NFAT5 regulates macrophage MHCII expression by controlling the transcription of its coactivator Ciita through a remote enhancer. This mechanism differs from those previously found in DCs and B lymphocytes and distinguishes macrophages from these APC lineages.
Abstract
MHCII in antigen-presenting cells (APCs) is a key regulator of adaptive immune responses. Expression of MHCII genes is controlled by the transcription coactivator CIITA, itself regulated through cell type–specific promoters. Here we show that the transcription factor NFAT5 is needed for expression of Ciita and MHCII in macrophages, but not in dendritic cells and other APCs. NFAT5-deficient macrophages showed defective activation of MHCII-dependent responses in CD4+ T lymphocytes and attenuated capacity to elicit graft rejection in vivo. Ultrasequencing analysis of NFAT5-immunoprecipitated chromatin uncovered an NFAT5-regulated region distally upstream of Ciita. This region was required for CIITA and hence MHCII expression, exhibited NFAT5-dependent characteristics of active enhancers such as H3K27 acetylation marks, and required NFAT5 to interact with Ciita myeloid promoter I. Our results uncover an NFAT5-regulated mechanism that maintains CIITA and MHCII expression in macrophages and thus modulates their T lymphocyte priming capacity.
Introduction
Presentation of antigen-derived peptides by MHC class II (MHCII) to CD4+ T lymphocytes is crucial for activating adaptive immune responses (DeSandro et al., 1999; Reith and Mach, 2001). Macrophages and dendritic cells (DCs) are antigen-presenting cells (APCs) that constitutively express MHCII in steady-state conditions (Reith et al., 2005). Also, when blood monocytes infiltrate tissues such as the gut, they acquire MHCII expression progressively as they mature to macrophages (Bain et al., 2014; Jakubzick et al., 2017). MHCII expression in macrophages and DCs is markedly enhanced by IFNγ, a cytokine produced by activated CD4 and CD8 T lymphocytes and various innate lymphoid cell subsets. IFNγ not only enhances MHCII expression in immune cells, but early works showed that it is a potent inducer of MHCII in nonimmune cells such as endothelial cells and fibroblasts, allowing them to acquire antigen presentation capacity (Collins et al., 1984). Macrophages are promoters of tolerance in tissues (Soroosh et al., 2013; Shouval et al., 2014), and their expression of MHCII is considered part of a mechanism that samples local signals such as host and commensal microbial antigens that are presented by MHCII to CD4+ T lymphocytes for activating tissue tolerance. Nonetheless, MHCII in tissue macrophages can also activate specific effector CD4+ T cells to mount potent inflammatory adaptive responses by presenting antigens from necrotic cells or pathogens. In this context, a positive feedback loop is established between macrophages and IFNγ-producing lymphoid populations by which MHCII-mediated antigen presentation and cytokines produced by macrophages stimulate T lymphocytes to produce IFNγ, which in turn enhances MHCII expression in the macrophage. MHCII-mediated communication between macrophages and lymphocytes occurs in diverse inflammation settings, for instance in obesity, where adipose tissue macrophages activated by stressed adipocytes drive CD4+ T cell activation and trigger obesity-induced inflammation and insulin resistance (Morris et al., 2013; Cho et al., 2014). Another example is provided by allogeneic graft rejection, where macrophages from the graft and those infiltrating from the host proliferate locally, release proinflammatory mediators, and ingest dead cells from the graft to present their antigens to T cells that mediate cytotoxic antigraft responses (Grau et al., 1998; Underhill et al., 1999; Breloer et al., 2002; Wyburn et al., 2005). These examples illustrate how the ability of macrophages to express moderate levels of MHCII is important to ensure immune tolerance while simultaneously allowing them to conduct local surveillance as long as homeostatic conditions prevail. However, upon disruption of tissue homeostasis, macrophages will up-regulate MHCII expression and antigen presentation capacity as they acquire a proinflammatory profile. Moderate expression of MHCII in steady-state macrophages distinguishes them from DCs, which express much higher levels of MHCII even in homeostatic conditions. In this regard, macrophages and myeloid DCs are thought to share common transcriptional mechanisms controlling MHCII, but differences in MHCII levels between both cell types as well as between homeostatic and inflammatory macrophages raise the question of whether macrophages might use specific mechanisms to regulate steady-state expression of MHCII.
Transcription of MHCII genes is controlled by a group of ubiquitously expressed factors that includes cAMP-responsive element binding protein (CREB1), regulatory factor X (RFX), and nuclear factor Y (NFY) proteins, all acting in concert with the MHCII transactivator (CIITA, also known as MHC2TA; Boss, 1997). The relevance of these transcription regulators is illustrated by bare lymphocyte syndrome, a severe immunodeficiency caused by mutations in CIITA or the RFX factors, all of which are essential for MHCII expression (DeSandro et al., 1999; Reith and Mach, 2001). The expression of MHCII in different populations of APCs is determined by cell lineage–specific mechanisms that control CIITA transcription (Boss and Jensen, 2003; Reith et al., 2005). Ciita promoter IV regulates its expression in nonhematopoietic APCs, promoter III drives it in cells of lymphoid origin such as B lymphocytes, and promoter I is the common regulator of CIITA expression in macrophages and conventional DCs, both in homeostasis and upon IFNγ stimulation (Muhlethaler-Mottet et al., 1997; Piskurich et al., 1998; Boss and Jensen, 2003; Reith et al., 2005).
NFAT5 is a transcription factor that shares structural and functional properties with NF-κB and NFATc proteins (Lopez-Rodríguez et al., 1999; López-Rodríguez et al., 2001). NFAT5 regulates gene expression in immune cells in different contexts, for instance during macrophage polarization and in response to pathogen-sensing receptors (Buxadé et al., 2012; Tellechea et al., 2018), during pre-TCR–induced T lymphocyte development (Berga-Bolaños et al., 2013), and in mature T cells (Berga-Bolaños et al., 2010; Alberdi et al., 2017). Apart from its ability to respond to specific immune receptors, NFAT5 is activated by hypertonicity, and indeed its antipathogen function can be enhanced under hypertonic conditions such as those present in wounds (Aramburu et al., 2006; Jantsch et al., 2015). Previous works had shown that NFAT5-deficient mice had reduced T cell responses to alloantigens in vivo (Go et al., 2004; Berga-Bolaños et al., 2010), a defect that was not observed in conditional KO mice in which only T cells were deficient in NFAT5 (Berga-Bolaños et al., 2010). In addition, a recent article reported a human patient with NFAT5 haploinsufficiency who suffered impaired adaptive immune responses and evidence of a primary immunodeficiency disorder (Boland et al., 2015). While these observations could be explained in part by the ability of NFAT5 to regulate macrophage polarization, they also raised the question of whether this factor could play a direct role in the capacity of APCs to contact with and induce antigen-dependent responses in T cells. This question had not been addressed in earlier works, so we analyzed the transcriptomes of wild-type and NFAT5-deficient macrophages to search for potential differences in the expression of genes regulating antigen presentation and macrophage–lymphocyte contact. We show that expression of MHCII and its transcription coactivator Ciita are impaired in NFAT5-deficient macrophages and identify a remote NFAT5-dependent Ciita enhancer that regulates CIITA expression. Lack of NFAT5 attenuated the ability of macrophages to activate MHCII-dependent CD4 T cell responses in vitro and delayed the rejection of incompatible skin grafts in vivo. The dependence of macrophages on NFAT5 for expressing CIITA and MHCII distinguishes them from other APC lineages such as DCs and B lymphocytes.
Results
NFAT5 regulates expression of MHCII genes in macrophages
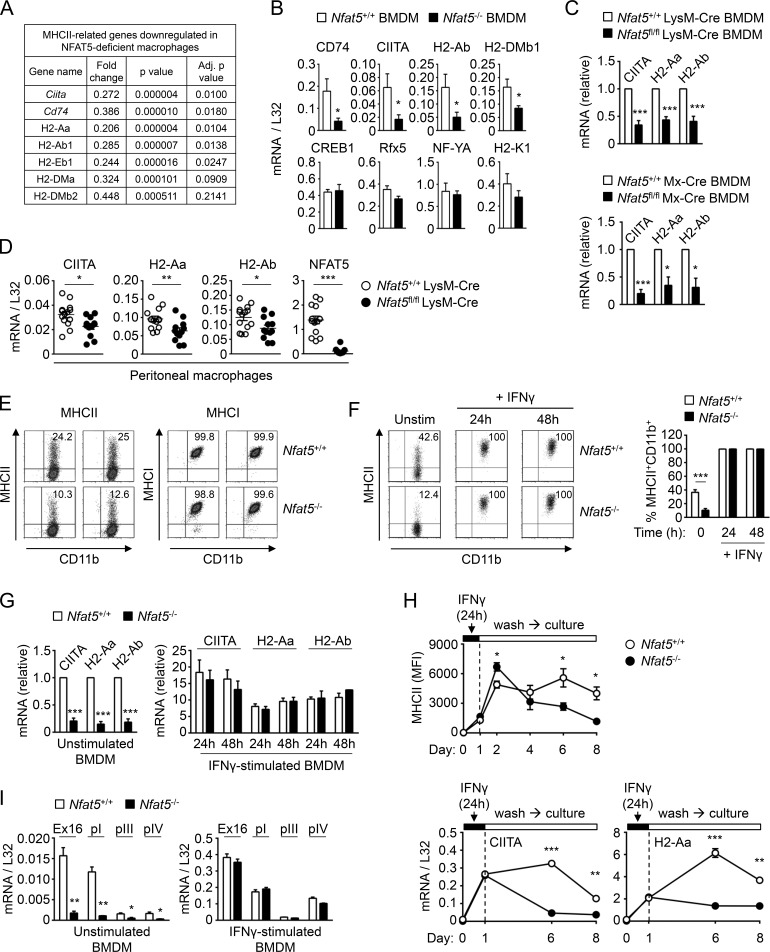
Global gene expression analysis of NFAT5-deficient bone marrow–derived macrophages (BMDMs) uncovered an extensive defect in the expression of a set of genes related to MHCII, the surface complex specialized in presenting antigenic peptides to CD4+ T lymphocytes (Fig. 1 A and Table S1). This defect was evidenced by a two- to fivefold decrease in mRNA levels of various MHCII genes, the invariant MHCII polypeptide CD74, and the MHCII transactivator CIITA, whereas genes encoding for other transcription regulators of MHCII, such as CREB1, RFX, and NFY factors, molecules coregulated with MHCII, adhesion, and costimulatory molecules, and MHC class I (MHCI) were not affected in NFAT5-deficient macrophages (Fig. 1 A and Table S1). We validated the findings from the transcriptome analysis for Ciita, H2-Aa, and H2-Ab by real-time quantitative PCR (RT-qPCR) in independent BMDM samples from NFAT5-deficient mice (Nfat5−/−), myeloid-specific NFAT5-deficient mice (Nfat5fl/fl LysM-Cre; Tellechea et al., 2018), and in inducible NFAT5-deficient mice (Nfat5fl/fl Mx-Cre; Tellechea et al., 2018; Fig. 1, B and C). We also confirmed the NFAT5-dependent expression of CD74 and the MHCII gene H2-DMb1, whereas expression of transcription factors that regulate MHCII genes (Rfx5, Nfya, and Creb1) and the MHCI gene H2-K1 did not require NFAT5 (Fig. 1 B). The dependence of CIITA and MHCII on NFAT5 was also observed in freshly isolated peritoneal macrophages from myeloid-specific NFAT5-deficient mice (Fig. 1 D). Flow cytometry analysis confirmed that impaired mRNA expression of MHCII genes in NFAT5-deficient macrophages was reflected in a lower proportion of MHCII-positive cells, whereas their MHCI expression was unaltered (Fig. 1 E). Intriguingly, we found that NFAT5-deficient DCs, either derived from bone marrow precursors (BMDCs) with GM-CSF or Flt3 ligand (Flt3L) or directly isolated from spleen as conventional myeloid and plasmacytoid DCs, had no significant defect in their expression of Ciita and MHCII (Fig. S1, A–C). B and T lymphocytes also expressed these genes in an NFAT5-independent manner (Fig. S1 D). Our finding that the closely related lineages of macrophages and myeloid DCs differed in their dependence on NFAT5 for CIITA and MHCII expression was unexpected, since until now both populations had been considered to regulate these genes through the same mechanisms. Basal expression of MHCII in APCs allows them to present antigens and prime CD4+ T lymphocytes, boosting IFNγ production by T cells. In turn, IFNγ enhances MHCII expression in macrophages, thus reinforcing a positive feedback loop. We asked whether lack of NFAT5 also affected the induction of CIITA and MHCII by IFNγ and found that acute stimulation with this cytokine strongly enhanced their expression in an NFAT5-independent manner (Fig. 1, F and G). However, after washing out IFNγ, NFAT5-deficient macrophages lost CIITA and MHCII expression faster than wild-type cells (Fig. 1 H). These results indicated that lack of NFAT5 in macrophages affected not only basal steady-state expression of CIITA and MHCII but also their sustained expression after transient stimulation with IFNγ. Finally, we analyzed the effect of NFAT5 in the expression of the different isoforms of Ciita (Muhlethaler-Mottet et al., 1997; Morris et al., 2013) in untreated and IFNγ-treated macrophages. Ciita expression in BMDMs was for the most part driven by promoter I in an NFAT5-dependent manner (Fig. 1 I). Promoters III and IV made a minor contribution to Ciita expression, which also seemed to be NFAT5-dependent. Acute stimulation with IFNγ induced Ciita expression from promoters I and IV, and, consistent with our previous result (Fig. 1 G), this was independent of NFAT5 (Fig. 1 I). This result indicated that NFAT5 is a relevant factor to control Ciita expression in macrophages through its myeloid promoter I. Altogether, our analysis of primary myeloid and lymphoid APCs uncovered a selective requirement of NFAT5 in macrophages for expressing CIITA and MHCII.
Figure 1.
MHCII expression in wild-type and NFAT5-deficient macrophages. (A) Reduced mRNA expression of MHCII-related genes in NFAT5-deficient BMDMs identified by microarray analysis (see Table S1 for additional genes). (B) mRNA expression of CIITA, H2-Ab, CD74, and H2-DMb1; MHCII regulatory factors Rfx5, NF-YA, and CREB1; and the MHCI gene H2-K1 in wild-type (Nfat5+/+) and NFAT5-deficient (Nfat5−/−) BMDMs. Values show the mean ± SEM of four independent experiments, each comparing BMDMs from one NFAT5-deficient mouse and a wild-type littermate. (C) CIITA and MHCII mRNA expression was analyzed by RT-qPCR in BMDMs from conditional NFAT5 deletion models. Values show the mean ± SEM of three independent experiments for LysM-Cre and six for Mx-Cre NFAT5 deletion models. Each experiment compared BMDMs from one NFAT5-deficient mouse and a wild-type littermate. Data for each respective mRNA are shown normalized to the wild-type sample, which was given a value of 1. (D) CIITA and MHCII mRNA expression in peritoneal macrophages from wild-type (Nfat5+/+ LysM-Cre) and Nfat5fl/fl LysM-Cre conditional KO. Each circle represents one individual mouse. Data are from 14 wild-type and 12 Nfat5fl/fl LysM-Cre conditional KO mice compiled through six independent experiments. (E) Flow cytometry analysis of MHCII and MHCI protein expression in BMDM from Nfat5+/+ LysM-Cre (wild-type, Nfat5+/+) and Nfat5fl/fl LysM-Cre mice (KO, Nfat5–/–) in two independent pairs of littermates. (F) Left: Representative flow cytometry analysis of MHCII expression in wild-type (Nfat5+/+) and NFAT5-deficient (Nfat5−/−) BMDMs. Right: Percentage of MHCII+ macrophages from BMDM cultures of littermate wild-type and NFAT5-deficient mice, either left unstimulated or treated with IFNγ (400 U/ml) for 24 or 48 h. Data are from six independent experiments with unstimulated cells and four more including unstimulated as well as IFNγ-treated macrophages. (G) mRNA levels of CIITA and MHCII genes in wild-type (Nfat5+/+) and NFAT5-deficient (Nfat5−/−) BMDMs, either left untreated (left) or stimulated with IFNγ (400 U/ml) for 24 or 48 h (right). Values for IFNγ-stimulated cells are represented relative to unstimulated cells, set as 1 for each respective gene in wild-type macrophages. Results are the mean ± SEM of three independent experiments, each comparing BMDMs from one NFAT5-deficient mouse and a wild-type littermate. (H) Expression of MHCII in wild-type and NFAT5-deficient BMDMs analyzed in basal conditions, after 24 h of IFNγ stimulation (100 U/ml), and at different days after washing out IFNγ. The upper panel shows the analysis of MHCII mean fluorescence intensity (MFI) in three independent pairs of wild-type and NFAT5-deficient BMDMs (mean ± SEM). Bottom panels show mRNA expression of CIITA and H2-Aa in the same experiment (mean ± SEM, n = 3). (I) Expression of CIITA transcripts from its different promoters (pI, pIII, and pIV) in wild-type (Nfat5+/+) and NFAT5-deficient (Nfat5–/–) BMDMs, either left unstimulated or stimulated with IFNγ for 24 h. CIITA Ex16 corresponds to an mRNA region common to all CIITA transcripts, which spans exons 16 to 18 in Ciita transcript NM_001243760.2 transcribed from promoter I. Values show the mean ± SEM of three independent experiments, each comparing BMDMs from one NFAT5-deficient mouse and a wild-type littermate. Statistical significance in B, D, F, H, and I was determined with an unpaired t test, and in C and G with a one-sample t test using the respective wild-type cells as reference with a value of 1. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Impaired activation of MHCII-dependent CD4+ T cell responses by NFAT5-deficient macrophages
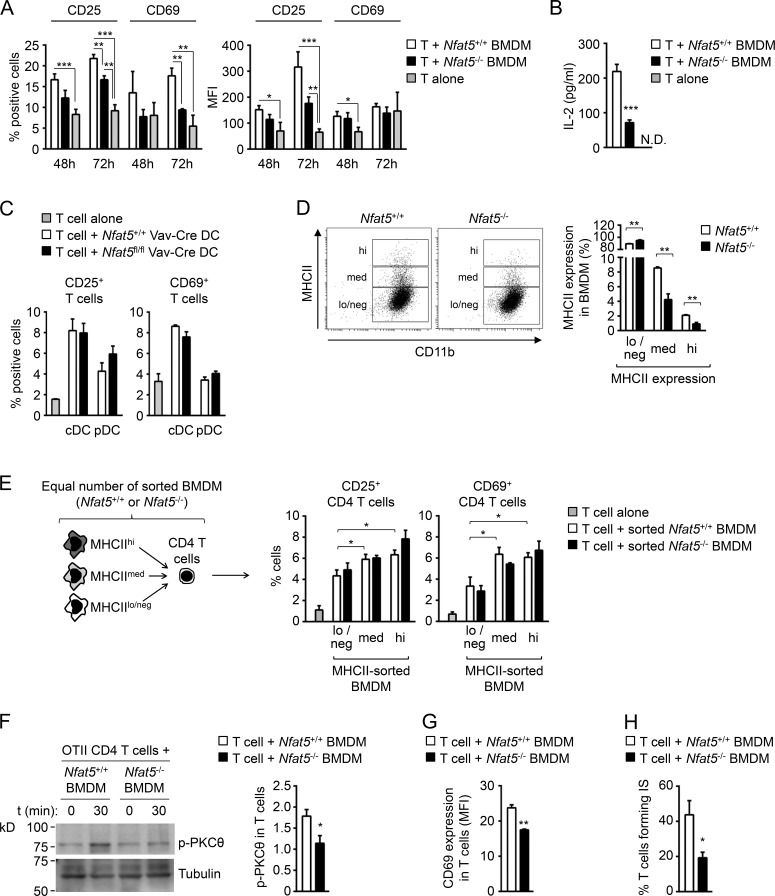
MHCII presentation of antigenic peptides by APCs is central for priming adaptive immune responses in CD4+ lymphocytes. We therefore analyzed the capacity of NFAT5-deficient macrophages (H2-Db) to activate allogeneic CD4+ T lymphocytes (isolated from BALB/c mice, H2-Dd) and observed that they were poorer inducers of T cell activation markers CD25, CD69, and IL-2 than wild-type macrophages (Fig. 2, A and B). We also analyzed NFAT5-deficient conventional myeloid and plasmacytoid DCs sorted from spleen, and they activated allogeneic CD4+ T cells to the same extent as wild-type DCs (Fig. 2 C). We observed that NFAT5-deficient macrophages presented a reduced proportion of cells expressing medium or high levels of MHCII protein (Fig. 2 D). In view of this, we used sorted NFAT5-deficient and control macrophages expressing the same level of MHCII (low, medium, or high) and found that they activated allogeneic T cells comparably (Fig. 2 E). This result indicated that the defective capacity of populations of NFAT5-deficient macrophages to activate allogeneic T cells was due to their having fewer cells expressing high MHCII levels. We then analyzed the capacity of NFAT5-deficient macrophages to form immunological synapses and activate early signaling events in TCR transgenic OTII CD4+ T lymphocytes upon MHCII-dependent OVA peptide presentation. NFAT5-deficient macrophages induced significantly lower early protein kinase C theta (PKCθ) phosphorylation and later CD69 expression in OTII CD4+ T cells than wild-type macrophages (Fig. 2, F and G). In these experiments, we also observed a reduced percentage of OTII T cells that remained bound to peptide-loaded NFAT5-deficient macrophages 18 h after their initial contact (Fig. 2 H), showing an impaired capacity of NFAT5-deficient macrophages to form synapses with CD4+ T cells. Altogether, these results showed that reduced MHCII expression in NFAT5-deficient macrophages impaired their capacity to activate TCR-mediated responses in CD4+ lymphocytes.
Figure 2.
Activation of allogeneic CD4+ T lymphocytes by NFAT5-deficient macrophages and DCs. (A and B) Activation of allogeneic CD4+ T cells (BALB/c mice, H2-Dd) in a mixed leukocyte reaction (MLR; BMDM-to-T-cell ratio of 1:1) with wild-type and NFAT5-deficient BMDM (129/sv mice, H2-Db). (A) CD25 and CD69 induction (percentage of positive cells and mean fluorescence intensity [MFI]) in CD4+ T cells was analyzed by flow cytometry. Statistical significance was determined by an unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. In the right panel, the difference for CD25 MFI between wild-type and NFAT5-deficient BMDM at 72 h was near significant, P = 0.07. (B) IL-2 production by CD4+ T cells cocultured with wild-type and NFAT5-deficient macrophages was analyzed in culture supernatants at 72 h. N.D., not detected. Results in A and B show the mean ± SEM from four independent experiments, each comparing BMDMs from one NFAT5-deficient mouse and a wild-type littermate. Statistical significance was determined by an unpaired t test. ***, P < 0.001. (C) Induction of CD25 and CD69 in allogeneic CD4 T cells after 48 h of culture with sorted conventional myeloid DCs (cDCs) or plasmacytoid DCs (pDCs; DC-to-T-cell ratio, 0.7:1) was analyzed by flow cytometry. DCs correspond to the sorting experiment shown in Fig. S1 C. The experiment analyzed DCs from three independent pairs of wild-type and Nfat5fl/fl Vav-Cre littermates. (D) Percentages of wild-type and NFAT5-deficient BMDM with high (hi), medium (med), or low/negative (lo/neg) expression of MHCII. Results show the mean ± SEM from three independent experiments, each comparing BMDMs from one NFAT5-deficient mouse and a wild-type littermate. Statistical significance was determined by an unpaired t test. **, P < 0.01. (E) Induction of CD25 and CD69 in allogeneic CD4 T cells was analyzed by flow cytometry after a 72-h MLR (BMDM-to-T-cell ratio of 1:1) with equal numbers of wild-type and NFAT5-deficient BMDMs sorted for high, medium, or low/negative MHCII surface expression. Results show the mean ± SEM from three independent BMDM cultures of each genotype analyzed in two independent MLRs. *, P < 0.05. (F–H) MHCII-dependent activation of OVA peptide-specific OTII CD4+ T cells and synapse formation by wild-type and NFAT5-deficient BMDMs. (F) Phospho-PKCθ was measured after 30 min of macrophage–T cell contact. The left panel shows one representative Western blot, and bars represent the quantification as fold-change relative to basal levels at t = 0 (which was given an arbitrary value of 1). (G) CD69 expression in T cells was analyzed at 14 h by flow cytometry. CD69 expression is shown as MFI. (H) Percentage of T cells forming immunological synapses (IS) with wild-type or NFAT5-deficient macrophages was determined by confocal microscopy after 18 h of culture. Results in F–H show the mean ± SEM from four to five independent BMDM cultures of each genotype, wild-type, and NFAT5-deficient, analyzed in three independent experiments. Statistical significance was determined by an unpaired t test. *, P < 0.05; **, P < 0.01.
Delayed graft rejection in a mouse model of myeloid-specific NFAT5-deficient skin transplant
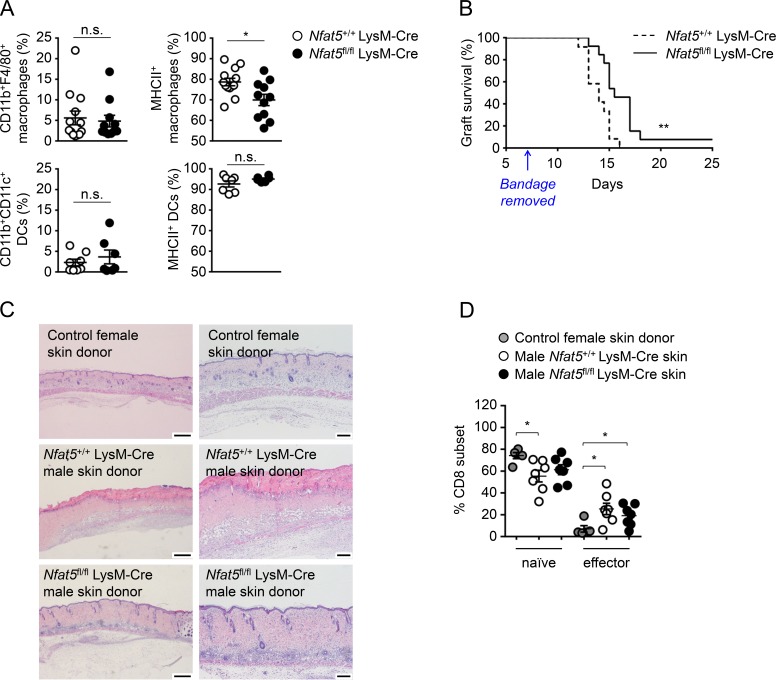
Given the central role of antigen presentation by MHCII in allograft rejection (Rosenberg and Singer, 1992), we tested whether lack of NFAT5 in macrophages improved graft acceptance in a mouse model of skin transplant. Engraftment of allogeneic skin in mice is a quite stringent experimental model for determining immune tolerance and reflects the challenges that exist to achieve prolonged survival of human solid organ grafts (Rosenberg and Singer, 1992; Auchinloss et al., 1999). First, we confirmed that skin macrophages (CD11b+ F4/80+) from Nfat5fl/fl LysM-Cre mice had a lower percentage of MHCII+ cells, whereas skin DCs (CD11b+ CD11c+) did not show this defect (Fig. 3 A), which was consistent with our previous observations (Figs. 1 and S1, A–C). We then performed sex-mismatched transplants of skin from myeloid-specific NFAT5-deficient (Nfat5fl/fl LysM-Cre) or control (Nfat5+/+ LysM-Cre) male donor mice in wild-type syngeneic (C57BL/6J) female recipients (Fig. S2 A). In this model, male skin grafts are rejected by female recipient T cells reacting to MHCII-presented male HY antigens (Rosenberg and Singer, 1992; Simpson et al., 1997). Skin grafts whose myeloid cells lacked NFAT5 survived longer (median graft survival of 15.5 d, between days 13 and 18, with one graft persisting for longer than 25 d) than those from littermate wild-type mice (median survival of 14 d, between days 12 and 16; Figs. 3 B and S2 B). Histological analysis between days 8 and 11 after transplant showed that skin grafts from Nfat5fl/fl LysM-Cre male donors, despite presenting detectable inflammation in subcutaneous tissues (hypodermis, panniculus carnosus, and adventitia), had better reepithelialization of both epidermis and dermis with clear vascularization and hair follicles (Fig. 3 C). By contrast, skin grafts of Nfat5+/+ LysM-Cre males exhibited chronic severe inflammation concentrated in the hypodermis and poor reepithelialization with extended necrosis in both the epidermis and dermis. Control transplants of wild-type female skin grafts in wild-type female recipients were included as quality control of the surgical procedure and showed healthy reepithelialization and vascularization (Fig. 3 C). These experiments also showed that mice transplanted with male wild-type skin presented a significant increase in the proportion of splenic effector CD8 T cells with a concomitant decrease in naive cells, whereas mice grafted with Nfat5fl/fl LysM-Cre skin showed an attenuated CD8+ T response (Fig. 3 D). For CD4+ T cells, we observed a comparable proportion of effector cells in both transplant groups (Fig. S2 C). As CD4 cell activation during graft rejection precedes and controls that of CD8 cells (Rosenberg and Singer, 1992; Auchinloss et al., 1999), it is possible that our analysis at the onset of rejection picked up CD8 cells still progressing to effectors, whereas CD4 effector cells had already reached comparable levels in both genotypes of skin grafts. Altogether, results from the transplant experiments were consistent with the in vitro T cell activation assays (Fig. 2) and suggested that reduced MHCII expression in NFAT5-deficient macrophages attenuated T cell activation and delayed graft rejection in vivo.
Figure 3.
Rejection of myeloid-specific NFAT5-deficient skin transplants. (A) Percentage of macrophages (CD11b+ F4/80+) and DCs (CD11b+ CD11c+) and their MHCII expression in skin biopsies of Nfat5+/+ LysM-Cre (wild-type) and Nfat5fl/fl LysM-Cre mice. Statistical significance was determined by an unpaired t test. *, P < 0.05. n.s., not significant. (B) Survival of skin grafts from Nfat5+/+ LysM-Cre and Nfat5fl/fl LysM-Cre male donor mice transplanted in wild-type female recipients. Results correspond to 10 independent transplant experiments, in each of which separate recipient female mice were respectively transplanted with skin from Nfat5+/+ LysM-Cre (wild-type) and Nfat5fl/fl LysM-Cre male mice. Transplant rejection was monitored after day 7, when the protective postsurgery bandage was removed (indicated by the arrow). Median survival for skin grafts of Nfat5+/+ LysM-Cre male mice (n = 12) was 14 d, and median survival for skin grafts of Nfat5fl/fl LysM-Cre male mice (n = 13) was 15.5 d (see Fig. S2 for representative pictures illustrating the time course of graft rejection). P = 0.0023, calculated with a Mantel–Cox log-rank test. (C) Histological analysis (hematoxylin and eosin staining) of skin grafts from a female wild-type donor (as quality control for the surgical procedure) and male Nfat5+/+ LysM-Cre and Nfat5fl/fl LysM-Cre donors 10 d after transplant in female recipients. Photographs are representative of histopathology analyses done in four control female skin transplants, six Nfat5+/+ LysM-Cre male skin transplants, and five Nfat5fl/fl LysM-Cre male skin transplants. Scale bar is 500 µm for the photographs in the left column, and 200 µm for the enlarged images in the right column. (D) Proportion of naive and effector CD8+ T cells in the spleens of transplanted mice. CD8+ T cells were analyzed in five independent transplant experiments, four of which included parallel controls with female mice transplanted with skin of a wild-type female donor (as shown in C). Recipient mice were sacrificed on the day when clear rejection was observed for wild-type Nfat5+/+ LysM-Cre male skin grafts (between days 12 and 16 after transplant). Results in the graphics are the mean ± SEM. *, P < 0.05. Significance was determined by a Mann–Whitney test.
NFAT5 binds to regulatory regions of Ciita and MHCII genes in macrophages
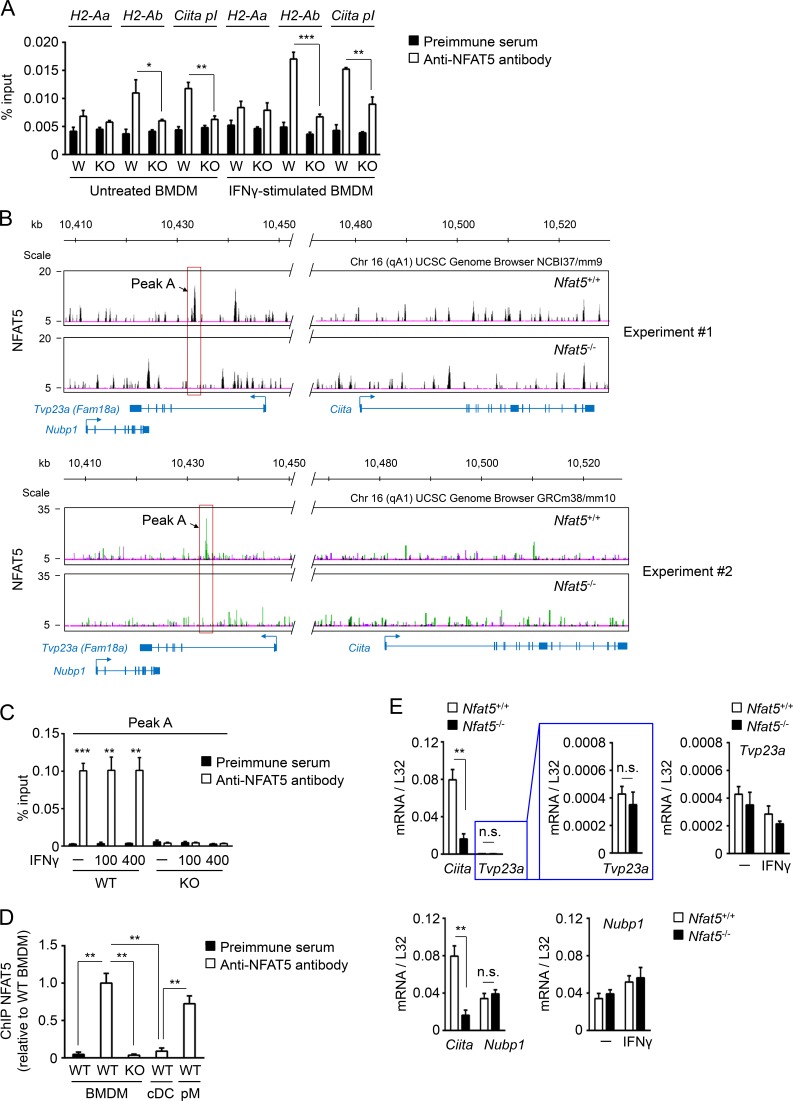
Sequences that match the consensus NFAT5-binding site (Lopez-Rodríguez et al., 1999) are present in the proximal promoter region of genes encoding for components of the MHCII complex, as well as in Ciita myeloid promoter I (Muhlethaler-Mottet et al., 1997; Piskurich et al., 1998; Fig. S3 A). We therefore asked whether NFAT5 bound these regulatory regions. Quantitative chromatin immunoprecipitation (qChIP) with a combination of two polyclonal antibodies to NFAT5 detected its specific binding to promoter I of Ciita and the promoter of H2-Ab both in untreated macrophages as well as in macrophages treated with IFNγ (Fig. 4 A), but we did not detect NFAT5-specific binding to the promoter region of H2-Aa. To obtain a wider coverage of MHCII and Ciita genomic regions, we decided to analyze NFAT5-immunoprecipitated chromatin by ultrasequencing. This approach did not discern specific NFAT5-binding peaks in a 200-kb region of mouse chromosome 17 that encompassed MHCII genes (Fig. S3 B); however, it identified one clear NFAT5-specific peak (peak A) −47 kb upstream of the Ciita locus in mouse chromosome 16 (Fig. 4 B). Peak A contained consensus NFAT5-binding sites (5′-GGAAA-3′), and we confirmed specific binding of NFAT5 to it by qChIP analysis of wild-type against NFAT5-deficient macrophages (Fig. 4 C). This binding was constitutive and occurred in steady-state conditions in unstimulated macrophages and was not affected by IFNγ (Fig. 4 C). A second NFAT5 peak at −38 kb was observed in one of the ChIP-seq experiments (Fig. 4 B, upper panel), and although it could be detected by qChIP, its signal was weaker than for peak A (data not shown). We then asked whether NFAT5 also bound to peak A in conventional myeloid DCs but did not detect its binding, which suggests that peak A recruits NFAT5 in macrophages but not DCs (Fig. 4 D). Because peak A was embedded within the first intron of the Tvp23a gene (also known as Fam18a), we asked whether NFAT5 might affect its expression in macrophages. This was not the case, as Tvp23a mRNA was detected at nearly background levels in macrophages, and its very low expression was independent of NFAT5 and IFNγ (Fig. 4 E, upper panels). Nubp1, the gene upstream of Tvp23a, was expressed at higher levels, but it was also IFNγ and NFAT5 independent (Fig. 4 E, bottom panels). In sum, these results identify a new NFAT5-bound element in macrophages that is located distally upstream of Ciita.
Figure 4.
Binding of NFAT5 to regulatory regions of MHCII-related genes and identification of a remote NFAT5-bound region upstream Ciita. (A) qChIP analysis of NFAT5 binding to promoters of MHCII genes and promoter I of Ciita in untreated or IFNγ-stimulated (400 U/ml, 4 h) wild-type (W) and NFAT5-deficient (KO) BMDMs (n = 3). Results shown are the mean ± SEM of three independent experiments with BMDMs from wild-type and NFAT5-deficient littermate mice. (B) ChIP-seq analysis of wild-type and NFAT5-deficient BMDMs showing the position of NFAT5-binding site peak A 47 kb upstream of the Ciita locus. Two independent experiments are shown. For experiment 1 (accession no. GSE107948), sequences were aligned to mouse chromosome 16 sequence in UCSC Genome Browser NCBIm37/mm9, and for experiment 2 (accession no. GSE107950), the sequence used was mouse chromosome 16 in UCSC Genome Browser GRCm38/mm10. Positions of genes Nubp1 and Tvp23a (Fam18a; the latter overlapping with Ciita peak A) are shown. (C) qChIP analysis of NFAT5 binding to peak A upstream of Ciita in macrophages left untreated (–) or stimulated with IFNγ (100 or 400 U/ml, 4 h; n = 5, except n = 3 for IFNγ 100 U/ml). Results show the mean ± SEM of four independent experiments comparing five pairs of BMDM cultures from wild-type and NFAT5-deficient littermate mice, all including unstimulated cells and treatment with 400 U/ml IFNγ, and three of them also including IFNγ 100 U/ml. (D) qChIP analysis of NFAT5 binding to peak A in conventional myeloid DCs (cDCs) in comparison with BMDMs and peritoneal macrophages (pMs). Binding of NFAT5 to peak A region in cDCs and pMs is represented as relative to its binding in wild-type BMDMs. qChIP with anti-NFAT5 antibodies in NFAT5-deficient BMDMs and with a preimmune serum are shown as negative controls. Results show the mean ± SEM of three independent experiments. (E) mRNA expression of Ciita, Tvp23a, and Nubp1 in wild-type and NFAT5-deficient BMDMs. RNA samples were extracted from three independent pairs of BMDM cultures from wild-type and NFAT5-deficient littermates. The left panels show the comparison between Ciita and Tvp23a or Nubp1 mRNA expression levels, and the right panels show the lack of responsiveness of Tvp23a and Nubp1 to IFNγ stimulation (400 U/ml, 24 h). Statistical significance in A and C–E was determined by an unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant.
Peak A (–47 kb Ciita) has NFAT5-dependent characteristics of an active enhancer
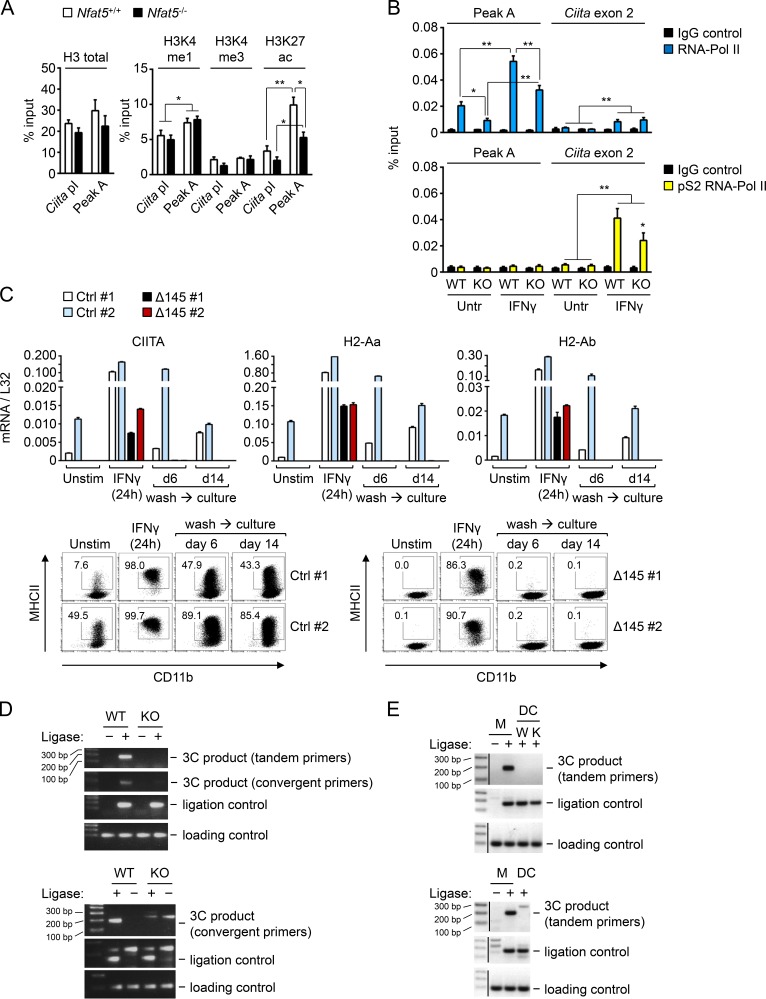
We then analyzed peak A and Ciita promoter I by qChIP in NFAT5-deficient or control macrophages for different histone modifications that are enriched in gene enhancers (H3K4me1) or promoters (H3K4me3) and a modification that marks transcribed regions and active enhancers (H3K27ac; Tie et al., 2009; Creyghton et al., 2010). We observed that peak A region had a slightly higher content in H3K4me1 compared with Ciita promoter I, and this was NFAT5 independent (Fig. 5 A). However, peak A had a greater density of H3K27ac than Ciita promoter I, and this accumulation was NFAT5 dependent (Fig. 5 A). The finding that peak A was enriched in H3K27ac, a mark of transcriptionally active regions, but was embedded within a gene (Tvp23a) that was not being transcribed suggested that it might act as an enhancer element for a different gene. Enhancers are also characterized by being enriched in RNA polymerase II (RNA Pol-II), although this enhancer-associated form is not coupled to transcription elongation and therefore is not phosphorylated in serine 2 of its carboxy terminal domain (CTD) repeat YSPTSPS. By contrast, promoters and the body of transcribed genes are marked by elongation-competent RNA Pol-II phosphorylated in serine 2 of its CTD (p-S2 RNA Pol-II; Koch et al., 2011). We found that peak A in unstimulated macrophages was enriched in total RNA Pol-II compared with Ciita exon 2, and this enrichment was dependent on NFAT5 (Fig. 5 B, upper panel). Abundance of total RNA Pol-II in both regions was enhanced by IFNγ, and this was NFAT5 dependent only in peak A. Regarding p-S2 RNA Pol-II, this form was detected only upon IFNγ stimulation in Ciita exon 2 but not in peak A, and its accumulation was partially dependent on NFAT5 (Fig. 5 B, bottom panel). These results are consistent with the interpretation that NFAT5 contributed both to the acquisition of enhancer-associated marks in peak A and to the recruitment of active RNA Pol II to Ciita transcribed regions and suggest that peak A functioned as an NFAT5-regulated enhancer for Ciita promoter I in macrophages.
Figure 5.
Characterization of NFAT5-binding site peak A upstream Ciita. (A) qChIP assays with antibodies specific for histone H3 and its indicated modifications in peak A and Ciita promoter I (Ciita pI) in wild-type and NFAT5-deficient BMDMs. Results show the mean ± SEM of four independent experiments, each comprising BMDMs from one NFAT5-deficient mouse and a wild-type littermate. *, P < 0.05; **, P < 0.01. (B) qChIP assays comparing the binding of RNA-Pol II and its transcription elongation-associated form (phosphorylated serine 2 of its CTD) to peak A and exon 2 of Ciita in wild-type (WT) and NFAT5-deficient (KO) BMDMs left untreated or stimulated with IFNγ (400 U/ml, 4 h). Results show the mean ± SEM of four independent experiments, each comparing one pair of BMDM cultures from wild-type and NFAT5-deficient littermate mice. Statistical significance was determined by an unpaired t test. *, P < 0.05; **, P < 0.01. (C) Effect of deletion of peak A on CIITA and MHCII expression in iBMDMs. Upper panels show mRNA expression of CIITA, H2-Aa, and H2-Ab in two control clones (Ctrl) and two clones with deletion of a 145-bp region in peak A comprising the two consensus NFAT5 binding sites (Δ145), following the CRISPR-Cas9 approach described in Fig. S4. Lower panels show the time course of MHCII protein expression analyzed in parallel by flow cytometry. Results shown are from one experiment comparing the respective clones in basal conditions, 24 h after IFNγ stimulation, and 6 and 14 d after washing out IFNγ. Reduced expression of MHCII expression in Δ145 clones was confirmed in two additional independent flow cytometry analyses (data not shown). (D) Two independent 3C experiments (representative of four independently performed experiments) showing the NFAT5-dependent intrachromosomal looping between peak A and Ciita promoter I in macrophages, detected by PCR amplification with diagnostic convergent and tandem primers. See Fig. S5 for a diagram of the 3C assay and sequencing analysis of the amplified 3C bands. (E) Two independent 3C experiments comparing the formation of intrachromosomal looping between peak A and Ciita promoter I in BMDMs (M) and myeloid conventional DCs. The upper panel includes wild-type (W) and NFAT5-deficient DCs (K).
Peak A (–47 kb Ciita) regulates Ciita and MHCII expression
To test whether peak A regulated the expression of Ciita, we used a CRISPR-Cas9 gene editing strategy for deleting a fragment of 145 base pairs from peak A, which included the two NFAT5 binding sites present in this region (5′-AGGAAAATT-3′ and 5′-AGGAAATT-3′; Fig. S4 A). For this approach, we used immortalized BMDMs (iBMDMs), which were transfected with a single vector that included expression cassettes for the Cas9 endonuclease, two guide RNA (gRNA) sequences to direct the deletion of peak A, and GFP for flow cytometry cell sorting (see Fig. S4 A for the deletion strategy). After limiting dilution and expansion of sorted cells, we obtained clones with an intact peak A region (control clones, Ctrl) or with a deletion of peak A (Δ145; see Fig. S4, B and C, for PCR analysis of genomic DNA in iBMDM clones). Analysis of Δ145 peak A clones showed a substantial reduction in their expression of CIITA and MHCII mRNA as well as MHCII protein compared with control nontargeted clones (Fig. 5 C). As we had observed that NFAT5-deficient macrophages induced robust MHCII expression upon acute IFNγ stimulation but lost it as they returned to basal conditions (Fig. 1 H), we analyzed whether Δ145 peak A cells exhibited a similar response. Δ145 peak A cells induced CIITA and MHCII expression upon acute IFNγ stimulation, although slightly less than control cells, and after IFNγ removal they were unable to sustain their expression (Fig. 5 C). These results showed that deletion of a small region containing the NFAT5 binding sites in this enhancer resembled the effect of suppressing NFAT5 by decreasing CIITA and MHCII expression and impairing macrophage ability to maintain their prolonged induction after transient stimulation with IFNγ. These results support the role of NFAT5-regulated peak A as a nonredundant enhancer element that regulates Ciita expression in macrophages.
Peak A (–47 kb Ciita) interacts with Ciita promoter I in native chromatin in an NFAT5-dependent manner
One mechanism by which enhancers stimulate transcription is by looping together remote regulatory elements and promoters. We used chromatin conformation capture (3C) analysis to test whether there was an intrachromosomal association between peak A and Ciita promoter I in primary macrophages. A basic diagram illustrating the 3C assay, as well as the positions of convergent and tandem primers used to analyze the association between both DNA regions, is shown in Fig. S5. PCR analysis of the 3C assay products with convergent primers yielded the 189-bp fragment predicted for a loop between peak A and Ciita promoter I only in samples from wild-type macrophages but not with NFAT5-deficient cells (Fig. 5 D). Similarly, tandem primers only produced the correct 163-bp amplicon in samples from wild-type macrophages (Fig. 5 D). Respective ligation and loading controls yielded the same amplified products in NFAT5-deficient and control macrophages. Sequencing of the PCR bands amplified with convergent and tandem primers in wild-type macrophages confirmed that they corresponded to the correct 3C product comprising a fusion between peak A and Ciita promoter I upon religation of the AflII restriction site (Fig. S5 C). By contrast, this interaction was not detected in conventional myeloid DCs (Fig. 5 E). These results indicated that NFAT5 facilitated the formation of a loop that brought the remote enhancer peak A into contact with Ciita promoter I in macrophages. Altogether, our identification of an NFAT5-regulated Ciita enhancer that controls MHCII expression in macrophages reveals a distinct mechanism that distinguishes them from other myeloid and lymphoid APCs.
Discussion
Expression of MHCII genes and their transcription coactivator CIITA is primarily regulated at the transcriptional level. Ciita expression in APCs is tightly controlled by the activity of three different cell type–specific promoters, with promoter I driving expression of Ciita and consequently MHCII genes in myeloid cells. Here we identify a distinct remote enhancer for Ciita in primary macrophages that is controlled by the Rel family transcription factor NFAT5. This enhancer formed an NFAT5-dependent intrachromosomal contact with Ciita myeloid promoter I and was needed to sustain CIITA and MHCII expression in macrophages in steady-state conditions and after stimulation with the MHCII-inducing cytokine IFNγ. The dependence of macrophages on NFAT5 for expressing MHCII distinguishes them from their closely related myeloid lineage of conventional DCs, which also transcribe Ciita from promoter I and until now were thought to use the same mechanisms to express these genes.
Consistent with the role of NFAT5 supporting MHCII expression in macrophages, we observed delayed rejection of skin grafts and reduced induction of effector T cells in mice transplanted with skin from myeloid-specific NFAT5-deficient mice. Although graft rejection could also involve other myeloid APCs, such as DCs, our observation that NFAT5-deficient skin DCs expressed normal levels of MHCII suggested that their antigen presentation capacity would not be impaired by the lack of NFAT5. Results in the transplant experiments also agree with our coculture assays showing a weaker capacity of NFAT5-deficient macrophages to directly activate MHCII-dependent T lymphocyte responses. The role of NFAT5 in macrophages during stimulation of T lymphocyte responses may not be limited to its control of MHCII expression. In this regard, we had previously shown that NFAT5 enhances proinflammatory polarization of macrophages, which would be consistent with promoting type 1 immune responses that contribute to graft rejection (Wyburn et al., 2005; Tellechea et al., 2018). Although this dual contribution of NFAT5 cannot be easily dissociated apart when analyzing T cell responses during graft rejection, our finding that an early MHCII-dependent TCR signaling event, PKCθ phosphorylation, was weaker in T cells stimulated in vitro by NFAT5-deficient macrophages strongly suggested a direct defect in MHCII-mediated TCR activation preceding later effects by macrophage-secreted cytokines. Therefore, the global effect of NFAT5 in an immune response would be determined by different aspects, including its roles in macrophage MHCII expression and proinflammatory macrophage polarization (Buxadé et al., 2012; Tellechea et al., 2018). Because NFAT5 may well influence other players in inflammatory responses such as DCs and T lymphocytes (Alberdi et al., 2017), the different NFAT5-mediated contributions could be interconnected, as inflammatory type 1 responses and MHCII expression positively regulate each other. In this regard, the need for NFAT5 in MHCII expression in steady-state macrophages suggests that NFAT5 could take part in an activation loop between macrophages and T cells (Morris et al., 2013; Cho et al., 2014). Reduced MHCII in NFAT5-deficient macrophages would decrease activation of CD4+ T lymphocytes and limit their production of factors such as IFNγ that in turn reinforce classic macrophage polarization, including enhanced transcription of CIITA and MHCII (Biswas and Mantovani, 2010). Our finding that IFNγ-stimulated NFAT5-deficient macrophages, as well as those lacking the NFAT5-regulated Ciita peak A enhancer, rapidly down-regulated MHCII after removing IFNγ also suggests that NFAT5 would be relevant for sustaining MHCII expression in macrophages in inflammatory environments where stimulatory cytokines delivered locally by T lymphocytes might fluctuate.
Our results show that NFAT5 can regulate Ciita transcription through a remote enhancer 47 kb upstream of the myeloid Ciita promoter I. This remote element exhibited NFAT5-dependent active enhancer marks, formed a loop with Ciita promoter I in an NFAT5-dependent manner, and its deletion in macrophages resembled the effect of reduced CIITA and MHCII expression observed in NFAT5-deficient macrophages. Still, differences in the expression of CIITA and MHCII were more drastic in iBMDM lacking Ciita enhancer than NFAT5-deficient macrophages, which could be due to the presence of transcription regulators other than NFAT5 that control this enhancer. For instance, among other factors that act in myeloid cells, the deleted element contains potential binding sites for PU.1 (5′-AGTTCCTCTT-3′) or Maf (5′-GGGTGGTGACATCGCTGTA-3′) factors, which play relevant roles in the control of macrophage enhancers (Ghisletti et al., 2010; Soucie et al., 2016). Distal elements forming intrachromosomal loops with Ciita promoters had been found in nonhematopoietic cell lines and B cells. In the carcinoma cell lines SW13 and HeLa, IFNγ-induced expression of CIITA was boosted by the chromatin remodeler BRG1, which facilitated interactions between distal regions and Ciita promoter IV (Ni et al., 2008). In human and mouse B cells, another set of distal elements was shown to interact with Ciita promoter III (Lohsen et al., 2014), and at least one of them required the transcription factor PU.1 to mediate this interaction (Yoon and Boss, 2010). None of these elements coincided with the −47-kb NFAT5-regulated Ciita enhancer identified in our current study. Our characterization in macrophages of an NFAT5-controlled Ciita regulatory mechanism different from those described in other cell types, together with the finding that NFAT5 was not required for CIITA and MHCII expression in DCs and B cells, suggests a remarkable diversity in mechanisms regulating MHCII expression in different APCs. Because differentiation and activation of immune cells is accompanied by extensive remodeling of chromatin architecture (Lin et al., 2012; Hakim et al., 2013; Bunting et al., 2016), the use of different distal elements to control cell type–specific Ciita promoters could likely be adapted to the chromatin topology of different types of APCs. Given that CIITA is critical for MHCII expression, it is tempting to speculate that evolution might have favored a diversification of strategies for controlling Ciita transcription in different APC lineages, so that mutations or dysfunctions that disrupted its expression in one type of APC would not affect others. In this regard, several questions arise, for instance how different APC lineages may acquire specific MHCII expression mechanisms during their ontogeny and how they regulate these mechanisms in response to environmental cues and immunological challenges. Unveiling the mechanistic basis of these processes might contribute to a better understanding of APC functional diversity and reveal as-yet-unsuspected pathological consequences of their deregulation.
Materials and methods
Mice
Nfat5+/− heterozygous mice were maintained in a pure 129/sv background and were crossed to obtain Nfat5−/− mice and control Nfat5+/+ littermates as described previously (López-Rodríguez et al., 2004). Nfat5-floxed mice (pure C57BL/6 background) have been described previously (Berga-Bolaños et al., 2010). For inducible deletion of NFAT5, Nfat5 floxed mice were crossed with mice carrying the Cre recombinase under the control of the Myxovirus resistance-1 (Mx-Cre) gene promoter (Kühn et al., 1995). Mx-Cre mice were a gift of M. Schmidt-Supprian (Technische Universität München, München, Germany). To induce Mx-Cre expression in vivo, Mx-Cre Nfat5fl/fl and control Mx-Cre Nfat5+/+ mice were injected intraperitoneally with poly(I):poly(C) (pIC, 15 mg/kg) three times, every other day. Bone marrow was extracted 12 d later and cultured to obtain wild-type and NFAT5-deficient BMDMs. Mice lacking Nfat5 in myeloid cells were obtained by crossing Nfat5-floxed mice with LysM-Cre mice (Clausen et al., 1999), purchased from the Jackson Laboratory. Mice lacking Nfat5 in blood cell lineages were obtained by crossing Nfat5-floxed mice with Vav-Cre mice (Stadtfeld and Graf, 2005), provided by T. Graf (Center for Genomic Regulation, Barcelona, Spain). OTII mice with a transgenic Vα2Vβ5 TCR specific for OVA323−339 peptide epitope in the context of IAb (Barnden et al., 1998) were housed at the animal facility of Centro de Investigaciones Biológicas (Madrid, Spain) and used for obtaining MHCII-restricted OVA-specific CD4 T lymphocytes. All experiments were performed using 6–10-wk-old NFAT5-deficient and control littermate mice that were bred and maintained in our animal facility in specific pathogen–free conditions. Animal handling and experiments were in accordance with protocols approved by the respective Animal Care and Use Committees of the Barcelona Biomedical Research Park (PRBB)/Universitat Pompeu Fabra (UPF; Barcelona, Spain) and Centro de Investigaciones Biológicas (for OTII mice) and were performed in accordance with the Declaration of Helsinki and the European Communities Council Directive (86/609/EEC).
Reagents
pIC was from InvivoGen (tlrl-pic), formaldehyde and glycine were from Sigma-Aldrich, LPS was from Sigma-Aldrich (Escherichia coli 055:B5; L2880), and recombinant mouse IFN-γ was purchased from ImmunoTools (12343534). Trizma base, glycine, EDTA, β-glycerophosphate, PMSF, leupeptin, pepstatin A, aprotinin, SDS, and Triton X-100 were from Sigma-Aldrich. Sodium fluoride (NaF) was from Merck. CellTracker Green 5-chloromethylfluorescein diacetate was from Thermo Fisher Scientific (C2925). Polyornithine, Hoechst 33342, β-mercaptoethanol, and phalloidin-tetramethyl-rhodamine B isothiocyanate were from Sigma-Aldrich.
Cell isolation and culture
BMDMs were generated as previously described (Buxadé et al., 2012). Briefly, mice were sacrificed and the femoral and tibial marrow was flushed from the bones with DMEM supplemented with 2 mM glutamine, 50 µM β-mercaptoethanol, and 1 mM sodium pyruvate plus penicillin/streptomycin (incomplete medium). Cells were then resuspended in complete DMEM (incomplete medium supplemented with 10% heat-inactivated FBS) with 25% (vol/vol) L929-conditioned medium (as the source of M-CSF) and incubated for 7 d in polystyrene dishes at 37°C in 5% CO2 atmosphere. Differentiated macrophages were harvested with PBS plus 5 mM EDTA by gentle pipetting, washed with PBS, and plated in tissue culture plates (2 × 106 cells/3 ml per well). iBMDMs have been described (Baroja-Mazo et al., 2014) and were provided by P. Pelegrín (Biomedical Research Institute of Murcia, University Clinical Hospital Virgen de la Arrixaca, Murcia, Spain). iBMDMs used for CRISPR-Cas9–mediated gene editing were confirmed to be mycoplasma negative and expressed basal surface MHCII as well as MHCII and CIITA mRNA levels comparable to fresh BMDM. They were maintained in complete DMEM and harvested by vigorous pipetting. BMDCs were obtained by culturing bone marrow cell suspensions in GM-CSF–supplemented medium for 10 d (Zal et al., 1994). Mature BMDCs were induced by stimulating GM-CSF–derived BMDCs with LPS (1 µg/ml) during the last 48 h of culture. BMDCs were also generated by culturing bone marrow cell suspensions with 200 ng/ml Flt3L (ImmunoTools; 12343305) for 10 d and sorting them as CD11c+ B220− cells. Peritoneal macrophages used for mRNA analysis were harvested by two consecutive lavages with 5 ml of ice-cold PBS, and then isolated with magnetic beads coated with anti-CD11b antibody (M1/70.15 hybridoma supernatant). Conventional DCs (CD11c+ B220−) and plasmacytoid DCs (CD11c+ B220+) used for mRNA or qChIP analysis were obtained from spleens by flow cytometry cell sorting excluding dead cells and neutrophils (Ly6G+). Thioglycollate-elicited peritoneal macrophages used for qChIP were isolated with magnetic beads coated with anti-CD11b antibody (M1/70.15 hybridoma supernatant) from peritoneal lavages of mice that had been injected with 3% Brewer thioglycollate medium (1 ml/mouse) 5 d earlier. B and T lymphocytes for mRNA analysis were isolated from spleen cell suspensions with magnetic beads coated with anti–mouse Ig and anti-CD4 antibodies, respectively. Skin macrophages and DCs used for flow cytometry analysis were obtained by excising a 2-cm2 piece of mouse back skin, mincing it with a scalpel, and digesting it with 0.5 mg/ml collagenase A (Roche; 10103578001) and 0.01% DNaseI (Sigma-Aldrich; D4263-5VL) in complete DMEM medium without β-mercaptoethanol during 1 h at 37°C with rotation. Samples were then filtered through a 70-µm cell strainer, and the filter was washed with 20 ml of DMEM. Filtered cells were then centrifuged for 8 min at 1,200 rpm (330 g), and pellets were resuspended in 200 µl of PBS containing 10% FBS and 0.1% sodium azide. Cells were then analyzed by flow cytometry to determine the percentage of MHCII+ cells in skin macrophages (CD11b+ F4/80+) and DCs (CD11b+ CD11c+).
mRNA analysis by RT-qPCR
Total RNA was isolated using the High Pure RNA Isolation kit (Roche; 11828665001) and quantified in a NanoDrop spectrophotometer (ND-1000). Typically, between 0.5 and 1 µg total RNA for BMDMs or 50–100 ng for peritoneal macrophages was retrotranscribed to cDNA using Transcriptor First Strand cDNA Synthesis Kit reverse transcription system (Roche; 04897030001). For RT-qPCR, LightCycler 480 SYBR Green I Master (Roche; 4887352001), LightCycler 480 Multiwell Plate (Roche; 04729749001), and the LightCycler 480 Real-Time PCR System (Roche) were used according to the instructions provided by the manufacturer. Samples were normalized to L32 ribosomal protein mRNA levels using LightCycler software, version 1.5. Primers used for RT-qPCR analysis are listed in Table S2.
Flow cytometry
Macrophage and DC suspensions were blocked for 20 min in PBS containing 10% FBS, 0.1% sodium azide, and an antibody to Fcγ receptors CD16/CD32 (1 µg antibody/106 cells; eBioscience; 14-0161). Cells were then incubated with fluorochrome-labeled isotype control antibodies or surface marker–specific antibodies (1 µg antibody/106 cells) and analyzed with FACSCalibur or LSRII flow cytometers and FACSDiva software (BD Biosciences). Flow cytometry analysis of CD69 induction during T cell activation by peptide-loaded macrophages was done as described (Riol-Blanco et al., 2009) using a Coulter Epics XL cytofluorometer with CXP Analysis software (Beckman Coulter). The following antibodies were used: FITC anti-mouse CD11b (clone M1/70, eBioscience; 11-0112-85); APC–eFluor 480 anti-mouse F4/80 (clone BM8, eBioscience; 47-4801-80); PE anti-mouse MHCII (I-A/I-E; clone M5/114.15.2, eBioscience; 12-5321-82); APC anti-mouse MHCII (I-A/I-E; clone M5/114.15.2, eBioscience; 17-5321); PE anti-mouse MHCI (28-14-8, eBioscience; 12-5999-82); FITC anti-mouse CD11c (N418, eBioscience; 11-0114-81); PE anti-mouse B220 (RA3-6B2, eBioscience; 12-0452-83); PE/Cy7 anti-mouse Ly6G (1A8, BioLegend; 127618); PE/Cy7 anti-mouse CD3ε (clone 145-2C11, BioLegend; 100320); PE-Cyanine5 anti-mouse CD4 (clone RM4-5, eBioscience; 15-0042-83); APC-Cy7 anti-mouse CD8a (clone 53-6.7, BD PharMingen; 561967); PE anti-mouse CD44 (clone IM7, eBioscience; 12-0441-83); APC anti-mouse CD62L (clone MEL-14, BD PharMingen; 553152); FITC anti-mouse CD25 (clone 7D4, BD PharMingen; 553072); PE anti-mouse CD69 (clone H1.2F3, eBioscience; 12-0691-82); and FITC anti-mouse CD69 (clone H1.2F3, BioLegend; 104505).
Western blot
Conjugates of OTII CD4 T cells and BMDMs were solubilized by boiling in SDS sample buffer (100 mM Tris HCl, pH 6.8, 0.05 mM sodium orthovanadate, 0.5 mM EDTA, 3% SDS, 1 mM EDTA, 2% β-mercaptoethanol, and 5% glycerol). After extraction, samples were separated by SDS-PAGE and electrotransferred to nitrocellulose membranes. After blocking with 5% nonfat milk in TBS, membranes were incubated with primary antibodies diluted in TBS plus 0.05% Tween 20 solution and 5% BSA. Subsequently, the membranes were incubated with suitable peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology), and immunoreactive bands were visualized using enhanced chemiluminescence reagents (Pierce). Anti-phospho-PKCθ (Thr538; Santa Cruz Biotechnology; sc-33885) and anti–α-tubulin (H-300, Santa Cruz Biotechnology; sc5546) antibodies were used. PKCθ is expressed in T lymphocytes but not in BMDMs (unpublished data; https://www.immgen.org/). The intensity of the immunoblot bands was quantified by densitometry using Multi Gauge software (Fujifilm).
Mixed leukocyte reaction
Wild-type and NFAT5-deficient 129/sv BMDMs (H-2Db) were seeded in 48-well plates (0.5 × 106 cells/well) before adding allogeneic CD4+ T cells from Balb/c mice (H-2Dd). T cells were isolated from the spleen and peripheral lymph nodes by positive selection using Dynabeads Mouse CD4 (Invitrogen; 114.45D) and DETACHaBEAD Mouse CD4 (Invitrogen; 124.06D) according to the manufacturer’s instructions. CD4+ T cells were added to the BMDM cultures in a 1:1 ratio. IL-2 production was measured in 72-h culture supernatants (eBioscience; BMS820FFSA), and T cell activation was analyzed after 48 and 72 h of coculture by flow cytometry using anti-CD4, anti-CD69, and anti-CD25 fluorescently labeled antibodies.
MHCII-restricted activation of OTII CD4 T lymphocytes and analysis of immunological synapses
CD4 OTII TCR transgenic CD4 T lymphocytes were obtained from spleens of 8-wk-old mice by negative immunomagnetic cell sorting (Miltenyi; 130-104-454), and immunological synapses were analyzed as previously described (Riol-Blanco et al., 2009). Briefly, wild-type or NFAT5-deficient BMDMs were first incubated for 30 min with 10 µg/ml OVA323-339-peptide (ISQAVHAAHAEINEAGR, GenScript; RP10610) in serum-free RPMI. Subsequently, OVA-peptide–loaded BMDMs and OTII CD4 T cells were mixed (ratio of 1 BMDM to 5 CD4 T cells) in RPMI supplemented with 10% FCS (complete medium). The conjugates were spun (50 g, 5 min) in a conical tube, maintained in complete medium at 37°C in 5% CO2 atmosphere for the times indicated in the legend of Fig. 2 F, and analyzed by Western blot for phospho-PKCθ (Thr538) induction (30 min after conjugate formation), flow cytometry for CD69 expression (14 h after conjugate formation), and confocal microscopy for synapse formation (18 h after conjugate formation). Confocal microscopy analysis of synapses was done as previously described (Riol-Blanco et al., 2009). Briefly, CD4 T cells were labeled for 30 min at 37°C with the fluorescent cell tracker 5-chloromethylfluorescein diacetate (5 µM) in 0.1% BSA in PBS and then extensively washed in PBS. Conjugates of 5-chloromethylfluorescein diacetate–labeled CD4 T cells and BMDM were fixed in 4% paraformaldehyde in PBS (15 min at room temperature) and then plated for 30 min onto coverslips coated with polyornithine (20 µg/ml). Subsequently, conjugates were permeabilized with 0.2% Triton X-100 (10 min at room temperature). Cells were first treated with 0.1% BSA (15 min) to block unspecific binding and then stained with phalloidin-tetramethylrhodamine and Hoechst 33342. Before mounting, samples were extensively washed with PBS and distilled water. Coverslips were mounted in fluorescent mounting medium, and representative fields were photographed through an oil-immersion lens. Laser-scanning confocal microscopy was performed with argon and helium/neon laser beams attached to an Ultra-spectral Leica TCS-SP2-AOBS inverted epifluorescence microscope using oil-immersion objectives. Image analysis was performed using Photoshop 7.0 (Adobe Systems) and Image software (Sánchez-Sánchez et al., 2004). For statistical analysis, 50–100 single and synapse-forming BMDMs were examined.
Mouse skin transplantation
The method used for full-thickness skin grafting was adapted from McKay et al. (2006) and Rosenberg (2001) and approved by the institutional Animal Ethical Committee (PRBB/UPF Animal Care and Use Committee). Donor and recipient mice were shaved the day before transplant. For this, the anterior part of the back was first shaved with a battery-operated razor and then chemically depilated with shaving cream (Vichy) to ensure complete hair removal. The day of transplantation, the donor tissue was first harvested from CO2-euthanized mice. The skin was sampled from the depilated dorsal part of the mouse using a 1.5-cm-diameter biopsy punch. The tissue with epidermis and dermis was placed in PBS until grafting to recipient mice. The recipient mouse was anesthetized with isoflurane, and a skin disc of the same size as the graft was surgically removed with the biopsy punch. The donor skin was then placed over the graft bed and secured in place with 6/0 silk sutures. The skin graft was covered with sterile Vaseline-impregnated gauze, and the recipient mouse was wrapped in a sterile bandage fixed with 4/0 silk sutures to avoid premature removal while allowing limb motion and ambulation. Transplanted animals were individually placed in clean cages under a heating lamp until they recovered from the anesthesia. 7 d after transplant, the bandage was carefully removed, and grafts were monitored daily for signs of rejection. Complete rejection was determined by necrosis affecting ≥90% of the graft. Histopathology analysis of skin grafts was done between days 8 and 11 after transplant. Briefly, tissue samples from euthanized mice were fixed overnight in 10% neutral buffered formalin, embedded in paraffin, sectioned in 3-µm-thick slabs, and dried. Slides were dewaxed and rehydrated through successive washes in graded ethanol to water before staining with hematoxylin and eosin. Microphotographs were taken with an Olympus DP73 digital camera. Assessment of naive and effector T lymphocytes in transplanted animals was done on the day when clear rejection of male Nfat5+/+ LysM-Cre skin grafts was observed, which ranged from day 12 to 16. Mice were sacrificed, their spleens were removed, and splenocytes were isolated after red blood cell lysis with ACK lysing buffer (Lonza; 10-548E). T lymphocytes were analyzed by flow cytometry using PE/Cy7 anti-CD3ε and APC-Cy7 anti-CD8a or PE-Cyanine5 anti-CD4 antibodies for gating CD8+ and CD4+ T lymphocytes, respectively. PE anti-CD44 and APC anti-CD62L antibodies were used to identify naive (CD62L+ CD44−) or effector (CD62L− CD44+) T cells.
Microarray analysis
Published microarray data of differential gene expression in wild-type and NFAT5-deficient BMDMs (Buxadé et al., 2012), with Gene Expression Omnibus (GEO) accession number GSE26343, were used for identifying NFAT5-regulated MHC-related genes as well as genes encoding for surface receptors and adhesion molecules mediating macrophage–T lymphocyte interactions.
qChIP
BMDMs cultured in 15-cm-diameter polystyrene dishes (18–20 × 106 cells), either left untreated or stimulated with IFNγ if indicated, were fixed with 0.75% formaldehyde for 10 min at room temperature. Formaldehyde was then quenched with glycine (final concentration 0.26 M) for 5 min. After the plates were washed twice with cold PBS, cells were collected with cell scrapers and lysed in 0.5 ml lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8, 1 mM PMSF, 5 µg/ml leupeptin/aprotinin, 1 µg/ml pepstatin A, 10 mM NaF, 10 mM sodium orthovanadate, and 10 mM β-glycerophosphate) for 30 min rotating at room temperature. Lysates were sonicated using the Bioruptor sonication system (Diagenode; Bioruptor UCD-200TM-EX). Each sample was divided in 2 × 1.5-ml tubes containing 250 µl lysate and sonicated for six cycles (30 s ON/OFF) at the high power setting to obtain DNA fragments between 500 and 1,000 bp. After sonication, samples were centrifuged to remove insoluble debris, supernatants were collected, and 5% of each sample was separated to use as a measure of chromatin input for normalization. The rest of the sample was diluted 1/10 in ChIP dilution buffer (1% Triton X-100, 20 mM Tris-HCl, pH 8, 2 mM EDTA, 150 mM NaCl, 1 mM PMSF, 5 µg/ml leupeptin, 5 µg/ml aprotinin, 1 µg/ml pepstatin A, 10 mM NaF, 10 mM sodium orthovanadate, and 10 mM β-glycerophosphate) for immunoprecipitation. Samples were precleared with protein A Sepharose beads (GE Healthcare; 17-0780-01) that were previously preadsorbed with fish sperm DNA (Roche; 11 467 140 001) and BSA (New England Biolabs; B9001S) for 1 h at 4°C. Specific antibodies were added to the lysates after removing the preclearing beads and incubated overnight at 4°C. Preadsorbed protein A Sepharose beads were then added, incubated for 1 h at 4°C, and washed three times with ChIP washing buffer (0.1% SDS, 1% Triton X-100, 20 mM Tris-HCl, pH 8, 2 mM EDTA, and 150 mM NaCl) and once with final washing buffer (0.1% SDS, 1% Triton X-100, 20 mM Tris-HCl, pH 8, 2 mM EDTA, and 500 mM NaCl). To elute the DNA, beads were incubated with elution buffer (1% SDS and 100 mM NaHCO3) for 15 min at room temperature. To reverse the cross-linking, samples were incubated overnight at 65°C with 5 ng/µl RNase (Roche; 11 119 915 001), and DNA was purified using the Qiagen PCR purification system. Antibodies used for ChIP were as follows: for NFAT5, a mixture of two rabbit polyclonal antibodies to the amino-terminal or DNA binding domain regions (Lopez-Rodríguez et al., 1999) was used, and preimmune serum served as control; for RNA Pol-II, anti-RNA polymerase II CTD repeat YSPTSPS antibody (Abcam; ab817) was used and a normal mouse IgG (Santa Cruz; sc-2025) included as a control; for RNA Pol-II Ser2, anti-RNA polymerase II CTD repeat YSPTSPS (phospho S2) antibody (Abcam; ab5095) and a normal rabbit IgG (Santa Cruz; sc-2027) were used. For histone modifications, anti-histone H3 monomethyl K4 antibody (Abcam; ab8895), anti-histone H3 trimethyl K4 antibody (Abcam; ab8580), and anti-histone H3 acetyl K27 antibody (Abcam; ab4729) were used, with a normal rabbit IgG (Santa Cruz; sc-2027) included as a control. Immunoprecipitated chromatins and their respective inputs were analyzed by RT-qPCR using the primers listed in Table S2. Immunoprecipitated DNA from each sample was normalized to its respective chromatin input.
Ultrasequencing of immunoprecipitated chromatin (ChIP-seq)
Lysates from formaldehyde-fixed wild-type and NFAT5-deficient BMDMs (fourteen 15-cm-diameter polystyrene dishes with 18–20 × 106 cells per plate) were obtained as described above for ChIP assays, except that sonication used two rounds of 10 cycles (30 s ON/OFF) at high power setting to obtain DNA fragments of 200–300 bp. After sonication, samples were centrifuged for 5 min at 13,000 rpm (15,700 g) at room temperature, and supernatants were collected together. Chromatin aliquots from the wild-type and NFAT5-deficient BMDM samples were separated to use as a measure of chromatin input. 900 µg sonicated chromatin from each genotype was diluted 1/10 in the same ChIP dilution buffer used above for conventional ChIP and precleared with 200 µl protein A agarose beads (Diagenode; kch-503-008) previously preadsorbed with BSA (Diagenode blocker for ChIP-Ab binding beads; kch-bloCKR-200) for 1 h at 4°C. For NFAT5 immunoprecipitation, a mixture of two rabbit polyclonal antibodies specific for NFAT5 amino-terminal or DNA binding domain regions (Lopez-Rodríguez et al., 1999) was added to the precleared lysates and incubated overnight at 4°C. Preadsorbed protein A agarose beads were added, incubated for 1 h at 4°C, and washed three times with ChIP washing buffer and once with final washing buffer. To elute the DNA, beads were incubated with elution buffer (1% SDS and 100 mM NaHCO3) for 15 min at room temperature. To reverse the cross-linking, samples were incubated with 5 ng/µl RNase (Roche) for 1 h at 37°C and 350 µg/ml proteinase K (Roche; 03115801001) for 2 h at 56°C and were left overnight at 65°C. Samples were purified by phenol extraction using the Phase Lock Gel system (5 Prime; 2302810). Finally, samples were resuspended in 30 µl Qiagen PCR purification elution buffer and quantified using PicoGreen and a Qubit fluorometer (Invitrogen). 20 ng of each sample was sequenced with an Illumina Genome Analyzer II platform (Illumina) at the Genomics Core Facility of the Center for Genomic Regulation (Barcelona, Spain). For sequencing, libraries were prepared using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs; E7370) according to the manufacturer’s protocol from 5–10 ng starting material. Final libraries were analyzed using the Agilent Bioanalyzer with a DNA High Sensitivity chip to estimate the quantity and check size distribution and were then quantified by qPCR using the KAPA Library Quantification Kit (KapaBiosystems; KK4835) before sequencing with Illumina TruSeq v3 chemistry on a HiSeq 2000. Reads produced by the ChIP-seq experiment GSE107948 were mapped using Bowtie aligner (v.0.12.7) to the reference genome NCBIM37.57 (mm9) with the option --best. Peaks were called by using MACS v.1.3.7.1 without creating the model and using the NFAT5-deficient macrophage sample (KO) as control. No input tracks were sequenced in the GSE107948 experiment. Reads from the second experiment, GSE107950, which included sequencing of the input DNA, were aligned using Bowtie2 (v.2.2.8) to the reference mm10 and fed to MACS (v.1.4.1) for peak calling again without building the model. Profiles were then created as bedgraph format by using BEDTools (v.2.25.0). The FastQC tool was used to check read quality. ENSEMBL annotation (versions 63 and 75) was used to annotate the peaks and detect the closest transcription start site (TSS). ChIP-seq signals around TSSs were calculated using BEDTools over TSS overlapping peaks and normalizing the coverage for the total number of mapped reads. After MACS analysis for peak calling, peak A in the first experiment (GSE107948) did not achieve a significant enrichment in wild-type macrophages, whereas in the second experiment (GSE107950), it showed a statistically significant value (−LOG10 [q value] = 19.116; q value = 7.66 × 10−20) in wild-type macrophages. Two independent experiments were performed and analyzed, and their respective GEO accession numbers are GSE107948 and GSE107950.
3C analysis
BMDMs cultured in 15-cm-diameter polystyrene dishes (18–20 × 106 cells) were fixed with 1% formaldehyde for 10 min at room temperature. Formaldehyde was then quenched with glycine (final concentration 0.125 M) for 5 min at room temperature. After 15 min on ice, plates were washed with cold PBS, and cells were collected with cell scrapers and 0.5 ml fresh lysis buffer (10 mM Tris HCl, pH 8, 10 mM NaCl, 0.2% NP-40, 1 mM PMSF, 5 µg/ml leupeptin/aprotinin, 1 µg/ml pepstatin A, 10 mM NaF, 10 mM sodium orthovanadate, and 10 mM β-glycerophosphate). Cells were then lysed on ice with a pestle B dounce homogenizer (15 strokes followed by 1 min on ice, repeated 15 times). After 30 min on ice, cell lysates were centrifuged for 5 min at 2,500 g at room temperature, and the supernatant was discarded. The pellet was washed twice with 500 µl of 1× restriction enzyme buffer and split into two 1.5-ml tubes with 362 µl per tube to separate “digested-not-ligated” and “digested-and-ligated” samples. Next, 38 µl of 1% SDS was added to each sample, heated at 65°C for 10 min, and digested overnight at 37°C with 400 U AflII restriction enzyme (New England Biolabs; R0520). After the first digestion, fresh AflII (200 U) was added and incubated for an additional 2 h. Finally, the enzyme was inactivated by adding 86 µl of 10% SDS and heating at 65°C for 30 min. Samples were then transferred to a 15-ml tube and ligated as follows: first, 745 µl of 10% Triton X-100 and 745 µl of 10× ligation buffer (500 mM Tris HCl, pH 7.5, 100 mM MgCl2, and 100 mM dithiothreitol) were added, and samples were incubated at 37°C for 30 min; then 80 µl of 10 mg/ml BSA, 80 µl of 100 mM ATP, 5,960 µl of milliQ H2O, and 4,000 units of T4 ligase (Thermo Fisher Scientific; EL0013) were added and incubated at 16°C for 4–5 h. After ligation, samples were treated with 37 µl of 22 mg/ml proteinase K (Roche) overnight at 65°C and purified by phenol/chloroform extraction. Samples were then treated with 1 µl of 10 mg/ml RNase A for 15 min at 37°C, and phenol extraction was repeated. Purified DNA was resuspended in 75 µl Qiagen PCR purification elution buffer and stored at −20°C or directly analyzed by PCR using the primers listed in Table S2. PCR products were run in 2% agarose gels, and Sanger sequencing was performed on purified bands from the gel (Genomics Facility, UPF).
CRISPR-Cas9 gene editing
A 145-bp region between positions 10,433,585 and 10,433,748 of mouse chromosome 16 (Chr 16 [qA1], UCSC Genome Browser; GRCm38/mm10), comprising two putative NFAT5 binding sites in peak A Ciita enhancer (described in this paper), was chosen for CRISPR-Cas9–mediated gene editing (illustrated in the schematic diagram of Fig. S4 A). Complementary DNA sequences for two 19-nucleotide gRNAs flanking the target region were subcloned in the CRISPR-Cas9 vector pSpCas9 (BB)-2A-GFP (PX458; Addgene; 48138). The resulting vector was introduced in iBMDM by electroporation (5 µg plasmid/10 × 106 cells in 0.4-cm gap cuvettes (Bio-Rad) at 320 V, 975 µF in a Bio-Rad Gene Pulser, and 24 h later, cells were sorted by flow cytometry based on GFP expression. Sorted cells were plated (5 cells/well) in 96-well round-bottom tissue culture plates, expanded, and cloned by limiting dilution to obtain individual iBMDM cell lines with either an intact peak A or a deletion (Δ145 peak A). Deletion of peak A was confirmed by PCR analysis of genomic DNA with diagnostic primers. gRNA sequences and diagnostic primers for deletion analysis are listed in Table S2.
Statistical analysis
All data are presented as mean ± SEM. Data were analyzed with GraphPad Prism 6 software. Normality (Gaussian distribution) of samples was determined by a D’Agostino–Pearson normality test before calculating statistical significance with an unpaired t test (for sets of samples with a Gaussian distribution) or Mann–Whitney test (samples with a non-Gaussian distribution). A one-sample t test was used when samples were compared with a reference control sample (set to an arbitrary value of 1). Specific statistics analyses, number of samples, and independent experiments done are indicated in each respective figure legend.
Accession numbers
ChIP-sequencing raw data of this study has been deposited under a controlled data access at the GEO, accession numbers GSE107948 and GSE107950.
Online supplemental material
Fig. S1 (related to Fig. 1) includes the expression of MHCII-related genes in NFAT5-deficient DCs and B and T lymphocytes. Fig. S2 (related to Fig. 3) shows a diagram of the skin graft assays, representative examples of graft progression, and the analysis of the activation of spleen CD4+ T cells from all experimental groups by the time of rejection. Fig. S3 (related to Fig. 4) illustrates the potential NFAT5 binding sites in the promoters of Ciita, H2-A2, and H2-Ab and also shows the results obtained in the ChIP-seq analysis in the MHCII locus. Fig. S4 (related to Fig. 5) shows the strategy used for the CRISPR-Cas9–induced deletion and the primers used for screening the clones obtained. It also includes the PCR results obtained with the diagnostic primers. Fig. S5 (related to Fig. 5) illustrates the 3C assay and the result obtained by sequencing the AflII-cut and ligated products comprising the myeloid promoter of Ciita and peak A. Table S1 (related to Fig. 1) shows the expression of genes encoding for molecules involved in the activation and communication between innate immune cells and T lymphocytes in NFAT5-deficient and control BMDMs. Table S2 lists the sequences of all primers used in this study classified by different technical approaches.
Supplementary Material
Acknowledgments
We are grateful to Pablo Pelegrín (Biomedical Research Institute of Murcia, University Clinical Hospital Virgen de la Arrixaca) for the iBMDM cell line, Jordi Sintes and Jordi Pou (Instituto Hospital del Mar de Investigaciones Médicas) for help with macrophage transfection protocols, Antonio Castrillo (Spanish National Research Council) for guidance with ChIP-seq methodology, Mª Dolores López-Maderuelo for advice with the skin transplant protocol, and Sara Capdevilla and Begoña Peñalba (Animal Facility, PRBB) for advice with experimental procedures in mice. Jochen Hoecht, Heinz Himmelbauer, and Anna Ferrer (Genomics Unit, Centre for Genomic Regulation [CRG]) are acknowledged for performing the ultrasequencing of the ChIP-seq experiments, and Luca Cozzuto (Bioinformatics Unit, CRG) for the bioinformatics analysis. We thank Carlo Carolis (Biomolecular Screening & Protein Technologies Unit, CRG) for guidance with the CRISPR-Cas9 strategy, Roger Anglada (Genomics Core Facility, UPF) for Sanger sequencing, and Oscar Fornas (Flow Cytometry Core Facility, UPF) for assistance with cell sorting. We also thank María García Belando and Mireia Viñas (UPF) for technical support with mouse genotyping, and members of our group for stimulating and helpful discussions.
This work was supported by the Spanish Ministry of Economy and Competitiveness (MINECO), Agencia Estatal de Investigación, and European Regional Development Fund (SAF2012-36535, SAF2015-71363-R, and BFU2016-77961-P), and Fundació la Marató de TV3 (1225-30 and 201619-30). We also acknowledge funding support from Generalitat de Catalunya (2014SGR1153, 2017SGR888, and 2017SGR702) and MINECO through the “Unidad de Excelencia María de Maeztu” funded by MINECO (MDM-2014-0370). H. Huerga Encabo was supported by a predoctoral fellowship of the Spanish Ministerio de Educación, Cultura y Deporte (FPU13/01798). M. Tellechea was supported by fellowships from Fundació Catalunya-La Pedrera (2011) and Generalitat de Catalunya (FI-DGR program 2013). C. López-Rodríguez is a recipient of an ICREA Acadèmia award from Institució Catalana de Recerca i Estudis Avançats (ICREA, Generalitat de Catalunya)
The authors declare no competing financial interests.
Author contributions: M. Buxadé performed and analyzed most of the experimental work and prepared the figures. H. Huerga Encabo performed and analyzed the mRNA and FACS experiments with DCs and the IFNγ withdrawal assays with BMDM and iBMDM, and prepared one ChIP-seq experiment. M. Riera-Borrull performed and analyzed the qChIP assays in DCs. L. Quintana-Gallardo prepared one ChIP-seq experiment. P. López-Cotarelo and J.L. Rodríguez-Fernández performed and analyzed the synapsis experiments. M. Tellechea isolated and analyzed mouse peritoneal macrophages. S. Martínez-Martínez and J.M. Redondo provided expertise with the skin transplants. J. Martín-Caballero and J.M. Flores performed and interpreted the histology analysis of skin grafts. E. Bosch contributed the analysis of genomic databases. J. Aramburu contributed with experiments design, manuscript writing, and figure preparation. C. López-Rodríguez designed and supervised the work, analyzed the results and interpreted the data, prepared the figures, and wrote the manuscript.
References
- Alberdi M., Iglesias M., Tejedor S., Merino R., López-Rodríguez C., and Aramburu J.. 2017. Context-dependent regulation of Th17-associated genes and IFNγ expression by the transcription factor NFAT5. Immunol. Cell Biol. 95:56–67. 10.1038/icb.2016.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramburu J., Drews-Elger K., Estrada-Gelonch A., Minguillón J., Morancho B., Santiago V., and López-Rodríguez C.. 2006. Regulation of the hypertonic stress response and other cellular functions by the Rel-like transcription factor NFAT5. Biochem. Pharmacol. 72:1597–1604. 10.1016/j.bcp.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Auchinloss H.J., Sykes M., and Sachs D.H.. 1999. Transplantation Immunology. In Fundamental Immunology. Paul W.E., editor. Lippincott-Raven, Philadelphia: pp. 1175–1235. [Google Scholar]
- Bain C.C., Bravo-Blas A., Scott C.L., Perdiguero E.G., Geissmann F., Henri S., Malissen B., Osborne L.C., Artis D., and Mowat A.M.. 2014. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 15:929–937. 10.1038/ni.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnden M.J., Allison J., Heath W.R., and Carbone F.R.. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34–40. 10.1046/j.1440-1711.1998.00709.x [DOI] [PubMed] [Google Scholar]
- Baroja-Mazo A., Martín-Sánchez F., Gomez A.I., Martínez C.M., Amores-Iniesta J., Compan V., Barberà-Cremades M., Yagüe J., Ruiz-Ortiz E., Antón J., et al. 2014. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat. Immunol. 15:738–748. 10.1038/ni.2919 [DOI] [PubMed] [Google Scholar]
- Berga-Bolaños R., Drews-Elger K., Aramburu J., and López-Rodríguez C.. 2010. NFAT5 regulates T lymphocyte homeostasis and CD24-dependent T cell expansion under pathologic hypernatremia. J. Immunol. 185:6624–6635. 10.4049/jimmunol.1001232 [DOI] [PubMed] [Google Scholar]
- Berga-Bolaños R., Alberdi M., Buxadé M., Aramburu J., and López-Rodríguez C.. 2013. NFAT5 induction by the pre-T-cell receptor serves as a selective survival signal in T-lymphocyte development. Proc. Natl. Acad. Sci. USA. 110:16091–16096. 10.1073/pnas.1215934110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S.K., and Mantovani A.. 2010. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 11:889–896. 10.1038/ni.1937 [DOI] [PubMed] [Google Scholar]
- Boland B.S., Widjaja C.E., Banno A., Zhang B., Kim S.H., Stoven S., Peterson M.R., Jones M.C., Su H.I.I., Crowe S.E., et al. 2015. Immunodeficiency and autoimmune enterocolopathy linked to NFAT5 haploinsufficiency. J. Immunol. 194:2551–2560. 10.4049/jimmunol.1401463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss J.M. 1997. Regulation of transcription of MHC class II genes. Curr. Opin. Immunol. 9:107–113. 10.1016/S0952-7915(97)80166-5 [DOI] [PubMed] [Google Scholar]
- Boss J.M., and Jensen P.E.. 2003. Transcriptional regulation of the MHC class II antigen presentation pathway. Curr. Opin. Immunol. 15:105–111. 10.1016/S0952-7915(02)00015-8 [DOI] [PubMed] [Google Scholar]
- Breloer M., Moré S.H., Osterloh A., Stelter F., Jack R.S., and Bonin Av A.. 2002. Macrophages as main inducers of IFN-gamma in T cells following administration of human and mouse heat shock protein 60. Int. Immunol. 14:1247–1253. 10.1093/intimm/dxf090 [DOI] [PubMed] [Google Scholar]
- Bunting K.L., Soong T.D., Singh R., Jiang Y., Béguelin W., Poloway D.W., Swed B.L., Hatzi K., Reisacher W., Teater M., et al. 2016. Multi-tiered reorganization of the genome during B cell affinity maturation anchored by a germinal center-specific locus control region. Immunity. 45:497–512. 10.1016/j.immuni.2016.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxadé M., Lunazzi G., Minguillón J., Iborra S., Berga-Bolaños R., Del Val M., Aramburu J., and López-Rodríguez C.. 2012. Gene expression induced by Toll-like receptors in macrophages requires the transcription factor NFAT5. J. Exp. Med. 209:379–393. 10.1084/jem.20111569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K.W., Morris D.L., DelProposto J.L., Geletka L., Zamarron B., Martinez-Santibanez G., Meyer K.A., Singer K., O’Rourke R.W., and Lumeng C.N.. 2014. An MHC II-dependent activation loop between adipose tissue macrophages and CD4+ T cells controls obesity-induced inflammation. Cell Reports. 9:605–617. 10.1016/j.celrep.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen B.E., Burkhardt C., Reith W., Renkawitz R., and Förster I.. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8:265–277. 10.1023/A:1008942828960 [DOI] [PubMed] [Google Scholar]
- Collins T., Korman A.J., Wake C.T., Boss J.M., Kappes D.J., Fiers W., Ault K.A., Gimbrone M.A. Jr., Strominger J.L., and Pober J.S.. 1984. Immune interferon activates multiple class II major histocompatibility complex genes and the associated invariant chain gene in human endothelial cells and dermal fibroblasts. Proc. Natl. Acad. Sci. USA. 81:4917–4921. 10.1073/pnas.81.15.4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A., et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 107:21931–21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSandro A., Nagarajan U.M., and Boss J.M.. 1999. The bare lymphocyte syndrome: molecular clues to the transcriptional regulation of major histocompatibility complex class II genes. Am. J. Hum. Genet. 65:279–286. 10.1086/302519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S., Barozzi I., Mietton F., Polletti S., De Santa F., Venturini E., Gregory L., Lonie L., Chew A., Wei C.L., et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 32:317–328. 10.1016/j.immuni.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Go W.Y., Liu X., Roti M.A., Liu F., and Ho S.N.. 2004. NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc. Natl. Acad. Sci. USA. 101:10673–10678. 10.1073/pnas.0403139101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau V., Herbst B., and Steiniger B.. 1998. Dynamics of monocytes/macrophages and T lymphocytes in acutely rejecting rat renal allografts. Cell Tissue Res. 291:117–126. 10.1007/s004410050985 [DOI] [PubMed] [Google Scholar]
- Hakim O., Sung M.-H., Nakayamada S., Voss T.C., Baek S., and Hager G.L.. 2013. Spatial congregation of STAT binding directs selective nuclear architecture during T-cell functional differentiation. Genome Res. 23:462–472. 10.1101/gr.147652.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick C.V., Randolph G.J., and Henson P.M.. 2017. Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 17:349–362. 10.1038/nri.2017.28 [DOI] [PubMed] [Google Scholar]
- Jantsch J., Schatz V., Friedrich D., Schröder A., Kopp C., Siegert I., Maronna A., Wendelborn D., Linz P., Binger K.J.J., et al. 2015. Cutaneous Na+ storage strengthens the antimicrobial barrier function of the skin and boosts macrophage-driven host defense. Cell Metab. 21:493–501. 10.1016/j.cmet.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F., Fenouil R., Gut M., Cauchy P., Albert T.K., Zacarias-Cabeza J., Spicuglia S., de la Chapelle A.L., Heidemann M., Hintermair C., et al. 2011. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 18:956–963. 10.1038/nsmb.2085 [DOI] [PubMed] [Google Scholar]
- Kühn R., Schwenk F., Aguet M., and Rajewsky K.. 1995. Inducible gene targeting in mice. Science. 269:1427–1429. 10.1126/science.7660125 [DOI] [PubMed] [Google Scholar]
- Lin Y.C., Benner C., Mansson R., Heinz S., Miyazaki K., Miyazaki M., Chandra V., Bossen C., Glass C.K., and Murre C.. 2012. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat. Immunol. 13:1196–1204. 10.1038/ni.2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohsen S., Majumder P., Scharer C.D., Barwick B.G., Austin J.W., Zinzow-Kramer W.M., and Boss J.M.. 2014. Common distal elements orchestrate CIITA isoform-specific expression in multiple cell types. Genes Immun. 15:543–555. 10.1038/gene.2014.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rodríguez C., Aramburu J., Rakeman A.S., and Rao A.. 1999. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc. Natl. Acad. Sci. USA. 96:7214–7219. 10.1073/pnas.96.13.7214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Rodríguez C., Aramburu J., Jin L., Rakeman A.S., Michino M., and Rao A.. 2001. Bridging the NFAT and NF-kappaB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity. 15:47–58. 10.1016/S1074-7613(01)00165-0 [DOI] [PubMed] [Google Scholar]
- López-Rodríguez C., Antos C.L., Shelton J.M., Richardson J.A., Lin F., Novobrantseva T.I., Bronson R.T., Igarashi P., Rao A., and Olson E.N.. 2004. Loss of NFAT5 results in renal atrophy and lack of tonicity-responsive gene expression. Proc. Natl. Acad. Sci. USA. 101:2392–2397. 10.1073/pnas.0308703100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D., Shigeoka A., Rubinstein M., Surh C., and Sprent J.. 2006. Simultaneous deletion of MyD88 and Trif delays major histocompatibility and minor antigen mismatch allograft rejection. Eur. J. Immunol. 36:1994–2002. 10.1002/eji.200636249 [DOI] [PubMed] [Google Scholar]
- Morris D.L., Cho K.W., Delproposto J.L., Oatmen K.E., Geletka L.M., Martinez-Santibanez G., Singer K., and Lumeng C.N.. 2013. Adipose tissue macrophages function as antigen-presenting cells and regulate adipose tissue CD4+ T cells in mice. Diabetes. 62:2762–2772. 10.2337/db12-1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlethaler-Mottet A., Otten L.A., Steimle V., and Mach B.. 1997. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 16:2851–2860. 10.1093/emboj/16.10.2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Abou El Hassan M., Xu Z., Yu T., and Bremner R.. 2008. The chromatin-remodeling enzyme BRG1 coordinates CIITA induction through many interdependent distal enhancers. Nat. Immunol. 9:785–793. 10.1038/ni.1619 [DOI] [PubMed] [Google Scholar]
- Piskurich J.F., Wang Y., Linhoff M.W., White L.C., and Ting J.P.. 1998. Identification of distinct regions of 5′ flanking DNA that mediate constitutive, IFN-gamma, STAT1, and TGF-beta-regulated expression of the class II transactivator gene. J. Immunol. 160:233–240. [PubMed] [Google Scholar]
- Reith W., and Mach B.. 2001. The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol. 19:331–373. 10.1146/annurev.immunol.19.1.331 [DOI] [PubMed] [Google Scholar]
- Reith W., LeibundGut-Landmann S., and Waldburger J.M.. 2005. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 5:793–806. 10.1038/nri1708 [DOI] [PubMed] [Google Scholar]
- Riol-Blanco L., Delgado-Martín C., Sánchez-Sánchez N., Alonso-C L.M., Gutiérrez-López M.D., Del Hoyo G.M., Navarro J., Sánchez-Madrid F., Cabañas C., Sánchez-Mateos P., and Rodríguez-Fernández J.L.. 2009. Immunological synapse formation inhibits, via NF-kappaB and FOXO1, the apoptosis of dendritic cells. Nat. Immunol. 10:753–760. 10.1038/ni.1750 [DOI] [PubMed] [Google Scholar]
- Rosenberg A.S. 2001. Skin allograft rejection. Curr. Protoc. Immunol. Chapter 4:Unit 4.4 [DOI] [PubMed] [Google Scholar]
- Rosenberg A.S., and Singer A.. 1992. Cellular basis of skin allograft rejection: an in vivo model of immune-mediated tissue destruction. Annu. Rev. Immunol. 10:333–358. 10.1146/annurev.iy.10.040192.002001 [DOI] [PubMed] [Google Scholar]
- Sánchez-Sánchez N., Riol-Blanco L., de la Rosa G., Puig-Kröger A., García-Bordas J., Martín D., Longo N., Cuadrado A., Cabañas C., Corbí A.L., et al. 2004. Chemokine receptor CCR7 induces intracellular signaling that inhibits apoptosis of mature dendritic cells. Blood. 104:619–625. 10.1182/blood-2003-11-3943 [DOI] [PubMed] [Google Scholar]
- Shouval D.S., Biswas A., Goettel J.A., McCann K., Conaway E., Redhu N.S., Mascanfroni I.D., Al Adham Z., Lavoie S., Ibourk M., et al. 2014. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. 40:706–719. 10.1016/j.immuni.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E., Scott D., and Chandler P.. 1997. The male-specific histocompatibility antigen, H-Y: a history of transplantation, immune response genes, sex determination and expression cloning. Annu. Rev. Immunol. 15:39–61. 10.1146/annurev.immunol.15.1.39 [DOI] [PubMed] [Google Scholar]
- Soroosh P., Doherty T.A., Duan W., Mehta A.K., Choi H., Adams Y.F., Mikulski Z., Khorram N., Rosenthal P., Broide D.H., and Croft M.. 2013. Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J. Exp. Med. 210:775–788. 10.1084/jem.20121849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucie E.L., Weng Z., Geirsdóttir L., Molawi K., Maurizio J., Fenouil R., Mossadegh-Keller N., Gimenez G., VanHille L., Beniazza M., et al. 2016. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science. 351:aad5510 10.1126/science.aad5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., and Graf T.. 2005. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development. 132:203–213. 10.1242/dev.01558 [DOI] [PubMed] [Google Scholar]
- Tellechea M., Buxadé M., Tejedor S., Aramburu J., and López-Rodríguez C.. 2018. NFAT5-regulated macrophage polarization supports the proinflammatory function of macrophages and T lymphocytes. J. Immunol. 200:305–315. 10.4049/jimmunol.1601942 [DOI] [PubMed] [Google Scholar]
- Tie F., Banerjee R., Stratton C.A., Prasad-Sinha J., Stepanik V., Zlobin A., Diaz M.O., Scacheri P.C., and Harte P.J.. 2009. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 136:3131–3141. 10.1242/dev.037127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill D.M., Bassetti M., Rudensky A., and Aderem A.. 1999. Dynamic interactions of macrophages with T cells during antigen presentation. J. Exp. Med. 190:1909–1914. 10.1084/jem.190.12.1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyburn K.R., Jose M.D., Wu H., Atkins R.C., and Chadban S.J.. 2005. The role of macrophages in allograft rejection. Transplantation. 80:1641–1647. 10.1097/01.tp.0000173903.26886.20 [DOI] [PubMed] [Google Scholar]
- Yoon H., and Boss J.M.. 2010. PU.1 binds to a distal regulatory element that is necessary for B cell-specific expression of CIITA. J. Immunol. 184:5018–5028. 10.4049/jimmunol.1000079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zal T., Volkmann A., and Stockinger B.. 1994. Mechanisms of tolerance induction in major histocompatibility complex class II-restricted T cells specific for a blood-borne self-antigen. J. Exp. Med. 180:2089–2099. 10.1084/jem.180.6.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.