Neutrophils enter tissues to mediate immunity and inflammation. Casanova-Acebes et al. show that migration into naive tissues is equally important. In the intestine, they provide remote support to hematopoietic niches, whereas in the lungs they regulate circadian transcription and metastasis.
Abstract
Immune protection relies on the capacity of neutrophils to infiltrate challenged tissues. Naive tissues, in contrast, are believed to remain free of these cells and protected from their toxic cargo. Here, we show that neutrophils are endowed with the capacity to infiltrate multiple tissues in the steady-state, a process that follows tissue-specific dynamics. By focusing in two particular tissues, the intestine and the lungs, we find that neutrophils infiltrating the intestine are engulfed by resident macrophages, resulting in repression of Il23 transcription, reduced G-CSF in plasma, and reinforced activity of distant bone marrow niches. In contrast, diurnal accumulation of neutrophils within the pulmonary vasculature influenced circadian transcription in the lungs. Neutrophil-influenced transcripts in this organ were associated with carcinogenesis and migration. Consistently, we found that neutrophils dictated the diurnal patterns of lung invasion by melanoma cells. Homeostatic infiltration of tissues unveils a facet of neutrophil biology that supports organ function, but can also instigate pathological states.
Graphical Abstract
Introduction
It has been long appreciated that infectious or physical insults are followed by rapid infiltration of tissues by circulating neutrophils, a process that is critical for the elimination of pathogens and tissue repair (Phillipson and Kubes, 2011) and is considered a hallmark of inflammation. Infected tissues or those affected by sterile stress can, however, be damaged as a consequence of the toxic activity of neutrophils (Phillipson and Kubes, 2011; Kruger et al., 2015), a property that has led to the common belief that healthy tissues are generally devoid of neutrophils. This prevailing belief explains why their roles in naive, unperturbed tissues remain poorly defined.
This gap in knowledge is paradoxical if one considers that neutrophils are the most abundant myeloid leukocytes in mammals and that they are equipped with one of the most efficient migratory capabilities among somatic cells (de Oliveira et al., 2016). In the absence of inflammatory signals, mature neutrophils are released from the bone marrow (BM) and circulate in blood for ∼12 h before abruptly disappearing from the bloodstream at defined times of the day (i.e., with circadian frequency; Furze and Rankin, 2008; Adrover et al., 2016). Classical studies in humans in which ex vivo radiolabeled neutrophils were infused back into healthy volunteers, as well as transfer studies in mice, suggested that the clearance of neutrophils from the bloodstream could be explained by infiltration of the BM, liver, and spleen, in which they are thought to be eliminated by phagocytosis (Saverymuttu et al., 1985; Suratt et al., 2001). This limited lifetime has further contributed to the assumption that they do not participate in normal tissue function, yet this view contrasts with the growing appreciation that innate immune cells, such as monocytes and macrophages, perform important functions in the steady-state (Auffray et al., 2007; Wynn et al., 2013).
Notwithstanding this restricted view of their biology, several studies suggest that neutrophils have support functions within healthy tissues. For example, neutrophils in the spleen support B cell maturation and antibody production (Puga et al., 2012), and in the BM, infiltrating neutrophils inhibit hematopoietic niches and trigger the release of hematopoietic progenitor cells (HPCs) into the bloodstream (Casanova-Acebes et al., 2013). In undefined tissues, phagocytosis of apoptotic neutrophils by dendritic cells and macrophages inhibits expression of Il23, a process that in turn controls granulopoiesis by regulating G-CSF production (Stark et al., 2005). Although these studies suggested that neutrophils can enter tissues to perform homeostatic (i.e., noninflammatory) roles, this contention has not been systematically explored. It remains unknown, for example, which tissues are actually infiltrated, the dynamics of this process, and more importantly, to what extent infiltrating neutrophils contribute to normal tissue physiology.
Here, we have examined the fate of neutrophils as they disappear from the circulation. We show that neutrophils infiltrate the vast majority of naive tissues in mice, often following strict diurnal patterns. Importantly, we find that infiltration of the intestinal mucosa and other extramedullary tissues supports hematopoietic niche activity in the BM, while diurnal infiltration of the lungs influences transcription and accounts for oscillatory metastatic invasion. Thus, neutrophils are endowed with important, nonimmune regulatory functions as they traffic through healthy tissues.
Results
Neutrophils infiltrate naive tissues with distinct rates and dynamics
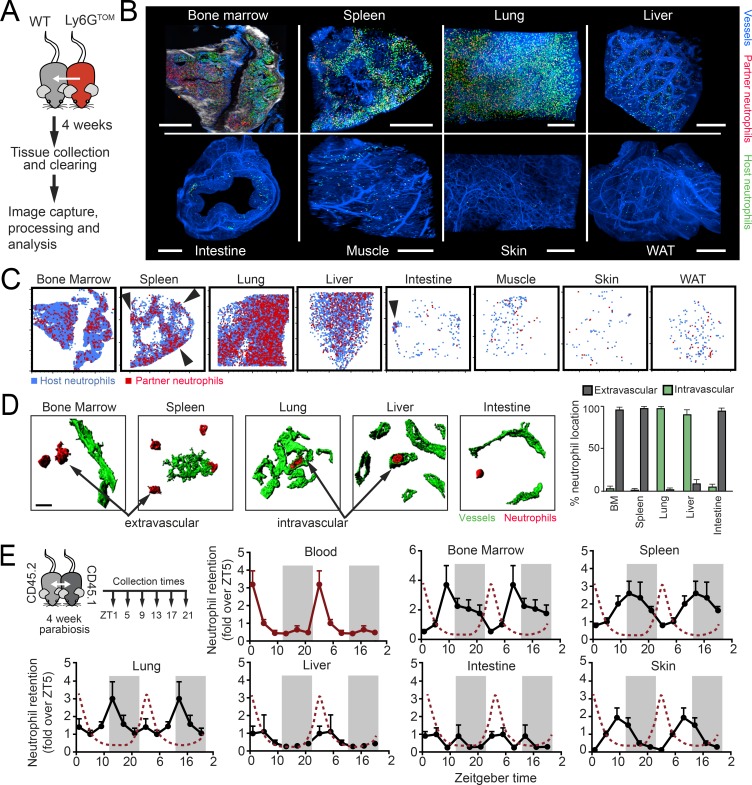
Neutrophils disappear from the bloodstream at the end of the animal’s resting period (Casanova-Acebes et al., 2013), but the fate of the cleared cells from blood is poorly defined. We performed an organism-wide search for the presence of neutrophils across multiple tissues of healthy mice housed under specific pathogen–free conditions. Using a criteria for detection of cells by both cytometric and immunofluorescence analyses with the neutrophil marker Ly6G in Lyz2GFP reporter mice (Faust et al., 2000), we found neutrophils in the majority of analyzed tissues (Fig. S1, A and B). Although they were most abundant in the BM, spleen, and lung tissues in which infiltration had been previously reported, neutrophils were also present in the liver, intestine, white adipose tissue (WAT), skin, and skeletal muscle (Fig. 1 A). Because these analyses could not discriminate between neutrophils produced locally and those that come from the circulation, we examined tissues of WT mice that had been exposed to the circulation of Lyz2GFP mice after 1 mo in parabiosis. We confirmed that, under this setting, the majority of partner-derived neutrophils in tissues originated from the circulation, rather than from HPCs potentially crossing between the parabiotic partners (see Materials and methods). We found that most tissues were infiltrated by blood-borne neutrophils (Fig. 1 B and Fig. S1 C), with partner-derived GFP+ neutrophils present in the same tissues as above, as well as in peripheral lymph nodes, kidneys, and heart (Fig. S1 D). We only failed to find neutrophils in the ovaries, testes, and brain (Fig. S1 D), a finding consistent with the immune-privileged status of these organs. The number of infiltrating neutrophils varied widely among tissues (Fig. 1 C). Quantitative estimations in the parabionts revealed that about one fourth of all detectable neutrophils in a mouse had already transited through the circulation (∼37.5 out of 155 × 106 cells, including the BM), implying that infiltration of tissues is quantitatively significant and does not necessarily lead to immediate elimination. Of note, while neutrophils trafficked through healthy tissues in a regular manner, the extent of this infiltration through tissues such as WAT, muscle, and intestine was still orders of magnitude lower than in organs such as the BM, spleen, and lung, which are known to have a high content of neutrophils at steady-state. Notably, the ratio of partner to host neutrophils in most tissues remained comparable to those in blood (Fig. 1 D), indicating constant replenishment from the circulation and transit through healthy tissues. The only tissue in which the ratio was skewed toward host neutrophils was the BM (Fig. 1 D), as expected from the active granulopoiesis found within this organ.
Figure 1.
Multi-organ infiltration of neutrophils in the steady-state. (A) Micrographs of tissues illustrating the presence of neutrophils (green; shown by yellow arrowheads) in Lyz2GFP reporter mice. These GFPHI cells also stained for Ly6G (see Fig. S1 B). Insets show details of neutrophils within each tissue. Bars: 100 µm (main images); 10 µm (inset images). (B) Neutrophils infiltrate multiple organs from the circulation, as observed in parabiotic pairs (left scheme). Representative images of Lyz2GFP neutrophils present in tissues of the nonfluorescent WT partner mouse. Bars: 50 µm (main images); 10 µm (inset images). (C) Quantification of partner-derived neutrophils per organ (except for blood, which represents 1 ml; for BM, one femur; for skin and muscle, 100 mg); n = 8–18 from three independent experiments. (D) Neutrophil chimerism in tissues of the nonfluorescent partner from WT with Lyz2GFP parabiotic pairs; n = 20–22 from two independent experiments. Note that chimerism in tissues equilibrates with that in blood, except for the BM. Error bars show mean ± SEM values.
To additionally define the distribution of infiltrating neutrophils within tissues, we next used multiphoton and light-sheet microscopy (LSM) of optically cleared organs of WT mice in parabiosis with Ly6GCRE; Rosa26TOM mice (Ly6GTOM, a neutrophil-specific reporter mouse; Hasenberg et al., 2015; Fig. 2 A), followed by image processing and analysis (Fig. 2 B, Fig. S2 A, and Videos S1–S8). In most organs, Ly6GTOM+ partner-derived neutrophils displayed what appeared to be a random distribution (Fig. 2 C), with host- and partner-derived cells showing overlapping localization. Only in the large intestine and in the spleen did we find areas of preferred concentration of neutrophils (Fig. 2 C): the marginal zone and red pulp in spleens (in agreement with previous reports; Puga et al., 2012; Video 2) and isolated patches in the intestine (Video 5). Additional image analyses revealed that partner-derived neutrophils preferentially localized within the tissue parenchyma (i.e., outside blood vessels) in the BM, spleen, and intestine (Fig. 2 D and Videos S1–S8). In heavily vascularized tissues such as liver and lungs, in contrast, neutrophils localized preferentially within vessels (Fig. 2 D). We confirmed these findings by flow cytometry (Fig. S2 B). The intravascular distribution in the liver and lungs is consistent with the long-known presence of a marginated neutrophil pool in these organs (Worthen et al., 1989; Gebb et al., 1995), and the infiltration of the BM and spleen aligns with previous studies (Puga et al., 2012; Scheiermann et al., 2012; Casanova-Acebes et al., 2013). Importantly, however, our findings show that homeostatic migration of neutrophils extends to virtually every tissue.
Figure 2.
Distribution and dynamics of neutrophils in tissues. (A) Experimental scheme and imaging of Ly6GTOM neutrophils in tissues of the WT parabiotic partner. (B) Distribution of neutrophils in optically cleared organs by multiphoton microscopy (for BM; white is bone captured by second harmonic generation) or LSM; n = organs from 2–3 mice. Blue, vessels; green, host-derived neutrophils; red, partner-derived neutrophils. Bars, 500 µm. See also Videos 1–8 showing partner-derived neutrophils within the tissue. (C) Positioning plots showing the distribution of host- (blue) and partner-derived (red) neutrophils in two-dimensional maps of each tissue. Arrows show areas where neutrophils cluster. Plots obtained from the tissues shown in B. (D) Confocal reconstruction of neutrophils (red) proximal to blood vessels (green) in representative tissues from parabionts as in A. Partner-derived neutrophils were preferentially found in the parenchyma (extravascular) or within vessels (intravascular), depending on the tissues, as quantified in the right bar graph. Bars show mean ± SEM values from the analysis 31–73 neutrophils per tissue from three different animals. (E) Left, experimental design. Curves show the number of partner-derived neutrophils as determined by flow cytometry from the tissues of parabiotic pairs collected at the indicated ZTs. Values are normalized to ZT5 and duplicated over two complete days for better visualization. Periods of darkness are shown by the shaded rectangles, and the red dotted lines show the oscillations in blood for reference. Values are mean ± SEM from 10–12 mice per time and tissue from two experiments. Intestine refers to the large intestine (colon).
Because neutrophil levels in blood oscillate with circadian frequency (Casanova-Acebes et al., 2013), we assessed the dynamics of infiltration in multiple tissues. We set up parabiotic pairs of congenic CD45.1 and CD45.2 WT mice and analyzed the frequency of partner-derived neutrophils in tissues collected in the morning (zeitgeber time 5 [ZT5], i.e., 5 h after lights on) or in the evening (ZT13), relative to their levels in blood (Fig. S2 C). We found elevated retention of neutrophils in the evening in most tissues, with the exception of the intestine, liver, and WAT, in which neutrophil numbers did not show rhythmic patterns (Fig. S2 C). To gain resolution in the temporal dynamics of infiltration, we repeated the analyses by sampling several of these tissues every 4 h during a full diurnal cycle. We found distinct diurnal, in-phase oscillations in the majority of tissues, with an acrophase at the beginning of the animals’ activity period (from ZT9 to ZT13, depending on the tissue; Fig. 2 E). Oscillations in these tissues were in anti-phase with those in the circulation such that numbers within tissues increased as they decreased from blood, a finding that extends earlier observations in the BM and muscle (Scheiermann et al., 2012). Conspicuous exceptions to these diurnal patterns were the intestine and liver, in agreement with our initial analyses (Fig. S2 C). Altogether, these observations revealed that neutrophils actively infiltrate most healthy tissues in numbers and with dynamics that are specific for each tissue.
Extramedullary infiltration of neutrophils supports BM niches
Given the finding that neutrophil infiltration in the steady-state occurs in multiple tissues, we examined the potential functions of neutrophils in some of these tissues by analyzing mouse models with defective migration under basal conditions.
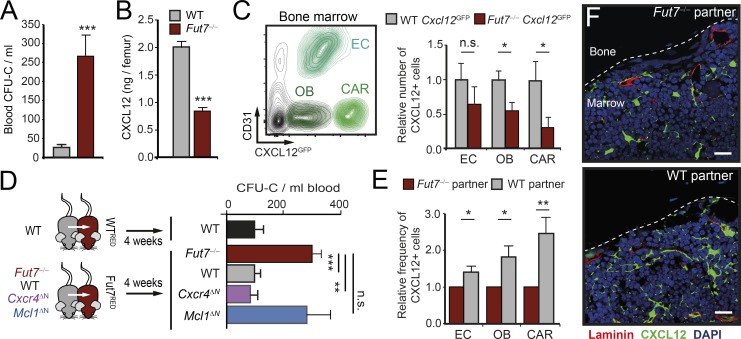
Endothelial selectins are essential for recruitment during inflammation, but they are also expressed in naive tissues. We chose to study mice lacking Fucosyltransferase 7 (Fut7−/− mice), in which leukocytes cannot synthesize selectin ligands and therefore display general defects in neutrophil trafficking (Malý et al., 1996). Notably, blood analysis of Fut7−/− mice revealed markedly increased numbers of HPCs in the bloodstream, which correlated with reduced levels in the BM of the chemokine CXCL12, the key retention factor for hematopoietic precursors (Fig. 3, A and B; Ding and Morrison, 2013). Consistently, Fut7−/− mice encoding for a reporter for CXCL12 expression (Fut7−/−; Cxcl12GFP mice) revealed a reduced number of BM stromal cells that produce this chemokine compared with Fut7-expressing Cxcl12GFP mice (Fig. 3 C), altogether indicating dysfunction of the BM in these migration-deficient mice. To determine whether these defects specifically originated from defective neutrophil infiltration, we set parabiotic pairs of Fut7−/− mice with WT partners or with mice that lacked neutrophils in blood (MRP8CRE; Mcl1flox/flox; Fig. S3 A; Dzhagalov et al., 2007). We reasoned that this approach would allow de novo entry of WT leukocytes into tissues of Fut7−/− mice and potential correction of the hematopoietic defects. Indeed, we found that only WT partners, but not those that were neutropenic, corrected the levels of HPCs in blood (Fig. 3 D), indicating that defective neutrophil trafficking was the origin of the hematopoietic disturbances in Fut7−/− mice. In agreement with this finding, exposure to WT neutrophils also caused elevations in CXCL12 protein and expression of niche-related genes in the BM of Fut7−/− mice (Fig. S3, B and C). Similarly, parabiosis of Fut7−/−; Cxcl12GFP mice with WT partners increased the number of CXCL12-producing cells in the BM (Fig. 3, E and F), without altering their numbers in extramedullary tissues (Fig. S3 D), indicating marrow-specific effects.
Figure 3.
Control of BM homeostasis by extramedullary neutrophils. (A) Number of CFUs in culture (CFU-C) at ZT5 in the blood of WT and Fut7−/− mice; n = 6 mice from two experiments. (B) Levels of CXCL12 protein in the BM of WT and Fut7−/− mice; n = 6–10 mice from two experiments. (C) Reduced number of CXCL12-producing cells in the BM of Fut7−/−; Cxcl12GFP mice, as measured by flow cytometry; n = 5–8 mice from two experiments. Density plot at left shows the gating strategy to identify endothelial cells (EC), osteoblasts (OB), and reticular cells (CAR). (D) Experimental design (left) and number of WTRED or Fut7RED-derived circulating progenitors (CFU-C) after 1 mo in parabiosis with WT or the indicated mutant mice. For comparison, the levels of circulating HPCs in WT control parabionts is shown; n = 3–20 mice. Cxcr4ΔN and Mcl1ΔN are neutrophil-specific mutants (characterized in Fig. S3). (E) Relative number of CXCL12-producing cells in the BM of Fut7−/−; Cxcl12GFP mice after 1 mo in parabiosis with Fut7−/− or WT mice; n = 8–9 mice from three independent experiments. (F) Micrographs showing GFP+ niche cells (green) in the BM of Fut7−/−; Cxcl12GFP mice after parabiosis with Fut7−/− or WT mice. Dashed lines outline the bone surface. Data from three mice per group. Bars, 20 µm. Data shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant, as determined by unpaired Student’s t test (A, B, C, and E), or ANOVA with Tukey’s multigroup correction (D).
To additionally determine whether the hematopoietic-correcting activity occurred locally, by neutrophils infiltrating the marrow, we analyzed Fut7−/− mice that were set in parabiosis with partners in which neutrophils do not express CXCR4 (MRP8CRE; Cxcr4flox/flox mice; Fig. S3 E). Neutrophils from these mice do not infiltrate the BM (Casanova-Acebes et al., 2013), but can migrate into other tissues (Fig. S3 F). Notably, CXCR4-deficient neutrophils corrected the hematopoietic defects in Fut7−/− mice as efficiently as WT cells (Fig. 3 D), indicating that the niche-supportive activity of neutrophils occurred outside of the BM. In combination, these data in Fut7−/− mice revealed that infiltration of one or more extramedullary tissues by neutrophils supports BM activity.
Phagocytosis of neutrophils inhibits cytokine production by intestinal macrophages
We next asked in which tissue(s) neutrophils exerted their BM-regulatory functions. We again took advantage of the parabiosis strategy to compare the differential infiltration of WT versus Fut7−/− neutrophils across multiple tissues by flow cytometry. Of all the tissues analyzed, Fut7−/− neutrophils presented impaired migration in the skin, muscle, and intestine (Fig. 4, A and B), pointing to these tissues as potential sites for niche regulation outside the BM.
Figure 4.
Regulatory functions of neutrophils in the intestine. (A) Experimental strategy and cytometry plots. Dot plots, gated on CD45+ CD11b+ Ly6G+ cells, show neutrophils in tissues of Fut7RED mice after parabiosis with Fut7−/− control or WT partners. Numbers indicate mean ± SEM of partner-to-host ratios corrected by ratios in blood and normalized to the Fut7−/−; Fut7−/− parabiotic pairs (see Materials and methods). (B) Radial plots showing the relative infiltration of partner-derived and Fut7−/− neutrophils in different tissues. Polygons show the mean ± SEM ratio of partner to Fut7RED neutrophils in each tissue, which are shown in the plots in A. Data from four to five pairs per group from three independent experiments. (C) Experimental approach to estimate phagocytosis of partner-derived fluorescent leukocytes by tissue macrophages using flow cytometry. (D) Percentage of macrophages that engulf partner-derived cells in organs of Fut7−/− parabionts paired with WTRED or Fut7RED partners; n = 5–6 mice from two independent experiments. (E) Number of circulating progenitors in Fut7−/− mice after 1 mo in parabiosis with Fut7−/− or WT partners or Fut7−/−; Cd169DTR mice paired with WT mice after macrophage depletion by treatment with DT; n = 6–12 mice from three independent experiments. (F) Whole-mount staining of the large intestine of Lyz2GFP mice showing GFP+ foci in the mucosal surface. Bar, 1 mm. (G) Detail of an ILF showing the distribution of CD169+ macrophages, MRP14+ neutrophils, and the microvasculature (laminin; white). Bar, 100 µm (see also Video 9). (H) Representative plot showing intestinal macrophages that phagocytose (red) or not (black) partner-derived DsRed+ cells, as in C and D. Bar graph at right shows expression of Il1b and Il23 in phagocytic macrophages relative to non-phagocytic cells; data are from cells sorted from three individual mice. (I) Scheme of the IL-23–IL-17–G-CSF pathway. (J) Number of circulating CFU-C in Fut7−/− mice after treatment with isotype or blocking antibodies against the cytokines shown in I; n = 5–6 mice from two (left) and one independent experiments. Data shown as mean ± SEM. *, P < 0.05; **, P < 0.001; ***, P < 0.001; n.s., not significant, as determined by unpaired t test analysis (B, D, H, and J) or one-way ANOVA analysis with Tukey’s multigroup test (E).
Previous studies showed that enhanced granulopoiesis in mice deficient in endothelial selectins is due to unleashed production of cytokines by phagocytes that fail to engulf senescent neutrophils (Stark et al., 2005), and we previously demonstrated that the transcriptional profile of tissue-resident macrophages is regulated by phagocytosis of infiltrating leukocytes (A-González et al., 2017). We therefore predicted a role for macrophage-mediated phagocytosis in BM regulation and tested for potential defects in phagocytosis in the identified tissues. To this end, we tracked the phagocytosis of fluorescent leukocytes from WTRED mice (WT mice expressing DsRed) in parabiosis with Fut7−/− partners following a previously described strategy (A-González et al., 2017) and compared it with the phagocytosis of leukocytes from Fut7RED mice in parabiosis with Fut7−/− partners (Fig. 4 C). We found that only intestinal macrophages presented defects in engulfment of Fut7−/− leukocytes, whereas macrophages in other tissues phagocytosed normally, including macrophages in the skin and muscle (Fig. 4 D). This finding was also consistent with the close proximity of neutrophils to CD169+ macrophages in the intestine (mean distance 5.3 ± 1 µm compared with >50 µm in the spleen; P < 0.001; Fig. S4 A). Together, these data identified the intestine as a potential site for BM regulation and suggested that macrophages might be active mediators of the niche-regulating effect of neutrophils. Consistent with this possibility, depletion of macrophages in Fut7−/− mice expressing the deleter CD169DTR allele, which depleted macrophages from the intestine (among other tissues; Fig. S4, B and C; Asano et al., 2015), abolished the capacity of WT neutrophils to correct HPC numbers in blood (Fig. 4 E).
To dissect potential regulatory mechanisms in the intestine, we performed whole-mount imaging of this tissue in Lyz2GFP mice. We found that neutrophils did not distribute homogeneously in the intestinal mucosa, but rather clustered in discrete areas enriched in B lymphocytes that resembled isolated lymphoid follicles (ILF; Pearson et al., 2012; Fig. 4 F and Fig. S4 C), an observation that was consistent with the clusters found in our initial distribution analyses (Fig. 2 C). High-resolution imaging revealed that neutrophils localized in the periphery of these follicles, proximal to CD169+ macrophages that also surrounded the ILF (Fig. 4 G and Video S9). These observations were intriguing because macrophages and lymphoid cells in the ILF have been reported to produce cytokines, including IL-1β and GM-CSF (Mortha et al., 2014), that could potentially regulate BM niches (Fig. S4 D). In support of this possibility, intestinal macrophages that phagocytosed DsRed+ leukocytes in the parabiosis model expressed lower levels of Il1b compared with non-phagocytic macrophages (Fig. 4 H), suggesting that perhaps repression of Il1b expression, and downstream GM-CSF production controlled niche activity and HPC release in blood in our model. We found, however, that mice deficient in IL-1Ra (the receptor for IL-1β), that lacked GM-CSF–producing lymphoid cells (Rorct−/−; Eberl et al., 2004), or deficient in GM-CSF (Csf2−/−), displayed normal elevations in circulating HPCs upon inhibition of selectins (to replicate the Fut7−/− phenotype; Fig. S4 E). Likewise, blockade of GM-CSF failed to reduce the number of HPCs in Fut7−/− mice (Fig. S4 F), altogether indicating that the IL-1β/GM-CSF pathway did not regulate the BM.
In our transcriptional analyses in Fut7−/− mice, we also noted strong reductions in Il23 expression by phagocytic macrophages (Fig. 4 H). IL-23 promotes expression of IL-17 by lymphoid cells, which in turn regulates expression of G-CSF (Stark et al., 2005), a cytokine that inhibits CXCL12 production by hematopoietic niche cells in the BM and mobilizes HPCs into blood (Christopher et al., 2009; Méndez-Ferrer et al., 2010; Fig. 4 I). We found that Fut7−/− mice exposed to neutrophils from WT partners displayed a trend toward less Il17-expressing T cells in the intestine (Fig. S4 G), as well as reduced levels of G-CSF in plasma (Fig. S4 H). These responses were consistent with regulation of this pathway by infiltrating neutrophils; consistently, blockade of IL-23, IL-17, or G-CSF resulted in marked reductions in the number of circulating HPCs in Fut7−/− mice (Fig. 4 J), indicating that dysregulation of this pathway in Fut7−/− mice caused the hematopoietic alterations. Of note, the observation that Rorct−/− mice, which have reduced IL-17–producing T cells, did not display anomalies in HPC mobilization suggests that other types of IL-17–producing cells distinct from RORγ-dependent T cells (Takahashi et al., 2008; Li et al., 2010; Bermejo et al., 2013) also participate in this pathway.
Collectively, these data show that intestinal macrophages respond to neutrophil infiltration by suppressing IL-23 production and suggest that regulation in the intestine could potentially control systemic G-CSF levels and BM activity. Consistent with this possibility, treatment of Fut7−/− mice with clodronate liposomes, which depleted macrophages in multiple organs but not in the intestine, still allowed normalization of circulating HPC levels in the parabiosis model (Fig. S4 I). Given the unavailability of intestine-specific models, however, we cannot rule out additional regulation emanating from other tissues, such as the lungs, in which interstitial macrophages displayed a trend toward reduced phagocytosis in Fut7−/− mice (Fig. S4 J).
When considering the temporal dynamics, steady infiltration of neutrophils in the intestine (Fig. 2 E) was at odds with the oscillatory pattern of circulating HPCs (Méndez-Ferrer et al., 2008; Casanova-Acebes et al., 2013). Analyses of circulating HPCs, however, revealed normal circadian patterns in Fut7−/− mice (Fig. S4 K) and in CD169DTR mice depleted of macrophages (Fig. S4 L), which suggested that infiltration and phagocytosis in extramedullary tissues controlled absolute numbers, but not diurnal rhythms, of circulating HPCs. Interestingly, mice in which neutrophils lacked CXCR4 (and therefore cannot infiltrate the BM; Fig. S3, E and F) were arrhythmic for circulating HPCs (Fig. S4 K). These observations suggested that neutrophils infiltrating different tissues have distinct effects on BM niches; in extramedullary tissues they regulate overall BM activity, whereas in the BM they regulate diurnal patterns, in agreement with a previous report (Casanova-Acebes et al., 2013).
Neutrophils regulate diurnal transcription in the lung
The identification of diurnal patterns of neutrophils in multiple tissues (Fig. 2 E) raised the question of whether neutrophils could influence circadian properties in tissues. We focused on the lung because neutrophils presented a marked diurnal pattern, were present in high numbers in this tissue (Fig. 1 C), and because little is known about the nonimmune roles of neutrophils in naive lungs. To search for possible circadian processes influenced by neutrophils, we collected lungs in the morning and in the evening (ZT4 and ZT16, respectively) from control mice, or from mice depleted of neutrophils using anti-Ly6G antibody for two consecutive days, and performed transcriptomic analysis by RNA sequencing (Fig. 5 A). Antibody treatment was effective at depleting pulmonary neutrophils (Fig. S5 A), as expected from their intravascular distribution (Fig. 2 D) and exposure to the circulating antibody, and had the additional advantage that short-term depletion prevented complications arising from sustained neutropenia (e.g., infections).
Figure 5.
Diurnal regulation of the lung transcriptome by neutrophils. (A) Experimental scheme for neutrophil depletion and collection of lungs at different diurnal times for RNA sequencing; n = 7 replicates per condition and time from two experiments. (B) Distribution of pulmonary transcripts: total genes (green; 100%) and genes that display diurnal changes in transcription (purple; 9.2%), a fraction of which loses diurnal oscillations when neutrophils are depleted (orange; representing 26.7% of diurnally oscillating or 2.5% of total transcripts). (C) Heat map of diurnally expressed genes that lose their diurnal variation in neutrophil-depleted lungs, with several representative genes indicated at right. Only genes with an adjusted P value <0.05 in a moderated t test in the non-depleted group were included; n = 7 mice per group. (D) Major biological functions affected by the neutrophil-regulated diurnal genes shown in C. The functions were identified from the pathways indicated by numbers (1–21) and listed in Fig. S5 C and Table S1. The total number of unique genes in the list was calculated for each biological function, and the color scale reflects the proportion of the genes in each pathway that contribute to the specific function.
Analysis of the transcriptomic data revealed that about one tenth (1,770 out of 19,184) of all pulmonary transcripts presented diurnal oscillations (similar to previous reports; Zhang et al., 2014) and that about one fourth of these gene transcripts specifically lost their oscillatory behavior upon neutrophil depletion (Fig. 5 B). Among the affected genes, many could be categorized in specific biological processes: some like Cept1 or Tbxas1 are needed for the biosynthesis of lipids (e.g., PAF or thromboxane A2), which have been involved in inflammation, cell growth, and metastasis (Im et al., 1996; Wculek and Malanchi, 2015); genes with functions in DNA repair, cell transformation, or cell growth, including Polq, Fgfr1, Akt2, Scube3, or Gata4; a small group of circadian regulators, including Atf5, Hcrtr2, and Bhlh341; and finally genes like Col6a6, Tgfb1i1, Cd44, Itga9, Ccl4, or Mmp9 that are associated with tissue structure and cell adhesion (Fig. 5 C). We could confirm lack of diurnal oscillation in these genes in an independent model of neutrophil depletion induced by diphtheria toxin (DT) in Mrp8CRE; iDTR mice (Figs. S5, A and B). Interrogation of canonical pathways using the Ingenuity package revealed that neutrophil depletion affected functions related to carcinogenesis, cell survival, and migration (Fig. 5 D, Fig. S5 C, and Table S1). When considered as a whole, these findings indicated that neutrophils regulate a diurnal program in the lungs and suggested potential effects in cell migration and susceptibility to cancer.
Metastatic invasion of the lungs is temporally controlled by neutrophils
To validate the transcriptomic analyses, we used a well-established model of metastasis in which B16F1 melanoma cells expressing the luciferase reporter are injected intravenously (Peinado et al., 2012). In this model, the injected cells that gain access to the lungs grow into metastatic lesions that can be scored by bioluminescence or histological analysis after 2 wk (Fig. 6 A). If diurnal invasion of the lungs by metastatic cells was influenced by neutrophils, however, one would expect that the number of metastatic foci would naturally oscillate depending on the time of injection of the tumoral cells. Indeed, we found a clear diurnal pattern in B16F1-induced metastasis, with more foci forming when the cells were injected in the morning and virtually no lesions when they were injected in the evening (Fig. 6 B). We confirmed this pattern both by bioluminescence at the time of lung collection (Fig. 6 B), as well as by histological analysis (Fig. 6 C).
Figure 6.
Diurnal metastasis of melanoma cells in lungs. (A) Experimental design to assess the effect of time of day in metastatic invasion of lungs by B16F1Luc melanoma cells. (B) Number of metastatic lesions 14 d after injection of B16F1Luc melanoma cells, as determined by the luminescence of lung explants, shown at bottom. Values are from a representative of two experiments, each with n = 3–5 mice per time point and experiment. (C) Representative H&E-stained sections of lungs from each time point. Bar, 100 µm. (D) Bioluminescence counts reflecting metastatic burden of lungs, 14 d after injection of B16F1Luc cells in the morning (AM, corresponding to ZT4) or in the evening (PM, corresponding to ZT16), in control mice or mice previously depleted of neutrophils. Values are normalized to AM; n = 9 mice from two independent experiments. Data are shown as mean ± SEM. *, P < 0.05; n.s., not significant, as determined by unpaired t test analysis.
To determine whether, and to what extent, this circadian behavior was regulated by neutrophils, we repeated the experiment at ZT4 and ZT16 (the times of maximal and minimal metastasis, respectively) in mice in which neutrophils had been depleted for 2 d. In these mice, the number of other immune cells was largely unaffected, except for moderate increases in dendritic cell numbers (not shown). Remarkably, the formation of metastatic foci lost its diurnal variability in the absence of neutrophils (Fig. 6 D). Given that the time of injection was determinant for the long-term metastatic spread in the lungs, we hypothesized that initial retention and seeding of tumoral cells in this organ was the key step regulated by neutrophils. Indeed, we found that neutrophil depletion resulted in loss of diurnal homing of melanoma cells to the lungs (Fig. 7, A–C). Remarkably, when we used mice in which we genetically disabled diurnal infiltration of neutrophils in lungs by neutrophil-specific deletion of the clock gene Bmal1 (Bmal1ΔN mice; Fig. 7 B), the homing of melanoma cells also lost its diurnal rhythmicity (Fig. 7 C).
Figure 7.
Homing of metastatic cells to lung is regulated by neutrophils. (A) Disrupted diurnal infiltration of Bmal1ΔN neutrophils into lungs. Experimental design. (B) Relative number of partner-derived neutrophils in the lungs of partner mice at AM or PM (ZT4 and 16, respectively), determined by flow cytometry; n = 4 (for Bmal1ΔN) to 20 (for WT) mice. ND, not determined for neutrophil-depleted (1A8 antibody) mice. (C) Relative homing of B16F1Luc cells to the lungs of control, neutrophil-depleted mice and Bmal1ΔN mice, as determined by flow cytometry 24 h after injection; n = 3–5 mice (control and depleted) or 9 (Bmal1ΔN) per time and experiment from at least two independent experiments. Values are normalized to AM. Values show mean ± SEM. *, P < 0.05; n.s., not significant, comparing AM versus AM or PM versus PM values, as determined by Student’s two-tailed unpaired t test analysis. (D) Schematic of neutrophil dynamics during homeostasis and physiological roles in intestine and lungs. Neutrophils released from the BM into blood infiltrate multiple tissues with distinct circadian dynamics (boxes next to tissue names). Most tissues display rhythms in anti-phase with blood, while infiltration in liver, WAT, and intestine are arrhythmic. Illustration of remote control of hematopoietic niches through modulation of Il23 transcription within intestinal ILF (bottom left box). In the lung, neutrophils regulated diurnal transcription (bottom right box), thereby affecting the susceptibility to metastatic invasion by B16F1 melanoma cells (right; see also Video 10).
Altogether, these data indicated that diurnal infiltration of neutrophils in lungs can control the secondary migration of other cells types, which in the case of circulating tumoral cells can create temporal windows of susceptibility to disease.
Discussion
It is paradoxical that despite their abundance and well-defined roles during infection and inflammation, much remains enigmatic about the dynamics, functions, and fate of neutrophils in healthy tissues. Here we show that far from being silently eliminated in dedicated organs (spleen, liver, and BM; Furze and Rankin, 2008), neutrophils are endowed with the capacity to infiltrate most tissues (Video 10). We show this to be a temporally gated behavior in many of these tissues, with preferential infiltration or intravascular retention during the early evening. Indeed, our findings confirm previously reported circadian migration into tissues like the BM, as well as the presence of neutrophil subsets in the red pulp of the naive spleen, and margination in the lungs and liver (Gebb et al., 1995; Chosay et al., 1997; Puga et al., 2012; Scheiermann et al., 2012). Importantly, our data extend the migratory capacity of neutrophils to many other tissues in the absence of stress and report distinct temporal patterns of infiltration (steady or diurnal) across the organism (Fig. 7 D). While the cues (temporal and spatial) that drive this behavior are currently unknown, circadian increases in vascular adhesion molecules and chemokines (Scheiermann et al., 2012) could provide the substrate and signals that guide neutrophils into tissues in the absence of inflammation.
Perhaps a more fundamental finding is that neutrophil infiltration in tissues regulates basal physiology. In the intestine, we present evidence suggesting that phagocytosis of incoming neutrophils by resident macrophages regulates cytokine production and participates in the remote regulation of BM niches through a process similar to the “neutrostat” model known to regulate granulopoiesis (Stark et al., 2005). Interestingly, additional mechanisms of immune-driven regulation of the BM exist. A relevant example is local inhibition of hematopoietic niches by BM-infiltrating neutrophils, a process that follows temporal patterns (Scheiermann et al., 2012; Casanova-Acebes et al., 2013) that align with the diurnal infiltration dynamics in the marrow (Fig. 2 E). Thus, neutrophils emerge as global regulators of hematopoietic niches by acting both locally in the marrow (to inhibit) or remotely (to enhance). Together with the reported role of splenic neutrophils in supporting B cell maturation and immunoglobulin production (Puga et al., 2012), a more general interpretation of the data is that neutrophils can adapt to the physiological demands of the organs that they infiltrate.
Among the tissues that displayed diurnal patterns of infiltration, the lung was of particular interest, given the high number of neutrophils compared with other tissues. In this sense, it is remarkable that the lung also displays one of the highest rates of circadian transcription, with a time of peak amplitude that roughly matches the time of infiltration by neutrophils, during the transition from day to night (Zhang et al., 2014). The neutrophil depletion experiments demonstrated that, among all the genes displaying circadian expression in the lung, a significant fraction (∼26%) was entrained by neutrophils. At present, we do not know how neutrophils influence circadian expression in lungs; they may alter the cellular composition of the tissue by facilitating infiltration of other cells. Alternatively, they could release soluble mediators that modulate transcription of pulmonary cells or produce proteases that penetrate into and activate neighboring cells, as shown in other tissues (Talukdar et al., 2012; Nicolás-Ávila et al., 2017).
Beyond these mechanistic considerations, our findings provide the first example of transcriptional programming of a tissue by infiltrating neutrophils and have important implications to understand the impact of myeloid cells in nonimmune physiology. Indeed, among the neutrophil-controlled genes, our transcriptomic analyses uncovered functions related to carcinogenesis and cell migration, which we validated in a model of pulmonary metastasis. These results are consistent with reports showing circadian variations in metastatic spread (Hrushesky et al., 1999), with the reported role of neutrophils in lung metastasis (Wculek and Malanchi, 2015; Coffelt et al., 2016) and with the higher incidence and severity of cancer in humans subjected to frequent shift-work or jetlag (Filipski et al., 2004; Scheiermann et al., 2013). Because the time of injection was critical for the outcome of metastasis, we propose that initial tissue seeding by tumoral cells is the critical step at which neutrophils influence metastatic spread. Further, although intrinsic expression of circadian genes in tumoral cells is known to influence tumorigenesis (Papagiannakopoulos et al., 2016), our data suggest that extrinsic temporal cues can also be important regulators of tumor progression. Beyond the effects on metastatic spread described here, it is likely that neutrophils that accumulate diurnally in naive lungs contribute to protect against potential infections (Yipp et al., 2017).
The varying infiltration dynamics of neutrophils among different tissues is intriguing, but the mechanisms behind these distinct tissue-associated patterns remain unknown. One possibility is that they arise from differential properties of microvascular cells in different organs (Nolan et al., 2013). In any case, a concept that emerges from our data are that the patterns of infiltration can instruct temporal functions in each tissue: constitutive (i.e., nonoscillatory) entry in the intestine could support niche function, whereas circadian infiltration of the BM entrains circadian release of HPCs (Casanova-Acebes et al., 2013), and in the lung, it determines diurnal patterns of transcription and organ invasion. It is therefore tempting to speculate that neutrophils influence the diurnal timing of multiple physiological processes across different tissues, a possibility that we are currently exploring.
In summary, our study demonstrates that neutrophils can accomplish important homeostatic functions after their lifetime in the circulation, and we provide relevant examples of how infiltration of the intestine and intravascular accumulation in the lungs influence important aspects of BM and pulmonary physiology (Fig. 7 D). More generally, our findings suggest the need to revise our ideas about the biology of neutrophils in the steady-state, including characterization of their transcriptional properties, lifespan, and homeostatic and pathological functions within tissues.
Materials and methods
Mice
All experiments were performed on 6–12-wk-old C57BL/6 male and female mice. Chow and water were available ad libitum. Mice deficient in Fut7, in both P- and E-selectins (encoded by Selp and Sele, respectively; PEdKO), have already been reported (Frenette et al., 1996; Malý et al., 1996). The following knock-in or transgenic mice were also used: Cxcr4flox/flox (Nie et al., 2004), hMRP8CRE (Passegué et al., 2004), Lyz2GFP (Faust et al., 2000), Mcl1flox/flox (Dzhagalov et al., 2007), Cxcl12GFP (Sugiyama et al., 2006), Rosa26iDTR (Buch et al., 2005), Ly6gCRE; Rosa26tdTomato (Hasenberg et al., 2015), and IL1r1−/− (Mortha et al., 2014). Mice with neutrophil-specific deficiency in CXCR4 or Bmal1 were generated by crossing hMRP8CRE with Cxcr4flox/flox mice (Cxcr4ΔN mice) or Arntlflox/flox (Bmal1ΔN mice), and neutropenic mice were generated by crossing hMRP8CRE with Mcl1flox/flox mice (Mcl1ΔN mice) or with Rosa26iDTR plus DT injection. Mice with high levels of tdTomato fluorescence in neutrophils were generated by crossing Ly6gCRE with mice expressing the Cre-inducible tdTomato in the Rosa26 locus as in (referred to here as Ly6GTOM; Hasenberg et al., 2015). The Fut7−/−; Cd169DTR, Fut7−/−; Il17GFP, Fut7RED, and Fut7−/−; Cxcl12GFP lines were generated by crossing Fut7-deficient with previously described Cd169DTR mice (Saito et al., 2001; Miyake et al., 2007; Chow et al., 2011; Esplugues et al., 2011), with transgenic mice expressing DsRed under the control of the β-actin promoter (WTRED; Vintersten et al., 2004) or with Cxcl12GFP mice, respectively. B6.129P2(Cg)-Rorctm2Litt/J (Rorc+/EGFP), generated as described (Eberl et al., 2004), were provided by D. Littmann (Skirball Institute, New York, NY). Csf2−/− mice were a gift from M. Manz (University Hospital, Zurich, Switzerland). All mice were in the C57BL/6 background and were maintained on a 12-h light/12-h darkness schedule. For some experiments, control WT mice of the same background were obtained from the animal facility at CNIC. Mice were housed in a specific pathogen–free facility at Fundación CNIC or Mount Sinai School of Medicine. Experimental procedures were approved by the Animal Care and Ethics Committee CNIC, Mount Sinai School of Medicine, and University of Toronto.
Parabiosis
To generate parabiotic pairs, we followed our previously described procedure (Casanova-Acebes et al., 2013). Anesthetized mice were shaved at the corresponding lateral aspects and matching skin incisions were made from the olecranon to the knee joint of each mouse, and the subcutaneous fascia was bluntly dissected to create ∼0.5 cm of free skin. The olecranon and knee joints were attached by a single 5-0 polypropylene suture and tie, and the dorsal and ventral skins were approximated by continuous suture. A single dose of flunixin-meglumine (Schering-Plough) was injected subcutaneously in each partner at the end of the surgical procedure (1 mg/kg). 1 mo after surgery, samples were obtained from each of the partners for analyses. To ensure that partner-derived neutrophils in tissues derived from the circulation rather than from partner progenitor cells that could have also crossed to the host mouse, we generated parabionts that were separated after 1 mo. In the separated mice we confirmed a rapid decline of partner-derived neutrophils which was reduced from 15.5% to 4% in blood and tissues after 4 d, which indicated that the majority of neutrophils found in tissues of the host mice derived from the circulation.
Isolation of tissue neutrophils for flow cytometry
8–12-wk-old C57BL/6 WT and Lyz2GFP mice were analyzed after 1 mo in parabiosis. The blood of each parabiont was collected and used to calculate input ratios for neutrophil exchange between the different partners. To normalize values between the different parabiotic pairs, we corrected the ratios of host versus partner-derived neutrophils in each tissue with the ratios present in blood for each animal.
Skin, large intestine (colon), skeletal muscle, lung, heart, kidney, inguinal lymph nodes, and WAT were digested in HBSS with liberase (1 U/ml; Roche) and DNase I (1 mU/ml; Sigma) for 30 min at 37°C. For BM and spleen, undigested tissues were mechanically dissociated to prepare single-cell suspensions. Enrichment of leukocytes from liver and skeletal muscle was performed with Percoll 36 and 40%, respectively. Large intestines were preincubated with HBSS containing 5 mM EDTA for 45 min at 37°C before digestion to remove epithelial cells.
Single-cell suspensions were incubated with antibodies against CD45, CD11b, and Ly6G (clone 1A8; Table S2). Samples were acquired in a LSRII Fortessa (BD Biosciences). We used GFP or DsRed expression in the CD45+ CD11b+ Ly6GHI population to discriminate host- versus partner-derived neutrophils (for Lyz2GFP or WTRED partners, respectively). Doublets and DAPI+ cells were excluded from analyses using the FlowJo software (Flowjo LLC).
Immunofluorescence of tissue-resident neutrophils
WT and Lyz2GFP were sacrificed with CO2 and perfused with PBS. Tissues were fixed in 4% paraformaldehyde (PFA) for 18 h at 4°C and subsequently cryopreserved with a sucrose gradient and frozen in optimal cutting temperature compound (OCT; Tissue Tek) in liquid nitrogen. Tissues were cut in 5-µm sections in a cryostat (CM1850; Leica). Neutrophil detection by immunofluorescence was performed in tissues blocked 1 h with 10% BSA and 2% goat serum. Tissues were then incubated with a rabbit anti-laminin antibody (1:300; Sigma) and biotinylated Ly6G (1:50) for 1 h at room temperature. Tissues were washed three times in PBS and incubated with secondary antibodies anti-rabbit Alexa Fluor 635 (Life Technologies) and streptavidin Alexa Fluor 546 (Invitrogen), both diluted 1:500. Tissues were washed in PBS and nuclei were stained with DAPI (1:1,000) before mounting with Mowiol (Sigma).
Imaging and quantification of neutrophil and vessels in tissues
We analyzed partner-derived neutrophils distribution in tissues of WT mice that were maintained in parabiosis with Ly6GTOM for 8 wk. Tissues were collected and frozen in OCT, and 30-µm slices were obtained in a cryostat (CM1850; Leica). Tissue sections were incubated with rat anti-endomucin (Santa Cruz) and rat anti-CD31 (eBioscience), followed by incubation with DAPI (Life Technologies), the endothelial-binding lectin IB4 conjugated with AF647 (Thermo Fisher), and a secondary chicken anti-rat AF647 antibody (Molecular Probes). Images from Ly6GTOM+ neutrophils were obtained using an SPE confocal microscope (Leica). 3D images of neutrophils and vessels were reconstructed with Imaris 8.0 (Bitplane AG). Quantification of intra- and extravascular neutrophils was done manually.
Quantification of absolute numbers of cells per tissue using fluorescent beads
Truecount beads (Truecount absolute counting tubes; BD Biosciences) were prepared at a concentration of 10,000 beads per milliliter of PBS buffer containing DAPI. 500 µl of the bead suspension were added to single-cell suspensions stained for flow cytometry. 300–500 beads were acquired per tube to ensure accuracy for estimation of absolute number of cells. For some tissues, neutrophil counts per milligram of tissue were calculated as follows:
Values were then corrected by the weight of the tissue fragment analyzed. For blood and BM, the absolute number of cells is presented as cells per ml of blood or per one femur. Blood counts were analyzed in an automated hemocounter.
Estimation of neutrophil number in tissues
We analyzed the blood, BM, spleen, liver, lung, lymph nodes, skin, large intestine, skeletal muscle, heart, kidney, and WAT of mice set in parabiosis with fluorescent (Lyz2GFP) or congenic CD45.1 partners, after 1 mo of circulatory exchange as indicated. We used Truecount beads to estimate absolute numbers per milliliter of blood, milligram of tissue, one femur, or total organ.
For estimation of total cells in the full organisms we assumed that the number of cells in one femur represents ∼5.9% (or 1 in 16.9) of the total BM (Chervenick et al., 1968), and therefore, we multiplied the number of partner-derived neutrophils by 16.9 for estimation of total cells cleared in the BM. We assumed a total blood volume of 2.5 ml. Because the number of partner-derived cells in WAT and lymph nodes was negligible in absolute terms compared with the other organs, we did not include these in our estimation of cleared neutrophils in tissues. We used these estimations to obtain the absolute number of neutrophils in all organs, as well as the absolute number of those originating from blood (partner derived) in one mouse. We also considered that the chimerism of partner-derived neutrophils in blood was 22.1 ± 2% (n = 18) and thus estimated that the total number of cleared neutrophils would be 4.5-fold (= 100/22.1) higher than that found for partner-derived cells only. From these calculations we obtained an estimation of the number of total neutrophils in all tissues analyzed and, of these, the number of those that had infiltrated from blood.
To estimate relative numbers of DsRed+ and DsRedNEG neutrophils in tissues of Fut7RED; WT and Fut7RED; Fut7−/− parabionts (see Fig. 4 A) we performed flow cytometry of blood and multiple tissues from the DsRed+ fluorescent partner, as indicated. Because we assumed that tissue neutrophils are derived from blood, the partner-to-host ratios for each tissue were corrected by the ratios in blood to eliminate experimental variations between different pairs. Finally, we corrected the ratios from Fut7RED; WT pairs by the ratios in Fut7RED; Fut7−/− pairs (which served as reference control since neutrophils from both partners have the same genotype) to obtain an estimation of the enrichment of WT over Fut7−/− neutrophils in each tissue.
Quantification of IL-17–producing cells
CD45.1 animals were irradiated and transplanted with WT, Fut7−/−, or Fut7−/−; Il17GFP. After 8 wk of reconstitution, the transplanted animals were set in parabiosis (as indicated above) with WT or Fut7−/− that had also been generated by BM transplantation. Mice were sacrificed after 4 wk, and the large intestine was processed as indicated above for staining with anti-CD45 conjugated with PerCP-Cy5.5 (clone 30-F11; Biolegend), anti-CD3E conjugated with APC (clone 145-2C11; BioLegend), and anti-CD4 conjugated with PECy7 (clone GK1.5; Tonbo). For the analysis of IL-17–producing cells in the intestine, female C57BL/6J or Rorc−/− mice age 8–12 wk were injected on days −5, −3, and −1 with 25 µg control Ig or anti-P and anti-E selectin antibodies (at ZT2). Blood was collected using cheek puncture immediately before sacrifice, and organs were harvested, and spleen was meshed through a 70-µm cell strainer. Intestinal tissues were removed, opened longitudinally, and rinsed in cold PBS. Epithelial cells were removed, and the remaining lamina propria was minced and digested using Collagenase and DNase. Live hematopoietic cells were enriched using a 40–80% Percoll gradient. Cells were then washed and resuspended in complete RPMI containing Brefeldin A, PMA, and Ionomycin. Following a 4 h incubation time at 37°C in a humidified incubator at 5% CO2, cells were washed and stained with anti-CD45. Cells were then fixed and stained intracellular with anti–IL-17A or isotype antibodies. Samples were analyzed on a BD Biosciences LSR Fortessa II and analyzed using FlowJo software.
RNA isolation, reverse transcription, and real-time quantitative PCR
Total RNA was prepared with RNA Extraction RNeasy Plus Mini- or Micro-kit (QIAGEN) for isolation from macrophages or with TRIzol (Sigma) for total tissue RNA. RNA was reverse transcribed with High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s protocol. Real-time quantitative PCR (SYBR-green; Applied Biosystems) assays were performed with an Applied Biosystems 7900HT Fast Real-Time PCR System sequencer detector. Expression was normalized to Hprt and/or 36b4 expression. Primer sequences are listed in Table S3.
Quantification of CXCL12 in BM
CXCL12 was measured from total BM extracellular fluid content obtained after flushing BM from femurs in 1 ml of PBS using commercially available ELISA reagents (R&D Systems). Samples were diluted 1:5 in PBS.
Progenitor assays in culture and hematological values
Blood was collected by punction of the facial vein, and 100 µl of blood was placed over a Lympholyte bed (Cedarlane Labs) and centrifuged to obtain mononuclear cells. Cells were added to semisolid media containing 1.25% methylcellulose (Sigma), 30% FBS (StemCell Technologies), 1% deionized BSA, 100 µM 2-mercaptoethanol, and conditioned medium (12.7% vol/vol) from the WEHI3 cell line (containing IL-3), HM-5 cell line (containing GM-CSF), and BHK/MKL cell line stably transfected to produce the secreted form of murine kit Ligand/SCF. Cultures were plated in duplicates in 35-mm culture dishes (NUNC A/S) and incubated at 37°C in 5% CO2. The numbers of CFU-Cs were scored on day 6 or 7 using an inverted microscope. Partner or endogenous-derived CFU-C were discriminated based on DsRed fluorescence. For total leukocyte counts, blood was obtained by maxillary puncture and leukocyte counts were analyzed using an Abacus automated counter (Diatron).
Flow cytometry
Flow cytometry was performed using established protocols as indicated above, at the Cellomics Unit of Fundación CNIC. Samples were acquired in a LSRII Fortessa or FACS Canto analyzer (BD Biosciences). Doublets and DAPI+ cells were excluded from analyses using the FlowJo software (Flowjo LLC). Antibodies used are listed in Table S2.
Macrophage and neutrophil depletion
Depletion of resident macrophages in either Cd169DTR or Fut7−/−; Cd169DTR mice was performed by intraperitoneal injection of DT (10 µg/kg; Sigma) at days −7, −5, and −2 before analysis. Alternatively, macrophages were depleted by a single intravenous injection of clodronate-loaded liposomes (100 µl, intravenously) 5 d before analysis. For neutrophil depletion, mice were injected subcutaneously with 3 µg/g mouse body weight of either a neutrophil-depleting antibody (rat anti-mouse Ly6G, clone 1A8; BioXcell) or control rat IgG (I4131; Sigma) for two consecutive days. Alternatively, Mrp8CRE; iDTR mice were injected subcutaneously with DT (10 ng/g mouse body weight) for three consecutive days before analysis.
Identification of CXCL12-producing cells in tissues
Identification of CXCL12-producing cells was performed in tissues digested as indicated before from Cxcl12GFP reporter mice. Single-cell suspensions were incubated during 15 min at 4°C in darkness with biotinylated antibodies against Ter119 and CD45, CD31-APC, and streptavidin Alexa Fluor 405. In the BM, GFP fluorescence intensity was used to discriminate CXCL12-abundant reticular cells from CD45NEGTer119NEG osteoblasts and CD45NEGTer119NEGCD31NEGGFPLOW populations. All other tissues analyzed showed no intermediate levels of CXCL12 expression, and therefore the populations were classified as endothelial (CD31+GFP+ and CD31+GFPNEG) and nonendothelial (CD31NEGGFP+ and CD31NEGGFPNEG). All samples were collected and analyzed by flow cytometry as indicated above.
For identification of CXCL12-producing cells in BM sections, mice were perfused with PBS and femurs were fixed during 18 h in 2% PFA at 4°C. Then, a decalcification process was done with PBS containing 0.25 M EDTA for 1 wk, followed by rehydration in 30% sucrose and freezing in OCT. After blocking, BM slices were incubated with a rabbit anti-mouse antibody for laminin (L9393; Sigma) for 1 h at room temperature. Samples were washed and incubated with an Alexa Fluor 647–labeled chicken secondary antibody against rabbit (A-21472; Life Technologies). Slides were mounted in Vectashield with DAPI (Vector Laboratories), and images were acquired using a Leica SP8 X or SP5 confocal microscope.
Identification and isolation of tissue macrophages that engulf partner-derived DsRed+ leukocytes
Preparation of tissue-resident macrophages was performed in tissues that were digested as indicated above. Macrophages in most tissues (spleen, skeletal muscle, liver, WAT, skin, and large intestine) were identified based on the expression of CD45, CD11b, and F4/80, and BM macrophages were identified as Gr1NEG F4/80+. Engulfment of leukocytes was based on DsRed signal acquisition by tissue-resident macrophages in the Fut7−/− nonfluorescent partner in Fut7RED; Fut7−/− and WTRED; Fut7−/− parabionts. Non-parabiotic WT mice were used as controls to set the appropriate gates. In some experiments engulfing and nonengulfing macrophages from the large intestine were sort-purified for transcriptomic analyses using a FACS Aria sorter (BD Biosciences).
In vivo blocking of G-CSF, GM-CSF, IL-23, IL-17, and P- and E-selectins
Fut7−/− mice were intravenously injected with control rIgG, a monoclonal anti–G-CSF (MAB414; R&D Systems) antibody for 3 d (daily dose of 50 µg/mouse) injected at ZT13, or monoclonal anti–GM-CSF (MAB415; R&D) injected once a day at ZT13 for two consecutive days. Leukocyte counts and blood CFU-C were analyzed the next day at ZT5 and ZT13.
For IL-23 or IL-17 blockade, Fut7−/− mice were intravenously injected with 200 µg isotype control rIgG, a monoclonal anti–IL-23 antibody (MAB1887, clone 320234; R&D Systems) or a monoclonal anti–IL-17 (MAB421, clone 50104; R&D Systems) antibody at ZT13. Leukocyte counts and blood CFU-C were analyzed the next day at ZT5.
To induce defective migration in various mutant mice, we injected mice intravenously with 25 µg of control rIgG or anti–P- and anti–E-selectin antibodies (clones RB40.34 and 9A9 from BioXcell, respectively) at days −5, −3, and −1 at ZT2.
Determination of diurnal rhythms
We used the Cosinor analysis (Nelson et al., 1979) to test for cyclical rhythmicity for a defined period of 24 h using the Graphpad Prism software. The method uses least squares for nonlinear fit of a cosine waveform based on the formula y = baseline + amplitude × cos (frequency × T + phase shift), where baseline is the mean of all values, amplitude is 1/2 × (maximum-minimum values), frequency is 2π/24 (0.2618), T is time, and phase shift is the time of acrophase.
Immunofluorescence and tissue clearing for LSM
6-wk-old male Ly6GTOM mice were placed under parabiosis with age- and sex-matched WT C57BL/6 mice. Parabionts were separated after 4 wk and euthanized by CO2 asphyxiation. The WT mouse was immediately transcardially perfused with 10 ml PBS with 2 mM EDTA (pH 7.4), followed by 10 ml of 4% PFA through the left ventricle. The spleen, lungs, liver, colon (intestine), quadriceps (muscle), ears (skin), and femur (BM) were dissected, and the cut ears were additionally treated with hair removal cream (Veet) according to manufacturer instructions. Organs were then fixed in 4% PFA overnight at 4°C, followed by extensive washing with PBS (3 × 1 h). For the femurs, decalcification was performed with 0.5 M EDTA (pH 8.0) overnight at room temperature, before embedding in OCT (TissueTek) and cryostat removal of part of the bone to expose the BM. Shaved bones were thawed and washed thoroughly in PBS to remove traces of OCT. With the exception of the mouse ears, the organs were permeabilized with graded methanol solutions (vol/vol in PBS) of 50% (30 min), 70% (30 min), 95% (30 min) and 100% (1 h), followed by 20% DMSO/80% methanol (1 h). Tissue was rehydrated back to PBS with 45 min incubation at room temperature. The ear skin was split into dorsal and ventral sides, and the cartilage was removed. Samples were then blocked overnight in blocking buffer (2.5% goat serum, 2.5% donkey serum, 5% BSA, 0.02% gelatin, 20% DMSO, and 0.3% Triton X-100 in PBS); for spleens, 40% DMSO was used instead of 20%), washed extensively with PBS (4 × 45 min), and incubated with primary antibodies in dilution buffer (2.5% goat serum, 2.5% donkey serum, 0.2% BSA, 0.02% gelatin, 5% DMSO, and 0.3% Triton X-100 in PBS) for three nights at 37°C. Rat anti-mouse MRP14 (clone 2B10, Abcam), hamster anti-mouse CD31 (clone 2H8, Millipore), and polyclonal rabbit anti-RFP (600–401-379, Rockland) were used at 1:300 dilution for all samples. Samples were washed extensively with washing buffer (3% NaCl + 0.3% Triton X-100 in PBS; 6 × 30 min) and PBS (2 × 1 h). The BM was incubated with anti-rat Alexa Fluor 568 (1:500), anti-hamster Alexa Fluor 647 (1:500) and anti-rabbit Alexa Fluor 680 (1:500) for 36 h at 4°C. All other organs were incubated with anti-rat Alexa Fluor 568 (1:300), anti-hamster Alexa Fluor 647 (1:300) and anti-rabbit Alexa Fluor 750 (1:400; Life Technologies) in dilution buffer for 36 h at 37°C. Organs were then washed with washing buffer (6 × 30 min) and PBS (2 × 1 h), and cleared using the BABB (1:2 of Benzyl alcohol/ Benzyl benzoate) protocol. The sample was dehydrated with graded methanol solutions (vol/vol in distilled water) of 50% (30 min), 70% (30 min), 95% (30 min), and 100% (3 × 30 min), equilibrated overnight with 50% BABB in methanol, and refractive index matched with 100% BABB for at least 30 min or until clear by visual inspection. Cleared organs were imaged using the light sheet Ultramicroscope (LaVision BioTec GmbH, Bielefeld, Germany) in BABB with 2×/0.5 objective (with zoom factor of 2.0), using a white light supercontinuum laser, except for the BM, which was imaged by tile scanning using a hybrid two-photon and conventional laser scanning confocal microscope (LSM 780, Zeiss) with 20×/1.0 water objective.
Analyses of LSM data
Post-acquisition, high resolution raw LSM data were smoothed with a median filter and down-sampled using Imaris 8 (Bitplane AG). Neutrophils were identified using the MRP14 channel by the “Spots” module of Imaris software, and their statistics were exported to comma-separated value (csv) files. These statistics included information such as their identifying indices, x, y, and z coordinates, as well as their mean channel intensities. The data were compiled into a single csv file compatible with the Flow Cytometry Standard (fcs) format using a custom Python 3.5 script and further converted into a Flow Cytometry Standard file by use of the flow cytometry software, FlowJo X (Treestar). Subsequently, RFP-positive neutrophils (from parabiont partners) were easily distinguished from RFP-negative neutrophils (host derived) by standard scatterplot gating in FlowJo, and their relative positioning were visualized and exported as images. The RFP classification data were exported as csv index lists and reimported back to Imaris software for visualization and verification using custom Python scripts and the Imaris “Xtension” module. In our imaging, we found the existence of cells that were RFP positive (i.e., partner derived) but were MRP14 negative (i.e., not neutrophils). Thus, to avoid confusion, for the figure images and videos, we performed masking of the MRP14 and RFP channels in Imaris to display only the underlying raw intensities that exist within the “Spots” surfaces, thereby removing autofluorescence and other RFP signals (see Fig. S2 A). The CD31 channel was not modified.
For the tiled confocal data of the BM, uneven image brightness was corrected using custom Python scripts and stitched using FIJI is Just ImageJ software (Preibisch et al., 2009; Schindelin et al., 2012). Individual cells were identified using the “Spots” module of Imaris software and imported into FlowJo as described earlier. Because there was significant spillover among the various red dyes, spillover coefficients were calculated, and compensation was applied to the appropriate channels before gating and subsequent verification checks. Pixel-based channel unmixing was performed on the images using the calculated spillover coefficients using custom Python scripts, and the results were visualized with Imaris. No masking was applied in this case.
Whole-mount staining of the large intestine (colon)
For whole-mount, large intestine colons were excised and flushed with cold PBS and opened by the mesenteric border. Colons exposing the lumen upwards were placed in HBSS with 5 mM EDTA in agitation for 30 min at 37°C. Epithelial cells were removed by several washes of the intestinal surface with HBSS/EDTA. Colons were washed in cold PBS and fixed in PFA 4% at 4°C overnight. Fixed intestines were washed three times in PBS containing 0.5% Triton-X 100 (PBST) and blocked overnight with PBST with 25% FBS at 4°C with shaking. Staining of neutrophils was performed with a biotinylated anti-Mrp14 antibody (clone 2B10 provided by N. Hogg, Cancer Research UK, London, England, UK), macrophages with an anti-CD169 (conjugated with Alexa Fluor 647; BD Biosciences) or anti-F4/80 (conjugated with Alexa Fluor 647; clone CI:A3-1; AbD Serotech) antibody and the basal membrane with a rabbit anti-laminin antibody (Sigma), in 25% FBS-PBST overnight at 4°C with shaking. Colons were washed and incubated with secondary antibody (goat anti-rabbit Alexa Fluor 405; Life Technologies) and Alexa Fluor 488–conjugated Streptavidin in 25% FBS-PBST for 4 h at room temperature. Intestines were washed and mounted in Mowiol. Imaging of whole-mount intestines was performed at the Microscopy & Dynamic Imaging Unit of the CNIC using a Leica SP8 X confocal microscopy system coupled to a DMI6000 inverted microscope, with 10× (HC PL Fluotar 10×/0.3 Dry) or 63× (HC PL Apo CS2 63×/1.4 OIL) magnification objectives.
Experimental lung metastasis
We used a previously described model (Peinado et al., 2012). 8–10-wk-old male C57BL/6 mice were intravenously injected at the indicated ZT times with 100,000 sub-confluent B16F1Luc cells (which express the luciferase gene) under isoflurane anesthesia. For in vivo imaging system visualization, after 14 d, mice were injected intraperitoneally with 100 µl (30 mg/ml in PBS) of XenoLight d-luciferin (Perkin Elmer) 10 min before sacrifice. Subsequently, their lungs were quickly harvested and subjected to IVIS imaging using the Xenogen IVIS 200 (Perkin Elmer), and their peak intensities were determined with manufacturer software (Living Image; Perkin Elmer). After IVIS imaging, the complete left lobes of the lungs were fixed in 4% PFA and embedded in paraffin. Lungs were serially sectioned (3 µm), and every 40 sections were stained with H&E. Four H&E-stained sections per lung were dedicated to histopathological examination and metastatic lesion quantification. Images were captured with an Olympus AX70 microscope applying cellSens software.
Homing of B16F1 cells to lungs
For the premetastatic homing assays, we used control WT, neutrophil-depleted mice and Bmal1ΔN mice. B16F1Luc cells were labeled with DiIC18 (D7757; Molecular Probes) according to the manufacturer instructions, and 100,000 cells were injected intravenously at the indicated ZT times into each mouse, under isoflurane anesthesia. For these experiments, mice were housed in standard room and light cabinet with a 12-h phase shift to allow injection of the same cell preparation into each group of mice. Mice were then sacrificed 24 h later for lung harvest. The whole left lobes of lungs were minced with scalpels and incubated in 2 ml of freshly prepared DMEM containing 0.8 mg/ml dispase (Gibco) and 1.5 mg/ml collagenase P (Roche) at 37°C in agitation for 15 min. After incubation, 10 ml of DMEM was added and tissue fragments, and cell suspensions were spun at 1,200 rpm for 5 min. Pellets were incubated with 0.05% trypsin-EDTA at 37°C for 2 min. Single-cell suspensions were filtered through 70-µm nylon filter and washed in PBS with 2 mM EDTA and 1% BSA. Cells were blocked with purified anti-CD16/CD32 (BD Biosciences) and stained with CD45-FITC (BD Biosciences) for flow cytometry, and absolute counts of CD45-negative DiIC18-positive cells were obtained with the use of commercial counting beads (Trucount; BD Biosciences).
Immune characterization of lungs
To quantify the different leukocyte subsets in the lungs of control or neutrophil-depleted mice, including T and B lymphocytes, NK cells, Ly6Chi and Ly6Clo monocytes, CD11b-positive and -negative dendritic cells, alveolar and interstitial macrophages, and eosinophils, we followed previously reported protocols (Yu et al., 2016). Mice were perfused before lung collection to avoid contamination from blood.
RNA sequencing analysis of lungs
Mice were treated with 3 µg/g mouse bodyweight of either anti-Ly6G (neutrophil depleted) or control IgG (nondepleted) antibodies for two consecutive days, in normal light-controlled mouse facility or 12 h inverted light cabinets. In the morning of the third day, mice were bled from the cheek just before the lung harvest to obtain blood samples for confirmation of neutrophil depletion using a standard hemocytometer (ABX Pentra 80; Horiba), as well as flow cytometry. In the Mrp8CRE; iDTR model, mice were treated with DT for 3 d as indicated above, and tissues were collected on the fourth day. Whole intact left lung lobes were harvested after perfusion of mice with saline at the same time for all the groups (which corresponds to ZT4 and ZT16, respectively, for “AM” and “PM” mice) and snap-frozen in 1 ml of TRIzol each. Whole RNA was obtained subsequently using mechanical disruption (Polytron PT 6100; Kinematica) and chloroform extraction and cleaned up using silica-based spin columns (Qiagen) according to manufacturer instructions. RNA quality was checked using capillary electrophoresis (Agilent). Five mice per group were experimented upon for two batches of experiments, but only the best quality RNA from three (batch 1) or four (batch 2) of the mice from each group was submitted for whole RNA next generation sequencing in the Genomics Unit of CNIC. 200 ng of total RNA was used to generate barcoded RNA sequencing libraries using the NEBNext Ultra RNA Library preparation kit (New England Biolabs). Libraries were sequenced with HiSeq2500 (Illumina) to generate 50-nucleotide single reads, with minimum of eight million reads per sample. FastQ files for each sample were obtained using CASAVA v1.8 software (Illumina). Reads were further processed using the CASAVA package (Illumina) to demultiplex reads according to adapter indexes and to produce FastQ files. Read quality was determined with the application FastQC. For data analysis, sequencing adaptor contaminations were removed from reads using Cutadapt v1.7.1, and the resulting reads were mapped on the transcriptome (GRCm38 Ensembl gene-build 84) and quantified using RSEM v1.2.20 and Kallisto v0.43.1. Only genes with at least 20 counts per million across all samples were considered for statistical analysis with limma, and genes with over five estimated counts in at least 47% of the samples were considered in sleuth. The data normalization and analysis was performed differently depending on the quantification program combination. Sleuth v0.29.0 R package was used to process Kallisto’s pseudocounts and to perform the Likelihood Ratio Test. The R package limma v3.32.2 was used to normalize the estimated counts from RSEM and to test differential expression using the moderated t statistics as in Smyth (2004). Raw and Benjamini-Hochberg adjusted P values (false discovery rate) were calculated for each of the tests. An adjusted P value of <0.05 was considered statistically significant. Differentially expressed genes after false discovery rate correction were used to identify overrepresented pathways using Ingenuity Pathway Analysis (IPA; Qiagen). Raw data for the RNA sequencing analyses, as well as the normalized counts and TPMs, can be publicly accessed at the Gene Expression Omnibus (GEO; NCBI) with GEO accession no. GSE106349.
Statistical analysis
Data from experiments are represented as mean values ± SEM. All parameters analyzed followed normal distribution as tested by D’Agostino-Pearson test. Statistics on the transcriptome analyses are detailed in the “RNA sequencing analysis of lungs” section. Paired t test was used when two groups were compared. All statistical analysis were performed using Excel or GraphPad software, and P values ≤0.05 were considered statistically significant (*). P values P ≤ 0.01 (**) and P ≤ 0.001 (***), as well as nonsignificant differences (n.s.), are indicated accordingly.
Online supplemental material
Fig. S1 shows cytometric and imaging analyses of neutrophils in multiple tissues, both in single and in parabiosed mouse systems. Fig. S2 shows the workflow for whole-mount tissue imaging, as well as intra- versus extravascular distribution of neutrophils and circadian patterns of accumulation in various tissues. Fig. S3 presents the characterization of Mcl1ΔN and CXCR4ΔN mice and the levels of niche-associated cells and factors across different tissues. Fig. S4 shows the distribution and function of neutrophils in the intestinal mucosa. Fig. S5 shows the efficiency of neutrophil depletion in the antibody and Mrp8CRE; iDTR models. The supplemental material also contains three tables. Table S1 shows the canonical pathways identified from the lung transcriptomic comparison of control versus neutropenic mice, and Tables S2 and S3 show the antibodies and primers used for this study, respectively. Finally, the supplemental material includes 10 videos. Videos 1–8 show three-dimensional (3D) reconstructions of eight different organs with the distribution of neutrophils within them. Video 9 shows the distribution of neutrophils and macrophages in intestinal ILF, and Video 10 shows a summary depiction of the main findings of this paper.
Supplementary Material
Acknowledgments
We thank I. Cossío for help with HSC analyses; M. Mazariegos, I. Ortega, J.M. Ligos, and Y.R. Tan for technical support; K. Kohno, M. Tanaka, L. Chen, and S. Lira for the generous gift of mice; J. Garaude, C. Scheiermann, and D. Lucas-Alcaraz for insightful comments; and V. Lavilla for video editing.
This study was supported by Deutsche Forschungsgemeinschaft grants CRC914-B08 and CRC1123-B04 to O. Soehnlein and C. Weber; intramural funding from A*STAR to S. Devi and L.G. Ng; Ministerio de Economía, Inovación, y Universidades (MCIU) grant no. SAF2012-31142 and Comunidad de Madrid grants S2010/BMD-2314 and SAF2013-49662-EXP to A. Hidalgo; MCIU BES grant 2010-032828 to M. Casanova-Acebes, grant SVP-2014-068595 to J.A. Nicolás-Ávila, grant BES-2013–065550 to J.M. Adrover, grant JCI-2012-14147 to L.A. Weiss, grant JCI-2012-12659 to N. A-González, and grant MSCA-IF-EF-748381 to I. Ballesteros from the H2020 Program from the EU. A. Mortha is the Tier 2 Canadian Research Chair in Mucosal Immunology and supported by an Operating Grant from the Canadian Institutes of Health Research (CIHR 388337). The CNIC is supported by the MCIU and the Pro CNIC Foundation, and is a Severo Ochoa Center of Excellence (MCIU award SEV-2015-0505).
The authors declare no competing financial interests.
Author contributions: M. Casanova-Acebes, J.A. Nicolás-Ávila, J.L. Li., S. García-Silva, A. Balachander, L.A. Weiss, J.M. Adrover, K. Burrows, N. A-González, S. Devi, A. Mortha, J.A. Quintana, G. Crainiciuc, M. Leiva, and I. Ballesteros performed experiments; A. Rubio-Ponce performed transcriptomic analyses; M. Gunzer, C. Weber, T. Nagasawa, O. Soehnlein, and M. Merad contributed essential reagents; M. Casanova-Acebes, L.G. Ng, H. Peinado, and A. Hidalgo designed and supervised experiments; and A. Hidalgo designed and supervised the study. M. Casanova-Acebes and A. Hidalgo wrote the manuscript, which was edited by all authors.
References
- Adrover J.M., Nicolás-Ávila J.A., and Hidalgo A.. 2016. Aging: A Temporal Dimension for Neutrophils. Trends Immunol. 37:334–345. 10.1016/j.it.2016.03.005 [DOI] [PubMed] [Google Scholar]
- A-González N., Quintana J.A., García-Silva S., Mazariegos M., González de la Aleja A., Nicolás-Ávila J.A., Walter W., Adrover J.M., Crainiciuc G., Kuchroo V.K., et al. 2017. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 214:1281–1296. 10.1084/jem.20161375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Takahashi N., Ushiki M., Monya M., Aihara F., Kuboki E., Moriyama S., Iida M., Kitamura H., Qiu C.H., et al. 2015. Intestinal CD169(+) macrophages initiate mucosal inflammation by secreting CCL8 that recruits inflammatory monocytes. Nat. Commun. 6:7802 10.1038/ncomms8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C., Fogg D., Garfa M., Elain G., Join-Lambert O., Kayal S., Sarnacki S., Cumano A., Lauvau G., and Geissmann F.. 2007. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science. 317:666–670. 10.1126/science.1142883 [DOI] [PubMed] [Google Scholar]
- Bermejo D.A., Jackson S.W., Gorosito-Serran M., Acosta-Rodriguez E.V., Amezcua-Vesely M.C., Sather B.D., Singh A.K., Khim S., Mucci J., Liggitt D., et al. 2013. Trypanosoma cruzi trans-sialidase initiates a program independent of the transcription factors RORγt and Ahr that leads to IL-17 production by activated B cells. Nat. Immunol. 14:514–522. 10.1038/ni.2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T., Heppner F.L., Tertilt C., Heinen T.J., Kremer M., Wunderlich F.T., Jung S., and Waisman A.. 2005. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods. 2:419–426. 10.1038/nmeth762 [DOI] [PubMed] [Google Scholar]
- Casanova-Acebes M., Pitaval C., Weiss L.A., Nombela-Arrieta C., Chèvre R., A-González N., Kunisaki Y., Zhang D., van Rooijen N., Silberstein L.E., et al. 2013. Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell. 153:1025–1035. 10.1016/j.cell.2013.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervenick P.A., Boggs D.R., Marsh J.C., Cartwright G.E., and Wintrobe M.M.. 1968. Quantitative studies of blood and bone marrow neutrophils in normal mice. Am. J. Physiol. 215:353–360. [DOI] [PubMed] [Google Scholar]
- Chosay J.G., Essani N.A., Dunn C.J., and Jaeschke H.. 1997. Neutrophil margination and extravasation in sinusoids and venules of liver during endotoxin-induced injury. Am. J. Physiol. 272:G1195–G1200. [DOI] [PubMed] [Google Scholar]
- Chow A., Lucas D., Hidalgo A., Méndez-Ferrer S., Hashimoto D., Scheiermann C., Battista M., Leboeuf M., Prophete C., van Rooijen N., et al. 2011. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J. Exp. Med. 208:261–271. 10.1084/jem.20101688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher M.J., Liu F., Hilton M.J., Long F., and Link D.C.. 2009. Suppression of CXCL12 production by bone marrow osteoblasts is a common and critical pathway for cytokine-induced mobilization. Blood. 114:1331–1339. 10.1182/blood-2008-10-184754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffelt S.B., Wellenstein M.D., and de Visser K.E.. 2016. Neutrophils in cancer: neutral no more. Nat. Rev. Cancer. 16:431–446. 10.1038/nrc.2016.52 [DOI] [PubMed] [Google Scholar]
- de Oliveira S., Rosowski E.E., and Huttenlocher A.. 2016. Neutrophil migration in infection and wound repair: going forward in reverse. Nat. Rev. Immunol. 16:378–391. 10.1038/nri.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., and Morrison S.J.. 2013. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 495:231–235. 10.1038/nature11885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhagalov I., St. John A., and He Y.W.. 2007. The antiapoptotic protein Mcl-1 is essential for the survival of neutrophils but not macrophages. Blood. 109:1620–1626. 10.1182/blood-2006-03-013771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G., Marmon S., Sunshine M.J., Rennert P.D., Choi Y., and Littman D.R.. 2004. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5:64–73. 10.1038/ni1022 [DOI] [PubMed] [Google Scholar]
- Esplugues E., Huber S., Gagliani N., Hauser A.E., Town T., Wan Y.Y., O’Connor W. Jr., Rongvaux A., Van Rooijen N., Haberman A.M., et al. 2011. Control of TH17 cells occurs in the small intestine. Nature. 475:514–518. 10.1038/nature10228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust N., Varas F., Kelly L.M., Heck S., and Graf T.. 2000. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood. 96:719–726. [PubMed] [Google Scholar]
- Filipski E., Delaunay F., King V.M., Wu M.W., Claustrat B., Gréchez-Cassiau A., Guettier C., Hastings M.H., and Francis L.. 2004. Effects of chronic jet lag on tumor progression in mice. Cancer Res. 64:7879–7885. 10.1158/0008-5472.CAN-04-0674 [DOI] [PubMed] [Google Scholar]
- Frenette P.S., Mayadas T.N., Rayburn H., Hynes R.O., and Wagner D.D.. 1996. Susceptibility to infection and altered hematopoiesis in mice deficient in both P- and E-selectins. Cell. 84:563–574. 10.1016/S0092-8674(00)81032-6 [DOI] [PubMed] [Google Scholar]
- Furze R.C., and Rankin S.M.. 2008. The role of the bone marrow in neutrophil clearance under homeostatic conditions in the mouse. FASEB J. 22:3111–3119. 10.1096/fj.08-109876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebb S.A., Graham J.A., Hanger C.C., Godbey P.S., Capen R.L., Doerschuk C.M., and Wagner W.W. Jr. 1995. Sites of leukocyte sequestration in the pulmonary microcirculation. J. Appl. Physiol. 79:493–497. 10.1152/jappl.1995.79.2.493 [DOI] [PubMed] [Google Scholar]
- Hasenberg A., Hasenberg M., Männ L., Neumann F., Borkenstein L., Stecher M., Kraus A., Engel D.R., Klingberg A., Seddigh P., et al. 2015. Catchup: a mouse model for imaging-based tracking and modulation of neutrophil granulocytes. Nat. Methods. 12:445–452. 10.1038/nmeth.3322 [DOI] [PubMed] [Google Scholar]
- Hrushesky W.J., Lester B., and Lannin D.. 1999. Circadian coordination of cancer growth and metastatic spread. Int. J. Cancer. 83:365–373. [DOI] [PubMed] [Google Scholar]
- Im S.Y., Ko H.M., Kim J.W., Lee H.K., Ha T.Y., Lee H.B., Oh S.J., Bai S., Chung K.C., Lee Y.B., et al. 1996. Augmentation of tumor metastasis by platelet-activating factor. Cancer Res. 56:2662–2665. [PubMed] [Google Scholar]
- Kruger P., Saffarzadeh M., Weber A.N., Rieber N., Radsak M., von Bernuth H., Benarafa C., Roos D., Skokowa J., and Hartl D.. 2015. Neutrophils: Between host defence, immune modulation, and tissue injury. PLoS Pathog. 11:e1004651 10.1371/journal.ppat.1004651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Huang L., Vergis A.L., Ye H., Bajwa A., Narayan V., Strieter R.M., Rosin D.L., and Okusa M.D.. 2010. IL-17 produced by neutrophils regulates IFN-gamma-mediated neutrophil migration in mouse kidney ischemia-reperfusion injury. J. Clin. Invest. 120:331–342. 10.1172/JCI38702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malý P., Thall A., Petryniak B., Rogers C.E., Smith P.L., Marks R.M., Kelly R.J., Gersten K.M., Cheng G., Saunders T.L., et al. 1996. The alpha(1,3)fucosyltransferase Fuc-TVII controls leukocyte trafficking through an essential role in L-, E-, and P-selectin ligand biosynthesis. Cell. 86:643–653. 10.1016/S0092-8674(00)80137-3 [DOI] [PubMed] [Google Scholar]
- Méndez-Ferrer S., Lucas D., Battista M., and Frenette P.S.. 2008. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 452:442–447. 10.1038/nature06685 [DOI] [PubMed] [Google Scholar]
- Méndez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma’ayan A., Enikolopov G.N., and Frenette P.S.. 2010. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 466:829–834. 10.1038/nature09262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y., Asano K., Kaise H., Uemura M., Nakayama M., and Tanaka M.. 2007. Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell-associated antigens. J. Clin. Invest. 117:2268–2278. 10.1172/JCI31990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortha A., Chudnovskiy A., Hashimoto D., Bogunovic M., Spencer S.P., Belkaid Y., and Merad M.. 2014. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 343:1249288 10.1126/science.1249288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W., Tong Y.L., Lee J.K., and Halberg F.. 1979. Methods for cosinor-rhythmometry. Chronobiologia. 6:305–323. [PubMed] [Google Scholar]
- Nicolás-Ávila J.A., Adrover J.M., and Hidalgo A.. 2017. Neutrophils in Homeostasis, Immunity, and Cancer. Immunity. 46:15–28. 10.1016/j.immuni.2016.12.012 [DOI] [PubMed] [Google Scholar]
- Nie Y., Waite J., Brewer F., Sunshine M.J., Littman D.R., and Zou Y.R.. 2004. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J. Exp. Med. 200:1145–1156. 10.1084/jem.20041185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan D.J., Ginsberg M., Israely E., Palikuqi B., Poulos M.G., James D., Ding B.S., Schachterle W., Liu Y., Rosenwaks Z., et al. 2013. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev. Cell. 26:204–219. 10.1016/j.devcel.2013.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannakopoulos T., Bauer M.R., Davidson S.M., Heimann M., Subbaraj L., Bhutkar A., Bartlebaugh J., Vander Heiden M.G., and Jacks T.. 2016. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 24:324–331. 10.1016/j.cmet.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegué E., Wagner E.F., and Weissman I.L.. 2004. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 119:431–443. 10.1016/j.cell.2004.10.010 [DOI] [PubMed] [Google Scholar]
- Pearson C., Uhlig H.H., and Powrie F.. 2012. Lymphoid microenvironments and innate lymphoid cells in the gut. Trends Immunol. 33:289–296. 10.1016/j.it.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Peinado H., Alečković M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., García-Santos G., Ghajar C., et al. 2012. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18:883–891. 10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillipson M., and Kubes P.. 2011. The neutrophil in vascular inflammation. Nat. Med. 17:1381–1390. 10.1038/nm.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S., Saalfeld S., and Tomancak P.. 2009. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 25:1463–1465. 10.1093/bioinformatics/btp184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga I., Cols M., Barra C.M., He B., Cassis L., Gentile M., Comerma L., Chorny A., Shan M., Xu W., et al. 2012. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nat. Immunol. 13:170–180. 10.1038/ni.2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Iwawaki T., Taya C., Yonekawa H., Noda M., Inui Y., Mekada E., Kimata Y., Tsuru A., and Kohno K.. 2001. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat. Biotechnol. 19:746–750. 10.1038/90795 [DOI] [PubMed] [Google Scholar]
- Saverymuttu S.H., Peters A.M., Keshavarzian A., Reavy H.J., and Lavender J.P.. 1985. The kinetics of 111indium distribution following injection of 111indium labelled autologous granulocytes in man. Br. J. Haematol. 61:675–685. 10.1111/j.1365-2141.1985.tb02882.x [DOI] [PubMed] [Google Scholar]
- Scheiermann C., Kunisaki Y., Lucas D., Chow A., Jang J.E., Zhang D., Hashimoto D., Merad M., and Frenette P.S.. 2012. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity. 37:290–301. 10.1016/j.immuni.2012.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiermann C., Kunisaki Y., and Frenette P.S.. 2013. Circadian control of the immune system. Nat. Rev. Immunol. 13:190–198. 10.1038/nri3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:e3 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Stark M.A., Huo Y., Burcin T.L., Morris M.A., Olson T.S., and Ley K.. 2005. Phagocytosis of apoptotic neutrophils regulates granulopoiesis via IL-23 and IL-17. Immunity. 22:285–294. 10.1016/j.immuni.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Kohara H., Noda M., and Nagasawa T.. 2006. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 25:977–988. 10.1016/j.immuni.2006.10.016 [DOI] [PubMed] [Google Scholar]
- Suratt B.T., Young S.K., Lieber J., Nick J.A., Henson P.M., and Worthen G.S.. 2001. Neutrophil maturation and activation determine anatomic site of clearance from circulation. Am. J. Physiol. Lung Cell. Mol. Physiol. 281:L913–L921. 10.1152/ajplung.2001.281.4.L913 [DOI] [PubMed] [Google Scholar]
- Takahashi N., Vanlaere I., de Rycke R., Cauwels A., Joosten L.A., Lubberts E., van den Berg W.B., and Libert C.. 2008. IL-17 produced by Paneth cells drives TNF-induced shock. J. Exp. Med. 205:1755–1761. 10.1084/jem.20080588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar S., Oh D.Y., Bandyopadhyay G., Li D., Xu J., McNelis J., Lu M., Li P., Yan Q., Zhu Y., et al. 2012. Neutrophils mediate insulin resistance in mice fed a high-fat diet through secreted elastase. Nat. Med. 18:1407–1412. 10.1038/nm.2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vintersten K., Monetti C., Gertsenstein M., Zhang P., Laszlo L., Biechele S., and Nagy A.. 2004. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 40:241–246. 10.1002/gene.20095 [DOI] [PubMed] [Google Scholar]
- Wculek S.K., and Malanchi I.. 2015. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature. 528:413–417. 10.1038/nature16140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthen G.S., Schwab B. III, Elson E.L., and Downey G.P.. 1989. Mechanics of stimulated neutrophils: cell stiffening induces retention in capillaries. Science. 245:183–186. 10.1126/science.2749255 [DOI] [PubMed] [Google Scholar]
- Wynn T.A., Chawla A., and Pollard J.W.. 2013. Macrophage biology in development, homeostasis and disease. Nature. 496:445–455. 10.1038/nature12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yipp B.G., Kim J.H., Lima R., Zbytnuik L.D., Petri B., Swanlund N., Ho M., Szeto V.G., Tak T., Koenderman L., et al. 2017. The Lung is a Host Defense Niche for Immediate Neutrophil-Mediated Vascular Protection. Sci. Immunol. 2:eaam8929 10.1126/sciimmunol.aam8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.R., O’Koren E.G., Hotten D.F., Kan M.J., Kopin D., Nelson E.R., Que L., and Gunn M.D.. 2016. A Protocol for the Comprehensive Flow Cytometric Analysis of Immune Cells in Normal and Inflamed Murine Non-Lymphoid Tissues. PLoS One. 11:e0150606 10.1371/journal.pone.0150606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Lahens N.F., Ballance H.I., Hughes M.E., and Hogenesch J.B.. 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. USA. 111:16219–16224. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.