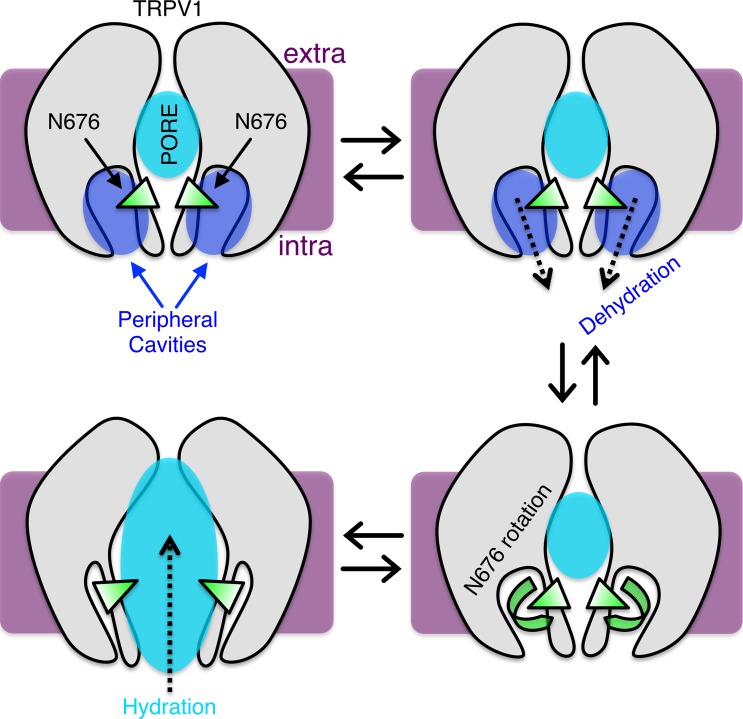
TRPV1 channels comprise four subunits containing six transmembrane segments (S1–S6) that surround a central pore. Kasimova et al. hypothesize that channel opening involves rotation of an S6 asparagine residue toward the pore, as well as associated pore hydration and external cavity dehydration.
Abstract
The transient receptor potential channel vanilloid type 1 (TRPV1) is activated by a variety of endogenous and exogenous stimuli and is involved in nociception and body temperature regulation. Although the structure of TRPV1 has been experimentally determined in both the closed and open states, very little is known about its activation mechanism. In particular, the conformational changes that occur in the pore domain and result in ionic conduction have not yet been identified. Here we suggest a hypothetical molecular mechanism for TRPV1 activation, which involves rotation of a conserved asparagine in S6 from a position facing the S4–S5 linker toward the pore. This rotation is associated with hydration of the pore and dehydration of the four peripheral cavities located between each S6 and S4–S5 linker. In light of our hypothesis, we perform bioinformatics analyses of TRP and other evolutionary related ion channels, evaluate newly available structures, and reexamine previously reported water accessibility and mutagenesis experiments. These analyses provide several independent lines of evidence to support our hypothesis. Finally, we show that our proposed molecular mechanism is compatible with the prevailing theory that the selectivity filter acts as a secondary gate in TRPV1.
Introduction
Transient receptor potential (TRP) channel vanilloid type 1 (TRPV1) is a member of the TRP family, which promotes nonselective cation current across cell membranes in response to heat, low pH, and inflammatory agents (Caterina et al., 1997; Tominaga et al., 1998; Hayes et al., 2000). Activation of this channel induces burning pain: the action potential triggered by TRPV1 in sensory ganglia and small sensory C and Aδ fibers is transmitted through the second-order projection neurons in the spinal cord to the thalamus and specific higher order brain areas, where it is perceived as pain (Julius and Basbaum, 2001; Carnevale and Rohacs, 2016).
TRPV1 is a homotetramer with each subunit composed of the transmembrane and cytoplasmic regions (Cao et al., 2013b; Liao et al., 2013; Gao et al., 2016). The transmembrane region has six helical segments (S1–S6); four of them (S1–S4) constitute a domain structurally homologous to the voltage sensor domain of voltage-gated ion channels. The remaining two helices (S5 and S6) assemble into the pore domain. Cryo-EM structures of TRPV1 in different functional states have revealed key elements of its activation mechanism (Cao et al., 2013b; Liao et al., 2013; Gao et al., 2016). For instance, it became apparent that the gate is located at the crossing of the S6 helical bundle. The gate is closed in the absence of an activating stimulus, while at room temperature it opens upon binding of the agonist capsaicin between S4 and S5 (Cao et al., 2013b). The molecular mechanism of this conformational change has been in part clarified by mutagenesis experiments (Yang et al., 2015; Gao et al., 2016): capsaicin forms a hydrogen bond with the S4–S5 linker and pulls it in the outward direction; as for voltage-gated ion channels, this motion relieves the constriction exerted by the linker on the S6 helical bundle.
However, the cryo-EM structure of the capsaicin-bound state is closed at cryogenic temperatures: the gate is narrower than that of the open state (Cao et al., 2013b), and in simulations it is dehydrated (Kasimova et al., 2018). Consistently, Jara-Oseguera et al. (2016) have shown that TRPV1 activation becomes increasingly less probable as the temperature is decreased even when capsaicin is present in large concentrations. Thus, the capsaicin-bound state is closed at cryogenic temperatures and opens as temperature is increased, with the midpoint of activation located approximately at 300°K.
We recently used molecular dynamics and free energy calculations to study the molecular mechanism of the closed-to-open transition in the capsaicin-bound state (Kasimova et al., 2018). Our major finding was that the conserved asparagine N676 in S6 adopts two alternative conformations in the open and closed states, projecting its side chain toward either the pore or four peripheral cavities located between the S6 helix and the S4–S5 linker, respectively. When TRPV1 is in the closed state, these cavities are connected to the intracellular solution and host several water molecules. Dehydration of the peripheral cavities correlates with the rotation of N676 from these cavities toward the pore. In this rotated conformation, N676 promotes pore hydration and thus permeability to ions (Fig. 1).
Figure 1.
A possible molecular mechanism for TRPV1 activation. The channel is shown in gray; water in the pore and peripheral cavities in cyan and blue, respectively; the conserved asparagine N676 in S6 in green; the membrane in purple (extra and intra denote extra- and intracellular solutions). In the capsaicin-bound closed state, the peripheral cavities host several water molecules, while the pore is partially dehydrated; N676 projects its side chain toward the peripheral cavities. Upon application of an activating stimulus, the peripheral cavities dehydrate, an event that correlates with the rotation of N676 toward the pore. The presence of a hydrophilic side chain inside the pore promotes its hydration and hence permeability to ions.
Here, we will briefly summarize our previous computational findings and discuss several independent pieces of experimental evidence that corroborate our hypothesis. We will show how these lend credibility to the rotation of N676 and the presence of the peripheral cavities. Finally, we will present an analysis of our previously reported simulations suggesting that the selectivity filter acts as a secondary gate.
Materials and methods
Molecular dynamics simulations
The molecular dynamics trajectories of the capsaicin-bound closed and open states were taken from our previous study (Kasimova et al., 2018). Briefly, the structure of the TRPV1 capsaicin-bound state (Cao et al., 2013b) with four capsaicin molecules (Elokely et al., 2016) was embedded in a hydrated 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) bilayer and surrounded by 150 mM NaCl solution. Four to six water molecules were added inside each peripheral cavity in one case (this system further relaxed to the closed state), while these cavities were left empty in the other case (this system further relaxed to the open state; Kasimova et al., 2018). The CHARMM36 force field (Mackerell et al., 2004) was used for the protein and the POPC lipids, the parameters derived in (Elokely et al., 2016) were used for capsaicin, and the TIP3P model (Jorgensen et al., 1983) was used for water molecules. The molecular dynamics simulations were performed following a multistep protocol (for the details, see Kasimova et al., 2018) using NAMD 2.10 (Phillips et al., 2005). The pressure and temperature were kept constant at 1 atm and 300°K, respectively, using Langevin dynamics. A cutoff of 11 Å with a switching function between 8 and 11 Å was used for the van der Waals interactions. A cutoff of 11 Å was considered for the short-range component of electrostatic interactions, and the long-range component was computed using particle mesh Ewald (Darden et al., 1993). The overall length of the molecular dynamics trajectory for each state was 750 ns. In addition, we performed the molecular dynamics simulations of the TRPV1 apo state (Liao et al., 2013) using an analogous setup. The overall length of the molecular dynamics trajectory for this state was 200 ns.
Analysis of sequence conservation
The multiple sequence alignments of the TRP and voltage-gated sodium and calcium channels were taken from our previous studies (Palovcak et al., 2015; Kasimova et al., 2016). To analyze the sequence conservation for these families, we computed the Shannon entropy profile using the logos website (Crooks et al., 2004).
Estimation of the accessibility of S6 residues to the intracellular solution
To estimate the accessibility of S6 residues to the intracellular solution, we considered the second halves of the molecular dynamics trajectories. We separated the intracellular solution from the rest applying the following procedure. We first calculated the three-dimensional map of water occupancy using the Volmap tool of VMD (Humphrey et al., 1996). Based on this, we generated a binary map by setting each voxel to either 0 or 1 depending on whether or not the local value of water occupancy overcomes a predefined threshold. After that, we used the gmx cluster of Gromacs 5.0.4 (Abraham et al., 2015) to group the voxels. This allowed us to identify the cluster containing the intracellular solution and all the regions connected to it. Finally, as a measure of accessibility, we calculated the overlap between the occupancy of the intracellular solution and that of each S6 residue.
Results and discussion
The π-bulge in the S6 helix is evolutionarily conserved across the TRP family
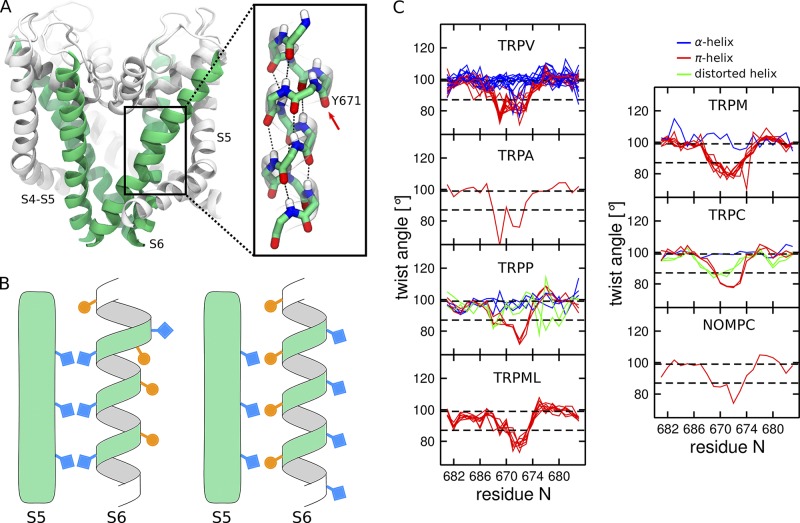
We start by recalling a remarkable feature of the capsaicin-bound structure (Cao et al., 2013b): the presence of a π-helical segment between Y671 and N676 (Fig. 2 A). α- and π-helices show an important difference: while in α-helices, hydrogen bonds join main chain groups that are four residues apart (i–i+4), this spacing changes to five residues in π-helices (i–i+5). Therefore, the presence of a π-helical segment in an α-helix implies a mismatch in the pattern of hydrogen bonds, which results in unpaired carbonyl and amino groups. In practice, one residue does not participate in the network of hydrogen bonds and bulges out of the α-helix, forming a so-called π-bulge (Fig. 2 A). The π-bulge is mobile: the bulging residue can “reabsorb” by forming hydrogen bonds at the expense of a neighboring residue, which, as a result, bulges out of the helix.
Figure 2.
π-helical segment in the S6 helix of TRP channels. (A) Cartoon representation of the TRPV1 pore domain (Cao et al., 2013b). The S6 helices are colored in green, and the rest of the pore domain in white. The inset shows the π-helical segment and the π-bulge located on the Y671 residue (highlighted with an arrow). The atoms are colored by element type: C in green, N in blue, O in red, and H in white. (B) Schematic representation of an α-helix (right) and an α-helix with a π-bulge (left). Note that the residues facing S5 in the α-helix are lining the pore when the π-bulge is introduced. (C) Per residue twist angle in the S6 helices of the available TRP structures estimated using HELANAL (Bansal et al., 2000). The structures with the π-bulge are shown in red (TRPV1, TRPV3, TRPV5, TRPV6, TRPA1, TRPM2, TRPM4, TRPM7, TRPC4, TRPP2, NOMPC, TRPML1, and TRPML3), and without the π-bulge in blue (TRPV2, TRPV3, TRPV4, TRPV5, TRPV6, TRPM8, TRPC3, and TRPP2). The structures in which S6 is distorted and cannot be assigned to either the α-helix or the α-helix with π-bulge are shown in green (TRPC3, TRPC6, and TRPP2). For the PDB codes of the structures, see Table 1. The twist angle is 99.4° and 86.6° for idealized α-helix and π-helix, respectively (highlighted by dashed black lines).
In one of our previous investigations, we hypothesized, on the basis of the sole TRPV1 structure, that this structural motif is conserved across the entire family of TRP channels (Palovcak et al., 2015). To test this hypothesis, we analyzed patterns of correlations in a large multiple sequence alignment containing ∼3,000 distinct sequences of genes encoding for TRP channels. The presence of a π-helical segment changes the register of the downstream α-helix: it offsets by one position all the subsequent residues, effectively imparting a rotation of approximately 100° to the downstream α-helix (Fig. 2 B). The change of the register results in a specific pattern of contacts between S6 and the neighboring S5. We detected this pattern in the multiple sequence alignment as strong correlations between S5 and S6 residues; importantly, these pairs of residues are in spatial proximity only with the “correct” register of S6, i.e., if the π-bulge is present (Palovcak et al., 2015). The conservation of this specific pattern of contacts between S5 and S6 across the entire TRP family suggests that the π-bulge is conserved as well.
Since then, several experimental structures of TRP channels have confirmed our hypothesis about the structural conservation of the π-bulge. The structures of TRPV1, TRPV3, TRPV5, TRPV6, TRPA1, TRPM2, TRPM4, TRPM7, TRPC4, TRPP2, TRPN-like channel NOMPC, TRPML1, and TRPML3 all show a π-helical segment in S6 (Fig. 2 C and Table 1; Cao et al., 2013b; Liao et al., 2013; Barad et al., 2015; Paulsen et al., 2015; Gao et al., 2016; Shen et al., 2016; Chen et al., 2017; Grieben et al., 2017; Guo et al., 2017; Hirschi et al., 2017; Jin et al., 2017; Schmiege et al., 2017; Winkler et al., 2017; Zhou et al., 2017; Autzen et al., 2018; Duan et al., 2018a,b; Hughes et al., 2018b; McGoldrick et al., 2018; Singh et al., 2018a,b; Su et al., 2018a; Vinayagam et al., 2018; Zhang et al., 2018). Interestingly, structures without the π-bulge have also been determined for some of these TRP channels (TRPV3, TRPV5, TRPV6, and TRPP2; Saotome et al., 2016; Singh et al., 2017, 2018a,b; Hughes et al., 2018a; Hulse et al., 2018; McGoldrick et al., 2018; Su et al., 2018b; Zheng et al., 2018), suggesting that a transition between the π- and α-helical conformations might be part of the functional cycle (Fig. 2 C and Table 1). Finally, only TRPV2, TRPV4, TRPM8, and TRPC3 do not show a π-helical segment in S6 (Huynh et al., 2016; Zubcevic et al., 2016, 2018; Deng et al., 2018; Fan et al., 2018; Hughes et al., 2018a; Yin et al., 2018). Either these TRP channels lack altogether the π-helical conformation or, alternatively, this conformation has not been experimentally captured yet.
Table 1. Structures of TRP channels.
| TRP channel | PDB ID | Resolution (Å) | Residue forming the gate | R (Å) | π-Bulge? | Conserved asparagine | State | Reference |
|---|---|---|---|---|---|---|---|---|
| TRPV1 | 5IRZ | 2.95 | I679 | 0.7 | Yes | N676 | In | Gao et al., 2016 |
| TRPV1 | 5IS0 | 3.43 | I679 | 0.7 | Yes | N676 | In | Gao et al., 2016 |
| TRPV1 | 3J5P | 3.275 | I679 | 0.8 | Yes | N676 | In | Liao et al., 2013 |
| TRPV1 | 3J5R | 4.2 | I679 | 1.8 | Yes | N676 | Out | Cao et al., 2013b |
| TRPV1 | 5IRX | 2.95 | I679 | 2.8 | Yes | N676 | In | Gao et al., 2016 |
| TRPV1 | 3J5Q | 3.8 | I679 | 3.0 | Yes | N676 | In | Cao et al., 2013b |
| TRPV2 | 5AN8 | 3.8 | M643 | 0.9 | No | N637 | Out | Zubcevic et al., 2016 |
| TRPV2 | 5HI9 | 4.4 | M645 | 1.1 | No | N639 | Out | Huynh et al., 2016 |
| TRPV2 | 6BWM | 3.9 | M643 not resolved | — | No | N637 | Out | Zubcevic et al., 2018 |
| TRPV2 | 6BWJ | 3.1 | M643 partially resolved | — | No | N637 | Out | Zubcevic et al., 2018 |
| TRPV3 | 6DVW | 4.3 | M677 | 0.9 | No | N671 | Out | Singh et al., 2018a |
| TRPV3 | 6DVY | 4.0 | M677 | 0.9 | No | N671 | Out | Singh et al., 2018a |
| TRPV3 | 6DVZ | 4.24 | I674 | 2.4 | Yes | N671 | In | Singh et al., 2018a |
| TRPV4 | 6BBJ | 3.8 | M714 | 0.8 | No | N708 | Out | Deng et al., 2018 |
| TRPV5 | 6B5V | 4.8 | M578 | 0.8 | No | N572 | Out | Hughes et al., 2018a |
| TRPV5 | 6DMU | 3.9 | I575 | 3.3 | Yes | N572 | In | Hughes et al., 2018b |
| TRPV6 | 6BO8 | 3.6 | I575 | 2.9 | Yes | N572 | In | McGoldrick et al., 2018 |
| TRPV6 | 6BO9 | 4.0 | I575 | 3.1 | Yes | N572 | In | McGoldrick et al., 2018 |
| TRPV6 | 6BOB | 3.9 | M577 | 0.8 | No | N571 | Out | McGoldrick et al., 2018 |
| TRPV6 | 6BOA | 4.2 | M578 | 0.8 | No | N572 | Out | McGoldrick et al., 2018 |
| TRPV6 | 5IWK | 3.247 | M577 | 0.6 | No | N571 | Out | Saotome et al., 2016 |
| TRPV6 | 5WO6 | 3.31 | M577 | 0.7 | No | N571 | Out | Singh et al., 2017 |
| TRPV6 | 6D7S | 4.34 | I575 | 2.7 | Yes | N572 | In | Singh et al., 2018b |
| TRPV6 | 6D7T | 4.44 | M578 | 0.8 | No | N572 | Out | Singh et al., 2018b |
| TRPA1 | 3J9P | 4.24 | I957 | 1.4 | Yes | N954 | In | Paulsen et al., 2015 |
| TRPM2 | 6CO7 | 3.07 | I1082 | 0.9 | Yes | N1079 | In | Zhang et al., 2018 |
| TRPM4 | 6BQV | 3.1 | I1040 | 0.7 | Yes | N1037 | In | Autzen et al., 2018 |
| TRPM4 | 6BQR | 3.2 | I1040 | 0.8 | Yes | N1037 | In | Autzen et al., 2018 |
| TRPM4 | 5WP6 | 3.8 | I1040 | 1.0 | Yes | N1037 | In | Winkler et al., 2017 |
| TRPM4 | 6BCJ | 3.14 | I1036 | 0.9 | Yes | N1033 | In | Guo et al., 2017 |
| TRPM4 | 6BCL | 3.54 | I1036 | 0.8 | Yes | N1033 | In | Guo et al., 2017 |
| TRPM4 | 6BCO | 2.88 | I1036 | 0.9 | Yes | N1033 | In | Guo et al., 2017 |
| TRPM4 | 6BCQ | 3.25 | I1036 | 0.9 | Yes | N1033 | In | Guo et al., 2017 |
| TRPM7 | 6BWD | 3.7 | I748/I931 | 0.8 | Yes | N745/N928 | In | Duan et al., 2018b |
| TRPM8 | 6BPQ | 4.1 | L973 | 0.8 | No | N967 | Out | Yin et al., 2018 |
| TRPC3 | 6CUD | 3.3 | I658 | 0.7 | No | N652 | Out | Fan et al., 2018 |
| TRPC3 | 5ZBG | 4.36 | I655 | 1.8 | S6 distorted | N652 | In | Tang et al., 2018 |
| TRPC4 | 5Z96 | 3.28 | I617 | 0.9 | Yes | N614 | In | Duan et al., 2018b |
| TRPC4 | 6G1K | 3.6 | I617 | 0.8 | Yes | N614 | In | Vinayagam et al., 2018 |
| TRPC6 | 5YX9 | 3.8 | I724 | 1.5 | S6 distorted | N721 | In | Tang et al., 2018 |
| TRPP2 | 5MKE | 4.3 | L677 | 1.9 | S6 distorted | N674 | In | Wilkes et al., 2017 |
| TRPP2 | 5MKF | 4.2 | L677 | 1.0 | S6 distorted | N674 | Out | Wilkes et al., 2017 |
| TRPP2 | 5T4D | 3.0 | L677 | 0.6 | Yes | N674 | In | Shen et al., 2016 |
| TRPP2 | 5K47 | 4.22 | L677 | 0.7 | Yes | N674 | In | Grieben et al., 2017 |
| TRPP2 | 6D1W | 3.54 | I680 | 2.3 | No | N674 | Out | Zheng et al., 2018 |
| TRPP2 | 5Z1W | 3.38 | I560 | 2.6 | No | N554 | Out | Su et al., 2018b |
| TRPP2 | 6DU8 | 3.11 | I560 | 0.5 | No | N554 | Out | Hulse et al., 2018 |
| TRPP2 | 6A70 | 3.6 | L677 | 0.9 | Yes | N674 | In | Su et al., 2018a |
| NOMPC | 5VKQ | 3.55 | I1554 | 0.8 | Yes | Q1551 | In | Jin et al., 2017 |
| TRPML1 | 5WJ5 | 3.72 | I514 | 0.9 | Yes | S511 | In | Schmiege et al., 2017 |
| TRPML1 | 5WJ9 | 3.49 | I514 | 2.4 | Yes | S511 | In | Schmiege et al., 2017 |
| TRPML1 | 5WPQ | 3.64 | I514 | 0.9 | Yes | S511 | In | Chen et al., 2017 |
| TRPML1 | 5WPT | 3.75 | I514 | 0.8 | Yes | S511 | In | Chen et al., 2017 |
| TRPML1 | 5WPV | 3.59 | I514 | 0.6 | Yes | S511 | In | Chen et al., 2017 |
| TRPML3 | 6AYE | 4.06 | I498 | 0.8 | Yes | S495 | In | Zhou et al., 2017 |
| TRPML3 | 6AYF | 3.62 | I498 | 3.3 | Yes | S495 | In | Zhou et al., 2017 |
| TRPML3 | 5W3S | 2.94 | I498 | 1.0 | Yes | S495 | In | Hirschi et al., 2017 |
Columns contain PDB ID, resolution, residue forming the gate, pore radius at the level of the gate, presence/absence of the π-bulge in the S6 helix, conserved asparagine, and conformational state of the conserved asparagine (inside or outside the pore). The pore radius at the level of the gate was estimated using HOLE (Smart et al., 1996). For TRPV2 6BWM and 6BWJ structures, the side chain of the residue forming the gate was either not resolved or resolved partially; the pore radius was not estimated for these structures.
The π-bulge causes conformational flexibility of neighboring residues
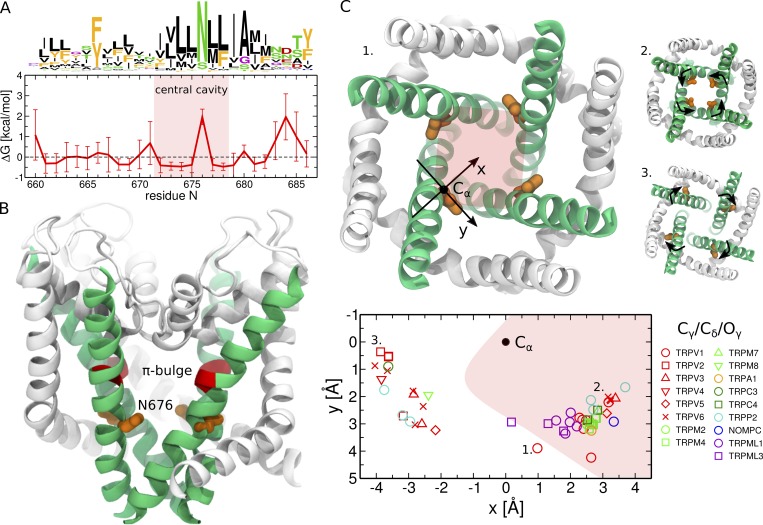
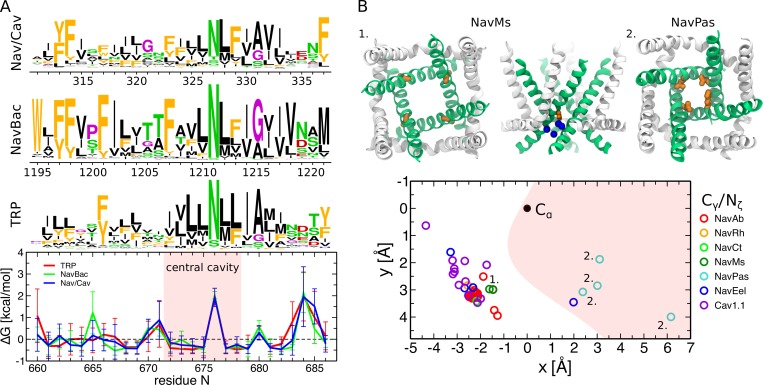
Analysis of a comprehensive multiple sequence alignment of TRP channels revealed that the pore-lining residues are all hydrophobic except for one that in most of the sequences is an asparagine (N676 in TRPV1, Fig. 3, A and B; Palovcak et al., 2015). This feature is strictly conserved in the two evolutionarily divergent families of TRPP and TRPML channels as well. In TRPML channels, instead of an asparagine, some sequences have a serine; also in these cases, serine is the only hydrophilic pore-lining residue. The conserved asparagine is located only ∼1.5 helical turns away from the π-bulge, which potentially endows this residue with conformational flexibility. Structures of different TRP channels (Cao et al., 2013b; Liao et al., 2013; Barad et al., 2015; Paulsen et al., 2015; Gao et al., 2016; Huynh et al., 2016; Saotome et al., 2016; Shen et al., 2016; Zubcevic et al., 2016, 2018; Chen et al., 2017; Grieben et al., 2017; Guo et al., 2017; Hirschi et al., 2017; Jin et al., 2017; Schmiege et al., 2017; Singh et al., 2017, 2018a,b; Winkler et al., 2017; Zhou et al., 2017; Autzen et al., 2018; Deng et al., 2018; Duan et al., 2018a,b; Fan et al., 2018; Hughes et al., 2018b,a; Hulse et al., 2018; McGoldrick et al., 2018; Su et al., 2018a,b; Vinayagam et al., 2018; Yin et al., 2018; Zhang et al., 2018; Zheng et al., 2018) indeed show that the orientation of N676 (or the corresponding amino acid) with respect to the pore can differ by as much as 180° between two structures (Fig. 3 C and Table 1). For instance, in the TRPV subfamily, the conserved asparagine points toward the pore in TRPV1 and some structures of TRPV3, TRPV5, and TRPV6 (Cao et al., 2013b; Liao et al., 2013; Barad et al., 2015; Gao et al., 2016; McGoldrick et al., 2018; Singh et al., 2018a,b ; Hughes et al., 2018b); points in the opposite direction (toward the S4–S5 linker) in TRPV2, TRPV4, and the remaining TRPV3, TRPV5, and TRPV6 structures (McGoldrick et al., 2018; Singh et al., 2018a,b ; Huynh et al., 2016; Saotome et al., 2016; Zubcevic et al., 2016, 2018; Singh et al., 2017; Deng et al., 2018; Hughes et al., 2018a); and, finally, lies just outside the pore in the TRPV1 capsaicin-bound structure (Cao et al., 2013b). Similar conformations are observed in other TRP families including TRPM, TRPC, and TRPP (Shen et al., 2016; Grieben et al., 2017; Guo et al., 2017; Winkler et al., 2017; Autzen et al., 2018; Duan et al., 2018b,a; Fan et al., 2018; Hulse et al., 2018; Su et al., 2018a,b; Vinayagam et al., 2018; Yin et al., 2018; Zhang et al., 2018; Zheng et al., 2018). Importantly, the structures with the conserved asparagine inside the pore or just outside of it all have the π-bulge; by contrast, those in which this residue points to the S4–S5 linker do not show any π-helical segment in S6 (Table 1).
Figure 3.
Conserved asparagine adopts a range of conformations from pointing toward the pore to pointing toward the S4–S5 linker. (A) Sequence logos and per-position hydrophobicity plot of the S6 helix across the TRP family (numbering based on the TRPV1 sequence; Palovcak et al., 2015). In the sequence logo, the height of each residue at a given position is proportional to its frequency at this position, and the height of the overall stack of residues decreases linearly with Shannon entropy (Crooks et al., 2004). Aromatic residues (F, W, Y) are shown in orange; hydrophobic not aromatic (A, C, I, L, M, V) in black; hydrophilic (N, Q, S, T) in green; positively (H, K, R) and negatively (D, E) charged in blue and red, respectively; and G and P in purple. Note that in a few cases, the conserved asparagine can be substituted by a serine (TRPML channels) or glutamine (TRPN-like channel NOMPC). In the hydrophobicity plot, positions with positive are hydrophilic, and those with negative are hydrophobic. The error bars represent SD. The red shaded area highlights the residues lining the central cavity (between Y671 and I679). (B) Cartoon representation of the TRPV1 pore domain (Cao et al., 2013b). The S6 helices are shown in green, and the rest of the pore domain in white. The π-bulge and the neighboring conserved asparagine are colored in red and orange, respectively. (C) Orientation of the conserved asparagine with respect to the pore in different TRP structures (Cao et al., 2013b; Liao et al., 2013; Barad et al., 2015; Paulsen et al., 2015; Gao et al., 2016; Huynh et al., 2016; Saotome et al., 2016; Shen et al., 2016; Zubcevic et al., 2016, 2018; Chen et al., 2017; Grieben et al., 2017; Guo et al., 2017; Hirschi et al., 2017; Jin et al., 2017; Schmiege et al., 2017; Singh et al., 2017, 2018a,b; Winkler et al., 2017; Zhou et al., 2017; Autzen et al., 2018; Deng et al., 2018; Duan et al., 2018a,b; Fan et al., 2018; Hughes et al., 2018b,a; Hulse et al., 2018; McGoldrick et al., 2018; Su et al., 2018a,b; Vinayagam et al., 2018; Yin et al., 2018; Zhang et al., 2018; Zheng et al., 2018). The side chain position in the plane perpendicular to the pore axis is shown: the Cα-atom is represented as a black dot and is located at (0,0), and the terminal Cγ-atom (Cδ and Oγ in NOMPC and TRPML channels, respectively) is shown as a colored symbol (a different one for each TRP channel). The x-axis is aligned with the vector connecting the Cα-atom and the center of the pore. The red shaded area highlights the pore region. The three insets show the pore domains of the TRPV1 capsaicin-bound structure (1), the TRPV6 open structure (2), and TRPV2 (3): the conserved asparagine points toward the S4–S5 linker in TRPV2, points toward the center of the pore in the TRPV6 open structure, and lies just outside the pore in the TRPV1 capsaicin-bound structure. In TRPV6 and TRPV2 (the two extreme cases), the black arrows highlight the direction of the asparagine rotation between the conformations inside and outside the pore.
This structural heterogeneity is in itself striking: evolutionary conservation points to functional relevance, yet the residue lacks a well defined conformation. Is the role of the conserved asparagine related to its flexibility? In our previous study (Kasimova et al., 2018), we have shown that in the TRPV1 capsaicin-bound structure, the rotation of N676 inside or outside the pore significantly affects the polarity of the latter and hence ionic transport: only when N676 is exposed to the pore, the latter is continuously hydrated and the free energy barrier for ionic transport at the level of the gate is ∼5.2 kcal/mol. By contrast, when N676 is interacting with the S4–S5 linker, the free energy barrier is larger than 12 kcal/mol, thereby preventing ionic transport. Consistent with these predictions, recently determined experimental structures of TRPV3, TRPV5, and TRPV6 show that the conserved asparagine faces the pore in the open conformations and the S4–S5 linker in the closed ones (Saotome et al., 2016; Singh et al., 2017, 2018a,b; Hughes et al., 2018a,b; McGoldrick et al., 2018).
It is important to note that the location of the conserved asparagine side chain inside the pore may not be the only determinant of channel opening. The radius of the pore should be, in any case, large enough to permit the passage of hydrated ions. Accordingly, several structures of TRP channels showing the conserved asparagine in the pore-facing configuration and the pore radius smaller than 2 Å are likely representatives of the closed states (Table 1).
Y671 motion affects gating at the selectivity filter and is coupled to N676 rotation
Are there functional data showing that the rotation of S6 underlies TRP channels’ gating? Steinberg et al. (2017) recently reported that TRPV1 activation correlates with a conformational change at the level of the π-bulge where a nonnatural amino acid bearing a coumarin side chain was introduced. Coumarin is a fluorophore with exquisite sensitivity to environmental polarity (Wang et al., 2006; Wagner, 2009); when genetically encoded into proteins, it can be used as reporter of changes in local hydration and conformational rearrangements. In the TRPV1 channel with Y671 substituted for coumarin, activation by capsaicin was found to cause an increase in both the photon counts and optical fluctuations. This effect suggests that coumarin undergoes a conformational transition from a more hydrated state to a less hydrated one (Steinberg et al., 2017).
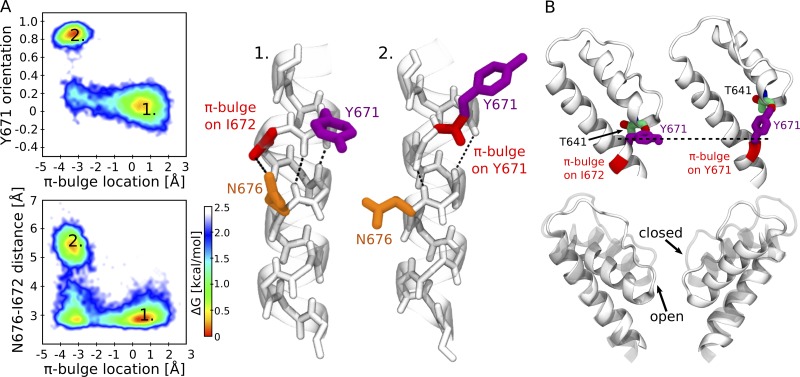
While the exact conformational change cannot be inferred from these experiments, molecular dynamics simulations performed on the TRPV1 wild type suggested a possible explanation for the observed changes in fluorescence (Steinberg et al., 2017). The simulations showed that the surface area of Y671 accessible to solvent decreases from 30% to 15% on passing from the closed to open states, in agreement with the experimental data for coumarin. The reason for this change is the conformational rearrangement of Y671 coupled to gating through N676. In the TRPV1 open state, Y671 is oriented perpendicularly with respect to the pore axis, and it is shielded from water molecules by the pore helices. The N676 side chain is located inside the pore, where it forms a hydrogen bond with the I672 carbonyl group, which stabilizes the π-bulge on I672 (Fig. 4 A). In the closed state, the N676 side chain moves toward the S4–S5 linker and no longer donates a hydrogen bond to the main chain. As a consequence, the π-bulge is found preferentially on Y671 (Fig. 4 A). The displacement of the π-bulge from I672 to Y671 causes a change of Y671 orientation with respect to the pore: it goes from being perpendicular to the pore axis in the open state to being parallel to it in the closed state (Fig. 4, A and B). In the conformation parallel to the pore axis, Y671 becomes less shielded by the pore helices and hence more accessible to water molecules (Fig. 4 B).
Figure 4.
Two alternative conformations of the S6 helix in the TRPV1 closed and open states. (A) Coupling between N676 and Y671. The top left panel shows Y671 orientation (the cosine of the angle between the Cα-Cγ vector and the pore axis) and the π-bulge position (the difference between the two distances: I672 carbonyl oxygen–N676 amine hydrogen and Y671 carbonyl oxygen–N676 amine hydrogen; positive values indicate the location of the π-bulge on I672, while negative ones indicate that the π-bulge is on Y671). Only two conformations are observed: open state (1), Y671 is perpendicular to the pore axis, and the π-bulge is on I672; and closed state (2), Y671 is parallel to the pore axis, and the π-bulge is on Y671. The bottom left panel shows the distance between N676 carboxamide carbon and I672 carbonyl oxygen and the π-bulge position. The energetically most favorable conformations are open state (1), a hydrogen bond between N676 carboxamide and I672 carbonyl oxygen is present, and the π-bulge is on I672; and closed state (2), the hydrogen bond between N676 carboxamide and I672 carbonyl oxygen is absent, and the π-bulge is on Y671. The right panel shows the cartoon representations of the two conformations. Y672 is colored in purple, the π-bulge in red, and N676 in orange. The dashed lines show the network of hydrogen bonds. (B) A change of Y671 orientation results in a displacement of the pore helix. The top panel shows the conformations of Y671 and of the adjacent pore helix in the open (left) and closed (right) states. The bottom panel shows the superposition between the pore helix in the open (solid) and closed (transparent) states.
Interestingly, the molecular dynamics simulations also suggest that the observed reorientation of Y671 is coupled to the conformational change of the selectivity filter. In particular, when the Y671 side chain is parallel to the pore axis, it donates a hydrogen bond to the T641 carbonyl group on the pore helix. This interaction displaces the pore helix toward the extracellular solution, resulting in a slight constriction of the selectivity filter. In our previous study, we have shown that this constriction is sufficient to increase the free energy barrier for ionic transport from ∼1.5 in the open state to ∼5.0 kcal/mol in the closed one (Kasimova et al., 2018). Thus our findings suggest that in TRPV1, the selectivity filter can act as a gate whose opening is coupled to the conformation of Y671 and N676. Can this coupling mechanism be conserved in other TRP channels? The analysis of the multiple sequence alignment argues against this hypothesis by showing a lack of conservation at position 671 (Fig. 3 A). Moreover, recent structural data indicate that in TRPML3, the selectivity filter adopts the same conformation in the closed and open states (Hirschi et al., 2017; Zhou et al., 2017).
Are the peripheral cavities the missing piece of the puzzle?
The molecular mechanism suggested here agrees with the one emerging from the TRPV3, TRPV5, and TRPV6 structures (Saotome et al., 2016; Singh et al., 2017, 2018a,b; Hughes et al., 2018a,b; McGoldrick et al., 2018) and complements it with a detailed description of the conformational rearrangements involved in activation: the rotation of the conserved asparagine increases the hydrophilicity of the pore surface, thereby promoting the hydration of this compartment. An interesting finding provided by the molecular dynamics simulations and not anticipated by these structures is the factor stabilizing the closed state, in which the asparagine is located outside the pore. Our simulations have shown that in the closed state of TRPV1, N676 is accommodated inside a small hydrophobic cavity located between the S4–S5 linker and S6 and connected to the intracellular solution (Kasimova et al., 2018). This cavity, which we call peripheral in contrast to the central cavity along the ionic pathway, hosts several water molecules forming hydrogen bonds with N676 carboxamide. Importantly, the hydration of the peripheral cavity and the N676 rotation are correlated (Kasimova et al., 2018): when the peripheral cavity dehydrates, N676 rotates inside the pore. Based on this observation, we suggested that at least for some stimuli, activation of TRPV1 is triggered by the dewetting of the peripheral cavities (Kasimova et al., 2018). Is there any experimental evidence that supports the existence of these peripheral cavities? Overall, we found three independent observations, which can be retrospectively interpreted in light of this suggested molecular mechanism: (i) the experiments probing S6 accessibility to intracellular solution (Salazar et al., 2009), (ii) the site-directed mutagenesis experiments targeting S6 residues (Susankova et al., 2007), and (iii) the structural studies of the evolutionary related voltage-gated sodium and calcium channels (Payandeh et al., 2011, 2012; Zhang et al., 2012; Tsai et al., 2013; Tang et al., 2014, 2016; Ahuja et al., 2015; Wu et al., 2015, 2016; Lenaeus et al., 2017; Shen et al., 2017; Sula et al., 2017; Yan et al., 2017; Irie et al., 2018).
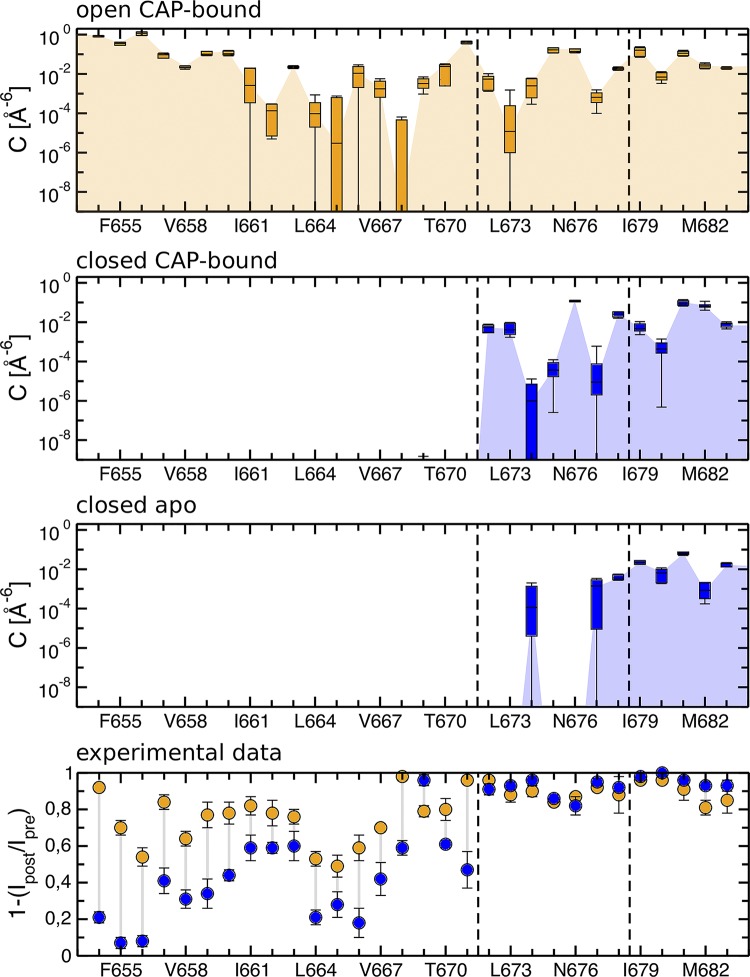
In a milestone paper by Salazar et al. (2009), the authors probed the accessibility of the TRPV1 pore residues to the intracellular solution; they mutated, one by one, all the S6 residues to cysteine and assessed their accessibility to thiol-modifying reagents in the open and closed states. Their results showed that modification of D654-Y671 is more prominent in the open state than in the closed one, and that of M682-G683 is state-independent. Interestingly, the rest of S6, namely the I672-L681 segment, showed different accessibility only when a bulky reagent was used; application of small reagents in the open or closed states, in contrast, resulted in a similar effect. Based on these data, the authors suggested the presence of lower and upper gates located at the levels of L681 and Y671, respectively; both gates are open in the open state, while in the closed state, the upper gate is closed and the lower gate is partially open (Salazar et al., 2009). This elegant explanation, however, turned out to be inconsistent with the structural information subsequently obtained via cryo-EM on the apo state (Liao et al., 2013), in which the gate at the level of the S6 bundle crossing was found to be tightly constricted with a radius of ∼1.0 Å, i.e., impermeable to any solute.
Can the presence of the peripheral cavities solve this apparent contradiction? To answer this question, we characterized the accessibility of the TRPV1 pore residues in the open and two closed states, including capsaicin-bound closed and apo closed (Fig. 5). In agreement with Salazar et al. (2009), we found that all of the S6 residues are accessible to the intracellular solution in the capsaicin-bound open state. Moreover, we found that the same set of residues, namely the I672-G683 segment, is accessible in the capsaicin-bound closed state. Despite the constriction at the level of I679, the residues up to and including I672 do interact with the intracellular solution. However, the interacting water molecules are not located in the pore, but rather in the peripheral cavities. Therefore, considering that the pore residues can be accessible to the intracellular solution through the peripheral cavities, one can resolve the contradiction between the water accessibility experiments (Salazar et al., 2009) and the structural information (Liao et al., 2013).
Figure 5.
Accessibility of the S6 residues to the intracellular solution in the TRPV1 open and closed states. The open and closed states data are shown in orange and blue, respectively. The three top panels show the computational results, while the bottom panel shows the experimental data extracted and digitalized from (Salazar et al., 2009). In the simulations, water accessibility is calculated as the overlap between the atomic density of a given S6 residue and that of the intracellular water (the values shown correspond to a sum over all four channel subunits). The open state data correspond to the results obtained for the capsaicin-bound (CAP-bound) open state (Kasimova et al., 2018); the closed state data correspond to the results obtained for the capsaicin-bound closed state and the apo closed state (Liao et al., 2013; Kasimova et al., 2018). The box plots report median and interquartile range with the whiskers representing the SD. The threshold for water accessibility was chosen to be zero. In the experiments, accessibility to the intracellular solution is measured as an effect on current after application of a thiol-modifying reagent. The dashed lines highlight the hypothetical upper gate according to Salazar et al. (2009) and I679 (the gate at the level of the S6 bundle crossing).
Interestingly, several pore residues above the gate are also accessible in the apo closed state, and similar to the capsaicin-bound closed state, they interact with the water molecules present in the peripheral cavities. However, in the apo state, the peripheral cavities are significantly smaller and host fewer water molecules. We hypothesize that this difference is the result of the S4–S5 linker movement: upon binding, capsaicin pulls the S4–S5 linker in the outward direction, thus expanding the space between the S4–S5 linker and S6 (Yang et al., 2015; Gao et al., 2016). The smaller size of the peripheral cavities in the apo closed state explains why only some of the I672-G683 residues are accessible to the intracellular solution.
Another piece of evidence lending credibility to our hypothesis comes from the alanine-scanning mutagenesis of the S6 residues performed by Susankova et al. (2007). The authors have identified a clear periodic pattern of the functional effects of mutations, which is consistent with the α-helical structure of S6 (Susankova et al., 2007). Subsequently obtained cryo-EM structures of TRPV1 showed that the residues whose mutation has the most significant effect on activation are located outside the pore and line the surface of the peripheral cavities (Fig. 6). Importantly, the response of the corresponding mutants to capsaicin, voltage, and heat was significantly reduced, indicating stabilization of the closed state versus the open one. A possible interpretation for these data is that the substitution of hydrophobic residues (I672, L673, L674, L678, and M682) with a less hydrophobic one (alanine) promotes the hydration of the peripheral cavities and thus stabilizes the closed state.
Figure 6.
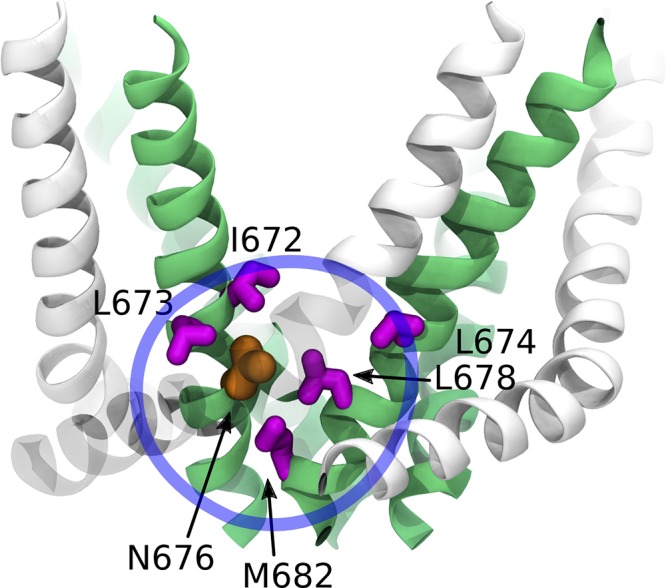
Mutagenesis of S6 residues lining the peripheral cavities. The residues mutated by Susankova et al. (2007) are shown in magenta, the conserved asparagine in orange, and the S6 helices and the rest of the pore domain in green and white, respectively. The peripheral cavity is highlighted with a blue circle.
Finally, we analyzed the families of voltage-gated sodium and calcium channels (Nav and Cav) that are structurally homologous and evolutionarily related to TRP channels (Payandeh et al., 2011, 2012; Zhang et al., 2012; Tsai et al., 2013; Tang et al., 2014, 2016; Ahuja et al., 2015; Wu et al., 2015, 2016; Lenaeus et al., 2017; Shen et al., 2017; Sula et al., 2017; Yan et al., 2017; Irie et al., 2018). Interestingly, these families also show a predominantly hydrophobic pore with the only exception of a conserved asparagine at the position corresponding to N676 (Fig. 7 A). In NaChBac, mutagenesis of this residue into several other amino acids yielded functional channels only in the case of the N225D mutant, and even in that case, gating of N225D was drastically affected (O’Reilly et al., 2017). Furthermore, the structures of Nav and Cav channels show that the conserved asparagine can adopt both the pore inward-facing (NavPas, all subunits; NavEel, one subunit out of four) and outward-facing conformations (Cav1.1, all subunits; NavAb, NavRh, NavMs, and NavEel, three subunits out of four; Fig. 7, B and C; Payandeh et al., 2011, 2012; Zhang et al., 2012; Tsai et al., 2013; Tang et al., 2014, 2016; Ahuja et al., 2015; Wu et al., 2015, 2016; Lenaeus et al., 2017; Shen et al., 2017; Sula et al., 2017; Yan et al., 2017; Irie et al., 2018). Do these channels have the peripheral cavities? A recent high-resolution structure of the bacterial sodium channel NavMs shows the presence of four cavities, whose location coincides with that of the peripheral ones in TRPV1 (Sula et al., 2017). Moreover, similar to the peripheral cavities, they host the side chain of the conserved asparagine and, importantly, few water molecules interacting with it.
Figure 7.
Structurally homologous and evolutionarily related families of voltage-gated sodium and calcium channels show a conserved asparagine in S6 and the presence of four hydrated cavities between S6 and the S4–S5 linker. (A) Sequence logos and per-position hydrophobicity plots of the S6 helix for voltage-gated sodium and calcium channels (Nav/Cav), bacterial voltage-gated sodium channels (NavBac), and TRP channels (TRP; Palovcak et al., 2015; Kasimova et al., 2016). In the sequence logos, the height of each residue at a given position is proportional to its frequency at this position, and the height of the overall stack of residues decreases linearly with Shannon entropy (Crooks et al., 2004). Aromatic residues (F, W, Y) are shown in orange; hydrophobic not aromatic (A, C, I, L, M, V) in black; hydrophilic (N, Q, S, T) in green; positively (H, K, R) and negatively (D, E) charged in blue and red, respectively; and G and P in purple. In the hydrophobicity plots, positions with positive are hydrophilic, and those with negative are hydrophobic. The error bars represent SD. The red shaded area highlights the residues lining the central cavity (between Y671 and I679). (B) Orientation of the conserved asparagine with respect to the pore in different Nav and Cav structures (Payandeh et al., 2011, 2012; Zhang et al., 2012; Tsai et al., 2013; Tang et al., 2014, 2016; Ahuja et al., 2015; Wu et al., 2015, 2016; Lenaeus et al., 2017; Shen et al., 2017; Sula et al., 2017; Yan et al., 2017; Irie et al., 2018). The side chain position in the plane perpendicular to the pore axis is shown: the Cα-atom is represented as a black dot and is located at (0,0), and the terminal Cγ-atom (Nζ in one of the NavPas subunits where the conserved asparagine is substituted for a lysine) is shown as colored symbols (different for each Nav/Cav channel). The x-axis is aligned with the vector connecting the Cα-atom and the center of the pore. The red shaded area highlights the pore region. The two insets show the pore domains of the NavMs structure (1, top and side views) and of the NavPas structure (2, top view). The conserved asparagine points toward the S4–S5 linker in NavMs and toward the pore in NavPas. In NavMs, the water molecules inside the cavity located between the S6 helix and the S4–S5 linker are shown in blue.
Conclusions
Despite two decades of extensive investigations, the molecular mechanism of TRPV1 activation is only partially understood. Catching an ion channel in the act of gating is, in general, a complex task requiring techniques with both high temporal and high spatial resolution. NMR spectroscopy and fluorescence-/luminescence-based approaches are typically the tools of choice, but they cannot easily probe the motions of an ion channel when embedded in a cell membrane. This limitation is particularly severe in the case of TRPV1, whose activity shows exquisite dependency on numerous environmental factors, such as lipid composition (Chuang et al., 2001; Cao et al., 2013a; Lukacs et al., 2013; Poblete et al., 2015; Rohacs, 2015; Gao et al., 2016), extracellular concentration of Na+ (Jara-Oseguera et al., 2016), and the presence of cholesterol or fatty acids (Liu et al., 2006; Matta et al., 2007; Picazo-Juárez et al., 2011; Morales-Lázaro et al., 2016). Accordingly, advanced experimental techniques, such as cell unroofing (Gordon et al., 2016; Zagotta et al., 2016) and incorporation of novel unnatural amino acids (Zagotta et al., 2016; Steinberg et al., 2017), are being constantly developed to overcome these limitations and introduce only minimal perturbations to the native environment of the channel. Despite the great progress, these techniques have focused so far only on specific aspects of TRPV1 activation. A global picture of this molecular process is still lacking, and it is unlikely that a single experimental technique will provide it in the foreseeable future.
Here, prompted by our previous molecular dynamics simulations (Kasimova et al., 2018) and bioinformatics analyses (Palovcak et al., 2015), we propose a molecular mechanism for TRPV1 activation. More than definite answers, our contribution provides testable predictions and an overall framework to rationalize a wealth of existing experimental data. Indeed, despite being consistent with the results of several independent studies, our hypothesis is still in part speculative. Most importantly, our work was only focused on the capsaicin-bound state, and the transition between that and the apo one was not investigated. It remains unclear how the conformation of the peripheral cavities changes along this transition and how capsaicin binding triggers this change. Based on the previous studies (Yang et al., 2015; Gao et al., 2016), we surmise that capsaicin promotes the expansion of the peripheral cavities between S6 and the S4–S5 linker. Enhanced sampling simulations starting from the apo state could provide answers to this question and also assess possible temperature dependency of the apo to capsaicin-bound transition.
Further research will be required to validate the molecular mechanism proposed here, including design of novel experiments able to discriminate among multiple alternative interpretations. For instance, incorporating unnatural amino acids in TRPV1 may allow for a fine-tuning of the polar properties of the pore and the peripheral cavities without perturbing their geometry too much. The same approach could be used to investigate the coupling between the selectivity filter and the gate. According to our model, this coupling involves the interaction between the side chain of N676 and the S6 helix main chain. The latter could be modified by introducing appropriately designed unnatural amino acids. From the computational point of view, long unbiased simulations could be used to sample gating events and provide direct confirmation of the activation mechanism. The Anton 2 supercomputer developed by D. E. Shaw Research (Shaw et al., 2009, 2014) and available through the Pittsburgh Supercomputing Center provides the opportunity to reach timescales comparable to those required for TRPV1 activation and thus to test the hypothesized chain of events. These simulations ought to involve, ideally, both the apo and capsaicin-bound states.
In closing, it is interesting to comment on an intriguing possibility emerging from our analyses: we found that the key structural features supporting our hypothesized mechanism are present also in the evolutionarily related families of voltage-gated sodium and calcium channels. In particular, the conserved asparagine in the S6 helix, the π-bulge, and the peripheral cavities are seemingly conserved beyond the family of TRP channels. This raises the possibility that voltage-gated sodium and calcium channels might share with TRP channels important aspects of the activation mechanism.
However, the sequence and structure conservation of S6 across TRP and voltage-gated sodium and calcium channels highlights a potentially problematic aspect of our hypothesis: in our previous computational investigation (Kasimova et al., 2018), we suggested that the rotation of N676 underlies temperature sensitivity in TRPV1. The evolutionary conservation of this asparagine cannot be easily reconciled with the fact that several TRP and voltage-gated sodium and calcium channels are insensitive to temperature. Future investigations ought to improve our understanding of the temperature sensitivity of ion channels and thus contribute to solving this conundrum.
Acknowledgments
This work was partially supported by the National Institutes of Health through grants R01GM093290, S10OD020095, NS055159, and P01GM055876 and the National Science Foundation through grants ACI-1614804 and CNS-1625061.
The authors declare no competing financial interests.
Author contributions: M.A. Kasimova and V. Carnevale designed the study. M.A. Kasimova collected the data and carried out analyses and simulations. M.A. Kasimova and D. Granata developed computational code to analyze molecular dynamics trajectories. M.A. Kasimova, A.T. Yazici, Y. Yudin, D. Granata, M.L. Klein, T. Rohacs, and V. Carnevale analyzed and interpreted the outcome of bioinformatics analyses and molecular dynamics simulations. T. Rohacs and V. Carnevale secured funding for the project. M.A. Kasimova and V. Carnevale wrote the paper with input from the other coauthors.
José D. Faraldo-Gómez served as editor.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., and Lindahl E.. 2015. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1–2:19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Ahuja S., Mukund S., Deng L., Khakh K., Chang E., Ho H., Shriver S., Young C., Lin S., Johnson J.P. Jr., et al. 2015. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science. 350:aac5464 10.1126/science.aac5464 [DOI] [PubMed] [Google Scholar]
- Autzen H.E., Myasnikov A.G., Campbell M.G., Asarnow D., Julius D., and Cheng Y.. 2018. Structure of the human TRPM4 ion channel in a lipid nanodisc. Science. 359:228–232. 10.1126/science.aar4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M., Kumar S., and Velavan R.. 2000. HELANAL: a program to characterize helix geometry in proteins. J. Biomol. Struct. Dyn. 17:811–819. 10.1080/07391102.2000.10506570 [DOI] [PubMed] [Google Scholar]
- Barad B.A., Echols N., Wang R.Y.-R., Cheng Y., DiMaio F., Adams P.D., and Fraser J.S.. 2015. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods. 12:943–946. 10.1038/nmeth.3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E., Cordero-Morales J.F., Liu B., Qin F., and Julius D.. 2013a TRPV1 channels are intrinsically heat sensitive and negatively regulated by phosphoinositide lipids. Neuron. 77:667–679. 10.1016/j.neuron.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E., Liao M., Cheng Y., and Julius D.. 2013b TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 504:113–118. 10.1038/nature12823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevale V., and Rohacs T.. 2016. TRPV1: A Target for Rational Drug Design. Pharmaceuticals (Basel). 9:E52 10.3390/ph9030052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina M.J., Schumacher M.A., Tominaga M., Rosen T.A., Levine J.D., and Julius D.. 1997. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 389:816–824. 10.1038/39807 [DOI] [PubMed] [Google Scholar]
- Chen Q., She J., Zeng W., Guo J., Xu H., Bai X.-C., and Jiang Y.. 2017. Structure of mammalian endolysosomal TRPML1 channel in nanodiscs. Nature. 550:415–418. 10.1038/nature24035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang H.H., Prescott E.D., Kong H., Shields S., Jordt S.E., Basbaum A.I., Chao M.V., and Julius D.. 2001. Bradykinin and nerve growth factor release the capsaicin receptor from PtdIns(4,5)P2-mediated inhibition. Nature. 411:957–962. 10.1038/35082088 [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.-M., and Brenner S.E.. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., and Pedersen L.. 1993. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- Deng Z., Paknejad N., Maksaev G., Sala-Rabanal M., Nichols C.G., Hite R.K., and Yuan P.. 2018. Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms. Nat. Struct. Mol. Biol. 25:252–260. 10.1038/s41594-018-0037-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J., Li J., Zeng B., Chen G.-L., Peng X., Zhang Y., Wang J., Clapham D.E., Li Z., and Zhang J.. 2018a Structure of the mouse TRPC4 ion channel. Nat. Commun. 9:3102 10.1038/s41467-018-05247-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J., Li Z., Li J., Hulse R.E., Santa-Cruz A., Valinsky W.C., Abiria S.A., Krapivinsky G., Zhang J., and Clapham D.E.. 2018b Structure of the mammalian TRPM7, a magnesium channel required during embryonic development. Proc. Natl. Acad. Sci. USA. 115:E8201–E8210. 10.1073/pnas.1810719115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elokely K., Velisetty P., Delemotte L., Palovcak E., Klein M.L., Rohacs T., and Carnevale V.. 2016. Understanding TRPV1 activation by ligands: Insights from the binding modes of capsaicin and resiniferatoxin. Proc. Natl. Acad. Sci. USA. 113:E137–E145. 10.1073/pnas.1517288113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Choi W., Sun W., Du J., and Lu W.. 2018. Structure of the human lipid-gated cation channel TRPC3. eLife. 7:e36852 10.7554/eLife.36852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Cao E., Julius D., and Cheng Y.. 2016. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 534:347–351. 10.1038/nature17964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.E., Senning E.N., Aman T.K., and Zagotta W.N.. 2016. Transition metal ion FRET to measure short-range distances at the intracellular surface of the plasma membrane. J. Gen. Physiol. 147:189–200. 10.1085/jgp.201511530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieben M., Pike A.C.W., Shintre C.A., Venturi E., El-Ajouz S., Tessitore A., Shrestha L., Mukhopadhyay S., Mahajan P., Chalk R., et al. 2017. Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2). Nat. Struct. Mol. Biol. 24:114–122. 10.1038/nsmb.3343 [DOI] [PubMed] [Google Scholar]
- Guo J., She J., Zeng W., Chen Q., Bai X.-C., and Jiang Y.. 2017. Structures of the calcium-activated, non-selective cation channel TRPM4. Nature. 552:205–209. 10.1038/nature24997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes P., Meadows H.J., Gunthorpe M.J., Harries M.H., Duckworth D.M., Cairns W., Harrison D.C., Clarke C.E., Ellington K., Prinjha R.K., et al. 2000. Cloning and functional expression of a human orthologue of rat vanilloid receptor-1. Pain. 88:205–215. 10.1016/S0304-3959(00)00353-5 [DOI] [PubMed] [Google Scholar]
- Hirschi M., Herzik M.A. Jr., Wie J., Suo Y., Borschel W.F., Ren D., Lander G.C., and Lee S.-Y.. 2017. Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Nature. 550:411–414. 10.1038/nature24055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T.E.T., Lodowski D.T., Huynh K.W., Yazici A., Del Rosario J., Kapoor A., Basak S., Samanta A., Han X., Chakrapani S., et al. 2018a Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM. Nat. Struct. Mol. Biol. 25:53–60. 10.1038/s41594-017-0009-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T.E.T., Pumroy R.A., Yazici A.T., Kasimova M.A., Fluck E.C., Huynh K.W., Samanta A., Molugu S., Zhou Z.H., Carnevale V., et al. 2018b Structural insights on TRPV5 gating by endogenous modulators. Nature Communications. 9:4198 10.1038/s41467-018-06753-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse R.E., Li Z., Huang R.K., Zhang J., and Clapham D.E.. 2018. Cryo-EM structure of the polycystin 2-l1 ion channel. eLife. 7:e36931 10.7554/eLife.36931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., and Schulten K.. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Huynh K.W., Cohen M.R., Jiang J., Samanta A., Lodowski D.T., Zhou Z.H., and Moiseenkova-Bell V.Y.. 2016. Structure of the full-length TRPV2 channel by cryo-EM. Nat. Commun. 7:11130 10.1038/ncomms11130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K., Haga Y., Shimomura T., and Fujiyoshi Y.. 2018. Optimized expression and purification of NavAb provide the structural insight into the voltage dependence. FEBS Lett. 592:274–283. 10.1002/1873-3468.12955 [DOI] [PubMed] [Google Scholar]
- Jara-Oseguera A., Bae C., and Swartz K.J.. 2016. An external sodium ion binding site controls allosteric gating in TRPV1 channels. eLife. 5:e13356 10.7554/eLife.13356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P., Bulkley D., Guo Y., Zhang W., Guo Z., Huynh W., Wu S., Meltzer S., Cheng T., Jan L.Y., et al. 2017. Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature. 547:118–122. 10.1038/nature22981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., and Klein M.L.. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Julius D., and Basbaum A.I.. 2001. Molecular mechanisms of nociception. Nature. 413:203–210. 10.1038/35093019 [DOI] [PubMed] [Google Scholar]
- Kasimova M.A., Granata D., and Carnevale V.. 2016. Voltage-Gated Sodium Channels: Evolutionary History and Distinctive Sequence Features. Curr. Top. Membr. 78:261–286. 10.1016/bs.ctm.2016.05.002 [DOI] [PubMed] [Google Scholar]
- Kasimova M.A., Yazici A., Yudin Y., Granata D., Klein M.L., Rohacs T., and Carnevale V.. 2018. Ion Channel Sensing: Are Fluctuations the Crux of the Matter? J. Phys. Chem. Lett. 9:1260–1264. 10.1021/acs.jpclett.7b03396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaeus M.J., Gamal El-Din T.M., Ing C., Ramanadane K., Pomès R., Zheng N., and Catterall W.A.. 2017. Structures of closed and open states of a voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA. 114:E3051–E3060. 10.1073/pnas.1700761114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Cao E., Julius D., and Cheng Y.. 2013. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 504:107–112. 10.1038/nature12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Huang W., Wu D., and Priestley J.V.. 2006. TRPV1, but not P2X, requires cholesterol for its function and membrane expression in rat nociceptors. Eur. J. Neurosci. 24:1–6. 10.1111/j.1460-9568.2006.04889.x [DOI] [PubMed] [Google Scholar]
- Lukacs V., Rives J.-M., Sun X., Zakharian E., and Rohacs T.. 2013. Promiscuous activation of transient receptor potential vanilloid 1 (TRPV1) channels by negatively charged intracellular lipids: the key role of endogenous phosphoinositides in maintaining channel activity. J. Biol. Chem. 288:35003–35013. 10.1074/jbc.M113.520288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerell A.D. Jr., Feig M., and Brooks C.L. III. 2004. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25:1400–1415. 10.1002/jcc.20065 [DOI] [PubMed] [Google Scholar]
- Matta J.A., Miyares R.L., and Ahern G.P.. 2007. TRPV1 is a novel target for omega-3 polyunsaturated fatty acids. J. Physiol. 578:397–411. 10.1113/jphysiol.2006.121988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick L.L., Singh A.K., Saotome K., Yelshanskaya M.V., Twomey E.C., Grassucci R.A., and Sobolevsky A.I.. 2018. Opening of the human epithelial calcium channel TRPV6. Nature. 553:233–237. 10.1038/nature25182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Lázaro S.L., Llorente I., Sierra-Ramírez F., López-Romero A.E., Ortíz-Rentería M., Serrano-Flores B., Simon S.A., Islas L.D., and Rosenbaum T.. 2016. Inhibition of TRPV1 channels by a naturally occurring omega-9 fatty acid reduces pain and itch. Nat. Commun. 7:13092 10.1038/ncomms13092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly A.O., Lattrell A., Miles A.J., Klinger A.B., Nau C., Wallace B.A., and Lampert A.. 2017. Mutagenesis of the NaChBac sodium channel discloses a functional role for a conserved S6 asparagine. Eur. Biophys. J. 46:665–674. 10.1007/s00249-017-1246-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palovcak E., Delemotte L., Klein M.L., and Carnevale V.. 2015. Comparative sequence analysis suggests a conserved gating mechanism for TRP channels. J. Gen. Physiol. 146:37–50. 10.1085/jgp.201411329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen C.E., Armache J.-P., Gao Y., Cheng Y., and Julius D.. 2015. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 520:511–517. 10.1038/nature14367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N., and Catterall W.A.. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358. 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J., Gamal El-Din T.M., Scheuer T., Zheng N., and Catterall W.A.. 2012. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 486:135–139. 10.1038/nature11077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., and Schulten K.. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picazo-Juárez G., Romero-Suárez S., Nieto-Posadas A., Llorente I., Jara-Oseguera A., Briggs M., McIntosh T.J., Simon S.A., Ladrón-de-Guevara E., Islas L.D., and Rosenbaum T.. 2011. Identification of a binding motif in the S5 helix that confers cholesterol sensitivity to the TRPV1 ion channel. J. Biol. Chem. 286:24966–24976. 10.1074/jbc.M111.237537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poblete H., Oyarzún I., Olivero P., Comer J., Zuñiga M., Sepulveda R.V., Báez-Nieto D., González Leon C., González-Nilo F., and Latorre R.. 2015. Molecular determinants of phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) binding to transient receptor potential V1 (TRPV1) channels. J. Biol. Chem. 290:2086–2098. 10.1074/jbc.M114.613620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohacs T. 2015. Phosphoinositide regulation of TRPV1 revisited. Pflugers Arch. 467:1851–1869. 10.1007/s00424-015-1695-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar H., Jara-Oseguera A., Hernández-García E., Llorente I., Arias-Olguín I.I., Soriano-García M., Islas L.D., and Rosenbaum T.. 2009. Structural determinants of gating in the TRPV1 channel. Nat. Struct. Mol. Biol. 16:704–710. 10.1038/nsmb.1633 [DOI] [PubMed] [Google Scholar]
- Saotome K., Singh A.K., Yelshanskaya M.V., and Sobolevsky A.I.. 2016. Crystal structure of the epithelial calcium channel TRPV6. Nature. 534:506–511. 10.1038/nature17975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiege P., Fine M., Blobel G., and Li X.. 2017. Human TRPML1 channel structures in open and closed conformations. Nature. 550:366–370. 10.1038/nature24036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw D.E., Dror R.O., Salmon J.K., Grossman J.P., Mackenzie K.M., Bank J.A., Young C., Deneroff M.M., Batson B., Bowers K.J., et al. 2009. Millisecond-scale Molecular Dynamics Simulations on Anton. In Proceedings of the Conference on High Performance Computing Networking, Storage and Analysis. ACM, New York: 39:1–39:11. [Google Scholar]
- Shaw D.E., Grossman J.P., Bank J.A., Batson B., Butts J.A., Chao J.C., Deneroff M.M., Dror R.O., Even A., Fenton C.H., et al. 2014. Anton 2: Raising the Bar for Performance and Programmability in a Special-Purpose Molecular Dynamics Supercomputer. In SC14: International Conference for High Performance Computing, Networking, Storage and Analysis. 41–53. [Google Scholar]
- Shen H., Zhou Q., Pan X., Li Z., Wu J., and Yan N.. 2017. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science. 355:eaal4326 10.1126/science.aal4326 [DOI] [PubMed] [Google Scholar]
- Shen P.S., Yang X., DeCaen P.G., Liu X., Bulkley D., Clapham D.E., and Cao E.. 2016. The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs. Cell. 167:763–773.e11. 10.1016/j.cell.2016.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Saotome K., and Sobolevsky A.I.. 2017. Swapping of transmembrane domains in the epithelial calcium channel TRPV6. Sci. Rep. 7:10669 10.1038/s41598-017-10993-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., McGoldrick L.L., and Sobolevsky A.I.. 2018a Structure and gating mechanism of the transient receptor potential channel TRPV3. Nat. Struct. Mol. Biol. 25:805–813. 10.1038/s41594-018-0108-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Saotome K., McGoldrick L.L., and Sobolevsky A.I.. 2018b Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB. Nat. Commun. 9:2465 10.1038/s41467-018-04828-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart O.S., Neduvelil J.G., Wang X., Wallace B.A., and Sansom M.S.P.. 1996. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14:354–360: 376. 10.1016/S0263-7855(97)00009-X [DOI] [PubMed] [Google Scholar]
- Steinberg X., Kasimova M.A., Cabezas-Bratesco D., Galpin J.D., Ladron-de-Guevara E., Villa F., Carnevale V., Islas L., Ahern C.A., and Brauchi S.E.. 2017. Conformational dynamics in TRPV1 channels reported by an encoded coumarin amino acid. eLife. 6:e28626 10.7554/eLife.28626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q., Hu F., Ge X., Lei J., Yu S., Wang T., Zhou Q., Mei C., and Shi Y.. 2018a Structure of the human PKD1-PKD2 complex. Science. 361:eaat9819. doi:. 10.1126/science.aat9819 [DOI] [PubMed] [Google Scholar]
- Su Q., Hu F., Liu Y., Ge X., Mei C., Yu S., Shen A., Zhou Q., Yan C., Lei J., et al. 2018b Cryo-EM structure of the polycystic kidney disease-like channel PKD2L1. Nat. Commun. 9:1192 10.1038/s41467-018-03606-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sula A., Booker J., Ng L.C.T., Naylor C.E., DeCaen P.G., and Wallace B.A.. 2017. The complete structure of an activated open sodium channel. Nat. Commun. 8:14205 10.1038/ncomms14205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susankova K., Ettrich R., Vyklicky L., Teisinger J., and Vlachova V.. 2007. Contribution of the putative inner-pore region to the gating of the transient receptor potential vanilloid subtype 1 channel (TRPV1). J. Neurosci. 27:7578–7585. 10.1523/JNEUROSCI.1956-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Gamal El-Din T.M., Payandeh J., Martinez G.Q., Heard T.M., Scheuer T., Zheng N., and Catterall W.A.. 2014. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature. 505:56–61. 10.1038/nature12775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Gamal El-Din T.M., Swanson T.M., Pryde D.C., Scheuer T., Zheng N., and Catterall W.A.. 2016. Structural basis for inhibition of a voltage-gated Ca2+ channel by Ca2+ antagonist drugs. Nature. 537:117–121. 10.1038/nature19102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Guo W., Zheng L., Wu J.-X., Liu M., Zhou X., Zhang X., and Chen L.. 2018. Structure of the receptor-activated human TRPC6 and TRPC3 ion channels. Cell Res. 28:746–755. 10.1038/s41422-018-0038-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M., Caterina M.J., Malmberg A.B., Rosen T.A., Gilbert H., Skinner K., Raumann B.E., Basbaum A.I., and Julius D.. 1998. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron. 21:531–543. 10.1016/S0896-6273(00)80564-4 [DOI] [PubMed] [Google Scholar]
- Tsai C.-J., Tani K., Irie K., Hiroaki Y., Shimomura T., McMillan D.G., Cook G.M., Schertler G.F.X., Fujiyoshi Y., and Li X.-D.. 2013. Two alternative conformations of a voltage-gated sodium channel. J. Mol. Biol. 425:4074–4088. 10.1016/j.jmb.2013.06.036 [DOI] [PubMed] [Google Scholar]
- Vinayagam D., Mager T., Apelbaum A., Bothe A., Merino F., Hofnagel O., Gatsogiannis C., and Raunser S.. 2018. Electron cryo-microscopy structure of the canonical TRPC4 ion channel. eLife. 7:e36615 10.7554/eLife.36615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner B.D. 2009. The use of coumarins as environmentally-sensitive fluorescent probes of heterogeneous inclusion systems. Molecules. 14:210–237. 10.3390/molecules14010210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Xie J., and Schultz P.G.. 2006. A genetically encoded fluorescent amino acid. J. Am. Chem. Soc. 128:8738–8739. 10.1021/ja062666k [DOI] [PubMed] [Google Scholar]
- Wilkes M., Madej M.G., Kreuter L., Rhinow D., Heinz V., De Sanctis S., Ruppel S., Richter R.M., Joos F., Grieben M., et al. 2017. Molecular insights into lipid-assisted Ca2+ regulation of the TRP channel Polycystin-2. Nat. Struct. Mol. Biol. 24:123–130. 10.1038/nsmb.3357 [DOI] [PubMed] [Google Scholar]
- Winkler P.A., Huang Y., Sun W., Du J., and Lü W.. 2017. Electron cryo-microscopy structure of a human TRPM4 channel. Nature. 552:200–204. 10.1038/nature24674 [DOI] [PubMed] [Google Scholar]
- Wu J., Yan Z., Li Z., Yan C., Lu S., Dong M., and Yan N.. 2015. Structure of the voltage-gated calcium channel Cav1.1 complex. Science. 350:aad2395 10.1126/science.aad2395 [DOI] [PubMed] [Google Scholar]
- Wu J., Yan Z., Li Z., Qian X., Lu S., Dong M., Zhou Q., and Yan N.. 2016. Structure of the voltage-gated calcium channel Ca(v)1.1 at 3.6 Å resolution. Nature. 537:191–196. 10.1038/nature19321 [DOI] [PubMed] [Google Scholar]
- Yan Z., Zhou Q., Wang L., Wu J., Zhao Y., Huang G., Peng W., Shen H., Lei J., and Yan N.. 2017. Structure of the Nav1.4-β1 Complex from Electric Eel. Cell. 170:470–482.e11. 10.1016/j.cell.2017.06.039 [DOI] [PubMed] [Google Scholar]
- Yang F., Xiao X., Cheng W., Yang W., Yu P., Song Z., Yarov-Yarovoy V., and Zheng J.. 2015. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat. Chem. Biol. 11:518–524. 10.1038/nchembio.1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wu M., Zubcevic L., Borschel W.F., Lander G.C., and Lee S.-Y.. 2018. Structure of the cold- and menthol-sensing ion channel TRPM8. Science. 359:237–241. 10.1126/science.aan4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta W.N., Gordon M.T., Senning E.N., Munari M.A., and Gordon S.E.. 2016. Measuring distances between TRPV1 and the plasma membrane using a noncanonical amino acid and transition metal ion FRET. J. Gen. Physiol. 147:201–216. 10.1085/jgp.201511531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ren W., DeCaen P., Yan C., Tao X., Tang L., Wang J., Hasegawa K., Kumasaka T., He J., et al. 2012. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 486:130–134. 10.1038/nature11054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Tóth B., Szollosi A., Chen J., and Csanády L.. 2018. Structure of a TRPM2 channel in complex with Ca2+ explains unique gating regulation. eLife. 7:e36409 10.7554/eLife.36409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Yang X., Hu R., Cai R., Hofmann L., Wang Z., Hu Q., Liu X., Bulkey D., Yu Y., et al. 2018. Hydrophobic pore gates regulate ion permeation in polycystic kidney disease 2 and 2L1 channels. Nat. Commun. 9:2302 10.1038/s41467-018-04586-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Li M., Su D., Jia Q., Li H., Li X., and Yang J.. 2017. Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states. Nat. Struct. Mol. Biol. 24:1146–1154. 10.1038/nsmb.3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubcevic L., Herzik M.A. Jr., Chung B.C., Liu Z., Lander G.C., and Lee S.-Y.. 2016. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat. Struct. Mol. Biol. 23:180–186. 10.1038/nsmb.3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubcevic L., Le S., Yang H., and Lee S.-Y.. 2018. Conformational plasticity in the selectivity filter of the TRPV2 ion channel. Nat. Struct. Mol. Biol. 25:405–415. 10.1038/s41594-018-0059-z [DOI] [PMC free article] [PubMed] [Google Scholar]