Sox4 is shown to be essential for the development of natural killer T cells that function as innate-like first responders. Existence of specific gene circuits to tailor TCR signal-dependent thymic selection into the innate-like αβ T cell lineage is revealed.
Abstract
Natural killer T (NKT) cells expressing the invariant T cell receptor (iTCR) serve an essential function in clearance of certain pathogens and have been implicated in autoimmune and allergic diseases. Complex effector programs of these iNKT cells are wired in the thymus, and upon thymic egress, they can respond within hours of antigenic challenges, classifying iNKT cells as innate-like. It has been assumed that the successful rearrangement of the invariant iTCRα chain is the central event in the divergence of immature thymocytes to the NKT cell lineage, but molecular properties that render the iTCR signaling distinct to permit the T cell lineage diversification remain obscure. Here we show that the High Mobility Group (HMG) transcription factor (TF) SOX4 controls the production of iNKT cells by inducing MicroRNA-181 (Mir181) to enhance TCR signaling and Ca2+ fluxes in precursors. These results suggest the existence of tailored, permissive gene circuits in iNKT precursors for innate-like T cell development.
Introduction
Lymphocytes of the immune system are broadly divided into the fast acting innate (or innate-like, within hours of infections) and slow responding (within days) adaptive cell lineages. Lymphocytes expressing clonal antigen receptors generated by the recombination-activating gene (RAG) recombinase were conventionally classified as adaptive immune cells. However, fundamentally different activities and origins of innate B1 cells (Herzenberg and Herzenberg, 1989), thymic programming of γδTCR+ and αβTCR+ thymocytes into discrete functional effectors before their egress into peripheral tissues (Ribot et al., 2009; Narayan et al., 2012; Lee et al., 2013), and the discoveries of TCR-negative innate lymphoid cells (ILCs) with a similar repertoire of effector function as T cell subsets have emphasized the insufficiency of clonal antigen receptor expression alone as the defining characteristics of immune cell types. Instead, the timing (before or after pathogen exposure) of establishment of effector subset defining gene networks has been put forth as the more cohesive parameter for lymphocyte subset classification (Kang and Malhotra, 2015).
γδ T cells, CD8α+, intraepithelial T cells, TCRαβ+ invariant natural killer T (iNKT) cells, and mucosa-associated invariant T (MAIT) cells are the four major thymic cell types that express TCRs, but still exhibit prominent innate immune cell characteristics. Effector subsets of each cell type differentiate in the thymus before being exported to the peripheral tissues. Unlike γδ T cells that develop from CD4−CD8− double-negative (DN) thymic precursors, iNKT and MAIT cells primarily arise from immature CD4+CD8+ double-positive (DP) thymocytes that productively rearrange clonotypic Tcra genes (Godfrey et al., 2010; Koay et al., 2016). However, a liver-tropic iNKT subset can arise from DN thymocytes (Dashtsoodol et al., 2017). The most common and well-studied innate-like murine αβ T cells are iNKT cells that are characterized by the expression of invariant Vα14-Jα18 TCR chain (Godfrey et al., 2010; Engel et al., 2016). iNKT cells recognize lipids in association with the MHC Class I–like molecule CD1d. Until recently, NKT cell development was predominantly studied by probing phenotypically confined sequential maturational intermediates from Stages 0–3 (characterized by usage of CD24, CD44, and NK1.1 markers). Stage 3 NKT cells produce only Th1 cytokines and thus referred to as NKT1, while Stages 1 and 2 contained heterogeneous pools of IL-4 and IL-17 (Th2- and Th17-like), producing NKT2 and NKT17 cells, respectively (Lee et al., 2013).
Newly devised segregation of thymic NKT effectors based on transcription factors (TFs) has constructed an alternative perspective to deduce molecular events in intrathymic NKT cell differentiation. Previously, several TFs known to be required for conventional αβ T cell development were also shown to be necessary for progressing through distinct stages of the thymic iNKT maturational stages. For example, RORγt mediates NKT cell development via its ability to prolong the survival of DP cells and permit Vα14 TCR gene rearrangements (Guo et al., 2002). Various other TFs that mediate signaling, selection, or survival of DP cells, such as E-box family member HEB, EGR2, RUNX1, and c-MYC, were also shown to be involved in the generation of iNKT cells (D’Cruz et al., 2010; Godfrey et al., 2010). Akin to γδ T effector subset programming in the thymus (Narayan et al., 2012), iNKT thymic subsets segregate based on differential TF activities with T-bet (Tbx21), PLZF (Zbtb16), and RORγt (Rorc), directing NKT1, NKT2, and NKT17 subset specification, respectively (Lee et al., 2013). These results suggested that innate-like T cell subtype development is commonly guided by gene regulatory circuits to program innate properties, regardless of antigen receptor types expressed. Nevertheless, TCR signal strength has been linked to selective iNKT effector differentiation (Lee et al., 2013; Cruz Tleugabulova et al., 2016), leaving open the possibility that both γδTCR and αβTCR can transmit similar cell fate instructive signals to program innate T cell subsets in the thymus.
To determine whether iNKT cells are regulated by TCR signaling-independent TFs, we focused on the High Mobility Group (HMG) box TFs. We showed that the HMG box TFs SOX4 and SOX13 are required for the thymic development of innate dermal γδ T cells programmed to produce IL-17 (Tγδ17; Malhotra et al., 2013). Here, we show that SOX4 is also necessary for the development of iNKT and MAIT cells, but appears not to impact adaptive αβ T cell differentiation. SOX4 regulates the expression of MicroRNA-181 (Mir181) family members in DP thymocytes that tunes TCR signaling (Li et al., 2007). SOX13 also controls iNKT cell generation, but its impact is primarily on NKT17 cells, mirroring the indispensable function of SOX13 in the generation Tγδ17 cells (Malhotra et al., 2013). Thus, both αβ and γδ innate T cell development is reliant on the HMG box TF network, supporting the shared properties of T cells contingent on thymic programming of effector function, rather than TCR types per se.
Results
SOX4 is required for Vα14 TCR+ iNKT cell development
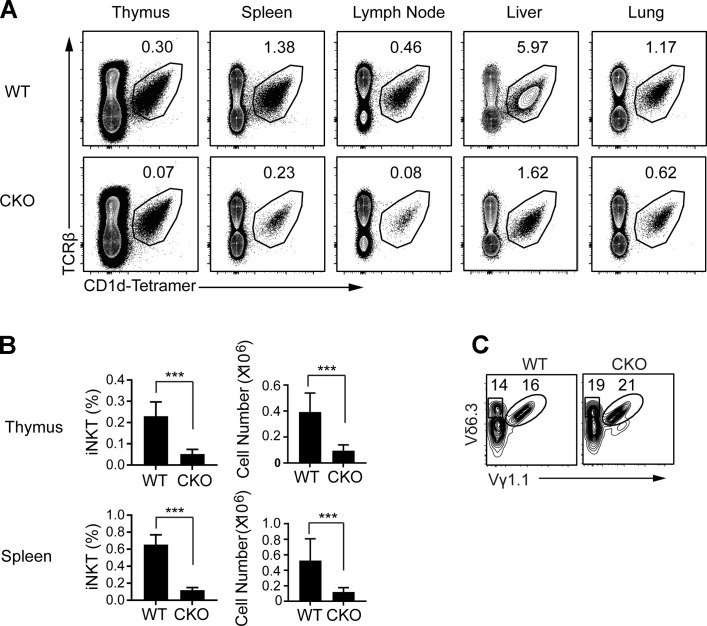
Sox4−/− mice are embryonic lethal (Schilham et al., 1996, 1997). SOX4 is a regulator of stem cell differentiation (Sinner et al., 2007; Novershtern et al., 2011), and Sox4−/− fetal liver stem cells are impaired in T and B cell generation (Schilham et al., 1996, 1997). In the thymus, Sox4 expression is highest in DN thymic precursors. Immature (CD24+) conventional αβTCR+ thymocytes (CD4+CD8+ DP) and immature innate thymocytes expressing γδTCR and those fated to become invariant Vα14 TCR+ NKT cells also express Sox4 (Narayan et al., 2012; Cohen et al., 2013; Immunological Genome Project Consortium, 2018). Upon transition to the mature state (CD24neg), the expression is extinguished in γδTCR+ and iNKT cells and decreased in conventional CD4 or CD8 single-positive thymocytes (Immunological Genome Project Consortium, 2018). To determine SOX4 function during intrathymic αβ T cell development, we bred Sox4 “floxed” mice to Cd2-Cre, Cd4-Cre, and Rorc-Cre mice so that all NKT cells that developed from thymic DN or DP precursor cells had undergone Cre-mediated recombinations (Fig. S1 A and data not shown) and lacked Sox4 transcripts (data not shown). We denote these mice as T cell–restricted Sox4-null allelic variants (T-Sox4−/−; conditional gene knocked out [CKO] in the figures). We previously showed that Tγδ17 cells are dependent on Sox4 for development (Malhotra et al., 2013), whereas function of Sox4 in αβ T cell development has not been established in detail. The proportions and cellularity of conventional T cell precursor subsets (DN subsets 1–4), DP, single-positive T cells, and FOXP3+ regulatory T cells were not affected in the absence of Sox4 (Fig. S1 B and data not shown). However, there was a striking decrease in the development of αβ iNKT cells in the absence of Sox4. The lack of iNKT cells was also observed in the peripheral tissues, in particular in LNs and the liver, where a large number of NKT cells normally reside (Fig. 1, A and B). The frequency of NFIL3-dependent adipose iNKT cells (Lynch et al., 2015) was reduced in T-Sox4−/−, but the difference was not statistically significant (Fig. S1 C). Similar iNKT cell developmental defects were observed in Cd4-Cre; T-Sox4−/− mice, indicating that SOX4 is primarily required at the DP stage of thymocyte differentiation (data not shown). Data generated from Cd2-Cre mice will be presented as the representative T-Sox4−/− mice herein.
Figure 1.
iNKT cell development is severely impaired in mice lacking Sox4 in hematopoietic cells. (A) Representative flow cytometric analyses show decreased frequencies of iNKT cells in the thymus, spleen, peripheral LNs, liver, and lung of WT (Cd2-iCre; Sox4+/+) and Cd2-iCre;Sox4fl/fl CKO mice. One of three independent experiments (minimum of three mice per genotype) is shown. (B) Summary of frequencies and cell numbers of iNKT cells in the thymus and spleen, with statistical significance denoted (***, P < 0.001, Student’s t test). Error bars denote SD. (C) γδTCR+ NKT cells (Vγ1.1+Vδ6.3+) as shown among gated γδTCR+ cells are not altered in CKO mice. Representative plots from one of two independent experiments.
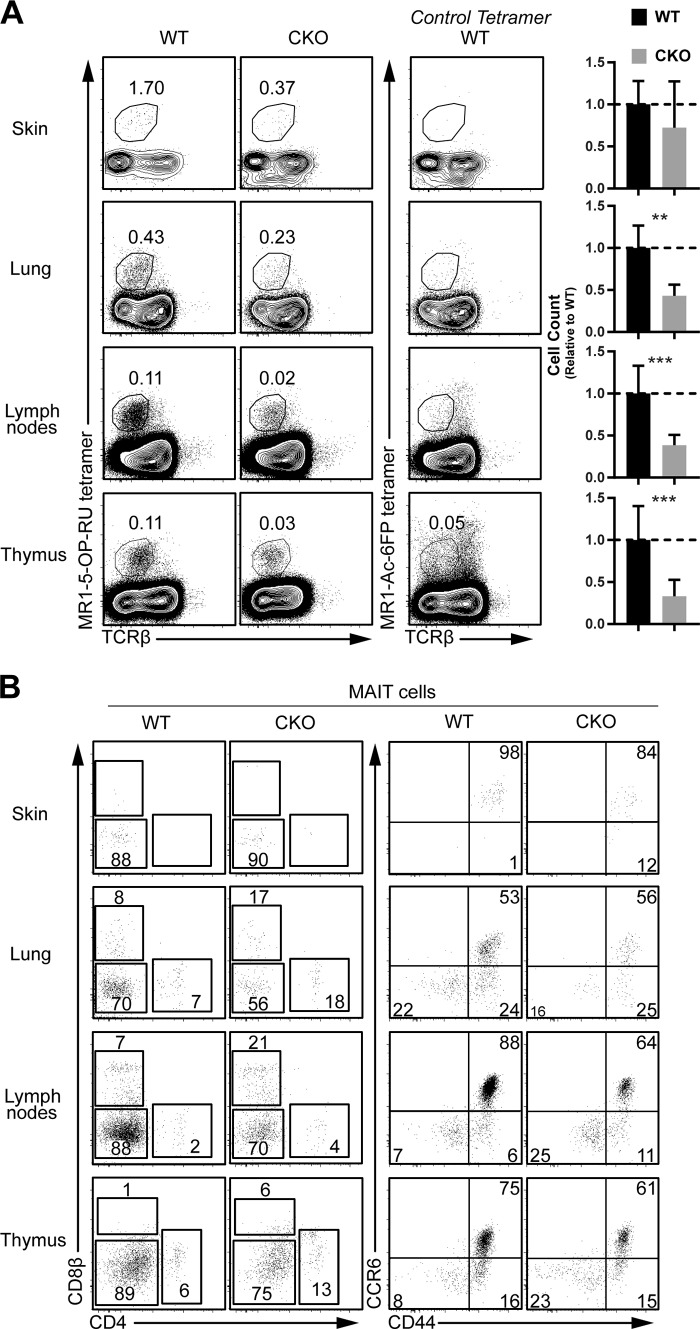
We also assessed development of other innate lymphoid effector subsets related to iNKT cells. CD1d-restricted but “diverse” type 2 NKT cells, identified as TCRβ+ NK1.1+ CD1d-PBS57neg cells, were also reduced in the thymus and spleen but not LN of T-Sox4−/− mice compared with WT controls (Fig. S1 D). γδTCR+ (Vγ1.1+Vδ6.3+) NKT cells and Vα18+ MAIT cells are two other PLZF-dependent innate T cell subsets in mice (Kovalovsky et al., 2008; Savage et al., 2008; Narayan et al., 2012; Pereira and Boucontet, 2012). TCR ligand for γδNKT cells is unknown, while MAIT cells recognize vitamin metabolites in the context of nonclassical MHC Class I molecule MR1 (Kjer-Nielsen et al., 2012). While SOX4 was not necessary for the development and/or maintenance of γδNKT cells (Fig. 1 C), it was required for the generation of MAIT cells in most tissues where two- to fourfold reduction in numbers was observed (Fig. 2 A). Of the residual MAIT cells that developed without SOX4, phenotypic analyses revealed no substantive alterations. In tissues other than the skin, there was a modest overrepresentation of CD4+ and CD8+ subsets over the predominant DN subset found in WT mice, and in the LN and thymus, “unactivated” CD44neg cells increased (Fig. 2 B). These results indicate that SOX4 controls the development of multiple innate T cell subsets while having no apparent impact on the production of adaptive αβ T cells.
Figure 2.
Defective innate MAIT cell development in Sox4 CKO mice. (A) Representative flow cytometric analyses of MAIT (TCRβintermediateMR1-5-OP1RU tetramer+) cells in indicated tissues from WT and Sox4 CKO mice are shown along with the tissue iNKT cellularities (normalized to WT cell counts; **, P < 0.01; ***, P < 0.001, Student’s t test) from three independent experiments for all tissues and of five experiments for thymus and LNs. Thymic profiles are gated on CD8neg thymocytes due to relatively high background staining of the control tetramer on DP thymocytes. Overall cellularity in tissues of WT and Sox4 CKO mice was not significantly altered and differences in proportions of MAIT cells reflect differences in cell numbers. For ears, there was a high variability, with one out of three of the Sox4-deficient mice analyzed exhibiting WT phenotype, leading to the nonsignificant difference. Error bars denote SD. (B) T cell coreceptor (CD4 and CD8; left panels), activation (CD44), and CCR6 chemokine receptor expression on WT and CKO MAIT cells.
Intrathymic iNKT cell developmental stages regulated by Sox4
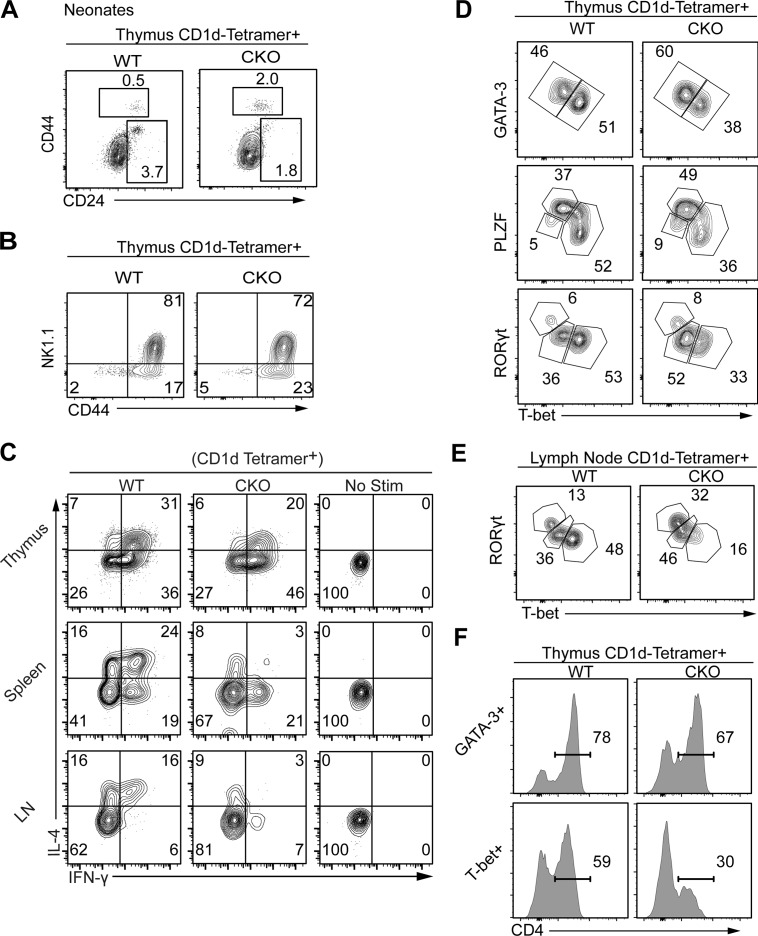
To pinpoint when during iNKT cell differentiation SOX4 exerts its function, we assessed thymic iNKT cells of T-Sox4−/− mice by tracking maturation markers and iNKT effector subset specific TFs. In neonates, when iNKT cells first arise (Gapin et al., 2001), the residual Sox4-deficient iNKT cells were proportionally enriched for the most immature (CD24+) iNKT cells (DP, CD1d-PBS-tetramer+CD24+; WT, 0.3 ± 0.2% vs. CKO, 2.5 ± 0.8%; means ± SEM; n = 5/genotype; Fig. 3 A). However, there was no significant difference in the number of these early iNKT subsets, suggesting that SOX4 is required for the transition from the DP stage to subsequent phases of iNKT cell maturation. As the mice aged, residual iNKT cells in T-Sox4−/− mice (range: ∼12–20% of the WT mice) exhibited relatively normal distribution of maturational intermediates (Fig. 3 B and Fig. S2 A) and marginally enhanced proliferation (Fig. S2 B) relative to WT mice. However, Sox4-deficient thymic and peripheral iNKT cells were impaired in IL-4 production (Fig. 3 C). In the thymus of T-Sox4−/− mice, the defect was more pronounced for IFNγ+IL-4+ dual producers, whereas in the periphery there was paucity of all IL-4–producing cells. Sox4-deficient iNKT cells maintained the capacity to produce IFNγ in the thymus and spleen. However, LN iNKT cells were defective in IFNγ production, correlating with an altered T-bet expression (below).
Figure 3.
Remnants of iNKT cells in Sox4 CKO mice are selectively depleted of NKT1 cells. (A) Representative flow cytometric analyses show a proportional enrichment for DP (CD8α+) CD1d-tetramer+ cells in neonatal CKO mice. Note that a general trend for decreased CD24 expression in Sox4 CKO DP thymocytes obscures the CD24+ NKT0 cells. (B) iNKT maturation marker CD44 and NK1.1 were analyzed in thymic iNKT cells of WT and CKO mice. Representative profiles from five independent experiments. (C) Programmed production of IL-4 and IFNγ from remnants of Sox4 CKO iNKT cells from indicated tissues was compared with normal iNKT cells (with no stimulation profiles). Representative profiles from four independent experiments. (D and E) iNKT effector subsets identified by the expression of indicated TFs in iNKT cells show decreased production of T-bet+ NKT1 cells in remnants of Sox4 CKO iNKT cells. Frequencies of each subset were assessed in the thymus (D) and peripheral LNs (E). Representative profiles from three independent experiments. (F) Proportions of CD4 iNKT cells within thymic GATA3+ or T-bet+ iNKT cells are shown. Representative plots from three independent experiments.
To determine whether the functional defect of residual Sox4-deficient iNKT cells was attributable to impaired expression of TFs that direct effector cytokine production, we assessed PLZF, GATA3, T-bet, and RORγt expression in Sox4-deficient iNKT cells. Starting in the thymus, these effector function programming TFs can be used to identify distinct effector subtypes (Lee et al., 2013; Engel et al., 2016). IL-4 production from iNKT cells has been shown to be regulated by a trio of TFs: PLZF, GATA3, and cMAF (Godfrey et al., 2010). Expression of all three TFs was not significantly altered in residual iNKT cells of T-Sox4−/− mice (Fig. 3 D and data not shown), indicating that the deficit in IL-4 production in the absence of Sox4 is not attributable to changes in the expression of the known regulators. Further, the expression of receptors for cytokines important in iNKT cell homeostasis, namely IL-2, IL-7, and IL-15, was not substantially and consistently altered in the residual peripheral iNKT cells in T-Sox4−/− mice, although some variations were observed in select lymphoid tissues (Fig. S2 C), suggesting that cytokine-mediated iNKT cell maintenance is unlikely to account for the deficit of effector programmed peripheral iNKT cells. Given that SOX4 functionally interacts with proteins controlling type 2 cytokine production (Kuwahara et al., 2012), the loss of these and other undocumented interactions may underpin the alterations in the capacity to make effector cytokines.
Another striking change in Sox4-deficient iNKT subsets was a decrease in T-bet+ NKT1 cells that was observed in all tissues examined (Fig. 3, D and E). In the thymus, the ratio of T-bet+ (NKT1):GATA3+ (NKT2) NKT cells was ∼1:2 in T-Sox4−/− mice, while in control mice, there was consistently more NKT1 cells (Fig. 3 D). GATA3+ iNKT cells are normally PLZFhi, and this pattern was maintained in T-Sox4−/− thymic residual iNKT cells (data not shown). Consistent with the impaired capacity of peripheral LN iNKT cells to produce IFNγ (Fig. 3 C), the reduction in residual T-bet+ iNKT cells was pronounced in the LNs, with a reciprocal enhancement of RORγt+ NKT17 cells (Fig. 3 E). Within the residual T-bet+ NKT1 population, the frequency of IFNγ+ cells was similar between WT and T-Sox4−/− mice (Fig. S2 D). Similar results were observed for IL-17A production by RORγt+ NKT17 cells, while IL-4 production was reduced approximately twofold among CD24–NK1.1– iNKT cells, which contains NKT2 cells. Additionally, more than half of T-bet+PLZFlo iNKT cells express CD4 normally, but Sox4-deficient T-bet+ counterparts were specifically depleted of the CD4+ subset (Fig. 3 F). These results establish the following hierarchy of Sox4 dependency among iNKT subsets: NKT1 > NKT2 > NKT17 > adipose iNKT.
It has been shown recently that TCF1 and LEF1 that are absolutely necessary for conventional αβ T cell development also mediate central functions during iNKT cell differentiation. While mutations in either TF curtail iNKT cell generation to the similar extent as T-Sox4−/− mice, TCF1 and LEF1 have more apparent impacts on NKT2/17 and NKT2 cells, respectively (Carr et al., 2015). T-Sox4−/− mice did not show altered TCF1 and LEF1 expression in DP thymocytes or iNKT cell subsets (Fig. S2 E). Compound mutations in Sox4 and Tcf7 led to a more severe depletion of iNKT cell compared with Sox4-deficient mice (Fig. S2 F), but interpretations of the data are challenging as there are general T cell developmental defects in Tcf7−/− mice. Nevertheless, unlike T-Sox4−/− iNKT cells, residual iNKT cells in double-deficient mice accumulated NK1.1+CD44− cells at the expense of NK1.1+CD44+ cells (Fig. S2 G), suggesting that TCF1 can perform a function distinct from SOX4. Together, these results show that the trio of HMG TFs (SOX4, TCF1, and LEF1) mediates nonredundant function in iNKT cell development, individually contributing to numerically normal output and mediating NKT subset–specific programming.
Deficient iNKT cell responses to cognate ligand in vivo in T-Sox4−/− mice
We next assessed whether the defects in both iNKT cell numbers and functional programming in the absence of SOX4 also resulted in deficient responses to the cognate iNKT cell TCR ligand, α-galactosylceramide (αGC), in vivo. T-Sox4−/− and littermate controls were injected with αGC and analyzed for a variety of iNKT cell-dependent processes. iNKT cells are activated to produce cytokines within hours after αGC injection; WT iNKT cells robustly produced IFNγ and IL-4 by 2 h after αGC administration, whereas cytokine production by T-Sox4−/− iNKT cells was significantly diminished (Fig. S3 A). Initial activation of iNKT cells by αGC leads to “transactivation” of NK cells (Carnaud et al., 1999); consistent with an early deficit in iNKT cell cytokine production, NK cell transactivation as assessed by IFNγ production and CD69 up-regulation was significantly reduced in T-Sox4−/− mice compared with WT mice (Fig. S3 B). After initial activation, iNKT cells down-regulate surface TCR followed by dramatic expansion and reexpression of TCR. Early down-modulation of the TCR (as assessed by CD1d-PBS57 staining) was similar when comparing iNKT cells from αGC-injected WT and T-Sox4−/− mice (Fig. S3 C), and we did not observe significant cell death of iNKT cells in either WT or T-Sox4−/− mice after αGC injection (Fig. S3 D). Interestingly, 3 d after αGC injection, T-Sox4−/− iNKT cells strongly expanded in a manner similar to iNKT cells from αGC-injected littermate controls (Fig. S3 E). Together these data indicate that SOX4 controls initial iNKT cell population size (Fig. 1) and effector function, but not peripheral iNKT cell proliferative responses after activation by a strong cognate ligand.
Sox4 is required in a DP thymocyte-intrinsic manner to generate iNKT cells
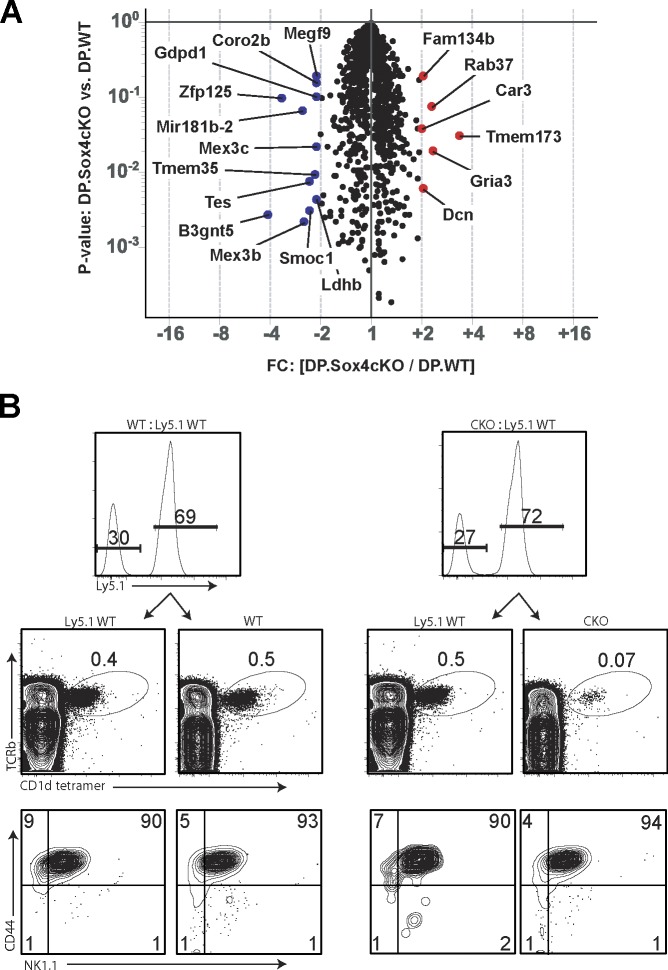
DP–DP cellular interactions generate iNKT cells and Sox4 is highly expressed in DP thymocytes, but at reduced amounts in most iNKT cells (Engel et al., 2016; Lee et al., 2016; Immunological Genome Project Consortium, 2018), suggesting that the initial SOX4-dependent programming of iNKT cell development is occurring in DP cells. Thus, to trace the origin of iNKT cell defect in T-Sox4−/− mice we first determined phenotypic and functional changes in Sox4-deficient DP thymocytes. DP thymocyte survival in T-Sox4−/− mice was identical to that in WT mice (Fig. S4 A) and Vα14-Jα18 TCR gene rearrangement, which is rare and requires normal DP cell survival (Guo et al., 2002), was unchanged (Fig. S4 B), indicating that the deficit in iNKT cells in T-Sox4−/− mice is not linked to a reduced rate of Vα14-Jα18 recombination events. Global differences in transcriptomes were also constrained, but some candidate molecules under direct or indirect control of SOX4 could be identified (Fig. 4 A). Expression of the two cell surface molecules on DP thymocytes, CD24 and CD1d, was reduced on Sox4-deficient DP cells (Fig. S4 C), and the array data indicated significant differences that did not meet the fold change criteria at the transcript level. HMG box TFs have been suggested to control CD24 and CD1d expression in cell lines (Shulewitz et al., 2006; Chen et al., 2010), but TCF1 and LEF1 do not impact CD24 or CD1d expression on thymocytes (Carr et al., 2015 and data not shown), raising the possibility that SOX4 may assume a more central role in vivo. Antibody (Ab)-mediated blockade of CD24 in WT mice (Askew and Harding, 2008) did not lead to altered iNKT cell differentiation (data not shown), suggesting that the reduced CD24 expression is unlikely to account for the iNKT developmental block in T-Sox4−/− mice.
Figure 4.
SOX4 is a T cell–intrinsic factor required for iNKT cell development from DP thymocytes. (A) Volcano plot shows genes that are expressed at higher (red) or lower (blue) in CKO DP thymocytes relative to WT counterparts. Genes whose expression was different by at least twofold on average with indicated P values are shown (FC, fold change). Expression values were determined using microarrays of three sorted DP thymocyte replicates for each genotype. Cd1d, Cd24, and Mir181a/b were all decreased in expression, but are not denoted on the plot as they were decreased 1.7–1.9-fold on average in transcript amounts as detected using microarrays. (B) Impaired iNKT cell development from Sox4-deficient precursors cannot be rescued in trans by codifferentiating WT T cells in the thymus. Flow cytometric analysis of thymocytes from Rag1−/− mice that were reconstituted with an ∼1:1 mix of WT:WT or WT:CKO BM cells ∼8–10 wk prior. Partner WT cells were from congenic C57BL/6 (Ly5.1) mice for tracking purposes. Shown are contributions from partner precursors (top panels; range for CKO reconstitution: 38–76% relative to partner WT cells, with no correlation between the extent of defective iNKT cell generation and relative reconstitution frequencies), iNKT cell frequencies (middle), and iNKT cell maturation profiles (bottom). Representative plots from two independent experiments with a minimum of five mixed BM chimeras/group.
The altered CD1d expression on Sox4-deficient thymocytes raised a possibility that the defective iNKT cell development was caused by decreased availability of the agonist CD1d complex for the TCR. To test this we generated mixed bone marrow (BM) chimeras consisting of congenically marked WT and T-Sox4−/− BM cells (1:1 ratio of CD45.1+ WT:CD45.2+ T-Sox4−/−) such that Sox4-deficient thymocytes would have access to normal CD1d on WT DP thymocytes. If the reduced expression of CD1d on mutant DP cells were the root cause of impaired iNKT cell generation the deficit from iNKT cell development should be restored in mixed chimeric mice. As in T-Sox4−/− mice, iNKT cells did not develop efficiently from mutant precursors in the mixed BM chimeras (Fig. 4 B; WT, 0.5 ± 0.1% iNKT cells; CKO, 0.04 ± 0.02%, n = 12). The few iNKT cells developing from Sox4-deficient precursors displayed relatively similar distribution of thymic iNKT subsets as WT progenitors (Fig. 4 B). These results demonstrated that the presence of WT DP thymocytes and normal amounts of CD1d cannot rescue the Sox4-deficient iNKT cell generation and that the iNKT defect in CKO mice arose from cell-intrinsic defects in Sox4-deficient iNKT precursors.
Impaired Ca2+ signaling in Sox4-deficient thymocytes
Analysis of TCRβ chain repertoire showed that the few iNKT cells that are found in the LNs in T-Sox4−/− mice were comparable to WT iNKT cells (Fig. S4 D). Remnants of Sox4-deficient thymic NKT1 cells, however, consistently expressed lowest amounts of CD5, a proxy for TCR signal strength (Azzam et al., 1998), compared with WT controls (Fig. S4 E). This raised the possibility that SOX4 modulates TCR signaling for NKT1 cell development. Consistent with this, Sox4-deficient DP thymocytes were impaired in Ca2+ signaling as indicated by the decreased Ca2+ flux upon TCR cross-linking in vitro (Fig. 5 A). The reduced expression of a Ca2+-regulated NFAT target gene Egr2 in Sox4-deficient DP thymocytes (Fig. S4 F) further supported the likelihood that the loss of iNKT cells in T-Sox4−/− is linked to abnormal TCR signaling. Mice lacking STIM1 and STIM2, which control store-operated Ca2+ entry, and Egr2-deficient mice also show selective defects in iNKT development, as conventional T cell generation is largely intact in these mutants (Lazarevic et al., 2009; Oh-hora et al., 2013).
Figure 5.
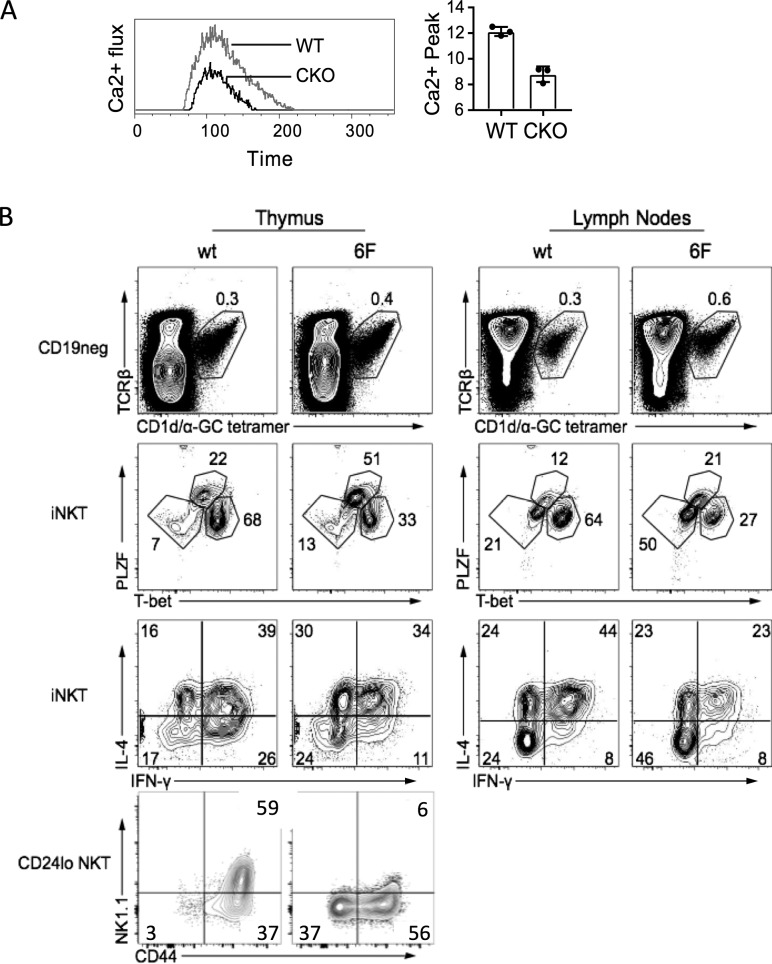
Evidence for impaired TCR signaling in Sox4-deficient thymocytes. (A) Impaired Ca2+ signaling in thymocytes lacking SOX4. Left, kinetic analysis of Ca2+ flux (ratios of Fluo 3:Fura red) in DP thymocytes from WT and Sox4 CKO mice after stimulation with CD3ε cross-linking. Representative plots from four independent experiments. Right, averages of peak flux (n = 4/genotype). Error bars denote SD. (B) Thymocytes from Cd2476F/6F (6F) mice with compromised TCR-signaling capacity also show impaired NKT1 cell generation and function. Shown from top to bottom panels are iNKT cell frequencies, iNKT cell subset distributions, effector cytokine production, and iNKT cell maturation profiles in the thymus and LNs of WT and Cd2476F/6F mice. Representative plots from three independent experiments using mice of age ranging from 4 to 6 wk old (thymus) or 8 to 12 wk old (LN).
Flow cytometric analyses of proxy for TCR signaling above suffers historically from very limited signal separations, necessitating additional experimental supports for SOX4-mediated control of TCR signaling. To determine whether mice with impaired TCR signaling show similar defects as T-Sox4−/− mice, we analyzed iNKT subset development in mice with signaling defects emanating from the CD3ζ chain, an obligate signal transducer of TCR. In these mice (6F: Cd2476F/6F) tyrosine phosphorylation sites in the six endogenous ITAMs of CD3ζ were substituted with phenylalanine (F), rendering them incapable of signal transduction and reducing the CD3 complex signal capacity by ∼60% (CD3ε and CD3δ contribute four ITAMs; Hwang et al., 2012). 6F mice have relatively mild perturbations in conventional αβ T cell development, but like T-Sox4−/− mice, production of select innate T cell subsets, including Tγδ17 cells (Spidale et al., 2018), is compromised. 6F mice had marginally increased frequencies of iNKT cells in the thymus and LNs that did not translate into significant differences in cellularity (data not shown), but the NKT1 (NK1.1+CD44+ or T-bet+) subset was highly underrepresented, and IFNγ production was reduced (Fig. 5 B). Although the NKT1 cell defect is more severe in T-Sox4−/− mice, the specific deficit in NKT1 cells observed in 6F mice support the reduced TCR signaling capacity in Sox4-deficient DP thymocytes as one component contributing to the iNKT cell developmental block.
SOX4 controls miR181 expression to regulate iNKT cell development
Selective requirement for TCR signaling apparatus for innate T cells has been linked to higher threshold of signals that need to be breached for their development. Global transcriptome analysis suggested a deficit in miR181 expression in Sox4-deficient DP thymocytes (Fig. 4 A), a finding confirmed by quantitative real-time PCR assay (Fig. 6 A). Mir181a/b-1 has been shown to be a positive regulator of TCR signaling and Ca2+ flux, and mice lacking Mir181a/b-1 have specific defects in iNKT cell development, with grossly normal conventional αβ T cell development, similar to T-Sox4−/− mice (Henao-Mejia et al., 2013; Ziętara et al., 2013). Interestingly, Sox4 expression has been reported to be diminished in Mir181a/b-1–deficient DP thymocytes, compared with WT controls (Henao-Mejia et al., 2013). SOX4 can directly control Mir181a/b-1 expression as chromatin immunoprecipitation (ChIP) assay using Abs to SOX4 showed that SOX4 is docked onto the regulatory region of Mir181a/b-1 in DP thymocytes at a HMG box consensus motif near the transcription start site (Fig. 6 B). Further, using anti-Histone ChIP, Histone 3 methylation pattern at the Mir181a/b-1 locus in WT DP thymocytes showed enrichment for active chromatin marks (H3K4me3), whereas in T-Sox4−/− DP thymocytes repressive marks (H3K27me3) dominated (Fig. 6 B). These results are consistent with SOX4 acting as a TF imposing a transcription permissive chromatin state of Mir181 genes.
Figure 6.
SOX4 regulates Mir181 expression in thymocytes. (A) Mir181a-1 and Mir181b-1 transcripts in DP thymocytes of WT and CKO mice were quantified by real-time PCR. One of three experiments is shown. (B) ChIP analysis for SOX4 docking and epigenetic modifications at the Mir181 locus of DP thymocytes. Each graph shows control Ab (matching the species of origin of experimental Ab) for the protein–chromatin complex precipitation followed by the chromatin states probed with Abs to indicated markers in sorted WT and Sox4 CKO DP thymocytes. The consensus DNA binding motif for SOX4 is located ∼2.3 kb upstream of the transcription start site of Mir181a-1 and denoted on the schematic (right; not to scale), which also shows the region assessed by quantitative PCR (arrows). Active (K4me3), poised (K9me3), and suppressed (K27me3) histone modifications in the region are shown. Statistical significance based on Student’s t test denoted. (C and D) iNKT cell development from Sox4-deficient precursors can be rescued by enforced Mir181a-1 expression. (C) Representative profiles of the rescued iNKT cell development among mature (CD24neg) thymocytes showing frequencies of iNKT cells from the Mir181a-1 retrovirus transduced (GFP+) and nontransduced (GFP−) precursors in the same mouse. Transduction studies using WT and CKO BM cells are shown. Representative plots from one of two independent experiments. (D) Summary of thymic iNKT cell frequencies from retroviral reconstitution experiments using infected WT (blank circles) and CKO (filled circles) BM cells. Only mice with >1% GFP+ cells among mature thymocytes were included in the calculation. Denoted significance in graphs was based on Student’s t test. Error bars denote SD.
To determine whether the diminished Mir181a-1 expression in Sox4-deficient thymocytes is in part responsible for the iNKT cell developmental block, we assessed the ability of retroviral Mir181a/b-1 expressing Sox4-deficient BM progenitors to generate iNKT cells in BM chimeras. Retrovirus-transduced cells were marked by GFP reporter expression. iNKT cells that developed from retrovirus-transduced Sox4-deficient progenitors developed similar frequencies of iNKT cells as those from WT progenitors (Fig. 6, C and D). Enforced Mir181a/b-1 expression restored normal NKT cell development from Sox4-deficient progenitors, including normal distribution of NKT subsets (data not shown). These results position SOX4 as an upstream regulator of the Mir181a/b-1 modulated TCR signaling pathway (Henao-Mejia et al., 2013; Ziętara et al., 2013) and that SOX4-regulated Mir181a/b-1 expression is critical for normal NKT cell development. Given the reciprocal reduction in the expression of Sox4 and Mir181 in DP thymocytes lacking Mir181a/b-1 and Sox4, respectively (Henao-Mejia et al., 2013), they are modeled to operate in a positive feedback loop to set TCR signal thresholds.
iNKT17 cell production also requires Sox13
A quartet of HMG box TFs, SOX4, SOX13, LEF1, and TCF1, controls Tγδ17 cell differentiation (Malhotra et al., 2013), and Sox4, Lef1, and Tcf7 have now been shown to be essential for iNKT cell generation. We next investigated whether Sox13 is also involved in iNKT cell development. In hematopoietic cells, SOX13 was identified as the first TF expressed distinctly in developing γδ versus αβ T cell lineages, active in γδTCR+ thymocytes, but absent in conventional αβTCR+ cells (Melichar et al., 2007). However, ECFP reporter driven by Sox13 promoter in mice marked early stages of iNKT cell development (Spidale et al., 2018). We confirmed the result using anti-GFP intracellular staining, which further showed that, in the thymus, ∼30% of RORγt+ iNKT cells expressed the Sox13 promoter-driven reporter, with reduced expression in RORγtneg iNKT cells (Fig. S5, A and B). This proxy for Sox13 expression pattern was supported by a global transcriptome analysis of sorted iNKT cell subsets (Cohen et al., 2013) and by single-cell transcriptome analysis of iNKT cells, which indicated a more restricted Sox13 transcription in a subset of individual NKT17 cells (Engel et al., 2016).
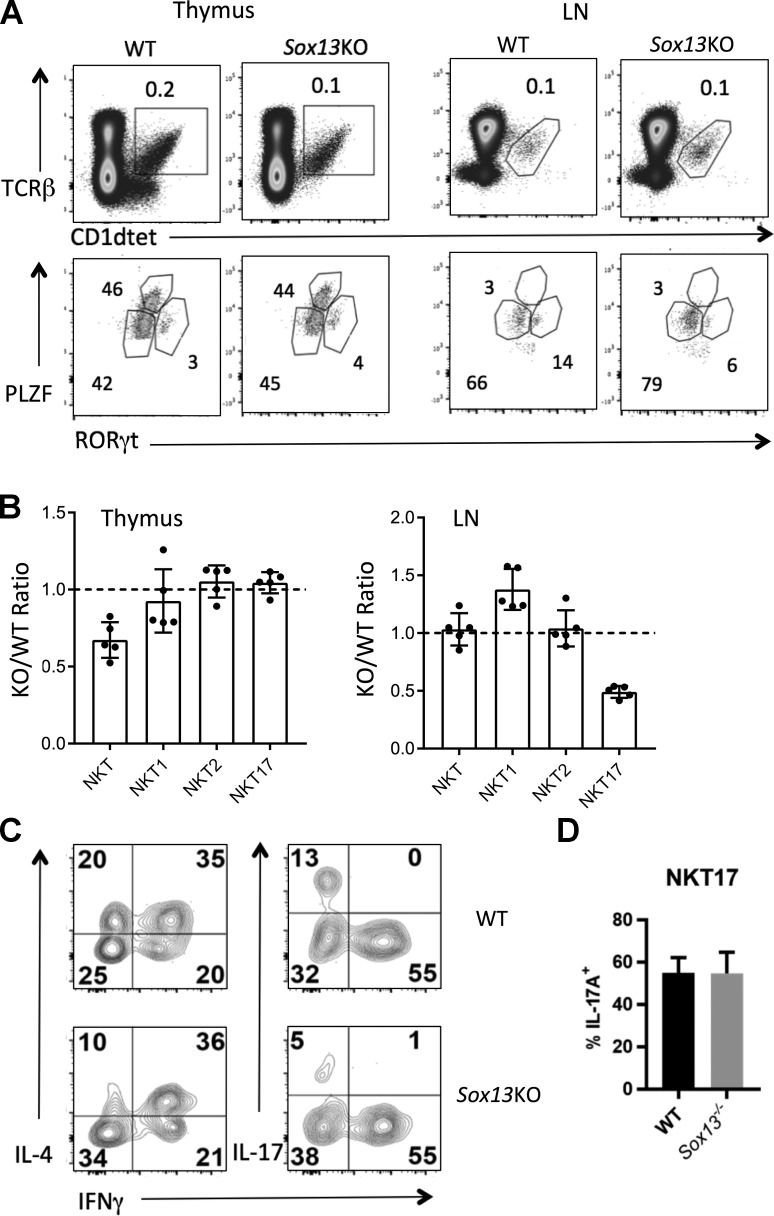
Sox13−/− (129/J) mice generated an approximately twofold-decreased number of total iNKT cells in the thymus (Fig. 7, A and B), but the numbers equilibrated in the LNs. Given that SOX13 institutes the gene network of Tγδ17 cells, a similar role was considered for NKT17 cells. A focused analysis of published NKT single-cell transcriptome data (Engel et al., 2016) for the expression of Tγδ17 cell gene signatures, such as Blk, Scart2 (5830411N06Rik), Aqp3, Camk2d, Gpr183, and Sox13, revealed a strong overlap between Tγδ17 and NKT17 cells (Fig. S5 C). Sox13−/− mice did not have a biased defect in NKT17 or RORγt expression in the thymus, but there was a twofold decrease in the number of RORγt+ NKT17 cells in peripheral LNs (Fig. 7, A and B) and the skin (data not shown) and concomitant decrease in IL-17 production in LN iNKT cells (Fig. 7 C). Similar to the case for IFNγ production by residual NKT1 cells in T-Sox4−/− mice (Fig. S2 D), IL-17A production by residual RORγt+ NKT17 cells in Sox13−/− mice was comparable to controls (Fig. 7 D). Combined with the expression of Sox13 in developing iNKT17 cells in the thymus, but not extrathymically (Immunological Genome Project Consortium, 2018; data not shown), results suggest that the reduction in the frequency of IL-17A+ iNKT cells originates from the thymus. While IFNγ production from iNKT cells was normal in the absence of Sox13, there was a twofold decrease in the cells that can produce IL-4 only (Fig. 7 C). This partial, effector subtype dependency on Sox13 could be specific to iNKT cells among innate-like αβ T cell subsets, as there were no significant alterations in MAIT cells in various tissues of Sox13−/− mice (data not shown). These results indicate that some NKT17 cells programmed to express Sox13 may not populate or expand in the extrathymic tissues. Hence, development of the two innate-like T cell lineages, αβTCR+ iNKT cells and γδ T cell subsets, is characterized by the shared requirement for the quartet of HMG box TFs, but the gene regulatory domains of the quartet in the T cell lineages are only partially overlapping, exhibiting considerable lineage specificity.
Figure 7.
SOX13 regulates NKT17 cell development. (A) Representative profiles showing the reduction of thymic iNKT cells and LN RORγt+ NKT17 cells in Sox13−/− (KO, ∼3–4 wk old, 129/J background) and aged-matched 129/J WT mice. One of two experiments. (B) Summary of the frequencies of total iNKT cells and NKT cell subsets defined by TFs in the thymus and LN of KO relative to WT mice. Each dot represents an individual mouse. Total tissue cellularity of KO and WT mice was indistinguishable. Data are combined from two independent experiments. (C) Representative profiles of effector cytokine (IL-4, IL-17A, and IFNγ) production by LN iNKT cells from WT and KO mice as measured by intracellular staining. One of two experiments with a minimum of n = 3/genotype is shown. (D) Thymic iNKT cells were stimulated with PMA and Ionomycin for simultaneous analysis of TFs and cytokine production. NKT17 cells were gated as live, B220/CD19−, TCRβ+, mCD1d-PBS57+, and RORγt+T-bet−. Data are summarized from two independent assays. Error bars denote SD.
Discussion
While the TF network driving intrathymic iNKT and MAIT cell after TCR-dependent selection is well characterized, there is less known about TFs in DP thymocytes that direct the differentiation of innate-like αβ T cells, in particular those that do not impact the development conventional αβ T cells. RORγt and RUNX1 have been identified as the earliest TFs necessary for iNKT cell generation from DP cells (Guo et al., 2002; Egawa et al., 2005; Thapa et al., 2017). Data here indicate that Sox4 also functions in DP cells to regulate iNKT cell development, in part by controlling Mir181 expression and tuning TCR signaling. Unlike most NKT effector lineage determining TFs, Sox4 expression is relatively high in DP thymocytes and severely down-regulated in CD1dTetramer+CD24neg cells (Fig. S5 C; Engel et al., 2016; Immunological Genome Project Consortium, 2018), supporting its primary function during the effector acquisition stage.
Recent studies (Narayan et al., 2012; Cohen et al., 2013; Jojic et al., 2013; Yin et al., 2013) have established that iNKT cells are more related in transcriptomes and function to innate NK and γδ T cells than to conventional αβ T cells. The shared operational domain of the HMG box TFs with γδ T cells elaborated here can partly account for the lineage relatedness at the gene expression level. However, the mechanistic basis for this relatedness remains to be determined. In its most streamlined model, the mechanism has been linked to the instructive function of TCR to specify T cell lineage choice. To date, most developmental studies on iNKT cells have focused on gene networks found in conventional T cell development. This was a natural progression, given the importance of the invariant TCR-mediated intrathymic selection of iNKT cells. Most molecules identified that block iNKT maturation are involved in strong TCR signaling (e.g., CD3 components, ZAP70, ITK [Arase et al., 1995; Iwabuchi et al., 2001; Felices and Berg, 2008], and EGR2 [Lazarevic et al., 2009]) or accessory molecules mediating DP–DP cell interactions (e.g., SLAM-SAP; Griewank et al., 2007). Hence, the acquisition of innate lymphoid properties of iNKT cells was linked to the unique TCR-mediated positive selection. With the identification of PLZF as the essential relay between the TCR and iNKT and MAIT effector cytokine signatures (Kovalovsky et al., 2008; Savage et al., 2008), the TCR-driven lineage divergence became coupled to induction of TFs that can impose functions characteristic of innate lymphocytes.
However, PLZF expression is skewed toward progenitors arising during murine fetal to neonatal stages that express Lin28b (Yuan et al., 2012). This regulator of MicroRNAs is expressed only in fetal hematopoietic progenitors and is responsible for turning on PLZF and other molecules associated with innate lymphocyte function. Moreover, the T cell lineage specification HMG TF TCF1 (Weber et al., 2011) represses Zbtb16 transcription, and PLZF+TCRneg thymic precursors do exist (data not shown), further supporting the uncoupling of Zbtb16 expression from TCR signaling per se. PLZF is selectively expressed in a subset of γδTCR+ NKT cells that express a near canonical receptor (Vγ1.1+Vδ6.3+) and share overlapping function and global gene expression profiles with αβ iNKT cells (Felices et al., 2009; Kreslavsky et al., 2009; Narayan et al., 2012; Cohen et al., 2013). Given the overall similarity between the subsets, it can be predicted that the quality of the NKT γδTCR and αβTCR signaling is convergent, if the TCR signaling primarily dictates cell lineage choice. However, TCR signaling requirements for the subsets are bipolar: αβ iNKT cells are absent or reduced in Cd247 (CD3ζ)−/−, Fyn−, and Itk−/− mice, but γδ NKT cells are increased in number in the mutants (Arase et al., 1995; Gadue et al., 1999; Felices and Berg, 2008; Felices et al., 2009; Yin, C., personal communication) or not altered compared with WT. Similarly, Sox4 or Mir181 deficiency impairs αβ iNKT, but not γδNKT cell differentiation (Sandrock et al., 2015). Although some of these molecules (e.g., FYN) may not exclusively convey TCR signals, these results are nevertheless incompatible with the “convergent TCR signaling” model to PLZF+ innate-like T cell differentiation from precursors.
An alternate mechanistic model of innate T cell development proposes that divergent instructive inputs lead to convergent outputs such that distinct cell-extrinsic cues can turn on specific T cell effector gene circuitries. As an illustration, distinct TCR signaling quality and/or quantity can be binned into on/off states of TFs that define T cell effector subtypes, but one signal input (e.g., TCR) can be substituted by other inputs (e.g., cytokines), if the underlying gene circuitry is already at different basal set points in distinct T cell types. The Tγδ17 gene program may represent one end of the spectrum where prewired gene programs dictate T cell lineage specification in the thymus, while conventional adaptive T cell development lies at the opposite end where TCR signaling is the ultimate arbiter. In this scheme, iNKT cells may hew more closely to the middle, where gene programs centered on the HMG TFs bias cells’ effector fates, but the activity of the TFs is integrated with TCR signaling. One consequence of this integration is that the shared core TF networks do not operate the same in different cell types. For illustration, SOX4 is dispensable for Rorc transcription in DP cells or NKT17 cells, whereas it is essential for Rorc transcription in γδ T cells (Malhotra et al., 2013). Conversely, SOX4 inhibits Th2 cell differentiation (Kuwahara et al., 2012), whereas SOX4 is necessary for IL-4 production from iNKT cells.
SOX4 is a unique TF that can modify TCR signaling during development by inducing Mir181a/b-1 and controlling other genes essential for iNKT cell selection (e.g., Cd1d) in DP cells. Mir181 has been previously demonstrated to enhance TCR signals in DP cells, and its germline deletion is detrimental for iNKT cell development (Li et al., 2007; Henao-Mejia et al., 2013; Ziętara et al., 2013). That a defect in Mir181 expression in T-Sox4−/− mice does not lead to perceptible alterations in conventional αβ T cell selection can be accounted for by two distinct processes. In conventional adaptive T cell development, the expansive breadth of TCR repertoire may tolerate altered TCR signaling capacity, while the iNKT’s constrained repertoire is not capable of the same compensation. Alternatively, iNKT cells may primarily arise from DP cells that stochastically express highest levels of SOX4 and Mir181. This notion of a preexisting clonal heterogeneity among DP cells, and attendant cell intrinsic variability in TCR signaling sensitivity that biases some DP cells toward specific effector subtypes, will be tested using Sox4 reporter mice we have generated.
In addition to the overall relatedness of NKT cells and γδ T cells, there is a remarkable conservation of gene expression signatures between Tγδ17 and NKT17 cells, including the expression of genes such as Sox13, Blk, and Scart2 that were previously thought to be γδ T cell lineage restricted. While the possibility that the T–T interactions underpinning NKT cell thymic selection mimic some aspects of preprogrammed Tγδ17 cell differentiation cannot be formally ruled out, we have recently discovered that there exists a dedicated, TCR signaling–independent progenitor subset that gives rise to Tγδ17 cells (Spidale et al., 2018). Thus, it is possible that some NKT17 cells originate from a distinct precursor cell type than the rest of NKT cells that arise from DP cells. Recently, it has been shown that there exist DN precursors of iNKT cells that are liver tropic and produce IFNγ (Dashtsoodol et al., 2017). Hence, developmental origin of innate-like iNKT cell subsets may be more complex than currently understood, in part accounting for the specific genetic requirements distinct from those for conventional adaptive T cells.
Materials and methods
Mice
Sox4fl/fl, Tcf7−/−, and Cd2476f/6f (all C57BL/6 background) mice were described previously (Hwang et al., 2012; Malhotra et al., 2013). Rorc-Cre and Rag1−/− mice were purchased from Jackson Laboratories. Sox13 promoter–ECFP “knock-in” mice were generated in house, and its characterization is included in Spidale et al. (2018). Mice were generally analyzed at 4–6 wk for thymic developmental studies and 8–12 wk for peripheral iNKT cell persistence and function. Mice were always age and sex matched within an experiment. All experiments were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee.
Flow cytometry
Abs to the following cell surface markers and cytokines were purchased from BD Biosciences or eBioscience: CD3, CD19, CD11b, Gr1, NK1.1, CD4, CD8, CD25, CD44, c-Kit, TCRβ, TCRδ, Vδ6.3, CCR6, CD24, IL-7R, CD27, CD45.1, IL-4, IL-17A, IFNγ, streptavidin-APC, streptavidin-APCcy7, and streptavidin-PE-Cy7. CD1d-PBS57 or αGC tetramers and control unloaded CD1d tetramers and MR1 tetramers loaded with 5-A-RU and methylglyoxal (resulting in the ligand 5-OP-RU) and control MR1 tetramers loaded with 6-FP were obtained from the National Institutes of Health (NIH) Tetramer Core Facility. Intranuclear staining was performed using the FOXP3 staining kit (eBioscience) for the following Abs: TCF1 and LEF1 (both from Cell Signaling); EGR2, T-bet, and GATA3 (all from eBioscience); Ki67 (BD Biosciences); RORγt (eBioscience); PLZF (Santa Cruz Biotechnology); and GFP Tag (Thermo). Anti-rabbit FITC Ab purchased from Molecular Probe (Invitrogen) was used for staining of LEF1 and TCF1. For PLZF staining, anti-mouse FITC or APC Abs purchased from eBioscience were used. Donkey anti-rabbit IgG (Jackson ImmunoResearch) was used as the secondary for anti-GFP Ab. Secondary Abs were screened based on no spurious staining of cells. Thymocyte viability was assessed using Annexin V (Leinco Technologies) and propidium iodide (Invitrogen) staining. Cells were labeled with a fixable viability dye (Invitrogen) to facilitate exclusion of dead cells during analysis. Data were acquired on a BD LSRII cytometer and were analyzed using FlowJo (Treestar). NKT cells were routinely identified by gating on forward scatter/side scatter lymphocyte population, doublet exclusion by forward scatter area versus height, viability dye−, B220/CD19−, TCRβ+, and CD1d-PBS57+. To detect cytokines, ex vivo thymic and LN cells were cultured (2 × 106/well) with PMA (10 ng/ml) and ionomycin (1 µg/ml) for 4 h at 37°C, with Golgi Stop and Golgi Plug (BD Biosciences) added after 1 h, according to the manufacturer’s protocol. After stimulation, cells were stained for cell surface markers and intracellular cytokine production after permeabilization using the Cytofix/Cytoperm kit (BD Biosciences) or, for simultaneous detection of TFs and cytokines, using eBiosicence Transcription Factor Staining Buffer Set. Gates for cytokine+ cells were always based on nonstimulated control cells stained with anti-cytokine Abs.
In vivo functional assays
αGC (KRN7000) was purchased from Cayman Chemical and dissolved in DMSO, and aliquots were stored at −20°C. Thawed aliquots were not refrozen or reused beyond the day of thawing. Mice were injected intravenously with 2 µg αGC diluted in PBS or Vehicle (Veh; 1% DMSO in PBS), and then splenocytes were analyzed at the time points after injection indicated in the figure legends. For assessment of iNKT cell and NK cell cytokine production after in vivo αGC administration, spleens were processed rapidly on ice and stained immediately without additional culture before fixation/permeabilization and intracellular staining as described above.
BM chimeras and retroviral transduction
5 × 106 mixed BM cells from congenic WT (Ly5.1/CD45.1):Sox4 CKO mice (ratios ranging from 4:1 to 1:4) or control Ly5.1 WT:Ly5.2 WT mice were used to reconstitute sublethally irradiated Rag1−/− or lethally irradiated C57BL/6 (Ly5.2) mice. Chimeras were sacrificed 10–14 wk after BM cell transfer. Mir181a-1 coding sequence (a gift from Q.-J. Li, Duke University, Durham, NC) was cloned into MSCV-IRES-GFP plasmid and used to infect BM progenitor enriched cells (by 5-FU injection 3 d before cell isolation) from CD45.1 congenic C57BL/6 mice per standard protocol. Infected BM cells were used to reconstitute sublethally irradiated Rag1−/− or lethally irradiated C57BL/6 (CD45.2) mice. Similar results were observed in both types of host. Reconstitution was monitored by the emergence of GFP+ cells in the blood, and mice were sacrificed 8–16 wk after BM cell transfer. Thymocytes and LN cells from WT and Sox4-deficient BM cells with or without exogenous Mir181a-1 expression were assessed for iNKT cell differentiation by flow cytometry.
Ca2+ flux measurement
Intracellular Ca2+ flux was determined by first activating thymocytes using biotinylated αCD3 Ab, followed by cell surface staining for CD4 and CD8 and loading with 16 µM Fluo-3 (FL-1) and 16 µM Fura Red (FL-3). Baseline Ca2+ was taken for 2 min before stimulation via the TCR cross-linking with 20 µg/ml streptavidin for 8 min. Intracellular [Ca2+] was determined by gating on DP thymocytes and calculating the mean fluorescence ratio of FL1:FL3 using FlowJo.
TCR rearrangement in genomic DNA
For analysis of Vα14-Jα18 TCR gene rearrangement in genomic DNA, DP thymocytes were sorted and genomic DNA isolated using the PureLink Genomic DNA Mini kit (Invitrogen). PCR was performed using Taq polymerase (Invitrogen) and the following thermal cycling conditions: 94°C, 3 min; 35 cycles of 94°C, 30 s; 55°C, 30 s; and 72°C, 30 s. PCR products were immediately electrophoresed on a 1.5% agarose gel and DNA visualized by ethidium bromide staining. The primer sequences used were Va14 F, 5′-GTTGTCCGTCAGGGAGAGAA-3′; Ja281 R, 5′-TCCCTAAGGCTGAACCTCTATC-3′; β-Actin F, 5′-ATGAAGATCCTGACCGAGCG-3′; and β-Actin R, 5′-TACTTGCGCTCAGGAGGAGC-3′.
Gene expression analysis and data visualization
DP thymocytes from three pooled male mice per genotype were sorted to >99% purity in three independent experiments. RNA processing from sorted thymocyte subsets and microarray analysis using the Gene1.0 ST array (Affymetrix) was performed at the ImmGen processing center (SOP, standard operating procedure; Immunological Genome Project Consortium, 2018). Microarray data analysis was performed using GenePattern modules (Broad Institute) as described in depth previously (Narayan et al., 2012). In brief, Multiplot (GenePattern) was used to identify differentially regulated genes and to generate Volcano plots. Genes were considered differentially regulated if they differed in expression by more than twofold, had a coefficient of variation (cv) among replicates of <0.5, and had a Student’s t test P value of <0.1. Raw data are available in the NCBI GEO database (GSE120477). For iNKT single-cell RNaseq data reanalysis, 203 single-cell sequencing data (GSE74597) were analyzed using R (package DEseq2). Heat map was read counts normalized from 203 single cells. Row-wise clustering and column-wise scale was applied using R package “pheatmap.” The gene list was based on the Tγδ17 gene signature at population (Narayan et al., 2012) and sc level (Spidale et al., 2018). Expression of select genes in iNKT subsets was illustrated as dot plots, with each dot representing one cell.
ChIPs
ChIP analysis was performed on total thymocytes (1–5 × 106) using the ChIP Assay kit (Millipore) following the manufacturer’s protocol. In brief, after de–cross-linking, DNA from the DNA–protein bead complexes was eluted and detected by qPCR (iQ SYBR Green Supermix and Bio-Rad iCycler) with specific primers for each gene target regulatory regions. Abs used were anti (α)-SOX4 (SCB; Santa Cruz Biotechnology), αH3K4me3 (17-614; Millipore), αH3K9me3 (17-625; Millipore), and αH3K27me3 (ab-6002; Abcam). Specificities of Abs against TFs were verified in T cell lines expressing transduced or endogenous target genes compared with nonexpressors. Reference genome was the mouse mm9/NCBI37 assembly. PCR primer locations were based on two TF search tools (Computational Biology Research Consortium and Genomatix) with assessment of the homology among species by www.decode.org and designed in Primer-Blast (National Center for Biotechnology Information).
Online supplemental material
Fig. S1 describes the efficiency of Cre-mediated recombination in T-Sox4−/− mice and additional details on T and NKT cell phenotype of T-Sox4−/− mice. Fig. S2 includes further characterization of residual iNKT cells in T-Sox4−/− mice, based on classical NKT cell developmental stages and cytokine production. Functional overlap with TCF1 and SOX4 in iNKT cells is also delved into. Fig. S3 focuses on residual function of iNKT cells in T-Sox4−/− mice after αGC treatments. Fig. S4 tests candidate molecules of iNKT cell development and TCR signaling that are modulated by SOX4. Fig. S5 shows Sox13 reporter expression in iNKT cells and reanalysis of published single-cell RNaseq data, centered on the Tγδ17 cell gene signature.
Supplementary Material
Acknowledgments
We are grateful to Qi-Jing Li (Duke University, Durham, NC) for Mir181 expression vectors, Takeshi Egawa (University of Washington, St. Louis, MO) for experiments to demonstrate that Sox4 expression is unaltered in c-Myc–deficient mice, and Paul Love (NIH, Bethesda, MD) for 6F mice.
This work was supported by NIH grants R21 AI07551 and RO1 AI01301 (to J. Kang).
The authors declare no competing financial interests.
Author contributions: N. Malhorta performed foundational experiments, analyzed data, prepared figures, and wrote the paper; Y. Qi performed experiments, analyzed data, and prepared figures; N.A. Spidale performed experiments, analyzed data, and prepared figures; M. Frascoli performed experiments, analyzed data, and prepared figures; B. Miu performed experiments, analyzed data, and prepared figures; O. Cho performed ChIP experiments, analyzed data, and prepared figures; K. Sylvia performed experiments, analyzed data, and prepared figures; J. Kang analyzed data, wrote the paper, and initiated and directed the study.
References
- Arase H., Ono S., Arase N., Park S.Y., Wakizaka K., Watanabe H., Ohno H., and Saito T.. 1995. Developmental arrest of NK1.1+ T cell antigen receptor (TCR)-alpha/beta+ T cells and expansion of NK1.1+ TCR-gamma/delta+ T cell development in CD3 zeta-deficient mice. J. Exp. Med. 182:891–895. 10.1084/jem.182.3.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew D., and Harding C.V.. 2008. Antigen processing and CD24 expression determine antigen presentation by splenic CD4+ and CD8+ dendritic cells. Immunology. 123:447–455. 10.1111/j.1365-2567.2007.02711.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam H.S., Grinberg A., Lui K., Shen H., Shores E.W., and Love P.E.. 1998. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188:2301–2311. 10.1084/jem.188.12.2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnaud C., Lee D., Donnars O., Park S.-H., Beavis A., Koezuka Y., and Bendelac A.. 1999. Cutting edge: Cross-talk between cells of the innate immune system: NKT cells rapidly activate NK cells. J. Immunol. 163:4647–4650. [PubMed] [Google Scholar]
- Carr T., Krishnamoorthy V., Yu S., Xue H.H., Kee B.L., and Verykokakis M.. 2015. The transcription factor lymphoid enhancer factor 1 controls invariant natural killer T cell expansion and Th2-type effector differentiation. J. Exp. Med. 212:793–807. 10.1084/jem.20141849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.Y., Zhang T., Pincus S.H., Wu S., Ricks D., Liu D., Sun Z., Maclaren N., and Lan M.S.. 2010. Human CD1D gene expression is regulated by LEF-1 through distal promoter regulatory elements. J. Immunol. 184:5047–5054. 10.4049/jimmunol.0901912 [DOI] [PubMed] [Google Scholar]
- Cohen N.R., Brennan P.J., Shay T., Watts G.F., Brigl M., Kang J., and Brenner M.B.. ImmGen Project Consortium . 2013. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat. Immunol. 14:90–99. 10.1038/ni.2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz Tleugabulova M., Escalante N.K., Deng S., Fieve S., Ereño-Orbea J., Savage P.B., Julien J.P., and Mallevaey T.. 2016. Discrete TCR Binding Kinetics Control Invariant NKT Cell Selection and Central Priming. J. Immunol. 197:3959–3969. 10.4049/jimmunol.1601382 [DOI] [PubMed] [Google Scholar]
- D’Cruz L.M., Knell J., Fujimoto J.K., and Goldrath A.W.. 2010. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat. Immunol. 11:240–249. 10.1038/ni.1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashtsoodol N., Shigeura T., Aihara M., Ozawa R., Kojo S., Harada M., Endo T.A., Watanabe T., Ohara O., and Taniguchi M.. 2017. Alternative pathway for the development of Vα14+ NKT cells directly from CD4-CD8- thymocytes that bypasses the CD4+CD8+ stage. Nat. Immunol. 18:274–282. 10.1038/ni.3668 [DOI] [PubMed] [Google Scholar]
- Egawa T., Eberl G., Taniuchi I., Benlagha K., Geissmann F., Hennighausen L., Bendelac A., and Littman D.R.. 2005. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 22:705–716. 10.1016/j.immuni.2005.03.011 [DOI] [PubMed] [Google Scholar]
- Engel I., Seumois G., Chavez L., Samaniego-Castruita D., White B., Chawla A., Mock D., Vijayanand P., and Kronenberg M.. 2016. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat. Immunol. 17:728–739. 10.1038/ni.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felices M., and Berg L.J.. 2008. The Tec kinases Itk and Rlk regulate NKT cell maturation, cytokine production, and survival. J. Immunol. 180:3007–3018. 10.4049/jimmunol.180.5.3007 [DOI] [PubMed] [Google Scholar]
- Felices M., Yin C.C., Kosaka Y., Kang J., and Berg L.J.. 2009. Tec kinase Itk in gammadeltaT cells is pivotal for controlling IgE production in vivo. Proc. Natl. Acad. Sci. USA. 106:8308–8313. 10.1073/pnas.0808459106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadue P., Morton N., and Stein P.L.. 1999. The Src family tyrosine kinase Fyn regulates natural killer T cell development. J. Exp. Med. 190:1189–1196. 10.1084/jem.190.8.1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapin L., Matsuda J.L., Surh C.D., and Kronenberg M.. 2001. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat. Immunol. 2:971–978. 10.1038/ni710 [DOI] [PubMed] [Google Scholar]
- Godfrey D.I., Stankovic S., and Baxter A.G.. 2010. Raising the NKT cell family. Nat. Immunol. 11:197–206. 10.1038/ni.1841 [DOI] [PubMed] [Google Scholar]
- Griewank K., Borowski C., Rietdijk S., Wang N., Julien A., Wei D.G., Mamchak A.A., Terhorst C., and Bendelac A.. 2007. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 27:751–762. 10.1016/j.immuni.2007.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Hawwari A., Li H., Sun Z., Mahanta S.K., Littman D.R., Krangel M.S., and He Y.W.. 2002. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat. Immunol. 3:469–476. 10.1038/ni791 [DOI] [PubMed] [Google Scholar]
- Henao-Mejia J., Williams A., Goff L.A., Staron M., Licona-Limón P., Kaech S.M., Nakayama M., Rinn J.L., and Flavell R.A.. 2013. The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity. 38:984–997. 10.1016/j.immuni.2013.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzenberg L.A., and Herzenberg L.A.. 1989. Toward a layered immune system. Cell. 59:953–954. 10.1016/0092-8674(89)90748-4 [DOI] [PubMed] [Google Scholar]
- Hwang S., Song K.D., Lesourne R., Lee J., Pinkhasov J., Li L., El-Khoury D., and Love P.E.. 2012. Reduced TCR signaling potential impairs negative selection but does not result in autoimmune disease. J. Exp. Med. 209:1781–1795. 10.1084/jem.20120058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immunological Genome Project Consortium 2018. Available at: immgen.org (accessed September 1, 2018).
- Iwabuchi K., Iwabuchi C., Tone S., Itoh D., Tosa N., Negishi I., Ogasawara K., Uede T., and Onoé K.. 2001. Defective development of NK1.1+ T-cell antigen receptor alphabeta+ cells in zeta-associated protein 70 null mice with an accumulation of NK1.1+ CD3- NK-like cells in the thymus. Blood. 97:1765–1775. 10.1182/blood.V97.6.1765 [DOI] [PubMed] [Google Scholar]
- Jojic V., Shay T., Sylvia K., Zuk O., Sun X., Kang J., Regev A., Koller D., Best A.J., Knell J., et al. Immunological Genome Project Consortium . 2013. Identification of transcriptional regulators in the mouse immune system. Nat. Immunol. 14:633–643. 10.1038/ni.2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., and Malhotra N.. 2015. Transcription factor networks directing the development, function, and evolution of innate lymphoid effectors. Annu. Rev. Immunol. 33:505–538. 10.1146/annurev-immunol-032414-112025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L., Patel O., Corbett A.J., Le Nours J., Meehan B., Liu L., Bhati M., Chen Z., Kostenko L., Reantragoon R., et al. 2012. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 491:717–723. 10.1038/nature11605 [DOI] [PubMed] [Google Scholar]
- Koay H.F., Gherardin N.A., Enders A., Loh L., Mackay L.K., Almeida C.F., Russ B.E., Nold-Petry C.A., Nold M.F., Bedoui S., et al. 2016. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat. Immunol. 17:1300–1311. 10.1038/ni.3565 [DOI] [PubMed] [Google Scholar]
- Kovalovsky D., Uche O.U., Eladad S., Hobbs R.M., Yi W., Alonzo E., Chua K., Eidson M., Kim H.J., Im J.S., et al. 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9:1055–1064. 10.1038/ni.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavsky T., Savage A.K., Hobbs R., Gounari F., Bronson R., Pereira P., Pandolfi P.P., Bendelac A., and von Boehmer H.. 2009. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc. Natl. Acad. Sci. USA. 106:12453–12458. 10.1073/pnas.0903895106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara M., Yamashita M., Shinoda K., Tofukuji S., Onodera A., Shinnakasu R., Motohashi S., Hosokawa H., Tumes D., Iwamura C., et al. 2012. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-β and suppresses T(H)2 differentiation. Nat. Immunol. 13:778–786. 10.1038/ni.2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V., Zullo A.J., Schweitzer M.N., Staton T.L., Gallo E.M., Crabtree G.R., and Glimcher L.H.. 2009. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat. Immunol. 10:306–313. 10.1038/ni.1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Holzapfel K.L., Zhu J., Jameson S.C., and Hogquist K.A.. 2013. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14:1146–1154. 10.1038/ni.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Starrett G.J., Lee S.T., Yang R., Henzler C.M., Jameson S.C., and Hogquist K.A.. 2016. Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst γδ T, Innate Lymphoid, and Th Cells. J. Immunol. 197:1460–1470. 10.4049/jimmunol.1600643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.J., Chau J., Ebert P.J., Sylvester G., Min H., Liu G., Braich R., Manoharan M., Soutschek J., Skare P., et al. 2007. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 129:147–161. 10.1016/j.cell.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Lynch L., Michelet X., Zhang S., Brennan P.J., Moseman A., Lester C., Besra G., Vomhof-Dekrey E.E., Tighe M., Koay H.F., et al. 2015. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nat. Immunol. 16:85–95. 10.1038/ni.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra N., Narayan K., Cho O.H., Sylvia K.E., Yin C., Melichar H., Rashighi M., Lefebvre V., Harris J.E., Berg L.J., and Kang J.. Immunological Genome Project Consortium . 2013. A network of high-mobility group box transcription factors programs innate interleukin-17 production. Immunity. 38:681–693. 10.1016/j.immuni.2013.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar H.J., Narayan K., Der S.D., Hiraoka Y., Gardiol N., Jeannet G., Held W., Chambers C.A., and Kang J.. 2007. Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science. 315:230–233. 10.1126/science.1135344 [DOI] [PubMed] [Google Scholar]
- Narayan K., Sylvia K.E., Malhotra N., Yin C.C., Martens G., Vallerskog T., Kornfeld H., Xiong N., Cohen N.R., Brenner M.B., et al. Immunological Genome Project Consortium . 2012. Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes. Nat. Immunol. 13:511–518. 10.1038/ni.2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N., Subramanian A., Lawton L.N., Mak R.H., Haining W.N., McConkey M.E., Habib N., Yosef N., Chang C.Y., Shay T., et al. 2011. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 144:296–309. 10.1016/j.cell.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh-hora M., Komatsu N., Pishyareh M., Feske S., Hori S., Taniguchi M., Rao A., and Takayanagi H.. 2013. Agonist-selected T cell development requires strong T cell receptor signaling and store-operated calcium entry. Immunity. 38:881–895. 10.1016/j.immuni.2013.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira P., and Boucontet L.. 2012. Innate NKTγδ and NKTαβ cells exert similar functions and compete for a thymic niche. Eur. J. Immunol. 42:1272–1281. 10.1002/eji.201142109 [DOI] [PubMed] [Google Scholar]
- Ribot J.C., deBarros A., Pang D.J., Neves J.F., Peperzak V., Roberts S.J., Girardi M., Borst J., Hayday A.C., Pennington D.J., and Silva-Santos B.. 2009. CD27 is a thymic determinant of the balance between interferon-gamma- and interleukin 17-producing gammadelta T cell subsets. Nat. Immunol. 10:427–436. 10.1038/ni.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrock I., Ziętara N., Łyszkiewicz M., Oberdörfer L., Witzlau K., Krueger A., and Prinz I.. 2015. MicroRNA-181a/b-1 Is Not Required for Innate γδ NKT Effector Cell Development. PLoS One. 10:e0145010 10.1371/journal.pone.0145010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., and Bendelac A.. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403. 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilham M.W., Oosterwegel M.A., Moerer P., Ya J., de Boer P.A., van de Wetering M., Verbeek S., Lamers W.H., Kruisbeek A.M., Cumano A., and Clevers H.. 1996. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 380:711–714. 10.1038/380711a0 [DOI] [PubMed] [Google Scholar]
- Schilham M.W., Moerer P., Cumano A., and Clevers H.C.. 1997. Sox-4 facilitates thymocyte differentiation. Eur. J. Immunol. 27:1292–1295. 10.1002/eji.1830270534 [DOI] [PubMed] [Google Scholar]
- Shulewitz M., Soloviev I., Wu T., Koeppen H., Polakis P., and Sakanaka C.. 2006. Repressor roles for TCF-4 and Sfrp1 in Wnt signaling in breast cancer. Oncogene. 25:4361–4369. 10.1038/sj.onc.1209470 [DOI] [PubMed] [Google Scholar]
- Sinner D., Kordich J.J., Spence J.R., Opoka R., Rankin S., Lin S.C., Jonatan D., Zorn A.M., and Wells J.M.. 2007. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol. Cell. Biol. 27:7802–7815. 10.1128/MCB.02179-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spidale N.A., Sylvia K., Narayan K., Miu B., Frascoli M., Melichar H.J., Zhihao W., Kisielow J., Palin A., Serwold T., et al. 2018. Interleukin-17 producing γδ T cells originate from SOX13+ progenitors that are independent of γδTCR signaling. Immunity. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa P., Manso B., Chung J.Y., Romera Arocha S., Xue H.H., Angelo D.B.S., and Shapiro V.S.. 2017. The differentiation of ROR-γt expressing iNKT17 cells is orchestrated by Runx1. Sci. Rep. 7:7018 10.1038/s41598-017-07365-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B.N., Chi A.W., Chavez A., Yashiro-Ohtani Y., Yang Q., Shestova O., and Bhandoola A.. 2011. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 476:63–68. 10.1038/nature10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C.C., Cho O.H., Sylvia K.E., Narayan K., Prince A.L., Evans J.W., Kang J., and Berg L.J.. 2013. The Tec kinase ITK regulates thymic expansion, emigration, and maturation of γδ NKT cells. J. Immunol. 190:2659–2669. 10.4049/jimmunol.1202531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Nguyen C.K., Liu X., Kanellopoulou C., and Muljo S.A.. 2012. Lin28b reprograms adult BM hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 335:1195–1200. 10.1126/science.1216557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziętara N., Łyszkiewicz M., Witzlau K., Naumann R., Hurwitz R., Langemeier J., Bohne J., Sandrock I., Ballmaier M., Weiss S., et al. 2013. Critical role for miR-181a/b-1 in agonist selection of invariant natural killer T cells. Proc. Natl. Acad. Sci. USA. 110:7407–7412. 10.1073/pnas.1221984110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.