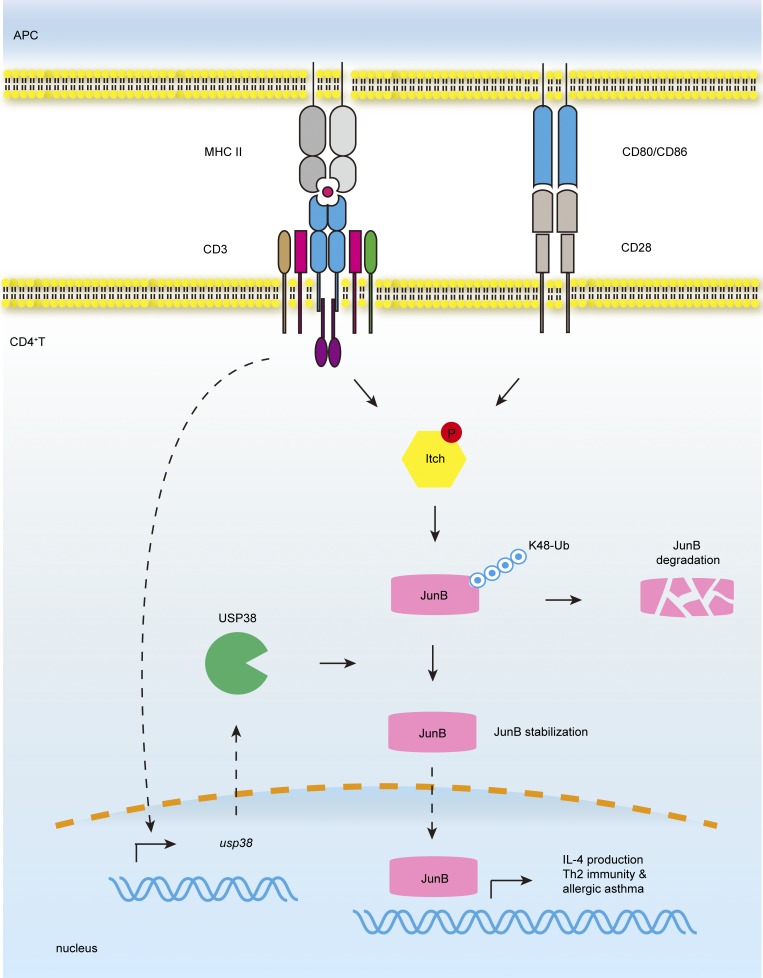
Although usp38 has recently been reported to be in a chromosome locus associated with human asthma in a GWAS study, its potential pathological role remains unknown. Chen et al. now demonstrate that usp38 is essential for asthmatic pathogenesis. USP38 is induced by TCR signaling and in turn promotes JunB stabilization to specifically regulate Th2 cell differentiation.
Abstract
Th2 immune response is critical for allergic asthma pathogenesis. Molecular mechanisms for regulating Th2 immunity are still not well understood. Here we report that the ubiquitin-specific protease USP38 is crucial for Th2-mediated allergic asthma. TCR stimulation up-regulated the USP38 level, and USP38 in turn mediated the protein stabilization of JunB, a transcription factor specific for Th2 development. Consequently, USP38 was specifically required for TCR-induced production of Th2 cytokines and Th2 development both in vitro and in vivo, and USP38-deficient mice were resistant to asthma pathogenesis induced by OVA or HDM. Mechanistically, USP38 directly associated with JunB, deubiquitinated Lys-48–linked poly-ubiquitination of JunB, and consequently blocked TCR-induced JunB turnover. USP38 represents the first identified deubiquitinase specifically for Th2 immunity and the associated asthma.
Graphical Abstract
Introduction
Asthma is a common pulmonary disease characterized by airway hyper-responsiveness and chronic inflammation (Lambrecht and Hammad, 2015). Th2 cells play a critical role in the pathogenesis of allergic diseases, including asthma, through producing characteristic cytokines IL-4, IL-5, and IL-13 (Fahy, 2015; Nakayama et al., 2017). These cytokines induce Th2 differentiation, eosinophil infiltration, and mucus production, respectively, to promote the airway pathophysiology (Takatsu and Nakajima, 2008; Gour and Wills-Karp, 2015). TCR recognition of cognate antigens trigger its signaling for downstream activation of several transcription factors to induce genes for T cell differentiation and function (Zhu et al., 2010; Brownlie and Zamoyska, 2013; Yamane and Paul, 2013). JunB, one of the TCR-activated transcription factors, plays an essential and specific role for Th2 development through promoting Il4 gene transcription (Li et al., 1999; Hartenstein et al., 2002). However, how the TCR pathway is regulated for Th2 development is not well understood.
Ubiquitination is an important protein modification to regulate signal transduction in T cell activation and differentiation (Hu and Sun, 2016). Some E3 ubiquitin ligases, including Cbl family, GRAIL, and Itch, play critical roles in T cell anergy and tolerance by regulating ubiquitination and degradation of key TCR signaling components (Heissmeyer et al., 2004; Mueller, 2004; Nurieva et al., 2010; Venuprasad, 2010). Itch, a member of Nedd4 family, also regulates Th2 differentiation and function through targeting the transcription factors JunB and c-Jun for ubiquitin-mediated degradation (Fang et al., 2002). JNK-mediated Itch phosphorylation is essential for its E3 ubiquitin ligase activity in the TCR signaling (Gao et al., 2004). Nedd4 family interacting protein-1 (Ndfip1) and Ndfip2 are also involved in JunB ubiquitination and degradation likely through activating the Nedd4 family E3 ligases Itch and Nedd4-2 (Oliver et al., 2006; O’Leary et al., 2016).
Protein ubiquitination is a reversible process tightly regulated by deubiquitinases (DUBs; Nijman et al., 2005). Compared with E3 ubiquitin ligases, the roles of DUBs in the regulation of TCR signaling and function are poorly characterized. Several DUBs, including A20 and CYLD, have been shown to be crucial for T cell activation and function (Reiley et al., 2006; Düwel et al., 2009). So far, there is no report of any DUBs involved in Th2 function. While the Nedd4 family members like Itch and Nedd4-2 are shown to be critical for ubiquitin-mediated degradation of JunB to shut off Th2 immunity (Fang et al., 2002; Heikamp et al., 2014), it is still not yet known whether the JunB ubiquitination and turnover is reversible by DUB. Here we found that TCR activation induced expression of ubiquitin-specific peptidase 38 (USP38), whose gene has been recently reported to be in a chromosome locus associated with human asthma in a genome-wide association study (GWAS; Hirota et al., 2011). We demonstrated that USP38 directly associated with JunB and removed its poly-ubiquitination to block JunB degradation in TCR signaling, thus initiating Th2 differentiation and driving allergic asthma.
Results
USP38 is required for allergic asthma induction
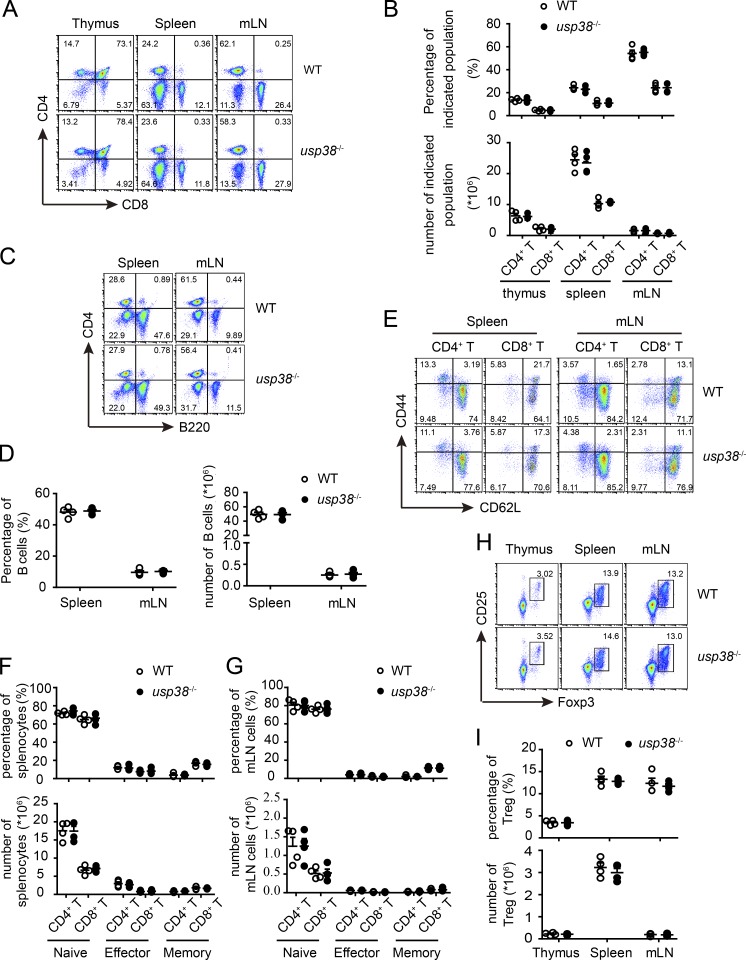
USP38 is a functionally not-characterized DUB (Hanpude et al., 2015) whose gene has been reported in a chromosome locus associated with adult asthma in a GWAS study (Hirota et al., 2011). To study its potential pathophysiological roles, we generated USP38-deficient mice by breeding usp38-floxed mice with E2a-Cre+/− mice (Fig. S1, A and B) and confirmed its complete knockout efficiency by determining its mRNA level by quantitative PCR (qPCR) and its protein level by Western blot (Fig. S1, C and D). The knockout mice were viable and fertile without any observable abnormality. USP38 deficiency did not affect the development of T cells and B cells in thymus, spleen, and mesenteric lymph node during homeostasis (Fig. 1, A–D). The deficiency also did not affect activation status of CD4+ and CD8+ T cells and T reg development (Fig. 1, E–I). Furthermore, we found that the T cell profiles including CD4+ and CD8+ T cell numbers, as well as their activation status in lungs, were not changed by USP38 deficiency (Fig. S2, A–D), indicating that USP38 does not affect the migratory ability of T cells into lungs and their activation there during homeostasis.
Figure 1.
USP38 deficiency does not affect homeostatic immune cell development. All the experiments below were carried on 6–10-wk-old littermates. (A and B) Cytoflow analysis (A) and quantification (B) of CD4- or CD8-positive populations in the indicated tissues out of usp38−/− or WT mice. (C and D) Cytoflow analysis (C) and quantification (D) of B cells in the indicated tissues out of usp38−/− or WT mice. (E–G) Cytoflow analysis (E) and quantification (F and G) of naive (CD62L+CD44−), effector (CD62L−CD44+), and memory (CD62L+CD44+) CD4+ T cell populations, as well as CD8+ T cells, in the indicated tissues out of usp38−/− or WT mice. (H and I) Cytoflow analysis (H) and quantification (I) of T reg cells in the indicated tissues out of usp38−/− or WT mice. Data are representative of four (A–I) independent experiments. Statistical significance was determined by Student’s t test. Error bars indicate the mean ± SEM.
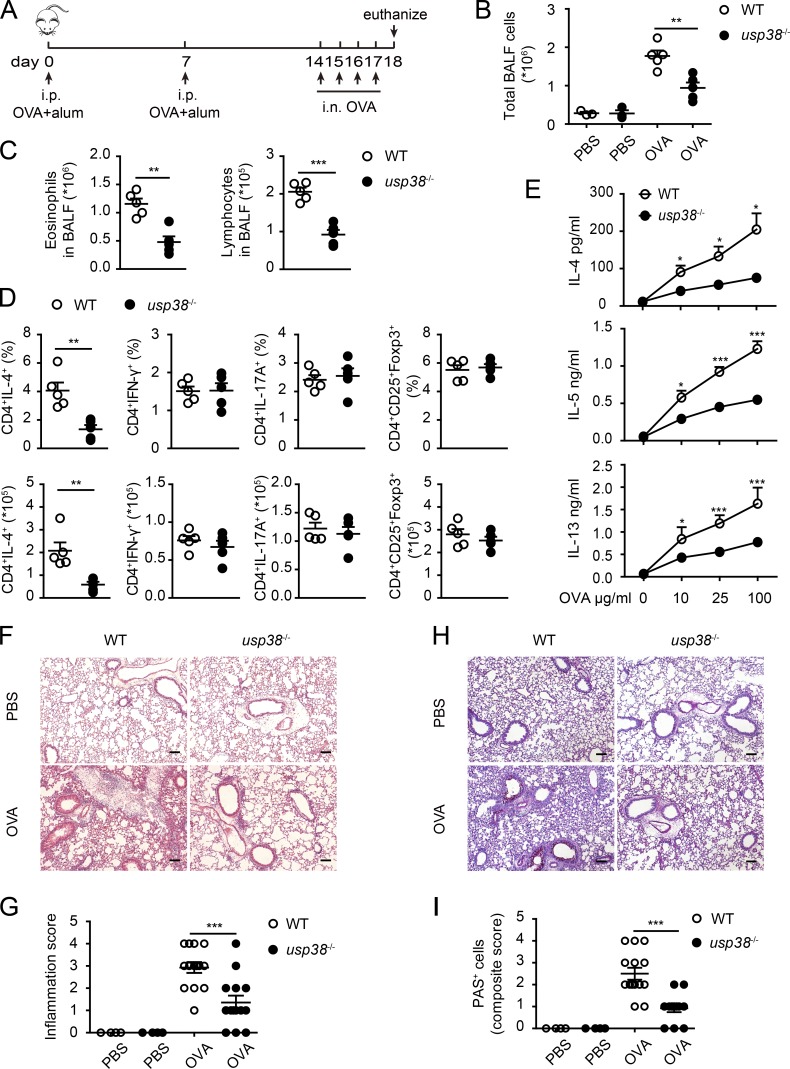
To explore if USP38 has any potential role in asthma pathogenesis, we made use of the OVA + Alum–induced allergic asthma model with the standard induction protocol (Fig. 2 A). USP38 deficiency resulted in marked reduction of total bronchoalveolar lavage fluid (BALF) cells (Fig. 2 B), as well as fewer eosinophils and lymphocytes in the BALF (Fig. 2 C), in the OVA model. To further evaluate T lymphocyte subpopulations, pulmonary mediastinal lymph node cells were collected and stimulated by Ionomycin and PMA, and then analyzed by cytoflow with markers for Th1, Th2, Th17, and T reg populations. We found that USP38 deficiency led to dramatic reduction of the percentage and absolute number of Th2 cells, but did not affect those of Th1, Th17, and T reg cell populations (Fig. 2 D). We then stimulated the pulmonary mediastinal lymph node cells with OVA and checked Th2 cytokines by ELISA. We found that the production of Th2 cytokines IL-4, IL-5, and IL-13 were markedly reduced in the USP38-deficient cells (Fig. 2 E), consistent with the reduced Th2 cells (Fig. 2 D). H&E staining showed that OVA-induced leukocyte infiltration and inflammation were much reduced in the USP38-deficient lungs (Fig. 2, F and G). Similarly, periodic acid-Schiff base (PAS) staining showed that USP38-deficient lungs had significantly decreased OVA-induced goblet cell hyperplasia (Fig. 2, H and I). These data suggest that USP38 is required for OVA-induced allergic asthma.
Figure 2.
USP38 critically promotes OVA-induced asthma. (A–I) 6–8-wk-old female usp38−/− mice and their littermate controls (WT) were analyzed individually. PBS group (n = 3); OVA group (n = 5). (A) A schematic diagram of OVA-induced allergic asthma. (B) Total number of BALF cells. (C) Numbers of eosinophils or lymphocytes in BALF by flow analysis. (D) Percentages and absolute numbers of Th1 (CD4+IFN-γ+), Th2 (CD4+IL-4+), Th17 (CD4+IL-17A+), and T reg (CD4+CD25+Foxp3+) cells in mediastinal lymph nodes (medLNs) from OVA-treated mice. (E) Levels of Th2 cytokines determined by ELISA from culture supernatants of medLNs cells treated with the indicated concentrations of OVA protein for 72 h. (F) H&E staining of lung sections from PBS- or OVA-treated mice. Original magnification is 10×. Bars, 100 µm. (G) Pathological score of perivascular and peribronchiolar inflammation as shown in F. (H) PAS staining of lung sections from PBS- or OVA-treated mice. Original magnification is 10×. Bars, 100 µm. (I) Quantification of goblet cell hyperplasia as shown in H. Data are representative of four (A–C) or two (D–I) independent experiments. Statistical significance was determined by Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate the mean ± SEM.
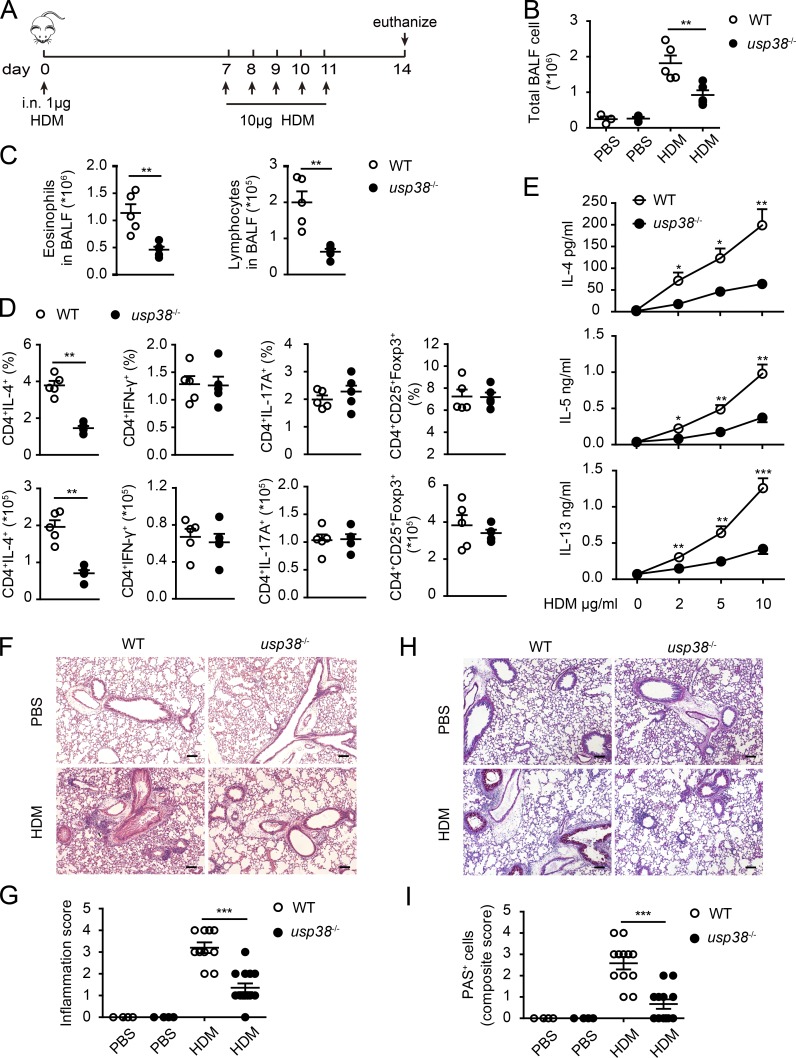
Next, we used house dust mite (HDM) extract, a natural allergen, to induce asthma and evaluate the role of USP38 in the HDM-induced asthmatic pathogenesis with the standard protocol as depicted in Fig. 3 A. Similar to the results in the OVA-induced asthma model (Fig. 2, B and C), the total BALF cells, as well as the eosinophils and lymphocytes in the BALF, were dramatically reduced in USP38-deficient mice in the HDM-induced asthma model (Fig. 3, B and C). Similarly, Th2 cell population, but not the other T cell populations checked (Th1, Th17, and T reg), was affected by USP38 deficiency (Fig. 3 D). Consistently, the secretion of Th2 cytokines (IL-4, IL-5, and IL-13) was dramatically lowered in the USP38-deficient cells (Fig. 3 E). The pathological staining with H&E or PAS also showed much reduced leukocyte infiltration and inflammation, as well as decreased severity of goblet cell hyperplasia, in USP38-deficient lungs in the HDM model (Fig. 3, F–I). Together, these results indicate that USP38 is also essential for HDM-induced allergic asthma.
Figure 3.
USP38 is crucial for HDM-induced asthma. (A–I) 6–8-wk-old female usp38−/− mice and their littermate controls were analyzed individually. PBS group (n = 3); HDM group (n = 5). (A) A schematic diagram of HDM-induced asthma. (B) Total number of BALF cells. (C) Cell counts of eosinophils or lymphocytes in BALF determined by flow analysis. (D) Percentages and absolute numbers of the indicated T cell subpopulations in medLNs. (E) Levels of Th2 cytokines determined by ELISA from Th2 ex vivo recall response. (F) H&E staining of lung sections from PBS- or HDM-treated mice. Original magnification is 10×. Bars, 100 µm. (G) Pathological score of perivascular and peribronchiolar inflammation as shown in F. (H) PAS staining of lung sections from PBS- or HDM-treated mice. Original magnification is 10×. Bars, 100 µm. (I) Quantification of goblet cell hyperplasia as shown in H. Data are representative of four (A–C) or two (D–I) independent experiments. Statistical significance was determined by Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate the mean ± SEM.
USP38 has a T cell intrinsic role in promoting allergic asthma
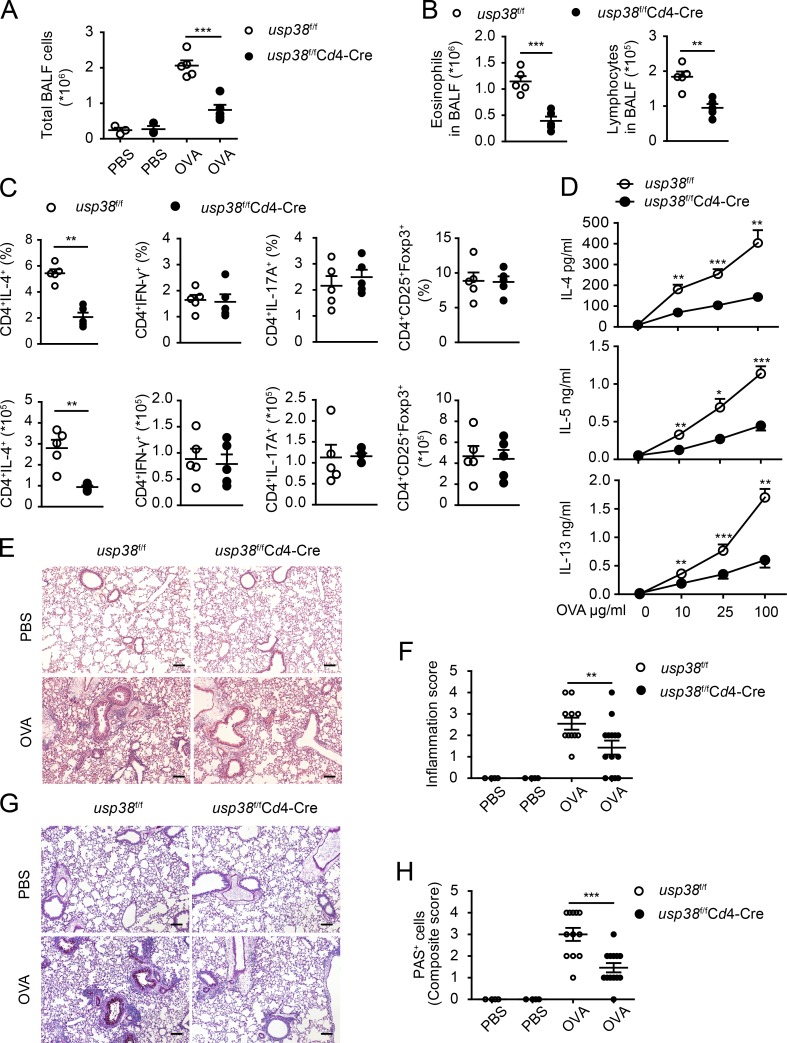
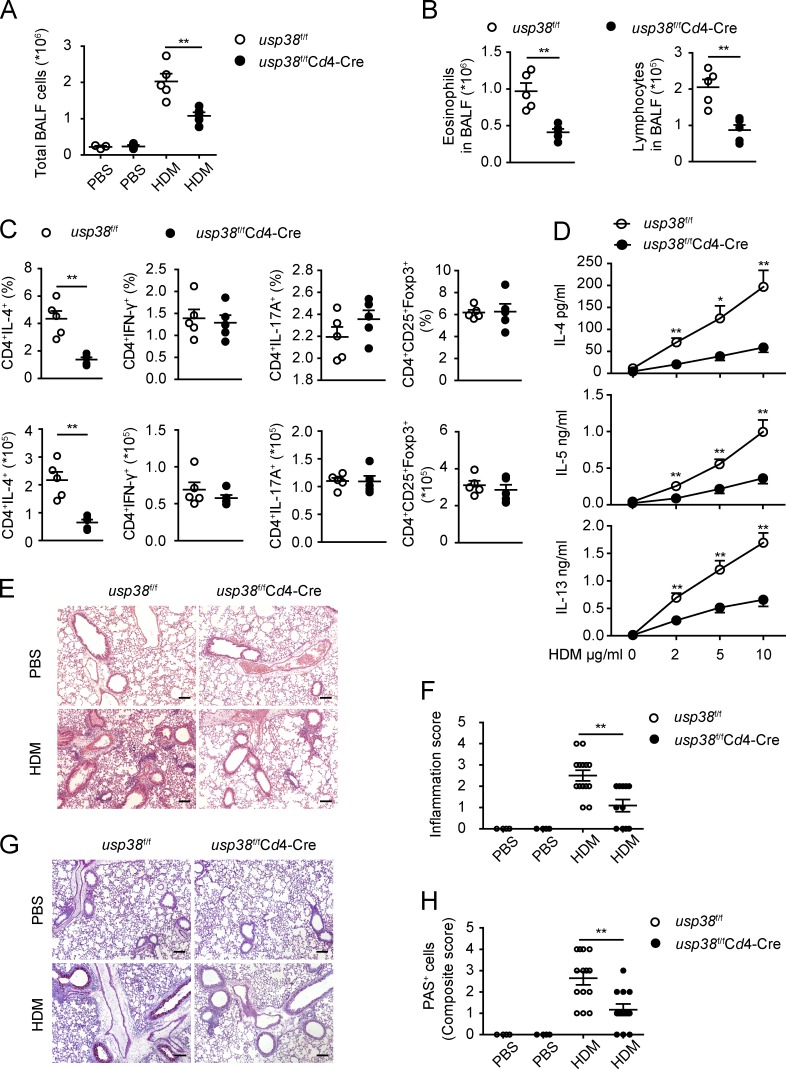
Th2 cells are critical for allergic asthma pathogenesis (Licona-Limón et al., 2013; Nakayama et al., 2017). We observed that USP38 deficiency resulted in dramatic reduction of Th2 cells in both OVA- and HDM-induced asthma models (Fig. 2 D and Fig. 3 D). To assess whether the critical promoting role of USP38 in asthma is T cell intrinsic, we generated USP38 deficiency specifically in T cells by breeding usp38-floxed mice with Cd4-Cre+/− mice (Fig. S1 A). USP38 was efficiently deleted specifically in T cells as determined by qPCR and Western blot (Fig. S1, E and F). The homeostatic immune cell development and activation were not altered in the T cell–specific USP38-deficient mice (Fig. S2, E–L). Similar to the phenotypes in the usp38 complete knockout mice (Fig. 2 and Fig. 3), USP38 deficiency in T cells led to markedly reduced number of total BALF cells, as well as the numbers of eosinophils and lymphocytes, in the OVA-induced asthma model (Fig. 4, A and B). Similarly, the T cell–specific USP38 deficiency dramatically reduced Th2 cells, but did not affect Th1, Th17, and T reg populations (Fig. 4 C). Consistent with the reduced Th2 cells, the production of Th2 cytokines was significantly decreased in the USP38-deficient T cells (Fig. 4 D). Accordingly, the leukocyte infiltration and inflammation were lowered in the T cell–specific USP38-deficient lungs in the OVA model (Fig. 4, E and F). The severity of goblet cell hyperplasia was also much reduced in the T cell–specific knockout mice (Fig. 4, G and H). These results support the T cell intrinsic role of USP38 in OVA-induced asthmatic pathogenesis. We then applied the HDM-induced model to the T cell–specific USP38-deficient mice and found similar results to the OVA-induced model. USP38 deficiency in T cells resulted in dramatically reduced numbers of total BALF cells (Fig. 5 A), eosinophils and lymphocytes (Fig. 5 B), decreased Th2 cells (Fig. 5 C), and reduced secretion of Th2 cytokines (Fig. 5 D). Leukocyte infiltration and inflammation, as well as goblet cell hyperplasia, were all reduced in the T cell–specific knockout lungs compared with WT lungs (Fig. 5, E–H). All these data demonstrate that USP38 expression in T cells is critical for its promoting role in asthmatic pathogenesis in both OVA- and HDM-induced asthma models.
Figure 4.
T cell–derived USP38 mediates the pathogenesis of OVA-induced asthma. (A–H) Mice with T cell–specific deletion of usp38 (usp38f/fCd4-Cre) and their littermate controls (usp38f/f) were used for OVA-induced asthma model. PBS group (n = 3); OVA group (n = 5). (A) Total number of BALF cells. (B) Quantification of eosinophils or lymphocytes in BALF determined by flow analysis. (C) Percentages and absolute numbers of the indicated T cell subpopulations in medLNs. (D) Levels of Th2 cytokines determined by ELISA from Th2 ex vivo recall response. (E) H&E staining of lung sections from PBS- or OVA-treated mice. Original magnification is 10×. Bar, 100 µm. (F) Pathological score of perivascular and peribronchiolar inflammation as shown in E. (G) PAS staining of lung sections from PBS- or OVA-treated mice. Original magnification is 10×. Bars, 100 µm. (H) Quantification of goblet cell hyperplasia as shown in H. Data are representative of four (A and B) or two (C–H) independent experiments. Statistical significance was determined by Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate the mean ± SEM.
Figure 5.
T cell–derived USP38 is essential for HDM-induced asthmatic pathogenesis. (A–H) Usp38f/fCd4-Cre and their littermate controls (usp38f/f) were used for HDM-induced asthma model. PBS group (n = 3); HDM group (n = 5). (A) Total number of BALF cells. (B) Numbers of eosinophils or lymphocytes in BALF determined by flow analysis. (C) Percentages and absolute numbers of the indicated T cell subpopulations in medLNs. (D) Levels of Th2 cytokines determined by ELISA from Th2 ex vivo recall response. (E) H&E staining of lung sections from PBS- or HDM-treated mice. Original magnification is 10×. Bars, 100 µm. (F) Pathological score of perivascular and peribronchiolar inflammation as shown in E. (G) PAS staining of lung sections from PBS- or HDM-treated mice. Original magnification is 10×. Bars, 100 µm. (H) Quantification of goblet cell hyperplasia as shown in H. Data are representative of four (A and B) or two (C–H) independent experiments. Statistical significance was determined by Student’s t test; *, P < 0.05; **, P < 0.01. Error bars indicate the mean ± SEM.
USP38 is specifically required for TCR-induced Th2 cytokine production and Th2 differentiation
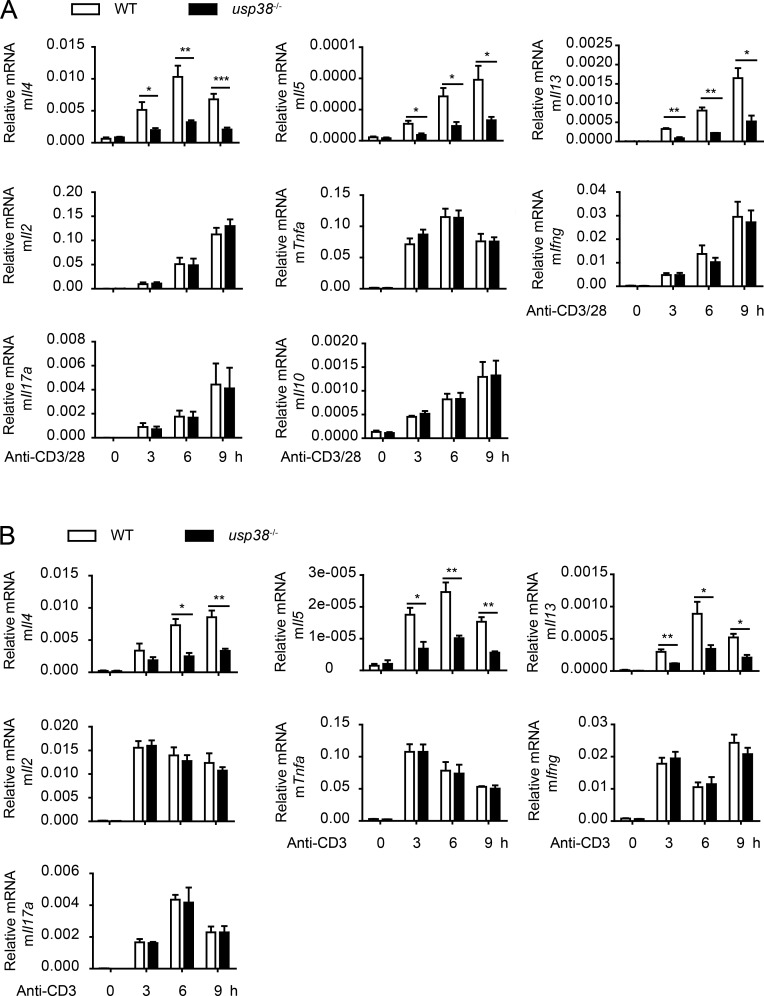
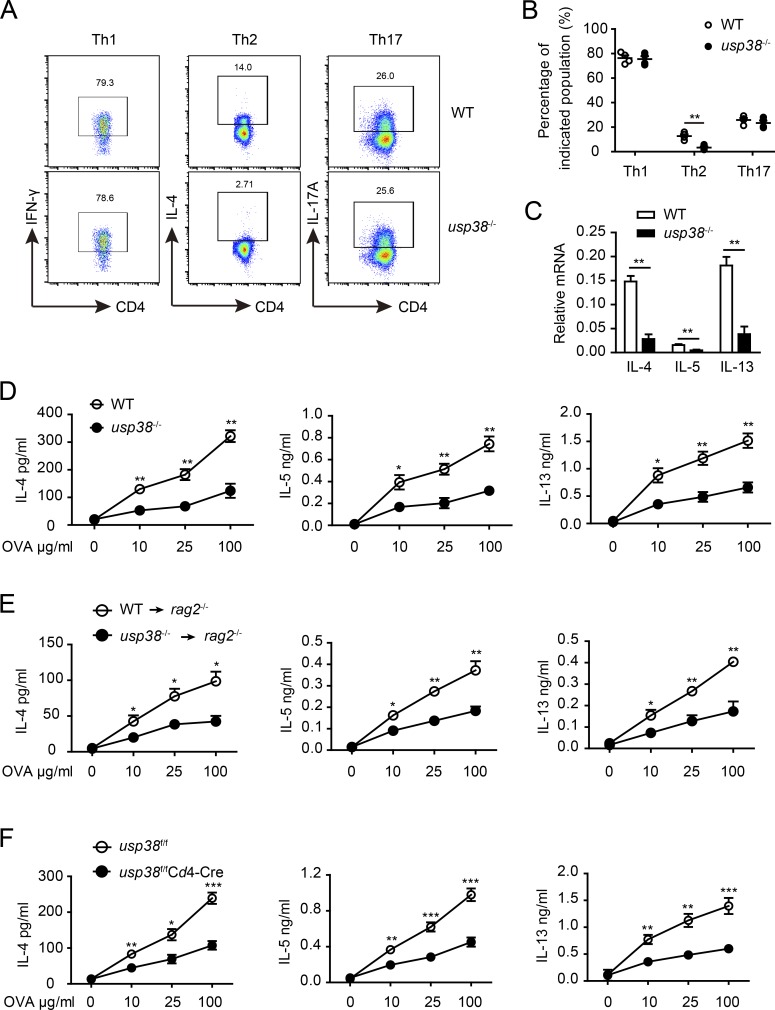
We showed above the T cell intrinsic role of USP38 in the two allergic asthma induction models (Fig. 4 and Fig. 5). To determine how USP38 affects T cell functions, we checked into T cell activation–induced genes and found that anti-CD3/CD28-induced production of Th2 cytokine genes (Il4, Il5, and Il13) was dramatically reduced, while the other checked genes, including Il2, Tnfa, Ifng, Il17a, and Il10, were not changed in USP38-deficient CD4+ T cells compared with WT control cells (Fig. 6 A). Furthermore, the Th2 cytokine genes induced by anti-CD3 alone were specifically and greatly reduced upon USP38 deficiency (Fig. 6 B), indicating that USP38 is involved in TCR signaling instead of CD28 costimulation signaling for the induction of Th2 genes. TCR-induced early IL-4 is critical for Th2 differentiation (Paul, 2010). We thus set up in vitro T cell differentiation assays and indeed found that USP38 was essential for Th2 cell differentiation, but not important for Th1 or Th17 cell differentiation (Fig. 7, A and B). Accordingly, USP38 deficiency dramatically reduced the production of the Th2 genes under Th2 differentiation condition (Fig. 7 C). We then checked into Th2 differentiation in vivo by preimmunization in mice with OVA + Alum intraperitoneally and then determination of Th2 cytokines by ELISA from OVA-rechallenged splenocytes ex vivo as described previously (Yang et al., 2009). Consistent with the results from Th2 differentiation in vitro (Fig. 7, A–C), the secretion of Th2 cytokines IL-4, IL-5, and IL-13 was greatly reduced from splenocytes of USP38-deficient mice after Th2 priming in vivo (Fig. 7 D). To avoid potential effects of USP38 deficiency in other cell types other than T cells, we purified CD4+ T cells from USP38-deficient and WT control spleens and then adoptively transferred the CD4+ T cells together with WT B cell into rag2−/− mice. Similarly, the production of the Th2 cytokines was dramatically decreased from the mice transferred with USP38-deficient T cells compared with the mice transferred with WT control T cells (Fig. 7 E). To further confirm the T cell intrinsic role of USP38 in in vivo Th2 differentiation, we used Cd4-cre–mediated USP38 deficiency in T cells and indeed found that USP38 expression in T cells was critical for Th2 cytokines production from the in vivo system (Fig. 7 F). Together, these data demonstrate that USP38 is essential for TCR-induced Th2 cytokines production, thus promoting Th2 differentiation in vitro and in vivo.
Figure 6.
USP38 positively regulates TCR-induced production of Th2 cytokines. (A) mRNA levels of the indicated genes determined by qPCR in the usp38−/− or WT CD4+ T cells that were treated by plated anti-CD3 (1 µg/ml) and plated anti-CD28 (1 µg/ml) for the indicated times. (B) mRNA levels of the indicated genes assessed by qPCR in the usp38−/− and WT CD4+ T cells that were stimulated by plated anti-CD3 (1 µg/ml) alone for the indicated time points. Data are representative of five (A) or three (B) independent experiments. Statistical significance was determined by Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate the mean ± SEM.
Figure 7.
USP38 is required for Th2 differentiation both in vitro and in vivo. (A) Flow chart of the indicated helper T cell subpopulations from cultures of usp38−/− or WT naive CD4+ T cells that were differentiated under Th1-, Th2-, or Th17-skewing conditions for 5 d. (B) Percentages of T cell subsets as shown in A. (C) mRNA levels of Il4, Il5, and Il13 in the Th2 cells obtained from A. (D) Concentrations of IL-4, IL-5, and IL-13 determined by ELISA in the supernatants from 3 d OVA-stimulated splenocytes out of WT or usp38−/− mice that were immunized with OVA in alum intraperitoneally for 7 d. (E) Concentrations of IL-4, IL-5, and IL-13 determined by ELISA in the supernatants from 3-d OVA-stimulated splenocytes out of the rag2−/− recipient mice that were transferred with WT or usp38−/− CD4+ T cells mixed with WT B cells at ratio of 1:1 and were immunized the next day with OVA in alum for 7 d. (F) Concentrations of IL-4, IL-5, and IL-13 determined by ELISA in the supernatants from 3-d OVA-stimulated splenocytes out of usp38f/fCd4-Cre and littermate control mice (usp38f/f) that were immunized as D. Data are representative of four (A–C), three (D and F), or two (E) independent experiments. Statistical significance was determined by Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate the mean ± SEM.
USP38 is required for TCR-induced JunB protein stability
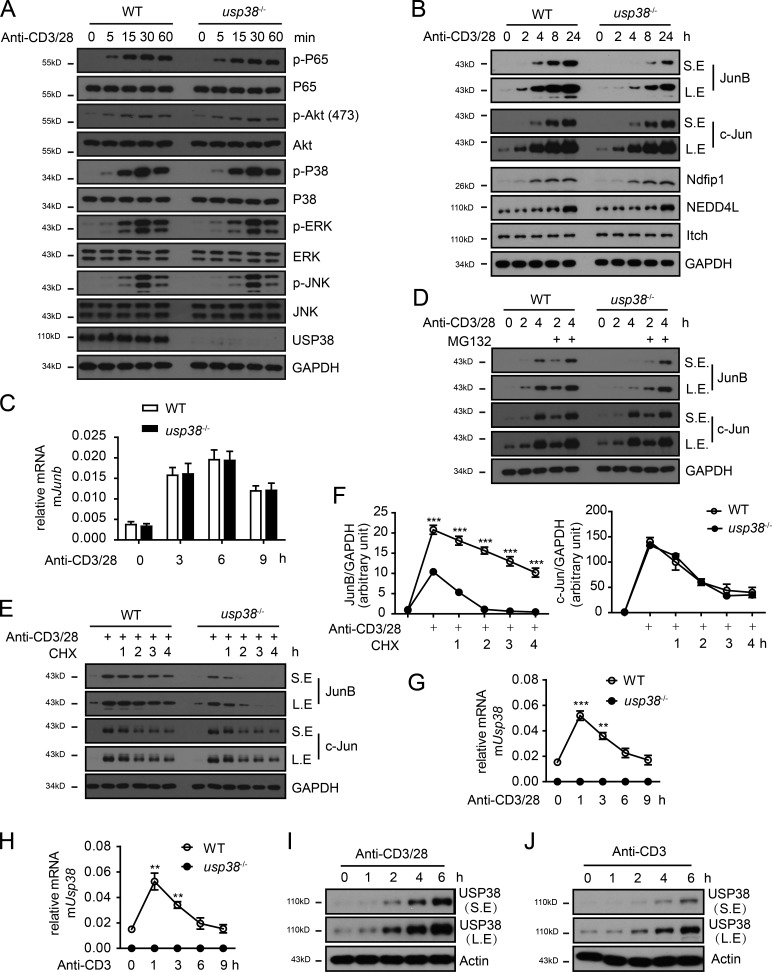
As USP38 was specifically required for TCR-induced Th2 cytokines production (Fig. 6), we next determined TCR signaling and found that USP38 deficiency did not affect TCR-induced activation of NF-κB, Akt, and MAPKs (JNK, ERK, and P38; Fig. 8 A). Interestingly, we observed that TCR-induced accumulation of JunB, but not that of c-Jun, was greatly prohibited in USP38-deficient T cells compared with WT control cells (Fig. 8 B). We further found that USP38 was not required for TCR-induced mRNA of Junb (Fig. 8 C), indicating that USP38 may contribute to JunB protein stability. To explore this point, we pretreated cells with MG132 to block proteasome-mediated protein degradation and indeed found that MG132 restored the TCR-induced accumulation defect of JunB protein in USP38-deficient T cells (Fig. 8 D). We also performed a chase experiment after anti-CD3/CD28 stimulation and found that JunB was degraded with a more rapid rate in USP38-deficient CD4+ T cells compared with WT control T cells in the presence of cycloheximide (CHX), while the rate of degradation of c-Jun is not affected by USP38 deficiency (Fig. 8, E and F).
Figure 8.
USP38 is essential for TCR-induced JunB protein stability. (A and B) Immunoblotting analyses of cell lysates from usp38−/− or WT CD4+ T cells that were stimulated by plated anti-CD3 (1 µg/ml) and plated anti-CD28 (1 µg/ml) for the indicated times. (C) mRNA level of Junb determined by qPCR in usp38−/− or WT CD4+ T cells that were stimulated by plated anti-CD3 (1 µg/ml) and plated anti-CD28 (1 µg/ml) for the indicated times. (D) Immunoblotting analyses of cell lysates from usp38−/− or WT CD4+ T cells that were pretreated with or without MG132 and then stimulated with anti-CD3/28 for the indicated time points. (E) Immunoblotting analyses of cell lysates from usp38−/− or WT CD4+ T cells that were stimulated by anti-CD3/28 for 6 h and then treated with CHX for the indicated period. (F) The relative abundance of JunB or c-Jun quantified by densitometric analysis that were normalized to that of GAPDH as showed in E. (G and H) mRNA levels of usp38 in the WT and usp38−/− CD4+ T cells that were stimulated by anti-CD3/28 (G) or anti-CD3 alone (H) for the indicated time points. (I and J) The protein levels of USP38 analyzed by immunoblotting analyses of cell lysates from WT CD4+ T cells that were stimulated with anti-CD3/28 (I) or anti-CD3 alone (J) for the indicated time points. (B, D, E, I, and J) S.E., short exposure; L.E., long exposure. All the data are representative of three independent experiments. Statistical significance was determined by Student’s t test, **, P < 0.01; ***, P < 0.001. Error bars indicate the mean ± SEM.
Since USP38 was required for TCR-induced JunB accumulation, we checked into USP38 expression regulation in TCR signaling. We observed that either anti-CD3/CD28 or anti-CD3 alone induced usp38 mRNA (Fig. 8, G and H). Similarly, USP38 protein was induced by anti-CD3/CD28 or anti-CD3 (Fig. 8, I and J), indicating that TCR signaling instead of CD28 signaling is responsible for USP38 induction. We further found that the mRNA level of usp38 did not show statistic difference in different CD4+ T cell subsets (Fig. S3 A) and was not induced by the subset-specific cytokines, including IL-2, IL-4, IL-6, IFN, and TGF-β in primary CD4+ T cells (Fig. S3 B), suggesting that usp38 expression is directly regulated by TCR signal rather than by cytokines downstream of TCR signaling. Given that a LacZ cassette is inserted in the usp38 gene locus in the targeting strategy (Fig. S1 A), we used β-Gal staining on lung section to visualize the USP38 expression. Consistent with the TCR-induced USP38 expression in vitro (Fig. 8, G–J), we found that USP38 was strongly induced in the infiltrated immune cells around the bronchia and blood vessels after OVA challenge (Fig. S3 C). We also noticed that USP38 was constitutively expressed and up-regulated after OVA treatment in bronchial epithelial cells (Fig. S3 C). It remains to be determined if the epithelial-derived USP38 expression has any potential pathophysiological functions. We did not observe any induction of usp38 by triggers of innate sensors like TLRs, RIG-I, AIM2, and cGAS in macrophages (Fig. S3, D–J). Accordingly, USP38 deficiency did not affect those innate sensor-induced production of downstream genes (Tnfa and Ifnb; Fig. S3 K). To our knowledge, it may not be feasible to operate β-Gal staining for LacZ together with immunofluorescence staining against a T cell marker like CD4. Considering that USP38 was induced by TCR signaling in T cells, but not induced in macrophages by the innate sensor triggers, we hypothesize that the observed USP38 expression in the infiltrated immune cells is likely due to its induction in T cells after OVA challenge.
Apart from its essential role in TCR-induced JunB stability, we found that USP38 was dispensable for the cytokine (IL-4, IL-2, and IL-6)-induced signaling in T cells (Fig. S4, A–C). Considering that USP38 had low constitutive expression in rest CD4+ T cells and was induced after TCR activation, we used retrovirus-mediated USP38 overexpression in WT CD4+ T cells and found that increased USP38 expression did not affect the cytokines-triggered signaling (Fig. S4, D–F). Similarly, the increased USP38 level did not affect TCR-induced pathways of NF-κB and MAPKs (Fig. S4 G). Altogether, our data suggest that TCR-induced USP38 specifically blocks JunB protein turnover, thus promoting Th2 cytokines production.
USP38 directly associates with JunB and removes TCR-induced JunB poly-ubiquitination
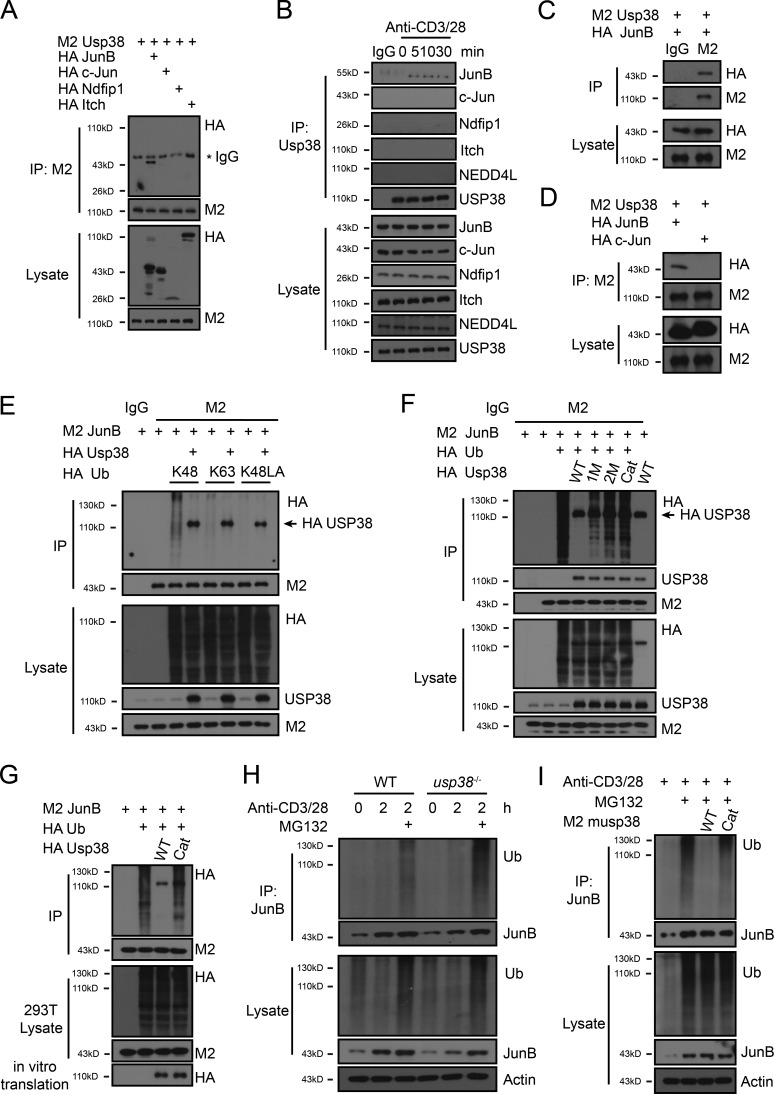
Since JunB is poly-ubiquitinated by Itch for degradation in TCR signaling (Fang et al., 2002; Gao et al., 2004), and we found that USP38 was required for TCR-induced JunB stability (Fig. 8 B), we hypothesized that USP38 may counteract the JunB ubiquitin system. To test this hypothesis, we first explored if USP38 interacts with JunB and found that USP38 specifically associated with JunB, but not c-Jun, in 293T overexpression system (Fig. 9 A), consistent with the specific role of USP38 for JunB, but not c-Jun accumulation in TCR signaling (Fig. 8 B). We then checked into the endogenous system and found that USP38 bound to JunB, but not c-Jun, in a TCR stimulation–dependent manner (Fig. 9 B). USP38 did not associate with Itch, nor its activator Ndfip1, in either 293T overexpression system or TCR-induced endogenous system (Fig. 9, A and B), indicating that USP38 may directly associate with JunB for its deubiquitination. Indeed, we found that USP38 interacted with JunB when both proteins were expressed through in vitro translation system (Fig. 9 C). Furthermore, USP38 directly associated with JunB, but not c-Jun (Fig. 9 D). In consistent with JunB ubiquitination for degradation, we observed that JunB was mainly Lys-48 poly-ubiquitinated in 293T overexpression system (Fig. 9 E). We then found that JunB poly-ubiquitination was removed by overexpression of WT USP38 in the 293T system (Fig. 9 F). To determine whether the DUB activity of USP38 is responsible for its effect on the removal of JunB poly-ubiquitination, we generated three DUB mutants of USP38 and found that the three sites mutation (C454S/H857A/D918N, named as Cat) had the most effect in blockage of the DUB activity in term of removal of the JunB poly-ubiquitination (Fig. 9 F). One potential reason for the differential effects of the three mutants could be that overexpressed USP38 with additional mutations may be more effective to function as dominant negative against the endogenous USP38 in the overexpression system. Although the reasons why C454S mutation did not totally block USP38 enzymatic activity are not clear, our mutation results are consistent with a recent report that additional mutations in the catalytic triad are necessary to render USP38 catalytic inactiveness (Lin et al., 2016). We further found that the poly-ubiquitination of JunB was removed in vitro by WT USP38 but not its enzyme activity mutant which were both expressed through the in vitro translation system (Fig. 9 G), indicating USP38 directly deubiqutinates JunB. We next moved to endogenous system and found that TCR-induced JunB poly-ubiquitination was greatly increased in USP38-deficient T cells only after MG132 treatment (Fig. 9 H), indicating that USP38 deubiqutinates the JunB ubiquitination to block its degradation in TCR signaling. We then immunoprecipitated the TCR-induced poly-ubiquitinated JunB and incubated it with WT USP38 or its enzyme activity mutant from the in vitro translation system. Indeed, we found that USP38 but not its mutant directly removed the ubiquitination of endogenous JunB from TCR signaling (Fig. 9 I). All these data suggest that USP38 directly associates with JunB, and its enzyme activity removes JunB ubiquitination in TCR signaling.
Figure 9.
USP38 deubiquitinates TCR-induced K48-linked poly-ubiquitination of JunB. (A) Co-immunoprecipitation from lysates of HEK293T cells that were transfected with the indicated plasmids. Whole lysates were immunoprecipitated (IP) with anti-M2 and followed by immunoblotting (WB) with anti-HA and anti-M2. (B) Co-immunoprecipitation from lysates of CD4+ T cells that were treated with anti-CD3/28 for the indicated time points. Whole lysates were IP with anti-USP38 and followed by WB with the indicated antibodies. (C and D) Co-immunoprecipitation of proteins that were synthesized by in vitro translation system. The product mixture was IP with IgG (C) or anti-M2 (D) and followed by WB with anti-HA or anti-M2. (E and F) Immunoblot analyses of ubiquitinated JunB in HEK293T cells that were transfected with the indicated plasmids. Cell lysates were IP with anti-M2 and WB with anti-HA, anti-M2, or anti-USP38. K48, only Lys48 site left in ubiquitination. K63, only Lys63 site left in ubiquitination. K48LA, only Lys48 mutated into Ala in ubiquitination. USP38 mutants: 1M (C454S), 2M (C454S/H857A), and Cat (C454S/H857A/D918N). (G) Immunoblot analyses of USP38-mediated removal of JunB ubiquitination. Whole lysates of HEK293T cells transfected with the indicated plasmids were IP with anti-M2. The immunoprecipitates were then mixed with products from in vitro translation system containing WT USP38 or its DUB mutant (Cat) and followed by WB with the indicated antibodies. (H) Immunoblot analyses of ubiquitinated JunB in CD4+ T cells that were pretreated with or without MG132 and then treated with anti-CD3/28 for 2 h. Whole lysates were IP with anti-JunB and followed by WB with the indicated antibodies. (I) Immunoblot analyses of USP38-mediated removal of endogenous JunB ubiquitination. CD4+ T cells were pretreated with or without MG132 and stimulated with anti-CD3/CD28 for 2 h. Cell lysates were IP with anti-JunB. The immunoprecipitates were then mixed with WT mUSP38 or its DUB mutant from in vitro translation system and followed by WB with the indicated antibodies. Data are representative of three (A–D, F, and H) or two (E, G, and I) independent experiments.
USP38 DUB activity is essential for TCR-induced JunB stability and Th2 differentiation
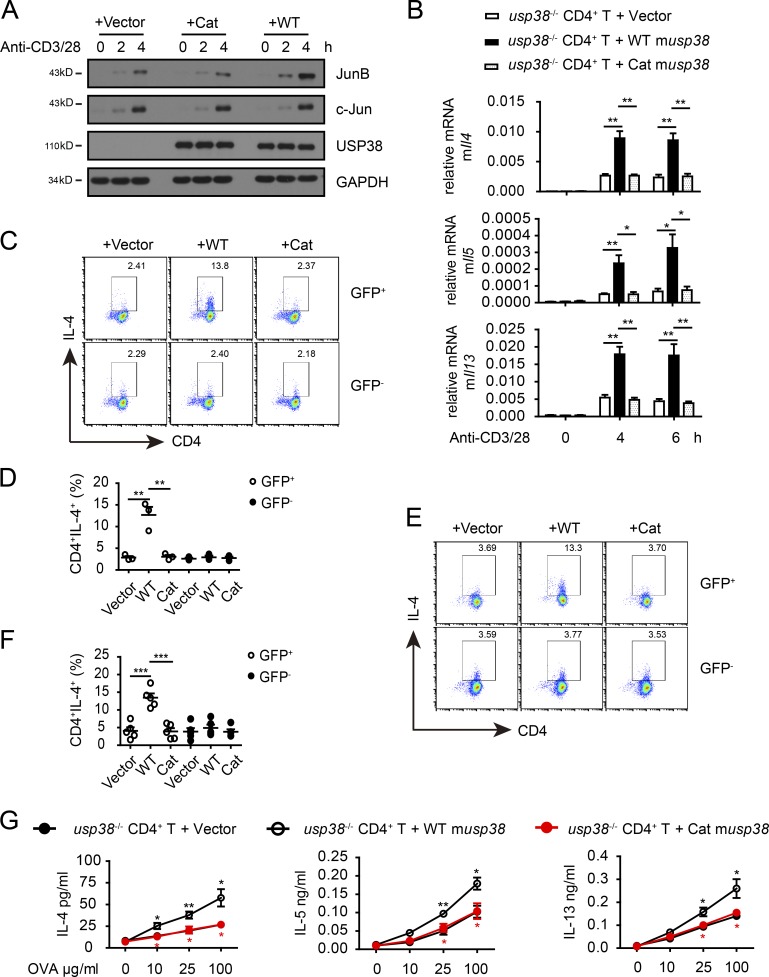
We showed above that USP38 directly associated with JunB to remove TCR-induced JunB ubiquitination (Fig. 9). Consistently, we found that retrovirus-mediated restoration of WT USP38 into USP38-deficient T cells recovered JunB protein accumulation by TCR signaling, while the USP38 enzyme activity mutant had no observable effect, although both WT USP38 and its mutant had similar expression levels (Fig. 10 A and Fig. S5 A), indicating USP38 DUB activity is totally responsible for its role in TCR-induced JunB protein stability. The restoration study also confirmed the specificity of USP38 in TCR-induced JunB instead of c-Jun stability (Fig. 10 A). Furthermore, putting WT USP38, but not its enzyme activity mutant, back into USP38-deficient T cells recovered TCR-induced production of Th2 genes, including Il4, Il5, and Il13 (Fig. 10 B). Both USP38 and its mutant had no effects on TCR-induced other genes, including Il2, Tnfa, Ifng, and Il17a (Fig. S5 B), further confirming the specific role of USP38 in TCR-induced production of Th2 genes. We then used the restoration system to confirm our results on the role of USP38 in Th2 differentiation out of WT and USP38-deficient T cells (Fig. 7). Indeed, restoration of WT USP38 into USP38-deficient T cells recovered the defect in Th2 differentiation in vitro (Fig. 10, C and D). Importantly, the enzyme activity mutant of USP38 had no effect on the recovery of the Th2 differentiation defect (Fig. 10, C and D), indicating that USP38 DUB activity is required for TCR-induced Th2 differentiation. Similarly in vivo, putting WT USP38, but not its enzyme activity mutant, back into USP38-deficient T cells restored Th2 differentiation and the production of Th2 cytokines (Fig. 10, E–G; and Fig. S5, C and D). Together, our data demonstrate that the DUB activity of USP38 is totally responsible for its effects on TCR-induced JunB protein stability and consequently Th2 cytokine production and Th2 differentiation.
Figure 10.
The DUB activity of USP38 is essential for its regulation of TCR-induced JunB stability and Th2 development. (A–G) Usp38−/− CD4+ T cells were restored with retrovirus containing empty vector or vectors encoding WT mouse USP38 or its DUB activity mutant (Cat). (A) Immunoblotting of cell lysates from the restored cells that were stimulated with anti-CD3 and anti-CD28 for the indicated times. (B) mRNA levels of the Th2 cytokine genes determined by qPCR in the restored cells that were stimulated with anti-CD3 and anti-CD28 for the indicated time points. (C and D) Flow chart (C) and quantification (D) of Th2 cell percentages by flow analysis in the restored cells that were cultured in Th2-skewing condition for 5 d. GFP positive stands for the virus-restored cells. (E and F) Flow chart (E) and quantification (F) of Th2 cell percentage. Rag2−/− recipient mice were transferred with the indicated restored cells (GFP, WT, and Cat) together with WT B cells at the ratio of 1:1 mixture and immunized the next day with OVA in alum for 7 d. The splenocytes were in vitro restimulated with OVA for 3 d in the presence of protein transport inhibitor for the last 6 h and then analyzed by cytoflow. (G) Concentrations of IL-4, IL-5, and IL-13 in the supernatants from the 3-d OVA-stimulated splenocytes out of the mice treated as E and F. Data are representative of three (A–D) or two (E–G) independent experiments. Statistical significance was determined by Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate the mean ± SEM.
Discussion
A usp38-containing genomic locus has recently been reported to be associated with adult asthma in a GWAS study (Hirota et al., 2011). A potential pathophysiological role of USP38 is still unknown. Here we show that USP38 is essential for asthmatic pathogenesis. USP38-deficient mice were resistant to allergic asthma induction by OVA or HDM. Through Cd4-Cre–mediated USP38 deficiency in T cells, we demonstrate the T cell intrinsic role of USP38 in asthma pathology. Furthermore, we found that USP38 was specifically required for TCR-induced production of Th2 genes (Il4, Il5, and Il13). Consequently, USP38 was essential for Th2 differentiation and the associated functions both in vitro and in vivo, while it was dispensable for Th1 or Th17 differentiation.
For the mechanistic insight into the specific role of USP38 in TCR-induced Th2 genes, we demonstrate that USP38 is specifically required for the protein, but not mRNA induction of Junb, a key essential and specific transcription factor for Th2 differentiation and function (Li et al., 1999; Hartenstein et al., 2002). The block of JunB protein induction in the USP38-deficient T cells was restored by the proteasome inhibitor MG132, and likewise, the degradation rate of JunB is more rapid in usp38−/− CD4+ T cells in the presence of CHX after TCR stimulation, suggesting that USP38 stabilizes JunB protein in TCR signaling. USP38 associated with JunB directly as well as in a TCR signal–dependent manner. USP38 de-ubiquitinated TCR-induced Lys-48–linked poly-ubiquitination of JunB and thus consequently blocked proteasome-mediated JunB degradation. The deubiquitinase activity of USP38 was responsible for its effects on TCR-induced JunB turnover and the consequent Th2 differentiation both in vitro and in vivo. Thus, our study identifies USP38 as a critical DUB, specifically for TCR-induced JunB protein stability and Th2 immunity.
Itch, the HECT-type E3 ubiquitin ligase of Nedd4 family, ubiquitinates JunB for proteasome-mediated degradation, thus suppressing Th2 immunity (Fang et al., 2002). Itch phosphorylation by JNK is required for its E3 ligase activity to target JunB turnover (Gao et al., 2004). Here we demonstrate that USP38 plays a counteractive role to Itch by deubiquitinating JunB for its stability. While Itch protein level was not regulated by TCR activation, we found that USP38 was induced with the kinetics similar to that of JunB. These results suggest that TCR ligation activates the JNK–Itch signaling axis to keep JunB levels in check to wait for further instruction, and USP38 induction is the signal to break the checkpoint for boosting Th2 response. While the JNK–Itch axis also targets c-Jun, another transcription factor downstream of TCR signaling (Gao et al., 2004), USP38 did not associate with c-Jun and was not required for c-Jun accumulation. In addition to Th2 response, Itch also regulates other T cell functions, such as T reg and Th17 (Venuprasad, 2010; Jin et al., 2013; Kathania et al., 2016). To our knowledge, there is so far no report of E3 ubiquitin ligase specific for TCR-induced Th2 differentiation and function. DUBs could be another way for controlling specificity of different T cell responses. As in our case, USP38 specifically boosts Th2 immunity likely through its expression regulation and its substrate binding specificity.
JunB has recently been reported to promote Th2 and Th17 development and suppress Th1 and T reg differentiation through analyses of Junb knockout mice (Carr et al., 2017), while our data showed that USP38 deficiency specifically affected Th2 development. The differential effects of JunB and USP38 on T cell subset differentiation is likely due to the potential differential roles of different JunB levels. As reported by Li et al. (1999), both mRNA and protein levels of Th2 cytokines were increased, while those of Th1 cytokine IFN-γ were not changed in the JunB transgenic T cells, compared with WT control T cells under Th1 differentiation condition. Analogous to the quantitative change of JunB level in the condition of the transgenic mice (Li et al., 1999), our data showed that USP38 only regulated JunB protein stability and was not required for Junb mRNA induction, and thus, there was still JunB induction in USP38-deficient CD4+ T cells upon TCR stimulation. Based on these transgenic and knockout studies as well as our data, we hypothesize that the change of JunB level (quantity) specifically regulates Th2 cell development, while a certain level of JunB is required and may be enough for its cooperation with other transcription factors to regulate other T cell subsets. It is also possible that total JunB deficiency could lead to changed cell state due to potential epigenetic changes for cell homeostasis balance. The remained level of JunB expression by its mRNA induction in USP38-deficient T cells may be enough for its regulation of other T cell subsets.
The mammalian genome encodes a large family of DUBs (near 100 members), most of which are not functionally well characterized (Nijman et al., 2005). Compared with E3 ubiquitin ligases, the potential roles of DUBs are even less defined in T cell functions (Gao et al., 2017). Recent studies have shown the important roles of several DUBs in regulating T cell development (Reiley et al., 2006; Dufner et al., 2015), T cell anergy (Soares et al., 2004), T cell activation (Düwel et al., 2009; Naik et al., 2014; Hu et al., 2016), and T cell differentiations (Liu et al., 2013; van Loosdregt et al., 2013; Zou et al., 2015). However, there is still no identification of DUBs in regulating Th2 immunity. Here we identify USP38 as the first DUB to regulate Th2 response. USP38 is an almost uncharacterized DUB with only one functional report showing that USP38 inhibits TBK1-mediated type 1 interferon signaling in macrophages (Lin et al., 2016). However, we did not find a role of USP38 in gene induction by innate sensors like RIG-I and cGAS in macrophages, and we found that USP38 was induced in T cells but not in macrophages. The inconsistent results on the role of USP38 in macrophage could be due to the differential experimental systems or the different knockout targeting strategies, as usp38 was targeted by the traditional homogenous recombination in our mice, while that report used TALEN technology (Lin et al., 2016). The specific regulation of Th2 immunity makes USP38 an attractive pharmacological target to develop small molecules that inhibit its enzyme activity for treating Th2-mediated inflammatory diseases such as asthma.
Materials and methods
Mice
The sperm carrying the loxp-flanked usp38 mutant gene was gotten from the European Conditional Mouse Mutagenesis Consortium (B6NTac;B6N-Usp38<tm1a(EUCOMM)Hmgu>/H (EM:05481), and used to generate usp38f/+ mice by Shanghai Research Center For Model Organisms (Biomodel). flp+/−, E2a-Cre+/−, Cd4-Cre+/−, and rag2−/− mice were all purchased from Biomodel. All the mice mentioned above were on C57BL/6 background. Usp38f/+ were mated with E2a-Cre+/− mice to remove the neomycin cassette and the targeted exon. The obtained heterozygous usp38+/− mice were intercrossed to generate usp38 knockout mice (usp38−/−) and its littermate WT control mice. To generate conditional knockout mice of usp38 in T cells, usp38f/+ mice were bred with flp+/− mice and then with Cd4-Cre+/− mice. The flp and E2a-Cre genes were eliminated through continuous passages. All mice were maintained under specific pathogen-free conditions, and 6–10-wk-old mice were used for experiments. All animal experimental protocols and performance were approved by the Institutional Biomedical Research Ethics Committee of the Shanghai Institutes for Biological Sciences (Chinese Academy of Sciences).
Reagents and constructs
Antibodies to USP38 (17767-1-AP), JunB (10486-1-AP), c-Jun (10024-2-AP), or Nedd4L (13690-1-AP) were from Proteintech. Antibody to Itch (611198) was from BD Biosciences. Antibodies to phosphorylated p65 (3033S), Akt (4060S), p38 (9211S), or JNK (9251S) were from Cell Signaling. Antibody to phosphorylated ERK (sc-7383) was from Santa Cruz Biotechnology. Anti-Flag and anti-GAPDH were from Sigma. Antibody to hemagglutinin was from Covance. Ionomycin (I0634-1MG) and PMA (P1585-1MG) were from Sigma.
The cDNAs of human Usp38, JunB, c-Jun, Itch, and Ndfip1 were generated from real-time PCR (RT-PCR) and then cloned into the pcDNA 3.0 vector. The hemagglutinin-tagged human Usp38 were then used to generate its DUB activity mutant plasmids (C454S, H857A, and D918N). The cDNA of mouse usp38 was similarly generated by RT-PCR and cloned into the pcDNA 3.0 vector. Then, the WT mouse usp38 and its DUB activity mutant were cloned into the pMSCV vector.
Preparation of primary CD4+ T cells and in vitro differentiation
Primary CD4+ T cells were isolated from the splenocytes of age- and sex-matched usp38−/− and WT mice using anti-CD4 microbeads (L3T4; Miltenyi Biotec). The cells were activated by plate-bound 1 µg/ml anti-CD3 (130-092-973; Miltenyi Biotec) and anti-CD28 (130-093-182; Miltenyi Biotec) or anti-CD3 alone for various periods.
For in vitro differentiation, 1 × 106 naive CD4+ T cells/ml were activated by plated anti-CD3 and anti-CD28 (1 µg/ml) and 10 ng/ml IL-2 with different skewing conditions for 48 h: Th1, 10 ng/ml IL-12 + 10 µg/ml anti–IL-4 (11B11); Th2, 10 ng/ml IL-4 + 10 μg/ml anti–IFN-γ (XMG1.2); and Th17, 10 ng/ml IL-6, and 5 ng/ml TGF-β with 10 ng/ml anti–IFN-γ and anti–IL-4. Then the cells were transferred into the same fresh culture mediums without anti-CD3 and anti-CD28 for another 72 h. All the cytokines used above were purchased from Peprotech. Anti–IL-4 and anti–IFN-γ were purchased from eBioscience.
Asthma induction
For OVA-induced allergic asthma, mice were immunized with OVA (100 µg/mouse) mixed in aluminum hydroxide intraperitoneally at days 0 and 7, and then the mice were challenged by 50 µg OVA dissolved in 50 µl PBS intranasally at days 14–17. 24 h later, the mice were sacrificed, and BALF was collected.
For HDM-induced allergic asthma, mice were immunized by 1 µg HDM dissolved in 50 µl PBS intranasally, and then the mice were challenged by 10 µg HDM dissolved in 50 µl saline intranasally from days 7–11. 3 d later, the mice were sacrificed and BALF were collected.
For ex vivo recall experiments, mediastinal lymph nodes cells were collected and restimulated by various concentration of OVA protein or HDM extract, and then cytokines in the supernatants were checked by ELISA. Alternatively, mediastinal lymph node cells were stimulated by PMA and ionomycin for 4 h in the presence of Golgiplug, and then the populations of Th1, Th2, Th17, and T reg cells were checked by cytoflow.
β-Gal staining
Lung tissues embedded in optimal cutting-temperature compound were prepared for frozen sections at a thickness of 10 µm. The sections were stained in the buffer containing 2 mM MgCl2, 0.01% sodium deoxycholate, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 20 mg/ml X-Gal overnight at 37°C in dark. Then, the sections were rinsed with PBS for three times and counterstained with eosin.
Isolation of lung lymphocytes
Lungs were harvested and cut into 1-mm pieces by surgical scissors and then incubated in 2 ml of complete DMEM containing 0.4 mg/ml of collagenase type VIII (Sigma) and 100 U/ml of DNase I (Sigma) at 37°C under agitation (250 rpm) for 90 min. The mixture was centrifuged at 500 g for 5 min at 4°C, and the precipitate was resuspended in 40% Percoll (GE Healthcare), followed by density gradient centrifugation with 80% Percoll at 2,500 rpm for 20 min. Lymphocytes were then collected from the interface between 40 and 80% Percoll.
In vivo Th2 differentiation and Elisa
Mice were immunized by OVA + Alum as described above. 7 d later, splenocytes were collected and restimulated by OVA protein for 3 d, and cytokines in the supernatant were checked by ELISA. For adoptive transfer, rag2−/− recipient mice were transferred with CD4+ T cells from WT or usp38−/− mice together with WT B cells at a 1:1 ratio of mixture. The recipient mice were then immunized with OVA as described above on the next day. CD4+ T cells and B cells were purified with anti-CD4 and anti-B220 beads (Miltenyi Biotec), respectively.
Concentrations of mouse IL-4, IL-5, and IL-13 were determined by Ready-Set-Go ELISA kits (eBioscience). Analyses were read out using a microplate reader set at 450 nm according to the manufacturer’s protocol.
Tissue histology
Lung tissues were fixed in 4% paraformaldehyde. Paraffin-embedded tissue sections were then stained with H&E. Each section was observed by upright microscope (Imager A2; Zeiss) and evaluated for the severity of peribronchiolar and perivascular inflammation by a semi-quantitative criteria-based method with a score from 0 to 4: 0, absent inflammation; 1, <25% of bronchials and vasculature were affected by inflammatory cell infiltration; 2, ∼25–50% of bronchials and vasculature were affected; 3, ∼50–75% of bronchials and vasculature were affected; and 4, >75% of bronchials and vasculature were affected or dense aggregations of infiltrated cells were observed.
Paraffin-embedded tissue section were also stained with PAS for scoring the severity of goblet cell hyperplasia by a similar way: 0, no PAS-positive cells; 1, <10% PAS-positive cells; 2, between 10 to 25% PAS-positive cells; 3, between 25 and 50% PAS-positive cells; 4, >50% PAS-positive cells within a certain visual field.
Flow cytometry
The following antibodies were used for the different cell populations: eosinophils, CD11b+(Apc)SiglecF+(PE)MHCII–(PE-Cy7); lymphocytes, CD45+(FITC)CD3e+(PE-Cy7); naive T cells, CD62L+(PE)CD44–(Apc); effector T cells, CD62L–(PE)CD44+(Apc); memory T cells, CD62L+(PE)CD44+(Apc); B cells, CD45+(FITC)B220+(PE-Cy7); Th1, CD45+(FITC)CD4+(PerCy5.5)IFN-γ+(PE); Th2, CD45+(FITC)CD4+(PerCy5.5)IL-4+(Apc); Th17, CD45+(FITC)CD4+(PerCy5.5)IL-17a+(Apc); and T reg, CD45+(FITC)CD4+(PerCy5.5)CD25+(Apc)Foxp3+(PE).
All the antibodies used above were from eBioscience. For intracellular cytokine staining, Cytofix/Cytoperm plus kit was from BD (55508). For Foxp3 staining, Foxp3-staining buffer set was purchased from eBioscience (00-5523-00). All the manipulations were followed by the manufacturer’s protocols.
RNA isolation and qRT-PCR
Total RNA was extracted from cells with TRIzol reagent according to the manufacturer’s instructions (Invitrogen). For cDNA synthesis, RNA was reverse-transcribed with a PrimeScript RT Reagent kit (Takara), and the cDNA was then amplified by RT-PCR using a SYBR Premix Extaq kit (Takara) on ABI VIIA7 system. The relative mRNA levels of target genes was normalized to expression of housekeeping gene Rpl13a. The primers are Il2, forward: 5′-CACTCCTCACAGTGACCTCAAG-3′ and reverse: 5′-GGGCAAGTAAAATTTGAAGGTG-3′; Tnfa, forward: 5′-TCTTCTCATTCCTGCTTGTGG and reverse: 5′-GGTCTGGGCCATAGAACTGA-3′; Ifng, forward: 5′-ATGAACGCTACACACTGCATC-3′ and reverse: 5′-CCATCCTTTTGCCAGTTCCTC-3′; Il4, forward: 5′-ACTTGAGAGAGATCATCGGCA-3′ and reverse: 5′-AGCTCCATGAGAACACTAGAGTT-3′; Il5, forward: 5′-CGCTCACCGAGCTCTGTTG-3′ and reverse: 5′-CCAATGCATAGCTGGTGATTTTT-3′; Il13, forward: 5′-GCTTATTGAGGAGCTGAGCAACA-3′ and reverse: 5′-GGCCAGGTCCACACTCCATA-3′; Il17a, forward: 5′-CAGGGAGAGCTTCATCTGTGT-3′ and reverse: 5′-GCTGAGCTTTGAGGGATGAT-3′; Junb, forward: 5′-GCACTGGGGACTTTGAGG-3′ and reverse: 5′-CGTCGCTTCCCTCAGTTC-3′; Usp38, forward: 5′-TCATCAGGAGCCTAACCACC-3′ and reverse: 5′-TCAGGAGAGCAATTACCCACG-3′.
In vitro translation, immunoprecipitation, and immunoblot analysis
WT human and mouse USP38 as well as their DUB activity mutants, human JunB and c-Jun were in vitro translated using TNT T7 Quick Coupled Transcription/Translation Systems (L1170; Promega) according to the manufacturer’s instructions. For co-immunoprecipitation, lysates were mixed with the indicated antibodies (1 µg each) for 4 h and followed by the addition of 40 µl of protein A Plus-Sepharose beads (Sigma) for additional 2 h at 4°C. For in vitro DUB activity against endogenous poly-ubiquitination–modified JunB, WT mouse USP38 and its mutant from in vitro translation system were immunoprecipitated by anti-Flag M2 Magnetic Beads (M8823; Sigma) and then eluted from the beads according to the manufacturer’s instruction. Immunoprecipitates were washed three times by Triton X-100 lysis buffer, pH 7.6, and boiled in 20 µl of 3× loading buffer. For immunoblot analysis, the detailed protocols were followed as previously described (Yao et al., 2012).
Retroviral transduction
WT mouse usp38 and its DUB activity mutant were cloned into pMSCV vector. Empty vector or vectors encoding WT or mutant usp38 were cotransfected with pCL-eco into platE cells, which stably expressed gagpol and VSVG. The culture supernatants rich in packaged viruses were collected at 48 h and used to infect primary usp38−/− or WT CD4+ T cells that were activated by anti-CD3 and anti-CD28 for 16 h. 2 h later, viruses were withdrawn and CD4+ T cells were cultured for another 24 h in the presence of anti-CD3 and anti-CD28 followed by the second infection using platE culture supernatants collected at 72 h. The successfully infected cells are CD4+GFP+ in cytoflow analysis. For TCR activation, the virus-infected CD4+ T cells (containing both CD4+GFP+ and CD4+GFP− cells) were cultured in complete 1640 medium with 10 ng/ml mIL-2 overnight and then stimulated by anti-CD3 and anti-CD28 for various periods after IL-2 removal. For adoptive transfer, the prepared virus-infected usp38−/− CD4+ T cells were transferred into rag2−/− mice together with WT B cells at a 1:1 ratio of mixture.
Preparation of bone marrow–derived macrophages (BMDMs)
BMDMs were generated as described previously (Yao et al., 2017). In brief, after removal of red blood cells, bone marrow cells were cultured at 3 × 106 cells per well in 6-well plates in complete DMEM containing 30% L929 conditioned medium. Fresh medium was added every 2 d. On day 6, cells were collected for further experiments.
Statistical analysis
Statistical analyses were performed using with GraphPad Prism software. A two-tailed Student’s t test was used for analysis of differences between the groups. P value of 0.05 or less was considered as significant.
Online supplemental material
Fig. S1 shows the strategy for generation of usp38 complete deficient or T cell–specific deficient mice. Fig. S2 indicates that USP38 deficiency does not affect homeostatic immune cell development. Fig. S3 depicts that usp38 is not induced by subset-specific cytokines in T cells nor by innate sensor triggers in BMDMs. Fig. S4 illustrates that USP38 does not affect the cytokines-induced signaling and TCR-induced activation of NF-κB and MAPKs. Fig. S5 documents that the DUB activity of USP38 is essential for Th2 development.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grants 81430036, 91542119, and 91429307), National Key Research and Development Program of China (grant 2018YFA0507402) and the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB19000000).
The authors declare no competing financial interests.
Y. Qian and S. Chen designed the experiments and wrote the manuscript; S. Chen conducted the experiments and analyzed the data. Y. Yao, F. Yun, M. Cao, Y. Zhang, J. Wang, and X. Song helped with experiments or manuscript. Y. Qian supervised the study.
References
- Brownlie R.J., and Zamoyska R.. 2013. T cell receptor signalling networks: branched, diversified and bounded. Nat. Rev. Immunol. 13:257–269. 10.1038/nri3403 [DOI] [PubMed] [Google Scholar]
- Carr T.M., Wheaton J.D., Houtz G.M., and Ciofani M.. 2017. JunB promotes Th17 cell identity and restrains alternative CD4+ T-cell programs during inflammation. Nat. Commun. 8:301 10.1038/s41467-017-00380-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufner A., Kisser A., Niendorf S., Basters A., Reissig S., Schönle A., Aichem A., Kurz T., Schlosser A., Yablonski D., et al. 2015. The ubiquitin-specific protease USP8 is critical for the development and homeostasis of T cells. Nat. Immunol. 16:950–960. 10.1038/ni.3230 [DOI] [PubMed] [Google Scholar]
- Düwel M., Welteke V., Oeckinghaus A., Baens M., Kloo B., Ferch U., Darnay B.G., Ruland J., Marynen P., and Krappmann D.. 2009. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J. Immunol. 182:7718–7728. 10.4049/jimmunol.0803313 [DOI] [PubMed] [Google Scholar]
- Fahy J.V. 2015. Type 2 inflammation in asthma--present in most, absent in many. Nat. Rev. Immunol. 15:57–65. 10.1038/nri3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D., Elly C., Gao B., Fang N., Altman Y., Joazeiro C., Hunter T., Copeland N., Jenkins N., and Liu Y.C.. 2002. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 3:281–287. 10.1038/ni763 [DOI] [PubMed] [Google Scholar]
- Gao M., Labuda T., Xia Y., Gallagher E., Fang D., Liu Y.C., and Karin M.. 2004. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 306:271–275. 10.1126/science.1099414 [DOI] [PubMed] [Google Scholar]
- Gao S.F., Zhong B., and Lin D.. 2017. Regulation of T helper cell differentiation by E3 ubiquitin ligases and deubiquitinating enzymes. Int. Immunopharmacol. 42:150–156. 10.1016/j.intimp.2016.11.013 [DOI] [PubMed] [Google Scholar]
- Gour N., and Wills-Karp M.. 2015. IL-4 and IL-13 signaling in allergic airway disease. Cytokine. 75:68–78. 10.1016/j.cyto.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanpude P., Bhattacharya S., Dey A.K., and Maiti T.K.. 2015. Deubiquitinating enzymes in cellular signaling and disease regulation. IUBMB Life. 67:544–555. 10.1002/iub.1402 [DOI] [PubMed] [Google Scholar]
- Hartenstein B., Teurich S., Hess J., Schenkel J., Schorpp-Kistner M., and Angel P.. 2002. Th2 cell-specific cytokine expression and allergen-induced airway inflammation depend on JunB. EMBO J. 21:6321–6329. 10.1093/emboj/cdf648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikamp E.B., Patel C.H., Collins S., Waickman A., Oh M.H., Sun I.H., Illei P., Sharma A., Naray-Fejes-Toth A., Fejes-Toth G., et al. 2014. The AGC kinase SGK1 regulates TH1 and TH2 differentiation downstream of the mTORC2 complex. Nat. Immunol. 15:457–464. 10.1038/ni.2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissmeyer V., Macián F., Im S.H., Varma R., Feske S., Venuprasad K., Gu H., Liu Y.C., Dustin M.L., and Rao A.. 2004. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 5:255–265. 10.1038/ni1047 [DOI] [PubMed] [Google Scholar]
- Hirota T., Takahashi A., Kubo M., Tsunoda T., Tomita K., Doi S., Fujita K., Miyatake A., Enomoto T., Miyagawa T., et al. 2011. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat. Genet. 43:893–896. 10.1038/ng.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., and Sun S.C.. 2016. Ubiquitin signaling in immune responses. Cell Res. 26:457–483. 10.1038/cr.2016.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Wang H., Xiao Y., Jin J., Chang J.H., Zou Q., Xie X., Cheng X., and Sun S.C.. 2016. Otud7b facilitates T cell activation and inflammatory responses by regulating Zap70 ubiquitination. J. Exp. Med. 213:399–414. 10.1084/jem.20151426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H.S., Park Y., Elly C., and Liu Y.C.. 2013. Itch expression by Treg cells controls Th2 inflammatory responses. J. Clin. Invest. 123:4923–4934. 10.1172/JCI69355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathania M., Khare P., Zeng M., Cantarel B., Zhang H., Ueno H., and Venuprasad K.. 2016. Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-γt ubiquitination. Nat. Immunol. 17:997–1004. 10.1038/ni.3488 [DOI] [PubMed] [Google Scholar]
- Lambrecht B.N., and Hammad H.. 2015. The immunology of asthma. Nat. Immunol. 16:45–56. 10.1038/ni.3049 [DOI] [PubMed] [Google Scholar]
- Li B., Tournier C., Davis R.J., and Flavell R.A.. 1999. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 18:420–432. 10.1093/emboj/18.2.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licona-Limón P., Kim L.K., Palm N.W., and Flavell R.A.. 2013. TH2, allergy and group 2 innate lymphoid cells. Nat. Immunol. 14:536–542. 10.1038/ni.2617 [DOI] [PubMed] [Google Scholar]
- Lin M., Zhao Z., Yang Z., Meng Q., Tan P., Xie W., Qin Y., Wang R.F., and Cui J.. 2016. USP38 Inhibits Type I Interferon Signaling by Editing TBK1 Ubiquitination through NLRP4 Signalosome. Mol. Cell. 64:267–281. 10.1016/j.molcel.2016.08.029 [DOI] [PubMed] [Google Scholar]
- Liu X., Li H., Zhong B., Blonska M., Gorjestani S., Yan M., Tian Q., Zhang D.E., Lin X., and Dong C.. 2013. USP18 inhibits NF-κB and NFAT activation during Th17 differentiation by deubiquitinating the TAK1-TAB1 complex. J. Exp. Med. 210:1575–1590. 10.1084/jem.20122327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D.L. 2004. E3 ubiquitin ligases as T cell anergy factors. Nat. Immunol. 5:883–890. 10.1038/ni1106 [DOI] [PubMed] [Google Scholar]
- Naik E., Webster J.D., DeVoss J., Liu J., Suriben R., and Dixit V.M.. 2014. Regulation of proximal T cell receptor signaling and tolerance induction by deubiquitinase Usp9X. J. Exp. Med. 211:1947–1955. 10.1084/jem.20140860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Hirahara K., Onodera A., Endo Y., Hosokawa H., Shinoda K., Tumes D.J., and Okamoto Y.. 2017. Th2 Cells in Health and Disease. Annu. Rev. Immunol. 35:53–84. 10.1146/annurev-immunol-051116-052350 [DOI] [PubMed] [Google Scholar]
- Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., and Bernards R.. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell. 123:773–786. 10.1016/j.cell.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Nurieva R.I., Zheng S., Jin W., Chung Y., Zhang Y., Martinez G.J., Reynolds J.M., Wang S.L., Lin X., Sun S.C., et al. 2010. The E3 ubiquitin ligase GRAIL regulates T cell tolerance and regulatory T cell function by mediating T cell receptor-CD3 degradation. Immunity. 32:670–680. 10.1016/j.immuni.2010.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary C.E., Riling C.R., Spruce L.A., Ding H., Kumar S., Deng G., Liu Y., Seeholzer S.H., and Oliver P.M.. 2016. Ndfip-mediated degradation of Jak1 tunes cytokine signalling to limit expansion of CD4+ effector T cells. Nat. Commun. 7:11226 10.1038/ncomms11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver P.M., Cao X., Worthen G.S., Shi P., Briones N., MacLeod M., White J., Kirby P., Kappler J., Marrack P., and Yang B.. 2006. Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity. 25:929–940. 10.1016/j.immuni.2006.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul W.E. 2010. What determines Th2 differentiation, in vitro and in vivo? Immunol. Cell Biol. 88:236–239. 10.1038/icb.2010.2 [DOI] [PubMed] [Google Scholar]
- Reiley W.W., Zhang M., Jin W., Losiewicz M., Donohue K.B., Norbury C.C., and Sun S.C.. 2006. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat. Immunol. 7:411–417. 10.1038/ni1315 [DOI] [PubMed] [Google Scholar]
- Soares L., Seroogy C., Skrenta H., Anandasabapathy N., Lovelace P., Chung C.D., Engleman E., and Fathman C.G.. 2004. Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat. Immunol. 5:45–54. 10.1038/ni1017 [DOI] [PubMed] [Google Scholar]
- Takatsu K., and Nakajima H.. 2008. IL-5 and eosinophilia. Curr. Opin. Immunol. 20:288–294. 10.1016/j.coi.2008.04.001 [DOI] [PubMed] [Google Scholar]
- van Loosdregt J., Fleskens V., Fu J., Brenkman A.B., Bekker C.P., Pals C.E., Meerding J., Berkers C.R., Barbi J., Gröne A., et al. 2013. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity. 39:259–271. 10.1016/j.immuni.2013.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuprasad K. 2010. Cbl-b and itch: key regulators of peripheral T-cell tolerance. Cancer Res. 70:3009–3012. 10.1158/0008-5472.CAN-09-4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H., and Paul W.E.. 2013. Early signaling events that underlie fate decisions of naive CD4(+) T cells toward distinct T-helper cell subsets. Immunol. Rev. 252:12–23. 10.1111/imr.12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.O., Angkasekwinai P., Zhu J., Peng J., Liu Z., Nurieva R., Liu X., Chung Y., Chang S.H., Sun B., and Dong C.. 2009. Requirement for the basic helix-loop-helix transcription factor Dec2 in initial TH2 lineage commitment. Nat. Immunol. 10:1260–1266. 10.1038/ni.1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Wang Y., Chen F., Huang Y., Zhu S., Leng Q., Wang H., Shi Y., and Qian Y.. 2012. NLRC5 regulates MHC class I antigen presentation in host defense against intracellular pathogens. Cell Res. 22:836–847. 10.1038/cr.2012.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Chen S., Cao M., Fan X., Yang T., Huang Y., Song X., Li Y., Ye L., Shen N., et al. 2017. Antigen-specific CD8+ T cell feedback activates NLRP3 inflammasome in antigen-presenting cells through perforin. Nat. Commun. 8:15402 10.1038/ncomms15402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yamane H., and Paul W.E.. 2010. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 28:445–489. 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q., Jin J., Xiao Y., Zhou X., Hu H., Cheng X., Kazimi N., Ullrich S.E., and Sun S.C.. 2015. T Cell Intrinsic USP15 Deficiency Promotes Excessive IFN-γ Production and an Immunosuppressive Tumor Microenvironment in MCA-Induced Fibrosarcoma. Cell Reports. 13:2470–2479. 10.1016/j.celrep.2015.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.