ABSTRACT
Several human diseases are thought to evolve due to a combination of host genetic mutations and environmental factors that include alterations in intestinal microbiota composition termed dysbiosis. Although in some cases, host genetics may shape the gut microbiota and enable it to provoke disease, experimentally disentangling cause and consequence in such host-microbe interactions requires strict control over non-genetic confounding factors. Mouse genetic studies previously proposed Nlrp6/ASC inflammasomes as innate immunity regulators of the intestinal ecosystem. In contrast, using littermate-controlled experimental setups, we recently showed that Nlrp6/ASC inflammasomes do not alter the gut microbiota composition. Our analyses indicated that maternal inheritance and long-term separate housing are non-genetic confounders that preclude the use of non-littermate mice when analyzing host genetic effects on intestinal ecology. Here, we summarize and discuss our gut microbiota analyses in inflammasome-deficient mice for illustrating the importance of littermate experimental design in studying host-microbiota interactions.
KEYWORDS: Nlrp6, ASC, caspase-1, inflammasomes, interleukin, DSS colitis, gut microbiota, dysbiosis, innate immunity, littermates
Need for controlled experimental design when evaluating cause and consequence in microbiota impacts on host health
The symbiotic relationship between the host and its intestinal commensal microbes is of crucial importance for immune homeostasis and overall health of the host.1,2 Sustained shifts in gut microbiota composition are termed dysbiosis, which are often characterized by a lower phylogenetic diversity and an over- and/or under-representation of particular microbial taxa. In humans, dysbiosis is observed in multiple immune and metabolic diseases, including inflammatory bowel disease (IBD) and obesity.3-5 The proposed causes for such diseases include host genetic factors in combination with external cues, such as environmental exposures and diet.6 These environmental factors could be a cause of dysbiosis, which then takes advantage of the host genetic susceptibility to provoke disease. Alternatively, the association of host genetic as well as microbiota composition changes with disease development suggests that in some cases there could be a linear chain of events, in which host genetic factors first alter the gut microbiota composition, which in turn causes or contributes to disease pathogenesis. In addition, rather than being a cause, dysbiosis could be a consequence of on-going inflammation and immune responses in patients, especially when concerning intestinal disorders such as IBD. In humans with established disease, disentangling the host versus microbiota cause and consequence in disease development is challenging. Therefore, mouse genetic studies in tightly controlled environmental conditions provide an elegant approach to investigate how host-microbiota interactions impact on intestinal ecology and disease susceptibility.7
Inflammasomes are oligomeric complexes in which particular cytosolic Pattern Recognition Receptors (PRRs) sense pathogen- or danger-associated cellular stress leading to caspase-1-dependent maturation of the cytokines interleukin (IL)-1β and IL-18.8 Human and mouse genetic observations suggested inflammasome deregulation as a potential host factor capable of provoking dysbiosis-driven intestinal disease. Indeed, polymorphisms in the gene encoding one of the inflammasome-activating PRRs, NLR nucleotide-binding domain, leucine-rich and Pyrin containing 3 (Nlrp3), are associated with an increased risk for IBD development in humans.9-11 Later, studies using genetic mouse models proposed inflammasomes as major regulators of the intestinal microbiota.12,13 Indeed, deletion of apoptosis-associated speck-like protein containing a CARD (ASC, encoded by Pycard), an adaptor protein needed for activating the Nlrp3 and also other inflammasomes, resulted in intestinal dysbiosis when compared to non-littermate separately housed C57BL/6 mice.12 Identical observations in Nlrp6 knockout (KO) mice suggested that the Nlrp6/ASC inflammasome was a key regulator of the gut microbiota.12 Moreover, a later study showed that also mice lacking Nlrp3 displayed a similar shift in the gut microbiota composition when compared with non-littermate separately housed wild-type (WT) mice.13 Importantly, the dysbiotic gut microbiota observed in Nlrp6- or ASC-deficient mice enhanced colitis development in WT mice. Indeed, WT mice co-housed with these inflammasome-deficient mice developed more severe Dextran Sodium Sulphate (DSS)-induced colitis than single housed wild-types, an effect attributed to horizontal transfer of colitogenic microbiota derived from the Nlrp6- or ASC-deficient mice.12 Likewise, horizontal microbiota transfer from inflammasome-deficient mice was proposed to underlie more severe hepatic steatosis and obesity in co-housed WT mice.13 Together, these studies suggested the possibility of host genetic inflammasome defects acting as initial culprits creating a microbiota composition that is subsequently able to drive inflammatory and metabolic diseases.
Using littermate-controlled experimental setups with mice deficient in Nlrp6 or ASC, we recently showed that Nlrp6 and ASC inflammasomes do not perform such a primary causative role in eliciting dysbiosis and its associated diseases.14 Detailed analyses of the fecal microbiota in these set-ups, as summarized below, highlighted the need for carefully designed experiments minimizing non-genetic confounders such as maternal inheritance and long-term separate housing when evaluating the impact of host immunity on intestinal ecosystems.
Littermate-controlled experiments reveal that mother and cage covariates trump host inflammasomes in shaping the commensal gut microbiota composition
In our study, we first tested the previously employed experimental set-up analyzing mice originating from distinct homozygous WT or Nlrp6-deficient breeding pairs. The fecal microbiota composition of these separately housed Nlrp6−/− and non-littermate C57BL/6J mice in our animal facility did not recapitulate the reported differences in Prevotellaceae levels, but did reveal several other differentially represented bacterial taxa, such as Porphyromonodaceae and Bacteroidaceae.14 However, despite these and other individual taxa differences, the microbiota populations did not cluster according to the Nlrp6−/− and C57BL/6J genotypes. Accordingly, distance-based redundancy analyses indicated that between host genetics, mother and cage covariates, only the latter two significantly contributed to the overall observed gut microbiota variation. Together, these data indicated that the fecal microbiota variation observed throughout non-littermate Nlrp6−/− and C57BL/6J mice in our animal facility had been driven by maternal inheritance and long-term housing separation rather than by host genetics.14
In order to minimize the impact of these non-genetic mother and cage drivers of microbiota variation, we next applied a setup in which littermates from Nlrp6+/− intercrosses were separated according to their genotype upon weaning (Fig 1A, left panel). In this approach, the uniform Nlrp6+/− genotype of the mothers allowed for control over the maternal inheritance of the microbiota, while the limited time spent in separate cages minimized the non-genetic housing confounder that was observed in the non-littermate set-up. However, although reducing the impact of these non-genetic confounders was expected to more easily detect true host genetic effects on the gut microbiota, we could not observe any influence of Nlrp6 deletion on the fecal microbiota composition in this littermate-controlled set-up. Indeed, no alterations in bacterial diversity or in the microbial community were observed in Nlrp6−/− mice compared to their Nlrp6+/+ littermates even after physical separation up to 1 year of age.14 Even though these observations using littermate controls indicated that Nlrp6 does not impact on the gut microbiota, we took our analyses a step further by generating mice in which Nlrp6 deletion could exert its presumed microbiota-shifting activity also during the pre-weaning time that is important for microbiota colonization of the intestine. For this purpose, the microbiota composition of F2 homozygous offspring from separately housed Nlrp6−/− and Nlrp6+/+ littermates was analyzed, since these F2 mice were separated also during the pre-weaning time (Fig 1A, right panel). Strikingly, F2 Nlrp6−/− and F2 Nlrp6+/+ mice harbored no differences in their fecal microbiota communities, indicating that Nlrp6 deficiency did not alter the intestinal microbial composition even when given a lifetime of potential impact.14
Figure 1.
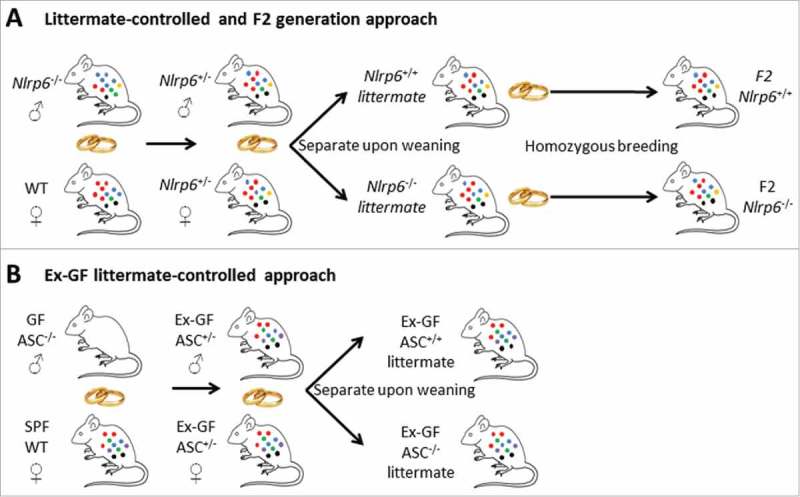
Littermate-controlled, F2 and ex-GF experimental approaches to evaluate host genetic impacts on the gut microbiota. (A) Breeding scheme used for generating Nlrp6−/− and Nlrp6+/+ littermates from non-littermate Nlrp6−/− and WT mice with a suspected Nlrp6-imposed dysbiosis, with generation of F2 mice for evaluating potential host genetic effects during pre-weaning time. Neither the Nlrp6−/− and Nlrp6+/+ littermates, nor the F2 Nlrp6−/− and Nlrp6+/+ mice reproduced the dysbiosis observed in non-littermate Nlrp6−/− and WT mice. (B) Breeding scheme used to generate ex-GF ASC−/− and ASC+/+ littermates from a GF ASC−/− male and an SPF WT female, in which the latter was the sole gut microbiota donor. Ex-GF ASC−/− and ASC+/+ littermates were allowed to colonize their intestines naturally after birth, and did not show differential gut microbiota compositions.
Finally, in a parallel series of experiments performed in an independent animal facility, we used separately housed littermates obtained from ASC+/− intercrosses to evaluate whether overall inflammasome deficiency affected the gut microbiota composition. In this breeding scheme, the ASC+/− parents derived from a germ-free (GF) ASC−/− male and a Specific Pathogen Free (SPF) WT C57BL/6J female (Fig 1B), which allowed analysis of the downstream impact of ASC on a gut microbiota community that originated solely from one C57BL/6J mouse. Like in our previous Nlrp6 experiments, ASC−/− and ASC+/− littermates separated as long as 15 weeks after weaning did not display any genotype-specific alterations in the fecal bacterial diversity or community structure.14 These analyses showed that when carefully controlling for maternal inheritance and housing effects through littermate-controlled set-ups, Nlrp6 and ASC inflammasomes did not affect the commensal gut microbiota composition.
NLRP6/ASC inflammasomes do not shape the commensal gut microbiota
Our above described results obtained with littermate-controlled experiments contrast to the previously suggested roles of Nlrp6 and ASC in shaping the intestinal microbiota composition.12,13,15-17 However, each of these earlier reports was based on fecal microbiota comparisons between non-littermate or externally sourced WT and KO mice that were separately housed for multiple generations. Several studies demonstrated animal facility-dependent microbiota differences in otherwise genetically identical mice,7,18-20 indicating that the gut microbiota composition can be influenced by multiple non-genetic factors. The latter could include also stochastic events that initiate random alterations in a given microbiota community, a novel concept that was recently coined the ‘Anna Karenina principle’.21 The variety of distinct microbial profiles arising due to stochastic triggers can later mistakenly be interpreted as ‘dysbiosis’ when transmitted further by maternal inheritance and preserved due to prolonged colony isolation, a phenomenon known as the legacy effect.22 In fact, we also encountered a legacy effect in our animal facility, as none of the littermate-controlled or F2 Nlrp6 analyses reproduced the bacterial taxa differences that we had initially observed in the non-littermate Nlrp6−/− versus C57BL/6J mice.14 As we identified mother and cage covariates as significant contributors to the gut microbiota variation in the latter set-up, microbial differences observed in these non-littermate mice likely represented legacy effects.
In contrast to our results, a recent study using littermate controls still found that the Nlrp6 inflammasome affected the gut microbiota.23 This study investigated the effect of Nlrp6 deficiency on a colitogenic IL-10-deficient background and observed altered gut microbiota composition in Il10−/−/Nlrp6−/− versus Il10−/−/Nlrp6+/+ littermates.23 However, the authors also detected differential intestinal inflammation between these Il10−/−/Nlrp6−/− and Il10−/−/Nlrp6+/+ littermates that could underlie the observed effects on intestinal ecology, as intestinal inflammation is known to influence the composition of the gut microbiota.24 Therefore, Seregin et al. performed re-colonization experiments in adult GF WT and Nlrp6−/− mice to assess the role of Nlrp6 on the gut microbiota composition in the absence of intestinal inflammation. Strikingly, fecal microbiota analyses of these ex-GF mice showed an impact of Nlrp6 on intestinal ecology within two weeks.23 These observations were similar to a previous study using an adult ex-GF approach, showing that the gut microbiota communities of ex-GF WT and Nlrp6−/− mice diverged at three weeks after re-colonisation.15 However, while this latter ex-GF study did not specify whether Prevotellaceae alterations previously observed in non-littermate WT and Nlrp6−/− mice were reproduced,15 Seregin et al. did not observe those reported effects on Prevotellaceae and rather found an increase in Akkermansia muciniphila in ex-GF Nlrp6−/− mice.23 These discordant observations in ex-GF mice illustrate that the nature of the donor SPF microbiota could determine potential host genetic effects on the gut microbiota in these experiments. Likewise, factors such as diet and the nature of the resident SPF microbiota composition may influence the impact of a host gene on the gut microbiota when investigated in different animal facilities. However, while the effects of a host gene on the microbiota may fluctuate across different facilities, littermate-controlled experiments within a given animal facility are first needed to exclude non-genetic confounders and as such to reveal whether this gene really impacts on the gut microbiota.
Nevertheless, the question arises whether re-colonizing non-littermate adult GF mice as in the Nlrp6 studies mentioned above,15,23 is an alternative valid approach for investigating host genetic impacts on the gut microbiota. In our ASC−/− study, we designed an ex-GF approach that resulted in analyzing WT and KO littermates that had all colonized their gastrointestinal tract in a natural manner after birth (Fig 1B).14 In contrast, the ex-GF studies showing Nlrp6 effects on the gut microbiota used adult mice that were re-colonized by housing in SPF conditions.15,23 Whereas natural colonization after birth is a gradual process in which niches are occupied by sequential bacterial colonizers, whole microbiota re-colonization in adult mice is likely to be influenced by colonization resistance. In addition, as re-colonizing adult GF mice is accompanied by changes in intestinal physiology and maturation of the host immune system, also these host processes could differentially affect the colonization success of various gut microbes. For instance, it is known that the mucus layer of GF mice is more permeable for bacterial penetration than that of SPF mice, a feature that is reverted upon intestinal microbial colonisation.25 In this respect, it is interesting to note that the Akkermansia muciniphila found to be enriched in adult ex-GF Nlrp6−/− mice are mucus-resident bacteria. Given the reported role for the Nlrp6 inflammasome in secreting mucus from sentinel goblet cells,26 it is plausible that re-colonizing adult NLRP6-deficient mice could be associated with differential repair of the ex-GF mucus layer, which could provoke Akkermansia colonization differences when compared with ex-GF WT mice. Therefore, it would be interesting to evaluate whether Nlrp6+/+ and Nlrp6−/− littermates derived from the ex-GF mice that were re-colonized as adults would yield similar Akkermansia abundance differences when compared with direct colonization of adult GF mice. Similar to our ex-GF approach with ASC−/− mice, such a combined littermate-based ex-GF approach would allow intestinal development and gut microbiota colonization to happen in a physiological manner after birth, and therefore is less likely to be influenced by experimental confounders.
Interestingly, our results showing the inability of inflammasomes to shape the commensal gut microbiota were recently confirmed by a study using an alternative approach to normalize the initial gut microbiota community. While not using littermates, Błażejewski et al. performed embryo transfers of WT C57BL/6N and isogenic caspase-1−/− mice to foster mothers harboring identical SPF intestinal microbial populations.27 This approach yielded WT and caspase-1−/− mice that colonized their gastrointestinal tract in a physiological manner after birth and that acquired normalized SPF gut microbiota compositions in these genotypes. In an impressive effort, the authors performed these embryo transfer experiments in two distinct SPF environments. The normalized fecal microbiota compositions of the WT and caspase-1−/− mice did not diverge over time in either of the SPF animal facilities. Thus, in accordance with our data obtained with ASC-deficient mice, Caspase-1 deletion in this study showed that overall inflammasome signaling does not impact on the commensal gut microbiota composition.
Inflammasomes do not protect from DSS colitis when normalizing the gut microbiota composition
Adding to the hypothesis that inflammasomes could be primary regulators of dysbiosis-associated diseases, Elinav et al. had shown that Nlrp6 deletion conferred increased susceptibility to DSS colitis secondary to intestinal microbial changes provoked by Nlrp6 deficiency.12 However, given our finding that Nlrp6 does not influence the gut microbiota composition, the host genetic effect of Nlrp6 on DSS colitis needed re-evaluation. Using the F2 Nlrp6−/− and F2 Nlrp6+/+ mice that showed no differences in gut microbiota composition, we could not observe any difference in DSS-induced colitis development between these genotypes.14 As such, our study showed that Nlrp6 neither affects intestinal microbiota composition nor predisposes mice to higher susceptibility to DSS-induced colitis. Therefore, whilst it is clear that microbial dysbiosis in some cases can provoke increased colitis, inflammasome deficiency does not drive a microbial dysbiosis capable of doing this. In addition, our DSS colitis results emphasize the importance of normalizing the gut microbiota between genotypes in order to reveal the physiological effect of a given host gene in DSS colitis.14,28
In this respect, also Błażejewski et al. investigated the influence of inflammasomes on DSS colitis susceptibility after normalizing the intestinal microbiota composition by embryo transfer. These DSS colitis experiments, performed in two distinct SPF facilities, showed that caspase-1 deletion in mice reduced DSS-induced colitis severity when compared with WT mice harboring an identical SPF gut microbiota.27 This reduced DSS severity phenotype of caspase-1−/− mice was accompanied with diminished intestinal levels of the inflammasome-activated cytokine IL-18. Indeed, littermate-controlled DSS experiments using epithelial IL-18-deficient mice (IL-18IEC-KO) had previously shown a detrimental role for IL-18 production during DSS colitis development.29 As Nlrp6 deficiency in our gut microbiota normalized DSS experiments did not exhibit protective effects, other caspase-1 inflammasomes presumably can produce sufficient amounts of mature IL-18 in the absence of Nlrp6. As it is plausible that the activation of these Nlrp6-independent inflammasomes depends on the nature and the activity of the intestinal microbiota, it will be interesting to identify the ligands and the inflammasomes responsible for this IL-18 production in future studies.
Nevertheless, the above DSS studies using gut microbiota normalized Nlrp6−/−, caspase-1−/− or IL-18IEC-KO mice (by embryo transfer or by littermate-controlled experiments) are all in contrast with multiple prior studies suggesting that mice lacking inflammasomes or IL-18 develop more severe DSS colitis when compared to non-littermate controls.12,30-32 Thus, these DSS colitis studies highlight the importance of eliminating non-genetic impacts on the gut microbiota - such as by using littermate controls – when studying host genetic effects on physiological processes that can be affected by the microbiota.28
Conclusion
Our recent report studying the impact of Nlrp6/ASC inflammasomes on intestinal ecology clearly showed the utmost importance to control for non-genetic mother and cage covariate-driven observations. We showed that littermate-controlled experimental design minimizes these confounders and we thereby showed that Nlrp6/ASC inflammasomes – in contrast to prior belief stemming from non-littermate controlled studies – do not shape the commensal gut microbiota composition.14
Inflammasomes were not the first innate immunity examples that created confusion with respect to regulating the intestinal microbiota. Indeed, although Toll-like receptor (TLR) as well as Nod2 signaling had been implicated in modelling the intestinal ecosystem,33,34 subsequent studies using littermate comparisons showed that these pathways did not exert the previously described effects on the gut microbiota composition.20,35 Hence, these studies confirmed using littermates as the appropriate experimental setup when investigating host genetic effects in intestinal microbiota regulation. Importantly however, while none of the above TLR, Nod2 and inflammasome signaling studies revealed impacts of host genetics on the gut microbiota using a littermate approach, it should be noted that physical littermate separation upon weaning is capable of detecting such effects. Indeed, studies investigating intestinal epithelial-specific TLR5 deficiency or full-body ablation of Card9 or IL-33 showed that weaning these genetic mouse models and their WT littermates in separate cages resulted in differential gut microbiota communities after 4–6 weeks of separation.36-38 Together, these several mouse genetic studies show that analyzing separately housed littermates is a well-controlled and valid approach for dissecting host genetic effects on intestinal ecosystems.
Overall, there is clear need for better understanding of the interplay between microbiota and the different players of host innate immune system. We hope that our recent study illustrated the need for standardized littermate-controlled experimental design in this research field, as we are convinced that this approach will minimize experimental discrepancies and will help to identify the causes and consequences in host-microbiota metagenomic observations.
Funding Statement
Fund for Scientific Research-Flanders (FWO) ID: Odysseus grant G.0C49.13N; Funding Source: Swiss National Science Foundation (SNSF) ID: SNSF310030_156022; Funding Source: EC | European Research Council (ERC) ID: 281785
Acknowledgments
K.D.M was supported by a grant from the SNSF (SNSF310030_156022) and the European Research Council (ERC, FP/2007-2013)/ERC grant Agreement no 281785). F.R. was supported by a Marie Heim-Vögtlin fellowship from SNSF and an ECCO Grant. A.W. was supported by the Odysseus grant G.0C49.13N from the Fund for Scientific Research-Flanders, and is a post-doctoral fellow with the Fund for Scientific Research-Flanders.
Disclosure of potential conflicts of interest
None of the authors have conflicts of interest or financial disclosures to report.
References
- 1.Belkaid Y, Harrison OJ. Homeostatic Immunity and the Microbiota. Immunity 2017; 46:562–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature 2016; 535:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, et al.. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 2016; 535:94–103. [DOI] [PubMed] [Google Scholar]
- 4.Halfvarson J, Brislawn CJ, Lamendella R, Vazquez-Baeza Y, Walters WA, Bramer LM, et al.. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2017; 2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez KB, Leone V, Chang EB. Western diets, gut dysbiosis, and metabolic diseases: Are they linked? Gut microbes 2017; 8:130–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumberg RS. Environment and Genes: What Is the Interaction? Dig Dis 2016; 34:20–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stappenbeck TS, Virgin HW. Accounting for reciprocal host-microbiome interactions in experimental science. Nature 2016; 534:191–9. [DOI] [PubMed] [Google Scholar]
- 8.Dubois H, Wullaert A, Lamkanfi M. General Strategies in Inflammasome Biology. Curr Top Microbiol Immunol 2016; 397:1–22. [DOI] [PubMed] [Google Scholar]
- 9.Cummings JR, Cooney RM, Clarke G, Beckly J, Geremia A, Pathan S, et al.. The genetics of NOD-like receptors in Crohn's disease. Tissue Antigens 2010; 76:48–56. [DOI] [PubMed] [Google Scholar]
- 10.Schoultz I, Verma D, Halfvarsson J, Torkvist L, Fredrikson M, Sjoqvist U, et al.. Combined polymorphisms in genes encoding the inflammasome components NALP3 and CARD8 confer susceptibility to Crohn's disease in Swedish men. The American journal of gastroenterology 2009; 104:1180–8. [DOI] [PubMed] [Google Scholar]
- 11.Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, et al.. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nature genetics 2009; 41:71–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al.. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011; 145:745–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al.. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012; 482:179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mamantopoulos M, Ronchi F, Van Hauwermeiren F, Vieira-Silva S, Yilmaz B, Martens L, et al.. Nlrp6- and ASC-Dependent Inflammasomes Do Not Shape the Commensal Gut Microbiota Composition. Immunity 2017; 47:339-48 e4. [DOI] [PubMed] [Google Scholar]
- 15.Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman-Schapira G, Mahdi JA, et al.. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 2015; 163:1428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anand PK, Malireddi RK, Lukens JR, Vogel P, Bertin J, Lamkanfi M, et al.. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature 2012; 488:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu B, Elinav E, Huber S, Strowig T, Hao L, Hafemann A, et al.. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci U S A 2013; 110:9862–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rausch P, Basic M, Batra A, Bischoff SC, Blaut M, Clavel T, et al.. Analysis of factors contributing to variation in the C57BL/6J fecal microbiota across German animal facilities. International journal of medical microbiology: IJMM 2016; 306: 343–55. [DOI] [PubMed] [Google Scholar]
- 19.Rogers GB, Kozlowska J, Keeble J, Metcalfe K, Fao M, Dowd SE, et al.. Functional divergence in gastrointestinal microbiota in physically-separated genetically identical mice. Scientific reports 2014; 4:5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, et al.. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med 2012; 209:1445–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaneveld JR, McMinds R, Vega Thurber R. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol 2017; 2:17121. [DOI] [PubMed] [Google Scholar]
- 22.Kostic AD, Howitt MR, Garrett WS. Exploring host-microbiota interactions in animal models and humans. Genes & development 2013; 27:701–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seregin SS, Golovchenko N, Schaf B, Chen J, Pudlo NA, Mitchell J, et al.. NLRP6 Protects Il10-/- Mice from Colitis by Limiting Colonization of Akkermansia muciniphila. Cell reports 2017; 19:733–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology 2008; 134:577–94. [DOI] [PubMed] [Google Scholar]
- 25.Johansson ME, Jakobsson HE, Holmen-Larsson J, Schutte A, Ermund A, Rodriguez-Pineiro AM, et al.. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host Microbe 2015; 18:582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birchenough GM, Nystrom EE, Johansson ME, Hansson GC. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 2016; 352:1535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blazejewski AJ, Thiemann S, Schenk A, Pils MC, Galvez EJC, Roy U, et al.. Microbiota Normalization Reveals that Canonical Caspase-1 Activation Exacerbates Chemically Induced Intestinal Inflammation. Cell reports 2017; 19:2319–30. [DOI] [PubMed] [Google Scholar]
- 28.McCoy KD, Geuking MB, Ronchi F. Gut Microbiome Standardization in Control and Experimental Mice. Curr Protoc Immunol 2017; 117:23 1 1- 1 13. [DOI] [PubMed] [Google Scholar]
- 29.Nowarski R, Jackson R, Gagliani N, de Zoete MR, Palm NW, Bailis W, et al.. Epithelial IL-18 Equilibrium Controls Barrier Function in Colitis. Cell 2015; 163:1444–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 2010; 32:379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, et al.. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity 2010; 32:367–78. [DOI] [PubMed] [Google Scholar]
- 32.Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, et al.. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med 2010; 207:1045–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al.. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 2008; 455:1109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, et al.. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A 2009; 106:15813–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson SJ, Zhou JY, Geddes K, Rubino SJ, Cho JH, Girardin SE, et al.. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut microbes 2013; 4:222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malik A, Sharma D, Zhu Q, Karki R, Guy CS, Vogel P, et al.. IL-33 regulates the IgA-microbiota axis to restrain IL-1alpha-dependent colitis and tumorigenesis. J Clin Invest 2016; 126: 4469–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chassaing B, Ley RE, Gewirtz AT. Intestinal Epithelial Cell Toll-like Receptor 5 Regulates the Intestinal Microbiota to Prevent Low-Grade Inflammation and Metabolic Syndrome in Mice. Gastroenterology 2014; 147: 1363–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, et al.. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016; 22: 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]


