ABSTRACT
Age-related macular degeneration (AMD) is a leading cause of blindness world-wide. Although the etiology of AMD is multifactorial, diet and nutrition have strong epidemiologic associations with disease onset and progression. Recent studies indicate a role for gut microbiota in development of AMD in mouse models and in some forms of human AMD. We previously found that consuming lower glycemia diets is associated with protection against AMD in humans and switching from higher to lower glycemia diets arrests AMD phenotypes in mice. Gut microbiota populations and circulating microbial cometabolites were altered in response to dietary carbohydrates, indicating a gut-retina axis. Here we explore additional gut microbiota-AMD interactions that point toward pathogenic roles for some gut microbiota families, including Ruminococcaceae and Lachnospiraceae, and individual members of Turicibacteraceae, Clostridiaceae, and Mogibacteriaceae. We also speculate on potential mechanisms by which gut microbiota influence AMD, with the objective of devising new AMD diagnoses and treatments.
KEYWORDS: Age-related macular degeneration, glycemic index, gut microbiome, microbial cometabolite, biomarker
Introduction
There are two forms of AMD: dry and wet. Both lead to visual impairment and potentially blindness due to death of photoreceptors. The latter, also called neovascular AMD, occurs when the choroidal blood supply in back of the eye invades into the photoreceptor layer. The vast majority of AMD is of the dry type. Our understanding of AMD has been greatly enhanced by epidemiologic studies that indicated important roles for environment, genetics, and diet in AMD morbidity.1,2 Strikingly, many of the risk factors for AMD like adiposity, smoking, and consumption of Western-style high glycemic index dietary patterns are also strong risk factors for CVD and diabetes, albeit they involve different tissues and often present at different times of life.3-5 The importance of dietary glycemic index has been validated in wildtype C57BL/6J mouse models of AMD wherein mice fed high glycemic index (HG) diets develop hallmarks of AMD, while mice fed low glycemic index (LG) diets do not.6,7 HG and LG diets used resemble standard rodent chow and are isocaloric, varying only in the type of starches used. The HG dietary starch consists of 100% amylopectin, a rapidly digested starch, while the LG dietary starch consists of 70% amylose/30% amylopectin, which is slowly digested. The effective glycemic index difference between these diets is approximately 55%, mirroring differences between the HG and LG breakfast cereals Cornflakes® and All-bran®.8,9 We recently demonstrated that switching diets from HG to LG diets midway through aging was able to prevent development of AMD and reverse accumulation of damage in the retina.7 Mice fed LG diets efficiently maintained their weights and were normoglycemic, whereas mice fed HG diets gained weight and become hyperglycemic and glucose intolerant.
We evaluated gut microbiota in LG and HG mice by 16S rDNA pyrosequencing and discovered alterations to gut microbiota populations that were mediated by diet and reversible by dietary change.7 We also evaluated the plasma and urine metabolomes and identified a large number of ‘microbial cometabolites’, metabolites whose levels change in response to microbiota, that were enriched in LG mice and that associated with protection against AMD features. We hypothesized that such metabolites, which included serotonin, hippurate, 4-hydroxyphenylacetate, tryptophan, trimethylamine, tyrosine, and indoxylsulfate, may alone, or in combination, mediate retinal neuroprotection, involving the neuroretina and the supporting retinal pigmented epithelial cells. By analogy with the gut-brain axis, we referred this signaling pathway as the gut-retina axis, with the recognition that it may not be distinct from the gut-brain axis, as the retina is a part of the central nervous system.
The gut microbiome in AMD
Using statistical approaches, we evaluated microbiota at multiple taxonomic levels to determine whether they showed associations with AMD, with diets, or with dietary change from HG to LG.7 As expected, some microbiota showed strong associations with diet. Enrichment with Bacteroidetes was associated with LG diet, in particular the S24–7 family and the Bacteroidia class, whereas high levels of Firmicutes were associated with HG diet, particularly the Clostridia class. We also showed that many groups of microbiota were associated with AMD, some of them in a diet-independent fashion. The associations were both positive and negative. Elevated populations with Clostridia and Bacilli classes were associated with greater risk for AMD. More Bacteroidia and Erysipelotrichi were associated with protection against AMD.
While these results were being prepared for publication, two additional studies were published that further enhanced our understanding about the connection between gut microbiota and AMD. The first was a mouse laser-induced model of neovascular AMD (wet AMD), in association with a high fat diet.10 The authors determined that high fat diets exacerbated AMD in a gut microbiota-dependent fashion. Furthermore, mice fed high fat diets showed increased gut permeability and systemic inflammation. Increased inflammation exacerbates choroidal neovascularization in the eye, which is what the authors found with high fat diets. The increased gut permeability associated with high fat diet was not mitigated via antibiotic treatment, but fecal transplantation from mice fed normal diets to mice fed the high fat diets, in conjunction with altering the gut microbiota, attenuated the neovascular response to laser injury. The major groups of microbiota that responded to diet and fecal transplantation were members of Bacteroidetes, which were higher in normal diets and members of Firmicutes and Proteobacteria, which were higher in high fat diets, consistent with previous literature and with the trends seen in our HG and LG mice.
Although mouse models of AMD have greatly enhanced our understanding of mechanisms of disease pathogenesis, including the role of gut microbiota, each model has limitations.11 In the case of our work and the study described above, diets were critical components and drivers of disease. Since the diet exerts such a strong and only partially reversible effect on gut microbiota, it may be impossible to definitively tease apart the influence of gut microbiota on AMD from other dietary effects. Furthermore, mouse microbiota and diets are quite dissimilar from those in humans. Therefore it is critical to both explore new mouse models of AMD, preferably in gnotobiotic mice, where the microbiome can be fully controlled and humanized. It is also essential to explore the gut microbiome in human AMD, which is just now emerging.12
In a first description of the gut microbiome in human AMD, 12 individuals with neovascular AMD were compared to 11 control individuals for gut microbiota composition.12 Even with this small sample size, principal component analysis demonstrated differences in microbiota composition between cases and controls. Further taxonomic associations identified bacteria enriched in cases or enriched in controls. Control individuals had higher amounts of Bacteroides, with the species Bacteroides eggerthii being significantly associated with controls. This finding is consistent with our finding of Bacteroidia being associated with LG diet and protection against AMD.7 AMD individuals had higher amounts of Anaerotruncus and Oscillibacter and at the species level Ruminococcus torques and Eubacterium ventriosum.12 All of these species or genera belong to the order of Clostridiales, which we observed to be associated with HG diet and AMD in our animal studies.7
At this time, it is premature to make generalizations about microbiome differences in AMD patients versus control individuals. The above study was quite small and the subjects were recruited from a single hospital in Switzerland. Future cohorts that encompass other forms of AMD will likely yield different results with different sets of limitations. Only after extensive phylogenetic and metagenomics studies are reported will we have a more definitive sense of how changes to the gut microbiota influence risk, progression, and phenotypes of AMD in people.
Associations of HG-induced AMD with individual taxonomic units
In our study of AMD related to HG and LG diets, as in many other microbiome studies, we grouped phylogenetically related taxa together and assessed association with AMD.7 This analytic approach is desirable because we sought to identify a consensus notion for the kinds of bacteria that work through similar functional pathways to influence AMD, while minimizing the number of multiple comparisons for statistical purposes. A shortcoming of this approach is that our analysis missed out on identification of microbiota that may have prominent effects, but that are negated by other highly-related members that have no effect or an opposite effect. As we learn more about the human microbiome, it has become clear that bacteria in closely-related species, or even different strains of the same species, can carry out very different functions within their host.13
We performed an ROC analysis using the same binary classifications of affected versus unaffected that we did in the original study at the level of individual metabolites (affected = retina damage score > 3). We included all of the OTUs identified in our sequencing analysis, using a false discovery rate cutoff of 0.1. The full list, grouped by taxonomic identification is shown in Table 1. Our analysis indicated multiple OTUs that could efficiently categorize affected from unaffected mice.
Table 1.
OTUs that significantly categorize AMD from non-AMD mice or non-AMD from AMD, grouped taxonomically, and ranked by area under the ROC curve or asymptotic P value (cutoff of false discovery rate < 0.1).
| OTU | Taxonomy | AUC | P value |
|---|---|---|---|
| OTU 11 | c_Bacilli;o_Turicibacterales;f_Turicibacteraceae | .983 | 3.09E-06 |
| OTU 28 | c_Clostridia;o_Clostridiales;f_Clostridiaceae | .972 | 5.02E-06 |
| OTU 126 | c_Clostridia;o_Clostridiales;f_[Mogibacteriaceae] | .941 | 2.03E-05 |
| OTU 418 | c_Clostridia;o_Clostridiales;f_Ruminococcaceae | .917 | 5.65E-05 |
| OTU 132 | .910 | 7.50E-05 | |
| OTU 266 | .863 | 4.54E-04 | |
| OTU 189 | .844 | 8.93E-04 | |
| OTU 187 | .840 | 1.01E-03 | |
| OTU 207 | .840 | 1.01E-03 | |
| OTU 324 | .835 | 1.20E-03 | |
| OTU 264 | .809 | 2.82E-03 | |
| OTU 260 | .802 | 3.51E-03 | |
| OTU 18 | .792 | 4.82E-03 | |
| OTU 258 | .783 | 6.24E-03 | |
| OTU 555 | c_Clostridia;o_Clostridiales | .885 | 1.95E-04 |
| OTU 52 | .868 | 3.75E-04 | |
| OTU 203 | .859 | 5.14E-04 | |
| OTU 384 | .847 | 7.91E-04 | |
| OTU 209 | .842 | 9.48E-04 | |
| OTU 65 | .833 | 1.27E-03 | |
| OTU 110 | .816 | 2.26E-03 | |
| OTU 208 | .816 | 2.26E-03 | |
| OTU 82 | .809 | 2.82E-03 | |
| OTU 399 | .807 | 2.98E-03 | |
| OTU 314 | .792 | 4.82E-03 | |
| OTU 86 | c_Clostridia;o_Clostridiales;f_Lachnospiraceae | .851 | 7.01E-04 |
| OTU 128 | .847 | 7.91E-04 | |
| OTU 155 | .845 | 8.41E-04 | |
| OTU 206 | .844 | 8.93E-04 | |
| OTU 235 | .842 | 9.48E-04 | |
| OTU 163 | .840 | 1.01E-03 | |
| OTU 115 | .819 | 2.02E-03 | |
| OTU 137 | .806 | 3.15E-03 | |
| OTU 499 | .785 | 5.93E-03 | |
| OTU 141 | c_Bacilli;o_Lactobacillales;f_Enterococcaceae | .839 | 1.07E-03 |
| OTU 345 | Unknown | .830 | 1.43E-03 |
| OTU 15 | c_Actinobacteria;o_Bifidobacteriales;f_Bifidobacteriaceae | .208 | 4.82E-03 |
| OTU 195 | c_Clostridia;o_Clostridiales | .108 | 1.49E-04 |
| OTU 13 | c_Erysipelotrichi;o_Erysipelotrichales;f_Erysipelotrichaceae | .104 | 1.30E-04 |
Shown in Figure 1 are examples of the top 3 OTUs associated with AMD and the top 2 OTUs associated with the non-disease state. These examples include 4 different families of microbiota and are representative of the remaining OTUs. Of the 39 OTUs that met statistical cutoffs, all but 3 were associated with AMD, and every categorized OTU belonged to the Firmicutes phylum. Several OTUs distinguished AMD from non-AMD nearly perfectly.
Figure 1.
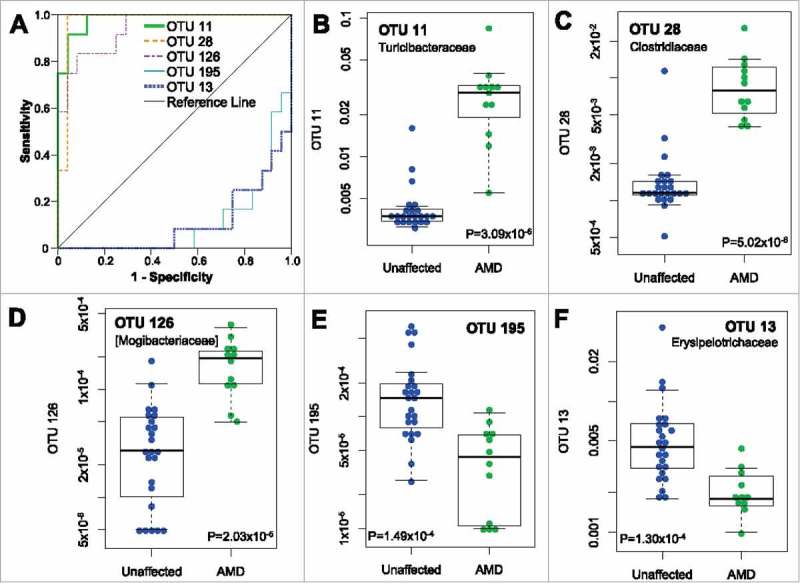
Categorization of AMD status by individual OTUs. (A) ROC curves shown for top 3 scoring and bottom 2 scoring OTUs. AUC values are listed in Table 1. An AUC of 1 indicates perfect categorization of AMD from unaffected while an AUC of 0 indicates perfect categorization of unaffected from AMD. (B-F) Boxplots for individual OTUs showing relative abundance of each OTU in unaffected mouse samples (n = 24) or affected mice samples (AMD, retina damage score >3 as in Rowan et al., n = 12).7
Grouping these OTUs taxonomically revealed an apparent enrichment for two particular families of the Clostridiales order, Ruminococcaceae and Lachnospiraceae. We applied enrichment analysis using the hypergeometric distribution to determine whether these groupings were likely chance events or not. As shown in Table 2, Ruminococcaceae were significantly enriched among AMD-predictive OTUs and Lachnospiraceae just missed statistical significance. The significant enrichment for Clostridiales with AMD was similar to our previous association analysis. However, the large number of OTUs that belonged to Ruminococcaceae and Lachnospiraceae was surprising, given that in aggregate neither family was significantly associated with AMD, even though they had some association with HG diets.7 Considering the large number of Ruminococcaceae and Lachnospiraceae OTUs that did not show significant association with AMD, one possible explanation is that these particular family members have distinct functions not shared by other family members, or might be related to one particular species like Ruminococcus torques, which is reported to be associated with human AMD.12 Alternatively, there could be a competitive interaction for a similar niche between highly related beneficial and pathogenic bacteria.
Table 2.
Taxonomic enrichment of OTUs that classify AMD from non-AMD mice (AMD-ROC).
| OTUs | p_Firmicutes | o_Clostridiales | f_Ruminococcaceae | f_Lachnospiraceae | |
|---|---|---|---|---|---|
| AMD-ROC | 36 | 35 | 33 | 11 | 9 |
| Total | 592 | 442 | 403 | 84 | 94 |
| P value | 2.45E-04 | 5.43E-04 | 0.00504 | 0.0552 |
Other OTUs associated with AMD or the non-AMD state were reflected in the larger taxonomic analysis from our previous analysis. For example, the Bacilli class association with AMD is accounted for by OTU 11 and OTU 141 and the Erysipelotrichales order association with protection from AMD is accounted for by OTU 13. OTU 13 and OTU 11 represented the highest and lowest AUCs from our ROC analysis and neither demonstrated strong dietary associations. Further elucidation of the species and functions represented by these OTUs will yield insights into the nature of their AMD associations.
Prospects of microbial cometabolites as indicators of AMD status
Gut microbiota carry out a broad array of activities in the body that affect our systematic physiology including effects on inflammation, metabolism, and the CNS.14-16 Although we are still unraveling the multiple mechanisms involved, it is clear from metabolomic studies that gut microbiota influence the synthesis and levels of many metabolites, which we collectively refer to as microbial cometabolites.17-19 Some cometabolites are only produced by gut bacteria (e.g. short-chain fatty acids).17,20 Other cometabolites reflect bacterial-specific modifications of host metabolites (e.g. trimethylamine is produced by gut microbiota from host choline or L-carnitine).21,22 Yet other cometabolites are largely synthesized by our host cells in response to bacterial signals (e.g. serotonin in response to bile acids).23 Bacterial production of cometabolites may utilize metabolic biosynthetic pathways that are distinct from the synthetic pathway that is exploited to make the same metabolite in mammals (e.g. chorismate, a precursor of tryptophan).24 Systematic measurements of microbial cometabolites reveal a biological readout of gut microbiota function independently from standard microbiome metagenomics analyses that may provide novel insights into host-microbiota functional interactions.
In our studies evaluating HG-induced AMD, we identified seven microbial cometabolite that negatively associated with AMD and were all present at higher abundance in LG-fed mice than HG mice.7 Six of these metabolites were identified in the urine samples taken several months before evaluation of AMD. Therefore, each metabolite represents a prospective biomarker for non-invasively monitoring AMD development. We are determining if these associations would translate to different mouse AMD models, which might impact gut microbiota in a different fashion, or in human AMD. Each metabolite offers its own potential link to metabolic pathways that may alter risk for AMD incidence or progress, as well as to the gut bacteria and gut bacterial gene products that contribute to its production. It is also possible that a metric based upon combinations of these metabolomic and microbiome data will provide even greater predictive capability.
In our metabolomics study, we did not identify any microbial cometabolites that were present at higher abundance in HG mice or that positively associated with AMD. However the analysis presented above certainly implicates HG-enriched microbiota in having specific pathogenic roles in AMD. We believe these mechanisms are distinct from the neuroprotective function of LG-enriched microbiota or their associated metabolites, although we can't rule out as yet unidentified AMD-associated microbial cometabolites. Pathogenic mechanisms may include increased inflammation and gut permeability, as suggested by high fat-diet exacerbated neovascular AMD.10 The mechanisms may be indirect via secondary effects on host metabolism, such as an obesogenic function or altered insulin signaling.25 Ongoing experiments using specific depletion methods and microbiota transfer are interrogating roles for microbiota, their products, and interactions with host in the etiology and treatment of AMD.
Funding Statement
This work was supported by the HHS | NIH | National Eye Institute (NEI) (EY13250) U.S. Department of Agriculture (USDA) (58-1950-0-014) U.S. Department of Agriculture (USDA) (58–1950-4–003) USDA | National Institute of Food and Agriculture (NIFA) (2015-05470) HHS | NIH | National Eye Institute (NEI) (EY021212) HHS | NIH | National Eye Institute (NEI) (EY026979).
Financial disclosures
None.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Sobrin L, Seddon JM. Nature and nurture- genes and environment- predict onset and progression of macular degeneration. Prog Retin Eye Res. 2014;40:1–15. doi: 10.1016/j.preteyeres.2013.12.004. PMID:24374240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weikel KA, Chiu CJ, Taylor A. Nutritional modulation of age-related macular degeneration. Mol Aspects Med. 2012;33:318–375. doi: 10.1016/j.mam.2012.03.005. PMID:22503690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams MK, Simpson JA, Aung KZ, Makeyeva GA, Giles GG, English DR, Hopper J, Guymer RH, Baird PN, Robman LD. Abdominal obesity and age-related macular degeneration. Am J Epidemiol. 2011;173:1246–1255. doi: 10.1093/aje/kwr005. PMID:21422060. [DOI] [PubMed] [Google Scholar]
- 4.Chiu CJ, Chang ML, Zhang FF, Li T, Gensler G, Schleicher M, Taylor A. The relationship of major American dietary patterns to age-related macular degeneration. Am J Ophthalmol. 2014;158:118-127 e111. doi: 10.1016/j.ajo.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu CJ, Taylor A. Dietary hyperglycemia, glycemic index and metabolic retinal diseases. Prog Retin Eye Res. 2011;30:18–53. doi: 10.1016/j.preteyeres.2010.09.001. PMID:20868767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weikel KA, Fitzgerald P, Shang F, Caceres MA, Bian Q, Handa JT, Stitt AW, Taylor A. Natural history of age-related retinal lesions that precede AMD in mice fed high or low glycemic index diets. Invest Ophthalmol Vis Sci. 2012;53:622–632. doi: 10.1167/iovs.11-8545. PMID:22205601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowan S, Jiang S, Korem T, Szymanski J, Chang ML, Szelog J, Cassalman C, Dasuri K, McGuire C, Nagai R, et al.. Involvement of a gut-retina axis in protection against dietary glycemia-induced age-related macular degeneration. Proc Natl Acad Sci U S A. 2017;114:E4472–E4481. doi: 10.1073/pnas.1702302114. PMID:28507131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pawlak DB, Kushner JA, Ludwig DS. Effects of dietary glycaemic index on adiposity, glucose homoeostasis, and plasma lipids in animals. Lancet. 2004. 2008;364:78–785. doi: 10.1016/S0140-6736(04)16937-7. PMID:15337404. [DOI] [PubMed] [Google Scholar]
- 9.Atkinson FS, Foster-Powell K, Brand-Miller JC. International tables of glycemic index and glycemic load values Diabetes Care. 2008;31:2281–2283. doi: 10.2337/dc08-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andriessen EM, Wilson AM, Mawambo G, Dejda A, Miloudi K, Sennlaub F, Sapieha P. Gut microbiota influences pathological angiogenesis in obesity-driven choroidal neovascularization. EMBO Mol Med. 2016;8:1366–1379. doi: 10.15252/emmm.201606531. PMID:27861126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pennesi ME, Neuringer M, Courtney RJ. Animal models of age related macular degeneration. Mol Aspects Med. 2012;33:487–509. doi: 10.1016/j.mam.2012.06.003. PMID:22705444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zinkernagel MS, Zysset-Burri DC, Keller I, Berger LE, Leichtle AB, Largiadèr CR, Fiedler GM, Wolf S. Association of the Intestinal Microbiome with the Development of Neovascular Age-Related Macular Degeneration. Sci Rep. 2017;7:40826. doi: 10.1038/srep40826. PMID:28094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG, et al.. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–66. PMID:28953883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeder BO, Backhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22:1079–1089. doi: 10.1038/nm.4185. PMID:27711063. [DOI] [PubMed] [Google Scholar]
- 15.Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31:69–75. doi: 10.1097/MOG.0000000000000139. PMID:25394236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: An integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. PMID:22424233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yap IK, Li JV, Saric J, Martin FP, Davies H, Wang Y, Wilson ID, Nicholson JK, Utzinger J, Marchesi JR, et al.. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res. 2008;7:3718–3728. doi: 10.1021/pr700864x. PMID:18698804. [DOI] [PubMed] [Google Scholar]
- 18.Lees HJ, Swann JR, Wilson ID, Nicholson JK, Holmes E. Hippurate: the natural history of a mammalian-microbial cometabolite. J Proteome Res. 2013;12:1527–1546. doi: 10.1021/pr300900b. PMID:23342949. [DOI] [PubMed] [Google Scholar]
- 19.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. PMID:19234110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, Bultman SJ. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. PMID:21531334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, et al.. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature medicine. 2013;19:576–585. doi: 10.1038/nm.3145. PMID:23563705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.al-Waiz M., Mikov M., Mitchell S.C. & Smith R.L. The exogenous origin of trimethylamine in the mouse. Metabolism. 1992;41:135–136. doi: 10.1016/0026-0495(92)90140-6. PMID:1736035. [DOI] [PubMed] [Google Scholar]
- 23.Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. PMID:25860609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diaz E, Ferrandez A, Prieto MA, Garcia JL. Biodegradation of aromatic compounds by Escherichia coli. Microbiol Mol Biol Rev. 2001;65:523–569, table of contents. doi: 10.1128/MMBR.65.4.523-569.2001. PMID:11729263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. PMID:27383980. [DOI] [PMC free article] [PubMed] [Google Scholar]


