Abstract
The spread of antimicrobial resistance has become a serious public health concern, making once treatable diseases deadly again and undermining breakthrough achievements of modern medicine 1,2. Drug combinations can aid in fighting multi-drug resistant (MDR) bacterial infections, yet they are largely unexplored and rarely used in clinics. To identify general principles for antibacterial drug combinations and understand their potential, we profiled ~3,000 dose-resolved combinations of antibiotics, human-targeted drugs and food additives in 6 strains from three Gram-negative pathogens, Escherichia coli, Salmonella Typhimurium and Pseudomonas aeruginosa. Despite their phylogenetic relatedness, more than 70% of the detected drug-drug interactions are species-specific and 20% display strain specificity, revealing a large potential for narrow-spectrum therapies. Overall, antagonisms are more common than synergies and occur almost exclusively between drugs targeting different cellular processes, whereas synergies are more conserved and enriched in drugs targeting the same process. We elucidate mechanisms underlying this dichotomy and further dissect the interactions of the food additive, vanillin. Finally, we demonstrate that several synergies are effective against MDR clinical isolates in vitro and during infections of Galleria mellonella larvae, with one reverting resistance to the last-resort antibiotic, colistin.
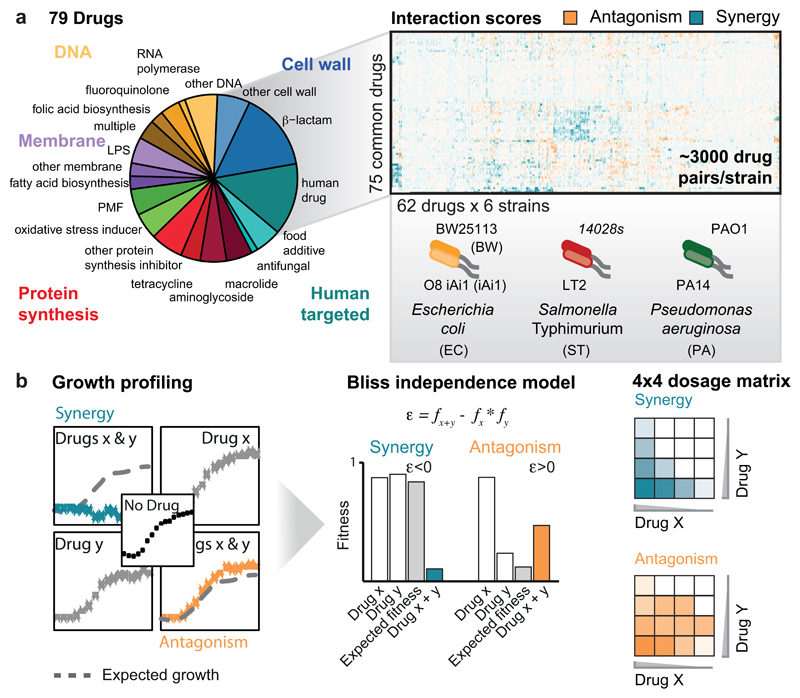
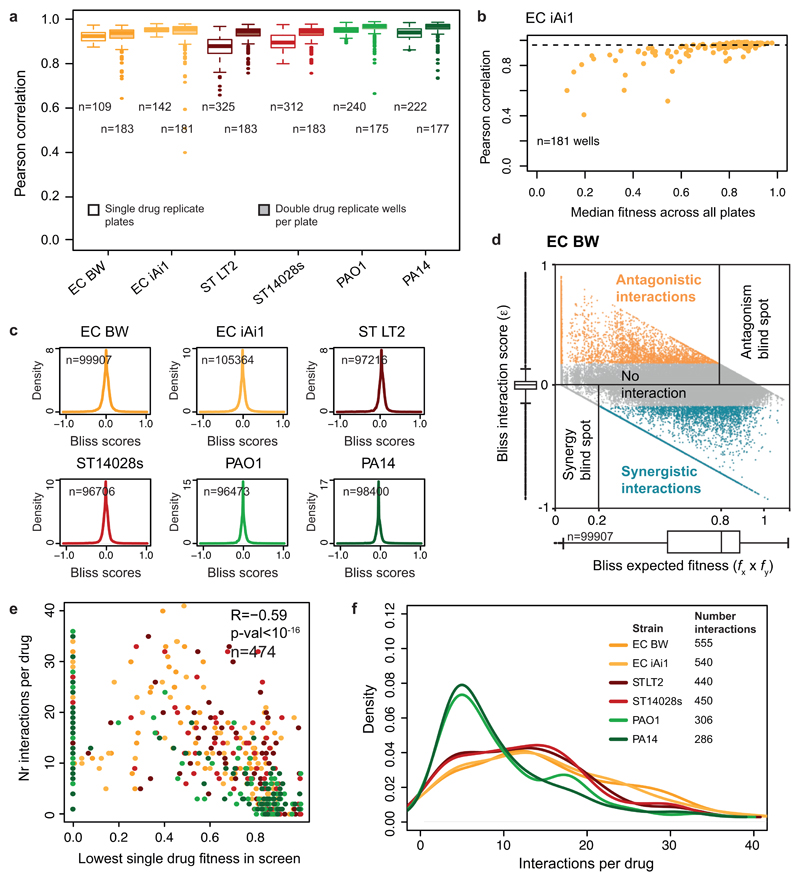
To study the characteristics and conservation of drug-drug interactions in bacteria, we selected three γ-proteobacterial species, E. coli, Salmonella enterica serogroup Typhimurium, and P. aeruginosa, all belonging to the highest risk groups for antibiotic resistance 3. We used model lab strains rather than MDR isolates to derive general principles behind drug-drug interactions without being confounded by horizontally transferred antibiotic resistance elements, and to facilitate follow-up experiments and comparisons with results from others. To further assess whether drug responses vary between strains of the same species, we included two strains per species (ED Fig. 1a), probing each in up to 79 compounds alone and in pairwise combinations. The compounds comprised 59% antibiotics (all major classes), 23% human-targeted drugs and food additives, most with reported antibacterial/adjuvant activity 4,5, and 18% of other compounds with known bacterial targets or genotoxic effects – e.g. proton motive force (PMF) inhibitors or oxidative damage agents, due to their potential relevance for antibiotic activity and/or uptake 6,7 (ED Fig. 1a; Supplementary Table 1). Altogether, we profiled up to 2,883 pairwise drug combinations in each of the 6 strains (17,050 in total). We assessed each drug combination in a 4x4 tailored dose matrix (Methods, Supplementary Table 1), using optical density as growth readout, and calculated fitness as the growth ratio between drug-treated and -untreated cells (ED Fig. 1-2, Methods). All experiments were done at least twice and on average 4x, with high replicate correlation (average Pearson Correlation = 0.93; ED Fig. 3a-b).
We quantified all drug-drug interactions using the Bliss independence model (ED Fig. 1b, Methods). Consistent with its null hypothesis, interaction scores were zero-centered for all species (ED Fig. 3c). From all the scores (ε) obtained per combination (4x4 dose matrix), we derived a single interaction score ranging from -1 to 1 (Methods). Synergies and antagonisms were considered significant if p-value < 0.05 (Benjamini-Hochberg corrected, 10,000 repetitions of a two-sided Wilcoxon rank-sum test). Strong interactions had an additional effect size requirement for || > 0.1, whereas weak interactions could satisfy the effect size threshold for one of the two strains of the same species, but be slightly below for the other (|| > 0.06; Methods). In total we detected ~19% interactions (synergies and antagonisms) for E. coli, ~16% for S. Typhimurium, and ~11% for P. aeruginosa (Supplementary Table 2). This is in between the >70% hit rate for 21 antibiotics tested in E. coli 8 and the <2% for a larger set of combinations tested in different fungi 9. Discrepancies are likely due to: (i) drug selection biases, (ii) single drug concentrations used in previous studies (which increase false negative and positive rates), and (iii) different data analysis. For example, we observed that drugs lacking antibacterial activity engage in fewer interactions (ED Fig. 3e). Robbins et al. screened pairwise combinations of 6 antifungals with 3,600 drugs, most of which had no antifungal activity 9, thus explaining the low number of interactions detected, whereas Yeh et al. profiled only bioactive antibiotics 8. Out of 79 drugs tested here, all had at least one interaction and a median of 5-13 interactions in the different strains (ED Fig. 3f).
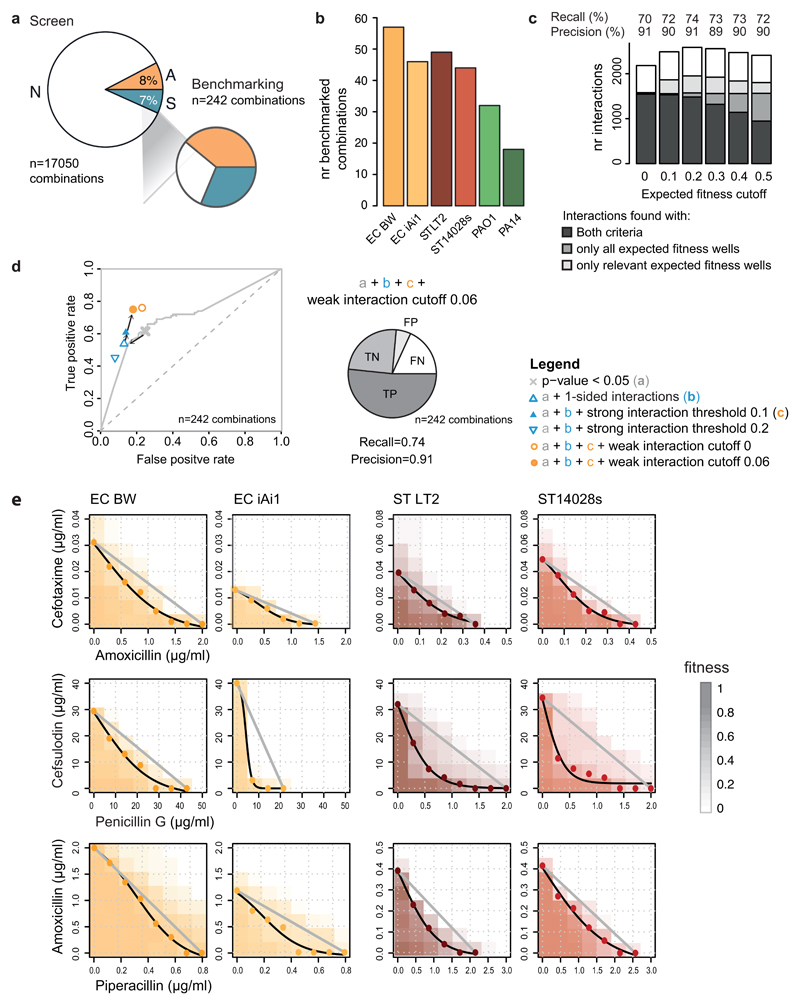
Since drug combinations have not been systematically probed in bacteria before, we lacked a ground truth for benchmarking our dataset. To overcome this limitation, we selected 242 combinations and created a validation set using higher-precision 8x8 checkerboard assays (ED Fig. 4a-b, Supplementary Table 3, Methods). We used this validation set to both assess the performance of our interaction identification approach and to benchmark our screen (ED Fig. 4c-d). Overall, we had precision and recall of 91% and 74%, respectively. The slightly lower recall can be partially explained by the larger coverage of drug concentration range in the validation experiments, which improves our ability to detect interactions (ED Fig. 5). We confirmed 90% of all weak interactions we probed in the validation set (n=46; Supplementary Table 3, ED Fig. 6), supporting the rationale of our interaction identification approach. Indeed, including weak interactions in our hits increases the recall (ED Fig. 4d). For a handful of the synergies observed between antibiotics of the same class (β-lactams), we confirmed the interactions using the Loewe additivity model (ED Fig. 4e), which is more suitable for assessing interactions between drugs with the same target.
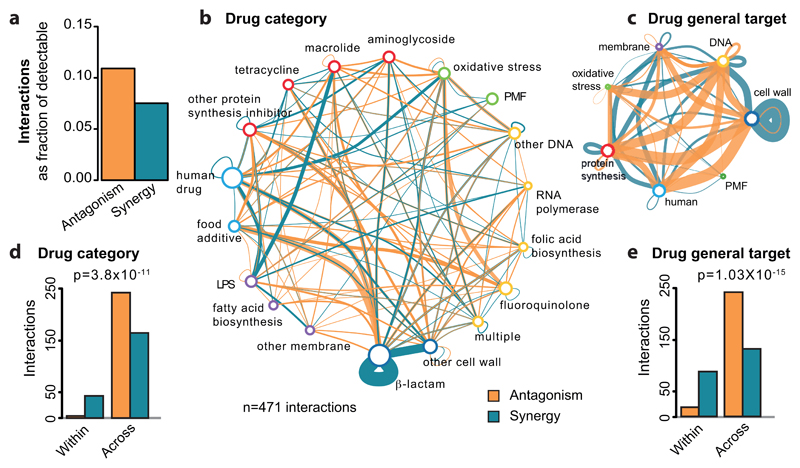
Overall, we detected 1354 antagonistic and 1230 synergistic interactions. Although this suggests that the two occur with similar frequencies, antagonisms are nearly 50% more prevalent than synergies, when correcting for the ability to detect both types of interactions (Fig. 1a). This is because we can detect antagonisms only for 75% of combinations (when at least one drug inhibits growth; ED Fig. 3d, Methods), whereas synergies are detectable for nearly all combinations. Higher prevalence of antagonisms has also been reported for antifungals 10.
Figure 1. Principles of drug-drug interaction networks.
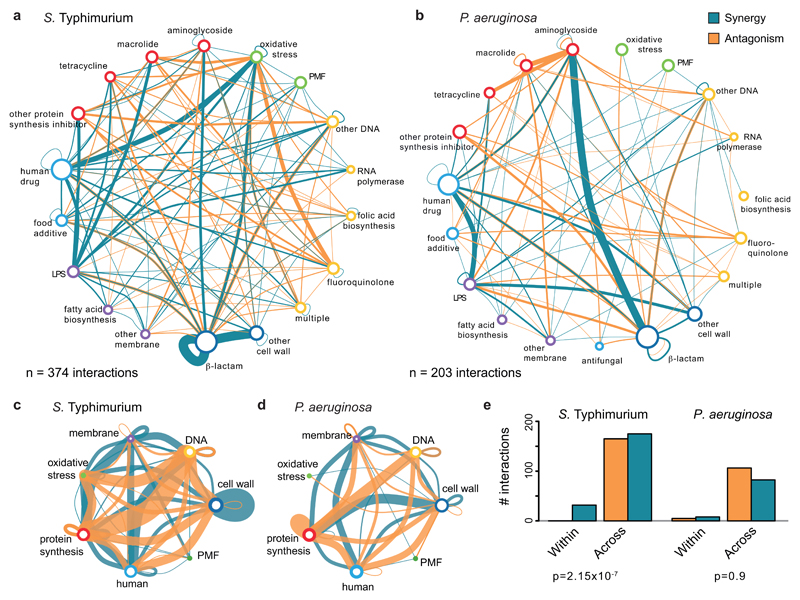
a) Antagonism is more prevalent than synergy. Fraction of observed over detectable interactions for the 6 strains. We detect more antagonistic (1354) than synergistic (1230) interactions, although our ability to detect antagonisms is lower than synergy: 12,778 versus 16,920 combinations. b & d) Drug-drug interaction networks in E. coli. Nodes represent either drug categories (b) or drugs grouped according to the general cellular process they target (d). Node color is as ED Fig. 1a and node size reflects the number of drugs within category. Edges represent synergy (blue) and antagonism (orange); thickness reflects number of interactions. Interactions between drugs of the same category/general cellular target are represented by self-interacting edges. Conserved interactions, including weak, are shown. c & e) Antagonisms occur almost exclusively between drugs belonging to different categories (c) or targeting different cellular processes (e), whereas synergies are also abundant between drugs within the same category (c) or targeting the same process (e). Quantification and Chi-squared test p-values from E. coli drug-drug interactions are shown in b and d, respectively.
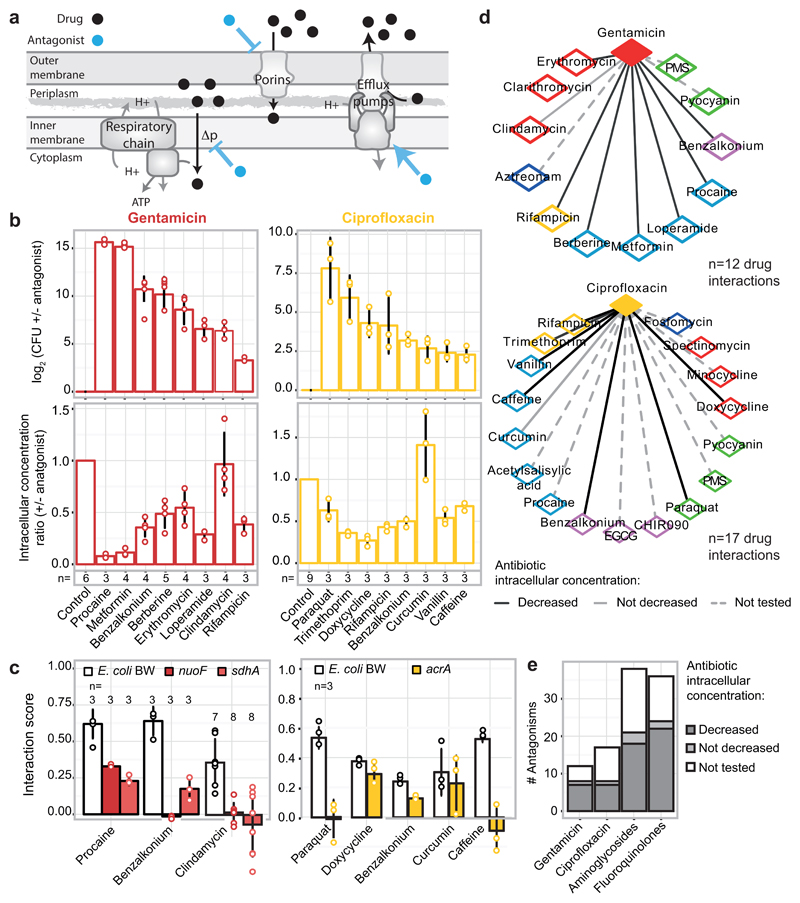
Strikingly, antagonisms and synergies exhibited a clear dichotomy in our data. Antagonism occurred almost exclusively between drugs targeting different cellular processes, while synergies were also abundant for drugs of the same class or targeting the same process (Fig. 1b-e, ED Fig. 7). Mechanistically, antagonism can be explained by interactions at the drug target level, with the two inhibitors helping the cell to buffer the distinct processes perturbed. DNA and protein synthesis inhibitors act this way in bacteria (Fig. 1b) 11. Consistent with this being a broader phenomenon, in genome-wide genetic interactions studies in yeast, alleviating interactions (antagonisms) are enriched between essential genes (the targets of anti-infectives), which are part of different functional processes 12. However, antagonism can also occur at the level of intracellular drug concentrations (ED Fig. 8a). We tested 16 antagonistic interactions of different drugs with gentamicin or ciprofloxacin in E. coli to investigate to what extent this occurs. Although initially detected at a growth inhibition level, all antagonisms held true at a killing level, with 14/16 decreasing the intracellular gentamicin or ciprofloxacin concentrations (ED Fig. 8b). In several cases tested, this likely occurred because the second drug decreased the PMF-energized uptake of gentamicin or increased AcrAB-TolC-dependent efflux of ciprofloxacin, as antagonisms were neutralized in the respective mutant backgrounds (ED Fig. 8c). Overall, our results suggest that a large fraction of antagonisms is due to modulation of intracellular drug concentrations, rather than due to direct interactions of the primary drug targets (ED Fig. 8d-e).
Unlike antagonistic interactions, synergies often occurred between drugs targeting the same cellular process (Fig. 1b-e, ED Fig. 7). In fact, synergies are significantly enriched within drugs of the same category across all three species (p-value < 10-16, Fisher’s exact test), given that there are ~15-fold more possible drug combinations across than within drug categories in our dataset. Mechanistically, targeting the same functional process at different steps could bypass its redundancy. For example, β-lactams have different affinities to the numerous and often redundant penicillin-binding-proteins (PBPs), likely explaining the many synergies between them (Fig. 1b, ED Fig. 4e & 7a-b).
Like antagonisms, synergies can also occur due to modulation of intracellular drug concentrations. Consistent with a general permeabilization role of membrane-targeting compounds in many organisms 9,13,14, and with drug uptake being a major bottleneck for Gram-negative pathogens, one fourth of all detected synergies contain at least one out of eight membrane-targeting drugs in our screen (two-sided Wilcoxon rank-sum test, p-value=0.06). However, membrane-targeting compounds account also for ~18% of antagonisms, suggesting that perturbations in membrane integrity can also decrease intracellular drug concentrations. Consistently, benzalkonium decreases the intracellular concentration of both gentamicin and ciprofloxacin, likely by interfering with their active import in the cell (ED Fig. 8b-c).
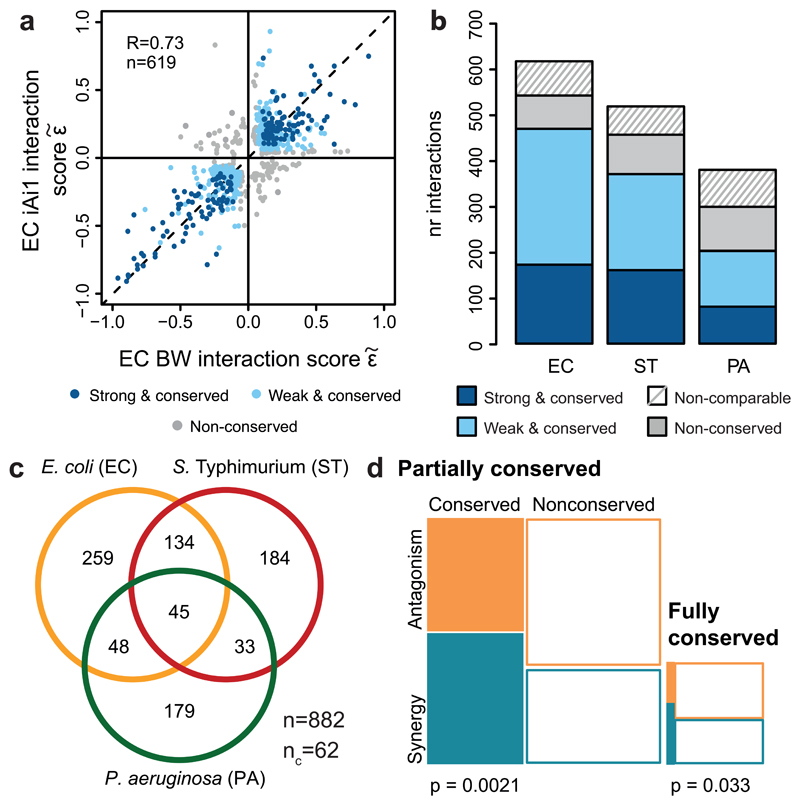
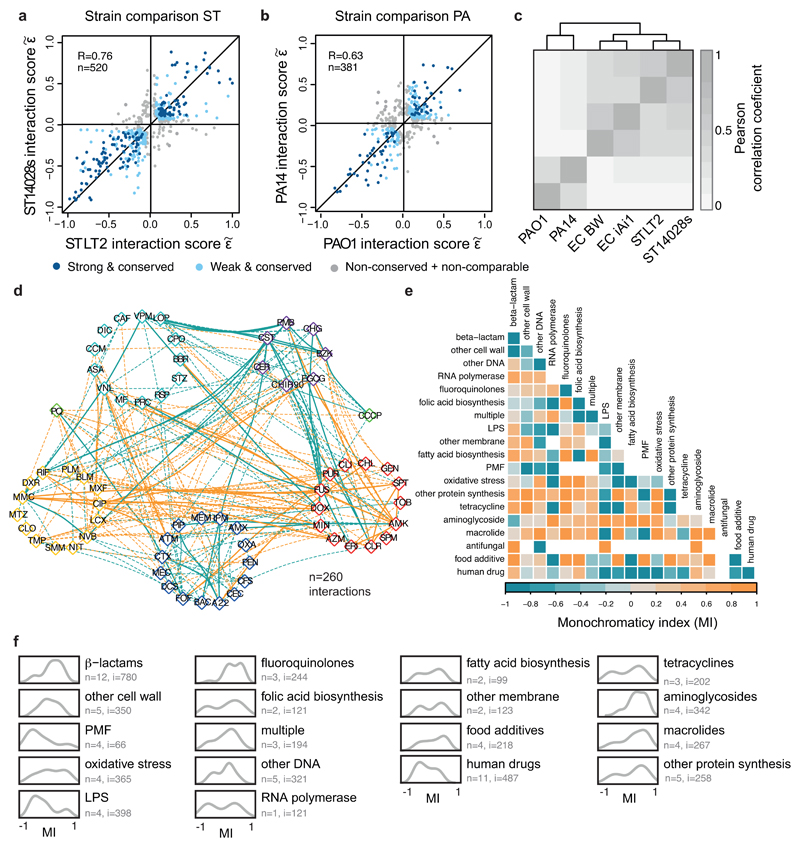
We next examined the conservation of drug-drug interactions. Interactions within species were highly conserved (Fig. 2a, ED Fig. 9a-b): 53-76%, depending on the species (Fig. 2b). Conservation is actually higher (68-87%, and on average 80%), if we disregard the non-comparable interactions for which the concentration range tested preclude us from detecting synergy or antagonism for both strains (Fig. 2b, ED Fig. 3d). High conservation of drug-drug interactions within species is consistent with the finding that such interactions are generally robust to simple genetic perturbations 15. Despite this high-degree conservation within species, 13-32% of the interactions remained strain-specific, with the majority being neutral in the second strain. Very few drug combinations synergized for one strain and antagonized for the other (16 interactions), but such strain differences held in our validation set (Supplementary Table 2).
Figure 2. Drug-drug interaction conservation.
a) Drug-drug interactions are conserved in E. coli. Scatter plot of interaction scores from the two E. coli strains; significant interactions for at least one of the strains are shown. Dark blue: strong and conserved interactions in both strains; light blue: strong interactions in one strain and concordant behavior in other (weak and conserved); grey: interactions occurring exclusively in one strain or conflicting between strains (non-conserved). R denotes the Pearson correlation, n the number interactions plotted. b) Drug-drug interactions are highly conserved within species. Colors as in a; non-comparable refers to combinations that have significantly different single drug dose responses between strains (Methods). c) Drug-drug interactions are largely species-specific; n = total number of interactions; nc = conflicting interactions between species, not accounted for in Venn diagram. d) Synergies are more conserved than antagonisms. Mosaic plots and Chi-squared test p-values show the quantification of synergy and antagonism among conserved (fully and partially) and non-conserved interactions between species.
While conservation is relatively high within species, it is very low across species (Fig. 2c, ED Fig. 9c). The majority (70%) of interactions occurred in one species, and only 5% were conserved in all three phylogenetically close-related species. Since conservation is much higher at the single-drug level for the three species (sharing resistance/sensitivity to 73% of the drugs; Supplementary Table 1, Methods), this indicates that drug combinations can impart species specificity to the drug action. Such specificities can be beneficial for creating narrow spectrum therapies with low collateral damage, by using synergies specific for pathogens and antagonisms specific for abundant commensals.
Moreover, we found that synergies are significantly more conserved than antagonisms (Fig. 2d), despite being less prevalent (Fig. 1a). This is presumably because: i) synergies are enriched between drugs of the same category, and interactions within functional processes are conserved across evolution 16; ii) membrane-targeting drugs have a general potentiation effect in Gram-negative bacteria; and iii) antagonisms often depend on drug import/uptake (ED Fig. 8), which are controlled by less conserved envelope machineries.
Exploring the network of conserved drug-drug interactions across the three species (ED Fig. 9d) exposed potential Achilles heels of Gram-negative bacteria, such as the strong synergy of colistin with macrolides 17, but also revealed that known antibiotic classes often behave non-uniformly. For example, the well-known synergy between β-lactams and aminoglycosides is confined to potent aminoglycosides used in our screen (amikacin and tobramycin) and β-lactams that target specifically the cell-division related PBPs (piperacillin, aztreonam, cefotaxime), in agreement with previous reports 18. To address whether pairwise drug interactions are Mode of Action (MoA)-driven (i.e. drug classes interacting purely synergistic or antagonistic with each other) 8, we calculated a monochromaticity index (MI) for all drug category pairs, across all species (Methods). For highly monochromatic category pairs, MI approaches 1 and -1 for antagonism and synergy, respectively. MI is overall high, especially between well-defined antibiotic classes. Yet, a number of them, including β-lactams, tetracyclines and macrolides, have mixed antagonisms and synergies with other antibiotic classes (ED Fig. 9e). While β-lactams have diverse affinities to their multiple PBP targets (potentially explaining the mixed interactions with other classes), the same does not apply to protein synthesis inhibitors, which have unique targets. In this case, non-uniform class behavior may be due to different chemical properties of the class members, and thus different dependencies on uptake and efflux systems. Aggregating the MI per drug category reinforced the view that broader categories exhibit less concordant interactions (ED Fig. 9f). Besides membrane targeting drugs, human-targeted drugs were the category exhibiting the most synergies, suggesting that many may act as adjuvants.
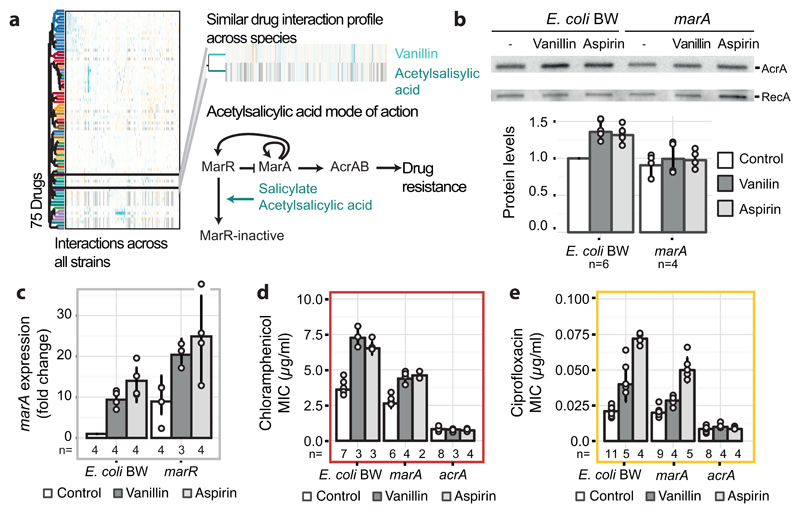
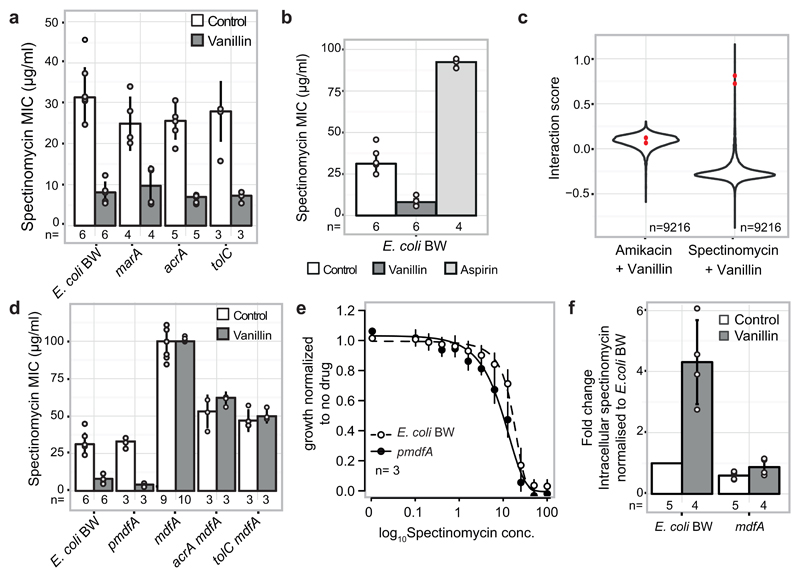
Since antibiotic classes interacted largely monochromatically, clustering drugs according to their interactions recapitulated the class groupings (ED Fig. 10). For example, cell-wall inhibitors grouped together, with further subdivisions being reflective of target specificity. Yet, exceptions were also evident, such as the macrolides, which split. Azithromycin, the only dibasic macrolide separates from its class co-members and clusters with two other basic antibiotics, bleomycin and phleomycin. Azithromycin interacts with and crosses the outer membrane (OM) of Gram-negative bacteria distinctly to other macrolides 17,19, and has also different binding kinetics to the peptide exit tunnel of the 50S ribosomal subunit 20. For drugs with unknown or less-well defined targets, clustering hinted towards possible MoA’s. Among them, we selected the flavoring compound vanillin, which clusters together with the structurally related acetylsalicylic acid (aspirin). Salicylate and aspirin induce the expression of the major efflux pump in enterobacteria, AcrAB-TolC via binding and inactivating the transcriptional repressor MarR 21 (Fig. 3a). Consistent with a similar action, vanillin treatment increased AcrA protein levels in E. coli, due to marA overexpression (Fig. 3b-c). Higher AcrA levels upon vanillin or aspirin treatment led to higher chloramphenicol and ciprofloxacin MICs (Fig. 3d-e). As previously reported for salicylate 22, vanillin exerts an additional minor effect on drug resistance in a MarR/A-independent manner, presumably via the MarA homologue, Rob (Fig. 3c-e).
Figure 3. Vanillin induces a multi-antibiotic-resistance (mar) phenotype.
a) Vanillin and aspirin (acetylsalicylic acid) have similar drug-drug interaction profiles (see ED Fig. 10), suggesting similar MoA’s. A schematic representation of the mar response induction via deactivation of the MarR repressor by salicylate/aspirin 21 is illustrated. b) Vanillin increases AcrA levels in a marA-dependent manner. A representative immunoblot of exponentially growing cells (all blots shown in Supplementary Fig. 1) after treatment with solvent, vanillin (150µg/ml) or aspirin (500µg/ml) is shown - loading controlled by cell density and constitutively expressed RecA. Barplots depict AcrA protein level quantification; c) marA expression levels upon vanillin (150µg/ml) or aspirin (500µg/ml) treatment are stronger in wildtype than in ΔmarR mutant. Expression is measured by RT-qPCR and normalized to no-drug treatment in wildtype; d & e) Vanillin (150 µg/ml) and aspirin (500µg/ml) increase the MIC of chloramphenicol (d) or ciprofloxacin (e). Antagonism is weaker and abolished in ΔmarA and ΔacrA mutants, respectively. n = number of independent biological replicates and error bars depict standard deviation (b-e).
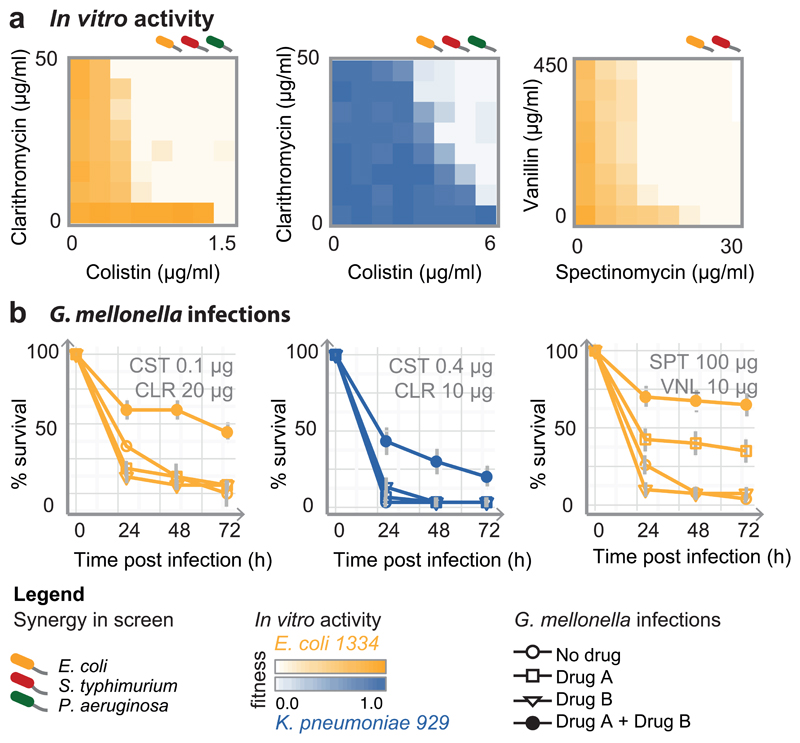
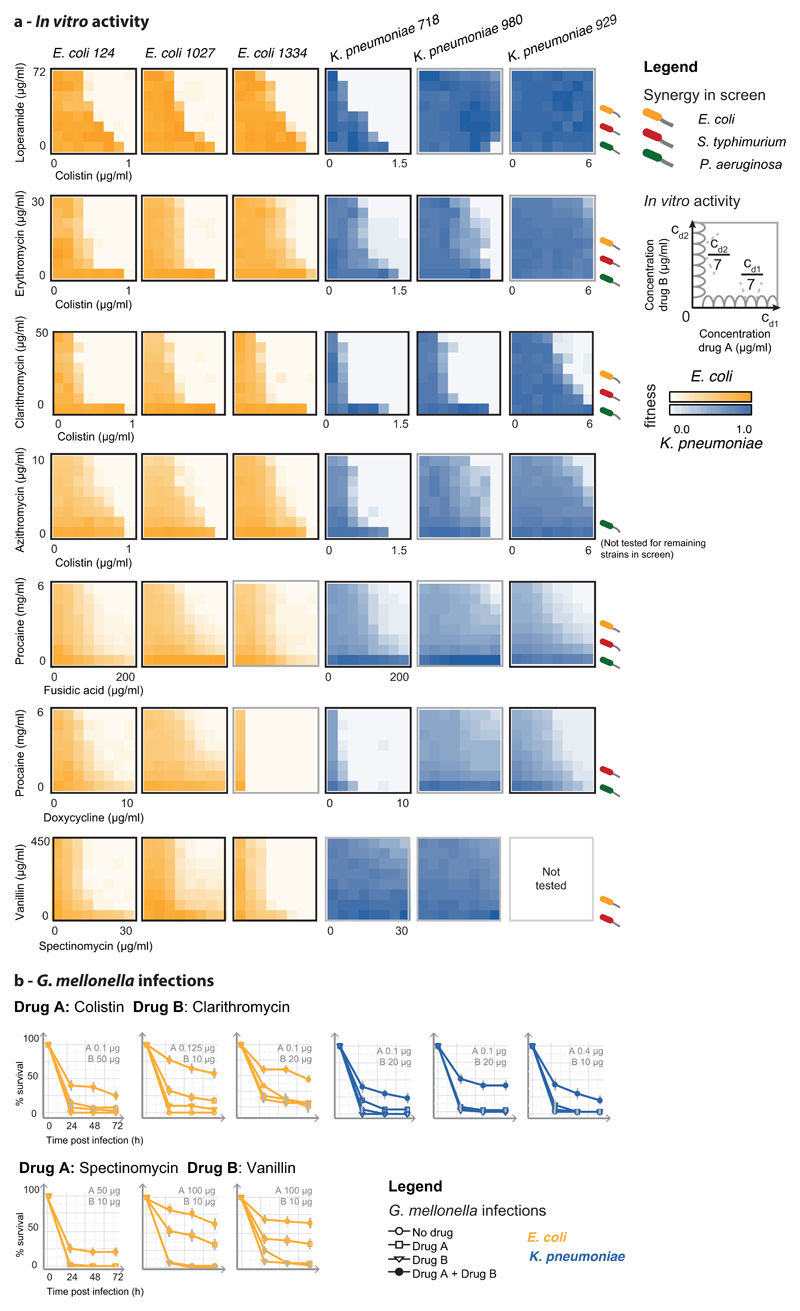
To test whether detected interactions are relevant for resistant isolates, we selected seven strong and conserved synergies, comprising antibiotics, human-targeted drugs or food additives, and assessed their efficacy against six MDR and XDR E. coli and Klebsiella pneumoniae clinical isolates. All strains were recovered from infected patients, belonging to successfully spread clonal lineages harboring extended spectrum β-lactamase (ESBL) resistance and various highly prevalent carbapenemases 23,24. One K. pneumoniae strain (929) is also resistant to the last-resort antibiotic, colistin, due to a chromosomal mutation (Supplementary Table 4). All drug pairs acted synergistically in most of the strains tested (Fig. 4a, ED Fig. 11a). We further tested colistin-clarithromycin and spectinomycin-vanillin, with an established infection model for evaluating antibacterial activity, using larvae the greater wax moth, Galleria mellonella. Both combinations also acted synergistically in vivo by increasing G. mellonella survival during infection (Fig. 4b & ED Fig. 11b).
Figure 4. Potent synergistic combinations against Gram-negative MDR clinical isolates.
a) In vitro synergies, shown as 8x8 checkerboards, for 3 MDR strains (more strains and synergies in ED Fig. 11). One of two biological replicates is shown. b) Drug synergies against the same MDR strains in the Galleria mellonella infection model (see also ED Fig. 11). Larvae were infected by E. coli and K. pneumoniae MDR isolates (106 and 104 CFU, respectively) and left untreated, or treated with single drugs or combination. % larvae survival was monitored at indicated intervals after infection – n=10 larvae per treatment. The average of 4 biological replicates are shown; error bars depict standard deviation.
The strongest of these synergies is between colistin and different macrolides (Fig. 4, ED Fig. 11). Although other polymyxins are known to help macrolides cross the OM of Gram-negative bacteria 17, this particular synergy occurred at low colistin concentration (< 0.3 µg/ml) and was active even for the intrinsically colistin-resistant strain (Fig. 4, K. pneumoniae 929), implying that macrolides may also potentiate colistin via a yet unknown mechanism. Similar resensitization of colistin-resistant pathogens to colistin by macrolides was recently reported for plasmid-borne colistin resistance 25, indicating that this synergy is independent of the resistance mechanism. In addition to antibiotic pairs, combinations of human-targeted drugs or food additives with antibiotics were also effective against MDR isolates, even when the former lacked antibacterial activity on their own (ED Fig. 11).
Finally, vanillin potentiated the activity of spectinomycin in E. coli MDR isolates. This was intriguing, since vanillin antagonizes many other drugs, including other aminoglycosides (Supplementary Table 2). We confirmed that this interaction is specific to spectinomycin and vanillin, and not to other aminoglycosides or aspirin, and thus independent of the vanillin effect on AcrAB-TolC (ED Fig. 12a-c). We then probed a genome-wide E. coli gene knockout library 26 to identify mutants that abrogate the vanillin-spectinomycin interaction, but do not influence the amikacin (another aminoglycoside)-vanillin interaction. One of the top hits was mdfA, which encodes for a Major Facilitator Superfamily transporter, exporting both charged and neutral compounds 27 (ED Fig. 12c). Consistent with MdfA modulating spectinomycin uptake, ΔmdfA cells were more resistant to spectinomycin and not responsive to vanillin (ED Fig. 12d), whereas cells overexpressing mdfA were more sensitive to spectinomycin (ED Fig. 12e, not visible at the MIC level in ED Fig. 12d), as previously reported 28, with vanillin further exacerbating this effect (ED Fig. 12d). Vanillin addition also increased the intracellular spectinomycin concentration in an mdfA-dependent manner (ED Fig. 12e). At this point, it is unclear how MdfA, which is known to export compounds out of the cell, facilitates spectinomycin import in the cell. However, the phylogenetic occurrence of mdfA is concordant with the species-specificity of this interaction, as we detected the synergy in E. coli and S. Typhimurium, but not in the phylogenetically more distant, P. aeruginosa and K. pneumoniae isolates, which lack mdfA. This synergy underlines the importance of exploring the role of food additives in combinatorial therapies 5.
In summary, we generated a comprehensive resource of pairwise drug combinations in Gram-negative bacteria, illuminating key principles of drug-drug interactions and providing a framework for assessing their conservation across organisms or individuals (Supplementary Discussion). Such information can nucleate similar screens in other microbes, studies investigating the underlying mechanism of pairwise drug combinations 11,15,29 and computational predictions of their outcomes 30,31. Some of the herein identified principles may go beyond anti-infectives and microbes 32. For antibacterial drug therapies, our study highlights the promise that non-antibiotic drugs hold as adjuvants, offers a new path for narrow spectrum therapies and identifies effective synergies against MDR clinical isolates (Supplementary Discussion). Further experimentation is required to address whether such synergies have clinical relevance.
Methods
Strains, plasmids and drugs
For each of the three Gram-negative species profiled in this study, we used two common sequenced lab strains for each species: Escherichia coli K-12 BW25113 and O8 IAI1, Salmonella enterica serovar Typhimurium LT2 and 14028s, Pseudomonas aeruginosa PAO1 and PA14. To validate selected synergies, we profiled 6 MDR clinical Enterobacteriaceae isolates recovered from human patient specimens: E. coli 124, 1027, 1334 and Klebsiella pneumoniae 718, 929 and 980 (see Supplementary Table 4 for details on antibiotic resistance determinants). For follow-up experiments, we used two closely-related E. coli K-12 model strains, BW25113 and MG1655.
All mutants used in this study were made using the E. coli Keio Knockout Collection 26 - after PCR-confirming and retransducing the mutation to wildtype BW25113 with the P1 phage (Supplementary Table 5). The kanamycin resistance cassette was excised when necessary using the plasmid pCP20 33. The plasmid used for mdfA overexpression was obtained from the mobile E. coli ORF library 34.
Drugs used in this study were purchased from Sigma Aldrich, except for metformin hydrochloride (TCI Chemicals), clindamycin and bleomycin (Applichem), CHIR-090 (MedChemtronica) and vanillin (Roth). Stocks were prepared according to supplier recommendations (preferably dissolved in water).
Minimal Inhibitory Concentration (MIC) calculation
We defined MIC as the lowest concentration required to inhibit growth of a microorganism after 8 hours of incubation in Lysogeny Broth (LB) at 37°C with shaking (384 wells plates, starting OD595nm 0.01). MICs of all drugs were computed using a logistic fit of growth (OD595nm for 8h) over 2-fold serial dilutions of the antibiotic concentrations for all strains used for the high-throughput screening and follow-up experiments.
High-throughput screening of pairwise drug interactions
For all drug combination experiments, drugs were diluted in LB to the appropriate working concentrations in transparent 384-well plates (Greiner BioOne GmbH), with each well containing 30µl in total. After the addition of drugs, cells were inoculated at initial OD595nm ~0.01 from an overnight culture. The same inoculum was used for all strains. All liquid handling (drug addition, cell mixing) was done with a Biomek FX liquid handler (Beckman Coulter). Plates were sealed with breathable membranes (Breathe-Easy®) and incubated at 37°C in a humidity-saturated incubator (Cytomat 2, Thermo Scientific) with continuous shaking and without lids to avoid condensation. OD595nm was measured every 40 min for 12 hours in a Filtermax F5 multimode plate reader (Molecular Devices).
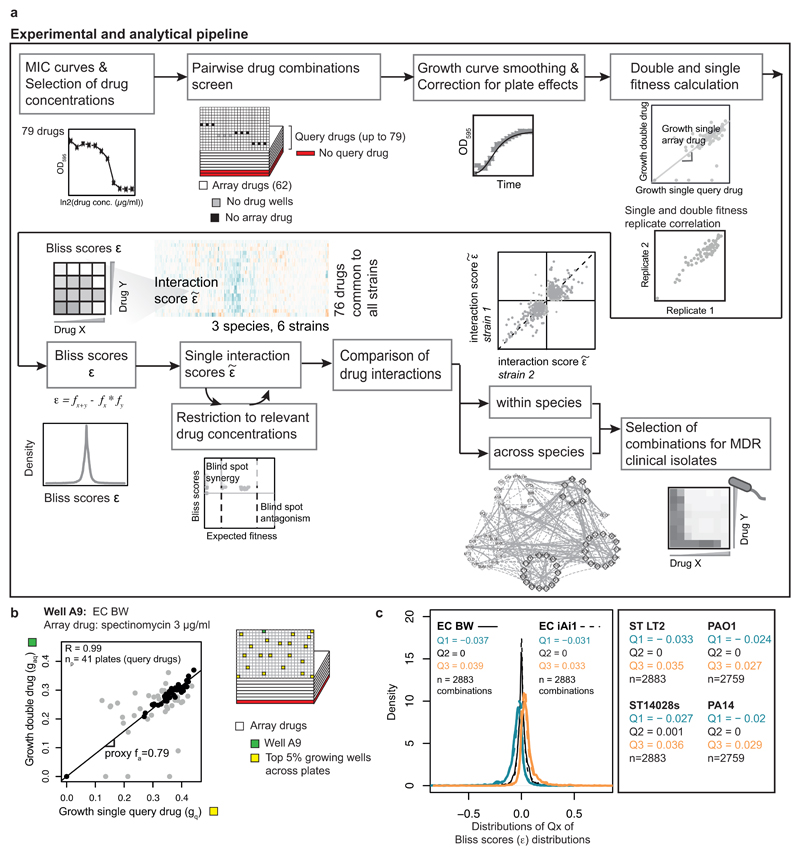
A flowchart of the experimental and analytical pipeline is shown in ED Fig. 2a. Data analysis was implemented with R and networks were created with Cytoscape 35.
Experimental Pipeline
The drug-drug interaction screen was performed using 4x4 checkerboards. 62 drugs were arrayed in 384 well plates with the different concentrations in duplicates (array drugs). Each plate contained 12 randomly distributed wells without arrayed drug: 9 wells containing only the query drug, and 3 wells without any drug. One query drug at a single concentration was added in all wells of the 384-well plate, except for the 3 control wells. All drugs were queried once per concentration, occasionally twice. We used 78 drugs as query in E. coli and S. Typhimurium, and 76 in P. aeruginosa. In total 79 query drugs were screened, out of which 75 were common for all three species (Supplementary Table 1). The 62 array drugs were a subset of the 79 query drugs. The same drug concentrations were used in both query and array drugs (Supplementary Table 1). Three drug concentrations (2-fold dilution series) were selected based on the MIC curves, tailored to the strain and drug. We targeted for nearly full, moderate, and mild/no growth inhibition –on average, corresponding to 50-100%, 25-50% and 0-25% of the MIC, respectively. The highest drug concentration and the lowest fitness obtained per single drug are listed in Supplementary Table 1. For drugs that do not inhibit growth on their own, we selected concentrations according to sensitivity of other strains/species or to their use in clinics or for research. E. coli and S. Typhimurium exhibited largely similar single drug dose responses within species, thus the same drug concentrations were used for both strains of each species. For P. aeruginosa, MICs often differed by several fold, thus drug concentrations were adjusted between the two strains (Supplementary Table 1).
Growth curves analysis
The Gompertz model was fitted to all growth curves (when growth was observed) by using the R package grofit version 1.1.1-1 for noise reduction. Quality of fit was assessed by Pearson correlation (R), which was > 0.95 for ~95% of all growth curves. R < 0.95 was indicative of either non-sigmoidal-shaped growth curves, typical of some drugs such as fosfomycin, or noisy data. In the former case, the original data was kept for further analysis. In the latter case, noisy data was removed from further analysis. Plate effects were corrected by fitting a polynomial to the median growth of each row and column. Background signal from LB was removed by subtracting the median curve of the non-growing wells from the same plate. These were wells in which either the single or the double drug treatments fully inhibited growth; each plate contained at least three such wells. Data was processed per strain and per batch to correct for systematic effects.
Fitness estimation
We used a single time-point OD595nm measurement (growth) for assessing fitness. This corresponded to the transition to stationary phase for cells grown without perturbation, as this allows us to capture the effect of drugs on lag-phase, growth rate or maximum growth. Thus, we used OD595nm at 8 hours for E. coli BW25113 and both P. aeruginosa strains, at 7 hours for the fast-growers E. coli iAi1 and S. Typhimurium 14028s, and at 9 hours for the slower growing S. Typhimurium LT2.
We used the Bliss model to assess interactions, as it can accommodate drugs that have no effect alone, but potentiate the activity of others (adjuvants) 36. This feature is especially relevant here, since we probed intrinsically antibiotic-resistant microbes (P. aeruginosa and MDR clinical isolates), and human-targeted drugs or food additives lacking antibacterial activity. According to the Bliss independence model 37 and assuming that drug-drug interactions are rare, for most drug combinations the fitness of arrayed drugs (fa) equals the fitness in the presence of both drugs (faq) divided by the fitness of the query drug alone (fq):
| (Eq. 1) |
| (Eq. 2) |
where ε denotes the Bliss score, f denotes fitness, g denotes growth, a denotes an arrayed drug, q denotes a query drug and 0 denotes no drug. The fitness in the presence of both drugs (faq) was calculated by dividing the growth in the presence of both drugs (gaq) by the median of the growth of drug-free wells from the same plate (g0). The fitness of the single query drugs (fq) was obtained by dividing the top 5% growing wells across each batch by the median of the growth of drug-free wells of each plate (g0). This metric is more robust to experimental errors than using only the 9 wells containing the query drug alone. Nevertheless, both estimators for fq yield very similar results (Pearson correlation = 0.98). In line with Eq. 2, the fitness of arrayed drugs (fa) was estimated by the slope of the line of best fit between gaq and gq across all plates (query drugs) within a batch:
| (Eq. 3) |
for given array drug m (am) across n query drugs q within a batch (ED Fig. 2b).
For array drugs with Pearson correlation between gaq and gq below 0.7, fa was estimated using only the query drugs corresponding to the interquartile range of gaq/gq (minimum n = 18 query drugs, ED Fig. 2b). Wells where r was still below 0.7, even after restricting the number of plates, were removed from further analysis due to high noise (~2%). For wells exhibiting no growth for > 75% of the plates within a batch fa was deemed as zero.
Interaction scores
Bliss independence
Bliss scores (ε) were calculated for each well as described above (Eq. 1). At least 3 x 3 drug concentrations x 2 (duplicates) x 2 (query and array drugs) = 36, or 18 (drugs used only as query) scores were obtained per drug pair. Drug-drug interactions were inferred based on the Bliss independence model in three steps: a) strong interactions based on complete ε distributions, b) strong interactions based on ε distributions restricted to relevant drug concentrations and c) weak and conserved interactions within species. Cross-species comparison, drug-drug interaction networks and monochromaticity analysis shown in this study include all drug-drug interactions.
a) Strong drug-drug interactions based on complete ε distributions
Strong drug-drug interactions were statistically assigned using a re-sampling approach. 10,000 repetitions of a two-sided Wilcoxon rank-sum test (per drug pair, per strain) were performed, in order to sample a representative set of ε for a given strain. For every repetition, the ε distribution of a given combination was compared to a ε distribution of the same size randomly sampled from the complete ε set for a given strain. P-values were calculated as follows:
| (Eq. 4) |
where N is the total number of repetitions (10,000) and pn is the p-value of the Wilcoxon rank-sum test obtained for the nth repetition. Strong drug-drug interactions were assigned to those drug pairs simultaneously satisfying two criteria: i) 1st or 3rd quartile of the ε distribution below -0.1 or higher than 0.1, for synergies or antagonisms respectively, and ii) p < 0.05 (after correcting for multiple testing, Benjamini-Hochberg). Only one-sided drug interactions were taken into account, thus those very few interactions satisfying the criteria concurrently for synergy and antagonism were re-assigned as neutral (only n=1 for > |0.1|). The highest absolute ε value between 1st and 3rd quartile was used as single interaction score () to reflect the strength of the drug-drug interactions.
b) Strong drug-drug interactions based on ε distributions restricted to relevant drug concentrations
Because drug interactions are concentration dependent, the same statistical procedure was repeated after restricting the drug concentration ratios to those relevant for either synergy or antagonism. This constraint was added by excluding ε values corresponding to concentration ratios where the expected fitness (product of the fitness on single drugs, fx*fy) was below 0.2 for synergy and above 0.8 for antagonism – blind spots for either interaction type (ED Fig. 3d). These interactions are described by their p-value and obtained with restricted drug concentration ratios. Although most interactions were detected based on both full and restricted ε distributions, each of the different methods uniquely identified interactions (ED Fig. 4c). With the expected fitness cutoff of 0.2, we identified the highest number of strong interactions (1950) with 90 uniquely identified interactions from full ε distributions and 379 from restricted (see also sensitivity analysis section in methods).
Restricting ε values based on expected fitness also allows defining whether synergy or antagonism is detectable for any given drug pair. No significant p-value was found for drug pairs with less than 5 ε scores within the relevant expected fitness space, as their sample size is insufficient. Synergy and antagonism could not be detected for 1% and 25% of all drug combinations, respectively.
c) Weak and conserved drug-drug interactions within species
For drug pairs with a strong drug-drug interaction in only one of the two strains per species, the criteria for assigning interactions for the second strain was relaxed to |second strain| > 0.06, provided that the interaction sign was the same. Interactions assigned with this approach are termed weak and conserved.
Loewe Additivity
For combinations between β-lactams for which high-resolution 8x8 checkerboards with sufficient growth inhibition was available in the validation dataset, Loewe additivity 38 was used to confirm the interactions. Drug-drug interactions were inferred by the shape of the isoboles (lines of equal growth) in two-dimensional drug concentration plots. Unless stated otherwise, all isoboles correspond to 50% growth inhibition (IC50) and were obtained by fitting a logistic model – with lines representing isoboles and dots IC50 interpolated concentrations. To interpolate IC50 concentrations (or other ICn%), a logistic model was used to fit the growth for each concentration of the first drug across different concentrations of the second drug. The null-hypothesis of this model is represented by the additivity line: a linear isobole connecting equal individual IC’s of the two drugs.
Sensitivity analysis
We confirmed the adequacy of the main statistical parameters used to assign interactions by performing a sensitivity analysis. Several expected fitness (fx*fy) cutoffs were tested, while keeping the other parameters constant (ED Fig. 4c). The added value of restricting the ε distributions to relevant drug concentrations (based on expected fitness) was strongly supported by the proportion of strong drug-drug interactions found exclusively using this criterion (~19% with our selected cutoff). The selected cutoff (0.2; disregarding wells with fx*fy < 0.2 for synergies and with fx*fy > 0.8 for antagonisms) resulted in the largest number of total interactions assigned, and the highest precision (91%) and recall (74%) after benchmarking against the validation dataset (ED Fig. 4c).
The suitability of the thresholds applied to define strong (| | > 0.1) and weak (| | > 0.06) interactions was assessed by their impact on the true and false positive rates (TPR and FPR respectively, ED Fig. 4d). A threshold of | | > 0.1 is beneficial, as it imposes a minimum strength to assign interactions. 0.1 corresponds to ~3 times the median of the 1st and 3rd quartiles across all ε distributions (ED Fig. 2c). Lowering this threshold results in lower TPR, because several drug pairs are reassigned to neutral due to ambiguity in calling interaction (we do not allow interactions to be both a synergy and an antagonism). Increasing this threshold lowers the TPR, because only very strong interactions will be assigned (ED Fig. 4d). Drug-drug interactions are highly conserved within species, exhibiting high correlation of observed for all species (Fig. 2a and ED Fig. 9a-b). This motivated us to relax the interaction strength threshold for the second strain if the interaction score | | was above 0.1 in the first strain, dubbing these interactions weak and conserved. Including weak and conserved interactions in our analysis increased the TPR by 15%. Adding a threshold for weak interactions of | | > 0.06 (~2 times the median of the 1st and 3rd quartiles of all distributions) is key for maintaining low FPR (ED Fig. 4d).
Benchmarking & clinical isolates checkerboard assays
8x8 checkerboard assays were performed for validating our screen (242 drug combinations - benchmarking dataset, Supplementary Table 3), as well as to test 7 selected synergies against 6 MDR clinical isolates (Fig. 4 & ED Fig. 11). As in the screen, growth was assessed based on OD595nm at the transition to stationary phase for the no drug controls. The timepoints used in the screen were used again for the validation set, whereas 8 hours were used for all E. coli and K. pneumoniae MDR isolates. Fitness was calculated by dividing OD595nm after single or double drug treatment by no drug treatment for each individual checkerboard. Bliss scores (ε) were calculated as before, resulting in 49 ε values per drug pair. Drug combinations were analyzed based on ε distributions, after removing wells in which one of the drugs alone and its subsequent combinations with the second drug completely inhibited growth. Antagonism was assigned when the median of the ε distribution was above 0.1 or the 3rd quartile was above 0.15. Similarly, synergies were assigned when the median of the ε distribution was below -0.1 or the 1st quartile was below -0.15. All experiments were done in biological duplicates, and interactions were considered effective when duplicates were consistent (vast majority of cases).
Assessing conservation of drug-drug interactions
Conservation of drug-drug interactions between strains of the same species was assessed by Pearson correlation of the interactions scores, . For potentially non-conserved drug-drug interactions, the expected fitness distributions of the two strains were taken into account. When the two distributions were significantly different according to a two-sided Wilcoxon rank-sum test (p-value < 0.05 after BH correction for multiple testing), the drug pairs were deemed as non-comparable between the two strains.
To assess the cross-species conservation of drug-drug interactions, we took into account only drug pairs that were probed in all three species. Drug-drug interactions were defined as being detected within a species, when detected in at least one of the two strains and no change of interaction sign was observed for the other strain. Interactions were then compared across the three species. Cases in which an interaction between drugs changed from synergy to antagonism or vice versa across species (conflicting interactions; ~7% of all interactions -Supplementary Table 2) were excluded from the comparative “across-species” Venn diagram (Fig. 2c). Note that with current analysis a given drug-drug interaction may be conserved across species, but not conserved within the species.
Conservation at the single drug level was defined based on shared resistance and sensitivity (Supplementary Table 1). A strain was considered sensitive to a given drug if one of the drug concentrations resulted in at least 30% growth inhibition. In line with conservation of drug-drug interactions across species, single drug responses are conserved across species when at least one strain of both species has the same sign (sensitive or resistant).
Monochromaticity index
The monochromaticity index (MI) between drug pairs was defined as in Szappanos et al. 39:
| (Eq. 5) |
where rij denotes the ratio of antagonism to all interactions between drugs from classes i and j, and b denotes the ratio of antagonism to all interactions. We set a minimum of 2 interactions between drugs from classes i and j in order to calculate the MI. MI equals 1 if only antagonisms occur between drugs from classes i and j, and -1 if only synergies occur. MI equals zero if the fraction of antagonism reflects the background ratio b. Both strong and weak drug interactions were taken into account across all species, in order to obtain one MI index per drug category pair.
Assessment of drug combinations in the Galleria mellonella infection model
Larvae of the greater wax moth (Galleria mellonella) at their final instar larval stage were used as an in vivo model to assess efficacy of drug combinations. Larvae were purchased from UK Waxworms (Sheffield, UK) and TZ-Terraristik (Cloppenburg, Germany). Stock solutions of vanillin (in 20% DMSO), spectinomycin, colistin and clarithromycin (20% DMSO/0.01% glacial acetic acid) were freshly prepared and diluted in PBS to the required concentration. Drugs and bacterial suspensions were administered by injection of 10 µL aliquots into the hemocoel via the last left (drugs) and right (antibiotic) proleg using Hamilton precision syringes. Controls included both uninfected larvae, and larvae which were injected into both last prolegs with the solvent used for the drugs. Drug toxicity was pre-evaluated by injection of serial dilutions of either single drugs or drug combination, and drugs were used at amounts that caused little/no toxicity. To identify an optimal inoculum, time-kill curves were generated by inoculating larvae with 10 µl of serial diluted bacterial suspensions (1x102 to 1x107 colony forming units [CFU]). For final experiments, groups of ten larvae were injected per strain/drug combination and placed into Petri dishes and incubated at 37 °C. Larvae were infected with a sublethal dose of 106 and 104 CFU for E. coli and K. pneumoniae isolates, respectively, and subsequently injected with indicated drugs, 1-hour post infection. Larvae survival was monitored at the indicated time points by two observers independently. Each strain/drug combination was evaluated in 4 independent experiments.
Cell viability assays and intracellular antibiotic concentration
Ciprofloxacin
Overnight cultures of E. coli BW25113 were diluted 1:1,000 into 50 ml LB and grown at 37°C to OD595nm ~0.5. Paraquat (50 µg/ml), Vanillin (150 µg/ml), Benzalkonium (5 µg/ml), Caffeine (200 µg/ml), Doxycycline (0.5 µg/ml), Rifampicin (5 µg/ml), Trimethoprim (5 µg/ml) or Curcumin (100 µg/ml), were added to the cultures and incubated at 37°C for 30 minutes prior to the addition of 2.5 µg/ml final concentration ciprofloxacin. The cultures were incubated at 37°C for 1 hour in the presence of both drugs. Cell viability was determined by counting CFUs after 16 hours incubation of washed cell pellets plated onto LB agar petri dishes. Intracellular ciprofloxacin was quantified using liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS), as previously described 40,41. Non-washed cell pellets 42 were directly frozen and lysed with 350 µl of acetonitrile, followed by three freeze-thaw cycles (thawing was performed in an ultrasonic bath for 5 min). Cell debris was pelleted at 16,000 g and the supernatant was filtered through a 0.22 µm syringe filter prior to injection. Chromatographic separation was achieved on a Waters BEH C18 column (2.1 × 50 mm; 1.7 µm) at 40 °C, with a 2 min gradient with flow rate of 0.5 mL/min: (i) 0–0.5 min, 1% mobile phase B; (ii) 0.5–1.2 min, linear gradient from 1 to 95% mobile phase B; (iii) 1.2–1.6 min, 95% mobile phase B; and (iv) 1.6–1.7 min, return to initial conditions (mobile phase A consisted of 0.1% formic acid in water, and mobile phase B consisted of 0.1% formic acid in acetonitrile). Samples were kept at 4 °C until analysis. Sample injection volume was 5 µL. Detection of ciprofloxacin was performed on a Waters Q-Tof premier instrument with electrospray ionization in positive mode. The transition 332>314 was monitored, with cone voltage set at 8 and collision energy set at 20. Intracellular ciprofloxacin was normalized to CFU at the time of ciprofloxacin addition.
Gentamicin
Intracellular gentamicin was quantified by measuring [3H]-gentamicin (1 mCi/ml; Hartmann Analytic Corp.), as previously described 7. Overnight cultures of E. coli MG1655 (the parental strain of BW25113) were diluted 1:100 into 5 ml LB and grown to OD595nm ~0.1. [3H]-gentamicin was diluted in cold gentamicin to obtain a 5 mg/ml (0,1 mCi/ml) stock solution, which was then added to the culture at a final concentration of 5 μg/ml (0,1 μCi/ml) together with the second drug: Berberine (200 µg/ml), Erythromycin (15 µg/ml), Metformin (13000 µg/ml), Procaine (6000 µg/ml), Loperamide (400 µg/ml), Benzalkonium (5 µg/ml), Rifampicin (5 µg/ml) or Clindamycin (200 µg/ml). Cultures were then incubated at 37°C on a rotary shaker. At 0, 0.5, 1, 1.5 and 2h time-points, 500 μl aliquots were removed and applied to a 0.45 μm-pore-size HAWP membrane filter (Millipore) pretreated with 1 ml of unlabeled gentamicin (250 μg/ml). Filters were washed with 10 ml of 1.5% NaCl, placed into counting vials, and dried for 30 min at 52°C. 8 ml of liquid scintillation were then added to the dried filters and vials were incubated overnight at room temperature before being counted for 5 min. Gentamicin uptake efficiency is expressed as total accumulation of gentamicin (ng) per 108 cells, and plotted here for the final time point (2h). Cell viability was determined by CFUs.
Spectinomycin
Intracellular spectinomycin was quantified by measuring [3H]-spectinomycin (1 μCi/mg; Hartmann Analytic Corp.). Overnight cultures of E. coli BW25113 were diluted 1:1,000 into 1 ml LB with and without vanillin (150 µg/ml) and grown to OD595nm ~0.5. 50 µg/ml [3H]-spectinomycin:spectinomycin 1:100 was added and the cultures were incubated for 1 h. Cultures were pelleted, washed twice with PBS with 50 μg/ml non-labeled spectinomycin, re-suspended in 1% SDS and incubated for 20 min at 85°C. The lysate was mixed with 8 ml liquid scintillation (Perkin Elmer ULTIMA Gold) and counted for 1 min using a Perkin Elmer Tri-Carb 2800TR. Measured radioactivity was normalized to cell number as measured by OD595nm.
RNA isolation, cDNA preparation and Quantitative RT-PCR
Overnight cultures of E. coli BW25113 and the marR deletion mutant (ΔmarR) were diluted 1:2,000 into 20 ml LB and grown at 37°C to OD595nm ~0.2. Aspirin or vanillin were added to the cultures to 500 and 150 µg/ml final concentration, respectively (DMSO was added in the control), followed by a 30 min incubation period at 37°C with agitation. Cells were harvested and RNA was extracted using the RNeasy Protect Bacteria Mini Kit (Qiagen). cDNA was prepared for qRT-PCR using SuperScript™ III Reverse Transcriptase (Thermo Fisher Scientific). marA expression levels were estimated by quantitative RT-PCR using SYBR™ Green PCR master mix following the manufacturer's instructions (Thermo Fisher Scientific). Primer sequences for marA and recA were previously described 29. All experiments were conducted in at least three biological replicates, and relative expression levels were estimated according to Livak et al. 43, using recA expression as reference.
Immunoblot analysis for protein quantification
Overnight cultures of E. coli BW25113 and the marA deletion mutant (ΔmarA) were diluted 1:1,000 into 50 ml LB containing 500 μg/ml aspirin, 150 μg/ml vanillin or DMSO (solvent control), followed by growth with agitation at 37°C to OD595nm ~0.5. Cells were washed in PBS containing corresponding drugs or DMSO, then resuspended to match OD595nm = 1. Cell pellets were resuspended in Laemmli buffer and heated to 95°C for 3 minutes followed by immunoblot analysis with α-AcrA polyclonal antiserum (gift from K.M. Pos) at 1:200,000 dilution. Primary antiserum was detected using anti-rabbit HRP (A0545 Sigma) at 1:5,000 dilution. Cell loading was controlled with the anti-RecA antibody (rabbit, ab63797 Abcam). For densitometry analysis, pixel intensity of AcrA bands from cell density normalised samples were quantified using ImageJ. At least four different biological replicates were blotted and summarized by their mean and standard deviation. Each biological replicate was run and blotted twice (technical replicates). Relative AcrA levels per biological replicate correspond to the average intensities of the technical replicates. All blots can be seen in Supplementary Fig. 1.
Screening the E. coli Keio Knockout Collection for identifying MoA of drug interactions
The E. coli Keio Knockout Collection 26 (two independent clones per mutant) was arrayed in 1536-format in LB agar plates using a Rotor HDA (Singer Instruments) as previously described 29. The growth of each mutant was estimated by colony opacity 44 after 13 hours incubation at 37°C in the absence and presence of vanillin (200 µg/ml), spectinomycin (4 µg/ml), and their combination. All plates were imaged under controlled lighting conditions (spImager S&P Robotics) using an 18-megapixel Canon Rebel T3i (Canon). Experiments were done in biological triplicates. Fitness of each mutant was calculating by dividing the growth in condition (vanillin, spectinomycin or both) by the growth in LB, after correcting for outer-frame plate effects 44. Bliss scores were calculated as per Eq. 1 per replicate and then averaged (Supplementary Table 7)
Extended Data
Extended Data Figure 1. High-throughput profiling of pairwise drug combinations in Gram-negative bacteria.
a) Drug and species selection for screen. The 79 drugs used in the combinatorial screen are grouped to categories (Supplementary Table 1). Antibacterials are grouped by target with the exception of antibiotic classes for which enough representatives were screened (>2) to form a separate category: β-lactams, macrolides, tetracyclines, fluoroquinolones and aminoglycosides. Classification of human-targeted drugs and food additives is not further refined, because the MoA is unclear for most. A subset of 62 arrayed drugs was profiled against the complete set of 79 drugs in 6 strains. Strains are color coded according to species. Strain colors and abbreviations are used in all main and ED figures. b) Quantification of drug-drug interactions. Growth was profiled by measuring optical density (OD595nm) over time in the presence of no, single and both drugs. Interactions were defined according to Bliss independence. Significantly lower or higher fitness than expectation (fx*fy) indicates synergy or antagonism, respectively. Synergy and antagonism were assessed by growth in 4x4 checkerboards (Methods).
Extended Data Figure 2. Data analysis pipeline.
a) Flowchart of the data analysis pipeline. b) Estimating single drug fitness of arrayed drugs. As drug-drug interactions are rare, the slope of the line of best fit between gaq (growth with double drug) and gq (growth with query drug alone – deduced from average of the top 5% growing wells across plates within a batch) across np query drugs (plates) corresponds to a proxy of the fitness of the arrayed drug alone, fa (Methods, Eq 3). R denotes the Pearson correlation coefficient between gaq and gq across np plates. Well A9 from E. coli BW25113 containing 3µg/ml spectinomycin is shown as an example of arrayed drugs with several interactions; several query drugs (plates) deviate from the expected fitness (light grey points), therefore only half of the plates corresponding to the interquartile range of gag/gq were used to estimate fa. c) Density distributions of quartiles 1, 2 and 3 of Bliss scores (ε) distributions for E. coli. Q1, Q2 and Q3 denote the median of quartiles 1, 2 and 3 of ε distributions, respectively. n denotes the number of drug combinations used.
Extended Data Figure 3. Data quality control.
a) High replicate correlation for single and double drug treatments. Transparent boxplots contain Pearson correlation coefficients between plates of the same batch containing arrayed drugs only (LB was used instead of the second drug). n represents the total number of correlations. Full boxplots contain Pearson correlation coefficients between double drug replicate wells within the same plate, across all plates. n represents the number of wells used for correlation, nmax = (62 drugs + 1 LB) x 3 concentrations = 189. Only wells with median growth above 0.1 were taken into account for this correlation analysis (see panel b). For all box plots the center line, limits, whiskers and points correspond to the median, upper and lower quartiles, 1.5x IQR and outliers, respectively. b) Wells with lower median growth have lower replicate correlation. The double drug correlation coefficients used to generate the boxplot from a are plotted as a function of the median growth of all wells across all plates for E. coli iAi1. Wells with overall lower growth (due to strong inhibition of arrayed drug) are less reproducible due to a combination of the lower spread of growth values and the sigmoidal nature of the drug dose response curves. c) Drug-drug interactions are rare. Density distributions of all Bliss scores (ε) obtained per strain. d) The ability to detect synergies and antagonisms depends on the effects of single drug treatments. Bliss scores (ε) are plotted as function of expected fitness (fx*fy) for all drug concentration ratios for all combinations in E. coli BW (example). Boxplots summarizing both variables are shown besides the axes (n=99,907 Bliss scores, center line, limits, whiskers and points correspond to the median, upper and lower quartiles, 1.5x IQR and outliers, respectively). Blind spots for detecting antagonism and synergy are indicated; they are both based on the expected fitness (see also ED Fig. 4c-d) and thus dependent on the growth of the strain with the single drugs. The number of drug combinations falling in the blind spot for antagonism is larger, due to the number of drugs used in the screen that do not inhibit E. coli on their own. e) Scatter plot of number of interactions per drug versus the minimum fitness of the drug alone (as obtained in screen, Supplementary Table 1). Strong and weak interactions are represented. n denotes the total number of interactions and R is the Pearson correlation coefficient. Strains are color coded as above. f) Density distributions of the number of interactions per drug for all strains.
Extended Data Figure 4. Benchmarking & sensitivity analysis.
a) Validation set is enriched in synergies and antagonisms to assess better true and false positives. Comparison of the interaction fractions between the screen and validation set. Both strong and weak interactions (Fig. 2b) are accounted for the screen tally. b) Number of benchmarked interactions per strain. c & d) Sensitivity analysis of the statistical thresholds for calling interactions. c) Total amount of interactions as function of the expected fitness (fx*fy) cutoff used for restricting the ε distributions to relevant drug concentrations. Strong drug-drug interactions are classified according to the ε distribution where they were significant: complete distribution only (i.e. all expected fitness wells), relevant wells only (i.e. all wells with fx*fy > cutoff for synergies and all wells with fx*fy < (1-cutoff) for antagonisms), or in both. Weak drug-drug interactions are independently assigned and represented in white. We selected an expected fitness cutoff of 0.2, as it resulted in the largest number of total interactions detected, with the highest precision and recall (91 and 74% respectively) after benchmarking against the validation dataset. d) Receiver operating characteristic (ROC) curve for the screen across different p-value thresholds (two-sided permutation test of Wilcoxon rank-sum) as a unique criterion for assigning interactions. The selected p-value (0.05) for screen threshold is indicated by a grey cross. Sensitivity to additional parameters for calling hits is shown: allowing interactions to be either antagonisms or synergies but not both (1-sided); strong and weak interaction thresholds. True and false positive rates were estimated based on the validation dataset. Precision and recall for the final and best performing set of parameters are shown: one-sided interactions, p < 0.05, fx*fy cutoff = 0.2 and |ε|>0.1 for strong interactions, |ε| > 0.06 for weak interactions. TP, TN, FP and FN stand for True Positives, True Negatives, False Positives and False Negatives, respectively. n indicates the total number of benchmarked drug combinations (Supplementary Table 3). e) Synergies between β-lactams according to Loewe additivity interaction model. The results of 8x8 checkerboards for 3 combinations between β-lactams in 4 strains are shown. The grey line in each plot represents null hypothesis in the Loewe additivity model, whereas the black line corresponds to the IC50 isobole, estimated by fitting a logistic curve to the interpolated drug concentrations (colored dots, Methods). Piperacillin did not reach 50% growth inhibition in E. coli, thus IC20 and IC40 isoboles were used for the amoxicillin + piperacillin combination in E. coli BW and E. coli iAi1, respectively.
Extended Data Figure 5. Benchmarking of non-comparable drug-drug interactions.
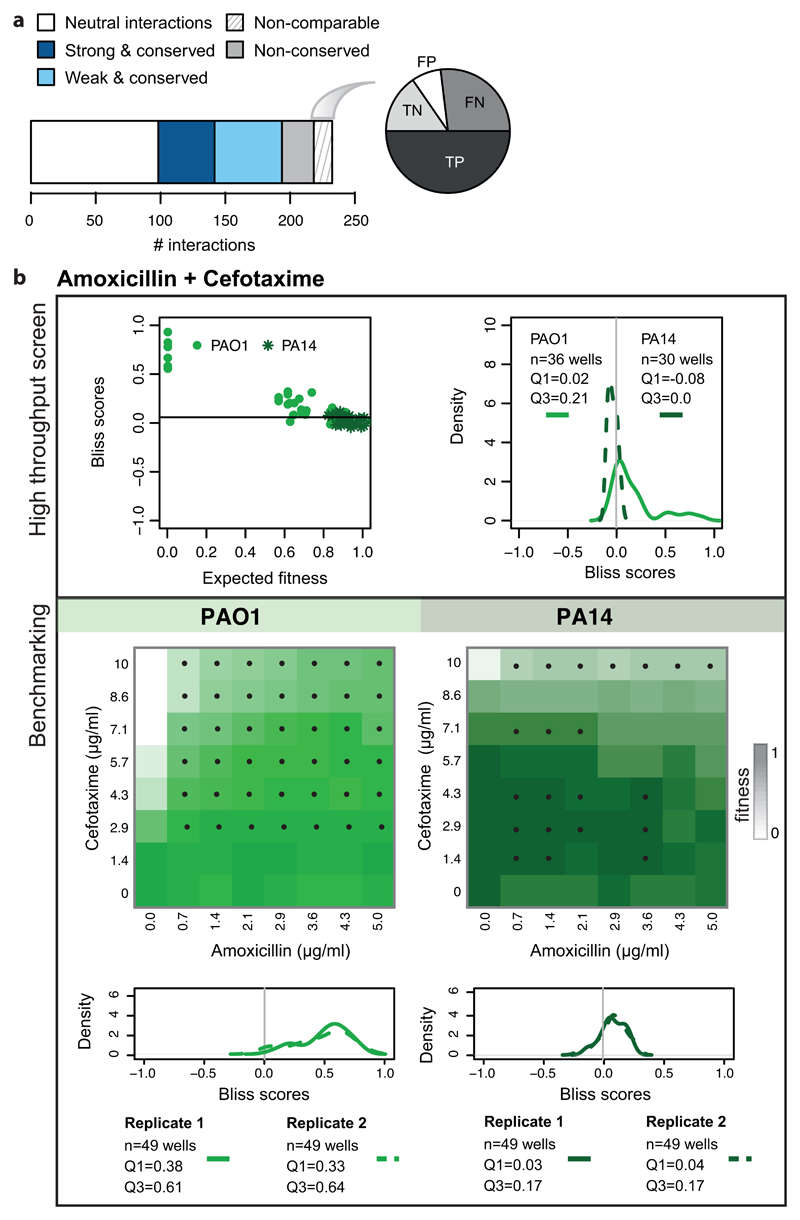
a) The barplot illustrates the division of benchmarked drug combinations according to their degree of conservation within species. The pie chart shows the proportion of False & True Positive (FP & TP) and False and True Negatives (FN & TN) within non-comparable drug-drug interactions. b) Combination of amoxicillin with cefotaxime in P. aeruginosa: an example of a non-comparable drug-drug interaction. The results of the screen are presented on the upper box. Bliss scores as function of expected fitness for both strains are presented on the left hand side, while a density distribution of the Bliss scores is shown on the right hand side. n denotes the total number of Bliss scores, Q1 and Q3 indicate the Bliss score for quartiles 1 and 3, respectively. Antagonism was detected only for PAO1 (Q3 > 0.1). PA14 was resistant to both drugs at concentrations screened (upper left panel), rendering the detection of antagonism impossible. The benchmarking results indicate that interaction is antagonistic in both strains (lower box), albeit weaker at PA14 and visible mostly at higher concentrations. Color on checkerboard reflects fitness and black dots correspond to drug-ratios where the Bliss score is above 0.1.
Extended Data Figure 6. Benchmarking of weak conserved drug-drug interactions.
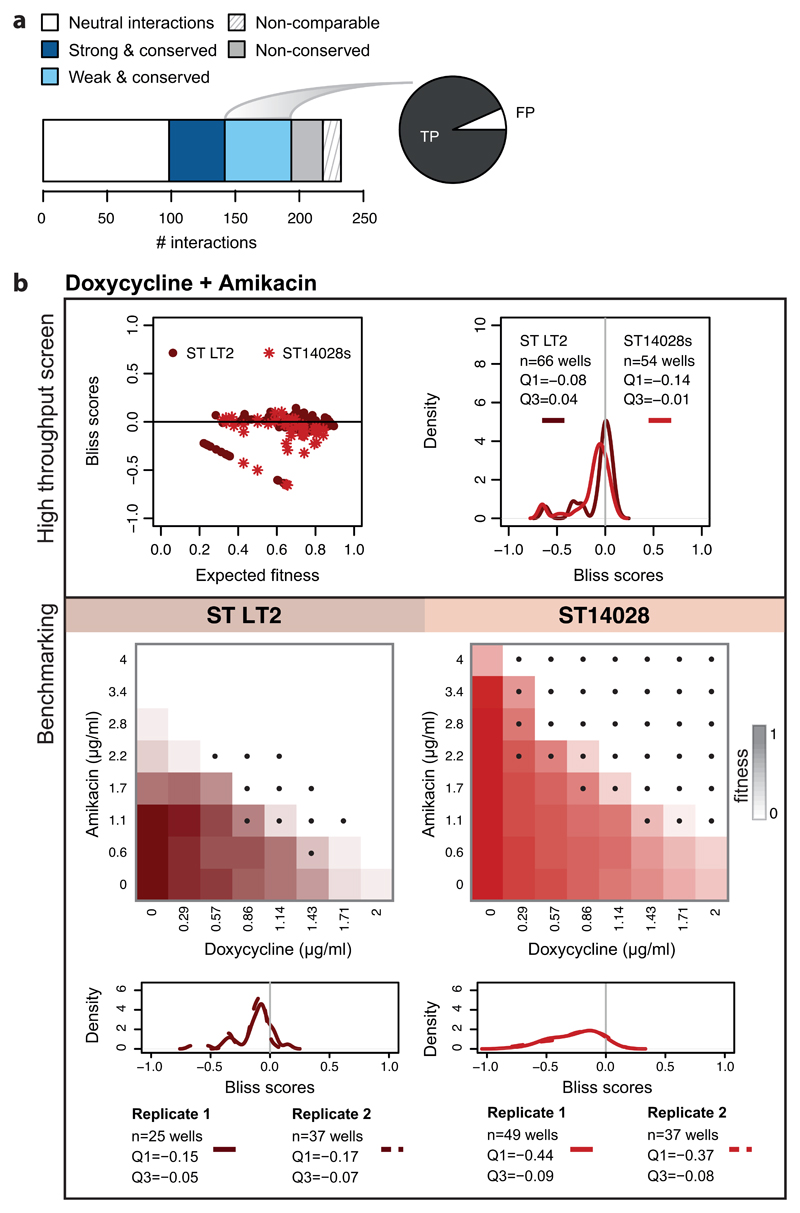
a) The barplot illustrates the division of benchmarked drug combinations as in ED Fig. 5a. The pie chart shows the proportion False Positives (FP) and True Positives (TP) within weak conserved interactions. b) Combination of doxycycline with amikacin in S. Typhimurium: an example of a weak conserved drug-drug interaction. The results of the screen are presented on the upper box. Bliss scores as function of expected fitness for both strains are presented on the left hand side, while a density distribution of the Bliss scores is shown on the right hand side. n denotes the total number of Bliss scores, Q1 and Q3 indicate the Bliss score for quartiles 1 and 3, respectively. A strong synergy was detected only for ST14028 (Q1 < -0.1), and then a weak conserved synergy was assigned afterwards to ST LT2 (Q1 < -0.06). The benchmarking results, presented on the box below, confirm that the interaction is synergistic in both strains. Color on checkerboard reflects fitness and black dots correspond to drug-ratios where the Bliss score is below -0.1.
Extended Data Figure 7. Salmonella and Pseudomonas drug-drug interaction networks.
a & b) Drug category interaction networks. Nodes represent drug categories according to ED Fig. 1a, and plotted as in Fig. 1b. Conserved interactions, including weak conserved, are shown here. One of the most well-known and broadly used synergies is that of aminoglycosides and β-lactams 45. Consistent with its use against P. aeruginosa in clinics, we detected multiple strong synergies between specific members of the two antibiotic classes in P. aeruginosa, but fewer interactions in the other two species. c & d) Drug-drug interactions across cellular processes. Representation as in a & b, but drug categories targeting the same general cellular process are grouped here. e) Quantification of synergy and antagonism in the networks from a & b, and the corresponding Chi-squared test p-value. As in E. coli, antagonism occurs more frequently than synergy and almost exclusively between drugs belonging to different categories in S. Typhimurium and P. aeruginosa. In P. aeruginosa, there are very few interactions occurring between drugs of the same category.
Extended Data Figure 8. Drug antagonisms are often due to decrease in intracellular drug concentrations.
a) Cartoon of possible MoAs for drug-drug interactions that function via modulation of the intracellular drug concentration. A drug (antagonist; blue) inhibits the uptake or promotes the efflux of another one (black), and thus decreases its intracellular concentration. b) Different antagonists (see methods for concentrations) of gentamicin (red – 5 µg/ml) and ciprofloxacin (gold – 2.5 µg/ml) identified in our screen for E. coli BW also rescue the killing effect of the two bactericidal drugs in the same strain or its parental MG1655 (top right and top left panel, respectively). With the exception of clindamycin (for gentamicin) and curcumin (for ciprofloxacin) all other antagonists decrease the intracellular concentration of their interacting drug (bottom panels) – gentamicin detected by using radiolabeled compound and ciprofloxacin with LC-MS/MS (Methods). The degree of rescue (upper panel) in many cases follows the decrease of intracellular concentration (lower panel), implying that most of these interactions depend at least partially on modulating the intracellular concentration of the antagonized drug. c) Antagonisms are resolved in E. coli BW mutants lacking key components controlling the intracellular concentration of the antagonized drug. Aminoglycosides depend on PMF-energized uptake and thus respiratory complexes 7,46; ciprofloxacin is effluxed by AcrAB-TolC 29,47. For gentamicin, most interactions are resolved when respiration is defected, even the one with clindamycin (not modulating intracellular gentamicin concentration- panel b) presumably because MoA and import of aminoglycosides are linked in a positive feedback loop 7,48. For ciprofloxacin, antagonisms with paraquat and caffeine are resolved in the ΔacrA mutant, implying that both compounds induce the AcrAB-TolC pump (known for paraquat). In contrast, interactions with curcumin, benzalkonium and doxycycline remain largely intact in the ΔacrA mutant. The first interaction is expected as curcumin does not modulate intracellular ciprofloxacin concentration (see panel b). In the other two cases, other component(s) besides AcrAB-TolC may be responsible for the altered ciprofloxacin import/export; for example, ciprofloxacin uses OmpF to enter the cell 49. Ciprofloxacin and gentamicin concentrations were adjusted in all strains according to MIC (70% and 100% MIC for ciprofloxacin and gentamicin, respectively; all drug concentrations are listed in Supplementary Table 6). Bliss interaction scores (ε) were calculated as in screen. Barplots and error bars in c & d represent the average and standard deviation, respectively, across n independent biological replicates. d) Gentamicin and ciprofloxacin antagonism networks for E. coli BW. Nodes represent drugs colored according to targeted cellular process (as ED Fig. 1a). Full and dashed edges represent antagonistic drug-drug interactions for which intracellular antibiotic concentration was and was not measured, respectively. Drug interactions that result in decreased intracellular concentration of the antagonized drug are represented by black edges. e) Quantification of antagonistic drug-drug interactions from the networks in (d). The bars for fluoroquinolones and aminoglycosides account for an extrapolation of antagonistic interactions to all other members of the two classes, assuming they behave the same as ciprofloxacin and gentamicin, respectively.
Extended Data Figure 9. Drug-drug interactions are largely conserved within species and only partially MoA-driven.
a & b) Drug-drug interactions are conserved in S. Typhimurium (a) and P. aeruginosa (b). Scatter plot of interaction scores in the two strains of each species; only significant interactions for at least one strain are shown. Colors and grouping as in Fig. 2a. R denotes the Pearson correlation and n the total number interactions plotted. Lower correlation in P. aeruginosa is presumably due to fewer and weaker interactions in total. c) Drug interaction profiles are phylogeny-driven. Clustering of strains based on Pearson correlation of their drug interaction profiles (taking into account all pairwise drug combinations; n=2759-2883, depending on the species). Strains of the same species cluster together, with the two enterobacterial species, E. coli and S. Typhimurium, behaving more similar to each other than to the phylogenetically more distant P. aeruginosa. d) Conserved drug-drug interaction network. Nodes represent individual drugs grouped and colored by targeted cellular process (as in ED Fig. 1a). Drug names are represented by 3 letter codes (Supplementary Table 1). Dashed and full edges correspond to conserved interactions between two or three species, respectively. Many of the human-targeted drugs, such as loperamide, verapamil and procaine exhibit a general potentiating effect, similar to that of membrane-targeting drugs. This suggests that they may also facilitate drug uptake or impair efflux, consistent with previous reports on the role of loperamide in E. coli and verapamil in Mycobacterium tuberculosis 4,50. e) Monochromaticity between all drug categories. The monochromaticity index (MI) reflects whether interactions between drugs of two categories are more synergistic (MI=-1) or antagonistic (MI=1) than the background proportion of synergy and antagonism. MI equals zero when interactions between two drug categories have the same proportion of synergy and antagonism as all interactions together. (Methods). MI was calculated using all interactions from the 6 strains for all category pairs that had at least 2 interactions. White cells in the heat map correspond to category pairs for which no (or an insufficient number of) interactions were observed. f) Human-targeted drugs, and LPS or PMF inhibitors are strong and promiscuous adjuvants. Density distributions of the MIs per drug category from panel e are shown. n denotes the amount of drugs in category involved in i interactions.
Extended Data Figure 10. Hierarchical clustering of drugs according to their interaction profiles.
Rows depict the 75 drugs common to all strains (colored according to drug category – ED Fig. 1a), and columns account for their interactions with other drugs in all six strains tested. Clustering was done using the median of the ε distributions, uncentered correlation and average linkage.
Extended Data Figure 11. Active synergies against Gram-negative MDR clinical isolates in vitro and in G. mellonella infection model.
Both human-targeted drugs (lately found to have an extended impact on bacteria 51) and food additives can promote the action of antibiotics in MDR strains, indicating that their use as antibacterial adjuvants should be explored further in the future. a) Drug combinations active against MDR E. coli and K. pneumoniae clinical isolates (related to Fig. 4). Interactions are shown as 8x8 checkerboards and synergies have a black bold border. Drug pairs are the same per line and indicated at the first checkerboard. The species in which the interaction was detected in the screen are indicated after the last checkerboard. Concentrations increase on equal steps per drug (see legend); only minimal and maximal concentrations are shown for the first strain of each species. Apart from colistin, the same concentration ranges were used for all E. coli and K. pneumoniae MDR strains. One of two replicates is shown. b) Drug synergies against the same MDR strains in the Galleria mellonella infection model. Larvae were infected by E. coli and K. pneumoniae MDR isolates (106 and 104 CFU, respectively) and left untreated, or treated with single drugs or in combination. % larvae survival was monitored at indicated intervals after infection – n=10 larvae per treatment. The averages of 4 biological replicates are plotted; error bars depict standard deviation.
Extended Data Figure 12. Mode of Action for the vanillin-spectinomycin synergy.
a) Spectinomycin MIC decreases upon addition of 100 µg/ml vanillin in the wildtype E. coli BW, as well as single-gene knockouts of members of the AcrAB-TolC efflux pump or its MarA regulator. Thus, the vanillin-spectinomycin synergy is independent of the effect of vanillin on AcrAB-TolC (Fig. 3). b) Synergy is specific to vanillin-spectinomycin, as spectinomycin is antagonized by 500 µg/ml of the vanillin-related compound, aspirin, thereby increasing the MIC ~3-fold. c) Profiling the vanillin-spectinomycin combination in the E. coli BW Keio collection 26 to deconvolute its MoA. Violin plots of the drug-drug interaction scores ε of all mutants (n=9216; Methods) are presented for the vanillin-spectinomycin combination (synergy) and as control, for the combination of vanillin with another aminoglycoside, amikacin (antagonism). The interaction scores of the two mdfA deletion clones present in the Keio library are indicated by red dots. The vanillin-spectinomycin synergy is lost in the absence of mdfA, whereas the vanillin-amikacin antagonism remains unaffected, indicating that the vanillin-spectinomycin synergy depends specifically on MdfA. d) Deletion of mdfA leads to increased spectinomycin MIC and abolishes the synergy with vanillin, independent of the presence or absence of AcrAB-TolC. Mild overexpression of mdfA from a plasmid (pmdfA - methods) further enhances the synergy with vanillin, decreasing the spectinomycin MIC by ~2-fold (compared to the MIC of the combination in the wildtype). Thus, MdfA levels are directly correlated to the degree of the spectinomycin-vanillin synergy. e) Overexpression of mdfA leads to increased spectinomycin sensitivity, even though MIC does not change. The growth of E. coli BW and pmdfA was measured (OD595nm after 8h) over 2-fold serial dilutions of spectinomycin and normalized to the no-drug growth of the corresponding strain (white and black dots represent the average of n=3 independent biological replicates, error bars represent standard deviation). Spectinomycin dose response was computed using a logistic fit of the averaged data points (MICs are calculated by fitting individual replicates first and then averaging). Fitted curves are represented by full and dashed lines for pmdfA and E. coli BW respectively. f) Vanillin leads to accumulation of spectinomycin in the cell in an mdfA-dependent manner. Intracellular spectinomycin is measured with the tritiated compound (Methods). Barplots and error bars in a, b, d & f represent the average and standard deviation, respectively, across n independent biological replicates.
Supplementary Material
Acknowledgements
We thank Pedro Beltrao (EBI) and Tobias Bollenbach (University of Cologne) for providing feedback on manuscript. We thank Klaas Martinus Pos (Goethe University, Frankfurt) for the α-AcrA antibody; Dominic Helm and the EMBL Proteomics Core Facility for help with MS experiments; the EMBL GBCS and the Centre for Statistical Analysis for advice on data analysis; Sara Riedel-Christ for help with Galleria mellonella experiments; and the Typas lab members for discussions. This work was partially supported by EMBL internal funding, the Sofja Kovalevskaja Award of the Alexander von Humboldt Foundation to ATy, the JPIAMR Combinatorials grant to FB (ANR) and ATy (BMBF), and the DFG (FOR 2251) to SG. AM and JS are supported by a fellowship from the EMBL Interdisciplinary Postdoc (EIPOD) program under Marie Curie Actions COFUND.
Footnotes
Data availability statement
All data supporting the findings of this study are included in this article as supplementary files.
Code availability
The code used for data analysis is available from https://github.com/AnaRitaBrochado/DrugInteractionsPipeline.
Author contributions
ARB and ATy conceived and designed the study. ARB, ATe and JB performed the screen; ARB, ATe and NN the validation screen; and ARB, MB, AM, JS, SB, MZ and JZB the mechanistic follow-up work. SG characterized the clinical isolates. ARB, ATe and SF performed the clinical isolate checkerboards, and EH and SG the G. mellonella infection experiments. ARB analyzed all data. BP, FB, SG and ATy supervised different parts of this study; BE, MS and PB provided advice. ARB and ATy wrote the paper with input from MS, PB and SG. All authors approved the final version.
References
- 1.Draft political declaration of the high-level meeting of the United Nations General Assembly on antimicrobial resistance. 2016 ( http://www.un.org/pga/71/2016/09/21/press-release-hl-meeting-on-antimicrobial-resistance/)
- 2.Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 3.Tacconelli E, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 4.Ejim L, et al. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat Chem Biol. 2011;7:348–350. doi: 10.1038/nchembio.559. [DOI] [PubMed] [Google Scholar]
- 5.Brown D. Antibiotic resistance breakers: can repurposed drugs fill the antibiotic discovery void? Nat Rev Drug Discov. 2015;14:821–832. doi: 10.1038/nrd4675. [DOI] [PubMed] [Google Scholar]
- 6.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 7.Ezraty B, et al. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science. 2013;340:1583–1587. doi: 10.1126/science.1238328. [DOI] [PubMed] [Google Scholar]
- 8.Yeh P, Tschumi AI, Kishony R. Functional classification of drugs by properties of their pairwise interactions. Nat Genet. 2006;38:489–494. doi: 10.1038/ng1755. ng1755 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Robbins N, et al. An Antifungal Combination Matrix Identifies a Rich Pool of Adjuvant Molecules that Enhance Drug Activity against Diverse Fungal Pathogens. Cell Rep. 2015;13:1481–1492. doi: 10.1016/j.celrep.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cokol M, et al. Large-scale identification and analysis of suppressive drug interactions. Chem Biol. 2014;21:541–551. doi: 10.1016/j.chembiol.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bollenbach T, Quan S, Chait R, Kishony R. Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell. 2009;139:707–718. doi: 10.1016/j.cell.2009.10.025. S0092-8674(09)01315-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costanzo M, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353 doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farha MA, Brown ED. Chemical probes of Escherichia coli uncovered through chemical-chemical interaction profiling with compounds of known biological activity. Chem Biol. 2010;17:852–862. doi: 10.1016/j.chembiol.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Stokes JM, et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.28. 17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chevereau G, Bollenbach T. Systematic discovery of drug interaction mechanisms. Mol Syst Biol. 2015;11:807. doi: 10.15252/msb.20156098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan CJ, et al. Hierarchical modularity and the evolution of genetic interactomes across species. Mol Cell. 2012;46:691–704. doi: 10.1016/j.molcel.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaara M. Outer membrane permeability barrier to azithromycin, clarithromycin, and roxithromycin in gram-negative enteric bacteria. Antimicrob Agents Chemother. 1993;37:354–356. doi: 10.1128/aac.37.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giamarellou H, Zissis NP, Tagari G, Bouzos J. In vitro synergistic activities of aminoglycosides and new beta-lactams against multiresistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1984;25:534–536. doi: 10.1128/aac.25.4.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamura Y, et al. Azithromycin exhibits bactericidal effects on Pseudomonas aeruginosa through interaction with the outer membrane. Antimicrob Agents Chemother. 2005;49:1377–1380. doi: 10.1128/AAC.49.4.1377-1380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petropoulos AD, et al. Time-resolved binding of azithromycin to Escherichia coli ribosomes. J Mol Biol. 2009;385:1179–1192. doi: 10.1016/j.jmb.2008.11.042. [DOI] [PubMed] [Google Scholar]
- 21.Hao Z, et al. The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli. Nat Chem Biol. 2014;10:21–28. doi: 10.1038/nchembio.1380. [DOI] [PubMed] [Google Scholar]
- 22.Chubiz LM, Glekas GD, Rao CV. Transcriptional cross talk within the mar-sox-rob regulon in Escherichia coli is limited to the rob and marRAB operons. J Bacteriol. 2012;194:4867–4875. doi: 10.1128/JB.00680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Göttig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-beta-lactamase with increased carbapenemase activity. J Antimicrob Chemother. 2013;68:1737–1740. doi: 10.1093/jac/dkt088. [DOI] [PubMed] [Google Scholar]
- 24.Göttig S, Gruber TM, Stecher B, Wichelhaus TA, Kempf VA. In vivo horizontal gene transfer of the carbapenemase OXA-48 during a nosocomial outbreak. Clin Infect Dis. 2015;60:1808–1815. doi: 10.1093/cid/civ191. [DOI] [PubMed] [Google Scholar]
- 25.MacNair CR, et al. Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat Commun. 2018;9:458. doi: 10.1038/s41467-018-02875-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yardeni EH, Zomot E, Bibi E. The fascinating but mysterious mechanistic aspects of multidrug transport by MdfA from Escherichia coli. Res Microbiol. 2017 doi: 10.1016/j.resmic.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Bohn C, Bouloc P. The Escherichia coli cmlA gene encodes the multidrug efflux pump Cmr/MdfA and is responsible for isopropyl-beta-D-thiogalactopyranoside exclusion and spectinomycin sensitivity. J Bacteriol. 1998;180:6072–6075. doi: 10.1128/jb.180.22.6072-6075.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nichols RJ, et al. Phenotypic Landscape of a Bacterial Cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wildenhain J, et al. Prediction of Synergism from Chemical-Genetic Interactions by Machine Learning. Cell Systems. 2015;1:383–395. doi: 10.1016/j.cels.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandrasekaran S, et al. Chemogenomics and orthology-based design of antibiotic combination therapies. Mol Syst Biol. 2016;12:872. doi: 10.15252/msb.20156777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehar J, et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat Biotechnol. 2009;27:659–666. doi: 10.1038/nbt.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Nat Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saka K, et al. A complete set of Escherichia coli open reading frames in mobile plasmids facilitating genetic studies. DNA Res. 2005;12:63–68. doi: 10.1093/dnares/12.1.63. [DOI] [PubMed] [Google Scholar]
- 35.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeh PJ, Hegreness MJ, Aiden AP, Kishony R. Drug interactions and the evolution of antibiotic resistance. Nat Rev Microbiol. 2009;7:460–466. doi: 10.1038/nrmicro2133. nrmicro2133 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bliss CI. The toxicity of poisons applied jointly. Ann Appl Biol. 1939;26:585–615. [Google Scholar]
- 38.Loewe S. Die quantitativen Probleme der Pharmakologie. Ergeb Physiol. 1928;27:47–187. [Google Scholar]
- 39.Szappanos B, et al. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nat Genet. 2011;43:656–662. doi: 10.1038/ng.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mateus A, et al. Prediction of intracellular exposure bridges the gap between target- and cell-based drug discovery. Proc Natl Acad Sci U S A. 2017;114:E6231–E6239. doi: 10.1073/pnas.1701848114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richter MF, et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature. 2017;545:299–304. doi: 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piddock LJ, Jin YF, Ricci V, Asuquo AE. Quinolone accumulation by Pseudomonas aeruginosa, Staphylococcus aureus and Escherichia coli. J Antimicrob Chemother. 1999;43:61–70. doi: 10.1093/jac/43.1.61. [DOI] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Kritikos G, et al. A tool named Iris for versatile high-throughput phenotyping in microorganisms. Nat Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.14. 17014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Safdar N, Handelsman J, Maki DG. Does combination antimicrobial therapy reduce mortality in Gram-negative bacteraemia? A meta-analysis. Lancet Infect Dis. 2004;4:519–527. doi: 10.1016/S1473-3099(04)01108-9. [DOI] [PubMed] [Google Scholar]
- 46.Taber HW, Mueller JP, Miller PF, Arrow AS. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev. 1987;51:439–457. doi: 10.1128/mr.51.4.439-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mazzariol A, Tokue Y, Kanegawa TM, Cornaglia G, Nikaido H. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob Agents Chemother. 2000;44:3441–3443. doi: 10.1128/aac.44.12.3441-3443.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis BD, Chen LL, Tai PC. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc Natl Acad Sci U S A. 1986;83:6164–6168. doi: 10.1073/pnas.83.16.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandes F, Neves P, Gameiro P, Loura LM, Prieto M. Ciprofloxacin interactions with bacterial protein OmpF: modelling of FRET from a multi-tryptophan protein trimer. Biochim Biophys Acta. 2007;1768:2822–2830. doi: 10.1016/j.bbamem.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 50.Machado D, et al. Ion Channel Blockers as Antimicrobial Agents, Efflux Inhibitors, and Enhancers of Macrophage Killing Activity against Drug Resistant Mycobacterium tuberculosis. PloS one. 2016;11:e0149326. doi: 10.1371/journal.pone.0149326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maier L, et al. Extensive impact of non-antibiotic drugs on human gut commensals. Nature. 2018;555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.