection process that favours the emergence of clones with a Warburg effect required for tumor growth: BAP1+/− cells constitutionally derive a large part of their metabolism through aerobic glycolysis. Thus when these cells develop mutations they are already primed for cancer growth [158].
In summary, BAP1 modulates Ca2+-dependent apoptosis and cell death via deubiquitylation of IP3R3 and, by regulating IP3R3 stability, and BAP1 also regulates cellular metabolic pathways. The cytoplasmic BAP1 activities, in concert with nuclear BAP1, modulate the high incidence of cancer in BAP1+/− carriers (100% cancer incidence during lifetime) and increase BAP1 null cancer cells resistance to chemotherapy [159].
Recent studies have elucidated how BRCA1 mutations cause breast and ovarian carcinomas (85). It was found that the interaction of BRCA1 with its partner BARD1 is necessary to recruit the exact genetic sequence needed to repair breaks in DNA caused by endogenous stress and environmental insults such as radiation exposure.
Tumour heterogeneity as the basis for acquired resistance in non-small cell lung cancer and glioblastoma: anti-cancer treatments as novel environmental interactors
Non-small cell lung cancer (NSCLC) is one of the most complex malignancies, largely due to tobacco-induced carcinogenesis. In NSCLC, TP53 mutations are very frequent, and the involvement of G × E interactions and p53 mutation spectrum in human lung cancer has long been studied [160]. Studies of G × E interactions in NSCLC have also identified other SNPs modulating the risk of tobacco-induced lung (and other) cancers [161–163]. Besides its association with tobacco, NSCLC is also well–recognized for a variety of treatable single driver oncogene-related subsets. Most prominent are activating mutations in the epidermal growth factor receptor (EGFR) and gain of function fusions in the anaplastic lymphoma kinase (ALK) or ROS1 genes, all of which are treatable with the corresponding tyrosine kinase inhibitors (TKIs) [164, 165]. Moreover, NSCLC is characterized by a remarkable degree of inter- and intra-patient tumour heterogeneity [166].
Tumour heterogeneity contributes to difficulty in initial diagnostic testing and in determining mechanisms of de novo and acquired resistance after oncogene-targeted therapies. In the subset of patients with the most treatable NSCLC (EGFR-, ALK- and ROS1-driven) response rates in each of these categories approximate 60–70% at most [164]. In the remainder of cases, de novo (primary) resistance is present, despite harbouring identical sensitizing mutations or fusions as in the responding cases. In the case of EGFR–mutated lung cancer, randomized trials against chemotherapy have demonstrated both higher response rates and improved disease control rates [167–171]. However, despite clear benefits for TKI therapy over chemotherapy in these settings, the eventual emergence of acquired resistance and progressive disease is universal. Furthermore, complete responses, typically thought of as the first step toward cure, are rarely achieved even with these targeted therapies.
Glioblastoma (GBM) is another prototype of multiclonal, highly heterogeneous tumour in which activating mutations or amplifications of Receptor Tyrosine Kinases (RTKs), such as EGFR, have major roles in the development and progression of the malignancy. Notably the degree of heterogeneity in GBM includes also key genetic alternations, such as the oncogenic EGFR variant III (EGFRvIII) [172, 173]. Oncogene amplification in extrachromosomal DNA (ecDNA) is a recently discovered mechanism adopted by GBM to maintain the cell-to-cell genetic variability that drives tumour progression and drug resistance [174]. Amplification of driver oncogenes in ecDNA results in increased transcript level. Mathematical modelling predicted that ecDNA amplification increases oncogene copy number variation compared to chromosomal amplification and effectively favours intratumoural heterogeneity and plasticity. Dynamic modulation of ecDNA context provides a powerful ability to the tumour to readapt to, and survive, oncogene-targeting therapies.
Recently, improved methodologies for distinguishing mechanisms of resistance, such as next-generation sequencing (NGS) of tumour tissue and/or plasma cell-free DNA, have become commercially available and clinical applicability has been established [166, 175, 176]. Further, these tools have been incorporated into large-scale clinical trials employing NGS in order to match patients with genotypic therapies targeting abnormalities within their tumours, such as the Lung-MAP study (NCT02154490). Moreover, determination of secondary mutations and/or bypass track mechanisms of acquired resistance are transforming the therapeutic landscape and directly influencing treatment decision-making [176]. The implementation of these techniques at the level of single cell resolution will allow a full characterization of the events associated with tumour heterogeneity and plasticity.
The rapid translation of new technologies into the clinic offers the potential for truly transformative approaches to cancer therapy consistent with the goals of personalized or precision medicine. In order to accomplish this, a deeper understanding of the relational complexity among germline genetic variants, acquired cancer mutations, ‘non-iatrogenic’ environmental factors and ‘iatrogenic’ environmental factors such as anti-cancer treatments is required.
Carbon source metabolism in acute myeloid leukaemia: gene × micro-environment interactions
G × E interaction studies have been proposed in some haematological malignancies [177]. Despite the absence of well defined G × E interactions, recent findings indicate that acute myeloid leukaemia (AML) may represent a valid study case for G × micro-E interactions, and help design novel specific therapies.
There are a variety of AML subtypes and several gene mutations are involved in the leukaemogenesis process. These include class I mutations which induce cellular proliferation, class II mutations which compromise normal differentiation [178], and a third class of genes encoding epigenetic modifiers, such as DNMT3A, IDH1, IDH2, and TET2 [178, 179]. Recent studies showed that these mutations are critically linked to aberrant metabolic phenotypes in AML pathogenesis and progression [180, 181]. A hypoxic microenvironment promotes the metabolic transformation to glycolytic phenotype of normal non-proliferating cells via the HIF-1 pathway, and progression of AML has been linked to the expansion of hypoxia in the subendosteal bone marrow niche relative to normal bone marrow [182]. AML cells display a significantly enhanced glycolytic metabolism [183], which was linked to poor survival in patients via contribution to Ara-C resistance in a clinical metabolomics study [181]. Increased glycolytic flux in AML cells accelerates glucose consumption and leads to glucose insufficiency in the bone marrow. Such a low-glucose microenvironment triggers a metabolic reprogramming in AML cells, with substantially increased expression of the fructose transporter, GLUT5 (SLC2A5), and thus, the ability of utilizing fructose to promote tumour cell growth relative to normal cells [184]. Fructose is the second most abundant blood sugar in human beings with a physiologically normal range of 0.5-1.0 mM [185, 186]. Fructose has previously been reported to alter the glycan structures on the cell surface of tumour cells and increase their proliferative and invasive properties when compared to glucose in vitro [187].
These evidences demonstrate that G × micro-E interactions play a critical role in accommodating energy stress and programming a new metabolic phenotype with enhanced fructose utilization in AML, leading to exacerbated leukemogenesis, as well as poor clinical outcomes regardless of the concentration of glucose present. Targeting the fructose pathway may lead to novel specific therapies for AML [184].
Obesity, metabolic syndrome and cancer
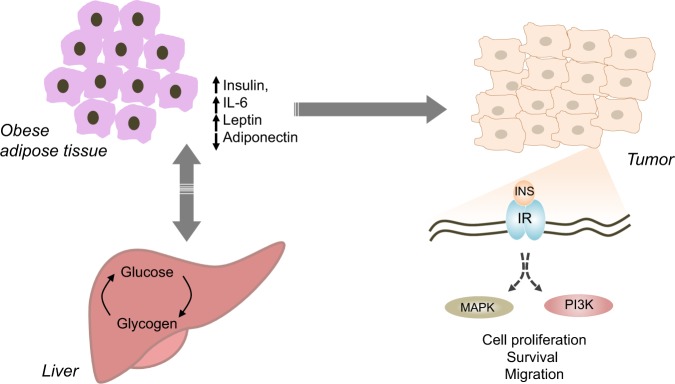
Lifestyle influences type and extent of exposure to extrinsic factors (Fig. 3). Reduced physical activity and high caloric intake have been recognised as risk factors for several malignancies [188]. In Western countries obesity is an epidemic, with one in three adult classified as obese. Despite epidemiological evidence indicating a clear relationship between obesity and oncologic conditions; the underlying molecular aspects determining the interaction between genetic predisposition, lifestyle, and metabolism have not been elucidated.
Adipose tissue contributes to maintenance of energetic balance in the organism engaging a complex network of systemic signalling that coordinate nutrient homeostasis. The excess of lipid storage alters the physiological metabolic balance of central and peripheral organs promoting constitutive signalling that communicates nutrient excess. The result of this process can include chronic hyperinsulinemia and aberrant adipokine signalling that eventually result in lower physiological defences against tumour development and progression. Hyperinsulinemia correlates with higher tumour incidence in humans [189, 190], and mouse studies have demonstrated that a high level of insulin triggers tumour progression in models such as c-Myc driven breast cancer [191]. Obesity can directly influence tumour growth, providing excess of energy and, also directly sustaining mitogenic signalling. The phosphatidylinositol 3-kinase (PI3-K) pathway is a component of the cellular insulin signalling transduction, physiologically contributing to an increase in glucose uptake in fat, liver and muscle cells by activating transcription, translation and membrane translocation of glucose transporter [192, 193]. Hyperactivation of insulin signalling leads to an unbalanced PI3-K pathway that in turn sustains aberrant cellular proliferation influencing cancer cell properties (Fig. 6) [194]. As a consequence the mutational status of this pathway in cancer cells enhances the ability of serum insulin to stimulate tumour growth, thereby playing a critical role in the response of the tumour to the obese status [195]. PI3-K inhibitors acutely induce insulin resistance in liver and muscle and result in elevation in serum glucose, and consequent elevation in serum insulin. Therapies that lower serum glucose and insulin, especially a ketogenic diet, could conceivably improve responses to PI3-K inhibitors, even in tumours carrying no mutations in the pathway.
Fig. 6.
Obesity and hyperinsulinemia influence cancer development and progression. Accumulation of adipose tissue in obesity systemically alters metabolic homeostasis, affecting endocrine signalling between organs and leading to condition such as chronic hyperinsulinemia. Insulin activates signalling pathways, including PI3-K and MAPK, which are generally required to sustain cancer cells. Overall these mechanisms might both contribute to reduction of physiological barriers against tumour ormation and promote cancer progression
There is substantial evidence in support of an implication of obesity in human cancer development [188], however it is still difficult to define a clear causative connection between the events. Many aspects associated with obesity, such as hyperinsulinemia, inflammation, endocrine signalling, have been independently associated to cancer [196]. It will be important to define whether obesity per se represents a risk factor or the hypercaloric diet, and whether the composition of the diet is associated with increased susceptibility to cancer.
Myelodysplastic syndromes, haematological malignancies and post-transcriptional control of gene expression
Chronic lymphocytic leukaemia (CLL) is the most common human leukaemia. Contributing environmental risk factors have not yet been identified yet; however, genetic predisposition appears very likely [197]. The loss of microRNA (miR)-15/16 on human chromosome 13q14 is the most common genetic alteration in CLL [198]. Physiologically, miR-15/16 regulates the expression of BCL2 [199], which plays an important part of the p53 signalling pathway, and that when overexpressed causes follicular cell lymphoma and CLL. Loss of miR-15/16 leads to BCL2 overexpression and CLL.
In addition to loss of miR-15/16 in CLL, deregulation of expression and processing of microRNAs has been observed in cancer. During transformation, cancer cells acquire clusters of enhancers, called super-enhancers (SEs). This class of regulatory elements is able to drive higher levels of transcription than typical enhancers. The acquisition of SEs boosts expression of oncogenes, favouring the establishment of the tumour phenotype. SE constituents act cooperatively and facilitate Drosha/DGCR8 recruitment and pri-miRNA processing to boost cell-specific miRNA production. During cancer progression SEs form at the miRNAs genes, enabling a co-transcriptional Drosha/DGCR8-dependent microRNA processing [200, 201].
Myelodysplastic syndromes (MDS) and leukaemia present a relatively high frequency of mutations in genes encoding proteins that function in, or regulate, splicing of mRNA precursors [202]. Cancer cells take advantage of deregulated mRNA splicing to produce aberrant proteins with added, deleted, or altered functional domains that contribute to malignancy, or else splicing errors can lead to protein loss-of-function. CRISPR/Cas9 introduction of the MDS-associated P95H mutation in the serine/arginine-rich splicing factor 2 (SRSF2) in K562 leukemia cells influences about 1% of total splicing events, very similar to the extent of splicing deregulation in MDS patients with the same mutation. Mutant SRSF2 does not lose its functionality but rather binds more tightly than its wild-type counterpart to RNA sites containing UCCAG while binding less tightly to UGGAG sites, thereby increasing or decreasing, respectively, inclusion of target exons [203]. Hence, mutations of spliceosomal genes contribute to the cancer phenotype, resulting in deregulated or aberrant splicing of specific mRNA precursors.
Precision medicine and targeted therapies
It may take a very long time for a research finding in the laboratory to make its way to the patient. For example, it took 32 years from Dr. Croce’s discovery of the critical role of the BCL2 gene in CLL to FDA approval in 2016 of an anti-Bcl2 drug for CLL. The anti-Bcl2, BH3 mimic, ABT199 or venetoclax, induces massive lysis of CLL cells: 80% of CLL patients respond to the drug with 85% showing responses lasting over 1 year, including complete remissions without evidence of minimal residual disease [204, 205].
Conventional cancer therapy kills cancer cells by causing DNA damage. However cancer cells are often deficient at least in one of the DNA repair mechanisms. Profiling the DNA repair pathways in patients might help in predicting radiation/chemotherapy responsiveness and therefore would help in designing personalised treatments. The Fanconi Anemia Gene D1, BRCA2, participates in homologous recombination (HR) repair and it is also linked to familial breast and ovarian cancer [206]. BRCA2-deficient epithelial ovarian cancers (EOCs) contain upregulated DNA polymerase θ (Polθ), a DNA polymerase, which promotes a compensatory Alternative end joining (Alt-EJ) DNA break repair. Synthetic lethality in mice between the Polθ and HR pathway and hypersensitivity of HR-deficient tumour to Polθ inhibition indicate a potential novel therapeutic approach for cancer with HR-inactivation, such as familial BRCA2 [207]. A successful example of exploiting synthetic lethality in cancer therapy is the development of inhibitors of poly ADP-ribose polymerases (PARPs), which selectively kill cancer cells with defective DNA repair, especially in breast and ovarian cancer patients with mutations in BRCA1 and BRCA2 [208].
While DNA repair deficiency contributes to cancer development by causing more genetic mutations, such deficiency can be harnessed for cancer immunotherapy because these mutations are expected to generate more neoantigens for recognition by the immune system. For example, patients with mismatch repair deficiency showed dramatic responses to PD-1 antibody treatments [209]. Such dramatic results have led to the recent approval by FDA of immune checkpoint blockade therapies for any solid tumors based on molecular diagnosis of mismatch repair deficiency (microsatellite instability) rather than tumor types.
The design of targeted therapies often clashes with the difficulty of targeting novel protein complexes, but computational tools have been developed to identify druggable binding sites. In the human proteome the number of potentially druggable protein–protein interactions is extremely wide and largely exceed the human kinome. The generally large, flat, and relatively featureless binding
sites of protein complexes make drug design very difficult by conventional approaches. Methods that integrate conventional methods to identify druggable areas at the protein surfaces together with sequence co-evolution tools used to identify protein–protein surfaces are now making it possible to overcome this challenge [210]. Success of this approach has been demonstrated on how the NEET family, a relatively new class of three related [2Fe-2S] proteins (CISD1-3), important in human health and disease, particularly cancer [211]. Protein levels of NAF-1 (CISD2), mitoNEET (CISD!) are elevated in human epithelial breast cancer cells, and their suppression by shRNA results in significantly reduced cell proliferation and tumour growth [212]. Structural studies and computational approaches led to identification of a small molecule named MAD-28 as an inhibitor of NAF-1 and mitoNEET. MAD-28 has been proven to selectively kill breast cancer cells, without apparent effect on normal epithelial cells and represents a potential alternative anti-cancer approach [213].
Conclusion
On the lengthy issue of cancer origin, with the ‘bad-luck’ vs ‘toxic insults’ contest, it should be considered that the exact evaluation of the number of stem cells and the mathematical model used are crucial points [3]. In fact, the number of stem cells may well vary during life, and their evaluation has been accounted for using different methods that are not directly comparable. On the other hand, if we accept that extrinsic risk factors are the main drivers of cancer, we should question the yet unidentified extrinsic risk factors that drive the cancer types for which such factors have not been identified [4].
The initiating events should be further investigated, for example spontaneous mutations and stem cell markers in different tissues at different ages. Indeed, despite a stable balance between the degree of DNA damage and the subsequent repair being maintained throughout life, there is a constant accumulation of genetic mutations with age in human adult stem cells, with a rate of circa 40 mutations/year, mostly unrelated to the large variation of cancer incidence in a specific tissue [214]. On the other hand, human cancers show very different rates and patterns of somatic mutations, which could be categorised in 21 general signatures [215]. An unbiased forward genetic approach using a mouse mutagenic program, along the lines as the one developed by Dr. Beutler that led to the identification of the Toll-like receptors (TLRs) as well as many other molecules with non-redundant function in the immune response, may help us identify mutations responsible for both quantitative and qualitative phenotypes to understand the mechanism by which certain mutations contribute to cancer development [216].
A landmark advance to evaluate the lifelong susceptibility to cancer has been allowed by the use of selective Cre-recombination of conditional lineage tracing in stem cells of mouse organs. By using CreER2-recombinase and LacZ from the endogenous Prom1 (Cd133) locus, activated with a tamoxifen-induced Rosa-ZsGreen fluorescence protein lineage tracing allele, Zhu et al. showed that tumour incidence is determined by the lifelong generative capacity of mutated cells, rather than the initiating mutation or the proliferative capacity: a combination of stem cell mutagenesis and extrinsic factors leading to a ‘perfect storm’ from which cancer arises [217]. A mutation analysis shows that only 18% of mutations occurred in oncogene/tumour suppressors, whilst 82% of mutations were in other genes, including genes regulating immune-surveillance and epigenetic modifiers consistent with random mutation theory [217].
A recent paper by Tomasetti et al. tried to clarify the contribution of random replication errors, environmental factors and heredity, emphasising the difference between aetiology of cancer mutations and cancer prevention [218]. These authors proposed that most cancers are preventable, as mutations dependent on environmental factors –even though numerically less prevalent than those dependent on random replication errors or hereditary—still contribute to cancer development, and their absence would result in prevention of a large number of cancer cases.
The current hypothesis is that while intrinsic random DNA errors are the most frequent type of mutations in all tissues and cells, including stem-cells, development of cancer is often facilitated and accelerated by the combinatory effect of these “inevitable” errors—as cells accumulate about 3 new mutations/cell division, together with those caused by exposure to mutagenic environmental carcinogens and hereditary mutations.
Acknowledgements
The meeting was made possible by a generous donation from the Barry and Virginia Weinman Foundation and by the generous support of the International Association for the Study of Lung Cancer (IASLC). M.C. research is supported by 1R01CA198138-01; DoD Translational Team Award; DoD Idea Award; University of Hawaii Foundation (through the Melohn Endowed Chair in Cancer Biology and Genetics; and through unrestricted donations from Honeywell International and from UNITED-FOR-A-CURE); H.Y research is supported by U01CA214195-01; DoD Translational Team Award; DoD Idea Award; H.I.P. is supported by U01CA214195-01, DoD Translational Team Award; J.B. is supported by P50CA196516, R01CA175754 and RP180192.
Compliance with ethical standards
Conflict of interest
M.C. has pending patent applications on BAP1; H.I.P., has patents pending for use of fibulin-3 for diagnosis of mesothelioma; M.C., H.Y and H.I.P. have patents pending for use of HMGB1 for diagnosis of mesothelioma; M.C. and H.Y. have patents pending for use of HMGB1 and its isoforms for diagnosis of mesothelioma; M.C. provides cost-free consultation for MM expertise and diagnosis to patients and colleagues and paid medical-legal expertise. The remaining authors declare that they have no conflict of interest.
Footnotes
Edited by: R.A. Knight.
Contributor Information
Michele Carbone, Email: mcarbone@cc.hawaii.edu.
Gerry Melino, Email: gm89@le.ac.uk.
References
- 1.Olson JS. The history of cancer: an annotated bibliography. New York: Greenwood Press; 1989. [Google Scholar]
- 2.Cardiff RD, Kenney N. A compendium of the mouse mammary tumor biologist: from the initial observations in the house mouse to the development of genetically engineered mice. Cold Spring Harb Perspect Biol 2011;3. [DOI] [PMC free article] [PubMed]
- 3.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu S, Powers S, Zhu W, Hannun YA. Substantial contribution of extrinsic risk factors to cancer development. Nature. 2016;529:43–47. doi: 10.1038/nature16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roushdy-Hammady I, Siegel J, Emri S, Testa JR, Carbone M. Genetic-susceptibility factor and malignant mesothelioma in the Cappadocian region of Turkey. Lancet. 2001;357:444–5. doi: 10.1016/S0140-6736(00)04013-7. [DOI] [PubMed] [Google Scholar]
- 6.Carbone M, Emri S, Dogan AU, Steele I, Tuncer M, Pass HI, et al. A mesothelioma epidemic in Cappadocia: scientific developments and unexpected social outcomes. Nat Rev Cancer. 2007;7:147–54. doi: 10.1038/nrc2068. [DOI] [PubMed] [Google Scholar]
- 7.Emri SA. The Cappadocia mesothelioma epidemic: its influence in Turkey and abroad. Ann Transl Med. 2017;5:239. doi: 10.21037/atm.2017.04.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonds NI, Ghazarian AA, Pimentel CB, Schully SD, Ellison GL, Gillanders EM, et al. Review of the Gene-Environment Interaction Literature in Cancer: What Do We Know? Genet Epidemiol. 2016;40:356–65. doi: 10.1002/gepi.21967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudolph A, Chang-Claude J, Schmidt MK. Gene-environment interaction and risk of breast cancer. Br J Cancer. 2016;114:125–33. doi: 10.1038/bjc.2015.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr SR, Akerley W, Hashibe M, Cannon-Albright LA. Evidence for a genetical contribution to non-smoking-related lung cancer. Thorax. 2015;70:1033–9. doi: 10.1136/thoraxjnl-2014-206584. [DOI] [PubMed] [Google Scholar]
- 11.Schottenfeld D, Beebe-Dimmer JL, Buffler PA, Omenn GS. Current perspective on the global and United States cancer burden attributable to lifestyle and environmental risk factors. Annu Rev Public Health. 2013;34:97–117. doi: 10.1146/annurev-publhealth-031912-114350. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Closas M, Malats N, Silverman D, Dosemeci M, Kogevinas M, Hein DW, et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366:649–59. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis SJ, Smith GD. Alcohol, ALDH2, and esophageal cancer: a meta-analysis which illustrates the potentials and limitations of a Mendelian randomization approach. Cancer Epidemiol Biomark Prev. 2005;14:1967–71. doi: 10.1158/1055-9965.EPI-05-0196. [DOI] [PubMed] [Google Scholar]
- 14.Thomas D. Gene--environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11:259–72. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moyerbrailean GA, Richards AL, Kurtz D, Kalita CA, Davis GO, Harvey CT, et al. High-throughput allele-specific expression across 250 environmental conditions. Genome Res. 2016;26:1627–38. doi: 10.1101/gr.209759.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cairns J, Lyon JL, Skolnick M. Cancer incidence in defined populations. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 17.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 18.Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, Collins N, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265:2088–90. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 19.Kamb A, Shattuck-Eidens D, Eeles R, Liu Q, Gruis NA, Ding W, et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet. 1994;8:23–26. doi: 10.1038/ng0994-22. [DOI] [PubMed] [Google Scholar]
- 20.Churpek JE, Marquez R, Neistadt B, Claussen K, Lee MK, Churpek MM, et al. Inherited mutations in cancer susceptibility genes are common among survivors of breast cancer who develop therapy-related leukemia. Cancer. 2016;122:304–11. doi: 10.1002/cncr.29615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pijpe A, Andrieu N, Easton DF, Kesminiene A, Cardis E, Nogues C, et al. Exposure to diagnostic radiation and risk of breast cancer among carriers of BRCA1/2 mutations: retrospective cohort study (GENE-RAD-RISK) BMJ. 2012;345:e5660. doi: 10.1136/bmj.e5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cannon-Albright LA, Thomas A, Goldgar DE, Gholami K, Rowe K, Jacobsen M, et al. Familiality of cancer in Utah. Cancer Res. 1994;54:2378–85. [PubMed] [Google Scholar]
- 23.Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, Sulem P, Kristjansson K, Arnason S, et al. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 2004;1:e65. doi: 10.1371/journal.pmed.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemminki K, Rawal R, Chen B, Bermejo JL. Genetic epidemiology of cancer: from families to heritable genes. Int J Cancer. 2004;111:944–50. doi: 10.1002/ijc.20355. [DOI] [PubMed] [Google Scholar]
- 25.Cannon-Albright LA, Dintelman S, Maness T, Backus S, Thomas A, Meyer LJ. Creation of a national resource with linked genealogy and phenotypic data: the Veterans Genealogy Project. Genet Med. 2013;15:541–7. doi: 10.1038/gim.2012.171. [DOI] [PubMed] [Google Scholar]
- 26.Comertpay S, Pastorino S, Tanji M, Mezzapelle R, Strianese O, Napolitano A, et al. Evaluation of clonal origin of malignant mesothelioma. J Transl Med. 2014;12:301. doi: 10.1186/s12967-014-0301-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carbone M, Ly BH, Dodson RF, Pagano I, Morris PT, Dogan UA, et al. Malignant mesothelioma: facts, myths, and hypotheses. J Cell Physiol. 2012;227:44–58. doi: 10.1002/jcp.22724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berry G, Reid A, Aboagye-Sarfo P, de Klerk NH, Olsen NJ, Merler E, et al. Malignant mesotheliomas in former miners and millers of crocidolite at Wittenoom (Western Australia) after more than 50 years follow-up. Br J Cancer. 2012;106:1016–20. doi: 10.1038/bjc.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumann F, Buck BJ, Metcalf RV, McLaurin BT, Merkler DJ, Carbone M. The Presence of Asbestos in the Natural Environment is Likely Related to Mesothelioma in Young Individuals and Women from Southern Nevada. J Thorac Oncol. 2015;10:731–7. doi: 10.1097/JTO.0000000000000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumann F, Ambrosi JP, Carbone M. Asbestos is not just asbestos: an unrecognised health hazard. Lancet Oncol. 2013;14:576–8. doi: 10.1016/S1470-2045(13)70257-2. [DOI] [PubMed] [Google Scholar]
- 31.Napolitano A, Antoine DJ, Pellegrini L, Baumann F, Pagano I, Pastorino S, et al. HMGB1 and Its Hyperacetylated Isoform are Sensitive and Specific Serum Biomarkers to Detect Asbestos Exposure and to Identify Mesothelioma Patients. Clin Cancer Res. 2016;22:3087–96. doi: 10.1158/1078-0432.CCR-15-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Rivera Z, Jube S, Nasu M, Bertino P, Goparaju C, et al. Programmed necrosis induced by asbestos in human mesothelial cells causes high-mobility group box 1 protein release and resultant inflammation. Proc Natl Acad Sci USA. 2010;107:12611–6. doi: 10.1073/pnas.1006542107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caputa G, Zhao S, Criado AE, Ory DS, Duncan JG, Schaffer JE. RNASET2 is required for ROS propagation during oxidative stress-mediated cell death. Cell Death Differ. 2016;23:347–57. doi: 10.1038/cdd.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simeonova PP, Luster MI. Iron and reactive oxygen species in the asbestos-induced tumor necrosis factor-alpha response from alveolar macrophages. Am J Respir Cell Mol Biol. 1995;12:676–83. doi: 10.1165/ajrcmb.12.6.7539275. [DOI] [PubMed] [Google Scholar]
- 35.Croce A, Allegrina M, Rinaudo C, Gaudino G, Yang H, Carbone M. Numerous Iron-Rich Particles Lie on the Surface of Erionite Fibers from Rome (Oregon, USA) and Karlik (Cappadocia, Turkey) Microsc Microanal. 2015;21:1341–7. doi: 10.1017/S1431927615014762. [DOI] [PubMed] [Google Scholar]
- 36.Yang H, Pellegrini L, Napolitano A, Giorgi C, Jube S, Preti A, et al. Aspirin delays mesothelioma growth by inhibiting HMGB1-mediated tumor progression. Cell Death Dis. 2015;6:e1786. doi: 10.1038/cddis.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pellegrini L, Xue J, Larson D, Pastorino S, Jube S, Forest KH, et al. HMGB1 targeting by ethyl pyruvate suppresses malignant phenotype of human mesothelioma. Oncotarget. 2017;8:22649–61. doi: 10.18632/oncotarget.15152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bononi A, Napolitano A, Pass HI, Yang H, Carbone M. Latest developments in our understanding of the pathogenesis of mesothelioma and the design of targeted therapies. Expert Rev Respir Med. 2015;9:633–54. doi: 10.1586/17476348.2015.1081066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshikawa Y, Emi M, Hashimoto-Tamaoki T, Ohmuraya M, Sato A, Tsujimura T, et al. High-density array-CGH with targeted NGS unmask multiple noncontiguous minute deletions on chromosome 3p21 in mesothelioma. Proc Natl Acad Sci USA. 2016;113:13432–7. doi: 10.1073/pnas.1612074113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nasu M, Emi M, Pastorino S, Tanji M, Powers A, Luk H, et al. High Incidence of Somatic BAP1 alterations in sporadic malignant mesothelioma. J Thorac Oncol. 2015;10:565–76. doi: 10.1097/JTO.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carbone M, Shimizu D, Napolitano A, Tanji M, Pass HI, Yang H, et al. Positive nuclear BAP1 immunostaining helps differentiate non-small cell lung carcinomas from malignant mesothelioma. Oncotarget. 2016;7:59314–21. doi: 10.18632/oncotarget.10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Napolitano A, Carbone M. Malignant Mesothelioma: Time toTranslate? Trends Cancer. 2016;2:467–74. doi: 10.1016/j.trecan.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–5. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carbone M, Flores EG, Emi M, Johnson TA, Tsunoda T, Behner D, et al. Combined Genetic and Genealogic Studies Uncover a Large BAP1 Cancer Syndrome Kindred Tracing Back Nine Generations to a Common Ancestor from the 1700s. PLoS Genet. 2015;11:e1005633. doi: 10.1371/journal.pgen.1005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baumann F, Flores E, Napolitano A, Kanodia S, Taioli E, Pass H, et al. Mesothelioma patients with germline BAP1 mutations have 7-fold improved long-term survival. Carcinogenesis. 2015;36:76–81. doi: 10.1093/carcin/bgu227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rusch A, Ziltener G, Nackaerts K, Weder W, Stahel RA, Felley-Bosco E. Prevalence of BRCA-1 associated protein 1 germline mutation in sporadic malignant pleural mesothelioma cases. Lung Cancer. 2015;87:77–79. doi: 10.1016/j.lungcan.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 47.Napolitano A, Pellegrini L, Dey A, Larson D, Tanji M, Flores EG, et al. Minimal asbestos exposure in germline BAP1 heterozygous mice is associated with deregulated inflammatory response and increased risk of mesothelioma. Oncogene. 2016;35:1996–2002. doi: 10.1038/onc.2015.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Betti M, Casalone E, Ferrante D, Romanelli A, Grosso F, Guarrera S, et al. Inference on germline BAP1 mutations and asbestos exposure from the analysis of familial and sporadic mesothelioma in a high-risk area. Genes Chromosomes Cancer. 2015;54:51–62. doi: 10.1002/gcc.22218. [DOI] [PubMed] [Google Scholar]
- 49.Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. BAP1 and cancer. Nat Rev Cancer. 2013;13:153–9. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–9. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pena-Llopis S, Christie A, Xie XJ, Brugarolas J. Cooperation and antagonism among cancer genes: the renal cancer paradigm. Cancer Res. 2013;73:4173–9. doi: 10.1158/0008-5472.CAN-13-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu YF, Cohn S, Christie A, McKenzie T, Wolff N, Do QN, et al. Modeling renal cell carcinoma in mice: Bap1 and Pbrm1 inactivation drive tumor grade. Cancer Discov. 2017;7:900–17. doi: 10.1158/2159-8290.CD-17-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Popova T, Hebert L, Jacquemin V, Gad S, Caux-Moncoutier V, Dubois-d’Enghien C, et al. Germline BAP1 mutations predispose to renal cell carcinomas. Am J Hum Genet. 2013;92:974–80. doi: 10.1016/j.ajhg.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt LS, Linehan WM. Genetic predisposition to kidney cancer. Semin Oncol. 2016;43:566–74. doi: 10.1053/j.seminoncol.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farley MN, Schmidt LS, Mester JL, Pena-Llopis S, Pavia-Jimenez A, Christie A, et al. A novel germline mutation in BAP1 predisposes to familial clear-cell renal cell carcinoma. Mol Cancer Res: MCR. 2013;11:1061–71. doi: 10.1158/1541-7786.MCR-13-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melkonian SC, Daniel CR, Ye Y, Tannir NM, Karam JA, Matin SF, et al. Gene-environment interaction of genome-wide association study-identified susceptibility loci and meat-cooking mutagens in the etiology of renal cell carcinoma. Cancer. 2016;122:108–15. doi: 10.1002/cncr.29543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scelo G, Riazalhosseini Y, Greger L, Letourneau L, Gonzalez-Porta M, Wozniak MB, et al. Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nat Commun. 2014;5:5135. doi: 10.1038/ncomms6135. [DOI] [PubMed] [Google Scholar]
- 58.Beckerman R, Prives C. Transcriptional regulation byp53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Charni M, Molchadsky A, Goldstein I, Solomon H, Tal P, Goldfinger N, et al. Novel p53 target genes secreted by the liver are involved in non-cell-autonomous regulation. Cell Death Differ. 2016;23:509–20. doi: 10.1038/cdd.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prives C, Bargonetti J, Farmer G, Ferrari E, Friedlander P, Wang Y, et al. DNA-binding properties of the p53 tumor suppressor protein. Cold Spring Harb Symp Quant Biol. 1994;59:207–13. doi: 10.1101/sqb.1994.059.01.025. [DOI] [PubMed] [Google Scholar]
- 61.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 62.Charni M, Aloni-Grinstein R, Molchadsky A, Rotter V. p53 on the crossroad between regeneration and cancer. Cell Death Differ. 2017;24:8–14. doi: 10.1038/cdd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lowe JM, Nguyen TA, Grimm SA, Gabor KA, Peddada SD, Li L, et al. The novel p53 target TNFAIP8 variant 2 is increased in cancer and offsets p53-dependent tumor suppression. Cell Death Differ. 2017;24:181–91. doi: 10.1038/cdd.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lopez I, Tournillon AS, Prado Martins R, Karakostis K, Malbert-Colas L, Nylander K, et al. p53-mediated suppression of BiP triggers BIK-induced apoptosis during prolonged endoplasmic reticulum stress. Cell Death Differ. 2017;24:1717–29. doi: 10.1038/cdd.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 66.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–17. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aggarwal M, Saxena R, Sinclair E, Fu Y, Jacobs A, Dyba M, et al. Reactivation of mutant p53 by a dietary-related compound phenethyl isothiocyanate inhibits tumor growth. Cell Death Differ. 2016;23:1615–27. doi: 10.1038/cdd.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170:1062–78. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Y, Chen C, Xu Z, Scuoppo C, Rillahan CD, Gao J, et al. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature. 2016;531:471–5. doi: 10.1038/nature17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weissmueller S, Manchado E, Saborowski M, Morris JPt, Wagenblast E, Davis CA, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell. 2014;157:382–94. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 72.Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–41. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 73.Kehrloesser S, Osterburg C, Tuppi M, Schafer B, Vousden KH, Dotsch V. Intrinsic aggregation propensity of the p63 and p73 TI domains correlates with p53R175H interaction and suggests further significance of aggregation events in the p53 family. Cell Death Differ. 2016;23:1952–60. doi: 10.1038/cdd.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–58. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alexandrova EM, Moll UM. Depleting stabilized GOF mutant p53 proteins by inhibiting molecular folding chaperones: a new promise in cancer therapy. Cell Death Differ. 2017;24:3–5. doi: 10.1038/cdd.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stracquadanio G, Wang X, Wallace MD, Grawenda AM, Zhang P, Hewitt J, et al. The importance of p53 pathway genetics in inherited and somatic cancer genomes. Nat Rev Cancer. 2016;16:251–65. doi: 10.1038/nrc.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sarkar J, Dominguez E, Li G, Kusewitt DF, Johnson DG. Modeling gene-environment interactions in oral cavity and esophageal cancers demonstrates a role for the p53 R72P polymorphism in modulating susceptibility. Mol Carcinog. 2014;53:648–58. doi: 10.1002/mc.22019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Costanzo A, Pediconi N, Narcisi A, Guerrieri F, Belloni L, Fausti F, et al. TP63 and TP73 in cancer, an unresolved “family” puzzle of complexity, redundancy and hierarchy. FEBS Lett. 2014;588:2590–9. doi: 10.1016/j.febslet.2014.06.047. [DOI] [PubMed] [Google Scholar]
- 79.Gebel J, Luh LM, Coutandin D, Osterburg C, Lohr F, Schafer B, et al. Mechanism of TAp73 inhibition by DeltaNp63 and structural basis of p63/p73 hetero-tetramerization. Cell Death Differ. 2016;23:1930–40. doi: 10.1038/cdd.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Nostrand JL, Bowen ME, Vogel H, Barna M, Attardi LD. The p53 family members have distinct roles during mammalian embryonic development. Cell Death Differ. 2017;24:575–9. doi: 10.1038/cdd.2016.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Amelio I, Melino G. The p53 family and the hypoxia-inducible factors (HIFs): determinants of cancer progression. Trends Biochem Sci. 2015;40:425–34. doi: 10.1016/j.tibs.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 82.Wu Q, Shi Y, Ge L, Ma D, Zhang H, Wang J. Relationship of p73 gene polymorphism and additional gene-smoking and gene-obesity interaction with non-small cell lung cancer risk. Oncotarget. 2017;8:34423–34428. doi: 10.18632/oncotarget.16257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G. The p53 family: guardians of maternal reproduction. Nat Rev Mol Cell Biol. 2011;12:259–65. doi: 10.1038/nrm3086. [DOI] [PubMed] [Google Scholar]
- 84.Memmi EM, Sanarico AG, Giacobbe A, Peschiaroli A, Frezza V, Cicalese A, et al. p63 Sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc Natl Acad Sci USA. 2015;112:3499–504. doi: 10.1073/pnas.1500762112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Viticchie G, Agostini M, Lena AM, Mancini M, Zhou H, Zolla L, et al. p63 supports aerobic respiration through hexokinase II. Proc Natl Acad Sci USA. 2015;112:11577–82. doi: 10.1073/pnas.1508871112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes & Dev. 2008;22:2677–91. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tomasini R, Tsuchihara K, Tsuda C, Lau SK, Wilhelm M, Ruffini A, et al. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc Natl Acad Sci USA. 2009;106:797–802. doi: 10.1073/pnas.0812096106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Amelio I, Inoue S, Markert EK, Levine AJ, Knight RA, Mak TW, et al. TAp73 opposes tumor angiogenesis by promoting hypoxia-inducible factor 1alpha degradation. Proc Natl Acad Sci USA. 2015;112:226–31. doi: 10.1073/pnas.1410609111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Amelio I, Markert EK, Rufini A, Antonov AV, Sayan BS, Tucci P, et al. p73 regulates serine biosynthesis in cancer. Oncogene. 2014;33:5039–46. doi: 10.1038/onc.2013.456. [DOI] [PubMed] [Google Scholar]
- 90.Rufini A, Niklison-Chirou MV, Inoue S, Tomasini R, Harris IS, Marino A, et al. TAp73 depletion accelerates aging through metabolic dysregulation. Genes & Dev. 2012;26:2009–14. doi: 10.1101/gad.197640.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Velletri T, Romeo F, Tucci P, Peschiaroli A, Annicchiarico-Petruzzelli M, Niklison-Chirou MV, et al. GLS2 is transcriptionally regulated by p73 and contributes to neuronal differentiation. Cell Cycle. 2013;12:3564–73. doi: 10.4161/cc.26771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.D’Alessandro A, Amelio I, Berkers CR, Antonov A, Vousden KH, Melino G, et al. Metabolic effect of TAp63alpha: enhanced glycolysis and pentose phosphate pathway, resulting in increased antioxidant defense. Oncotarget. 2014;5:7722–33. doi: 10.18632/oncotarget.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA. 2010;107:7455–60. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Amelio I, Antonov AA, Catani MV, Massoud R, Bernassola F, Knight RA, et al. TAp73 promotes anabolism. Oncotarget. 2014;5:12820–934. doi: 10.18632/oncotarget.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Solomon H, Brauning B, Fainer I, Ben-Nissan G, Rabani S, Goldfinger N, et al. Post-translational regulation of p53 function through 20S proteasome-mediated cleavage. Cell Death Differ. 2017;24:2187–98. doi: 10.1038/cdd.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nemajerova A, Amelio I, Gebel J, Dotsch V, Melino G, Moll UM. Non-oncogenic roles of TAp73: from multiciliogenesis to metabolism. Cell death Differ. 2017;25:144–153. doi: 10.1038/cdd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Amelio I, Cutruzzola F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39:191–8. doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Antonov A, Agostini M, Morello M, Minieri M, Melino G, Amelio I. Bioinformatics analysis of the serine and glycine pathway in cancer cells. Oncotarget. 2014;5:11004–13. doi: 10.18632/oncotarget.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sharif T, Ahn DG, Liu RZ, Pringle E, Martell E, Dai C, et al. The NAD(+) salvage pathway modulates cancer cell viability via p73. Cell Death Differ. 2016;23:669–80. doi: 10.1038/cdd.2015.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Clendening JW, Pandyra A, Boutros PC, El Ghamrasni S, Khosravi F, Trentin GA, et al. Dysregulation of the mevalonate pathway promotes transformation. Proc Natl Acad Sci USA. 2010;107:15051–6. doi: 10.1073/pnas.0910258107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ingallina E, Sorrentino G, Bertolio R, Lisek K, Zannini A, Azzolin L, et al. Mechanical cues control mutant p53 stability through a mevalonate-RhoA axis. Nat Cell Biol. 2018;20:28–35. doi: 10.1038/s41556-017-0009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Parrales A, Ranjan A, Iyer SV, Padhye S, Weir SJ, Roy A, et al. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat Cell Biol. 2016;18:1233–43. doi: 10.1038/ncb3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ahern TP, Pedersen L, Tarp M, Cronin-Fenton DP, Garne JP, Silliman RA, et al. Statin prescriptions and breast cancer recurrence risk: a Danish nationwide prospective cohort study. J Natl Cancer Inst. 2011;103:1461–8. doi: 10.1093/jnci/djr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Clendening JW, Pandyra A, Li Z, Boutros PC, Martirosyan A, Lehner R, et al. Exploiting the mevalonate pathway to distinguish statin-sensitive multiple myeloma. Blood. 2010;115:4787–97. doi: 10.1182/blood-2009-07-230508. [DOI] [PubMed] [Google Scholar]
- 105.Mikoshiba K. IP3 receptor/Ca2+channel: from discovery to new signaling concepts. J Neurochem. 2007;102:1426–46. doi: 10.1111/j.1471-4159.2007.04825.x. [DOI] [PubMed] [Google Scholar]
- 106.Mikoshiba K. Role of IP3 receptor signaling in cell functions and diseases. Adv Biol Regul. 2015;57(Supplement C):217–27. doi: 10.1016/j.jbior.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 107.Berridge MJ. The inositol trisphosphate/calcium signaling pathway in health and disease. Physiol Rev. 2016;96:1261–96. doi: 10.1152/physrev.00006.2016. [DOI] [PubMed] [Google Scholar]
- 108.Furuichi T, Yoshikawa S, Miyawaki A, Wada K, Maeda N, Mikoshiba K. Primary structure and functional expression of the inositol 1,4,5-trisphosphate-binding protein P400. Nature. 1989;342:32–38. doi: 10.1038/342032a0. [DOI] [PubMed] [Google Scholar]
- 109.Mignery GA, Newton CL, Archer BT, 3rd, Sudhof TC. Structure and expression of the rat inositol 1,4,5-trisphosphate receptor. J Biol Chem. 1990;265:12679–85. [PubMed] [Google Scholar]
- 110.Sudhof TC, Newton CL, Archer BT, 3rd, Ushkaryov YA, Mignery GA. Structure of a novel InsP3 receptor. EMBO J. 1991;10:3199–206. doi: 10.1002/j.1460-2075.1991.tb04882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Blondel O, Takeda J, Janssen H, Seino S, Bell GI. Sequence and functional characterization of a third inositol trisphosphate receptor subtype, IP3R-3, expressed in pancreatic islets, kidney, gastrointestinal tract, and other tissues. J Biol Chem. 1993;268:11356–63. [PubMed] [Google Scholar]
- 112.Yamamoto-Hino M, Sugiyama T, Hikichi K, Mattei MG, Hasegawa K, Sekine S, et al. Cloning and characterization of human type 2 and type 3 inositol 1,4,5-trisphosphate receptors. Recept & Channels. 1994;2:9–22. [PubMed] [Google Scholar]
- 113.Yamada N, Makino Y, Clark RA, Pearson DW, Mattei MG, Guénet JL, et al. Human inositol 1,4,5-trisphosphate type-1 receptor, Ins<em>P</em>3R1: structure, function, regulation of expression and chromosomal localization. Biochem J. 1994;302:781–90. doi: 10.1042/bj3020781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.White C, Li C, Yang J, Petrenko NB, Madesh M, Thompson CB, et al. The endoplasmic reticulum gateway to apoptosis by Bcl-X(L) modulation of the InsP3R. Nat Cell Biol. 2005;7:1021–8. doi: 10.1038/ncb1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Eckenrode EF, Yang J, Velmurugan GV, Foskett JK, White C. Apoptosis protection by Mcl-1 and Bcl-2 modulation of inositol 1,4,5-trisphosphate receptor-dependent Ca2+signaling. J Biol Chem. 2010;285:13678–84. doi: 10.1074/jbc.M109.096040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Szado T, Vanderheyden V, Parys JB, De Smedt H, Rietdorf K, Kotelevets L, et al. Phosphorylation of inositol 1,4,5-trisphosphate receptors by protein kinase B/Akt inhibits Ca2+release and apoptosis. Proc Natl Acad Sci. 2008;105:2427–32. doi: 10.1073/pnas.0711324105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boehning D, Patterson RL, Sedaghat L, Glebova NO, Kurosaki T, Snyder SH. Cytochrome c binds to inositol (1,4,5) trisphosphate receptors, amplifying calcium-dependent apoptosis. Nat Cell Biol. 2003;5:1051–61. doi: 10.1038/ncb1063. [DOI] [PubMed] [Google Scholar]
- 118.Zhang S, Mizutani A, Hisatsune C, Higo T, Bannai H, Nakayama T, et al. Protein 4.1N is required for translocation of inositol 1,4,5-trisphosphate receptor type 1 to the basolateral membrane domain in polarized Madin-Darby canine kidney cells. J Biol Chem. 2003;278:4048–56. doi: 10.1074/jbc.M209960200. [DOI] [PubMed] [Google Scholar]
- 119.Kawaai K, Hisatsune C, Kuroda Y, Mizutani A, Tashiro T, Mikoshiba K. 80K-H interacts with inositol 1,4,5-trisphosphate (IP3) receptors and regulates IP3-induced calcium release activity. J Biol Chem. 2009;284:372–80. doi: 10.1074/jbc.M805828200. [DOI] [PubMed] [Google Scholar]
- 120.Tang TS, Tu H, Chan EYW, Maximov A, Wang Z, Wellington CL, et al. Huntingtin and Huntingtin-Associated Protein 1 Influence Neuronal Calcium Signaling Mediated by Inositol-(1,4,5) Triphosphate Receptor Type 1. Neuron. 2003;39:227–39. doi: 10.1016/s0896-6273(03)00366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sung PJ, Tsai FD, Vais H, Court H, Yang J, Fehrenbacher N, et al. Phosphorylated K-Ras limits cell survival by blocking Bcl-xL sensitization of inositol trisphosphate receptors. Proc Natl Acad Sci. 2013;110:20593–8. doi: 10.1073/pnas.1306431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang S, Hisatsune C, Matsu-Ura T, Mikoshiba K. G-protein-coupled receptor kinase-interacting proteins inhibit apoptosis by inositol 1,4,5-triphosphate receptor-mediated Ca2+signal regulation. J Biol Chem. 2009;284:29158–69. doi: 10.1074/jbc.M109.041509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hedgepeth SC, Garcia MI, Wagner LE, Rodriguez AM, Chintapalli SV, Snyder RR, et al. The BRCA1 Tumor Suppressor Binds to Inositol 1,4,5-Trisphosphate Receptors to Stimulate Apoptotic Calcium Release. J Biol Chem. 2015;290:7304–13. doi: 10.1074/jbc.M114.611186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kuchay S, Giorgi C, Simoneschi D, Pagan J, Missiroli S, Saraf A, et al. PTEN counteracts FBXL2 to promote IP3R3- and Ca2+-mediated apoptosis limiting tumour growth. Nature. 2017;546:554–8. doi: 10.1038/nature22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bononi A, Giorgi C, Patergnani S, Larson D, Verbruggen K, Tanji M, et al. BAP1 regulates IP3R3-mediated Ca2+flux to mitochondria suppressing cell transformation. Nature. 2017;546:549–53. doi: 10.1038/nature22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hamada K, Miyatake H, Terauchi A, Mikoshiba K. IP3-mediated gating mechanism of the IP3 receptor revealed by mutagenesis and X-ray crystallography. Proc Natl Acad Sci USA. 2017;114:4661–6. doi: 10.1073/pnas.1701420114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rizzuto R, Brini M, Murgia M, Pozzan T. Microdomains with high Ca2+close to IP3-sensitive channels that are sensed by neighboring mitochondria. Sci (New Y, NY) 1993;262:744–7. doi: 10.1126/science.8235595. [DOI] [PubMed] [Google Scholar]
- 128.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–78. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 129.Gomez L, Thiebaut PA, Paillard M, Ducreux S, Abrial M, Crola Da Silva C, et al. The SR/ER-mitochondria calcium crosstalk is regulated by GSK3beta during reperfusion injury. Cell Death Differ. 2016;23:313–22. doi: 10.1038/cdd.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol. 2012;13:607–25. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vance JE. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem. 1990;265:7248–56. [PubMed] [Google Scholar]
- 132.Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, et al. Close Contacts with the Endoplasmic Reticulum as Determinants of Mitochondrial Ca2+2+Responses. Sci (New Y, NY) 1998;280:1763–6. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 133.Csordás G, Renken C, Várnai P, Walter L, Weaver D, Buttle KF, et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–21. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Szabadkai G, Bianchi K, Várnai P, De Stefani D, Wieckowski MR, Cavagna D, et al. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+2+channels. J Cell Biol. 2006;175:901–11. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.De Stefani D, Bononi A, Romagnoli A, Messina A, De Pinto V, Pinton P, et al. VDAC1 selectively transfers apoptotic Ca2+signals to mitochondria. Cell Death Differ. 2012;19:267–73. doi: 10.1038/cdd.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–40. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–5. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Khan AA, Soloski MJ, Sharp AH, Schilling G, Sabatini DM, Li SH, et al. Lymphocyte apoptosis: mediation by increased type 3 inositol 1,4,5-trisphosphate receptor. Sci (New Y, NY) 1996;273:503–7. doi: 10.1126/science.273.5274.503. [DOI] [PubMed] [Google Scholar]
- 139.Jayaraman T, Marks AR. T cells deficient in inositol 1,4,5-trisphosphate receptor are resistant to apoptosis. Mol Cell Biol. 1997;17:3005–12. doi: 10.1128/mcb.17.6.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sugawara H, Kurosaki M, Takata M, Kurosaki T. Genetic evidence for involvement of type 1, type 2 and type 3 inositol 1,4,5-trisphosphate receptors in signal transduction through the B-cell antigen receptor. EMBO J. 1997;16:3078–88. doi: 10.1093/emboj/16.11.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Joseph SK, Hajnoczky G. IP3 receptors in cell survival and apoptosis: Ca2+release and beyond. Apoptosis: Int J Program Cell death. 2007;12:951–68. doi: 10.1007/s10495-007-0719-7. [DOI] [PubMed] [Google Scholar]
- 142.Rasola A, Bernardi P. Mitochondrial permeability transition in Ca(2+)-dependent apoptosis and necrosis. Cell Calcium. 2011;50:222–33. doi: 10.1016/j.ceca.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 143.Boehning D, van Rossum DB, Patterson RL, Snyder SH. A peptide inhibitor of cytochrome c/inositol 1,4,5-trisphosphate receptor binding blocks intrinsic and extrinsic cell death pathways. Proc Natl Acad Sci USA. 2005;102:1466–71. doi: 10.1073/pnas.0409650102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Greenberg EF, Lavik AR, Distelhorst CW. Bcl-2 regulation of the inositol 1,4,5-trisphosphate receptor and calcium signaling in normal and malignant lymphocytes: potential new target for cancer treatment. Biochim Biophys Acta. 2014;1843:2205–10. doi: 10.1016/j.bbamcr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vervloessem T, Kerkhofs M, La Rovere RM, Sneyers F, Parys JB, Bultynck G Bcl-2 inhibitors as anti-cancer therapeutics: the impact of and on calcium signaling. Cell Calcium 2018;70:102–116. [DOI] [PubMed]
- 146.Ando H, Mizutani A, Matsu-ura T, Mikoshiba K. IRBIT, a novel inositol 1,4,5-trisphosphate (IP3) receptor-binding protein, is released from the IP3 receptor upon IP3 binding to the receptor. J Biol Chem. 2003;278:10602–12. doi: 10.1074/jbc.M210119200. [DOI] [PubMed] [Google Scholar]
- 147.Ando H, Mizutani A, Kiefer H, Tsuzurugi D, Michikawa T, Mikoshiba K. IRBIT suppresses IP3 receptor activity by competing with IP3 for the common binding site on the IP3 receptor. Mol Cell. 2006;22:795–806. doi: 10.1016/j.molcel.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 148.Ando H, Mizutani A, Mikoshiba K. An IRBIT homologue lacks binding activity to inositol 1,4,5-trisphosphate receptor due to the unique N-terminal appendage. J Neurochem. 2009;109:539–50. doi: 10.1111/j.1471-4159.2009.05979.x. [DOI] [PubMed] [Google Scholar]
- 149.Kawaai K, Ando H, Satoh N, Yamada H, Ogawa N, Hirose M, et al. Splicing variation of Long-IRBIT determines the target selectivity of IRBIT family proteins. Proc Natl Acad Sci USA. 2017;114:3921–6. doi: 10.1073/pnas.1618514114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ando H, Kawaai K, Mikoshiba K. Biochimica et biophysica acta. IRBIT: a regulator of ion channels and ion transporters; 2014. pp. 2195–204. [DOI] [PubMed] [Google Scholar]
- 151.Kawaai K, Mizutani A, Shoji H, Ogawa N, Ebisui E, Kuroda Y, et al. IRBIT regulates CaMKIIalpha activity and contributes to catecholamine homeostasis through tyrosine hydroxylase phosphorylation. Proc Natl Acad Sci USA. 2015;112:5515–20. doi: 10.1073/pnas.1503310112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ando H, Hirose M, Gainche L, Kawaai K, Bonneau B, Ijuin T, et al. IRBIT Interacts with the Catalytic Core of Phosphatidylinositol Phosphate Kinase Type Ialpha and IIalpha through Conserved Catalytic Aspartate Residues. PLoS One. 2015;10:e0141569. doi: 10.1371/journal.pone.0141569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bonneau B, Ando H, Kawaai K, Hirose M, Takahashi-Iwanaga H, Mikoshiba K IRBIT controls apoptosis by interacting with the Bcl-2 homolog, Bcl2l10, and by promoting ER-mitochondria contact. Elife 2016, 5. pii:e19896. [DOI] [PMC free article] [PubMed]
- 154.Arnaoutov A, Dasso M. Enzyme regulation. IRBIT Is a Nov Regul ribonucleotide reductase High eukaryotes Sci. 2014;345:1512–5. doi: 10.1126/science.1251550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ando H, Kawaai K, Bonneau B, Mikoshiba K Remodeling of Ca(2+) signaling in cancer: Regulation of inositol 1,4,5-trisphosphate receptors through oncogenes and tumor suppressors. Advances in Biological Regulation 2018;6:64–76. [DOI] [PubMed]
- 156.Yu H, Pak H, Hammond-Martel I, Ghram M, Rodrigue A, Daou S, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci USA. 2014;111:285–90. doi: 10.1073/pnas.1309085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ismail IH, Davidson R, Gagne JP, Xu ZZ, Poirier GG, Hendzel MJ. Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res. 2014;74:4282–94. doi: 10.1158/0008-5472.CAN-13-3109. [DOI] [PubMed] [Google Scholar]
- 158.Bononi A, Yang H, Giorgi C, Patergnani S, Pellegrini L, Su M, et al. Germline BAP1 mutations induce a Warburg effect. Cell Death Differ. 2017;24:1694–704. doi: 10.1038/cdd.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Amelio I. Genes versus Environment: cytoplasmic BAP1 determines the toxic response to environmental stressors in mesothelioma. Cell death & Dis. 2017;8:e2907. doi: 10.1038/cddis.2017.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bennett WP, Hussain SP, Vahakangas KH, Khan MA, Shields PG, Harris CC. Molecular epidemiology of human cancer risk: gene-environment interactions and p53 mutation spectrum in human lung cancer. J Pathol. 1999;187:8–18. doi: 10.1002/(SICI)1096-9896(199901)187:1<8::AID-PATH232>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 161.Zhang R, Chu M, Zhao Y, Wu C, Guo H, Shi Y, et al. A genome-wide gene-environment interaction analysis for tobacco smoke and lung cancer susceptibility. Carcinogenesis. 2014;35:1528–35. doi: 10.1093/carcin/bgu076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Haugen A, Ryberg D, Mollerup S, Zienolddiny S, Skaug V, Svendsrud DH. Gene-environment interactions in human lung cancer. Toxicol Lett. 2000;112-3:233–7. doi: 10.1016/s0378-4274(99)00275-1. [DOI] [PubMed] [Google Scholar]
- 163.Wu X, Zhao H, Suk R, Christiani DC. Genetic susceptibility to tobacco-related cancer. Oncogene. 2004;23:6500–623. doi: 10.1038/sj.onc.1207811. [DOI] [PubMed] [Google Scholar]
- 164.Thomas A, Liu SV, Subramaniam DS, Giaccone G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol. 2015;12:511–26. doi: 10.1038/nrclinonc.2015.90. [DOI] [PubMed] [Google Scholar]
- 165.Yu T, Chen X, Zhang W, Liu J, Avdiushko R, Napier DL, et al. KLF4 regulates adult lung tumor-initiating cells and represses K-Ras-mediated lung cancer. Cell Death Differ. 2016;23:207–15. doi: 10.1038/cdd.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 167.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 168.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–8. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 169.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–42. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 170.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–46. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 171.Yang JC, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015;16:141–51. doi: 10.1016/S1470-2045(14)71173-8. [DOI] [PubMed] [Google Scholar]
- 172.Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15:302–10. doi: 10.1038/nrc3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Piyush T, Chacko AR, Sindrewicz P, Hilkens J, Rhodes JM, Yu LG. Interaction of galectin-3 with MUC1 on cell surface promotes EGFR dimerization and activation in human epithelial cancer cells. Cell Death Differ. 2017;24:1937–47. doi: 10.1038/cdd.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–5. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early stage lung cancer evolution. Nature 2017;545:446–451. [DOI] [PMC free article] [PubMed]
- 176.Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med. 2017;376:629–40. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Brisson GD, Alves LR, Pombo-de-Oliveira MS. Genetic susceptibility in childhood acute leukaemias: a systematic review. Ecancermedicalscience. 2015;9:539. doi: 10.3332/ecancer.2015.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet. 2013;381:484–95. doi: 10.1016/S0140-6736(12)61727-9. [DOI] [PubMed] [Google Scholar]
- 179.Wang J, Wang H, Wang LY, Cai D, Duan Z, Zhang Y, et al. Silencing the epigenetic silencer KDM4A for TRAIL and DR5 simultaneous induction and antitumor therapy. Cell Death Differ. 2016;23:1886–96. doi: 10.1038/cdd.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, et al. R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–5. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen WL, Wang JH, Zhao AH, Xu X, Wang YH, Chen TL, et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood. 2014;124:1645–54. doi: 10.1182/blood-2014-02-554204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tabe Y, Konopleva M. Advances in understanding the leukaemia microenvironment. Br J Haematol. 2014;164:767–78. doi: 10.1111/bjh.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Herst PM, Howman RA, Neeson PJ, Berridge MV, Ritchie DS. The level of glycolytic metabolism in acute myeloid leukemia blasts at diagnosis is prognostic for clinical outcome. J Leukoc Biol. 2011;89:51–55. doi: 10.1189/jlb.0710417. [DOI] [PubMed] [Google Scholar]
- 184.Chen WL, Wang YY, Zhao A, Xia L, Xie G, Su M, et al. Enhanced fructose utilization mediated by SLC2A5 Is a unique metabolic feature of acute myeloid leukemia with therapeutic potential. Cancer Cell. 2016;30:779–91. doi: 10.1016/j.ccell.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu H, Huang D, McArthur DL, Boros LG, Nissen N, Heaney AP. Fructose induces transketolase flux to promote pancreatic cancer growth. Cancer Res. 2010;70:6368–76. doi: 10.1158/0008-5472.CAN-09-4615. [DOI] [PubMed] [Google Scholar]
- 186.Barone S, Fussell SL, Singh AK, Lucas F, Xu J, Kim C, et al. Slc2a5 (Glut5) is essential for the absorption of fructose in the intestine and generation of fructose-induced hypertension. J Biol Chem. 2009;284:5056–66. doi: 10.1074/jbc.M808128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Monzavi-Karbassi B, Hine RJ, Stanley JS, Ramani VP, Carcel-Trullols J, Whitehead TL, et al. Fructose as a carbon source induces an aggressive phenotype in MDA-MB-468 breast tumor cells. Int J Oncol. 2010;37:615–22. doi: 10.3892/ijo_00000710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hopkins BD, Goncalves MD, Cantley LCObesity. and Cancer Mechanisms: Cancer Metabolism. J Clin Oncol: Off J Am Soc Clin Oncol. 2016;34:4277–83. doi: 10.1200/JCO.2016.67.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hammarsten J, Hogstedt B. Hyperinsulinaemia: a prospective risk factor for lethal clinical prostate cancer. Eur J Cancer. 2005;41:2887–95. doi: 10.1016/j.ejca.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 190.Yoon YS, Keum N, Zhang X, Cho E, Giovannucci EL. Hyperinsulinemia, insulin resistance and colorectal adenomas: A meta-analysis. Metab: Clin Exp. 2015;64:1324–33. doi: 10.1016/j.metabol.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 191.Ferguson RD, Novosyadlyy R, Fierz Y, Alikhani N, Sun H, Yakar S, et al. Hyperinsulinemia enhances c-Myc-mediated mammary tumor development and advances metastatic progression to the lung in a mouse model of type 2 diabetes. Breast Cancer Res: BCR. 2012;14:R8. doi: 10.1186/bcr3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lien EC, Lyssiotis CA, Cantley LC. Metabolic Reprogramming by the PI3K-Akt-mTOR Pathway in Cancer. Recent Results Cancer Res Fortschr der Krebsforsch Progres dans Les Rech sur Le Cancer. 2016;207:39–72. doi: 10.1007/978-3-319-42118-6_3. [DOI] [PubMed] [Google Scholar]
- 193.Huang S, Czech MP. The GLUT4 glucose transporter. Cell Metab. 2007;5:237–52. doi: 10.1016/j.cmet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 194.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K Pathway in Human Disease. Cell. 2017;170:605–35. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kalaany NY, Sabatini DM. Tumours with PI3K activation are resistant to dietary restriction. Nature. 2009;458:725–31. doi: 10.1038/nature07782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Matassa DS, Amoroso MR, Lu H, Avolio R, Arzeni D, Procaccini C, et al. Oxidative metabolism drives inflammation-induced platinum resistance in human ovarian cancer. Cell Death Differ. 2016;23:1542–54. doi: 10.1038/cdd.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Slager SL, Caporaso NE, de Sanjose S, Goldin LR. Genetic susceptibility to chronic lymphocytic leukemia. Semin Hematol. 2013;50:296–302. doi: 10.1053/j.seminhematol.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Suzuki HI, Young RA, Sharp PA. Super-enhancer-mediated rna processing revealed by integrative microrna network analysis. Cell. 2017;168:1000–14 e1015. doi: 10.1016/j.cell.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kang H, Kim C, Lee H, Rho JG, Seo JW, Nam JW, et al. Downregulation of microRNA-362-3p and microRNA-329 promotes tumor progression in human breast cancer. Cell Death Differ. 2016;23:484–95. doi: 10.1038/cdd.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013;3:1228–37. doi: 10.1158/2159-8290.CD-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhang J, Lieu YK, Ali AM, Penson A, Reggio KS, Rabadan R, et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci USA. 2015;112:E4726–4734. doi: 10.1073/pnas.1514105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–8. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 205.Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JF, et al. Targeting BCL2 with Venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med. 2016;374:311–22. doi: 10.1056/NEJMoa1513257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–9. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 207.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518:258–62. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Konecny GE, Kristeleit RS. PARP inhibitors for BRCA1/2-mutated and sporadic ovarian cancer: current practice and future directions. Br J Cancer. 2016;115:1157–73. doi: 10.1038/bjc.2016.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–13. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bai F, Morcos F, Cheng RR, Jiang H, Onuchic JN. Elucidating the druggable interface of protein-protein interactions using fragment docking and coevolutionary analysis. Proc Natl Acad Sci USA. 2016;113:E8051–E8058. doi: 10.1073/pnas.1615932113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lipper CH, Karmi O, Sohn YS, Darash-Yahana M, Lammert H, Song L, et al. Structure of the human monomeric NEET protein MiNT and its role in regulating iron and reactive oxygen species in cancer cells. Proc Natl Acad Sci USA. 2018;115:272–7. doi: 10.1073/pnas.1715842115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sohn YS, Tamir S, Song L, Michaeli D, Matouk I, Conlan AR, et al. NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc Natl Acad Sci USA. 2013;110:14676–81. doi: 10.1073/pnas.1313198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bai F, Morcos F, Sohn YS, Darash-Yahana M, Rezende CO, Lipper CH, et al. The Fe-S cluster-containing NEET proteins mitoNEET and NAF-1 as chemotherapeutic targets in breast cancer. Proc Natl Acad Sci USA. 2015;112:3698–703. doi: 10.1073/pnas.1502960112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–4. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Moresco EM, Li X, Beutler B. Going forward with genetics: recent technological advances and forward genetics in mice. Am J Pathol. 2013;182:1462–73. doi: 10.1016/j.ajpath.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhu L, Finkelstein D, Gao C, Shi L, Wang Y, Lopez-Terrada D, et al. Multi-organ mapping of cancer risk. Cell. 2016;166:1132–46 e1137. doi: 10.1016/j.cell.2016.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tomasetti C, Li L, Vogelstein B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science. 2017;355:1330–4. doi: 10.1126/science.aaf9011. [DOI] [PMC free article] [PubMed] [Google Scholar]