Philip and Harrison introduce research from Condon et al., who use high resolution imaging to reveal novel surface structures involved in macropinocytosis.
Abstract
Macropinocytosis or “cell drinking” involves the elaboration of membrane ruffles that enclose and internalize extracellular fluids. Using lattice light sheet microscopy, Condon et al. (2018. J. Cell Biol. https://doi.org/10.1083/jcb.201804137) reveal the presence of parallel membrane protrusions termed “tent poles” that flank and direct membrane ruffle formation.
Macrophages are sentinel cells that constantly sample the extracellular milieu for antigens and nutrients. Macrophages use constitutively active surface protrusions called membrane ruffles that close to produce a fluid-filled organelle termed the macropinosome. While heterogeneous in nature, membrane ruffle formation is amplified in macrophages after exposure to growth factors and bacterial products. Elegant mechanistic work spearheaded by Joel Swanson’s group has led to a detailed understanding of how phospholipids and F-actin remodeling proteins build a membrane ruffle (1–4). To date, phase-contrast and confocal imaging of macrophages have described macropinosome formation as the induction of broad, linear ruffles that circularize into a cup. The cups close and fuse with the plasma membrane, presumably at multiple contact sites (3, 5).
In this issue, Condon et al. shed additional light on the mechanism of macropinosome formation by imaging activated macrophages with lattice light sheet microscopy (LLSM). With LLSM, 2D optical lattices create an ultrathin light sheet that can be scanned through the specimen to create a 3D image (6). The decoupling of the illumination and detection pathways in LLSM allow for improved axial resolution. This, combined with negligible photobleaching, allows for unprecedented 4D observations of highly dynamic biological processes (6).
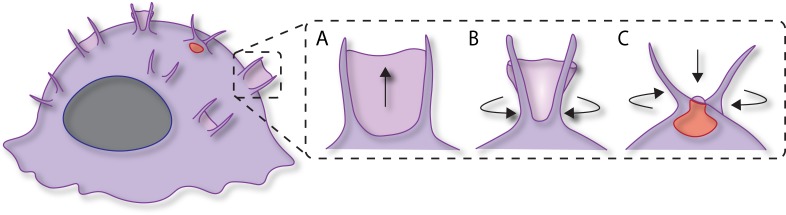
Macropinosome formation was investigated by Condon et al. (7) using the RAW264.7 macrophage cell line that was stimulated with bacteria lipopolysaccharide (LPS) and stably transfected with GFP-LifeAct to image F-actin. LLSM allowed live cell imaging of the complete volume of transfected macrophages every 1.4 s for periods up to several minutes. Imaging large fields of view of activated macrophages showed sites of recurring membrane ruffles or “hotspots” on the macrophage cell surface. Most striking within these regions was the presence of long filopodial-like F-actin extensions, termed “tent poles,” which were associated with over 80% of membrane ruffles. Using high temporal resolution dissection, the authors divided macropinosome formation into tent pole extension, membrane/F-actin veil formation between the tent poles, and ruffle circularization corresponding to convergence of the tent poles, followed by collapse of the membrane veil (Fig. 1). Particularly fascinating was the crossing over and twisting of the tent poles at the time of nascent macropinosome formation (Fig. 1), detected by the accumulation of early endosome markers Halo-Rab5 and -2xFYVE on the descending vacuole. How the tent pole structures twist is not yet known but the authors suggest an intriguing parallel with the similar rotational movements at the base of filopodia that are driven by myosin V (8).
Figure 1.
Macropinosome formation by tent pole ruffling. (A) Erected tent poles support the formation of membrane/F-actin veils. (B) The meeting of tent poles results in circularization of the ruffles. (C) The collapse of the membrane veil coincident with tent pole crossing over and twisting leads to macropinosome formation.
Condon et al. (7) provide imaging evidence that tent pole twisting causes membrane constriction coincident with macropinosome formation. The collapse of the membrane/F-actin veil that occurs when tent poles first crossover could restrict the membrane fusion sites to a single, confined location. The mechanism of membrane fusion, scission, and sealing to create a discrete intracellular organelle has remained enigmatic for macropinocytosis as well as another large-scale internalization process, phagocytosis. During clathrin-mediated endocytosis, membrane fusion requires contact of the membranes, directed by BAR domain–containing proteins (9). Due to the inherent curvature of these proteins, their sequential recruitment progressively constricts the membranes together until membrane scission occurs via dynamin. The twisting of tent poles may provide the required constriction of membranes that become closely apposed when tent poles interact and crossover. Presumably, a purse string–type of contraction would also occur to coalesce the membrane veils for fusion at a single aperture, if endocytic mechanisms are used. The next steps will be to identify the molecular players driving lipid bilayer scission and resealing to form a discrete macropinosome.
To probe for a mechanism behind LPS-induced macropinosome formation, Condon et al. (7) performed an exhaustive Rab screen, since several Rab GTPases are known to be integral components of membrane ruffles. Rab13 showed the most prominent enrichment and activation in ruffles by LPS, determined using a novel Förster resonance energy transfer biosensor and 3D-structured illumination microscopy. Finally, the authors used Rab13 siRNA and developed Rab13 CRISPR knockout cells to determine a functional role for Rab13 in tent pole ruffles and macropinocytosis in macrophages. LPS-induced membrane ruffling was attenuated and specifically the production of large macropinosomes was impaired in the absence of Rab13. Most compelling was the impact of Rab13 knockdown on tent pole ruffle formation and dynamics, which was assessed using LLSM. Tent poles were still present but the elaboration of F-actin veils between them was reduced. Furthermore, LPS-induced ruffle circulation and twisting of the tent poles was attenuated in Rab13 knockout macrophages. This comprehensive set of experiments showed that Rab13 was necessary for tent pole-veil elaboration and tent pole dynamics that are causal steps in large macropinosome formation in LPS-stimulated macrophages.
As demonstrated by the striking LLSM videos in Condon et al. (7), a large surface area of plasma membrane must be devoted to the formation of a large macropinosome. The authors speculate that tent poles may serve as structural props to erect these elaborate membrane structures. This is consistent with membrane ruffle hotspots on macrophages, which may reflect sites where core structures of tent poles are maintained or recycled. How tent poles interact and spatially coordinate expansion and constriction of the ruffle body remains unclear, and the data presented by Condon et al. (7) offer some directions to be further investigated. Ultrastructurally, are the tent poles bona fide filopodia, and what regulates their formation? How are the lipids and F-actin in tent poles coordinated with the broad ruffle sheets they support? Tent poles may define the diffusion barrier believed to aggregate signaling complexes within the membrane ruffle (5), which is a tantalizing prospect for future investigation. Finally, what drives the upward expansion and downward collapse of the membrane veils between the tent poles? Evidence by Condon et al. (7) suggests that Rab13 is a major player for the interactions of tent poles with the membrane veils and for ruffle circularization. Identification of other key players involved in these processes will allow loss-of-function analysis to tease out the precise contributions of tent poles in erecting and directing ruffle morphogenesis and macropinosome formation.
The newly described tent pole ruffles by Condon et al. (7) has provided a fascinating new perspective in the field of macropinocytosis. Additionally, these findings have opened up important questions and highlighted areas in need of more investigation. Macrophage dorsal ruffles have largely been observed using epifluorescence and confocal imaging in growth factor–stimulated macrophages (3, 5) and whether the absence of tent pole structures in these studies are due to stimuli differences or imaging modality remains to be determined. Finally, the advent of LLSM technology will help deduce at high spatiotemporal resolution the relative roles of known players in membrane ruffle formation, such as Rac1 and phosphoinositide 3-kinase (1, 10), and their potential contributions in tent pole–directed macropinosome formation in macrophages.
Acknowledgments
We thank Drs. Nicolas Touret and Mauricio Terebiznik for scientific discussions and critical reading of the manuscript.
R.E. Harrison is supported by a Natural Science and Engineering Research Council grant (RGPIN 298538-09).
The authors declare no competing financial interests.
References
- 1.Araki N., et al. 1996. J. Cell Biol. 135:1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levin R., et al. 2015. Biochim. Biophys. Acta. 1851:805–823. [DOI] [PubMed] [Google Scholar]
- 3.Swanson J.A. 2008. Nat. Rev. Mol. Cell Biol. 9:639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swanson J.A., and Watts C.. 1995. Trends Cell Biol. 5:424–428. [DOI] [PubMed] [Google Scholar]
- 5.Welliver T.P., et al. 2011. J. Cell Sci. 124:4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen B.C., et al. 2014. Science. 346:1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condon N.D., et al. J. Cell Biol. 2018 doi: 10.1083/jcb.201804137. [DOI] [Google Scholar]
- 8.Tamada A., et al. 2010. J. Cell Biol. 188:429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarsch I.K., et al. 2016. J. Cell Biol. 214:375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nobes C., and Marsh M.. 2000. Curr. Biol. 10:R739–R741. [DOI] [PubMed] [Google Scholar]