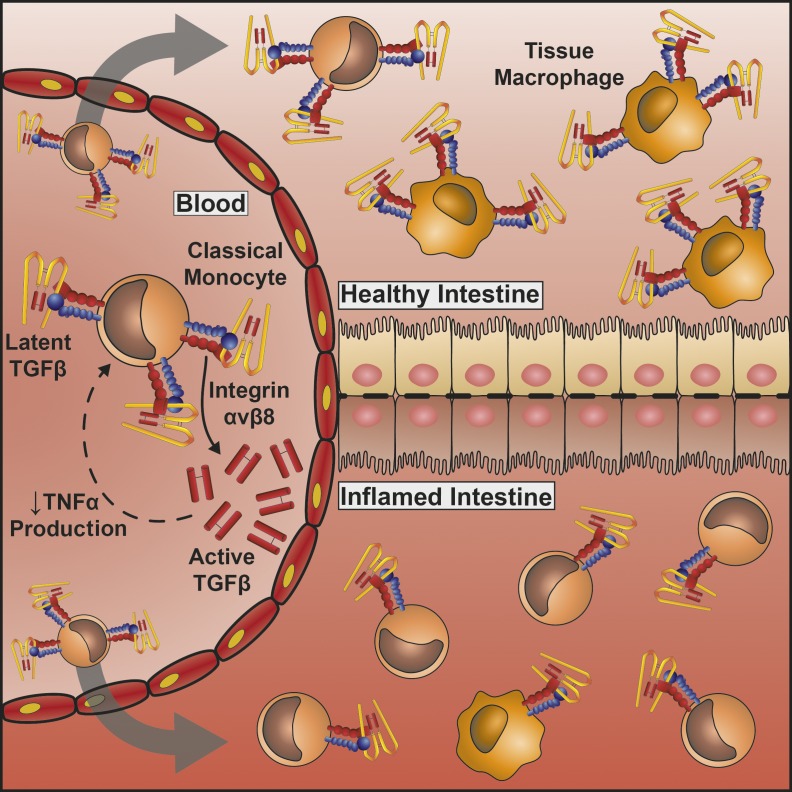
TGFβ is a crucial immune regulator and attractive therapeutic target but needs to be activated to function. Kelly et al. show that human monocytes and macrophages activate TGFβ via expression of an integrin, αvβ8, which dampens pro-inflammatory cytokine production and is disrupted in inflammatory bowel disease.
Abstract
Monocytes are crucial immune cells involved in regulation of inflammation either directly or via differentiation into macrophages in tissues. However, many aspects of how their function is controlled in health and disease are not understood. Here we show that human blood monocytes activate high levels of the cytokine TGFβ, a pathway that is not evident in mouse monocytes. Human CD14+, but not CD16+, monocytes activate TGFβ via expression of the integrin αvβ8 and matrix metalloproteinase 14, which dampens their production of TNFα in response to LPS. Additionally, when monocytes differentiate into macrophages, integrin expression and TGFβ-activating ability are maintained in anti-inflammatory macrophages but down-regulated in pro-inflammatory macrophages. In the healthy human intestine, integrin αvβ8 is highly expressed on mature tissue macrophages, with these cells and their integrin expression being significantly reduced in active inflammatory bowel disease. Thus, our data suggest that integrin αvβ8–mediated TGFβ activation plays a key role in regulation of monocyte inflammatory responses and intestinal macrophage homeostasis.
Graphical Abstract
Introduction
Monocytes are short-lived bone marrow–derived immune cells that play an important role in orchestrating inflammation but also promoting tolerance (Jakubzick et al., 2017). These cells are involved in phagocytosis, cytokine secretion, and antigen presentation but are probably best known for their role as a source of tissue macrophages. While not all macrophages are monocyte derived, with many being seeded at birth and maintained during life (Ginhoux and Guilliams, 2016), the intestine appears to be particularly reliant on peripheral monocytes as a source of intestinal macrophages (Bain et al., 2013, 2014). Although extensive work has been performed on pathways regulating murine monocyte and macrophage function, how these cells are regulated in humans and how such pathways are altered in inflammatory disease are less well understood.
TGFβ is a key multifunctional cytokine that is currently being targeted in a number of human diseases such as cancer and inflammatory disorders (Akhurst and Hata, 2012; Akhurst, 2017). An important function of TGFβ is its ability to regulate immunity, particularly T cell responses (Travis and Sheppard, 2014). While TGFβ is proposed to drive some effector T cell responses, specifically differentiation of Th9 (Dardalhon et al., 2008; Veldhoen et al., 2008) and Th17 cells (Korn et al., 2009), it is best known for its tolerogenic function (Kelly et al., 2017; Sanjabi et al., 2017). For example, TGFβ is important for promoting regulatory T cells (Chen et al., 2003; Davidson et al., 2007; Zheng et al., 2007; Liu et al., 2008) and blocking differentiation and function of Th1 (Gorham et al., 1998; Gorelik et al., 2002; Laouar et al., 2005) and Th2 cells (Gorelik et al., 2000; Heath et al., 2000; Kuwahara et al., 2012). Indeed, a key role for TGFβ in maintaining T cell homeostasis is shown by the observation that the TGFβ1 knockout mouse dies from CD4+ T cell–driven multi-organ inflammation early in life (Shull et al., 1992; Kulkarni et al., 1993). Although much is known about how TGFβ regulates T cell responses, much less is known about how TGFβ regulates innate immune responses, including regulation of monocytes and macrophages, and how such pathways are altered in disease.
TGFβ is expressed ubiquitously, but is always secreted as a latent complex, consisting of the latency-associated peptide (LAP) and active TGFβ moieties (Gleizes et al., 1997; Munger et al., 1997). Both LAP and active TGFβ are encoded by the TGFB1 gene, are cleaved from each other intracellularly but remain noncovalently bound, and are secreted with LAP folded around active TGFβ to mask its TGFβ receptor binding sites (Munger et al., 1997). Thus, the function of TGFβ in the regulation of immune responses is controlled by mechanisms that regulate latent TGFβ activation. However, with many of the previous studies being performed in mice, how TGFβ is activated to control human immunity and how this is altered in disease settings are poorly understood.
Here we show that human monocytes and macrophages are key activators of latent TGFβ, with such pathways not present in mice. Mechanistically, this ability to activate TGFβ is dependent on expression of an integrin, αvβ8, with this activation important in dampening pro-inflammatory cytokine production by monocytes. Furthermore, we find that this pathway is highly expressed on human intestinal macrophages, which are reduced in inflammatory bowel disease (IBD) and replaced by incoming pro-inflammatory monocytes/macrophages that lack expression of αvβ8. Our data therefore highlight a new pathway by which human monocyte/macrophage function is regulated, which could be therapeutically targeted to modulate TGFβ-mediated control of innate immunity in inflammatory disease.
Results and discussion
Human CD14+ monocytes activate TGFβ via expression of the integrin αvβ8
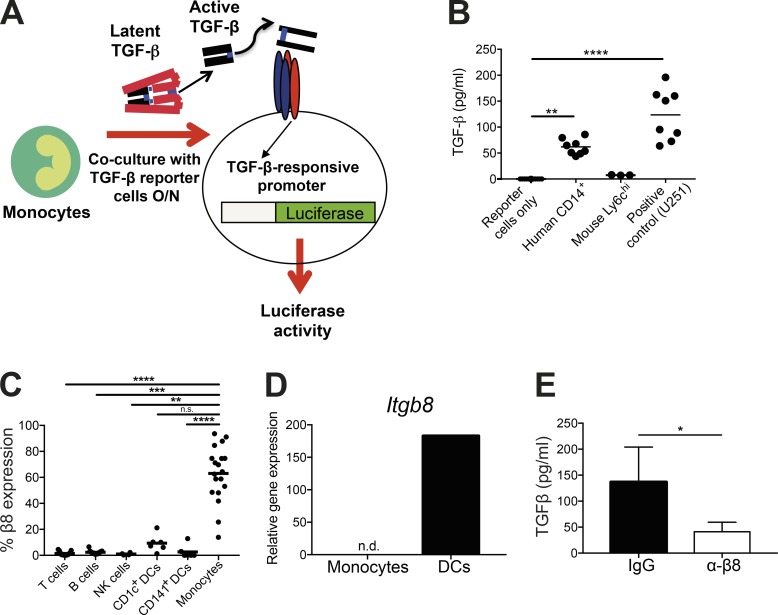
Monocytes represent the major mononuclear phagocyte cell population in the periphery with a crucial role in mediating inflammatory responses via regulation of adaptive immunity and maturation into tissue macrophages. Given its major role in controlling immune responses, we hypothesized that TGFβ activation may be important for regulation of monocyte function. We therefore first assessed the ability of CD14+ monocytes from human blood to activate TGFβ via co-culture with an active TGFβ reporter cell line (Abe et al., 1994; Fig. 1 A). As a positive control for the assay, we used U251 cells, which we have previously shown to activate high levels of TGFβ via integrin αvβ8 (Fenton et al., 2017). We found that human CD14+ monocytes were potent activators of TGFβ (Fig. 1 B). Surprisingly, this ability was not present on murine monocytes, which did not activate any TGFβ above the baseline of reporter cells alone (Fig. 1 B), suggesting that this pathway is specific to humans. The failure to detect TGFβ activation by mouse monocytes was not due to an inability of the reporter cells to respond to murine TGFβ, as a near identical response was observed in the presence of recombinant active mouse and human TGFβ1 (Fig. S1 A). Additionally, the lack of TGFβ activation by mouse monocytes could not be explained by their inability to produce latent TGFβ, which was shown to be equivalent to human monocytes (Fig. S1 B).
Figure 1.
Human peripheral blood monocytes activate TGFβ in an integrin αvβ8–dependent manner. (A) Illustration of TGFβ activation assay whereby cells of interest are co-cultured with active TGFβ reporter cells (Abe et al., 1994) expressing a TGFβ-inducible promoter fused to luciferase. (B) Human CD14+ blood monocytes (n = 8) and murine splenic Ly6c+ monocytes (n = 3, each data point representing two pooled mice) were isolated and co-cultured with active TGFβ reporter cells overnight, before measuring luciferase activity (active TGFβ concentration calculated from standard curve). Reporter cells alone were used as a negative control (n = 8), and U251 cells (n = 8; Fenton et al., 2017) were used as a positive control. (C) Healthy human PBMCs were analyzed by flow cytometry. Following gating on single, live cells, integrin β8 expression was identified on lymphocyte populations (CD3 for T cells, CD19 for B cells, and CD56 for NK cells), DCs (lineage-negative [CD3, CD14, CD15, CD16, CD19, CD20, and CD56] HLA-DR+ CD11c+ with subsets identified by CD1c and CD141 staining), and monocytes (HLA-DR+, CD14+, and CD16+) following gating on forward and side scatter (n = 4–19). (D) RNA from murine spleen Ly6chi monocytes (lineage-negative [TCRβ, NK1.1, B220, CD19, Ter119, Ly6G, and Siglec F], CD11b+, Ly6c+) was analyzed for Itgb8 expression by real-time PCR, normalizing to the housekeeping gene Hprt (n = 6, each from two pooled mice). Bone marrow–derived DCs were used as a positive control. (E) TGFβ activation of human CD14+ monocytes (n = 6) was measured in the presence of an isotype control antibody or an integrin β8 blocking antibody. Bars show mean and SD. n.s., not significant; n.d., not detected; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; P values were determined by Kruskal–Wallis with Dunn’s multiple comparisons test (B and C) and Wilcoxon matched-pairs signed rank test (E).
Next, we looked to identify mechanisms responsible for the ability of human CD14+ monocytes to activate TGFβ. As we and others have recently shown that an integrin, αvβ8, is a key activator of TGFβ in the murine immune system (Lacy-Hulbert et al., 2007; Travis et al., 2007; Acharya et al., 2010; Païdassi et al., 2011; Worthington et al., 2011, 2013, 2015), we assessed expression of integrin αvβ8 on human peripheral immune cells by flow cytometry. As recently described (Fenton et al., 2017), integrin αvβ8 was expressed by CD1c+ but not CD141+ dendritic cells (DCs; Fig. 1 C). However, integrin αvβ8 was most highly expressed by blood monocytes, with ∼70% of these cells expressing the integrin (Fig. 1 C). In contrast, minimal expression of integrin αvβ8 was observed on lymphocyte populations (Fig. 1 C). In agreement with their lack of ability to activate TGFβ (Fig. 1 B), expression of integrin αvβ8 on murine Ly6chi monocytes isolated from spleen was undetectable, in contrast to expression on murine DCs (Fig. 1 D), which we have previously shown use integrin αvβ8 to activate TGFβ (Travis et al., 2007; Worthington et al., 2011).
To directly test whether expression of integrin αvβ8 is responsible for the ability of human blood monocytes to activate TGFβ, we used a recently described integrin β8 functional blocking antibody (Fenton et al., 2017). Treatment of monocytes with the integrin-blocking antibody almost completely ablated their ability to activate TGFβ (Fig. 1 E). These results show that human but not murine monocytes can activate high levels of the cytokine TGFβ, which is dependent on expression of the integrin αvβ8.
Previous results have shown that both mouse and human DCs can activate TGFβ via integrin αvβ8, suggesting some level of conservation in this pathway (Worthington et al., 2011; Fenton et al., 2017). However, our data now suggest that there is a new cellular pathway that regulates TGFβ activity in the human immune system, via monocyte expression of integrin αvβ8. Although our data show that murine monocytes are not capable of activating TGFβ, we cannot rule out that, in some biological contexts such as infection or inflammation, certain mouse monocyte subsets could activate TGFβ via integrin αvβ8–dependent or –independent pathways. However, the species-specific differences observed are in agreement with studies suggesting significant gene expression (Ingersoll et al., 2010) and functional (Gaidt et al., 2016) differences between human and mouse monocytes. Given the great interest in targeting TGFβ function in human diseases such as cancer and inflammation (Akhurst and Hata, 2012), it is crucial to understand human-specific pathways controlling the function of TGFβ.
All human monocyte subsets express integrin αvβ8, but only CD14+ classical and intermediate monocytes activate TGFβ
In humans, distinct monocyte subsets can be identified based on expression of the surface proteins CD14 and CD16 (Ziegler-Heitbrock, 2015; Collin and Bigley, 2016). Thus, CD14+CD16− classical monocytes represent 70–80% of blood monocytes, with CD14+CD16+ intermediate and CD14−CD16+ nonclassical monocyte subsets also existing, with recent data suggesting that human monocytes differentiate in a sequential manner from classical to intermediate and nonclassical monocytes (Patel et al., 2017). The different subsets are proposed to have distinct functions; for example, classical monocytes can produce pro-inflammatory cytokines and migrate to tissue sites where they differentiate into macrophages (Epelman et al., 2014; Ginhoux and Jung, 2014), whereas nonclassical CD16+ monocytes are thought to patrol the endothelium and be involved in anti-viral responses (Auffray et al., 2007; Cros et al., 2010). Therefore, we next determined whether there was heterogeneity in the ability of different human monocyte subsets to activate TGFβ.
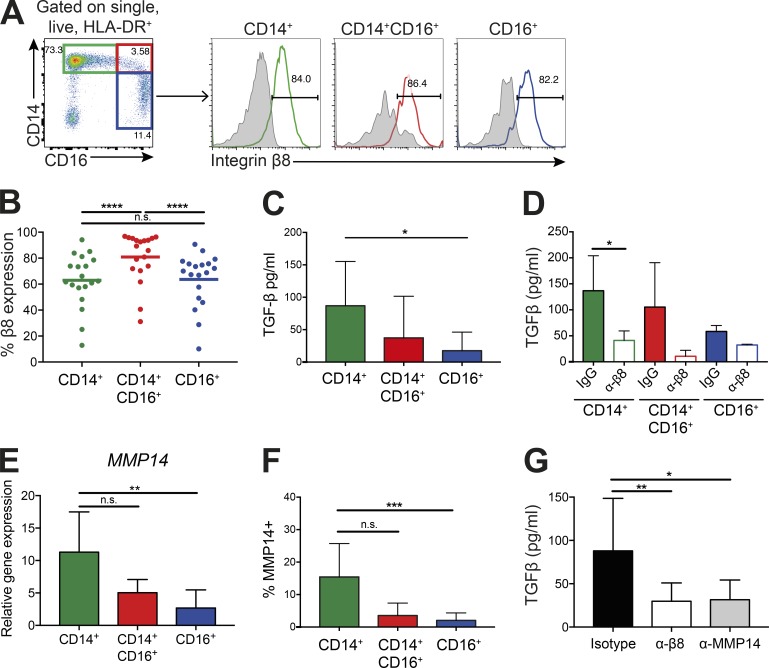
We first assessed integrin αvβ8 expression levels on different human monocyte subsets. Expression was similarly high on classical, intermediate, and nonclassical monocytes, with a slightly but significantly higher expression present on intermediate monocytes (Fig. 2, A and B), suggesting that all human monocyte subsets would be capable of activating TGFβ. However, although classical monocytes activated high levels of TGFβ, which was inhibited by an integrin αvβ8 functional blocking antibody, activation was almost completely absent by nonclassical monocytes, with intermediate levels of TGFβ activation observed by the intermediate monocyte subset (Fig. 2, C and D). Differences in TGFβ activation between monocytes could not be explained by differences in their ability to produce latent TGFβ, with all subsets expressing equivalent levels of latent TGFβ1 (Fig. S1 C). These data suggest that expression of integrin αvβ8 promotes TGFβ activation by classical monocytes, but that expression of the integrin is not sufficient to activate TGFβ in all monocyte subsets.
Figure 2.
The CD14+ classical monocyte subset activates high levels of TGFβ via integrin αvβ8 and MMP14. (A and B) PBMCs were gated on single, live HLA-DR+ cells, then as CD14+CD16− classical monocytes (green gate), CD14+CD16+ intermediate monocytes (red gate), and CD14−CD16+ nonclassical monocytes (blue gate). Levels of integrin β8 were measured on each subset (colored histogram, integrin β8 antibody; gray histogram, isotype control). Representative flow cytometry plots (A) and quantification of integrin β8 levels (B); n = 19 healthy donors. (C and D) Monocyte subsets were isolated by flow cytometry sorting and co-cultured overnight with a TGFβ reporter cell line (C; n = 10–13 donors) in the presence of an isotype or anti–integrin β8 antibody (D; n = 3–6 donors). Bars show mean and SD. (E and F) MMP14 expression was measured by real-time PCR, normalizing to the housekeeping gene B2M (E; n ≥ 5 for each subset) and flow cytometry (F; n = 8 for each subset) in monocyte subsets. Bars show mean and SD. (G) Active TGFβ production by CD14+ monocytes in the presence of an isotype control, integrin β8 blocking, or MMP14 blocking antibody (n = 6 donors). Bars show mean and SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n.s., not significant; P values were determined by Friedman test with Dunn’s multiple comparisons post-test (B, F, and G), Wilcoxon matched-pairs signed rank test (D), and Kruskal–Wallis with Dunn’s multiple comparisons post-test (C and E).
To determine mechanisms that promote differential TGFβ activation by monocyte subsets, despite all expressing high integrin αvβ8 levels, we focused on the expression of matrix metalloproteinase 14 (MMP14, also known as MT1-MMP), which has been suggested to synergize with integrin αvβ8 to promote TGFβ activation in some cells (Mu et al., 2002). Expression levels of MMP14 correlated with the ability of monocyte subsets to activate TGFβ via integrin αvβ8, with the highest levels seen in classical monocytes, and almost 10-fold lower levels in nonclassical monocytes (Fig. 2, E and F). To more directly assess the role for MMP14 in the ability of monocytes to activate TGFβ, we analyzed the ability of CD14+ monocytes to activate TGFβ in the presence or absence of an MMP14-blocking antibody. Inhibition of MMP14 resulted in a significant decrease in TGFβ activation, to similar levels observed in the presence of an integrin β8–blocking antibody (Fig. 2 G). Thus, overall, these data show an association between the expression of MMP14 and the ability of monocyte subsets to activate TGFβ, with classical monocytes requiring both integrin αvβ8 and MMP14 activity to promote TGFβ activation. Activation of TGFβ by other integrins, such as αvβ6, does not require MMP14 (Munger et al., 1999), and the ability of integrin αvβ8 to activate TGFβ in some cell types—for example, regulatory T cells (Stockis et al., 2017)—appears to be MMP14-independent. Here, our data suggest that expression of MMP14 at high levels on CD14+ monocytes likely cooperates with integrin αvβ8 to activate TGFβ via its enzymatic activity, a process that is lacking in other monocyte subsets.
Integrin αvβ8–mediated TGFβ activation dampens pro-inflammatory cytokine production by human blood classical monocytes
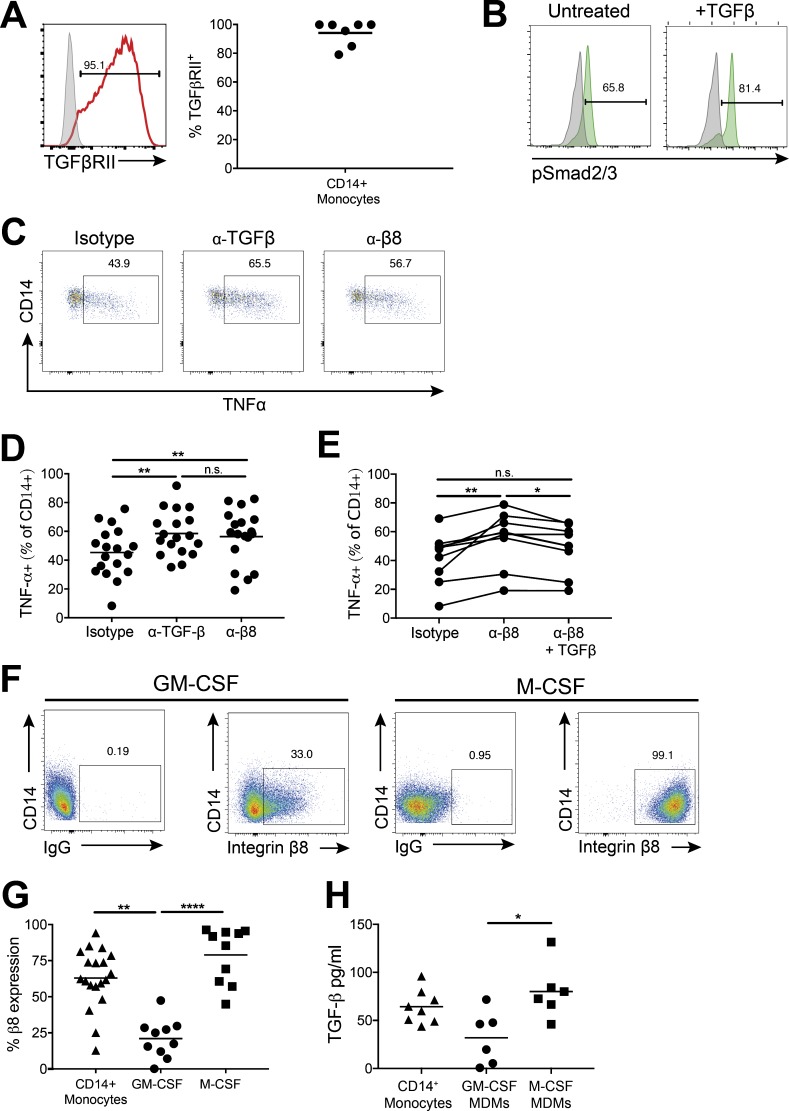
We next investigated the functional importance of TGFβ activation by human monocytes, focusing on CD14+ classical monocytes, given that they activate the highest levels of TGFβ via integrin αvβ8 and MMP14. In support of a potential autocrine function for TGFβ activation by classical monocytes, these cells express high levels of the TGFβ receptor (Fig. 3 A), which is functionally active, as shown by phosphorylation of the TGFβ signaling protein Smad2/3 in response to active TGFβ (Fig. 3 B).
Figure 3.
Integrin αvβ8–mediated TGFβ activation reduces monocyte TNFα production in response to LPS and is down-regulated upon differentiation to pro-inflammatory macrophages. (A) TGFβ receptor chain II (TGFβRII, red histogram) levels were assessed by flow cytometry on CD14+ monocytes (isotype control, gray histogram; graph shows cumulative data of n = 7 donors). (B) Levels of the TGFβ signaling molecule pSmad2/3 were measured in CD14+ classical monocytes (n = 3) after treatment of PBMCs with 5 ng/ml TGFβ for 30 min (isotype control shown in gray). Representative flow cytometry plots are displayed. (C and D) TNFα production by CD14+ monocytes was measured by intracellular flow cytometry, following PBMC culture overnight in the presence of an isotype control, anti-TGFβ, or anti–integrin β8 blocking antibody followed by activation with 10 ng/ml LPS for 4 h in the presence of a protein transport inhibitor; C shows representative flow cytometry plots; D shows cumulative data (n = 18). (E) TNFα production by CD14+ monocytes as in C and D, with addition of 5 ng/ml active TGFβ in the presence of anti–integrin β8 antibody (n = 9). (F and G) CD14+ monocytes were cultured for 6 d with GM-CSF or M-CSF to differentiate to macrophages and integrin β8 expression measured by flow cytometry; F displays representative flow cytometry plots for macrophage expression of integrin β8; G shows pooled data for monocyte and macrophage expression of integrin β8 from 10–19 donors. (H) Monocytes and MDMs were co-cultured overnight with active TGFβ reporter cells (n = 6–8 donors). *, P < 0.05; **, P < 0.01; ****, P < 0.0001, n.s., not significant; P values were determined by Friedman test with Dunn’s multiple comparisons post-test (D and E) or Kruskal–Wallis with Dunn’s multiple comparisons post-test (G and H).
As monocytes are key, short-lived innate effector cells that can produce pro-inflammatory cytokines in response to pathogen challenge, and TGFβ can regulate cytokine production in other immune cells such as T cells (Kelly et al., 2017), we hypothesized that integrin αvβ8–mediated TGFβ activation may modulate the ability of monocytes to produce cytokines. To test this possibility, cells were cultured in the presence of blocking antibodies against TGFβ or integrin αvβ8 and then stimulated with LPS. In the presence of a TGFβ-blocking antibody, monocytes produced significantly increased levels of the pro-inflammatory cytokine TNFα (Fig. 3, C and D). Importantly, blockade of integrin αvβ8 yielded a similar increase in TNFα production by monocytes (Fig. 3, C and D), with this increase significantly reduced by addition of active TGFβ (Fig. 3 E).
Thus, these results indicate that integrin-mediated TGFβ activation normally functions to restrict CD14+ monocyte pro-inflammatory cytokine secretion. Such a function may act to potentiate over-activation of monocytes to prevent excessive inflammation. However, the functional importance of such a feedback loop requires further attention, especially in the light of recent data suggesting that, at least in the mouse, TNFα is an important survival factor for monocytes in steady state and inflammation (Wolf et al., 2017).
Integrin αvβ8–mediated TGFβ activation is up-regulated on anti-inflammatory macrophages and down-regulated on pro-inflammatory macrophages
An important function of monocytes is the migration from the bone marrow and peripheral blood to tissue sites where they can differentiate into macrophages during homeostasis and inflammation (Ginhoux and Guilliams, 2016). We therefore tested whether the ability of monocytes to activate TGFβ was altered after differentiation into macrophages. To this end, we differentiated monocytes to macrophages in vitro, using the cytokines GM-CSF and M-CSF, which have been shown previously to give rise to pro-inflammatory and anti-inflammatory phenotypes, respectively (Murray et al., 2014). Indeed, in agreement with previous findings (Akagawa, 2002; Akagawa et al., 2006), we showed that GM-CSF–derived macrophages produced lower levels of IL-10 and were less phagocytic than M-CSF–derived macrophages (Fig. S2, A and B).
We found that pro-inflammatory GM-CSF–differentiated macrophages down-regulated expression of integrin αvβ8 compared with CD14+ monocytes, whereas monocytes differentiated to anti-inflammatory macrophages in the presence of M-CSF expressed high levels of integrin αvβ8, displaying close to 100% expression (Fig. 3, F and G). Additional polarization of GM-CSF–treated cells with IFN-γ and M-CSF–treated cells with IL-4, shown to further promote pro- and anti-inflammatory phenotypes, respectively (Murray et al., 2014), showed similar changes in integrin αvβ8 expression levels to GM-CSF and M-CSF treatment alone (Fig. S2, C–E). Expression of MMP14 was uniformly higher on both GM-CSF– and M-CSF–differentiated macrophages versus monocytes (Fig. S2 F), suggesting that signals received during the differentiation caused up-regulation of MMP14 expression.
These results suggest that macrophage populations may have differential abilities to activate TGFβ. Indeed, M-CSF–differentiated macrophages activated similar levels of TGFβ to CD14+ monocytes, whereas GM-CSF–differentiated macrophages showed a reduced ability to activate TGFβ (Fig. 3 H). Thus, our data show that anti-inflammatory macrophages are able to activate TGFβ, whereas this ability is down-regulated on more pro-inflammatory macrophage populations. Indeed, these results fit with previous findings suggesting that TGFβ is an important “M2” anti-inflammatory macrophage marker (Italiani and Boraschi, 2014).
Human intestinal macrophages express high levels of integrin αvβ8 and are reduced in patients with IBD
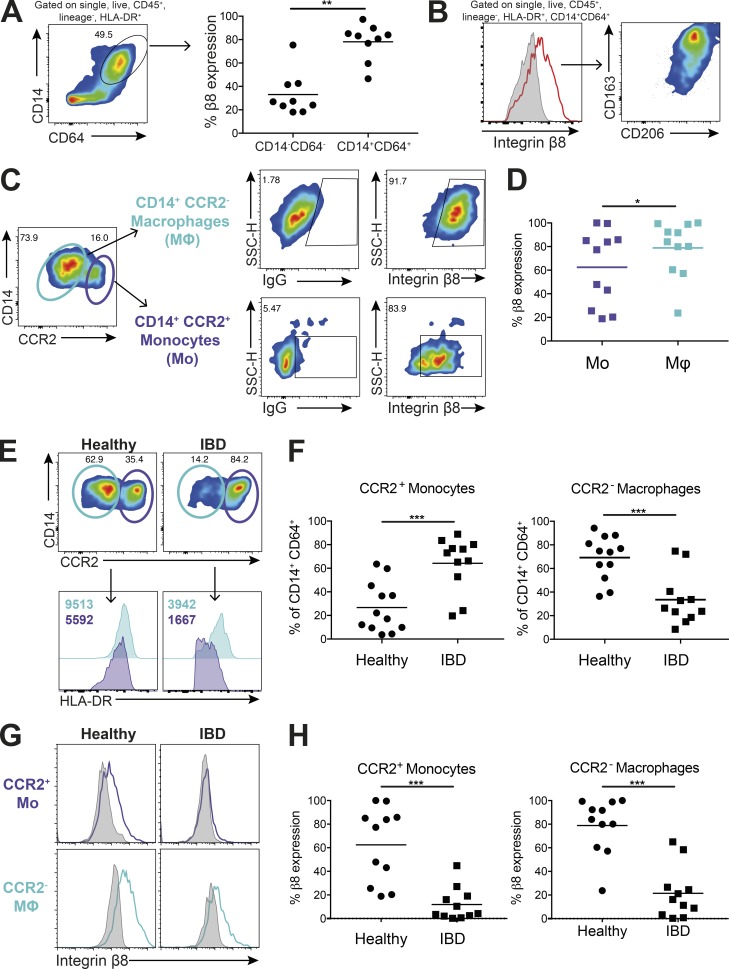
While in vitro–derived macrophages are a useful model, macrophages exist in multiple different phenotypes in vivo (Xue et al., 2014). In healthy tissue, anti-inflammatory macrophages capable of maintaining homeostasis are common, whereas a switch to pro-inflammatory monocytes/macrophages occurs during infection and inflammation (Varol et al., 2015). Given the proposed importance of tolerogenic macrophages in the intestine and the fact that intestinal macrophages derive predominantly from blood monocytes (Bain et al., 2014), coupled with data suggesting that TGFβ is a crucial cytokine in maintenance of immune homeostasis in the gut (Konkel and Chen, 2011; Kelly et al., 2017), we next assessed expression of integrin αvβ8 on human intestinal tissue macrophages. We found that intestinal macrophages, identified by gating on CD45+ lineage-negative, HLA-DR+CD14+CD64+ cells (Fig. S3 A), expressed high levels of integrin αvβ8 compared with CD64− cells (Fig. 4 A). These integrin αvβ8+ intestinal macrophages also express CD163 and CD206 (Fig. 4 B), which have been proposed as markers of anti-inflammatory intestinal macrophages (Bain et al., 2013). Notably, murine intestinal macrophages, like murine monocytes, do not express integrin αvβ8 (Fig. S3 B), again highlighting an important species-specific difference in pathways that activate TGFβ.
Figure 4.
Integrin αvβ8 is highly expressed on intestinal macrophages and is decreased in patients with IBD. (A) Integrin β8 levels were analyzed by flow cytometry on human colonic monocyte/macrophage populations, gated as single, live, CD45+, lineage-negative (CD3, CD15, CD19, CD20, and CD56), HLA-DR+CD14+CD64+ cells. Expression of integrin αvβ8 was compared with equivalent CD14−CD64− cells (n = 9). (B) Integrin β8+ monocytes/macrophages were analyzed for expression of the tissue macrophage markers CD163 and CD206 by flow cytometry (representative image from n = 9 donors). (C and D) Human monocyte/macrophage populations, identified as described in A, were gated for expression of the monocyte marker CCR2 and expression of integrin β8 on monocytes (CD14+CCR2+) and macrophages (CD14+CCR2−) analyzed. Representative flow cytometry plots (C) and cumulative data (D) from 11 donors are depicted. SSC-H, side scatter height. (E) Monocyte and macrophage populations in the intestinal tissue of healthy versus inflamed IBD patients were analyzed by flow cytometry (gated as described in C and D) with histograms showing levels of HLA-DR expression on monocytes (purple) and macrophages (blue) in healthy tissue compared with IBD. Representative plots shown, with F showing cumulative data for CCR2+ monocyte and CCR2– macrophage levels from 11–12 healthy or IBD donors. (G and H) Expression levels of integrin αvβ8 were measured on monocyte/macrophage populations from healthy patients and patients with IBD. Representative histograms (G) and cumulative data (H) from 11 healthy and IBD donors are depicted. Bars show mean and SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; P values were determined by Wilcoxon matched-pairs signed rank test (A and D) or Mann–Whitney (F and H).
We also gated cells on the basis of CCR2, with CCR2+ cells proposed to represent recently emigrated intestinal monocytes that are yet to differentiate to macrophages (Bain and Mowat, 2014). Such cells are a minor fraction of total intestinal HLA-DR+CD14+CD64+ cells in the healthy human intestine (Fig. 4 C) and display low forward scatter/side scatter levels compared with CCR2– cells (Fig. S3 C), further supporting that such cells represent a monocyte/immature macrophage population. We found that both CD14+CCR2+ monocyte and CD14+CCR2− macrophage populations in the intestine expressed high integrin αvβ8 levels, with significantly higher levels found on tissue macrophages (Fig. 4, C and D). Thus, together these data suggest that integrin αvβ8 is highly expressed on monocytes that enter the healthy intestine, and remains high as cells develop into macrophages.
Having established that integrin αvβ8 is conserved between human peripheral CD14+ monocytes and intestinal macrophages in the healthy intestine, we next addressed whether the pathway was altered in individuals with IBD. We observed a shift in the balance of monocytes and macrophages in the inflamed versus healthy intestine, with a significant increase in a CD14hi HLA-DRdim CCR2+ population (Fig. 4, E and F), proposed from previous work in mouse and humans to represent an inflammatory monocyte population (Kamada et al., 2008; Bain et al., 2013; Ogino et al., 2013; Thiesen et al., 2014; Magnusson et al., 2016). This influx of monocytes substantially reduces the proportion of tissue macrophages present (Fig. 4, E and F). Importantly, whereas both the monocyte population and tissue macrophages display clear expression of integrin αvβ8 in the healthy intestine, expression of the integrin is significantly down-regulated on inflammatory monocytes and remaining tissue macrophages in IBD (Fig. 4, G and H). MMP14 expression was also present in both intestinal monocytes and macrophages in healthy tissue, but was not altered in cells from patients with IBD (Fig. S3 D). When patients were stratified into those with Crohn’s disease or ulcerative colitis, significant reductions in monocyte and macrophage integrin β8 expression were observed in both disorders (Fig. S3, E and F). Thus, our results show that integrin αvβ8 is highly expressed on monocyte/macrophage populations in the steady-state intestine, but during inflammation, populations of integrin αvβ8–negative monocytes predominate.
Previous data have shown that, although levels of total latent TGFβ are increased in the intestine in IBD (Babyatsky et al., 1996), TGFβ signaling is in fact decreased (Monteleone et al., 2001). This apparently paradoxical finding was proposed to be due to enhanced expression of the TGFβ signaling inhibitor Smad7 in IBD, with targeting of Smad7 promoting enhanced TGFβ signaling (Monteleone et al., 2001). Our data now suggest that a reduced ability to activate TGFβ in the inflamed intestine, via down-regulation of integrin αvβ8–expressing monocytes/macrophages, may contribute to the reduced ability to trigger TGFβ signaling in the inflamed intestine. Together with our data (Fig. 3, C–E) showing that integrin αvβ8 expression on monocytes is required for the dampening of pro-inflammatory cytokine expression, our work suggests that down-regulation of this autocrine regulatory pathway in inflammatory monocyte/macrophage populations may promote inflammation. Additionally, as recent work in mouse models has suggested that TGFβ can promote intestinal macrophage differentiation in the steady state (Schridde et al., 2017), reduction of TGFβ activation pathways during inflammation may help explain the reduced proportions of intestinal macrophage populations observed in IBD patients.
To conclude, we have demonstrated that human but not murine monocytes and macrophages are specialized to activate TGFβ via expression of integrin αvβ8, which plays a regulatory role in dampening LPS-induced pro-inflammatory cytokine responses. This pathway is present on intestinal monocyte/macrophage populations during health, but is dysregulated during intestinal inflammation, suggesting that the pathway may represent a key immune regulatory mechanism in inflammation. Thus, promoting monocyte/macrophage ability to activate TGFβ via integrin αvβ8 may be a valid therapeutic strategy in inflammatory disorders such as IBD. To achieve this, it will be important in the future to determine signals that lead to the down-regulation of integrin αvβ8 during inflammation, which could then be targeted to promote the pathway. Targeting integrin αvβ8 (via small molecule inhibitors or blocking antibodies) is also attractive in the treatment in fibrosis (given the pro-fibrotic properties of TGFβ) and cancer (given the anti-inflammatory properties of TGFβ in the tumor microenvironment). However, our new data now highlight previously unappreciated cell types that will also be targeted by blocking integrin αvβ8 in humans, which will need to be considered in future therapeutic strategies.
Materials and methods
Samples and ethics
Human samples were obtained according to the principles expressed in the Declaration of Helsinki and under local ethical guidelines, and approved by the North West National Ethics Service (reference no. 15/NW/0007). Intestinal tissue samples were obtained via the Central Manchester University Hospitals National Health Service (NHS) Foundation Trust Biobank from patients undergoing surgery or endoscopy for chronically active Crohn’s disease or ulcerative colitis. The diagnosis of IBD was confirmed by established clinical, radiological, and histological criteria. Specimens were collected from healthy areas of colorectal cancer patients or patients undergoing endoscopy as noninflammatory controls. All patients provided written, informed consent for the collection of tissue samples and subsequent analysis. Healthy blood was obtained from donors recruited locally (according to the University of Manchester ethics and guidelines) or from leukocyte cones sourced from the blood bank (National Blood Service, Manchester, UK). Patient demographic and clinical information is listed in Table 1.
Table 1. Demographic and clinical data for patients who donated intestinal samples analyzed in the study.
| Sample type | Gender | Tissue | Age (yr) | Diagnosis | IBD medication |
|---|---|---|---|---|---|
| Healthy (non-IBD) samples | |||||
| Resections | Male | Colon | 73 | Sigmoid colon cancer | N/A |
| Male | Colon | 73 | Concurrent bladder and rectal cancer | N/A | |
| Female | Colon | 74 | Sigmoid adenocarcinoma | N/A | |
| Male | Colon | 75 | Colorectal cancer with liver metastases | N/A | |
| Female | Colon | 63 | Caecal tumor and bladder tumor | N/A | |
| Female | Colon | 73 | Adenocarcinoma | N/A | |
| Male | Colon | 64 | Adenocarcinoma | N/A | |
| Male | Colon | 64 | Adenocarcinoma | N/A | |
| Female | Colon | 51 | Colon cancer with metastases | N/A | |
| Male | Colon | 40 | Rectal carcinoma with metastases | N/A | |
| Male | Rectum | 67 | Rectal carcinoma | N/A | |
| Female | Colon | 57 | Colon cancer with metastases | N/A | |
| Female | Colon | 61 | Colonic polyp | N/A | |
| Biopsies | Male | Colon | 46 | Healthy (screen for cancer) | N/A |
| Female | Colon | 61 | Healthy (screen for cancer) | N/A | |
| Male | Colon | 79 | Healthy (screen for cancer) | N/A | |
| IBD patients | |||||
| Resections | Female | Colon | 31 | Active Crohn’s disease | Azathioprine |
| Female | Colon | 21 | Active Crohn’s disease | Vedolizumab, prednisolone | |
| Male | Colon | 25 | Active Crohn’s disease | Infliximab, methotrexate | |
| Female | Colon | 47 | Active Crohn's disease | Prednisolone, simethicone | |
| Female | Colon | 59 | Active Crohn’s disease | Azathioprine, adalimumab, methotrexate | |
| Male | Colon | 51 | Active ulcerative colitis | Adalimumab | |
| Male | Colon | 25 | Active ulcerative colitis | Azathioprine, mesalazine | |
| Biopsies | Male | Rectum | 25 | Active Crohn’s disease | Adalimumab, methotrexate |
| Female | Colon | 23 | Active Crohn’s disease | Infliximab | |
| Male | Rectum | 17 | Active ulcerative colitis | Infliximab | |
| Male | Rectum | 57 | Active ulcerative colitis | Prednisone, omeprazole, mesalazine | |
N/A, not applicable.
Peripheral blood mononuclear cells (PBMCs) and monocyte isolation
PBMCs were separated from healthy donor blood by layering over Ficoll Paque Plus (GE Healthcare) before centrifugation. Monocytes were isolated from PBMCs by magnetic bead separation using CD14 microbeads (Miltenyi Biotech), as per the manufacturer’s instructions. Monocyte purity was consistently >95%. Monocyte subsets were isolated by pre-enrichment using a Pan-Monocyte Isolation Kit or HLA-DR bead isolation kit (both from Miltenyi Biotech) followed by sorting by flow cytometry based on CD14 and CD16 expression.
Monocyte-derived macrophages (MDMs)
MDMs were differentiated in RPMI plus l-glutamine (Sigma-Aldrich) with 10% FCS (Life Technologies, Thermo Fisher Scientific) and 1% penicillin/streptomycin (Sigma-Aldrich) in 12-well plates at 0.5 × 106/ml for 6–7 d. Recombinant human GM-CSF or M-CSF (Peprotech) was added at a concentration of 50 ng/ml. Fresh media and cytokines were added on day 3.
Murine monocyte isolation
Monocytes were isolated from the spleens of wild-type C57BL/6 mice. Briefly, spleens were pressed through a 40-µM cell strainer and washed with RPMI. Red cells were lysed using red cell lysis buffer (Sigma-Aldrich), and cells pre-enriched using a Monocyte Isolation Kit (BM; Miltenyi Biotech) as per the manufacturer’s instructions. Fc receptor block of enriched cells, using TruStain FcX (BioLegend), was performed, followed by staining and sorting of cells using the FACS Aria (BD Biosciences) via gating on lineage-negative (TCRβ, NK1.1, B220, CD19, Ter119, Ly6G, and Siglec F), CD11b+, Ly6c+ monocytes. 7-AAD (BioLegend) was used to discriminate dead cells (see Table S1 for antibody details).
Isolation of lamina propria mononuclear cells (LPMCs)
LPMCs were isolated from tissue by first removing muscle and fat and then washing in HBSS, penicillin-streptomycin, G418 (Melford), and 1 mM dithiothreitol (Sigma-Aldrich). Tissue was then washed three times in HBSS, penicillin-streptomycin, G418, and 1 mM EDTA (Sigma-Aldrich) to remove epithelial cells. The tissue was then cut into small pieces and digested in Collagenase A (Roche) in 10% RPMI, penicillin-streptomycin, and G418 with 60 U/ml DNase I (Roche) for 1 h in a shaking incubator at 37°C. After this time, the tissue was filtered through a 40-µM filter, and cells were analyzed by flow cytometry.
Flow cytometry surface staining
PBMCs, monocytes, MDMs, and LPMCs were stained in PBS with 1% FCS and 0.1% sodium azide (FACS buffer) by first preincubating in 2% mouse serum (Invitrogen), followed by staining for 20 min on ice (see Table S1 for details of all antibodies used). Cells were analyzed using a LSR Fortessa cytometer (BD Biosciences). Data were analyzed using Flowjo v10.1 (TreeStar).
Phosphorylated (p)Smad2/3 intracellular staining
For pSmad2/3 staining, 100 µl of whole blood (100 µl/test) was either untreated or stimulated with TGFβ (Peprotech) for 30 min and then fixed by adding 1 ml of prewarmed 1X Lyse/Fix Buffer (BD Biosciences). Surface staining was performed for 60 min at room temperature using HLA-DR, CD14, and CD16, followed by washing and permeabilization using ice-cold Perm buffer III (BD Biosciences) for 30 min on ice. Following permeabilization, intracellular staining was performed using pSmad2/3 antibody (BD Biosciences) for 60 min at room temperature. Samples were washed in FACS buffer and acquired on the LSR Fortessa flow cytometer on the same day.
Monocyte intracellular cytokine staining
Isolated PBMCs were cultured at 2 × 106/ml in 10% RPMI in a 96-well flat-bottomed plate overnight in the presence of mouse IgG (40 µg/ml; Sigma-Aldrich) and isotype control antibody (40 µg/ml; clone MOPC-21; BioXCell), TGFβ blocking antibody (40 µg/ml; clone 1D11; BioXCell), or integrin β8 blocking antibody (20 µg/ml; clone ADWA16; Fenton et al., 2017). In some experiments, 5 ng/ml active TGFβ (Peprotech) was also added to cultures overnight. Cells were then stimulated with 10 ng/ml LPS (L4391; Sigma-Aldrich) for 4 h in the presence of GolgiPlug protein transport inhibitor (BD Biosciences). Following stimulation, cells were stained with lineage markers (CD3, CD15, CD19, CD20, and CD56), HLA-DR, CD14, and CD16, permeabilized with 0.5% saponin (Sigma-Aldrich), and then stained for intracellular TNFα. Cells were washed in saponin, resuspended in FACS buffer, and acquired on the LSR Fortessa on the same day.
TGFβ activation assay
Transformed mink lung epithelial cells transfected with a plasmid containing firefly luciferase complementary DNA downstream of a TGFβ-sensitive promoter (a gift from D. Rifkin, New York University, New York, NY; Abe et al., 1994) were seeded in 96-well flat-bottomed plates in DMEM containing 10% FCS for at least 3 h. Monocytes or MDMs were added in the same medium in the presence of mouse IgG (40 µg/ml; Sigma-Aldrich) and isotype control antibody (40 µg/ml; clone MOPC-21; BioXCell), TGFβ blocking antibody (40 µg/ml; clone 1D11; BioXCell), integrin β8 blocking antibody (20 µg/ml; clone ADWA16) as described previously (Worthington et al., 2011; Fenton et al., 2017), or MMP14 blocking antibody (MAB3328; Merck Millipore). Cells were co-cultured for 16–20 h, and luciferase was then detected using the Luciferase Assay System (Promega) using an Infinite M200 Pro instrument (Tecan). A standard curve of active TGFβ (Peprotech) was generated and used to calculate levels of active TGFβ from luminescence intensity observed. TGFβ activity was determined as sample values with anti-TGFβ antibody background values subtracted. For comparison of mouse and human active TGFβ, recombinant proteins were purchased from BioLegend.
Real-time PCR
RNA was isolated using RNeasy Mini or Micro kits (Qiagen). cDNA was synthesized using a High-Capacity RNA-to-cDNA kit (Applied Biosystems), and PCR was performed with Taqman Universal Mastermix II (Applied Biosystems/Thermo Fisher Scientific) on a QuantStudio 12K Flex instrument (Applied Biosystems). Primers used were as follows (all from Applied Biosystems): mouse Itgb8 primers (Mm00623991_m1), mouse Tgfb1 primers (Mm01178820_m1), human MMP14 primers (Hs01037003_g1), and human TGFB1 primers (Hs00998133_m1). Expression levels were calculated relative to the control genes B2M (Hs00984230_m1) for human and Hprt (Mm01545399_m1) for mouse.
IL-10 ELISA
96-well Nunc Maxisorp plates (eBioscience) were coated with anti–IL-10 capture antibody overnight (eBioscience). Following a blocking step with ELISA/ELISPOT diluent (eBioscience), supernatants from day 7 MDM cultures were incubated in wells followed by addition of a biotinylated IL-10 detection antibody (eBioscience). Avidin-HRP (eBioscience) was added and 3,3′,5,5′-tetramethylbenzidine (TMB) solution was used to detect peroxidase activity, followed by TMB stop solution (BioLegend). Absorbance readings were measured using an Infinite M200 Pro plate reader (Tecan).
Phagocytosis assay
On day 7 of culture, MDM supernatants were removed and replaced with fresh media. pHrodo Red E. Coli Bioparticles Conjugate for Phagocytosis (Life Technologies/Thermo Fisher Scientific) was added and incubated for 1 h, with pretreatment of cells with 5 µg/ml of Cytochalasin D (Sigma-Aldrich) for 30 min before addition of pHrodo particles used as a negative control. Cells were washed, removed by gentle scraping, fixed in 2% formaldehyde (Sigma-Aldrich), and analyzed by flow cytometry using an LSR II flow cytometer (BD Biosciences).
Statistical analysis
Data were analyzed using GraphPad Prism version 7.0 for Mac. Statistical differences were tested as described in figure legends. Data are displayed as mean or mean and SD, as described in the figure legends.
Online supplemental material
Fig. S1 shows active TGFβ reporter cell responses to mouse and human TGFβ1, and expression of TGFβ1 by human and mouse monocytes, and human monocyte subsets. Fig. S2 shows functional characterization of different MDM models and integrin αvβ8 and MMP14 expression on MDMs differentiated with various cytokines. Fig. S3 shows the gating strategy for monocyte/macrophage populations in human intestinal tissue, expression of integrin β8 on murine intestinal macrophages, MMP14 expression by monocytes and macrophages in health versus IBD, and expression of integrin β8 in Crohn’s disease and ulcerative colitis patients. Table S1 is a list of monoclonal antibodies used in the study.
Supplementary Material
Acknowledgments
We thank Amha Atakilit and Prof. Dean Sheppard (University of California, San Francisco) for providing anti–integrin β8 antibody reagents, Dr. Gareth Howell and Mike Jackson in the Faculty of Biology, Medicine and Health Flow Cytometry Core Facility for help with flow cytometry, and members of the Biological Service Facility at the University of Manchester for help with animal work. We thank Dr. Jay Brown, Yasmin Ahmed, and other members of the Manchester University NHS Foundation Trust Biobank for help in acquisition of human intestinal samples, and all staff and patients at Manchester University NHS Foundation Trust who aided with collection and donated tissue samples. We thank Prof. Judith Allen, Prof. Richard Grencis, Prof. Dave Thornton, Prof. Daniel Davis, Dr. Anu Goenka, and Dr. John Grainger for critical reading of our manuscript, Dr. Joshua Casulli for creation of the graphical abstract, and Dr. Tovah Shaw for assistance with sample processing.
Work was funded by a Manchester Collaborative Centre for Inflammation Research award and the Medical Research Council (MR/M00242X/1; M.A. Travis), the Wellcome Trust (grant no. 209741/Z/17/Z; M.A. Travis), and a Wellcome Trust clinical PhD training fellowship (108746/Z/15/Z; E.E. Shuttleworth). The Wellcome Centre for Cell-Matrix Research, University of Manchester, is supported by core funding from the Wellcome Trust (grant no. 203128/Z/16/Z).
The authors declare no competing financial interests.
Author contributions: A. Kelly and M.A. Travis conceived the project; M.A. Travis, A. Kelly, C.P. McEntee, S. Gunaltay, E.E. Shuttleworth, and S.A. Houston designed experiments; A. Kelly, C.P. McEntee, S. Gunaltay, E.E. Shuttleworth, C. Smedley, S.A. Houston, T.M. Fenton, and E.R. Mann performed experiments; A. Kelly and M.A. Travis wrote the manuscript, and all other authors provided critical review of the manuscript; and E.E. Shuttleworth and S. Levison provided essential patient samples and clinical analysis.
References
- Abe M., Harpel J.G., Metz C.N., Nunes I., Loskutoff D.J., and Rifkin D.B.. 1994. An assay for transforming growth factor-beta using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal. Biochem. 216:276–284. 10.1006/abio.1994.1042 [DOI] [PubMed] [Google Scholar]
- Acharya M., Mukhopadhyay S., Païdassi H., Jamil T., Chow C., Kissler S., Stuart L.M., Hynes R.O., and Lacy-Hulbert A.. 2010. αv Integrin expression by DCs is required for Th17 cell differentiation and development of experimental autoimmune encephalomyelitis in mice. J. Clin. Invest. 120:4445–4452. 10.1172/JCI43796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagawa K.S. 2002. Functional heterogeneity of colony-stimulating factor-induced human monocyte-derived macrophages. Int. J. Hematol. 76:27–34. 10.1007/BF02982715 [DOI] [PubMed] [Google Scholar]
- Akagawa K.S., Komuro I., Kanazawa H., Yamazaki T., Mochida K., and Kishi F.. 2006. Functional heterogeneity of colony-stimulating factor-induced human monocyte-derived macrophages. Respirology. 11(suppl 1):S32–S36. 10.1111/j.1440-1843.2006.00805.x [DOI] [PubMed] [Google Scholar]
- Akhurst R.J. 2017. Targeting TGF-β Signaling for Therapeutic Gain. Cold Spring Harb. Perspect. Biol. 9:a022301 10.1101/cshperspect.a022301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhurst R.J., and Hata A.. 2012. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 11:790–811. 10.1038/nrd3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C., Fogg D., Garfa M., Elain G., Join-Lambert O., Kayal S., Sarnacki S., Cumano A., Lauvau G., and Geissmann F.. 2007. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science. 317:666–670. 10.1126/science.1142883 [DOI] [PubMed] [Google Scholar]
- Babyatsky M.W., Rossiter G., and Podolsky D.K.. 1996. Expression of transforming growth factors alpha and beta in colonic mucosa in inflammatory bowel disease. Gastroenterology. 110:975–984. 10.1053/gast.1996.v110.pm8613031 [DOI] [PubMed] [Google Scholar]
- Bain C.C., and Mowat A.M.. 2014. The monocyte-macrophage axis in the intestine. Cell. Immunol. 291:41–48. 10.1016/j.cellimm.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain C.C., Scott C.L., Uronen-Hansson H., Gudjonsson S., Jansson O., Grip O., Guilliams M., Malissen B., Agace W.W., and Mowat A.M.. 2013. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 6:498–510. 10.1038/mi.2012.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain C.C., Bravo-Blas A., Scott C.L., Perdiguero E.G., Geissmann F., Henri S., Malissen B., Osborne L.C., Artis D., and Mowat A.M.. 2014. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 15:929–937. 10.1038/ni.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Jin W., Hardegen N., Lei K.J., Li L., Marinos N., McGrady G., and Wahl S.M.. 2003. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198:1875–1886. 10.1084/jem.20030152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin M., and Bigley V.. 2016. Monocyte, Macrophage, and Dendritic Cell Development: the Human Perspective. Microbiol. Spectr. 4 10.1128/microbiolspec.MCHD-0015-2015 [DOI] [PubMed] [Google Scholar]
- Cros J., Cagnard N., Woollard K., Patey N., Zhang S.Y., Senechal B., Puel A., Biswas S.K., Moshous D., Picard C., et al. 2010. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 33:375–386. 10.1016/j.immuni.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardalhon V., Awasthi A., Kwon H., Galileos G., Gao W., Sobel R.A., Mitsdoerffer M., Strom T.B., Elyaman W., Ho I.C., et al. 2008. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nat. Immunol. 9:1347–1355. 10.1038/ni.1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson T.S., DiPaolo R.J., Andersson J., and Shevach E.M.. 2007. Cutting Edge: IL-2 is essential for TGF-beta-mediated induction of Foxp3+ T regulatory cells. J. Immunol. 178:4022–4026. 10.4049/jimmunol.178.7.4022 [DOI] [PubMed] [Google Scholar]
- Epelman S., Lavine K.J., and Randolph G.J.. 2014. Origin and functions of tissue macrophages. Immunity. 41:21–35. 10.1016/j.immuni.2014.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton T.M., Kelly A., Shuttleworth E.E., Smedley C., Atakilit A., Powrie F., Campbell S., Nishimura S.L., Sheppard D., Levison S., et al. 2017. Inflammatory cues enhance TGFβ activation by distinct subsets of human intestinal dendritic cells via integrin αvβ8. Mucosal Immunol. 10:624–634. 10.1038/mi.2016.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidt M.M., Ebert T.S., Chauhan D., Schmidt T., Schmid-Burgk J.L., Rapino F., Robertson A.A., Cooper M.A., Graf T., and Hornung V.. 2016. Human Monocytes Engage an Alternative Inflammasome Pathway. Immunity. 44:833–846. 10.1016/j.immuni.2016.01.012 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., and Guilliams M.. 2016. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity. 44:439–449. 10.1016/j.immuni.2016.02.024 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., and Jung S.. 2014. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14:392–404. 10.1038/nri3671 [DOI] [PubMed] [Google Scholar]
- Gleizes P.E., Munger J.S., Nunes I., Harpel J.G., Mazzieri R., Noguera I., and Rifkin D.B.. 1997. TGF-beta latency: biological significance and mechanisms of activation. Stem Cells. 15:190–197. 10.1002/stem.150190 [DOI] [PubMed] [Google Scholar]
- Gorelik L., Fields P.E., and Flavell R.A.. 2000. Cutting edge: TGF-beta inhibits Th type 2 development through inhibition of GATA-3 expression. J. Immunol. 165:4773–4777. 10.4049/jimmunol.165.9.4773 [DOI] [PubMed] [Google Scholar]
- Gorelik L., Constant S., and Flavell R.A.. 2002. Mechanism of transforming growth factor beta-induced inhibition of T helper type 1 differentiation. J. Exp. Med. 195:1499–1505. 10.1084/jem.20012076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorham J.D., Güler M.L., Fenoglio D., Gubler U., and Murphy K.M.. 1998. Low dose TGF-beta attenuates IL-12 responsiveness in murine Th cells. J. Immunol. 161:1664–1670. [PubMed] [Google Scholar]
- Heath V.L., Murphy E.E., Crain C., Tomlinson M.G., and O’Garra A.. 2000. TGF-beta1 down-regulates Th2 development and results in decreased IL-4-induced STAT6 activation and GATA-3 expression. Eur. J. Immunol. 30:2639–2649. [DOI] [PubMed] [Google Scholar]
- Ingersoll M.A., Spanbroek R., Lottaz C., Gautier E.L., Frankenberger M., Hoffmann R., Lang R., Haniffa M., Collin M., Tacke F., et al. 2010. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood. 115:e10–e19. 10.1182/blood-2009-07-235028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiani P., and Boraschi D.. 2014. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front. Immunol. 5:514 10.3389/fimmu.2014.00514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick C.V., Randolph G.J., and Henson P.M.. 2017. Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 17:349–362. 10.1038/nri.2017.28 [DOI] [PubMed] [Google Scholar]
- Kamada N., Hisamatsu T., Okamoto S., Chinen H., Kobayashi T., Sato T., Sakuraba A., Kitazume M.T., Sugita A., Koganei K., et al. 2008. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J. Clin. Invest. 118:2269–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A., Houston S.A., Sherwood E., Casulli J., and Travis M.A.. 2017. Regulation of Innate and Adaptive Immunity by TGFβ. Adv. Immunol. 134:137–233. 10.1016/bs.ai.2017.01.001 [DOI] [PubMed] [Google Scholar]
- Konkel J.E., and Chen W.. 2011. Balancing acts: the role of TGF-β in the mucosal immune system. Trends Mol. Med. 17:668–676. 10.1016/j.molmed.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Oukka M., and Kuchroo V.K.. 2009. IL-17 and Th17 Cells. Annu. Rev. Immunol. 27:485–517. 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- Kulkarni A.B., Huh C.G., Becker D., Geiser A., Lyght M., Flanders K.C., Roberts A.B., Sporn M.B., Ward J.M., and Karlsson S.. 1993. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc. Natl. Acad. Sci. USA. 90:770–774. 10.1073/pnas.90.2.770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara M., Yamashita M., Shinoda K., Tofukuji S., Onodera A., Shinnakasu R., Motohashi S., Hosokawa H., Tumes D., Iwamura C., et al. 2012. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-β and suppresses T(H)2 differentiation. Nat. Immunol. 13:778–786. 10.1038/ni.2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy-Hulbert A., Smith A.M., Tissire H., Barry M., Crowley D., Bronson R.T., Roes J.T., Savill J.S., and Hynes R.O.. 2007. Ulcerative colitis and autoimmunity induced by loss of myeloid alphav integrins. Proc. Natl. Acad. Sci. USA. 104:15823–15828. 10.1073/pnas.0707421104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laouar Y., Sutterwala F.S., Gorelik L., and Flavell R.A.. 2005. Transforming growth factor-beta controls T helper type 1 cell development through regulation of natural killer cell interferon-gamma. Nat. Immunol. 6:600–607. 10.1038/ni1197 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang P., Li J., Kulkarni A.B., Perruche S., and Chen W.. 2008. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat. Immunol. 9:632–640. 10.1038/ni.1607 [DOI] [PubMed] [Google Scholar]
- Magnusson M.K., Brynjólfsson S.F., Dige A., Uronen-Hansson H., Börjesson L.G., Bengtsson J.L., Gudjonsson S., Öhman L., Agnholt J., Sjövall H., et al. 2016. Macrophage and dendritic cell subsets in IBD: ALDH+ cells are reduced in colon tissue of patients with ulcerative colitis regardless of inflammation. Mucosal Immunol. 9:171–182. 10.1038/mi.2015.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone G., Kumberova A., Croft N.M., McKenzie C., Steer H.W., and MacDonald T.T.. 2001. Blocking Smad7 restores TGF-beta1 signaling in chronic inflammatory bowel disease. J. Clin. Invest. 108:601–609. 10.1172/JCI12821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu D., Cambier S., Fjellbirkeland L., Baron J.L., Munger J.S., Kawakatsu H., Sheppard D., Broaddus V.C., and Nishimura S.L.. 2002. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J. Cell Biol. 157:493–507. 10.1083/jcb.200109100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J.S., Harpel J.G., Gleizes P.E., Mazzieri R., Nunes I., and Rifkin D.B.. 1997. Latent transforming growth factor-beta: structural features and mechanisms of activation. Kidney Int. 51:1376–1382. 10.1038/ki.1997.188 [DOI] [PubMed] [Google Scholar]
- Munger J.S., Huang X., Kawakatsu H., Griffiths M.J., Dalton S.L., Wu J., Pittet J.F., Kaminski N., Garat C., Matthay M.A., et al. 1999. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 96:319–328. 10.1016/S0092-8674(00)80545-0 [DOI] [PubMed] [Google Scholar]
- Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T., et al. 2014. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 41:14–20. 10.1016/j.immuni.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T., Nishimura J., Barman S., Kayama H., Uematsu S., Okuzaki D., Osawa H., Haraguchi N., Uemura M., Hata T., et al. 2013. Increased Th17-inducing activity of CD14+ CD163 low myeloid cells in intestinal lamina propria of patients with Crohn’s disease. Gastroenterology. 145:1380–91.e1. 10.1053/j.gastro.2013.08.049 [DOI] [PubMed] [Google Scholar]
- Païdassi H., Acharya M., Zhang A., Mukhopadhyay S., Kwon M., Chow C., Stuart L.M., Savill J., and Lacy-Hulbert A.. 2011. Preferential expression of integrin αvβ8 promotes generation of regulatory T cells by mouse CD103+ dendritic cells. Gastroenterology. 141:1813–1820. 10.1053/j.gastro.2011.06.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.A., Zhang Y., Fullerton J.N., Boelen L., Rongvaux A., Maini A.A., Bigley V., Flavell R.A., Gilroy D.W., Asquith B., et al. 2017. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J. Exp. Med. 214:1913–1923. 10.1084/jem.20170355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi S., Oh S.A., and Li M.O.. 2017. Regulation of the Immune Response by TGF-β: From Conception to Autoimmunity and Infection. Cold Spring Harb. Perspect. Biol. 9:a022236 10.1101/cshperspect.a022236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schridde A., Bain C.C., Mayer J.U., Montgomery J., Pollet E., Denecke B., Milling S.W.F., Jenkins S.J., Dalod M., Henri S., et al. 2017. Tissue-specific differentiation of colonic macrophages requires TGFβ receptor-mediated signaling. Mucosal Immunol. 10:1387–1399. 10.1038/mi.2016.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull M.M., Ormsby I., Kier A.B., Pawlowski S., Diebold R.J., Yin M., Allen R., Sidman C., Proetzel G., Calvin D., et al. 1992. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 359:693–699. 10.1038/359693a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockis J., Liénart S., Colau D., Collignon A., Nishimura S.L., Sheppard D., Coulie P.G., and Lucas S.. 2017. Blocking immunosuppression by human Tregs in vivo with antibodies targeting integrin αVβ8. Proc. Natl. Acad. Sci. USA. 114:E10161–E10168. 10.1073/pnas.1710680114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiesen S., Janciauskiene S., Uronen-Hansson H., Agace W., Högerkorp C.M., Spee P., Håkansson K., and Grip O.. 2014. CD14(hi)HLA-DR(dim) macrophages, with a resemblance to classical blood monocytes, dominate inflamed mucosa in Crohn’s disease. J. Leukoc. Biol. 95:531–541. 10.1189/jlb.0113021 [DOI] [PubMed] [Google Scholar]
- Travis M.A., and Sheppard D.. 2014. TGF-β activation and function in immunity. Annu. Rev. Immunol. 32:51–82. 10.1146/annurev-immunol-032713-120257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis M.A., Reizis B., Melton A.C., Masteller E., Tang Q., Proctor J.M., Wang Y., Bernstein X., Huang X., Reichardt L.F., et al. 2007. Loss of integrin alpha(v)beta8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 449:361–365. 10.1038/nature06110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C., Mildner A., and Jung S.. 2015. Macrophages: development and tissue specialization. Annu. Rev. Immunol. 33:643–675. 10.1146/annurev-immunol-032414-112220 [DOI] [PubMed] [Google Scholar]
- Veldhoen M., Uyttenhove C., van Snick J., Helmby H., Westendorf A., Buer J., Martin B., Wilhelm C., and Stockinger B.. 2008. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 9:1341–1346. 10.1038/ni.1659 [DOI] [PubMed] [Google Scholar]
- Wolf Y., Shemer A., Polonsky M., Gross M., Mildner A., Yona S., David E., Kim K.W., Goldmann T., Amit I., et al. 2017. Autonomous TNF is critical for in vivo monocyte survival in steady state and inflammation. J. Exp. Med. 214:905–917. 10.1084/jem.20160499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington J.J., Czajkowska B.I., Melton A.C., and Travis M.A.. 2011. Intestinal dendritic cells specialize to activate transforming growth factor-β and induce Foxp3+ regulatory T cells via integrin αvβ8. Gastroenterology. 141:1802–1812. 10.1053/j.gastro.2011.06.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington J.J., Klementowicz J.E., Rahman S., Czajkowska B.I., Smedley C., Waldmann H., Sparwasser T., Grencis R.K., and Travis M.A.. 2013. Loss of the TGFβ-activating integrin αvβ8 on dendritic cells protects mice from chronic intestinal parasitic infection via control of type 2 immunity. PLoS Pathog. 9:e1003675 10.1371/journal.ppat.1003675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington J.J., Kelly A., Smedley C., Bauché D., Campbell S., Marie J.C., and Travis M.A.. 2015. Integrin αvβ8-Mediated TGF-β Activation by Effector Regulatory T Cells Is Essential for Suppression of T-Cell-Mediated Inflammation. Immunity. 42:903–915. 10.1016/j.immuni.2015.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J., Schmidt S.V., Sander J., Draffehn A., Krebs W., Quester I., De Nardo D., Gohel T.D., Emde M., Schmidleithner L., et al. 2014. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 40:274–288. 10.1016/j.immuni.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.G., Wang J., Wang P., Gray J.D., and Horwitz D.A.. 2007. IL-2 is essential for TGF-beta to convert naive CD4+CD25- cells to CD25+Foxp3+ regulatory T cells and for expansion of these cells. J. Immunol. 178:2018–2027. 10.4049/jimmunol.178.4.2018 [DOI] [PubMed] [Google Scholar]
- Ziegler-Heitbrock L. 2015. Blood Monocytes and Their Subsets: Established Features and Open Questions. Front. Immunol. 6:423 10.3389/fimmu.2015.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.