Abstract
Autophagy of mitochondria (mitophagy) is essential for maintaining muscle mass and healthy skeletal muscle. Patients with heritable phosphatidic acid phosphatase lipin-1–null mutations present with severe rhabdomyolysis and muscle atrophy in glycolytic muscle fibers, which are accompanied with mitochondrial aggregates and reduced mitochondrial cytochrome c oxidase activity. However, the underlying mechanisms leading to muscle atrophy as a result of lipin-1 deficiency are still not clear. In this study, we found that lipin-1 deficiency in mice is associated with a marked accumulation of abnormal mitochondria and autophagic vacuoles in glycolytic muscle fibers. Our studies using lipin-1–deficient myoblasts suggest that lipin-1 participates in B-cell leukemia (BCL)-2 adenovirus E1B 19 kDa protein–interacting protein 3 (Bnip3)–regulated mitophagy by interacting with microtubule-associated protein 1A/1B-light chain (LC)3, which is an important step in the recruitment of mitochondria to nascent autophagosomes. The requirement of lipin-1 for Bnip3–mediated mitophagy was further verified in vivo in lipin-1–deficient green fluorescent protein-LC3 transgenic mice (lipin-1−/−-GFP-LC3). Finally, we showed that lipin-1 deficiency in mice resulted in defective mitochondrial adaptation to starvation-induced metabolic stress and impaired contractile muscle force in glycolytic muscle fibers. In summary, our study suggests that deregulated mitophagy arising from lipin-1 deficiency is associated with impaired muscle function and may contribute to muscle rhabdomyolysis in humans.—Alshudukhi, A. A., Zhu, J., Huang, D., Jama, A., Smith, J. D., Wang, Q. J., Esser, K. A., Ren, H. Lipin-1 regulates Bnip3–mediated mitophagy in glycolytic muscle.
Keywords: fld, mitochondrial autophagy, contractile force, rhabdomyolysis, LC3
Skeletal muscle is an extremely plastic tissue that can rapidly adapt to changes in energetic demands. Most skeletal muscles are a mixture of oxidative slow-twitch (type I) and fast-twitch (type IIA) fibers and glycolytic fast-twitch fibers (type IIB/IIX) (1, 2). Skeletal muscle atrophy, induced by aging (sarcopenia), prolonged fasting states (starvation), cancer cachexia, sepsis, chronic heart failure, or diabetes, is nutrient related and predominantly restricted to the glycolytic muscle fibers (3).
Autophagy is essential for muscle plasticity; it degrades protein aggregates and damaged organelles (4–7). This process involves the sequestration of cytoplasmic components and organelles in double-membraned autophagosomes (8, 9), which subsequently fuse with lysosomes, where their cargoes are degraded and recycled. The efficient removal of dysfunctional organelles and aberrant protein aggregates is critical for the maintenance of muscle metabolic homeostasis.
Autophagy is generally a nonselective degradation process in which autophagosomes nondiscriminately engulf cytosolic materials; however, damaged mitochondria can be selectively removed by autophagy (i.e., mitophagy) (10–14). Although mitophagy uses a common core autophagic machinery, it must have a mechanism to ensure selective elimination of specific cargos, mitochondria. Studies in yeast have identified the genes necessary for mitophagy (15–17). Moreover, Kristensen et al. (17) found that during serum and amino acid starvation, cytosolic proteins are degraded early on, whereas mitochondria are degraded at a much later time, suggesting that the degradation of proteins and organelles are distinct processes that occur during starvation.
To date, two categories of mitophagy have been described: 1) phosphatase and tensin homolog-induced putative kinase-1/Parkin, and 2) mitochondrial receptor-mediated mitophagy (6, 9, 11, 12). B-cell leukemia (BCL)-2/adenovirus E1B 19 kDa protein–interacting protein 3 (Bnip3), a mitophagy receptor, plays a key role in the autophagic removal of mitochondria in skeletal muscle (18, 19). This protein interacts with the autophagy protein microtubule-associated protein 1A/1B– light chain 3 (LC3) via a conserved LC3 interaction region, bringing mitochondria into autophagosomes during mitophagy (20, 21).
Failure of general autophagy or mitophagy is particularly detrimental in postmitotic tissues such as skeletal muscle (22–25). Autophagy blockages induced by the skeletal muscle–specific deletion of autophagy-related protein-7 result in profound muscle atrophy and an age-dependent decrease in muscle contractile force (22). Autophagy-related protein-7–null muscles show an accumulation of swollen and irregular-shaped mitochondria (22). In patients with Pompe disease, a failure of productive autophagy in muscle tissue contributed significantly to muscle weakness (26).
Lipin-1 (encoded by the LPIN1 gene) is a phosphatidic acid phosphatase that catalyzes the conversion of phosphatidic acid to diacylglycerol, which is involved in the de novo synthesis of phospholipids and triglycerides (27–31). Human studies have found that patients with heritable LPIN1–null mutations often develop muscle atrophy accompanied by mitochondrial aggregates and reduced mitochondrial cytochrome c oxidase activity in glycolytic muscle fibers (32, 33). Selective knockout of Lpin-1 gene in skeletal muscle of MCK-Lpin-1Δ115 mice in Dr. Schweitzer’s collected data (Schweitzer et al., unpublished results) leads to an increased occurrence of swollen mitochondria with impaired mitochondrial function, suggesting defective mitochondria clearance. Zhang et al. (34) reported that lipin-1 phosphatidic acid phosphatase activity is necessary for autophagosome maturation and that lipin-1 deficiency in myoblasts leads to impaired autophagy. However, the role of lipin-1 in the mitophagy process has not been fully explored.
In the current study the glycolytic extensor digitorum longus (EDL) muscle of lipin-1–deficient fatty liver dystrophy (fld) mice exhibited significantly increased mitochondrial aggregation and autophagic vacuole formation. Lipin-1 regulated the autophagic clearance of damaged mitochondria through Bnip3–mediated mitophagy. Furthermore, lipin-1 deficiency induced a defect in mitophagy associated with reduced mitochondrial energy potential and muscle force. Therefore, our study provides new pathophysiologic insights into the potential roles of lipin-1 and subsequently mitophagy in rhabdomyolysis.
MATERIALS AND METHODS
Animals
BALB/cByJ-Lpin-1fld/J mice carrying a spontaneous mutation in the Lpin-1 gene (fld) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). CAG promoter–driven green fluorescent protein (GFP)-LC3 transgenic mice in C57BL/6 background were provided as a generous gift from Dr. Noboro Mizushima (University of Tokyo, Tokyo, Japan) (35, 36). We generated Lipin-1–deficient GFP-LC3 transgenic mice (lipin-1−/−-GFP-LC3) by crossing fld mice with GFP-LC3 transgenic mice. Because lipin-1−/−-GFP-LC3 compound mice have a mixed C57BL/6xBALB/cByJ background, we used their GFP-LC3 lipin wild-type (WT) littermates as positive controls. The fld and WT mice were deprived of food for 16 h with free access to drinking water. GFP-LC3 control and lipin-1−/−-GFP-LC3 transgenic mice were subjected to intraperitoneal injection with chloroquine diphosphate (50 mg/kg body weight; MilliporeSigma, Burlington, MA, USA) every 24 h for 10 d (37), followed by another 16 h of starvation. All animal experiments were performed in accordance with relevant guidelines and regulations approved by the Animal Care and Use Committee of Wright State University.
Generation of lipin-1–deficient C2C12 myoblasts
Lipin-1 deficient C2C12 myoblasts were generated by the genome-editing clustered regularly interspaced short palindromic repeat (CRISPR)/caspase (Cas)-9 system. Lipin-1 gRNA (5′-CACGTCCGCTTCGGCAAGAT-3′) was generated with the CRISPR design tool (Massachusetts Institute of Technology, Cambridge, MA, USA; http://crispr.mit.edu) and was ligated into pSpCas9 (BB)-2A-Puro (PX495) v.2.0 (plasmid 62988; Addgene, Cambridge, MA, USA) using the method described by Ran et al. (38). Successful cloning of gRNA into PX495 was verified by sequencing through GeneWiz. The pSpCas9 (BB)-2A-puro-lipin-1gRNA plasmid was transfected into C2C12 myoblasts. One day after transfection, the cells were enriched with puromycin-containing medium (3 µg/ml) to select and generate a stable lipin-1–knockout C2C12 cell line. Depletion of lipin-1 in this stable cell line was verified and monitored by both Western blot and mRNA analysis.
Muscle force measurements
EDL muscle fiber bundles isolated from WT and fld mice were fixed by tethers of braided silk sutures (4-0) at the proximal and distal tendons before removal from the legs. To measure muscle force, the distal end was attached to a platinum wire hook inside a temperature-controlled muscle bath (Radnoti, Monrovia, CA, USA) of oxygenated Krebs-Ringer solution (137 mM NaCl, 5 mM KCl, 1 mM MgSO4, 1 mM NaH2PO4, 24 mM NaHCO3, and 2 mM CaCl2; pH 7.3) perfused with 95% O2-5% CO2 at room temperature. The proximal end was mounted on a 300C-LR lever arm (Aurora Scientific, Aurora, ON, Canada). The muscle was positioned between platinum electrodes and subjected to electrical field stimulation (supramaximum voltage, 0.5 ms pulse duration) using a 701C stimulator (Aurora Scientific). Force output was measured and recorded with 610A Dynamic Muscle Control software (Aurora Scientific). Bath temperature was maintained at 25°C throughout the protocol. Optimal length was determined by adjusting the EDL position to maximize twitch force at 1 Hz, and length was measured with digital calipers. After an initial 300 Hz tetanic stimulation, the force–frequency relationship was determined by using contractions at 1-min intervals with stimulus frequencies of 1, 15, 30, 50, 80, 120, 150, and 200 Hz and a pulse duration of 500 ms. After each experiment, the muscle was removed from the apparatus, gently blotted dry, and weighed. Physiologic cross-sectional area (CSA) was estimated according to Brooks and ;Faulkner (39) and used to calculate specific force (force/CSA), which was expressed as newtons per square centimeter. Data were analyzed by using repeated-measures ANOVA and Bonferroni post hoc comparison. Differences of P < 0.05 were considered significant.
Electron microscopy
Muscle specimens from mice were fixed in 3.5% glutaraldehyde in 0.1 M cacodylate buffer (pH7.4) at 4°C for 2 h followed by buffer rinse and postfixation in 1% osmium tetroxide for 1 h. Muscle sections were then dehydrated sequentially by 50, 70, 80, 90, and 100% ethanol, infiltrated with graded mixtures of propylene oxides, then embedded in fresh resin and incubated at 60°C for 48 h. Ultrathin sections (60 nm thick) were cut on an Ultracut E Microtome (Reichert-Jung; Leica Microsystems, Wetzlar Germany) with a diamond knife (Diatome, Hatfield, PA, USA), collected on copper grids, and stained with uranyl acetate and lead citrate. Electron micrographs were obtained with a Tecnai Biotwin 12-Electron Microscope (Thermo Fisher Scientific, Waltham, MA, USA) equipped with a 4K digital camera (Gatan, Pleasanton, CA, USA).
Western blot analysis
EDL, soleus, or primary myoblasts were homogenized with a glass homogenizer in 20 volumes of lysis buffer containing 20 mM Tris HCl (pH 7.5) and 2% SDS. Relative protein expression levels were examined by immunoblot analysis with anti-Bnip3 (1:1000; ab10433; Abcam, Cambridge, MA, USA), anti-Tom20 (1:500; 17764; Santa Cruz Biotechnology, Dallas, TX, USA), anti-Parkin (1:1000; 2132; Cell Signaling Technology, Danvers, MA, USA), anti-LC3 (1:1000; 3868; Cell Signaling Technology), anti-p62 (1:1000; P0067; MilliporeSigma, Burlington, MA, USA), anti-cytochrome c oxidase subunit IV (COX-IV) (1:1000; 4850; Cell Signaling Technology), and anti-α-tubulin (1:1000; T5168; MilliporeSigma) primary antibodies, followed by incubation with goat anti-mouse IgG, goat anti-rabbit IgG IR Dye 680 or 800 (Li-Cor Biosciences, Lincoln, NE, USA), or goat anti-mouse and anti-rabbit IgG-horseradish peroxidase (Promega, Madison, WI, USA) secondary antibodies. The blots were imaged with an infrared scanner (Odyssey Infrared Imaging System; Li-Cor Biosciences) or Amersham Imager 600 (GE Healthcare, Wilmington, MA, USA).
Immunoprecipitation
For immunoprecipitation, WT and stable lipin-1–deficient C2C12 myoblasts were cotransfected with GFP-Bnip3 and mCherry-LC3. After 24 h, transfected cells were either not treated or serum starved for 4 h and lysed in Pierce IP lysis buffer (1861603; Thermo Fisher Scientific). An equal amount of protein lysate was incubated with mouse anti-Bnip3 mAbs (1:100; ab10433; Abcam) overnight at 4°C, followed by incubation with 50 µl Pierce protein A/G argose beads (20421; Thermo Fisher Scientific) for 4 h. The immune complexes were subjected to Western blot analysis with rabbit anti-LC3 antibody (1:1000; Cell Signaling Technology) and rabbit phospho-serine antibody (1:1000; ab9332; Abcam). Total protein lysates were also subjected to Western blot analyses with anti-Bnip3 and anti-LC3 antibodies.
Immunofluorescence and microscopy, image processing, and morphometric analyses
Muscle biopsies collected from GFP-LC3 control and lipin-1−/−-GFP-LC3 transgenic mice were frozen in an isopentane/liquid nitrogen slurry. Muscle biopsies were cut into 7 μm–thick slices at −23°C in a cryostat microtome (CM1850; Leica Microsystems). The cryosections were air dried for 1 h, washed with PBS for 5 min, and fixed in 4% paraformaldehyde for 7 min at room temperature. After a wash in PBS, the sections were blocked in PBS containing 5% fetal bovine serum for 1 h at room temperature, followed by incubation with anti-Bnip3 (1:100; ab10433; Abcam) in 1% blocking buffer overnight at 4°C. The sections were then incubated with goat anti-mouse AlexaFluor 488 (1:200; Thermo Fisher Scientific) and goat anti-rabbit AlexaFluor 555 (1:200; Thermo Fisher Scientific) in amplification diluents. After DAPI incubation and washes, the slides were mounted in Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA, USA).
For imaging cells, WT or lipin-1–deficient myoblasts were seeded onto 13 mm round glass coverslips. The cells were transfected with GFP-Bnip3 and mCherry-LC3 plasmids. After 24 h, they were either not treated, serum starved for 2 h, or serum starved for 2 h followed by treatment with 200 nM bafilomycin A1 for 2 h. The cells were fixed in 4% paraformaldehyde. Colocalization of GFP-Bnip3 and mCherry-LC3 was examined with an inverted microscope (IX70; Olympus, Tokyo, Japan) equipped with a DFC7000T camera (Leica Microsystems). All digital microscopic images were acquired at room temperature. GFP-Bnip3 was a generous gift from Dr. Wen-xing Ding (University of Kansas Medical Center, Kansas City, KS, USA) (40).
Preparation of mitochondria-enriched fraction
Mitochondria were enriched from mouse skeletal muscle by a method described by Garcia-Cazarin et al. (41). In brief, EDL and soleus muscles from fld and WT mice were collected and sequentially washed with prechilled PBS, PBS containing 10 mM EDTA, and isolation buffer [containing 10 mM EDTA, 215 mM d-mannitol, and 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; pH 7.4), 0.1% bovine serum albumin, and 75 mM sucrose]. After homogenization in a glass homogenizer, the tissue homogenates were transferred to 2 ml microcentrifuge tubes and centrifuged at 700 g for 10 min at 4°C. The supernatant was transferred to a fresh, prechilled microcentrifuge tube and centrifuged at 10,500 g for 10 min at 4°C to pellet the mitochondria-enriched fraction. The pellet was again washed with ice-cold isolation buffer and lysed in RIPA buffer for Western blot analyses.
Mitochondrial membrane potential of myoblasts
Primary myoblasts isolated from WT or fld mice were seeded onto 13 mm round glass coverslips. After 24 h, the cells were either not starved or starved in HBSS medium for 6 h followed by incubation with 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1) dye (5 μg/ml; Thermo Fisher Scientific) in HBSS, containing 10 mM HEPES (pH 7.4), for 20 min at 37°C. After extensive washing with HBSS, the JC-1 monomer and aggregate fluorescence in the myoblasts was monitored by a confocal microscope. Laser line intensity, photometric gain, photomultiplier setting, filter attenuation, and pinhole size were kept constant throughout the experiments. Confocal images were processed with an image analysis system (KS 300; Zeiss GmbH, Jena, Germany). The size, number of pixels, and ratios between fluorescence intensity in the red and green channels in each region of interest were calculated and compared according to the same procedures with NIS-Elements BR 3.10 image analyzer software (Nikon, Melville, NY, USA).
Statistical analysis
In each graph, unless noted otherwise, data represent means ± se of independent experiments. Statistical significance was calculated by a 2-tailed Student’s t test, unless noted otherwise. Values of P < 0.05 were considered statistically significant.
RESULTS
Abnormal mitochondrial aggregation and autophagic vacuole accumulation in EDL muscle of fld mice
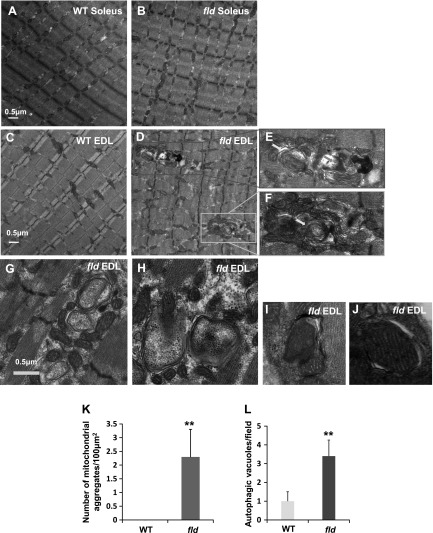
Patients with compound heterozygous loss of lipin-1 function exhibit abnormal mitochondrial aggregates with functional defects, as well as muscle wasting in glycolytic muscle fibers (32). To determine whether lipin-1–deficient fld mice have similar abnormal muscle morphology, we examined the soleus (Fig. 1A, B) and EDL (Fig. 1C–J) muscles from WT and fld mice using negative staining electron microscopy. Although WT EDL muscle exhibited normal ultrastructure in terms of the distribution and localization of mitochondria and other organelles (Fig. 1C), lipin-1 deficiency in EDL muscle resulted in abnormal aggregation of mitochondria, often spanning more than one z line (Fig. 1D–F). A mean of 2.3 mitochondrial aggregates per 100 μm2 was observed in fld EDL muscle, whereas no mitochondrial aggregate was observed in WT EDL muscle (Fig. 1K). Within the aggregates of mitochondria, we observed autophagic vacuoles (Fig. 1E–J), including those containing mitochondria (Fig. 1H–J). The morphology of some of the vacuoles resemble multilamellar bodies, which are membrane-bound organelles composed of concentric membrane layers (Fig. 1E–H). The number of autophagic vacuoles was also increased in fld EDL muscle (Fig. 1L). In contrast, mitochondrial aggregates were less obvious in the soleus muscle of either WT (Fig. 1A) or fld (Fig. 1B) mice.
Figure 1.
Lipin-1 deficiency results in mitochondrial aggregation and an increase in autophagic vacuoles. Representative electron micrographs of soleus of WT (A) and fld (B), and EDL of WT (C) and fld (D–J) mice. The morphology of some of the vacuoles resembles autophagosomes (E, F, yellow arrows) and autolysosomes (E, blue arrows). G–J) Representative micrographs showing mitochondrion-containing autophagic vacuoles. K) Quantification of the number of mitochondrial aggregates per 100 μm2. L) Quantification of the number of autophagic vacuoles per view field. **P < 0.01.
Lipin-1 deficiency is associated with impaired mitophagy
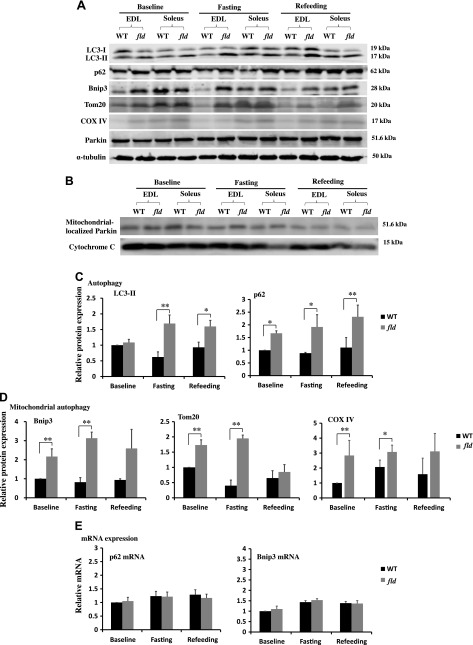
Because similar morphologic defects such as abnormal mitochondrial aggregation and autophagic vacuole accumulation have been described in autophagy–deficient skeletal muscle (25, 42), we examined the role of lipin-1 in mitophagy in EDL and soleus muscles by detecting the expression of representative autophagy and mitochondrial proteins during food deprivation and refeeding. LC3-II is associated with autophagosome membranes and thus is a commonly used marker for autophagosomes (43). Adaptor protein p62 facilitates the autophagic turnover of ubiquitinated protein aggregates by binding directly to both ubiquitin and LC3 (44). Our data show that LC3-II levels in fld EDL were higher than those in WT EDL in food-deprived animals and remained at high levels even after refeeding (Fig. 2A, C). Increased LC3-II levels in fld EDL as compared to WT EDL, at both basal levels and in response to food deprivation may be related to autophagy induction or impaired autophagic flux. To distinguish these two possibilities, we further examined the levels of p62, which is degraded by autophagy. We observed higher levels of p62 in fld EDL than in WT EDL under basal, food deprivation for 16 h, and then refed for 5 h (Fig. 2A, C). Of note, the mRNA expression level of p62 was not affected by lipin-1 deficiency (Fig. 2E), suggesting that the increased p62 protein level in lipin-1–deficient EDL was caused by blocked clearance rather than increased synthesis. This result is consistent with a blockade of autophagic flux in fld EDL and in agreement with Zhang et al. (34).
Figure 2.
Lipin-1 deficiency inhibits mitophagy. A) Representative immunoblots of key proteins involved in autophagy and mitophagy from EDL and soleus muscles of WT and fld mice fed with chow diet, deprived of food for 16 h (fasting) or food deprived for 16 h followed by refeeding for 5 h (refeeding). B) Western blot of Parkin in mitochondria-enriched fractions prepared from EDL and soleus muscles of WT and fld mice in basal, unfed, and refeeding states. The expression levels of cytochrome c in mitochondrial fraction were used as the loading control. C) Densitometry of LC3-II and p62 immunoblots of EDL muscles in WT and fld mice in the basal, unfed, and refeeding states. D) Densitometry of Bnip3, Tom20, and COX-IV immunoblots of EDL muscles in WT and fld mice in the basal, unfed, and refeeding states. E) Real-time RT-PCR of mRNA levels of p62 and Bnip3 in EDL muscle of WT and fld mice in the basal, unfed, and refeeding states. Data represent means ± se (n = 3/group). *P < 0.05, **P < 0.01. Experiments were repeated for at least 3 times.
We also examined the degradation of outer mitochondrial membrane protein, Tom20, and an inner membrane protein, COX-IV, and found higher levels of both Tom20 and COX-IV in fld EDL than in WT EDL in mice left unfed and after refeeding (Fig. 2A, D), suggesting that lipin-1 is important for mitophagy. As mitophagy can be regulated by either the Pink/Parkin pathway or the mitochondrial receptor-mediated pathways (9, 11), we examined the expression of Parkin in EDL and the soleus muscle of fld and WT mice and found no difference in Parkin levels in the basal, unfed, and refed states (Fig. 2A). We also isolated mitochondrial fractions of WT and fld EDL muscle in the basal, unfed, and refed states and found no obvious lipin-1 deficiency–related differences in Parkin protein levels in these mitochondrial fractions (Fig. 2B). This result suggests that a deficiency in lipin-1 does not affect the recruitment of Parkin to mitochondria.
Next, we examined the expression levels of Bnip3, a mitochondrial receptor. Bnip3 resides primarily on mitochondrial outer membranes (45) and has been implicated as a regulator of mitophagy (11, 21). The levels of Bnip3 protein expression in EDL muscles of fld mice were significantly higher than those in WT EDL muscle under basal conditions, increased further in response to food deprivation, and remained high after refeeding (Fig. 2A, D). Similar to p62, Bnip3 mRNA levels were similar between WT and fld mice (Fig. 2E) suggesting that the Bnip3 increase in fld was related to defective clearance by mitophagy. Together, our data suggest that lipin-1 is necessary for Bnip3–, but not pink/Parkin–mediated mitophagy in mouse EDL muscle.
Lipin-1 deficiency results in impaired Bnip3–LC3 interaction
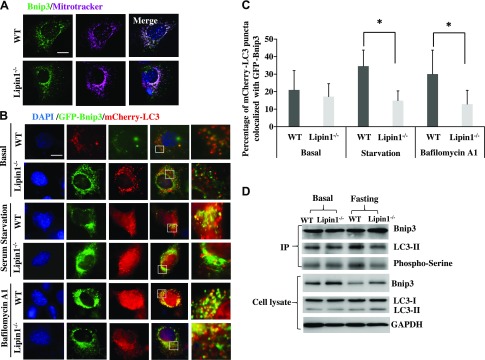
Because impaired mitophagy was mainly observed in EDL muscle of fld mice, the following experiments focused on comparing fld EDL with WT EDL muscle. To further understand how lipin-1 affects Bnip3–mediated mitophagy in mouse EDL muscle, we used lipin-1–deficient myoblasts isolated from fld mice to determine the mechanism by which lipin-1 deficiency impairs Bnip3–mediated mitophagy. Bnip3 is a BH3-only protein that is localized to the outer mitochondrial membrane in response to cellular stress, including starvation; it binds to LC3 to recruit damaged mitochondria to autophagosomes (21, 46). We first transfected WT and lipin-1–deficient myoblasts with GFP-Bnip3 plasmids and examined the subcellular localization of GFP-Bnip3 in response to 6 h of serum deprivation. Bnip3 protein was highly colocalized with mitochondria, which was detected by Mitotracker (Thermo Fisher Scientific) in both WT and lipin-1–deficient myoblasts (Fig. 3A).
Figure 3.
Lipin-1 contributes to LC3-Bnip3–mediated autophagosome recruitment. A) Primary myoblasts isolated from WT and fld mice were transfected with GFP-Bnip3. Twenty-four hours after transfection, mitochondria were stained by Mitotracker dye. Subcellular colocalization of Bnip3 and mitochondria in the presence (WT) and absence (Lipin-1−/−) of lipin-1 were identified. Scale bar, 50 μm. B) WT and Lipin-1−/− myoblasts were transfected with GFP-Bnip3 and mCherry-LC3 plasmids. Twenty-four hours after transfection, cells were not treated, serum starved for 2 h, or serum starved for 2 h followed by 2 h of bafilomycin A1 (200 nM) treatment. Higher magnification images represent the regions enclosed in white squares. C) Quantification of the average percentage of mCherry-LC3 puncta colocalized with GFP-Bnip3. Data are means ± se. *P < 0.05. D) Lipin-1 deficiency suppressed the interaction of Bnip3 and LC3, likely through inhibition of Bnip3 phosphorylation. Lipin-1–deficient (Lipin-1−/−) and control (WT) myoblasts, transfected with GFP-Bnip3 and mcherry-LC3, were either not treated or treated with serum-free medium for 4 h. The cell lysates were subjected to immunoprecipitation with mouse anti-Bnip3 mAb. The immunoprecipitates were immunoblotted with rabbit anti-LC3 to evaluate the Bnip3-LC3 interaction or rabbit anti-phosphoserine antibody to assess Bnip3 phosphorylation.
We next cotransfected primary WT and lipin-1–deficient myoblasts with plasmids expressing GFP-Bnip3 and mCherry-LC3. We evaluated the colocalization of GFP-Bnip3 and mCherry-LC3 puncta to examine whether mitochondria were sequestered by autophagosomes after no treatment, 2 h of serum deprivation, or 2 h of serum deprivation followed by 2 h of treatment with bafilomycin A1 (lysosomal inhibitor, 200 nM). Quantification of Bnip3–containing autophagosomes was performed by calculating the percentage of GFP-Bnip3+mCherry-LC3-labeled autophagic structures including autophagosomes and autolysosomes. We observed that in response to serum starvation, extensive mitophagy occurred in GFP-Bnip3- and mCherry-LC3–cotransfected WT cells, where 34.6% mCherry-LC3–labeled autophagosomes/autolysosomes colocalized with GFP-Bnip3. However, the percentage of LC3+/Bnip3+ costained autophagosomes/autolysosomes was significantly lower in lipin-1–deficient myoblasts, as only 14.9% mCherry-LC3–labeled autophagosomes/autolysosomes colocalized with GFP-Bnip3, suggesting that lipin-1 deficiency significantly inhibited autophagosome recruitment of mitochondria by Bnip3 (Fig. 3B, C). Similar results were observed in bafilomycin A1-treated cells (30% in WT vs. 12.8% in lipin-1−/−).
We further examined the interaction between Bnip3 and LC3 by using immunoprecipitation. Lipin-1–deficient and WT myoblasts were cotransfected with GFP-Bnip3 and mcherry-LC3. Transfected cells were either not treated or serum starved for 4 h. The cell lysates were then subjected to immunoprecipitation with the mouse anti-Bnip3 mAb. The immunoprecipitates were blotted with rabbit anti-LC3 to evaluate the Bnip3-LC3 interaction. Our results showed less LC3 interaction with Bnip3 in the absence of lipin-1 (Fig. 3D). LC3-I is cytosolic (47), and we detected LC3-I in the total cell lysate but not in the immunoprecipitates.
Previous study suggested that Bnip3 phosphorylation at serine residues 17 and 24 in the LC3-interacting region is critical for the recognition of LC3 (20). However, phospho-Bnip3 antibody is not available. To evaluate whether lipin-1 promotes Bnip3 activation through phosphorylation, we also immunoblotted the Bnip3 immunoprecipitates with mouse anti-phosphoserine antibody to assess Bnip3 phosphorylation. Our results showed that Bnip3 serine phosphorylation was reduced in the absence of lipin-1 under food-deprivation conditions (Fig. 3D), suggesting that lipin-1 is necessary for Bnip3 serine phosphorylation and thus LC3 binding.
Together, these results confirmed the requirement of lipin-1 in myoblasts for the starvation-induced interaction between Bnip3 and LC3, which is a critical step during recruitment of cargos (i.e., mitochondria) to autophagosomes. We did not observe any obvious differences in Bnip3-LC3 interaction at basal levels (Fig. 3B).
Requirement of lipin-1 in Bnip3–mediated mitophagy was confirmed in vivo
To verify the effect of lipin-1 deficiency on mitophagy processes in vivo, we generated lipin-1–deficient GFP-LC3 transgenic mice (lipin-1−/−–GFP-LC3) by crossing lipin-1–deficient fld mice with GFP-LC3 transgenic mice. We monitored GFP-LC3, Bnip3, and p62 protein expression levels in the lipin-1−/−-GFP-LC3 mice in basal and starvation states, and compared results with those from the lipin-1–deficient fld mice without GFP-LC3. We found that lipin-1 deficiency induced accumulation of Bnip3 and LC3 proteins in lipin-1−/−-GFP-LC3 mice, consistent with findings in fld mice (data not shown).
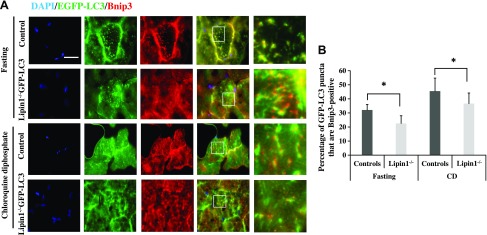
To address the effect of lipin-1 deficiency on autophagic flux in vivo, we withheld food from GFP-LC3-expressing (WT control) and lipin-1−/−–GFP-LC3 mice after no treatment or treatment with chloroquine diphosphate (a lysosomal inhibitor, 50 mg/kg body weight, every 24 h for 10 d; MilliporeSigma) (Fig. 4). Consistent with the results in cell culture (Fig. 2), 16 h of food deprivation induced increased punctate GFP expression in lipin-1−/−–GFP-LC3 mice compared with GFP-LC3 controls, but the colocalization of Bnip3 and GFP-LC3 was less in lipin-1−/−-GFP-LC3 mice than in GFP-LC3 controls (Fig. 4A, B). Chloroquine diphosphate treatment increased the number of punctate GFP-LC3, and Bnip3-GFP-LC3 colocalization, but lipin-1−/−-GFP-LC3 mice still had a smaller percentage of Bnip3+ autophagosomes/autolysosomes than GFP-LC3 controls. Together, our results suggest that lipin-1 deficiency impairs Bnip3–mediated mitophagy in vivo.
Figure 4.
Lipin-1 is essential in Bnip3–mediated mitophagy in vivo. A) Immunofluorescence detection of Bnip3 and GFP-LC3 in GFP-LC3 control and lipin-1−/−-GFP-LC3 transgenic mice that were food deprived for 16 h, or were treated with chloroquine diphosphate (50 mg/kg body weight, intraperitoneal injection for 10 d) followed by food deprivation for 16 h (n = 3 for each group). Higher magnification images represent the regions enclosed in white squares. EDL muscles were cryosectioned and stained with anti-Bnip3 antibody. Scale bar, 50 μm. Subcellular localization of Bnip3 (red) and GFP-LC3 (green) in GFP-LC3 control (Control) and lipin-1−/−-GFP-LC3 (Lipin-1−/−) were quantified in (B) as the percentage of GFP-LC3 puncta that are Bnip3+. Significant differences between groups were determined with Student’s t test. *P < 0.05.
Mitophagy defect impacts mitochondrial membrane potential and muscle force
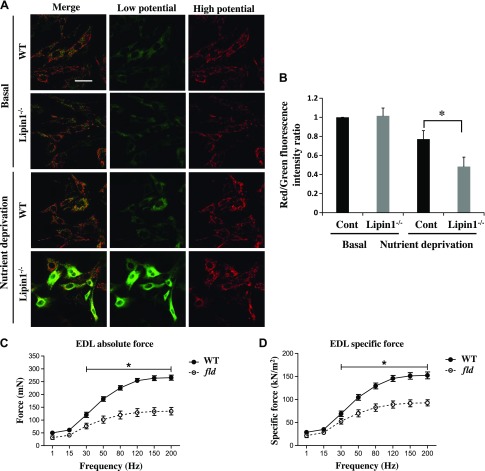
Last, we determined the impact of lipin-1 deficiency–induced blockade of mitophagy on cellular energy demand and muscle physiology. We first measured mitochondrial membrane potential, an important parameter of mitochondrial function and an indicator of cell health, in primary myoblasts isolated from the EDL muscle of fld and WT mice. Mitochondrial membrane potential was detected by a unique fluorescent cationic dye, JC-1, which alters its fluorescence properties at different membrane potentials. At low membrane potentials, JC-1 produces a green fluorescence; whereas at high membrane potentials, it forms “J aggregates” with red fluorescence (48, 49). Mitochondrial depolarization is indicated by a decrease in red:green fluorescence intensity ratios. We found that there was no difference in mitochondrial membrane potential between WT and fld myoblasts under basal conditions. However, in response to nutrient starvation for 6 h, lipin-1 deficiency caused a significantly reduced mitochondrial membrane potential in myoblasts isolated from EDL of fld mice, as compared with WT mice (Fig. 5A, B).
Figure 5.
Lipin-1 deficiency leads to reduced mitochondrial energy potential and impaired muscle force. A) Lipin-1 deficiency in primary myoblasts isolated from fld mice (compared with WT mice) led to reduced mitochondrial energy potential, as probed by JC-1 dye. Primary myoblasts isolated from WT and fld mice were either untreated or starved with an HBSS medium containing 10 mM HEPES for 6 h at 37°C followed by incubation with 5 μg/ml JC-1 dye at 37°C for 20 min. After extensive washing with HBSS, the medium was switched to HBSS before immediate live imaging. All images were acquired under the same instrument settings. Scale bar, 50 μm. B) The ratio of JC-1 red to green staining was subjected to image analysis to measure the mitochondrial membrane potential. Data are means ± se. *P < 0.05. Absolute force (C) and force per CSA (specific force) (D) in EDL of WT (n = 4) and fld (n = 4) mice. Tetanic contractions were elicited at 1 min intervals with stimulus frequencies of 1, 15, 30, 50, 80, 120, 150, and 200 Hz. *P < 0.05, by 1-way repeated-measures ANOVA and Bonferroni post hoc comparison.
It has been reported that impaired mitochondrial clearance contributes to reduced contractile force in skeletal muscle (22). To further understand the functional consequences of mitophagy defects from lipin-1 deficiency, we evaluated absolute and specific contractile force in isolated EDL muscles from fld and WT mice at 3–5 mo of age (Fig. 5C, D). Maximum absolute isometric tetanic force was determined from the plateau obtained in a frequency-force experiment. Specific force was calculated by normalizing absolute force to the muscle fiber CSA. We found that EDL muscle from fld mice displayed a 46% reduction in absolute force and a 40% decrease in specific force compared to WT EDL muscle with a stimulus frequency of 200 Hz (Fig. 5C, D).
DISCUSSION
In our study, we found that lipin-1 is involved in Bnip3–mediated sequestration of mitochondria by autophagosomes in EDL muscle. Lipin-1-deficiency resulted in both impaired mitochondrial clearance and loss of membrane potential as well as contributed to muscle weakness in the EDL muscle of fld mice.
We investigated the impact of a lipin-1 deficiency on mitochondrial autophagy. Compared to WT muscle, we found that levels of Bnip3 protein in EDL muscles of fld mice were higher than in WT mice in basal conditions, and further increased in response to food deprivation and remained elevated after refeeding in fld but not in WT mice, indicating mitophagy blockage as a result of the lipin-1 deficiency. This finding suggests a mechanism for mitochondrial aggregate accumulation in muscle from fld mice. Specifically, we found that, other than facilitating the fusion of autophagosomes to lysosomes as previously found (34), lipin-1 may also promote Bnip3–mediated cargo recruitment in mitophagy. Therefore, it is likely that in the absence of lipin-1, the inhibition of both mitophagy and general autophagy contributes to mitochondrial abnormality. Although inhibition of mitophagy slows down the formation of mitochondria-containing autophagosomes, defective general autophagy prevents their degradation.
Lipin-1 could be involved in Bnip3-LC3 interactions. This notion is supported by our findings that levels of Bnip3 expression in EDL muscles of fld mice were higher under basal conditions, increased further in response to food deprivation, and remained high after refeeding, as compared to WT muscle, whereas Bnip3 mRNA levels were very similar between WT and fld mice. Further evidence from in vitro and in vivo experiments showed that the colocalization of Bnip3 and LC3 was affected by lipin-1 deficiency. Previous study suggested that Bnip3 phosphorylation at serines 17 and 24 is critical for its binding to LC3 (20). It is possible that lipin-1 mediates Bnip3-LC3 interaction through Bnip3 phosphorylation. Indeed, our data show reduced Bnip3 serine phosphorylation in lipin-1–deficient myoblasts. The kinase that regulates Bnip3 phosphorylation is still unknown. Lipin-1 catalyzes the biosynthesis of diacylglycerol which is an important signaling molecule regulating PKC and -D activation (34). Therefore, we hypothesize that PKC or PKD, activated by diacylglycerol generated by lipin-1, could phosphorylate Bnip3. Further studies are needed to identify whether Bnip3 can be phosphorylated by PKC or PKD. In addition, although lipin-1 is involved in Bnip3–mediated mitophagy, we cannot rule out the possibility that other proteins and lipids, such as fibrillin unique N-terminal–domain containing -1 (50), small mothers against decapentaplegic homolog (SMAD) ubiquitination regulatory-1 (51), and cardiolipin (52) can also serve as additional mitophagy receptors. Further studies are needed to evaluate their contributions.
Previous studies showed that the impairment of basal mitophagy or autophagy is deleterious to muscle homeostasis and contributes to muscle atrophy and weakness (22–24, 53). We evaluated the functional consequences of defective mitophagy by measuring mitochondrial membrane potential and muscle force. Physiologic measurements of muscle force in EDL muscle of fld mice showed muscle weakness which is consistent with previous reports on impaired mitophagy/autophagy. Mitochondria are essential for ATP generation, and in our study, the data showed that lipin-1 deficiency reduced mitochondrial membrane potential. Therefore, it is possible that lipin-1 deficiency impairs mitochondrial membrane potential, leading to muscle weakness. In addition, ultrastructural studies of lipin-1–deficient EDL muscle display a marked increase in mitochondrial aggregates spanning from one to the next z line, accompanied by an increased number of autophagic vacuoles. It is also possible that mitochondrial aggregates and autophagic vacuoles alter the normal disposition of sarcomeric and contractile proteins and impair normal actin–myosin interactions, further contributing to muscle weakness. Further studies are needed to determine the mechanisms of how impaired mitophagy, induced by lipin-1 deficiency, leads to muscle weakness. Our observation is consistent with Dr. Schweitzer’s recent work (Schweitzer et al., unpublished results) in which the selective knockout of Lpin-1 gene in the skeletal muscle of MCK-Lpin-1Δ115 mice displayed an increased occurrence of swollen mitochondria with impaired mitochondrial function and progressive myopathy.
In response to nutrient deprivation, lipin-1–deficient myoblasts had reduced mitochondrial membrane potential, suggesting that lipin-1 deficiency is detrimental to mitochondrial function under metabolic stress. If the degree of mitochondrial interconnection becomes incompatible with the lack of autophagic removal of damaged mitochondria, the mitochondrial network collapses. Lipin-1 deficiency is associated with massive rhabdomyolysis episodes in humans which are typically triggered by fasting or intense exercise (54, 55). Because fasting and exercise also trigger autophagy (32), it is possible that impaired autophagy, including mitophagy, contributes to the muscle pathophysiologies in lipin-1–deficient patients under metabolic stress.
In our study, we found that EDL muscle is more subject to mitophagy defects than is oxidative soleus muscle, which is consistent with previous studies showing that the glycolytic fibers are more vulnerable under diabetes, chronic heart failure, aging, and cancer cachexia-induced muscle atrophy conditions compared to oxidative muscle fibers (56). One explanation is that protein synthesis and degradation occur more rapidly in oxidative muscle fiber than in glycolytic muscle fiber (56). It has also been reported that fiber-specific atrophy is attributable to different signaling pathways. For example, muscle wasting is associated with a Fyn oncogene/signal transducer and activator of transcription -3/vacuolar protein sorting-34 (Fyn/STAT3/Vps34) signaling pathway in glycolytic muscles, but not in oxidative muscles (57). This detailed mechanism should be further studied.
In summary, we found that lipin-1 plays an important role in Bnip3–mediated mitophagy by altering its interactions with LC3. Inhibition of mitophagy induced by lipin-1 deficiency was more pronounced in glycolytic muscle fibers, as compared to oxidative muscle fibers. Furthermore, lipin-1 deficiency led to abnormal mitochondrial aggregation and mitochondrial dysfunction, as indicated by decreased mitochondrial membrane potential, which may greatly affect mitochondrial adaptation to metabolic stress. Our study suggests that dysregulated mitophagy resulting from lipin-1 deficiency is associated with impaired muscle function. Our study contributes to the understanding of the link between lipin-1 deficiency and muscle wasting and weakness in lipin-1–deficient patients, and in patients with chronic heart failure, cancer cachexia, sepsis, diabetes, age-related muscle wasting (sarcopenia), or disuse atrophy.
ACKNOWLEDGMENTS
The authors thank Dr. Noboro Mizushima (University of Tokyo, Tokyo, Japan) for the GFP-LC3 mice, Dr. Wen-Xing Ding (University of Kansas Medical School, Kansas City, KS, USA) for the GFP-Bnip3 plasmid, and Dr. Feng Zhang (McGovern Institute for Brian Research, Massachusetts Institute of Technology, Cambridge, MA, USA) for pSpCas9 (BB)-2A-Puro (PX495) v.2.0 (plasmid 62988; Addgene, Cambridge, MA, USA). This work was supported by startup funds from Wright State University (to H.R.), U.S. National Institutes of Health Center of Biomedical Research Excellence on Obesity and Cardiovascular Diseases Grant P20 GM103527-06, and Beginning Grant-in-Aid 11BGIA7710059 and Scientist Development Grant 12SDG12050697 from the American Heart Association (to H.R.). The authors declare no conflicts of interest.
Glossary
- Bnip3
B-cell leukemia (BCL)-2/adenovirus E1B 19 kDa protein-interacting protein 3
- CSA
cross-sectional area
- COX-IV
cytochrome c oxidase subunit IV
- CRISPR/Cas9
clustered regularly interspaced short palindromic repeat/caspase-9
- EDL
extensor digitorum longus
- fld
fatty liver dystrophy
- GFP
green fluorescent protein
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- JC-1
5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide
- LC3
microtubule-associated protein 1A/1B-light chain 3
- WT
wild type
AUTHOR CONTRIBUTIONS
A. A. Alshudukhi, J. Zhu, D. Huang, A. Jama, J. D. Smith, and H. Ren performed studies and analyzed data; H. Ren designed and supervised the work and wrote the manuscript; and Q. J. Wang and K. A. Esser provided intellectual contribution to the experimental design and data interpretation and edited the manuscript.
REFERENCES
- 1.Schiaffino S., Reggiani C. (2011) Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531 [DOI] [PubMed] [Google Scholar]
- 2.Goodman C. A., Kotecki J. A., Jacobs B. L., Hornberger T. A. (2012) Muscle fiber type-dependent differences in the regulation of protein synthesis. PLoS One 7, e37890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandri M. (2013) Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Int. J. Biochem. Cell Biol. 45, 2121–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He C., Klionsky D. J. (2009) Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi A. M., Ryter S. W., Levine B. (2013) Autophagy in human health and disease. N. Engl. J. Med. 368, 1845–1846 [DOI] [PubMed] [Google Scholar]
- 6.Ravikumar B., Sarkar S., Davies J. E., Futter M., Garcia-Arencibia M., Green-Thompson Z. W., Jimenez-Sanchez M., Korolchuk V. I., Lichtenberg M., Luo S., Massey D. C., Menzies F. M., Moreau K., Narayanan U., Renna M., Siddiqi F. H., Underwood B. R., Winslow A. R., Rubinsztein D. C. (2010) Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 90, 1383–1435 [DOI] [PubMed] [Google Scholar]
- 7.Kliosnky D., Abdelmohsen K., Abe A., et al. (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eskelinen E. L., Kovács A. L. (2011) Double membranes vs. lipid bilayers, and their significance for correct identification of macroautophagic structures. Autophagy 7, 931–932 [DOI] [PubMed] [Google Scholar]
- 9.Kubli D. A., Gustafsson A. B. (2012) Mitochondria and mitophagy: the yin and yang of cell death control. Circ. Res. 111, 1208–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J. (2013) Autophagy and mitophagy in cellular damage control. Redox Biol. 1, 19–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding W. X., Yin X. M. (2012) Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol. Chem. 393, 547–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim I., Rodriguez-Enriquez S., Lemasters J. J. (2007) Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hönscher C., Mari M., Auffarth K., Bohnert M., Griffith J., Geerts W., van der Laan M., Cabrera M., Reggiori F., Ungermann C. (2014) Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell 30, 86–94 [DOI] [PubMed] [Google Scholar]
- 14.Reggiori F., Komatsu M., Finley K., Simonsen A. (2012) Autophagy: more than a nonselective pathway. Int. J. Cell Biol. 2012, 219625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto K., Kondo-Okamoto N., Ohsumi Y. (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97 [DOI] [PubMed] [Google Scholar]
- 16.Xie Z., Klionsky D. J. (2007) Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 17.Kristensen A. R., Schandorff S., Høyer-Hansen M., Nielsen M. O., Jäättelä M., Dengjel J., Andersen J. S. (2008) Ordered organelle degradation during starvation-induced autophagy. Mol. Cell. Proteomics 7, 2419–2428 [DOI] [PubMed] [Google Scholar]
- 18.Rikka S., Quinsay M. N., Thomas R. L., Kubli D. A., Zhang X., Murphy A. N., Gustafsson A. B. (2011) Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 18, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonaldo P., Sandri M. (2013) Cellular and molecular mechanisms of muscle atrophy. Dis. Model. Mech. 6, 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Y., Massen S., Terenzio M., Lang V., Chen-Lindner S., Eils R., Novak I., Dikic I., Hamacher-Brady A., Brady N. R. (2013) Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J. Biol. Chem. 288, 1099–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanna R. A., Quinsay M. N., Orogo A. M., Giang K., Rikka S., Gustafsson A. B. (2012) Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J. Biol. Chem. 287, 19094–19104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masiero E., Agatea L., Mammucari C., Blaauw B., Loro E., Komatsu M., Metzger D., Reggiani C., Schiaffino S., Sandri M. (2009) Autophagy is required to maintain muscle mass. Cell Metab. 10, 507–515 [DOI] [PubMed] [Google Scholar]
- 23.Raben N., Hill V., Shea L., Takikita S., Baum R., Mizushima N., Ralston E., Plotz P. (2008) Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum. Mol. Genet. 17, 3897–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masiero E., Sandri M. (2010) Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy 6, 307–309 [DOI] [PubMed] [Google Scholar]
- 25.Wu J. J., Quijano C., Chen E., Liu H., Cao L., Fergusson M. M., Rovira I. I., Gutkind S., Daniels M. P., Komatsu M., Finkel T. (2009) Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany N.Y.) 1, 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shea L., Raben N. (2009) Autophagy in skeletal muscle: implications for Pompe disease. Int. J. Clin. Pharmacol. Ther. 47 (Suppl 1), S42–S47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Csaki L. S., Reue K. (2010) Lipins: multifunctional lipid metabolism proteins. Annu. Rev. Nutr. 30, 257–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reue K., Brindley D. N. (2008) Thematic Review Series. Glycerolipids: multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J. Lipid Res. 49, 2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reue K., Dwyer J. R. (2009) Lipin proteins and metabolic homeostasis. J. Lipid Res. 50 (Suppl), S109–S114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren H., Federico L., Huang H., Sunkara M., Drennan T., Frohman M. A., Smyth S. S., Morris A. J. (2010) A phosphatidic acid binding/nuclear localization motif determines lipin1 function in lipid metabolism and adipogenesis. Mol. Biol. Cell 21, 3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang W., Zhu J., Zhuang X., Zhang X., Luo T., Esser K. A., Ren H. (2015) Lipin1 regulates skeletal muscle differentiation through extracellular signal-regulated kinase (ERK) activation and cyclin D complex-regulated cell cycle withdrawal. J. Biol. Chem. 290, 23646–23655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michot C., Hubert L., Romero N. B., Gouda A., Mamoune A., Mathew S., Kirk E., Viollet L., Rahman S., Bekri S., Peters H., McGill J., Glamuzina E., Farrar M., von der Hagen M., Alexander I. E., Kirmse B., Barth M., Laforet P., Benlian P., Munnich A., JeanPierre M., Elpeleg O., Pines O., Delahodde A., de Keyzer Y., de Lonlay P. (2012) Study of LPIN1, LPIN2 and LPIN3 in rhabdomyolysis and exercise-induced myalgia. J. Inherit. Metab. Dis. 35, 1119–1128 [DOI] [PubMed] [Google Scholar]
- 33.Michot C., Hubert L., Brivet M., De Meirleir L., Valayannopoulos V., Müller-Felber W., Venkateswaran R., Ogier H., Desguerre I., Altuzarra C., Thompson E., Smitka M., Huebner A., Husson M., Horvath R., Chinnery P., Vaz F. M., Munnich A., Elpeleg O., Delahodde A., de Keyzer Y., de Lonlay P. (2010) LPIN1 gene mutations: a major cause of severe rhabdomyolysis in early childhood. Hum. Mutat. 31, E1564–E1573 [DOI] [PubMed] [Google Scholar]
- 34.Zhang P., Verity M. A., Reue K. (2014) Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab. 20, 267–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuma A., Mizushima N. (2008) Chromosomal mapping of the GFP-LC3 transgene in GFP-LC3 mice. Autophagy 4, 61–62 [DOI] [PubMed] [Google Scholar]
- 37.Grumati P., Coletto L., Sabatelli P., Cescon M., Angelin A., Bertaggia E., Blaauw B., Urciuolo A., Tiepolo T., Merlini L., Maraldi N. M., Bernardi P., Sandri M., Bonaldo P. (2010) Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat. Med. 16, 1313–1320 [DOI] [PubMed] [Google Scholar]
- 38.Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brooks S. V., Faulkner J. A. (1988) Contractile properties of skeletal muscles from young, adult and aged mice. J. Physiol. 404, 71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding W. X., Ni H. M., Li M., Liao Y., Chen X., Stolz D. B., Dorn G. W., II, Yin X. M. (2010) Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J. Biol. Chem. 285, 27879–27890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Cazarin M. L., Snider N. N., Andrade F. H. (2011) Mitochondrial isolation from skeletal muscle. J. Vis. Exp. 49:2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nemazanyy I., Blaauw B., Paolini C., Caillaud C., Protasi F., Mueller A., Proikas-Cezanne T., Russell R. C., Guan K. L., Nishino I., Sandri M., Pende M., Panasyuk G. (2013) Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease. EMBO Mol. Med. 5, 870–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanida I., Ueno T., Kominami E. (2008) LC3 and autophagy. Methods Mol. Biol. 445, 77–88 [DOI] [PubMed] [Google Scholar]
- 44.Komatsu M., Ichimura Y. (2010) Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 584, 1374–1378 [DOI] [PubMed] [Google Scholar]
- 45.Ray R., Chen G., Vande Velde C., Cizeau J., Park J. H., Reed J. C., Gietz R. D., Greenberg A. H. (2000) BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J. Biol. Chem. 275, 1439–1448 [DOI] [PubMed] [Google Scholar]
- 46.Shi R. Y., Zhu S. H., Li V., Gibson S. B., Xu X. S., Kong J. M. (2014) BNIP3 interacting with LC3 triggers excessive mitophagy in delayed neuronal death in stroke. CNS Neurosci. Ther. 20, 1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mizushima N., Yoshimori T. (2007) How to interpret LC3 immunoblotting. Autophagy 3, 542–545 [DOI] [PubMed] [Google Scholar]
- 48.Salvioli S., Ardizzoni A., Franceschi C., Cossarizza A. (1997) JC-1, but not DiOC6(3) or rhodamine 123, is a reliable fluorescent probe to assess delta psi changes in intact cells: implications for studies on mitochondrial functionality during apoptosis. FEBS Lett. 411, 77–82 [DOI] [PubMed] [Google Scholar]
- 49.Green D. R., Reed J. C. (1998) Mitochondria and apoptosis. Science 281, 1309–1312 [DOI] [PubMed] [Google Scholar]
- 50.Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P., Ma Q., Zhu C., Wang R., Qi W., Huang L., Xue P., Li B., Wang X., Jin H., Wang J., Yang F., Liu P., Zhu Y., Sui S., Chen Q. (2012) Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14, 177–185 [DOI] [PubMed] [Google Scholar]
- 51.Orvedahl A., Sumpter R., Jr., Xiao G., Ng A., Zou Z., Tang Y., Narimatsu M., Gilpin C., Sun Q., Roth M., Forst C. V., Wrana J. L., Zhang Y. E., Luby-Phelps K., Xavier R. J., Xie Y., Levine B. (2011) Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chu C. T., Ji J., Dagda R. K., Jiang J. F., Tyurina Y. Y., Kapralov A. A., Tyurin V. A., Yanamala N., Shrivastava I. H., Mohammadyani D., Wang K. Z. Q., Zhu J., Klein-Seetharaman J., Balasubramanian K., Amoscato A. A., Borisenko G., Huang Z., Gusdon A. M., Cheikhi A., Steer E. K., Wang R., Baty C., Watkins S., Bahar I., Bayir H., Kagan V. E. (2013) Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 15, 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lokireddy S., Wijesoma I. W., Teng S., Bonala S., Gluckman P. D., McFarlane C., Sharma M., Kambadur R. (2012) The ubiquitin ligase Mul1 induces mitophagy in skeletal muscle in response to muscle-wasting stimuli. Cell Metab. 16, 613–624 [DOI] [PubMed] [Google Scholar]
- 54.Bergounioux J., Brassier A., Rambaud C., Bustarret O., Michot C., Hubert L., Arnoux J. B., Laquerriere A., Bekri S., Galene-Gromez S., Bonnet D., Hubert P., de Lonlay P. (2012) Fatal rhabdomyolysis in 2 children with LPIN1 mutations. J. Pediatr. 160, 1052–1054 [DOI] [PubMed] [Google Scholar]
- 55.Zeharia A., Shaag A., Houtkooper R. H., Hindi T., de Lonlay P., Erez G., Hubert L., Saada A., de Keyzer Y., Eshel G., Vaz F. M., Pines O., Elpeleg O. (2008) Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am. J. Hum. Genet. 83, 489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y., Pessin J. E. (2013) Mechanisms for fiber-type specificity of skeletal muscle atrophy. Curr. Opin. Clin. Nutr. Metab. Care 16, 243–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamada E., Bastie C. C., Koga H., Wang Y., Cuervo A. M., Pessin J. E. (2012) Mouse skeletal muscle fiber-type-specific macroautophagy and muscle wasting are regulated by a Fyn/STAT3/Vps34 signaling pathway. Cell Reports 1, 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]