Nuclear speckles (NSs) store splicing factors. Wang et al. show that many naturally intronless mRNAs associate with NSs and that speckle association enhances their export by facilitating TREX recruitment, suggesting that trafficking to NSs could be an important quality control step in intronless mRNA export.
Abstract
Nuclear speckles (NSs) serve as splicing factor storage sites. In this study, we unexpectedly found that many endogenous intronless mRNAs, which do not undergo splicing, associate with NSs. These associations do not require transcription, polyadenylation, or the polyA tail. Rather, exonic splicing enhancers present in intronless mRNAs and their binding partners, SR proteins, promote intronless mRNA localization to NSs. Significantly, speckle targeting of mRNAs promotes the recruitment of the TREX export complex and their TREX-dependent nuclear export. Furthermore, TREX, which accumulates in NSs, is required for releasing intronless mRNAs from NSs, whereas NXF1, which is mainly detected at nuclear pores, is not. Upon NXF1 depletion, the TREX protein UAP56 loses speckle concentration but coaccumulates with intronless mRNAs and polyA RNAs in the nucleoplasm, and these RNAs are trapped in NSs upon UAP56 codepletion. We propose that the export-competent messenger RNP assembly mainly occurs in NSs for intronless mRNAs and that entering NSs serves as a quality control step in mRNA export.
Introduction
Nuclear speckles (NSs; also known as splicing-factor compartments, interchromatin granules, or SC35 domains) are dynamic nuclear structures located in mammalian cells. Although it has been >50 yr since their initial discovery (Swift, 1959), functions of NSs are still unclear (Lamond and Spector, 2003; Spector and Lamond, 2011). Currently, the only widely accepted function of NSs is that of the storage/modification sites of splicing factors (Spector and Lamond, 2011). Multiple studies have demonstrated that splicing is required for the association of mRNAs with NSs (Johnson et al., 2000; Melčák et al., 2001; Ishihama et al., 2008; Funatsu, 2009; Dias et al., 2010). Although it remains highly controversial whether splicing occurs in NSs, accumulating evidence has suggested their involvement in splicing regulation. Splicing was thought to occur at perichromatin fibrils surrounding NSs (Fu and Maniatis, 1990; Spector et al., 1991; Cmarko et al., 1999). Different from this view, there are also studies suggesting that splicing occurs directly in NSs (Johnson et al., 2000; Melčák et al., 2001; Hall et al., 2006; Ishihama et al., 2008; Funatsu, 2009; Dias et al., 2010). More recently, using antibodies that specifically detect active spliceosomes, Girard et al. (2012) reported that both of these views are true. Their data indicate that 80% of splicing events occur cotranscriptionally at the periphery of NSs, whereas 20% of them occur posttranscriptionally within these subnuclear structures (Girard et al., 2012).
Except for splicing factors, other important mRNA metabolic factors such as mRNA export factors and components of the exon–junction complex are also enriched in NSs (Mayeda et al., 1999; Kataoka et al., 2000; Zhou et al., 2000; Gatfield et al., 2001; Masuda et al., 2005). In the nuclei of mammalian cells, a significant portion of polyA RNAs is present in NSs (Carter et al., 1991; Visa et al., 1993; Huang et al., 1994; Dias et al., 2010). When components of the TREX complex that serves as a key nuclear export adaptor are depleted, polyA RNAs as well as mRNAs derived from intron-containing reporter genes are almost exclusively accumulated in these subnuclear structures (Sträßer et al., 2002; Dias et al., 2010; Chi et al., 2013). Considering that the vast majority of splicing events occur at speckle surrounding sites, these results suggest that a significant fraction of spliced mRNAs might enter NSs after splicing. Consistent with this possibility, it has been shown that the COL1A1 mRNA is almost entirely spliced before entering NSs (Johnson et al., 2000). Why do spliced mRNAs enter NSs? One possibility is that these spliced mRNAs might be assembled into export-competent messenger RNPs (mRNPs) in these domains. In disagreement with this possibility, it was reported that speckle-localized polyA RNAs are trapped in this foci and not to be released to the cytoplasm (Huang et al., 1994). However, to date, direct evidence that NSs are involved in mRNA export is still lacking.
Approximately 3% of protein-coding genes do not have introns. Although they represent the minority in the human genome, intronless genes mostly encode proteins with fundamental functions such as signal transduction factors and regulatory proteins important for growth, proliferation, and development (Grzybowska, 2012). Since splicing does not occur to naturally intronless mRNAs, they are thought not to pass through NSs (Johnson et al., 2000; Melčák et al., 2001; Hall et al., 2006; Ishihama et al., 2008; Funatsu, 2009; Dias et al., 2010; Lei et al., 2011). Consistent with this view, a previous study reported that three naturally intronless mRNAs, including HSPB3, IFN-α1, and IFN-β1, do not associate with NSs (Lei et al., 2011). Intronless mRNAs are exported to the cytoplasm by using the same machinery as spliced ones (Palazzo et al., 2007; Lei et al., 2011; Akef et al., 2013; Chi et al., 2014). The lack of association of naturally intronless mRNAs with NSs does not support the possibility that mRNA export factors are recruited in these domains. However, more recent studies reported that some cis-acting elements that were known to function in mRNA export could also promote speckle association of intronless reporter transcripts. For example, the signal sequence coding region (SSCR) from the mouse major histocompatibility complex H2kb gene promotes both speckle association and nuclear export of a reporter cDNA transcript (Palazzo et al., 2007; Akef et al., 2013). Further, influenza viral unspliced M1 mRNA is also targeted to NSs (Mor et al., 2016). However, currently, it remains unclear whether naturally intronless mRNAs associate with NSs or where they are assembled into export-competent mRNPs.
In this study, we unexpectedly found that many naturally intronless mRNAs, including eight exogenously expressed and four endogenous ones, associate with NSs. Mechanistically, exonic splicing enhancers (ESEs) present in intronless mRNAs and their binding partners SR proteins promote their speckle association. Functionally, speckle association of intronless mRNAs enhances mRNA export by facilitating recruitment of the TREX complex. Further, we provide evidence supporting the notion that TREX recruitment mainly occurs in NSs. Given that the current view for NSs is that they are the sites for storage of splicing factors and regulation of splicing, our study expands the role of these subnuclear regions in assembling export-competent mRNPs and suggests that trafficking to NSs could be an important quality control step in mRNA export.
Results
Exogenous naturally intronless mRNAs enter NSs rapidly after transcription
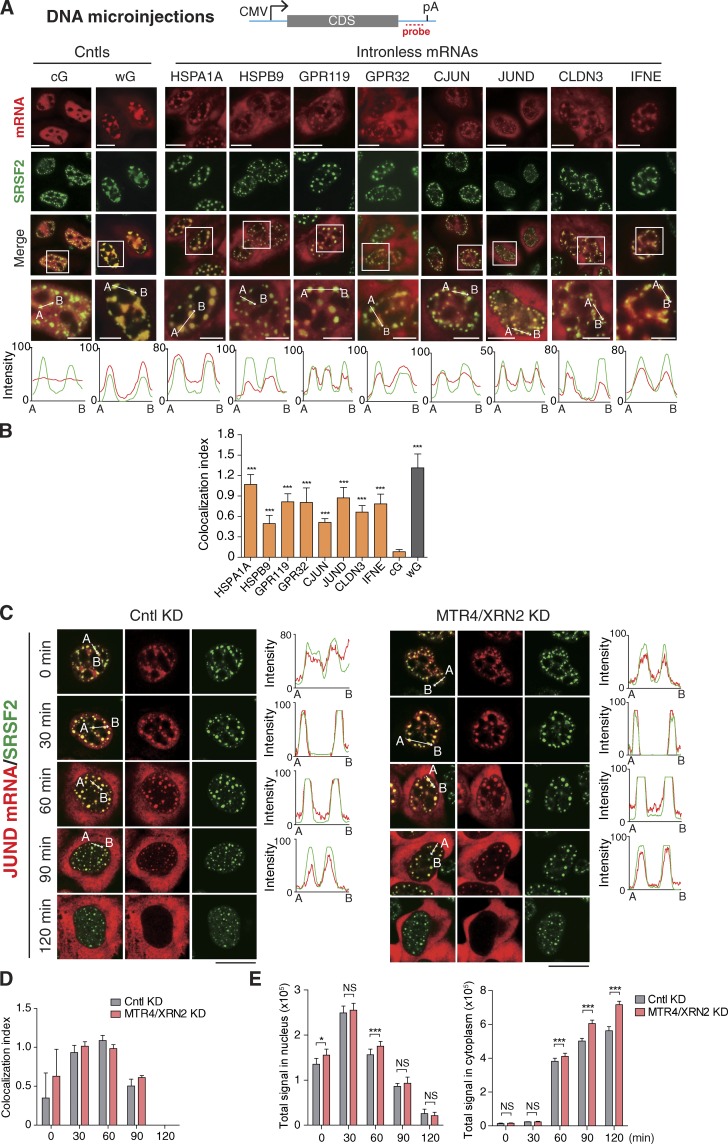
To initially obtain some clue where naturally intronless mRNAs distribute in the nucleus before nuclear export, we performed DNA microinjection experiments. We randomly picked eight intronless genes, including HSPA1A, HSPB9, GPR119, GPR32, CJUN, JUND, CLDN3, and IFNE, and cloned the coding regions into reporter constructs. These reporter DNA constructs were microinjected into the nuclei of HeLa cells, followed by FISH analysis to detect the corresponding mRNAs. For controls (Cntls), the wG (β-globin pre-mRNA) and the cG (β-globin cDNA) constructs were also microinjected. 20 min after microinjection, α-amanitin was added to inhibit transcription. To avoid extensive cG mRNA degradation, we took the images at 30 min after transcription inhibition when the levels of the two reporter mRNAs seemed similar (Valencia et al., 2008). Consistent with a previous study (Dias et al., 2010), the wG mRNA was largely distributed in nuclear foci that colocalized with SRSF2, a standard marker for NSs, whereas the cG mRNA mostly diffused in the nucleoplasm (Fig. 1 A, Cntls). For naturally intronless mRNAs, different time points were taken to observe their nuclear distribution as well as cytoplasmic accumulation. Specifically, FISH images are shown at 1 h (CJUN and JUND), 2 h (HSPA1A), or 4 h (all the others) after the addition of α-amanitin (Fig. 1 A). Significantly, in the nucleus, all of these intronless mRNAs are localized in specific foci that generally colocalized with SRSF2 (Fig. 1 A), and the colocalization index was >0.5 (Fig. 1 B). These data indicate that many naturally intronless mRNAs, but not artificial cDNA transcript, derived from reporter DNA constructs associate with NSs.
Figure 1.
Exogenously expressed naturally intronless mRNAs associate with NSs. (A) Top: Schematic of reporter constructs. Sequences from the vector and the genes of interest are indicated as a cyan line and gray bar, respectively. The positions of the promoter, polyA (pA) site, and FISH probe are indicated. Bottom: Equal amounts of reporter constructs were microinjected into HeLa nuclei, and α-amanitin (4 µg/ml) was added 20 min after injection. At 30 min (cG and wG), 1 h (CJUN and JUND), 2 h (HSPA1A), or 4 h (others) after the addition of α-amanitin, FISH with the 3′ vector probe and IF with the SRSF2 antibody were performed. Higher magnification of the boxed regions is shown. The red and green lines in the graphs show the intensity of the FISH and SRSF2 IF signal along the freely positioned arrow indicated from A to B, respectively. CMV, cytomegalovirus; CDS, coding sequence. (B) Colocalization index of mRNA foci with SRSF2 speckles in each group shown in A. (C–E) Confocal microscopic images showing the JUND mRNA transcribed from the microinjected reporter construct and SRSF2 in HeLa cells treated with Cntl or MRT4/XRN2 siRNA for 72 h. The red and green lines in the graphs show the intensity of the FISH and SRSF2 IF signal, respectively (C). Colocalization indexes of the JUND FISH foci and SRSF2 dots at each time point are shown in D. Quantifications of nuclear and cytoplasmic JUND mRNA FISH signals are shown in E. Data represent the mean ± SD from three independent experiments; n = 10. Bars: 10 µm (magnification); 20 µm (others). Statistical analysis was performed using an unpaired t test. *, P < 0.05; ***, P < 0.01. KD, knockdown.
To investigate the kinetics of speckle targeting of intronless mRNAs and to examine whether speckle-associated intronless mRNAs could be exported to the cytoplasm, time courses with the use of FISH and immunofluorescence (IF) were performed for the JUND and HSPA1A constructs (Figs. 1 C and S1 A). Representative confocal images of a cell at each time point are shown. Significantly, for the JUND mRNA, the FISH signal was apparently detected in NSs as early as 0 min after the addition of α-amanitin (20 min after injection; Fig. 1, C and D), indicating that intronless mRNAs enter NSs rapidly after transcription. The mRNA was still mostly concentrated at NSs at 30 min. Since 60 min, the JUND mRNA started to accumulate in the cytoplasm, accompanied by a gradually reduced signal in NSs. At 120 min, the mRNA was completely detected in the cytoplasm. This result suggests that the JUND mRNA distributed in NSs could be released to the cytoplasm. However, it was also possible that the mRNA localized in NSs had been degraded rather than released to the cytoplasm. To examine this possibility, we inhibited nuclear mRNA degradation pathways by depleting the 5′-to-3′ ribonuclease XRN2 as well as MTR4, an essential cofactor of the nuclear exosome harboring both endo- and 3′-to-5′ exoribonuclease activity (Fig. 1, C–E; and Fig. S1, B–F; Mitchell et al., 1997; Allmang et al., 1999; West et al., 2004; Schilders et al., 2007; Schneider et al., 2007; Lebreton et al., 2008; Fasken et al., 2011; Lubas et al., 2011). The overall JUND FISH signal was increased ∼1.25-fold, and the levels of exosome targets were elevated in these cells, evident of inhibited degradation (Figs. 1 E and S1, C–F; Fan et al., 2017). However, the kinetics of JUND mRNA speckle exit was similar to that of Cntl cells, supporting the notion that speckle-localized JUND mRNA was not degraded but was released to the cytoplasm (Fig. 1, C–E). Importantly, a similar observation was obtained with the HSPA1A mRNA (Fig. S1, A–C and F), although both the entry into and the exit from NSs was slower than the JUND mRNA. We noted that upon JUND mRNA export completion, most of SRSF2 lost its regular speckled pattern but diffused in the nucleoplasm of all injected cells (Figs. 1 C and S1 G). A similar phenomenon was observed when rapidly exported spliced mRNAs were introduced to the cells (Dias et al., 2010; Shi et al., 2015). This is consistent with the notion that NS components dynamically traffic between these domains and the nucleoplasm (Misteli, 2001). Together, these data indicate that naturally intronless mRNAs enter NSs rapidly after transcription and that speckle-associated intronless mRNAs could be exported to the cytoplasm.
Endogenous naturally intronless mRNAs are detected within NSs
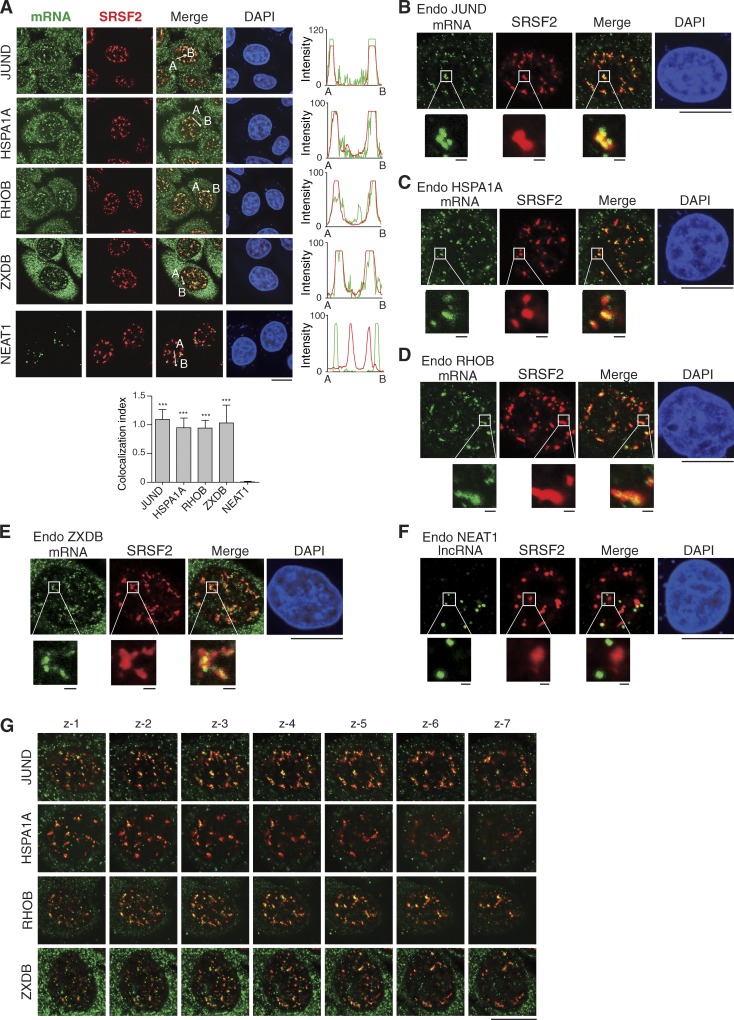
We next examined whether endogenous naturally intronless mRNAs associate with NSs. Four relatively abundant endogenous intronless mRNAs in HeLa cells, including JUND, HSPA1A, RHOB, and ZXDB, were chosen for this analysis. The long noncoding RNA NEAT1 was used as a Cntl not localizing in NSs (Bond and Fox, 2009; Chen and Carmichael, 2009). Fields of multiple cells are shown in Fig. 2 A, and a representative cell is shown in Fig. 2 (B–F). Apparent nuclear FISH signals were detected for these intronless mRNAs, probably due to the lack of the coupling of mRNA export to splicing. Confocal microscopic analysis revealed that all four endogenous intronless mRNAs generally colocalized with SRSF2 in the nucleus, whereas NEAT1 did not (Fig. 2, A–F). Higher magnification of the images showed that most of these intronless mRNAs localized in the center of NSs, although some of them were detected at the periphery (Fig. 2, B–E). Z stack images and 3D video reconstruction further confirmed the localization of intronless mRNAs within NSs (Fig. 2 G and Videos 1, 2, 3, and 4). Together, our data suggest that many endogenous naturally intronless mRNAs associate with NSs.
Figure 2.
Endogenous intronless mRNAs associate with NSs. (A) Confocal microscopic images to show distributions of endogenous (Endo) RNAs and NSs detected by FISH with transcript-specific probes and IF with the SRSF2 antibody. To clearly visualize the NEAT1 foci, a shorter exposure is shown. The green and red lines in the graphs demonstrate the RNA and SRSF2 IF signal intensity, respectively. The colocalization index of RNA foci with SRSF2 speckles is shown in the bottom. Data represent the mean ± SD from three independent experiments; n = 10. Statistical analysis was performed using an unpaired t test. ***, P < 0.01. (B) High magnification of a representative cell with the JUND mRNA and SRSF2 detected is shown. The further higher magnification of the boxed region is shown. (C–F) Same as in B except that the HSPA1A mRNA, RHOB mRNA, ZXDB mRNA, and NEAT1 lncRNA were detected. (G) The top to bottom sections of Z-stacked confocal images were taken in steps of 0.5 µm. Bars: 1 µm (magnifications); 20 µm (others).
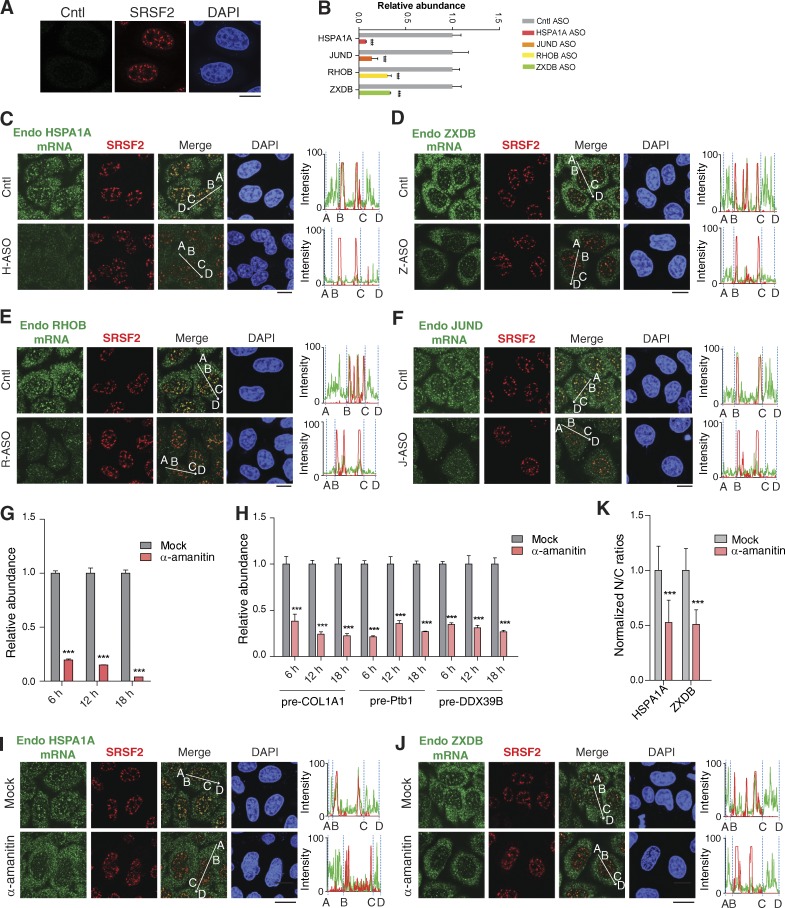
Notably, a nonspecific probe did not detect any apparent signal, and the FISH signals were apparently reduced in cells treated with transcript-specific antisense oligonucleotides (ASOs), demonstrating the specificities of the FISH signals (Fig. 3, A–F). Furthermore, when transcription was inhibited with α-amanitin, FISH signals were apparently reduced in NSs, and most mRNAs were detected in the cytoplasm (Fig. 3, G–K), further supporting the notion that intronless mRNAs localized at NSs could be released to the cytoplasm.
Figure 3.
Endogenous intronless mRNAs are specifically detected in NSs. (A) FISH signals with a negative Cntl probe targeting the antisense transcript of the mouse long noncoding RNA Gm14635, SRSF2 IF, and DAPI staining are shown. (B) RT-PCRs to examine the knockdown efficiencies of endogenous intronless mRNAs in cells electronically transfected with ASOs for 48 h. (C–F) FISH signals of endogenous (Endo) intronless mRNAs in HeLa cells were microinjected with a Cntl ASO or ASO targeting the corresponding intronless mRNAs for 10 h. Note that the same exposures were taken for the cells treated with Cntl or transcript-specific ASO. (G and H) RT-qPCRs to examine the levels of the wG mRNA derived from microinjected constructs (G) or endogenous mRNAs (H) in HeLa cells mock-treated or treated with α-amanitin (4 µg/ml) for 6, 12, and 18 h. The graphs show the relative abundance of wG mRNA to DNA or relative abundance of COL1A1, PTB1, and DDX39B pre-mRNAs to 18S rRNA. Data represent the mean ± SEM; n = 3. (I–K) Confocal microscopic images show the distribution of endogenous HSPA1A (I) and ZXDB (J) mRNAs and NSs in HeLa cell mock-treated or treated with α-amanitin (4 µg/ml) for 18 h. The same exposures were taken for the cells treated with Cntl or α-amanitin. The average N/C ratios of each intronless mRNA are shown in the graph (K). The data represent the mean ± SD from three independent experiments; n = 10. Bars, 20 µm. The green and red lines in the graphs demonstrate the mRNA and SRSF2 IF signal intensity, respectively. Statistical analysis was performed using an unpaired t test. ***, P < 0.01.
Transcription, polyadenylation, or the polyA tail is not required for naturally intronless mRNAs entering NSs
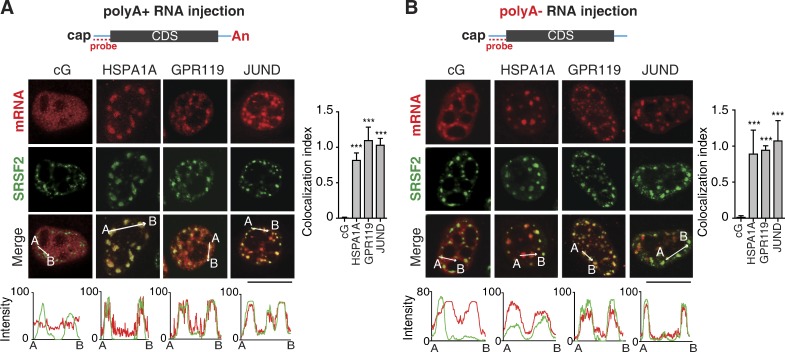
How could naturally intronless mRNAs be targeted to NSs? As all intronless mRNAs we tested associated with NSs, we first considered whether some common processes such as transcription and polyadenylation might facilitate these associations. If true, one would expect that mRNAs synthesized and polyadenylated in vitro do not enter NSs. To investigate this possibility, we in vitro transcribed HSPA1A, GRP119, JUND, and cG (Cntl) mRNAs using T7 polymerase, followed by in vitro polyadenylation. When these mRNAs were microinjected into HeLa nuclei, at 30 min after injection, the HSPA1A, GRP119, and JUND mRNAs, but not the cG mRNA, clearly colocalized with SRSF2 (Fig. 4 A). Thus, neither transcription nor polyadenylation is required for naturally intronless mRNAs entering NSs.
Figure 4.
In vitro–transcribed naturally intronless mRNAs without a polyA tail are accumulated in NSs. (A) Top: Schematic of reporter constructs. Sequences from the vector and the genes of interest are indicated as cyan line and gray bar, respectively. The positions of the cap, polyA tail (An), and FISH probe are indicated. Confocal microscopic images show the distribution of microinjected cG, HSPA1A, GPR119, and JUND mRNAs as well as NSs. FISH with the 5′ vector probe and SRSF2 IF were performed 30 min after microinjection. Bar, 20 µm. The red and green lines in the graphs demonstrate the mRNA and SRSF2 IF signal intensity, respectively. The colocalization indexes are shown on the right. Data represent the mean ± SD from three independent experiments; n = 10. Statistical analysis was performed by using the unpaired Student’s t test. ***, P < 0.01. CDS, coding sequence. (B) Same as in A, except that in vitro–transcibed but not polyadenylated intronless mRNAs were used for microinjection.
Another common feature of intronless mRNAs is the polyA tail. To examine its potential roles, we injected in vitro–transcribed HSPA1A, GPR119, JUND, and cG mRNAs without a polyA tail. As shown in Fig. 4 B, 30 min after injection, polyA- naturally intronless mRNAs but not the cG mRNA were apparently accumulated in NSs, indicating that speckle targeting of intronless mRNAs does not depend on the polyA tail.
Cis-acting elements facilitate intronless mRNA association with NSs
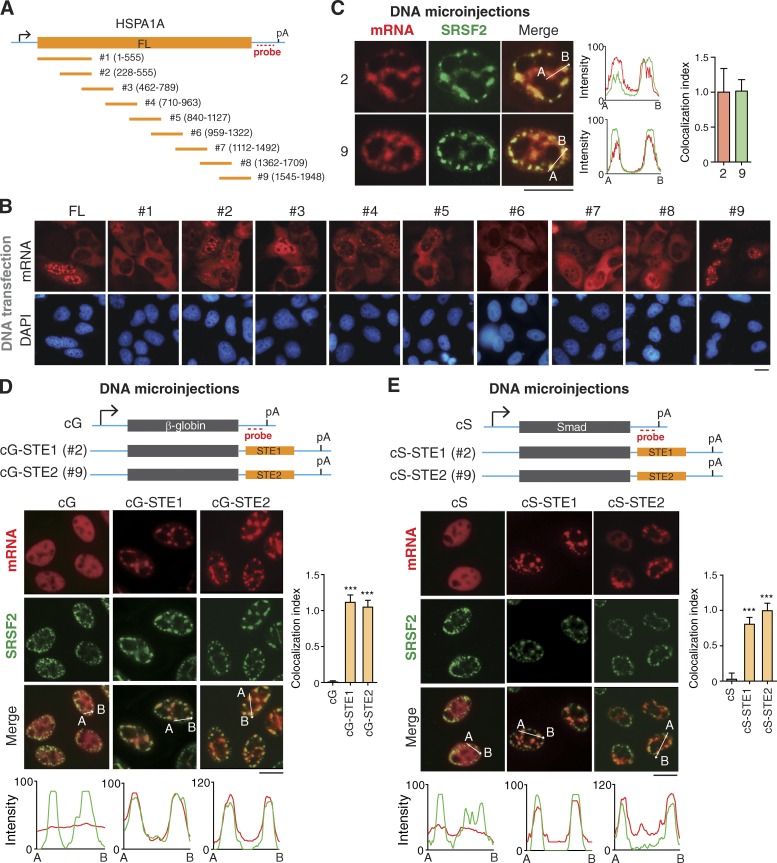
We next considered the possibility that some cis-acting elements might function in targeting intronless mRNAs to NSs. Among the eight exogenous intronless mRNAs we tested, HSPA1A showed the most significant speckle-association phenotype. We thus separated the HSPA1A gene into nine overlapping fragments of 200–300 bp in length and cloned them into an expression plasmid (Fig. 5 A). When these plasmids were transfected, the no. 2 and no. 9 fragments apparently localized in speckled nuclear foci, whereas others were mostly cytoplasmic at 24 h after transfection (Fig. 5 B). We confirmed speckle associations of the no. 2 and no. 9 fragments with microinjection experiments (Fig. 5 C). These data suggest that these HSPA1A fragments might contain speckle-targeting signals.
Figure 5.
Multiple fragments in naturally intronless mRNAs facilitate speckle targeting. (A) Schematic of the HSPA1A constructs. Sequences from the vector and HSPA1A gene are indicated as a cyan line and an orange bar, respectively. The positions of the promoter, polyA (pA) signal, and FISH probe are marked. FL, full length. (B) Fluorescence microscopic images show the distribution of mRNA fragments transcribed from transfected DNA constructs shown in A. FISH with the 3′ vector probe was performed at 24 h after transfection. (C) Confocal microscopic images show the distribution of HSPA1A fragment mRNAs derived from microinjected DNA construct and NSs. FISH with the 3′ vector probe and SRSF2 IF were performed at 1 h after the addition of α-amanitin. The colocalization index is shown on the right. (D) Top: Schematic of cG constructs. Sequence from the vector is indicated as cyan line, and the regions of the cG and STEs are indicated as a gray and an orange bar, respectively. The positions of promoter, polyA site, and FISH probe are marked. Bottom: Fluorescence microscopic images show the distribution of cG, cG-STE1, and cG-STE2 mRNAs transcribed from microinjected DNA constructs and NSs. α-Amanitin (4 µg/ml) was added 20 min after microinjection. FISH with the 3′ probe and IF with the SRSF2 antibody were performed at 30 min after the addition of α-amanitin. The colocalization index is shown on the right. Data represent the mean ± SD from three independent experiments; n = 10. (E) Same as in D, except that the Smad cDNA transcript (cS) was used. Bars, 20 µm. The red and green lines in the graphs demonstrate the mRNA and SRSF2 IF signal intensities, respectively. Statistical analysis was performed by using the unpaired t test. ***, P < 0.01.
To examine whether no. 2 and no. 9 fragments can facilitate mRNA association with NSs, we inserted them into the 3′ of the β-globin and Smad cDNA transcripts (cG and cS mRNA, respectively), which otherwise do not associate with NSs (Fig. 5, D and E; Dias et al., 2010). Significantly, with either fragment, the cG and cS mRNAs were now apparently colocalized with SRSF2 (Fig. 5, D and E). We thus named no. 2 and no. 9 fragments as speckle-targeting element (STE) 1 and 2, respectively. Together, these data indicate that specific intronless mRNA sequences function in targeting them to NSs.
It was possible that the cytoplasmic localized HSPA1A fragments could have passed through NSs before nuclear export. Indeed, when another three randomly picked fragments, no. 1, no. 5, and no. 6, were microinjected, they also apparently associated with NSs at 30 min after injection (Fig. S2, A and B). The observation that multiple RNA fragments show speckle localization raised the possibility that speckle-targeting sequences are interspersed, rather than localized in a specific region, in intronless mRNAs. Consistent with this possibility, when the JUND coding region was separated into three fragments, all of them associated with NSs to a different extent (Fig. S2, C and D).
ESEs function in speckle targeting of intronless mRNAs via SR proteins
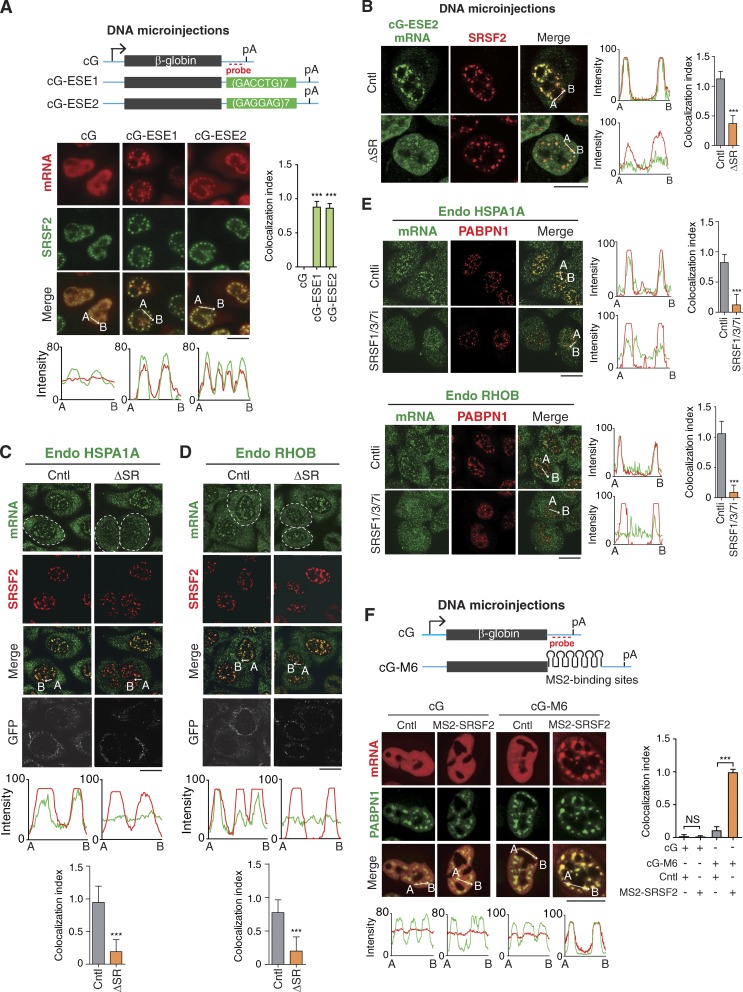
What could be the interspersed speckle targeting sequences in intronless mRNAs? ESEs are purine-rich RNA sequences that stimulate splicing by recruiting SR proteins (Tacke and Manley, 1995; Liu et al., 1998, 2000; Blencowe, 2000; Wu et al., 2005; Cáceres and Hurst, 2013). Recently, a specific subgroup of ESEs was reported to be overrepresented in intronless genes more than in intergenic sequences (Savisaar and Hurst, 2016). We found that on average, each intronless gene contains >30 copies of these ESEs (Fig. S3, A and B). This led us to speculate that ESEs might function in targeting intronless mRNAs to NSs. The two STEs we identified do contain multiple ESEs (Fig. S3 C). We chose two of the most enriched ESEs in the HSPA1A gene, 5′-GACCTG-3′ and 5′-GAGGAG-3′, for further functional study (Fig. S3 D). Significantly, insertion of seven tandem repeats of either of these ESEs resulted in the apparent association of the cG mRNA with NSs (Fig. 6 A). This speckle targeting was not due to cryptic splicing (Fig. S3 E). These data suggest that ESEs are indeed sufficient for driving intronless mRNAs to NSs.
Figure 6.
ESEs and SR proteins function in speckle association of intronless mRNAs. (A) Top: Schematic of cG constructs. Sequence from the vector is indicated as the cyan line, and the regions of the cG and ESEs are indicated as gray and green bars, respectively. The positions of the promoter, polyA (pA) signal, and FISH probe are marked. Bottom: Fluorescence microscopic images show the distribution of cG, cG-ESE1, and cG-ESE2 mRNAs transcribed from microinjected DNA constructs and NSs. α-Amanitin (4 µg/ml) was added 20 min after microinjection. FISH with the 3′ probe and IF with the SRSF2 antibody were performed at 2 h after the addition of α-amanitin. The colocalization index is shown on the right. The red and green lines in the graphs demonstrate the mRNA and SRSF2 IF signal intensities, respectively. (B) Confocal fluorescence microscopic images to show the distribution of the cG-ESE2 mRNA coexpressed with the Cntl or ΔSR RNAs and NSs. α-Amanitin (4 µg/ml) was added 20 min after coinjection of the cG-ESE2 construct (30 ng/µl), together with the Cntl or the ΔSR construct (100 ng/µl). FISH with the β-globin probe and IF with the SRSF2 antibody were performed at 2 h after the addition of α-amanitin. The colocalization index is shown on the right. The green and red lines in the graphs demonstrate the mRNA and SRSF2 IF signal intensities, respectively. The colocalization index is shown on the right. (C and D) Confocal fluorescence microscopic images show the distribution of the endogenous HSPA1A (C) and RHOB (D) mRNAs and NSs in cells expressing the Cntl or ΔSR RNA. To allow the release of mRNAs that had entered NSs before SR depletion, 8 h after microinjection of the Cntl or ΔSR construct, FISH with transcript-specific probes, SRSF2 IF, and GFP IF indicating the microinjected cells (circles) were performed. Note that to avoid signal leakage between channels, only a tiny amount of GFP construct was injected. The green and red lines in the graphs demonstrate the mRNA and SRSF2 IF signal intensities, respectively. The colocalization index is shown at the bottom. (E) Confocal fluorescence microscopic images show the distribution of the endogenous HSPA1A and RHOB mRNAs and NSs in cells treated with Cntl or SRSF1/3/7 siRNAs for 72 h. The green and red lines in the graphs demonstrate the mRNA and PABPN1 IF signal intensities, respectively. The colocalization index is shown on the right. (F) Top: Schematic of cG and cG-M6 constructs. Sequences from the vector and cG are indicated as cyan line and gray bars, respectively. The positions of the promoter, polyA (pA) signal, and FISH probe are marked. Bottom: Confocal fluorescence microscopic images show the distribution of cG and cG-M6 transcribed from microinjected DNA constructs and NSs in cells coexpressing GFP-MS2 (Cntl) or MS2-SRSF2. α-Amanitin (4 µg/ml) was added 20 min after microinjection. FISH with the 3′ probe and IF with the PABPN1 antibody were performed at 2 h after the addition of α-amanitin. The colocalization index is shown on the right. The red and green lines in the graphs demonstrate the mRNA and PABPN1 IF signal intensities, respectively. Data represent the mean ± SD from three independent experiments; n = 10. Bars, 20 µm. Statistical analysis was performed using an unpaired t test. ***, P < 0.01.
By considering the prevalent binding of ESEs with SR proteins, it was possible that they promote speckle association of intronless mRNAs via SR proteins. To investigate this possibility, we examined whether depletion of SR proteins affects speckle association of intronless mRNAs. Many SR proteins share common binding motifs and often functionally substitute for one another (Chandler et al., 1997). Thus, we used the well-characterized FP RNA element (referred to in this study as ΔSR RNA) that binds to multiple SR proteins and can titrate SR proteins (Sun et al., 1993; Dirksen et al., 2000; Li et al., 2000). The FPD RNA element (referred to in this study as the Cntl RNA), which does not bind to SR proteins (Sun et al., 1993; Dirksen et al., 2000; Li et al., 2000), was used as a Cntl. No apparent effect on speckle formation was detected with either RNA element. Significantly, when the ΔSR RNA, but not the Cntl RNA, was expressed, speckle association of the cG-ESE2 mRNA was apparently weakened (Fig. 6 B), indicating that SR proteins are required for ESE-mediated speckle targeting.
We next examined the effect of the ΔSR RNA on the distribution of endogenous intronless mRNAs including HSPA1A and RHOB. No apparent difference was detected in speckle association of these mRNAs in cells treated or not treated with the Cntl RNA (Fig. 6, C and D, Cntl; comparing circled cells with noncircled cells). In contrast, in cells expressing the ΔSR RNA, speckle association was significantly weakened compared with that in the neighboring untreated cells (Fig. 6, C and D, ΔSR; comparing circled cells with noncircled cells). These results indicate that speckle targeting of endogenous intronless mRNAs depends on SR proteins. To further corroborate the role of SR proteins in intronless mRNA speckle targeting, we next knocked down a panel of SR proteins and assessed speckle localization of the endogenous HSPA1A and RHOB mRNAs (Figs. 6 E and S4, A–C). Considering that SRSF2 was knocked down in some of the samples, to give a fair comparison of the effect of different SRSF proteins, we used PABPN1 as the NSs marker in this experiment (Krause et al., 1994). Significantly, co-knockdown of SRSF1/3/7 apparently abolished speckle localization of these mRNAs, and co-knockdown of SRSF1/6/7 also had a moderate effect (Figs. 6 E and S4, B and C). Together, these data demonstrate that SR proteins are required for targeting intronless mRNAs to NSs.
To examine these roles of SR proteins in a more direct way, we tethered the MS2-tagged SR protein SRSF2 to the cG mRNA containing six MS2-binding sites (cG-M6). MS2-GFP was used as a negative Cntl. As shown in Fig. 6 F, tethering MS2-SRSF2 but not MS2-GFP resulted in the apparent association of the cG-M6 mRNA with NSs. Note that the cG mRNA without MS2-binding site did not show speckle localization. Together, these data indicate that ESEs present in intronless mRNAs promote their speckle association via SR proteins.
Speckle association of intronless mRNAs promotes TREX recruitment and mRNA export
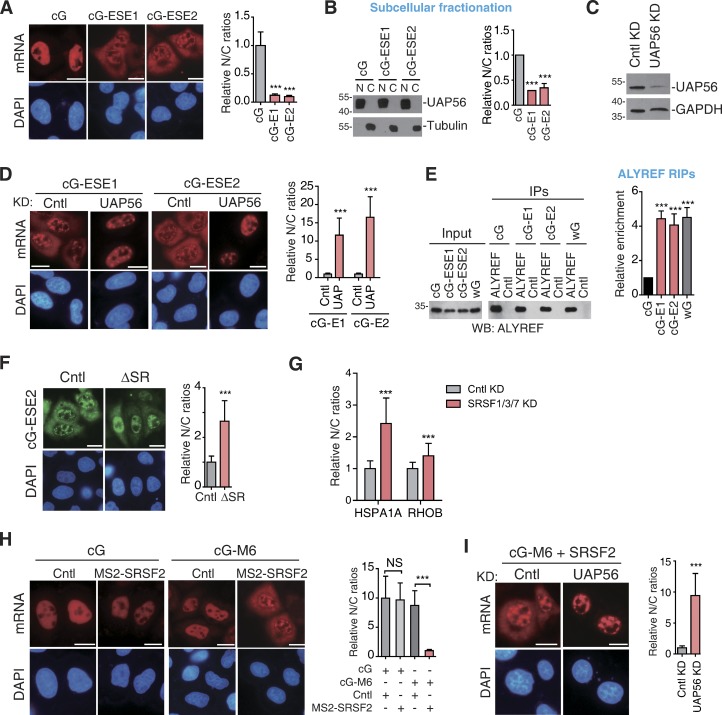
We next sought to investigate the functional relevance for intronless mRNA associating with NSs. Considering that mRNA export factors are also enriched in NSs (Zhou et al., 2000; Gatfield et al., 2001; Masuda et al., 2005), we speculated that this association might facilitate the assembly of export-competent mRNPs. If true, targeting cDNA transcripts, which cannot efficiently recruit TREX and are mostly retained in the nucleus, to NSs might promote TREX recruitment and mRNA export. To examine this possibility, we microinjected cG, cG-ESE1, and cG-ESE2 reporter constructs and examined their nucleocytoplasmic (N/C) distribution 4 h after injection. As previously reported (Valencia et al., 2008; Dias et al., 2010), the cG mRNA was largely retained in the nucleus (Fig. 7 A). Significantly, apparent cytoplasmic accumulation was observed with the cG-ESE1 and cG-ESE2 mRNAs (Fig. 7 A), suggesting that ESEs indeed promote mRNA export. To further verify this, we next transfected these constructs into HeLa cells and performed subcellular fractionation. Western analysis confirmed the purities of nuclear and cytoplasmic fractions (Fig. 7 B). Reverse transcription–quantitative PCR (RT-qPCR) data revealed that with ESEs, the N/C ratios of the cG mRNA apparently decreased (Fig. 7 B). Together, these results indicate that ESEs promotes intronless mRNA export.
Figure 7.
Speckle association promotes TREX recruitment and TREX-dependent intronless mRNA export. (A) Fluorescence microscopic images show the distribution of the cG, cG-ESE1, and cG-ESE2 mRNA derived from equal amounts of microinjected cG, cG-ESE1, and cG-ESE2 constructs. α-Amanitin (4 µg/ml) was added at 20 min after injection, and 4 h later, FISH with the 3′ vector probe was performed. DAPI staining marks the nucleus. Relative N/C ratios are shown on the right. Data represent the mean ± SD from three independent experiments; n = 30. (B) Equal amounts of cG, cG-ESE1, and cG-ESE2 constructs were transfected into HeLa cells. 24 h later, N/C fractionation was performed. Western blotting (WB) was used to examine the purities of the nuclear and cytoplasmic fraction by using UAP56 and tubulin as markers (left). Ratios of nuclear to cytoplasmic β-globin mRNAs after normalization to NEAT1 (nucleus) and GAPDH (cytoplasm) in each fraction are presented in the bar graph (right). Data represent the mean ± SEM; n = 3. (C) Western analysis was used to examine the knockdown (KD) efficiency of UAP56. (D) Similar to A, except that Cntl or UAP56 knockdown cells were used. Note that to achieve efficient mRNA nuclear retention and to avoid secondary effect, the cells were treated with UAP56 and URH49 siRNAs for 48 h. Data represent the mean ± SD from three independent experiments; n = 30. (E) RIPs with UV cross-linking were performed with an ALYREF antibody or IgG (see details in Materials and methods). Western analysis was performed by using the ALYREF antibody (left). The relative IP efficiencies of cG mRNAs to the cS mRNA were quantified and are indicated in the graph (right). Data represent the mean ± SEM; n = 3. (F) Fluorescence microscopic images show the distribution of the cG-ESE2 mRNA derived from the microinjected cG-ESE2 construct. α-Amanitin (4 µg/ml) was added at 20 min after injection, and 4 h later, FISH with the β-globin probe was performed. DAPI staining marks the nucleus. Relative N/C ratios are shown on the right. Data represent the mean ± SD from three independent experiments; n = 30. (G) The graph shows the average N/C ratios of the endogenous HSPA1A and RHOB mRNAs in cells shown in Fig. 6 E. Data represent the mean ± SD from three independent experiments; n = 30. (H) Fluorescence microscopic images show the distribution of the cG or cG-M6 mRNAs derived from microinjected constructs in cells coexpressing GFP-MS2 or MS2-SRSF2. FISH with the 3′ vector probe was performed at 4 h after injection. The graph shows the relative N/C ratios. Data represent the mean ± SD from three independent experiments; n = 30. (I) Similar to H, except that the cG-M6 and MS2-SRSF2 were coinjected into Cntl or UAP56 knockdown cells (48 h knockdown). Data represent the mean ± SD from three independent experiments; n = 30. Bars, 20 µm. Statistical analysis was performed using an unpaired t test. ***, P < 0.01. Molecular masses are given in kilodaltons.
Considering that multiple SR proteins interact with the mRNA export receptor NXF1 and serve as NXF1 adaptors (Huang et al., 2003, 2004; Müller-McNicoll et al., 2016), it was possible that ESE-mediated mRNA export was a result of direct recruitment of NXF1 by SR proteins. If true, TREX, which serves as another nuclear export adaptor, would not be required for ESE-mediated mRNA export. However, this is not the case. Knockdown of the TREX component UAP56 apparently inhibited ESE-mediated mRNA export (Fig. 7, C and D), indicating that the effect of ESEs on mRNA export was not due to SR-mediated NXF1 recruitment.
To examine whether ESE-mediated export promotion was a result of enhanced TREX recruitment, UV cross-linking RNA immunoprecipitations (IPs; RIPs) with an antibody to the TREX component ALYREF, an RNA-binding protein, were performed. The cG and wG mRNAs were used as the negative and the positive Cntl for TREX recruitment, respectively (Masuda et al., 2005; Fan et al., 2017). As expected, ALYREF associated with the wG mRNA apparently more than the cG mRNA (Fig. 7 E). Significantly, compared with the cG mRNA, ALYREF associations with the cG-ESE1 and cG-ESE2 mRNAs were strengthened up to fourfold (Fig. 7 E), suggesting that ESE-mediated speckle association indeed promoted TREX recruitment.
It was also possible that ESE-mediated export promotion was a result of sequence-dependent TREX recruitment, rather than speckle association. In this case, depleting SR proteins should not affect ESE-mediated mRNA export. However, expression of the ΔSR RNA but not the Cntl RNA significantly inhibited the cytoplasmic accumulation of the cG-ESE2 mRNA (Fig. 7 F), suggesting that ESE-mediated speckle association, but not the ESE itself, was important for mRNA export promotion. Consistent with this view, co-knockdown of SRSF1/3/7 also resulted in nuclear retention of endogenous HSPA1A and RHOB mRNAs (Figs. 6 E and 7 G). The lack of apparent effect of the ΔSR RNA on the cytoplasmic accumulation of endogenous intronless mRNAs (Fig. 6, C and D) was likely because the mRNAs that had been exported to the cytoplasm before SR titration were still detectable.
To further investigate the effect of speckle association on mRNA export, we next examined whether tethering SRSF2, which neither interacts with NXF1 nor has a direct role in mRNA export (Huang et al., 2003, 2004; Müller-McNicoll et al., 2016), promoted mRNA cytoplasmic accumulation. As shown in Fig. 7 H, 4 h after injection, the cG-M6 mRNA but not the cG mRNA was apparently detected in the cytoplasm when coinjected with MS2-SRSF2. Similar results were obtained when these constructs were transfected and followed by subcellular fractionation and RT-qPCRs (Fig. S4 D). Importantly, export promotion by SRSF2 tethering was also dependent on TREX as depletion of UAP56 eliminated this effect (Fig. 7 I). Together, these data indicate that speckle association promotes intronless mRNA export by facilitating TREX recruitment.
Evidence that TREX recruitment mainly occurs in NSs
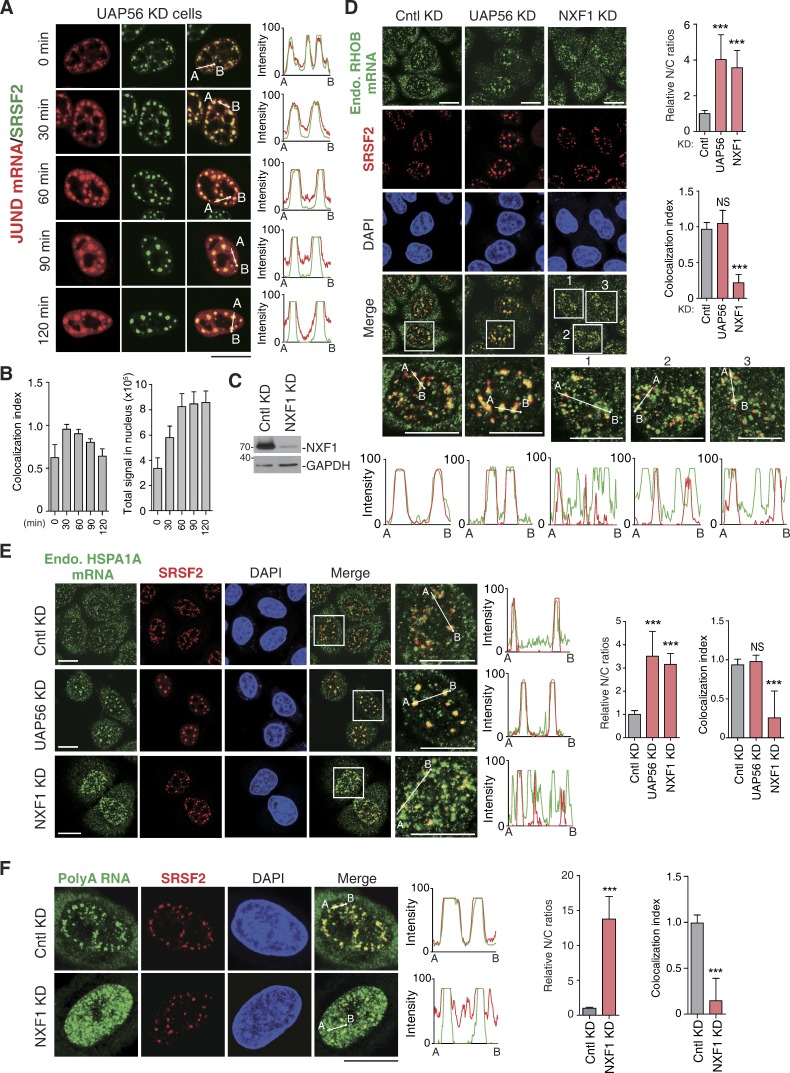
Considering that TREX components are concentrated at NSs (Zhou et al., 2000; Gatfield et al., 2001; Masuda et al., 2005) and speckle targeting promotes TREX binding, it is possible that TREX recruitment mainly occurs in these domains. In support of this possibility, TREX depletion results in retention of polyA RNAs and mRNAs derived from intron-containing reporter genes in NSs (Dias et al., 2010; Chi et al., 2013). To further investigate this possibility, we examined how intronless mRNA distribution was affected by depletion of the TREX component UAP56. Significantly, the exogenously expressed JUND and HSPA1A mRNAs were largely detected in NSs without apparently reduced levels up to 120 and 300 min, respectively, when they were completely released to the cytoplasm in normal cells (Figs. 1 C and 8, A and B; and Figs. S1 A and S4 E). Importantly, exclusive accumulation of the endogenous RHOB, HSPA1A, and ZXDB mRNAs in NSs was also observed in UAP56-depleted cells (Fig. 8, D and E; and Fig. S4 F). Note that this accumulation was not a result of altered mRNA levels as among three endogenous mRNA we examined, only the level of HSPA1A mRNA was affected (Fig. S4 G). These results are consistent with the notion that TREX recruitment occurs in NSs.
Figure 8.
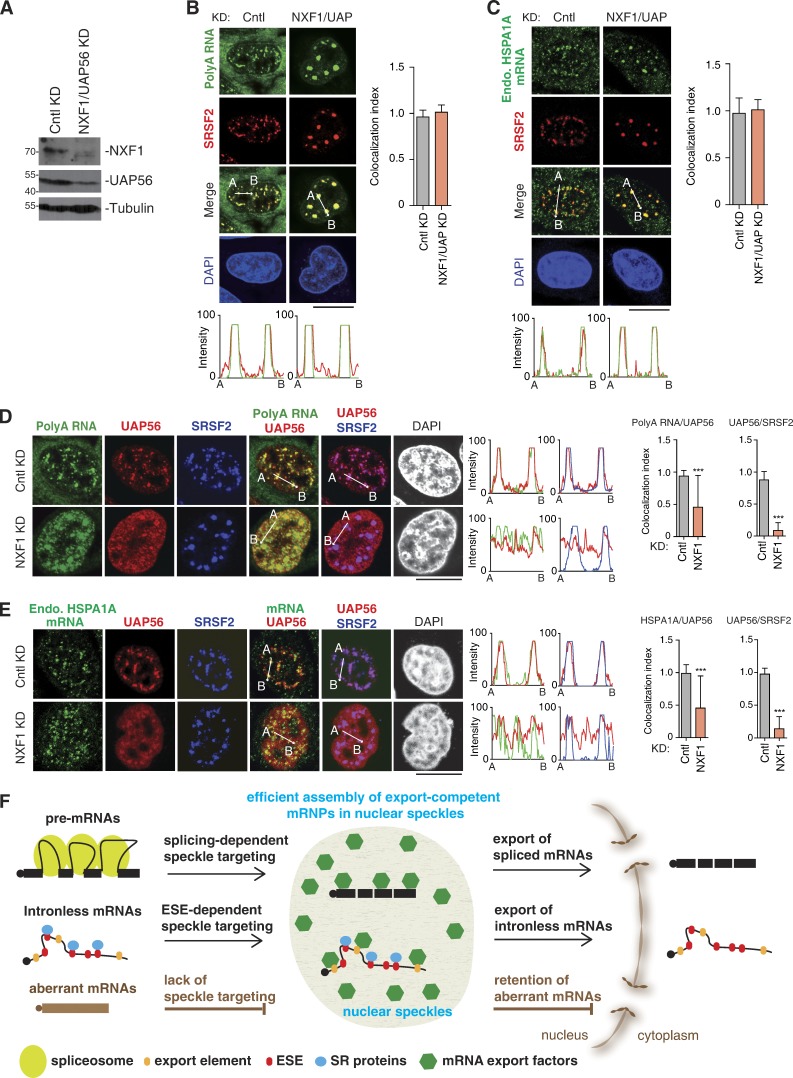
NXF1 depletion results in nuclear mRNA retention both inside and outside of NSs. (A and B) Confocal microscopic images show the JUND mRNA transcribed from the microinjected reporter construct and SRSF2 in HeLa cells treated with UAP56/URH49 siRNA for 48 h. The red and green lines in the graphs show the intensity of the FISH and SRSF2 IF signal, respectively (A). Colocalization indexes of the JUND FISH foci and SRSF2 dots (left) and total FISH signal quantification (right) at each time point are shown in B. (C) Western analysis was used to examine the knockdown (KD) efficiency of NXF1. (D) Confocal microscopic images show the endogenous RHOB mRNA and NSs (SRSF2) in HeLa cells treated with Cntl, UAP56/URH49, or NXF1siRNAs for 48 h. Higher magnification of the boxed regions is shown. The green and red in the line scan graphs demonstrate the intensities of the mRNA and the SRSF2 IF signals, respectively. The bar graphs show the relative N/C ratios (top) and colocalization index (bottom) in each treatment group. Data represent the mean ± SD from three independent experiments; n = 10. (E) Same as in D, except that the endogenous HSPA1A mRNA was examined. (F) Same as in D, except that the bulk polyA RNAs were examined. Bars, 20 µm. Statistical analysis was performed by using the unpaired Student’s t test. ***, P < 0.01. Molecular masses are given in kilodaltons.
However, it was also possible that TREX recruitment might take place outside of NSs, and when UAP56 was knocked down, intronless mRNAs were trapped in NSs due to their associations with SR proteins. In this case, one would expect that inhibiting mRNA export in other ways would result in similar speckle accumulation of intronless mRNAs. However, although depletion of NXF1, which mainly localizes at nuclear pores (Segref et al., 1997; Forler et al., 2004), resulted in nuclear export inhibitions, the RHOB, HSPA1A, and ZXDB mRNAs were detected not only inside but also apparently outside of NSs (Fig. 8, C–E; and Fig. S4 F). Importantly, similar accumulation pattern was also observed with total polyA RNAs (Fig. 8 F). These data suggest that TREX, but not NXF1, is required for mRNA release from NSs.
Had mRNAs accumulated in the nucleoplasm of NXF1 depleted cells passed through NSs and recruited TREX? If they had, these mRNAs would have been blocked in these domains in NXF1 and UAP56 co-knockdown cells. Indeed, both polyA RNAs and the endogenous HSPA1A mRNA were mainly detected in NSs in these cells (Fig. 9, A–C). As previously reported, UAP56 is mainly detected in NSs in Cntl cells (Fig. 9, D and E, Cntl KD; Gatfield et al., 2001). Intriguingly, upon NXF1 depletion, UAP56 apparently lost its speckle concentration but mostly diffused throughout the nucleoplasm and partially colocalized with polyA RNAs and HSPA1A mRNA (Fig. 9, D and E, NXF1 KD). These results indicate that the mRNAs accumulated outside of NSs upon NXF1 depletion had passed through NSs and recruited TREX. Together, these results from every angle strongly support the notion that TREX recruitment mainly occurs in NSs.
Figure 9.
Nucleoplasmic accumulated mRNAs upon NXF1 depletion had passed through NSs and recruited export factors. (A) Western analysis was used to examine the knockdown (KD) efficiency of NXF1 and UAP56. Note that to achieve efficient mRNA nuclear retention and to avoid extensive cell death, the cells were treated with UAP56 and URH49 siRNAs for 36 h. (B and C) Confocal microscopic images show the distribution of polyA RNAs (B) or the endogenous HSPA1A mRNA (C) and NSs (SRSF2) in HeLa cells treated with Cntl or UAP56/URH49/NXF1 siRNAs for 36 h. The green and red lines in the graphs demonstrate the RNA and SRSF2 IF signal intensities, respectively. Colocalization indexes are shown on the right. (D and E) Confocal microscopic images show the distribution of polyA RNAs (D) or the endogenous (Endo.) HSPA1A mRNA (E), UAP56, and NSs (SRSF2) in HeLa cells treated with Cntl or NXF1 siRNA for 48 h. The green, red, and blue lines in the graphs demonstrate the RNA, UAP56, and SRSF2 signal intensities, respectively. Colocalization indexes are shown on the right. Data represent the mean ± SD from three independent experiments; n = 10. (F) mRNAs derived from intron-containing genes associate with NSs dependent on splicing, whereas naturally intronless mRNAs enter these subnuclear structures via SR proteins that are bound on ESEs. Aberrant mRNAs such as cDNA transcripts cannot enter NS due to lack of splicing. In NSs, mRNA export factors are efficiently recruited due to their high concentration, resulting in efficient nuclear export of spliced and intronless mRNAs. Aberrant mRNAs have a limited chance to get access to mRNA export factors and are mostly retained in the nucleus. Bars, 20 µm. Statistical analysis was performed using an unpaired t test. ***, P < 0.01. Molecular masses are given in kilodaltons.
Discussion
As a general rule, nuclear bodies serve to concentrate proteins involved in similar processes in a confined space, probably to enhance reaction efficiency and facilitate regulation. NSs have long been known for their relevance in splicing (Lerner et al., 1981; Spector et al., 1983, 1991; Fu and Maniatis, 1990; Fu, 1993; Lamond and Spector, 2003; Spector and Lamond, 2011). Except for splicing factors, mRNA export factors are also enriched in these subnuclear structures (Zhou et al., 2000; Gatfield et al., 2001; Masuda et al., 2005). Depletion of mRNA export factors such as TREX proteins results in retention of polyA RNAs and spliced reporter mRNAs in NSs (Dias et al., 2010; Chi et al., 2013; Shi et al., 2015). Based on previous studies and our data, we propose that NSs are sites for the assembly of many export-competent mRNPs (Fig. 9 F). mRNAs derived from intron-containing genes associate with NSs dependent on splicing (Johnson et al., 2000; Melčák et al., 2001; Ishihama et al., 2008; Dias et al., 2010), whereas intronless mRNAs enter these domains via ESEs where they probably recruit mRNA export factors using specific cis-acting elements, e.g., cytoplasmic accumulation regions (CARs; Lei et al., 2011, 2013). On one hand, recruitment of mRNA export factors in NSs is efficient, probably due to their enrichment. On the other hand, aberrant mRNAs that cannot be targeted to NSs, such as cDNA transcripts and defectively processed mRNAs, could be limited for the access of mRNA export factors and thus are mostly retained and degraded in the nucleus. Our study adds a new layer of function to NSs in the assembly of export-competent mRNPs and supports the notion that entering NSs might be an important quality control step in mRNA export. We note that since mRNAs can be diverse and differences in cell types may influence RNA processing, the generality of the function of NSs in mRNA export remains to be studied.
Although it is controversial whether cotranscriptional splicing occurs in the center of NSs or at surrounding sites, multiple studies have shown that posttranscriptional splicing occurs in these domains (Girard et al., 2012; Boutz et al., 2015). Splicing promotes mRNA export by enhancing recruitment of mRNA export factors, i.e., TREX components (Zhou et al., 2000; Masuda et al., 2005; Chi et al., 2013). This conclusion was drawn mainly based on the observation that compared with cDNA transcripts, TREX recruitment to the corresponding spliced mRNAs was apparently enhanced. This enhancement has been thought to be due to physical interactions between splicing factors and mRNA export factors. Although several potential functional interactions have been identified, solid evidence is still lacking (Chi et al., 2013; Gromadzka et al., 2016). We found that speckle targeting of β-globin cDNA transcript, which otherwise diffuse and are retained in the nucleoplasm, in two different ways significantly enhanced mRNA export (Fig. 7, A, B, and H; and Fig. S4 D). This finding raised the possibility that the effect of splicing on promoting mRNA export might in part be due to speckle association. Consistent with this possibility, the sequence of SSCR that promotes nuclear export of cDNA transcripts also facilitates speckle targeting (Palazzo et al., 2007; Akef et al., 2013). Further, depletion of ALYREF resulted in the trapping of the viral unspliced M1 mRNA into NSs and enhanced its splicing, suggesting cross-talk between NSs and intronless mRNA export (Mor et al., 2016). In addition, we previously found that a subelement in the posttranscriptional regulatory element (SEP1) of hepatitis B viral RNAs promoted nuclear export of cDNA transcripts (Chi et al., 2014). We revisited the localization of the cG-SEP1 mRNA and found that it also traffics into NSs (Fig. S5). We cannot exclude the possibility that speckle targeting and export promotion resulted from both SSCR and SEP1 are independent events. However, together with the findings that ESE and SR protein-mediated speckle association enhances TREX recruitment and mRNA export, we speculate that many export-promoting elements might execute their roles partially by facilitating speckle targeting. A previous study reported that insertion of an intron in U snRNAs resulted in their export via mRNA export pathway (Ohno et al., 2002). It is possible that this export pathway switch was due to the association of RNAs with different nuclear bodies.
On average, >30 ESEs are present in each intronless gene. This explains why multiple RNA fragments show speckle associations. Further, we note that since we studied only two ESEs, it remains possible that different ESEs have varied ability in facilitating mRNA speckle targeting. This, and the fact that intronless genes contain different numbers of ESEs, might explain why different intronless mRNAs associate with NSs to a varied extent. Although we detected the apparent association of intronless mRNAs with NSs, it remains possible that speckle targeting of these mRNAs is not very efficient and those that cannot traffic into these domains are degraded in the nucleoplasm. In support of this possibility, compared with spliced mRNAs, intronless mRNAs more tend to be degraded by the nuclear exosome (Fan et al., 2017). Further, speckle association does not seem to be a definite requisite for intronless mRNA export as intronless mRNAs that do not apparently associate with NSs could be exported (Lei et al., 2011). In addition, replication-dependent histone mRNAs that are known to mainly localize in histone locus bodies are exported via directly recruiting the mRNA export receptor NXF1 (Huang and Steitz, 2001; Huang et al., 2003; Marzluff et al., 2008). We speculate that intronless mRNAs that do not enter NSs might evolve specific mechanisms for efficiently recruiting mRNA export factors diffusing in the nucleoplasm. Further studies are required to understand these mechanisms.
Materials and methods
Plasmids and antibodies
To construct naturally intronless reporter plasmids, the coding sequence of HSPA1A, HSPB9, GPR119, GPR32, CJUN, JUND, CLDN3, and IFNE were inserted to pcDNA3 (Invitrogen). HSPA1A and JUND fragments were cloned into the pcDNA3. cG, cG-M6, wG, and cS constructs were described previously (Valencia et al., 2008; Shi et al., 2015). The sequence of cG was inserted into the STE1 or STE2 construct to make the cG-STE1 or cG-STE2 plasmid. Seven tandem repeats of 5′-GACCTG-3′ or 5′-GAGGAG-3′ were inserted into the cG construct to make the cG-ESE1 and cG-ESE2 constructs. To construct GFP-MS2 plasmid, the coding sequence of MS2 was inserted to pEGFP-C1 (Takara Bio Inc.). To construct MS2-SRSF2-HA plasmid, the coding sequence of MS2-SRSF2 was cloned into pEGFP-N3 (Takara Bio Inc.) in which GFP was replaced by HA. To construct the Cntl and ΔSR RNA expression plasmid, the Cntl or ΔSR sequence was inserted into pcDNA3. The pEGFP-C3 (Takara Bio Inc.) was used as a transfection Cntl.
The antibodies against ALYREF and UAP56 were described previously (Chi et al., 2014). The SRSF2 (1:2,000, mouse; Sigma-Aldrich), XRN2 (1:500, rabbit; ProteinTech), MTR4 (1:1,000, rabbit; ABclonal), GAPDH (1:1,000, mouse; Sigma-Aldrich), digoxin (1:200, sheep; Roche), PABPN1 (1:1,000, rabbit; ABclonal), tubulin (1:1,000, mouse; Sigma-Aldrich), GFP (1:2,000, rabbit; Sigma-Aldrich), and NXF1 (1:500, rabbit; ProteinTech) antibodies were purchased. IgG was purchased from Santa Cruz. Alexa Fluor 546–, Alexa Fluor 488–, or Alexa Fluor 647–conjugated secondary antibodies were purchased from Life Technologies.
Cell culture, RNAi, and transfection
HeLa cells were cultured in DMEM supplemented with 10% FBS (Biochrom). siRNA and DNA transfections were performed with RNAi MAX (Invitrogen) and Lipofectamine 2000 (Invitrogen), respectively, according to the manufacturer’s protocols. ASOs were introduced to HeLa cells by using electroporation (NEPA21) according to the manufacturer’s recommendations. Targeting sequences of siRNAs and ASOs are shown in Table S1. Note that UAP56 and its homolog URH49 must be co–knocked down to observe an obvious export block (Kapadia et al., 2006).
Microinjections, FISH, and IF
DNA microinjection and FISH were performed as previously described (Shi et al., 2015). DNA (100 ng/µl) was microinjected into HeLa cells plated on a fibronectin-coated 20-mm coverslip at the bottom of 35-mm dishes, followed by incubation at 37°C. For each experiment, 300–400 cells were microinjected. After 20 min incubation, transcription was terminated with 4 µg/ml α-amanitin (Sigma-Aldrich), and incubation was continued for the desired period of time before fixation with 4% PFA. After incubation with 0.1% Triton X-100, the cells were hybridized with the FISH probes overnight at 37°C. After being washed with 1× SSC/50% formamide four times, the cells were stained with DAPI. The FISH probes were labeled at the 5′ end with Alexa Fluor 546 or Alexa Fluor 488 and HPLC purified. For RNA injection, the RNA (1 µg/µl) was coinjected with FITC-conjugated 70-kD dextran as an injection marker. The sequences of the vector probes and β-globin probe are shown in Table S2. For IF, SRSF2 (1:2,000), GFP (1:2,000), PABPN1 (1:1,000), and UAP56 (1:200) antibodies were used. After primary antibody incubation, cells were washed three times in 1× PBS and incubated for 1 h at room temperature with Alexa Fluor 488–, Alexa Fluor 546–, or Alexa Fluor 647–conjugated secondary antibodies 1:1,000 in blocking buffer (1× PBS, 0.1% Triton X-100, and 2 mg/ml BSA).
To detect the endogenous RNAs with SRSF2 simultaneously, HeLa cells were fixed with 3.6% formaldehyde plus 10% acetic acid in 1× PBS for 20 min, followed by three washes with 1× PBS and permeabilization with 1× PBS/0.1% Triton X-100/2mM vanadyl ribonucleoside complex (VRC) for 15 min. The cells were incubated with specific probes that were labeled with digoxin at 50°C for 16 h The targeting sequences of the transcript-specific probes are shown in Table S2. After extensive wash, cells were incubated with the digoxin and SRSF2 antibody diluted in blocking buffer for 1 h. After three washes with 1× PBS and incubation with the Alexa Flour 488–labeled anti–sheep antibody (1:1,000) for 1 h, the cells were washed with 1× PBS three times and incubated with the Alexa Fluor 546–labeled anti–mouse antibody for another 1 h, followed by DAPI staining and three washes with 1× PBS.
Confocal imaging was performed on a FLUOVIEW FV1000 microscope (Olympus) by using a 60× 1.35 NA objective lens and acquired with FV10-ASW software (Olympus) or was performed using an A1plus laser-scanning confocal microscope with a 60× 1.40 NA objective lens and acquired with NIS Elements (Nikon) software. Fluorescence microscopic images were captured with a DP72 charge-coupled device camera on IX71 microscope by using DP-BSW software (Olympus). All imaging was performed at room temperature, and all samples were covered with 1× PBS. 3D reconstitutions were performed with FV10-ASW software (Olympus).
To ensure that the colocalization index could reflect the extent of colocalization, the data were analyzed in three steps. First, it was judged whether there was a focus by the comparison of the intensity of the focus signal with that of the nucleoplasm signal (>20% was considered positive). Then, colocalization of two foci was judged by comparing the peak locations of intronless mRNAs and NSs markers. The intronless mRNA peaks localizing within the central region (±5%) of the speckle peaks were considered positive. Finally, we calculated the colocalization index using the area of intronless mRNA signals divided by the area of speckle focus signal. Notably, when counting the signals of endogenous FISH, the obvious noise signals were discarded. The intensity of signals and the area under the curves were counted with ImageJ 1.33u software (National Institutes of Health). N/C ratios were calculated as described (Valencia et al., 2008). The area (AW) and the average signal intensity of the whole cell (IW) were determined separately by using the ImageJ software. Similarly, the area (AN) and the average signal intensity of the nucleus (IN) were determined. The background intensity (BIW for the whole cell, and BIN for the nucleus) was determined by using a cell without a FISH signal. The total cell signal (W) was calculated as AW × (IW − BIW). Similarly, the nuclear signal (N) was calculated as AN × (IN − BIN), and the cytoplasmic signal (C) was equal to W − N. The N/C ratio was equal to N/(W −N).
Subcellular fractionation
Subcellular fractionation was performed as described (Shi et al., 2017). Briefly, 4 × 105 HeLa cells were suspended in 1 ml lysis buffer (10 mM Tris, pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% IGEPAL, and 2 mM VRC) and incubated for 5 min on ice. The lysate was centrifuged at 1,000 g for 3 min at 4°C, and the pellet and the supernatant were kept as the nuclei and the cytoplasmic fraction, respectively. The supernatant fraction was further centrifuged at 13,000 rpm for 10 min at 4°C and then collected carefully to a new tube. The nuclear pellets were subjected to one additional wash with 1 ml lysis buffer containing 0.5% deoxycholic acid and one additional wash with lysis buffer. Finally, the purified nuclei were resuspended in 100 µl lysis buffer. Nuclear or cytoplasmic RNAs were extracted by using TRI reagent (Sigma-Aldrich).
RIPs
RIPs with UV cross-linking were performed as described with some modification (Yeo et al., 2009). Different β-globin constructs were cotransfected with the cS construct into HeLa cells. 4 h after transfection, cells were washed with cold 1× PBS and irradiated at 150 mJ/cm2 with UV. The cells were then suspended in 1 ml RIP buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40, 0.25% sodium deoxycholate, 0.05% SDS, 1 mM PMSF, 2 mM VRC, and protease inhibitor cocktail), followed by 10 rounds of sonication (10-s work with 60-s rest) on ice. After centrifugation at 13,000 rpm at 4°C, the supernatants were precleared with 50 µl Dynabeads A (Invitrogen) and 20 µg/ml yeast tRNA at 4°C for 1 h. The precleared cell lysate was incubated with Dynabeads A, which were coated with the ALYREF antibody or IgG at 4°C for 4 h. The beads were washed three times with buffer I (50 mM Tris-HCl, pH 7.5, 1 M NaCl, 0.5% NP-40, 0.25% sodium deoxycholate, 0.05% SDS, and 2 mM VRC) and three times with buffer II (50 mM Tris-HCl, pH 7.5, 1 M NaCl, 0.5% NP-40, 0.25% sodium deoxycholate, 0.05% SDS, 2 mM VRC, and 1 M urea). The immunoprecipitated RNA was extracted by using TRI reagent following proteinase K treatment. RNAs were then treated with the RNase-free RQ DNase I (Promega) for 2 h at 37°C, and cDNAs were synthesized from 1 µg RNAs with random primer by using M-MLV reverse transcription (Promega). For qPCRs, cDNAs were amplified by using GoTaq qPCR Master Mix (Promega) according to the manufacturer’s protocol. Primer sequences are listed in Table S3. For data quantification, we first calculated IP efficiency for each mRNA using immunoprecipitated mRNA divided by the corresponding input. Then, the β-globin IP efficiency/cS IP efficiency for each different β-globin construct was calculated. Further, the final relative IP efficiency of the cG mRNA was set at 1, and the ratios of other β-globin mRNAs to the cG mRNA were calculated and shown in the bar graph.
In vitro transcription
Capped naturally intronless mRNAs were in vitro transcribed by using the mMessage mMachine Kit (Ambion), and the polyA tail was added with the Poly(A) tailing kit (Ambion) using the mMessage mMachine Kit reaction products. The mRNAs were purified with the MEGAclear Kit (Ambion). The polyA mRNAs were prepared in the same way, with the polyA-tailing step being omitted.
DNA extraction
The DNA microinjected into the HeLa cells was extracted with TRI reagent with some modification based on the manufacturer’s protocols. After the RNA isolation, the interphase was removed to a new column. The DNA was precipitated with 3 M NaAc and 100% ethanol. The DNA pellet was washed twice in 0.1 M trisodium citrate and 10% ethanol solution. Following one more wash in 75% ethanol, the DNA pellet was dissolved in 8 mM NaOH. To amplify DNA by PCR, the solution was adjusted to pH 8.4 using 0.1 M Hepes.
Statistics
All results were repeated at least three times in independent experiments. Data are presented as mean ± SD or mean ± SEM, and statistical significance was determined by using the unpaired Student’s t test, which was two-sided. The variance was similar between the groups that were being statistically compared. Datasets were analyzed for statistical significance by using GraphPad Prism software (version 6). P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows the evidence that speckle-localized HSPA1A mRNAs were released to the cytoplasm rather than being degraded. Fig. S2 shows multiple fragments of HSPA1A and JUND mRNAs associating with NSs. Fig. S3 shows the presence of ESEs in naturally intronless genes and that introducing ESEs does not cause cryptic splicing events of the cG mRNA. Fig. S4 shows the roles of SR and TREX proteins in speckle localization and nuclear export of intronless mRNAs and the distribution of exogenous HSPA1A or endogenous ZXDB mRNA in cells treated with UAP56 or NXF1 siRNAs. Fig. S5 shows SEP1 facilitates speckle association of cDNA transcripts. Table S1 shows siRNA and ASO targeting sequences. Table S2 shows probe sequences. Table S3 shows primer sequences. Videos 1, 2, 3, and 4 show colocalization of JUND (Video 1), HSPA1A (Video 2), RHOB (Video 3), and ZXDB (Video 4) mRNAs and SRSF2 with 3D reconstitutions.
Supplementary Material
Acknowledgments
We thank Cheng Lab members for useful suggestions and discussions.
This work was supported by grants from National Key R&D Program of China (2017YFA0504400) and the National Natural Science Foundation of China (31570822, 91540104, and 31770880).
The authors declare no competing financial interests.
Author contributions: K. Wang and H. Cheng conceived the project. K. Wang performed most of the experiments and analysis. L. Wang made JUND fragment constructs and cultured the cells. J. Wang made cG-STE1, cG-STE2, cS-STE1, and cS-STE2 constructs and carried out parts of the microinjection, IF, and FISH experiments. S. Chen made HSPA1A fragment constructs. M. Shi performed parts of Western blotting. K. Wang and H. Cheng wrote the manuscript. All authors discussed the results and commented on the manuscript.
References
- Akef A., Zhang H., Masuda S., and Palazzo A.F.. 2013. Trafficking of mRNAs containing ALREX-promoting elements through nuclear speckles. Nucleus. 4:326–340. 10.4161/nucl.26052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., and Mitchell P.. 1999. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev. 13:2148–2158. 10.1101/gad.13.16.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B.J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106–110. 10.1016/S0968-0004(00)01549-8 [DOI] [PubMed] [Google Scholar]
- Bond C.S., and Fox A.H.. 2009. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 186:637–644. 10.1083/jcb.200906113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz P.L., Bhutkar A., and Sharp P.A.. 2015. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 29:63–80. 10.1101/gad.247361.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres E.F., and Hurst L.D.. 2013. The evolution, impact and properties of exonic splice enhancers. Genome Biol. 14:R143 10.1186/gb-2013-14-12-r143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter K.C., Taneja K.L., and Lawrence J.B.. 1991. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J. Cell Biol. 115:1191–1202. 10.1083/jcb.115.5.1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler S.D., Mayeda A., Yeakley J.M., Krainer A.R., and Fu X.D.. 1997. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc. Natl. Acad. Sci. USA. 94:3596–3601. 10.1073/pnas.94.8.3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.L., and Carmichael G.G.. 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 35:467–478. 10.1016/j.molcel.2009.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi B., Wang Q., Wu G., Tan M., Wang L., Shi M., Chang X., and Cheng H.. 2013. Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic Acids Res. 41:1294–1306. 10.1093/nar/gks1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi B., Wang K., Du Y., Gui B., Chang X., Wang L., Fan J., Chen S., Wu X., Li G., and Cheng H.. 2014. A Sub-Element in PRE enhances nuclear export of intronless mRNAs by recruiting the TREX complex via ZC3H18. Nucleic Acids Res. 42:7305–7318. 10.1093/nar/gku350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmarko D., Verschure P.J., Martin T.E., Dahmus M.E., Krause S., Fu X.D., van Driel R., and Fakan S.. 1999. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell. 10:211–223. 10.1091/mbc.10.1.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias A.P., Dufu K., Lei H., and Reed R.. 2010. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat. Commun. 1:97 10.1038/ncomms1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen W.P., Li X., Mayeda A., Krainer A.R., and Rottman F.M.. 2000. Mapping the SF2/ASF binding sites in the bovine growth hormone exonic splicing enhancer. J. Biol. Chem. 275:29170–29177. 10.1074/jbc.M001126200 [DOI] [PubMed] [Google Scholar]
- Fan J., Kuai B., Wu G., Wu X., Chi B., Wang L., Wang K., Shi Z., Zhang H., Chen S., et al. . 2017. Exosome cofactor hMTR4 competes with export adaptor ALYREF to ensure balanced nuclear RNA pools for degradation and export. EMBO J. 36:2870–2886. 10.15252/embj.201696139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasken M.B., Leung S.W., Banerjee A., Kodani M.O., Chavez R., Bowman E.A., Purohit M.K., Rubinson M.E., Rubinson E.H., and Corbett A.H.. 2011. Air1 zinc knuckles 4 and 5 and a conserved IWRXY motif are critical for the function and integrity of the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) RNA quality control complex. J. Biol. Chem. 286:37429–37445. 10.1074/jbc.M111.271494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forler D., Rabut G., Ciccarelli F.D., Herold A., Köcher T., Niggeweg R., Bork P., Ellenberg J., and Izaurralde E.. 2004. RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export. Mol. Cell. Biol. 24:1155–1167. 10.1128/MCB.24.3.1155-1167.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.D. 1993. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 365:82–85. 10.1038/365082a0 [DOI] [PubMed] [Google Scholar]
- Fu X.D., and Maniatis T.. 1990. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 343:437–441. 10.1038/343437a0 [DOI] [PubMed] [Google Scholar]
- Funatsu T. 2009. [Single-molecule imaging and quantification of mRNAs in a living cell]. Yakugaku Zasshi. 129:265–272. 10.1248/yakushi.129.265 [DOI] [PubMed] [Google Scholar]
- Gatfield D., Le Hir H., Schmitt C., Braun I.C., Köcher T., Wilm M., and Izaurralde E.. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11:1716–1721. 10.1016/S0960-9822(01)00532-2 [DOI] [PubMed] [Google Scholar]
- Girard C., Will C.L., Peng J., Makarov E.M., Kastner B., Lemm I., Urlaub H., Hartmuth K., and Lührmann R.. 2012. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 3:994 10.1038/ncomms1998 [DOI] [PubMed] [Google Scholar]
- Gromadzka A.M., Steckelberg A.L., Singh K.K., Hofmann K., and Gehring N.H.. 2016. A short conserved motif in ALYREF directs cap- and EJC-dependent assembly of export complexes on spliced mRNAs. Nucleic Acids Res. 44:2348–2361. 10.1093/nar/gkw009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzybowska E.A. 2012. Human intronless genes: functional groups, associated diseases, evolution, and mRNA processing in absence of splicing. Biochem. Biophys. Res. Commun. 424:1–6. 10.1016/j.bbrc.2012.06.092 [DOI] [PubMed] [Google Scholar]
- Hall L.L., Smith K.P., Byron M., and Lawrence J.B.. 2006. Molecular anatomy of a speckle. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 288:664–675. 10.1002/ar.a.20336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., and Steitz J.A.. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 7:899–905. 10.1016/S1097-2765(01)00233-7 [DOI] [PubMed] [Google Scholar]
- Huang S., Deerinck T.J., Ellisman M.H., and Spector D.L.. 1994. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J. Cell Biol. 126:877–899. 10.1083/jcb.126.4.877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Gattoni R., Stévenin J., and Steitz J.A.. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 11:837–843. 10.1016/S1097-2765(03)00089-3 [DOI] [PubMed] [Google Scholar]
- Huang Y., Yario T.A., and Steitz J.A.. 2004. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. USA. 101:9666–9670. 10.1073/pnas.0403533101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y., Tadakuma H., Tani T., and Funatsu T.. 2008. The dynamics of pre-mRNAs and poly(A)+ RNA at speckles in living cells revealed by iFRAP studies. Exp. Cell Res. 314:748–762. 10.1016/j.yexcr.2007.10.023 [DOI] [PubMed] [Google Scholar]
- Johnson C., Primorac D., McKinstry M., McNeil J., Rowe D., and Lawrence J.B.. 2000. Tracking COL1A1 RNA in osteogenesis imperfecta. splice-defective transcripts initiate transport from the gene but are retained within the SC35 domain. J. Cell Biol. 150:417–432. 10.1083/jcb.150.3.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia F., Pryor A., Chang T.H., and Johnson L.F.. 2006. Nuclear localization of poly(A)+ mRNA following siRNA reduction of expression of the mammalian RNA helicases UAP56 and URH49. Gene. 384:37–44. 10.1016/j.gene.2006.07.010 [DOI] [PubMed] [Google Scholar]
- Kataoka N., Yong J., Kim V.N., Velazquez F., Perkinson R.A., Wang F., and Dreyfuss G.. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell. 6:673–682. 10.1016/S1097-2765(00)00065-4 [DOI] [PubMed] [Google Scholar]
- Krause S., Fakan S., Weis K., and Wahle E.. 1994. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp. Cell Res. 214:75–82. 10.1006/excr.1994.1235 [DOI] [PubMed] [Google Scholar]
- Lamond A.I., and Spector D.L.. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4:605–612. 10.1038/nrm1172 [DOI] [PubMed] [Google Scholar]
- Lebreton A., Tomecki R., Dziembowski A., and Séraphin B.. 2008. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 456:993–996. 10.1038/nature07480 [DOI] [PubMed] [Google Scholar]
- Lei H., Dias A.P., and Reed R.. 2011. Export and stability of naturally intronless mRNAs require specific coding region sequences and the TREX mRNA export complex. Proc. Natl. Acad. Sci. USA. 108:17985–17990. 10.1073/pnas.1113076108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H., Zhai B., Yin S., Gygi S., and Reed R.. 2013. Evidence that a consensus element found in naturally intronless mRNAs promotes mRNA export. Nucleic Acids Res. 41:2517–2525. 10.1093/nar/gks1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E.A., Lerner M.R., Janeway C.A. Jr., and Steitz J.A.. 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA. 78:2737–2741. 10.1073/pnas.78.5.2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Shambaugh M.E., Rottman F.M., and Bokar J.A.. 2000. SR proteins Asf/SF2 and 9G8 interact to activate enhancer-dependent intron D splicing of bovine growth hormone pre-mRNA in vitro. RNA. 6:1847–1858. 10.1017/S1355838200000674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.X., Zhang M., and Krainer A.R.. 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 12:1998–2012. 10.1101/gad.12.13.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.X., Chew S.L., Cartegni L., Zhang M.Q., and Krainer A.R.. 2000. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 20:1063–1071. 10.1128/MCB.20.3.1063-1071.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M., Christensen M.S., Kristiansen M.S., Domanski M., Falkenby L.G., Lykke-Andersen S., Andersen J.S., Dziembowski A., and Jensen T.H.. 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell. 43:624–637. 10.1016/j.molcel.2011.06.028 [DOI] [PubMed] [Google Scholar]
- Marzluff W.F., Wagner E.J., and Duronio R.J.. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9:843–854. 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S., Das R., Cheng H., Hurt E., Dorman N., and Reed R.. 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 19:1512–1517. 10.1101/gad.1302205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Badolato J., Kobayashi R., Zhang M.Q., Gardiner E.M., and Krainer A.R.. 1999. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J. 18:4560–4570. 10.1093/emboj/18.16.4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melčák I., Melcáková S., Kopský V., Vecerová J., and Raska I.. 2001. Prespliceosomal assembly on microinjected precursor mRNA takes place in nuclear speckles. Mol. Biol. Cell. 12:393–406. 10.1091/mbc.12.2.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science. 291:843–847. 10.1126/science.291.5505.843 [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Shevchenko A., Mann M., and Tollervey D.. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 91:457–466. 10.1016/S0092-8674(00)80432-8 [DOI] [PubMed] [Google Scholar]
- Mor A., White A., Zhang K., Thompson M., Esparza M., Muñoz-Moreno R., Koide K., Lynch K.W., García-Sastre A., and Fontoura B.M.. 2016. Influenza virus mRNA trafficking through host nuclear speckles. Nat. Microbiol. 1:16069 10.1038/nmicrobiol.2016.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-McNicoll M., Botti V., de Jesus Domingues A.M., Brandl H., Schwich O.D., Steiner M.C., Curk T., Poser I., Zarnack K., and Neugebauer K.M.. 2016. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 30:553–566. 10.1101/gad.276477.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M., Segref A., Kuersten S., and Mattaj I.W.. 2002. Identity elements used in export of mRNAs. Mol. Cell. 9:659–671. 10.1016/S1097-2765(02)00454-9 [DOI] [PubMed] [Google Scholar]
- Palazzo A.F., Springer M., Shibata Y., Lee C.S., Dias A.P., and Rapoport T.A.. 2007. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 5:e322 10.1371/journal.pbio.0050322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savisaar R., and Hurst L.D.. 2016. Purifying Selection on Exonic Splice Enhancers in Intronless Genes. Mol. Biol. Evol. 33:1396–1418. 10.1093/molbev/msw018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G., van Dijk E., and Pruijn G.J.. 2007. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 35:2564–2572. 10.1093/nar/gkm082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., Anderson J.T., and Tollervey D.. 2007. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol. Cell. 27:324–331. 10.1016/j.molcel.2007.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma K., Doye V., Hellwig A., Huber J., Lührmann R., and Hurt E.. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256–3271. 10.1093/emboj/16.11.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Zhang H., Wang L., Zhu C., Sheng K., Du Y., Wang K., Dias A., Chen S., Whitman M., et al. . 2015. Premature Termination Codons Are Recognized in the Nucleus in A Reading-Frame Dependent Manner. Cell Discov. 1:15001 10.1038/celldisc.2015.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Zhang H., Wu X., He Z., Wang L., Yin S., Tian B., Li G., and Cheng H.. 2017. ALYREF mainly binds to the 5′ and the 3′ regions of the mRNA in vivo. Nucleic Acids Res. 45:9640–9653. 10.1093/nar/gkx597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.L., and Lamond A.I.. 2011. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3:a000646 10.1101/cshperspect.a000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.L., Schrier W.H., and Busch H.. 1983. Immunoelectron microscopic localization of snRNPs. Biol. Cell. 49:1–10. 10.1111/j.1768-322X.1984.tb00215.x [DOI] [PubMed] [Google Scholar]
- Spector D.L., Fu X.D., and Maniatis T.. 1991. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 10:3467–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondón A.G., Aguilera A., Struhl K., Reed R., and Hurt E.. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 417:304–308. 10.1038/nature746 [DOI] [PubMed] [Google Scholar]
- Sun Q., Mayeda A., Hampson R.K., Krainer A.R., and Rottman F.M.. 1993. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 7:2598–2608. 10.1101/gad.7.12b.2598 [DOI] [PubMed] [Google Scholar]
- Swift H. 1959. Studies on nuclear fine structure. Brookhaven Symp. Biol. 12:134–152. [PubMed] [Google Scholar]
- Tacke R., and Manley J.L.. 1995. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14:3540–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia P., Dias A.P., and Reed R.. 2008. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. USA. 105:3386–3391. 10.1073/pnas.0800250105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N., Puvion-Dutilleul F., Harper F., Bachellerie J.P., and Puvion E.. 1993. Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp. Cell Res. 208:19–34. 10.1006/excr.1993.1218 [DOI] [PubMed] [Google Scholar]
- West S., Gromak N., and Proudfoot N.J.. 2004. Human 5′ --> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 432:522–525. 10.1038/nature03035 [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhang Y., and Zhang J.. 2005. Distribution of exonic splicing enhancer elements in human genes. Genomics. 86:329–336. 10.1016/j.ygeno.2005.05.011 [DOI] [PubMed] [Google Scholar]
- Yeo G.W., Coufal N.G., Liang T.Y., Peng G.E., Fu X.D., and Gage F.H.. 2009. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 16:130–137. 10.1038/nsmb.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Luo M.J., Straesser K., Katahira J., Hurt E., and Reed R.. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 407:401–405. 10.1038/35030160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.