Abstract
The CRISPR system provides adaptive immunity against mobile genetic elements in prokaryotes, utilising small CRISPR RNAs which direct effector complexes to degrade invading nucleic acids1–3. Type III effector complexes were recently demonstrated to synthesise a novel second messenger, cyclic oligoadenylate (cOA), on binding target RNA4,5. cOA in turn binds to and activates ribonucleases and other factors via a CARF (CRISPR associated Rossman Fold) domain, inducing an antiviral state in the cell that is important for immunity. The mechanism of the “off-switch” that resets the system is not understood. Here, we report the identification of the nuclease that degrades these cOA ring molecules. The “Ring nuclease” is itself a CARF family protein with a metal independent mechanism, which cleaves cA4 rings to generate linear di-adenylate species and switches off the antiviral state. The identification of Ring nucleases adds an important insight to the CRISPR system.
Cyclic oligoadenylate (cOA, with a ring size of 4 or 6 AMP monomers) has emerged as a key second messenger signalling the presence of invading mobile genetic elements in prokaryotes harbouring type III (Csm/Cmr) CRISPR systems4,5. cOA is synthesised by the Cyclase domain of the Cas10 subunit, which is activated by the binding of target RNA. cOA in turn activates a range of proteins with CARF (CRISPR associated Rossman Fold) domains including HEPN (Higher Eukaryotes and Prokaryotes Nucleotide binding) domain ribonucleases (Csm6/Csx1) and transcription factors6–8, precipitating an antiviral state in infected cells that may enhance viral clearance, result in dormancy, or cell death9. There are interesting parallels with the cGAS-cGAMP-STING pathway in eukaryotes, where detection of DNA in the cytoplasm leads to synthesis of a cyclic nucleotide that activates the host immune response10. The HEPN ribonucleases are important for CRISPR-based immunity in vivo11–13. cOA is therefore a potent signalling molecule that must be tightly controlled if cells are to survive a viral infection. cOA synthesis is switched off when type III effectors cleave and release viral RNA targets5,14. However, this will not remove extant cOA, potentially leading to untrammelled ribonuclease activity and cell death. Other cyclic nucleotide signalling molecules in prokaryotes, such as di-cAMP, are degraded by specific phosphodiesterases15,16, but the enzyme specific for cOA is unknown. Here, we report the identification and characterisation of a family of enzymes that degrade cA4, providing a mechanism for the deactivation of the antiviral state induced by cOA in cells harbouring a type III CRISPR system.
To identify the enzyme responsible for the degradation of cOA, we undertook a classical biochemical approach. Starting with a cell lysate from the crenarchaeote Sulfolobus solfataricus (Sso), we noted the presence of an activity that converted radioactively labelled cA4 into a form (X) that migrated more slowly in denaturing gel electrophoresis (Fig. 1). We fractionated the cell lysate through three chromatography steps (phenyl-sepharose, size exclusion and heparin) and followed the activity. The final purification step was followed by assay of each fraction and SDS-PAGE analysis (Fig. 1d), revealing that the activity correlated with a single protein that was identified by mass spectrometry as Sso2081 – a member of the CARF domain-containing protein family17. To confirm that Sso2081 was the nuclease responsible for degradation of cA4, we expressed and purified the recombinant protein in E. coli. The enzyme converted cA4 to a slower-migrating species on gel electrophoresis, as seen for the activity from S. solfataricus, in a reaction independent of divalent metal ions (Fig. 1e). We also noted that the Csx1 nuclease (Sso1389; PDB 2I71)14 is found close to an uncharacterised CARF domain protein of known structure, Sso1393 that is homologous to Sso2081 (Extended Data Fig. 1 & 2). Pure recombinant Sso1393 also exhibits cA4 degradation activity, whilst Csx1 does not (Fig. 1e). In contrast, of the three enzymes, only Csx1 displays cA4-stimulated ribonuclease activity on a linear RNA substrate (Fig. 1f). Sso2081 and Sso1393 degrade cA4 with single-turnover rate constants of 0.23 ± 0.01 and 0.024 ± 0.0004 min-1, respectively (Fig. 2a and Extended Data Fig. 3). The ten-fold higher specific activity of Sso2081, combined with its higher expression levels in S. solfataricus18, suggest that it is the major cA4-degrading enzyme in this organism.
Figure 1. Purification and identification of the enzyme that degrades cA4.
S. solfataricus cell lysate was fractionated by a, phenyl-sepharose, b, size exclusion and c, heparin chromatography. At each stage, fractions were assayed for cA4 conversion activity using radioactive cA4, and active fractions generating product X (indicated by shaded boxes) were pooled for the next stage. d, Following the heparin column, each fraction was assayed and analysed by SDS-PAGE. The band corresponding to the peak of activity (arrowed) was excised from the gel and identified by mass spectrometry as Sso2081, a CARF domain protein. e, Purified recombinant Sso2081 and 1393 degrade cA4, but Csx1 does not. f, Only Csx1 degrades linear RNA in the presence of cA4. Panels a-f are representative of at least duplicate experiments.
Figure 2. Kinetics and products of Ring nuclease activity.
a, Single turnover kinetic analysis of cA4 cleavage by Sso2081 and Sso1393. Sso2081 is a 10-fold faster enzyme. Data points are the means of triplicate measurements, with standard deviation shown, and are technical replicates representative of duplicate experiments. b, TLC analysis of substrates and products. Lane 1 shows cA4 synthesised by the Csm complex. Lane 2 shows the product (X) of conversion by Sso2081 after 20 min. Lane 3 shows product (X) following phosphorylation by PNK. Lanes 4-7 show the products (X and Y) of cA4 conversion by Sso1393 after 20 and 120 min, respectively, before and after PNK treatment. Lanes 8 and 9 are markers for P-A2>P and P-A4>P generated by the MazF nuclease. This gel is representative of duplicate experiments. c, d, LC-HRMS analysis of cA4 cleavage by Sso2081 and Sso1393, respectively, showing ion chromatograms extracted for m/z 657.10 ± 0.5 corresponding to cA4-2, A4>P-2 and A2>P-1. The control is cA4 incubated for 150 min without enzyme. Traces for the conversion of cA4 by Sso2081 after 60 min, and by Sso1393 after 60 and 150 min are shown. Products X and Y are indicated. The bottom traces show the linear oligoadenylates A2>P and A4>P, generated by MazF, as standards. The data are representative of at least duplicate experiments.
Metal independent ribonucleases and ribozymes, including the Cas6 nuclease family19, share a common mechanism: activation of the 2’-hydroxyl of the ribose sugar as the nucleophile that attacks the phosphodiester bond targeted for cleavage, leading to products with a 2’,3’-cyclic phosphate and a 5’-hydroxyl moiety20. To determine the degradation products of cA4 treated with Sso2081 and 1393, we analysed radioactively labelled species by thin layer chromatography (TLC) (Fig. 2b and Extended Data Fig 4). Sso2081 converted cA4 into a faster-migrating form (X) that could be phosphorylated by treatment with polynucleotide kinase (PNK), showing that it was a linear product with a 5’-OH group (lanes 2 and 3). For Sso1393 an linear intermediate product (Y) with a 5’-OH group that converted over time into the final product (X) was also observed (lanes 4-7). Lanes 8 and 9 show standards generated by cleavage of RNA oligonucleotides by the MazF ribonuclease14 to generate the species P-A2>P (5’-phospho-ApAp with a cyclic 2’,3’ phosphate) and P-A4>P, respectively. The final products of Sso2081 and 1393 run at the same position as the P-A2>P standard once phosphorylated by PNK, whilst the intermediate (Y) observed for Sso1393 runs at the same position as the P-A4>P standard.
To identify products X and Y, we incubated Sso2081 and 1393 individually with cA4 and analysed the reaction products by liquid chromatography-high resolution mass spectrometry (LC-HRMS, Fig. 2c and d). Upon 60 min incubation with recombinant Sso2081, the peak corresponding to cA4 had almost completely disappeared in favour of the main product (X) with a retention time of 4.4 min alongside a trace of compound Y eluting at 11.9 min (Fig. 2c and Extended Data Fig. 5). Linear A2 and A4 species with a 2',3’-cyclic phosphate, generated using the MazF toxin14, eluted with comparable retention times as products X and Y, respectively. Mass spectrometry revealed product X to have a neutral mass of 658.104 amu, consistent with linear A2>P. The mass of product Y (1316.208 amu), on the other hand, was consistent with A4>P. The same products were observed for Sso1393 (Fig. 2d and Extended Data Fig. 6). Again, A2>P (X) was the main product, with A4>P (Y) more visible at early time points, consistent with the slower catalytic rate of Sso1393. Thus, these enzymes break the cA4 ring using a metal-independent mechanism, generating linear A4>P intermediate and A2>P products with 5’-OH and 2’,3’-cyclic phosphate termini. We tested the specific activity of Sso2081 against available cyclic nucleotide molecules: cA6, cA4, cyclic di-AMP, cyclic di-GMP and cyclic GMP-AMP (Extended Data Fig. 7) confirming that the enzyme is highly specific for degradation of cA4, with a very low level of activity directed against cA6 and no detectable degradation of the cyclic di-nucleotides. We propose the collective term “Ring nucleases” for this family of phosphodiesterases.
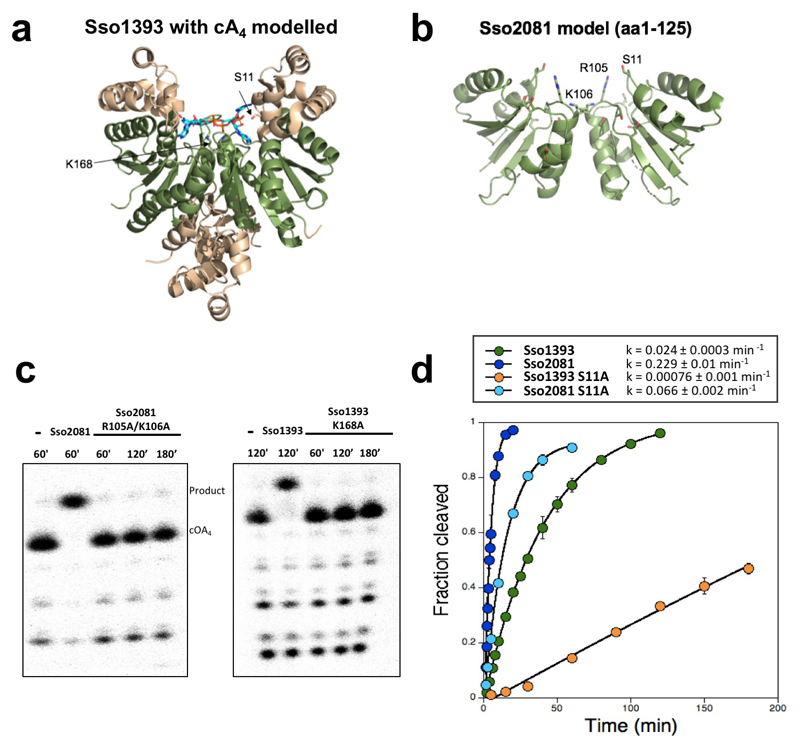
The structure of Sso1393 (PDB 3QYF, unpublished) reveals a canonical CARF domain formed by a homodimeric subunit arrangement, with a C-terminal extension. To elucidate the mechanism of Ring nucleases, we docked cA4 into the CARF domain of Sso1393 (Fig. 3a and Extended Data Fig. 8). This simple model without energy minimisation allows hypotheses concerning the enzyme mechanism to be drawn. Using structure-guided multiple sequence alignment of Sso1393, Sso2081 and homologues (Extended Data Fig.2), we identified conserved residues positioned close to the cA4 binding site that might play a role in catalysis (Fig. 3). We noted the conservation of a lysine (K168 in Sso1393) predicted to lie in the centre of the cA4 binding site, which could play a role in catalysis. Sso2081 has two basic residues (R105/K106) in this position. The K168A variant of Sso1393 was catalytically inactive, as was the Sso2081 R105A/K106A variant, confirming an important role in catalysis, possibly by stabilising the pentacovalent phosphorous generated in the transition state20 (Fig. 3b, Extended Data Fig.8). The conserved residue S11 is also implicated in catalysis. The S11A variant had a catalytic rate constant reduced by 3.5- and 32-fold for Sso2081 and Sso1393, respectively (Fig. 3b, Extended Data Fig.8). Consistent with the absence of Ring nuclease activity, Csx1 lacks these key active site residues.
Figure 3. Structure and mechanism of Ring nucleases.
a, Structure of the CARF domain of Sso1393 with cA4 docked. The active site residues K168 and S11 are shown. b, Kinetic analysis of Sso1393 (wild-type, S11A and K168A variants) and Sso2081 (wild-type, S11A and R105A/K106A variants). Catalytic rate constants under single turnover conditions are plotted. The data points are derived from exponential fits to the triplicate rate measurements presented in Extended Data Fig. 8, with the standard errors derived from curve fitting shown. c, Cartoon showing the reaction scheme for conversion of cyclic to linear A4>P and A2>P. S11 may participate in the correct positioning of the 2’-OH group of the ribose to facilitate nucleophilic attack, whilst the basic residue K168 (and R105/K106 in Sso2081) may stabilise the penta-covalent phosphorus formed in the transition state.
Based on the evidence available, we propose that cleavage of cA4 is catalysed by binding the molecule in an orientation that allows the 2’-hydroxyl of the ribose to attack the bridging phosphorus, coupled with stabilisation of the developing transition state by the basic residue(s) at the base of the binding pocket (Fig. 3c). The single turnover rate constant for Sso2081, at below 1 min-1, is comparable to the Cas6 nuclease family, which use a similar mechanism19,21. Given the dimeric organisation of the CARF domain, there is the possibility for two active sites acting on opposite sides of the cA4 ring, consistent with the observation of an A2>P product. For the slower Sso1393 enzyme, appreciable levels of A4>P intermediate were observed, suggesting that the two active sites need not function in a concerted manner.
We next wished to reconstitute the cOA signalling system in vitro to confirm the function of the Ring nucleases. We first tested the ability of Ring nucleases to deactivate Csx1, observing that as cA4 is converted to A2>P, the activation of the Csx1 ribonuclease is abrogated (Extended Data Fig. 9). We next set up a reaction containing the Csm effector complex, 0.5 mM ATP and a variable concentration of target RNA from 10 to 0.01 nM, to activate cA4 synthesis. After incubation for 1 h at 70 °C in the presence or absence of 2.5 µM Sso2081, the HEPN ribonuclease Csx1 (0.5 µM) together with radioactively labelled Csx1 substrate RNA were added. Transcript levels of the sso2081 gene are about 2-fold higher than those of the csx1 gene in uninfected S. solfataricus cells18, and large changes in expression of neither gene was observed on infection with the STIV virus22. Target RNA binding by Csm activates the Cyclase domain, switching on cA4 production. The Cyclase domain is deactivated when target RNA is cleaved and dissociates from Csm, leaving the complex free to bind to further targets14. Higher target RNA concentrations thus result in higher levels of cA4 and stronger activation of Csx1, which degrades a labelled substrate RNA (Fig. 4a). The presence of Sso2081 abrogated the activation of Csx1 in a target RNA concentration-dependent manner. This recapitulates the situation likely to prevail in cells infected with a virus, where viral RNA load determines the extent of cOA production and speed of subsequent clearance by the Ring nuclease (Fig. 4b).
Figure 4. Reconstitution of the cOA signalling pathway.
a, The Csm effector generates cA4 in proportion to the amount of viral target RNA present 14, activating the HEPN nuclease Csx1. The presence of Sso2081 (2.5 µM) for 1 h prior to the addition of Csx1 (0.5 µM) for 20 min at 70 °C reversed Csx1 activation partially when 10 nM target RNA was present, and fully when lower amounts of RNA were used. Control c shows the experiment in the presence of Sso2081 and absence of target RNA. Levels of cA4 and A2>P were monitored by TLC. The data are representative of three separate experiments. b, Schematic of the cOA signalling system. Type III effectors loaded with crRNA bind to viral target RNA, activating the HD nuclease, which targets viral DNA, and the Cyclase domain. cOA is synthesised, activating the Csx1 ribonuclease. Target RNA cleavage and subsequent dissociation deactivates the Cyclase domain, stopping synthesis of cOA. The Ring nucleases complete the deactivation of the antiviral state by degrading extant cOA.
Thus, we have identified a family of CARF-domain containing Ring nucleases as the enzymes responsible for the degradation of cA4, which is synthesised by type III CRISPR systems in response to detection of viral RNA. The Ring nucleases act as the “off-switch” for the system, limiting the damage caused by the HEPN ribonucleases activated by cOA once invading RNA has been cleared from the cell. The relatively slow kinetics of cA4 degradation by Ring nucleases are probably consistent with the requirement to maintain the antiviral response for an appropriate period, but we cannot rule out the possibility that other factors increase the cOA clearance rate under certain circumstances. Clearly, it will be important to follow up these biochemical studies with genetic analyses to elucidate the function of Ring nucleases in the antiviral cOA-signalling pathway in vivo.
Many type III CRISPR systems have multiple associated CARF-domain containing proteins, some of which may be specialised for the degradation of cOA. Ring nucleases are hard to identify using bioinformatic analysis, as they involve the “grafting” of a minimal ribonuclease catalytic site on to an extensive cOA binding site in the CARF domain. In a minimal system, it is possible that a single enzyme has a C-terminal HEPN family ribonuclease coupled to an N-terminal CARF family Ring nuclease. This would allow cOA binding to rapidly switch on the ribonuclease activity and then slowly auto-deactivate by cOA cleavage. However, specialisation of CARF proteins as Ring nucleases would yield the advantage of allowing levels of cOA-activated ribonuclease and cOA-degrading Ring nuclease activity to be controlled independently. It is also possible that cA6 rings such as those generated in Streptococcus thermophilus will be processed differently, as they are likely to have more conformational flexibility than cA4. The recent identification of a wide range of as-yet uncharacterised CARF domain proteins across the prokaryotes8 suggests that we have only scratched the surface of this system. These are fruitful areas for future studies.
Methods
Purification of cOA degrading enzyme from S. solfataricus cellular extract
S. solfataricus (Sso) P2 was grown as previously described23 and cells pelleted by centrifugation at 4000 rpm (Beckman Coulter Avanti JXN-26; JLA8.1 rotor) for 15 min at 4 °C. Cells were suspended in buffer A containing 100 mM sodium phosphate pH 7.0 and 1.5 M ammonium sulphate with one EDTA-free protease inhibitor tablet (Roche). Cells were lysed by sonicating six times for 1 min with 1 min rest intervals on ice, and the lysate was ultracentrifuged at 40000 rpm (Beckman Coulter Optima L-90K; 70 Ti rotor) and 4 °C for 45 min before filtering and loading onto a phenyl-sepharose column (GE Healthcare) pre-equilibrated with buffer A. Protein was eluted with a linear gradient of buffer B containing 100 mM sodium phosphate pH 7.0 across 10 column volumes (CV). Each fraction was assayed for cyclic tetra-adenosine (cA4) degradation activity and fractions displaying cA4 degradation were pooled and concentrated using a 10 kDa molecular weight cut-off ultracentrifugal concentrator (Amicon Millipore). Concentrated protein was then further separated by size-exclusion chromatography (S200; 26/60 GE Healthcare) in buffer containing 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (HEPES) pH 7.5 and 150 mM KCl. Fractions were assayed for cA4 degradation, and active fractions pooled and concentrated as previously, exchanging into 10 mM 2-(N-morpholino)ethanesulfonic acid (MES) pH 6.0 buffer during concentration. Concentrated protein was then loaded on to a 5 ml HiTrap heparin column (GE Healthcare) equilibrated with 10 mM MES pH 6.0 buffer, washing unbound protein with 8% buffer C containing 10 mM MES pH 6.0 and 1.0 M NaCl for 2 CV. Protein was eluted by a linear gradient to 30% buffer C across 5 CV followed by a gradient to 100% buffer C across 4 CV. After assaying for cA4 degradation, fractions of interest were visualised by sodium-dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE). A protein band of ~18 kDa, the presence and abundance of which corresponded to a peak in cA4 degradation was excised, trypsin digested and identified by mass spectrometry.
Cloning, expression and purification of Sso2081, Sso1393 and variants
The cloning, expression and purification of Sso1389 (Csx1) has been described previously14. A synthetic gene encoding Sso2081 was purchased from Integrated DNA Technologies, Coralville, IA, United States (IDT), while Sso1393 was PCR amplified from S. solfataricus P2 genomic DNA. Genes encoding Sso2081 and Sso1393 were cloned into the pEHisTEV vector24, and transformed into E. coli DH5α competent cells. Sso2081 S11A, Sso2081 R105A/K106A, Sso1393 S11A and Sso1393 K168A variants were generated by site directed mutagenesis using the QuickChange Site-Directed Mutagenesis protocol (Agilent Technologies), with DNA primers purchased from IDT. Sequence verified constructs were transformed into C43 (DE3) E. coli cells for protein expression.
Expression of recombinant Sso2081 and variants was induced with 0.4 mM isopropyl β-D-1-thiogalactopyranoside at an OD600 of ~0.8, and cells incubated at 16 °C overnight with shaking at 180 rpm before harvesting by centrifugation at 4000 rpm (JLA8.1 rotor) at 4 °C for 15 min. Cells were suspended in lysis buffer (50 mM Tris-HCl pH 8.0, 0.5 M NaCl, 10 mM imidazole and 10% glycerol) with 1 mg/ml chicken egg lysozyme (Sigma-Aldrich) and one EDTA-free protease inhibitor tablet. Cells were sonicated six times for 1 min with 1 min rest intervals on ice at 4 °C. Cell lysate was then ultracentrifuged at 40000 rpm (70 Ti rotor) for 45 min at 4 °C, filtered and loaded onto a 5 ml HisTrap FF column (GE Healthcare) equilibrated with buffer D containing 50 mM Tris-HCl pH 8.0, 0.5 M NaCl, 30 mM imidazole and 10% glycerol. After washing unbound protein with 20 CV buffer D, recombinant Sso2081 and mutants were eluted with a linear gradient (0-20%) of buffer D supplemented with 0.5 M imidazole across 15 CV, then holding at 20% for 4 CV. Pooled fractions were concentrated as described previously, and the hexa-histidine affinity tag was removed by incubating protein with Tobacco etch virus (TEV) protease (10:1) for 4 h at 37 °C. His-tag cleaved Sso2081 was isolated from TEV by affinity chromatography as detailed above, eluting with buffer D prior to further purification by size-exclusion chromatography (S200 26/60; GE Healthcare) in buffer containing 20 mM Tris-HCl pH 8.0, 0.5 M NaCl and 1 mM DTT.
Expression of recombinant Sso1393 (wild-type and variants) was induced as above, except cells were grown at 16 °C overnight before harvest. Cells were lysed and protein purified as for Sso2081. All proteins were aliquoted, flash frozen with liquid nitrogen, and stored at -80 °C.
Cyclic oligoadenosine nuclease assays and kinetic analysis
32P-α-ATP incorporated cyclic oligoadenylate was generated using the Csm complex as previously described14. Briefly, the Csm complex was incubated for 2 h at 70 ºC in 100 µl final reaction volume in a pH 5.5 buffer containing 20 mM MES, 100 mM potassium glutamate and 1 mM DTT supplemented with 2 mM MgCl2, 1 mM ATP, 3 nM 32P-α-ATP and 100 nM A26 target RNA: (5’-AGGGUCGUUGUUAAGAACGACGUUGUUAGAAGUUGGGUAUGGUGGAGA). The reaction was stopped by phenol-chloroform extraction followed by chloroform extraction. Sso1393, Sso2081, and their mutants were assayed for labelled cA4 degradation by incubating 2 μM protein dimer with 1/300 diluted Csm generated 32P labelled cOA (0.33 µl per 100 µl reaction) in buffer E (20 mM 2-amino-2-(hydroxymethyl)-1,3-propanediol pH 8.0, 100 mM NaCl, 1 mM EDTA and 1 mM DTT) at 70 °C. At desired time points, a 10 μl aliquot was removed, and the reaction quenched by adding to chilled phenol-chloroform (Ambion). Subsequently, 5 μl of deproteinised reaction product was extracted into 5 μl 100% formamide for denaturing PAGE. Control reactions include cOA incubated in buffer E without protein at 4 °C and at 60 °C up to the endpoint of each experiment (typically 20, 120 or 180 min) phenol-chloroform extracted as above. All experiments were carried out in triplicate. cA4 degradation was visualised by phosphorimaging following denaturing PAGE (7M Urea, 20% acrylamide, 1x TBE). For kinetic analysis, cA4 cleavage was quantified using the Bio-Formats plugin25 of ImageJ26 as distributed in the Fiji package27 and fitted to a single exponential curve using Kaleidagraph (Synergy Software), as described previously28.
HEPN nuclease deactivation assays
For degradation of cA4, Csm generated cOA was incubated with 2 μM Sso2081 dimer or 4 μM Sso1393 dimer in buffer E for 60 min and 120 min, respectively. Subsequently, the reaction was deproteinised by phenol-chloroform extraction and diluted two-fold in RNase free water. As a control reaction, cOA was mock treated in buffer with water in place of protein and deproteinised as before. 500 nM Csx1 dimer was incubated with 50 nM A1 RNA and either no cOA activator, 1/100 diluted untreated Csm cOA, 1/100 mock-treated cOA or 1/100 Ring nuclease treated cOA in buffer containing 20 mM MES pH 5.5, 100 mM K-glutamate and 1 mM DTT for 60 min at 70 °C. Reactions were quenched by the addition of a reaction volume equivalent of 100% formamide, and RNA cleavage was assessed by phosphorimaging following denaturing PAGE.
Reconstitution of the cOA signalling pathway
The Csm complex (70 nM carrying the crRNA targeting A26) was incubated for 1 h at 70 ºC in presence of 2.5 µM of Sso2081 dimer and various concentrations of target A26 ssRNA in a reaction containing 20 mM MES pH 6.0, 100 mM NaCl, 1 mg/ml BSA, 2 mM MgCl2, 0.5 mM ATP and a 5’-labeled A1 ssRNA that is not recognised by the Csm complex (5’-AGGGUAUUAUUUGUUUGUUUCUUCUAAACUAUAAGCUAGUUCUGGAGA). After 1 h incubation, 500 nM of Csx1 dimer was added to the reaction for a further 20 min. Reactions were quenched by deproteination with phenol-chloroform extraction and run on a 20% acrylamide 7 M urea denaturing gel prior to phosphorimaging to visualise A1 RNA cleavage. The same experiment was carried out in the presence of 3 pM 32P-α-ATP and unlabelled A1 RNA to visualise the formation and degradation of cA4, and 1 µl of the reaction was run on TLC prior to phosphorimaging.
Generation of standards using the MazF nuclease
The E. coli toxin-antitoxin MazEF was purified as previously described14. Active MazF was liberated by either trypsin (Promega) digestion (1600:1) at 37 °C for 15 min or by incubation with Factor X Activated (0.1 unit per 1 mg of protein; Sigma-Aldrich) in FXa buffer containing 10 mM Tris-HCl pH 8.0 and 1 mM DTT. For generating linear oligoadenylates A2>P (5’ hydroxyl-Ap-Ap with a 2’,3’-cyclic phosphate) and A4>P, 30 μM A2 (AAACAUCAG) or A4 (AAAAACAUCAG) RNA was incubated with MazF in FXa buffer for 1 h at 37 °C. RNA was deproteinised by phenol-chloroform extraction followed by chloroform extraction. For use as standards, A2>P and A4>P linear oligoadenylates were 5’-end labelled using 32P-γ-ATP and T4 Polynucleotide Kinase (PNK; Thermo Fisher Scientific) via its forward reaction.
Thin-layer chromatography
Experiments were performed using silica gel on a 20 x 20 cm TLC plate (Supelco Sigma-Aldrich) containing a fluorescent indicator, allowing visualisation of non-radioactive products by UV shadowing. Prior to loading, all samples were deproteinised by phenol-chloroform followed by chloroform extraction. Non-radioactive or radioactive samples (from 0.1 µl to 2 µl) were loaded 1 cm above the bottom of the TLC plate. The TLC plate was then placed in a sealed glass chamber pre-warmed at 30 °C and containing 0.5 cm of a running buffer composed of H2O (30 %), ethanol (70 %) and ammonium bicarbonate (0.2 M), pH 9.3. The buffer was allowed to rise along the plate through capillary action until the migration front reached 15 cm. The plate was dried and radioactive sample migration visualised by phosphorimaging while non-radioactive sample migration was pictured under UV light (254 nm).
Liquid chromatography-high resolution mass spectrometry (LC-HRMS)
LC-HRMS analysis was performed on a Thermo Scientific Velos Pro instrument equipped with HESI source and Dionex UltiMate 3000 chromatography system. Samples were deproteinised as described for TLC. Compounds were separated on a Kinetex 2.6 µm EVO C18 column (2.1 ? 100 mm, Phenomenex) using a linear gradient of acetonitrile (B) against 20 mM ammonium bicarbonate (A): 0 – 5 min 2% B, 5 – 33 min 2 – 15% B, 33 – 35 min 15 – 98% B, 35 – 40 min 98% B, 40 – 41 min 98 – 2% B, 41 – 45 min 2% B. The flow rate was 350 µl min-1 and column temperature was 40 °C. UV data were recorded at 254 nm. Mass data were acquired on the FT mass analyzer in negative ion mode with scan range m/z 150 – 1500 at a resolution of 30,000. Source voltage was set to 3.5 kV, capillary temperature was 350 °C, and source heater temperature was 250 °C. Data were analysed using Xcalibur (Thermo Scientific). Extracted ion chromatograms were smoothed using the Boxcar function at default settings.
Data Availability Statement
All electrophoretic separation and kinetic data are included as source data and supplementary information. Raw MS data are available on request from M.F.W.
Extended Data
Extended Data Figure 1. Genome organisation of the CRISPR-Cas locus of S. solfataricus.
The type I-A, III-B and III-D effector complex operons are depicted, along with genes encoding adaptation proteins and the position of the six CRISPR loci (A-F). Genes outlined in blue encode proteins with CARF domains. The structures of three CARF family proteins are shown with CARF domains coloured green, along with the structure of cA4.
Extended Data Figure 2. Structure-guided sequence alignment of Sso1393, Sso2081 and orthologues.
Multiple sequence alignment showing Sso1393, together with three homologues, aligned with Sso2081 and three homologues. Secondary structure is shown above the alignment, based on the structure of Sso1393 (PDB 3QYF). Ser-11 and Lys-168 are indicated by asterisks. Conserved residues are shaded. Sequences aligned are from S. solfataricus (Sso1393, Sso2081); S. islandicus REY15A (SiRE_0811, SiRE_0455); S. islandicus M.16.4 (M164_0884); S. acidocaldarius (Saci_1063) and S. tokodaii (Stk_17430).
Extended Data Figure 3. Single-turnover kinetic analysis of cA4 degradation by a, Sso2081 and b, Sso1393.
Representative phosphorimages of denaturing polyacrylamide gel electrophoresis (PAGE) analysing the reaction of radioactively labelled cA4 with a, 2 μM Sso2081 or b, 2 μM Sso1393 at 60 °C over time. Triplicate experiments were carried out, quantified by phosphorimaging and the fraction of cA4 cleaved plotted in Figure 3b.
Extended Data Figure 4. Sso2081 and Sso1393 cA4 degradation mechanism investigated by TLC.
Csm generated cOA (lane 1) was incubated with 2 μM Sso2081 dimer at 60 °C to determine the intermediate (Y) and final (X) reaction product over time (lanes 2-10). Lanes 11-13 show reaction product seen in lanes 2, 4 and 6, 5’-end phosphorylated using T4 polynucleotide kinase (PNK) for identification of reaction intermediates and products by comparison to 5’-end phosphorylated MazF nuclease generated HO-A2>P and HO-A4>P standards. Lanes 14 & 15 show the reaction products of 2 μM Sso1393 dimer incubated with cOA at 70 °C for 20 and 120 min, respectively. Reaction product from lanes 14 & 15 are 5’-end phosphorylated by PNK for comparison to P-A2>P and P-A4>P standards. Comparison of PNK treated reaction product to standards showed the presence of a low amount of intermediate (P-Y) during the Sso2081 cA4 cleavage reaction, which migrated similarly to the P-A4>P standard and did not change in abundance over time, whereas the abundance of the final product (P-X) increased over time. In contrast, comparison of Sso1393 PNK treated 20 min and 120 min reaction products showed a decrease of the intermediate (P-Y) over time and increase of product (P-X).
Extended Data Figure 5. LC-MS analysis of Sso2081 reactions.
a, Ion chromatograms extracted for m/z 657.1 (cA2-1, A2>P-1, cA4-2, A4>P-2). Individual lanes are labelled. cOAs (mainly cA4), derived from reaction of Csm with ATP, was incubated with Sso2081 for 60 and 150 min. Linear oligoadenylates with 2’,3’-cyclic phosphate were derived from hydrolysis of A4 RNA oligonucleotide with MazF.
b, UV traces at 254 nm. Peaks that change in intensity over the course of the enzymatic reaction are indicated by arrows. The three peaks that decreased or increased over the course of the reaction all match the changes in abundance of the m/z 657.1 species. No changes are observed after 60 min reaction time. The broad peak at 4.9 min is an unknown contaminant likely resulting from the phenol-chloroform extraction. Shifts in retention time are possibly due to matrix effects.
c, Mass spectra for cA4 and product X. Calculated for C20H23N10O12P2-1 (cA2 / A2>P) m/z 657.0978, found 657.0966 (δm -1.8 ppm); calculated for C40H46N20O24P4-2 (cA4 / A4>P) m/z 657.0978, found 657.0967 (δm -1.6 ppm). The data presented are representative of duplicate experiments.
Extended Data Figure 6. LC-MS analysis of Sso1393 reactions.
a, Ion chromatograms extracted for m/z 657.1 (cA2-1, A2>P-1, cA4-2, A4>P-2). Individual lanes are labeled. cOAs (mainly cA4), derived from reaction of Csm with ATP, was incubated with Sso1393 for 60 and 150 min or without Ring nuclease for 150 min. A control in which the ATP from the Csm cyclase reaction had been omitted was also analysed as control. Linear oligoadenylates with 2’,3’-cyclic phosphate were derived from hydrolysis of suitable DNA oligonucleotide subtrates with the toxin MazF; whereas cA2 was a commercially available standard. The traces show clearly the difference in retention time between the linear and cyclic isomers.
b, UV traces at 254 nm. Peaks that change in intensity over the course of the enzymatic reaction are indicated by arrows. The three peaks that decreased or increased over the course of the reaction all match the changes in abundance of the m/z 657.1 species. The broad peak at 4.1 – 6 min is an unknown contaminant likely resulting from the phenol-chloroform extraction.
c, Mass spectra for species X and Y. Calculated for C20H23N10O12P2-1 (cA2 / A2>P) m/z 657.0978, found 657.0968 (δm -1.4 ppm); calculated for C40H46N20O24P4-2 (cA4 / A4>P) m/z 657.0978, found 657.0967 (δm -1.6 ppm). The data presented are representative of duplicate experiments.
Extended Data Figure 7. Specificity of Sso2081 for cyclic nucleotide substrates.
Various cyclic di- and oligonucleotides were incubated in the absence (ctrl) or presence of Sso2081 as described in Materials and Methods. Dinucleotides and cA6 were obtained from BIOLOG Life Science Institute (Bremen), cA4 was obtained from an enzymatic reaction with S. solfataricus Csm. Protein-free extracts were analyzed by LC-HRMS essentially as before but using a shorter column and gradient. EICs for substrate and expected products are shown in each panel. No reaction was observed for any of the cyclic dinucleotides. cA4, was completely converted to linear A2>P by Sso2081 whilst only a small percentage of cA6 was converted, demonstrating a clear preference of Sso2081 for its physiological substrate cA4. The data presented are representative of duplicate experiments.
Extended Data Figure 8. Structure and kinetic analysis of Sso1393 and Sso2081.
a, Structure of Sso1393 (PDB 3QYF), with cA4 docked at the active site. This is an orthogonal view to that shown in Fig. 3. The CARF domain is coloured green. The side chains of Ser-11 and Lys-168 are shown.
b, Model of the Sso2081 structure. Only the CARF domain (aa1-125) is modelled. Conserved residues Ser-11, Arg-105 and Lys-106 are labelled. Model generated using Phyre29.
c, Representative phosphorimages of denaturing PAGE assessing the cA4 degradation activity of Sso2081 compared to its catalytically inactive R105A/K106A mutant over time (left hand panel), and Sso1393 activity compared to its K168A mutant (right hand panel). All reactions were carried out at 70 °C with 2 μM protein dimer. The data presented are representative of triplicate experiments.
d, Single-turnover kinetics of Sso2081 and Sso1393 plotted alongside their S11A active site mutants and fitted to an exponential equation. Rate constants are displayed with the legend (Sso2081; 0.23 ± 0.01 min-1, Sso2081 S11A; 0.066 ± 0.002 min-1, Sso1393; 0.024 ± 0.0003 min-1, Sso1393 S11A; 0.00076 ± 0.001 min-1). Experiments were carried out in triplicate with means and standard deviation shown. The data presented are technical replicates and are representative of duplicate experiments.
Extended Data Figure 9. Ring nucleases abrogate activation of the Csx1 nuclease by converting cA4 to A2>P.
a, In the absence of cA4, Csx1 had no ribonuclease activity (lane c), but addition of cA4 resulted in degradation of the substrate RNA molecule. When cA4 was pre-incubated for 1 h at 70 °C with Sso2081 in buffer E before addition to the assay, Csx1 ribonuclease activity was abolished. Mock treatment using buffer instead of Sso2081 (denoted ‘*’) had no effect.
b, As for a, but pre-incubated with Sso1393 for 2 h at 70 °C.
c, Using radioactive cA4, the deactivation of Csx1 by Sso2081 and Sso1393 was observed to correlate with the conversion of cA4 into A2>P. Results shown are representative of at least triplicate experiments.
Supplementary Material
Acknowledgements
This work was funded by grants from the Biotechnology and Biological Sciences Research Council (REF BB/M000400/1 and BB/M021017/1). M.F.W. is a Wolfson Research Merit Award holder. We acknowledge the contribution of the Mass Spectrometry Unit of the University of St Andrews to this work.
Footnotes
Author contributions J.S.A. carried out chromatography to identify the major cA4 degrading enzyme, carried out enzyme assays and analysis, and developed HEPN nuclease deactivation assays; C.R. carried out TLC to identify cA4 degradation products and reconstituted the cOA signaling pathway; S.Grü. carried out and analysed the mass spectrometry; S.Gra. generated expression plasmids and purified proteins; M.F.W. oversaw the work, analysed the data and wrote the manuscript. All authors contributed to data analysis and writing.
Competing interests The authors declare no competing interests.
References
- 1.Mojica FJ, Rodriguez-Valera F. The discovery of CRISPR in archaea and bacteria. FEBS J. 2016;283:3162–3169. doi: 10.1111/febs.13766. [DOI] [PubMed] [Google Scholar]
- 2.Makarova KS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nature Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang F, Doudna JA. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol. 2015;30:100–111. doi: 10.1016/j.sbi.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niewoehner O, et al. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017;548:543–548. doi: 10.1038/nature23467. [DOI] [PubMed] [Google Scholar]
- 5.Kazlauskiene M, Kostiuk G, Venclovas C, Tamulaitis G, Siksnys V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017;357:605–609. doi: 10.1126/science.aao0100. [DOI] [PubMed] [Google Scholar]
- 6.Makarova KS, Anantharaman V, Grishin NV, Koonin EV, Aravind L. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front Genet. 2014;5:102. doi: 10.3389/fgene.2014.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burroughs AM, Zhang D, Schaffer DE, Iyer LM, Aravind L. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucl Acids Res. 2015;43:10633–10654. doi: 10.1093/nar/gkv1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shmakov SA, Makarova KS, Wolf YI, Severinov KV, Koonin EV. Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc Natl Acad Sci USA. 2018 doi: 10.1073/pnas.1803440115. 1803440115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koonin EV, Makarova KS. Discovery of Oligonucleotide Signaling Mediated by CRISPR-Associated Polymerases Solves Two Puzzles but Leaves an Enigma. ACS Chem Biol. 2018;13:309–312. doi: 10.1021/acschembio.7b00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 11.Deng L, Garrett RA, Shah SA, Peng X, She Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol. 2013;87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- 12.Hatoum-Aslan A, Maniv I, Samai P, Marraffini LA. Genetic Characterization of Antiplasmid Immunity through a Type III-A CRISPR-Cas System. J Bacteriol. 2014;196:310–317. doi: 10.1128/JB.01130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang W, Samai P, Marraffini LA. Degradation of Phage Transcripts by CRISPR-Associated RNases Enables Type III CRISPR-Cas Immunity. Cell. 2016;164:710–721. doi: 10.1016/j.cell.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouillon C, Athukoralage JS, Graham S, Grüschow S, White MF. Control of cyclic oligoadenylate synthesis in a type III CRISPR system. eLife. 2018;7:e36734. doi: 10.7554/eLife.36734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huynh TN, Woodward JJ. Too much of a good thing: regulated depletion of c-di-AMP in the bacterial cytoplasm. Curr Opin Microbiol. 2016;30:22–29. doi: 10.1016/j.mib.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dey RJ, et al. Inhibition of innate immune cytosolic surveillance by an M. tuberculosis phosphodiesterase. Nat Chem Biol. 2017;13:210–217. doi: 10.1038/nchembio.2254. [DOI] [PubMed] [Google Scholar]
- 17.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nature Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wurtzel O, et al. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–141. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haurwitz RE, Sternberg SH, Doudna JA. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 2012;31:2824–2832. doi: 10.1038/emboj.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sokolowski RD, Graham S, White MF. Cas6 specificity and CRISPR RNA loading in a complex CRISPR-Cas system. Nucl Acids Res. 2014;42:6532–6541. doi: 10.1093/nar/gku308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortmann AC, et al. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J Virol. 2008;82:4874–4883. doi: 10.1128/JVI.02583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albers SV, et al. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl Environ Microbiol. 2006;72:102–111. doi: 10.1128/AEM.72.1.102-111.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, Naismith JH. A simple and efficient expression and purification system using two newly constructed vectors. Prot Expr Purif. 2009;63:102–111. doi: 10.1016/j.pep.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linkert M, et al. Metadata matters: access to image data in the real world. J Cell Biol. 2010;189:777–782. doi: 10.1083/jcb.201004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Meth. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–672. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All electrophoretic separation and kinetic data are included as source data and supplementary information. Raw MS data are available on request from M.F.W.