The mechanisms underlying the differentiation of T follicular helper (Tfh) cell subsets are poorly understood. Here, Fang et al. show that the NKG2Dhigh Tfh cells in germinal centers with a history of T-bet expression represent the IFN-γ–producing Tfh subset.
Abstract
T follicular helper (Tfh) cells express transcription factor BCL-6 and cytokine IL-21. Mature Tfh cells are also capable of producing IFN-γ without expressing the Th1 transcription factor T-bet. Whether this IFN-γ–producing Tfh population represents a unique Tfh subset with a distinct differentiation pathway is poorly understood. By using T-bet fate–mapping mouse strains, we discovered that almost all the IFN-γ–producing Tfh cells have previously expressed T-bet and express high levels of NKG2D. DNase I hypersensitivity analysis indicated that the Ifng gene locus is partially accessible in this “ex–T-bet” population with a history of T-bet expression. Furthermore, multicolor tissue imaging revealed that the ex–T-bet Tfh cells found in germinal centers express IFN-γ in situ. Finally, we found that IFN-γ–expressing Tfh cells are absent in T-bet–deficient mice, but fully present in mice with T-bet deletion at late stages of T cell differentiation. Together, our findings demonstrate that transient expression of T-bet epigenetically imprints the Ifng locus for cytokine production in this Th1-like Tfh cell subset.
Introduction
T follicular helper (Tfh) cells are considered as a distinct subset of CD4 T helper (Th) cells, in parallel with classical type 1 Th (Th1), type 2 Th (Th2), and IL-17–producing Th (Th17) cells (King, 2009; Zhu et al., 2010; Crotty, 2011, 2014). However, while Tfh cells mainly produce IL-21 as their signature cytokine, several studies have also shown that some Tfh cells are capable of expressing Th1- or Th2-signature cytokines, IFN-γ or IL-4, both of which contribute to the regulation of different B cell Ig isotype switching (Snapper and Paul, 1987; Johnston et al., 2009; Reinhardt et al., 2009; Lu et al., 2011). Overproduction of IFN-γ by Tfh cells also contributes to autoimmune disease lupus-associated pathology (Lee et al., 2012). However, whether IFN-γ–producing Tfh cells represent a unique subset of Tfh cells or all the Tfh cells have the capacity to produce low amounts of IFN-γ is unknown.
The transcription factor BCL-6 is the master regulator for the differentiation and functions of Tfh cells (Johnston et al., 2009; Nurieva et al., 2009; Hatzi et al., 2015) and inhibits the expression of T-bet, a crucial transcription factor for differentiation of IFN-γ–producing Th1 cells (Szabo et al., 2000; Nurieva et al., 2009; Qi, 2016). Conversely, T-bet inhibits Tfh cell commitment by diverting BCL-6 from its target genes and/or by repressing BCL-6 expression (Nakayamada et al., 2011; Oestreich et al., 2011, 2012). Consistent with the idea of mutual repression between BCL-6 and T-bet, it has been shown that mature Tfh cells that express BCL-6 do not express T-bet (Nurieva et al., 2008). However, a balance between BCL-6 and T-bet may also be achieved with their coexpression under certain circumstances, and thus, mature Tfh cells generated in vivo in response to bacterial or viral infections uniformly express low levels of T-bet (Pepper et al., 2011; Hale et al., 2013; Weinstein et al., 2018). Nevertheless, whether such low levels of T-bet expression are sufficient to induce IFN-γ production is not clear. It has been shown that although T-bet expression at low levels in a regulatory T (T reg) subset is sufficient to induce chemokine receptor CXCR3 expression, such low amounts of T-bet are not sufficient to induce IFN-γ production (Yu et al., 2015). Therefore, how Tfh cells with low or no T-bet expression can produce IFN-γ is still not known.
Interestingly, some studies have shown that BCL-6 and T-bet may be coexpressed at high levels by some CD4 T cells at early stage of infections (Fahey et al., 2011; Kitano et al., 2011; Nakayamada et al., 2011; Pepper et al., 2011; Hale et al., 2013; Schmitt et al., 2016; Vella et al., 2017; Weinstein et al., 2018). It has been suggested that BCL-6/T-bet coexpressing early “Th1” cells may become mature Th1 cells by down-regulating BCL-6 during Th1 differentiation (Nakayamada et al., 2011). However, the relationship between these BCL-6/T-bet coexpressing cells and mature Tfh cells is not clear. It is possible that some CD4 T cells may initially express high levels of T-bet with or without BCL-6 expression and undergo chromatin remodeling at the Ifng locus, and during the process of these cells becoming BCL-6–expressing Tfh cells and migrating to B cell follicle, T-bet expression would be extinguished by BCL-6. Nevertheless, in germinal centers (GCs), these mature Tfh cells that have previously expressed T-bet (referred to as ex–T-bet cells hereafter) may epigenetically memorize their potential to produce IFN-γ.
Here we used a T-bet reporter and T-bet fate–mapping mouse strain to test this intriguing hypothesis. We found that ex–T-bet cells in the steady-state enriched for genes that are preferentially expressed by Tfh cells. Fully developed Tfh cells generated upon immunization in GC did not express T-bet; however, a substantial proportion of Tfh cells consisted of ex–T-bet cells. Among the Tfh cells found in GC, the ex–T-bet population represented the major IFN-γ–producing population in situ. Antigen-specific ex–T-bet Tfh cells had remodeled the Ifng locus and T-bet was essential for IFN-γ production by these Tfh cells. Finally, genome-wide analysis of Tfh cell subsets revealed that cell surface marker NKG2D was preferentially expressed by the ex–T-bet Tfh subset.
Results and discussion
Tfh signature genes are preferentially expressed by ex–T-bet cells in steady-state
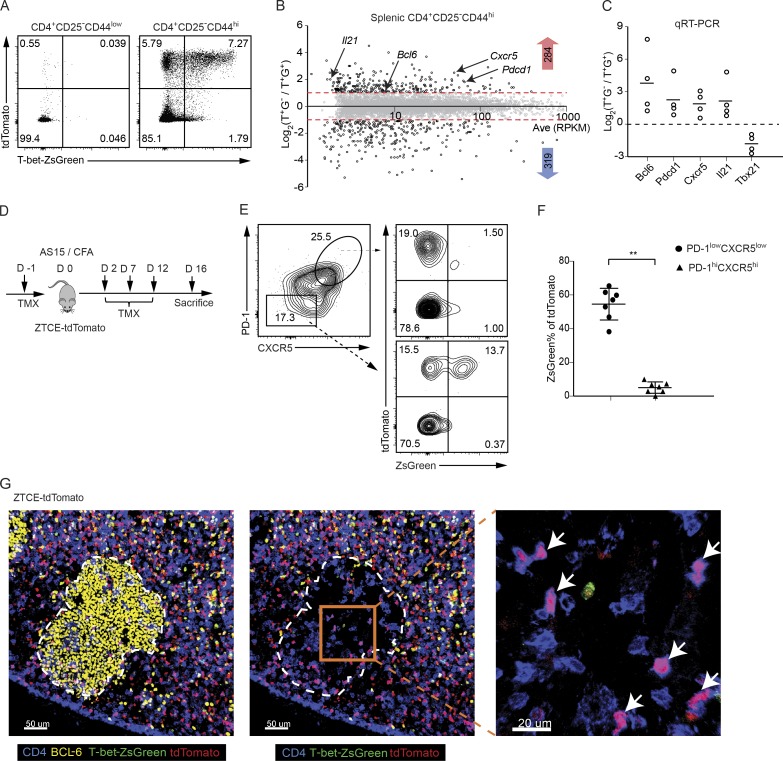
Although “one transcription factor, one cell fate” is a useful model to simply describe terminally differentiated CD4 T cells, accumulating evidence indicates that one “homogenous” cell type, particularly during their differentiation, may be quite heterogeneous based on dynamic coexpression of master transcription factors at the single-cell level (Fang and Zhu, 2017). We have previously reported that T-bet and GATA3 expression by T reg cells are dynamic, but transient expression of T-bet does not seem to be limited within the T reg population (Yu et al., 2015). To track the fate of T-bet–expressing effector CD4 T cells, we further studied the T-bet fate–mapping mouse strain (Fig. S1 A), T-bet-ZsGreen-T2A-CreERT2-Rosa26-loxP-STOP-loxP-tdTomato (ZTCE-tdTomato), in which cells that have expressed T-bet during the period of tamoxifen (TMX) treatment are permanently marked by the expression of a fluorescent protein, tdTomato, even after they have turned off T-bet expression, which can be determined by the expression of another fluorescent protein, ZsGreen.
Naive CD4 T cells from the ZTCE-tdTomato T-bet fate–mapping mice were cultured under Th1 or Th17 conditions in vitro with or without 4-hydroxytamoxifen (4-OHT) treatment. Almost all the 4-OHT–treated Th1 cells, but not Th17 cells, expressed both ZsGreen and tdTomato, indicating the faithfulness of this reporter system (Fig. S1 B). The effectiveness of T-bet fate–mapping was also confirmed in vivo since most ZsGreen-expressing splenic CD4 T effector cells within the memory-like population (CD4+CD44hiCD25−) were also tdTomato-positive in the ZTCE-tdTomato mice injected with TMX, while the naive CD4 T cells (CD4+CD44lowCD25−) in the same animal did not express either ZsGreen or tdTomato (Fig. 1 A). We also observed that a substantial proportion of tdTomato-positive memory-like CD4 T cells did not express ZsGreen (Fig. 1 A); based on the properties of the reporter system, these are presumably cells that had transiently expressed T-bet during the period of exposure to TMX (ex–T-bet cells). To verify the T-bet protein levels in T-bet–expressing and ex–T-bet cells, we sorted CD4+CD44highCD25−tdTomato+ZsGreen+ and CD4+CD44highCD25−tdTomato+ZsGreen− cells as shown in Fig. 1 A and then performed intracellular staining. Consistent with T-bet fate–mapping system, T-bet protein was detected in all the purified CD4+CD44highCD25−tdTomato+ZsGreen+ T-bet–expressing cells; however, the vast majority of CD4+CD44highCD25−tdTomato+ZsGreen− cells did not express T-bet and thus were ex–T-bet cells (Fig. 1 A and Fig. S1 C).
Figure 1.
Tfh cells in GCs do not express T-bet but some have a history of T-bet expression. (A) ZTCE-tdTomato mice were treated (i.p.) with TMX on days 0, 2, and 5. After 10 d, the expression of ZsGreen and tdTomato by the splenic naive CD4 T cells (CD4+CD44lowCD25−) and memory-like CD4 T effector cells (CD4+CD44hiCD25−) cells was assessed by flow cytometry. The plot represents a typical profile from more than five experiments. (B) Splenic CD4+CD44highCD25−tdTomato+ZsGreen− (T+G−) and CD4+CD44highCD25−tdTomato+ZsGreen+ (T+G+) cells sorted from TMX-treated naive ZTCE-tdTomato mice were used for RNA-Seq analysis. Samples are in biological duplicates. (C) Relative gene expression levels of Tfh-related genes in CD4+CD44highCD25−tdTomato+ZsGreen− (T+G−) and CD4+CD44highCD25−tdTomato+ZsGreen+ (T+G+) populations were measured by qRT-PCR (n = 4). Data are representative of three independent experiments. (D) Experimental procedure of immunizing ZTCE-tdTomato mice with AS15/CFA. (E and F) ZTCE-tdTomato mice were immunized with AS15/CFA for 16 d and treated with TMX as shown in D. dLN cells were analyzed by flow cytometry. The percentage of ZsGreen+ cells among the tdTomato+ cells was calculated for the PD-1lowCXCR5low non-Tfh and PD-1hiCXCR5hi Tfh populations (mean ± SD; n = 7; *, P < 0.0001). Data are representative of three independent experiments. (G) ZTCE-tdTomato mice were treated as in D. Different cell types in GC were analyzed by multicolor tissue imaging. GC (yellow, BCL-6); ex–T-bet-Tfh cell (pink, CD4-blue and tdTomato-red); T-bet-ZsGreen-green. Arrows indicate ex–T-bet-Tfh cells in GC. Data are representative of two independent experiments with two animals in each experiment.
We then performed RNA sequencing (RNA-Seq) analysis to determine whether these ex–T-bet cells have specific gene expression profiles. Comparing to the T-bet–expressing (tdTomato+ZsGreen+, T+G+) population, we identified 284 genes that were up-regulated and 319 genes that were down-regulated in the ex–T-bet (tdTomato+ZsGreen−, T+G−) population (Fig. 1 B and Table S1). To determine what potential CD4 T subsets are within the ex–T-bet population, we checked the 284 up-regulated genes for transcription factors and cytokines that are important for different CD4 T subsets and found that transcription factors Bcl6, Rorc, and Foxp3, and cytokines Il21 and Tgfb3 were enriched in the ex–T-bet population (Fig. S1, D and E). GO cellular component analysis showed that cell surface molecules Cxcr5 and Pdcd1 were highly expressed in the ex–T-bet population (Fig. S1 F). Cxcr5, Pdcd1, Bcl6, and Il21 mRNA levels were also confirmed by quantitative reverse-transcription PCR (qRT-PCR; Fig. 1 C). Collectively, these data indicate that T-bet may be dynamically expressed by several subsets of splenic CD4 T cells, including a T reg subset as we reported previously (Yu et al., 2015), and that the ex–T-bet cell population also contains Tfh-like cells since the expression of Tfh signature genes including Cxcr5, Pdcd1, Bcl6, and Il21 was enriched in this population.
A subset of Tfh cells induced by immunization has a history of T-bet expression
Since an ex–T-bet population within splenocytes from naive mice displays a Tfh-like phenotype, we wondered whether T-bet is transiently expressed during Tfh cell differentiation in vivo. Some reports using infection models have shown that T-bet is expressed by all the Tfh cells, but at low levels (Pepper et al., 2011; Hale et al., 2013; Weinstein et al., 2018); this is probably due to a strong Th1 response in these models.
To investigate the heterogeneity of Tfh cells in vivo, we adopted an immunization protocol known to generate a robust antigen-specific Tfh cell response without biasing toward a Th1 response (Grover et al., 2012; Riteau et al., 2016). ZTCE-tdTomato mice were immunized with the 15-mer peptide, AS15, whose sequence is derived from a protein of Toxoplasma gondii, as an antigen emulsified in CFA. To make sure that TMX was delivered to cells when animals were immunized, we pretreated mice with TMX 1 d before immunization (Fig. 1 D). Since Tfh-like cells were found in naive mice, we used tetramer staining to track newly differentiated antigen-specific Tfh cells in our study (Fig. S2 A). After 16 d of immunization, the vast majority of the antigen-specific CXCR5highPD-1high Tfh cells in the draining lymph node (dLN) did not express the T-bet-ZsGreen reporter (Fig. 1, E and F), which is consistent with an antagonistic relationship between transcription factor T-bet and BCL-6. However, a substantial proportion of these Tfh cells (∼20%) displayed an ex–T-bet cell phenotype (tdTomato+ZsGreen−). In contrast, even though ex–T-bet cells were also found in the CXCR5lowPD-1low non-Tfh cell population, about half of these tdTomato+ cells also expressed high levels of ZsGreen (Fig. 1 E). Interestingly, on day 6 after immunization, a substantial proportion (∼10–15%) of CXCR5highPD-1high cell population expressed T-bet-ZsGreen, but the frequency of T-bet-ZsGreen–expressing cells, as well as mean fluorescence intensity of T-bet-ZsGreen within the CXCR5highPD-1high Tfh cell population, dramatically reduced on day 16 (Fig. S2 B). These results indicate that T-bet is transiently expressed during the differentiation of a Tfh subset.
Although CXCR5 is wildly used to identify Tfh cells, the function of mature Tfh cells depends on their location (Victora and Nussenzweig, 2012; Qi, 2016), and they reside in B cell follicle with some within GC. BCL-6 is highly expressed by GC B cells and Tfh cells and widely used to identify GC. We used multiplex immunofluorescence confocal imaging of the dLN to localize the ex–T-bet CD4 T cells. Consistent with the flow cytometry data, a substantial fraction of CD4 T cells found in GC were ex–T-bet (CD4+tdTomato+, shown as pink in the image), but not T-bet–expressing (CD4+ZsGreen+), cells (Fig. 1 G). Thus, using T-bet fate–mapping mice, we found T-bet is transiently expressed during differentiation of a Tfh cell subset, and ex–T-bet Tfh cells can be found within GC in vivo after immunization.
The ex–T-bet Tfh cells in GCs are potent IFN-γ–producing cells
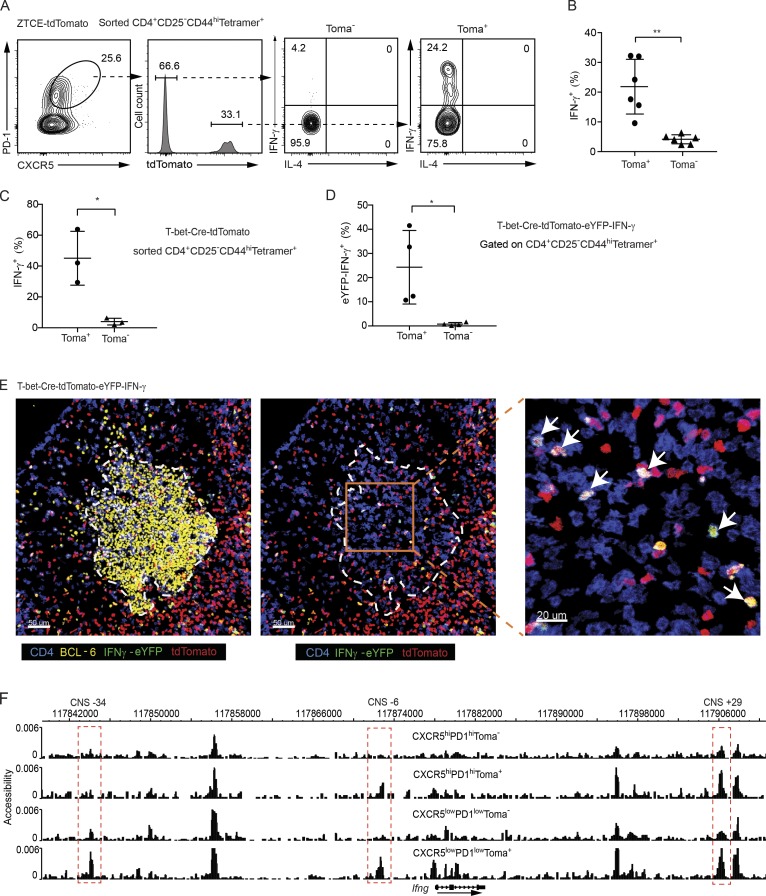
To test whether the AS15-specific Tfh cells can express IFN-γ, and if so, to understand the relationship between IFN-γ–producing Tfh cells and ex–T-bet Tfh cells, we purified and studied the CD4+CD44highCD25−Tetramer+ cells from the AS15/CFA-immunized ZTCE-tdTomato mice. Strikingly, among the Tfh compartment, only the ex–T-bet (tdTomato+) population represented potent IFN-γ–producing cells (Fig. 2, A and B). By using another T-bet fate–mapping mouse strain, T-bet-Cre-tdTomato (Fig. S2, C and D), in which all cells that are currently expressing or have previously expressed T-bet are labeled by tdTomato expression in the absence of TMX, we confirmed that all the cells capable of IFN-γ production were within the population that had expressed T-bet (Fig. 2 C and Fig. S2 E). To further confirm IFN-γ production without in vitro restimulation and to visualize IFN-γ–expressing ex–T-bet Tfh cells in situ, we made an IFN-γ-eYFP-T-bet-Cre-tdTomato mouse strain. Flow cytometry analysis showed that all eYFP+ IFN-γ–producing cells were within the tdTomato+ population (Fig. 2 D and Fig. S2 F). Imaging of the draining lymph node of the immunized mice also showed that almost all the IFN-γ–producing Tfh cells (eYFP+CD4+) in the GC were tdTomato+ (Fig. 2 E). Collectively, our data indicate that, although Tfh cells do not express T-bet in GC, the IFN-γ–expressing Tfh cells in GC represent a subset of Tfh cells that have a history of T-bet expression.
Figure 2.
Ex–T-bet Tfh cells display a partial accessibility at the Ifng locus and have the capability to express IFN-γ. (A–C) Sorted CD4+CD44highCD25−Tetramer+ cells from the immunized TMX-treated ZTCE-tdTomato mice (A and B) or TMX-untreated T-bet-Cre-tdTomato mice (C) were cultured in IL-2–supplemented media overnight. Cells were then stimulated with PMA and ionomycin for 4 h. Cytokine production was assessed by intracellular staining. Percentages of IFN-γ–producing cells were calculated (mean ± SD; in B: **, P < 0.005, n = 6; in C: *, P < 0.05, n = 3). Data are representative of two independent experiments (A–C). (D) Cells from dLN of immunized IFN-γ-eYFP-T-bet-Cre-tdTomato mice were stained with cell surface markers. Percentage of eYFP+ cells among the CD4+CD44highCD25−Tetramer+ tdTomato+ or tdTomato− Tfh (CXCR5highPD-1high) population was calculated (mean ± SD; *, P < 0.05, n = 4). Data are representative of three independent experiments. (E) IFN-γ-eYFP-T-bet-Cre-tdTomato mice were immunized with AS15/CFA. dLN was harvest for imaging analysis. GC (yellow, BCL-6); IFN-γ–expressing ex–T-bet-Tfh cell (shown in white; CD4 shown in blue; tdTomato shown in red; and IFN-γ-eYFP shown in green). Arrows indicate IFN-γ–expressing ex–T-bet-Tfh cells. Data are representative of two independent experiments with two animals in each experiment. (F) CD4+CD44highCD25−Tetramer+-CXCR5highPD-1hightdTomato−, CXCR5highPD-1hightdTomato+, CXCR5lowPD-1lowtdTomato−, and CXCR5lowPD-1lowtdTomato+ cells were sorted from AS15/CFA-immunized T-bet-Cre-tdTomato mice. DHS analysis was performed through scDNase-Seq, and the Ifng gene locus was viewed by the Washington University genome browser. Peaks at the CNS-34, CNS-6 and CNS+29 sites that displayed differential accessibility among the samples are highlighted in red. Cell samples are in biological duplicates.
The Ifng locus is accessible in ex–T-bet Tfh but not in other Tfh cells
Epigenetic information often predicts a development fate and “memorializes” the prior effect of a transcription factor during cell differentiation (Rothenberg, 2013), which can be reflected by “open chromatin regions” identified through detection of DNase I hypersensitivity sites (DHSs; Boyle et al., 2008; Thurman et al., 2012). It has been reported that T-bet is involved in chromatin remodeling at the Ifng locus during Th1 cell differentiation (Mullen et al., 2001; Zhu et al., 2012). However, it is not known whether transient expression of T-bet is sufficient to remodel the Ifng locus. To evaluate the accessibility of Ifng locus in the antigen-specific ex–T-bet Tfh cells, we applied a sensitive DHS assay (scDNase-Seq; Jin et al., 2015) with tdTomato− and tdTomato+ Tfh (CXCR5highPD-1high) as well as tdTomato− and tdTomato+ non-Tfh (CXCR5lowPD-1low) populations within the CD4+CD44highCD25−Tetramer+ cells from the AS15/CFA-immunized T-bet-Cre-tdTomato mice. Interestingly, the Ifng locus was accessible at the CNS-6 and CNS+29 regions in both ex–T-bet Tfh cells (CXCR5highPD-1hightdTomato+) and CXCR5lowPD-1lowtdTomato+ population that presumably contains conventional Th1 cells, whereas these regions were inaccessible or much less accessible in the tdTomato− Tfh and non-Tfh cells (Fig. 2 F). In contrast, the CNS-34 site was primarily accessible in CXCR5lowPD-1lowtdTomato+ population. Thus, the Ifng locus is partially accessible in ex–T-bet Tfh cells compared with the conventional Th1 cells and these results suggest that at early stage of Tfh cell differentiation, transient expression of T-bet may epigenetically imprint the Ifng locus for its later expression in a subset of Tfh cells.
Transient T-bet expression is required for IFN-γ production by Tfh cells
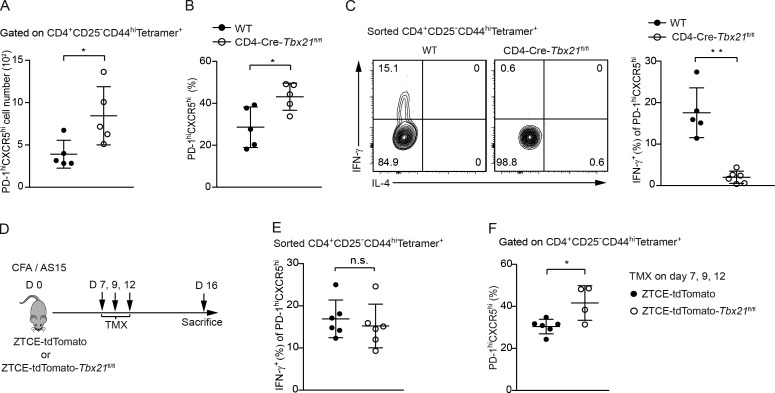
The above results showed that ex–T-bet Tfh cells had the capability to express IFN-γ. However, whether transient T-bet expression during development is necessary for Tfh cells to produce IFN-γ is unknown. We first immunized Tbx21fl/fl-CD4-Cre and wild-type control mice. The percentage, as well as the total cell numbers, of antigen-specific CXCR5highPD-1high Tfh cells were higher in T-bet–deficient mice than in wild-type control mice (Fig. 3, A and B). This is consistent with the function of T-bet in inhibiting BCL-6 activity as previously reported (Nakayamada et al., 2011). Nevertheless, IFN-γ expression by Tfh cells was abolished in the absence of T-bet (Fig. 3 C). To test whether T-bet is required at late stages of T cell differentiation, we treated ZTCE-tdTomato and ZTCE-tdTomato-Tbx21fl/fl mice with TMX after 7 d of immunization (Fig. 3 D and Fig. S1 A). Interestingly, the Tfh cells from the ZTCE-tdTomato-Tbx21fl/fl mice (T-bet late–knock-out mice) could still express normal levels of IFN-γ (Fig. 3 E). However, the percentage of antigen-specific CXCR5highPD-1high Tfh cells were higher in ZTCE-tdTomato-Tbx21fl/fl mice treated with TMX (Fig. 3 F). Thus, transient but not continuous T-bet expression is essential for Tfh cells to acquire and maintain their ability to produce IFN-γ.
Figure 3.
Early but not late expression of T-bet is necessary for IFN-γ production in Tfh cells. (A and B) CD4-Cre-Tbx21fl/fl and wild-type (WT) control mice were immunized with AS15/CFA for 16 d. The total cell number (A) and percentage (B) of CD4+CD44highCD25−Tetramer+CXCR5highPD-1high population was calculated (mean ± SD; *, P < 0.05, n = 5). Data are representative of three independent experiments. (C) CD4+CD44highCD25−Tetramer+ cells from immunized CD4-Cre-Tbx21fl/fl and wild-type control mice were sorted and cultured in IL-2 supplemented media overnight. Cytokine production was assessed by intracellular staining 4 h after stimulation with PMA and ionomycin. Percentages of IFN-γ–producing cells were calculated (mean ± SD; **, P < 0.0005; n ≥ 5). Data are representative of two independent experiments. (D) Experimental procedure of immunizing the ZTCE-tdTomato-Tbx21fl/fl mice with AS15/CFA while inducing late T-bet deletion. (E) ZTCE-tdTomato and ZTCE-tdTomato-Tbx21fl/fl mice were immunized with AS15/CFA for 16 d and treated with TMX on days 7, 9, and 12 as shown in D. CD4+CD44highCD25−Tetramer+ cells were sorted and cultured in IL-2–supplemented media overnight. Cells were then stimulated with PMA and ionomycin for 4 h. Cytokine production was assessed by intracellular staining. Percentages of IFN-γ–producing cells were calculated (mean ± SD; n.s.; not statistically significant, n = 6). Data are representative of two independent experiments. (F) Mice were treated as in D. The percentage of CD4+CD44highCD25−Tetramer+CXCR5highPD-1high population was calculated (mean ± SD; *, P < 0.05; n ≥ 4). Data are representative of two independent experiments.
NKG2D is preferentially expressed by ex–T-bet Tfh cells
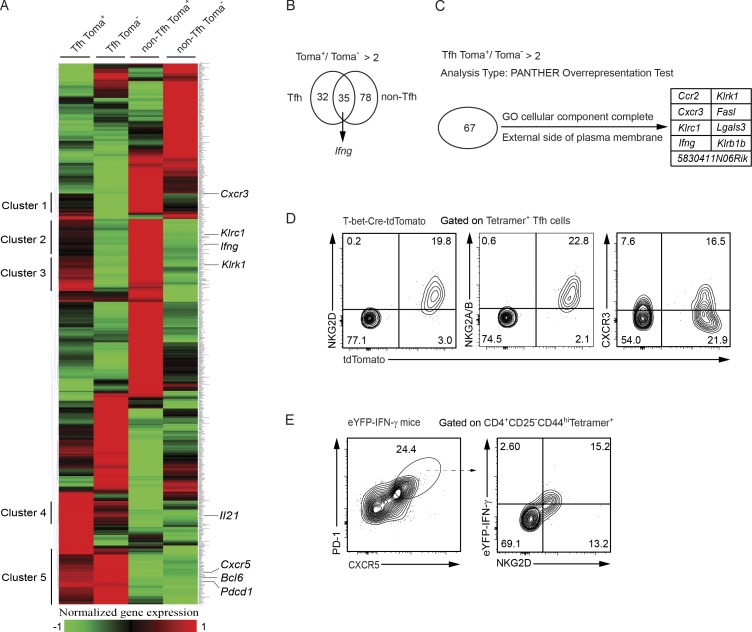
Given that transient T-bet expression is essential for Tfh cells to acquire their ability to produce IFN-γ, finding specific surface protein(s) to distinguish ex–T-bet Tfh cells from other Tfh cells will be helpful to track IFN-γ–producing Tfh cells without using the T-bet fate–mapping tools. We thus performed RNA-Seq analysis on the same cell subsets that were used for scDNaseI-Seq analysis. Clustering of all differentially expressed genes among these four groups (Fig. 4 A and Table S2) revealed T-bet fate–mapping–associated gene clusters (Clusters 1, 2, and 3) and Tfh cell-specific gene clusters (Clusters 4 and 5). Within the Cluster 2, a subset of genes, including Ifng, was expressed at higher levels in tdTomato+ Tfh cells than in tdTomato− Tfh cells as expected. Interestingly, genes in Cluster 3, such as Klrk1, were highly expressed in both Tfh and non-Tfh cells that were tdTomato+. There was a substantial overlap between the 67 genes that were higher in tdTomato+ Tfh cells than in their tdTomato− counterparts, and the 113 genes that were higher in tdTomato+ non-Tfh cells than in their tdTomato− counterparts (Fig. 4 B). Although Ifng expression was increased in both tdTomato+ Tfh and non-Tfh population, higher levels of Ifng transcripts were detected in tdTomato+ non-Tfh cells compared with tdTomato+ Tfh cells (Table S2), which is consistent with the DHS results shown in Fig. 2 F. Within the 67 genes, there were 9 differentially expressed genes that encode proteins located at plasma membrane (Fig. 4 C). Indeed, high protein expression levels of NKG2D (encoded by Klrk1) and NKG2A/B (encoded by Klrc1) were found in tdTomato+, but not tdTomato− antigen-specific Tfh cells (Fig. 4 D). On the other hand, a well-known Th1-specific protein CXCR3, which is also a T-bet target gene, was expressed only by a fraction of tdTomato+ Tfh cells (Fig. 4 D). Furthermore, by using eYFP-IFN-γ reporter mice, we found that all the antigen-specific IFN-γ–producing Tfh cells were NKG2D positive (Fig. 4 E). Thus, NKG2D serves as a useful cell surface marker to identify the IFN-γ–producing Tfh subset.
Figure 4.
Unique gene expression profile of the ex–T-bet Tfh cells reveals cell surface markers for identifying this population. (A) RNA-Seq analysis was performed using CD4+CD44highCD25−Tetramer+-CXCR5highPD-1hightdTomato+, CXCR5highPD-1hightdTomato−, CXCR5lowPD-1lowtdTomato+, and CXCR5lowPD-1lowtdTomato− antigen-specific cell populations from dLN of AS15/CFA-immunized T-bet-Cre-tdTomato mice. Differentially expressed genes were clustered. Cell samples are in biological duplicates. (B) Overlapping of the 67 and 113 genes that were expressed at higher levels in tdTomato+ cells compared with tdTomato− counterparts in Tfh and non-Tfh compartment, respectively. (C) tdTomato+ Tfh cell–specific genes (compared with the tdTomato− Tfh cells) that encode cell surface molecules. (D) Antigen-specific Tfh cells from the AS15/CFA-immunized T-bet-Cre-tdTomato mice were stained with anti-NKG2D, anti-NKG2A/B, or anti-CXCR3. Data are representative of three independent experiments. (E) NKG2D and IFN-γ-eYFP expression in antigen-specific Tfh cells from immunized IFN-γ-eYFP mice were measured. Data are representative of two independent experiments with five animals in each experiment.
In conclusion, we have identified a subset of Tfh cells with a history of T-bet expression and epigenetic imprints on the Ifng locus; these cells express NKG2D and have the capability to express IFN-γ in GC. Early but not late T-bet expression is required for the development of IFN-γ–producing Tfh cells. In fact, T-bet expression at late stages of T cell differentiation is inhibitory for the generation of this subset. Thus, manipulating the duration of T-bet expression in vivo may regulate the balance between IFN-γ–producing Th1 cells and IFN-γ–producing Tfh cells.
Tfh cells and T reg cells have been regarded as parallel CD4 T cell subsets to conventional Th1, Th2, and Th17 cells (O’Shea and Paul, 2010). However, it has been recently shown that the master regulators for conventional effector T helper cells, T-bet (for Th1), GATA3 (for Th2), and RORγt (for Th17) can be expressed by different subsets of T reg cells (Campbell and Koch, 2011; Yu et al., 2015; Levine et al., 2017). For Tfh cells, our finding that a subset of Tfh cells have a history of T-bet expression may have a broader implication since some Tfh cells are capable of expressing the Th2 signature cytokine IL-4, and some Tfh cells that induce antigen-specific IgA responses in the gut are derived from IL-17–producing, presumably RORγt-expressing, cells (Reinhardt et al., 2009; Liang et al., 2011; Lu et al., 2011; Hirota et al., 2013). It has been reported that while GATA3 is critical for the acquisition of IL-4–producing capacity, it is no longer essential for IL-4 production after cell fate commitment (Zhu et al., 2004). It has also been previously shown that during an in vivo response to LCMV infection, BCL-6–expressing Tfh cells 5 d after infection express high levels of T-bet; however, the mature Tfh cells 15 d after infection express very low levels of T-bet (Weinstein et al., 2018). It is likely that high levels of T-bet expression during early Tfh cell differentiation determine the capacity of these pre-Tfh cells to produce IFN-γ, whereas low levels of T-bet expression in mature Tfh cells may not be functionally relevant.
Based on our current study and these previous reports discussed above, we propose that transient induction of a particular effector master regulator, including T-bet, GATA3 and RORγt may be a common feature for the development of distinct IFN-γ–, IL-4–, or IL-17–producing Tfh subsets in GCs. Thus, dynamic coexpression of multiple antagonizing transcription factors could be a plausible mechanism that results in heterogeneity and plasticity of CD4 T helper cells. In a striking parallel with T reg cells, Tfh cells should also be considered to exist in distinct differentiated states similar to conventional effector T helper cells.
Materials and methods
Mice
T-bet-ZsGreen-T2A-CreERT2 (ZTCE)-Rosa26-loxP-STOP-loxP-tdTomato (ZTCE-tdTomato) mice on C57BL/6 background was previously described (Yu et al., 2015), and ZTCE-tdTomato mice were bred to Tbx21fl/fl mice to generate ZTCE-tdTomato-Tbx21fl/fl. T-bet-Cre mice (Haddad et al., 2013) (JAX 024507) were bred onto Rosa26-loxP-STOP-loxP-tdTomato (Madisen et al., 2010) (JAX 7914) for six generations to generate T-bet-Cre-tdTomato mouse strain. T-bet-Cre-tdTomato mice were further crossed with IFN-γ-eYFP (Reinhardt et al., 2009; JAX 017580) mice to generate IFN-γ-eYFP-T-bet-Cre-tdTomato. T-bet-ZsGreen mice were previously generated (Zhu et al., 2012) and maintained at the National Institute of Allergies and Infectious Diseases (NIAID)–Taconic repository (Line 8419). Tbx21fl/fl (Intlekofer et al., 2008) were bred to CD4-Cre mice (Lee et al., 2001) to generate CD4-Cre-Tbx21fl/fl mice. All mice were 8–12 wk of age when they were used under a protocol approved by the NIAID Animal Care and Use Committee. Mice were bred and/or maintained in the NIAID specific pathogen-free animal facilities.
Mouse manipulation
Naive ZTCE-tdTomato mice were administrated with 3 mg TMX (T5648; Sigma) in corn oil (C8267; Sigma) through i.p. injection on days 0, 2, and 5. 10 d after TMX injection, splenic cells were harvested for sorting and followed by RNA-Seq analysis. In all immunization assays, mice were immunized with 20-µg peptide AS15 (AVEIHRPVPGTAPPS; synthesized by Peptide Synthesis and Analysis Unit, Research Technologies Branch, NIAID) emulsified in the oil and CFA (F5506, Sigma) by subcutaneous s.c. injection at two sites along the back (50 µl/site). After 16 d, inguinal draining lymph nodes (dLN) were harvested to make single-cell suspension for staining or fixed for imaging. ZTCE-tdTomato mice were treated with TMX on days −1, 2, 7, and 12 and immunized with AS15/CFA on day 0. Cells were stained at 37°C for 1 h with Tetramer I-Ab-AVEIHRPVPGTAPPS-APC or -BV421 provided by the National Institutes of Health Tetramer Core Facility. To delete T-bet at a late stage, ZTCE-tdTomato-Tbx21fl/fl mice were injected with TMX on days 7, 9, and 12 after they were immunized with AS15/CFA.
Cell culture and purification
Naive CD4 T cells (CD4+CD44lowCD62LhighCD25−) were sorted from peripheral lymph nodes by using FACSAria (BD Biosciences). Sorted cells were mixed with T cell–depleted splenocytes (used as antigen-presenting cells) and cultured under Th1 conditions (anti-CD3, 1 µg/ml; anti-CD28, 3 µg/ml; anti–IL-4, 10 µg/ml; IL-12, 10 ng/ml; and IL-2, 100 U/ml) or Th17 conditions (anti-CD3, 1 µg/ml; anti-CD28, 3 µg/ml; anti–IL-4, 10 µg/ml; anti–IFN-γ, 10 µg/ml; anti–IL-12, 10 µg/ml; TGFβ1, 1 ng/ml; IL-6, 10 ng/ml; and IL-1β, 10 ng/ml) for 4 d. 4-OHT was added at the beginning of the culture. FACS-sorted CD4+CD44highCD25−Tetramer+ cells from dLN were cultured in resting condition (IL-2, 50 U/ml) overnight before restimulation. The basic culturing media is the RPMI medium 1640 (Invitrogen) supplemented with 10% FBS (Hyclone), 200 mM glutamine, 100 mM sodium pyruvate (Gibco), 50 µM β-mercaptoethanol (Sigma), 100 U/ml penicillin, and 100 µg/ml streptomycin.
Immunofluorescent staining
Mice were immunized with AS15/CFA, and dLNs were harvested after 16 d. Samples were treated with the fixation and permeabilization solution (554722; BD Bioscience) for 12 h. Samples were then dehydrated in 30% sucrose and embedded in optimal cutting temperature freezing media (Sakura Finetek). CM3050S cryostat (Leica) and Superfrost Plus slides (VWR) were used to make sections, which were further permeabilized and blocked in PBS (0.1% Triton X-100 and 10% normal mouse serum) and then stained with antibodies in PBS (0.01% Triton X-100 and 5% normal mouse serum). Fluormount G (Southern Biotech) and Leica TCS SP8 confocal microscope were used to collect signaling from slides. Data were analyzed by Imaris software (Bitplane). Antibodies against CD4 (RM4-5), BCL-6 (K112-91), and B220 (RA3-6B2) were purchased from BD Bioscience; antibody against GFP/YFP (A-21311) was purchased from Thermo Fisher.
Flow cytometry analysis and reagents
Unless otherwise stated, cell staining was performed at 4°C. Single-cell suspensions were first incubated with anti-CD16/32 (2.4G2) for 15 min to block antibody Fc receptors. To detect intracellular cytokine production in vitro, cells were stimulated with 10 ng/ml phorbol 12-myristate 13-acetate (PMA) and 500 nM ionomycin for 4 h in the presence of monensin (00-4505-41; eBioscience). Cells were fixed in 4% paraformaldehyde for 15 min at room temperature and permeabilized with 0.5% Triton X-100. For transcription factors staining, the Foxp3 Staining Buffer Set (00-5523-00; eBioscience) was used according to the manufacturer’s instruction. Flow cytometry data were collected from the LSR II or FORTESSA (BD Biosciences), and the results were analyzed with the FlowJo 10 software (Tree Star). Antibodies were purchased from several commercial sources indicated below. Antibodies against CD4 (RM4-5), CD25 (PC61.5), CD44 (IM7), CD62L (MEL-14), and PD-1 (J43) were from eBioscience; anti-CXCR5 (2G8), IFN-γ (XMG1.2), T-bet (O4-46), CD138 (281–2), CD19 (1D3), B220 (RA3-6B2), NKG2D (9C11G4), and NKG2A/C/E (20d5) from BD Bioscience; anti-IL-4 (11B11) was from BioLegend; 2.4G2 (PUR001) was from Harlan; and Fixable Viability Dye eFluor 506 was from eBioscience.
qRT–PCR
RNeasy Mini kit (74104; Qiagen) was used to harvest total RNAs from sorted cell samples. The diluted RNAs were reverse transcribed with PrimeScript RT Master Mix (RR036A; TaKaRa) and analyzed with 7900HT Fast Real-Time PCR system (Applied Biosystems) using FastStart Universal SYBR Green Master (Rox; 04913850001; Roche). Expression of specific gene transcripts was measured by using the following primer pairs: Il21, 5′-GGACCCTTGTCTGTCTGGTAG-3′ and 5′-TGTGGAGCTGATAGAAGTTCAGG-3′; Cxcr5, 5′-ATGAACTACCCACTAACCCTGG-3′ and 5′- TGTAGGGGAATCTCCGTGCT-3′; Bcl6, 5′- CCGGCACGCTAGTGATGTT-3′ and 5′-TGTCTTATGGGCTCTAAACTGCT-3′; Pdcd1, 5′-ACCCTGGTCATTCACTTGGG-3′ and 5′-CATTTGCTCCCTCTGACACTG-3′; Tbx21, 5′-AGCAAGGACGGCGAATGTT-3′ and 5′-GGGTGGACATATAAGCGGTTC-3′. Hprt was used for normalization purposes.
RNA-Seq and scDNase-Seq analysis
CD4+CD44highCD25− tdTomato+ZsGreen− and CD4+CD44highCD25− tdTomato+ZsGreen+ cells from naive mice after TMX treatment were used for RNA-Seq experiment. CD4+CD44highCD25−Tetramer+-CXCR5highPD-1hightdTomato+, CXCR5highPD-1hightdTomato−, CXCR5lowPD-1lowtdTomato+, and CXCR5lowPD-1lowtdTomato− cells from AS15/CFA-immunized T-bet-Cre-tdTomato mice were used for RNA-Seq and scDNase-Seq analysis. The total RNAs were harvested and purified by using Qiagen’s miRNeasy micro kit (217084; Qiagen). Qiagen’s DNase set (79254; Qiagen) was used for on-column DNase digestion. PolyA-tailed RNAs were purified from total RNA by using Dynabeads mRNA DIRECT kit (61012; Ambion Life Technologies). scDNase-Seq was performed according to a protocol described previously (Jin et al., 2015). The RNA-Seq and scDNase-Seq libraries were sequenced with Illumina HiSeq system, and 50 bp reads were generated by the National Heart, Lung, and Blood Institute DNA Sequencing and Computational Biology Core. Sequence reads were mapped to mouse genome (mm9) by using bowtie 2 with default settings (Langmead and Salzberg, 2012), and reads mapped to multiple positions (MAPQ < 10) were discarded. In the DHS analysis, only one read for genomic site with multiple alignment hits were retained. Gene expression levels were measured by RPKM (reads per kilobase of exon per million reads; Mortazavi et al., 2008). Differentially expressed genes were identified by edgeR 3 (Robinson et al., 2010) with the following criteria: FDR (false discovery rate) < 0.01, FC (fold change) log2 ≥ 1, and RPKM ≥ 3. Differently expressed genes were clustered by using software MeV. SICER (Zang et al., 2009) was used to call peaks (window size is 200 bp, gap size is 400 bp, and others by defaults). The location of protein is analyzed by PANTHER Overrepresentation Test–GO cellular component complete.
Statistics
Differences between groups were determined by two-tailed unpaired or paired Student’s t test with Prism 7 software (GraphPad). Data are presented as mean ± SD. A P < 0.05 was considered statistically significant and is indicated as * or **; not statistically significant is indicated as n.s.
Accession code
The RNA-Seq and scDNAase-Seq datasets are available in the Gene Expression Omnibus database under the accession no. GSE102959.
Online supplemental material
Fig. S1 shows generation and characterization of inducible T-bet fate–mapping mouse strain and identification of Tfh-related gene expression in ex–T-bet population. Fig. S2 shows Tfh cell gating strategy and immunization of constitutive T-bet fate mapping mice. Table S1 is an Excel file of differentially expressed genes between CD4+CD44highCD25−tdTomato+ZsGreen+ and CD4+CD44highCD25−tdTomato+ZsGreen− cell populations. Table S2 is an Excel file of differentially expressed genes in CD4+CD44highCD25−Tetramer+-CXCR5highPD-1hightdTomato+, CXCR5highPD-1hightdTomato−, CXCR5lowPD-1lowtdTomato+, and CXCR5lowPD-1lowtdTomato− antigen-specific cell populations.
Supplementary Material
Acknowledgments
We thank Drs. R.N. Germain, J.J. O’Shea, and P.L. Schwartzberg for their critical reading of the paper. We thank K. Weng for cell sorting.
This work is supported by the Division of Intramural Research of the NIAID (U.S. National Institutes of Health grant 1ZIA-AI-001169).
The authors declare no competing financial interests.
Author contributions: D. Fang and J. Zhu conceived the project. D. Fang performed the majority of experiments. K. Cui contributed to RNA-Seq and scDNase-Seq experiments. G. Hu, and R. Li performed bioinformatics analysis. K. Mao contributed to immunofluorescence staining and analysis. N. Riteau and M. Zheng helped in some immunization experiments and data analysis. S.L. Reiner, A. Sher, and K. Zhao provided critical advice to the project and edited the manuscript. S.L. Reiner also contributed critical reagents. D. Fang and J. Zhu wrote the manuscript. J. Zhu supervised the project.
References
- Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., and Crawford G.E.. 2008. High-resolution mapping and characterization of open chromatin across the genome. Cell. 132:311–322. 10.1016/j.cell.2007.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D.J., and Koch M.A.. 2011. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. Immunol. 11:119–130. 10.1038/nri2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Crotty S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity. 41:529–542. 10.1016/j.immuni.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey L.M., Wilson E.B., Elsaesser H., Fistonich C.D., McGavern D.B., and Brooks D.G.. 2011. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J. Exp. Med. 208:987–999. 10.1084/jem.20101773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D., and Zhu J.. 2017. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J. Exp. Med. 214:1861–1876. 10.1084/jem.20170494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover H.S., Blanchard N., Gonzalez F., Chan S., Robey E.A., and Shastri N.. 2012. The Toxoplasma gondii peptide AS15 elicits CD4 T cells that can control parasite burden. Infect. Immun. 80:3279–3288. 10.1128/IAI.00425-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad R., Lanjuin A., Madisen L., Zeng H., Murthy V.N., and Uchida N.. 2013. Olfactory cortical neurons read out a relative time code in the olfactory bulb. Nat. Neurosci. 16:949–957. 10.1038/nn.3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale J.S., Youngblood B., Latner D.R., Mohammed A.U., Ye L., Akondy R.S., Wu T., Iyer S.S., and Ahmed R.. 2013. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 38:805–817. 10.1016/j.immuni.2013.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzi K., Nance J.P., Kroenke M.A., Bothwell M., Haddad E.K., Melnick A., and Crotty S.. 2015. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J. Exp. Med. 212:539–553. 10.1084/jem.20141380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Turner J.E., Villa M., Duarte J.H., Demengeot J., Steinmetz O.M., and Stockinger B.. 2013. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat. Immunol. 14:372–379. 10.1038/ni.2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer A.M., Banerjee A., Takemoto N., Gordon S.M., Dejong C.S., Shin H., Hunter C.A., Wherry E.J., Lindsten T., and Reiner S.L.. 2008. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 321:408–411. 10.1126/science.1159806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Tang Q., Wan M., Cui K., Zhang Y., Ren G., Ni B., Sklar J., Przytycka T.M., Childs R., et al. . 2015. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature. 528:142–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., and Crotty S.. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010. 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C. 2009. New insights into the differentiation and function of T follicular helper cells. Nat. Rev. Immunol. 9:757–766. 10.1038/nri2644 [DOI] [PubMed] [Google Scholar]
- Kitano M., Moriyama S., Ando Y., Hikida M., Mori Y., Kurosaki T., and Okada T.. 2011. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 34:961–972. 10.1016/j.immuni.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., et al. . 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774. 10.1016/S1074-7613(01)00227-8 [DOI] [PubMed] [Google Scholar]
- Lee S.K., Silva D.G., Martin J.L., Pratama A., Hu X., Chang P.P., Walters G., and Vinuesa C.G.. 2012. Interferon-γ excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity. 37:880–892. 10.1016/j.immuni.2012.10.010 [DOI] [PubMed] [Google Scholar]
- Levine A.G., Mendoza A., Hemmers S., Moltedo B., Niec R.E., Schizas M., Hoyos B.E., Putintseva E.V., Chaudhry A., Dikiy S., et al. . 2017. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature. 546:421–425. 10.1038/nature22360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H.E., Reinhardt R.L., Bando J.K., Sullivan B.M., Ho I.C., and Locksley R.M.. 2011. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat. Immunol. 13:58–66. 10.1038/ni.2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K.T., Kanno Y., Cannons J.L., Handon R., Bible P., Elkahloun A.G., Anderson S.M., Wei L., Sun H., O’Shea J.J., and Schwartzberg P.L.. 2011. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 35:622–632. 10.1016/j.immuni.2011.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. . 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., and Wold B.. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 5:621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Mullen A.C., High F.A., Hutchins A.S., Lee H.W., Villarino A.V., Livingston D.M., Kung A.L., Cereb N., Yao T.P., Yang S.Y., and Reiner S.L.. 2001. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 292:1907–1910. 10.1126/science.1059835 [DOI] [PubMed] [Google Scholar]
- Nakayamada S., Kanno Y., Takahashi H., Jankovic D., Lu K.T., Johnson T.A., Sun H.W., Vahedi G., Hakim O., Handon R., et al. . 2011. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 35:919–931. 10.1016/j.immuni.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Hwang D., Yang X.O., Kang H.S., Ma L., Wang Y.H., Watowich S.S., Jetten A.M., Tian Q., and Dong C.. 2008. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 29:138–149. 10.1016/j.immuni.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.H., and Dong C.. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005. 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea J.J., and Paul W.E.. 2010. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 327:1098–1102. 10.1126/science.1178334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich K.J., Huang A.C., and Weinmann A.S.. 2011. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J. Exp. Med. 208:1001–1013. 10.1084/jem.20102144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich K.J., Mohn S.E., and Weinmann A.S.. 2012. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat. Immunol. 13:405–411. 10.1038/ni.2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper M., Pagán A.J., Igyártó B.Z., Taylor J.J., and Jenkins M.K.. 2011. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 35:583–595. 10.1016/j.immuni.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H. 2016. T follicular helper cells in space-time. Nat. Rev. Immunol. 16:612–625. 10.1038/nri.2016.94 [DOI] [PubMed] [Google Scholar]
- Reinhardt R.L., Liang H.E., and Locksley R.M.. 2009. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 10:385–393. 10.1038/ni.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riteau N., Radtke A.J., Shenderov K., Mittereder L., Oland S.D., Hieny S., Jankovic D., and Sher A.. 2016. Water-in-Oil-Only Adjuvants Selectively Promote T Follicular Helper Cell Polarization through a Type I IFN and IL-6-Dependent Pathway. J. Immunol. 197:3884–3893. 10.4049/jimmunol.1600883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E.V. 2013. Epigenetic mechanisms and developmental choice hierarchies in T-lymphocyte development. Brief. Funct. Genomics. 12:512–524. 10.1093/bfgp/elt027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N., Liu Y., Bentebibel S.E., and Ueno H.. 2016. Molecular Mechanisms Regulating T Helper 1 versus T Follicular Helper Cell Differentiation in Humans. Cell Reports. 16:1082–1095. 10.1016/j.celrep.2016.06.063 [DOI] [PubMed] [Google Scholar]
- Snapper C.M., and Paul W.E.. 1987. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 236:944–947. 10.1126/science.3107127 [DOI] [PubMed] [Google Scholar]
- Szabo S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., and Glimcher L.H.. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 100:655–669. 10.1016/S0092-8674(00)80702-3 [DOI] [PubMed] [Google Scholar]
- Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B., et al. . 2012. The accessible chromatin landscape of the human genome. Nature. 489:75–82. 10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella L.A., Herati R.S., and Wherry E.J.. 2017. CD4+ T Cell Differentiation in Chronic Viral Infections: The Tfh Perspective. Trends Mol. Med. 23:1072–1087. 10.1016/j.molmed.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Weinstein J.S., Laidlaw B.J., Lu Y., Wang J.K., Schulz V.P., Li N., Herman E.I., Kaech S.M., Gallagher P.G., and Craft J.. 2018. STAT4 and T-bet control follicular helper T cell development in viral infections. J. Exp. Med. 215:337–355. 10.1084/jem.20170457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Sharma S., Edwards J., Feigenbaum L., and Zhu J.. 2015. Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat. Immunol. 16:197–206. 10.1038/ni.3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C., Schones D.E., Zeng C., Cui K., Zhao K., and Peng W.. 2009. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 25:1952–1958. 10.1093/bioinformatics/btp340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Min B., Hu-Li J., Watson C.J., Grinberg A., Wang Q., Killeen N., Urban J.F. Jr., Guo L., and Paul W.E.. 2004. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat. Immunol. 5:1157–1165. 10.1038/ni1128 [DOI] [PubMed] [Google Scholar]
- Zhu J., Yamane H., and Paul W.E.. 2010. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 28:445–489. 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Jankovic D., Oler A.J., Wei G., Sharma S., Hu G., Guo L., Yagi R., Yamane H., Punkosdy G., et al. . 2012. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 37:660–673. 10.1016/j.immuni.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.