Morita et al. identify the endoplasmic reticulum multispanning membrane protein TMEM41B as a factor required for autophagosome formation. TMEM41B interacts physically and functionally with VMP1, a structurally related autophagy protein.
Abstract
Macroautophagy is an intracellular degradation process that requires multiple autophagy-related (ATG) genes. In this study, we performed a genome-wide screen using the autophagic flux reporter GFP-LC3-RFP and identified TMEM41B as a novel ATG gene. TMEM41B is a multispanning membrane protein localized in the endoplasmic reticulum (ER). It has a conserved domain also found in vacuole membrane protein 1 (VMP1), another ER multispanning membrane protein essential for autophagy, yeast Tvp38, and the bacterial DedA family of putative half-transporters. Deletion of TMEM41B blocked the formation of autophagosomes at an early step, causing accumulation of ATG proteins and small vesicles but not elongating autophagosome-like structures. Furthermore, lipid droplets accumulated in TMEM41B-knockout (KO) cells. The phenotype of TMEM41B-KO cells resembled those of VMP1-KO cells. Indeed, TMEM41B and VMP1 formed a complex in vivo and in vitro, and overexpression of VMP1 restored autophagic flux in TMEM41B-KO cells. These results suggest that TMEM41B and VMP1 function together at an early step of autophagosome formation.
Introduction
Macroautophagy (hereafter autophagy) is an evolutionarily conserved intracellular protein degradation process (Mizushima and Komatsu, 2011; Lamb et al., 2013; Abada and Elazar, 2014). Autophagy targets both nonselective and selective substrates, while the ubiquitin–proteasome system degrades only selective substrates. During autophagy, a membrane sac called an isolation membrane or phagophore forms in the cytoplasm, elongates, and closes to form an autophagosome. The autophagosome then fuses with lysosomes, where its contents are degraded by lysosomal enzymes.
Autophagy involves multiple complex subprocesses and consequently requires many molecules. Currently, >40 autophagy-related (ATG) genes are known to be involved in various types of autophagy (Mizushima, 2018). Most of these genes were identified by genetic screens performed in the yeast Saccharomyces cerevisiae (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Harding et al., 1995). In addition, genes required for pexophagy (autophagic degradation of peroxisomes) were identified in Komagataella phaffii (previously known as Pichia pastoris; Strømhaug et al., 2001; van Dijk et al., 2001; Mukaiyama et al., 2002; Dunn et al., 2005). Genetic screens for autophagic factors have also been performed in Caenorhabditis elegans (Tian et al., 2010) and Schizosaccharomyces pombe (Sun et al., 2013). Several autophagy genes have been identified in mammals by nongenetic methods. FIP200 (also known as RB1CC1) and ATG101 were identified as an ULK-interacting protein (Hara et al., 2008) and an Atg13-interacting protein (Hosokawa et al., 2009a; Mercer et al., 2009), respectively. Vacuole membrane protein 1 (VMP1), which is induced in acute pancreatitis, is an ER-localized transmembrane protein that is required for autophagosome formation (Ropolo et al., 2007). S. cerevisiae lacks some of these factors including ATG101 (Hosokawa et al., 2009a; Mercer et al., 2009), VMP1 (Ropolo et al., 2007), EPG5 (Wang et al., 2016), and EI24 (Tian et al., 2010). Therefore, it is possible that more autophagy factors not revealed in yeast screens remain to be discovered in metazoans.
Until recently, techniques for genetically perturbing mammalian cells were limited, and genome-wide screens targeting autophagy could only be performed using siRNA-mediated gene silencing (Chan et al., 2007; Orvedahl et al., 2011; Hale et al., 2016; Jung et al., 2017). However, the recent emergence of the CRISPR-Cas9 system and the development of high-speed sequencing techniques have enabled knockout (KO)-based genome-wide screening in mammalian cells (Shalem et al., 2014; Wang, 2014). In the autophagy field, a CRISPR-based genome-wide screen identified the UFMylation pathway as a regulator of expression of SQSTM1/p62, a substrate of autophagy (DeJesus et al., 2016). Another group discovered an autophagy-independent lysosomal targeting pathway by a genome-wide screen using NCOA4, an adaptor involved in ferritin autophagy, as an indicator (Goodwin et al., 2017).
In this study, we performed an independent genome-wide screen using our novel autophagic flux reporter (Kaizuka et al., 2016) and identified TMEM41B as a novel autophagy-related gene. TMEM41B localizes on the ER and functions together with VMP1, a structurally related autophagy protein.
Results and discussion
A genome-scale CRISPR screen using an autophagic flux reporter
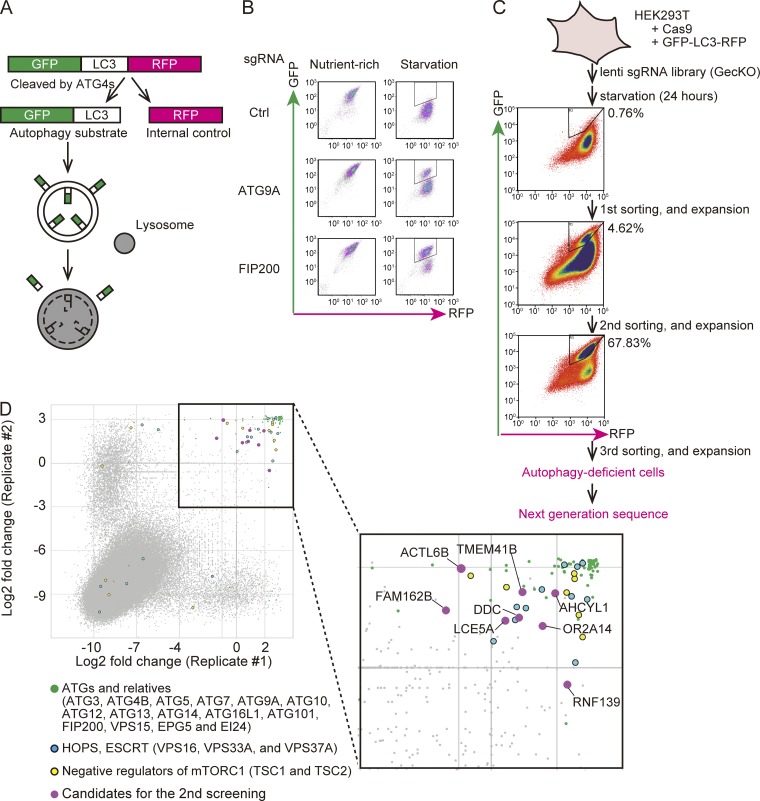
To perform CRISPR-based genome-wide screening, we used a novel autophagic flux reporter recently established by our group (Kaizuka et al., 2016). In this system, GFP-LC3-RFP is cleaved by endogenous ATG4 proteins to yield equimolar amounts of GFP-LC3 and RFP (Fig. 1 A). GFP-LC3 is then conjugated with phosphatidylethanolamine (PE) on isolation membranes and is quenched/degraded when delivered to the lysosome by autophagy. Meanwhile, RFP stays in the cytosol, serving as an internal control. Thus, the reduction in the GFP:RFP fluorescence ratio represents autophagic flux (Fig. 1 A).
Figure 1.
CRISPR-mediated genome-wide screen using an autophagic flux reporter. (A) Schematic representation of the autophagic flux reporter GFP-LC3-RFP. GFP-LC3-RFP is cleaved by endogenous ATG4 family proteins to yield equimolar amounts of GFP-LC3 (autophagy substrate) and RFP (internal control). Reduction in the GFP:RFP ratio indicates autophagic activity. (B) HEK293T cells expressing Cas9 and GFP-LC3-RFP were transduced with or without sgRNAs targeting ATG9A and FIP200 and selected with puromycin. The GFP and RFP intensities were determined by flow cytometry under nutrient-rich and starvation conditions. The autophagy-deficient population is indicated by the region of interest (ROI). (C) Schematic representation of the CRISPR-mediated genome-wide screen. An sgRNA library (GeCKO) was introduced to HEK293T cells expressing Cas9 and GFP-LC3-RFP. The cell population that did not respond to starvation (indicated by the ROI) was collected by FACS and expanded. After repeating this enrichment process three times, genomic DNA was extracted and subjected to next-generation sequencing. The proportion (%) of the autophagy-deficient population is indicated by the ROI. (D) Scatterplot of the results of two replicates. Data represent log2 (fold change) of read counts of individual sgRNAs before versus after enrichment. Enriched sgRNAs are shown in the separate panel. Canonical ATG genes and known autophagy-related genes (green), genes encoding HOPS and ESCRT components (blue), negative regulators of mTORC1 (yellow), and high-scoring genes not previously linked to autophagy (magenta) are indicated.
We established a HEK293T cell line stably expressing GFP-LC3-RFP and Cas9. When these cells were starved, the GFP signal decreased, while the RFP signal remained constant (Fig. 1 B). This specific reduction of the GFP signal was inhibited following introduction of single-guide RNAs (sgRNAs) targeting ATG9A and FIP200 (also known as RB1CC1), both of which are required for autophagy (Fig. 1 B), confirming that this cell line is suitable for detection of deficiencies in autophagy. For genome-wide screening, these cells were transduced with the human GeCKO library, a pooled lentiCRISPR library comprising 123,411 sgRNAs targeting 19,050 human genes (six sgRNAs per gene; Fig. 1 C; Sanjana et al., 2014; Shalem et al., 2014). 14 d after transduction, the cells were starved for 24 h and subjected to FACS to select cells that did not exhibit a starvation-induced reduction in the GFP:RFP ratio (i.e., autophagy-deficient cells). By repeating the culture and sorting steps three times, we enriched for autophagy-deficient cells (Fig. 1 C). Genomic DNA was extracted from cells before the first sort (control) and after the third sort (autophagy deficient) and subjected to next-generation sequencing analysis.
We performed the screen twice and plotted the abundance of each sgRNA (Fig. 1 D). Most nonredundant ATGs (ATG3, ATG4B, ATG5, ATG7, ATG9A, ATG10, ATG12, ATG13, ATG14, ATG16L1, ATG101, and FIP200) scored highly in both replicates, confirming that the screens were effective and almost saturated (Fig. 1 D and Table S1). In addition to canonical ATGs, genes encoding components of the homotypic fusion and protein sorting (HOPS) complex (VPS16 and VPS33A), negative regulators of mTORC1 (TSC1 and TSC2), and noncanonical ATGs (VPS15, EPG5, and EI24) were also enriched. Endocytosis- and lysosome-related genes were not enriched, with the exception of VPS37A, probably because these genes are required for normal growth and were eliminated during the enrichment procedure.
TMEM41B, a novel gene required for autophagy
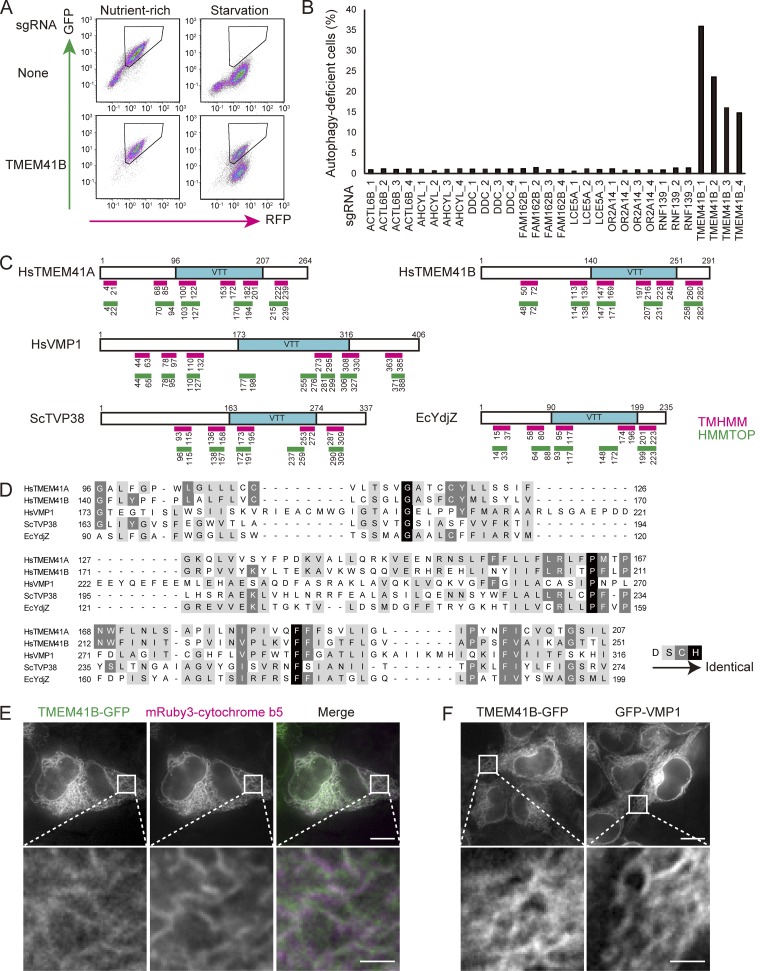
For the secondary screen, we selected eight genes for which at least one of the six corresponding sgRNAs were enriched in the primary screen: ACTL6B, AHCYL1, DDC, FAM162B, LCE5A, OR2A14, RNF139, and TMEM41B. HEK293T cells expressing GFP-LC3-RFP and Cas9 were transduced with three or four independent lentiviruses carrying sgRNAs against each gene, and the fraction of autophagy-deficient cells was determined. Autophagic flux was reduced in cells expressing any of the four sgRNAs against TMEM41B (Fig. 2, A and B) but not in those expressing sgRNAs against the other candidates (Fig. 2 B).
Figure 2.
TMEM41B, a novel ATG candidate. (A and B) Results of the secondary screen. HEK293T cells expressing Cas9 and GFP-LC3-RFP were transduced with sgRNAs (three or four for each candidate) and selected with puromycin. GFP and RFP intensities were determined by flow cytometry under nutrient-rich and starvation conditions (24 h). Representative plots of cells treated with TMEM41B sgRNA are shown in A. The proportion of the population that was autophagy deficient (indicated by ROI) was determined and shown in B. Data in B are representative of two independent experiments. (C) Domain structures of Homo sapiens (Hs) TMEM41A, HsTMEM41B, HsVMP1, S. cerevisiae (Sc) Tvp38, and E. coli (Ec) YdjZ. Presence of VTT domains (blue) was predicted by Pfam search. Positions of transmembrane domains were predicted by TMHMM (magenta) and HMMTOP (green) and are shown as lines. The VTT domain was previously called the SNARE-assoc domain. (D) Multiple sequence alignment of the VTT domains of HsTMEM41A, HsTMEM41B, HsVMP1, ScTvp38, and EcYdjZ. Positions are colored according to increasing sequence identity: white, not conserved; gray, similar; black, identical. (E) TMEM41B-GFP colocalized with mRuby3–cytochrome b5 (an ER marker) in HEK293T cells. (F) Neither TMEM41B-GFP nor GFP-VMP1 forms punctate structures under starvation conditions. Bars: 10 µm (main images); 2 µm (insets).
TMEM41B (also known as Stasimon) is a major target of spinal muscular atrophy–dependent U12 splicing and is required for normal synaptic transmission of motor neurons (Imlach et al., 2012). However, its molecular function remains unknown. TMEM41B was predicted by TMHMM software (Krogh et al., 2001) to have six transmembrane domains (Fig. 2 C). A Pfam domain search (Finn et al., 2016) revealed that TMEM41B has a SNARE-assoc domain (Fig. 2 C). This domain is also present in VMP1, another autophagy-related ER protein (Ropolo et al., 2007; Zhao et al., 2017), TMEM41A, yeast Tvp38 (and its mammalian homologue TMEM64), and the bacterial DedA family of proteins, e.g., Escherichia coli YdjZ (Fig. 2, C and D; Doerrler et al., 2013). Because there is no evidence that this domain associates with SNARE proteins, we renamed it the VMP1, TMEM41, and Tvp38 (VTT) domain. Notably, E. coli YdjX and YdjZ, which belong to the DedA family, are predicted to be half-transporters (Khafizov et al., 2010; Keller et al., 2014). This domain likely contains three transmembrane domains, but the TMHMM and HMMTOP (Tusnády and Simon, 2001) algorithms yielded different predictions (Fig. 2 C), suggesting that this domain forms complicated or discontinuous structures within the membrane. Outside the VTT domains, the sequences of TMEM41B and VMP1 are less well conserved (Fig. S1 A). Both TMEM41B and VMP1 have typical transmembrane domains before and after the VTT domain: TMEM41B has two upstream and one downstream, whereas VMP1 has three upstream and two downstream (Fig. 2 C). Like VMP1, TMEM41B is localized in the ER (Fig. 2 E) and does not form punctate structures under starvation conditions (Fig. 2 F).
Impaired autophagic activity caused by deletion of TMEM41B
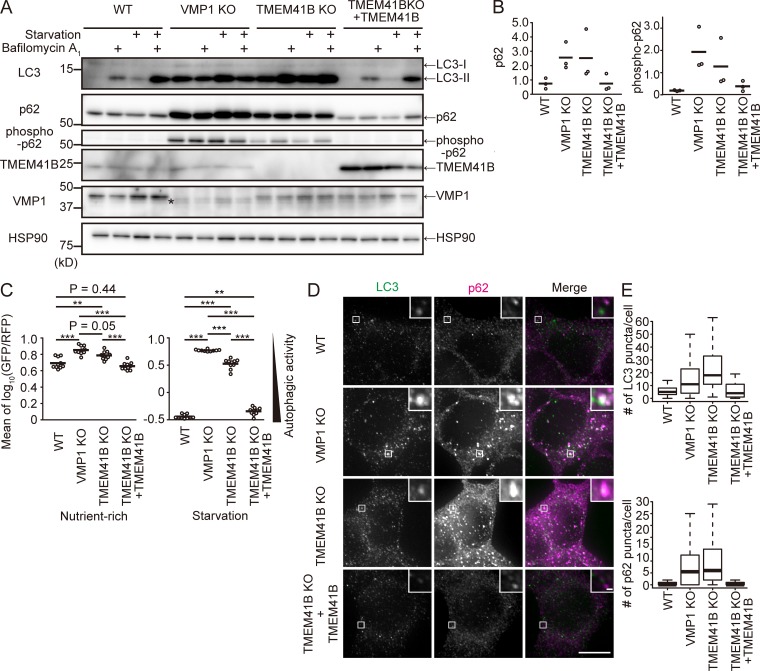
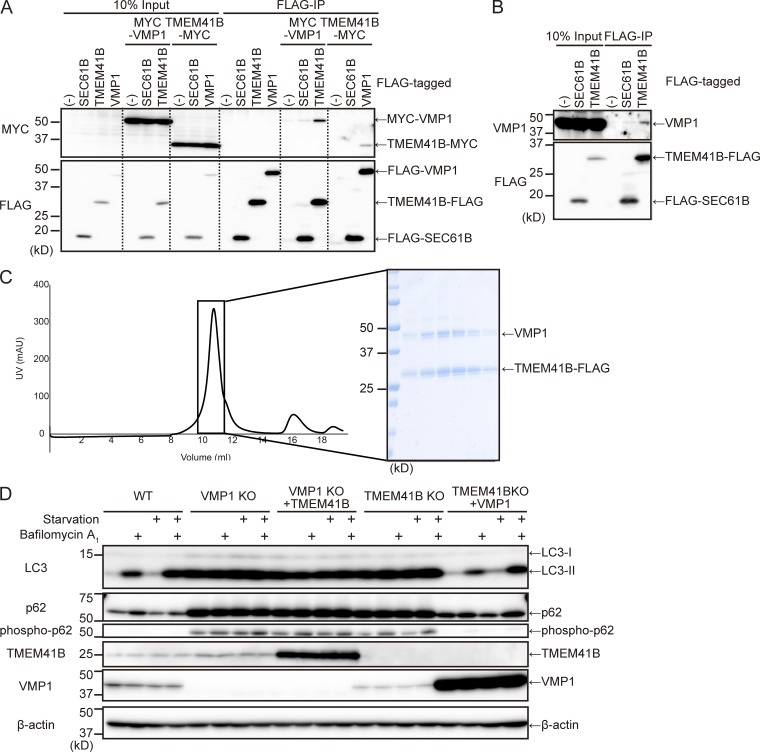
To further characterize the roles of these proteins in autophagy, we established TMEM41B-KO and VMP1-KO HEK293T cell lines using the CRISPR-Cas9 system (Fig. S1, B and C). In WT cells, the amount of LC3-II, the PE-conjugated form of the protein, increased under starvation conditions, suggesting that the number of autophagosomes increased (Fig. 3 A). Bafilomycin A1, a vacuolar-type H+-ATPase inhibitor, caused a further increase in the level of LC3-II, representing normal autophagic flux. By contrast, in VMP1 KO cells, the level of LC3-II was already high under growing conditions and did not further increase under starvation or bafilomycin A1 treatment, suggesting that autophagic flux was almost completely blocked (Fig. 3 A). Similarly, in TMEM41B-KO cells, autophagic flux was profoundly blocked (Fig. 3 A). p62/SQSTM1 and phospho-p62/SQSTM1, selective substrates of autophagy, accumulated in both TMEM41B-KO and VMP1-KO cells, but to a lesser extent in the former (Fig. 3, A and B). Indeed, quantitative measurements of autophagic activity using the GFP-LC3-RFP reporter revealed that TMEM41B-KO cells had a slightly milder defect than VMP1-KO cells (Fig. 3 C). All defects in TMEM41B-KO cells were restored by reexpression of exogenous TMEM41B, ruling out the possibility of an off-target effect (Fig. 3, A–C), but not by expression of TMEM41B lacking the VTT domain (Fig. S2 A). TMEM41A and TMEM41B were not redundant: expression of TMEM41A failed to rescue TMEM41B-KO cells (Fig. S2 A), and knockdown of TMEM41A did not further impair autophagic activity of TMEM41B-KO cells (Fig. S2 B). Immunofluorescence microscopy revealed that p62 accumulated as punctate structures in both TMEM41B-KO and VMP1-KO cells (Fig. 3, D and E). These structures often colocalized with LC3 (Fig. 3 D). Together, these data confirm that TMEM41B is required for autophagic flux.
Figure 3.
Defective autophagy in TMEM41B-KO and VMP1-KO cells. (A and B) Autophagic flux assay of TMEM41B-KO and VMP1-KO HEK293T cells. Cells were cultured with or without bafilomycin A1 under nutrient-rich or starvation conditions for 2 h. The asterisk indicates a nonspecific band detected by the anti-VMP1 antibody (A). Quantification of band intensities of p62 and phospho-p62 versus HSP90. Bars indicate means (B). (C) Autophagic flux measured using the GFP-LC3-RFP reporter under nutrient-rich and starvation conditions (24 h). Bars indicate means. Differences were statistically analyzed by Tukey-Kramer post hoc test. **, P < 0.01; ***, P < 0.001. (D) WT, TMEM41B-KO, VMP1-KO, and rescued TMEM41B-KO HEK293T cells were cultured under nutrient-rich conditions and subjected to immunofluorescence microscopy. Bars: 10 µm (main images); 500 nm (insets). (E) Numbers of LC3 and p62 puncta were quantified. Data were collected from >50 cells for each genotype. Solid bars indicate median, boxes the interquartile range (25th to 75th percentile), and whiskers 1.5 times the interquartile range.
Defective autophagosome formation in TMEM41B-KO cells
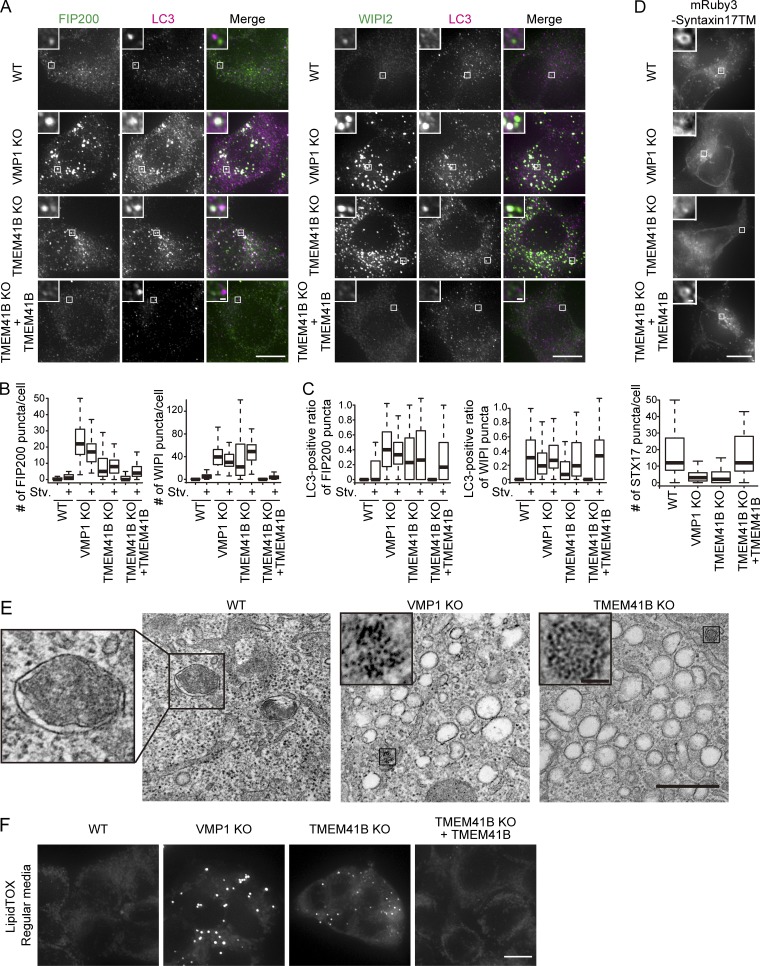
To identify the step at which autophagy was blocked in TMEM41B-KO cells, we monitored the recruitment of other ATG proteins. Depletion of VMP1 resulted in accumulation of puncta positive for FIP200 and WIPI2 under both nutrient-rich and starvation conditions (Fig. 4, A and B) as previously reported (Itakura and Mizushima, 2010; Zhao et al., 2017). Similarly, many puncta positive for FIP200 and WIPI2 were observed in TMEM41B-KO cells even under nutrient-rich conditions (Fig. 4, A and B). These structures often colocalized with LC3 (Fig. 4, A and C). Because FIP200 and WIPI2 are present on isolation membranes but not on mature autophagosomes, these data suggest that autophagosome formation is stalled in TMEM41B-KO and VMP1-KO cells.
Figure 4.
Defective autophagosome formation in TMEM41B-KO and VMP1-KO cells. (A) WT, TMEM41B-KO, VMP1-KO, and rescued TMEM41B-KO HEK293T cells were cultured under nutrient-rich and starvation (Stv) conditions for 2 h and subjected to immunofluorescence microscopy. Bars: 10 µm (main images); 500 nm (insets). (B) Numbers of FIP200 and WIPI2 puncta in cells under nutrient-rich and starvation conditions were quantified as in Fig. 3 E. Data were collected from >70 cells for each sample. (C) Quantification of colocalization between FIP200 and LC3 and between WIPI2 and LC3. Data were collected from >70 cells for each sample. (D) WT, TMEM41B-KO, VMP1-KO, and rescued TMEM41B-KO cells expressing mRuby3-STX17TM were starved for 2 h and observed by fluorescence microscopy. Data were collected from >40 cells and quantified as in Fig. 3 E. Bars: 10 µm (main images); 500 nm (insets). (E) Transmission EM of WT, VMP1-KO, and TMEM41B-KO cells under starvation conditions (2 h). Insets indicate an autophagosome (in WT) or ferritin clusters (in KO). Bars: 500 nm (main images); 50 nm (insets). (F) WT, TMEM41B-KO, VMP1-KO, and rescued TMEM41B-KO HEK293T cells were stained with LipidTOX. Bar, 10 µm.
To determine whether completed autophagosomes were formed, we used the autophagosomal SNARE syntaxin17 (STX17) as an autophagosome marker (Itakura et al., 2012). In WT cells, mRuby3-tagged STX17TM (a mutant of STX17 that contains only the transmembrane domain but acts similarly to full-length STX17) formed ring-shaped structures throughout the cytoplasm under starvation conditions, which represented completed autophagosomes (Fig. 4 D). By contrast, in TMEM41B-KO and VMP1-KO cells, such ring-shaped STX17 structures were absent, and only small puncta or clusters were observed, suggesting that autophagosomes were not formed (Fig. 4 D). Transmission EM revealed accumulation of small vesicles and ferritin clusters but not autophagosomes in TMEM41B-KO cells and VMP1-KO cells (Fig. 4 E). These aberrant structures were reminiscent of features previously observed in VMP1-knockdown cells (Kishi-Itakura et al., 2014). Taken together, these data suggest that in TMEM41B-KO and VMP1-KO cells, autophagosomal formation is blocked at or before the elongation step, causing precursor structures containing isolation membrane markers to accumulate.
Accumulation of lipid droplets in TMEM41B-KO cells
To further characterize the role for TMEM41B, we investigated the nonautophagic phenotype of TMEM41B-KO cells. Lipid droplets accumulated in VMP1-KO cells (Fig. 4 F; Tábara and Escalante, 2016; Zhao et al., 2017). Likewise, lipid droplets accumulated in TMEM41B-KO cells (Fig. 4 F), suggesting that TMEM41B has a function similar to that of VMP1 in nonautophagic pathways.
TMEM41B physically and functionally interacts with VMP1
Our observation that TMEM41B-KO and VMP1-KO cells had similar phenotypes coupled with the fact that both proteins reside in the ER implied that TMEM41B and VMP1 form a functional complex. Indeed, exogenously expressed VMP1 interacted with TMEM41B (Fig. 5 A). This interaction was specific as neither protein interacted with SEC61B, another ER membrane protein. Endogenous VMP1 also coimmunoprecipitated with TMEM41B-FLAG (Fig. 5 B). Interaction between TMEM41B and other core ATG proteins was not detected (Fig. S3). These data demonstrate that TMEM41B and VMP1 interact with each other in vivo.
Figure 5.
Interaction and functional relationships between TMEM41B and VMP1. (A and B) Coimmunoprecipitation of TMEM41B and VMP1. WT cells stably expressing indicated tagged proteins were immunoprecipitated with anti-FLAG antibody and detected with anti-FLAG and anti-MYC (A) or anti-VMP1 antibody (B). (C) Coexpression and purification of TMEM41B–VMP1 complex. Chromatogram of Superdex 200 Increase 10/300 gel filtration purification of the VMP1–TMEM41B complex. The main peak fractions were collected and subjected to SDS-PAGE. (D) WT, VMP1-KO, and TMEM41B-KO cells with or without overexpression of indicated proteins were subjected to autophagic flux assays. Cells were cultured with or without bafilomycin A1 under nutrient-rich or starvation conditions for 2 h.
To determine whether TMEM41B directly interacts with VMP1, we performed in vitro binding assays. TMEM41B-FLAG and VMP1-TEV-GFP-His were coexpressed in Sf9 cells and sequentially purified on TALON (Cobalt) metal affinity resin and anti-FLAG M2 affinity gel (Fig. S2 C). The eluate was evaluated by size-exclusion chromatography and SDS-PAGE. TMEM41B and VMP1 were codistributed as a single peak (Fig. 5 C). The band intensities imply that more than one TMEM41B molecule associates with each VMP1 molecule. These in vitro results suggest that TMEM41B and VMP1 directly interact with each other.
To explore the functional relationship between TMEM41B and VMP1, we investigated whether overexpression of either TMEM41B or VMP1 could rescue deficiency of the other. Overexpression of TMEM41B did not rescue the autophagy defect in VMP1-KO cells (Fig. 5 D). However, overexpression of VMP1 restored LC3 turnover and p62 degradation in TMEM41B-KO cells (Fig. 5 D). These results suggest that VMP1 is functionally interacted with TMEM41B.
In this study, we identified TMEM41B as a novel autophagy-related gene required for autophagosome formation. TMEM41B contains a VTT domain also found in VMP1, and the two proteins physically and functionally interact with each other (Fig. 5). Remarkably, the VTT domain is conserved in E. coli DedA family proteins such as YdjX and YdjZ. Although the precise function of these bacterial proteins remains unknown, they may be half-transporters (Khafizov et al., 2010; Keller et al., 2014). Accordingly, TMEM41B and VMP1 may also be half-transporters that together form a complete transporter. VMP1 has been proposed to regulate the activity of the Ca2+ pump SERCA (Zhao et al., 2017), but VMP1 itself may also be involved in ion homeostasis. The VTT domain is also present in other proteins such as TMEM41A and Tvp38 (and its mammalian homologue TMEM64; Fig. 2 D), but the molecular functions of these proteins also remain largely unknown. Determining the structures of the VTT domain and VTT domain–containing proteins will provide new insights into the autophagy pathway as well as more general cellular functions conserved in both prokaryotes and eukaryotes.
Materials and methods
Antibodies and reagents
The following antibodies were used for immunoblotting: mouse monoclonal antibodies against HSP90 (610419; BD) and β-actin (A2228; Sigma-Aldrich) and rabbit polyclonal antibodies against p62/SQSTM1 (PM045; MBL), phospho-p62 (PM074; MBL), TMEM41B (HPA014946; Sigma-Aldrich), VMP1 (PM072; MBL), TMEM41A (20768-1-AP; ProteinTech), Atg101 (SAB4200175; Sigma-Aldrich), Beclin1 (3738; Cell Signaling Technology), MYC (16282-AP; ProteinTech), and peroxidase-conjugated anti-FLAG antibody (015-22391; Wako). Rabbit polyclonal antibody against LC3 was described previously (Hosokawa et al., 2006). Peroxidase-conjugated anti-mouse and anti-rabbit immunoglobulins (111-035-144; Jackson ImmunoResearch Laboratories, Inc.) were used as secondary antibodies. The following antibodies were used for immunocytochemistry: rabbit polyclonal antibody against FIP200 (17250-1-AP; ProteinTech), rabbit polyclonal antibody against WIPI2 (SAB4200400; Sigma-Aldrich) and p62/SQSTM1 (PM045; MBL), and mouse monoclonal antibody against LC3 (CTB-LC3-2-IC; CosmoBio). Alexa Fluor 488–conjugated goat anti-mouse IgG and Alexa Fluor 568–conjugated goat anti-rabbit IgG (A-11029 and A-11036; Thermo Fisher Scientific) were used as secondary antibodies. LipidTOX (H34476; Thermo Fisher Scientific) was used for staining of lipid droplets. For transient expression, Fugene HD (VPE2311; Promega) was used.
Plasmids
LentiCas9-Blast (52962; Addgene) and human GeCKO v2 Library (2 Plasmid System; lentiGuide-Puro; 1000000049; Addgene) were provided by F. Zhang (Massachusetts Institute of Technology, Cambridge, MA). The GFP-LC3-RFP reporter (Kaizuka et al., 2016) was inserted into pMXs-IP (Kitamura et al., 2003), and the puromycin-resistant gene cassette (PuroR) was removed to generate pMXs-GFP-LC3-RFP. sgRNAs targeting FIP200 (5′-TATGTATTTCTGGTTAACAC-3′) and ATG9A (5′-CCTGTTGGTGCACGTCGCCG-3′) were inserted into lentiGuide-Puro. LentiCRISPR v2 was provided by F. Zhang through Addgene (52961; Sanjana et al., 2014). sgRNAs targeting ACTL6B (5′-GTCATGTCGCCCCTCAAGAA-3′, 5′-ATGAGCGGGGGCGTCTACGG-3′, 5′-AGTCCGCGCTGGGTACGCTG-3′, and 5′-ACTATGAGCGGGGGCGTCTA-3′), AHCYL1 (5′-GCCAGTTTTGGTGGGGAATT-3′, 5′-GCAGGCAGAATTTGGACGCC-3′, 5′-CGCCGAGAAGTACTCCTTCA-3′, and 5′-TCGAGCGAGACAAAGATCTT-3′), DDC (5′-ACGCCAGGTCTACCCTGACG-3′, 5′-GCTTGCGGACATGCTGTGCG-3′, 5′-GGTACCCGGGCTCCACGTCA-3′, and 5′-AGCAGCATGTTGTGGTCCCC-3′), FAM162B (5′-CGCGGGCTAACAGTCCGCTG-3′, 5′-AGGGCGTTTCAAATCGATGG-3′, 5′-GTGGCGTGAAGAAGCTGCAT-3′, and 5′-GACATGAATCCTTAACAAGT-3′), LCE5A (5′-GGGCATTTAGGGGTACATTT-3′, 5′-TGGAACTCTGAGGTCGGCGT-3′, and 5′-TGGGCATGGGGCTGAACACT-3′), OR2A14 (5′-GATTTCATGAGGCCCGCAGA-3′, 5′-ACCAACCTGGAATCGCGGCA-3′, 5′-TGATGTCTGTGATCCATGTC-3′, and 5′-CATAAGATTCGTCAGCATCT-3′), RNF139 (5′-GGTCTGGGCGGCGCTCGAAG-3′, 5′-CAAAAGATAAACATATCGTG-3′, and 5′-GTCTACTACGTTCGTTCAAC-3′), and TMEM41B (5′-GTCGCCGAACGATCGCAGTT-3′, 5′-GAAAAATCCTGGGTAGAAGC-3′, 5′-TATACTTACTCACTAAGCTG-3′, and 5′-CTGTATCAACTTACAACAGC-3′) were cloned to lentiCRISPR v2. sgRNAs targeting TMEM41B (5′-GTCGCCGAACGATCGCAGTT-3′) and VMP1 (5′-CTTTTGTATGCCTACTGGAT-3′) were cloned into pSpCas9(BB)-2A-GFP (PX458; gift from F. Zhang; 48138; Addgene). cDNAs encoding human TMEM41B (NP_055827.1), TMEM41A (NP_542383.1), SEC61B (Nishimura et al., 2017), VMP1 (Itakura and Mizushima, 2010), cytochrome b5 (Itakura and Mizushima, 2010), VPS34 (Itakura et al., 2008), and ATG9A (Nishimura et al., 2017) were inserted into pMRXIP (Saitoh et al., 2002), pMRXIN, or pMRXIZ together with EGFP, codon-optimized mRuby3 (modified from pKanCMV-mClover3-mRuby3; 74252; Addgene), 3×FLAG, or 3×MYC. WIPI2B (NP_057087.2) and LC3B (Kabeya et al., 2000) were inserted into p3×FLAG CMV10 (E7658; Sigma-Aldrich). pMRXIP-mRuby3-STX17TM was described previously (Matsui et al., 2018). pMRXIN and pMRXIZ were constructed by replacing PuroR of pMRXIP with a neomycin-resistant gene cassette or zeocin-resistant cassette. p3×FLAG CMV10-ATG13 (Hosokawa et al., 2009b) and p3×FLAG CMV10-ATG14 (Itakura et al., 2008) were previously described. Truncated constructs were prepared by PCR-mediated site-directed mutagenesis.
Cell culture
Cells were cultured in DMEM (D6546; Sigma-Aldrich) supplemented with 10% FBS (172012; Sigma-Aldrich), 50 U/ml penicillin, 50 µg/ml streptomycin (15070-063; Gibco), and 2 mM glutamine (25030-081; Gibco; regular medium) in a 5% CO2 incubator. For starvation treatment, cells were washed twice with PBS and cultured in Earle’s balanced salt solution (E2888; Sigma-Aldrich) or amino acid–free DMEM (048-33575; Wako) without FBS. For bafilomycin A1 treatment (B1793; Sigma-Aldrich), cells were cultured with 100 nM bafilomycin A1 for 2 h.
Preparation of lentivirus and retrovirus
For preparation of lentivirus, HEK293T cells were transiently transfected with a lentiviral vector together with pCMV-VSV-G (gift from R.A. Weinberg, Whitehead Institute for Biomedical Research, Cambridge, MA) and psPAX2 (gift from D. Trono, Ecole Polytechnique Federale de Lausanne, Lausanne, Switzerland) using Lipofectamine 2000 (11668019; Thermo Fisher Scientific). After 2–3 d culture, the supernatant was passed through a 0.45-µm syringe filter unit (SLHV033RB; EMD Millipore). For preparation of retrovirus, HEK293T cells were transiently transfected with a retroviral vector together with pCG-VSV-G and pCG-gag-pol (gifts from T. Yasui, Osaka University, Osaka, Japan) using Lipofectamine 2000, and virus was collected from the supernatant as described above.
Generation of stable cell lines
Cells were infected with retrovirus or lentivirus, and stable transformants were selected with puromycin (P8833; Sigma-Aldrich), blasticidin (022-18713; Wako), zeocin (R25005; Thermo Fisher Scientific), or geneticin (10131; Thermo Fisher Scientific).
Establishment of TMEM41B-KO and VMP1-KO cells
HEK293T cells were transfected with pX458-encoding sgRNAs targeting TMEM41B and VMP1 using ViaFect (E4981; Promega). 2 d after transfection, GFP-positive cells were isolated using a cell sorter (MoFlo Astrios EQ; Beckman Coulter), and single clones were obtained. Clones with mutations in both alleles were identified by PCR and confirmed by sequencing of genomic DNA.
RNAi
Stealth RNAi oligonucleotides were purchased from Thermo Fisher Scientific. The sequences used were as follows: luciferase siRNA antisense, 5′-CGCGGUCGGUAAAGUUGUUCCAUUU-3′, and sense, 5′-AAAUGGAACAACUUUACCGACCGCG-3′; and human TMEM41A siRNA antisense, 5′-ACAGCUUAAAGACAGUGUCCCAGGA-3′, and sense, 5′-UCCUGGGACACUGUCUUUAAGCUGU-3′. Stealth RNAi oligonucleotides were transfected into cells using Lipofectamine RNAiMAX (13778150; Thermo Fisher Scientific). After 2 d, the cells were transfected again with the same siRNAs and cultured for an additional 2 d before analysis.
Genome-wide screening
HEK293T cells expressing GFP-LC3-RFP and Cas9 were infected with the pooled lentiviral library at an MOI of 0.5–0.7 and selected with puromycin. 2 wk after infection, cells were starved for 24 h and subjected to FACS using MoFlo AstriosEQ (Beckman Coulter). After sorting, cells were cultured, expanded, and subjected to FACS again. Totally, the enrichment by FACS was performed three times.
Genomic DNA sequencing and data processing
Genomic DNA sequencing and data processing were performed as previously described (Shalem et al., 2014). Briefly, genomic DNA was extracted from 4 × 107 cells (>300× coverage over the GeCKO library) using the Blood & Cell Culture Midi kit (13343; QIAGEN). From 240 µg genomic DNA, sgRNA sequences were amplified by PCR using Herculase II Fusion DNA polymerase (600675; Agilent Technologies). A second PCR was then performed to attach Illumina adaptors and barcode samples. The primers used in the first PCR were as follows: F1, 5′-AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG-3′, and R1, 5′-CTTTAGTTTGTATGTCTGTTGCTATTATGTCTACTATTCTTTCC-3′. The primers used in the second PCR were as follows: F2, 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-(5-bp variable length sequence)-(6-bp barcode)-TCTTGTGGAAAGGACGAAACACCG-3′; R2, 5′-CAAGCAGAAGACGGCATACGAGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCTACTATTCTTTCCCCTGCACTGT-3′. The amplicons from the second PCR were gel extracted and subjected to DNA sequencing on a HiSeq 2500 (Illumina) sequencer. Samples were sequenced with the rapid run mode of the single-ended 71 bp. Sequence reads of control and test samples were distinguished by the presence of the following sequences: control, 5′-ATTGGCTCTTGTGGAAAGGACGAAACACCG-3′, and test, 5′-TACAAGTCTTGTGGAAAGGACGAAACACCG-3′. The target insertion sites were designated by the 20-bp sequences followed by the above sequences, allowing one or two mismatches. After trimming, the number of reads completely matching the GeCKO v2 library sequence was calculated.
Flow cytometry
Cells were treated with trypsin-EDTA (25300062; Gibco; 0.05% for analysis and 0.005% for sorting) for 1.5–2 min and collected in ice-cold PBS. After washing, cells were analyzed on an EC800 cell analyzer (Sony) equipped with 488-nm and 561-nm lasers. Data were processed with the Kaluza software (Beckman Coulter) and R software (3.2.4; https://www.r-project.org/). For sorting, cells were resuspended with HBSS containing penicillin, streptomycin, and EDTA and subjected to FACS on a cell sorter (MoFlo AstriosEQ; Beckman Coulter).
Immunoblotting
Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and Complete EDTA-free protease inhibitor cocktail [05056489001; Roche]) for 15 min on ice and then centrifuged at 12,000 g for 15 min. Supernatant was collected as cell lysate and boiled in sample buffer (46.7 mM Tris-HCl, pH 6.8, 5% glycerol, 1.67% SDS, 1.55% DTT, and 0.02% bromophenol blue). Samples were subsequently separated by SDS-PAGE and transferred to Immobilon-P polyvinylidene difluoride membranes (IPVH00010; EMD Millipore). Immunoblotting analysis was performed with the indicated antibodies. Super-Signal West Pico Chemiluminescent Substrate (1856135; Thermo Fisher Scientific) or Immobilon Western Chemiluminescent HRP Substrate (P90715; EMD Millipore) was used to visualize signals, which were detected on a Fusion System Solo 7S (M&S Instruments Inc.). Contrast and brightness adjustment, treatment, and quantification were performed in the Fiji software (ImageJ; National Institutes of Health; Schindelin et al., 2012) and Photoshop CS6 (Adobe).
Immunocytochemistry and fluorescence microscopy
Cells grown on coverslips were washed with PBS and fixed with 4% PFA in PBS for 20 min on ice. After permeabilization with 50 µg/ml digitonin in PBS for 5 min, cells were blocked with 3% BSA in PBS for 30 min, incubated with primary antibodies for 1 h, washed, and then incubated with secondary antibodies for 1 h. Fluorescence microscopy was performed on a DeltaVision Elite (GE Healthcare) equipped with a 60× Plan Apochromat oil-immersion objective lens (NA 1.42; Olympus) and a cooled charge-coupled device camera (CoolSNAP HQ2; Photometrics). All images were deconvoluted using the DeltaVision software (SoftWoRx; Applied Precision Ltd.) to remove out-of-focus material. All images except for those shown in Fig. 2 (E and F) and the insets in Fig. 4 D were z projected by Fiji. For visualization of STX17-positive autophagosomes, cells stably expressing mRuby3-tagged STX17TM were placed on a glass-bottomed dish (617870; Greiner Bio-one) in amino acid–free DMEM (048–33575; Wako) and observed on a DeltaVision Elite. During live-cell imaging, the dish was mounted in a chamber (INUB-ONI-F2; TOKAI HIT) to maintain incubation conditions at 37°C and 5% CO2. Quantitative analyses were performed on z-projected images using the Fiji and R software.
Immunoprecipitation (IP)
Cell lysates were prepared in lysis/IP buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 1% dodecyl maltoside [DDM], and 0.2% cholesteryl hemisuccinate [CHS]) for 15 min on ice and then centrifuged at 17,700 g for 15 min. The supernatants were incubated with anti-FLAG M2 affinity gel (A2220; Sigma-Aldrich) for 3 h at 4°C with gentle rotation. The beads were washed two times in wash buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 0.5% DDM, and 0.1% CHS), and the bound proteins were eluted with SDS-PAGE sample buffer.
EM
For EM analysis, cells were cultured on a cell-tight C-2 cell disk (MS-0113K; Sumitomo Bakelite). After 2 h culture under nutrient-rich or starvation conditions, cells were fixed with 2.5% glutaraldehyde (G015; TAAB) in 0.1 M phosphate buffer, pH 7.4, for 2 h on ice. After three washes with 0.1 M phosphate buffer, pH 7.4, the cells were postfixed with 1% osmium tetroxide in 0.1 M phosphate buffer, pH 7.4, for 2 h, dehydrated, and embedded in epon 812 following a standard procedure. Ultrathin sections were stained with uranyl acetate and lead citrate and observed on an H-7100 electron microscope (Hitachi).
Sequence alignment
Protein sequences were obtained from the NCBI protein database and aligned using Constraint-based Multiple Alignment Tool (COBALT). In figures, residues that are identical or similar in all sequences are shaded black or gray, respectively. Similar residues were determined by Molecular Evolutionary Genetics Analysis (MEGA; Kumar et al., 2018).
Coexpression and purification of TMEM41B–VMP1 complex
The human TMEM41B gene was subcloned into a modified pFastBac vector, with the resultant construct encoding a FLAG epitope tag (DYKDDDDK) at the C terminus. The human VMP1 gene was subcloned into a modified pFastBac vector, with the resultant construct encoding a TEV cleavage site followed by a GFP-His8 tag at the C terminus. Recombinant baculovirus was prepared using the Bac-to-Bac baculovirus expression system (10359016; Invitrogen). Sf9 insect cells were infected with viruses at a cell density of 4.0 × 106 per ml in Sf900II medium and grown for 48 h at 27°C. Harvested cells were suspended and solubilized in buffer A (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 10% glycerol, 1% DDM, and 0.2% CHS) for 1 h at 4°C. The supernatant was separated from the insoluble material by ultracentrifugation at 180,000 g for 1 h and then incubated with TALON resin (635504; Takara Bio Inc.) for 1 h at 4°C. The resin was washed with 10 column volumes of buffer B (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 0.03% DDM, 0.006% CHS, and 15 mM imidazole), and bound protein was eluted in buffer C (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 0.03% DDM, 0.006% CHS, and 200 mM imidazole). The eluate was incubated with anti-FLAG M2 affinity gel (A2220-25ML; Sigma-Aldrich) for 1.5 h at 4°C. The resin was washed with the 10-column volume of buffer D (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 0.03% DDM, and 0.006% CHS). The VMP1–TMEM41B complex was eluted in buffer E (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 0.03% DDM, 0.006% CHS, and 0.1 mg/ml Flag peptide). The eluate was treated with 1:10 (wt/wt) TEV protease and dialysis buffer F (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, and 2 mM 2-mercaptoethanol) overnight. The complex was concentrated using a centrifugal filter device (UFC805096; EMD Millipore; 50-kD molecular weight cutoff) and loaded onto a Superdex 200 Increase 10/300 size-exclusion column (28990944; GE Healthcare) and equilibrated in buffer G (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5% glycerol, 0.03% DDM, and 2 mM 2-mercaptoethanol). Fractions containing the proteins were analyzed by SDS-PAGE.
Statistical analysis
Multiple comparisons were performed by the Tukey-Kramer test using R software. Data distributions were assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows the similarity of VMP1 and TMEM41B and the positions of sgRNA-targeted sequences and resultant mutations in the human TMEM41B and VMP1 genes. Fig. S2 shows the requirement for the VTT domain of TMEM41B, the role of TMEM41A, and affinity purification of TMEM41B and VMP1. Fig. S3 shows no interaction between TMEM41B and core ATG proteins. Table S1 shows the results of the genome-wide pooled CRISPR screen.
Supplementary Material
Acknowledgments
We thank Hayashi Yamamoto for constructive discussion, Katsumori Segawa for helpful advice for genome-wide screening, Feng Zhang for the human GeCKO library and other CRISPR vectors, Toshio Kitamura for pMXs-IP, Shoji Yamaoka for pMRXIP, Teruhito Yasui for pCG-VSV-G and pCG-gag-pol, Robert A. Weinberg for pCMV-VSV-G, and Didier Trono for psPAX2.
This study was supported by a Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research on Innovative Areas (grant 25111005) and a Japan Science and Technology Agency (JST) Exploratory Research for Advanced Technology (ERATO; grant JPMJER1702) grant to N. Mizushima.
The authors declare no competing financial interests.
Author contributions: K. Morita and N. Mizushima designed the project. K. Morita performed most of the cell biology experiments with help from N. Tamura, K. Mimura, and H. Morishita. Y. Hama performed the phylogenetic analysis. T. Izume performed the coexpression and purification of the VMP1–TMEM41B complex with supervision by W. Shihoya, and O. Nureki. Y. Yamashita, T. Ueno, and H. Mano performed next-generation sequencing. Y. Sakamaki carried out the EM experiments. K. Morita, T. Izume, and N. Mizushima wrote the manuscript. All authors analyzed and discussed the results and commented on the manuscript.
References
- Abada A., and Elazar Z.. 2014. Getting ready for building: signaling and autophagosome biogenesis. EMBO Rep. 15:839–852. 10.15252/embr.201439076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.Y.W., Kir S., and Tooze S.A.. 2007. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 282:25464–25474. 10.1074/jbc.M703663200 [DOI] [PubMed] [Google Scholar]
- DeJesus R., Moretti F., McAllister G., Wang Z., Bergman P., Liu S., Frias E., Alford J., Reece-Hoyes J.S., Lindeman A., et al. 2016. Functional CRISPR screening identifies the ufmylation pathway as a regulator of SQSTM1/p62. eLife. 5:1–16. 10.7554/eLife.17290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerrler W.T., Sikdar R., Kumar S., and Boughner L.A.. 2013. New functions for the ancient DedA membrane protein family. J. Bacteriol. 195:3–11. 10.1128/JB.01006-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W.A. Jr., Cregg J.M., Kiel J.A.K.W., van der Klei I.J., Oku M., Sakai Y., Sibirny A.A., Stasyk O.V., and Veenhuis M.. 2005. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 1:75–83. 10.4161/auto.1.2.1737 [DOI] [PubMed] [Google Scholar]
- Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin J.M., Dowdle W.E., DeJesus R., Wang Z., Bergman P., Kobylarz M., Lindeman A., Xavier R.J., McAllister G., Nyfeler B., et al. 2017. Autophagy-independent lysosomal targeting regulated by ULK1/2-FIP200 and ATG9. Cell Reports. 20:2341–2356. 10.1016/j.celrep.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C.M., Cheng Q., Ortuno D., Huang M., Nojima D., Kassner P.D., Wang S., Ollmann M.M., and Carlisle H.J.. 2016. Identification of modulators of autophagic flux in an image-based high content siRNA screen. Autophagy. 12:713–726. 10.1080/15548627.2016.1147669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., and Mizushima N.. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181:497–510. 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding T.M., Morano K.A., Scott S.V., and Klionsky D.J.. 1995. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131:591–602. 10.1083/jcb.131.3.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Hara Y., and Mizushima N.. 2006. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 580:2623–2629. 10.1016/j.febslet.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., et al. 2009b Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 20:1981–1991. 10.1091/mbc.e08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Sasaki T., Iemura S., Natsume T., Hara T., and Mizushima N.. 2009a Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 5:973–979. 10.4161/auto.5.7.9296 [DOI] [PubMed] [Google Scholar]
- Imlach W.L., Beck E.S., Choi B.J., Lotti F., Pellizzoni L., and McCabe B.D.. 2012. SMN is required for sensory-motor circuit function in Drosophila. Cell. 151:427–439. 10.1016/j.cell.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., and Mizushima N.. 2010. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 6:764–776. 10.4161/auto.6.6.12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., and Mizushima N.. 2008. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 19:5360–5372. 10.1091/mbc.e08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Jung J., Nayak A., Schaeffer V., Starzetz T., Kirsch A.K., Müller S., Dikic I., Mittelbronn M., and Behrends C.. 2017. Multiplex image-based autophagy RNAi screening identifies SMCR8 as ULK1 kinase activity and gene expression regulator. eLife. 6:1–32. 10.7554/eLife.23063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., and Yoshimori T.. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizuka T., Morishita H., Hama Y., Tsukamoto S., Matsui T., Toyota Y., Kodama A., Ishihara T., Mizushima T., and Mizushima N.. 2016. An autophagic flux probe that releases an internal control. Mol. Cell. 64:835–849. 10.1016/j.molcel.2016.09.037 [DOI] [PubMed] [Google Scholar]
- Keller R., Ziegler C., and Schneider D.. 2014. When two turn into one: evolution of membrane transporters from half modules. Biol. Chem. 395:1379–1388. 10.1515/hsz-2014-0224 [DOI] [PubMed] [Google Scholar]
- Khafizov K., Staritzbichler R., Stamm M., and Forrest L.R.. 2010. A study of the evolution of inverted-topology repeats from LeuT-fold transporters using AlignMe. Biochemistry. 49:10702–10713. 10.1021/bi101256x [DOI] [PubMed] [Google Scholar]
- Kishi-Itakura C., Koyama-Honda I., Itakura E., and Mizushima N.. 2014. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell Sci. 127:4089–4102. 10.1242/jcs.156034 [DOI] [PubMed] [Google Scholar]
- Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., and Kumagai H.. 2003. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31:1007–1014 https://doi.org/S0301472X03002601. [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., and Sonnhammer E.L.L.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., and Tamura K.. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35:1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C.A., Yoshimori T., and Tooze S.A.. 2013. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14:759–774. 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- Matsui T., Jiang P., Nakano S., Sakamaki Y., Yamamoto H., and Mizushima N.. 2018. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J. Cell Biol. 10.1083/jcb.201712058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer C.A., Kaliappan A., and Dennis P.B.. 2009. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 5:649–662. 10.4161/auto.5.5.8249 [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2018. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 20:521–527. 10.1038/s41556-018-0092-5 [DOI] [PubMed] [Google Scholar]
- Mizushima N., and Komatsu M.. 2011. Autophagy: renovation of cells and tissues. Cell. 147:728–741. 10.1016/j.cell.2011.10.026 [DOI] [PubMed] [Google Scholar]
- Mukaiyama H., Oku M., Baba M., Samizo T., Hammond A.T., Glick B.S., Kato N., and Sakai Y.. 2002. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells. 7:75–90. 10.1046/j.1356-9597.2001.00499.x [DOI] [PubMed] [Google Scholar]
- Nishimura T., Tamura N., Kono N., Shimanaka Y., Arai H., Yamamoto H., and Mizushima N.. 2017. Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J. 36:1719–1735. 10.15252/embj.201695189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., Sumpter R. Jr., Xiao G., Ng A., Zou Z., Tang Y., Narimatsu M., Gilpin C., Sun Q., Roth M., et al. 2011. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 480:113–117. 10.1038/nature10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropolo A., Grasso D., Pardo R., Sacchetti M.L., Archange C., Lo Re A., Seux M., Nowak J., Gonzalez C.D., Iovanna J.L., and Vaccaro M.I.. 2007. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J. Biol. Chem. 282:37124–37133. 10.1074/jbc.M706956200 [DOI] [PubMed] [Google Scholar]
- Saitoh T., Nakano H., Yamamoto N., and Yamaoka S.. 2002. Lymphotoxin-β receptor mediates NEMO-independent NF-kappaB activation. FEBS Lett. 532:45–51. 10.1016/S0014-5793(02)03622-0 [DOI] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., and Zhang F.. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343:84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strømhaug P.E., Bevan A., and Dunn W.A. Jr. 2001. GSA11 encodes a unique 208-kDa protein required for pexophagy and autophagy in Pichia pastoris. J. Biol. Chem. 276:42422–42435. 10.1074/jbc.M104087200 [DOI] [PubMed] [Google Scholar]
- Sun L.L., Li M., Suo F., Liu X.M., Shen E.Z., Yang B., Dong M.Q., He W.Z., and Du L.L.. 2013. Global analysis of fission yeast mating genes reveals new autophagy factors. PLoS Genet. 9:e1003715 10.1371/journal.pgen.1003715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tábara L.-C., and Escalante R.. 2016. VMP1 establishes ER-microdomains that regulate membrane contact sites and autophagy. PLoS One. 11:e0166499 10.1371/journal.pone.0166499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., and Wolf D.H.. 1994. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349:275–280. 10.1016/0014-5793(94)00672-5 [DOI] [PubMed] [Google Scholar]
- Tian Y., Li Z., Hu W., Ren H., Tian E., Zhao Y., Lu Q., Huang X., Yang P., Li X., et al. 2010. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 141:1042–1055. 10.1016/j.cell.2010.04.034 [DOI] [PubMed] [Google Scholar]
- Tsukada M., and Ohsumi Y.. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169–174. 10.1016/0014-5793(93)80398-E [DOI] [PubMed] [Google Scholar]
- Tusnády G.E., and Simon I.. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics. 17:849–850. 10.1093/bioinformatics/17.9.849 [DOI] [PubMed] [Google Scholar]
- van Dijk R., Faber K.N., Hammond A.T., Glick B.S., Veenhuis M., and Kiel J.A.K.W.. 2001. Tagging Hansenula polymorpha genes by random integration of linear DNA fragments (RALF). Mol. Genet. Genomics. 266:646–656. 10.1007/s004380100584 [DOI] [PubMed] [Google Scholar]
- Wang T. 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 343:80–84. 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Miao G., Xue X., Guo X., Yuan C., Wang Z., Zhang G., Chen Y., Feng D., Hu J., and Zhang H.. 2016. The Vici syndrome protein EPG5 is a Rab7 Effector that determines the fusion specificity of autophagosomes with late endosomes/lysosomes. Mol. Cell. 63:781–795. 10.1016/j.molcel.2016.08.021 [DOI] [PubMed] [Google Scholar]
- Zhao Y.G., Chen Y., Miao G., Zhao H., Qu W., Li D., Wang Z., Liu N., Li L., Chen S., et al. 2017. The ER-Localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell. 67:974–989. 10.1016/j.molcel.2017.08.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.