Fan et al. have identified a population of recirculating Treg cells with greater suppressive ability and a unique tissue distribution. Using single-cell RNA-seq, they place these Treg cells at the apex of the Treg developmental trajectory and show that similar cells may exist in humans.
Abstract
Regulatory T (Treg) cells prevent autoimmunity by limiting immune responses and inflammation in the secondary lymphoid organs and nonlymphoid tissues. While unique subsets of Treg cells have been described in some nonlymphoid tissues, their relationship to Treg cells in secondary lymphoid organs and circulation remains unclear. Furthermore, it is possible that Treg cells from similar tissue types share largely similar properties. We have identified a short-lived effector Treg cell subset that expresses the α2 integrin, CD49b, and exhibits a unique tissue distribution, being abundant in peripheral blood, vasculature, skin, and skin-draining lymph nodes, but uncommon in the intestines and in viscera-draining lymph nodes. CD49b+ Treg cells, which display superior functionality revealed by in vitro and in vivo assays, appear to develop after multiple rounds of cell division and TCR-dependent activation. Accordingly, single-cell RNA-seq analysis placed these cells at the apex of the Treg developmental trajectory. These results shed light on the identity and development of a functionally potent subset of mature effector Treg cells that recirculate through and survey peripheral tissues.
Introduction
Lymphocytes are widely dispersed throughout the body and can be categorized on the basis of their migratory behaviors into recirculating or tissue-resident cells (von Andrian and Mackay, 2000; Fan and Rudensky, 2016). Importantly, these behaviors subserve immune effector functions and modes of immunosurveillance. For example, naive T cells and central memory T cells recirculate through secondary lymphoid organs (SLOs), scanning for their cognate peptide–MHC complexes, while tissue-resident memory T (TRM) cells embed in tissues to act as antigen-specific sentinels against recurrent infection. Among innate lymphocytes, recirculating natural killer (NK) cells inspect parenchymal cells for signs of infection or transformation, while tissue-resident innate lymphoid cells (ILCs) are early sources of pivotal cytokines. Thus, through the combined network of recirculating and tissue-resident lymphocytes, the mammalian immune system is able to efficiently monitor far-flung anatomical locations and meet diverse signals of perturbed homeostasis with the appropriate response.
Regulatory T (Treg) cells are a lineage of CD4+ T cells required throughout life to suppress autoreactive T cells that escape thymic selection (Josefowicz et al., 2012). Deficiency in Treg cell abundance, fitness, or function all lead to lethal multi-organ autoimmunity. Treg cells perform their suppressor function through a variety of mechanisms that include the production of immunoregulatory mediators, such as IL-10, TGF-β, and adenosine, and the depletion of IL-2 and down-modulation of costimulatory molecules. These mechanisms may play varying roles in different tissues. In the SLOs, recent studies have suggested that consumption of IL-2 by Treg cells in the proximity of conventional T cells is an important suppressor mechanism; Treg cells that lack the ability to consume IL-2 through the high-affinity IL-2 receptor are specifically unable to restrain CD8+ T cell expansion (Liu et al., 2015; Chinen et al., 2016). Besides SLOs, Treg cells can also be found in nonlymphoid tissues (NLTs), and a disruption of Treg cell trafficking to NLTs results in tissue-specific inflammation (Sather et al., 2007). NLT-localized Treg cells are transcriptionally distinct from their lymphoid tissue counterparts and may exhibit distinct functional modalities (Feuerer et al., 2009; Cipolletta et al., 2012; Schiering et al., 2014; Ohnmacht et al., 2015; Sefik et al., 2015). For example, production of IL-10 by Treg cells present at mucosal surfaces, foremost in the intestine, plays a nonredundant role in suppressing inflammation at these sites. Accordingly, mice lacking IL-10 in Treg cells exhibit selective mucosal inflammation rather than the systemic lymphoproliferation seen in mice lacking all Treg cells or in mice in which Treg cells are unable to consume IL-2 (Rubtsov et al., 2008). Furthermore, Treg cells also contribute to repair of NLTs after injury by producing amphiregulin, a ligand of the epidermal growth factor family (Burzyn et al., 2013; Arpaia et al., 2015).
Despite considerable progress in understanding the functional “geography” of Treg cells, the relationships between Treg cells in SLOs and those in NLTs remain unclear. Although parabiosis studies show that Treg cells in NLTs equilibrate more slowly than those in SLOs, suggesting a longer dwell time in NLTs (Kolodin et al., 2015), donor-derived cells do not persist in NLTs upon disconnection of parabiotic mice, indicating that most tissue Treg populations are continuously replenished from circulation (Luo et al., 2016). Tracking studies using photoactivatable tagging of cells show unexpectedly high egress of Treg cells from NLTs to their draining LNs in the steady-state, which increases during inflammation (Tomura et al., 2010; Morton et al., 2014; Ikebuchi et al., 2016). Consequently, Treg cells found in draining LNs share many properties with Treg cells from the tissues that they drain. Collectively, these results suggest that, unlike ILCs and tissue macrophages, Treg cells in adult mice have limited self-renewal capacity in NLTs or that a large recirculating Treg pool masks a small number of bona fide tissue-resident cells. While studies of adipose tissue and colonic Treg cells have led to the idea that Treg cells exhibit individual tissue-specific features and functions, the properties and ontogeny of Treg cells in other tissues have not been closely examined. It remained possible that common tissue characteristics, such as the extent of vascularization and oxygenation, or developmental origins may impart common features on Treg cells present in these tissues. In particular, highly vascularized tissues such as lung and liver contain Treg cells that transcriptionally cluster together with those from skin (Burzyn et al., 2013), but also with lymphoid organ Treg cells (Delacher et al., 2017). Thus, we sought to explore how Treg cells from lymphoid organs, circulation, and NLTs are related by investigating transcriptional and functional features of Treg cells present in highly vascularized tissues.
Results
CD49b+ Treg cells are enriched in vasculature and skin
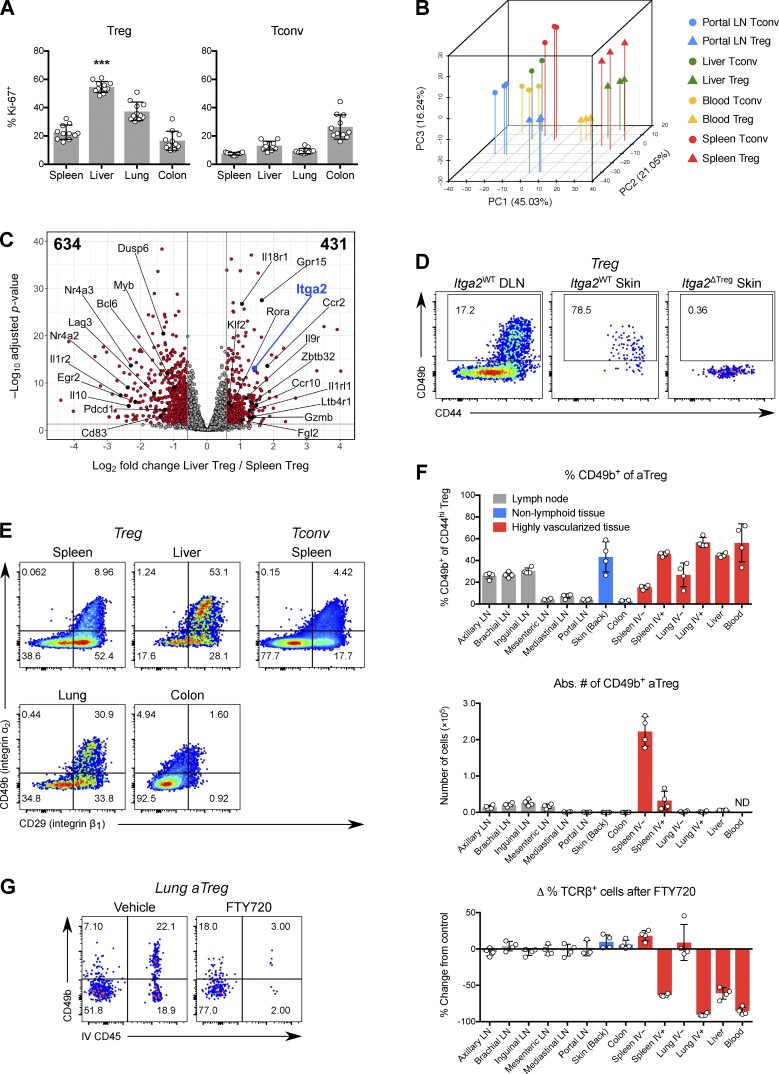
With the aim of understanding how Treg cells in highly vascularized tissues are related to Treg cells in other organs, we first used flow cytometry to characterize the phenotype of Treg cells from the liver, an organ with dual circulatory input and vital metabolic and detoxifying functions (Thomson and Knolle, 2010). Liver Treg cells contained a high percentage of Ki-67+ cells, even when compared with other tissues with predominantly activated Treg (aTreg) cells, such as the colon (Fig. 1 A and Fig. S1, A and B) and also when compared with liver CD4+ Foxp3− “conventional” T (Tconv) cells. To elucidate the transcriptional features associated with this phenotype, we performed RNA sequencing (RNA-seq) analysis of bulk Treg and Tconv cells isolated from liver, blood, spleen, and the liver-draining portal LN (Fig. S2 A). Principal component analysis (PCA) revealed a clear separation of cell transcriptomes by both tissue and Treg/Tconv cell identity, with cells from the liver falling between blood- and spleen-derived cells (Fig. 1 B). Compared with splenic Treg cells, hepatic Treg cells were enriched for genes related to proliferation and activation and genes encoding chemokine and cytokine receptors (Fig. 1 C).
Figure 1.
A subset of activated Treg cells expresses CD49b and is enriched in vasculature and skin. (A) Percentage of Ki-67+ cells among CD4+ Foxp3+ (Treg) cells or CD4+ Foxp3− (Tconv) cells in tissues of B6 mice. Data were pooled from three independent experiments (n = 11 total). ***, P < 0.001 compared with any other group by paired t test. (B) PCA, using the top 1,000 genes with highest variance, of Treg and Tconv cells from various mouse tissues and profiled by RNA-seq. (C) Volcano plot of RNA-seq results, comparing liver and spleen Treg cells. Genes with FC ≤ 0.67 or ≥ 1.5, and adjusted P value ≤ 0.05, are shown in red and enumerated in the top left and right corners, respectively. (D) Representative flow cytometric data showing expression of CD49b and CD44 in Treg cells from ear skin or draining (cervical) LN of Foxp3YFP-Cre Itga2+/+ (Itga2WT) or Foxp3YFP-Cre Itga2fl/fl (Itga2ΔTreg) mice. DLN, draining LN. (E) Expression of CD49b (integrin α2) and CD29 (integrin β1) in Treg cells from various tissues or Tconv cells from spleen. (F) Top, percentage of CD49b+ cells among CD4+ Foxp3+ CD44hi aTreg cells from tissues of B6 mice. Most liver and all blood lymphocytes are intravascular. Intravascular cells in LNs, skin, and colon are scarce (<5%) and were gated out in this analysis. Middle, absolute number of CD49b+ aTreg cells in tissues of B6 mice. Bottom, change in the percentage of TCRβ+ NK-1.1− CD1d-tetramer− cells (of CD45+ cells) 12 h after treatment with FTY720, compared with vehicle-treated mice. IV+, intravascular CD45 labeling+; IV−, extravascular; ND, not determined. (G) CD49b expression versus intravascular CD45 labeling of lung CD44hi aTreg cells 12 h after mice were treated with FTY720 or vehicle. Data in D–G are representative of three or more independent experiments with three to five mice per group. Bars depict means ± SD. See also Fig. S1 and Fig. S2.
One gene with significantly higher expression in liver and blood Treg cells was Itga2, encoding integrin α2, which is also known as CD49b or DX5 (Fig. 1 C). CD49b obligately heterodimerizes with CD29 (integrin β1) to form the very late antigen-2 (VLA-2) complex, a receptor for fibrillar collagen and other extracellular matrix (ECM) proteins (Madamanchi et al., 2014). CD49b is a well-known marker for recirculating mature NK cells, differentiating them from tissue-resident CD49a+ group 1 ILCs (Peng et al., 2013; Sojka et al., 2014; O’Sullivan et al., 2016). CD49b expression also defines subsets of memory CD4+ T cells with distinct cytokine profiles and has been implicated in the homing of memory T cells to bone marrow (Kassiotis et al., 2006; Tokoyoda et al., 2009). When we examined CD49b expression in Treg cells by flow cytometry, we noted that it marks a distinct population that is differentially abundant across tissues in a manner resembling the distribution of Ki-67+ Treg cells (Fig. 1, D and E). All CD49b+ Treg cells coexpressed CD29/integrin β1 and CD44, a marker of aTreg cells (Fig. 1, D and E). However, not all β1-expressing Treg cells expressed CD49b, as β1 can pair with other integrin α chains. While CD49b+ Treg cells were present at high frequencies in liver, lung, skin, and peripheral blood, they were scarce in the colon and other gut-associated tissues (Fig. 1, E and F). In both lung and spleen, CD49b+ Treg cells were enriched in vascular beds, as revealed by their efficient labeling upon the brief intravenous administration of a fluorescent CD45 antibody (Fig. 1, F and G; Anderson et al., 2014). Accordingly, T cells in this compartment were almost completely depleted 12 h after treatment with the S1PR1 agonist FTY720 (Fig. 1, F and G). Furthermore, CD49b+ Treg cells were enriched in skin-draining axillary, brachial, and inguinal LNs, but scarce in the mesenteric, mediastinal, and portal LNs draining the intestines, lung, and liver, respectively (Fig. 1 F). However, in absolute terms, CD49b+ Treg cells were numerically most abundant in the white pulp of the spleen (Fig. 1 F). Collectively, these data suggest that in healthy mice, CD49b marks a novel Treg population that appears to preferentially recirculate between skin, vasculature, and SLOs.
CD49b defines Treg cells with superior migratory ability
In light of our finding that a distinct CD49b+ Treg cell subset is enriched in liver and blood relative to spleen, we examined genes up-regulated in liver and blood Treg cells over spleen and portal LN cells in our RNA-seq data, with the goal of identifying genes associated with CD49b+ Treg cells independently of their tissue of origin (Fig. S1 C). As expected, one of the top genes identified in this approach was Itga2. Gene ontology (GO) terms related to migration, adhesion, maturation, and cytokine sensing were also overrepresented in liver and blood Treg cells (Fig. S1 C). To confirm that these genes were truly enriched in CD49b+ Treg cells, we performed RNA-seq of splenic CD49b+ and CD49b− Treg cells that were of otherwise similar activation status based on expression of CD44 and CD62L (Fig. 2, A–D; Fig. S2 B; and Fig. S1, D–G). This revealed that splenic CD49b+ Treg cells closely resemble hepatic Treg cells (Fig. S1 D) and express genes that facilitate recirculation through SLOs (e.g., Klf2 and S1pr1), as well as homing to skin (Ccr10, Ccr4, and Gpr15) and to sites of inflammation (Ccr2, Ltb4r1, and Cysltr1; Fig. 2, B–D; and Fig. S1 E). Splenic CD49b+ Treg cells were also enriched for the gene signatures of Treg cells that experience activation in an inflammatory environment (Arvey et al., 2014) or through constitutive STAT5 signaling (Chinen et al., 2016), but they were relatively depleted of transcripts that are TCR dependent in Treg cells (Fig. S1, F and G; Levine et al., 2017). These transcriptomic analyses, as well as the association of CD49b+ Treg cells with vasculature that we observed by flow cytometry, suggested that enhanced migration and circulation are defining properties of highly activated CD49b+ Treg cells.
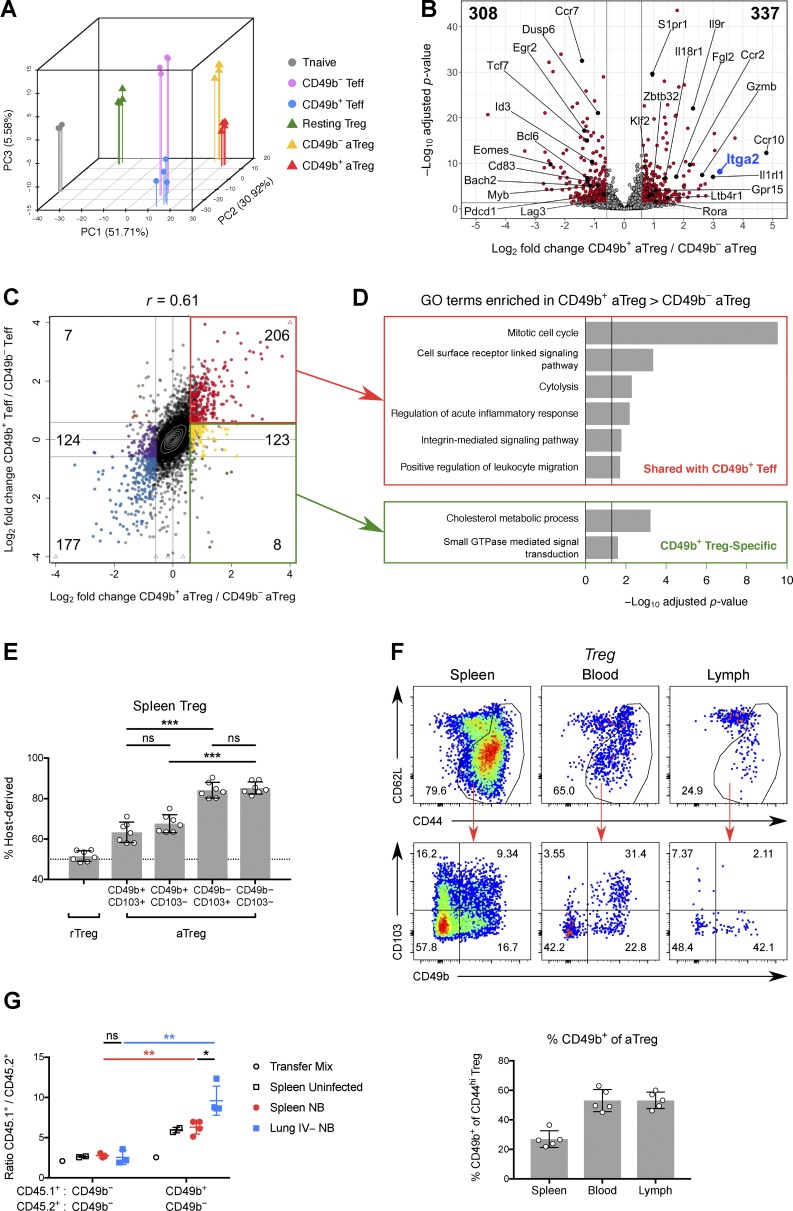
Figure 2.
CD49b+ Treg cells are recirculatory and traffic to sites of inflammation. (A) PCA, using the top 1,000 genes with highest variance, of mouse splenic Treg and Tconv cell subsets profiled by RNA-seq. (B) Volcano plot of RNA-seq results, comparing CD49b+ and CD49b− aTreg cells. Numbers in top left and right corners indicate numbers of genes down- or up-regulated in CD49b+ Treg cells, respectively. (C) Log2 FC in gene expression between CD49b+ and CD49b− splenic aTreg cells plotted against log2 FC between CD49b+ and CD49b− splenic Teff cells. Genes that are differentially expressed between the aTreg cell subsets (padj ≤ 0.05, FC ≥ 1.5 or ≤ 0.67) are further subdivided by their FC in Teff cells (FC ≥ 1.5; 0.67 ≤ FC ≤ 1.5; or FC ≤ 0.67) and enumerated. (D) GO terms significantly overrepresented among subgroups of differentially expressed genes from C. (E) Chimerism of splenic Treg subsets after 12 d of parabiosis between congenic WT mice. Each point represents the average host chimerism of an independent pair of mice. Data are pooled from two independent experiments (n = 7 pairs of mice). (F) Top, CD49b versus CD103 expression on aTreg cells from various tissues of B6 mice. Lymph was collected from the thoracic duct. Bottom, quantification. Data are representative of three independent experiments with three to five mice per group. (G) Mixtures of congenically marked aTreg cells were adoptively transferred into mice 5 d after infection with N. brasiliensis or mock infection. The ratios of CD45.1+ to CD45.2+ aTreg cells in the injection mix, spleen, and lung parenchyma (IV−) after 14 h are shown. Data are representative of two independent experiments with three mice per group. Bars depict means ± SD; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 by multiple t tests; ns, not significant. See also Fig. S1 and Fig. S2. NB, N. brasiliensis.
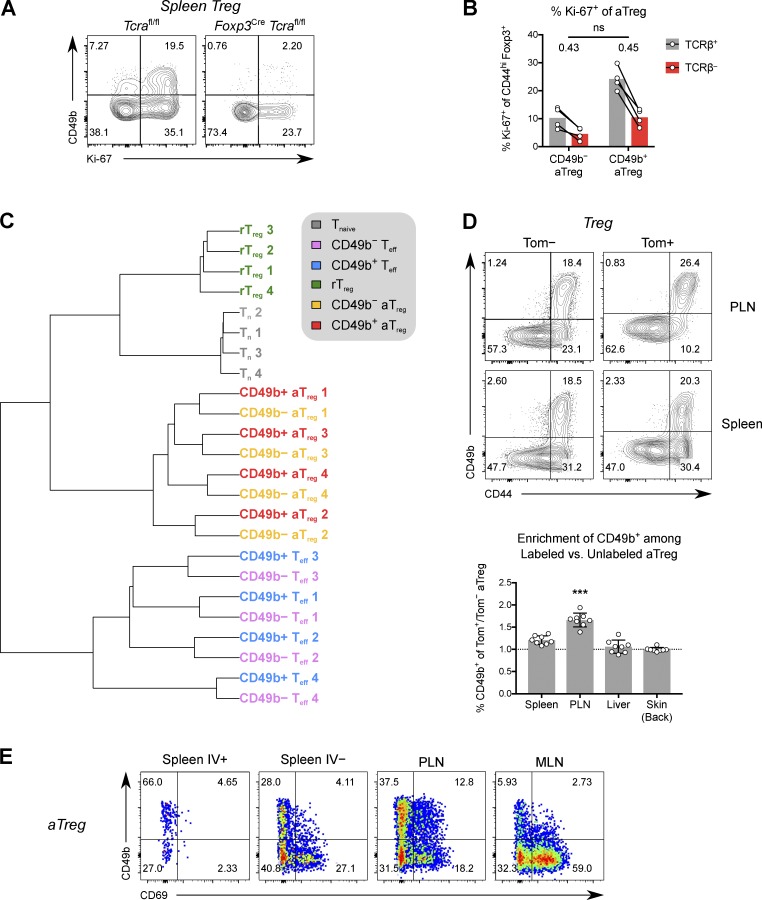
To experimentally test this possibility, we first examined parabiotic mice. After 2 wk of parabiosis, CD49b+ aTreg cells in spleen and the skin-draining peripheral LNs (PLNs) showed significantly more exchange between parabionts than CD49b− aTreg cells, although this was less than the extent seen for resting Treg (rTreg) cells (Fig. 2 E and Fig. S5 D). Previously, CD49b expression by a subset of CD103+ Treg cells had been noted (Stephens et al., 2007); CD103 marks an aTreg cell population enriched in NLTs (Huehn et al., 2004). We found, however, that despite partial overlap with CD103, only CD49b expression was predictive of faster exchange of splenic and PLN aTreg cells in parabionts (Fig. 2 E and Fig. S5 D).
The observed enrichment of CD49b+ Treg cells in skin-draining LNs suggested that skin is not their terminal destination. This was corroborated by flow cytometric analysis of cells in thoracic duct lymph. Although aTreg cells as a whole were relatively underrepresented in lymph compared with blood, we found that a similar fraction of the aTreg cells in lymph express CD49b as in blood (Fig. 2 F). Donor-derived CD49b+ Treg cells could be found in thoracic duct lymph of Lta−/− parabionts (data not shown), which lack SLOs, indicating that these cells can in fact return from NLTs and do not merely egress from efferent lymphatics (Gerlach et al., 2016). Although many aTreg cells in blood coexpressed CD49b and CD103, few lymph-derived aTreg cells expressed CD103 while CD49b expression was preserved (Fig. 2 F). Moreover, CD103 does not show the same distribution across tissue Treg cells as CD49b, being highly expressed in intestinal Treg cells (Korn et al., 2014; Schiering et al., 2014; data not shown). Thus, distinct signals likely induce the expression of these integrins on overlapping subsets of aTreg cells, but only CD49b marks cells enriched in skin, vasculature, and lymphoid tissues and possessing superior migratory ability.
To test the ability of CD49b+ Treg cells to home to a site of inflammation, we cotransferred CD49b+ or CD49b− aTreg cells along with congenically marked CD49b− aTreg cells at a fixed ratio into lymphoreplete mice infected with the helminth Nippostrongylus brasiliensis. After 14 h, we examined the ratio of CD49b+ to CD49b− Treg cells in the spleen and lungs of uninfected or infected mice. We observed enrichment of CD49b+ Treg cells in the spleen relative to the transfer mix and a much greater enrichment (∼10-fold over the transfer mix) in helminth-infected lung parenchyma (Fig. 2 G), demonstrating the superior ability of CD49b+ Treg cells to accumulate at sites of inflammation. Together with our RNA-seq analysis, these experiments suggested that CD49b+ Treg cells are a Treg population equipped for circulating immunosurveillance.
CD49b+ Treg cells are more suppressive than other Treg cells
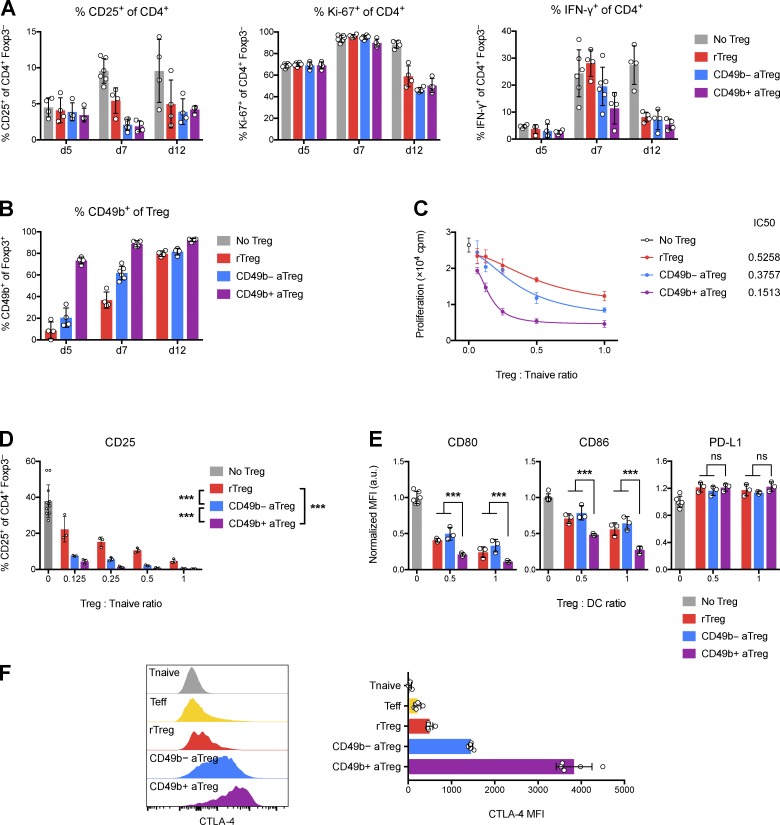
We next sought to assess the suppressive capacity of CD49b+ Treg cells in an adoptive transfer setting: we cotransferred effector CD4+ T (Teff) cells and CD49b+ aTreg, CD49b− aTreg, or rTreg cells into T cell–deficient mice and measured activation of the transferred Teff cells (Fig. 3 A). However, at time points when robust suppression of Teff cells could be observed, a large fraction of initially CD49b− rTreg or aTreg cells had become CD49b+ (Fig. 3 B). Transfer of Treg cells that were CD49b+ from the outset did not provide durably superior protection compared with Treg cells that became CD49b+ in vivo (Fig. 3 A). Thus, the steady generation of CD49b+ Treg cells from adoptively transferred CD49b− Treg cells masked a potential difference in suppressor capacity and confounded its assessment in an in vivo suppression assay, due to its extended duration. Therefore, we resorted to a short-term direct assessment of Treg suppressor function in vitro. We found that CD49b+ Treg cells were markedly more potent than resting or CD49b− aTreg cells in their ability to suppress in vitro proliferation and activation of naive T cells (Fig. 3, C and D). Furthermore, upon co-culture with dendritic cells only, CD49b+ Treg cells suppressed LPS-induced CD80 and CD86 up-regulation by dendritic cells more effectively than the other Treg populations, consistent with the higher expression of CTLA-4 on CD49b+ Treg cells (Fig. 3, E and F). These results suggest that CD49b+ Treg cells are highly effective suppressors and that lymphopenic and inflammatory conditions drive Treg cells of initially different functional states to converge on the potently suppressive CD49b+ state.
Figure 3.
CD49b+ Treg cells are more suppressive than other Treg cells in vitro, but all Treg populations converge on the CD49b+ state under inflammatory conditions in vivo. (A and B) Treg cells and preactivated Teff cells were cotransferred into T cell–deficient recipients. Shown is the expression of CD25, Ki-67, and IFN-γ in Teff cells and CD49b in Treg cells recovered from skin-draining PLNs at various times after transfer. (C) Treg-mediated suppression of naive T cell thymidine incorporation in vitro. (D) Treg-mediated suppression of naive T cell CD25 expression in vitro. (E) Treg-mediated suppression of dendritic cell costimulatory molecule expression in vitro. a.u., arbitrary units. (F) CTLA-4 expression by splenic Treg and Tconv cell subsets. Data are representative of two (A and B) or three (C–F) independent experiments. Bars depict means ± SD; ***, P ≤ 0.001 by two-way ANOVA; ns, not significant. See also Fig. S3.
In NK cells and neutrophils, CD49b has been shown to facilitate motility over ECM (Werr et al., 2000; Garrod et al., 2007). Thus, we asked whether CD49b itself was important for Treg function. However, we found that ablation of a conditional Itga2 allele (encoding CD49b) in Treg cells did not result in noticeable impairment of Treg cell function as manifested by spontaneous disease or exacerbated lung or skin inflammation upon challenge with N. brasiliensis or dinitrofluorobenzene, respectively (Fig. S3, A–C). CD49b-deficient Treg cells were also fully suppressive in vivo (Fig. S3 D) and were not further competitively disadvantaged in the presence of their CD49b-sufficient counterparts in mosaic heterozygous female Foxp3Cre/+ Itga2fl/fl mice (Fig. S3 E). Thus, CD49b expression identifies a Treg subset with greater migratory and potent suppressive ability, but CD49b itself is nonessential for Treg function.
Treg cells that have proliferated extensively express CD49b
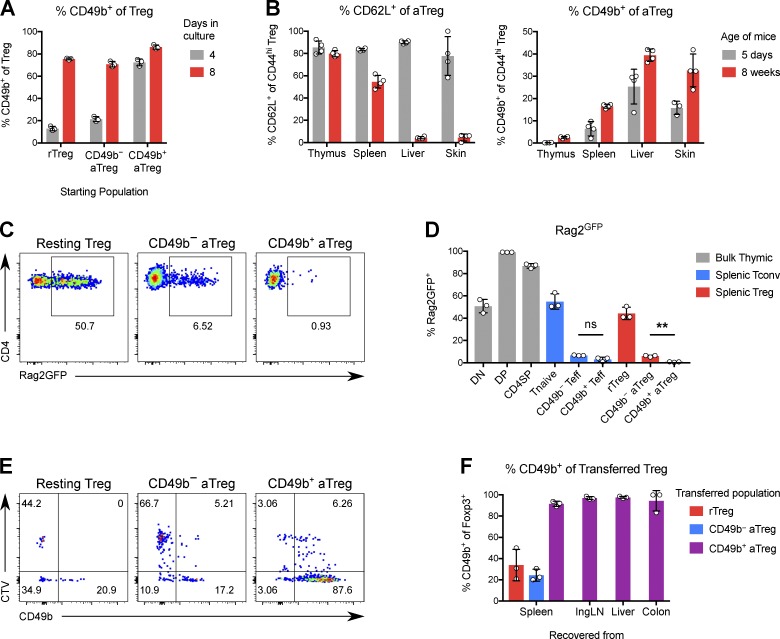
Adoptive transfer experiments showed that most Treg cells could become CD49b+ after a period of time in an inflammatory and proliferation-promoting environment. However, CD49b− aTreg cells were able to express CD49b earlier than resting Treg cells on average. To confirm this result in a setting where the transferred Treg cells do not influence the degree of inflammation, we adoptively transferred small numbers of each of the three populations (rTreg cells and CD49b± aTreg cells) into diphtheria toxin (DT)–treated Foxp3GFP-DTR mice, in which the transferred cells are also strongly driven to proliferate by inflammation and an empty Treg niche. Despite all three populations showing maximal induction of Ki-67, resting Treg cells induced CD49b to a significantly lesser degree than CD49b− aTreg cells (Fig. S5, A and B). Thus, neither inflammation nor proliferation per se is sufficient to fully drive CD49b expression in cells that are not fully mature.
VLA-2 derives its name from its expression on human T cells that have been stimulated in vitro for weeks (Hemler et al., 1985). We found that this held true for mouse Treg cells: ∼20% of CD49b− resting or activated Treg cells cultured with IL-2 and anti-CD3/CD28 turned on CD49b after 4 d, and this increased to >70% after 8 d (Fig. 4 A). In vivo, we observed that the percentage of CD49b+ Treg cells was two to three times higher in adult mice than in neonatal mice, further suggesting that multiple cell divisions are required for Treg cells to express CD49b (Fig. 4 B). Indeed, almost all CD49b+ Treg cells in Rag2GFP mice lack GFP, indicating that they have proliferated numerous times in the periphery and consequently lost GFP (Fig. 4, C and D). These results support a sequential model in which rTreg cells receive strong TCR stimulation to become aTreg cells, which do not yet express CD49b until they have undergone further extensive proliferation.
Figure 4.
Treg cells stably express CD49b in a maturation-dependent manner. (A) Percentage of CD49b+ cells among Treg cells stimulated in culture with α-CD3/CD28 and IL-2 for 4 or 8 d. (B) Percentage of CD62L+ or CD49b+ cells among aTreg cells in tissues of 5-d-old or 8-wk-old mice. (C) Expression of Rag2GFP in Treg subsets from 4-wk-old Rag2GFP mice. (D) Quantification of C. **, P ≤ 0.01 by paired t test; ns, not significant. (E) The indicated Foxp3GFP Treg subsets were separately transferred into sublethally irradiated congenic Foxp3GFP mice. Shown is the expression of CD49b versus dilution of CTV by transferred Treg cells recovered from spleen after 6 d. (F) Quantification of CD49b expression by transferred Treg cells. Data are representative of two independent experiments. Bars depict means ± SD.
Finally, we asked whether CD49b expression is stable in Treg cells. To test this, we isolated rTreg cells or CD49b+ or CD49b− aTreg cells and adoptively transferred them into sublethally irradiated congenic recipients, a setting which poses a much more stringent test of Treg phenotypic stability while ameliorating the poor take of Treg cells upon transfer into lymphoreplete hosts. We found that >90% of CD49b+ Treg cells remained CD49b+ regardless of the tissue from which the cells were recovered (Fig. 4, E and F). In the absence of genetic fate-mapping tools, we cannot tell whether the ∼10% of cells that “lost” CD49b expression reflects true instability of expression or is due to the imprecision of sorting based on CD49b, which is not as highly expressed in Treg cells as in NK cells (Bajénoff et al., 2006). Consistent with other measures of increased proliferation, CD49b+ Treg cells diluted their CellTrace violet (CTV) label more than the other Treg subsets, and some Treg cells that were sorted as CD49b− turned on CD49b in a manner associated with proliferation. We also observed that CD49b+ Treg cells showed more 5-ethynyl-2′-deoxyuridine (EdU) incorporation, as well as more rapid loss of incorporated EdU (Fig. S5 C). Thus, CD49b not only identifies Treg cells with a history of proliferation, but also predicts future proliferation and shorter half-life. Altogether, these data show that CD49b is a marker of Treg cell maturation and marks short-lived effector-like cells that are functionally mature and potent with respect to both migration and suppression.
Unbiased discovery of recirculating Treg population structure
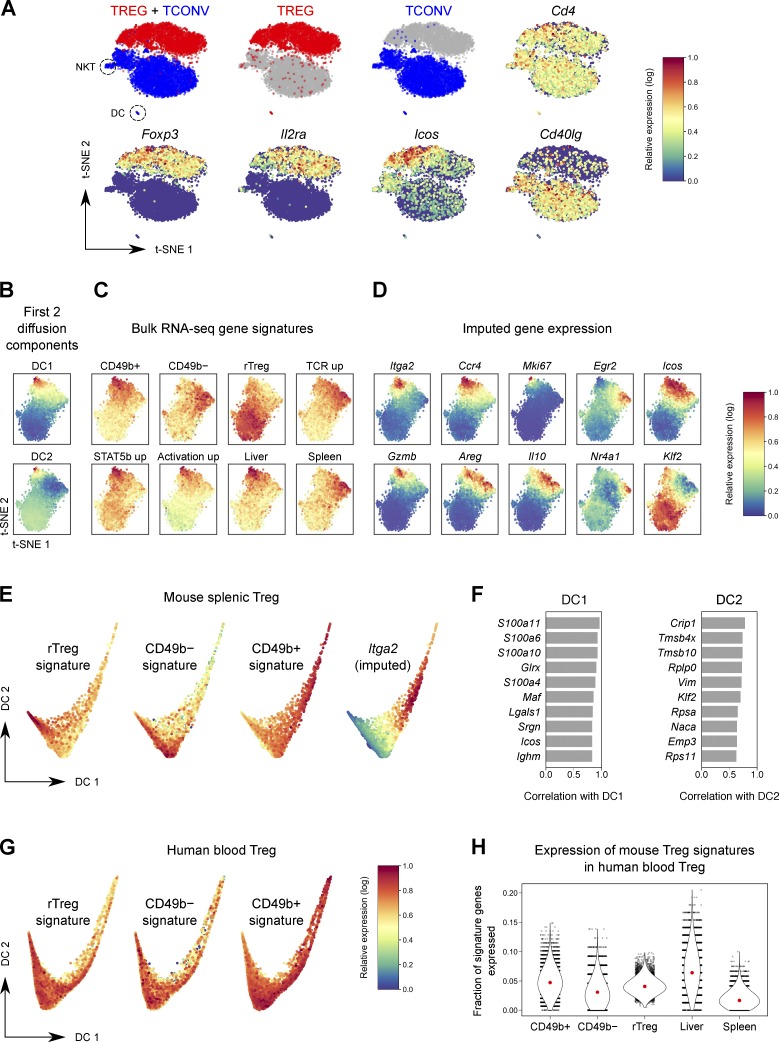
To situate the CD49b+ Treg subset within the larger Treg population hierarchy and estimate developmental trajectories of Treg cells in an unbiased fashion, we performed single-cell RNA-seq (scRNA-seq) of splenic Treg and Tconv cells (Fig. S2 C). When visualized by t-distributed stochastic neighbor embedding (t-SNE; van der Maaten and Hinton, 2008; Amir et al., 2013), Treg and Tconv cells formed distinct clusters on the basis of sort origin (Fig. 5 A and Fig. S4 A). 94% of cells in the Treg cluster were sorted as Treg cells, and 96% of cells in the Tconv cell cluster were sorted as Tconv cells; this error is within the range of sort impurity. A first-pass clustering of these data using PhenoGraph (Levine et al., 2015) segmented the Tconv and Treg cell clusters into subclusters driven by activation state (Fig. S4, B and C). As expected, unlike the discrete separation of Tconv cells from Treg cells, NK T cells, and (contaminating) dendritic cells in our data, the activation subclusters within the Tconv and Treg cell populations represented continuous rather than discrete states. This is consistent with the gradient of activation states seen in scRNA-seq data of T cells from human tumors (Azizi et al., 2018). Thus, instead of clustering, we performed further analysis using a diffusion map approach (Fig. 5, B, E, and F; and Fig. S4 E), as this has been shown to better represent populations that fall along continuous trajectories (Haghverdi et al., 2015).
Figure 5.
scRNA-seq situates CD49b+ Treg cells within the splenic Treg phenotypic landscape. (A) t-SNE plot of scRNA-seq data of splenic Tconv and Treg cells. Cells colored red were sorted as Treg cells; blue cells were sorted as Tconv cells. (B–D) t-SNE plot of Treg cells only, colored by the first two DCs (B), expression of bulk RNA-seq gene signatures (C), and imputed expression of selected genes (D). (E) Plot of the first two DCs of mouse splenic Treg cells, colored by expression of bulk RNA-seq gene signatures, or imputed expression of Itga2. (F) Top genes that correlate with DC1 and DC2. (G) Plot of the first two DCs of human blood Treg cells (Zheng et al., 2017), colored by expression of murine bulk RNA-seq gene signatures. (H) The fraction of genes in each signature expressed by human blood Treg cells. Each dot is a cell. Red dot, median. P < 10−10 by paired t test for comparisons between CD49b+/CD49b−/rTreg signatures or between liver and spleen Treg signatures.
The first diffusion component (DC), capturing the most prominent axis of variation in the data, correlated strongly with activation (Fig. 5, B and E; and Fig. S4 E). When we mapped our bulk RNA-seq gene signatures onto the t-SNE plot along with the first few DCs, we observed that the CD49b+ signature (as well as the related liver, activation, and STAT5b signatures) mapped to the cells highest for DC1, while the rTreg signature anti-correlated with DC1 (Fig. 5, C and E; and Fig. S4 E). The second DC seemed to delineate a Treg population that highly expresses the Treg TCR-dependent gene signature and was intermediate for activation-associated gene expression. Thus, the Treg population structure in the spleen seems to revolve around three poles corresponding to quiescence (rTreg cells), activation and proliferation (CD49b+ aTreg cells), and recent TCR stimulation (CD49b− aTreg cells; Fig. 5, C and E).
The superior recirculatory properties of CD49b+ Treg cells observed in parabiosis experiments correlated with expression of Klf2 in single-cell data (Fig. 5, D and F; and Fig. S4 D). KLF2 is a transcription factor that controls the expression of trafficking molecules such as S1PR1 and CD62L that facilitate recirculation through lymphoid tissues (Carlson et al., 2006; Sebzda et al., 2008). Down-regulation of KLF2 supports the restricted migration patterns of TRM cells and follicular helper T cells, while maintenance of KLF2 expression by Foxo1 is essential for the recirculation of naive T cells (Kerdiles et al., 2009; Skon et al., 2013; Lee et al., 2015; Weber et al., 2015). In our data, Klf2 was one of the genes most highly correlated to DC2, i.e., being expressed minimally in CD49b− Treg cells, which show the most limited exchange by parabiosis; at higher levels in CD49b+ Treg cells; and at the highest levels in “professional” recirculating rTreg cells, which equilibrate most rapidly. Thus, an unbiased transcriptional assessment of Treg population structure identifies phenotypic states underlain by differences in migratory behavior.
We next asked whether CD49b defines an equivalent aTreg cell population in human peripheral blood. Although human Treg cells also fall along a continuum of activation states like mouse Treg cells (Fig. 5 G), we found that CD49b marks a much smaller percentage of human blood Treg cells (Fig. S4 F), and we failed to find Itga2 among expressed genes in a publicly available scRNA-seq dataset of ∼10,000 CD25+ Treg cells from human blood (Zheng et al., 2017). This could be because CD49b, which we showed to be functionally redundant in mouse Treg cells, may not be conserved as a marker, though cells of an activation state analogous to CD49b+ Treg cells are present bearing other markers. Thus, we analyzed the aforementioned human blood Treg scRNA-seq dataset using gene signatures from our bulk RNA-seq data. Indeed, we found that the signatures of mouse CD49b+ and liver Treg cells were more highly expressed in human blood Treg cells than the signatures of CD49b− and splenic Treg cells, respectively (Fig. 5 H and Fig. S4 G). Thus, although CD49b itself exhibits more restricted expression in humans, the broader gene expression profile of CD49b+ Treg cells appears to be conserved between mice and humans.
Shared repertoires of CD49b+ and CD49b− aTreg cells
Bulk and scRNA-seq analyses of splenic Treg cells had both shown that CD49b+ cells express lower levels of TCR-dependent genes than CD49b− aTreg cells. Accordingly, we observed lower protein expression of CD69 (Fig. 6 E), a marker of recent TCR activation, in splenic CD49b+ Treg cells. These findings raised questions of the role of TCR signaling in CD49b+ Treg cell generation and whether CD49b+ Treg and Treg cells highly expressing TCR-dependent markers represent stable states of Treg cell differentiation instructed by distinct TCR repertoires. First, we examined Foxp3YFP-Cre Tcrafl/fl mice, in which maturation from resting to activated Treg cells is blocked, resulting in lethal autoimmunity (Levine et al., 2014). As expected, constitutive loss of the TCR abrogated all CD49b expression on Treg cells, in spite of the inflammatory environment (Fig. 6 A). However, cells did not completely lose the ability to proliferate and express Ki-67. Therefore, TCR signaling is absolutely required for Treg cells to express CD49b, and, as in the transfer setting, proliferation or activation per se is not sufficient. However, loss of TCR blocked development of all aTreg cells, not only those expressing CD49b. We next administered tamoxifen to Foxp3ERT2-Cre Tracfl/+ mice to acutely ablate the TCR in 50% of Treg cells. In these mice, which do not develop overt autoimmunity, loss of the TCR in Treg cells led to a decrease in Ki-67 percentage of similar magnitude in both CD49b+ and CD49b− aTreg cells (Fig. 6 B), showing that they are dependent on continuous TCR signaling to a similar degree. Thus, although CD49b+ Treg cells found in highly vascularized tissues showed less prominent TCR-dependent gene expression, they are no less reliant on TCR-driven signals than CD49b− aTreg cells.
Figure 6.
Similarity of TCR repertoires suggests continuous differentiation of CD49b− to CD49b+ aTreg cells. (A) Expression of Ki-67 and CD49b in splenic Treg cells from 2.5-wk-old Foxp3YFP-Cre Tcrafl/fl or Tcrafl/fl littermate control mice. (B) Foxp3ERT2-Cre Tcra+/fl mice were given tamoxifen. The percentages of Ki-67+ cells among splenic TCRβ± CD49b± aTreg cells on day 7 is shown. ns, P > 0.05 by paired t test comparing TCRβ−/TCRβ+ FCs in CD49b− and CD49b+ aTreg cells. (C) Hierarchical clustering on F2 pairwise similarity of TCRα chain amino acid CDR3 repertoires from Treg and Tconv cell subsets from Foxp3GFP Tcra+/− DO11.10-TCRβ–transgenic mice. (D) Representative flow cytometric data and ratio of CD49b+ frequency on tagged (Tom+) versus untagged (Tom−) aTreg cells 6 wk after 7-d-old mice were given 4-OHT. ***, P < 0.001 compared with any other group (multiple t tests). (E) Representative flow cytometric data of CD49b versus CD69 expression by aTreg cells in various tissues. Data are representative of three (A, B, and E) or two (D) independent experiments. Bars depict means ± SD.
To directly address the role of TCR specificity in CD49b+ Treg cell differentiation, we sequenced the TCRα chains of splenic rTreg cells and CD49b− and CD49b+ aTreg cells, and the corresponding Tconv cell populations, isolated from mice with a “fixed” transgene-encoded TCRβ chain (Fig. S2 B). Clustering the CD49b± TCR repertoires by the fraction of repertoire occupied by shared clonotypes (F2 similarity metric of VDJtools software; Shugay et al., 2015; Izraelson et al., 2018) revealed that CD49b+ and CD49b− aTreg cells cluster together by individual mouse (Fig. 6 C). After accounting for sampling stochasticity, there did not appear to be a pattern of clonotypes that was specific to either subset, suggesting that essentially all CD49b− Treg cells could give rise to CD49b+ cells, though likely at differing clonotype-dependent rates. These data indicate that differentiation into CD49b+ aTreg cells is not instructed by unique TCR specificities, but that CD49b− and CD49b+ aTreg cells are likely to be temporally related, with CD49b− aTreg cells generated first.
Because the progeny of Treg cells generated early in life are highly suppressive (Yang et al., 2015), it remained possible that the highly activated and suppressive CD49b+ Treg cells are generated predominantly from early precursors. To evaluate this hypothesis, we administered 4-hydroxytamoxifen (4-OHT) to Foxp3GFP-DTR Cd4ERT2-Cre Rosa26LSL-tdTomato “CD4 fate-mapping” mice at 7 d of age or at 6 wk of age, and examined the percentage of tagged cells that were CD49b+ 6 wk later. Indeed, a modestly higher percentage of day 7–tagged aTreg cells showed CD49b expression compared with aTreg cells tagged at 6 wk (Fig. S5 E), suggesting that the neonatal environment, which is relatively lymphopenic, favors CD49b+ Treg development.
Unexpectedly, we also observed that compared to untagged aTreg cells, after 6 wk, neonatally fate-mapped aTreg cells were highly enriched in CD49b+ Treg cells in the skin-draining PLNs, more so than in other lymphoid and NLTs, suggesting that CD49b+ aTreg cells represent a maturation state for PLN Treg cells (Fig. 6 D). A similar trend was seen in fate-mapped adult mice, although the enrichment was not as striking (Fig. S5 F). Additionally, CD49b+ Treg cells in PLNs showed higher expression of CD69 compared with their counterparts in spleen or mesenteric LN (Fig. 6 E), suggesting that CD49b+ aTreg cells preferentially receive stimulation in PLNs. Altogether, these results delineate a model in which CD49b+ aTreg cells arise from skin- and PLN-homing CD49b− aTreg cells that have undergone extensive rounds of proliferation in the physiological setting. This leads to transcriptional changes, including up-regulation of a battery of chemokine and cytokine receptor genes, which collectively mediate this population’s greater ability to recirculate and suppress.
Discussion
Our studies showed that in mice, a major aTreg cell population in blood, highly vascularized organs, and skin expresses CD49b and is developmentally and functionally mature in comparison to other Treg cell subsets. CD49b, which was previously shown to distinguish recirculating mature NK cells from immature NK cells and tissue-resident ILC1s (O’Sullivan et al., 2016), is expressed in Treg cells after multiple rounds of proliferation. scRNA-seq analysis of splenic Treg cells placed CD49b+ Treg cells at the apex of a gradient of activation, enriched for genes promoting recirculation and relatively depleted of genes reflecting recent TCR activation. However, differentiation of CD49b± aTreg cells does not represent a choice between alternative “fates” driven by distinct TCR specificities, and CD49b+ Treg cells are no less dependent on their TCR, but rather appear to preferentially receive stimulation in skin-draining PLNs. Our results thus suggest that under physiological conditions, recirculating CD49b+ Treg cells are a mature effector Treg population that continuously surveys skin, vasculature, and lymphoid tissues.
Integrins mediate migration, adhesion, force production, or sensing in many cell types (Wolfenson et al., 2013). CD103 (Itgae) or CD49a (Itga1) deficiency impairs retention or survival of CD8+ TRM cells (Ray et al., 2004; Casey et al., 2012; Mackay et al., 2013), and CD103 has been implicated in intestinal Treg cell retention (Suffia et al., 2005; Braun et al., 2015; Yuan et al., 2015). Although CD49b was thought to be important for platelet adhesion and for cell migration over ECM, Itga2-deficient mice are viable and fertile and do not show a major phenotype besides subtle differences in mammary gland branching morphogenesis and ex vivo platelet function (Chen et al., 2002; Holtkötter et al., 2002; Habart et al., 2013). Accordingly, we did not observe a nonredundant role for CD49b itself in Treg cell function or migration using Treg cell–specific gene ablation.
VLA-2 (integrin α2β1), the complex of CD49b and CD29, was initially identified through its expression on human T cells stimulated with IL-2 for weeks in vitro (Hemler et al., 1985). We found that in mouse Treg cells, CD49b expression also follows extensive proliferation. Consistent with this, we observed that while fewer CD49b+ Treg cells were found in neonates, Treg cells fate mapped in neonatal life were modestly more likely to become CD49b+ cells after 6 wk. However, CD49b expression is not simply a marker of proliferation per se, as resting Treg cells driven to proliferate upon adoptive transfer into Treg cell–depleted recipients are less capable of expressing CD49b than aTreg cells that exhibit similar proliferative activity based on Ki-67 expression. CD49b+ Treg cells are transcriptionally similar to Treg cells that express the active form of STAT5 and also to aTreg cells from mice recovering from transient Treg cell ablation, in which IL-2 is abundant. Because CD49b expression was induced slowly over time, even in the presence of excess IL-2, we suggest that persistent STAT5 signaling and inflammatory environments rich in IL-2 promote CD49b expression indirectly through stimulating persistent proliferation of Treg cells and rescuing otherwise short-lived effector-like cells.
Recent studies described several Treg subsets based on various markers. Our experiments suggest that aTreg cells can be further subdivided with respect to maturity and function, with CD49b marking cells that are more advanced for both. Although CD103 also marks Treg cells with heightened suppressive capacity and can be expressed in an overlapping manner with CD49b (Huehn et al., 2004; Stephens et al., 2007), CD103 shows a completely different tissue distribution in Treg cells. Furthermore, CD103 expression on its own does not mark highly recirculating cells—only when it coincides with CD49b expression—and is expressed in a TGF-β–dependent manner by a variety of tissue-resident cell types (Casey et al., 2012; Mackay et al., 2013; Zhang and Bevan, 2013). Thus, CD49b is likely a more definitive marker for a mature Treg cell subset with a migratory phenotype, given its consistently high expression across blood and lymph and its known expression conditional on extensive proliferative history.
CD49b+ Treg cells are able to traffic to and from skin, as indicated by their enrichment in skin and skin-draining PLNs and the expression of skin-homing receptor genes on splenic CD49b+ Treg cells. This observation agrees with a report showing that the majority of Treg cells in human blood express skin-homing receptors (Hirahara et al., 2006) and that Treg cells bearing a single TCR specificity for a likely skin antigen are sufficient to prevent most of the lymphoproliferation associated with Treg deficiency (Levine et al., 2017). Our fate-mapping data also indicated that CD49b− Treg cells in PLNs are particularly enriched in cells that will become CD49b+.
The seemingly mutual exclusivity observed for splenic CD49b+ aTreg cells and CD49b− aTreg cells highly expressing markers of recent TCR activation was unlikely to be due to the existence of distinct aTreg cell “fates” driven by unique TCR specificities. We found that the TCR repertoires of CD49b+ and CD49b− aTreg cells cluster by individual mouse, and the majority of aTreg cell clonotypes showed representation in both CD49b− and CD49b+ aTreg cells. Our TCR repertoire, fate-mapping, and adoptive transfer analyses thus argue in favor of a model in which most CD49b− aTreg cells can become CD49b+ with extensive proliferation, but early homeostatic proliferation by neonatal cells, perhaps driven in part by recognition of skin antigens, may initially bias the frequencies of clonotypes that become CD49b+. The relative absence of the CD49b+ Treg cell in intestine-associated tissues suggests that there are at least two major circuits of Treg maturation and migration: one culminating in CD49b+ Treg cells and centered in the skin and another involving peripherally induced Treg cells centered in the intestine.
On a per-cell basis, CD49b+ Treg cells exhibit more potent suppression of naive T cell proliferation and LPS-induced costimulatory molecule expression on dendritic cells. While multiple factors likely contribute to their greater suppressive potency, we speculate that a greater ability to interact with dendritic cells mostly accounts for this phenotype (Onishi et al., 2008; Chinen et al., 2016). Although prominent up-regulation of CD49b by initially CD49b− cells in our adoptive transfer studies confounded unequivocal comparison of their functionality in vivo, the onset of robust suppression coincided with the expression of CD49b by most transferred Treg cells. Moreover, in an in vivo migration assay, CD49b+ Treg cells showed superior ability to migrate to a site of inflammation. Intriguingly, in a study from the “pre-Foxp3” era, adoptive transfer of DX5+ (CD49b+) T cells was shown to protect mice expressing the diabetogenic BDC2.5 TCR from autoimmune diabetes (Gonzalez et al., 2001); our study would suggest that this protection was in fact mediated by mature effector CD49b+ Foxp3+ Treg cells.
In conclusion, CD49b+ Treg cells represent a differentiated population of Treg cells that have undergone many rounds of cell division. These cells are enriched in skin-homing cells, exhibit potent suppressor function and are more capable of homing to sites of inflammation. We suggest that CD49b+ Treg cells are the major recirculating effector Treg population that surveys for and responds to inflammation in mice.
Materials and methods
Mice
Foxp3GFP (Fontenot et al., 2005), Foxp3GFP-DTR (Kim et al., 2007), Foxp3Thy1.1 (Liston et al., 2008), Foxp3YFP-Cre (Rubtsov et al., 2008), Foxp3ERT2-Cre (Rubtsov et al., 2010), Cd4ERT2-Cre (Śledzińska et al., 2013), Il10GFP (Kamanaka et al., 2006), Tcrafl (Polic et al., 2001), TcrbKO (Mombaerts et al., 1992), and TcrdKO (Itohara et al., 1993) mice have been previously described and were bred in the specific pathogen–free animal facility of Memorial Sloan Kettering Cancer Center. C57BL/6J, CD45.1, and Itga2fl (B6.Cg-Itga2tm1.1Tkun/J; Habart et al., 2013) mice were obtained from Jackson Laboratories. All mice were used in accordance with institutional guidelines and under protocols approved by the Sloan Kettering Institutional Animal Care and Use Committee.
Reagent administration in mice
DT
To deplete Treg cells and activate conventional T cells, 50 µg/kg DT in PBS was administered on days 0 (retroorbitally) and 2 (intraperitoneally), and mice were used on day 7.
EdU
1 mg of EdU in 200 µl PBS was injected retroorbitally per mouse 3.5 or 1 d before analysis.
FTY720
FTY720 (reconstituted to 25 mg/ml in DMSO) was diluted in 2% wt/vol (2-hydroxypropyl)-β-cyclodextrin in PBS. Mice were injected intraperitoneally with 1 mg/kg FTY720 or vehicle 14 h before being euthanized for analysis.
Tamoxifen
80 mg of tamoxifen was vortexed in 100 µl of ethanol for 1 min. The resulting slurry was mixed with 1.9 ml of olive oil and heated at 55°C until totally dissolved. 200 µl of 40 mg/ml tamoxifen was administered to mice by oral gavage on days 0 and 1, and mice were analyzed on day 9.
4-OHT
Adult and neonatal mice were injected intraperitoneally with 75 mg/kg 4-OHT dissolved in DMSO.
N. brasiliensis infection
Mice were injected subcutaneously with 450 infective L3 larvae, passaged in a Wistar rat and cultured as previously described (Camberis et al., 2003).
Parabiosis
Female 6–8-wk-old CD45.1 and CD45.2 B6 mice underwent surgery to establish parabiosis, as previously described (Kamran et al., 2013). Under isoflurane anesthesia, corresponding lateral skin incisions were made from elbow to knee in each mouse, forelimbs and hindlimbs were tied together using nonabsorbable nylon suture, and the skin incisions were closed using stainless steel wound clips. For 2 wk after surgery, mice were maintained on a diet supplemented with trimethoprim-sulfamethoxazole for infection prophylaxis.
Generation of bone marrow chimeras
Bone marrow was isolated from mouse femurs by crushing the bones with a mortar and pestle. CD90+ T and TER-119+ erythroid cells were depleted from the bone marrow using the Dynabeads FlowComp Mouse Pan T kit (Invitrogen) with added biotinylated TER-119 antibody. 2.5 × 106 T- and erythroid-depleted bone marrow cells were injected retroorbitally into Tcrb−/− Tcrd−/− mice 1 d after irradiation (950 rads). Mice were administered 2 mg/ml neomycin in drinking water for 4 wk and were used 12 wk after transplant.
Isolation of single-cell suspensions from mouse tissues
After euthanasia by asphyxiation with CO2, mice were perfused with ice-cold PBS through the left ventricle until the liver, lungs, and kidneys turned completely pale. Liver and lung Treg percentages, numbers, and phenotypes were similar with or without perfusion (data not shown).
Lymphoid organs
Spleens and LNs were dissociated into single-cell suspensions using frosted glass slides and filtered through 100-µm nylon mesh. Spleens were additionally resuspended in ammonium-chloride-potassium (ACK) lysis buffer for 1 min at room temperature to lyse red blood cells.
Blood and lymph
Using a lancet, blood was collected from the submandibular vein into ACK lysis buffer. To collect thoracic duct lymph, mice were euthanized with CO2, and lymph was collected from the cisterna chyli using pulled glass microcapillary needles containing 10 µl of heparin solution.
Liver and lungs
The liver and lungs were manually minced using scissors and incubated for 45 min in collagenase solution (RPMI 1640 containing 1 mg/ml collagenase, 1 U/ml DNase I, 5% fetal calf serum, 1% l-glutamine, 1% penicillin-streptomycin, and 10 mM HEPES) with shaking at 37°C, in the presence of ceramic beads. After being passed through 100-µm strainers, liver suspensions were further purified with a discontinuous 44% over 67% Percoll gradient, while lungs were passed over a straight 40% Percoll gradient. Red blood cells were lysed in both using ACK lysis buffer.
Large intestine
Large intestines were flushed of their contents with ice-cold PBS, cut open lengthwise, and chopped transversely into 0.5-cm pieces. To dislodge the epithelial fraction, the pieces of intestine were incubated in DTT/EDTA solution (PBS containing 1 mM dithiothreitol, 1 mM EDTA, 5% FCS, 1% l-glutamine, 1% pen-strep, and 10 mM HEPES) at 37°C, with shaking, for 15 min. After washing out the supernatant containing the epithelial fraction, the remaining intestinal pieces were then incubated in collagenase solution for 40 min at 37°C with shaking and ceramic beads, passed through a 100-µm strainer, and purified using 40% Percoll.
Skin
Mice were euthanized after being shaved and depilated. Back skin was collected and scraped with forceps to remove subcutaneous fat and connective tissue. The skin was then minced finely using scissors, resuspended in collagenase solution, and shaken with ceramic beads for 1 h. The resulting single-cell suspension was passed through a 40-µm filter, washed, and centrifuged in 40% Percoll to remove cellular debris. Ear skin was treated similarly, except that it did not require depilation, but required an additional step in which the skin was separated from the underlying cartilage using forceps.
Intravascular cell labeling
Under isoflurane anesthesia, mice were injected retroorbitally with 3 µg of Alexa Fluor 647–, APC-, APC-eFluor 780–, or BV 421–conjugated anti-CD45 (clone 30-F11) or anti-CD45.2 (clone 104) antibody in 200 µl of PBS. After 3 min, mice were euthanized and then perfused with PBS to wash out (potential) excess unbound antibody from vascular spaces. In practice, no difference was observed in the percentage of IV-labeled CD4+ lymphocytes whether or not perfusion was performed (data not shown).
Ex vivo restimulation for intracellular cytokine staining
Single-cell suspensions were restimulated in U-bottom plates with 50 ng/ml PMA and 500 ng/ml ionomycin or 5 µg/ml soluble α-CD3 (clone 145-2C11; BioXCell) and 5 µg/ml soluble α-CD28 (clone 37.51; BioXCell), both in the presence of 1 µg/ml brefeldin A.
Flow cytometry
Single-cell suspensions were first incubated with 1:1,000 Ghost Dye violet 510 (Tonbo Biosciences) and 1:1,000 α-CD16/CD32 (clone 2.4G2; Tonbo) in PBS for 5 min. Cells were then stained for cell surface antigens in FACS buffer (PBS containing 0.05% sodium azide, 2% FCS, and 2 mM EDTA) for 30 min. After washing with FACS buffer, cells were either analyzed live or underwent fixation and intracellular staining using the Foxp3/transcription factor staining kit (eBioscience). For intracellular cytokine staining, the BD Cytofix/Cytoperm kit was used instead. Flow cytometric data were acquired on a BD LSR II and analyzed using FlowJo X software. CD1d tetramers loaded with PBS-57 were obtained from the National Institutes of Health (NIH) Tetramer Core Facility.
For detection of incorporated EdU, cells were stained for surface and intracellular antigens as described above and were additionally labeled through a click reaction using Alexa Fluor 647 azide (2 µM of azide in ddH2O with 0.5 µM CuSO4, 100 mM Tris-HCl, and 50 mM ascorbic acid).
Cell sorting
To isolate Treg and Tconv cells for in vivo and in vitro assays, pooled spleens and LNs were dissociated and then enriched for CD4+ cells using the Dynabeads FlowComp Mouse CD4 kit (Invitrogen). Cells were then stained for surface antigens and resuspended in isolation buffer (DMEM with 2% FCS, 1% pen-strep, 10 mM HEPES, and 2 mM EDTA). Cells were sorted on a BD FACSAria II.
Expansion and isolation of CD11c+ dendritic cells
To expand dendritic cells in vivo, CD45.1 congenic mice were injected subcutaneously with 106 Flt3L-expressing B16 melanoma cells. After 2 wk, the spleen was harvested and dissociated to a single-cell suspension. CD11c+ splenic dendritic cells were isolated using mouse CD11c MicroBeads (Miltenyi Biotec).
In vitro suppression of dendritic cells
104 Flt3L-expanded CD11c+ dendritic cells were co-cultured in U-bottom plates with titrated numbers of FACS-sorted Treg cells and 0.5 µg/ml α-CD3 antibody (clone 145-2C11), with or without 1 µg/ml LPS. After 48 h, cell numbers and costimulatory molecule expression were analyzed by flow cytometry.
In vitro suppression of naive T cell proliferation
CTV dilution
104 FACS-sorted naive T cells were labeled with CTV and co-cultured in 96-well U-bottom plates with 104 Flt3L-expanded CD11c+ dendritic cells, titrated numbers of FACS-sorted Treg cells, and 0.5 µg/ml α-CD3 antibody. After 72 h, cell numbers, CD25 expression, and CTV dilution were analyzed by flow cytometry.
Thymidine incorporation
4 × 104 FACS-sorted naive T cells were co-cultured in 96-well U-bottom plates with 105 irradiated (2,000 rads) CD4-depleted splenocytes, titrated numbers of FACS-sorted Treg cells, and 1 µg/ml α-CD3 antibody. After 72 h, [3H]-thymidine was added to each well to a final concentration of 5 nCi/µl for an additional 8 h. Cells were harvested to a glass fiber filter mat using a PerkinElmer cell harvester, and radioactivity was measured using a PerkinElmer TopCount scintillation counter.
In vivo migration assay
Treg cells from CD45.2 Foxp3GFP or Foxp3Thy1.1 Il10GFP mice were mixed at a ∼3:1 ratio with Treg cells from CD45.1/2 (heterozygous) Foxp3GFP mice and injected retroorbitally into CD45.1 congenic Foxp3GFP-DTR mice on day 5 of infection with N. brasiliensis larvae. A total of 3 × 105 Treg cells were injected per mouse. 14 h later, mice were euthanized after intravascular anti-CD45 labeling, and spleens and lungs were analyzed by flow cytometry for the ratio of CD45.2 to CD45.1/2 Treg cells.
In vivo suppression assay
CD45.1 Foxp3GFP-DTR mice were given DT on days 0 (intravenously) and 2 (intraperitoneally). On day 7, 1.05 × 106 CD44hi Foxp3− Tconv cells from pooled DT-treated CD45.1 Foxp3GFP-DTR mice, and 3.5 × 105 Foxp3+ Treg cells (3:1 ratio) from pooled Foxp3GFP mice were retroorbitally injected into Tcrb−/− Tcrd−/− mice. Mice were euthanized on days 5, 7, and 12, and activation marker expression, absolute cell numbers, and cytokine production upon ex vivo restimulation were determined for transferred Tconv cells from spleen and PLNs.
Bulk RNA-sequencing
Treg cells from liver and lymphoid tissues
Tissues from nine 5-mo-old male Foxp3Thy1.1 mice were divided into three pools of three, which were needed to obtain enough cells from liver and blood. Both liver and spleen were processed using the “Liver and lungs” cell isolation procedure described above, including collagenase digestion and Percoll purification. Portal LN and blood were processed as described for those respective tissues. Live CD45+ TCRβ+ NK-1.1− CD1d Tetramer− CD4+ CD8α− Foxp3+ Treg or Foxp3− Tconv cells (Fig. S2) were sorted from these tissues and resuspended in TRIzol.
Splenic naive, CD49b−, and CD49b+ Treg and Tconv cells
After Dynabeads CD4-positive selection, CD62Lhi CD44lo, CD44hi CD49b−, and CD44hi CD49b+ subsets of Foxp3+ Treg or Foxp3− Tconv cells were sorted from individual spleens of four 2-mo-old male Foxp3GFP mice (Fig. S2). Cells were first gated to be TCRβ+ CD4+ CD8α− NK-1.1− CD1d Tetramer−. Cells were double-sorted, and the second sort was directly into TRIzol LS.
Total RNA was extracted and underwent SMARTer amplification, poly(A) selection, and Illumina TruSeq paired-end library preparation following manufacturer’s protocols. Samples were sequenced on the Illumina HiSeq 2500 to a depth of 30–50 million 50-bp read pairs per sample. Read alignment and processing were performed as previously described (Anders et al., 2013; Chinen et al., 2016). Raw reads were trimmed using Trimmomatic v0.32 with standard settings to remove low-quality reads and adaptor contamination (Bolger et al., 2014). The trimmed reads were then aligned to the mouse genome (Ensembl assembly GRCm38) using TopHat2 v2.0.11 implementing Bowtie2 v2.2.2 with default settings (Langmead and Salzberg, 2012; Kim et al., 2013). Read alignments were sorted with SAMtools v0.1.19 (Li et al., 2009) before being counted to genomic features using HTSeq v0.6.1p1 (Anders et al., 2015). Differential gene expression was analyzed using DESeq2 1.6.3 in R version 3.1.0 (Love et al., 2014). For the liver RNA-seq data set, hepatocyte contamination was removed by excluding a cluster of genes that was highly expressed in bulk liver RNA-seq (Denzler et al., 2014), lowly expressed in liver Treg and Tconv cells, and showed high intra-sample correlation with albumin, the most highly expressed gene in bulk liver RNA-seq. Gene ontology analyses confirmed that the set of contaminating genes was highly enriched for genes encoding plasma proteins and genes involved in metabolic processes.
Bulk RNA-seq bioinformatic analyses
The distribution of read counts across all genes was bimodal in bulk RNA-seq data. The assumption that this corresponded to “expressed” and “non-expressed” genes was supported by examination of marker genes known to be expressed or not expressed in Treg and Tconv cells. The local minimum between the two peaks was chosen to be the threshold for expression.
Bulk RNA-seq gene signature definitions
Treg activation–dependent genes were up- or down-regulated (2× fold change [FC], padj < 0.01) in Treg cells from Foxp3DTR mice recovering from punctual regulatory T cell depletion (Arvey et al., 2014). STAT5b-regulated genes were up- or down-regulated (2× FC, padj < 0.05) in Treg cells expressing constitutively active STAT5b (Chinen et al., 2016). The signatures of genes regulated by the TCR in Treg cells had been defined as genes up- or down-regulated (padj < 0.05) in TCR-sufficient over TCR-deficient CD44hi Treg cells from healthy Foxp3ERT2-Cre Tcrafl/+ mice that remained significant (i.e., were not rescued by inflammation) in TCR-deficient Treg cells from sick Foxp3ERT2-Cre Tcrafl/fl mice (Levine et al., 2017). Liver Treg genes were up-regulated in liver over splenic Treg cells and vice versa for spleen Treg genes (2× FC, padj < 0.05). The signature of CD49b+ Treg cells was defined as genes up- or down-regulated in CD49b+ aTreg cells compared with both CD49b− aTreg cells and rTreg cells (1.5× FC, padj < 0.05); the signatures of rTreg cells and CD49b− aTreg cells were determined using the corresponding comparisons against the other two groups. Enrichment of bulk RNA-seq signatures was tested using the two-sided Kolmogorov-Smirnov test.
GO analyses
Biological process GO term enrichment was calculated using BiNGO v3.0.3 (Maere et al., 2005) in Cytoscape v3.2.1 (Shannon et al., 2003), using the hypergeometric test and applying a significance cutoff of false discovery rate (FDR)–adjusted P value ≤ 0.05. All expressed genes were entered as the reference set, and the GO and annotation files used were downloaded on October 25, 2015. The output from BiNGO was imported into EnrichmentMap v2.0.1 (Merico et al., 2010) in Cytoscape, and an EnrichmentMap was generated using a Jaccard similarity coefficient cutoff of 0.25, an FDR-adjusted P value cutoff of 0.05, and excluding gene sets that were represented by fewer than three genes. Non-redundant GO terms were manually selected from the EnrichmentMap.
TCR sequencing
After Dynabeads CD4-positive selection, CD62Lhi CD44lo, CD44hi CD49b−, and CD44hi CD49b+ subsets of Foxp3+ Treg or Foxp3− T onv cells were double-sorted from individual spleens of four 3-mo-old male Foxp3GFP Tcra+/− DO11.10-transgenic mice. Cells were first gated to be Vβ8+ CD4+ CD8α− NK-1.1− CD1d Tetramer−. After sorting, cells were lysed with Buffer RLT Plus (Qiagen), and RNA was purified using the RNeasy Micro kit (Qiagen). Purified RNA was used to prepare TCRα cDNA libraries using a previously published method (Feng et al., 2015). Libraries were loaded onto a MiSeq v2 paired-end cartridge and run on a MiSeq sequencer (Ilumina). Unique molecular identifier (UMI)–based grouping of sequencing reads covering the same starting cDNA molecules was performed using MIGEC software (Shugay et al., 2014). TCRα repertoire extraction was performed using MiXCR software (Bolotin et al., 2015). Dendrogram clustering using the F2 overlap metric was performed using VDJtools (Shugay et al., 2015).
scRNA-seq
After Dynabeads CD4-positive selection, CD4+ Foxp3+ Treg and CD4+ Foxp3− Tconv cells were sorted from the pooled spleens of three 8-wk-old male Foxp3GFP mice. Single cells were then isolated in microfluidic droplets with barcoded DNA primers and RT reagents using the 10x Genomics Chromium instrument and Chromium Single Cell 3′ Reagent kit (v2). The cDNA synthesis/barcoding was performed at 53°C for 45 min, followed by heat inactivation at 85°C for 5 min. The resulting barcoded cDNA was PCR-amplified and prepared for sequencing according to the Single Cell 3′ Reagent kit v2 User Guide (CG00052; Rev D). In total, 10,260 cells were barcoded and sequenced. Next-gen sequencing was performed on the Illumina HiSeq 2500 instrument, using the paired-end option (R1, 26 reads; R2, 98 reads; and index, 8 reads), which yielded ∼250 million reads per 10,000 single cells. Notably, the Treg and Tconv cell samples were run side by side on the 10x Chromium, underwent library preparation together, and were loaded onto one sequencing lane of the HiSeq. FASTQ files containing the transcriptome and barcoding metadata were de-multiplexed using the SEQC pipeline (Azizi et al., 2018), resulting in 3,450 UMIs per cell on average.
scRNA-seq bioinformatic analyses
Tconv and Treg cell datasets were merged by taking the union of genes in each set and filling in zeroes for cells with missing genes. PCA was performed on the merged Tconv and Treg cell dataset using as variables the number of genes, number of UMIs, and percent comprised of mitochondrial genes, per cell. Multivariate outliers were then identified and removed using the mvoutlier v2.0.9 R package (Filzmoser and Gschwandtner, 2018). Using this approach, 3% of Tconv cells and 15.8% of Treg cells were discarded, generally due to having low complexity and/or high mitochondrial content. (Similar results were obtained when simple cutoffs—thresholds of QC metric histograms—were used. However, the latter method retained more actively cycling cells which complicated library size normalization and subsequent diffusion map analysis, though the overall relationships between cell clusters were fully preserved [Fig. S4 A].) Genes expressed in fewer than five cells were discarded. The data were then normalized by the median library size, log transformed, and reduced to 20 principal components, beyond which additional PCs contributed negligible information. For visualization, t-SNE was performed on the principal components using a perplexity of 40 (van der Maaten and Hinton, 2008). As a first pass, the data were then clustered using PhenoGraph, which yielded fairly reproducible clustering at values of k between 20 and 80 (Levine et al., 2015), as assessed by the adjusted Rand index. This allowed us to identify a cluster of contaminating dendritic cells (expressing multiple antigen presentation genes) and another cluster of NK T cells (highly expressing Ifng, Il4, and Cxcr6 and to which most Zbtb16 transcripts, encoding PLZF, mapped), which were removed from downstream analyses.
To analyze the phenotypic landscape of Treg cells only, we used tools associated with the Wishbone package in Python (Setty et al., 2016). Data were normalized, log transformed, and reduced to 20 principal components as before. t-SNE was run on the PCs with a perplexity of 40, and t-SNE plots were colored by unimputed gene expression, gene expression imputed using MAGIC (van Dijk et al., 2017), summated expression of bulk RNA-seq gene signatures, and the first two DCs.
Data availability
Tissue Treg bulk RNA-seq, Treg subset bulk RNA-seq, scRNA-seq, and TCR-seq data have been deposited to the Gene Expression Omnibus under accession numbers GSE121480, GSE121482, GSE121034, and GSE121147, respectively.
Statistical analyses
Statistical tests for nongenomic data were performed in GraphPad Prism 7. Unless otherwise noted, differences between groups were analyzed using unpaired or paired two-tailed t tests with FDR correction for multiple hypotheses.
Online supplemental material
Fig. S1 shows additional analyses of Treg bulk RNA-seq data. Fig. S2 shows the gating strategies used for cell sorting and flow cytometry. Fig. S3 shows that CD49b itself is nonessential for Treg survival or function. Fig. S4 shows additional analyses of splenic Treg scRNA-seq data. Fig. S5 shows additional experiments related to Treg population dynamics.
Supplementary Material
Acknowledgments
We thank S. Lee, A. Chaudhry, and K. Wu for help with cell isolation; I. Masilionis and E. Azizi for help with scRNA-seq data acquisition and analysis, respectively; Y. Chen and D. Wesemann for providing Rag2GFP tissues; S. Dikiy and N. Arpaia for help with cell sorting; R. Franklin for help with parabiosis; B. Hoyos for maintenance of N. brasiliensis; B. Hoyos, A. Bravo, J. Verter, D. Konstantinovsky, and R. Fromme for general laboratory support; and members of the Rudensky laboratory for helpful discussions.
X. Fan was supported by NIH Medical Scientist Training Program grant T32GM07739 to the Weill Cornell/Rockefeller University/Sloan Kettering Tri-Institutional MD-PhD Program and NIH grant F30AI122721. A.Y. Rudensky was supported by NIH grant R37AI034206, NIH/NCI Cancer Center Support Grant P30CA008748, the Ludwig Center at Memorial Sloan Kettering Cancer Center, and the Hilton-Ludwig Cancer Prevention Initiative. A.Y. Rudensky is an investigator with the Howard Hughes Medical Institute. A.N. Davydov was supported by the Ministry of Education, Youth, and Sports of the Czech Republic under the project CEITEC 2020 LQ1601. D.M. Chudakov was supported by the Ministry of Education and Science of the Russian Federation grant 14.W03.31.0005.
The authors declare no competing financial interests.
Author contributions: conceptualization, X. Fan and A.Y. Rudensky; formal analysis, X. Fan, A.N. Davydov, R. Sharma, and D.M. Chudakov; investigation, X. Fan, B. Moltedo, A. Mendoza, M.B. Faire, and L. Mazutis; writing (original draft), X. Fan; writing, review, and editing, X. Fan, D. Pe’er, and A.Y. Rudensky; visualization, X. Fan and A.N. Davydov; supervision, project administration, and funding acquisition, A.Y. Rudensky.
References
- Amir A.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., and Pe’er D.. 2013. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31:545–552. 10.1038/nbt.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., McCarthy D.J., Chen Y., Okoniewski M., Smyth G.K., Huber W., and Robinson M.D.. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8:1765–1786. 10.1038/nprot.2013.099 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K.G., Mayer-Barber K., Sung H., Beura L., James B.R., Taylor J.J., Qunaj L., Griffith T.S., Vezys V., Barber D.L., and Masopust D.. 2014. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9:209–222. 10.1038/nprot.2014.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N., Green J.A., Moltedo B., Arvey A., Hemmers S., Yuan S., Treuting P.M., and Rudensky A.Y.. 2015. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell. 162:1078–1089. 10.1016/j.cell.2015.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvey A., van der Veeken J., Samstein R.M., Feng Y., Stamatoyannopoulos J.A., and Rudensky A.Y.. 2014. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 15:580–587. 10.1038/ni.2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi E., Carr A.J., Plitas G., Cornish A.E., Konopacki C., Prabhakaran S., Nainys J., Wu K., Kiseliovas V., Setty M., et al. 2018. Single-cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell. 174:1293–1308.e36. 10.1016/j.cell.2018.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajénoff M., Breart B., Huang A.Y.C., Qi H., Cazareth J., Braud V.M., Germain R.N., and Glaichenhaus N.. 2006. Natural killer cell behavior in lymph nodes revealed by static and real-time imaging. J. Exp. Med. 203:619–631. 10.1084/jem.20051474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., and Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., and Chudakov D.M.. 2015. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 12:380–381. 10.1038/nmeth.3364 [DOI] [PubMed] [Google Scholar]
- Braun A., Dewert N., Brunnert F., Schnabel V., Hardenberg J.-H., Richter B., Zachmann K., Cording S., Claßen A., Brans R., et al. 2015. Integrin αE(CD103) Is Involved in Regulatory T-Cell Function in Allergic Contact Hypersensitivity. J. Invest. Dermatol. 135:2982–2991. 10.1038/jid.2015.287 [DOI] [PubMed] [Google Scholar]
- Burzyn D., Kuswanto W., Kolodin D., Shadrach J.L., Cerletti M., Jang Y., Sefik E., Tan T.G., Wagers A.J., Benoist C., and Mathis D.. 2013. A special population of regulatory T cells potentiates muscle repair. Cell. 155:1282–1295. 10.1016/j.cell.2013.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberis M., Le Gros G., and Urban J. Jr. 2003. Animal model of Nippostrongylus brasiliensis and Heligmosomoides polygyrus. Curr. Protoc. Immunol. Chapter 19:19.12. [DOI] [PubMed] [Google Scholar]
- Carlson C.M., Endrizzi B.T., Wu J., Ding X., Weinreich M.A., Walsh E.R., Wani M.A., Lingrel J.B., Hogquist K.A., and Jameson S.C.. 2006. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 442:299–302. 10.1038/nature04882 [DOI] [PubMed] [Google Scholar]
- Casey K.A., Fraser K.A., Schenkel J.M., Moran A., Abt M.C., Beura L.K., Lucas P.J., Artis D., Wherry E.J., Hogquist K., et al. 2012. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188:4866–4875. 10.4049/jimmunol.1200402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Diacovo T.G., Grenache D.G., Santoro S.A., and Zutter M.M.. 2002. The α(2) integrin subunit-deficient mouse: a multifaceted phenotype including defects of branching morphogenesis and hemostasis. Am. J. Pathol. 161:337–344. 10.1016/S0002-9440(10)64185-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen T., Kannan A.K., Levine A.G., Fan X., Klein U., Zheng Y., Gasteiger G., Feng Y., Fontenot J.D., and Rudensky A.Y.. 2016. An essential role for the IL-2 receptor in Treg cell function. Nat. Immunol. 17:1322–1333. 10.1038/ni.3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolletta D., Feuerer M., Li A., Kamei N., Lee J., Shoelson S.E., Benoist C., and Mathis D.. 2012. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 486:549–553. 10.1038/nature11132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacher M., Imbusch C.D., Weichenhan D., Breiling A., Hotz-Wagenblatt A., Träger U., Hofer A.C., Kägebein D., Wang Q., Frauhammer F., et al. 2017. Genome-wide DNA-methylation landscape defines specialization of regulatory T cells in tissues. Nat. Immunol. 18:1160–1172. 10.1038/ni.3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R., Agarwal V., Stefano J., Bartel D.P., and Stoffel M.. 2014. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 54:766–776. 10.1016/j.molcel.2014.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., and Rudensky A.Y.. 2016. Hallmarks of Tissue-Resident Lymphocytes. Cell. 164:1198–1211. 10.1016/j.cell.2016.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., van der Veeken J., Shugay M., Putintseva E.V., Osmanbeyoglu H.U., Dikiy S., Hoyos B.E., Moltedo B., Hemmers S., Treuting P., et al. 2015. A mechanism for expansion of regulatory T-cell repertoire and its role in self-tolerance. Nature. 528:132–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M., Herrero L., Cipolletta D., Naaz A., Wong J., Nayer A., Lee J., Goldfine A.B., Benoist C., Shoelson S., and Mathis D.. 2009. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 15:930–939. 10.1038/nm.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filzmoser P., and Gschwandtner M.. 2018. mvoutlier: Multivariate Outlier Detection Based on Robust Methods. Available at: https://cran.r-project.org/web/packages/mvoutlier/index.html (accessed June 15, 2018).
- Fontenot J.D., Rasmussen J.P., Williams L.M., Dooley J.L., Farr A.G., and Rudensky A.Y.. 2005. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 22:329–341. 10.1016/j.immuni.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Garrod K.R., Wei S.H., Parker I., and Cahalan M.D.. 2007. Natural killer cells actively patrol peripheral lymph nodes forming stable conjugates to eliminate MHC-mismatched targets. Proc. Natl. Acad. Sci. USA. 104:12081–12086. 10.1073/pnas.0702867104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C., Moseman E.A., Loughhead S.M., Alvarez D., Zwijnenburg A.J., Waanders L., Garg R., de la Torre J.C., and von Andrian U.H.. 2016. The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity. 45:1270–1284. 10.1016/j.immuni.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A., Andre-Schmutz I., Carnaud C., Mathis D., and Benoist C.. 2001. Damage control, rather than unresponsiveness, effected by protective DX5+ T cells in autoimmune diabetes. Nat. Immunol. 2:1117–1125. 10.1038/ni738 [DOI] [PubMed] [Google Scholar]
- Habart D., Cheli Y., Nugent D.J., Ruggeri Z.M., and Kunicki T.J.. 2013. Conditional knockout of integrin α2β1 in murine megakaryocytes leads to reduced mean platelet volume. PLoS One. 8:e55094 10.1371/journal.pone.0055094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L., Buettner F., and Theis F.J.. 2015. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 31:2989–2998. 10.1093/bioinformatics/btv325 [DOI] [PubMed] [Google Scholar]
- Hemler M.E., Jacobson J.G., and Strominger J.L.. 1985. Biochemical characterization of VLA-1 and VLA-2. Cell surface heterodimers on activated T cells. J. Biol. Chem. 260:15246–15252. [PubMed] [Google Scholar]
- Hirahara K., Liu L., Clark R.A., Yamanaka K., Fuhlbrigge R.C., and Kupper T.S.. 2006. The majority of human peripheral blood CD4+CD25highFoxp3+ regulatory T cells bear functional skin-homing receptors. J. Immunol. 177:4488–4494. 10.4049/jimmunol.177.7.4488 [DOI] [PubMed] [Google Scholar]
- Holtkötter O., Nieswandt B., Smyth N., Müller W., Hafner M., Schulte V., Krieg T., and Eckes B.. 2002. Integrin α 2-deficient mice develop normally, are fertile, but display partially defective platelet interaction with collagen. J. Biol. Chem. 277:10789–10794. 10.1074/jbc.M112307200 [DOI] [PubMed] [Google Scholar]
- Huehn J., Siegmund K., Lehmann J.C.U., Siewert C., Haubold U., Feuerer M., Debes G.F., Lauber J., Frey O., Przybylski G.K., et al. 2004. Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4+ regulatory T cells. J. Exp. Med. 199:303–313. 10.1084/jem.20031562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikebuchi R., Teraguchi S., Vandenbon A., Honda T., Shand F.H.W., Nakanishi Y., Watanabe T., and Tomura M.. 2016. A rare subset of skin-tropic regulatory T cells expressing Il10/Gzmb inhibits the cutaneous immune response. Sci. Rep. 6:35002 10.1038/srep35002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itohara S., Mombaerts P., Lafaille J., Iacomini J., Nelson A., Clarke A.R., Hooper M.L., Farr A., and Tonegawa S.. 1993. T cell receptor delta gene mutant mice: independent generation of alpha beta T cells and programmed rearrangements of gamma delta TCR genes. Cell. 72:337–348. 10.1016/0092-8674(93)90112-4 [DOI] [PubMed] [Google Scholar]
- Izraelson M., Nakonechnaya T.O., Moltedo B., Egorov E.S., Kasatskaya S.A., Putintseva E.V., Mamedov I.Z., Staroverov D.B., Shemiakina I.I., Zakharova M.Y., et al. 2018. Comparative analysis of murine T-cell receptor repertoires. Immunology. 153:133–144. 10.1111/imm.12857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz S.Z., Lu L.-F., and Rudensky A.Y.. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30:531–564. 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamanaka M., Kim S.T., Wan Y.Y., Sutterwala F.S., Lara-Tejero M., Galán J.E., Harhaj E., and Flavell R.A.. 2006. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity. 25:941–952. 10.1016/j.immuni.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Kamran P., Sereti K.-I., Zhao P., Ali S.R., Weissman I.L., and Ardehali R.. 2013. Parabiosis in mice: a detailed protocol. J. Vis. Exp. 80:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassiotis G., Gray D., Kiafard Z., Zwirner J., and Stockinger B.. 2006. Functional specialization of memory Th cells revealed by expression of integrin CD49b. J. Immunol. 177:968–975. 10.4049/jimmunol.177.2.968 [DOI] [PubMed] [Google Scholar]
- Kerdiles Y.M., Beisner D.R., Tinoco R., Dejean A.S., Castrillon D.H., DePinho R.A., and Hedrick S.M.. 2009. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat. Immunol. 10:176–184. 10.1038/ni.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Rasmussen J.P., and Rudensky A.Y.. 2007. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8:191–197. 10.1038/ni1428 [DOI] [PubMed] [Google Scholar]
- Kolodin D., van Panhuys N., Li C., Magnuson A.M., Cipolletta D., Miller C.M., Wagers A., Germain R.N., Benoist C., and Mathis D.. 2015. Antigen- and cytokine-driven accumulation of regulatory T cells in visceral adipose tissue of lean mice. Cell Metab. 21:543–557. 10.1016/j.cmet.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn L.L., Hubbeling H.G., Porrett P.M., Yang Q., Barnett L.G., and Laufer T.M.. 2014. Regulatory T cells occupy an isolated niche in the intestine that is antigen independent. Cell Reports. 9:1567–1573. 10.1016/j.celrep.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Skon C.N., Lee Y.J., Oh S., Taylor J.J., Malhotra D., Jenkins M.K., Rosenfeld M.G., Hogquist K.A., and Jameson S.C.. 2015. The transcription factor KLF2 restrains CD4+ T follicular helper cell differentiation. Immunity. 42:252–264. 10.1016/j.immuni.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.G., Arvey A., Jin W., and Rudensky A.Y.. 2014. Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15:1070–1078. 10.1038/ni.3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.G., Hemmers S., Baptista A.P., Schizas M., Faire M.B., Moltedo B., Konopacki C., Schmidt-Supprian M., Germain R.N., Treuting P.M., and Rudensky A.Y.. 2017. Suppression of lethal autoimmunity by regulatory T cells with a single TCR specificity. J. Exp. Med. 214:609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir A.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R., et al. 2015. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 162:184–197. 10.1016/j.cell.2015.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A., Nutsch K.M., Farr A.G., Lund J.M., Rasmussen J.P., Koni P.A., and Rudensky A.Y.. 2008. Differentiation of regulatory Foxp3+ T cells in the thymic cortex. Proc. Natl. Acad. Sci. USA. 105:11903–11908. 10.1073/pnas.0801506105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Gerner M.Y., Van Panhuys N., Levine A.G., Rudensky A.Y., and Germain R.N.. 2015. Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature. 528:225–230. 10.1038/nature16169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C.T., Liao W., Dadi S., Toure A., and Li M.O.. 2016. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature. 529:532–536. 10.1038/nature16486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay L.K., Rahimpour A., Ma J.Z., Collins N., Stock A.T., Hafon M.-L., Vega-Ramos J., Lauzurica P., Mueller S.N., Stefanovic T., et al. 2013. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14:1294–1301. 10.1038/ni.2744 [DOI] [PubMed] [Google Scholar]
- Madamanchi A., Santoro S.A., and Zutter M.M.. 2014. Alpha2beta1 integrin. I Domain Integrins. Springer Netherlands, Dordrecht, Netherlands: pp. 41–60. [Google Scholar]
- Maere S., Heymans K., and Kuiper M.. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 21:3448–3449. 10.1093/bioinformatics/bti551 [DOI] [PubMed] [Google Scholar]
- Merico D., Isserlin R., Stueker O., Emili A., and Bader G.D.. 2010. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 5:e13984 10.1371/journal.pone.0013984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P., Clarke A.R., Rudnicki M.A., Iacomini J., Itohara S., Lafaille J.J., Wang L., Ichikawa Y., Jaenisch R., Hooper M.L., et al. 1992. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 360:225–231. 10.1038/360225a0 [DOI] [PubMed] [Google Scholar]
- Morton A.M., Sefik E., Upadhyay R., Weissleder R., Benoist C., and Mathis D.. 2014. Endoscopic photoconversion reveals unexpectedly broad leukocyte trafficking to and from the gut. Proc. Natl. Acad. Sci. USA. 111:6696–6701. 10.1073/pnas.1405634111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan T.E., Rapp M., Fan X., Weizman O.-E., Bhardwaj P., Adams N.M., Walzer T., Dannenberg A.J., and Sun J.C.. 2016. Adipose-Resident Group 1 Innate Lymphoid Cells Promote Obesity-Associated Insulin Resistance. Immunity. 45:428–441. 10.1016/j.immuni.2016.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht C., Park J., Cording S., Wing J.B., Atarashi K., Obata Y., Gaboriau-Routhiau V., Marques R., Dulauroy S., Fedoseeva M., et al. 2015. The microbiota regulates type 2 immunity through RORγt+ T cells. Science. 349:989–993. 10.1126/science.aac4263 [DOI] [PubMed] [Google Scholar]
- Onishi Y., Fehervari Z., Yamaguchi T., and Sakaguchi S.. 2008. Foxp3+ natural regulatory T cells preferentially form aggregates on dendritic cells in vitro and actively inhibit their maturation. Proc. Natl. Acad. Sci. USA. 105:10113–10118. 10.1073/pnas.0711106105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Jiang X., Chen Y., Sojka D.K., Wei H., Gao X., Sun R., Yokoyama W.M., and Tian Z.. 2013. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J. Clin. Invest. 123:1444–1456. 10.1172/JCI66381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polic B., Kunkel D., Scheffold A., and Rajewsky K.. 2001. How alpha beta T cells deal with induced TCR alpha ablation. Proc. Natl. Acad. Sci. USA. 98:8744–8749. 10.1073/pnas.141218898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S.J., Franki S.N., Pierce R.H., Dimitrova S., Koteliansky V., Sprague A.G., Doherty P.C., de Fougerolles A.R., and Topham D.J.. 2004. The collagen binding alpha1beta1 integrin VLA-1 regulates CD8 T cell-mediated immune protection against heterologous influenza infection. Immunity. 20:167–179. 10.1016/S1074-7613(04)00021-4 [DOI] [PubMed] [Google Scholar]
- Rubtsov Y.P., Rasmussen J.P., Chi E.Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W.R. Jr., et al. 2008. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 28:546–558. 10.1016/j.immuni.2008.02.017 [DOI] [PubMed] [Google Scholar]
- Rubtsov Y.P., Niec R.E., Josefowicz S., Li L., Darce J., Mathis D., Benoist C., and Rudensky A.Y.. 2010. Stability of the regulatory T cell lineage in vivo. Science. 329:1667–1671. 10.1126/science.1191996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather B.D., Treuting P., Perdue N., Miazgowicz M., Fontenot J.D., Rudensky A.Y., and Campbell D.J.. 2007. Altering the distribution of Foxp3(+) regulatory T cells results in tissue-specific inflammatory disease. J. Exp. Med. 204:1335–1347. 10.1084/jem.20070081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiering C., Krausgruber T., Chomka A., Fröhlich A., Adelmann K., Wohlfert E.A., Pott J., Griseri T., Bollrath J., Hegazy A.N., et al. 2014. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 513:564–568. 10.1038/nature13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebzda E., Zou Z., Lee J.S., Wang T., and Kahn M.L.. 2008. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat. Immunol. 9:292–300. 10.1038/ni1565 [DOI] [PubMed] [Google Scholar]
- Sefik E., Geva-Zatorsky N., Oh S., Konnikova L., Zemmour D., McGuire A.M., Burzyn D., Ortiz-Lopez A., Lobera M., Yang J., et al. 2015. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science. 349:993–997. 10.1126/science.aaa9420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M., Tadmor M.D., Reich-Zeliger S., Angel O., Salame T.M., Kathail P., Choi K., Bendall S., Friedman N., and Pe’er D.. 2016. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 34:637–645. 10.1038/nbt.3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., and Ideker T.. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugay M., Britanova O.V., Merzlyak E.M., Turchaninova M.A., Mamedov I.Z., Tuganbaev T.R., Bolotin D.A., Staroverov D.B., Putintseva E.V., Plevova K., et al. 2014. Towards error-free profiling of immune repertoires. Nat. Methods. 11:653–655. 10.1038/nmeth.2960 [DOI] [PubMed] [Google Scholar]
- Shugay M., Bagaev D.V., Turchaninova M.A., Bolotin D.A., Britanova O.V., Putintseva E.V., Pogorelyy M.V., Nazarov V.I., Zvyagin I.V., Kirgizova V.I., et al. 2015. VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLOS Comput. Biol. 11:e1004503 10.1371/journal.pcbi.1004503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skon C.N., Lee J.-Y., Anderson K.G., Masopust D., Hogquist K.A., and Jameson S.C.. 2013. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat. Immunol. 14:1285–1293. 10.1038/ni.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śledzińska A., Hemmers S., Mair F., Gorka O., Ruland J., Fairbairn L., Nissler A., Müller W., Waisman A., Becher B., and Buch T.. 2013. TGF-β signalling is required for CD4+ T cell homeostasis but dispensable for regulatory T cell function. PLoS Biol. 11:e1001674 10.1371/journal.pbio.1001674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka D.K., Plougastel-Douglas B., Yang L., Pak-Wittel M.A., Artyomov M.N., Ivanova Y., Zhong C., Chase J.M., Rothman P.B., Yu J., et al. 2014. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 3:e01659 10.7554/eLife.01659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens G.L., Andersson J., and Shevach E.M.. 2007. Distinct subsets of FoxP3+ regulatory T cells participate in the control of immune responses. J. Immunol. 178:6901–6911. 10.4049/jimmunol.178.11.6901 [DOI] [PubMed] [Google Scholar]
- Suffia I., Reckling S.K., Salay G., and Belkaid Y.. 2005. A role for CD103 in the retention of CD4+CD25+ Treg and control of Leishmania major infection. J. Immunol. 174:5444–5455. 10.4049/jimmunol.174.9.5444 [DOI] [PubMed] [Google Scholar]
- Thomson A.W., and Knolle P.A.. 2010. Antigen-presenting cell function in the tolerogenic liver environment. Nat. Rev. Immunol. 10:753–766. 10.1038/nri2858 [DOI] [PubMed] [Google Scholar]
- Tokoyoda K., Zehentmeier S., Hegazy A.N., Albrecht I., Grün J.R., Löhning M., and Radbruch A.. 2009. Professional memory CD4+ T lymphocytes preferentially reside and rest in the bone marrow. Immunity. 30:721–730. 10.1016/j.immuni.2009.03.015 [DOI] [PubMed] [Google Scholar]
- Tomura M., Honda T., Tanizaki H., Otsuka A., Egawa G., Tokura Y., Waldmann H., Hori S., Cyster J.G., Watanabe T., et al. 2010. Activated regulatory T cells are the major T cell type emigrating from the skin during a cutaneous immune response in mice. J. Clin. Invest. 120:883–893. 10.1172/JCI40926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L., and Hinton G.. 2008. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 9:2579–2605. [Google Scholar]
- van Dijk D., Nainys J., Sharma R., Kathail P., Carr A.J., Moon K.R., Mazutis L., Wolf G., Krishnaswamy S., and Pe’er D.. 2017. MAGIC: A diffusion-based imputation method reveals gene-gene interactions in single-cell RNA-sequencing data. https://www.biorxiv.org/content/early/2017/02/25/111591 (Preprint posted February 25, 2017).
- von Andrian U.H., and Mackay C.R.. 2000. T-cell function and migration. Two sides of the same coin. N. Engl. J. Med. 343:1020–1034. 10.1056/NEJM200010053431407 [DOI] [PubMed] [Google Scholar]
- Weber J.P., Fuhrmann F., Feist R.K., Lahmann A., Al Baz M.S., Gentz L.-J., Vu Van D., Mages H.W., Haftmann C., Riedel R., et al. 2015. ICOS maintains the T follicular helper cell phenotype by down-regulating Krüppel-like factor 2. J. Exp. Med. 212:217–233. 10.1084/jem.20141432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werr J., Johansson J., Eriksson E.E., Hedqvist P., Ruoslahti E., and Lindbom L.. 2000. Integrin alpha(2)beta(1) (VLA-2) is a principal receptor used by neutrophils for locomotion in extravascular tissue. Blood. 95:1804–1809. [PubMed] [Google Scholar]
- Wolfenson H., Lavelin I., and Geiger B.. 2013. Dynamic regulation of the structure and functions of integrin adhesions. Dev. Cell. 24:447–458. 10.1016/j.devcel.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Fujikado N., Kolodin D., Benoist C., and Mathis D.. 2015. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 348:589–594. 10.1126/science.aaa7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Dee M.J., Altman N.H., and Malek T.R.. 2015. IL-2Rβ-dependent signaling and CD103 functionally cooperate to maintain tolerance in the gut mucosa. J. Immunol. 194:1334–1346. 10.4049/jimmunol.1400955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., and Bevan M.J.. 2013. Transforming growth factor-β signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity. 39:687–696. 10.1016/j.immuni.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8:14049 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tissue Treg bulk RNA-seq, Treg subset bulk RNA-seq, scRNA-seq, and TCR-seq data have been deposited to the Gene Expression Omnibus under accession numbers GSE121480, GSE121482, GSE121034, and GSE121147, respectively.