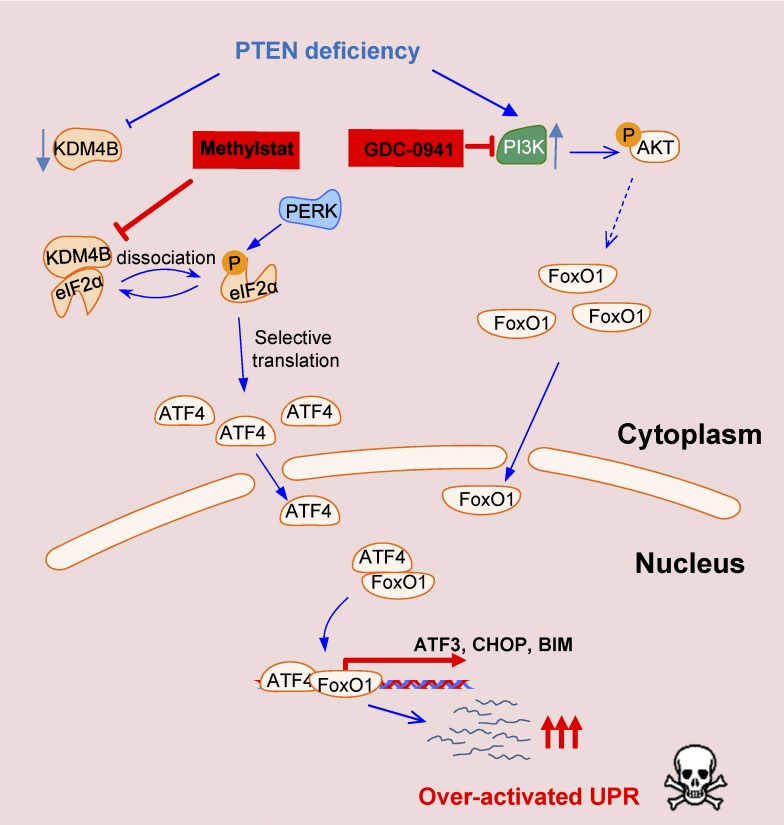
Wang et al. report an unexpected role of demethylase KDM4B in regulating unfolded protein response (UPR). A stepwise hyperactivation of UPR by co-targeting the KDM4B and PI3K pathway uncovers a therapeutic vulnerability of PTEN-deficient TNBC that otherwise would be resistant to PI3K inhibition.
Abstract
PTEN deficiency in breast cancer leads to resistance to PI3K–AKT inhibitor treatment despite aberrant activation of this signaling pathway. Here, we report that genetic depletion or small molecule inhibition of KDM4B histone demethylase activates the unfolded protein response (UPR) pathway and results in preferential apoptosis in PTEN-deficient triple-negative breast cancers (TNBCs). Intriguingly, this function of KDM4B on UPR requires its demethylase activity but is independent of its canonical role in histone modification, and acts through its cytoplasmic interaction with eIF2α, a crucial component of UPR signaling, resulting in reduced phosphorylation of this component. Targeting KDM4B in combination with PI3K inhibition induces further activation of UPR, leading to robust synergy in apoptosis. These findings identify KDM4B as a therapeutic vulnerability in PTEN-deficient TNBC that otherwise would be resistant to PI3K inhibition.
Graphical Abstract
Introduction
The PI3K–AKT pathway is frequently hyperactivated in breast cancer with genetic abnormalities in PIK3CA, PTEN, and other genes (Cancer Genome Atlas Network, 2012), and multiple PI3K inhibitors are currently in late-stage clinical development (Juric et al., 2017; Mayer et al., 2017). Intriguingly, breast cancer cells bearing the PIK3CA mutation, which are primarily found in luminal types of breast cancer, have shown a good response to PI3K/AKT inhibitors in vitro. However, PTEN-deficient cells, which are primarily found in triple-negative breast cancer (TNBC), appear to be resistant to PI3K/AKT inhibitors (Brachmann et al., 2009; Tanaka et al., 2011; Weigelt et al., 2011; Juric et al., 2015; Kim et al., 2017). TNBC is a heterogeneous and clinically aggressive disease for which there is no targeted therapy (Lehmann et al., 2011; Vaz-Luis et al., 2014). Given that PTEN deficiency occurs in up to 35% of TNBCs (Cancer Genome Atlas Network, 2012), it is important to develop therapeutic approaches targeting PTEN-deficient TNBC.
The unfolded protein response (UPR), often activated upon ER stress, is an adaptive pathway that protects cells from ER stress–mediated cell death (Kim et al., 2008). However, when ER stress is severe or prolonged, and ER homeostasis restoration fails, the UPR becomes “over-activated” and triggers cell death through constitutive activation of eIF2α/ATF4 and downstream apoptotic targets such as CHOP and BIM (Hetz, 2012; Wang and Kaufman, 2014). As tumors often have a high level of basal UPR (Wang and Kaufman, 2014; Urra et al., 2016), one hypothesis is that the hyperactive UPR can be exploited to trigger cancer apoptosis. Indeed, it is currently believed that activation of UPR can sensitize cells to ER stress, which has been shown to be a useful strategy in melanoma and lung cancer treatment (Feng et al., 2014; Jeon et al., 2015; Ramadori et al., 2015), but the mechanisms for UPR activation remains to be demonstrated, and whether this strategy could be applied to other cancer types remains unknown.
This study was undertaken to identify molecular vulnerabilities in PTEN-deficient TNBC in order to develop a therapeutic approach targeting PTEN deficiency in breast cancer. To this end, we identified a synthetic lethality in the context of PTEN deficiency that involves a noncanonical role of KDM4B in restricting the UPR, as well as functional cross-talk between UPR and PI3K signaling in controlling the activity of UPR and apoptosis induction. Accordingly, inhibiting KDM4B promoted UPR activation for apoptosis induction in PTEN-deficient TNBC cells, an action that became more robust when combined with a PI3K inhibitor.
Results
Synthetic lethality compound screening identifies the vulnerability of PTEN deficiency to KDM inhibitor Methylstat
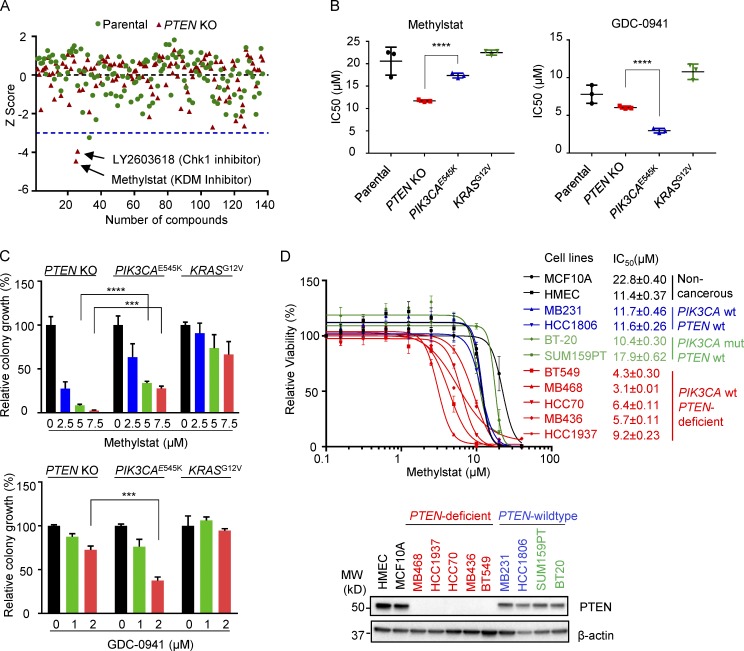
To identify the genetic vulnerability of PTEN deficiency and potential small molecules with selective activity against PTEN-deficient breast cancer, we performed a compound screening using noncancerous breast epithelial MCF10A cell lines in which PTEN is intact or genetically depleted (PTEN-KO). Among the 140 small molecule compounds that target pathways involved in growth, metabolism, and epigenetic modification, Methylstat, a KDM histone demethylase inhibitor (Luo et al., 2011), proved to best selectively impair the viability of PTEN-KO cells compared with parental MCF10A counterparts (using a threshold z-score −3 or less as a cutoff; Fig. 1 A). Another promising compound was an inhibitor of Chk1, the activity of which has previously been implicated in PTEN deficiency (Puc et al., 2005; Puc and Parsons, 2005) and was thus not pursued.
Figure 1.
Drug screening identifies KDM inhibitor Methylstat selectively impairing PTEN-deficient breast cancer cells. (A) MCF10A parental and PTEN-KO cells treated for 4 d with a compound library consisting of 140 small molecule inhibitors. Cell viability was assessed using a CellTiter-Glo Luminescent Cell Viability Assay. Shown is the scatter plot of z-scores normalized to DMSO control in MCF10A parental (green) and PTEN-KO cells (red). Blue dashed line indicates a z-score of –3 as the significance threshold. (B) IC50s of Methylstat or GDC-0941 in indicated isogenic MCF10A cells as determined by a CellTiter-Glo Luminescent Cell Viability Assay. (C) Colony formation assay was performed in indicated isogenic MCF10A cells in 0.5% methylcellulose containing the indicated concentration of Methylstat or GDC-0941 for 1 wk. Representative data of two independent experiments are shown. (D) IC50s of Methylstat in indicated breast cell lines with different PTEN and PIK3CA status. Top: Cells were treated with Methylstat for 3 d, and viability was assessed using a CellTiter-Glo Luminescent Cell Viability Assay. Bottom: Western blot analysis of PTEN in indicated breast cell lines. MW, molecular weight. See also Fig. S1. All data are representative of three independent experiments unless stated otherwise. Data are expressed as means ± SD. P values were determined by two-tailed unpaired Student’s t test; *** P ≤ 0.001, **** P ≤ 0.0001.
To verify the selectivity of Methylstat on PTEN deficiency, we further compared MCF10A cell lines with overexpression of oncogenic PIK3CAE545K or KRASG12V mutants. Although these isogenic lines showed similar capacities in cell proliferation when grown in monolayer or colony formation in soft agar (Fig. S1, A and B), they displayed differential responses to Methylstat and the PI3K inhibitor GDC-0941, respectively: PTEN-KO cells were most responsive to Methylstat, with a significantly lower IC50 compared with PIK3CAE545K or KRASG12V cells (Fig. 1 B). PIK3CAE545K cells, on the contrary, were most sensitive to GDC-0941, as expected, compared with the other lines (Fig. 1 B). Assessment with a colony formation assay also yielded similar results: Methylstat mainly impaired the colony growth of PTEN-KO cells, while GDC-0941 was more effective in PIK3CAE545K cells (Fig. 1 C). These findings establish the preferential selectivity of the histone KDM family inhibitor Methylstat against PTEN deficiency.
In a panel of TNBC cell lines with known PTEN and PIK3CA status, we further demonstrated that Methylstat preferentially affected the viability of PTEN-deficient TNBC cells compared with PTEN wild-type cells (Fig. 1 D). It is noteworthy that SUM159PT and BT-20 TNBC cells, known to harbor a PIK3CA-activating mutation, did not show a preferential response to Methylstat, consistent with our observations in MCF10A PIK3CAE545K cells. Our collective findings using various isogenic cell lines to model oncogenic events, as well as breast cancer cell lines with different PTEN and PIK3CA status, demonstrated that Methylstat preferentially affects TNBC cells with PTEN deficiency, but not PIK3CA mutations.
KDM inhibitor Methylstat induces UPR activation in PTEN-deficient TNBC
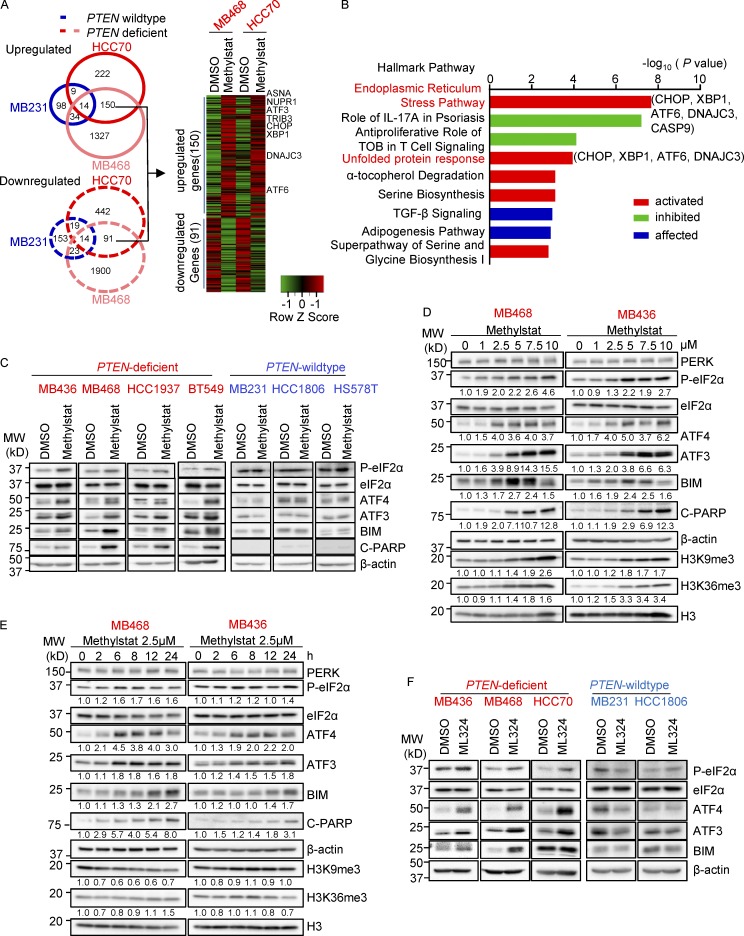
To determine the mechanism by which Methylstat favors PTEN-deficient TNBC cells, we performed a transcriptome analysis to identify genes with selective responsiveness to Methylstat in PTEN-deficient, but not PTEN wild-type, cells. Two PTEN-deficient TNBC lines, MDA-MB-468 (hereafter MB468) and HCC70, and a PTEN wild-type cell line MDA-MB-231 (hereafter MB231), were analyzed, and we identified 241 Methylstat-responsive genes, including 150 up-regulated and 91 down-regulated genes (using a 1.5-fold cutoff, P ≤ 0.05), selectively in PTEN-deficient cells (Fig. 2 A). Pathway analysis of the Methylstat-responsive transcriptome revealed significant enrichment for gene sets involved in ER stress and the UPR pathway (Fig. 2 B and Table S1), suggesting possible ER stress/UPR activation by Methylstat in PTEN-deficient TNBC cells. Consistently, ATF4, a key transcription factor in ER stress/UPR activation, was found to be the top upstream regulator (P = 4.05 × 10−17) under Methylstat treatment, which is in line with the up-regulation of its downstream targets, such as ATF3, CHOP (also known as DDIT3), and TRIB3 (Fig. 2 A and Table S2). Further analysis using gene set enrichment analysis (GSEA) supported this hypothesis, as Methylstat significantly induced gene sets known to be activated by two well-known ER stress inducers, thapsigargin (Tg) and tunicamycin (Tm; Koo et al., 2012; Fig. S1 C). As a control, the gene set known to be induced by the genotoxic drug doxorubicin (Flamant et al., 2012) was not induced by Methylstat (Fig. S1 D).
Figure 2.
Methylstat activates the UPR pathway in PTEN-deficient TNBC cells. (A) Venn diagram showing up- and down-regulated genes (>1.5 fold, P ≤ 0.05) by Methylstat (2.5 µM) for 24 h in PTEN-deficient MB468 and HCC70 and PTEN wild-type MB231 cells (left panel). Heat map is showing common Methylstat-responsive genes in PTEN-deficient MB468 and HCC70 (right panel). (B) IPA showing nine hallmark pathways exhibiting enrichment of Methylstat-responsive genes in MB468 and HCC70. Graph displays category scores as –log10 (P value) from Fisher’s exact test. TOB, transducer of ErbB2. (C) Western blot analysis of the UPR pathway in indicated TNBC cell lines treated with 2.5 µM Methylstat for 24 h. MW, molecular weight. (D) Western blot analysis of the UPR pathway in response to indicated concentrations of Methylstat for 24 h. (E) Western blot analysis of UPR in cells treated with 2.5 µM Methylstat for indicated hours. (F) Western blot analysis of UPR in indicated TNBC cells treated with 5 µM ML324 for 24 h. See also Fig. S1 and Tables S1 and S2. All data are representative of three independent experiments.
Western blot analysis of a panel of TNBC cell lines verified that Methylstat activated the UPR pathway, as evidenced by increased levels of eIF2α phosphorylation, ATF4, ATF3, and BIM in PTEN-deficient cells, but not in PTEN wild-type cells (Fig. 2 C). Similarly, Methylstat induced poly (ADP-ribose) polymerase (PARP) cleavage, indicating apoptosis in PTEN-deficient cells, but not in PTEN wild-type cells (Fig. 2 C). Dose response analysis showed that Methylstat treatment for 24 h activated UPR, PARP cleavage, and the histone methylation targets (H3K9me3 and H3K36me3) in a dose-dependent manner (Fig. 2 D). Notably, Methylstat treated at 2.5 µM was sufficient to activate UPR without inducing histone trimethylation on H3K9 and H3K36, the known histone targets of KDM4 (Klose et al., 2006; Whetstine et al., 2006; Fig. 2 D). A further time course analysis showed that Methylstat at 2.5 µM activated UPR as early as 6 h without affecting histone targets (Fig. 2 E). These observations indicated that Methylstat-induced UPR activation is a primary effect and is independent of its canonical role in chromatin modifications.
Methylstat is known to target KDM4 and KDM6 family histone demethylases (Luo et al., 2011). A KDM6-specific inhibitor, GSK-J4, included in the compound screening, however, did not show selective activity toward PTEN-KO cells (Fig. 1 A), arguing against KDM6B being a relevant target of Methylstat in this system. Moreover, ML324, a more specific KDM4 inhibitor, showed a similar effect as Methylstat in PTEN-deficient TNBC cells (Fig. 2 F). Taken together, these observations suggest that KDM4, rather than KDM6, accounts for the activity of Methylstat toward PTEN deficiency.
KDM4B is a relevant target of Methylstat and represses UPR activity in PTEN-deficient TNBC through cytoplasmic interaction with eIF2α, preventing eIF2α phosphorylation by protein kinase R–like ER kinase (PERK)
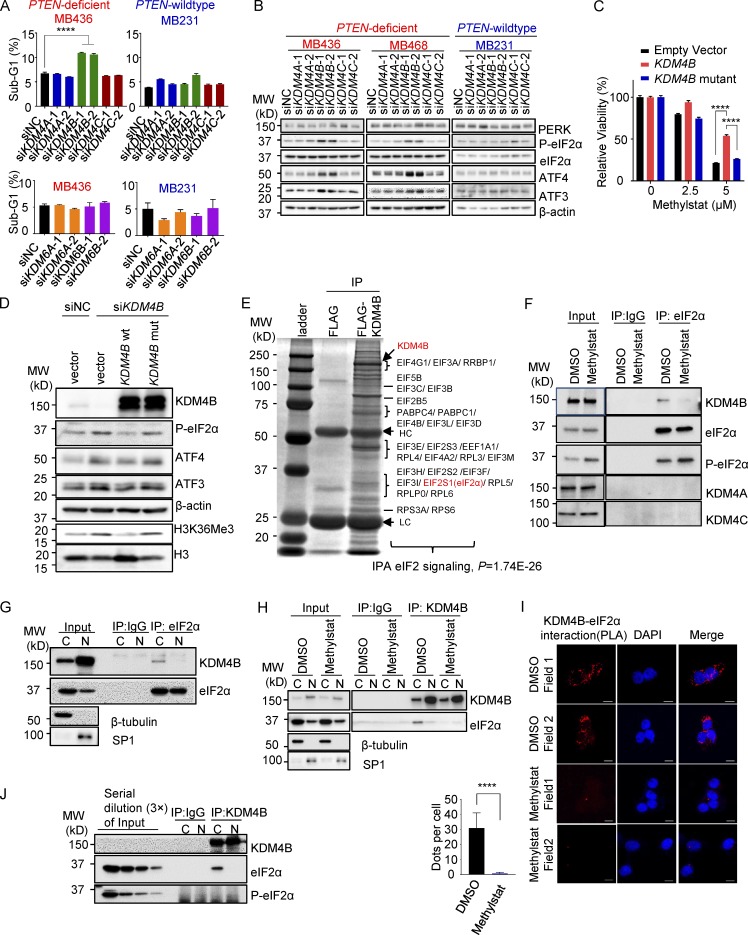
To determine which members of the KDM4/6 family are relevant targets of the Methylstat-induced phenotype, we first depleted the individual members of KDM4 (KDM4A, KDM4B, and KDM4C) and KDM6 (KDM6A and KDM6B) in TNBC and examined the corresponding effect on cell death and UPR activation. Upon individual knockdown of the KDM members using corresponding siRNAs (Fig. S2 A), we showed that only KDM4B silencing was able to mimic the Methylstat effect and induced significant cell death and UPR activation in PTEN-deficient TNBC cells (but not in PTEN wild-type cells; Fig. 3, A and B), ruling out the involvement of other KDM4/6 family members in the regulation of UPR in this setting.
Figure 3.
KDM4B represses UPR activity through cytoplasmic interaction with eIF2α. (A) Cell death determined by the percentage of a sub-G1 flow cytometry assay in indicated cell lines treated with indicated siRNAs for 48 h. (B) Western blot analysis of the UPR pathway in indicated cell lines treated with indicated siRNAs. MW, molecular weight. (C) Response to Methylstat in MB436 cell lines expressing empty vector, KDM4B wild-type, or KDM4B mutant plasmids. (D) Western blot analysis of MB436 cells transfected with indicated siRNAs and followed by plasmid transfection 24 h later. (E) Immunoaffinity purification of the KDM4B-containing protein complex. Cell extracts from MB436 cells stably expressing FLAG and FLAG-KDM4B were immunoprecipitated (IP) with anti-FLAG beads. All the immunoprecipitated proteins were resolved by SDS-PAGE and Coomassie blue staining. The protein bands were retrieved and analyzed by mass spectrometry. The interacting proteins were further analyzed by IPA analysis. (F) Coimmunoprecipitation study of endogenous KDM4B and eIF2α in MB436 cells treated with vehicle and 2.5 µM Methylstat for 24 h. (G) Coimmunoprecipitation study using eIF2α antibody in MB436 cells following cytoplasmic and nuclear fractionation. (H) Coimmunoprecipitation study of KDM4B and eIF2α in MB436 cells treated with vehicle and 2.5 µM Methylstat for 24 h following cytoplasmic and nuclear fractionation. (I) DMSO- and Methylstat-treated MDA-MB-436 cells were stained with specific antibodies against eIF2α (mouse) and KDM4B (rabbit) before PLA. The PLA signals between endogenous eIF2α and KDM4B are shown in the red channel; DAPI was used to stain the nuclei (blue); and the merged images show the overlay of the red and blue channels. Dots per cells were analyzed from two independent experiments. Scale bars, 10 µm. (J) Coimmunoprecipitation study using KDM4B antibody in MB436 cells following cytoplasmic and nuclear fractionation. See also Fig. S2. All data are representative of at least three independent experiments unless stated otherwise. Data are expressed as mean ± SD. P values were determined by two-tailed unpaired Student’s t test; **** P ≤ 0.0001.
Next, we demonstrated that Methylstat-induced loss of viability of MB436 cells was at least partially rescued by overexpressing the ectopic wild-type KDM4B but not the mutant KDM4B JmjC domain mutant (H189G/E191Q) known to be deficient in catalytic demethylase activity (Beyer et al., 2008; Fig. 3 C), supporting that KDM4B is a relevant target of Methylstat. Moreover, to verify the role of KDM4B in regulating the UPR pathway, “rescue” experiments were performed using the ectopic expression of a siRNA-resistant KDM4B or the H189G/E191Q KDM4B mutant. Western blot analysis showed that the UPR activation induced by KDM4B knockdown was abolished by overexpression of the wild-type KDM4B, but not the mutant KDM4B (Fig. 3 D), indicating an indispensable role of the demethylase activity of KDM4B in suppressing the UPR pathway. A detailed KDM4B domain structure activity analysis showed that although depletion of PHD1, PHD2, or Tudor domain reduced the ability of KDM4B to suppress the UPR pathway, they retained its histone demethylase activity toward H3K9Me3 and H3K36Me3 (Fodor et al., 2006; Fig. S2, B and C). Together, these studies provided evidence that KDM4B is a relevant target of Methylstat, and KDM4B represses UPR activity independently of its epigenetic activity on chromatin.
To investigate the molecular mechanism by which KDM4B regulates the UPR pathway, we sought to examine if KDM4B directly interferes with UPR signaling. Ectopic expression of a FLAG-tagged KDM4B (FLAG-KDM4B) in MB436 cells followed by immunoprecipitation with anti-FLAG antibody identified multiple components of the mRNA translation machinery (Fig. 3 E) by mass spectrometry analysis, including eIF2α (also known as EIF2S1), a key node of UPR regulation, suggesting that KDM4B may regulate UPR activity through physical interaction with eIF2α containing translation machinery complex. Further ingenuity pathway analysis (IPA) of the coimmunoprecipitated proteins also revealed a significant enrichment for proteins involved in eIF2 signaling (P = 1.74E-26; Fig. 3 E). We therefore speculate that eIF2α might be a key intersection point at which KDM4B interferes with the UPR pathway. Further validation through coimmunoprecipitation analysis showed that eIF2α indeed interacted with KDM4B, and this interaction was abolished by Methylstat (Fig. 3 F). Considering that the majority of KDM4B is localized in the nucleus, while eIF2α is mainly in the cytoplasm, we performed cell fractionation experiments to investigate the site of the interaction. The results showed that KDM4B, albeit more abundantly expressed in the nucleus, mainly interacted with eIF2α in the cytoplasm, which was abolished by Methystat (Fig. 3, G and H). As a separate validation, the proximity ligation assay (PLA), which detects the interaction of two proteins in situ in cells, confirmed the interaction between KDM4B and eIF2α in the cytoplasm which was also highly susceptible to Methylstat treatment (Fig. 3 I). Moreover, KDM4B seemed to interact only with nonphosphorylated eIF2α, as evidenced by the lack of phosphorylated eIF2α (P-eIF2α) detected in the KDM4B immunoprecipitation (Fig. 3 J). On the other hand, Methylstat treatment led to increased interaction between PERK and eIF2α (Fig. S2 D), and P-eIF2α pull-down did not detect KDM4B, but showed increased PERK upon Methylstat treatment (Fig. S2 E). These data suggest that Methylstat disrupted the KDM4B–eIF2α interaction while promoting the interaction between PERK and eIF2α, leading to increased eIF2α phosphorylation. Similarly, PERK depletion diminished Methylstat-induced eIF2α phosphorylation (Fig. S2 F). Based on these observations, we propose that KDM4B regulates UPR through direct interaction with eIF2α, resulting in reduced eIF2α phosphorylation by its upstream kinase PERK.
To determine which domain of KDM4B is important for binding to eIF2α, we constructed three KDM4B mutants lacking the N-terminus (amino acids 1–309, containing the jmjN and jmjC domain, the catalytic domain for demethylase activity; ΔN), middle part (amino acids 310–730, devoid of any defined structural motif but important for β-catenin binding; ΔM; Berry et al., 2014), and C-terminus (amino acids 731–1096, containing PHD the and TUDOR domain, which are involved in recognizing methylated histone residues; ΔC), respectively (Fig. S2 B), and performed coimmunoprecipitation experiments in cells expressing these constructs. We found attenuated interactions between eIF2α and two KDM4B mutants (KDM4BΔN and ΔC), which revealed that both the catalytic domain and C-terminus of KDM4B are required, at least partially, for KDM4B binding to eIF2α (Fig. S2 G). Similarly, a catalytic activity-dead KDM4B mutant (H189G/E191Q) also showed attenuated interaction with eIF2α (Fig. S2 H). Moreover, depletion of the middle part fully abolished the binding, suggesting an essential role of its domain (Fig. S2 G). Accordingly, while overexpression of wild-type KDM4B decreased the level of P-eIF2α, KDM4B mutants, especially jumonji domain deletion, acted as dominant negative mutants to increase the phosphorylation of eIF2α (Fig. S2 G). Next, we wondered whether two other KDM4 family members, KDM4A and KDM4C, could bind to eIF2α. It is worth noting that the middle domain of KDM4 family members was unconserved (Fig. S2 I) by the MultAlin analysis (Corpet, 1988). Consistent with our finding that the middle part of KDM4B is important to the interaction with eIF2α, no interactions were found between KDM4A/C and eIF2α (Fig. 3 F and Fig. S2 J), demonstrating the specificity of KDM4B interaction with eIF2α. Collectively, these data suggested that demethylase activity-competent KDM4B is required for regulation of eIF2α and the UPR activity, whereas the interaction between KDM4B and eIF2α is required but not sufficient for this regulation.
PTEN loss results in down-regulation of KDM4B and enhanced UPR activity
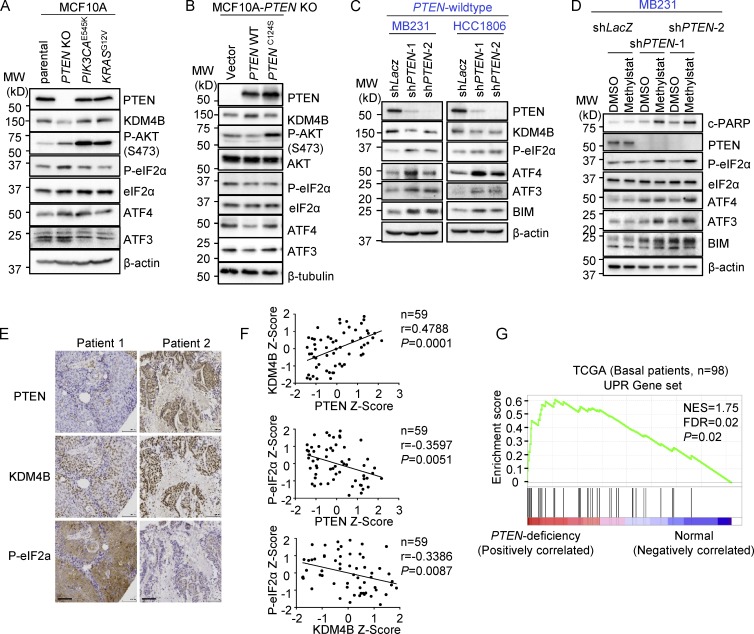
Given that PTEN deficiency confers susceptibility to Methylstat, we examined whether PTEN deficiency causes intrinsic activation of UPR, which results in cell sensitization to further stress. Interestingly, PTEN-KO MCF10A cells, when compared with PIK3CAE545K or KRASG12V cells, showed down-regulation of KDM4B, but enhanced UPR activity, as shown by increased P-eIF2α, ATF4, and ATF3 (Fig. 4 A). This finding suggests that KDM4B down-regulation and UPR activation are associated with PTEN loss. Furthermore, reconstitution of a wild-type PTEN, but not the phosphatase-deficient PTEN, in PTEN-KO cells restored expression of KDM4B and abolished UPR activation (Fig. 4 B). PTEN knockdown in PTEN wild-type MB231 and HCC1806 cells resulted in UPR activation (Fig. 4 C), which conferred sensitivity to further UPR activation, BIM induction, and PARP cleavage in response to Methylstat treatment (Fig. 4 D). To investigate how PTEN regulates the level of KDM4B, a wider panel of TNBCs with different PTEN status was studied. Although the down-regulation of KDM4B protein was founded in PTEN-deficient TNBC cells (Fig. S3 A), it was not because of differences in KDM4B stability, since the half-life of KDM4B remained the same when PTEN was knocked down/out (Fig. S3 B). Quantitative PCR analysis revealed that the mRNA of KDM4B was down-regulated after PTEN depletion (Fig. S3 C) as well as in PTEN-deficient TNBCs (Fig. S3 D). This effect of PTEN on KDM4B mRNA may be related to its chromatin regulation function (Chen et al., 2014b; Benitez et al., 2017).
Figure 4.
PTEN loss results in down-regulation of KDM4B and activation of UPR. (A) Western blot analysis of the UPR pathway in indicated isogenic MCF10A cell lines. MW, molecular weight. (B) Western blot analysis of UPR in PTEN-KO–MCF10A cells with ectopic expression of wild-type PTEN and PTEN-C124S proteins. (C) Western blot analysis of PTEN wild-type MB231- and HCC1806-expressing PTEN shRNAs. (D) Western blot analysis of the UPR pathway in MB231 cells expressing PTEN shRNAs treated with vehicle or Methylstat (2.5 µM) for 24 h. (E) Representative IHC staining of PTEN, KDM4B, and P-eIF2α in a TNBC tumor specimen. Scale bars, 100 µm. (F) Correlation analysis of IHC staining in tissue microarray between PTEN, KDM4B, and P-eIF2α in TNBC patients (n = 59). Pearson correlation coefficient and two-tailed P values are shown. (G) GSEA was performed with UPR gene signatures (which was implicated in protein folding or translational control and ER-associated protein degradation; see Table S3) on TCGA dataset where PTEN deficiency–related genes were ordered from largest to smallest in 98 basal patients. Basal patients with deep deletion, mutation, mRNA down-regulation, and/or protein down-regulation of PTEN were defined as PTEN deficient; the rest were defined as PTEN wild-type (PTEN deficiency, n = 27; PTEN wild-type, n = 71). NES, normalized enrichment score; FDR, false discovery rate. See also Fig. S3 and Table S3. All Western blot data are representative of three independent experiments.
To investigate the correlation among PTEN, KDM4B, and UPR in clinical breast cancer samples, we performed immunohistochemistry (IHC) analysis of a set of 59 TNBC tissue microarray samples. The results showed that loss of PTEN was often accompanied by reduced expression of KDM4B but elevated P-eIF2α (Fig. 4 E), with a strong positive correlation between PTEN and KDM4B and a reverse correlation with P-eIF2α (Fig. 4 F). Further GSEA of a cohort of TNBC-enriched basal breast patients from The Cancer Genome Atlas (TCGA) database consisting of 71 PTEN wild-type and 27 PTEN-deficient tumor samples revealed that the UPR gene signature was significantly enriched in PTEN-deficient tumors compared with PTEN wild-type tumors (Fig. 4 G). Together, these findings demonstrated that PTEN deficiency is associated with reduced KDM4B expression but augmented UPR activity, conferring enhanced susceptibility to further UPR activation upon Methylstat treatment.
KDM4B inhibition synergizes with a PI3K inhibitor to induce further UPR activation, leading to robust apoptosis of PTEN-deficient TNBC
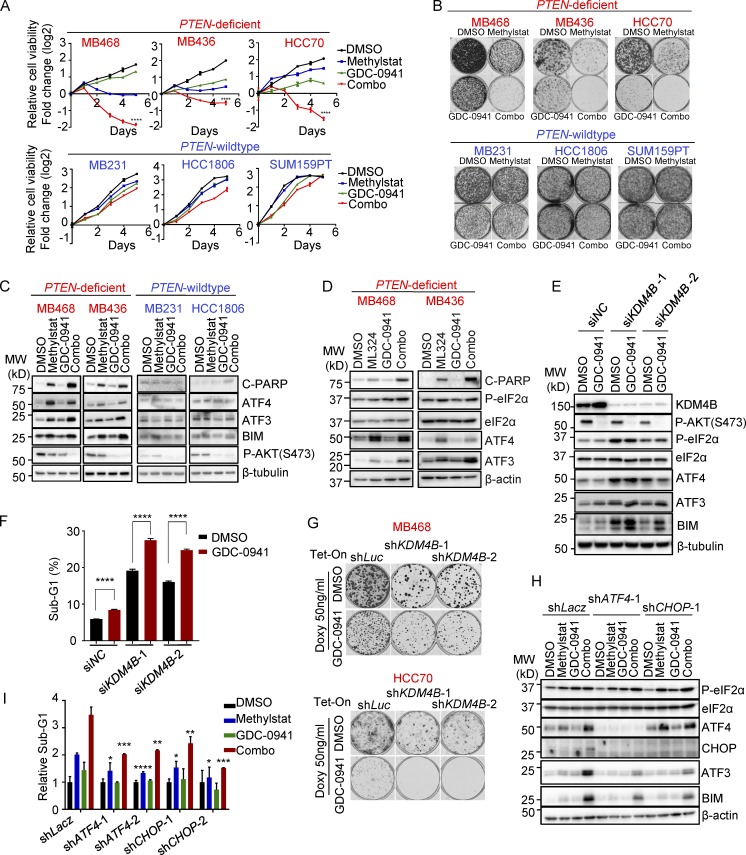
Since PTEN-deficient TNBC cells are susceptible to KDM4B inhibition but resistant to PI3K inhibitors, we tested the potential benefit of coinhibition of KDM4B and the PI3K pathway in these cells. As shown in multiple TNBC cell lines, the two drugs synergized, and the combination treatment consistently exhibited robust impairment of cell viability (Fig. 5 A) and colony formation (Fig. 5 B), as well as triggering cell death in PTEN-deficient TNBC cells (Fig. S4 A) but not PTEN wild-type TNBC cells. In accordance with the cellular phenotype, the combination treatment resulted in enhanced PARP cleavage and UPR activation compared with the single treatment in PTEN-deficient TNBC cells (Fig. 5 C), which was also supported by increased expression of UPR downstream target genes as assessed by RT-PCR (Fig. S4 B). ML324 exhibited a similar effect as Methylstat when combined with GDC-0941 (Fig. 5 D and Fig. S4 C), confirming that this robust combination effect in PTEN-deficient TNBC cells was not unique to Methylstat, but also applies to other KDM4B inhibitors. Similar to pharmacologic inhibition, we also confirmed that genetic depletion of KDM4B, when combined with GDC-0941, also yielded synergistic activation of UPR (Fig. 5 E), as well as cell death (Fig. 5 F) and impaired colony formation (Fig. 5 G and Fig. S4 D) selectively in PTEN-deficient TNBC cells. Interestingly, the synergistic effect of combination treatment was only found on the downstream targets of ATF4 (ATF3 and BIM), indicating that ATF4 is the key node to trigger the synergistic effect.
Figure 5.
Methylstat and GDC-0941 synergize to boost UPR and cell death in PTEN-deficient TNBC cells. (A) Cellular response to Methylstat (2.5 µM), GDC-0941 (1 µM), or both in indicated cell lines as determined by a CellTiter-Glo Luminescent Cell Viability Assay. Representative data are expressed as log2-fold change normalized to day 0 of three technical replicates. (B) Colony formation assay in cells treated as indicated. Cells were stained with crystal violet when DMSO-treated cells were confluent. (C) Western blot analysis of UPR in PTEN-deficient and PTEN wild-type TNBC cells treated with Methylstat and indicated drugs for 48 h. MW, molecular weight. (D) Western blot analysis of UPR in PTEN-deficient TNBC cells treated with ML324 and indicated drugs for 48 h. (E) Western blot analysis of UPR in MB436 cells transfected with indicated KDM4B siRNAs followed by treatment of GDC-0941 for 48 h. (F) Cell death determined by sub-G1 analysis in MB436 cells in E. (G) Colony formation assay in PTEN-deficient TNBC cells expressing inducible shRNAs. Cells growing in medium with doxycycline were treated with DMSO and GDC-0941 until control cells were confluent, and then colonies were stained with crystal violet. (H) Western blot analysis of MB436 cells expressing inducible shRNAs grown in medium containing doxycycline for 48 h followed by drug treatment for another 48 h. (I) Cell death presented as sub-G1 in MB436 cells, as in H. Drug treatment: 2.5 µM Methylstat, 1 µM GDC-0941, or in combination. See also Fig. S4. All data are representative of three independent experiments unless stated otherwise. Data are expressed as mean ± SD. P values were determined by two-tailed unpaired Student’s t test; * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001.
To confirm the role of UPR activation elicited by the combination treatment in killing PTEN-deficient cells, we showed that, indeed, inducible knockdown of ATF4 or CHOP, the two key factors mediating UPR-induced cell death, not only weakened downstream activation of ATF3 and BIM in response to the combination treatment (Fig. 5 H) but also significantly reduced apoptotic cell death (Fig. 5 I), reinforcing the pro-apoptotic role of UPR activation in this treatment. Collectively, our data revealed that the co-targeting KDM4B and PI3K provided a remarkable combinatorial effect on UPR activation, leading to marked apoptosis of PTEN-deficient TNBC cells.
ATF4 and FoxO1 nuclear translocation converge to induce UPR target gene activation for apoptosis upon combined Methylstat and GDC-0941 treatment
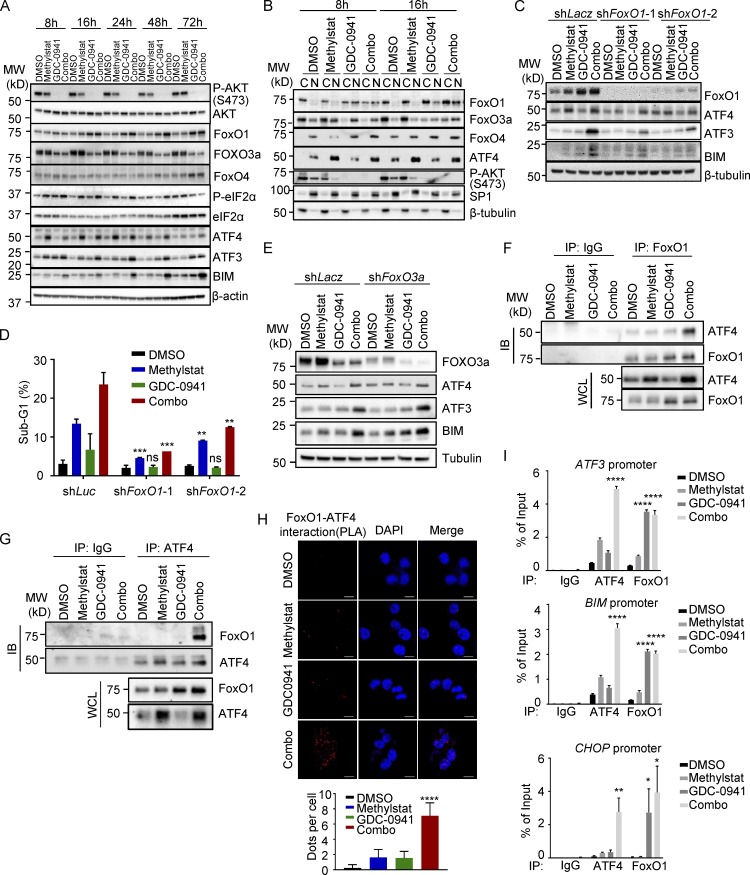
As discussed before, Western blot analysis of the UPR cascade revealed that the combination of Methylstat and GDC-0941 induced a synergistic effect on ATF3 and BIM but did not affect P-eIF2α and ATF4 (Fig. 5, C and E), indicating that the synergistic effect on UPR activation occurred downstream of ATF4. Hence, we hypothesized that a transcription factor responsive to PI3K–AKT inhibition might be engaged to cooperate with ATF4 to activate the UPR downstream genes. FoxO family members are among the most important transcription factors regulated by the PI3K–AKT pathway and are involved in anti-apoptosis and ER stress response (Martinez et al., 2008; Ghosh et al., 2012), and FoxO1 has been found to cooperate with ATF4 to regulate bone formation in osteoblasts (Rached et al., 2010). We thus monitored the dynamic changes of FoxO family members FoxO1, FoxO3a, and FoxO4 along with the UPR pathway in response to Methylstat, GDC-0941, or combination treatment for 8–72 h. FoxO1, but not other FoxO members, was the only one showing synergistic induction along with ATF3 and BIM upon combined treatment, though P-eIF2α and ATF4 were unchanged by treatment with GDC-0941 alone or combination treatment (Fig. 6 A). Moreover, FoxO1 displayed marked nuclear translocation and accumulation in response to GDC-0941 or combination treatment (Fig. 6 B). In contrast, although FoxO3a also showed nuclear translocation upon GDC-0941 treatment, it did not show increased accumulation upon combination treatment (Fig. 6 B), suggesting that FoxO1 may be the candidate transcription factor contributing to the induction of UPR downstream targets such as ATF3 and BIM. Indeed, FoxO1 knockdown effectively impaired expression of ATF3 and BIM, as well as cell death (Fig. 6, C and D), whereas FoxO3a knockdown failed to do so (Fig. 6 E), confirming that FoxO1 plays a crucial role in causing cell death in response to the combination treatment.
Figure 6.
FoxO1 and ATF4 cooperate to directly activate UPR pro-apoptotic gene expression. (A) Western blot analysis of MB436 treated with Methylstat, GDC-0941, or in combination for the indicated hours. MW, molecular weight. (B) Subcellular fractionation and Western blot analysis of MB436 treated with the indicated drugs for 8 and 16 h. (C) Western blot analysis of MB436 cells with induced knockdown of FoxO1 treated with Methylstat, GDC-0941, and in combination for 48 h. (D) Cell death presented as sub-G1 was assessed by flow cytometry in MB436 cells from C. (E) Western blot analysis of MB436 cells with induced knockdown of FoxO3a treated with Methylstat, GDC-0941, and in combination for 48 h. (F and G) Coimmunoprecipitation study of FoxO1 and ATF4 in MB436 treated with the indicated drugs for 24 h using FoxO1 (F) and ATF4 (G) antibody, respectively. IB, immunoblotting; WCL, whole cell lysate; IP, immunoprecipitation. (H) Indicated drug–treated MB-436 cells were stained with specific antibodies against ATF4 (mouse) and FoxO1 (rabbit) before PLA. The PLA signals between endogenous ATF4 and FoxO1 are shown in the red channel; DAPI was used to stain the nuclei (blue); and the merged images show the overlay of the red and blue channels. Dots per cells were analyzed from two independent experiments. Scale bars, 10 µm. (I) ChIP-PCR analysis of ATF4 and FoxO1 enrichments on ATF3, BIM, and CHOP promoters in MB436 cells treated with Methylstat (2.5 µM), GDC-0941 (1 µM), or in combination for 24 h. All data are representative of at least three independent experiments unless stated otherwise. P values were determined by two-tailed unpaired Student’s t test; ns P > 0.05, * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001.
Mechanistically, we further demonstrated that ATF4 was able to interact with FoxO1 through coimmunoprecipitation experiments, which was more evident under the conditions of the combination treatment (Fig. 6, F and G). Furthermore, a direct nuclear interaction between ATF4 and FoxO1 was supported by PLAs (Fig. 6 H). These data raised the possibility that ATF4 potentially cooperated with FoxO1 to directly bind to the UPR downstream target promoters such as ATF3, BIM, and CHOP. To validate this theory, we performed chromatin immunoprecipitation (ChIP) analysis using ATF4 or FoxO1 antibody in MB436 cells. As expected, ATF4 directly bound to the promoters of ATF3, BIM, and CHOP in response to Methylstat treatment, whereas FoxO1 bound these promoters mainly upon GDC-0941 treatment (Fig. 6 I). Of note, once FoxO1 was enriched, the binding of ATF4 to these promoters was boosted robustly under the combination treatment (Fig. 6 I), indicating that nuclear localization of FoxO1 facilitated the recruitment of ATF4 to these gene promoters. Indeed, the depletion of FoxO1 attenuated the binding of ATF4 to these promoters under GDC-0941 and combination treatment (Fig. S4 E). Taken together, our findings demonstrated that ATF4 induction by KDM4B inhibition cooperates with FoxO1, which is activated by PI3K inhibition to induce a robust activation of UPR downstream pro-apoptotic target genes, leading to strong apoptosis.
Co-targeting KDM4B and PI3K in PTEN-deficient TNBC xenograft tumor models
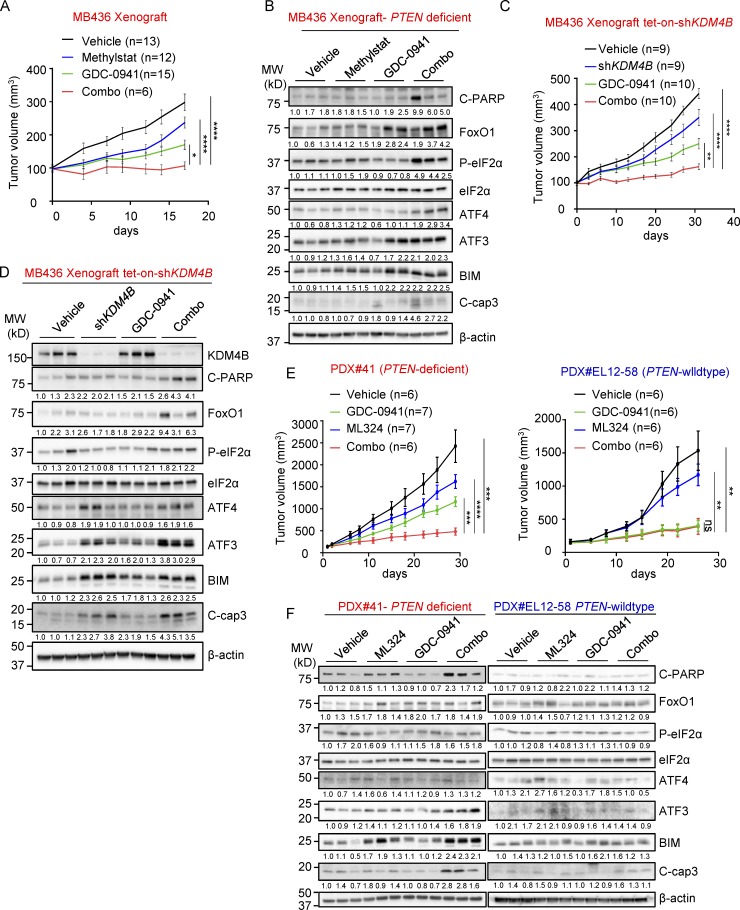
To assess the potential value of co-targeting KDM4B and PI3K in vivo, we tested Methylstat and GDC-0941 alone or in combination in mice engrafted with PTEN-deficient MB436 cells. Methylstat and GDC-0941 alone resulted in tumor suppression, an effect markedly enhanced by their combination (Fig. 7 A and Fig. S5, A and C). Consistent with tumor suppression, combination treatment resulted in prolonged UPR activation and induced apoptosis in MB436 xenografts (Fig. 7 B). Similarly, using MB436 cells with doxycycline-inducible knockdown of KDM4B, KDM4B knockdown upon doxycycline treatment also substantially suppressed tumor growth and decreased tumor mass when combined with GDC-0941 (Fig. 7 C and Fig. S5 D), accompanied by up-regulation of the UPR pathway and increased cell apoptosis in tumor samples (Fig. 7 D and Fig. S5 G). To expand the study, we also employed two patient-derived xenograft (PDX) models: PDX#41 (PTEN-deficient) and PDX#EL12-58 (PTEN wild-type), and tested them with ML324 and GDC-0941. The result showed the combinatory effect on PDX#41 as expected. However, PDX#EL12-58 was highly sensitive to GDC-0941, and therefore the combination treatment did not further reduce the tumor volume (Fig. 7 E and Fig. S5, B, E, and F). Further analysis showed that combination treatment led to an obvious UPR activation and apoptosis induction in PDX#41 but not in PDX#EL12-58 (Fig. 7 F and Fig. S5 G), which is in line with our findings in vitro. Taken together, these in vivo findings demonstrated that co-targeting KDM4B and PI3K could be a potential strategy in treating PTEN-deficient TNBC.
Figure 7.
The effect of Methylstat and GDC-0941 combination in vivo. (A) NOD-SCID mice bearing MB436 xenografts were treated with vehicle (n = 13) and Methylstat (30 mg/kg; n = 12) by intraperitoneal injection twice a week, GDC-0941 (100 mg/kg; n = 15) by oral gavage daily, or both drugs (n = 6) for 2 wk. Tumor volumes were measured twice a week. (B) Western blot analysis of representative tumors from each experimental group in A. MW, molecular weight. (C) NOD-SCID mice bearing MB436 (TetOn-shKDM4B) xenografts were fed with diets of doxycycline or treated with GDC-0941 (100 mg/kg) by oral gavage daily or in combination as indicted for 1 mo. Tumor volumes were measured twice a week. Vehicle (n = 9), doxycycline (n = 9), GDC-0941 (n = 10), combination (n = 10). (D) Western blot analysis of representative tumors from each experimental group in C. (E) NOD-SCID mice bearing PTEN-deficient PDX#41 were treated with vehicle (n = 6) and ML324 (60 mg/kg; n = 7) by intraperitoneal injection daily, GDC-0941 (100 mg/kg; n = 7) by oral gavage daily, or both drugs (n = 6) for 2 wk. NOD-SCID mice bearing PTEN wild-type PDX#EL12-58 were treated with vehicle (n = 6), ML324 (60 mg/kg; n = 6), GDC-0941 (100 mg/kg; n = 6), or both drugs (n = 6) for 2 wk. Tumor volumes were measured twice a week. (F) Western blot analysis of representative tumors from each experimental group in E. See also Fig. S5. Data are presented as mean ± SEM. P values were determined by two-tailed unpaired Student’s t test; ns P > 0.05, * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001.
Discussion
The discovery of effective targeted therapies for treating TNBCs remains an unresolved clinical challenge. PTEN-deficient tumors comprise a large proportion of TNBC. Thus, efforts to identify synthetic-lethal vulnerabilities of PTEN deficiency is of great importance in TNBC therapy (Mendes-Pereira et al., 2009; Mason et al., 2014; Zhao et al., 2017).
We have defined, through chemical screening and functional characterization, a pathway comprising two distinct molecular nodes that cooperate to generate effective induction of apoptosis in PTEN-deficient TNBC. This pathway centers on a convergent generation of UPR signals to achieve a pro-apoptotic effect. Not surprisingly, in cancer cells, the UPR pathway serves as an adaptive response pathway to sustain protein homeostasis and protect cells from an over-reactive stress environment, but when hyperactivated, this mechanism can be destructive. Consistent with our findings, it has been reported that some aggressive TNBCs show a consistently high level of UPR (Chen et al., 2014a). Therefore, the intrinsically higher level of UPR found in PTEN-deficient TNBC invariably causes increased vulnerability to further UPR inducers.
In contrast to the previously reported oncogenic functions of KDM4B relying on the nuclear role of histone regulation (Beyer et al., 2008; Kawazu et al., 2011; Shi et al., 2011; Young et al., 2013; Berry et al., 2014), our study shows a nonchromatin effect of KDM4B in tumorigenesis that is achieved by regulating UPR through cytoplasmic interactions with eIF2α. Recent studies have also suggested the cytoplasmic role of KDM4B in influencing androgen receptor degradation (Coffey et al., 2013) and the cytoplasmic role of KDM4A associated with translation machinery that affects the initiation and protein synthesis (Van Rechem et al., 2015). We show that KDM4B demethylase activity is critically required in this context, and its inhibition by Methylstat can elicit apoptosis through hyperactivation of UPR. This differs from previous reports indicating that targeting KDM4B inhibits tumorigenesis in hormonally responsive breast cancers (Kawazu et al., 2011; Shi et al., 2011), in which it acts as a cofactor to mediate expression of ER target genes through chromatin modification. However, it remains unclear whether eIF2α is a direct substrate of KDM4B demethylase. The mechanism by which KDM4B demethylase activity functions in this process needs to be further investigated.
Herein, we show that KDM4B inhibition produced marked apoptosis when combined with a PI3K inhibitor that targets aberrant activation of PI3K–AKT but yielded meager clinical benefit alone. Strikingly, the combination showed a significant suppressive effect on tumors both in vitro and in vivo due to its greatly enhanced lethal UPR signal. FoxO1, which was shown to contribute to the synergistic effect of the combination, provides an additional signal axis to boost ATF4-mediated UPR signal. FoxO family members are known to be targeted by the PI3K pathway and regulate apoptosis, in part through nuclear translation, where it induces genes involved in apoptosis (Martinez et al., 2008). Our study shows, for the first time, that FoxO1, but not other members of the FoxO family, participate in the UPR as a cofactor of ATF4 to regulate UPR downstream gene promoters, resulting in a boost of lethal UPR signal including CHOP, ATF3, and BIM. Thus, the discovery of the specific role of KDM4B-mediated UPR in the interplay with PI3K signaling provides a rationale for combination therapies with PI3K inhibitors in PTEN-deficient breast cancer.
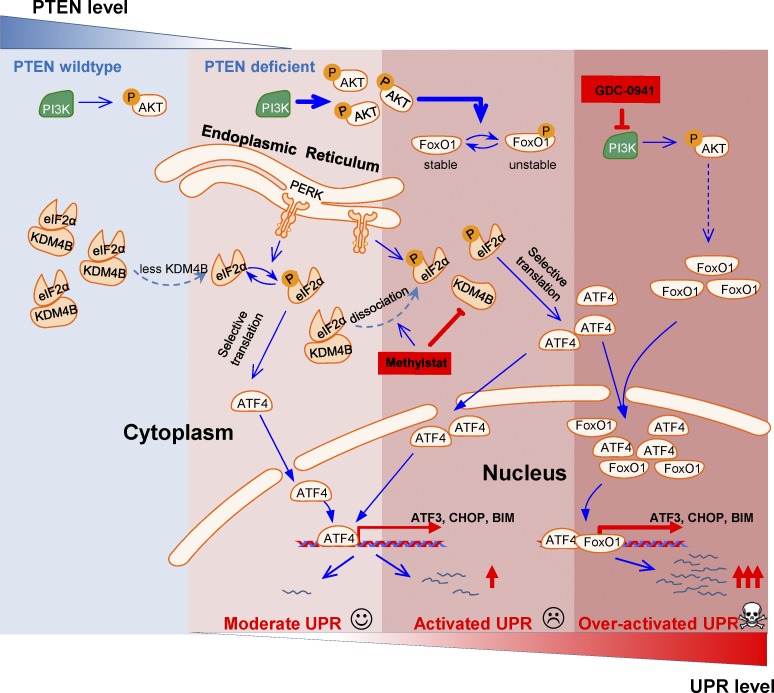
Our study suggests that the level of KDM4B might dictate the threshold of UPR activation for triggering apoptosis such that PTEN-deficient TNBC has an enhanced ability to undergo UPR-mediated apoptosis upon further inhibition of its activity. Moreover, in the context of PI3K inhibition, the UPR signal is further enhanced due to FoxO1 activation, leading to even higher susceptibility to apoptosis (Fig. 8). Thus, our results illustrate an approach for stepwise activation of UPR to trigger apoptosis in PTEN-deficient TNBC, which might be broadly exploited in other cancers with high frequencies of PTEN deficiency, such as prostate cancer and glioblastoma.
Figure 8.
Schematic representation of the proposed mechanisms of over-activation of UPR to trigger apoptosis. Stepwise activation of the UPR pathway by PTEN deficiency, KDM4B inhibition, and PI3K/AKT inhibition leads to cell death in breast cancer cells.
Materials and methods
Cell culture and drug treatments
All breast cancer cell lines were from ATCC. MCF10A parental (catalog no. HD PAR-021) and MCF10A PTEN-KO (catalog no. HD 101–006) cells were purchased from Horizon. MDA-MB-436, MB468, MB231, BT549, HS578T, and BT-20 cells were grown in DMEM with 10% FBS. HCC1806, HCC70, and HCC1937 were cultured in RPMI medium supplemented with 10% FBS. SUM159PT cells were cultured in Ham’s F12 medium complemented with 5% FBS, 1 µg/ml hydrocortisone, and 0.2 UI/ml insulin. Human mammary epithelial cells and MCF10A normal breast epithelial cell lines were cultured in DMEM/F12 with 5% horse serum, 20 ng/ml epidermal growth factor, 0.5 mg/ml hydrocortisone, 100 ng/ml cholera toxin, and 10 µg/ml insulin. All media were also supplemented with 5,000 U/ml penicillin/streptomycin. Cells were grown in a 37°C humidified, 5% CO2 atmosphere incubator.
Methylstat (Merck; 420303; or Abcam; ab144566), GDC-0941 (Axon Medchem; Axon-1377), and ML324 (Cayman Chemical; 1222800-79-4) were used at indicated concentrations.
Drug screening
MCF10A parental and MCF10A PTEN-KO cells were used for drug screening. 1,000 cells were seeded per well, in triplicate, in 96-well plates 24 h before drug treatment. A compound library containing 140 small molecules were used for the screening. After 4 d of treatment, cell viability was measured using a CellTiter-Glo Luminescent Cell Viability Assay (Promega; G7573). All values were normalized to that of DMSO treatment at day 4. The value of individual compounds was then transformed to z-score values by the formula
where X stands for normalized value, μ stands for mean, and σ stands for SD. The cutoff z-score was set at –3.
Gene expression profiling and quantitative PCR analysis
MB468, HCC70, and MB231 cells were harvested after 24 h of drug exposure (DMSO and 2.5 µM Methylstat). Total RNA was isolated and purified using the RNeasy Mini Kit (Qiagen). Microarray hybridization was performed using Human HT-12 V4.0 Expression Beadchip (Illumina). GeneSpring software (Agilent Technologies) was used for data analysis. Differential gene expression was determined by 1.5-fold cutoff and statistical analysis (P ≤ 0.05). Differentially expressed gene sets between control and Methylstat-treated groups were subjected to IPA for gene ontology analysis. RT-PCR assays were performed using a High Capacity cDNA Archive Kit and KAPA SyBr Fast qPCR Kit (KAPA Biosystems). Quantitative PCR was performed using the Applied Biosystems PRISM 7500 Fast Real-Time PCR system. For quantification of mRNA levels, β-actin was used as internal controls. Real-time primer sequences are shown in Table S4.
Immunoblotting
All immunoblotting analyses were performed following SDS-PAGE using standard methods. Antibodies used for immunoblotting were KDM4B (Cell Signaling; #8639, 1:1,000 dilution), cleaved PARP (Cell Signaling; #9541, 1:1,000 dilution), ATF4 (Cell Signaling; #11815, 1:1,000 dilution), p-AKT-S473 (Cell Signaling; #4058, 1:1,000 dilution), pan-AKT (Cell Signaling; #4691, 1:1,000 dilution), P-eIF2α (Cell Signaling; #9721, 1:1,000 dilution), PTEN (Cell Signaling; #9188, 1:1,000 dilution), CHOP (Cell Signaling; #2895, 1:500 dilution), eIF2α (Cell Signaling; #9722, 1:1,000 dilution), FoxO1 (Cell Signaling; #2880, 1:1,000 dilution), BIM (Cell Signaling; #2819, 1:1,000 dilution), ATF3 (Santa Cruz; sc-188, 1:1,000 dilution), H3K9Me3 (Millipore; #07-422, 1:2,000 dilution), H3K36Me3 (Cell Signaling; #9763, 1:2,000 dilution), H3 (Cell Signaling; #3638, 1:1,000 dilution), β-actin (Sigma-Aldrich; A1978, 1:20,000 dilution), SP1 (Millipore; #07-645, 1:1,000 dilution), and β-tubulin (Sigma-Aldrich; T4026, 1:2,000 dilution).
Colony formation assay
For monolayer cell colony formation assays, 3,000 cells were seeded on 6-well plates with DMSO or indicated drugs. Cells were grown for 2–3 wk until DMSO-treated cells were confluent. The surviving colonies were stained with 0.5% crystal violet after methanol fixation. For anchorage-independent growth, cells were suspended in the top layer of complete medium containing 0.3% agar or complete medium with 0.5% methylcellulose and were then layered onto complete medium containing 0.6% agar in 6-well plates. After 2 wk, colonies were stained with iodonitrotetrazolium chloride (Sigma-Aldrich) overnight.
siRNAs, shRNAs, viral infection, and overexpression
All siRNAs and negative controls were synthesized by Sigma-Aldrich. siRNA transfections were performed using Lipofectamine RNAimax (Thermo Fisher Scientific) following the manufacturer’s instructions. The target sequences of siRNAs are shown in Table S5. For shRNA delivery, pLV-H1-EF1α-RFP-Puro (Biosettia; SORT-B31) or pLV-H1TetO-RFP-Puro (Biosettia; SORT-B31) were used as backbones for constitutive knockdown or inducible knockdown, respectively. Anti-Luc shRNA was used as a negative control. Virus particles for individual shRNAs were produced in 293T cells. Briefly, pLV shRNA plasmids, together with psPAX2 plasmid (Addgene) and pMD2.G plasmid (Addgene; the ratio of pLV:psPAX2:pMD2.G is equal to 4:3:1), were transfected into HEK293T cells using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Medium containing transfection reagent was removed 6 h after transfection and replaced with fresh medium. For infection, indicated cells were seeded into 10-cm dishes and allowed to grow for 24 h. Lentiviral particles were added to cells along with polybrene (8 μg/ml, Sigma-Aldrich) and 48 h later selected with puromycin. For doxycycline treatment, inducible cells were treated with doxycycline to a final concentration of 50 ng/ml. The target sequences of shRNAs are shown in Table S5.
For gene overexpression, the pMN-GFP retroviral expression vector (a gift from Dr. Linda Penn, University of Toronto, Toronto, Canada) was used. To generate the KDM4B wild-type and mutants overexpressing plasmids, KDM4B and its mutants were amplified from cDNA of MCF10A cells by PCR and inserted into the pMN-GFP vector. Virus particles were produced in Plat A cells.
Cell proliferation assay and flow cytometry
Cell proliferation assays and flow cytometry were performed as described previously (Wee et al., 2015). Briefly, an optimal number of cells were seeded 24 h before drug treatment in triplicate. Cell viability was measured using a CellTiter-Glo Luminescent Cell Viability Assay (Promega; G7573) daily for 6 d. All values were normalized to the value of day 0 and then transformed to log2 values.
For flow cytometry, cells were washed with PBS buffer twice, fixed in 70% ethanol, and stained with propidium iodide (50 mg/ml) after RNase treatment at room temperature for 30 min. The resuspended cells were then subjected to FACS analysis by FACScalibur or LSRFortessa X-20 (BD Biosciences), and data were analyzed by FACSDiva software (BD Biosciences).
Cell fractionation
Cells were washed twice with PBS buffer and collected by centrifugation for 5 min at 600 g. Cell pellets were resuspended in hypotonic buffer (25 mM Tris-HCl, pH 7.4, 1 mM MgCl2, and 5 mM KCl) and kept on ice for 5 min. An equal volume of hypotonic buffer containing 1% NP-40 was added and kept on ice for another 5 min. Lysed cells were then centrifuged at 5,000 g for 5 min, and the supernatant containing the cytoplasmic fraction was collected. Nuclei were resuspended in nuclear resuspension buffer (20 mM Hepes, pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, and 1 mM PMSF) for further incubation at 4°C for 30 min. Nuclear fraction was collected after removing insoluble membrane debris by centrifugation at 12,000 g for 10 min. All buffers contained a protease inhibitor cocktail and phosphatase inhibitor cocktail (Roche Diagnostics).
Coimmunoprecipitation
For immunoprecipitation, the following antibodies were used: KDM4B (Cell Signaling; #8639, 1:200 dilution), eIF2α (Santa Cruz; sc-133132, 1:200 dilution), PERK (Cell Signaling; #5683, 1:200 dilution), and P-eIF2α (Cell Signaling; #3398, 1:200 dilution). Coimmunoprecipitation assays were performed as described previously (Wee et al., 2015) with a modified protocol using NETN buffer (25 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 10% glycerol, and 0.3% NP-40) containing protease inhibitor cocktail and phosphatase inhibitor cocktail (Roche Diagnostics) to extract whole-cell protein, or a cell fractionation protocol for cytoplasmic and nuclear fractionation. Cell lysates were incubated with the indicated antibodies at 4°C overnight. The protein complex was captured using protein A–agarose (for rabbit primary antibody) or protein G–agarose (for mouse primary antibody) beads (Roche Diagnostics) at 4°C for 4 h, and agarose beads were collected by centrifuge and washed three times with washing buffer. The precipitated proteins were dissolved in SDS sample buffer along with 3 mM dithiothreitol and subjected to Western blot analysis.
Liquid chromatography–tandem mass spectrometry analysis
The proteins that specifically interact with KDM4B were pulled down by flag-KDM4B in MB436 cells and were separated using SDS-PAGE followed by Coomassie blue staining (Thermo Fisher Scientific; #24615). Then, these proteins were identified by liquid chromatography–tandem mass spectrometry analysis, which was performed by PTM Biolabs Inc.
ChIP
For ChIP, the following antibodies were used: ATF4 (Santa Cruz; sc-200x; 1:100 dilution) and FoxO1 (Abcam; ab39670, 1:100 dilution). ChIP assay was performed as described previously (Lee et al., 2011). Briefly, 1 × 107 cells were cross-linked using 1% formaldehyde for 10 min, and reactions were quenched by addition of 0.125 M glycine for 5 min at room temperature. Cell pellets were washed three times with cold TBSE buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1 mM EDTA). Cells were lysed in SDS lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 1% SDS) for 5 min on ice. Chromatin fragmentation was performed using a Branson Digital Sonifier (25% amplitude, 10 s on and 20 s off for 16 cycles) to achieve a DNA shear length of 200–500 bp. Solubilized chromatin was diluted 10 times in dilution buffer (10 mM Tris-HCl, pH 7.4, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.01% SDS) and precleared with Dynabead (Life Technologies) for 4 h at 4°C. Precleared chromatin was then incubated with appropriate antibodies overnight at 4°C. The complexes of chromatin and antibodies were captured with Dynabead at 4°C for 2 h. Immune complexes were then washed three times with dilution buffer and once with TE buffer (10 mM Tris-HCl, pH 7.5, and 1 mM EDTA), sequentially. Elution and reverse cross-linking were performed in elution buffer (50 mM Tris-HCl, pH 7.5, 10 mM EDTA, and 1% SDS) at 68°C with vortex overnight followed by digestion with proteinase K (20 mg/ml) at 42°C for 2 h. Eluted DNA was purified using the E.Z.N.A. Cycle Pure Kit (Omega). Precipitated chromatin DNA was analyzed by real-time quantitative PCR using the KAPA SyBr Fast qPCR kit (KAPA Biosystems). Quantification of ChIP results was performed relative to the input amount. Primer sets are designed to amplify ∼150 bp around the indicated region shown in Table S4.
PLA
PLAs were performed with a Duolink In Situ Red Starter Kit (Sigma-Aldrich; DUO92101) according to the manufacturer’s instructions. The antibodies used for PLA were KDM4B (Cell Signaling; #8639, 1:100), eIF2α (Santa Cruz; sc-133132, 1:100), ATF4 (Santa Cruz; sc-390063, 1:100), and FoxO1 (Abcam; ab39670, 1:100).
IHC staining
Human breast cancer tissue microarray, BR10011a, was purchased from US Biomax. The antibodies used for IHC were KDM4B (Cell Signaling; #8639, 1:200 dilution, pH 9.0), PTEN (Cell Signaling; #9559, 1:200 dilution, pH 9.0), P-eIF2α (Cell Signaling; #3398, 1:50 dilution, pH 9.0), and ATF3 (Abcam; ab58668, 1:50 dilution, pH 9.0). IHC staining and image analysis were performed by the Histopathology Department of the Institute of Molecular and Cell Biology, Agency for Science, Technology, and Research (A*STAR), Singapore. Slides were scanned using a Leica SCN400 slide scanner (Leica Microsystems). Tissue microarray cores were analyzed using the Measure Stained Cells algorithm of Slidepath Tissue IA software (Leica Microsystems). IHC staining of individual proteins was quantified by the H-score method, in which 0, 1+, 2+, and 3+ indicate negative, weak, moderate, and strong staining, respectively. The percentage of cells at each staining was calculated, and the H-score was assigned by the following formula: [1 × (% cells 1+) + 2 × (% cells 2+) + 3 × (% cells 3+)]. The H score, ranging from 0 to 300, was transformed to z-score value by the formula
where X stands for H-score value, μ stands for mean, and σ stands for standard deviation.
GSEA
For analysis of Methylstat-treatment expression data, Tm, Tg, and doxycycline gene signatures were used as described previously (Feng et al., 2014). The top 100 genes induced by Tm (GEO accession no. GSE24500), Tg (GEO accession no. GSE24500), or doxorubicin (GEO accession no. GSE39042) were defined as Tm, Tg, and doxycycline gene signatures, respectively.
For analysis of PTEN-deficient gene expression data, all patient data (Cancer Genome Atlas Network, 2012) were downloaded from the Cancer Genome Browser. All basal patients were chosen for further analysis according to the clinical data regarding subtypes extracted from the TCGA BrCA dataset. The patients with deep deletion, missense mutation, inframe mutation, truncating mutation, protein down-regulation, and mRNA down-regulation of PTEN were defined as PTEN deficient. A UPR-related gene signature consisting of 39 genes responsible for protein folding, translational control, and ER-associated protein degradation was used (Lecca et al., 2005) for enrichment analysis. The gene list can be found in Table S3.
Animal experiments
NOD/SCID mice were purchased from InVivos. All animal studies were conducted in compliance with animal protocols approved by the A*STAR-Biopolis Institutional Animal Care and Use Committee of Singapore. 6–8-wk-old NOD/SCID mice were used for tumor engraftment. 1 × 106 MB436 cells or MB436 cells carrying doxycycline-inducible shKDM4B#1, suspended in PBS:Matrigel (BD Biosciences; catalog no. 354234) at a 1:1 ratio, were engrafted in the mammary fat pad of the mice. Treatment was started after the average of tumor volume reached 100 mm3, and mice were randomized.
For in vivo single and combinatorial drug tests, Methylstat (30 mg/kg) dissolved in DMSO:Cremophor:ethanol:PBS at a ratio of 1:2:2:5 was administered via intraperitoneal injection twice a week. ML324 (60 mg/kg) dissolved in DMSO:Cremophor:ethanol:PBS at a ratio of 1.5:2:2:4.5 was administered via intraperitoneal injection daily. GDC-0941 (100 mg/kg) dissolved in 10% DMSO, 5% Tween 20, and 85% water was given daily by oral gavage. For in vivo tumor suppression efficacy of KDM4B knockdown, GDC-0941, and in combination treatment, mice were given free access to diets with or without doxycycline (Bio-Serv; catalog no. S3888) or treated with GDC-0941 (100 mg/kg) daily or in combination as indicated. For the PDX model, each mouse was implanted with one PDX tumor (PDX#41 or PDX#EL12-58) on one side of the mammary fat pad. Treatment was started after the average of tumor volume reached 100 mm3, and mice were randomized.
Primary mammary tumors were measured by a vernier caliper twice a week, and tumor volume was calculated by the formula: V = W × W × L/2, where V is tumor volume, W is tumor width, and L is tumor length. Data are expressed as mean ± SEM. Mice were sacrificed by CO2 inhalation, and tumors were harvested for further analysis.
Statistical analysis
All statistical analyses were performed with GraphPad Prism version 7.0, and the differences were assessed by two-tailed Student’s t test. Correlation analyses were done using Spearman’s correlation coefficients in GraphPad Prism 7. In all statistical tests, the significance was accepted at P value ≤ 0.05. (ns P > 0.05, * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, and **** P ≤ 0.0001). For IPA, all ingenuity canonical pathways were included as customized pathways. A common gene list of up-regulated and down-regulated (1.5-fold cutoff, P ≤ 0.05) genes in both MB468 and HCC70, but not in MB231, upon Methylstat treatment generated from microarray was uploaded for IPA core analysis.
Accession nos.
The expression microarray dataset generated in this study has been deposited in the GEO repository under accession no. GSE103018.
Online supplemental material
Fig. S1 shows GSEA of Methylstat-induced UPR activation in PTEN-deficient TNBC cell lines. Fig. S2 shows that KDM4B regulates the UPR pathway through interaction with eIF2α. Fig. S3 shows that KDM4B down-regulation in PTEN-deficient cells is related to mRNA reduction but not protein stability. Fig. S4 shows that Methylstat and GDC-0941 synergize to induce cell death in PTEN-deficient TNBC cells through the UPR pathway. Fig. S5 shows that combination of KDM4B inhibition and PI3K inhibitor suppresses PTEN-deficient breast cancer growth in vivo. Table S1 contains a list of up- and down-regulated genes by Methylstat in MB468 and HCC70. Table S2 contains the most changed ATF4 targets under Methylstat treatment in HCC70 and MDA-MB468. Table S3 contains a gene list of UPR signature used in Fig. 4 G. Table S4 contains primers for quantitative PCR and ChIP-PCR used in this study. Table S5 contains siRNAs, shRNA constructs, and targeting sequences used in this study.
Supplementary Material
Acknowledgments
We thank the Histopathology Department from the Institute of Molecular and Cell Biology, Agency for Science, Technology and Research of Singapore (A*STAR), for their service in IHC staining and analysis.
This work was supported by A*STAR and the Singapore Ministry of Health’s National Medical Research Council Open Fund Individual Research Grants (NMRC/OFIRG/0023/2016 to Q. Yu). This work was also supported by an A*STAR Graduate Academy Singapore International Graduate Award scholarship to G. Oguz.
The authors declare no competing financial interests.
Author contributions: Q. Yu conceived and supervised the study. W. Wang contributed to the design, conduct, and interpretation of all experiments. G. Oguz performed bioinformatics and statistical analyses. W. Wang and P.L. Lee performed Western blot analyses. Y. Bao performed UPR analysis in MCF10A cells. P. Wang provided crucial reagents, and M.G. Terp and H.J. Ditzel provided the PTEN-deficient PDX model. W. Wang and Q. Yu wrote the manuscript with inputs from all co-authors.
References
- Benitez J.A., Ma J., D’Antonio M., Boyer A., Camargo M.F., Zanca C., Kelly S., Khodadadi-Jamayran A., Jameson N.M., Andersen M., et al. 2017. PTEN regulates glioblastoma oncogenesis through chromatin-associated complexes of DAXX and histone H3.3. Nat. Commun. 8:15223 10.1038/ncomms15223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry W.L., Kim T.D., and Janknecht R.. 2014. Stimulation of β-catenin and colon cancer cell growth by the KDM4B histone demethylase. Int. J. Oncol. 44:1341–1348. 10.3892/ijo.2014.2279 [DOI] [PubMed] [Google Scholar]
- Beyer S., Kristensen M.M., Jensen K.S., Johansen J.V., and Staller P.. 2008. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 283:36542–36552. 10.1074/jbc.M804578200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann S.M., Hofmann I., Schnell C., Fritsch C., Wee S., Lane H., Wang S., Garcia-Echeverria C., and Maira S.M.. 2009. Specific apoptosis induction by the dual PI3K/mTor inhibitor NVP-BEZ235 in HER2 amplified and PIK3CA mutant breast cancer cells. Proc. Natl. Acad. Sci. USA. 106:22299–22304. 10.1073/pnas.0905152106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490:61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Iliopoulos D., Zhang Q., Tang Q., Greenblatt M.B., Hatziapostolou M., Lim E., Tam W.L., Ni M., Chen Y., et al. 2014a XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 508:103–107. 10.1038/nature13119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.H., Zhu M., Yang J., Liang H., He J., He S., Wang P., Kang X., McNutt M.A., Yin Y., and Shen W.H.. 2014b PTEN interacts with histone H1 and controls chromatin condensation. Cell Reports. 8:2003–2014. 10.1016/j.celrep.2014.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey K., Rogerson L., Ryan-Munden C., Alkharaif D., Stockley J., Heer R., Sahadevan K., O’Neill D., Jones D., Darby S., et al. 2013. The lysine demethylase, KDM4B, is a key molecule in androgen receptor signalling and turnover. Nucleic Acids Res. 41:4433–4446. 10.1093/nar/gkt106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881–10890. 10.1093/nar/16.22.10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.X., Sokol E.S., Del Vecchio C.A., Sanduja S., Claessen J.H., Proia T.A., Jin D.X., Reinhardt F., Ploegh H.L., Wang Q., and Gupta P.B.. 2014. Epithelial-to-mesenchymal transition activates PERK-eIF2α and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 4:702–715. 10.1158/2159-8290.CD-13-0945 [DOI] [PubMed] [Google Scholar]
- Flamant L., Roegiers E., Pierre M., Hayez A., Sterpin C., De Backer O., Arnould T., Poumay Y., and Michiels C.. 2012. TMEM45A is essential for hypoxia-induced chemoresistance in breast and liver cancer cells. BMC Cancer. 12:391 10.1186/1471-2407-12-391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor B.D., Kubicek S., Yonezawa M., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., and Jenuwein T.. 2006. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20:1557–1562. 10.1101/gad.388206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A.P., Klocke B.J., Ballestas M.E., and Roth K.A.. 2012. CHOP potentially co-operates with FOXO3a in neuronal cells to regulate PUMA and BIM expression in response to ER stress. PLoS One. 7:e39586 10.1371/journal.pone.0039586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C. 2012. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13:89–102. 10.1038/nrm3270 [DOI] [PubMed] [Google Scholar]
- Jeon Y.J., Khelifa S., Ratnikov B., Scott D.A., Feng Y., Parisi F., Ruller C., Lau E., Kim H., Brill L.M., et al. 2015. Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stress-inducing chemotherapies. Cancer Cell. 27:354–369. 10.1016/j.ccell.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric D., Castel P., Griffith M., Griffith O.L., Won H.H., Ellis H., Ebbesen S.H., Ainscough B.J., Ramu A., Iyer G., et al. 2015. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature. 518:240–244. 10.1038/nature13948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric D., Krop I., Ramanathan R.K., Wilson T.R., Ware J.A., Sanabria Bohorquez S.M., Savage H.M., Sampath D., Salphati L., Lin R.S., et al. 2017. Phase I Dose-Escalation Study of Taselisib, an Oral PI3K Inhibitor, in Patients with Advanced Solid Tumors. Cancer Discov. 7:704–715. 10.1158/2159-8290.CD-16-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazu M., Saso K., Tong K.I., McQuire T., Goto K., Son D.O., Wakeham A., Miyagishi M., Mak T.W., and Okada H.. 2011. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One. 6:e17830 10.1371/journal.pone.0017830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Xu W., and Reed J.C.. 2008. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 7:1013–1030. 10.1038/nrd2755 [DOI] [PubMed] [Google Scholar]
- Kim S.-B., Dent R., Im S.-A., Espié M., Blau S., Tan A.R., Isakoff S.J., Oliveira M., Saura C., Wongchenko M.J., et al. LOTUS investigators . 2017. Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 18:1360–1372. 10.1016/S1470-2045(17)30450-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R.J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., and Zhang Y.. 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 442:312–316. 10.1038/nature04853 [DOI] [PubMed] [Google Scholar]
- Koo H.J., Piao Y., and Pak Y.K.. 2012. Endoplasmic reticulum stress impairs insulin signaling through mitochondrial damage in SH-SY5Y cells. Neurosignals. 20:265–280. 10.1159/000333069 [DOI] [PubMed] [Google Scholar]
- Lecca M.R., Wagner U., Patrignani A., Berger E.G., and Hennet T.. 2005. Genome-wide analysis of the unfolded protein response in fibroblasts from congenital disorders of glycosylation type-I patients. FASEB J. 19:240–242. 10.1096/fj.04-2397fje [DOI] [PubMed] [Google Scholar]
- Lee S.T., Li Z., Wu Z., Aau M., Guan P., Karuturi R.K., Liou Y.C., and Yu Q.. 2011. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol. Cell. 43:798–810. 10.1016/j.molcel.2011.08.011 [DOI] [PubMed] [Google Scholar]
- Lehmann B.D., Bauer J.A., Chen X., Sanders M.E., Chakravarthy A.B., Shyr Y., and Pietenpol J.A.. 2011. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121:2750–2767. 10.1172/JCI45014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Liu Y., Kubicek S., Myllyharju J., Tumber A., Ng S., Che K.H., Podoll J., Heightman T.D., Oppermann U., et al. 2011. A selective inhibitor and probe of the cellular functions of Jumonji C domain-containing histone demethylases. J. Am. Chem. Soc. 133:9451–9456. 10.1021/ja201597b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez S.C., Tanabe K., Cras-Méneur C., Abumrad N.A., Bernal-Mizrachi E., and Permutt M.A.. 2008. Inhibition of Foxo1 protects pancreatic islet beta-cells against fatty acid and endoplasmic reticulum stress-induced apoptosis. Diabetes. 57:846–859. 10.2337/db07-0595 [DOI] [PubMed] [Google Scholar]
- Mason J.M., Lin D.C., Wei X., Che Y., Yao Y., Kiarash R., Cescon D.W., Fletcher G.C., Awrey D.E., Bray M.R., et al. 2014. Functional characterization of CFI-400945, a Polo-like kinase 4 inhibitor, as a potential anticancer agent. Cancer Cell. 26:163–176. 10.1016/j.ccr.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Mayer I.A., Abramson V.G., Formisano L., Balko J.M., Estrada M.V., Sanders M.E., Juric D., Solit D., Berger M.F., Won H.H., et al. 2017. A Phase Ib Study of Alpelisib (BYL719), a PI3Kα-Specific Inhibitor, with Letrozole in ER+/HER2- Metastatic Breast Cancer. Clin. Cancer Res. 23:26–34. 10.1158/1078-0432.CCR-16-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes-Pereira A.M., Martin S.A., Brough R., McCarthy A., Taylor J.R., Kim J.S., Waldman T., Lord C.J., and Ashworth A.. 2009. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol. Med. 1:315–322. 10.1002/emmm.200900041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puc J., and Parsons R.. 2005. PTEN loss inhibits CHK1 to cause double stranded-DNA breaks in cells. Cell Cycle. 4:927–929. 10.4161/cc.4.7.1795 [DOI] [PubMed] [Google Scholar]
- Puc J., Keniry M., Li H.S., Pandita T.K., Choudhury A.D., Memeo L., Mansukhani M., Murty V.V., Gaciong Z., Meek S.E., et al. 2005. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell. 7:193–204. 10.1016/j.ccr.2005.01.009 [DOI] [PubMed] [Google Scholar]
- Rached M.T., Kode A., Xu L., Yoshikawa Y., Paik J.H., Depinho R.A., and Kousteni S.. 2010. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metab. 11:147–160. 10.1016/j.cmet.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadori G., Konstantinidou G., Venkateswaran N., Biscotti T., Morlock L., Galié M., Williams N.S., Luchetti M., Santinelli A., Scaglioni P.P., and Coppari R.. 2015. Diet-induced unresolved ER stress hinders KRAS-driven lung tumorigenesis. Cell Metab. 21:117–125. 10.1016/j.cmet.2014.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Sun L., Li Q., Liang J., Yu W., Yi X., Yang X., Li Y., Han X., Zhang Y., et al. 2011. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl. Acad. Sci. USA. 108:7541–7546. 10.1073/pnas.1017374108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Yoshida M., Tanimura H., Fujii T., Sakata K., Tachibana Y., Ohwada J., Ebiike H., Kuramoto S., Morita K., et al. 2011. The selective class I PI3K inhibitor CH5132799 targets human cancers harboring oncogenic PIK3CA mutations. Clin. Cancer Res. 17:3272–3281. 10.1158/1078-0432.CCR-10-2882 [DOI] [PubMed] [Google Scholar]
- Urra H., Dufey E., Avril T., Chevet E., and Hetz C.. 2016. Endoplasmic Reticulum Stress and the Hallmarks of Cancer. Trends Cancer. 2:252–262. 10.1016/j.trecan.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Van Rechem C., Black J.C., Boukhali M., Aryee M.J., Gräslund S., Haas W., Benes C.H., and Whetstine J.R.. 2015. Lysine demethylase KDM4A associates with translation machinery and regulates protein synthesis. Cancer Discov. 5:255–263. 10.1158/2159-8290.CD-14-1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz-Luis I., Ottesen R.A., Hughes M.E., Mamet R., Burstein H.J., Edge S.B., Gonzalez-Angulo A.M., Moy B., Rugo H.S., Theriault R.L., et al. 2014. Outcomes by tumor subtype and treatment pattern in women with small, node-negative breast cancer: a multi-institutional study. J. Clin. Oncol. 32:2142–2150. 10.1200/JCO.2013.53.1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., and Kaufman R.J.. 2014. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer. 14:581–597. 10.1038/nrc3800 [DOI] [PubMed] [Google Scholar]
- Wee Z.N., Yatim S.M., Kohlbauer V.K., Feng M., Goh J.Y., Bao Y., Lee P.L., Zhang S., Wang P.P., Lim E., et al. 2015. IRAK1 is a therapeutic target that drives breast cancer metastasis and resistance to paclitaxel. Nat. Commun. 6:8746 10.1038/ncomms9746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B., Warne P.H., and Downward J.. 2011. PIK3CA mutation, but not PTEN loss of function, determines the sensitivity of breast cancer cells to mTOR inhibitory drugs. Oncogene. 30:3222–3233. 10.1038/onc.2011.42 [DOI] [PubMed] [Google Scholar]
- Whetstine J.R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., and Shi Y.. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 125:467–481. 10.1016/j.cell.2006.03.028 [DOI] [PubMed] [Google Scholar]
- Young L.C., McDonald D.W., and Hendzel M.J.. 2013. Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following γ-irradiation. J. Biol. Chem. 288:21376–21388. 10.1074/jbc.M113.491514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Lu X., Wang G., Lan Z., Liao W., Li J., Liang X., Chen J.R., Shah S., Shang X., et al. 2017. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature. 542:484–488. 10.1038/nature21357 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.