Venkei and Yamashita summarize recent advances in our understanding of asymmetric stem cell division in tissue homeostasis.
Abstract
The asymmetric cell division of stem cells, which produces one stem cell and one differentiating cell, has emerged as a mechanism to balance stem cell self-renewal and differentiation. Elaborate cellular mechanisms that orchestrate the processes required for asymmetric cell divisions are often shared between stem cells and other asymmetrically dividing cells. During asymmetric cell division, cells must establish asymmetry/polarity, which is guided by varying degrees of intrinsic versus extrinsic cues, and use intracellular machineries to divide in a desired orientation in the context of the asymmetry/polarity. Recent studies have expanded our knowledge on the mechanisms of asymmetric cell divisions, revealing the previously unappreciated complexity in setting up the cellular and/or environmental asymmetry, ensuring binary outcomes of the fate determination. In this review, we summarize recent progress in understanding the mechanisms and regulations of asymmetric stem cell division.
Introduction
Asymmetric cell division is a widespread process, occurring in organisms ranging from prokaryotes to highly complex multicellular organisms (Pereira et al., 2001; Inaba and Yamashita, 2012). In multicellular organisms, asymmetric cell division is critical for fate diversification. Asymmetric division of stem cells creates one stem cell and one differentiating cell, a simple yet elegant way to balance stem cell self-renewal and differentiation (Morrison and Kimble, 2006; Knoblich, 2008; Inaba and Yamashita, 2012; Chen et al., 2016a). This balance in turn ensures long-term tissue homeostasis, a failure of which is speculated to lead to tumorigenesis and/or tissue degeneration (Morrison and Kimble, 2006; Chen et al., 2016a).
Asymmetric stem cell division involves a sequence of coordinated processes. Cell fate–determining factors are provided either cell extrinsically (Fig. 1 A) or intrinsically (Fig. 1 B) to stem cells in a polarized manner. By coordinating the division orientation with the position of polarized fate determinants, the daughters of stem cells acquire distinct fates: either to self-renew their stem cell identity or to commit to differentiation. Earlier work has revealed many of the basic fundamental mechanisms for asymmetric cell divisions, while recent progress has made it clear that asymmetric stem cell division involves many additional layers of regulation.
Figure 1.
Framework of asymmetric cell division. (A and B) Asymmetric cell division dictated by extrinsic (A) or intrinsic (B) fate determinants. (C) Asymmetric division of Drosophila male GSC. The hub cells provide the polarized source of fate determinants (self-renewal ligands Upd and Dpp), which are received by GSC receptor Dome and Tkv, respectively. GSCs are attached to the hub via adherens junctions, ensuring their retention in the niche. The mother centrosome anchors to the adherens junctions via astral MTs, instructing spindle orientation in mitosis. In parallel, the receptor Dome binds to Eb1 to capture MTs to orient the spindle. GSC division creates a gonialblast (GB), the differentiating daughter. (D) Drosophila NBs divide asymmetrically by segregating fate determinants (e.g., Miranda and Prospero) to GMCs (green crescent). Apical polarity complex (e.g., Par3–Par6–aPKC complex and Pins; brown crescent) captures MTs from the activated daughter centrosome to orient the spindle.
In this review, we will first briefly describe the framework of asymmetric stem cell division, although we refer the readers to recent reviews on the topic for a detailed discussion on these established frameworks. Then, we will focus on emerging mechanisms that reveal the complexity of regulation in achieving asymmetric stem cell division.
Framework of asymmetric cell division
The term “asymmetric cell division” ultimately refers to the asymmetry in cell fates, although many other forms of asymmetries accompany cell divisions, as will be discussed. Accordingly, in defining asymmetric cell division, the most critical asymmetry is that of fate-determining factors. Fate-determining factors can be provided in two ways: (1) extracellular environments that define cell fate may be presented to two daughter cells in an asymmetric manner, and (2) intracellular fate determinants may be polarized within a cell and segregated asymmetrically upon cell division (Fig. 1, A and B).
Extracellular environments that define stem cell identity are called stem cell niches. Niches typically present signaling molecules (such as ligands) to stem cells, which activate downstream transcriptional networks within stem cells to specify their identity (Morrison and Spradling, 2008; Losick et al., 2011). For example, Drosophila melanogaster male and female germline stem cells (GSCs) provide two of the best-characterized models of asymmetric stem cell division within the niche (Fuller and Spradling, 2007; Lehmann, 2012). In the Drosophila testes, postmitotic somatic hub cells function as a major constituent of the stem cell niche by secreting the critical self-renewal ligands Unpaired (Upd; a cytokine homologue) and Decapentaplegic (Dpp)/Glass bottom boat (Gbb; both of which are bone morphogenetic protein signaling pathway ligands; Fig. 1 C; Kiger et al., 2001; Tulina and Matunis, 2001; Shivdasani and Ingham, 2003; Kawase et al., 2004; Schulz et al., 2004). In the Drosophila ovary, terminal filament cells and cap cells constitute the niche by secreting Dpp ligand (Xie and Spradling, 2000).
Alternatively, stem cell identity can be determined by intrinsic fate determinants. In such a scenario, asymmetric division is achieved by polarizing fate determinants on one side of the cell, which are subsequently segregated into only one daughter of the division. The Drosophila neuroblast (NB) presents the best-understood example of this category (Doe and Bowerman, 2001; Yu et al., 2006; Prehoda, 2009; Gallaud et al., 2017). In mitotic Drosophila NBs, a number of fate-determining factors (Numb, brain tumor [Brat], Prospero, Miranda, Staufen and prospero mRNA) are polarized basally to be subsequently inherited by the differentiating daughter called ganglion mother cells (GMCs; Fig. 1 D; Reichert, 2011; Sousa-Nunes and Somers, 2013; Janssens and Lee, 2014; Gallaud et al., 2017). These fate-determining factors are produced by NBs, remain inactive in NBs, and then ensure differentiation upon segregation into GMCs.
By coordinating with the position of the fate-determining factors, stem cells polarize their cytoskeleton to orient their spindles in preparation for asymmetric stem cell division. Here, the ultimate goal is to orient the division plane such that the two daughter cells will adapt distinct cell fates. In general, a particular area of cell cortex is specified to anchor the spindle. Such a cortical area is formed in coordination with other landmarks of the cell such as the position of the stem cell niche or fate determinants. In short, the cell cortex anchors machineries that capture microtubule (MT) plus ends that emanate from the centrosome/spindle pole. When this anchoring is coupled with depolymerization of MTs, it generates pulling forces that can anchor the spindle perpendicular to the cortex. In many cases, dynein plays dual roles in capturing MT plus ends and generating pulling force by walking toward the minus end of the MTs (Grill and Hyman, 2005; di Pietro et al., 2016).
These basic mechanisms appear to be ubiquitously used in many asymmetrically dividing cells including nonstem cells. While this framework describes the fundamentals of asymmetric cell division, recent studies have revealed the necessity of more complex mechanisms to ensure asymmetric stem cell divisions. In the following sections, we will review these emerging mechanisms.
Coupling spindle orientation with the landmark of stem cell asymmetry
The most critical aspect of asymmetric stem cell division is polarization of fate determinants and division orientation. In Drosophila NBs, the Par3–Par6–atypical protein kinase C (aPKC) polarity complex forms the apical crescent, which marks one side of the NB to instruct both the spindle orientation and the position of the fate determinants at the other side of the NB as the basal crescent, thereby coordinating these two processes (Gallaud et al., 2017). Although these cell-intrinsic machineries are sufficient to achieve asymmetric NB division, external cues also instruct the overall cell polarity in the context of the tissue. When an embryonic NB is attached to epithelial cells (in the intact tissue as well as in a cell culture), the position of the apical crescent is determined and maintained with respect to the cell contact (Siegrist and Doe, 2006). This is mediated by activation of the G protein–coupled receptor Trapped in endoderm 1 (Tre1) in NBs. Tre1 in turn recruits Pins, a major organizer of the apical crescent, to the apical side defined by the presence of epithelial cells, thereby coordinating the position of the epithelial layer with the NB polarity (Fig. 1 D; Yoshiura et al., 2012).
When the cell fate determinants are provided extrinsically, the cells must sense from which side the signal is coming and convey this information to the machinery that orients the spindle. Drosophila male GSCs divide asymmetrically by orienting the spindle perpendicular to the hub cells (Fig. 1 C; Yamashita et al., 2003). This spindle orientation is precisely determined by the positioning of the centrosomes during interphase: the mother centrosome is typically anchored near the hub cells, whereas the daughter centrosome migrates toward the opposite side to prepare correct orientation of the spindle (Yamashita et al., 2007). Drosophila female GSCs also orient their spindles perpendicular to the cap cells to divide asymmetrically. The major mechanism that orients spindles toward the cap cells is the spectrosome, a germline-specific membranous organelle. The spectrosome is almost always positioned near the cap cells, and the spindle is anchored to the spectrosome (Deng and Lin, 1997). In addition, the centrosomes are also oriented with respect to the cap cells, contributing to the spindle orientation (Lu et al., 2012).
How GSCs sense the position of the niche cells to orient their spindle is of critical importance in achieving asymmetric divisions. In the male GSC niche, two parallel mechanisms appear to orient the spindle with respect to the hub. First, GSCs attach to the hub cells via adherens junctions to remain in the niche (Yamashita et al., 2003), which is also the case for female GSCs (Song et al., 2002), and this adhesion provides an ideal landmark for GSCs to determine which side is the hub/cap cell (Fig. 1 C). Adherens junctions anchor astral MTs emanating from the mother centrosome (Yamashita et al., 2007), and E-cadherin, a component of adherens junctions, plays a critical role in male GSC spindle orientation (Inaba et al., 2010). In parallel, male GSCs also use the self-renewal ligand Upd to directly sense the position of the niche cells. Upd secreted from the hub cells dictates the localization of its receptor Dome, which in turn directly binds to a major MT regulator, plus end–binding protein 1 (Eb1), to orient the spindle (Fig. 1 C; Chen et al., 2018).
Ligand–receptor interactions, as observed in male GSCs, may be universal mechanisms for cells to sense the position of signal-sending cells. For example, during early development of Caenorhabditis elegans embryos, the spindle in the endomesodermal precursor (EMS) cell is oriented toward the P2 cell (germ cell precursor). The P2 cell expresses the Wnt ligand MOM-2, whereas the EMS cell expresses its receptor Frizzled (MOM-5), regulating the spindle orientation in EMS cells (Goldstein et al., 2006). Mouse embryonic stem cells are shown to polarize toward the Wnt3a ligand, when it is presented on the beads, leading to polarization of the cell and spindle orientation toward the beads (Habib et al., 2013). Although this is a phenomenon observed in an in vitro setting, it is striking in that it demonstrates the ability of a single ligand species to induce the polarization of the entire cell. These results indicate that the cells can directly sense the position of the ligands (thus the position of ligand-expressing cells) via receptors, using these as cues to orient the spindles toward the ligand source.
Certain stem cells attach to the ECM/basement membrane, which provides a polarity cue toward which stem cell division orients. For example, in mouse epidermal stem cells, stem cells adhere to the basement membrane with integrin, and the spindle is oriented toward the basement membrane to divide asymmetrically (Lechler and Fuchs, 2005; Williams et al., 2011; Seldin et al., 2013), although other mechanisms such as cell migration might also contribute to the asymmetric outcome (Poulson and Lechler, 2012). Similar mechanisms involving the basement membrane as a polarity cue in spindle orientation/asymmetric stem cell division have been observed in several systems including Drosophila intestinal stem cells (Goulas et al., 2012), mouse spinal cord neuroepithelial stem cells (Farkas and Huttner, 2008; Loulier et al., 2009), and mouse muscle satellite cells (Kuang et al., 2007; Le Grand et al., 2009; Bentzinger et al., 2013), suggesting a universality of this mechanism.
In addition, recent studies illuminated the potential influence of physical properties of cells and their environments (such as membrane stiffness) on stem cells’ behavior. For example, in human hematopoietic stem/progenitor cells, myosin-IIb was observed to be asymmetrically inherited during their divisions (Shin et al., 2014). As myosin-IIb was shown to localize to where cells are mechanically stressed, it raises a possibility that the endogenous niche environment may use such characteristics to polarize stem cells to achieve asymmetric stem cell divisions. In another example, it was reported that asymmetric divisions of mouse satellite cells (muscle stem cells) are influenced by cellular geometry (Yennek et al., 2014): depending on the micropatterns to which cultured satellite cells are adhered, satellite cells underwent asymmetric versus symmetric divisions assessed by nonrandom sister chromatid segregation. Although these studies have not examined the spindle orientation as a mechanism to achieve observed asymmetries, it is tempting to speculate that the physical environment of the niche influences stem cell behaviors, thus regulating asymmetric cell divisions.
Establishing asymmetry for binary fate determination
As described, polarization of fate determinants and spindle orientation are two critical aspects of asymmetric stem cell divisions. However, recent studies have demonstrated that additional layers of regulation are required to ensure the binary outcome of asymmetric cell division. Much of the processes of cell division (proliferation), such as DNA replication/repair and chromosome segregation, are designed to generate two essentially identical cells with the same genetic information and similar sets of cellular components (organelles and cytoplasm). Also, two daughter cells are juxtaposed to each other at the completion of cell division, and thus it is not an easy task to present these two cells with drastically different microenvironments such that two cells would take different fates. Therefore, there must be mechanisms that strictly define the asymmetry and/or amplify the initial, possibly subtle asymmetries to achieve binary cell fates through a single cell division. Recent progress provides a glance at these mechanisms that ensure binary asymmetric fates: self-renewal versus differentiation.
Defining the niche space
Although asymmetric inheritance of niche interface may seem a simple way by which stem cells divide asymmetrically, additional layers of regulation are required to ensure that the two daughters of a stem cell division are exposed to different signaling environments. Considering that two daughters of a stem cell division are juxtaposed to each other and that the ligands from the niche can diffuse, defining the border of the niche between two juxtaposing cells requires elaborate mechanisms. For example, glypican-type transmembrane proteins, which stabilize ligands in the extracellular space (Yan and Lin, 2009; Sarrazin et al., 2011), contribute to defining the niche space. Hub cells in the male GSC niche and cap cells in the female GSC niche express the glypicans, dally-like and dally, respectively, and these glypican molecules are critical to maintaining GSCs by stabilizing Upd and Dpp within the niche and restricting its diffusion outside the niche (Fig. 2 A; Hayashi et al., 2009). In addition, Drosophila male GSCs use thin protrusions termed MT-based nanotubes (MT-nanotubes) to limit the niche signaling to GSCs (Inaba et al., 2015a). GSCs form MT-nanotubes, which extend into the invagination of hub cells. Thickveins (Tkv), the receptor for Dpp, is expressed in GSCs and specifically trafficked into MT-nanotubes, where it interacts with Dpp secreted from the hub cells. In this manner, the surface of MT-nanotubes is used as an exclusive platform for productive ligand (Dpp)–receptor (Tkv) interaction. This effectively excludes nonstem cells from the influence of the Dpp signaling. In this manner, the potential influence of diffusive ligands on nonstem cells is prevented, creating the sharp boundary of inside versus outside of the niche (Fig. 2 A). Similar thin protrusions called cytonemes are reported to deliver the Hh ligand from the cap cells to another type of somatic cells, escort cells, in creating the female GSC niche (Rojas-Ríos et al., 2012), suggesting that targeted delivery of signaling molecules is a critical means by which cells communicate without influencing nearby cells.
Figure 2.
Breaking symmetry and amplifying asymmetry for binary fate determination. (A) In Drosophila male GSCs, MT-nanotubes and glypican restrict the effective range of niche signaling. MT-nanotubes are extended from the GSC into the hub, where Tkv in the GSC is recruited and engages in signaling with Dpp secreted from the hub. Glypican binds to secreted ligands to maintain their effective concentration near the niche while preventing their diffusion. These mechanisms contribute to limit the niche signaling to GSCs while excluding nonstem cells. (B) Mother–daughter centrosome asymmetry may promote fate asymmetry in apical progenitor cells, the neural stem cells in the ventricle zone of the developing neocortex in mice. Upon mitotic entry, the mother centrosome retains a remnant of ciliary membrane, leading to faster ciliary growth in the next cell cycle. This may break the symmetry of two daughter cells as the cell with the mother centrosome might engage in signaling sooner than its sibling. (C) In Drosophila SOP cells, symmetry breaking on the central spindle leads to biased segregation of SARA endosomes to pIIa cells in addition to asymmetric segregation of cortically localized Numb. These two mechanisms together lead to high Notch activation in pIIa cells, resulting in fate asymmetry.
Breaking symmetry and amplifying asymmetry
As discussed, tissues may create two distinct microenvironments by limiting the diffusion of ligands such that two daughter cells are exposed to different signaling ligands. However, many types of asymmetric divisions require different mechanisms. For example, self-renewal ligands may be provided from a distant tissue under certain circumstances (e.g., endocrine-based signaling). For example, proliferation of mouse hematopoietic stem cells is known to be positively regulated by estrogen (Nakada et al., 2014). In such a scenario, alternative mechanisms are required to break the symmetry of the two daughter cells to make them adapt two distinct fates.
The difference between the mother versus daughter centrosome may provide such a mechanism to break symmetry, using the initial, possibly subtle asymmetry to generate fate asymmetry. Stereotypical inheritance of the mother versus daughter centrosomes have been observed in several systems. Such a phenomenon was first discovered in Drosophila male GSCs (Yamashita et al., 2007), where the mother centrosome is consistently inherited by the stem cells as described previously (Fig. 1 C). A similar pattern of inheritance, i.e., the mother centrosome being inherited by the stem cells, was observed in mouse radial glial progenitor cells (Wang et al., 2009). Interestingly, in Drosophila NBs and female GSCs (Conduit and Raff, 2010; Januschke et al., 2011; Salzmann et al., 2014), the stem cells consistently inherit the daughter centrosomes. NBs undergo elaborate processes to switch the MT-organizing center (MTOC) activity of mother versus daughter centrosomes to allow the inheritance of the daughter centrosome by NBs and/or the inheritance of the mother by GMCs (Fig. 1 D; Rusan and Peifer, 2007; Lerit and Rusan, 2013; Singh et al., 2014).
Although the significance of the difference between mother versus daughter centrosomes remains incompletely understood, it may provide an elegant way to amplify a subtle asymmetry. Following division, the cell that inherited the mother centrosome grows the primary cilia earlier than its sibling that inherited the daughter centrosome (Anderson and Stearns, 2009). This could bias the receptivity of the two sibling cells to signaling ligands such as Hedgehog, conferring them with distinct cell fates. In mouse radial glial progenitor cells, the mother centriole carries ciliary membrane when it is internalized before mitosis. This inheritance of ciliary membrane promotes a faster reassembly of primary cilia in the next cell cycle (Fig. 2 B; Paridaen et al., 2013). These results suggest that the difference between mother versus daughter centrosomes can trigger the cell fate asymmetry. Once triggered, two cells can signal to each other to further enhance distinct fates via juxtacrine signaling.
Another elegant example of how asymmetry is created and then amplified to generate two distinct cell fates is found in Drosophila sensory organ precursor cells (SOPs). SOPs reside in the single-layered epithelium of the Drosophila pupal thorax, where their asymmetric division generates two daughter cells, pIIa and pIIb, which eventually generate four different cells that constitute a single mechanosensory hair unit (hair cell, socket cell, neuron, and glia; Schweisguth, 2015). A SOP is planar polarized within the epithelial layer and divides along this axis to generate posterior pIIa and anterior pIIb cells (Fig. 2 C). Binary fate determination of pIIa and pIIb cells depends on Notch signaling between these two cells: pIIb produces Notch ligands (Dl and Ser), which are received by the pIIa cell to confer its identity. The asymmetry between pIIa and pIIb is created in two ways: Numb is asymmetrically segregated to pIIb cells during SOP division, which later functions as an antagonist of Notch signaling, repressing Notch activity in pIIb cells. In parallel, the internalized pool of Delta and Notch is associated with the Smad anchor for receptor activation (Sara) endosomes, which are trafficked to pIIa cells during cytokinesis, enhancing Notch activation in pIIa cells (Coumailleau et al., 2009). This biased segregation of Sara endosomes into pIIa is mediated by biased MT orientation at the central spindle, whereby more plus ends are oriented toward the pIIa side and the kinesin-like protein at 98A carries Sara endosomes toward the pIIa cell (Derivery et al., 2015; Loubéry et al., 2017). The Sara endosomes have been shown to segregate asymmetrically in other cell types as well, such as zebrafish neural precursor cells (Kressmann et al., 2015) and Drosophila intestinal stem cells (Montagne and Gonzalez-Gaitan, 2014), suggesting the conservation of this mechanism in generating fate asymmetry.
Checkpoint mechanisms to safeguard asymmetric division
Critical cellular processes are often under the regulation of checkpoint mechanisms, which ensure the completion of a certain process before a cell can proceed to the next step. For example, the DNA damage checkpoint ensures that DNA replication and repair is complete before mitotic entry (Löbrich and Jeggo, 2007), and the spindle assembly checkpoint verifies that all kinetochores have established bipolar attachments before anaphase (Musacchio, 2015). Lack of these checkpoints can cause mutations or chromosome missegregation. The failure of asymmetric stem cell division can have comparable deleterious consequences for cells, tissues, and organisms. Indeed, failure in asymmetric stem cell division has been speculated to lead to stem cell overproliferation due to symmetric self-renewal or to stem cell depletion due to symmetric differentiation (Morrison and Kimble, 2006). Although there is no definitive proof to date that the defective asymmetric stem cell division is the root cause of stem cell overproliferation or depletion, spindle misorientation has been shown to increase stem cell number in several contexts due to erroneous inheritance of fate-determining factors (Yamashita et al., 2003; Cabernard and Doe, 2009), highlighting the importance of ensuring successful asymmetric stem cell divisions.
In the unicellular budding yeast, a spindle position checkpoint mechanism monitors that the spindle orientation/position is correct during their divisions (Pereira et al., 2001; Seshan and Amon, 2004; Caydasi and Pereira, 2012). A few checkpoint or checkpoint-like phenomena in stem cells of multicellular organisms have been reported. In Drosophila NBs, spindle misorientation in metaphase is often corrected by a phenomenon called telophase rescue, where the spindle reorients correctly and divides asymmetrically (Siller and Doe, 2008). Also, in mitotic NBs mutant for centrosomal proteins Bld10/Cep135 or Wdr62, the metaphase spindle is often misoriented due to defective establishment of mother–daughter centrosome asymmetry, but this does not lead to symmetric division, pointing to the presence of a correction (thus checkpoint) mechanism (Singh et al., 2014; Ramdas Nair et al., 2016). Although the molecular identity of these phenomena and whether these represent a checkpoint mechanism remains unknown, tight temporal orders of two processes point to the presence of such mechanisms.
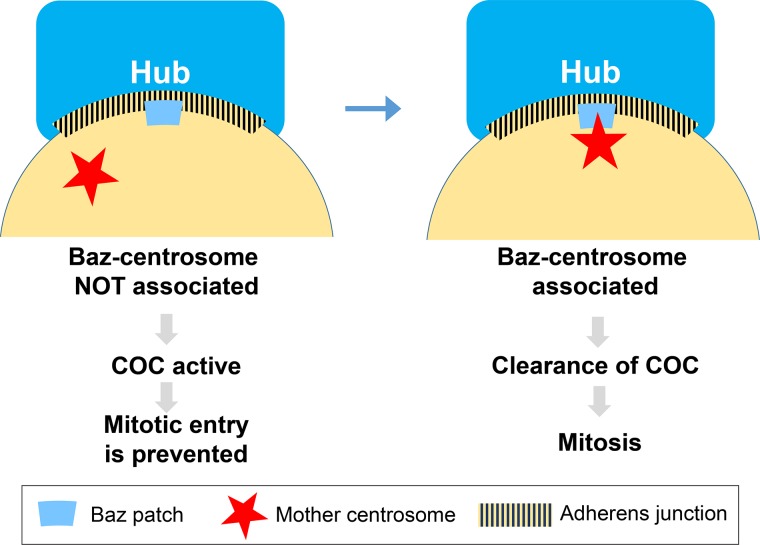
The Drosophila male GSCs possess a centrosome orientation checkpoint (COC) to ensure correct spindle orientation. The COC monitors centrosome orientation in interphase and halts mitotic entry if centrosomes are not correctly oriented (Fig. 3; Cheng et al., 2008; Pereira and Yamashita, 2011). COC activity is specific to stem cells, and differentiating spermatogonia (SGs), which divide symmetrically, do not exhibit this checkpoint activity, suggesting that COC is specifically designed to ensure asymmetric stem cell divisions (Venkei and Yamashita, 2015). To date, a few molecular players of the COC have been characterized. A polarity protein Bazooka (Baz)/Par-3 forms a small ∼2-µm Baz patch along the hub–GSC interface. The mother centrosome is closely associated with the Baz patch right before mitotic entry. That centrosome orientation is correct appears to be inferred by this association between the Baz patch and the centrosome, thus permitting mitotic entry of GSCs (Inaba et al., 2015b). Par-1, a kinase that phosphorylates Baz, is also a part of COC. Whereas Par-1–dependent phosphorylation of Baz is critical for COC activity, Par-1 also functions to sequester cyclin A to the spectrosome, a membranous, ER-like organelle specific to germ cells, to prevent mitotic entry (Yuan et al., 2012). It remains unknown how Par-1 phosphorylation of Baz and sequestration of Cyclin A are coordinated to regulate COC activity.
Figure 3.
The COC: A safeguard mechanism of spindle orientation in Drosophila male GSCs. In interphase Drosophila male GSCs, the polarity protein Baz (Par3) forms a small structure along the adherens junction (Baz patch), which becomes a docking point for the mother centrosome. GSCs interpret the Baz patch–centrosome interaction as indicating the correct centrosome orientation, permitting mitotic entry. This mechanism ensures that GSCs enter mitosis only when they are ready to orient the mitotic spindle perpendicular to the hub cells.
Interestingly, the COC is used in a different context: in response to poor nutrition, centrosomes become highly misoriented in GSCs, which in turn activates COC and thus reduces the rate of GSC proliferation (Roth et al., 2012). In this manner, the tissue adapts to poor nutrition by reducing resources for spermatogenesis. During aging, GSC centrosomes become highly misoriented, contributing to the decline in spermatogenesis during aging (Cheng et al., 2008). Centrosome misorientation during aging, however, is not a process of adaptation as is the case due to poor nutrition. Instead, GSC centrosome misorientation during aging is due to the increase in dedifferentiation (Cheng et al., 2008): although dedifferentiation provides a mechanism to maintain GSC pool during aging, dedifferentiated GSCs are not as potent as native ones, leading to an eventual decline in spermatogenesis.
Drosophila female GSCs appear to have a similar checkpoint that arrests GSCs in response to misoriented centrosomes. In this case, if centrosomes are not correctly oriented, GSCs enter mitosis and arrest in prophase (Lu et al., 2012). However, in female GSCs, the major mechanism that orients the spindle is the anchoring of the spindle pole to the spectrosome (Deng and Lin, 1997), and it remains unknown how spindle orientation may be monitored in female GSCs.
Asymmetric segregation of cellular components during stem cell divisions
In addition to the asymmetric segregation of cell fate determinants, asymmetric stem cell division involves many additional asymmetries, the implications of which are beginning to be understood.
Centrosomes
As described earlier, mother versus daughter centrosomes segregate in a stereotypical manner during asymmetric divisions of several stem cell systems. The two centrosomes in a cell are intrinsically asymmetric due to a semiconservative manner of centrosome duplication, and their age difference results in differential activity of mother versus daughter centrosomes as MTOC (Yamashita et al., 2007; Pelletier and Yamashita, 2012; Gasic et al., 2015; Loncarek and Bettencourt-Dias, 2018).
In addition to the roles of centrosome asymmetry in centrosome positioning/spindle orientation and symmetry breaking (Fig. 1, C and D), differential MTOC activities between mother versus daughter centrosomes can host a plethora of asymmetries. For example, fate-determining mRNAs have been observed to be associated with the centrosomes (Lambert and Nagy, 2002; Lécuyer et al., 2007; Ramat et al., 2017), raising a possibility that the mother (or daughter) centrosomes may be specifically associated with such mRNAs during asymmetric cell divisions. In addition, other asymmetries that will be discussed below have been reported to require functional centrosomes, pointing to the possibility that mother–daughter centrosome asymmetry orchestrates asymmetric cell division in its entirety.
The unique behavior of centrosomes in asymmetrically dividing stem cells has led to the idea that stem cell centrosomes are uniquely regulated, possibly containing stem cell–specific centrosomal components. On one hand, the repeated inheritance of the mother centrosome, which has higher MTOC activity, by Drosophila male GSCs (Yamashita et al., 2007) or mouse neural progenitor cells (Wang et al., 2009) may be interpreted as a default of anchoring one centrosome in a position for correct spindle orientation and may not necessarily represent special fate-determining properties. However, in some other stem cells such as Drosophila NBs (Conduit and Raff, 2010; Januschke et al., 2011) and female GSCs (Salzmann et al., 2014), daughter centrosomes are inherited by the stem cells. In particular, Drosophila NBs undergo elaborate processes of switching the MTOC activities between mother and daughter centrosomes, down-regulating the mother centrosome and up-regulating the daughter, pointing to the possibility that there are reasons for the mother or daughter centrosome to be inherited by specific cells. Several centrosomal proteins have been identified as being required for maintaining mother–daughter asymmetry and their accurate inheritance. For example, Centrobin (Cnb) specifically localizes to the daughter centrosome and is required for dominant MTOC activity of the daughter centrosome (Januschke et al., 2013). Wdr62 is required to maintain high MTOC activity of the daughter centrosome (Ramdas Nair et al., 2016). Bld10/Cep135 is required to down-regulate the mother (Singh et al., 2014). Several other genes including Plp and calmodulin have been shown to regulate mother–daughter centrosome inheritance (Schoborg et al., 2015). Defective mother–daughter asymmetry or inheritance in the mutants of these genes only impacts spindle orientation, and no drastic outcome in cell fate has been observed. Therefore, biological significance of mother–daughter centrosome inheritance beyond its role in spindle orientation remains unknown.
These elaborate regulatory mechanisms to ensure asymmetric behavior and inheritance of mother versus daughter centrosomes imply the presence of stem cell–specific regulation of centrosomes. Such stem cell–specific centrosome behaviors are likely regulated by stem cell–specific centrosomal proteins. Accordingly, identification of stem cell centrosome-specific proteins will be of particular interest in the future investigation. However, the difficulty in obtaining sufficient amounts/purities of stem cells for proteomic analysis has hampered such efforts. To date, only one such protein has been identified: Kinesin-like protein at 10A (Klp10A), a depolymerizing kinesin, has been identified as a protein that is enriched on the centrosomes of stem cells but not those of differentiating cells in the Drosophila male germline (Chen et al., 2016b). Interestingly, depletion of Klp10A led to abnormal elongation of the mother centrosome in stem cells without influencing the size of the daughter centrosome in GSCs or any centrosomes in nonstem cells. These observations indeed indicate that the stem cell (mother) centrosomes are under unique regulations. Components that specifically localize to the mother (or daughter) centrosomes of stem cells that determine/influence cell fates are yet to be identified.
Asymmetries in spindle and cell size
Asymmetric cell division is sometimes associated with the asymmetry in cell size. For example, the first division of the C. elegans zygote yields a larger anterior cell and a smaller posterior cell, which is mediated by spindle pulling force toward the posterior cortex (Grill et al., 2003; Grill and Hyman, 2005). Drosophila NB divisions also yield a larger NB and a smaller GMC (Fig. 1 D). In the case of NBs, asymmetric daughter cell size involves asymmetric spindle morphology: the apical half spindle is much larger than the basal half spindle. In addition, a specialized cytokinesis mechanism, mediated by regulation of cortical myosin, ensures that cleavage furrow is positioned basally to generate a smaller GMC (Cabernard et al., 2010; Roubinet et al., 2017; Tsankova et al., 2017). Drosophila male GSCs’ division normally yields two daughter cells of the same size. However, interestingly, when the mother centrosome becomes abnormally large in the absence of Klp10A, the spindle becomes asymmetric, yielding a larger GSC and a smaller GB (differentiating daughter of GSC), similar to NB divisions (Chen et al., 2016b). This leads to frequent death of small GBs, suggesting that cell size regulation is a critical aspect of asymmetric cell division.
Sister chromatids
Although sister chromatids are considered to be exact copies of each other, several hypotheses have been put forward to postulate that asymmetries in certain aspects associated with sister chromatids may underlie asymmetric cell divisions. The immortal strand hypothesis was one of the first hypotheses for this concept and proposes that long-living cells such as stem cells may avoid accumulation of replication-induced mutations by preferentially inheriting older DNA strands (Rando, 2007). Many studies to test this hypothesis with the heterogenous cell populations with low stem cell frequency have left ambiguity. Mouse satellite cells (muscle stem cells) and human/mouse embryonic stem cells exhibit a high frequency of biased sister chromatid segregation when old versus new sister chromatids are distinguished by pulse labeling of DNA with bromodeoxyuridine (Conboy et al., 2007; Rocheteau et al., 2012; Elabd et al., 2013). Old DNA strands exhibit a tendency to segregate to the less-differentiated cells during satellite cell divisions (Conboy et al., 2007; Rocheteau et al., 2012). In embryonic stem cells, the involvement of epigenetic information was implied, as the biased DNA strand segregation depends on the DNA methyltransferase Dnmt3 (Elabd et al., 2013). Drosophila male GSCs do not exhibit immortal strand segregation (Yadlapalli et al., 2011), although female GSCs are reported to exhibit immortal strand segregation (Karpowicz et al., 2009). Despite not segregating the immortal strands, male GSCs segregate sister chromatids of X and Y chromosomes with a striking bias in a manner dependent on the functional centrosome (Fig. 4 A; Yadlapalli and Yamashita, 2013). In all these cases, the underlying mechanisms remain largely elusive. Likewise, there is no evidence that the biased segregation functions to avoid accumulation of mutations, and accordingly, the meaning of nonrandom sister chromatid segregation is yet to be established.
Figure 4.

Biased segregation of cellular components during asymmetric stem cell division. (A) In Drosophila male GSCs, sister chromatids of X and Y chromosomes are segregated nonrandomly. Image courtesy of G. Watase (University of Michigan, Ann Arbor, MI; Yadlapalli and Yamashita, 2013). (B) Drosophila male GSCs preferentially retain old histone H3 (green), whereas new histone H3 (red) is preferentially segregated to GBs. Image courtesy of X. Chen (Johns Hopkins University, Baltimore, MD; Tran et al., 2012). (C) An example of asymmetric segregation of protein aggregates. Image adapted from Rujano et al. (2006), published in PLoS Biology under the Creative Commons Attribution License. (D) Old mitochondria labeled by photoactivated Omp25-paGFP are preferentially segregated to the differentiating daughter of human mammary stem-like cells. Image adapted from Katajisto et al. (2015) with the permission from the authors and the American Association for the Advancement of Science.
Having the same genetic information, distinct cell fates between two sister cells upon asymmetric cell division must be ultimately tracked down to distinct gene expression patterns, i.e., epigenetic information. Therefore, it has been hypothesized that epigenetic information such as DNA methylation and histone modification may be distinct between two sister chromatids, leading to different cell fates. Above-mentioned involvement of Dnmt3 in biased DNA segregation (Elabd et al., 2013) is consistent with this idea. In Drosophila male GSCs, it has been shown that old versus new canonical histone H3.1 exhibits asymmetric segregation between GSC and GB, suggesting that histone H3.1 might carry distinct epigenetic information during GSC division (Fig. 4 B; Tran et al., 2012; Xie et al., 2015). It remains unknown which genes are regulated by such distinct epigenetic information. Also, it remains unknown how this asymmetry in histone segregation is related to the nonrandom sister chromatid segregation of X and Y chromosomes.
Senescence-causing factors
Cells undergo replicative senescence, the state in which cells are alive yet permanently arrested in cell cycle, after a certain number of cell divisions (20–30 divisions) due to accumulation of factors/conditions that interfere with cell proliferation such as damaged proteins and unstable genome (Hernandez-Segura et al., 2018; Sapieha and Mallette, 2018). To maintain the proliferative potential of a cell, even unicellular organisms such as bacteria and yeasts undergo asymmetric cell divisions to partition undesirable characteristics such as damaged proteins and compromised ribosomal DNA (rDNA) loci to the old cell while rejuvenating the young cell (Aguilaniu et al., 2003; Ganley et al., 2009; Winkler et al., 2010; Liu et al., 2011; Kobayashi, 2014; Hill et al., 2016).
The stem cells of multicellular organisms appear to employ similar mechanisms as damaged protein has been observed to segregate asymmetrically in dividing mammalian cells including human embryonic stem cells (ESCs; Fig. 4 C; Rujano et al., 2006; Fuentealba et al., 2008; Moore et al., 2015). Chromosomal rDNA copy number decreases during the aging of Drosophila male GSCs and recovers in the subsequent generation (Lu et al., 2018), possibly implying the presense of a similar mechanism as observed in yeast. In mouse hematopoietic stem cells, the nucleolus accumulates replicative stress during aging, which may reflect instability of rDNA, although asymmetric segregation of rDNA damage was not examined in these cells (Flach et al., 2014). Old mitochondria are preferentially segregated to the differentiating daughter, whereas stem cells inherited fewer older mitochondria during the asymmetric division of human mammary stem-like cells, suggesting that stem cells might be protected from senescence by eliminating old, nonfunctional mitochondria (Fig. 4 D; Katajisto et al., 2015). In these stem cells, the old mitochondria accumulate around the nuclear periphery, while new mitochondria remain equally distributed, leading to biased segregation of old mitochondria to differentiating cells. While the details of the underlying mitotic machinery are not yet understood, mitochondrial fission and fusion appear to be essential for biased segregation of old mitochondria. Together, these studies suggest that asymmetric divisions may be used to protect stem cells from senescence by isolating the senescence factors to one cell to rejuvenate the other.
Concluding remarks
Asymmetric division is a critical mechanism that balances stem cell self-renewal and differentiation. In this review, we described recent advances that have added to our understanding of asymmetric stem cell divisions. Emerging studies have revealed complexities in mechanisms that ensure asymmetric outcome of divisions. We have also started to appreciate multitudes of asymmetries, which may impact daughter cell behavior even if they may not determine the cell fate per se. These new discoveries made us realize (yet again) that we have not understood much of the secrets of life. Nonetheless, we have begun to appreciate what might be missing from our knowledge, paving the way to deepening our understanding in future studies. In particular, it will be of great interest to understand the meaning of intriguing asymmetries with unknown functions/significance that have been observed in recent years.
Acknowledgments
We thank Drs. Xin Chen and George Watase for generously sharing images, and we thank the Yamashita laboratory members for discussions.
The research in the Yamashita laboratory is supported by the Howard Hughes Medical Institute and the National Institute of General Medical Sciences (R01GM118308 to Y.M. Yamashita).
The authors declare no competing financial interests.
References
- Aguilaniu H., Gustafsson L., Rigoulet M., and Nyström T.. 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 299:1751–1753. 10.1126/science.1080418 [DOI] [PubMed] [Google Scholar]
- Anderson C.T., and Stearns T.. 2009. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr. Biol. 19:1498–1502. 10.1016/j.cub.2009.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger C.F., Wang Y.X., von Maltzahn J., Soleimani V.D., Yin H., and Rudnicki M.A.. 2013. Fibronectin regulates Wnt7a signaling and satellite cell expansion. Cell Stem Cell. 12:75–87. 10.1016/j.stem.2012.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabernard C., and Doe C.Q.. 2009. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev. Cell. 17:134–141. 10.1016/j.devcel.2009.06.009 [DOI] [PubMed] [Google Scholar]
- Cabernard C., Prehoda K.E., and Doe C.Q.. 2010. A spindle-independent cleavage furrow positioning pathway. Nature. 467:91–94. 10.1038/nature09334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caydasi A.K., and Pereira G.. 2012. SPOC alert--when chromosomes get the wrong direction. Exp. Cell Res. 318:1421–1427. 10.1016/j.yexcr.2012.03.031 [DOI] [PubMed] [Google Scholar]
- Chen C., Fingerhut J.M., and Yamashita Y.M.. 2016a The ins(ide) and outs(ide) of asymmetric stem cell division. Curr. Opin. Cell Biol. 43:1–6. 10.1016/j.ceb.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Inaba M., Venkei Z.G., and Yamashita Y.M.. 2016b Klp10A, a stem cell centrosome-enriched kinesin, balances asymmetries in Drosophila male germline stem cell division. eLife. 5:e20977 10.7554/eLife.20977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Cummings R., Mordovanakis A., Hunt A.J., Mayer M., Sept D., and Yamashita Y.M.. 2018. Cytokine receptor-Eb1 interaction couples cell polarity and fate during asymmetric cell division. eLife. 7:e33685 10.7554/eLife.33685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Türkel N., Hemati N., Fuller M.T., Hunt A.J., and Yamashita Y.M.. 2008. Centrosome misorientation reduces stem cell division during ageing. Nature. 456:599–604. 10.1038/nature07386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy M.J., Karasov A.O., and Rando T.A.. 2007. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 5:e102 10.1371/journal.pbio.0050102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P.T., and Raff J.W.. 2010. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr. Biol. 20:2187–2192. 10.1016/j.cub.2010.11.055 [DOI] [PubMed] [Google Scholar]
- Coumailleau F., Fürthauer M., Knoblich J.A., and González-Gaitán M.. 2009. Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature. 458:1051–1055. 10.1038/nature07854 [DOI] [PubMed] [Google Scholar]
- Deng W., and Lin H.. 1997. Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila. Dev. Biol. 189:79–94. 10.1006/dbio.1997.8669 [DOI] [PubMed] [Google Scholar]
- Derivery E., Seum C., Daeden A., Loubéry S., Holtzer L., Jülicher F., and Gonzalez-Gaitan M.. 2015. Polarized endosome dynamics by spindle asymmetry during asymmetric cell division. Nature. 528:280–285. 10.1038/nature16443 [DOI] [PubMed] [Google Scholar]
- di Pietro F., Echard A., and Morin X.. 2016. Regulation of mitotic spindle orientation: an integrated view. EMBO Rep. 17:1106–1130. 10.15252/embr.201642292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C.Q., and Bowerman B.. 2001. Asymmetric cell division: fly neuroblast meets worm zygote. Curr. Opin. Cell Biol. 13:68–75. 10.1016/S0955-0674(00)00176-9 [DOI] [PubMed] [Google Scholar]
- Elabd C., Cousin W., Chen R.Y., Chooljian M.S., Pham J.T., Conboy I.M., and Conboy M.J.. 2013. DNA methyltransferase-3-dependent nonrandom template segregation in differentiating embryonic stem cells. J. Cell Biol. 203:73–85. 10.1083/jcb.201307110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas L.M., and Huttner W.B.. 2008. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr. Opin. Cell Biol. 20:707–715. 10.1016/j.ceb.2008.09.008 [DOI] [PubMed] [Google Scholar]
- Flach J., Bakker S.T., Mohrin M., Conroy P.C., Pietras E.M., Reynaud D., Alvarez S., Diolaiti M.E., Ugarte F., Forsberg E.C., et al. 2014. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 512:198–202. 10.1038/nature13619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba L.C., Eivers E., Geissert D., Taelman V., and De Robertis E.M.. 2008. Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc. Natl. Acad. Sci. USA. 105:7732–7737. 10.1073/pnas.0803027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller M.T., and Spradling A.C.. 2007. Male and female Drosophila germline stem cells: two versions of immortality. Science. 316:402–404. 10.1126/science.1140861 [DOI] [PubMed] [Google Scholar]
- Gallaud E., Pham T., and Cabernard C.. 2017. Drosophila melanogaster Neuroblasts: A Model for Asymmetric Stem Cell Divisions. Results Probl. Cell Differ. 61:183–210. 10.1007/978-3-319-53150-2_8 [DOI] [PubMed] [Google Scholar]
- Ganley A.R., Ide S., Saka K., and Kobayashi T.. 2009. The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol. Cell. 35:683–693. 10.1016/j.molcel.2009.07.012 [DOI] [PubMed] [Google Scholar]
- Gasic I., Nerurkar P., and Meraldi P.. 2015. Centrosome age regulates kinetochore-microtubule stability and biases chromosome mis-segregation. eLife. 4:e07909 10.7554/eLife.07909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B., Takeshita H., Mizumoto K., and Sawa H.. 2006. Wnt signals can function as positional cues in establishing cell polarity. Dev. Cell. 10:391–396. 10.1016/j.devcel.2005.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas S., Conder R., and Knoblich J.A.. 2012. The Par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell. 11:529–540. 10.1016/j.stem.2012.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S.W., and Hyman A.A.. 2005. Spindle positioning by cortical pulling forces. Dev. Cell. 8:461–465. 10.1016/j.devcel.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Grill S.W., Howard J., Schäffer E., Stelzer E.H., and Hyman A.A.. 2003. The distribution of active force generators controls mitotic spindle position. Science. 301:518–521. 10.1126/science.1086560 [DOI] [PubMed] [Google Scholar]
- Habib S.J., Chen B.C., Tsai F.C., Anastassiadis K., Meyer T., Betzig E., and Nusse R.. 2013. A localized Wnt signal orients asymmetric stem cell division in vitro. Science. 339:1445–1448. 10.1126/science.1231077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Kobayashi S., and Nakato H.. 2009. Drosophila glypicans regulate the germline stem cell niche. J. Cell Biol. 187:473–480. 10.1083/jcb.200904118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Segura A., Nehme J., and Demaria M.. 2018. Hallmarks of Cellular Senescence. Trends Cell Biol. 28:436–453. 10.1016/j.tcb.2018.02.001 [DOI] [PubMed] [Google Scholar]
- Hill S.M., Hao X., Grönvall J., Spikings-Nordby S., Widlund P.O., Amen T., Jörhov A., Josefson R., Kaganovich D., Liu B., and Nyström T.. 2016. Asymmetric Inheritance of Aggregated Proteins and Age Reset in Yeast Are Regulated by Vac17-Dependent Vacuolar Functions. Cell Reports. 16:826–838. 10.1016/j.celrep.2016.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M., and Yamashita Y.M.. 2012. Asymmetric stem cell division: precision for robustness. Cell Stem Cell. 11:461–469. 10.1016/j.stem.2012.09.003 [DOI] [PubMed] [Google Scholar]
- Inaba M., Yuan H., Salzmann V., Fuller M.T., and Yamashita Y.M.. 2010. E-cadherin is required for centrosome and spindle orientation in Drosophila male germline stem cells. PLoS One. 5:e12473 10.1371/journal.pone.0012473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M., Buszczak M., and Yamashita Y.M.. 2015a Nanotubes mediate niche-stem-cell signalling in the Drosophila testis. Nature. 523:329–332. 10.1038/nature14602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M., Venkei Z.G., and Yamashita Y.M.. 2015b The polarity protein Baz forms a platform for the centrosome orientation during asymmetric stem cell division in the Drosophila male germline. eLife. 4:e04960 10.7554/eLife.04960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens D.H., and Lee C.Y.. 2014. It takes two to tango, a dance between the cells of origin and cancer stem cells in the Drosophila larval brain. Semin. Cell Dev. Biol. 28:63–69. 10.1016/j.semcdb.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Llamazares S., Reina J., and Gonzalez C.. 2011. Drosophila neuroblasts retain the daughter centrosome. Nat. Commun. 2:243 10.1038/ncomms1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Reina J., Llamazares S., Bertran T., Rossi F., Roig J., and Gonzalez C.. 2013. Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 15:241–248. 10.1038/ncb2671 [DOI] [PubMed] [Google Scholar]
- Karpowicz P., Pellikka M., Chea E., Godt D., Tepass U., and van der Kooy D.. 2009. The germline stem cells of Drosophila melanogaster partition DNA non-randomly. Eur. J. Cell Biol. 88:397–408. 10.1016/j.ejcb.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Katajisto P., Döhla J., Chaffer C.L., Pentinmikko N., Marjanovic N., Iqbal S., Zoncu R., Chen W., Weinberg R.A., and Sabatini D.M.. 2015. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 348:340–343. 10.1126/science.1260384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase E., Wong M.D., Ding B.C., and Xie T.. 2004. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development. 131:1365–1375. 10.1242/dev.01025 [DOI] [PubMed] [Google Scholar]
- Kiger A.A., Jones D.L., Schulz C., Rogers M.B., and Fuller M.T.. 2001. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 294:2542–2545. 10.1126/science.1066707 [DOI] [PubMed] [Google Scholar]
- Knoblich J.A. 2008. Mechanisms of asymmetric stem cell division. Cell. 132:583–597. 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Kobayashi T. 2014. Ribosomal RNA gene repeats, their stability and cellular senescence. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 90:119–129. 10.2183/pjab.90.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressmann S., Campos C., Castanon I., Fürthauer M., and González-Gaitán M.. 2015. Directional Notch trafficking in Sara endosomes during asymmetric cell division in the spinal cord. Nat. Cell Biol. 17:333–339. 10.1038/ncb3119 [DOI] [PubMed] [Google Scholar]
- Kuang S., Kuroda K., Le Grand F., and Rudnicki M.A.. 2007. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 129:999–1010. 10.1016/j.cell.2007.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.D., and Nagy L.M.. 2002. Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature. 420:682–686. 10.1038/nature01241 [DOI] [PubMed] [Google Scholar]
- Lechler T., and Fuchs E.. 2005. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 437:275–280. 10.1038/nature03922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T.R., Tomancak P., and Krause H.M.. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 131:174–187. 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Le Grand F., Jones A.E., Seale V., Scimè A., and Rudnicki M.A.. 2009. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 4:535–547. 10.1016/j.stem.2009.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R. 2012. Germline stem cells: origin and destiny. Cell Stem Cell. 10:729–739. 10.1016/j.stem.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit D.A., and Rusan N.M.. 2013. PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J. Cell Biol. 202:1013–1022. 10.1083/jcb.201303141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Larsson L., Franssens V., Hao X., Hill S.M., Andersson V., Höglund D., Song J., Yang X., Öling D., et al. 2011. Segregation of protein aggregates involves actin and the polarity machinery. Cell. 147:959–961. 10.1016/j.cell.2011.11.018 [DOI] [PubMed] [Google Scholar]
- Löbrich M., and Jeggo P.A.. 2007. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer. 7:861–869. 10.1038/nrc2248 [DOI] [PubMed] [Google Scholar]
- Loncarek J., and Bettencourt-Dias M.. 2018. Building the right centriole for each cell type. J. Cell Biol. 217:823–835. 10.1083/jcb.201704093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick V.P., Morris L.X., Fox D.T., and Spradling A.. 2011. Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev. Cell. 21:159–171. 10.1016/j.devcel.2011.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubéry S., Daeden A., Seum C., Holtzer L., Moraleda A., Damond N., Derivery E., Schmidt T., and Gonzalez-Gaitan M.. 2017. Sara phosphorylation state controls the dispatch of endosomes from the central spindle during asymmetric division. Nat. Commun. 8:15285 10.1038/ncomms15285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulier K., Lathia J.D., Marthiens V., Relucio J., Mughal M.R., Tang S.C., Coksaygan T., Hall P.E., Chigurupati S., Patton B., et al. 2009. beta1 integrin maintains integrity of the embryonic neocortical stem cell niche. PLoS Biol. 7:e1000176 10.1371/journal.pbio.1000176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K.L., Nelson J.O., Watase G.J., Warsinger-Pepe N., and Yamashita Y.M.. 2018. Transgenerational dynamics of rDNA copy number in Drosophila male germline stem cells. eLife. 7:e32421 10.7554/eLife.32421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Casanueva M.O., Mahowald A.P., Kato M., Lauterbach D., and Ferguson E.L.. 2012. Niche-associated activation of rac promotes the asymmetric division of Drosophila female germline stem cells. PLoS Biol. 10:e1001357 10.1371/journal.pbio.1001357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne C., and Gonzalez-Gaitan M.. 2014. Sara endosomes and the asymmetric division of intestinal stem cells. Development. 141:2014–2023. 10.1242/dev.104240 [DOI] [PubMed] [Google Scholar]
- Moore D.L., Pilz G.A., Araúzo-Bravo M.J., Barral Y., and Jessberger S.. 2015. A mechanism for the segregation of age in mammalian neural stem cells. Science. 349:1334–1338. 10.1126/science.aac9868 [DOI] [PubMed] [Google Scholar]
- Morrison S.J., and Kimble J.. 2006. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 441:1068–1074. 10.1038/nature04956 [DOI] [PubMed] [Google Scholar]
- Morrison S.J., and Spradling A.C.. 2008. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 132:598–611. 10.1016/j.cell.2008.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A. 2015. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 25:R1002–R1018. 10.1016/j.cub.2015.08.051 [DOI] [PubMed] [Google Scholar]
- Nakada D., Oguro H., Levi B.P., Ryan N., Kitano A., Saitoh Y., Takeichi M., Wendt G.R., and Morrison S.J.. 2014. Oestrogen increases haematopoietic stem-cell self-renewal in females and during pregnancy. Nature. 505:555–558. 10.1038/nature12932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaen J.T., Wilsch-Bräuninger M., and Huttner W.B.. 2013. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell. 155:333–344. 10.1016/j.cell.2013.08.060 [DOI] [PubMed] [Google Scholar]
- Pelletier L., and Yamashita Y.M.. 2012. Centrosome asymmetry and inheritance during animal development. Curr. Opin. Cell Biol. 24:541–546. 10.1016/j.ceb.2012.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., and Yamashita Y.M.. 2011. Fly meets yeast: checking the correct orientation of cell division. Trends Cell Biol. 21:526–533. 10.1016/j.tcb.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Tanaka T.U., Nasmyth K., and Schiebel E.. 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 20:6359–6370. 10.1093/emboj/20.22.6359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulson N.D., and Lechler T.. 2012. Asymmetric cell divisions in the epidermis. Int. Rev. Cell Mol. Biol. 295:199–232. 10.1016/B978-0-12-394306-4.00012-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehoda K.E. 2009. Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harb. Perspect. Biol. 1:a001388 10.1101/cshperspect.a001388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramat A., Hannaford M., and Januschke J.. 2017. Maintenance of Miranda Localization in Drosophila Neuroblasts Involves Interaction with the Cognate mRNA. Curr. Biol. 27:2101–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdas Nair A., Singh P., Salvador Garcia D., Rodriguez-Crespo D., Egger B., and Cabernard C.. 2016. The Microcephaly-Associated Protein Wdr62/CG7337 Is Required to Maintain Centrosome Asymmetry in Drosophila Neuroblasts. Cell Reports. 14:1100–1113. 10.1016/j.celrep.2015.12.097 [DOI] [PubMed] [Google Scholar]
- Rando T.A. 2007. The immortal strand hypothesis: segregation and reconstruction. Cell. 129:1239–1243. 10.1016/j.cell.2007.06.019 [DOI] [PubMed] [Google Scholar]
- Reichert H. 2011. Drosophila neural stem cells: cell cycle control of self-renewal, differentiation, and termination in brain development. Results Probl. Cell Differ. 53:529–546. 10.1007/978-3-642-19065-0_21 [DOI] [PubMed] [Google Scholar]
- Rocheteau P., Gayraud-Morel B., Siegl-Cachedenier I., Blasco M.A., and Tajbakhsh S.. 2012. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 148:112–125. 10.1016/j.cell.2011.11.049 [DOI] [PubMed] [Google Scholar]
- Rojas-Ríos P., Guerrero I., and González-Reyes A.. 2012. Cytoneme-mediated delivery of hedgehog regulates the expression of bone morphogenetic proteins to maintain germline stem cells in Drosophila. PLoS Biol. 10:e1001298 10.1371/journal.pbio.1001298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth T.M., Chiang C.Y., Inaba M., Yuan H., Salzmann V., Roth C.E., and Yamashita Y.M.. 2012. Centrosome misorientation mediates slowing of the cell cycle under limited nutrient conditions in Drosophila male germline stem cells. Mol. Biol. Cell. 23:1524–1532. 10.1091/mbc.e11-12-0999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubinet C., Tsankova A., Pham T.T., Monnard A., Caussinus E., Affolter M., and Cabernard C.. 2017. Spatio-temporally separated cortical flows and spindle geometry establish physical asymmetry in fly neural stem cells. Nat. Commun. 8:1383 10.1038/s41467-017-01391-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujano M.A., Bosveld F., Salomons F.A., Dijk F., van Waarde M.A., van der Want J.J., de Vos R.A., Brunt E.R., Sibon O.C., and Kampinga H.H.. 2006. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 4:e417 10.1371/journal.pbio.0040417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan N.M., and Peifer M.. 2007. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177:13–20. 10.1083/jcb.200612140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann V., Chen C., Chiang C.Y., Tiyaboonchai A., Mayer M., and Yamashita Y.M.. 2014. Centrosome-dependent asymmetric inheritance of the midbody ring in Drosophila germline stem cell division. Mol. Biol. Cell. 25:267–275. 10.1091/mbc.e13-09-0541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapieha P., and Mallette F.A.. 2018. Cellular Senescence in Postmitotic Cells: Beyond Growth Arrest. Trends Cell Biol. 28:595–607. 10.1016/j.tcb.2018.03.003 [DOI] [PubMed] [Google Scholar]
- Sarrazin S., Lamanna W.C., and Esko J.D.. 2011. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3:a004952 10.1101/cshperspect.a004952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoborg T., Zajac A.L., Fagerstrom C.J., Guillen R.X., and Rusan N.M.. 2015. An Asp-CaM complex is required for centrosome-pole cohesion and centrosome inheritance in neural stem cells. J. Cell Biol. 211:987–998. 10.1083/jcb.201509054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C., Kiger A.A., Tazuke S.I., Yamashita Y.M., Pantalena-Filho L.C., Jones D.L., Wood C.G., and Fuller M.T.. 2004. A misexpression screen reveals effects of bag-of-marbles and TGF beta class signaling on the Drosophila male germ-line stem cell lineage. Genetics. 167:707–723. 10.1534/genetics.103.023184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F. 2015. Asymmetric cell division in the Drosophila bristle lineage: from the polarization of sensory organ precursor cells to Notch-mediated binary fate decision. Wiley Interdiscip. Rev. Dev. Biol. 4:299–309. 10.1002/wdev.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin L., Poulson N.D., Foote H.P., and Lechler T.. 2013. NuMA localization, stability, and function in spindle orientation involve 4.1 and Cdk1 interactions. Mol. Biol. Cell. 24:3651–3662. 10.1091/mbc.e13-05-0277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshan A., and Amon A.. 2004. Linked for life: temporal and spatial coordination of late mitotic events. Curr. Opin. Cell Biol. 16:41–48. 10.1016/j.ceb.2003.11.003 [DOI] [PubMed] [Google Scholar]
- Shin J.W., Buxboim A., Spinler K.R., Swift J., Christian D.A., Hunter C.A., Léon C., Gachet C., Dingal P.C., Ivanovska I.L., et al. 2014. Contractile forces sustain and polarize hematopoiesis from stem and progenitor cells. Cell Stem Cell. 14:81–93. 10.1016/j.stem.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani A.A., and Ingham P.W.. 2003. Regulation of stem cell maintenance and transit amplifying cell proliferation by tgf-beta signaling in Drosophila spermatogenesis. Curr. Biol. 13:2065–2072. 10.1016/j.cub.2003.10.063 [DOI] [PubMed] [Google Scholar]
- Siegrist S.E., and Doe C.Q.. 2006. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development. 133:529–536. 10.1242/dev.02211 [DOI] [PubMed] [Google Scholar]
- Siller K.H., and Doe C.Q.. 2008. Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Dev. Biol. 319:1–9. 10.1016/j.ydbio.2008.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Ramdas Nair A., and Cabernard C.. 2014. The centriolar protein Bld10/Cep135 is required to establish centrosome asymmetry in Drosophila neuroblasts. Curr. Biol. 24:1548–1555. 10.1016/j.cub.2014.05.050 [DOI] [PubMed] [Google Scholar]
- Song X., Zhu C.H., Doan C., and Xie T.. 2002. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science. 296:1855–1857. 10.1126/science.1069871 [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R., and Somers W.G.. 2013. Mechanisms of asymmetric progenitor divisions in the Drosophila central nervous system. Adv. Exp. Med. Biol. 786:79–102. 10.1007/978-94-007-6621-1_6 [DOI] [PubMed] [Google Scholar]
- Tran V., Lim C., Xie J., and Chen X.. 2012. Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science. 338:679–682. 10.1126/science.1226028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova A., Pham T.T., Garcia D.S., Otte F., and Cabernard C.. 2017. Cell Polarity Regulates Biased Myosin Activity and Dynamics during Asymmetric Cell Division via Drosophila Rho Kinase and Protein Kinase N. Dev. Cell. 42:143–155. [DOI] [PubMed] [Google Scholar]
- Tulina N., and Matunis E.. 2001. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 294:2546–2549. 10.1126/science.1066700 [DOI] [PubMed] [Google Scholar]
- Venkei Z.G., and Yamashita Y.M.. 2015. The centrosome orientation checkpoint is germline stem cell specific and operates prior to the spindle assembly checkpoint in Drosophila testis. Development. 142:62–69. 10.1242/dev.117044 [DOI] [PubMed] [Google Scholar]
- Wang X., Tsai J.W., Imai J.H., Lian W.N., Vallee R.B., and Shi S.H.. 2009. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 461:947–955. 10.1038/nature08435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.E., Beronja S., Pasolli H.A., and Fuchs E.. 2011. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 470:353–358. 10.1038/nature09793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J., Seybert A., König L., Pruggnaller S., Haselmann U., Sourjik V., Weiss M., Frangakis A.S., Mogk A., and Bukau B.. 2010. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 29:910–923. 10.1038/emboj.2009.412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T., and Spradling A.C.. 2000. A niche maintaining germ line stem cells in the Drosophila ovary. Science. 290:328–330. 10.1126/science.290.5490.328 [DOI] [PubMed] [Google Scholar]
- Xie J., Wooten M., Tran V., Chen B.C., Pozmanter C., Simbolon C., Betzig E., and Chen X.. 2015. Histone H3 Threonine Phosphorylation Regulates Asymmetric Histone Inheritance in the Drosophila Male Germline. Cell. 163:920–933. 10.1016/j.cell.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadlapalli S., and Yamashita Y.M.. 2013. Chromosome-specific nonrandom sister chromatid segregation during stem-cell division. Nature. 498:251–254. 10.1038/nature12106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadlapalli S., Cheng J., and Yamashita Y.M.. 2011. Drosophila male germline stem cells do not asymmetrically segregate chromosome strands. J. Cell Sci. 124:933–939. 10.1242/jcs.079798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y.M., Jones D.L., and Fuller M.T.. 2003. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 301:1547–1550. 10.1126/science.1087795 [DOI] [PubMed] [Google Scholar]
- Yamashita Y.M., Mahowald A.P., Perlin J.R., and Fuller M.T.. 2007. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 315:518–521. 10.1126/science.1134910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., and Lin X.. 2009. Shaping morphogen gradients by proteoglycans. Cold Spring Harb. Perspect. Biol. 1:a002493 10.1101/cshperspect.a002493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yennek S., Burute M., Théry M., and Tajbakhsh S.. 2014. Cell adhesion geometry regulates non-random DNA segregation and asymmetric cell fates in mouse skeletal muscle stem cells. Cell Reports. 7:961–970. 10.1016/j.celrep.2014.04.016 [DOI] [PubMed] [Google Scholar]
- Yoshiura S., Ohta N., and Matsuzaki F.. 2012. Tre1 GPCR signaling orients stem cell divisions in the Drosophila central nervous system. Dev. Cell. 22:79–91. 10.1016/j.devcel.2011.10.027 [DOI] [PubMed] [Google Scholar]
- Yu F., Kuo C.T., and Jan Y.N.. 2006. Drosophila neuroblast asymmetric cell division: recent advances and implications for stem cell biology. Neuron. 51:13–20. 10.1016/j.neuron.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Yuan H., Chiang C.Y., Cheng J., Salzmann V., and Yamashita Y.M.. 2012. Regulation of cyclin A localization downstream of Par-1 function is critical for the centrosome orientation checkpoint in Drosophila male germline stem cells. Dev. Biol. 361:57–67. 10.1016/j.ydbio.2011.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]