Condon et al. use lattice light-sheet microscopy to analyze live macrophages and define a new model of macropinosome formation and closure through tent pole ruffles. The ruffles, which are enhanced by LPS and regulated by Rab13, are erected and supported by F-actin tent poles that cross over and twist to constrict the forming macropinosomes.
Abstract
Pathogen-mediated activation of macrophages arms innate immune responses that include enhanced surface ruffling and macropinocytosis for environmental sampling and receptor internalization and signaling. Activation of macrophages with bacterial lipopolysaccharide (LPS) generates prominent dorsal ruffles, which are precursors for macropinosomes. Very rapid, high-resolution imaging of live macrophages with lattice light sheet microscopy (LLSM) reveals new features and actions of dorsal ruffles, which redefine the process of macropinosome formation and closure. We offer a new model in which ruffles are erected and supported by F-actin tent poles that cross over and twist to constrict the forming macropinosomes. This process allows for formation of large macropinosomes induced by LPS. We further describe the enrichment of active Rab13 on tent pole ruffles and show that CRISPR deletion of Rab13 results in aberrant tent pole ruffles and blocks the formation of large LPS-induced macropinosomes. Based on the exquisite temporal and spatial resolution of LLSM, we can redefine the ruffling and macropinosome processes that underpin innate immune responses.
Introduction
The macrophage cell surface is distinguished by its highly dynamic protrusions, constant movement, and its high rate of membrane turnover, all of which serve the role of macrophages as innate immune sentinels (Stow and Condon, 2016). Surface ruffling is a prevalent feature of immune cells and can give rise to macropinosomes. According to current dogma, linear ruffles transiently appear and then circularize (closing a C shape) to form the macropinocytic cup (Swanson, 2008); but how this open cup forms a closed macropinosome is yet to be fully discerned. Other cell types have much larger, more stable, so-called circular dorsal ruffles (Orth and McNiven, 2006). Macrophages ruffle and macropinocytose constitutively, but growth factors like colony-stimulating factor and contact with pathogens or activation of Toll-like receptors enhance ruffling and macropinocytosis (Patel and Harrison, 2008; Swanson, 2008; Canton et al., 2016; Wall et al., 2017). In these and other cells, macropinocytosis also supports cell growth and mTORC1 signaling through the uptake of macromolecules and amino acids, respectively (Welliver and Swanson, 2012; Commisso et al., 2013; Yoshida et al., 2015). Dorsal ruffles and the macropinosomes they generate are key receptor signaling domains, including for Toll-like receptors (Kagan et al., 2008; Luo et al., 2014). Dorsal ruffles are formed through the actions of actin polymerizing proteins, and they become enriched in membrane phospholipids, lipid kinases, signaling kinases, and small G proteins that support these as specialized sites for signaling (Swanson, 2008; Yoshida et al., 2009; Bohdanowicz et al., 2013).
Several Rab GTPases, including Rabs 8, 34, and 5 are associated with surface ruffles or macropinosomes (Sun et al., 2003; Schnatwinkel et al., 2004; Wall et al., 2017). Herein we also introduce Rab13 as a Rab functioning in this milieu. Rab13 has afore noted roles in post-Golgi trafficking (Nokes et al., 2008), Glut4-containing vesicle exocytosis (Sun et al., 2016), and epithelial junction regulation (Köhler et al., 2004), and it is found at the leading edge of cancer cells where it promotes migration and metastasis (Ioannou et al., 2015). The imaging performed in the current study serves to reveal a specific role for Rab13 in the assembly of large LPS-induced ruffles that give rise to enlarged macropinosomes in activated macrophages.
The formation of ruffles and macropinosomes has been studied using multiple imaging modalities, and previous studies have described ruffle formation, macropinosome closure, and subsequent scission from the plasma membrane (Welliver and Swanson, 2012; Levin et al., 2015). However, these events occur on a scale of minutes (Welliver and Swanson, 2012; Wall et al., 2017) and take place in a 3D space spanning micrometers in the x, y, and z dimensions, with protein transition events occurring at sub-minute intervals. In general, most imaging modalities lack the integrated temporal and spatial resolution to adequately capture these events, leaving open questions about the nature of how ruffles generate macropinosomes. In this study, we used lattice light sheet microscopy (LLSM) for live imaging of the macrophage surface, in 3D, with speeds and resolutions that have not previously been possible. This has produced new insights into ruffling and macropinosome formation, revealing new structural features of the ruffles and a new mode of ruffle closure that redefine this essential internalization pathway.
Results and discussion
A new type of dorsal ruffle formation revealed by LLSM
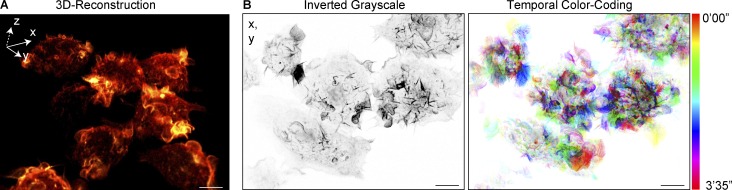
The surface of LPS activated macrophages is replete with prominent and highly dynamic F-actin–rich dorsal ruffles, which can be optimally viewed through the high-speed/high-resolution imaging generated by LLSM. RAW macrophage cell lines were stably transfected with GFP-LifeAct, and using LLSM, we could, for the first time, image large fields of view with multiple activated cells and at great temporal and spatial resolution. The video in Fig. 1 A (Video 1) shows the complete cell volume captured every 1.4 s, depicting the constant ruffling of the macrophage surface. A second example (Fig. 1 B), displays ruffles in a single frame of a time series (Video 2), and with temporal color coding, a series of 75 frames (∼3.5 min) illustrates the highly dynamic and transient nature of the ruffles themselves. Thus, this imaging highlights the constant motion of the macrophage cell surface and the highly dynamic nature of the dorsal ruffles themselves, which is important contextual information for events occurring on these membrane domains, such as fluid uptake, as well as receptor clustering, signaling, and endocytosis (Stow and Condon, 2016).
Figure 1.
LLSM of macrophage ruffling. RAW 264.7 macrophages stably expressing GFP-LifeAct and incubated in medium containing LPS were imaged using LLSM. (A) Single frame from LLSM recording (Video 1) visualized in 3D by tilting the field of view (FOV) to visualize the top and side of the cell. GFP-LifeAct fluorescence was visualized using the Orange-Hot Look Up Table (LUT) from the Amira data visualization software. (B) A second recording is depicted as an inverted grayscale MIP (Video 2) and then temporal color coding of 75 frames (1’45”) is illustrated in the right-hand panel. Bars, 10 µm.
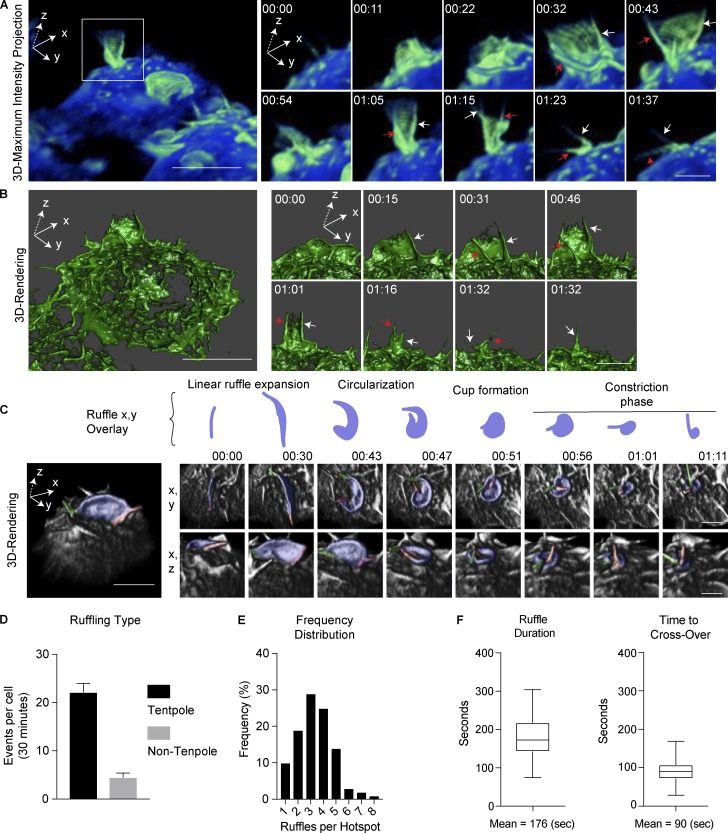
Previous studies have comprehensively mapped the formation of C-shaped dorsal ruffles and macropinocytic cups on macrophages and other cells (Swanson, 2008). However, at high spatial and temporal resolution with LLSM, a new type of dorsal ruffle emerges, and this is displayed in 3D volumetric data (Fig. 2 A and Video 3). Two prominent filopodial-like extensions (red and white arrows, Fig. 2 A) erected on the cell surface at the outer edges of the ruffle act as “tent poles” to raise up the large, F-actin sheet—the veil of the ruffle—between them. In Video 3, these tent poles can be seen dramatically crossing over in a twisting action. 3D surface rendering displays the ruffle structure and the dynamic movements of the tent pole pair within the sheet (red and white arrows, Fig. 2 B and Video 4). In this sequence, the ruffle is clearly seen constricting as it sinks into the cell or collapses, commensurate with the anticipated formation of a macropinosome. A further 3D-rendered cell shows the crossing over of the tent poles accompanying circularization of the membrane ruffle (Fig. 2 C and Video 5). The color overlays on these images depict typical tent poles functioning as pairs, with one drawn away from the other, stringing the ruffle between them. Then, the second pole circles back, conspicuously crossing over the first until the bases come together, and the tent poles twist together, ratcheting in the ruffle veil as a spiral as it simultaneously sinks into the cell (Fig. 2 C and Video 5). The top-down view shows the tent poles (red and green) twisting together in a clockwise fashion, coinciding with constriction of the ruffle aperture, followed by presumptive closure of a nascent macropinosome. Thus, the F-actin tent poles are a major new and integral feature of the ruffles. The tent poles are dynamic and difficult to visualize as part of the ruffles using other forms of live cell imaging, which is why they may not have been described previously. They have the outward appearance of filopodia, and whether they differ at a molecular level from other forms of filopodia on the cell surface is not yet known.
Figure 2.
LLSM imaging reveals tent pole ruffles. Macrophages stably expressing GFP-LifeAct incubated in medium containing LPS were imaged using LLSM. (A) Single frame from LLSM recording (Video 3) visualized by tilting the FOV on the x axis and using the Amira Blue-Green LUT. Boxed region shows 11-s intervals illustrating a full ruffling event. Tent poles at the ruffle margins are marked with red and white arrows. (B) A second example using a 3D-surface render of GFP-LifeAct (Video 4) to show details of tent poles within the F-actin sheath. Poles indicated by the red and white arrows switch sides during crossover. (C) 3D rendering of x,y and x,z ruffle event (Video 5). The marginal tent poles are overlaid with red and green. The ruffle veil is overlaid blue, and in the upper panel, it is extracted from the x,y plane to illustrate the linear ruffle circularization and constriction phases. Bars: 10 µm (main panels); 5 µm (inset panels). Time stamps, min:s. (D) Quantification of ruffle type from Videos 1 and 2. Data are represented as mean ± SEM; n = 6 cells with a total of 133 tent pole–associated and 27 non–tent pole–associated ruffles. (E) Formation of ruffles in the same location (hotspots) from a 30-min period in Videos 1 and 2, represented as a frequency distribution. Frequency distribution is calculated as a percentage from n = 10 cells, containing 104 hotspots and 345 ruffles. (F) Quantification of mean ruffle duration and mean time to crossover of tent poles. Data are represented as a min/max box whisker plot calculated using n = 115 events.
The movement and twisting of the tent poles implies the need for actin-based motors at the tent pole bases for both linear and rotational motion. Potential myosin motors, such as motors of the myosin I, II, and V subfamilies are found in dorsal ruffles (Edgar and Bennett, 1997). An interesting precedent for rotational movement is the myosin V–driven rotation at the base of filopodia, which happens in a circular motion and in the direction of a right handed screw (Tamada et al., 2010; Zidovska and Sackmann, 2011). Finally, the tent poles move to circumscribe the outer limits of the forming macropinocytic cup, and this could form the proposed diffusion barrier, which segregates and concentrates signaling components within the ruffle (Welliver et al., 2011).
Quantification based on extended videos (e.g., Videos 1 and 2) show that the majority (83%) of the ruffles appearing are associated with tent pole ruffles during their lifetimes, and the other 17% includes a variety of small and petal-shaped ruffles that are not further defined here (Fig. 2 D). It is notable that tent pole ruffles keep reforming in “hotspots” on the cell surface (Fig. 2 E), with examples even showing apparent reuse of tent poles to form successive ruffles, suggesting that there may be a sub-cortical concentration of machinery or membrane delivery that regenerates tent poles and ruffling. The tent pole ruffles are very dynamic and short-lived with a mean duration of 176 s, and 90 s of this time precedes the tent pole crossover that demarks the cup formation (Fig. 2 F). These attributes highlight the more rapid, transient nature of the tent pole ruffles and their different behavior, compared with prototypical circular dorsal ruffles on other cell types that can last up to 30 min (Krueger et al., 2003). Finally, while LLSM imaging of tent pole ruffles has been done largely on RAW macrophages, we have also observed tent pole ruffles on other cell types, including cancer cells (Fig. S1 A), showing that this ruffling mechanism is not limited to immune cells.
Formation of macropinosomes from tent pole ruffles
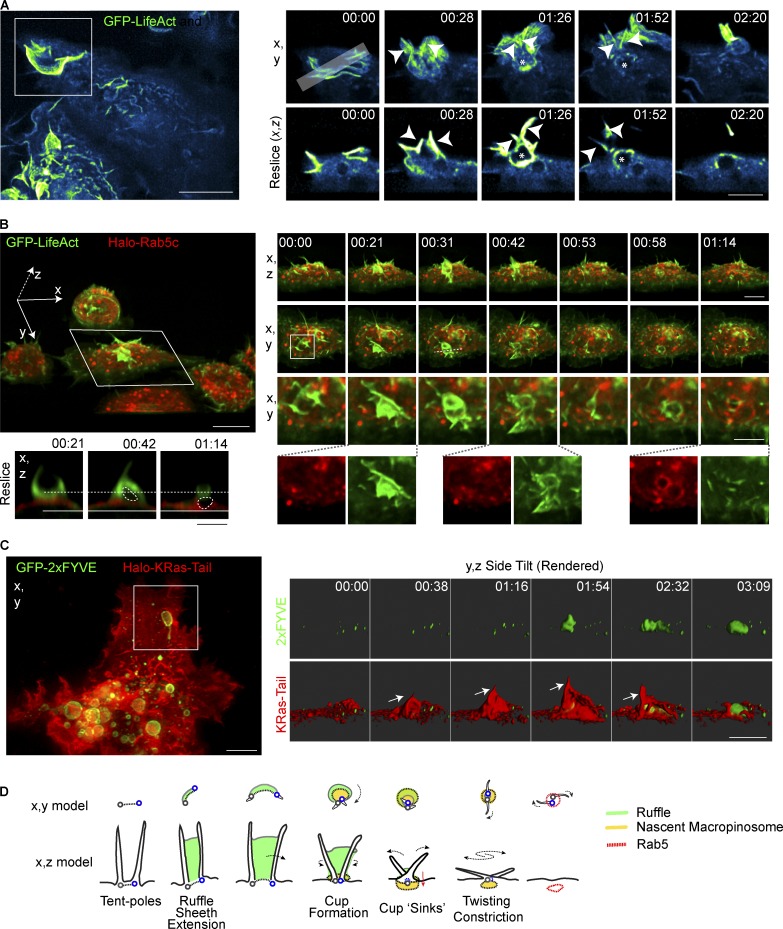
We next gathered evidence to prove that tent pole ruffles do indeed go on to form macropinosomes as they collapse. In imaging of ruffling events, an empty void appears in the subcortical space under GFP-LifeAct–labeled tent pole ruffles at the point of tent pole crossover (Fig. 3 A and Video 6). In Fig. 3 A, there is an existing void (a presumptive macropinosome) and a new void forming at the base of a tent pole ruffle (asterisk). These voids can also be seen forming beneath tent pole ruffles labeled simultaneously for F-actin and a membrane marker, Halo-KRas-Tail (Fig. S1 B and Video 7). The fate of these voids was then confirmed by imaging cells coexpressing GFP-LifeAct and Halo-Rab5c as an early macropinosome marker (Fig. 3 B and Video 8). Viewed from the top and the side, F-actin–rich dorsal ruffles form, twist, and collapse, followed by the recruitment of Halo-Rab5c on the underlying macropinosome. In key frames we show that the void underneath the F-actin ruffle appears before Rab5c labeling and it sinks down into the cytoplasm (Fig. 3 B; reslice). The final frames of the video then show that F-actin depolymerizes as the early macropinosomes acquire Rab5c, in accord with current knowledge (Schnatwinkel et al., 2004). A second example of a tent pole ruffle converting to a macropinosome is shown in cells expressing Halo-KRas-Tail and GFP-2×FYVE as marker of the early macropinosome phosphoinositide and a Rab5 product (Fig. 3 C and Video 9). Thus, the tent pole dorsal ruffles do indeed form macropinosomes from the membrane sheet that begins to sink at the same time as the tent poles begin to cross over and then twist to help close the macropinosome. This completes the tent pole ruffle-to-macropinosome life cycle, which is depicted as a model in Fig. 3 D. The model highlights that the action of the tent poles produces a single tight aperture for final sealing, in contrast to the current model for cup formation and closure, which entails sealing at multiple points or extended areas around the ruffle/cup margins (Swanson, 2008). While the nature of the sealing is still a matter of conjecture, this new model reduces this action to virtually a single point on the plasma membrane.
Figure 3.
Tent pole ruffles collapse to form macropinosomes. (A) Macrophages stably expressing GFP-LifeAct. Cells were treated with 100 ng/ml LPS and imaged using LLSM. GFP-LifeAct is represented using the Cyan Hot LUT from ImageJ. Key frames from Video 6. MIP (x,y) and an x,z resliced through the ruffling event is indicated by the gray bar in the top left inset frame. Arrowheads indicate the position of tent poles. Asterisks denotes the macropinosomal void. (B) Macrophages stably expressing GFP-LifeAct and Halo-Rab5c imaged in medium containing LPS. Cells were imaged using LLSM and Halo-tagged proteins were labeled with 10nM JF549. Left large panel shows whole FOV tilted on the x axis from Videos 6 and 7. Small panels show x,z and x,y of boxed cell with second inset box showing one tent pole ruffling event. Reslice of key frames shows the void of the presumptive macropinosome indicated on the merged image with the dotted circle. Separation of the void from the plasma membrane cannot be verified in these images. (C) LLSM of RAW 264.7 macrophages transiently expressing GFP-2×FYVE and Halo-KRas-Tail. Cells were stained with 10 nM JF549 and imaged in media containing LPS. Inset frames were rendered using Imaris and visualized on an x,z side-tilt. Arrow indicates tent pole position in ruffle. (D) Model of macropinosome formation incorporating tent pole crossover and twisting during the constriction phase. Green indicates the ruffle veil, the nascent macropinosome in yellow. The base of each pole is marked as a gray and blue circle. Bars: 10 µm (main panels); 5 µm (inset panels). Time stamps, min:s.
To further dissect the model of tent pole ruffles, it will now be important to make use of LLSM to reexamine, at high resolution, the well-described perturbations in ruffling and macropinocytosis induced by inhibition or genetic deletion of key regulators. These include the PI3K activity needed for macropinosome completion and closure (Araki et al., 1996) and the actions of phosphatidic acid (Bohdanowicz et al., 2013), ARF6 (Radhakrishna et al., 1999), Rac1 (Nobes and Marsh, 2000), SNX1 and SNX5 (Bryant et al., 2007; Lim et al., 2012), and F-actin assembly proteins in ruffle formation.
Identification of Rab13 as a ruffle-associated small GTPase in activated macrophages
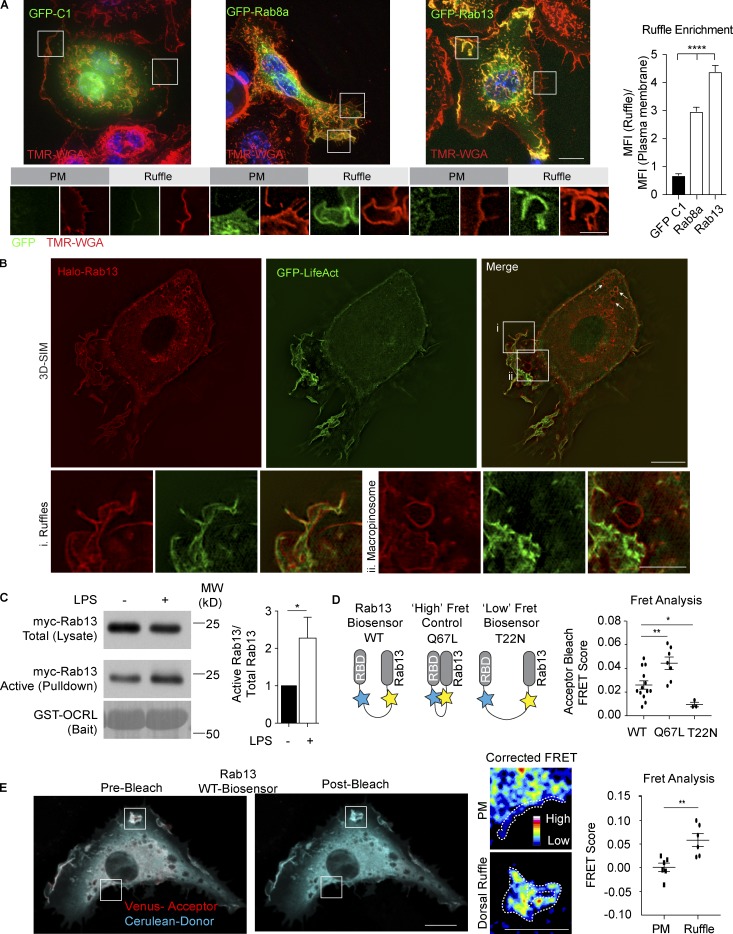
We next set out to identify molecules that define the tent pole ruffle structures on LPS-activated macrophages. To do so, we turned to Rab GTPases, which act as switches at various stages of ruffles and macropinosomes. Since dorsal ruffles are enriched in negatively charged lipids, which may contribute to membrane recruitment (Heo et al., 2006), our initial screen ranked Rabs by C-terminal charge, and a selection of Rabs were expressed in macrophages as GFP-fusion proteins to measure ruffle localization and enrichment. GFP-Rab13 emerged with the most pronounced enrichment in dorsal ruffles, compared with a panel of other Rabs, including Rab8a, which is transiently in macrophage ruffles (Luo et al., 2014; Wall et al., 2017; Fig. 4 A and Fig. S3 A). Although Rab13 has not previously been characterized in macrophages, its known attributes befit the context of ruffles, including its close relationship as a GTPase to Rab8 (Ioannou and McPherson, 2016) and its role in migration at the leading edge of cancer cells (Sun et al., 2010; Ioannou et al., 2015).
Figure 4.
Identification of active Rab13 in LPS-induced dorsal ruffles. (A) Representative images of macrophages transiently transfected with GFP-C1, GFP-Rab8a, and GFP-Rab13. Cell surfaces were stained using TMR-WGA (red) and cells counter stained with DAPI (blue). 3D stacks were taken using the DeltaVision deconvolution microscope. Inset images show examples of plasma membrane and ruffle regions used to produce the quantification shown for the relative ruffle enrichment by ratio of normalized mean fluorescence intensity (MFI) for both the ruffle and resting regions. Quantification of ruffle enrichment is displayed as mean ± SEM from n = 20 cells per Rab over three independent experiments. Each cell had five independent measurements for ruffles and for plasma membrane regions. (B) 3D-SIM of macrophages stably expressing Halo-Rab13 (red) and GFP-LifeAct (green). Cells were fixed before counterstaining with Halo-Ligand JF549 (10 nM), Rab13 localizes to ruffles (B i) and to peripheral macropinosome-like structures (B ii). (C) Rab13 activation assay. Cell lysates of myc-Rab13 expressing macrophages were analyzed for relative levels of active Rab13. Cells were untreated or treated for 30 min with 100 ng/ml LPS. Western blot is representative of three independent experiments. GST-OCRL (RBD) pulldowns were probed with anti-myc to detect active Rab13. Ponceau stain shows amount of GST-OCRL used as bait. Input cell lysate was run on separate lanes to show total Rab13. Quantification of Western blots are from three individual experiments and displayed as mean ± SEM; *, P < 0.05, where the data are normalized to nonstimulated GTP-Rab13 levels. (D) Schematics of Rab13 FRET biosensors and FRET measurements of whole cells expressing WT, dominant negative (T22N), and constitutively active (Q67L) Rab13 biosensors transiently expressed in macrophages. FRET acceptor bleaching score was calculated on n = 13, 7, and 3 for WT, Q67L, and T22N, respectively, from three independent transfections. Graphs are represented as mean ± SEM. (E) Macrophages transiently expressing the WT Rab13 FRET biosensor were imaged using a Zeiss 710 LSM confocal microscope. Boxed areas within bleached regions highlight ruffles and nonruffle plasma membrane. Within the boxed areas, dotted lines indicate example regions of interest used to calculate FRET scores. Data presented as mean ± SEM; n = 6 cells. Test for statistical significance was calculated using unpaired t tests, where *, P < 0.05 and **, P < 0.01. Bars: 10 µm (main panels); 5 µm (inset panels).
More detailed localization of Halo-Rab13 in activated macrophages was performed by 3D–structured illumination microscopy (3D-SIM) that revealed the presence of Halo-Rab13 on intracellular vesicular membranes and on the plasma membrane, where it is concentrated on F-actin–rich ruffles (Fig. 4 B, i). Perinuclear localization of Halo-Rab13 is consistent with previous reports of Rab13 acting in post-Golgi trafficking (Nokes et al., 2008). Halo-Rab13 labeling appears on F-actin labeled endosomes in the cell periphery (arrows, Fig. 4 B) and also on early macropinosome-like structures, which are mostly devoid of F-actin (Fig. 4 B ii). Recruitment of Rab13 was assessed during macropinocytosis by live cell imaging of mCherry-Rab13, coexpressed with macropinosome-associated Rab5a or Rab8a (Lanzetti et al., 2004; Schnatwinkel et al., 2004; Wall et al., 2017). In live cells, mCherry-Rab13 labeling is located on ruffles and macropinosomes, gradually diminishing as the macropinosomes mature and acquire GFP-Rab5a (Fig. S1 C). Rab8a is typically recruited to ruffles that transition to early macropinosomes (Wall et al., 2017), and we find Rab13 on ruffles overlapping with Rab8a during the transition to a Rab5a-positive macropinosome (Fig. S1 D). Thus, in fixed and live cells, Rab13 is prominent on large dorsal ruffles and on early-stage macropinosomal membranes in LPS-treated macrophages.
Stimulation of macrophages with LPS enhances ruffling and macropinocytosis (Patel and Harrison, 2008; Zanoni et al., 2011), and accordingly, it increases the number of Halo-Rab13–labeled ruffles on activated cells. To test whether LPS activates the GTPase Rab13, we used an activation assay based on capture of GTP-Rab13 by a Rab-binding domain (RBD) from the Inositol polyphosphate 5-phosphatase OCRL1, fused to GST (Wall et al., 2017). While there is a basal level of active (GTP-bound) Rab13 in macrophages, this was increased (nearly doubled) after LPS treatment, indicating that Rab13 nucleotide loading is part of the Toll-like receptor 4 (TLR4)–driven response in macrophages (Fig. 4 C). Rab13 associates with membranes in both a GDP- and GTP-bound state (Ioannou et al., 2015, 2016). To determine where Rab13 is most active after LPS stimulation we made use of our previously described, Rab13 FRET-based biosensor, consisting of full-length WT Rab13 coupled to the RBD of MICAL-L2 (Ioannou et al., 2015). Independent biosensors containing Rab13 T22N (inactive mutant, GDP-bound) and Q67L (active mutant, GTP bound) were used to demonstrate the dynamic range between inactive and active states (Fig. 4 D). FRET measurements calculated for both dorsal ruffles and other plasma membrane regions in LPS-treated cells reveal a significant elevation in Rab13 activity at dorsal ruffles (Fig. 4 E). Collectively, these results show that LPS activates Rab13 and enhances the accumulation of active GTP-loaded Rab13 in macrophage dorsal ruffles. This complements previous use of these biosensors, which reveal that in breast cancer cells, active Rab13 accumulates on membrane protrusions at the leading edge of migrating cells (Ioannou et al., 2015).
Rab13 regulates ruffling and formation of LPS-induced macropinosomes
To determine whether Rab13 has functional roles in ruffling and macropinocytosis, we established imaged-based quantitative assays for these functions. Dorsal ruffling was quantified as F-actin protrusions on the dorsal surface relative to dorsal and total cell areas of a large number of fixed cells in an analysis that revealed a 1.3-fold, LPS-induced increase in ruffling (Fig. S2, A and B). To demonstrate how this ruffling affects downstream macropinocytosis, the fluid phase tracer Alexa Fluor 555–Dextran was used to measure uptake (Fig. S2 C). Quantification of individual macropinosomes revealed a doubling of macropinocytic uptake over a 10-min period after addition of LPS (Fig. S2 D), representing enhanced capacity for membrane recycling and environmental sampling in activated macrophages. Macropinosome size is also notably increased in LPS treated cells, which have a higher proportion of large (designated as >1.3 µm2) versus small (<1.3 µm2) macropinosomes (Fig. S2 E). This increase in the number of large macropinosomes almost exclusively accounts for the overall increase in macropinocytic uptake, as the number of small macropinosomes does not change after LPS treatment (Fig. S2 E) and the number of cells with large macropinosomes increases (Fig. S2 F). The LPS-induced population of large macropinosomes can also be seen in primary bone marrow–derived macrophages (BMM) labeled with dextran (Fig. S2 G). These enlarged organelles are in line with similar enlarged macropinosomes that can be induced by growth factor stimulation of macrophages and that are distinct from smaller, calcium-dependent, constitutive macropinosomes (Canton et al., 2016).
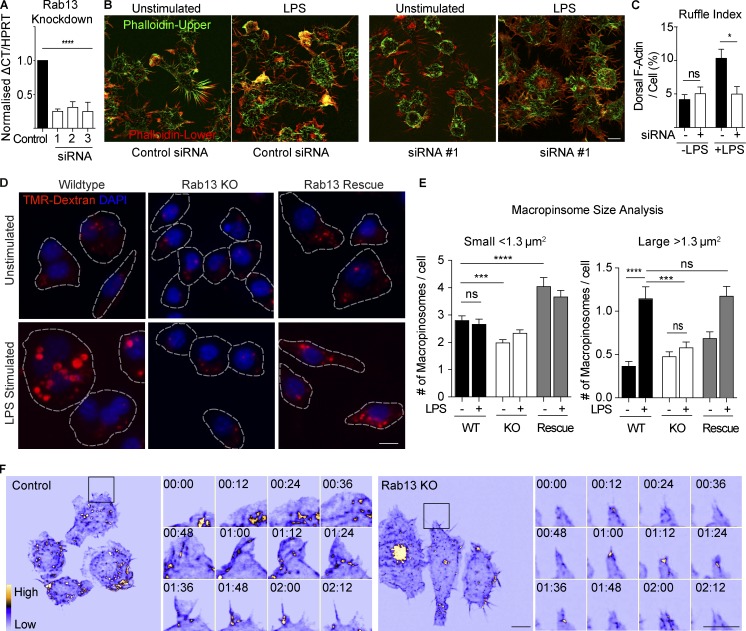
Ruffling assays were performed on siRNA treated cells where Rab13 was knocked down (by ∼75%) using three independent siRNAs (Fig. 5 A). In control cells, we observe a doubling in dorsal F-actin projections after LPS treatment, but in Rab13-depleted cells there is no LPS-induced increase in dorsal ruffles and little to no change in ruffling in untreated cells (Fig. 5, B and C). These results indicate a role for Rab13 in the formation of LPS-induced dorsal ruffles. To analyze the downstream effect on macropinocytosis, we generated CRISPR-mediated Rab13 knockout (KO; >90% reduction in Rab13) and rescue (reexpression of myc-tagged Rab13 in KO line) cell lines (Fig. S3, B–D). Before and after activation with LPS, cell lines were tested using dextran uptake into the previously designated populations of small (<1.3 µm2) versus large (>1.3 µm2) macropinosomes (Fig. 5, D and E). Again, we did not see any increase in the total number of small macropinosomes formed after LPS stimulation in control cells. The most dramatic effect is that abolition of the LPS-induced increase in large macropinosomes in Rab13 KO cells (Fig. 5 E). In the rescue cell lines, the number of large macropinosomes returns to control levels, attributing this effect specifically to Rab13. When calculated as total fluid uptake, the data show that LPS doubles fluid uptake by the cells, and this capacity is driven by Rab13-mediated large macropinosomes (Fig. S3 E). In Rab13 KO cells, there was also a smaller effect on constitutive macropinosomes and a small increase in the rescue cell lines, which is most likely due to some cells overexpressing Rab13 (Fig. S3, C and D). In these CRISPR/Cas9-mediated KO cell lines the marked and predominant loss of the LPS-induced large macropinosomes suggests an essential and selective role for Rab13 in the formation of ruffles and large LPS-induced macropinosomes.
Figure 5.
Rab13 regulates dorsal ruffle formation and macropinocytosis in LPS activated macrophages. (A) Real-time quantitative PCR (qRT-PCR) from Rab13 siRNA treated cells using three independent siRNA duplexes. Data are normalized to HPRT and represented as mean ± SEM; n = 3 individual experiments; ****, P < 0.0001. (B and C) Ruffle index assay described in the Materials and methods to quantify ruffling on control and Rab13 siRNA–treated macrophages with and without LPS for 30 min. F-Actin was stained with Alexa Fluor 488–phalloidin and 3D z stacks imaged using a DeltaVision deconvolution microscope. Threshold images of each channel were used for area measurements to generate the Ruffle Index. Data presented as mean ± SEM of ≥10 cells in multiple experiments. *, P < 0.05. (D) Macropinocytosis assay. CRISPR cell lines (WT, Rab13 KO, and Rab13 Rescue) were pretreated with or without 100 ng/ml LPS for 15 min before a 15-min incubation with Alexa Fluor 555–dextran (100 µg/ml). Cell membranes were stained with Alexa Fluor 488–WGA (not shown) to segment cells (dotted lines indicate cell borders), and nuclei were labeled with DAPI after fixation. (E) Macropinosome size analysis. Numbers of small (<1.3 µm2) versus large (>1.3 µm2) macropinosomes were calculated before and after LPS stimulation in each cell line. Data are displayed as mean ± SEM with n ≥ 400 macropinosomes per group. Tests for statistical significance were calculated using unpaired t tests. (F) Control and Rab13 KO cell lines stably expressing GFP-LifeAct were imaged by LLSM. Example frames are displayed as an MIP using the ICA LUT in ImageJ (Video 10). Inset region illustrates a full tent pole ruffling event for control and Rab13 KO cells displayed every 12 s. Tests for statistical significance was calculated using unpaired t tests. Bars, 10 µm. Time stamps, min:s.
We next used LLSM imaging of both siRNA-depleted and CRISPR KO cells, each expressing GFP-LifeAct to observe real time changes in ruffling induced by loss of Rab13. Loss of Rab13 did not block ruffling per se, but significant changes to ruffling behavior were evident (Fig. 5 F and Video 10). Whereas in activated control cells, there is typical tent pole ruffling (Fig. 5 F, inset panels), in Rab13 KO cells ruffling is defective. Specifically, tent poles are still formed on the cell surface but these structures were less likely to erect patent F-actin/membrane veils between them. Some tent poles erected incomplete or smaller veils, but subsequent events like ruffle circularization and tent pole twisting were seen much less often. To quantify these changes, we performed segmentation of the brightest ruffling events and tracked the ruffle center of mass to measure ruffle speed (average displacement/time), their displacement (total distance moved), and ruffle duration (total time; Fig. S3, F and G). In Rab13 depleted cells, a drop in both mean ruffle speed (from 4.4 to <3 µM/s2) and displacement (from 10 to 5 µM/s2) are parameters that are consistent with the formation of smaller and less active ruffles. Rab13-depleted cells still have ruffling and they retain most of their capacity for making constitutive (small) macropinosomes. Collectively, this evidence shows that Rab13 is required, not for tent pole formation, but for subsequent, optimal formation, and patent behavior of the tent pole ruffles, and it implicates these as causal steps in the specific loss of the large LPS-induced macropinosomes seen in Rab13 depleted cells.
Two potential mechanistic roles for Rab13 can be envisioned from these findings: (1) control of F-actin polymerization or branching to form the ruffle veil or (2) control of membrane trafficking to the plasma membrane at the site of ruffle formation, which could deliver membrane into the large ruffle veil. Rab13 has several effectors that could be involved in either of these functions and that indirectly mediate actin polymerization in different settings, including PKA/VASP at tight junctions in epithelial cells (Köhler et al., 2004) and MICAL-L2/actinin4 that supports Glut4 trafficking (Sun et al., 2016) and neurite outgrowth (Sakane et al., 2010). In breast cancer cells, Rab13 is activated by DENND2B in a complex with MICAL-L2 on leading edges for tumor metastasis (Ioannou et al., 2015; Ioannou and McPherson, 2016). Rab35-recruited MICAL-L1 coalesces the roles of Rab13 alongside Rabs 8 and 36 in neurite outgrowth (Kobayashi et al., 2014). In lymphocytes, Mst1 phosphorylates and activates DENND1C, and subsequently Rab13, to regulate integrin trafficking and cell motility (Nishikimi et al., 2014). Interestingly, MyD88-dependent TLR signaling also regulates the Mst1–hippo pathway (Hong et al., 2018; Zhang et al., 2018) and may provide a link to Rab13 activity during TLR4 signaling and possibly macropinocytosis. Thus, we anticipate that Rab13 will function on tent pole ruffles with a known or new effector, in a sequence of Rab GTPases, and the identities of these molecules are yet to be determined.
LLSM can now be further used to determine the universality of tent pole ruffles, beyond the macrophages and cancer cells shown here, where it may well be able to discern functional or cell type adaptions or differences in behavior. Interestingly, LLSM has been used in another study to look at ruffling and macropinosome cup formation in Dictyostelium discoideum (Veltman et al., 2016). In this organism, the macropinocytic cups were formed in a different manner, namely, from an outwardly spreading patch of active Ras, Rac, and PIP3 that finally constricted into a ring. This is distinct from the tent pole ruffles herein and from the circularization of erect linear ruffles described previously in immune cells (Welliver et al., 2011). However, it is notable that Dictyostelium LLSM images shown by Veltman et al. (2016) do appear to have ruffles with tent pole like structures. With the development of machine learning tools and software for big image data, LLSM holds the potential to develop mathematically defined 4D models to statistically interrogate molecular components and functions of tent pole ruffles and macropinosomes.
Materials and methods
Antibodies and reagents
A primary antibody against GAPDH (5174) was purchased from Cell Signaling Technology. Mouse anti-myc IgG was concentrated from a hybridoma supernatant (9E10). Alexa Fluor 488–phalloidin (A12379), wheat germ agglutinin (WGA) 488 (W11261) and tetramethylrhodamine (TMR; W849), DAPI (Dilactate; D3571), and 70,000 MW Oregon Green 488 (D7173) or 10,000 MW Alexa Fluor 555–Dextran (D34679) were purchased from Life Technologies. HRP-conjugated goat anti-mouse and anti-rabbit antibodies (81-6520) were obtained from Zymed Laboratories Inc. LPS purified from Salmonella enterica serotype Minnesota Re 595 was purchased from Sigma-Aldrich and used at 100 ng/ml unless otherwise stated. Gibson Assembly Master Mix was from New England Biolabs (E2611). The GeneArt genomic cleavage detection kit (A24372) was from Invitrogen (Thermo Fisher Scientific). JF549 coupled to Halo-ligand was used to label both live and fixed cells at 10 nM and was provided by L. Lavis (Janelia, Ashburn, VA; Grimm et al., 2015).
Cell culture
All experiments used the RAW 264.7 macrophage-like cell line from ATCC (-TIB71). Unless otherwise stated, cells were maintained and passaged in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% heat-inactivated FCS (Thermo Fisher Scientific) and 2 mM l-glutamine (Invitrogen) and kept at 37°C in humidified 5% CO2. Primary mouse BMMs used for dextran uptake assays, were obtained from femur-derived bone marrow cells from C57BL/6 mice. Cells were ex vivo differentiated for 7 d in RPMI medium supplemented with 10% FBS, 20 U/ml penicillin, 20 µg/ml streptomycin, and 100 ng/ml purified recombinant macrophage colony stimulating factor–1 (11343118; ImmunoTools GmbH).
Plasmids and stable cell lines
pEGFP-C1 vector (Clontech) containing the open reading frames of mouse Rab8a and 13, were gifts from M. Fukuda (Tohoku University, Sendai, Japan). Stable cell lines were generated using the pEF6/V5-His TOPO TA vector (K961020) from Life Technologies, and pEF1α-Halo-(neomycin) was generated by replacing the cytomegalovirus promoter in pHTN HaloTag cytomegalovirus-neo vector (G7721) from Promega with the EF-1α promoter by PCR and restriction ligation. pEF1α-Halo-Rab13 was generated by PCR and ligation reaction using the following primers: Halo-Rab13-EcoRI-F, 5′-GAATTCATGGCCAAAGCCTACGAC-3′; Halo-Rab13-EcoRV-R, 5′-AATTGGGCCCTCAGCCTAACAAGCACTTGT-3′. pEF1α-Halo-K-RasTail and pEF1a-Halo-Rab5c were generated by Gibson assembly using the Gibson assembly kit from NEB and Rab5c inserted by PCR ligation into the EcoRI and NotI sites in the multiple cloning site. Rab5c was amplified using the following primers: forward, 5′-ATATGAATTCATGGCGGGTCGAGGAGGT-3′; reverse, 5′-AATTGCGGCCGCTCAGTTGCTGCAGCACTG-3′. pEF6-myc-Rab13 was generated by restriction digestion of the mouse Rab13 ORF from peGFP-Rab13 using BsrGI and BamHI and replacing the Rab8a ORF from a previously generated pEF6-myc-Rab8a construct (unpublished data). pEF6-GFP-N1 was generated by cloning the GFP ORF from pEGFP-N1 (Clonetech) and inserting into the pEF6 backbone. peGFP-LifeAct and peGFP-2×FYVE were gifts from F. Meunier (University of Queensland, Brisbane, Australia), and GFP-LifeAct was subcloned into pEF6-GFP-N1 by restriction digestion. Cells were transfected with Lipofectamine 2000, and stable cell lines were generated using either G418/neomycin (100 µg/ml) and/or blasticidin (4 µg/ml). Forster resonance energy transfer (FRET) biosensor constructs were provided by P. McPherson and L. Hodgson (Ioannou et al., 2015). Three biosensor constructs consisted of WT Rab13, dominant negative mutant (T22N) Rab13, and constitutively active mutant (Q67L) Rab13, fused at the N-terminus to Venus (acceptor), a linker, mCer3 (donor), then the RBD of MICAL-L2.
siRNA knockdown and CRISPR/Cas9 gene KO
Knockdown of Rab13 by siRNA was performed using the Amaxa nucleofection system and reagent V as per the manufacturer’s instructions (V VCA-1003) with the Nucleofector 2b, program D-032. Stealth RNAi siRNA was from Invitrogen, nontargeting control siRNA was human HDAC1, 5′-GAACUACCCACUGCGAGACGGCAUU-3′; mouse siRNA Rab13 no. 1, 5′-CCGUGACAUCUUGCUCAAGACAGGA-3′; siRNA Rab13 no. 2, 5′-CCAAGAACGAUUCAAGACAAUAACU-3′; and siRNA Rab13 no. 3, 5′-GGCUCGAGAGCACAGAAUCCGAUU-3′. Cells were collected and resuspended in 100 µl Amaxa Reagent V at a density of 2.5 × 107/ml and 6 µl of 20 nM siRNA was used. Cells were seeded onto plates for experimentation the following day at a density of 0.2 × 106 /ml siRNA-treated cells.
CRISPR/Cas9 stable KO cell lines were developed using homology-directed repair as previously described (Wall et al., 2017; Fig. S2). The ∼1,500-bp 5′ (left) and 3′ (right) homology arms flanking the CRISPR targeting site used the following primers: Rab13_LA_F, 5′-TCGAGATTTAATTAAGATACGCGTGACGATACTGCCTTCTGTAAGCCTCTC-3′; Rab13_LA_R, 5′-GAATAGGAACTTCGGTAAAGCTTATCCGATGGGAAGAGCCAACACTGGTA-3′; Rab13_RA_F, 5′-CGGTATATATATATATATGTCGACATGGTTTCAAGTTGCTGCTCATCGGG-3′; Rab13_RA_R, 5′-AGATCTGCGATCGCAATCAATTGCTGGTTAAACTGAAAGTGGGCCACCA-3′. gRNA sequence was designed using the Zhang Lab CRISPR design tool. The gRNA for mouse Rab13 (5′-ACGACCACCTCTTCAAGT-3′) was chosen, and oligos were annealed and ligated into pSpCas9(BB)-2A-GFP (px458) from the Feng Zhang Lab at Massachusetts Institute of Technology (plasmid 48138; Addgene). Co-transfection of each plasmid (5 µg) was performed using Amaxa nucleofection (Lonza), and cells were selected with G418 (100 µg/ml) 24 h after transfection.
Rab13 activation assay
The Rab13 activation assay was previously described (Wall et al., 2017). In brief, a GST fusion protein of the RBD of OCRL (amino acids 539–901) was used to pull down active GTP-loaded Rab13 from a stable cell line expressing myc-tagged Rab13. MicroSpin columns (27-3565-01; GE Healthcare) were used for all of the pulldowns. Beads were washed with ice-cold lysis buffer (20 mM Tris-HCl, pH 7.2, 150 mM NaCl, 5 mM MgCl2, 1% NP-40, and 5% glycerol); cOmplete protease inhibitors (Roche Applied Science) and phosSTOP tablets (Roche Applied Science) were added to lysis buffer before use. Elution was achieved conventionally by boiling in 2× SDS-PAGE sample buffer for 5 min. The samples were subjected to immunoblots. Total and active Rab13 is detected using anti-myc antibody from input and pulldown extracts, respectively.
Cell culture for fluorescence microscopy
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) for antibody staining or labeling. For fixed cell assays, cells were seeded on 1.5 coverslips at 0.2 × 106/ml for next day experiments. For transfection the next day they were seeded at 0.1 × 106/ml. For live cell experiments, transfected cells were seeded onto glass-bottom 35-mm dishes (MatTek), and for LLSM experiments, cells were seeded on 5-mm diameter coverslips (64- 0700; Warner Instruments).
Microscopy
Betzig LLSM (Chen et al., 2014).
Custom built, lattice light sheet microscope by E. Betzig was housed at the Advanced Imaging Center (AIC; Howard Hughes Medical Institute Janelia Research Campus, Ashburn, VA). Excitation was achieved with 488-nm or 560-nm diode lasers (MPB Communications) at 20–50% acousto-optic tunable filter (AOTF) transmittance with 30-mW and 50-mW initial box power (respectively) through an excitation objective (Special Optics 0.65 NA 3.74-mm water-dipping lens) and was detected via a Nikon CFI Apo LWD 25× 1.1 NA water-dipping objective with a 2.5× tube lens. Live cells were imaged in 37°C-heated, water-coupled bath in L15 medium (21083027; Life Technologies) with 5% FBS and Pen/Strep (10 µg/ml) and were acquired with 2× Hamamatsu Orca Flash 4.0 V2 sCMOS cameras in custom written LabView Software. Post-image deskewing and deconvolution was performed using Janelia custom software and 10 iterations of the Richardson-Lucy algorithm.
3i LLSM.
The 3i lattice light sheet microscope was used at the Institute for Molecular Bioscience, The University of Queensland, Brisbane, Australia. Excitation was achieved with a Coherant Sapphire 500-mW, 488-nm laser, or 560-nm diode lasers (MPB Communications) at 2–5% AOTF transmittance through an excitation objective (Special Optics 28.6× 0.7 NA 3.74-mm water-dipping lens) and is detected via a Nikon CFI Apo LWD 25× 1.1 NA water-dipping objective with a 2.5× tube lens. Live cells were imaged in 37°C-heated, water-coupled bath in L15 medium (21083027; Life Technologies) with 5% FBS and Pen/Strep (10 µg/ml) and were acquired with 2× Hamamatsu Orca Flash 4.0 V2 sCMOS cameras in SlideBook. Post-image deskewing and deconvolution was performed using Microvolution Deconvolution software and 20 iterations of the Richardson-Lucy algorithm.
Applied precision personal Deltavision.
Olympus IX80 inverted microscope fitted with an Olympus 60× 1.4 NA Plan Apochromat objective, Lumencor 7 line LED light source, and Photometrics CoolSnap HQ2 camera running SoftWorx. Post image processing was performed using Applied Biosystems internal deconvolution software and 10 cycles of the conservative algorithm.
Lin Shao 3D-SIM (Gustafsson et al., 2008).
Custom Built 3D-SIM microscope using a Zeiss 100× 1.49 NA Plan-Fluar objective, with 488- and 560-nm lasers. Five phase-shifted images were taken per channel/z stack with the pattern generated with an spatial light modulator, and images of cells in L15 medium (21083027; Life Technologies) with 5% FBS and Pen/Strep (10 µg/ml) were acquired using 2× Orca Flash 4.0 V2 sCMOS cameras using custom acquisition software.
Zeiss LSM 710 confocal with external GaAsP.
Zeiss Axiovert stand with a 710 LSM scanhead, with 63× 1.4 C-Apochromat objective, and 458-, 488-, 514-, and 561-nm lasers, fitted with external twin GaAsP detectors, running Zen Black 2012 software. Cells were maintained at 37°C within a Peacon incubation chamber and imaged in CO2-independent medium (18045088; Thermo Fisher Scientific) with 5% FBS and Pen/Strep (10 µg/ml).
Zeiss LSM 880 confocal with Fast AiryScan.
Zeiss Axiovert stand with an 880 LSM scanhead with a 63× 1.4 NA Plan Apochromat objective, and 488-, 561-, and 613-nm lasers with a Fast AiryScan module, running Zen Black 2.3. Cells were imaged in CO2-independent medium (18045088; Thermo Fisher Scientific) with 5% FBS and Pen/Strep (10 µg/ml).
Zeiss Axioimager with Apotome 2.
Upright Zeiss AxioImager microscope with Apotome 2, with 40× 1.3 NA Plan Neofluar objective, coupled with HXP 120-W light source running Zen Blue 2 software.
Nikon TiE inverted wide field.
Custom designed, Nikon TiE inverted wide field system with 60× 1.4 NA Plan Apochromat objective, and Lumencor 7 line LED light source, with high-speed Sutter Instruments filter wheels and is housed in a 37°C OKO laboratories incubation chamber, running NIS Elements AR 4.3. Cells were imaged in CO2-independent medium (18045088; Thermo Fisher Scientific) with 5% FBS and Pen/Strep (10 µg/ml) using a Hamamatsu Orca Flash 4.0 sCMOS. Post-image processing was performed using Microvolution Deconvolution software and 20 cycles of the Richardson-Lucy algorithm.
Post-imaging visualization and analysis software
For LLSM data visualization and analysis, Amira version 4.6, Arivis Vision4D X64 and Imaris X64 version 9.2 software packages were used. For all other confocal and wide field microscopy, post-imaging analysis and visualization was performed using ImageJ (Fiji version 2.0.0-rc-68/1.52e).
Ruffling and macropinocytosis assays
For the ruffle index assay, RAW macrophages were incubated with or without LPS 100 ng/ml for 30 min before fixation. F-Actin was labeled with Alexa Fluor 488–phalloidin (Life Technologies), and nuclei were labeled with DAPI. 3D z stacks were acquired and analyzed using ImageJ software. The nuclear mid-point was selected (using DAPI), and the cell volumes below (cell area–F-actin) and above (dorsal ruffle F-actin) were subjected to a maximum intensity projection (MIP) and separated into red and green channels, respectively (see Fig. S2). Each channel was background subtracted, and a median filter of two pixels was applied. Each channel was then thresholded into a binary image, and area measurements performed using ImageJ. The ruffle index was calculated as the percentage of dorsal F-actin (pixel area)/cell (pixel area).
For the ruffle enrichment assay, RAW cells on glass coverslips were transiently transfected with selected GFP-Rabs and left overnight before LPS treatment and fixation. Plasma membrane was stained with TMR-WGA, and cells were counter stained with DAPI. Z stacks were acquired using the personal DeltaVision microscope. Fluorescence intensities of GFP-Rabs and TMR-WGA were assessed at both ruffles and “resting” regions of the plasma membrane and expressed as ratios. Macropinosomes were labeled during 15-min incubation with Alexa Fluor 555–dextran (100 µg/ml). Cells were fixed and counterstained with 488-WGA and DAPI. Individual macropinosomes >0.2 µm in diameter (Kerr and Teasdale, 2009) were counted, and their areas were measured per cell using ImageJ.
Acceptor photobleaching analysis (FRET)
Acceptor photobleaching to measure the FRET efficiency of the Rab13 biosensors has previously been described (Ioannou et al., 2015). In brief, Rab13 imaging was performed using a Zeiss 710 LSM confocal to acquire five images of the donor (Cerulean3) and acceptor (Venus) channels before bleaching of the acceptor (Venus) channel by ≥50% with the 514-nm laser. Five images were taken before and after a 10-iteration bleaching protocol. Mean pixel gray levels of Cerulean were measured before and after Venus bleaching to calculate the FRET efficiency according to the equation:
qRT-PCR
Quantification of qRT-PCR has been described (Luo et al., 2014). In brief, Rab13 expression levels were analyzed in siRNA-treated and CRISPR/Cas9 KO lines using the following primers: hypoxanthine phosphoribosyl transferase (HPRT), 5′-TCAGGCAGTATAATCCAAAGATGGT-3′ (forward) and 5′-AGTCTGGCTTATATCCAACACTTCG-3′ (reverse); mRab13, 5′-CGAACCGTGGACATAGAGGG-3′ (forward) and 5′-CCGCAGAGGCATTCTCTTTGA-3′ (reverse). RNA was extracted using TRIzol (Life Technologies) as per manufacturer’s protocol. 1 µg of purified RNA was used for cDNA synthesis using Superscript III (Life Technologies) as per manufacturer’s instructions. cDNA was diluted in 200 µl of ultrapure dH2O. PCR was completed using SYBR green (Applied Biosystems). ΔCT was calculated using Microsoft Excel and was quantified using GraphPad Prism6.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software package version 7.0. Data are presented as mean ± SEM unless otherwise stated. Datasets were subject to normality tests (Shapiro-Wilk and Chi Squared) before performing unpaired t test. Significance was calculated at P < 0.05 and in general *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Where indicated in figure legends, before statistical analysis, data were normalized to internal controls using calculations in Excel.
Online supplemental material
Fig. S1 shows (A) LLSM imaging of GFP-LifeAct in MDA-MB-231 cells and (B) GFP-LifeAct and Halo-Kras-Tail in RAW 264.7 cells and wide field imaging of mCherry-Rab13 with GFP-Rab5a and GFP-Rab8a. Fig. S2 contains the ruffle index assay methodology and macropinocytosis assay and quantification from RAW 264.7 macrophages and examples from primary BMMs. Fig. S3 contains the Rab mini-screen for ruffle enrichment, CRISPR-design strategy and quantification and imaging of Rab13 KO and recovery cell lines. Fig. S3 E shows total macropinocytic uptake in these cell lines. Fig. S3 (F and G) contains imaging and quantification of ruffling in control and Rab13 siRNA–treated cells. Videos 1 and 2 show LLSM of macrophage ruffling. Videos 3–5 show LLSM of tent pole ruffles. Video 6 shows LLSM of tent pole–associated ruffle conversion to a macropinosome. Video 7 show dual color LLSM of tent pole ruffle formation (Halo-KRasTail/GFP-LifeAct). Video 8 shows dual color LLSM of tent pole ruffle formation (Halo-Rab5c/GFP-LifeAct). Video 9 shows dual color rendering of LLSM of tent pole ruffle formation (Halo-KRasTail/GFP-2×FYVE). Video 10 shows LLSM of Rab13 CRISPR KO macrophages.
Supplementary Material
Acknowledgments
We thank T. Khromykh and D. Brown for expert technical assistance. Imaging was performed in Institute for Molecular Bioscience’s Australian Cancer Research Foundation–funded Cancer Biology Imaging and Cancer Ultrastructure and Function Facilities. Original LLSM and 3D-SIM imaging was performed at the Advanced Imaging Center (AIC) – Howard Hughes Medical Institute (HHMI) Janelia Research Campus. Our thanks to Eric Betzig for help and advice with imaging and for his interpretation of the tent pole ruffle data, and we thank Lin Shao, AIC for help with 3D-SIM imaging. The AIC is a jointly funded venture of the Gordon and Betty Moore Foundation and the HHMI. We thank Luke Lavis from HHMI for generously providing the Janelia Fluor – Halo Ligands.
Research funding was from the National Health and Medical Research Council of Australia (grants 1098710 and 1003021 to J.L. Stow), Australian Research Council (grants AAW LE170100206 and DP180101910 to J.L. Stow), and National Institutes of Health (grant CA205262 to L. Hodgson). PhD scholarship funding (N.D. Condon) was from the Australian government and Yulgilbar Alzheimer Research Program. P.S. McPherson is a James McGill Professor and is supported by a Foundation Grant from the Canadian Institutes of Health Research. L. Hodgson is an Irma T. Hirschl Career Scientist.
The authors declare no competing financial interests.
Author contributions: N.D. Condon, J.L. Stow, and A.A. Wall conceived the project, designed the experiments, analyzed the data, and wrote the manuscript. N.D. Condon and A.A. Wall performed the imaging and experiments. L. Luo helped with experiments and analysis for Fig. 4 C. P.S. McPherson, M.S. loannou, and L. Hodgson designed and supplied the Rab13 biosensor and contributed to interpretation of data. J.M. Heddleston and T.-L. Chew helped with experimental design, performing original LLSM imaging and post-image processing of data, and interpretation of the imaging data.
References
- Araki N., Johnson M.T., and Swanson J.A.. 1996. A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J. Cell Biol. 135:1249–1260. 10.1083/jcb.135.5.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohdanowicz M., Schlam D., Hermansson M., Rizzuti D., Fairn G.D., Ueyama T., Somerharju P., Du G., and Grinstein S.. 2013. Phosphatidic acid is required for the constitutive ruffling and macropinocytosis of phagocytes. Mol. Biol. Cell. 24:1700–1712: S1–S7. 10.1091/mbc.e12-11-0789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D.M., Kerr M.C., Hammond L.A., Joseph S.R., Mostov K.E., Teasdale R.D., and Stow J.L.. 2007. EGF induces macropinocytosis and SNX1-modulated recycling of E-cadherin. J. Cell Sci. 120:1818–1828. 10.1242/jcs.000653 [DOI] [PubMed] [Google Scholar]
- Canton J., Schlam D., Breuer C., Gütschow M., Glogauer M., and Grinstein S.. 2016. Calcium-sensing receptors signal constitutive macropinocytosis and facilitate the uptake of NOD2 ligands in macrophages. Nat. Commun. 7:11284 10.1038/ncomms11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.C., Legant W.R., Wang K., Shao L., Milkie D.E., Davidson M.W., Janetopoulos C., Wu X.S., Hammer J.A. III, Liu Z., et al. 2014. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 346:1257998 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C., Davidson S.M., Soydaner-Azeloglu R.G., Parker S.J., Kamphorst J.J., Hackett S., Grabocka E., Nofal M., Drebin J.A., Thompson C.B., et al. 2013. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 497:633–637. 10.1038/nature12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar A.J., and Bennett J.P.. 1997. Circular ruffle formation in rat basophilic leukemia cells in response to antigen stimulation. Eur. J. Cell Biol. 73:132–140. [PubMed] [Google Scholar]
- Grimm J.B., English B.P., Chen J., Slaughter J.P., Zhang Z., Revyakin A., Patel R., Macklin J.J., Normanno D., Singer R.H., et al. 2015. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods. 12:244–250: 3: 250. 10.1038/nmeth.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M.G., Shao L., Carlton P.M., Wang C.J., Golubovskaya I.N., Cande W.Z., Agard D.A., and Sedat J.W.. 2008. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94:4957–4970. 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo W.D., Inoue T., Park W.S., Kim M.L., Park B.O., Wandless T.J., and Meyer T.. 2006. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 314:1458–1461. 10.1126/science.1134389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Li X., Zhou D., Geng J., and Chen L.. 2018. Role of Hippo signaling in regulating immunity. Cell. Mol. Immunol. 10.1038/s41423-018-0007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou M.S., and McPherson P.S.. 2016. Regulation of Cancer Cell Behavior by the Small GTPase Rab13. J. Biol. Chem. 291:9929–9937. 10.1074/jbc.R116.715193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou M.S., Bell E.S., Girard M., Chaineau M., Hamlin J.N., Daubaras M., Monast A., Park M., Hodgson L., and McPherson P.S.. 2015. DENND2B activates Rab13 at the leading edge of migrating cells and promotes metastatic behavior. J. Cell Biol. 208:629–648. 10.1083/jcb.201407068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou M.S., Girard M., and McPherson P.S.. 2016. Rab13 Traffics on Vesicles Independent of Prenylation. J. Biol. Chem. 291:10726–10735. 10.1074/jbc.M116.722298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan J.C., Su T., Horng T., Chow A., Akira S., and Medzhitov R.. 2008. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 9:361–368. 10.1038/ni1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr M.C., and Teasdale R.D.. 2009. Defining macropinocytosis. Traffic. 10:364–371 10.1111/J.1600-0854.2009.00878.X. [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Etoh K., Ohbayashi N., and Fukuda M.. 2014. Rab35 promotes the recruitment of Rab8, Rab13 and Rab36 to recycling endosomes through MICAL-L1 during neurite outgrowth. Biol. Open. 3:803–814. 10.1242/bio.20148771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler K., Louvard D., and Zahraoui A.. 2004. Rab13 regulates PKA signaling during tight junction assembly. J. Cell Biol. 165:175–180. 10.1083/jcb.200312118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger E.W., Orth J.D., Cao H., and McNiven M.A.. 2003. A dynamin-cortactin-Arp2/3 complex mediates actin reorganization in growth factor-stimulated cells. Mol. Biol. Cell. 14:1085–1096. 10.1091/mbc.e02-08-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetti L., Palamidessi A., Areces L., Scita G., and Di Fiore P.P.. 2004. Rab5 is a signalling GTPase involved in actin remodelling by receptor tyrosine kinases. Nature. 429:309–314. 10.1038/nature02542 [DOI] [PubMed] [Google Scholar]
- Levin R., Grinstein S., and Schlam D.. 2015. Phosphoinositides in phagocytosis and macropinocytosis. Biochim. Biophys. Acta. 1851:805–823. 10.1016/j.bbalip.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Lim J.P., Teasdale R.D., and Gleeson P.A.. 2012. SNX5 is essential for efficient macropinocytosis and antigen processing in primary macrophages. Biol. Open. 1:904–914. 10.1242/bio.20122204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Wall A.A., Yeo J.C., Condon N.D., Norwood S.J., Schoenwaelder S., Chen K.W., Jackson S., Jenkins B.J., Hartland E.L., et al. 2014. Rab8a interacts directly with PI3Kγ to modulate TLR4-driven PI3K and mTOR signalling. Nat. Commun. 5:4407 10.1038/ncomms5407 [DOI] [PubMed] [Google Scholar]
- Nishikimi A., Ishihara S., Ozawa M., Etoh K., Fukuda M., Kinashi T., and Katagiri K.. 2014. Rab13 acts downstream of the kinase Mst1 to deliver the integrin LFA-1 to the cell surface for lymphocyte trafficking. Sci. Signal. 7:ra72 10.1126/scisignal.2005199 [DOI] [PubMed] [Google Scholar]
- Nobes C., and Marsh M.. 2000. Dendritic cells: new roles for Cdc42 and Rac in antigen uptake? Curr. Biol. 10:R739–R741. 10.1016/S0960-9822(00)00736-3 [DOI] [PubMed] [Google Scholar]
- Nokes R.L., Fields I.C., Collins R.N., and Fölsch H.. 2008. Rab13 regulates membrane trafficking between TGN and recycling endosomes in polarized epithelial cells. J. Cell Biol. 182:845–853. 10.1083/jcb.200802176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., and McNiven M.A.. 2006. Get off my back! Rapid receptor internalization through circular dorsal ruffles. Cancer Res. 66:11094–11096. 10.1158/0008-5472.CAN-06-3397 [DOI] [PubMed] [Google Scholar]
- Patel P.C., and Harrison R.E.. 2008. Membrane ruffles capture C3bi-opsonized particles in activated macrophages. Mol. Biol. Cell. 19:4628–4639. 10.1091/mbc.e08-02-0223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna H., Al-Awar O., Khachikian Z., and Donaldson J.G.. 1999. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J. Cell Sci. 112:855–866. [DOI] [PubMed] [Google Scholar]
- Sakane A., Honda K., and Sasaki T.. 2010. Rab13 regulates neurite outgrowth in PC12 cells through its effector protein, JRAB/MICAL-L2. Mol. Cell. Biol. 30:1077–1087. 10.1128/MCB.01067-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnatwinkel C., Christoforidis S., Lindsay M.R., Uttenweiler-Joseph S., Wilm M., Parton R.G., and Zerial M.. 2004. The Rab5 effector Rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol. 2:E261 10.1371/journal.pbio.0020261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow J.L., and Condon N.D.. 2016. The cell surface environment for pathogen recognition and entry. Clin. Transl. Immunology. 5:e71 10.1038/cti.2016.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P., Yamamoto H., Suetsugu S., Miki H., Takenawa T., and Endo T.. 2003. Small GTPase Rah/Rab34 is associated with membrane ruffles and macropinosomes and promotes macropinosome formation. J. Biol. Chem. 278:4063–4071. 10.1074/jbc.M208699200 [DOI] [PubMed] [Google Scholar]
- Sun Y., Bilan P.J., Liu Z., and Klip A.. 2010. Rab8A and Rab13 are activated by insulin and regulate GLUT4 translocation in muscle cells. Proc. Natl. Acad. Sci. USA. 107:19909–19914. 10.1073/pnas.1009523107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Jaldin-Fincati J., Liu Z., Bilan P.J., and Klip A.. 2016. A complex of Rab13 with MICAL-L2 and α-actinin-4 is essential for insulin-dependent GLUT4 exocytosis. Mol. Biol. Cell. 27:75–89. 10.1091/mbc.e15-05-0319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J.A. 2008. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 9:639–649. 10.1038/nrm2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada A., Kawase S., Murakami F., and Kamiguchi H.. 2010. Autonomous right-screw rotation of growth cone filopodia drives neurite turning. J. Cell Biol. 188:429–441. 10.1083/jcb.200906043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman D.M., Williams T.D., Bloomfield G., Chen B.C., Betzig E., Insall R.H., and Kay R.R.. 2016. A plasma membrane template for macropinocytic cups. eLife. 5:e20085 10.7554/eLife.20085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall A.A., Luo L., Hung Y., Tong S.J., Condon N.D., Blumenthal A., Sweet M.J., and Stow J.L.. 2017. Small GTPase Rab8a-recruited Phosphatidylinositol 3-Kinase γ Regulates Signaling and Cytokine Outputs from Endosomal Toll-like Receptors. J. Biol. Chem. 292:4411–4422. 10.1074/jbc.M116.766337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welliver T.P., and Swanson J.A.. 2012. A growth factor signaling cascade confined to circular ruffles in macrophages. Biol. Open. 1:754–760. 10.1242/bio.20121784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welliver T.P., Chang S.L., Linderman J.J., and Swanson J.A.. 2011. Ruffles limit diffusion in the plasma membrane during macropinosome formation. J. Cell Sci. 124:4106–4114. 10.1242/jcs.091538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Hoppe A.D., Araki N., and Swanson J.A.. 2009. Sequential signaling in plasma-membrane domains during macropinosome formation in macrophages. J. Cell Sci. 122:3250–3261. 10.1242/jcs.053207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Pacitto R., Yao Y., Inoki K., and Swanson J.A.. 2015. Growth factor signaling to mTORC1 by amino acid-laden macropinosomes. J. Cell Biol. 211:159–172. 10.1083/jcb.201504097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni I., Ostuni R., Marek L.R., Barresi S., Barbalat R., Barton G.M., Granucci F., and Kagan J.C.. 2011. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 147:868–880 10.1016/J.Cell.2011.09.051s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang H., and Zhao B.. 2018. Hippo Signaling in the Immune System. Trends Biochem. Sci. 43:77–80. 10.1016/j.tibs.2017.11.009 [DOI] [PubMed] [Google Scholar]
- Zidovska A., and Sackmann E.. 2011. On the mechanical stabilization of filopodia. Biophys. J. 100:1428–1437. 10.1016/j.bpj.2011.01.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.