ABSTRACT
Antibiotics offer an efficient means for managing diseases caused by bacterial pathogens. However, antibiotics are typically broad spectrum and they can indiscriminately kill beneficial microbes in body habitats such as the gut, deleteriously affecting the commensal gut microbiota. In addition, many bacteria have developed or are developing resistance to antibiotics, which complicates treatment and creates significant challenges in clinical medicine. Therefore, there is a real and urgent medical need to develop alternative antimicrobial approaches that will kill specific problem-causing bacteria without disturbing a normal, and often beneficial, gut microbiota. One such potential alternative approach is the use of lytic bacteriophages for managing bacterial infections, including those caused by multidrug-resistant pathogens. In the present study, we comparatively analysed the efficacy of a bacteriophage cocktail targeting Escherichia coli with that of a broad-spectrum antibiotic (ciprofloxacin) using an in vitro model of the small intestine. The parameters examined included (i) the impact on a specific, pre-chosen targeted E. coli strain, and (ii) the impact on a selected non-targeted bacterial population, which was chosen to represent a defined microbial consortium typical of a healthy small intestine. During these studies, we also examined stability of bacteriophages against various pH and bile concentrations commonly found in the intestinal tract of humans. The bacteriophage cocktail was slightly more stable in the simulated duodenum conditions compared to the simulated ileum (0.12 vs. 0.58 log decrease in phage titers, respectively). It was equally effective as ciprofloxacin in reducing E. coli in the simulated gut conditions (2–3 log reduction), but had much milder (none) impact on the commensal, non-targeted bacteria compared to the antibiotic.
KEYWORDS: phage, persistence, antibiotic, in vitro, small intestine
1. Introduction
Antibiotics are a type of antimicrobial drug used for either killing (bactericidal antibiotics; e.g., vancomycin) or inhibiting the growth (bacteriostatic antibiotics; e.g., tetracycline) of microorganisms. The discovery of antibiotics in the first half of the 20th century revolutionized medicine as it allowed treatment of bacterial infections that were essentially untreatable during the pre-antibiotic era; in addition to saving countless lives, it also fostered the development of many sophisticated medical procedures (e.g., organ transplantation surgeries) that were previously not possible due to, in part, the inability to manage bacterial infections that are unavoidable during such surgical procedures. However, extensive use (and sometimes misuse) of antibiotics since their discovery has also led to an emergence of antibiotic-resistant bacteria – bacteria that cannot be killed by commonly available antibiotics, and in some cases by any available antibiotics; e.g., multiple drug resistance (MDR), extensively drug resistant (XDR), and pan-drug-resistant (PDR) bacteria.
This emergence of drug-resistance is a very serious problem in modern medicine, which imposes a significant social and economic burden on the society, including the loss of lives.1,2 The problem is further exacerbated by the fact that most antibiotics are broad-spectrum. In practice this means that in addition to targeting disease-causing bacteria, antibiotic therapy can cause substantial collateral damage to the human microbiota by killing many other, non-targeted, and often beneficial bacteria. This collateral effect can, and often does, lead to dysbiosis,3,4 further promoting the emergence of resistant bacteria (via selective pressure), and may also facilitate horizontal transfer of resistance genes.5,6 Moreover, growing evidence suggests that microbiota disturbance caused by antibiotics may promote various other health problems such as obesity, asthma, inflammatory bowel disease, and diabetes.1-4 Therefore, there is an increasing interest to manage infections caused by antibiotic-resistant pathogens by selectively targeting the disease-causing bacteria, without disturbing the commensal microbiota of the human gastrointestinal (GI) tract. One intriguing concept in that regard is the use of lytic bacteriophages, to selectively and specifically kill disease-causing bacteria, including MDR, XDR, and PDR.
Bacteriophages (or “phages” for short) are bacteria-infecting viruses. Lytic phages have potent bactericidal activity against their host bacterial strains. During the lytic cycle the phage infects the cell, using the cell's replication and translation machinery to replicate and then lyses the cell releasing new phage particles into the environment. In cases where overwhelming concentrations of phage are applied “lysis from without” might occur as well.7,8 Phages are also very specific: they only attack their targeted bacterial hosts, and they cannot infect human or other eukaryotic cells. Even within bacterial taxa, and in clear contrast to broad-spectrum antibiotics, phages usually only lyse strains or a subgroup of strains within the bacterial species, making targeted bacterial therapy possible.
Increased presence of Adherent-Invasive Escherichia coli in the ileum has been associated with Ileal Crohn's disease (ICD)9-11 and ICD-associated E. coli has been found to manifest multidrug resistance.12 Therefore, phages targeting these bacteria might be an alternative for antibiotics and have potential therapeutic ability in ICD management. In order to lyse their targeted bacteria in the small or large intestine, orally-administered phages must pass through the harsh environment of the human GI tract, including the low pH in the stomach, presence of pancreatic enzymes, and bile salts in the small intestine.13-15 These factors may reduce phage viability/stability and render them less effective or ineffective; yet, despite the long history of therapeutic use of phages in humans, there is striking paucity of information on the pharmacokinetics of bacteriophages when they are administered orally.16-18 Thus, the goal of this study was to start to elucidate the persistence of bacteriophages in the human GI tact, by testing a phage cocktail targeting E. coli in a simulated human small intestinal model system. During these studies, we also compared the impact of the phage cocktail vs. a commonly used broad-spectrum antibiotic, ciprofloxacin, on the levels of E. coli, and other selected (non-targeted, representative of the “normal” microbiota) bacteria in the simulated small intestine system.
2. Materials and methods
2.1. Small intestine model system: Consortium of microorganisms and growth conditions
To simulate a normal, healthy, small intestine microbiome, a consortium of 7 bacterial species were selected to represent a healthy ileal microbiota19,20 (Table 1). All bacteria were acquired from the German Collection of Microorganisms and Cell cultures (DSMZ).21 Strains were propagated in Gifu Anaerobic Medium (GAM Broth, NISSUI) in an anaerobic bench (CoyLab, USA), under strictly anaerobic conditions using an Anaerobic jar (Merck) together with the AnaeroGen system (Oxoid). Before each experiment, all strains were cultured separately in 10 ml GAM broth for the incubation time optimal for each individual strain according to.23 After propagation, bacterial cells were centrifuged for 2 minutes at 4,000 g, the supernatant was discarded, and precipitated bacteria were resuspended in volumes of PBS (pH 7.4) adequate to obtain a final concentration of ca. 108 CFU/ml for each strain. After adjusting concentrations, all strains were mixed in an equal ratio and either used immediately or stored in liquid nitrogen until future use.
Table 1.
Bacterial strains, their source, culturing time and range on growth media. Taken from {Cieplak:Rpap5xgx}.
| Species | Strain source | Origin | Culture time [h] | Culture media |
|---|---|---|---|---|
| Escherichia coli | DSM 1058 | Human intestine | 24 | VRB, MCC, GAM |
| Streptococcus salivarius | DSM 20560 | Blood | 6 | M17, GAM |
| Streptococcus luteinensis | DSM 15350 | Human isolate | 24 | M17, GAM |
| Enterococcus faecalis | DSM 20478 | Human faeces | 24 | MCC, GAM |
| Bacteroides fragilis | DSM 2151 | Appendix abscess | 24 | GAM |
| Veillonella parvula | DSM 2008 | Human intestine | 48 | GAM |
| Flavonifractor plautii | DSM 6740 | Human faeces | 48 | GAM |
VRB: Violet Red Bile Agar, M17: M17 Agar, MCC: MacConkey Agar and GAM: Gifu Anaerobic Agar.
2.2. Bacteriophage preparation
For this pilot study, we specifically formulated a bacteriophage preparation to target one strain from the representative ileal consortium: E. coli DSM 1058. For the phage cocktail, individual monophages were selected from Intralytix's bacteriophage collection, based on their ability to lyse the DSM 1058 strain in the classical Spot Test assay.22 Various dilutions were used for the spot test to differentiate between lysis and inhibition (i.e., low dilutions resulting in plaques were tested as well). The resulting susceptibility data were analyzed using the PhageSelector™ program (proprietary program developed by Intralytix) to formulate bacteriophage cocktail Ec17B153DK1. The resulting cocktail is composed of 3 phages (ECML-363, ECML-122 and ECML-359) each having potent lytic activity against E. coli DSM 1058. Each component monophage was propagated separately in their respective E. coli host strains at 37°C, with Multiplicity of infections (MOIs) ranging from 2 × 10−4 to 1 × 10−1. Following propagation, each phage was harvested by filtering through a 0.2-micron filter and concentrated/buffer exchanged in a 0.85% saline solution. Following buffer exchange, the three monophages were combined in approximately equal concentrations to produce the phage cocktail Ec17B153DK1. The final cocktail was then sterile filtered through a 0.2-micron filter and stored refrigerated (2–8°C) until use.
The ability of each of the three phages included in the Ec17B153DK1 phage cocktail, and the phage cocktail itself, was also tested against an additional 607 E. coli strains (Fig. 1) and against the 6 non-E. coli strains (Table 1) used in our model system.
Figure 1.
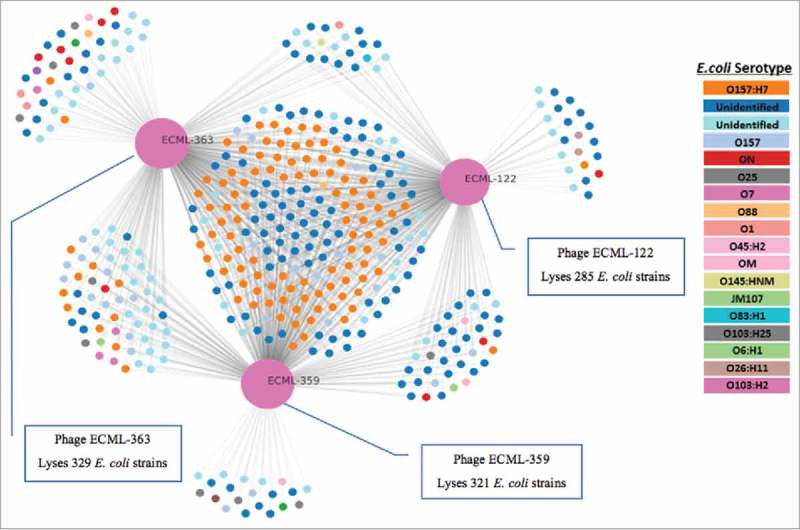
Host range of the Ec17B153DK1 component phages against 408 E. coli isolates. Host range and interaction network of the three monophages (ECML-122, ECML-363, ECML-359) included the Ec17B153DK1 phage cocktail. Phages are depicted as large pink circles. Results shown are for 408 susceptible E. coli strains (i.e., 199 resistant strains were not included in the chart for simplicity). Each small circle is an E. coli isolate. The gray lines connecting each phage to bacterial strains indicate the ability to infect and kill that given E. coli strain based on the Spot Test assay. Bacterial strains killed by more than one phage are grouped towards the canter of the chart. The graph was generated using proprietary PhageSelector™ program developed by Intralytix, Inc.
2.3. Small intestine in vitro simulation
To simulate passage of phages through the human small intestine (SI), we used a newly developed dynamic in vitro model (TSI).23 The TSI consists of 5 reactors with working volumes of 12 ml each. Each reactor simulates the small intestine of one individual. Parameters like pH, temperature, and bile salts concentration and pancreatic juice were monitored and maintained at physiologically relevant levels during simulations (4–10 mM bile salts; 40 – 100 U/ml pancreatic juice). Both bile salts and pancreatic juice originated from pigs. Reactors were prepared and run according to.23 Briefly, before addition of phages to the reactors, the pH was adjusted and confirmed to be 6.5 at which point 0.5 ml of the phage cocktail (10.81 log PFU/ml) or controls were added to the simulated small intestines. From there on the whole experiment is divided into 3 stages: duodenum, jejunum and ileum, each of them characterized by different conditions. During duodenum passage (2 hours), pH is elevated from 6.5 to 6.8 and pancreatic juice and bile salts are administrated. During the jejunum stage (4 hours), pH is raised from 6.8 to 7.2, while bile salts and small nutrients are re-absorped. Finally, during the ileum stage the small intestine microbial consortium / E. coli suspension is added and pH is stabilized at 7.2 during 2 hours as described in detail in.23
2.4. Bacteriophages persistence and impact on E. coli during small intestine passage
To test the persistence of the phage cocktail during small intestinal transit, and its efficacy against the targeted E. coli strain, four TSI reactors were inoculated with 0.5 ml of the bacteriophage cocktail (10.81 log PFU/ml), in the last reactor we added saline solution (0.5 ml, 0.9% NaCl) as control. Simulated small intestinal digestion was carried out as described in.23 Before the ileal step 1 ml of E. coli suspension (7 log CFU/ml), propagated in a similar manner as described in section 3.1. was added into each reactor. Persistence of the phage cocktail was tested before the duodenum (baseline), after duodenum and after the ilium stage by determining plaque forming units (PFU) counts24 in an E. coli host strain grown on VRB (Violet Red Bile Agar, Sigma-Aldrich) selective agar media. Efficiency of phage cocktail against E. coli was quantified at the end of the ileal stage by comparing bacteria counts before and after the ileal stage in phage-treated vs. untreated control samples, via colony plate counts on VRB agar medium.
2.5. Impact of bacteriophage cocktail versus ciprofloxacin on the microbiota
To compare the impact of the bacteriophage cocktail vs. an antibiotic on the ileal microbiota, a mixture of 7 bacterial strains of various spp. (Table 1) was co-incubated with the phage cocktail, the antibiotic (ciprofloxacin), or water (control), and the impact of each treatment on the overall bacterial counts was determined by plating on selective and non-selective substrates. Experiments were conducted for each preparation and feeding condition in duplicate. Briefly, 0.5 ml of phage cocktail (10.81 log PFU/ml) was added in two reactors and 0.5 ml of water was added to the other three reactors, of which two were also supplemented with 1 ml of ciprofloxacin (500 mg/l in final solution). Before the ileal phase of the experiment, 1 ml of microbial consortia (see Table 1), and 3 ml of fresh SIF consisting of bile salts and pancreatic juice were added to each reactor. After the experiment, samples were taken from each reactor and plated on the following 4 media, followed by incubation at 37 °C for 24 h: (1) VRB for enumeration of E. coli, (2) M17 Agar (Oxoid) for enumeration of Streptococcus sp., (3) MacConkey Agar (MCC, Sigma-Aldrich) for enumeration of Enterococcus feacalis, and (4) Gifu Anaerobic Agar (GAM, NISSUI) where all microbes from the small intestine consortium (Table 1) can grow.
2.6. Statistical analysis
All statistical analyses were performed using the Prism 7 v 7.0b software (GraphPad). Differences between groups were calculated using one-way ANOVA with Tukey's multiple comparisons test, with a single pooled variance. Significance was determined at P<0.05 level.
3. Results
3.1. Bacteriophage preparation: Target range and specificity
Bacteriophage cocktail Ec17B153DK1 was specifically formulated for this study. It was specifically designed to lyse E. coli DSM 1058, which is one of 7 strains included in the small intestine model consortia. Each of the three phages included in the Ec17B153DK1 cocktail was capable of lysing DSM 1058. We also tested monophages in the cocktail for their ability to lyse a panel of an additional 607 E. coli isolates, using the classical Spot Test assay; the cocktail lysed 408 (67%) of 607 E. coli strains examined (including DSM 1058) (Fig. 1). The cocktail was also tested against six strains of the non-targeted consortia included in our model system (Table 1), using the same Spot Test assay. None of the six non-E. coli strains included in our TSI model were susceptible to the cocktail (data not shown).
3.2. Bacteriophage stability in the small intestine in vitro model
We tested persistence of phages in the small intestine in the presence of Escherichia coli and in two different feeding conditions mimicking either a “fasted” small intestine (i.e., mimicking conditions before a meal; bile salts = 4mM; pancreatic Juice = 40 U/ml) or a “fed” small intestine (i.e., mimicking conditions after a meal; bile salts = 10 mM; pancreatic juice = 100 U/ml). The number of phages did not change significantly (P>0.05) under the fasted conditions for either the duodenum (10.59 ± 0.06 log PFU/ml) or the ileum simulated conditions remaining at ca. 10.32 ± 0.17 log PFU/ml). In the fed state, the concentration of phages remained stable until the end of duodenum stage (10.76 ± 0.14 log PFU/ml) and then slightly decreased at the end of the ileum stage (from 10.81 log PFU/ml to 10.17 log PFU/ml (±0.35); P = 0.048),.
3.3. Impact of the bacteriophage cocktail vs. antibiotic on the levels of E. coli in the small intestine in vitro model
The ability of our phage cocktail to reduce the levels of the targeted E. coli strain in the model system was evaluated by inoculating 1 ml of E. coli culture into the simulated ileal section (both fed and fasted) of the small intestine, and adding either the phage cocktail or sterile water (as control). For both fasted and fed conditions, we observed a significant decrease in E. coli cell counts (Fig. 2 Fasted: P <0.01, Fed: P <0.01). Specifically, addition of bacteriophages reduced the levels of the targeted E. coli cells by ca. 2.5 log (99,5%) in both fed and fasted conditions. There was no reduction of E. coli levels in the phage-untreated, control group; in fact, the E. coli population slightly increased in control samples under the fed conditions (Fig. 2). The reduction in the E. coli population was slightly more pronounced under the fed condition compared to fasted conditions, although the difference was not statistically significant.
Figure 2.
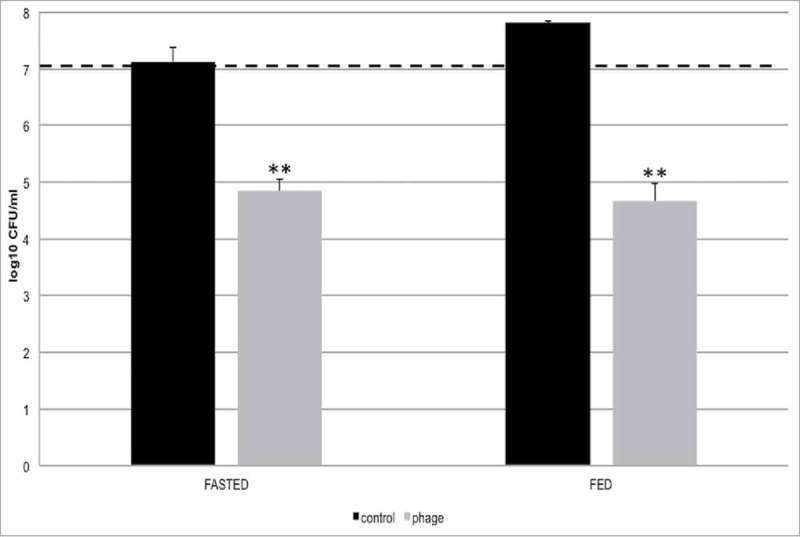
Lytic activity of the bacteriophage cocktail against targeted bacteria in the TSI model. Testing the lytic activity of the phage cocktail in two feeding conditions (fasted, fed). E. coli culture was added to the reactors at the ileum stage (ca. 7 log CFU/mL). The data shown are for the ileum compartment. The dotted line indicates the number of E. coli before treatment with the bacteriophage cocktail. All experiments were performed in triplicates (n = 3). *P < 0.05 – significant. **P < 0.01 – highly significant.
3.4. Impact of bacteriophage cocktail vs. antibiotic on the ileal bacterial community
The impact of a broad-spectrum antibiotic (ciprofloxacin) on the ileal bacterial consortia was compared to that of the bacteriophage cocktail (Fig. 3). The phage cocktail and ciprofloxacin showed similar efficacy in reducing E. coli, each yielding on average a 2.5 log (99.5%) reduction (Fig. 3). However, the phage cocktail was highly specific towards targeted E. coli species and did not significantly reduce any of the other species of the ileal consortium (p>0.05 for both Fasted state and Fed state) (Fig. 3). In contrast, ciprofloxacin reduced counts of all other bacteria on average of 1 log (90%) on all growth media for both fed and fasted states (Fig. 3).
Figure 3.
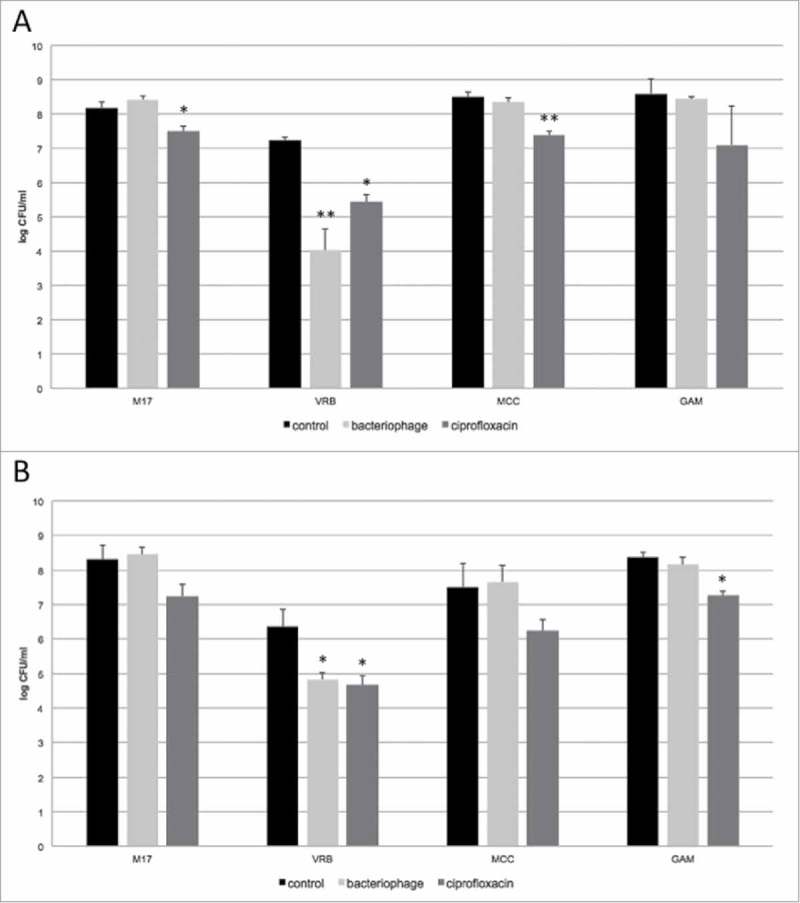
Impact of ciprofloxacin and bacteriophage cocktail on simulated small intestine microbiota in fasted (A) and fed (B) conditions (7 different bacterial spp., Table 1) in the TSI model. Survival of different bacteria species from the simulated small intestinal microbiota was tested on four different culturing media: Violet Red Bile Agar (VRB) for enumeration of E. coli, M17 Agar (M17) for enumeration of Streptococcus sp., MacConkey Agar (MCC) for enumeration of E. faecalis, and Gifu Anaerobic Agar (GAM) where all species from small intestinal consortium can be cultivated. All experiments were performed in duplicate (n = 2). *P < 0.05 – significant (difference between bacteriophage or ciprofloxacin treatment compared to control samples). **P < 0.01 – highly significant (difference between bacteriophage or ciprofloxacin treatment compared to control samples).
4. Discussion
4.1. Phages retain significant infectivity after passage through simulated small intestines
Most phage treatments are administrated orally, although phages have been applied to humans therapeutically via various other routes as well, including auricular, intravesical, intrapulmonary, rectal, topical, and intravenous (reviewed in25). For oral administration, it is generally believed that phages are deleteriously impacted by the low acidity of the stomach; hence, many oral applications involved administering phages together with bicarbonate water (to neutralize stomach acidity) or in a special enteric formulations (gel caps or tablets) that provide protection during passage through the acidic stomach environment (pH ranging from 1 – 2 up to 4 – 5) by dissolving and releasing phages only during less acidic sections of the intestinal track, such as the small intestine (pH 6 – 7.4), caecum (pH 5.7) or the rectum (pH 6.7).26 However, rigorous data on the persistence of phages in various GI tract sections are not available, and it is not well established how various physiological conditions that impact gut parameters (e.g., consumption of foods) may further influence this.
To begin addressing these questions, a TSI in vitro system23 was used to determine phage stability and efficacy by simulating various sections of the small intestine (duodenum, jejunum or ileum) both in “fasted” or “fed” conditions. In general, the varying environmental conditions (pH, bile) during simulated small intestinal passage did not affect the stability of the phages. There was slight decrease in phage titers in the ileum under the fed conditions; however, the reduction was relatively minor and the phage cocktail was still able to significantly reduce E. coli levels in our in vitro system. Specifically, the phage cocktail reduced E. coli levels by the same ca. 2.5 log both in fed and fasted conditions. This study's results diverge from the study by Ma et al., who reported larger (1.29 and 1.67 log units, respectively) loss of Salmonella phage Felix O1) titers in 1% and 2% bile solution.27 The negative impact of bile salts on phage viability has been reported by some other authors as well28-30; however, the topic is still not fully understood as other investigators – and this study – suggest that phages can be fairly stable and remain infectious when exposed to bile salts ranging from 4–10 mM).31 It is possible that the phage viability will be more profoundly reduced in vivo due to many additional factors (e.g., intestinal peristalsis, more complex microbiota, various diets, etc.) that may deleteriously impact phage particles in the GI tract. Also, different bacteriophages can have different stability against bile salts, pH, etc. Elucidating the underlying mechanisms could prove to be a fertile area for subsequent investigations, and the results could offer important insights into the basic phage biology as well as for designing optimal phage preparations for various oral phage therapy applications.,
4.2. Bacteriophages preserve representative small intestine microbiome
In our studies, the phage preparation and ciprofloxacin reduced the E. coli levels in our in vitro model system by the same ca. 2.5 log. However, the two antimicrobials had very different impact on the non-targeted microbiota. Specifically, phage administration had no detectable impact on the six non-E. coli bacterial species included in our model system; whereas, ciprofloxacin reduced the levels of all bacteria in the consortium (Fig. 3). Short and long-term use of antibiotics could lead to a wide range of undesired effects, such as diarrhea, dysbioses, and other indirect deleterious effects such as obesity or colitis. These deleterious effects are usually due to the antibiotic-caused non-specific reduction of beneficial gut microbes, and the associated general perturbance in the gut microbiota. In this context, ciprofloxacin is one of the most widely used antibiotics to treat E. coli infections in the gut.32,33 Unfortunately, it is also a major disruptor of the gut microbiota, and has been shown to profoundly alter the natural taxa in the GIT after only a single course of treatment, with some taxa never recolonizing.34 Our studies provide further support of this indirect action of the antibiotic, as it led to a 1 log-reduction of the representative SI consortia in this study. In contrast, the phage preparation only impacted the E. coli populations, and had no impact on any of the other six “commensal” bacterial species included in our model.
The specificity of bacteriophages may offer some important medical / health benefits. For example, it can help mitigate unnecessary bacterial diversity reduction (and potential dysbiosis) in the gut. While there is a plethora of knowledge on microbial dysbiosis as it relates to antibiotic treatments, there is less understanding on how phages may affect the microbiota. Of the few studies examining phage therapy, none have reported any adverse effects on human health, or major alterations of the microbiome.17,18,35,36 The present study corroborated those findings. Noteworthy, the bacteriophage cocktail did also lyse a large number of other E. coli strains in vitro; i.e., it was not solely lytic for one targeted E. coli strain. Thus, the use of such cocktail may still potentially impact some non-targeted E. coli in the small intestine. However, the use of a broad host range E. coli phage T4 (“broad host range” defined here as the ability of a given bacteriophage to lyse several strains of the same bacteria species) has been reported not to deleteriously change the normal E. coli microbiota of human volunteers.16 Additional studies, including larger scale trials in human volunteers, will be required to better elucidate the impact of phage administration on the commensal microbiota. Overall, our data support the idea that properly selected phage preparations could be potentially at least equally effective as a commonly prescribed antibiotic in reducing the E. coli levels in the small intestine, but may have a significantly milder impact on non-targeted, “normal microbiota” bacteria.
Funding Statement
The research leading to these results has received funding from the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme FP7/2007-2013/under REA grant agreement n° 606713 The people programme (Marie Curie Actions).
Disclosure of potential conflicts of interest
This manuscript has not been published elsewhere and has not been submitted simultaneously for publication elsewhere.
References
- 1.Blaser M. Antibiotic overuse: stop the killing of beneficial bacteria. Nature. Nature Publishing Group; 2011;476:393–4. doi: 10.1038/476393a. PMID:21866137. [DOI] [PubMed] [Google Scholar]
- 2.Pérez-Cobas AE, Artacho A, Knecht H, Ferrús ML, Friedrichs A, Ott SJ, Moya A, Latorre A, Gosalbes MJ. Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PLoS One. Public Library of Science; 2013;8:e80201. doi: 10.1371/journal.pone.0080201. PMID:24282523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dethlefsen Les Shmlsdar The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLoS Biol. Public Library of Science; 2008. November;6:2383–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewardson AJ, Huttner B, Harbarth S. At least it won't hurt: the personal risks of antibiotic exposure. Curr Opin Pharmacol. Elsevier; 2011;11:446–52. doi: 10.1016/j.coph.2011.06.011. PMID:21775205. [DOI] [PubMed] [Google Scholar]
- 5.US CFDCAP Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Office of Infectious Disease. Antibiotic resistance threats in the United States; 2013 April. Available at: https://www.cdc.gov/drugresistance/threatreport-2013/index.html [Google Scholar]
- 6.Vollaard EJ, Clasener HA. Colonization resistance. Antimicrobial Agents and Chemotherapy. American Society for Microbiology (ASM). 1994;38:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abedon ST. Lysis from without. Bacteriophage. 2011. January;1:46–49. doi: 10.4161/bact.1.1.13980. PMID:21687534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woolston J, Parks AR, Abuladze T, Anderson B, Li M, Carter C, Hanna LF, Heyse S, Charbonneau D, Sulakvelidze A. Bacteriophages lytic for Salmonella rapidly reduce Salmonella contamination on glass and stainless steel surfaces. Bacteriophage. 2013. July;3:e25697. doi: 10.4161/bact.25697. PMID:24228226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel J-F. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology. Elsevier; 1998;115:1405–13. doi: 10.1016/S0016-5085(98)70019-8. PMID:9834268. [DOI] [PubMed] [Google Scholar]
- 10.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser A-L, Barnich N, Bringer M-A, Swidsinski A, Beaugerie L, Colombel J-F. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004. July;127:412–21. doi: 10.1053/j.gastro.2004.04.061. PMID:15300573. [DOI] [PubMed] [Google Scholar]
- 11.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG, et al.. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 2007. September;1:403–18. doi: 10.1038/ismej.2007.52. PMID:18043660. [DOI] [PubMed] [Google Scholar]
- 12.Dogan B, Scherl E, Bosworth B, Yantiss R, Altier C, McDonough PL, Jiang Z-D, DuPont HL, Garneau P, Harel J, et al.. Multidrug resistance is common in Escherichia coli associated with ileal Crohn's disease. Inflamm Bowel Dis. 2013. January;19:141–50. doi: 10.1002/ibd.22971. PMID:22508665. [DOI] [PubMed] [Google Scholar]
- 13.Clark WA. Comparison of several methods for preserving bacteriophages. Appl Environ Microbiol. Am Soc Microbiol. 1962;10:466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabig M, Herman-Antosiewicz A, Kwiatkowska M, Los M, Thomas MS, Węgrzyn G. The cell surface protein Ag43 facilitates phage infection of Escherichia coli in the presence of bile salts and carbohydrates. Microbiology (Reading, Engl.). 2002. May;148:1533–42. doi: 10.1099/00221287-148-5-1533. PMID:11988528. [DOI] [PubMed] [Google Scholar]
- 15.Joerger RD. Alternatives to antibiotics: bacteriocins, antimicrobial peptides and bacteriophages. Poult Sci. Oxford University Press; 2003;82:640–7. doi: 10.1093/ps/82.4.640. PMID:12710486. [DOI] [PubMed] [Google Scholar]
- 16.Bruttin A, Brüssow H. Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy. Antimicrobial Agents and Chemotherapy. Am Soc Microbiol. 2005;49:2874–8. doi: 10.1128/AAC.49.7.2874-2878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarker SA, McCallin S, Barretto C, Berger B, Pittet A-C, Sultana S, Krause L, Huq S, Bibiloni R, Bruttin A, et al.. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology. 2012;434:222–32. doi: 10.1016/j.virol.2012.09.002. PMID:23102968. [DOI] [PubMed] [Google Scholar]
- 18.Sarker SA, Sultana S, Reuteler G, Moine D, Descombes P, Charton F, Bourdin G, McCallin S, Ngom-Bru C, Neville T, et al.. Oral Phage Therapy of Acute Bacterial Diarrhea With Two Coliphage Preparations: A Randomized Trial in Children From Bangladesh. EBioMedicine. 2016. February;4:124–37. doi: 10.1016/j.ebiom.2015.12.023. PMID:26981577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dlugosz A, Winckler B, Lundin E, Zakikhany K, Sandström G, Ye W, Engstrand L, Lindberg G. No difference in small bowel microbiota between patients with irritable bowel syndrome and healthy controls. Nature Scientific Reports. 2015. February;5:8508. doi: 10.1038/srep08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung C-S, Chang P-F, Liao C-H, Lee T-H, Chen Y, Lee Y-C, Wu M-S, Wang H-P, Ni Y-H. Differences of microbiota in small bowel and faeces between irritable bowel syndrome patients and healthy subjects. Scand. J. Gastroenterol. 2016;51(4):410–9. doi: 10.3109/00365521.2015.1116107. Epub 2015 Nov 23. [DOI] [PubMed] [Google Scholar]
- 21.Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). http://dsmz.de/.
- 22.Adams MH. Host specificity. Bacteriophages. London: Interscience Publishers Ltd. 1959;3(3):121–35. doi: 10.4161/bact.25697. [DOI] [Google Scholar]
- 23.Cieplak T, Wiese M, Nielsen S, Van de Wiele T, van den Berg F, Nielsen DS. The Smallest Intestine (TSI) – an in vitro model of the small intestine with increased throughput. bioRxiv doi: https://doi.org/ 10.1101/244764 2018. [DOI] [PubMed] [Google Scholar]
- 24.Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. Enumeration of bacteriophages by double agar overlay plaque assay Bacteriophages: Methods Mol Biol. 2009;501:69–76. doi: 10.1007/978-1-60327-164-6_7. PMID: 19066811.2395061 [DOI] [PubMed] [Google Scholar]
- 25.Sulakvelidze A, Kutter E. Bacteriophage therapy in humans. In: Kutter A, Sulakvelidze A (eds). Bacteriophages: Biology and Applications. CRC Press: Boca Raton, USA; 2005. p. 381–436. [Google Scholar]
- 26.Fallinborg J, Christensen LA, Ingemannielsen M, Jacobsen BA, Abildgaard K, Rasmussen HH, Rasmussen SN. Measurement of Gastrointestinal Ph and Regional Transit Times in Normal-Children. J. Pediatr. Gastroenterol. Nutr. 1990. August;11:211–4. doi: 10.1097/00005176-199008000-00010. PMID:2395061. [DOI] [PubMed] [Google Scholar]
- 27.Ma Y, Pacan JC, Wang Q, Xu Y, Huang X, Korenevsky A, Sabour PM. Microencapsulation of bacteriophage felix O1 into chitosan-alginate microspheres for oral delivery. Appl Environ Microbiol. 2008. July;74:4799–805. doi: 10.1128/AEM.00246-08. PMID:18515488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tóthová L, Bábíčková J, Celec P. Phage survival: The biodegradability of M13 phage display library in vitro. Biotechnol Appl Biochem. 2012. December;59:490–4. doi: 10.1002/bab.1050. PMID:23586959. [DOI] [PubMed] [Google Scholar]
- 29.Nobrega FL, Costa AR, Santos JF, Siliakus MF, van Lent JWM, Kengen SWM, Azeredo J, Kluskens LD. Genetically manipulated phages with improved pH resistance for oral administration in veterinary medicine. Sci Rep. 2016;6:1–12. Article ID: 39235. doi: 10.1038/srep39235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scanlan PD, Bischofberger AM, Hall AR. Modification of Escherichia coli-bacteriophage interactions by surfactants and antibiotics in vitro. FEMS Microbiol Ecol. 2017. January;93:1–9. doi: 10.1093/femsec/fiw211. PMID:27737900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koo J, DePaola A, Marshall DL. Effect of Simulated Gastric Fluid and Bile on Survival of Vibrio vulnificus and Vibrio vulnificus Phage. J Food Prot. 2000. December;63:1665–9. doi: 10.4315/0362-028X-63.12.1665. PMID:11131888. [DOI] [PubMed] [Google Scholar]
- 32.Johnson JR, Kuskowski MA, Menard M, Gajewski A, Xercavins M, Garau J. Similarity between human and chicken Escherichia coli isolates in relation to ciprofloxacin resistance status. J Infect Dis. Oxford University Press; 2006;194:71–78. doi: 10.1086/504921. PMID:16741884. [DOI] [PubMed] [Google Scholar]
- 33.World Health Organization The Selection and Use of Essential Medicines: Report of the WHO Expert Committee, 2015 (including the 19th WHO Model List of Essential Medicines and the 5th WHO Model List of Essential Medicines for Children). World Health Organization; 2016. [Google Scholar]
- 34.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. National Acad Sciences; 2011;108:4554–61. doi: 10.1073/pnas.1000087107. PMID:20847294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCallin S, Sarker SA, Barrett C, Sultana S, Berger B, Huq S, Krause L, Bibiloni R, Schmitt B, Reuteler G, et al.. Safety analysis of a Russian phage cocktail: From MetaGenomic analysis to oral application in healthy human subjects. Virology. 2013;443:187–96. doi: 10.1016/j.virol.2013.05.022. PMID:23755967. [DOI] [PubMed] [Google Scholar]
- 36.Mai V, Ukhanova M, Reinhard MK, Li M, Sulakvelidze A. Bacteriophage administration significantly reduces Shigella colonization and shedding by Shigella-challenged mice without deleterious side effects and distortions in the gut microbiota. Bacteriophage. 2015. October;5:e1088124. doi: 10.1080/21597081.2015.1088124. PMID:26909243. [DOI] [PMC free article] [PubMed] [Google Scholar]


