Cheng and Schorey identified a regulatory mechanism where cytosolic DNA and RNA sensing pathways synergistically regulate type I IFN production in macrophages and mice during a Mycobacterium tuberculosis infection and show that activation of mitochondrial antiviral signaling protein (MAVS) promotes M.tb survival in mice.
Abstract
RNA sensing pathways are key elements in a host immune response to viral pathogens, but little is known of their importance during bacterial infections. We found that Mycobacterium tuberculosis (M.tb) actively releases RNA into the macrophage cytosol using the mycobacterial SecA2 and ESX-1 secretion systems. The cytosolic M.tb RNA induces IFN-β production through the host RIG-I/MAVS/IRF7 RNA sensing pathway. The inducible expression of IRF7 within infected cells requires an autocrine signaling through IFN-β and its receptor, and this early IFN-β production is dependent on STING and IRF3 activation. M.tb infection studies using Mavs−/− mice support a role for RNA sensors in regulating IFN-β production and bacterial replication in vivo. Together, our data indicate that M.tb RNA is actively released during an infection and promotes IFN-β production through a regulatory mechanism involving cross-talk between DNA and RNA sensor pathways, and our data support the hypothesis that bacterial RNA can drive a host immune response.
Graphical Abstract
Introduction
Innate immunity is the first line of our host defense against microbial pathogens. Cells respond to microbial pathogen-associated molecular patterns through engagement of pattern recognition receptors (PRRs) resulting in the initiation of an immune response (Brubaker et al., 2015). Engagement of host PRRs triggers downstream signaling pathways resulting in the production of various cytokines and chemokines including type I IFNs. The type I IFN receptor (IFNAR), which is composed of two subunits, IFNAR1 and IFNAR2, regulates the transcription of a set of IFN-stimulated genes, many of which are important in promoting antiviral immunity (McNab et al., 2015). However, the relevancy of type I IFNs in an immune response to bacterial infections is less understood and, depending on the pathogen, may have beneficial or detrimental effects (Kovarik et al., 2016). In the context of a Mycobacterium tuberculosis (M.tb) infection, the production of type I IFNs appears to limit the protective immune response (O’Garra et al., 2013). For example, Ifnar1−/− mice show increased resistance to an M.tb infection when compared with WT mice (Stanley et al., 2007; Dorhoi et al., 2014). A link between increased expression of IFN-stimulated genes and active tuberculosis has also been observed in human genetic studies (Berry et al., 2010).
Since published reports indicate that the type I IFN response observed upon an M.tb infection plays an important role in tuberculosis pathogenesis and immunity, recent work has focused on what mycobacterial components and host pathways are involved. A number of studies indicate that host cytosolic DNA sensing pathways are crucial for M.tb-induced type I IFN production (Manzanillo et al., 2012; Collins et al., 2015; Dey et al., 2015; Wassermann et al., 2015; Watson et al., 2015). The cytosolic DNA sensor cyclic GMP–AMP synthase (cGAS) directly recognizes M.tb DNA, which is released through an ESX-1–dependent manner. The DNA-bound cGAS subsequently activates STING (stimulator of IFN genes) and the transcription factor IRF3, resulting in type I IFN production (Manzanillo et al., 2012; Collins et al., 2015; Wassermann et al., 2015; Watson et al., 2015). M.tb also releases a bacterial second messenger, cyclic-di-adenosine monophosphate (c-di-AMP), which stimulates IFN-β expression through a cGAS-independent but STING-dependent pathway (Dey et al., 2015). Although these published studies indicate that STING activation is required for type I IFN production in host cells upon an M.tb infection, there are likely additional pathways that intersect/amplify the host response.
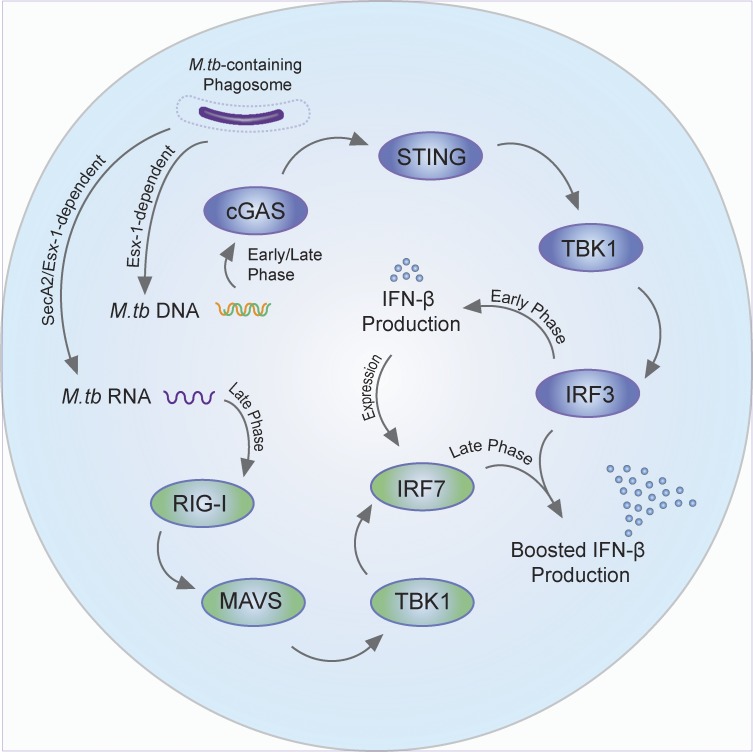
Our previous studies indicate that M.tb RNA is present in exosomes, which are endosome-derived membrane vesicles released from cells that function in intercellular communication (Schorey et al., 2015; Singh et al., 2015). The presence of M.tb RNA in exosomes suggests that the RNA is released from M.tb during an infection and therefore potentially detected by host endosomal or cytosolic RNA sensors. However, there is very limited information on the role of mycobacterial RNA in M.tb pathogenesis, and this is likely due to an assumed lack of accessibility of bacterial RNA to corresponding host PRRs. In the present study, we show that M.tb mRNA is released into the macrophage cytosol through a SecA2- and ESX-1–dependent mechanism and activates the retinoic acid–inducible gene (RIG-I)/mitochondrial antiviral signaling protein (MAVS)/tank-binding kinase 1 (TBK1)/IRF7 signaling pathway. Activation of this RNA sensing pathway requires prior STING activation and works synergistically with the DNA sensing pathway to stimulate IFN-β production in host cells during an M.tb infection. In addition, our findings demonstrate a role for MAVS in regulating an immune response to an M.tb infection in vivo.
Results
M.tb releases mRNAs through a SecA2-dependent pathway
M.tb RNA has been detected in mycobacterial culture supernatant previously (Obregón-Henao et al., 2012). However, it was unclear if this RNA was released by bacterial lysis or through an RNA transporter, or contained within outer membrane vesicles, which can also contribute to release of extracellular bacterial RNA. To begin defining the mechanisms of RNA release we cultured an M.tb H37Rv strain expressing DsRed in liquid broth until midexponential phase and analyzed for M.tb RNA in the culture filtrate. To remove any potential contamination from outer membrane vesicles, the culture filtrates were subject to ultracentrifugation before RNA extraction. Six different M.tb mRNAs were detected (Fig. S1 A), but no PCR products were detected in the absence of reverse transcription, excluding the contamination from genomic DNA (data not shown). The RNA was released from intact bacteria as we failed to detect DsRed mRNA (Fig. S1 A) in the culture filtrate. A similar result was observed in Mycobacterium bovis bacillus Calmette-Guérin (BCG) expressing DsRed (data not shown). Additionally, we measured the M.tb RNAP-β, a mycobacterial lysis marker (Reyna et al., 2016). As shown in Fig. S1 B, no RNAP-β was detected in the culture filtrate under the culture conditions used for RNA isolation. In contrast, the presence of EsxB, a known secreted protein, was readily detected in the culture supernatant. Similar to H37Rv, M.tb CDC1551 and Erdman strains both release mRNA, but not 16S ribosomal RNA (rRNA), into the culture media (Fig. S1, C and D).
The presence of M.tb mRNAs but not DsRed in the culture filtrate suggests the presences of an active RNA transporter. SecA2 was previously shown in Listeria monocytogenes to be required for release of RNA into the bacterial culture media (Abdullah et al., 2012). However, this study did not define any specific nucleic acids that were released by the Listeria. Mycobacteria also express a SecA2 secretion system, and therefore, we evaluated its potential role in mRNA release. As shown in Fig. 1 (A and B), we detected all six M.tb mRNAs in the culture filtrate of WT and the secA2 complemented strains but not for the ΔsecA2 mutant. In contrast, we detected all the transcripts in the cell lysates for each CDC1551 strain.
Figure 1.
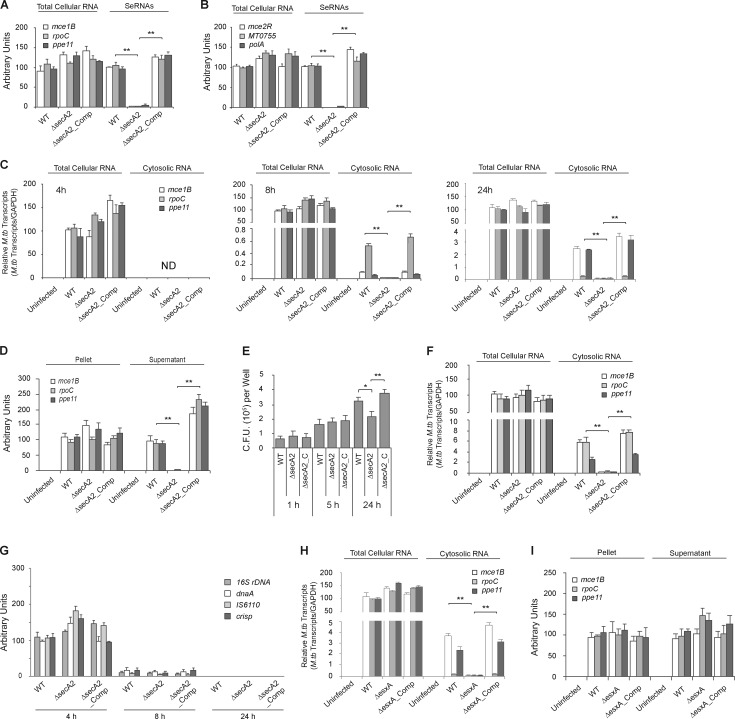
The M.tb SecA2 and Esx-1 secretion systems are important for delivery of mycobacterial mRNAs into the cytosol of infected macrophages. (A and B) Use of qRT-PCR to determine the abundance of M.tb mRNAs in total cellular RNA and seRNAs. 16S rRNA was used as an input control for total cellular RNA but was not detected in seRNAs. Each M.tb transcript abundance was expressed as the percentage relative to WT. (C) Use of qRT-PCR to determine M.tb mRNA abundance in macrophages (total cellular RNA) or the cytosol (cytosolic RNA) at the times indicated. All mRNAs were normalized to GAPDH. Each transcript was expressed as percentage relative to its abundance in total cellular RNA from macrophages infected with WT M.tb. (D) qRT-PCR analysis for M.tb mRNA in lysed Raw264.7 cells infected for 24 h with indicated M.tb CDC1551 strains. Lysed Raw264.7 cells were separated into M.tb cells (pellet) and Raw264.7 cell lysate (supernatant), and RNA was purified from both fractions. The data were normalized as described in A. (E) Survival of M.tb strains within BMMs infected at an MOI of 10. Results are mean CFUs ± SD of three independent infections at each time point (n = 3 independent infections per group). (F) Similar to C, but assayed for M.tb mRNA abundance in F4/80+ macrophages isolated from C57BL/6 mice (n = 3 mice per group) aerosol-infected for 2 wk with M.tb CDC1551 strains. (G) M.tb DNA (16S rDNA, dnaK, IS6110, and crisp) in the cytosol of infected macrophages was measured by PCR. (H) Raw264.7 cells were infected for 24 h with WT, ΔesxA, or ΔesxA_Comp (esxA complemented strain) M.tb Erdman strains. qRT-PCR was used to quantify the M.tb transcripts present in whole cells and cytosol. Data were normalized as described for C. (I) Similar to D, except cells were infected with the indicated M.tb Erdman strains. Data are mean ± SD of triplicate wells and representative of at least three independent experiments. * P < 0.05 and ** P < 0.01 by two-tailed Student’s t test. ND, not detected.
M.tb mRNA released through SecA2 is found in the cytosol of infected macrophages
To determine if SecA2 was also involved in release of M.tb RNA during a macrophage infection, we infected Raw264.7 cells with WT, ΔsecA2, and secA2 complemented CDC1551 strains and evaluated the cytosol at different times after infection for M.tb transcripts previously detected in culture filtrate. We detected by quantitative RT-PCR all the tested M.tb transcripts in the cytosol of WT CDC1551-infected macrophages at 8 and 24 h after infection (Fig. 1 C). However, we failed to detect or detected at low levels these transcripts in the cytosol of macrophages infected with ΔsecA2 CDC1551. Macrophages infected with the complemented strain showed comparable results to WT CDC1551 (Fig. 1 C). No M.tb rRNA was detected by qRT-PCR at all three time points (data not shown). Moreover, although we detected the M.tb transcripts at a similar level in total cellular RNA at all time points, we failed to detect M.tb RNA in the cytosol 4 h after infection (Fig. 1 C). It is interesting that the three RNAs show different kinetics in their presence in the host cytosol with rpoC more prevalent in the cytosol at 8 h, while mce18 and ppe11 were more abundant at 24 h. This suggests that there may be differences in the transport processes between the different RNAs. Nevertheless, the releases of all three RNAs were dependent on M.tb expressing SecA2. To determine if, during a bone marrow–derived macrophage (BMM) infection, the ΔsecA2 CDC1551 release RNA from the mycobacteria but the RNA fails to gain entry into the cytosol, we lysed the infected cells under conditions that permeabilized host membranes including phagosomes. Under these conditions, we failed to detect M.tb RNA in the infected cell lysate but could detect the RNA in the M.tb isolated from the infected BMM (Fig. 1 D). Since previous results indicate that M.tb secA2 mutants are attenuated within macrophages (Sullivan et al., 2012), we evaluated the bacterial load for the different CDC1551 strains during the first 24 h of infection. M.tb was present within infected macrophages for all strains, although there was a slight decrease in bacterial numbers in cells infected with the ΔsecA2 CDC1551 relative to WT 24 h after infection (Fig. 1 E). Cells isolated from the bronchoalveolar levage fluid of M.tb-infected mice are known to contain mycobacteria (Repasy et al., 2013). Therefore, we isolated F4/80+ macrophages from CDC1551-infected mice. Consistent with the results observed in BMMs, M.tb mRNAs were also detected in the cytosol of F4/80+ macrophages isolated from WT or secA2 complemented M.tb-infected mice, but the M.tb RNA was absent or present at low levels in macrophages isolated from ΔsecA2-infected mice (Fig. 1 F). Previous studies also indicate that M.tb DNA is present in the cytosol of infected macrophages (Manzanillo et al., 2012; Watson et al., 2015). Therefore, we evaluated whether the SecA2 secretion system was also required for release of M.tb DNA and served as a common transporter for mycobacterial nucleic acids. Testing for M.tb DNA species previously identified in the cytosol of infected macrophages (Watson et al., 2015), we observed no effect of SecA2 expression on the levels of M.tb DNA in the cytosol (Fig. 1 G). Further, in contrast to the M.tb RNA, we detected the highest levels of M.tb DNA in the cytosol at 4 h after infection with little or no detectable DNA at 8 and 24 h (Fig. 1 G).
Cytosolic localization of M.tb mRNA requires the ESX-1 secretion system
Previous studies have identified the mycobacterial ESX-1 secretion system is required for release of M.tb DNA into the cytosol of infected macrophages (Manzanillo et al., 2012; Watson et al., 2015). To evaluate its potential role in mRNA release, we infected macrophages with M. bovis BCG, which lacks a functional ESX-1 secretion system due to the loss of the RD1 region (Brodin et al., 2002). In BCG-infected cells, we failed to detect mycobacterial transcripts in the cytosol (Fig. S2, A and B). To further define ESX-1’s role in the transport of M.tb RNA into the cytosol, we infected macrophages with WT, ΔesxA, and esxA complemented Erdman strains. As observed for BCG, cytosolic M.tb mRNA was only detected in macrophages infected with Erdman strains that have a functional ESX-1 secretion system (Fig. 1 H). As expected, release of M.tb mRNA during broth culture was not dependent on ESX-1 (Fig. S2, C and D). Moreover, under conditions that lyse cellular compartments including phagosomes but not M.tb, we detected the M.tb RNA in the cell lysate of ΔesxA-infected cells (Fig. 1 I). However, using these same lysis conditions, we did not detect M.tb RNA in the lysate of cells infected with the ΔsecA2 mutant (Fig. 1 D).
To extend the role of SecA2 and ESX-1 secretion systems in the release of M.tb mRNAs beyond the few mRNAs tested by PCR, we developed a method as outlined in Fig. 2 to specifically quantify the M.tb RNA in the cytosol of macrophages infected with M.tb strains labeled with 5-ethynyl uridine (EU; Fig. 2 A). Using this method, we found that labeled M.tb RNA present in the macrophage cytosol is significantly reduced in cells infected with M.tb lacking SecA2 or EsxA compared with WT or complemented M.tb strains (Fig. 2, B–E). Additionally, two sharp rRNA peaks corresponding to mycobacterial 16S and 23S rRNA were detected in total cellular RNA (Fig. 2, B and D) but were absent in cytosolic RNA (Fig. 2, C and E). The M.tb RNA abundance was further determined by qRT-PCR on the total cellular RNA and cytosolic RNA before (Fig. 2, F and H) or after (Fig. 2, G and I) biotin-streptavidin purification. As seen in Fig. 2 (F and H), each M.tb RNA (mce1B, rpoC, ppe11, and 16S rRNA) showed a comparable abundance in total cellular RNA isolated from macrophages infected with various M.tb strains. However, the abundance of these mycobacterial mRNAs was significantly decreased or undetectable in the cytosol of macrophages infected with the ΔsecA2 or ΔesxA strains. When analyzing biotin-streptavidin–purified RNA from total cellular RNA, all M.tb RNA species showed a similar level of abundance in cells infected with various M.tb strains (Fig. 2, G and I). In contrast, after biotin-streptavidin purification, M.tb mRNA abundance diminished or was undetectable in the cytosol of macrophage infected with the ΔsecA2 or ΔesxA mutant (Fig. 2, G and I). In cells infected with M.tb strains that were not labeled with EU, no M.tb RNA was detected after biotin-streptavidin purification. As expected, only the M.tb mRNAs but not the 16S rRNA were detected in the cytosol of cells infected with M.tb before or after the biotin-streptavidin purification (Fig. 2, F–I). No macrophage GAPDH mRNA was detected in the RNA after the biotin-streptavidin purification.
Figure 2.
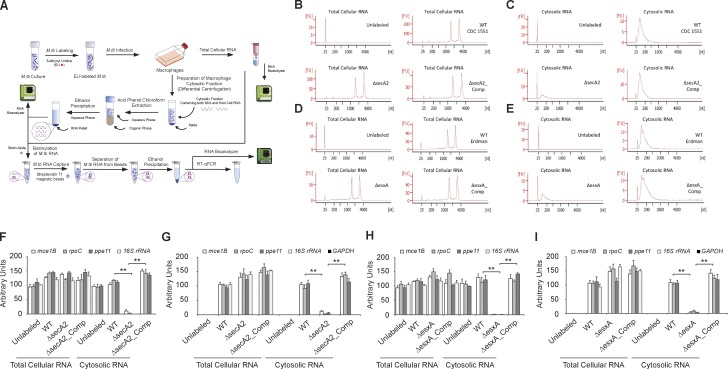
Mycobacterial SecA2 and ESX-1 secretion systems are required for delivery of M.tb RNA into the cytosol of macrophages. (A) Schematic diagram of the experiment design. (B–E) The RNA bioanalyzer results for the EU-labeled RNA from total cell lysate (B and D) and cytosolic fraction (C and E). (B and C) Cells infected with the M.tb CDC1551 strains. (D and E) Cells infected with the M.tb Erdman strains. (F and H) qRT-PCR of the M.tb transcripts (mce1B, rpoC, ppe11, and 16S rRNA) before pull-down. Each transcript was normalized to GAPDH. The abundance of each transcript was expressed relative to the M.tb transcript/GAPDH ratio defined for unlabeled WT M.tb-infected cells for both total cellular and cytosolic RNA. (G and I) qRT-PCR of the M.tb and host transcripts (mce1B, rpoC, ppe11, 16S rRNA, and GAPDH) after biotin-streptavidin purification. The abundance of each transcript was expressed relative to WT M.tb-infected cells for both total cellular and cytosolic RNA. Unlabeled indicates host cells infected with unlabeled WT M.tb. Data are representative of two independent experiments. (F–I) Data are mean ± SD of duplicate wells. ** P < 0.01 by two-tailed Student’s t test.
M.tb stimulates a SecA2- and MAVS-dependent IFN-β expression
The presence of mycobacterial mRNAs in the cytoplasm suggest a possible engagement of host cytosolic RNA sensors and activation of cytosolic surveillance pathways including the induction of type I IFNs (Wu and Chen, 2014; McNab et al., 2015). We first quantified the IFN-β transcript levels in macrophages infected with WT and ΔsecA2 as well as ΔsecA2 complemented CDC1551 strains. As shown in Fig. 3 A, there were increased IFN-β mRNA levels in M.tb-infected cells compared with uninfected cells at all time points tested after infection. A relatively low level of SecA2-independent IFN-β expression was observed in M.tb-infected cells at 4 h. In contrast, a significantly higher level of IFN-β transcript was observed at 8 and 24 h after infection, and this increase was SecA2 dependent (Fig. 3 A). The SecA2 secretion system was required for maximum IFN-β protein production as macrophages infected for 24 h with the SecA2-deficient strain secreted IFN-β at significantly lower levels compared with macrophages infected with WT or complemented strains (Fig. 3 B). Nevertheless, in macrophages infected with the SecA2-deficient CDC1551, we observed increased IFN-β expression compared with uninfected cells, indicating a SecA2-independent mechanism for induction of IFN-β expression (Fig. 3, A and B). In contrast to SecA2, the absences of EsxA resulted in a marked reduction in IFN-β at all times after infection including 4 h (Fig. 3 C).
Figure 3.
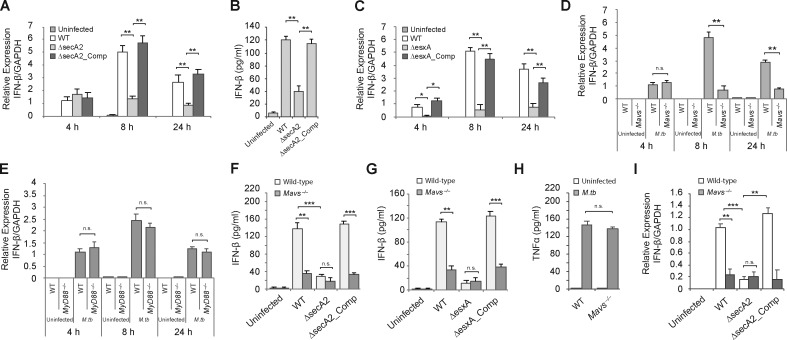
The SecA2 and ESX-1 secretion systems are required for IFN-β production by M.tb-infected BMMs, and this is mediated through a MAVS-dependent pathway. (A) WT BMMs were infected at a 10:1 MOI with the M.tb CDC1551 strains for 4, 8, or 24 h. The IFN-β mRNA levels were determined by qRT-PCR. (B) IFN-β in the BMM culture supernatants was defined by ELISA 24 h after infection. (C) Similar to A, but macrophages were infected with the indicated Erdman strains. (D) WT and Mavs−/− BMMs were infected with WT M.tb CDC1551 for the times indicated or left uninfected. The IFN-β mRNA levels were defined by qRT-PCR. (E) WT and MyD88−/− BMMs were left uninfected or infected with WT M.tb CDC1551 strain for 4, 8, and 24 h, and the IFN-β mRNA levels were defined by qRT-PCR. (F) WT and Mavs−/− BMMs were infected with the M.tb CDC1551 strains. IFN-β protein levels in the culture supernatants were quantified by ELISA 24 h after infection. (G) Same as in E, except BMMs were infected with the M.tb Erdman strains. (H) WT and Mavs−/− BMMs were infected with the WT M.tb CDC1551 strain, and the level of TNFα was measured by ELISA 24 h after infection. (I) qRT-PCR analysis for IFN-β in alveolar macrophages isolated from M.tb-infected WT and Mavs−/− mice 2 wk after infection. For qRT-PCR, the IFN-β mRNA levels were expressed as fold change relative to WT M.tb (A) and BMMs (C and D) 4 h after infection or WT mice infected with WT M.tb (H). mRNA levels were normalized to GAPDH. Data are mean ± SD of triplicate wells and representative of at least three independent experiments. n.s., not statistically significant; ** P < 0.01 and *** P < 0.001 by two-tailed Student’s t test.
The above data suggest that a mycobacterial component released through the SecA2 secretion system was important in stimulating IFN-β production. To determine if this component was M.tb RNA, we evaluated whether known RNA sensors were required for the SecA2-dependent induction of IFN-β. Cytosolic RNA sensors are known to induce IFN-β production through the mitochondrial membrane protein MAVS (also known as IPS-1, CARDIF, and VISA), which interacts with RNA sensors via its caspase recuitment domain and is an essential adaptor protein in the host cytosolic RNA sensing pathway (Wu and Chen, 2014). Therefore, we investigated the IFN-β expression in WT and Mavs−/− murine BMMs infected with WT CDC1551 M.tb. As shown in Fig. 3 D, the increased IFN-β transcript levels at 4 h after infection were MAVS independent. This correlated with our RNA localization data, as we failed to detect cytoplasmic M.tb mRNA at the 4-h time point. However, after an 8- and 24-h infection in which we detected M.tb mRNA in the cytosol, the induction of IFN-β was significantly dependent on MAVS (Fig. 3 D). In contrast, IFN-β production induced by an M.tb infection was independent of MyD88 (myeloid differentiation primary response gene 88), a critical adaptor protein in endosomal TLR-mediated RNA sensing pathway (Fig. 3 E; McNab et al., 2015). To evaluate the connection between MAVS and the SecA2 secretion system, Mavs−/− BMMs were infected for 24 h with the ΔsecA2 CDC1551 as well as WT and complemented strains, and the amount of secreted IFN-β was quantified. As shown in Fig. 3 F, the additional loss of MAVS function had no significant effect on IFN-β protein expression following an M.tb infection with the ΔsecA2-deficient strain, suggesting that the major effector released through the SecA2 secretion system functions through MAVS. Similar findings were observed when we infected Mavs−/− BMMs with the ΔesxA and parental Erdman strains (Fig. 3 G), indicating the ESX-1–dependent M.tb RNA release is an important driver for MAVS-regulated IFN-β induction in M.tb-infected macrophages. The cellular response was relatively specific as the loss of MAVS expression had no effect on the M.tb-induced TNF-α expression in BMMs (Fig. 3 H). As seen in BMM culture, IFN-β production was diminished in F4/80+ macrophages isolated from WT M.tb-infected Mavs−/− mice compared with macrophages from infected WT mice (Fig. 3 H). Furthermore, there was no significant difference in IFN-β production by F4/80+ macrophages isolated from WT and MAVS−/− mice when the mice were infected with the ΔsecA2 strain (Fig. 3 I).
Mycobacterial mRNAs are recognized by the cytosolic RNA sensor RIG-I
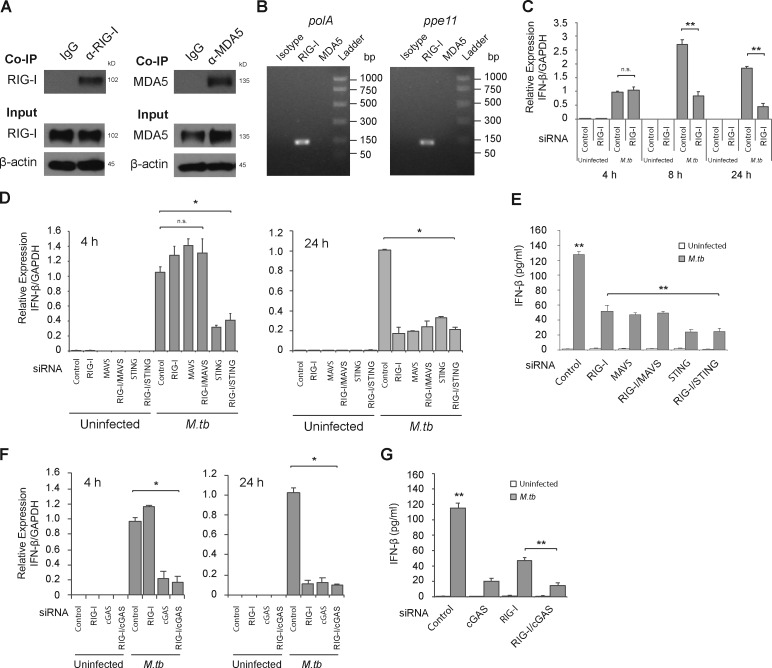
There are three well-characterized cytosolic RNA sensors in nucleated cells: RIG-I, LGP2, and MDA5 (Wu and Chen, 2014). Among them, RIG-I and MDA5 directly bind to MAVS via the caspase recruitment domain after activation. To determine whether cytosolic M.tb RNA is recognized by one of these RNA sensors, we performed immunoprecipitation experiments using antibodies against MDA5 and RIG-I. The immunoprecipitated RIG-I and MDA5 proteins were verified by Western blot (Fig. 4 A). Two M.tb-specific mRNAs, polA and ppe11, were coimmunoprecipitated with RIG-I but were not detected in the MDA5 pull-down (Fig. 4 B). The RT-PCR products were further confirmed by DNA sequencing (data not shown).
Figure 4.
Cytosolic RNA sensor RIG-I recognizes M.tb mRNAs and is required for IFN-β production by M.tb-infected cells. (A) Western blot of RIG-I and MDA5 before (input) and after immunoprecipitation with anti–RIG-I or anti-MDA5 antibodies. β-actin was used as a loading control. (B) The M.tb transcripts pulled down by the immunoprecipitation were defined by RT-PCR. (C) Control or RIG-I siRNA-treated BMMs were infected for 4, 8, and 24 h with WT M.tb CDC1551 and the IFN-β mRNA levels defined by qRT-PCR. (D) BMMs were first treated with an siRNA or a mixture of siRNAs as indicated and then infected for 4 and 24 h with WT M.tb CDC1551 strain, and the expression level of IFN-β was shown as a fold change relative to control siRNA-treated cells infected with WT M.tb for 4 h (C and D) or 24 h (D). mRNA levels were normalized to GAPDH expression. (E) IFN-β levels in BMM culture supernatants were quantified by ELISA 24 h after M.tb infection. (F and G) Similar to D and E, respectively, but BMMs were pretreated with cGAS or combined siRNA. Data are mean ± SD of triplicate wells and are representative of at least three independent experiments. n.s., not statistically significant; * P < 0.05 and ** P < 0.01 by two-tailed Student’s t test.
To further evaluate RIG-I’s role in type I IFN production, we used siRNA to knock down RIG-I expression in BMMs and measured IFN-β mRNA levels upon an M.tb infection. Analogous to what was observed in Mavs−/− BMMs, we observed no difference in IFN-β expression between control oligo and RIG-I siRNA at 4 h after infection but significantly lower expression of IFN-β mRNA in RIG-I siRNA-treated cells compared with controls at 8 and 24 h after infection (Fig. 4 C).
Cross-talk between STING-dependent and RIG-I/MAVS–dependent signaling pathways contributes to the late IFN-β expression after M.tb infection
The STING-dependent DNA sensing pathway is required for the induction of IFN-β in macrophages during an M.tb infection (Manzanillo et al., 2012; Collins et al., 2015; Wassermann et al., 2015; Watson et al., 2015). To evaluate the link between the RIG-I/MAVS–mediated production of IFN-β and that induced through STING, we generated siRNA knockdowns for the different sensors in combination and measured the levels of IFN-β transcripts at 4 and 24 h after M.tb infection. We were able to achieve ∼90% knockdown of gene expression in the BMMs for the different knockdowns (Fig. S3). As shown in Fig. 4 D, knockdown of RIG-I and/or MAVS had no effect on IFN-β mRNA levels at the 4-h infection time point. However, knockdown of STING resulted in a marked loss of IFN-β mRNA (Fig. 4 D). At 24 h after infection, again STING knockdown resulted in a loss in IFN-β mRNA. In contrast to the 4-h infection, knockdown of RIG-I and/or MAVS also resulted in a loss of IFN-β transcript at the 24-h time point and were comparable to what was observed in the STING knockdown (Figs. 4 D). Blocking both RIG-I/MAVS and STING had no additional effect compared with STING alone (Figs. 4 D). At the protein level, we observed an ∼65% decrease in cytokine released from the RIG-I and/or MAVS knockdown cells after a 24-h infection, while the decrease was slightly but significantly higher in STING knockdown cells (Fig. 4 E). Similar to STING, the siRNA knockdown of cGAS (Fig. S3 G), a critical cytosolic DNA sensor in M.tb DNA-driven IFN-β production in macrophages, also showed diminished IFN-β transcript levels at 4 and 24 h (Fig. 4 F) and protein levels (Fig. 4 G) at 24 h after M.tb infection. Furthermore, RIG-I knockdown also had no additional effect on IFN-β in cGAS knockdown BMMs after M.tb infection (Fig. 4 F).
Mycobacterial infection activates the RIG-I/MAVS/TBK1/IRF7 signaling pathway that drives IFN-β expression
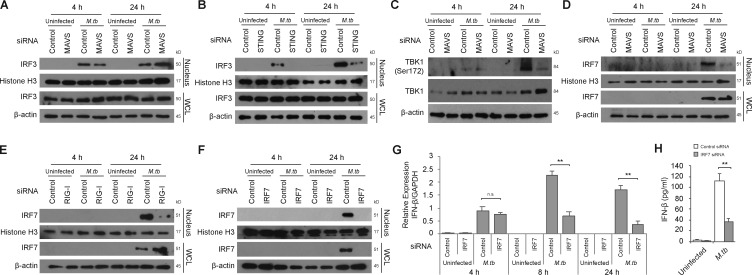
The above data indicate that STING is essential for the early and late IFN-β induction, while RIG-I/MAVS are functioning only at later times after infection. However, the tie between these two pathways was not clear. IRF3 and IRF7 are two central transcription factors for type I IFN production in response to cytosolic viral DNA and RNA in host cells (Honda et al., 2006; Tamura et al., 2008; McNab et al., 2015). These two transcription factors are phosphorylated and activated by TBK1, a downstream protein kinase shared by STING- and MAVS-mediated signaling pathways (Wu and Chen, 2014). Activated IRF3 and IRF7 are translocated into the nucleus to drive type I IFN expression (Honda et al., 2006). Unlike IRF3, IRF7 is not constitutively expressed but is induced by various mediators including IFN-β (Marié et al., 1998; Sato et al., 1998). IRF3 and IRF7 can function as homodimers or as heterodimers to drive IFN-β production (Honda et al., 2006; Tamura et al., 2008). Here, we hypothesized that IRF7 expression is induced upon an M.tb infection and is important in amplifying IFN-β production at later stages of an M.tb infection and that IRF7 activation is mediated through an M.tb RNA/RIG-I/MAVS pathway. We first confirmed the early activation of IRF3 and tested whether its activation was dependent on MAVS. As shown in Fig. 5 A, IRF3 was expressed in both infected and uninfected macrophages, but its nuclear translocation was only seen in M.tb-infected macrophages. IRF3 nuclear localization was observed at both 4 and 24 h after infection and was inhibited not by knockdown of MAVS (Fig. 5 A) but by STING knockdown (Fig. 5 B), as previously shown in Sting−/− BMMs (Manzanillo et al., 2012). The protein kinase TBK1 showed increased phosphorylation at Serine 172 in infected macrophages with the highest levels observed 24 h after infection (Fig. 5 C). This phosphorylation, which results in an active TBK1, was partially dependent on MAVS at 24 h after infection. We also evaluated IRF7 expression and found it expressed in macrophages 24 h but not 4 h after M.tb infection. In contrast to IFR3, its nuclear translocation was dependent on MAVS (Fig. 5 D) and RIG-I (Fig. 5 E). To determine if IRF7 was important in the M.tb-mediated induction of IFN-β, we used siRNA knockdown, which resulted in undetectable levels of IRF7 in cells by Western blot (Fig. 5 F), and observed a significant loss in IFN-β mRNA at 8 and 24 h after infection (Fig. 5 G). No effect of IRF7 knockdown on IFN-β mRNA abundance in macrophages was observed at 4 h after infection. The IRF7 knockdown also resulted in an ∼70% decrease in IFN-β protein production 24 h after infection (Fig. 5 H).
Figure 5.
IRF7 expression is induced during an M.tb infection and is required for the MAVS-dependent IFN-β expression. BMMs were treated with control, MAVS, or STING siRNA and either left uninfected or infected for 4 or 24 h with WT M.tb CDC1551 strain at an MOI of 10. Histone H3 and β-actin were used as loading controls for nuclear fraction and WCL, respectively. (A and B) Western blot for IRF3 in BMM WCLs or nuclear fractions (nucleus). (C) Western blot for TBK1 and Phospho-TBK1 (Ser172) in WCL. (D) Western blot for IRF7 in WCL and nuclear fraction of MAVS knockdown BMMs. (E) Similar to D, but using RIG-I-knockdown BMMs. (F) Similar to D, but using IRF7 knockdown BMMS. (G) Control or IRF7 knockdown BMMs were left uninfected or infected with the WT M.tb CDC1551 strain for the indicated times. The level of IFN-β mRNA expression was defined by qRT-PCR. (H) IFN-β protein levels in BMM culture supernatants were quantified by ELISA 24 h after infection. IFN-β levels are mean ± SD of triplicate wells and are representative of at least three independent experiments. n.s., not statistically significant; ** P < 0.01 by two-tailed Student’s t test.
Host cytosolic DNA and RNA sensing pathways synergistically contribute to the IFN-β expression in M.tb-infected macrophages
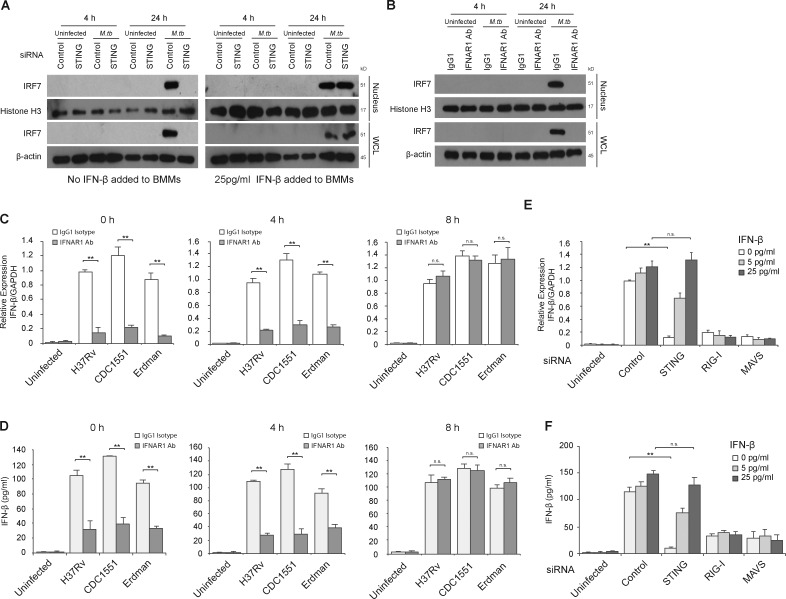
Previous studies have shown IRF7 expression during a viral infection to be regulated by an early production of IFN-β via a positive-feedback loop (Honda et al., 2006). To test if STING-dependent early IFN-β induction has a similar role during M.tb infection, we first measured IRF7 expression by Western blot in uninfected and M.tb-infected BMMs that were pretreated with control siRNA or siRNA to STING. As shown in Fig. 6 A, IRF7 expression was observed 24 h after infection and blocked in STING knockdown BMMs. This STING-dependent induction of IRF7 could be overcome by the addition of exogenous IFN-β to the infected macrophages (Fig. 6 A), suggesting an IFN-β autocrine-dependent IRF7 expression. However, exogenous IFN-β alone, at the concentrations used, failed to induce IRF7 production in uninfected BMMs. Additional studies indicated that IRF7 induction by IFN-β was dose dependent with a higher concentration of IFN-β required to induce IRF7 expression in uninfected compared with infected BMMs (Fig. S4 A). To further confirm the requirement for IFN-β in inducing IRF7 expression in M.tb-infected BMMs, the IFN-β signaling pathway was blocked using an IFNAR1-blocking antibody, which, when added to infected cells, resulted in the loss of IRF7 expression (Fig. 6 B).
Figure 6.
Early STING-dependent production of IFN-β is required for activation of the RIG-I/MAVS/IRF7 signaling pathway. (A) Western blot for IRF7 in BMM WCLs or nuclear fractions (nucleus). BMMs were treated with control or STING siRNA and either left uninfected or infected for 4 or 24 h with WT M.tb CDC1551 strain at an MOI of 10. For one set of samples, BMMs were also treated with 25 pg/ml of IFN-β at the time of infection. Histone H3 and β-actin were used as loading controls for nuclear fraction and WCL, respectively. (B) WT BMMs were infected with WT M.tb CDC1551 strain in the presence of an IFN-β receptor blocking antibody (IFNAR1 Ab) or isotype control (IgG1). (C) IFNAR1 blocking Ab or control IgG1 were added at different times after infection (0, 4, and 8 h). IFN-β mRNA levels in BMMs were defined by qRT-PCR 24 h after M.tb infection (H37Rv, CDC1551, and Erdman) or in uninfected cells. (D) As described for C, except IFN-β protein levels in the culture supernatant were quantified by ELISA. (E) Use of qRT-PCR to define IFN-β mRNA expression levels in control, STING, RIG-I, or MAVS siRNA-treated BMMs following a 24-h infection with the WT M.tb CDC1551 strain or in uninfected cells. At the time of infection, recombinant mouse IFN-β was added to BMMs at a final concentration of 5 or 25 pg/ml or was left untreated. (F) Same as E, except IFN-β protein levels in culture supernatant were quantified by ELISA. For qRT-PCR, the mRNA expression levels of IFN-β were expressed as fold change relative to WT H37Rv-infected cells treated with isotype antibody (C) or infected BMMs treated with control siRNA, no IFN-β (E). Levels were normalized to GAPDH. Data are representative of at least three independent experiments. Data are mean ± SD of triplicate wells. n.s., not statistically significant; ** P < 0.01 by two-tailed Student’s t test.
To determine if the IFN-β released during the early stages of an M.tb infection serves as an autocrine signal to promote the late IFN-β production in macrophages, BMMs were treated with the blocking antibody specific to IFNAR1 at different time points after infection. Blocking the IFN-β signaling pathway at the start or 4 h after infection resulted in a significant decrease in IFN-β transcription induced by all three WT M.tb strains (Fig. 6 C). However, IFNAR1-blocking antibody had no effect when added at 8 h after infection, suggesting that a STING-dependent early production of IFN-β is required for the subsequent IRF7-mediated IFN-β transcription (Fig. 6 C). Similar results were found for IFN-β protein production by M.tb-infected BMMs treated with the IFNAR1-blocking antibody (Fig. 6 D).
The experiments described above suggest that the STING-dependent production of IFN-β by M.tb-infected macrophages could be bypassed by the addition of exogenous IFN-β. As shown in Fig. 6 E, the addition of IFN-β to M.tb-infected STING knockdown BMMs rescued the IFN-β transcript levels to what was observed in control siRNA-treated and M.tb-infected cells. However, exogenous IFN-β failed to rescue the IFN-β response in RIG-I or MAVS knockdown BMMs infected with WT M.tb. Further, addition of exogenous IFN-β had no visible effect on IFN-β mRNA production in IRF7 knockdown BMMs infected with WT M.tb (Fig. S4 B). Similar results were observed at the protein level (Fig. 6 F and Fig. S4 C). Addition of exogenous IFN-β significantly increased IFN-β mRNA expression in M.tb-infected STING knockdown cells but not RIG-I, MAVS, or IRF7 knockdown cells. This functional rescue was specific to the IFN-β receptor-mediated signaling pathway as the addition of the IFNAR1 blocking antibody prevented the IFN-β production observed in STING knockdown cells primed with IFN-β (Fig. S4, D and E). Together, these results indicate that the initial IFN-β produced upon an M.tb infection is STING dependent. This early production of IFN-β works in an autocrine manner through its receptor IFNAR to stimulate IRF7 production, which is subsequently activated in a RIG-I/MAVS–dependent manner to significantly amplify IFN-β production in M.tb-infected host cells.
Host cytosolic RNA sensing pathway is required for in vivo IFN-β production and M.tb pathogenesis in mice
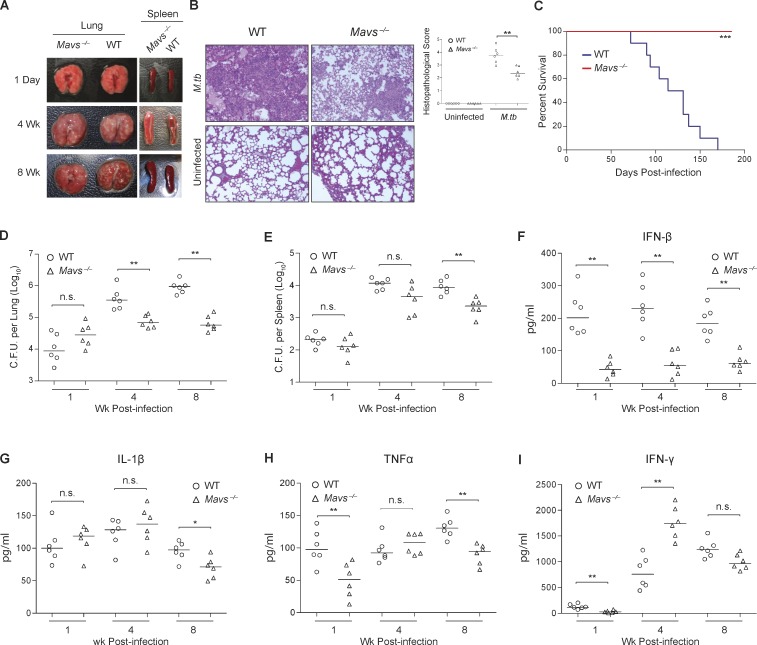
To address the importance of the RNA sensing pathway in the immune response to an M.tb infection, we infected Mavs−/− and congenic C57BL/6 mice using a low-dose aerosol route. The gross morphological and histopathological analysis indicates that at 4 wk after infection, there was an increased cellular infiltration within the lungs of WT infected mice when compared with infected Mavs−/− (Fig. 7, A and B). In addition, the spleens of infected Mavs−/− were enlarged relative to WT at 4 and 8 wk after infection (Fig. 7 A). Interestingly, Mavs−/− mice showed a significant increase in survival after a low-dose aerosol M.tb infection in comparison with WT C57BL/6 mice (Fig. 7 C). In addition, bacterial burden in the lung was significantly lower in Mavs-/– mice at 4 and 8 wk after infection compared with WT mice, while the spleens were statistically different only after 8 wk (Fig. 7, D and E). We evaluated the cytokine response by measuring the presence of IFN-β, IL-1β, TNF-α, and IFN-γ in serum of M.tb-infected Mavs−/− and WT mice after infection (Fig. 7, F–I). Consistent with the in vitro study using Mavs−/− BMMs, decreased IFN-β production was observed in the serum of M.tb-infected Mavs−/− compared with infected WT mice at all time points (Fig. 7 F). Elevated production of IL-1β and TNF-α was observed in the serum of WT mice 8 wk after infection, but this may be a consequence of a higher bacterial burden in WT infected mice (Fig. 7, G and H). In contrast, IFN-γ production was higher in the serum of M.tb-infected Mavs−/− mice but only at the 4-wk time point (Fig. 7 I).
Figure 7.
MAVS is required for M.tb-induced IFN-β production and facilitates bacterial survival in vivo. (A) Representative pictures of Mavs−/− and WT C57BL/6 mouse lungs 1 d and 2, 4, and 8 wk after infection. (B) Representative H&E staining of lung sections from Mavs−/− and WT C57BL/6 mice and histopathological scoring of multiple lung sections at 4 wk after infection. (C) Kaplan–Meier survival analysis of Mavs−/− and WT C57BL/6 mice infected with WT CDC1551 M.tb via the aerosol route (n = 10 per group). *** P < 0.001 by log-rank test. (D and E) Mycobacterial burdens in the lung (D) and spleen (E) of mice at different times after infection. (F–I) The ELISA analysis of IFN-β (F), IL-1 β (G), TNF-α (H), and IFN-γ (I) in the serum of mice infected with M.tb. n.s., not statistically significant; for D–I (n = 3 per group), * P < 0.05 and ** P < 0.01 by Mann–Whitney U test.
Discussion
The importance of bacterial RNA as an inducer of a host immune response remains relatively undefined. Most of the studies have focused on purified bacterial RNA and transfection of target cells to evaluate their roles (Eberle et al., 2009; Mancuso et al., 2009; Eigenbrod et al., 2012; Li and Chen, 2012; Oldenburg et al., 2012; Cervantes et al., 2013; Eigenbrod and Dalpke, 2015; Fieber et al., 2015). Koski et al. demonstrated that human dendritic cells (DCs) transfected with bacterial RNA but not eukaryotic RNA induce high levels of IL-12 (Koski et al., 2004), and more recent studies showed that group B Streptococcus RNA can induce IFN-β production in mouse DCs through TLR7 (Eberle et al., 2009; Mancuso et al., 2009). Obregón-Henao et al. found small–molecular weight RNA, present in the M.tb culture filtrate, which could stimulate human monocyte apoptosis in a caspase-8–dependent pathway (Obregón-Henao et al., 2012). However, few studies have focused on the contribution of RNA during a live bacterial infection. A recent publication by Fieber et al. has identified a role for TLR13 in TNF and IL-6 production by murine DCs upon a Streptococcus pyogenes infection (Li and Chen, 2012; Fieber et al., 2015). Our studies identified M.tb RNA in exosomes released from infected murine macrophages (Singh et al., 2015) suggesting that M.tb RNA is released during an active macrophage infection. The presences of extracellular RNA could stem from bacterial lysis and/or active transport. Our bacterial culture studies, however, using DsRed-expressing BCG and M.tb suggest a selectivity for which RNA species are released, although we cannot rule out that some of the extracellular RNA is due to cell lysis, particularly within an infected macrophage.
Extracellular release of bacterial DNA through the type IV secretion system has been well studied (Christie et al., 2014); however, little is known about mechanisms of RNA release. Studies with L. monocytogenes identified that bacterial RNA was released into bacterial culture supernatant in a SecA2-dependent way (Abdullah et al., 2012). Our study found M.tb RNA in the cytosol of infected macrophages, and this localization required both the bacterial SecA2 and Esx-1 secretion systems. Our studies support a role for SecA2 in the release of the M.tb RNA from the bacteria and Esx-1 for the release of the RNA into the cytosol. The SecA2 secretion system is found in a limited number of gram-positive bacteria including Listeria, Staphylococcus aureus, Streptococcus gordonii, and all tested mycobacterial species (Rigel and Braunstein, 2008). Although not required for bacterial survival and growth in culture media, the SecA2 M.tb mutant is attenuated in both cultured macrophages and mice due in part to the SecA2-dependent release of proteins involved in inhibiting phagosome maturation (Kurtz et al., 2006; Sullivan et al., 2012). The M.tb RNA-mediated production of IFN-β and SecA2’s involvement in the release of M.tb RNA during an infection may also play into SecA2’s role in virulence. How SecA2 mediates the transport of bacterial RNA through the M.tb cytoplasmic membrane and whether there is selectivity and regulation of this process is presently unknown. One possible mechanism involves RNA-binding proteins with chaperon-like function that capture and assist bacterial RNA transport via the SecA2 system (Miller et al., 2017). Interestingly, we did not see a SecA2-dependent release of M.tb DNA again, suggesting some specificity in binding/transport.
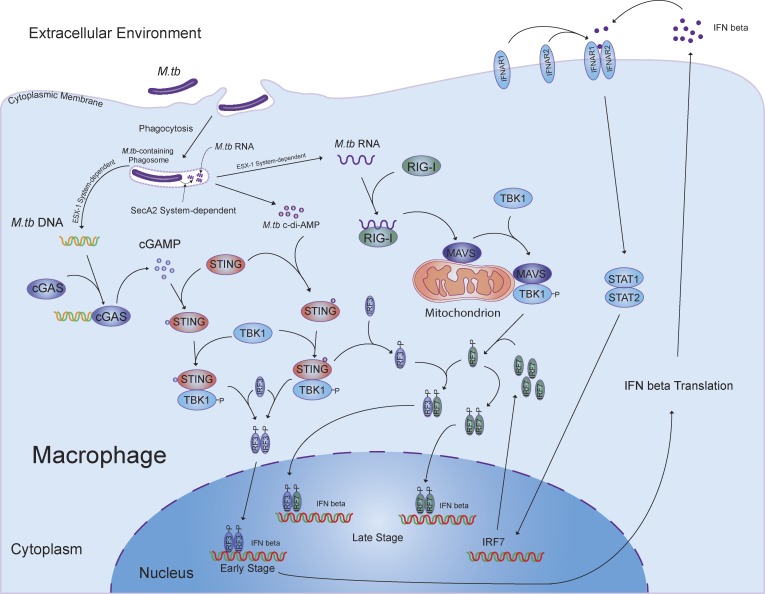
The secA2-deficient M.tb provides an excellent model to address the role of bacterial RNA in the innate immune response during an infection. We measured IFN-β production in host cells infected with secA2-deficient M.tb as well as WT and complemented CDC1551 strains. Although the initial production of IFN-β was not SecA2 dependent, the overall expression of IFN-β was significantly decreased in cells infected with the ΔsecA2 CDC1551 when measured 24 h after infection. This stimulation was M.tb RNA dependent as both RIG-I and MAVS, two well-known proteins involved in the host RNA sensing pathway, were required for this SecA2-dependent induction of IFN-β, and RIG-I directly recognized cytosolic M.tb mRNA. As observed with the SecA2-deficient CDC1551, the early induction of IFN-β production was not dependent on RIG-I or MAVS, analogous to the previous study which demonstrated a MAVS-independent IFN-β production at 3 h after infection (Manzanillo et al., 2012). Moreover, our studies as well as previous work demonstrate a requirement for the STING-dependent DNA sensing pathway in stimulating IFN-β production during an M.tb infection (Manzanillo et al., 2012; Collins et al., 2015; Dey et al., 2015; Wassermann et al., 2015; Watson et al., 2015). In these studies, activation of STING was found to be induced either by M.tb DNA or through M.tb c-di-AMP. Our current study shows that during an M.tb infection, IFN-β production is regulated via a mechanism involving both cytosolic DNA and RNA sensing pathways (Fig. 8). M.tb c-di-AMP appears to play a minimal role in our study as we observed a similar dependence on both cGAS and STING for induction of IFN-β upon an M.tb infection. The initial cGAS- and STING-dependent IFN-β production following an M.tb infection functions through autocrine binding to INFAR to induce expression of IRF7. IRF7, which is known to bind to the IFN-β promoter either as a homodimer or a heterodimer with IRF3 (Honda and Taniguchi, 2006), is subsequently activated in a RIG-I/MAVS/TBK1–dependent pathway to markedly enhance IFN-β production. Overall, the cGAS/STING-dependent DNA sensing pathway activated by M.tb DNA is critical for the type I IFN production during the course of M.tb infection, and the RIG-I/MAVS–dependent RNA sensing pathway participates at later times after infection as a second essential factor to initiate a robust production of type I IFNs in host cells. The M.tb DNA abundance and cGAMP levels in host cells decline over time during M.tb infection (Fig. 1 G; Collins et al., 2015), suggesting that cGAS may not promote IFN-β production at later times after infection. However, it is clearly required for the initial IFN-β production, which, as indicated above, is required for driving IRF7 expression and subsequent IFN-β production.
Figure 8.
Cross-talk between the host DNA and RNA signaling pathways contributes to an M.tb-induced IFN-β production in macrophages.
A dose-dependent response to IFN-β indicates that both infected and uninfected BMMs produce IRF7, although infected cells are stimulated to produce IRF7 at a lower concentration of IFN-β relative to uninfected cells. This suggests that engagement of other PRRs during an M.tb infection provides some costimulation, although which signaling pathways are involved is presently unclear. Both MAVS and STING are known to activate the kinase TBK1, which subsequently phosphorylates and activates the transcription factors IRF3 and IRF7 (McNab et al., 2015). However, the TBK1 activated by MAVS and STING may be spatially separated as previous studies using HEK293T cells and Sendai virus infection have shown that stable complexes are formed between STING, TBK1, and IRF3 as well as MAVS, TBK1, and IRF3 (Tanaka and Chen, 2012; Liu et al., 2015). These studies suggest that the activated forms of STING and MAVS function as scaffolds to recruit TBK1 and IRF3 and potentially IRF7 into a complex. Our results showed a diminished TBK1 activation in the MAVS knockdown BMMs, resulting in an undetectable IRF7 but no change in IRF3 nuclear localization. These results support a spatial separation of STING and MAVS activation of TBK1 and its phosphorylation of IRF3 and IRF7, respectively. However, confirmation of this hypothesis awaits further study.
The RIG-I/MAVS–dependent RNA signaling is required for immunity against a number of viral pathogens (Kumar et al., 2006; Sun et al., 2006). However, its importance in antibacterial immunity remains to be elucidated (Li et al., 2011). Our infection studies using Mavs−/− mice support a role for mycobacterial RNA in driving a host immune response, including IFN-β production, in vivo. Surprisingly, unlike the findings in cGAS−/− and STINGgt/gt mice (Collins et al., 2015; Watson et al., 2015), M.tb growth is significantly attenuated in Mavs−/− mice over time, as was observed in the Irf3−/− mice, partially supporting the hypothesis that type I IFN functions as a negative regulator during an M.tb infection (Manzanillo et al., 2012; McNab et al., 2015). Although cGAS and STING are crucial for the IFN-β production during an M.tb infection, they also contribute to other host cellular responses including autophagy and inflammasome activation that restrict M.tb survival. Diminished inflammasome activation associated with loss of cGAS and STING expression may mask the positive effect on host immunity associated with lower type I IFN production in these knockout mice (Wassermann et al., 2015; Watson et al., 2015). Besides the type I IFNs, other host factors driven by the MAVS activation might also play a role in the immune response during an M.tb infection. Interestingly, Mavs−/− mice show an increased level of IFN-γ in the serum of M.tb-infected mice, which might also contribute to the diminished bacterial load in the lung. Previous studies in Mycobacterium leprae indicate that IFN-γ production was preferentially expressed in self-healing tuberculoid lesions, while IFN-β and IL-10 were preferentially expressed in progressive lepromatous lesions (Teles et al., 2013). This study also showed that IFN-β and IL-10 inhibited the antimicrobial peptide response normally induced in macrophages upon IFN-γ stimulation. Our data support the hypothesis that differential production of IFNs may play a role in mycobacterial pathogenesis.
In summary, our studies indicate a role for M.tb RNA in driving an immune response and support a role for bacterial RNA as an important factor in the host recognition of a bacterial infection. Future studies will need to address the relative importance of this foreign RNA-driven immunity during an M.tb infection, particularly in the context of a human infection, as well as gain a better understanding of the mechanism by which RNA is released from the mycobacteria and whether other bacterial secretion systems are capable of transporting RNA across their cell membrane.
Materials and methods
Mice
Mavs−/− mice on a C57BL/6 background were obtained as a generous gift from Dr. Stanley Perlman (University of Iowa, Iowa City, IA; Suthar et al., 2012; Zhao et al., 2016). WT C57BL/6 mice were originally purchased from Jackson Laboratories and bred at the University of Notre Dame. Breeding and housing of all mice used for these experiments took place within the institutional animal facility under specific pathogen-free conditions. The M.tb infection was performed in the biosafety level 3 laboratory. The University of Notre Dame is accredited through the Animal Welfare Assurance (A3093-01). All animal experiments were approved by the Institutional Animal Care and Use Committees of University of Notre Dame.
Bacterial strains
All M.tb Erdman strains were the kind gifts of Dr. Patricia Champion (University of Notre Dame, Notre Dame, IN). WT M.tb CDC1551 was ordered from BEI Resources. The M.tb secA2 (MT1869) transposon insertion mutant strain (position of insertion: 1352) was from the Tuberculosis Animal Research and Gene Evaluation Taskforce. Before infection of macrophages, all M.tb strains were grown in MiddleBrook 7H9 broth (cat. 271310; BD) supplemented with 10% (vol/vol) Middlebrook oleic acid-albumin-dextrose-catalase (OADC; cat. 211886; BD) and 0.2% glycerol until midexponential phase, and washed three times with complete medium for BMMs or Raw264.7 cell when required.
Cell culture
BMMs were isolated from WT C57BL/6, Mavs−/−, or MyD88−/− mice (female, 6–8 wk) as described previously (Roach and Schorey, 2002), and cells were grown in DMEM (D7777; Sigma-Aldrich) supplemented with 10% (vol/vol) heat-inactivated FBS (SH30910.03; Hyclone), 20% L929 cell-conditional medium as a source of macrophage colony-stimulating factor, and 100 U/ml penicillin and 100 U/ml streptomycin (SV30010; HyClone) at 37°C and 5% CO2. The Raw264.7 cells were cultured in DMEM supplemented with 10% (vol/vol) heat-inactivated FBS.
siRNA transfection
Mouse BMMs (3 × 105 cells per well) were transfected with siRNA oligos (25 pmol/3 × 105 cells) listed in Table S1 in 24-well plates using Lipofectamine 2000 (cat. 11668-027; Invitrogen) following the manufacturer’s protocol in BMM complete medium for 24 h at 37°C and 5% CO2. AllStars Negative Control siRNA (cat. 1027280; Qiagen) was used as a negative control. During M.tb infection, siRNAs were always included in the cell culture.
Generation of ΔsecA2 complementary strain
To rescue M.tb CDC1551 secA2-inactivated mutant, the SecA2-encoding gene, MT1869, was cloned into an integrative vector pMH406-hyg, a generous gift from Dr. Patricia Champion (University of Notre Dame, Notre Dame, IN). Briefly, a 2,456-bp DNA fragment covering the secA2 open reading frame was amplified using two phosphorylated primers, forward (5′-CTGCAGAATTCAGGAGTCCAGCGTGAACGTGCACGGTTGTCC-3′) and reverse (5′-GCGTTAACTCAGCGGAACACCCCGGG-3′; Table S1); Phusion high-fidelity DNA polymerase (M0530; NEB); and M.tb CDC1551 genomic DNA as the template. The vector pMH406-hyg was digested with restriction endonuclease PvuII and HpaI (NEB), and the blunt ends were dephosphorylated with Shrimp Alkaline Phosphatase (M0371; NEB). The PCR products were purified using Qiagen PCR Purification Kit (cat. 28104; Qiagen) following the manufacturer’s protocol and then cloned into the linearized and dephosphorylated pMH406-hyg. The resulting plasmid named as pMH406-sA2 was verified by digestion with restriction endonuclease PvuII and HpaI, confirmed by DNA sequencing, and finally transformed into the ΔsecA2 strain of M.tb by electrotransformation as described previously (Larsen et al., 2007). The positive M.tb colonies were selected on Middlebrook 7H10 agar plates (cat. M199; HiMedia Laboratories) supplemented with 0.5% glycerol, 10% OADC, 50 µg/ml of hygromycin, and 20 µg/ml of kanamycin at 37°C. Three independent colonies were picked and confirmed by PCR using secA2-specific primers and Western blot of PhoS1 (data not shown), one substrate of the SecA2 secretion system (Feltcher et al., 2015).
Survival assay of M.tb strains in macrophages
The BMMs were infected with M.tb CDC1551 strains at a multiplicity of infection (MOI) of 10 for 1 h, and cells were washed with complete BMM medium as described above. Cells were incubated for another 1, 5, and 24 h at 37°C and 5% CO2 and subsequently washed with precold PBS three times and lysed in 0.05% SDS. A series of dilution of cell lysates in PBS (1×) were added onto 7H11 agar plates (cat. 7244A; Acumedia) supplemented with 10% (vol/vol) OADC and 0.2% glycerol. Plates were incubated at 37°C for 3–4 wk until counting.
RNA purification
To determine M.tb secreted/released RNAs (seRNAs) in mycobacterial culture supernatant, M.tb strains were recovered from frozen stocks at −80°C and grown in 7H9 broth supplemented with 10% (vol/vol) OADC until midexponential phase. 1 ml of mycobacterial culture was centrifuged at 2,000 rpm for 5 min at room temperature (RT) and washed with 7H9 broth three times before inoculation into a fresh 100-ml complete 7H9 liquid medium and grown until midexponential phase. The mycobacteria were inoculated into fresh media twice to minimize the presence of any dead bacteria. The M.tb cultures were spun at 2,000 rpm for 10 min at RT. The M.tb pellets were washed with TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) three times. Cells were disrupted using a beadbeater, and total cellular RNAs were purified using Qiagen RNeasy Plus Mini Kit. For seRNAs, M.tb culture supernatant from previous centrifugation was passed through a 0.2-micron filter (cat. 28145-501; VWR), followed by an ultracentrifugation at 100,000 xg for 2 h at 4°C. The resulting supernatant was mixed with 0.1 volume of NH4OAc (5 M) and one volume of precold isopropanol and incubated at −70°C for 30 min. The RNAs were spun down at 14,000 xg for 15 min at 4°C. The RNA pellets were resuspended in Buffer RLT Plus and further purified using Qiagen RNeasy Plus Mini Kit. To analyze M.tb transcripts in the cytosol of infected macrophages, Raw264.7 cells were infected with various M.tb strains, and cytosolic RNA was isolated as described in Purification of EU-labeled M.tb RNAs from infected host cells. Total cellular RNA was purified from infected cells with Qiagen RNeasy Plus Mini Kit. To measure IFN-β expression level in BMMs, cells were infected with M.tb strains at a MOI of 10 for various times as indicated. Total cell RNA was purified using Qiagen RNeasy Plus Mini Kit.
For isolation of RNA released from M.tb, phagosome-enriched material was isolated from M.tb-infected Raw264.7 cells 24 h after infection (MOI = 10), as described previously (Beatty et al., 2002). Briefly, cells were resuspended in homogenization buffer (20 mM Hepes [pH 7.4], 250 mM sucrose, 10 mM KCl, 2 mM MgCl2, and 1 mM EDTA) plus RNase Inhibitor (1 U/μl) and disrupted by multiple passage through a 3-ml syringe with a 25 G needle. Cell lysate was centrifuged at 300 xg for 10 min at 4°C and two subsequent postnuclear spins at 100 xg for 10 min at 4°C. The resulting supernatant was added onto a step gradient of 50 and 12% sucrose and spun at 800 xg for 45 min at 4°C. The 12%/50% interface was isolated, diluted 1:2 in homogenization buffer, layered on 10% Ficoll 70,000/150 mM sucrose, and centrifuged at 1,300 xg for 45 min at 4°C. The pellet was resuspended in homogenization buffer and layered on 12% Ficoll 400,000/150 mM sucrose, and spun at 1,500 xg for 45 min at 4°C. The resulting pellet containing phagosomes was gently lysed in the lysis buffer (10 mM Hepes [pH 7.8], 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, and 0.5% NP-40) containing RNase inhibitor (1 U/μl) in ice for 10–15 min, which differentially disrupted phagosome membrane but not M.tb cell wall. The lysates were centrifuged at 1,200 xg for 10 min at 4°C to pellet M.tb. The RNA in supernatant was purified as M.tb seRNAs using Qiagen RNeasy Plus Mini Kit, and RNA in M.tb pellet (total cellular RNA) was purified as described above.
qRT-PCR
The RNAs were first treated with DNase I (cat. 18068015; Invitrogen) following the manufacturer’s protocol. For M.tb seRNAs and cytosolic RNAs, cDNA was synthesized with avian myeloblastosis virus (AMV) reverse transcription (M0277; NEB) and one of the M.tb gene-specific primers as listed in Table S1. For IFN-β analysis, cDNA was synthesized using AMV reverse transcription and Oligo(dT)20 primer. Quantitative PCR using PerfeCTa SYBR Green SuperMix (cat. 95054; Quantabio) and the primers listed in Table S1 was performed on StepOnePlus Real-Time PCR System (Applied Biosystems).
Isolation of bacterial culture supernatant and host cytosolic DNA and PCR
For M.tb culture supernatant DNA isolation, M.tb was grown and culture supernatant was prepared as described above for M.tb seRNAs isolation. The DNA was precipitated with 100% ethanol (three volumes) and sodium acetate (3 M, pH 5.2, 0.1 volume) at RT for 30 min and further extracted in phenol-chloroform. Finally, extracellular DNA was precipitated again using 100% ethanol and sodium acetate. For host cytosolic DNA, Raw264.7 cells were infected with M.tb CDC1551 strains at an MOI of 10 for 4, 8, and 24 h at 37°C and 5% CO2. Cytosolic fraction of infected cells was prepared as described in Purification of EU-labeled M.tb RNAs from infected host cells, and extracted with one volume of phenol/chloroform/isoamyl alcohol (pH 6.6; FisherBiotech). Cytosolic DNA was precipitated by adding 0.1 volume of NaOAc (3 M, pH 5.5) and two volumes of 100% ethanol. To determine the level of M.tb DNA fragments, isolated DNA was treated with RNase A (cat. 12091021; Invitrogen) and then served as the template in PCR mix using GoTaq Green Master Mix and M.tb-specific primers described previously (Watson et al., 2015).
Purification of EU-labeled M.tb RNAs from infected host cells
The M.tb RNA was labeled and isolated using Click-iT Nascent RNA Capture Kit (C10365; Life Technologies) following the manufacturer’s instruction. Briefly, M.tb strains were labeled in 7H9 broth supplemented with 10% (vol/vol) OADC in the presence of EU (0.2 mM) for 24 h at 37°C. Labeled bacterial cells were washed with complete DMEM three times to remove extracellular EU. The Raw264.7 cells were infected with labeled M.tb strains at a MOI of 10 for 24 h at 37°C. Total cellular RNA was isolated from infected cells using Qiagen RNeasy Mini Kit (cat. 74136; Qiagen), and cytosolic RNA from cytosolic fraction by ethanol precipitation. For cytosolic RNA, cells were washed with precold PBS three times and then resuspended in lysis buffer (20 mM Hepes [pH 7.4], 250 mM sucrose, 10 mM KCl, 2 mM MgCl2, and 1 mM EDTA) plus RNase inhibitor (M0314; NEB) and disrupted with a syringe and 27 G needle by 25 strikes. Cell lysates were then centrifuged at 350 xg for 5 min at 4°C and subsequently at 12,000 xg for 10 min at 4°C, to remove cell debris and undisrupted cells. The supernatant was passed through a 0.2-micron filter and centrifuged at 100,000 xg for 2 h at 4°C. One volume of Acid-Phenol:Chloroform (pH 4.5; cat. AM9720; Ambion) was added into the resulting supernatant, vortexed for 30–60 s, and centrifuged at 12,000 xg for 5 min at 4°C to remove proteins and DNA. Aqueous phase was transferred into a new tube and mixed with 0.1 volume of 3 M NaOAc (pH 5.0) and three volumes of ice cold 100% ethanol. RNA was precipitated at −20°C for 1 h and centrifuged at 14,000 xg for 30 min at 4°C. The RNA pellet was washed with 0.5 ml ice cold 70% ethanol three times and resuspended in 100 μl nuclease-free water. The EU-labeled M.tb RNA was purified from prepared total cellular RNA and cytosolic RNA following the manufacturer’s protocol. Captured RNA was eluted from Dynabeads MyOne Streptavidin T1 magnetic beads in 20 μl of elution buffer (10 mM EDTA [pH 8.2] and 95% formamide) at 65°C for 2 min. Eluted solution was then mixed with two volumes (40 μl) of RNase-free water, 0.1 volume of 3 M NaOAc, and four volumes (240 μl) of 100% EtOH and incubated at −70 for 1 h. RNA was centrifuged at 14,000 xg for 30 min at 4°C and washed with 0.5 ml ice cold 70% ethanol three times and resuspended in 20 μl nuclease-free water.
Coimmunoprecipitation
The Raw264.7 cells were infected with WT M.tb CDC1551 at a MOI of 10 for 24 h at 37°C and then washed with precold PBS (1×) three times. Cells were lysed in NP-40 lysis buffer (50 mM Hepes [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, and 0.5 mM dithiothreitol) with 1 U/μl RNase inhibitor (M0314; NEB) in ice for 30 min. The cell lysates were centrifuged at 12,000 xg, 5 min, 4°C, and supernatant was used for immunoprecipitation. Protein A/G PLUS-agarose beads (20 μl; SC-2003; Santa Cruz Biotech) were washed with NP-40 lysis buffer three times and incubated with RIG-I (cat. 3743; Cell Signaling Technology) and MDA-5 (cat. 5321; Cell Signaling Technology) monoclonal antibodies overnight at 4°C with gentle rolling. Antibody-conjugated beads were incubated with prepared cell lysates for 5 h at 4°C with gentle rolling in the presence of 1 U/μl RNase inhibitor. Beads were washed with NP-40 lysis buffer three times and treated with proteinase K (cat. 97062-238; VWR) at 50°C for 30 min. Proteins were removed by Acid-Phenol:Chloroform (pH 4.5) extraction. The RNA was precipitated by adding 0.1 volume of 3 M NaOAC and one volume of precold isopropanol and resuspended in nuclease-free water as described above. Purified RNA was used for RT-PCR analysis. Before Acid-Phenol:Chloroform extraction, an aliquot of samples were stored for Western blot analysis. For RT-PCR, RNAs were treated with DNase I (cat. 18068015; Invitrogen), and cDNA were synthesized using AMV reverse transcription (M0277; NEB). PCR was performed with GoTaq Green Master Mix (M712B; Promega) and primers listed in Table S1.
Whole-cell lysates (WCLs) and nuclear fraction preparation
The siRNA-treated BMMs were infected for 4 or 24 h with the indicated M.tb strains at a MOI of 10 and washed with precold PBS three times. WCLs were prepared by adding WCL lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 1.0% Triton X-100) containing 1× protease inhibitor cocktail (P8340; Sigma-Aldrich) and incubated in ice for 30 min. Nuclear fraction was prepared as described previously (Flaherty et al., 2002). Briefly, cells were washed in precold PBS three times and with lysis buffer (10 mM Hepes [pH 7.8], 10 mM KCl, 2 mM MgCl2, and 0.1 mM EDTA) once. Cell pellets were then resuspended in 500 µl of lysis buffer containing 1× protease inhibitor cocktail and incubated in ice for 15 min, and then 25 µl of 10% NP-40 was added and mixed thoroughly. Cell lysates were centrifuged at 1,200 xg for 10 min at 4°C. The pellets (nuclear fraction) were washed with lysis buffer three times and resuspended in WCL lysis buffer and incubated in ice for 30 min before adding SDS-loading buffer (5×).
Immunoblotting
For Western blot analysis, prepared WCL and nuclear fraction were denatured at 95°C for 10 min, separated by 12.5% SDS-PAGE gel. Proteins were transferred onto nitrocellulose membranes and blotted with rabbit anti-IRF3 (cat. A303-384A; Bethyl Laboratories Inc.), anti-IRF7 (cat. 3941; Prosci Inc.), anti-TBK1 (cat. 3504; Cell Signaling Technology), anti–phospho-TBK1 (Ser172; cat. 5483; Cell Signaling Technology), anti–RIG-I (cat. 3743; Cell Signaling Technology), anti–MDA-5 (cat. 5321; Cell Signaling Technology), anti-cGAS (cat. Sc-515777; SCBT), anti–β-actin (cat. 4970; Cell Signaling Technology), anti-MAVS (cat. Sc-365333; SCBT), anti-STING (cat. NBP2-24683SS), and anti-histone H3 (cat. 9717; Cell Signaling Technology) antibodies, followed by goat anti-rabbit IgG-HRP (cat. 31460; Thermo Scientific).
For M.tb culture supernatant proteins, M.tb strains were grown in Sauton’s liquid medium until midexponential phase, and culture supernatant fraction was prepared from 50 ml bacterial culture as described previously (Reyna et al., 2016). 10 µg of sample was loaded into 12% SDS-PAGE gel, and M.tb RNA polymerase subunit β (RNAP-β) and EsxB (CFP-10) proteins were probed using mouse anti–RNAP-β antibody (ab12087; Abcam) and rabbit anti-EsxB antibody (NR-13801; BEI Resources), respectively.
IFNAR blocking and stimulating assays
For receptor blocking assay, siRNA-treated BMMs were infected with WT M.tb strains at an MOI of 10, and IFNAR1 blocking antibody (cat. 127303; BioLegend) was added into the cell culture at a final concentration of 2 µg/ml at 0, 4, and 8 h after infection. Mouse IgG1 isotype (cat. 401403; BioLegend) served as a negative control. For receptor stimulating assay, recombinant mouse IFN-β (cat. 8234-MB-010; R&D Systems) was added at the beginning of M.tb infection into the cell culture media at the final concentration indicated. The IFN-β expression level was determined by RT-qPCR and ELISA at 24 h after infection.
Mouse infection and survival assay
Mavs−/− and WT C57BL/6 mice (8–10 wk old, female) were aerosol-infected with WT M.tb (CDC1551) to a dose of ∼1,000 CFUs in the lung, using a Glas-Col Inhalation Exposure System (Glas-Col) as described previously (Cheng and Schorey, 2013). The M.tb input in the lung of mice was determined at day 1. 4 and 8 wk after infection, mouse serum was harvested via cardiac puncture and prepared using BD Microtainer Serum Separator Tube. Additionally, mouse lungs and spleens were harvested, homogenized, and plated onto Middlebrook 7H11 agar plates, and mycobacterial colonies were counted after 3–4 wk of incubation at 37°C and expressed as log10 CFUs per organ. For pathological analysis, mouse lung sections were prepared and stained with H&E at the Histology Core Facility of University of Notre Dame, and histopathological score was evaluated as described previously (Cheng and Schorey, 2013). For mouse survival experiment, Mavs−/− and WT C57BL/6 mice were infected as described above, and the severity of clinical signs was monitored. Morbid mice were killed according to Institutional Animal Care and Use Committees protocol and counted as a data point the day of sacrifice.
Alveolar macrophage isolation
Mice were infected with M.tb strains as described above, and mouse bronchoalveolar lavage was prepared as described previously (Gonzalez-Juarrero et al., 2003). Alveolar macrophages in bronchoalveolar lavage were further purified using Dynabeads Protein G (cat. 10003D; Invitrogen) conjugated with anti-mouse F4/80 antibody (cat. 123102; BioLegend). Cytosolic RNA and total cellular RNA were purified as described above.
ELISA
Macrophage culture supernatants were used to measure IFN-β and TNFα level at 24 h after M.tb infection. Culture supernatants were centrifuged at 1,000 xg for 10 min at 4°C and subsequently passed through 0.2-micron filter two times. The ELISA was performed according to the manufacturer’s protocol (eBioscience) using Avidin-HRP (cat. 18-4100-94; eBioscience) and TMB (cat. 00-4201-56; eBioscience). For TNFα, capture antibody (cat. 14-7341-85; eBioscience), detection antibody (cat. 13-7341-85; ebioscience), and TNFα standard (cat. 39-8321-60; eBioscience) were used. For IFN-β, capture antibody (purified anti-mouse IFN-β antibody, cat. 519202; BioLegend), detection antibody (biotin anti-mouse IFN-β antibody, cat. 508105; BioLegend), and IFN-β standard (cat. 581309; BioLegend) were used. Mouse IFN-γ and IL-1β were measured using IFN gamma (cat. 88-7314-22; eBioscience) and IL-1 β (cat. 88-7013-22; eBioscience) mouse ELISA kit, respectively.
Statistical analysis
Statistical analysis was performed to determine differences between groups by two-tailed paired Student’s t tests, Mann–Whitney U test, or Kaplan–Meier survival analysis using GraphPad Prism software (version 5.04). A P value < 0.05 was considered significant.
Online supplemental material
In Fig. S1, we show that M.tb releases mycobacterial but not DsRed transcripts into the bacterial culture supernatant. In Fig. S2, we demonstrate that the ESX-1 secretion system is important for delivery of mycobacterial mRNAs into the macrophage cytosol but not into the culture supernatant. In Fig. S3, we characterized the efficiency of the siRNA-mediated knockdown of MAVS, STING, RIG-I, and IRF7 in mouse BMMs. In Fig. S4, we show that addition of exogenous IFN-β to M.tb-infected macrophages did not rescue IFN-β production in IRF7 knockdown cells. Table S1 shows primer and siRNA sequences.
Supplementary Material
Acknowledgments
We are deeply grateful to Dr. Stanley Perlman from the University of Iowa for providing the MAVS–/– mice. We are also very appreciative to the Tuberculosis Animal Research and Gene Evaluation Taskforce Program at the John Hopkins University School of Medicine for providing us with the SecA2 CDC1551 mutant. We very much appreciate the gift of the M.tb Erdman strains from Dr. Patricia Champion at the University of Notre Dame.
Funds for this work were provided by the National Institute of Allergy and Infectious Diseases grant AI052439.
The authors declare no competing financial interests.
Author contributions: Y. Cheng was responsible for the experiments performed in this study and assisted in the writing of the manuscript. J.S. Schorey aided in the design of the experiments and wrote the manuscript.
References
- Abdullah Z., Schlee M., Roth S., Mraheil M.A., Barchet W., Böttcher J., Hain T., Geiger S., Hayakawa Y., Fritz J.H., et al. 2012. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 31:4153–4164. 10.1038/emboj.2012.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty W.L., Rhoades E.R., Hsu D.K., Liu F.-T., and Russell D.G.. 2002. Association of a macrophage galactoside-binding protein with Mycobacterium-containing phagosomes. Cell. Microbiol. 4:167–176. 10.1046/j.1462-5822.2002.00183.x [DOI] [PubMed] [Google Scholar]
- Berry M.P.R., Graham C.M., McNab F.W., Xu Z., Bloch S.A.A., Oni T., Wilkinson K.A., Banchereau R., Skinner J., Wilkinson R.J., et al. 2010. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 466:973–977. 10.1038/nature09247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P., Eiglmeier K., Marmiesse M., Billault A., Garnier T., Niemann S., Cole S.T., and Brosch R.. 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect. Immun. 70:5568–5578. 10.1128/IAI.70.10.5568-5578.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker S.W., Bonham K.S., Zanoni I., and Kagan J.C.. 2015. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 33:257–290. 10.1146/annurev-immunol-032414-112240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes J.L., La Vake C.J., Weinerman B., Luu S., O’Connell C., Verardi P.H., and Salazar J.C.. 2013. Human TLR8 is activated upon recognition of Borrelia burgdorferi RNA in the phagosome of human monocytes. J. Leukoc. Biol. 94:1231–1241. 10.1189/jlb.0413206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., and Schorey J.S.. 2013. Exosomes carrying mycobacterial antigens can protect mice against Mycobacterium tuberculosis infection. Eur. J. Immunol. 43:3279–3290. 10.1002/eji.201343727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P.J., Whitaker N., and González-Rivera C.. 2014. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta. 1843:1578–1591. 10.1016/j.bbamcr.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A.C., Cai H., Li T., Franco L.H., Li X.-D., Nair V.R., Scharn C.R., Stamm C.E., Levine B., Chen Z.J., and Shiloh M.U.. 2015. Cyclic GMP-AMP synthase is an innate immune DNA sensor for Mycobacterium tuberculosis. Cell Host Microbe. 17:820–828. 10.1016/j.chom.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B., Dey R.J., Cheung L.S., Pokkali S., Guo H., Lee J.-H., and Bishai W.R.. 2015. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat. Med. 21:401–406. 10.1038/nm.3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorhoi A., Yeremeev V., Nouailles G., Weiner J. III, Jörg S., Heinemann E., Oberbeck-Müller D., Knaul J.K., Vogelzang A., Reece S.T., et al. 2014. Type I IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. Eur. J. Immunol. 44:2380–2393. 10.1002/eji.201344219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle F., Sirin M., Binder M., and Dalpke A.H.. 2009. Bacterial RNA is recognized by different sets of immunoreceptors. Eur. J. Immunol. 39:2537–2547. 10.1002/eji.200838978 [DOI] [PubMed] [Google Scholar]
- Eigenbrod T., and Dalpke A.H.. 2015. Bacterial RNA: An underestimated stimulus for innate immune responses. J. Immunol. 195:411–418. 10.4049/jimmunol.1500530 [DOI] [PubMed] [Google Scholar]
- Eigenbrod T., Franchi L., Muñoz-Planillo R., Kirschning C.J., Freudenberg M.A., Núñez G., and Dalpke A.. 2012. Bacterial RNA mediates activation of caspase-1 and IL-1β release independently of TLRs 3, 7, 9 and TRIF but is dependent on UNC93B. J. Immunol. 189:328–336. 10.4049/jimmunol.1103258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltcher M.E., Gunawardena H.P., Zulauf K.E., Malik S., Griffin J.E., Sassetti C.M., Chen X., and Braunstein M.. 2015. Label-free quantitative proteomics reveals a role for the Mycobacterium tuberculosis SecA2 pathway in exporting solute binding proteins and Mce transporters to the cell wall. Mol. Cell. Proteomics. 14:1501–1516. 10.1074/mcp.M114.044685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieber C., Janos M., Koestler T., Gratz N., Li X.-D., Castiglia V., Aberle M., Sauert M., Wegner M., Alexopoulou L., et al. 2015. Innate immune response to Streptococcus pyogenes depends on the combined activation of TLR13 and TLR2. PLoS One. 10:e0119727 10.1371/journal.pone.0119727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty D.M., Monick M.M., Carter A.B., Peterson M.W., and Hunninghake G.W.. 2002. Oxidant-mediated increases in redox factor-1 nuclear protein and activator protein-1 DNA binding in asbestos-treated macrophages. J. Immunol. 168:5675–5681. 10.4049/jimmunol.168.11.5675 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Juarrero M., Shim T.S., Kipnis A., Junqueira-Kipnis A.P., and Orme I.M.. 2003. Dynamics of macrophage cell populations during murine pulmonary tuberculosis. J. Immunol. 171:3128–3135. 10.4049/jimmunol.171.6.3128 [DOI] [PubMed] [Google Scholar]
- Honda K., and Taniguchi T.. 2006. IRFs: Master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644–658. 10.1038/nri1900 [DOI] [PubMed] [Google Scholar]
- Honda K., Takaoka A., and Taniguchi T.. 2006. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 25:349–360. 10.1016/j.immuni.2006.08.009 [DOI] [PubMed] [Google Scholar]
- Koski G.K., Karikó K., Xu S., Weissman D., Cohen P.A., and Czerniecki B.J.. 2004. Cutting edge: Innate immune system discriminates between RNA containing bacterial versus eukaryotic structural features that prime for high-level IL-12 secretion by dendritic cells. J. Immunol. 172:3989–3993. 10.4049/jimmunol.172.7.3989 [DOI] [PubMed] [Google Scholar]
- Kovarik P., Castiglia V., Ivin M., and Ebner F.. 2016. Type I interferons in bacterial infections: A balancing act. Front. Immunol. 7:652 10.3389/fimmu.2016.00652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H., Kawai T., Kato H., Sato S., Takahashi K., Coban C., Yamamoto M., Uematsu S., Ishii K.J., Takeuchi O., and Akira S.. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 203:1795–1803. 10.1084/jem.20060792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., McKinnon K.P., Runge M.S., Ting J.P.-Y., and Braunstein M.. 2006. The SecA2 secretion factor of Mycobacterium tuberculosis promotes growth in macrophages and inhibits the host immune response. Infect. Immun. 74:6855–6864. 10.1128/IAI.01022-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M.H., Biermann K., Tandberg S., Hsu T., and Jacobs W.R.. 2007. Genetic manipulation of Mycobacterium tuberculosis. Curr. Protoc. Microbiol. Chapter 10:Unit 10A.2. doi:. 10.1002/9780471729259.mc10a02s6 [DOI] [PubMed] [Google Scholar]
- Li X.-D., and Chen Z.J.. 2012. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. eLife. 1:e00102 10.7554/eLife.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-D., Chiu Y.-H., Ismail A.S., Behrendt C.L., Wight-Carter M., Hooper L.V., and Chen Z.J.. 2011. Mitochondrial antiviral signaling protein (MAVS) monitors commensal bacteria and induces an immune response that prevents experimental colitis. Proc. Natl. Acad. Sci. USA. 108:17390–17395. 10.1073/pnas.1107114108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.-T., Grishin N.V., and Chen Z.J.. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 347:aaa2630 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- Mancuso G., Gambuzza M., Midiri A., Biondo C., Papasergi S., Akira S., Teti G., and Beninati C.. 2009. Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat. Immunol. 10:587–594. 10.1038/ni.1733 [DOI] [PubMed] [Google Scholar]
- Manzanillo P.S., Shiloh M.U., Portnoy D.A., and Cox J.S.. 2012. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 11:469–480. 10.1016/j.chom.2012.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marié I., Durbin J.E., and Levy D.E.. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660–6669. 10.1093/emboj/17.22.6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F., Mayer-Barber K., Sher A., Wack A., and O’Garra A.. 2015. Type I interferons in infectious disease. Nat. Rev. Immunol. 15:87–103. 10.1038/nri3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B.K., Zulauf K.E., and Braunstein M.. 2017. The Sec pathways and exportomes of Mycobacterium tuberculosis. Microbiol. Spectr. 5:TBTB2-0013-2016. 10.1128/microbiolspec.TBTB2-0013-2016 [DOI] [PubMed] [Google Scholar]
- O’Garra A., Redford P.S., McNab F.W., Bloom C.I., Wilkinson R.J., and Berry M.P.R.. 2013. The immune response in tuberculosis. Annu. Rev. Immunol. 31:475–527. 10.1146/annurev-immunol-032712-095939 [DOI] [PubMed] [Google Scholar]
- Obregón-Henao A., Duque-Correa M.A., Rojas M., García L.F., Brennan P.J., Ortiz B.L., and Belisle J.T.. 2012. Stable extracellular RNA fragments of Mycobacterium tuberculosis induce early apoptosis in human monocytes via a caspase-8 dependent mechanism. PLoS One. 7:e29970 10.1371/journal.pone.0029970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg M., Krüger A., Ferstl R., Kaufmann A., Nees G., Sigmund A., Bathke B., Lauterbach H., Suter M., Dreher S., et al. 2012. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 337:1111–1115. 10.1126/science.1220363 [DOI] [PubMed] [Google Scholar]
- Repasy T., Lee J., Marino S., Martinez N., Kirschner D.E., Hendricks G., Baker S., Wilson A.A., Kotton D.N., and Kornfeld H.. 2013. Intracellular bacillary burden reflects a burst size for Mycobacterium tuberculosis in vivo. PLoS Pathog. 9:e1003190 10.1371/journal.ppat.1003190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyna C., Mba Medie F., Champion M.M., and Champion P.A.. 2016. Rational engineering of a virulence gene from Mycobacterium tuberculosis facilitates proteomic analysis of a natural protein N-terminus. Sci. Rep. 6:33265 10.1038/srep33265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigel N.W., and Braunstein M.. 2008. A new twist on an old pathway--accessory Sec [corrected] systems. Mol. Microbiol. 69:291–302. 10.1111/j.1365-2958.2008.06294.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach S.K., and Schorey J.S.. 2002. Differential regulation of the mitogen-activated protein kinases by pathogenic and nonpathogenic mycobacteria. Infect. Immun. 70:3040–3052. 10.1128/IAI.70.6.3040-3052.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Hata N., Asagiri M., Nakaya T., Taniguchi T., and Tanaka N.. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441:106–110. 10.1016/S0014-5793(98)01514-2 [DOI] [PubMed] [Google Scholar]
- Schorey J.S., Cheng Y., Singh P.P., and Smith V.L.. 2015. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 16:24–43. 10.15252/embr.201439363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P.P., Li L., and Schorey J.S.. 2015. Exosomal RNA from Mycobacterium tuberculosis-infected cells is functional in recipient macrophages. Traffic. 16:555–571. 10.1111/tra.12278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S.A., Johndrow J.E., Manzanillo P., and Cox J.S.. 2007. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J. Immunol. 178:3143–3152. 10.4049/jimmunol.178.5.3143 [DOI] [PubMed] [Google Scholar]
- Sullivan J.T., Young E.F., McCann J.R., and Braunstein M.. 2012. The Mycobacterium tuberculosis SecA2 system subverts phagosome maturation to promote growth in macrophages. Infect. Immun. 80:996–1006. 10.1128/IAI.05987-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Sun L., Liu H.-H., Chen X., Seth R.B., Forman J., and Chen Z.J.. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 24:633–642. 10.1016/j.immuni.2006.04.004 [DOI] [PubMed] [Google Scholar]
- Suthar M.S., Ramos H.J., Brassil M.M., Netland J., Chappell C.P., Blahnik G., McMillan A., Diamond M.S., Clark E.A., Bevan M.J., and Gale M. Jr. 2012. The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity. 37:235–248. 10.1016/j.immuni.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Yanai H., Savitsky D., and Taniguchi T.. 2008. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26:535–584. 10.1146/annurev.immunol.26.021607.090400 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., and Chen Z.J.. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5:ra20 10.1126/scisignal.2002521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles R.M., Graeber T.G., Krutzik S.R., Montoya D., Schenk M., Lee D.J., Komisopoulou E., Kelly-Scumpia K., Chun R., Iyer S.S., et al. 2013. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science. 339:1448–1453. 10.1126/science.1233665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassermann R., Gulen M.F., Sala C., Perin S.G., Lou Y., Rybniker J., Schmid-Burgk J.L., Schmidt T., Hornung V., Cole S.T., and Ablasser A.. 2015. Mycobacterium tuberculosis differentially activates cGAS- and inflammasome-dependent intracellular immune responses through ESX-1. Cell Host Microbe. 17:799–810. 10.1016/j.chom.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Watson R.O., Bell S.L., MacDuff D.A., Kimmey J.M., Diner E.J., Olivas J., Vance R.E., Stallings C.L., Virgin H.W., and Cox J.S.. 2015. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe. 17:811–819. 10.1016/j.chom.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., and Chen Z.J.. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32:461–488. 10.1146/annurev-immunol-032713-120156 [DOI] [PubMed] [Google Scholar]
- Zhao J., Vijay R., Zhao J., Gale M. Jr., Diamond M.S., and Perlman S.. 2016. MAVS expressed by hematopoietic cells is critical for control of West Nile Virus infection and pathogenesis. J. Virol. 90:7098–7108. 10.1128/JVI.00707-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.