Abstract
Stable mutualism between a host and its resident bacteria requires a moderated immune response to control bacterial population size without eliciting excessive inflammation that could harm both partners. Little is known about the specific molecular mechanisms utilized by bacterial mutualists to temper their hosts’ responses and protect themselves from aggressive immune attack. Using a gnotobiotic larval zebrafish model, we identified an Aeromonas secreted immunomodulatory protein, AimA. AimA is required during colonization to prevent intestinal inflammation that simultaneously compromises both bacterial and host survival. Administration of exogenous AimA prevents excessive intestinal neutrophil accumulation and protects against septic shock in models of both bacterially and chemically induced intestinal inflammation. We determined the molecular structure of AimA, which revealed two related calycin-like domains with structural similarity to the mammalian immune modulatory protein, lipocalin-2. As a secreted bacterial protein required by both partners for optimal fitness, AimA is an exemplar bacterial mutualism factor.
Research organism: Zebrafish
eLife digest
Animals, including humans, harbor vast numbers of bacteria inside our digestive tracts. But rather than wage constant war, we have learned to coexist peacefully, and many of these bacteria are important to keep us healthy. Our immune system controls the number of bacteria, but in some diseases, this balance fails, and immune cells called neutrophils start a defense response. However, such attacks also cause inflammation in our guts, which can damage the tissues and organs.
Understanding the delicate balance between immune cells and individual bacteria in humans remains a challenge. Our gut bacteria live in complex communities with many species of microorganisms. Using animals like zebrafish can help to find out if gut bacteria are able to prevent such inflammation. These fish can grow under sterile conditions in the laboratory, allowing the study of added individual bacteria and the molecules they release. They are also transparent, making it easy to capture images of their organs under the microscope. In the wild, they live side-by-side with a type of bacteria called Aeromonas.
Here, Rolig et al. added Aeromonas bacteria to sterile zebrafish and observed how they interacted with the fish immune system. This revealed that the bacteria produce a protein to pacify immune cells, named 'Aeromonas immune modulator' (AimA). The sequence of this new protein did not look like any known molecules, but its 3D structure resembled a protein found in animals called lipocalin. AimA dampened inflammation in the gut.
When Aeromonas without AimA colonized the zebrafish, their neutrophils went to war. Their guts became inflamed and the bacteria started to die. But when the zebrafish had no neutrophils in their gut, nothing could stop the AimA-deficient bacteria and they grew too much. They produced toxic products, triggering septic shock and killing the fish. Adding AimA back into the fish stopped the inflammation and prevented septic shock, restoring balance. Both partners need AimA to survive. The bacteria need it to shield them from the immune response and the fish need it for protection against toxic products made by the bacteria.
Understanding proteins like AimA could help us to control bacteria and gut inflammation. Further work could reveal ways to use similar molecules to treat inflammation or to boost the growth of friendly bacteria.
Introduction
The vertebrate gastrointestinal tract harbors the highest density of microbes recorded for any microbial habitat (Whitman et al., 1998). These microbial communities are not only tolerated by their hosts, but are also required for normal host development and physiology, highlighting the intimacy of these mutualisms (McFall-Ngai et al., 2013). How these host-microbe mutualisms are established and maintained is unresolved. Understanding these processes will require identifying the molecular mechanisms by which resident microbes modulate the host immune system and defining the benefit these microbes derive from investing in such interactions. This knowledge is critical to dissect the microbial etiology of a range of diseases associated with excessive inflammation, including inflammatory bowel disease (IBD), diabetes, metabolic syndrome, and sepsis (Huttenhower et al., 2014; Turnbaugh et al., 2006; Wen et al., 2008). Furthermore, a thorough understanding of these host-microbe mutualisms will accelerate the development of effective treatments for these inflammatory diseases. Here, we explore the host-bacterial mutualism between zebrafish and their prominent colonizer, Aeromonas, and identify a secreted bacterial protein that promotes both bacterial colonization and host survival by preventing excessive inflammation.
Pathogenic bacterial species are often defined by unique virulence factors that enable host colonization and determine the severity of host disease symptoms. Much less well known are the unique factors produced by anti-inflammatory bacterial species, which likely play a key role in establishing host-bacterial mutualism and maintaining intestinal homeostasis. One such factor is the zwitterionic polysaccharide, PSA, produced by the human symbiont Bacteroidetes fragilis, that induces a toleragenic T cell profile and protects against intestinal inflammation (Mazmanian et al., 2008; Mazmanian et al., 2005; Round and Mazmanian, 2010). The anti-inflammatory properties of Lactobacillus rhamnosus have been attributed to secreted proteins p75 and p40 (Yan et al., 2007), and Faecalibacterium prausnitzii secretes an anti-inflammatory 15 kDa protein called microbial anti-inflammatory molecule or MAM (Yan et al., 2007; Quévrain et al., 2016; Sokol et al., 2008; Carlsson et al., 2013; Martín et al., 2014). These secreted proteins reduce inflammation in mouse models of colitis (Yan et al., 2011), which suggests their therapeutic potential, but we do not yet understand how these proteins aid the bacteria that produce them. Investigating the bacterial fitness benefits for producing anti-inflammatory activities is critical for developing approaches to promote membership of these immunoregulatory bacterial species in chronic inflammatory diseases like IBD and to use bacterial immune modulators as therapeutics to shape microbiota composition.
The investigation of microbiota-derived immunomodulatory factors is challenging because their effects on the host are subtler then those of disease-causing toxins and they are produced by complex and genetically intractable microbial consortia. We used the zebrafish, Danio rerio, as a high-throughput, gnotobiotic model vertebrate system to identify individual bacterial products with immunomodulatory properties and understand how those products promote host-bacterial mutualism. Zebrafish have long been used as a vertebrate model for developmental biology because of their optical transparency, high fecundity, and rapid early development (Grunwald and Eisen, 2002). Zebrafish embryos develop ex-utero within their protective chorions and first encounter environmental microbes when they hatch as larvae between 2 and 3 days post fertilization (dpf). By 5 dpf the animals have a fully functional digestive tract that is colonized with bacteria (Bates et al., 2006; Stephens et al., 2016; Wallace et al., 2005). The larvae are also equipped with a functional innate immune system with highly dynamic neutrophils and macrophages that serve as the primary cellular defenses before the emergence of adaptive immune cells at approximately 3 weeks of age (Renshaw and Trede, 2012). The zebrafish is an excellent model for studying the establishment of host-bacterial symbioses (Burns and Guillemin, 2017) because hundreds of zebrafish can be easily derived and maintained in a gnotobiotic state with defined microbial isolates during larval development (Melancon et al., 2017). The large population sizes of animals allow for the analysis of the subtle effects of resident microbes on host phenotypes. For example, we have shown that intestinal neutrophil populations in larval zebrafish range from an average of 3 cells in germ free (GF) animals to 7 cells in conventionally reared (CV) animals, a difference that can be readily detected in well-powered experiments despite extensive inter-host variation (Rolig et al., 2015). Our use of gnotobiology to assay the effect of individual microbes and their products on the host has been aided by the fact that many zebrafish bacterial isolates are culturable, have known genome sequences (Stephens et al., 2016), and are genetically tractable (Hill et al., 2016; Wiles et al., 2016). This gnotobiotic approach, however, fails to capture the complexity of naturally occurring vertebrate microbiota. Additionally, in the zebrafish we are largely limited to using gnotobiology to study the larval period because we have yet to develop adequate nutritional and husbandry approaches to promote the growth of GF zebrafish to adulthood.
In this study we focused on the mutualism established between larval zebrafish and their resident Aeromonas species. Aeromonas is an important symbiont of the zebrafish and the sole bacterial genus that we found present in 100% of individuals at all developmental time points in a comprehensive longitudinal study of zebrafish gut microbiomes across their lifespan (Stephens et al., 2016). Furthermore, we have shown that in mono-associations, representatives of the Aeromonas genus can fully rescue defects in glycan expression (Bates et al., 2006), epithelial cell proliferation (Cheesman et al., 2011), innate immune response (Rolig et al., 2015), and pancreatic beta cell expansion (Hill et al., 2016) that are characteristic phenotypes of GF fish. We have examined the dynamics of the Aeromonas-zebrafish mutualism, taking advantage of the optical transparency of the larvae to monitor both bacterial and host cells in vivo (Taormina et al., 2012; Jemielita et al., 2014). Here we specifically quantified the innate immune response against Aeromonas bacterial genetic variants, using GFP-expressing intestinal neutrophils as a metric of the host response (Renshaw et al., 2006). Neutrophils are a primary component of the initial inflammatory response and are responsive to both infecting pathogens and resident intestinal microbes (Harvie and Huttenlocher, 2015; Bates et al., 2007; Kanther et al., 2014). This responsiveness makes the intestinal neutrophil population an ideal readout of the host-microbe mutualism between zebrafish and Aeromonas.
Using the gnotobiotic zebrafish model, we identified a potent immune modulatory protein secreted by Aeromonas, which we named Aeromonas immune modulator A (AimA) because it reduces intestinal inflammation. To understand the molecular mechanism of AimA’s function, we determined the protein crystal structure, which revealed two related calycin-like domains that are structurally similar to the immune modulatory factor found in mammals, lipocalin-2 (LCN2). Consistent with AimA targeting the host immune response, as opposed to counteracting a specific bacterial product, we showed that purified AimA, and each of its subdomains, prevents chemically induced intestinal inflammation. Furthermore, we found that this activity is blocked by exogenous mouse LCN2 (mLCN2), implying a shared mechanism of action between these structurally similar bacterial and mammalian proteins. However, AimA does not appear to function like LCN2 as a binding protein for bacterial siderophores. To understand AimA’s function for Aeromonas, we generated mutants lacking aimA and its homologue aimB. In mono-associations with aim mutants, we found that both the host and bacterial partners suffered reduced viability, which could be rescued with exogenous AimA protein. We showed that the fitness cost to the bacteria was due to excessive inflammation because the aim mutant survival deficit was erased in an immunocompromised host. In parallel, we showed AimA’s benefit to the host extended to protection from lipopolysaccharide (LPS) intoxication. Together, these data provide an exemplar of a bacteria immune modulatory factor that is beneficial for both partners in a bacterial mutualism.
Results
Aeromonas secretes a protein that modulates the intestinal neutrophil response
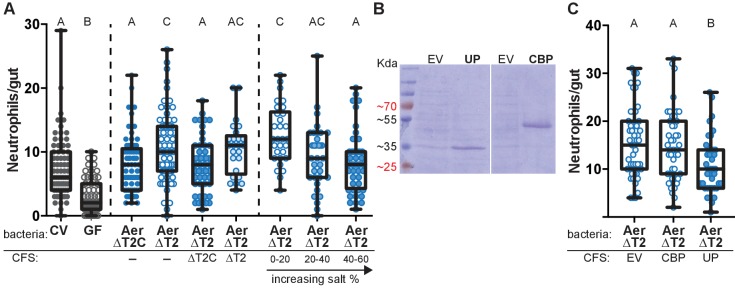
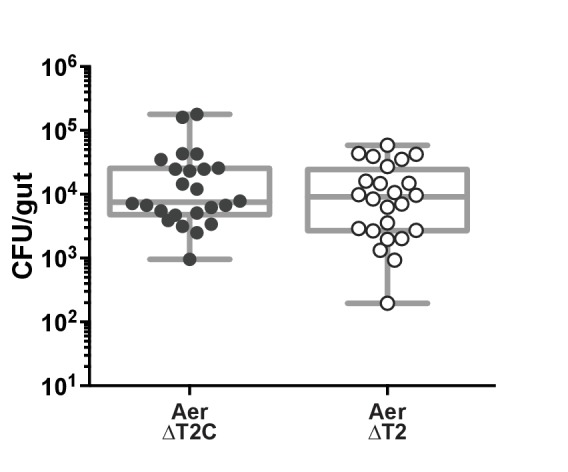
Based on the mutualism between Aeromonas and zebrafish (Stephens et al., 2016), we hypothesized that Aeromonas would use specific secreted factors to co-exist with its host. We tested this hypothesis by comparing the zebrafish intestinal neutrophil response to mono-association with an Aeromonas isolate, A. veronii strain Hm21 (Maltz and Graf, 2011), versus mono-association with an isogenic mutant lacking the type two secretion system (T2SS)(ΔT2)(Table 1), a major system for secretion in Gram-negative bacteria (Maltz and Graf, 2011). We colonized each bacterial strain in zebrafish from 4 dpf to 6 dpf and measured the number of intestinal neutrophils that accumulated in response to each colonizing bacterial strain. In these experiments we compared the ΔT2 strain to a genetically complemented strain, ΔT2C, previously shown to restore T2SS function (Maltz and Graf, 2011), which allowed us to rule out the possibility that phenotypes associated with the ΔT2 strain were due to polar or second site mutations. The ΔT2C and ΔT2 colonized the zebrafish intestine to similar levels in mono-associations (Figure 1—figure supplement 1), however the ΔT2 mutant induced a greater intestinal neutrophil response than the ΔT2C (Figure 1A). This result suggests that A. veronii strain Hm21 secretes a product that decreases the neutrophil response to its colonization. The increased neutrophil response to ΔT2 was rescued by concurrent treatment with cell-free supernatant (CFS) collected from the ΔT2C strain, but not CFS collected from ΔT2 (Figure 1A). To identify the factor produced by ΔT2C and not from the ΔT2 strain that resulted in fewer intestinal neutrophils, we performed mass-spectrophotometry on the CFS from these two strains and determined which proteins were enriched in the ΔT2C compared to the ΔT2 strain. This analysis resulted in a list of 22 proteins that were enriched by greater than 10 counts in the ΔT2C CFS (Supplemental file 1). We narrowed down the potential candidates further by performing ammonium sulfate fractionation on the ΔT2C CFS and adding fractions to fish mono-associated with ΔT2. We found neutrophil modulating activity primarily in the 40–60% salt fraction, but also saw some activity in the 20–40% salt fraction (Figure 1A). An SDS PAGE gel of these fractions revealed a similar banding pattern for each fraction, except the bands in the 20–40% fraction were considerably weaker. We observed two prominent bands at 33 kDa and 55 kDa. These sizes corresponded to two proteins on our list of proteins enriched in the ΔT2C CFS, an uncharacterized protein (UP) and a chitin binding protein (CBP), respectively (Supplemental file 1). To test whether either of these proteins modulated the neutrophil response to ΔT2 colonization, we cloned the sequence for each gene into an overexpression vector and induced expression in Escherichia coli, resulting in E. coli CFS that was heavily dominated by the protein of interest (Figure 1B). We treated zebrafish mono-associated with ΔT2 with the CFS resulting from overexpression of each protein of interest and found neutrophil modulating activity in the E. coli CFS with the 33 kDa UP and not with CBP or control CFS from E. coli containing an empty expression vector (Figure 1C). We named this neutrophil-modulating protein Aeromonas immune modulator A (AimA).
Table 1. Strain table.
| Strain | Characteristics | Ref. or source |
Manuscript abbreviation |
|---|---|---|---|
| Hm21S | Parent strain, SmR | Graf, 1999 | Aer |
| HE-1095 | Hm21S::interrupted exeM mTn5 KmR SmR | Maltz and Graf, 2011 | Aer ΔT2 |
| HEC-1344 | HE-1095::Tn7 containing TpR
exeMN + promoter region |
Maltz and Graf, 2011 | Aer ΔT2C |
| ASRC7 | Hm21S aimA::cmR | This study | Aer ΔaimA |
| ASRD5 | Hm21S ΔaimB | This study | Aer ΔaimB |
| ASRD4 | Hm21S aimA::cmR; ΔaimB | This study | Aer ΔAΔB |
| ZOR0001 | Zebrafish Aeromonas isolate | Stephens | ZF Aer |
| ASRC9 | ZOR0001 aimA::cmR | This study | ZF Aer ΔaimA |
Figure 1. Aeromonas secretes a protein that regulates intestinal neutrophil response to a general model of inflammation.
(A) Intestinal neutrophil response to conventional (CV), germ free (GF), and mono-associations of wild-type Aeromonas (Aer ΔT2C), and an isogenic mutant of the type II secretion system (ΔT2). ΔT2 induces a greater neutrophil response that is rescued by addition of cell free supernatant (CFS) from the wild-type Aeromonas strain. Ammonium sulfate fractionation narrowed potential candidates down to two proteins, an unidentified protein (UP) or chitin binding protein (CBP). (B) E. coli BL21 carrying pET21b expression vector overexpression of the candidate proteins of interest. (C) Addition of CFS containing UP rescued the increased neutrophil response induced by ΔT2. EV: empty vector control CFS. Letters denote p<0.05, ANOVA followed by Tukey’s post hoc test. Each box represents the first to third quartiles, center bar the median, and whiskers the maximum and minimum. Each dot represents one fish; data collected from at least two independent experiments; n ≥ 24.
Figure 1—figure supplement 1. Colonization of A. veronii strain Hm21 type two secretion mutant.
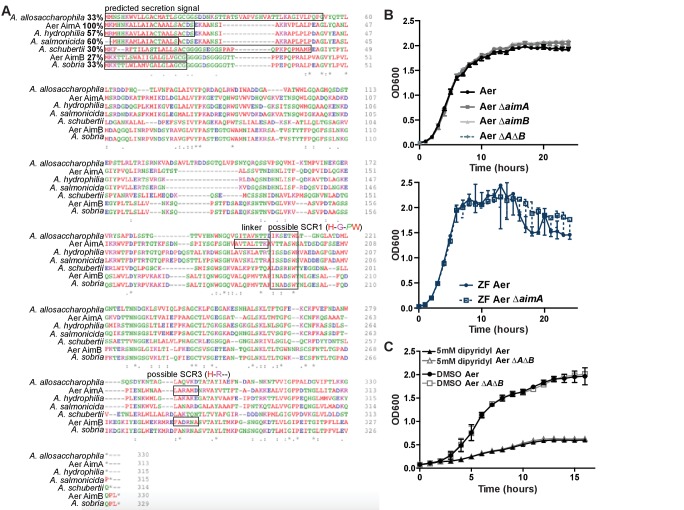
The AimA protein has two distinct calycin domains with related folds
DNA and amino acid homology searches revealed that no homologues to AimA exist outside of the Aeromonas genera. Further, analysis of the amino acid sequence of AimA with the N terminal amino acid secretion signal removed revealed a lack of sequence identity with known domains or structurally characterized proteins and therefore offered little insight into the structure or function of AimA. Thus, to gain insight into the mechanism of AimA, we constructed a His-tagged version that was purified from E. coli, and we crystallized the protein. We determined the molecular structure of AimA using a heavy atom derivative and Single-wavelength Anomalous Dispersion (SAD) phasing to a resolution of 2.3 Å (PDB 6B7L; Table 2).
Table 2. Data Collection and Refinement Statistics for Deposited Model.
| Data collection | Iodide | Native | ||||
|---|---|---|---|---|---|---|
| space group | P622 | P622 | ||||
| unit cell a, b, c (Å) | 161.4 | 161.4 | 66.6 | 160.5 | 160.5 | 66.2 |
| alpha, beta, gamma (degrees) | 90 | 90 | 120 | 90 | 90 | 120 |
| resolution (Å) | 50.0–2.7 (2.75–2.70) | 41.1–2.30 | ||||
| completeness (%) | 100 (100) | 100 (100) | ||||
| no. unique reflections | 14772 (711) | 22818 (3244) | ||||
| multiplicity | 37.7 (38.1) | 70.2 (71.6) | ||||
| I/sigma> | 44.2 (3.5) | 16.1 (1.4) | ||||
| CC1/2 | 1 | 0.861 | 1.0 (0.9) | |||
| CC1/2 anomolous | 1 | 0.873 | ||||
| R merge (%) | 19.6 (486) | 19.1 (574) | ||||
| Refinement | ||||||
| R work (%) | 17.2 | |||||
| R free (%) | 20.4 | |||||
| no. of molecules in the asymetric unit | 1 | |||||
| no. protein residues | 290 | |||||
| no. of waters | 69 | |||||
| rmsd for lengths (Å) | 0.008 | |||||
| rmsd for angles (deg) | 1.1 | |||||
| Ramachandran plot (%) | ||||||
| preferred | 96.2 | |||||
| allowed | 3.4 | |||||
| outliers | 0.4 | |||||
| Avg. B factor (Å^2) | ||||||
| mainchain* | 81 | |||||
| waters | 75 | |||||
| new PDB entry | 6B7L |
* Several loop regions display high mobility but have been modeled due to a visible chain path in the electron density, resulting in an increase in the average observed B-factors of the main chain.
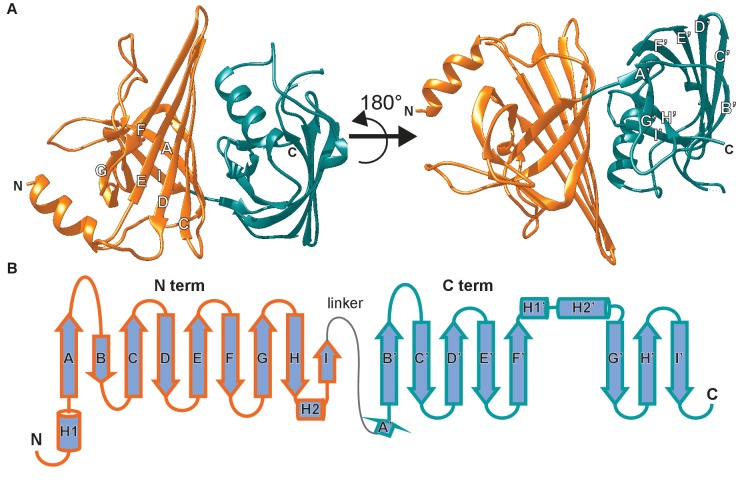
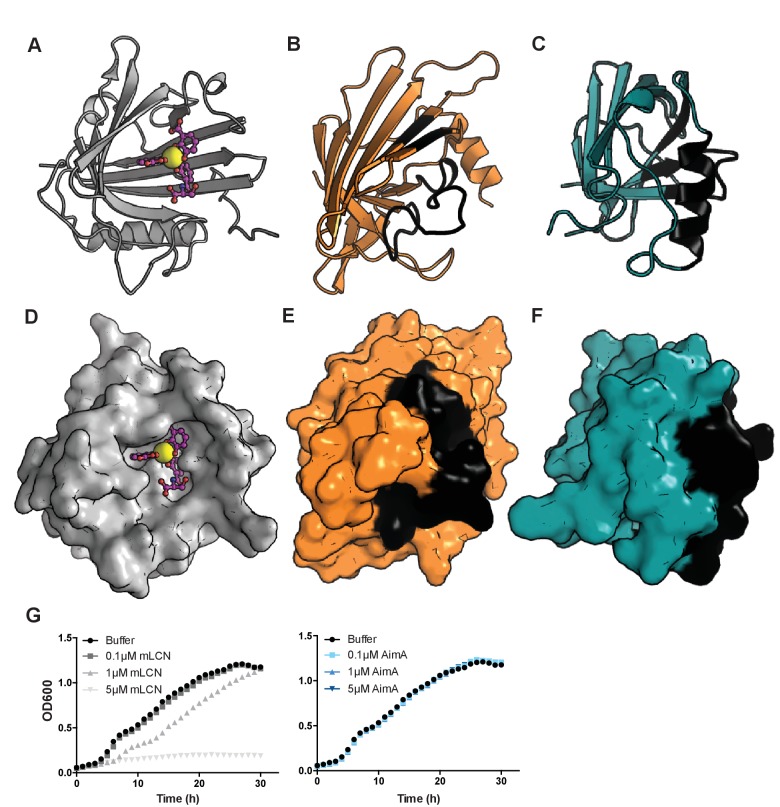
The structure of AimA revealed two domains connected by a short linker (Figure 2A,B). β-strands dominate each domain with the carboxy terminal (C-term) domain forming a complete β-barrel and the amino terminal (N-term) domain containing a curved β-sheet. Structural homology searches of full length AimA against all Protein Data Bank-deposited structures using PDBeFold resulted in structures that aligned to only one domain or the other, with the majority aligning with the C-term domain (Krissinel et al., 2004; Berman et al., 2000). Therefore, we performed structural homology searches against each domain separately, which revealed that both domains had similarity to proteins in the calycin superfamily (Supplemental file 2 and Figure 2—figure supplement 1).
Figure 2. The crystal structure of AimA reveals two calycin domains.
(A) The 2.3 Å structure of AimA displays two calycin domains (PDB ID 6B7L). (B) The amino terminal domain (orange) of AimA is connected by a short linker to the carboxy terminal domain (teal) and both contain an eight-stranded full (C-term) or partial (N-term) β-barrel.
Figure 2—figure supplement 1. Structural homology of the two domains of AimA to avidins.
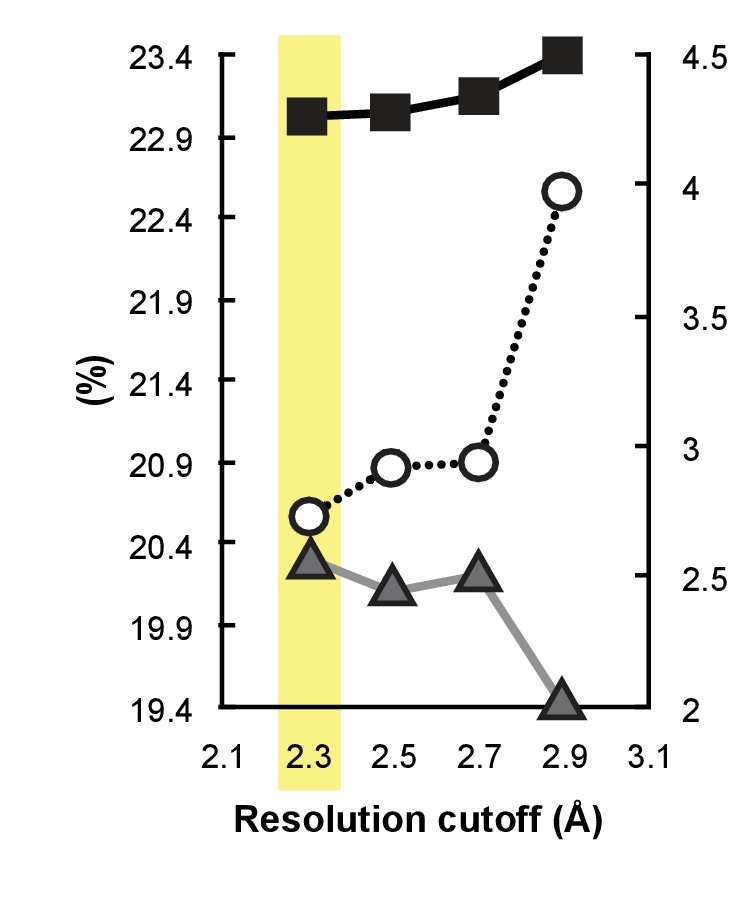
Figure 2—figure supplement 2. A paired refinement demonstrates that the extended resolution improves the quality of the model.
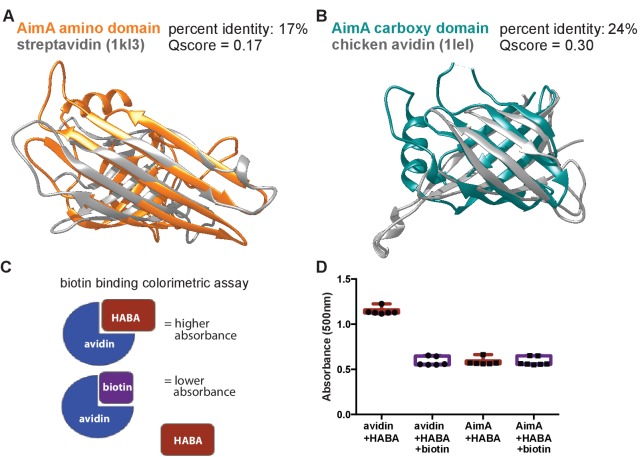
The calycin superfamily of proteins contains members from all domains of life and includes lipocalins, fatty acid binding proteins, and avidins. This superfamily is defined by an anti-parallel β-barrel with a repeated +1 topology (Flower, 1996). Notably, the calycin superfamily is known for structural conservation without high amino acid sequence conservation (Flower, 1996; Lakshmi et al., 2015), consistent with a lack of sequence homology between AimA and other calycin family members. The N-term domain of AimA has an incomplete β-barrel, but maintains some structural homology to streptavidin. Streptavidin binds biotin tightly, but we found no evidence that AimA binds biotin (Figure 2—figure supplement 1). The C-term domain of AimA has structural homology both to avidins and lipocalins (Supplemental file 2).
In a model of intestinal inflammation, AimA reduces neutrophil influx
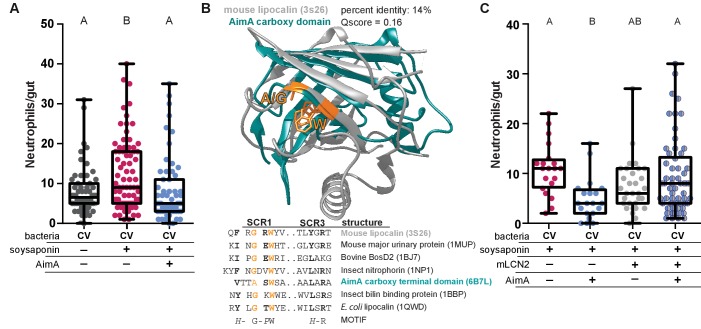
AimA’s structural similarity to lipocalin proteins was intriguing because of these proteins’ known roles in modulating neutrophil behavior (Moschen et al., 2017). Based on this structural similarity, we wondered whether AimA’s influence on intestinal neutrophil numbers was specific to Aeromonas colonization or whether it acts as a more general immunoregulatory molecule to influence neutrophil behavior. To test whether AimA modulates neutrophil numbers in response to stimuli other than Aeromonas mono-association, we employed the zebrafish model of soysaponin-induced inflammation. Farmed fish, such as salmon and carp, fed soybean meal as a protein source are well known to develop intestinal inflammation, and zebrafish are a good model of this irritation (Hedrera et al., 2013). We fed conventionally raised zebrafish larvae Zeiglers fish food with 0.3% soysaponin from 4 dpf to 6 dpf and saw a significant increase in the number of intestinal neutrophils in response to the soysaponin, as expected (Figure 3A). When we treated the fish with 100 ng/mL purified AimA concurrently with soysaponin, AimA prevented the increase in neutrophil influx in response to soysaponin (Figure 3A). This result demonstrates that AimA alone can inhibit pro-inflammatory signaling pathways elicited by stimuli other than Aeromonas.
Figure 3. AimA contains regions structurally conserved with the lipocalin family and reduces soysaponin induced inflammation.
(A) Feeding zebrafish soy saponin induces increased intestinal neutrophil response, and treating those fish with 100 ng/mL purified AimA prevents the increased intestinal neutrophil response. Letters indicate significantly different groups; ANOVA with multiple comparisons. Each dot represents one fish; data collected from at least two independent experiments; n ≥ 24. (B) Structural overlay of mouse lipocalin (PDB ID 3S26) and AimA C-term domain using PDBeFold. Qscore is a structural overlay quality score that takes into account both the root mean standard deviation (RMSD) of the Cα carbons and the alignment length. Qscore of 1 is perfect alignment, 0 is no alignment. The residues highlighted in orange are conserved in both the sequences and the structures (see table below). Displayed are the sequences of Structurally Conserved Regions (SCR) 1 and 3 of a representative set of kernel and outlier lipocalins, with PDB IDs in parenthesis. The C-term domain of AimA is included and contains a subset of conserved SCR residues. Bold residues are conserved across the sequences. Orange residues are conserved across the sequences and in the structures of mLCN and the C-term domain of AimA. (C) Treatment of conventionally raised (CV) fish with soysaponin and lipocalin prevents AimA from reducing the neutrophil response. Each dot represents one fish; n ≥ 20 from at least three independent experiments. Letters indicate significantly different groups; ANOVA with multiple comparisons.
Figure 3—figure supplement 1. AimA does not bind enterobactin.
Presence of mouse lipocalin-2 inhibits AimA function
Because AimA reduces neutrophil influx in a model of intestinal inflammation and the C-term domain has structural homology to lipocalin proteins (Figure 3A, Supplementary file 2), we hypothesized that the neutrophil reducing capacity of AimA was related to its lipocalin-like structure. Lipocalins are defined by three key structurally conserved regions, SCR1, SCR2, and SCR3; of these, SCR1 contains four key residues and is the most conserved (Figure 3B) (Flower, 1996; Lakshmi et al., 2015). The SCR1 in the C-term domain of AimA overlaps structurally with the SCR1 of mouse lipocalin-2 (mLCN2) and shares some related amino acids (Figure 3B). Given this structural similarity, we hypothesized that AimA may function in the same pathways as mLCN2. Thus, we asked whether the presence of mLCN2 protein would alter the capacity of AimA to reduce the neutrophil response to soysaponin. We found that the addition of mLCN2 concurrently with soysaponin did not influence the neutrophil response to soysaponin (Figure 3C); however, addition of mLCN2 in conjunction with AimA blocked the protective effect of AimA against soysaponin-induced inflammation. This result suggests that mLCN2 may interfere with AimA’s activity by direct binding, competing for the same host receptor, or acting in a competing pathway.
LCN2’s immunomodulatory activity is mediated, at least in part, by binding iron-sequestering siderophores, including enterobactin from E. coli (Xiao et al., 2017), thus we explored whether AimA had a similar enterobactin-binding activity. The original evidence for LCN2’s enterobactin binding came from the protein crystal structure of human LCN2, in which enterobactin was co-crystalized (Goetz et al., 2002). We did not find enterobactin in the AimA structure, which was expected because the E. coli strain, BL21 DE3, which we used to express AimA, does not produce enterobactin. However, in comparison to the wide enterobactin-binding cleft (calyx) of LCN2, the homologous regions of the N- and C-term domains of AimA are much narrower, are occluded by loops or a helix, and lack the enterobactin-interacting residues of LCN (Figure 3—figure supplement 1A–F). To test whether AimA binds enterobactin, we grew E. coli K12 strain (MG1655) under iron limiting conditions that make it dependent on the siderophore function of its secreted enterobactin, which can be sequestered by exogenously added LCN2 (Goetz et al., 2002). As predicted, when we grew E. coli K12 in the presence of the iron chelator dipyridyl to limit available iron, we observed a dose dependent inhibition of E. coli growth upon addition of purified mouse LCN2 protein (Figure 3—figure supplement 1G). In contrast, when we added the same range of concentrations of purified AimA protein to the E. coli cultures, we saw no growth inhibition (Figure 3—figure supplement 1G), suggesting that AimA does not bind and sequester enterobactin in a similar manner to LCN2.
Each individual domain of AimA retains neutrophil regulating capacity
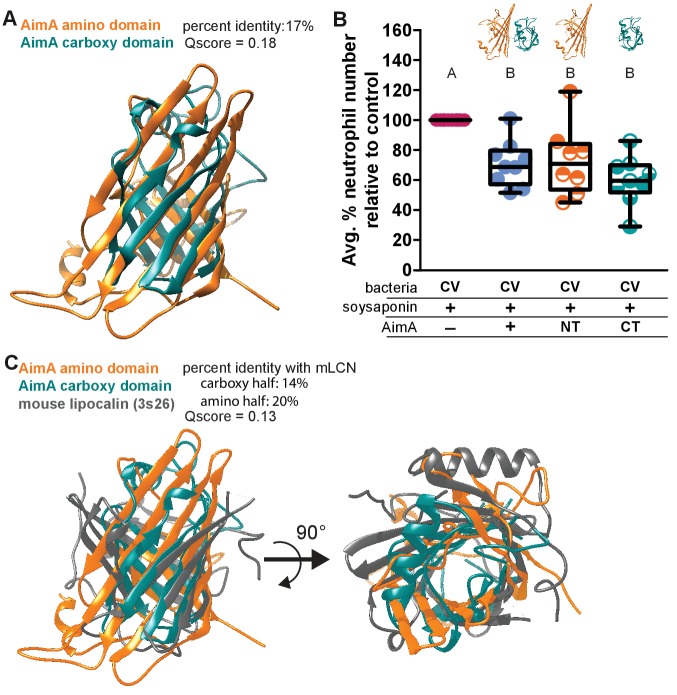
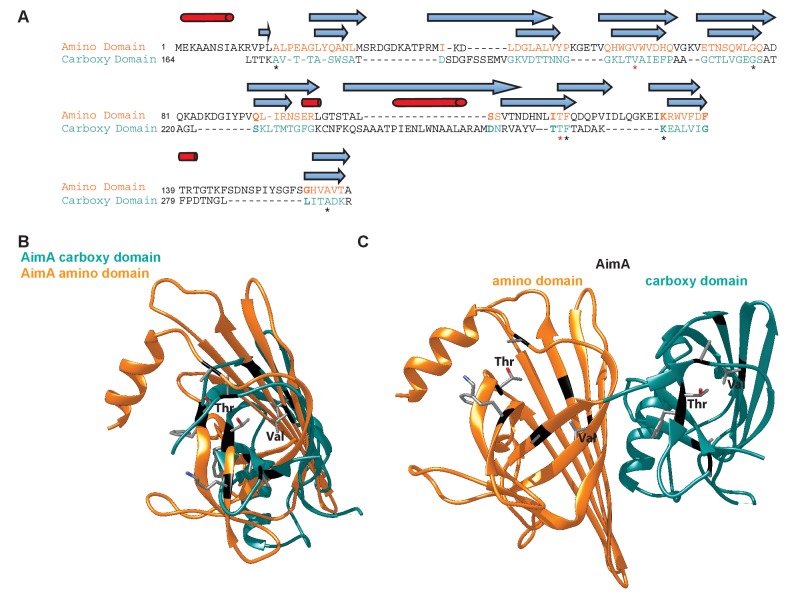
Human LCN2 exists both as a monomer and a homodimer, and while the functional distinction between the two forms is unknown, the homodimer is the major molecular form secreted by neutrophils (Cai et al., 2010). Both the C- and N-terminal domains of AimA have structural homology to proteins in the calycin superfamily. Although they share only 17% amino acid identity, a structural overlay between the two domains reveals some structural conservation (Figure 4A and Figure 4—figure supplement 1A). Because human LCN2 exists as both a monomer and homodimer and the two domains of AimA each have a lipocalin-like fold, we asked if each domain alone was sufficient to alter the intestinal neutrophil response. The individual domains were not as soluble as full-length AimA, but we were able to crudely purify each domain. Concurrently with soysaponin, we added approximately 100 ng/mL of the domain of interest to zebrafish from 4 to 6 dpf. We found that both the N- and C-term domains alone were sufficient to reduce the neutrophil response in the soysaponin model of intestinal inflammation (Figure 4B). An in depth analysis of the structural overlay between the N- and C-term domains revealed that seven out of eight β-strands in the barrels align well; however, only seven total residues are in analogous functional positions across the two domains (Figure 4—figure supplement 1). Of these seven residues, two—Val 60/201 and Thr 117/265—stand out as possible candidates to interact with a hydrophobic ligand that could bind inside the barrel cavity. The other five residues may overlap by coincidence or they may be positioned to interact with a promiscuous protein or ligand partner. Furthermore, given the overall structural similarity between the N- and C-term domains, it is possible that critical residues are in a flexible loop region that could become structured upon binding (Figure 4A and Figure 4—figure supplement 1A). Interestingly, both AimA domains have comparable structural similarity with mLCN2 (Figure 4C). While the mLCN2 binding cleft for enterobactin is not conserved in the domains of AimA, the majority of the β-strands in the barrels overlap in all three domains, which suggests these domains could interact with the same binding partner(s) (Figure 4C).
Figure 4. Both domains of AimA retain neutrophil modulating function.
(A) Overlay of C-term and N-term domains of AimA using PDBeFold. (B) Conventionally raised (CV) fish fed soysaponin (SS) and treated with either purified full-length AimA or purified N-term (NT) or C-term (CT). Each dot represents the average percent of neutrophil influx in a flask of 15 fish from the average neutrophil influx of a control flask (soysaponin only) of 15 fish. n ≥ 9 flasks from at least three independent experiments. Letters indicate significantly different groups; ANOVA with multiple comparisons. (C) Overlay of C- and N-term domains of AimA and mLCN using PDBeFold.
Figure 4—figure supplement 1. Structural comparison of N-term and C-term domains of AimA.
AimA controls host neutrophil response and promotes Aeromonas colonization
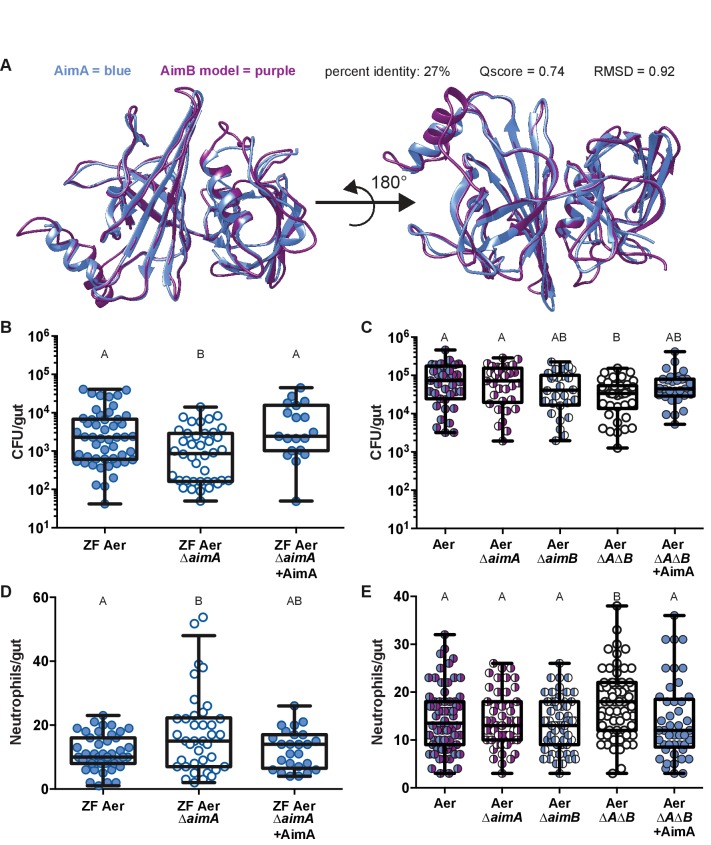
With a clear understanding that AimA controls the host intestinal neutrophil response, we next asked whether the activity of AimA also increases Aeromonas fitness, thus facilitating a mutualistic relationship with the host. We began by asking how prevalent AimA was across bacterial genomes. Knowing that proteins in the calycin superfamily have low sequence conservation, we were not surprised to find AimA homologues by sequence similarity only within the Aeromonas genus (Figure 5—figure supplement 1A). We found an almost identical homologue (99%) to AimA in a zebrafish intestinal Aeromonas isolate, ZOR0001, referred to here as ZF Aer (Table 1) (Stephens et al., 2016). Some Aeromonas species, including A. veronii strain Hm21 (Table 1), have both AimA and a second copy, which we named AimB. AimB is distantly related to AimA by amino acid sequence conservation (27%), yet a model generated by the Iterative Threading ASSEmbly Refinement (I-TASSER) program of the structure of AimB overlays directly on the structure of AimA (Figure 5A) (Yang et al., 2015; Roy et al., 2010; Zhang, 2008). A reexamination of our mass spectrometry analysis of the ΔT2C CFS uncovered peptides corresponding to AimB that were enriched in the sample from ΔT2C compared to the ΔT2 mutant, although the difference between the two samples was less than our original threshold (Supplemental file 1).
Figure 5. AimA reduces neutrophil influx and promotes colonization of Aeromonas.
(A) Structural overlay (PDBeFold) of AimA (blue) and the model of AimB (purple) generated by I-TASSER using AimA as a threading structure. (B) Colonization level of wild-type ZF Aeromonas and ZF Aer ΔAimA. The colonization defect of ZF Aer ΔAimA is rescued by treatment with 100 ng/mL purified AimA. (C) Colonization level of wild-type Aeromonas, ΔAimA, ΔAimB, and ΔAΔB. Each of the single mutants colonizes as well as wild type, while the double mutant has a significantly reduced colonization level. This phenotype is rescued by treatment with 100 ng/mL purified AimA. (D) Intestinal neutrophil response to ZF Aer and the colonization defect of ZF Aer ΔAimA. (E) Intestinal neutrophil response to wild-type Aeromonas, ΔAimA, ΔAimB, and ΔAΔB. Each of the single mutants induces a similar neutrophil response to wild type, while the double mutant induces significantly greater response. This phenotype is rescued by treatment with 100 ng/mL purified AimA for both Aeromonas isolates. For all graphs, each dot represents one fish; n ≥ 23 from at least three independent experiments. Letters indicate significantly different groups, ANOVA with multiple comparisons.
Figure 5—figure supplement 1. Homologues to AimA are found across the Aeromonas genus.
Figure 5—figure supplement 2. ZF Aer ΔAimA suffers the greatest colonization defect seven dpf.
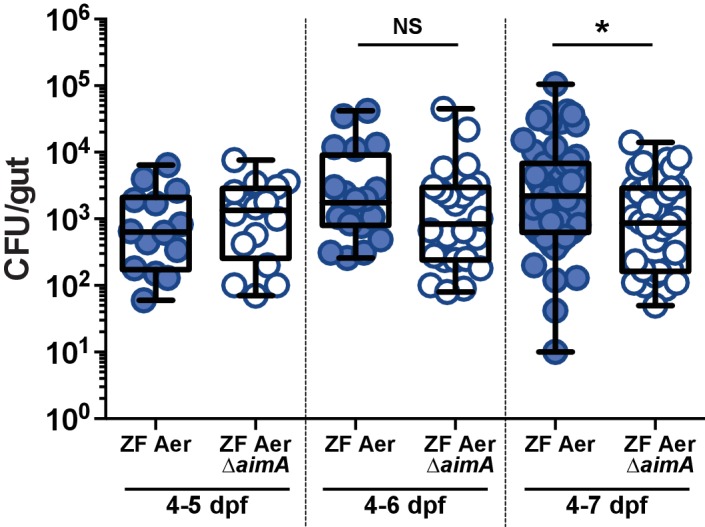
To determine whether the bioactive protein AimA and its homologue AimB benefit Aeromonas, we constructed deletion strains of each gene individually and of both genes in A. veronii strain Hm21 (Aer ΔaimA, Aer ΔaimB, and Aer ΔAΔB) and an AimA deletion in the ZF Aer background (ZF Aer ΔaimA). All of these mutants displayed normal growth in vitro in rich media (Figure 5—figure supplement 1B). Because AimA has structural homology to LCN2, which binds siderophores, and Aeromonas species make several siderophores, including enterobactin (Maltz et al., 2015), we asked whether AimA functions in iron acquisition for Aeromonas by testing the ability of wild type Hm21 and the Aer ΔAΔB mutant to grow in iron depleted media containing dipyridyl. We found that the two strains had identical growth curves when grown under iron starvation (Figure 5—figure supplement 1C), suggesting that AimA and AimB do not function in iron acquisition for Aeromonas.
We next tested whether the strains lacking aim genes exhibited growth defects in mono-associations in larval zebrafish. We found that when inoculated into GF larvae at 3 dpf, the ZF Aer ΔaimA mutant colonized gnotobiotic zebrafish to lower levels than its wild type counterpart (Figure 5B), with the colonization defect becoming apparent at 7 dpf (Figure 5—figure supplement 2). We observed a similar colonization defect at 7 dpf with the Aer ΔAΔB mutant relative to the wild-type strain (Figure 5C). However, the single gene deletion strains colonized to wild-type levels, suggesting that AimA and AimB provide redundant functions for Aeromonas growth within the larval intestine. We note that Aer ΔT2 did not exhibit a colonization defect (Figure 1—figure supplement 1) similar to the Aer ΔAΔB mutant, which could be due to other phenotypic consequences of its altered secretion profile.
We next tested whether colonization defects of the aim deficient strains could be rescued by supplementation of the flask media with purified AimA protein. Remarkably, addition of purified AimA protein at a concentration of 100 ng/mL to the flask water from 4 to 7 dpf completely restored the colonization efficiency of the aim deficient strains (Figure 5B,C).
Because purified AimA reduces the intestinal neutrophil response (Figure 3A,C), we hypothesized that the aim deletion strains would induce more inflammation, and thus upon mono-association could create a less hospitable intestinal environment for Aeromonas colonization. To test our hypothesis, we quantified intestinal neutrophil numbers in response to Δaim mono-associations at 7 dpf, corresponding to the time point when the bacterial colonization deficit for ZF Aer ΔaimA was apparent (Figure 5—figure supplement 2). We found that zebrafish mono-associated with ZF Aer ΔaimA exhibited significantly higher intestinal neutrophil counts as compared with those colonized with wild-type ZF Aer (Figure 5D), despite carrying a lower bacterial load (Figure 5B). Similarly, zebrafish colonized with the double mutant, Aer ΔAΔB, had significantly higher neutrophil counts than fish colonized with wild-type Aer (Figure 5E), despite being colonized at a lower level (Figure 5C). Notably, the Aer ΔaimA and Aer ΔaimB single mutants, which did not exhibit decreased colonization in mono-associations, also did not induce higher intestinal neutrophil numbers (Figure 5E). For both Aeromonas lineages, when exogenous AimA was added to the flasks of fish mono-associated with inflammation-inducing Δaim strains, the protein reduced neutrophil numbers back to normal levels (Figure 5D,E) and at the same time restored colonization levels (Figure 5B,C).
AimA promotes host survival of inflammatory challenge
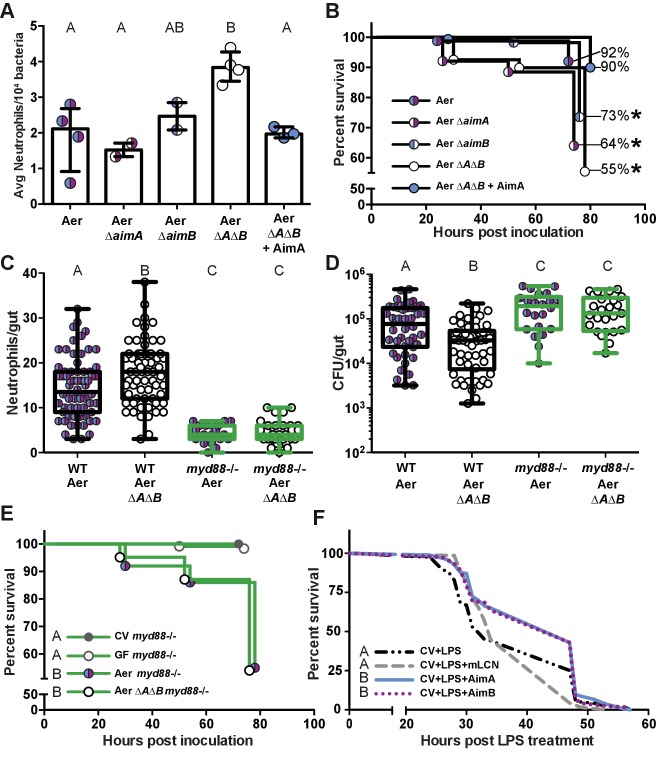
We previously showed that resident zebrafish bacterial species can vary dramatically in their per capita impact on host response to bacterial colonization (Rolig et al., 2015). By extrapolating the number of neutrophils elicited per 104 colonizing bacteria, we calculated that Aer ΔAΔB is much more immuno-stimulatory that the wild-type strain, recruiting nearly twice the number of neutrophils, on average, for the same number of bacteria (Figure 6A). This suggests that the Aim proteins function to reduce Aeromonas’ immune stimulation, allowing it to reach high colonization density without eliciting a strong inflammatory response.
Figure 6. The increased neutrophil response to Aeromonas ΔAΔB causes decreased survival rate.
(A) The per capita effect of wild-type Aeromonas, ΔAimA, ΔAimB, and ΔAΔB. Each dot represents the average neutrophil response from a flask of 15 fish divided by the average colonization level from a flask of 15 fish, normalized to 104. (B) Survival curve of zebrafish mono-associated with wild-type A. veronii (N = 163), ΔAimA (N = 139), ΔAimB (N = 174), ΔAΔB (N = 148), and ΔAΔB + AimA (N = 60). *indicates significant difference from the survival curve with wild-type Aeromonas, Mantel-Cox test. (C) myd88-/- transgenic fish lack a neutrophil response to bacteria. Each dot represents one fish. (D) ΔAΔB colonization is rescued to wild-type colonization levels in myd88-/- transgenic fish. Further, both wild-type Aeromonas and ΔAΔB reach significantly higher colonization levels in the myd88-/- transgenic fish compared to wild-type fish, indicating that the innate immune response limits commensal bacterial growth. (E) Survival curves of myd88-/- zebrafish over a 3 day infection. GF myd88-/- (N = 45), CV myd88-/- (N = 120), Aer myd88-/- (N = 100), ΔAΔB myd88-/- (N = 124). (F) Survival curve of conventionally raised (CV) fish treated with LPS (N = 163), with LPS and AimA (N = 163), with LPS and AimB (N = 121), or with LPS and mLCN (N = 69). Letters next to key indicate significant difference between the survival curves, Mantel-Cox test. For all graphs, letters indicate significance by ANOVA with multiple comparisons.
Knowing that loss of the Aim proteins resulted in both a significantly increased per capita effect and intestinal neutrophil response, we explored whether the increased neutrophil response led to other downstream consequences for the host by monitoring the survival rate of mono-associated fish. By 72 hr post-inoculation, the survival rate of zebrafish colonized with wild-type Aer was 92% (n = 163). We observed a significant decline in the survival of fish mono-associated with Aeromonas lacking Aim genes [Aer ΔaimA (n = 139), Aer ΔaimB (n = 174), and Aer ΔAΔB (n = 148)], with survival rates of 64%, 73%, and 55%, respectively (Figure 6B). Remarkably, this decreased survival rate was rescued back to 90% by the presence of purified AimA protein (n = 60; Figure 6B). These data demonstrate that the Aim proteins act to promote both bacterial colonization and host survival, identifying AimA as a key mediator of host-bacterial mutualism.
To test whether AimA benefits both the bacteria and the host by controlling the neutrophil response to bacterial colonization, we evaluated the importance of Aim proteins in a colonization model lacking intestinal neutrophils. We reasoned that in the absence of intestinal inflammation, bacteria possessing or lacking Aim proteins would have a similar capacity to survive in the intestine. For these experiments, we used myd88-/- mutant fish lacking the adaptor protein for inducing many toll-like receptor and IL-1 dependent pro-inflammatory pathways (Larsson et al., 2012); these fish have a severely attenuated intestinal neutrophil response (Figure 6C) that is indistinguishable from GF wild-type zebrafish (Figure 1A), as we have reported previously (Bates et al., 2007; Burns et al., 2017). We found that both Aer ΔAΔB and wild-type Aer colonized myd88-/- fish to equivalent levels, which were significantly higher than in wild-type fish (Figure 6D). These results suggest that the decreased colonization of Aer ΔAΔB compared to wild-type Aeromonas in wild-type fish is due to the increased inflammatory response to the mutant bacteria, and moreover suggest that the hosts’ innate immune response contributes to limiting bacterial colonization. We further found that myd88-/- fish inoculated with wild-type Aeromonas or Aer ΔAΔB died at the same rate (Figure 6E), suggesting that the increased inflammatory response of wild-type fish colonized with Aer ΔAΔB compared to wild-type Aeromonas accounts for the decreased fish survival rate with the mutant bacteria. Notably, the myd88-/- fish mono-associated with either wild-type or Aer ΔAΔB perished at higher rates than wild type fish, indicating that there is a host fitness cost to failing to control Aeromonas levels.
Given the increase in bacterial colonization and fish death in the myd88-/- fish, regardless of the colonizing strain, we hypothesized that these fish may be dying of septic shock as a result of bacterial overgrowth. Thus we asked whether AimA could delay death as a result of another zebrafish model of bacterial septic shock, which is induced by LPS intoxication (Bates et al., 2007; Philip and Wang, 2017). To test this, we treated fish with 100 ng/mL AimA on 4 dpf and then challenged them with 600 μg/mL LPS on 5 dpf and tracked their survival over the subsequent 60 hr. The median survival time post LPS treatment was 32 hr, which we found was extended to 47 hr in the presence of AimA (Figure 6F). Using the same treatment paradigm, we found that purified AimB also extended the median survival of LPS-exposed zebrafish to 47 hr (Figure 6F). In contrast, treatment with mLCN2 had no effect on zebrafish survival (median survival 34 hr, Figure 6F). Thus, AimA and AimB, but not mLCN2, significantly extend the survival of zebrafish in an LPS intoxication model.
Discussion
A universal feature of animals is their co-existence with resident microbial communities.
Appropriate modulation of host immune responses to resident microbes is critical for maintaining health, with too muted a response leaving hosts susceptible to microbial overgrowth (Dukowicz et al., 2007; Rolig et al., 2017) and too aggressive a response wreaking havoc on both host tissue and resident microbiota (Huttenhower et al., 2014). Fitness effects of host-bacterial interactions are often considered from the host perspective, yet it is essential to examine both sides of these partnerships to understand how these interactions evolve and persist. Here we describe the discovery of AimA, a bacterial immunomodulatory protein secreted by Aeromonas that is required during bacterial colonization of larval zebrafish to prevent an intestinal inflammatory response that is detrimental to both partners. We determined the molecular structure of AimA and show that it consists of two structurally similar lipocalin-like domains, each of which acts to suppress intestinal inflammation. We demonstrate that purified AimA protein is protective against host intestinal inflammation and LPS intoxication and simultaneously rescues Aeromonas growth in the context of inflammation. When the inflammatory response is attenuated in myd88 deficient hosts, then the bacterial colonization advantage associated with AimA production is eliminated, indicating that it is AimA’s anti-inflammatory activity that benefits Aeromonas.
All resident bacteria produce microbial associated molecular patterns (MAMPs) that are inherently pro-inflammatory, as we have shown in zebrafish larvae for LPS, which elicits intestinal neutrophil influx, proinflammatory cytokine gene expression, and endotoxemia in a dose-dependent manner (Bates et al., 2007). However, our analysis of dose response curves to resident bacteria revealed that few zebrafish microbiota members elicit dose-dependent inflammation (Rolig et al., 2015; Rolig et al., 2017). We did identify a few examples of bacteria with potent and dose-dependent inhibitory effects on intestinal neutrophil influx. These dose-dependent inhibitory effects were exploited to identify an immunoregulatory Shewanella strain that produces a secreted, heat-labile, anti-inflammatory factor (Rolig et al., 2015) and an Escherichia strain that reduces intestinal inflammation through a secretion-independent mechanism (Rolig et al., 2017). However, the majority of zebrafish gut bacteria we surveyed, including Aeromonas species, showed no correlation between their bacterial load and the number of neutrophils they elicited (Rolig et al., 2015; Rolig et al., 2017), suggesting that most gut bacteria posses both pro-inflammatory products, like LPS, along with neutralizing mechanisms to counteract the effects on their hosts. AimA, which decreases intestinal neutrophil influx and septic shock elicited by both Aeromonas and LPS, represents such a neutralizing activity. Future studies will determine whether AimA acts primarily by reducing the production of pro-inflammatory cytokines that recruit neutrophils to the intestine or by inhibiting the responsiveness of neutrophils to such cues.
Sequence-based searches for AimA-related proteins recovered only other Aeromonas homologues, including the highly divergent but functionally redundant AimB. Structural determination of AimA, however, revealed two linked domains of the lipocalin superfamily. Lipocalin domains are found across the tree of life, with bacterial lipocalin fold-containing proteins being only more recently identified (Bishop, 2000). In eukaryotes, proteins with lipocalin folds commonly bind small hydrophobic molecules such as odorant molecules, retinol, and fatty acids (Flower, 1996), but proteins with a lipocalin fold, especially those found in prokaryotes, can also take on a variety of other functions, such as inducing apoptosis (Tavares and Pathak, 2017), binding extracellular matrix (Parker et al., 2016), and binding hydrophobic antibiotics and antimicrobials (El-Halfawy et al., 2017; Sisinni et al., 2010).
We were particularly intrigued by AimA’s structural similarity to mammalian LCN2, a known regulator of intestinal inflammation (Moschen et al., 2017; Xiao et al., 2017). LCN2 was originally identified and purified from neutrophil granules and shown to play a critical role in neutrophil extravasation and migration—processes that AimA may also modulate. LCN2 was subsequently shown to be a sensitive fecal biomarker of intestinal inflammation in humans and mice (Chassaing et al., 2012). Inflammation-associated induction of LCN2 appears to be protective against pathology because Lcn2 deficient mice are sensitized to developing colitis (Toyonaga et al., 2016; Moschen et al., 2016; Singh et al., 2016). Further, Lactococcus lactis engineered to overexpress mLCN2 confers greater protection against colitis than a control strain of L. lactis in a mouse model of acute colitis (Saha et al., 2017). While it is clear that LCN2 protects against inflammation, the mechanisms by which it does so are not well understood. One possibility is that mLCN2’s capacity to sequester bacterial enterobactin-like siderophores may cause beneficial shifts in intestinal microbiota composition and reduce the toxic effects of iron in the inflamed environment (Moschen et al., 2016). Our data, however, suggest that AimA confers protection against inflammation by a distinct mechanism, because we found no evidence that AimA binds enterobactin or contributes to Aeromonas survival in iron limiting conditions. To test for a possible overlap between LCN2 and AimA function, we asked whether addition of mLCN2 to a model of soysaponin-induced intestinal inflammation would alter AimA’s efficacy. Indeed, the presence of mLCN2 inhibited the ability of AimA to reduce the neutrophil response. This result may indicate that the two proteins compete by acting on the same pathway governing neutrophil behavior. For example, AimA and mLCN2 may compete directly for binding to the same host receptor. LCN2 binds at least two known mammalian receptors, megalin and LCN2R, which have homologues in zebrafish (Kim et al., 2011), although the binding to these receptors has not been directly connected to the influence of LCN2 on neutrophil behavior (Moschen et al., 2017; Cabedo Martinez et al., 2016). Another possibility is that mLCN2 interferes with AimA’s activity by binding directly to AimA. mLCN2 is known to exist both as monomers and as dimers (Axelsson et al., 1995; Perduca et al., 2001; Niemi et al., 2015), and while little is known about the conditions under which each form is active, the dimeric form is associated with neutrophils (Cai et al., 2010). In AimA, the two lipocalin-like domains are tethered, but we found that each structural half can function independently to inhibit inflammation. Future biochemical studies will determine whether either of AimA’s lipocalin domains can homodimerize or bind to other lipocalin proteins and which oligomeric states of AimA confer its immune modulatory function.
Our analysis of aimA and aimB deficient Aeromonas revealed that the proteins’ anti-inflammatory activity on the host is inextricable from the fitness advantage it confers to the bacteria. In this regard, AimA represents a new class of bacterial effector proteins, which we refer to as mutualism factors. Whereas virulence factors promote the fitness of pathogens to the detriment of their hosts and are revealed by the decreased fitness of the pathogen mutants coupled to the increased fitness of the host, a mutualism factor like AimA confers mutual benefits, and both partners suffer when it is removed from the host-microbe symbiosis. Another example of a mutualism factor is the zwitterionic polysaccharide PSA produced by Bacteroidetes fragilis, which colonizes the murine colon. PSA induces T regulatory cells (T regs), which benefit its murine host by preventing excessive inflammation. The T regs in turn contribute to B. fragilis colonization, as demonstrated by the fact that in the absence of T regs, due either to genetic ablation or absence of PSA-induction, B. fragilis suffers a colonization defect (Round et al., 2011).
Given the constraints of bacterial-host coexistence in the vertebrate intestine, we speculate that intestinal microbiota are rich sources of novel anti-inflammatory factors that enable resident microbes to colonize this tissue at high density without eliciting excessive inflammation. Such anti-inflammatory factors offer promise for useful therapeutic applications, and the high throughput gnotobiotic larval zebrafish system provides an experimentally tractable platform for their discovery. These factors, however, may represent novel biological molecules that defy easy functional classification based purely on their gene sequences or the gene sequences of their biosynthetic machinery, thus additional approaches, such as protein structure determination, will be needed to further advance our understanding of their mechanisms of action. In the case of Aeromonas AimA, the crystal structure revealed it to belong to a large family of lipocalin proteins, which includes the mammalian immunomodulatory protein LCN2. Our discovery of AimA raises the possibility that AimA and LCN2 have overlapping functions as mutualism factors in the vertebrate intestine even though they are provisioned by different members of the partnership. Given the diversity of the microbial communities that are part of the complex host-microbe ecosystem, AimA is likely only the first of many unique bacterial proteins that facilitate mutualistic interactions critical for both host and resident microbes to thrive.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) |
BL21 DE3 | New England Biolabs | C2527 | |
| Strain, strain background (E. coli) |
K12 strain (MG1655) | doi: 10.1128/JB.188.3.928–933.2006 | Dr. Matthew Mulvey, Univ. of Utah |
|
| Strain, strain background (Aeromonas veronii) |
HM21S; Aer; Aeromonas Hm21; Hm21 |
doi:10.1128/AEM.01621–10 | Parent strain, SmR | |
| Strain, strain background (A. veronii) |
HE-1095; Aer DT2 | doi:10.1128/AEM.01621–10 | Hm21S::interrupted exeM mTn5 KmR SmR |
|
| Strain, strain background (A. veronii) |
HEC-1344; Aer DT2C | doi:10.1128/AEM.01621–10 | HE-1095::Tn7 containing TpR exeMN + promoter region |
|
| Strain, strain background (A. veronii) |
ASRC7; Aer DaimA | this study | Hm21S aimA::cmR | |
| Strain, strain background (A. veronii) |
ASRD5; Aer DaimB | this study | Hm21S DaimB | |
| Strain, strain background (A. veronii) |
ASRD4; Aer DADB |
this study | Hm21S aimA::cmR;
DaimB |
|
| Strain, strain background (A. veronii) |
ZOR0001; ZF Aer |
doi:10.1038/ismej.2015.140 | Zebrafish Aeromonas
isolate |
|
| Strain, strain background (A. veronii) |
ZOR0001; ZF Aer DaimA |
this study | ZOR0001 aimA::cmR | |
| Genetic reagent (Danio rerio) |
AB x Tu strain; wild type zebrafish |
UO Zebrafish facility | ||
| Genetic reagent (Danio rerio) |
myd88-/- | PMID: 28973938 | ||
| Genetic reagent (Danio rerio) |
Tg(BACmpx:GFP) i114; mpx:GFP |
doi:10.1182/blood -2006-05-024075 |
||
| Recombinant DNA reagent |
pET21B | Genscript | ||
| Chemical compound, drug |
LPS, E. Coli
O111:B4 |
Sigma | L2630 | |
| Peptide, recombinant protein |
Lipocalin 2, LCN2, Siderocalin |
Biolegend | 588002 | |
| Peptide, recombinant protein |
AimA | this study | NCBI hypothetical protein WP_021230730.1 |
see Materials and Methods |
| Peptide, recombinant protein |
AimB | this study | NCBI hypothetical protein WP_021230165.1 |
see Materials and Methods |
| Peptide, recombinant protein |
mLCN2 | this study | NCBI gene NM_008491.1 | see Materials and Methods |
Gnotobiotic zebrafish husbandry
All zebrafish experiments were performed following protocols approved by the University of Oregon Institutional Animal Care and Use Committee and followed standard zebrafish protocols (Westerfield, 2000). Conventionally-raised (CV) wild-type (AB x Tu strain), Tg(BACmpx:GFP)i114 (referred to as mpx:GFP) (Renshaw et al., 2006) and myd88-/- (Burns et al., 2017) were maintained as described (Westerfield, 2000). Zebrafish embryos were derived germ free (GF) as previously described (Melancon et al., 2017). Subsequently, 15 GF embryos were transferred to sterile tissue culture flasks (25 cm2, Techno Plastic Products, Trasadingen, Switzerland) with 15 mL embryo medium (EM) (Melancon et al., 2017). Monoassociated zebrafish were generated by inoculating flasks with 4 dpf GF zebrafish with 106 colony forming units (CFU)/mL of each bacterial strain. GF flasks were chosen at random for their respective treatment, and the researcher was blinded to the treatment group until after data collection. Bacteria used for monoassociations were zebrafish isolate Aeromonas ZOR0001 (Stephens et al., 2016), Aeromonas ZOR0001ΔaimA, Aeromonas ZOR0001 ΔaimA complement, Aeromonas HE-1095 (ΔT2) (Maltz and Graf, 2011), Aeromonas HEC-1344 (ΔT2C) (Maltz and Graf, 2011), Aeromonas Hm21S (Graf, 1999), Hm21S ΔaimA, Hm21S ΔaimB, and HM21S ΔaimAΔaimB. Control CV fish were prepared GF, as above, and inoculated with 1 mL of fish facility water on 4 dpf. All manipulations to the GF flasks were performed under a class II A/B3 biological safety cabinet. The flasks were kept at 28° C until analysis of fluorescent myeloperoxidase positive (MPX+) cells on 7 dpf.
Aeromonas mutants were created using plasmids and protocols as described in Wiles et al. (2018). Briefly, Aeromonas mutants were constructed using homologous recombination of 1 kb regions upstream and downstream of the aimA gene to replace aimA with a chloramphenicol resistance gene. The aimB mutant was constructed in a similar manner, except as a markerless deletion by first generating a merodiploid strain.
Histology and quantification of neutrophils
Zebrafish larvae were fixed in 4% paraformaldehyde (PFA) overnight. Whole larvae were stained with Myeloperoxidase kit (Sigma) following the manufacturer’s protocol and processed and analyzed as previously described (Bates et al., 2007), except that MPO +cells were quantified in whole dissected intestines rather than tissue sections. For analysis of neutrophils in mpx:GFP fish, GFP +cells in the intestine were quantified as previously described (Rolig et al., 2015). Briefly, the mpx:GFP zebrafish were anesthetized in Tricaine (Western Chemical, Inc., Ferndale, WA) and mounted in 4% methylcellulose (Fisher, Fair Lawn, NJ), and their intestines were sterilely dissected. The number of GFP-positive cells was quantified visually for each fish using a fluorescent microscope (SteREO Discovery.V8, Zeiss).
Microbiota quantification
To determine the CFU/intestine, dissected zebrafish intestines were placed in 100 μL sterile EM, homogenized in a bullet blender (Next Advance), diluted, and cultured on tryptic soy agar (TSA, BD, Sparks MD). The TSA plates were incubated at 30° C overnight and then colonies were counted.
Concentration and fractionation of cell-free supernatant
Aeromonas HEC-1344 or E. coli BL21 were grown overnight to stationary phase. Then 500 μL of the overnight culture was used to inoculate a 50 mL culture, which was kept shaking at 30° C for 2 hr, until the bacteria had grown through exponential and into stationary phase (OD of approximate 0.6). To overexpress proteins of interest, 1 mM of IPTG was added to the cultures during exponential growth phase and allowed to grow for an additional 2 hr at 30° C. To prepare the CFS, the 50 mL cultures were centrifuged at 7000 x g for 10 min at 4° C. Subsequently, the supernatant was filtered through a 0.22 μm sterile tube top filter (Corning Inc., Corning, NY). The sterile supernatant was concentrated at 4° C for 1 hr at 3000 x g with a centrifugal device that has a 10 kDa weight cut off (Pall Life Sciences, Ann Arbor, MI). The concentration of the supernatant was determined with a Nanodrop and inoculated into the flasks at a final concentration of 500 ng/mL.
Ammonium sulfate fractionation experiments were done as previously described (Hill et al., 2016). Briefly, unconcentrated CFS from a 50 mL overnight culture of Aeromonas HEC-1344 was fractionated by slowly adding cold 100% ammonium sulfate until solutions reached 20%, 40%, and 60% ammonium sulfate. Precipitated proteins were collected by centrifugation at 4° C, 15000 g for 15 min. Recovered proteins were resuspended in cold EM, dialyzed for 2–3 hr at 4° C, then added to mono-associated fish flasks at a concentration of 500 ng/mL.
Mass spectrophotometry
The protein constituents of concentrated cell free supernatants from Aeromonas HE-1095 and Aeromonas HEC-1344 (Maltz and Graf, 2011) were determined by analysis of peptide MS/MS spectra at the Proteomics Shared Resource Facility at Oregon Health and Sciences University in Portland, Oregon.
Inflammatory assays
Soysaponin (Sigma) was mixed with Zieglers fish food at a concentration of 0.3%. Ten CV zebrafish were maintained 10 mL EM in 60 × 15 mm petri dishes. Larval fish were fed once daily from 4 dpf to 6 dpf. During each feeding, the larvae had access to the food for 3–4 hr before being washed into fresh EM. For experiments with mLCN2, recombinant mouse LCN2 (Biolegend) was added to the fish EM at a concentration of 100 ng/mL after the soysaponin feeding on 4 dpf and 5 dpf after the fish were moved into fresh EM.
To induce LPS intoxication, CV zebrafish were treated with 600 μg/mL LPS (Sigma) on 5 dpf and monitored for survival for the following 2 days. In additional treatment groups, 100 ng/mL AimA, AimB, or mLCN (purified as described below) were added on 4 dpf.
AimA, AimB, and mLCN2 protein purification
The aimA gene was PCR amplified from gDNA excluding the 5’ 66 nucleotide (22 amino acid) secretion signal and cloned into pET21b using NdeI and XhoI restriction sites. The resultant gene product (NCBI hypothetical protein WP_021230730.1) expressed well in E. coli BL21 DE3 as a C-terminal 6X His tagged protein of 300 amino acids long (including His tag and linker).
The E. coli culture was grown at 37° C until OD600 0.4–0.6, then moved to 30° C and induced with 1 mM IPTG for 3–4 hr. All subsequent steps were performed at 4° C. 1–2 L of pelleted E. coli cells were lysed in lysis buffer (50 mM HEPES pH 7.9, 300 mM NaCl, 10 mM imidazole and 5 mM BME), sonicated, and debris pelleted. The supernatant was washed over 5 mL Ni-NTA resin in a gravity column that was pre-washed with lysis buffer. The resin was washed with 15X bed volume of lysis buffer, then 10X bed volume of lysis buffer with 30 mM imidazole, 10X bed volume of lysis buffer with 50 mM imidazole and finally eluted with 3x - 5x bed volume of lysis buffer plus 100–300 mM imidazole. The high absorbance (280 nm wavelength) fractions were pooled and dialyzed overnight into 150 mM NaCl, 50 mM HEPES pH 7.9 and 5 mM BME, concentrated, flash frozen in liquid nitrogen, and stored at −80° C.
The aimB gene, excluding the 5’ 57 nucleotide (19 amino acid) secretion signal, was cloned into pET21b using NdeI and XhoI restriction sites (GenScript). The resulting gene product (NCBI hypothetical protein WP_021230165.1) expressed well in E. coli BL21 DE3 as a C-terminal 6X His tagged protein of 321 amino acids long (including His tag and linker). AimB protein was expressed and purified similarly to AimA, above, with some changes. The lysate was run over HisTrap HP 5 mL column (GE healthcare) using an AKTA FPLC (GE healthcare) and eluted with a gradient from 10 to 500 mM imidazole. Fractions containing the purest AimB were pooled, dialyzed (150 mM NaCl, 50 mM HEPES pH 7.9 and 5 mM BME) and concentrated. Due to the difficulty of separating away contaminating E. coli proteins, AimB was only crudely purified to >50% pure by SDS PAGE.
The mouse lipocalin2 gene (NCBI gene NM_008491.1), excluding the 5’ 63 nucleotide (21 amino acids) secretion signal, was cloned into pET21b (GenScript), using Nde1 and Xho1 restriction sites. Mouse LCN2 with a C-term 6X His tag expressed well in E. coli BL21 DE3 using 0.25 mM IPTG at 18° C overnight. mLCN2 was purified using Ni-NTA agarose resin (Qiagen) in a gravity column with increasing concentrations of imidazole from 10 to 30 mM, and eluted with 50 mM imidazole. The resultant 22 kDa protein (189 amino acids including His tag and linker) was dialyzed into 300 mM NaCl, 50 mM potassium phosphate pH 7 and 5 mM BME. The dialyzed protein was concentrated and confirmed to be >95% pure by SDS PAGE.
AimA protein crystallization
Purified AimA with C-terminal 6x His tag was concentrated to 10.9 mg/mL and set up in hanging drops as 1 μL protein: 1 uL well solution at room temperature. AimA crystallized in thick hexagons in 3.5 M sodium formate, 75 mM NaCl, 25 mM HEPES pH 7.9 and 2.5 mM BME. One to two weeks after the crystals grew, they were transferred to wells containing 3.8 M sodium formate to toughen them up for approximately one week. The heavy atom derivative crystals were then transferred to drops with 3.8 M sodium formate, 0.5 M NaI (for iodide data set) and 15% glycerol (as a cryoprotectant) for several hours before being scooped and flash frozen in liquid nitrogen for data collection at the Advanced Light Source in Berkeley, CA, beamline 5.0.2 using the Pilatus detector at a wavelength of 1.0 Å. The native crystals were transferred to 3.8 M sodium formate and 15% PEG 200 briefly, then flash frozen in liquid nitrogen for data collection as described for the iodide soaked crystals.
Structure determination of AimA
Data processing
The heavy atom derivative was solved by the single-wavelength anomalous diffraction (SAD) method from a single crystal derivatized with I-, with data collected at wavelength λ = 1 Å. The data set was integrated and scaled to resolution 2.7 Å using HKL3000 (Minor et al., 2006) with the merging analysis indicating the P622 space group. Although data were isotropic, diffraction spots were smeared in a manner indicating the presence of order-disorder. In addition, the scaling B-factor increase of ~40 Å2, which is equivalent to a dose of ~40 MGy, indicated severe radiation damage. Therefore, it was necessary to apply the ‘automatic corrections’ computational procedure to optimize the error model (Borek et al., 2010; Borek et al., 2013; Borek et al., 2007), and this was essential for the success of the experimental phasing described below.
The estimated level of anomalous signal was ~3.6% of the native intensity. The search for heavy atom positions was performed to a resolution of 3.7 Å. The 30 positions of I- were identified using SHELXC/D (Sheldrick, 2008), run within HKL3000, with correlation coefficients: CCAll = 41.4%, CCWeak = 16.1%. The handedness of the best solution was determined with SHELXE. The heavy atom positions were refined to 2.7 Å with MLPHARE (Otwinowski, 1991), with the final Figure of Merit (FOM) reaching 0.14 for all observations. Solvent flattening was performed by DM (Cowtan and Main, 1998). The procedure produced an interpretable electron density map that was used for iterative automatic model building with BUCCANEER, Coot and REFMAC (Cowtan, 2006; Emsley and Cowtan, 2004; Murshudov et al., 1997; Murshudov et al., 1999) – all run within HKL3000 with the ‘HKL Builder’ option. That procedure resulted in 100% of the model being built with 90% of the side chains docked. At this point, the R and R-free factors were ~27% and~35%, respectively.
Two native AimA datasets from separate crystals were indexed and integrated with iMosflm 7.2.1 (Battye et al., 2011) and scaled using SCALA (Collaborative Computational Project, Number 4, 1994). The two datasets were found to be isomorphous, and were combined using POINTLESS (Collaborative Computational Project, Number 4, 1994). The high-resolution cutoff was determined by the method of Karplus and Diederichs (Karplus and Diederichs, 2012) using a CC1/2 of >0.3 and completeness of >50% in the highest resolution shell. This method has been utilized in numerous other studies (Perkins et al., 2016; Evans and Murshudov, 2013; Kern et al., 2013) and has been cited over 900 times since its publication in Science in 2012. Using this criteria the correlation between two halves of the data are used to determine the point at which signal falls away into noise (Evans and Murshudov, 2013), and Rmerg values can rise to values much higher than what has traditionally been thought of as allowable. Using this strategy we were able to extend the resolution from 2.9 Å (where the data would have been cut based on Rmerg ~0.6) to 2.3 Å. To further test the validity of using this noisy high-resolution data to refine the model, a series of paired refinements were conducted (Karplus and Diederichs, 2012). The model was first refined using data out to 2.9, 2.7, 2.5, or 2.3 Å and then, since R values are only comparable when calculated at the same resolution (Karplus and Diederichs, 2012), R and Rfree were calculated for each refined model at 2.9 Å (Figure 2—figure supplement 2). The extra resolution improves both R (higher value) and Rfree (lower value), showing that the model is improved in predictive quality and is less overfit using the extended resolution cutoff. In this case CC1/2 remains quite high at 0.9 in the high-resolution shell, and the <I/σ >at 1.4 is not far below a traditional cutoff of 2.0. Data statistics are summarized in Table 2.
Structure refinement
Manual model building was performed using Coot 0.8.1.6 (Emsley and Cowtan, 2004) and refinement was carried out using PHENIX 1.12–2829 (Adams et al., 2010). Initial rigid body refinement resulted in R/Rfree values of 26.5/28.3%. Using the extended resolution improved the electron density maps and allowed placement of additional water molecules, two formate molecules (present at 3.5 M in the crystallization buffer), N-terminal residues 1–8, and an alternate chain path for residues 153–165, improving R/Rfree to 20.9/24.8%. Residues 180–181 are at the tip of a disordered loop and were not modeled, and residues 293–294 and the C-terminal His-tag beyond it are not visible in the electron density. Electron density is weak in several regions including the N-terminus and several loops, but the chain path was clear enough to build at least the backbone atoms for these residues. In late stages of refinement, TLS was implemented using one group per chain, dropping R/Rfree to 17.9/20.9%. B-factor weights were optimized in the final refinement step, yielding final R/Rfree of 17.2/20.4% for the final AimA model (Table 2).
E.coli and Aeromonas growth curves
E. coli and Aeromonas cultures were grown overnight (37° C and 30° C, respectively) from a glycerol freezer stock in Luria Bertani (LB) broth to stationary phase. The cultures were back-diluted 1:100 the next morning in LB and loaded into a transparent 96-well plate. We used a total volume of 200 μL per well. 2,2’-dipyridyl (Sigma-Aldrich) titrations in DMSO were administered in 10 μL treatments for the Aeromonas growth curves. For the E. coli growth curves, AimA and mLCN titrations were administered in 20 μL treatments, with dipyridyl added to 300 μM in the LB 1:100 back-dilution. Controls for the Aeromonas and E. coli growth curves were treatments of DMSO or protein buffer (300 mM NaCl, 50 mM potassium phosphate pH 7, 5 mM BME), respectively. Growth (OD 600 nm) at 30° C was monitored using a FLUOStar Omega (BMG LABTECH) plate reader. Readings were taken every 1 hr for up to 36 hr. Replicate curves (three or four) were plotted with standard deviation using Prism (GraphPad).
Statistical analysis
The appropriate sample size for experiments quantifying intestinal neutrophils and bacterial colonization level was estimated a priori using a power of 84% and a significance level of 0.05. From previously published data on intestinal neutrophil quantification (Rolig et al., 2015) and zebrafish gut bacterial mono-association colonization (Rolig et al., 2015; Hill et al., 2016) we estimated a effect size of 0.24 for neutrophil influx and 0.35 for bacterial colonization. These parameters suggested using an n of 40 and 23 in order to detect significant changes between treatment groups for either neutrophil influx or bacterial colonization, respectively. Each experiment described herein contains about 10–15 biological replicates (individual fish per treatment group). These experiments of 10–15 fish were repeated multiple times (technical replicates), resulting in pooled data sets ranging from 20 to 70 biological replicates. Grubbs statistical outlier test was applied to the data and data points that met the criteria for a statistical outlier were removed. In the figures, these data are presented as box and whisker plots, which display the data median (line within the box), first and third quartiles (top and bottom of the box), and minimum and maximum values (whiskers). All data points that generate the box and whisker plots are presented as individual points within the plot. These pooled data were analyzed through the statistical software Prism Graphpad software. For experiments measuring a single variable with multiple treatment groups, a single factor ANOVA with post hoc means testing (Tukey) was utilized. A p-value of less than 0.05 was required to reject the null hypothesis that no difference existed between groups of data. For survival curves a Mantel-Cox test was used to determine significance.
Acknowledgements
We thank Rose Sockol and the UO Zebrafish Facility staff for fish husbandry. We thank Joerg Graf for providing the Aeromonas veronii HM21 strains and S James Remington for advice on crystallography.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Contributor Information
Karen Guillemin, Email: kguillem@uoregon.edu.
Wendy S Garrett, Harvard TH Chan School of Public Health, United States.
Xochitl Morgan, University of Otago, New Zealand.
Funding Information
This paper was supported by the following grants:
National Institute of General Medical Sciences P50GM098911 to Karen Guillemin.
National Institute of Diabetes and Digestive and Kidney Diseases F32DK098884 to Annah S Rolig.
Eunice Kennedy Shriver National Institute of Child Health and Human Development P01HD22486 to Karen Guillemin.
Additional information
Competing interests
Have two patent applications related to AimA pending in the United States, Europe, and Canada (US Patent App. 15/883,999 and 15/311,667).
No competing interests declared.
Karen Guillemin: Have two patent applications related to AimA pending in the United States, Europe, and Canada (US Patent App. 15/883,999 and 15/311,667).
Author contributions
Conceptualization, Formal analysis, Funding acquisition, Investigation, Visualization, Methodology, Writing—original draft.
Formal analysis, Investigation, Visualization, Writing—review and editing, Purified AimA and AimB, Determined the crystal structure of AimA.
Investigation, Performed genetic and functional characterizations of the Aim proteins.
Investigation, Performed genetic and functional characterizations of the Aim proteins.
Investigation, Made significant intellectual contributions to the analysis of crystallographic data.
Investigation, Characterized the secreted proteins from the type 2 secretion mutant and complemented strain of Aeromonas.
Investigation, Writing—review and editing, Performed genetic and functional characterizations of the Aim proteins.
Conceptualization, Resources, Supervision, Funding acquisition, Project administration, Writing—review and editing.
Ethics
Animal experimentation: All zebrafish experiments were performed following protocols approved by the University of Oregon Institutional Animal Care and Use Committee (#15-98) and following standard zebrafish protocols.
Additional files
Mass-spectrophotometry was performed on the CFS from ∆T2C and ∆T2 determined which proteins were enriched in the ΔT2C compared to the ΔT2 strain. This table lists the top 22 proteins that were enriched by greater than 10 counts in the ΔT2C CFS.
Structural homology searches performed against the N and C terminal domains of AimA separately revealed that both domains have similarity to proteins in the calycin superfamily, primarily avidins and lipocalins.
Data availability
Diffraction data have been deposited in PDB under the accession code 6B7L.
The following dataset was generated:
Rolig AS, Sweeney EG, Kaye LE, DeSantis MD, Perkins A, Banse AV, Hamilton MK, Guillemin K. 2018. Aeromonas Immune Modulator A Structure. RCSB Protein Data Bank. 6B7L
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson L, Bergenfeldt M, Ohlsson K. Studies of the release and turnover of a human neutrophil lipocalin. Scandinavian Journal of Clinical and Laboratory Investigation. 1995;55:577–588. doi: 10.3109/00365519509110257. [DOI] [PubMed] [Google Scholar]
- Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Developmental Biology. 2006;297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host & Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallographica. Section D, Biological Crystallography. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop RE. The bacterial lipocalins. Biochimica Et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 2000;1482:73–83. doi: 10.1016/S0167-4838(00)00138-2. [DOI] [PubMed] [Google Scholar]
- Borek D, Ginell SL, Cymborowski M, Minor W, Otwinowski Z. The many faces of radiation-induced changes. Journal of Synchrotron Radiation. 2007;14:24–33. doi: 10.1107/S0909049506046589. [DOI] [PubMed] [Google Scholar]
- Borek D, Cymborowski M, Machius M, Minor W, Otwinowski Z. Diffraction data analysis in the presence of radiation damage. Acta Crystallographica Section D Biological Crystallography. 2010;66:426–436. doi: 10.1107/S0907444909040177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borek D, Dauter Z, Otwinowski Z. Identification of patterns in diffraction intensities affected by radiation exposure. Journal of Synchrotron Radiation. 2013;20:37–48. doi: 10.1107/S0909049512048807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns AR, Guillemin K. The scales of the zebrafish: host-microbiota interactions from proteins to populations. Current Opinion in Microbiology. 2017;38:137–141. doi: 10.1016/j.mib.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns AR, Miller E, Agarwal M, Rolig AS, Milligan-Myhre K, Seredick S. Interhost dispersal alters microbiome assembly and can overwhelm host innate immunity in an experimental zebrafish model. Proc Natl Acad Sci. 2017 doi: 10.1073/pnas.1702511114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabedo Martinez AI, Weinhäupl K, Lee WK, Wolff NA, Storch B, Żerko S, Konrat R, Koźmiński W, Breuker K, Thévenod F, Coudevylle N. Biochemical and structural characterization of the interaction between the siderocalin NGAL/LCN2 (Neutrophil Gelatinase-associated Lipocalin/Lipocalin 2) and the N-terminal domain of its endocytic receptor SLC22A17. Journal of Biological Chemistry. 2016;291:2917–2930. doi: 10.1074/jbc.M115.685644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Rubin J, Han W, Venge P, Xu S. The origin of multiple molecular forms in urine of HNL/NGAL. Clinical Journal of the American Society of Nephrology. 2010;5:2229–2235. doi: 10.2215/CJN.00980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson AH, Yakymenko O, Olivier I, Håkansson F, Postma E, Keita AV, Söderholm JD. Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice DSS colitis. Scandinavian Journal of Gastroenterology. 2013;48:1136–1144. doi: 10.3109/00365521.2013.828773. [DOI] [PubMed] [Google Scholar]
- Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, Vijay-Kumar M. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS ONE. 2012;7:e44328. doi: 10.1371/journal.pone.0044328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the wnt pathway and microbial signaling via Myd88. PNAS. 2011;108 Suppl 1 doi: 10.1073/pnas.1000072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallographica Section D Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cowtan K, Main P. Miscellaneous algorithms for density modification. Acta Crystallographica Section D Biological Crystallography. 1998;54:487–493. doi: 10.1107/S0907444997011980. [DOI] [PubMed] [Google Scholar]
- Cowtan K. The buccaneer software for automated model building. 1. Tracing Protein Chains. Acta Crystallogr Sect D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Dukowicz AC, Lacy BE, Levine GM. Small intestinal bacterial overgrowth: a comprehensive review. Gastroenterology & Hepatology. 2007;3:112–122. [PMC free article] [PubMed] [Google Scholar]
- El-Halfawy OM, Klett J, Ingram RJ, Loutet SA, Murphy MEP, Martín-Santamaría S, Valvano MA. Antibiotic capture by bacterial lipocalins uncovers an extracellular mechanism of intrinsic antibiotic resistance. mBio. 2017;8:1–15. doi: 10.1128/mBio.00225-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallographica Section D Biological Crystallography. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower DR. The lipocalin protein family: structure and function. Biochemical Journal. 1996;318:1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Molecular Cell. 2002;10:1033–1043. doi: 10.1016/S1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- Graf J. Symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech: a novel model for digestive tract associations. Infection and Immunity. 1999;67:1–7. doi: 10.1128/iai.67.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald DJ, Eisen JS. Headwaters of the zebrafish -- emergence of a new model vertebrate. Nature Reviews Genetics. 2002;3:717–724. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- Harvie EA, Huttenlocher A. Neutrophils in host defense: new insights from zebrafish. Journal of Leukocyte Biology. 2015;98:523–537. doi: 10.1189/jlb.4MR1114-524R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrera MI, Galdames JA, Jimenez-Reyes MF, Reyes AE, Avendaño-Herrera R, Romero J, Feijóo CG. Soybean meal induces intestinal inflammation in zebrafish larvae. PLoS ONE. 2013;8:e69983. doi: 10.1371/journal.pone.0069983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JH, Franzosa EA, Huttenhower C, Guillemin K. A conserved bacterial protein induces pancreatic beta cell expansion during zebrafish development. eLife. 2016;5:e20145. doi: 10.7554/eLife.20145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40:843–854. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielita M, Taormina MJ, Burns AR, Hampton JS, Rolig AS, Guillemin K, Parthasarathy R. Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. mBio. 2014;5:1–8. doi: 10.1128/mBio.01751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, Tomkovich S, Xiaolun S, Grosser MR, Koo J, Flynn EJ, Jobin C, Rawls JF. Commensal microbiota stimulate systemic neutrophil migration through induction of serum amyloid A. Cellular Microbiology. 2014;16:1053–1067. doi: 10.1111/cmi.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern J, Alonso-Mori R, Tran R, Hattne J, Gildea RJ, Echols N, Glöckner C, Hellmich J, Laksmono H, Sierra RG, Lassalle-Kaiser B, Koroidov S, Lampe A, Han G, Gul S, Difiore D, Milathianaki D, Fry AR, Miahnahri A, Schafer DW, Messerschmidt M, Seibert MM, Koglin JE, Sokaras D, Weng TC, Sellberg J, Latimer MJ, Grosse-Kunstleve RW, Zwart PH, White WE, Glatzel P, Adams PD, Bogan MJ, Williams GJ, Boutet S, Messinger J, Zouni A, Sauter NK, Yachandra VK, Bergmann U, Yano J. Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature. Science. 2013;340:491–495. doi: 10.1126/science.1234273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lee S, Park HC, Lee WH, Lee MS, Suk K. Modulation of glial and neuronal migration by lipocalin-2 in zebrafish. Immune Network. 2011;11:342–347. doi: 10.4110/in.2011.11.6.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K, matching S-structure. SSM), a new tool for fast protein structure alignment in three dimensions. Acta crystallogr sect D biol crystallogr. International Union of Crystallography. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Lakshmi B, Mishra M, Srinivasan N, Archunan G. Structure-Based phylogenetic analysis of the lipocalin superfamily. Plos One. 2015;10:e0135507–e0135518. doi: 10.1371/journal.pone.0135507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE, Bäckhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltz M, Graf J. The type II secretion system is essential for erythrocyte lysis and gut colonization by the leech digestive tract symbiont Aeromonas veronii. Applied and Environmental Microbiology. 2011;77:597–603. doi: 10.1128/AEM.01621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltz M, LeVarge BL, Graf J. Identification of iron and heme utilization genes in Aeromonas and their role in the colonization of the leech digestive tract. Frontiers in Microbiology. 2015;6:1–14. doi: 10.3389/fmicb.2015.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín R, Chain F, Miquel S, Lu J, Gratadoux JJ, Sokol H, Verdu EF, Bercik P, Bermúdez-Humarán LG, Langella P. The commensal bacterium Faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflammatory Bowel Diseases. 2014;20:417–430. doi: 10.1097/01.MIB.0000440815.76627.64. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. Animals in a bacterial world, a new imperative for the life sciences. PNAS. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melancon E, Gomez De La Torre Canny S, Sichel S, Kelly M, Wiles TJ, Rawls JF, Eisen JS, Guillemin K. Best practices for germ-free derivation and gnotobiotic zebrafish husbandry. Methods in Cell Biology. 2017;138:61–100. doi: 10.1016/bs.mcb.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL -3000: the integration of data reduction and structure solution – from diffraction images to an initial model in minutes. Acta Crystallographica Section D Biological Crystallography. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Moschen AR, Gerner RR, Wang J, Klepsch V, Adolph TE, Reider SJ, Hackl H, Pfister A, Schilling J, Moser PL, Kempster SL, Swidsinski A, Orth Höller D, Weiss G, Baines JF, Kaser A, Tilg H. Lipocalin 2 protects from inflammation and tumorigenesis associated with gut Microbiota alterations. Cell Host & Microbe. 2016;19:455–469. doi: 10.1016/j.chom.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Moschen AR, Adolph TE, Gerner RR, Wieser V, Tilg H. Lipocalin-2: a master mediator of intestinal and metabolic inflammation. Trends in Endocrinology & Metabolism. 2017;28:388–397. doi: 10.1016/j.tem.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallographica Section D Biological Crystallography. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- Niemi MH, Rytkönen-Nissinen M, Miettinen I, Jänis J, Virtanen T, Rouvinen J. Dimerization of lipocalin allergens. Scientific Reports. 2015;5:13841. doi: 10.1038/srep13841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. Maximum likelihood refinement of heavy atom parameters. Pap Present CCP4 Study Weekend 1991 [Google Scholar]
- Parker ML, Houston S, Pětrošová H, Lithgow KV, Hof R, Wetherell C, Kao WC, Lin YP, Moriarty TJ, Ebady R, Cameron CE, Boulanger MJ. The structure of Treponema pallidum Tp0751 (Pallilysin) Reveals a Non-canonical lipocalin fold that mediates adhesion to extracellular matrix components and interactions with host cells. PLOS Pathogens. 2016;12:e1005919–e1005924. doi: 10.1371/journal.ppat.1005919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perduca M, Mancia F, Del Giorgio R, Monaco HL. Crystal structure of a truncated form of porcine odorant-binding protein. Proteins: Structure, Function, and Genetics. 2001;42:201–209. doi: 10.1002/1097-0134(20010201)42:2<201::AID-PROT70>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Perkins A, Parsonage D, Nelson KJ, Ogba OM, Cheong PH, Poole LB, Karplus PA. Peroxiredoxin catalysis at atomic resolution. Structure. 2016;24:1668–1678. doi: 10.1016/j.str.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip AM, Wang Y. Development of a zebrafish Sepsis model for High-Throughput drug discovery. Molecular Medicine. 2017;23:00188. doi: 10.2119/molmed.2016.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quévrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermúdez-Humarán LG, Pigneur B, Lequin O, Kharrat P, Thomas G, Rainteau D, Aubry C, Breyner N, Afonso C, Lavielle S, Grill JP, Chassaing G, Chatel JM, Trugnan G, Xavier R, Langella P, Sokol H, Seksik P. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in crohn's disease. Gut. 2016;65:415–425. doi: 10.1136/gutjnl-2014-307649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- Renshaw SA, Trede NS. A model 450 million years in the making: zebrafish and vertebrate immunity. Disease Models & Mechanisms. 2012;5:38–47. doi: 10.1242/dmm.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolig AS, Parthasarathy R, Burns AR, Bohannan BJ, Guillemin K. Individual members of the Microbiota disproportionately modulate host innate immune responses. Cell Host & Microbe. 2015;18:613–620. doi: 10.1016/j.chom.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolig AS, Mittge EK, Ganz J, Troll JV, Melancon E, Wiles TJ, Alligood K, Stephens WZ, Eisen JS, Guillemin K. The enteric nervous system promotes intestinal health by constraining Microbiota composition. PLOS Biology. 2017;15:e2000689. doi: 10.1371/journal.pbio.2000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK, a CT. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. PNAS. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y, Prediction F. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P, Chassaing B, Yeoh BS, Viennois E, Xiao X, Kennett MJ, Singh V, Vijay-Kumar M. Ectopic expression of innate immune protein, Lipocalin-2, in Lactococcus lactis protects against gut and environmental stressors. Inflammatory Bowel Diseases. 2017;23:1120–1132. doi: 10.1097/MIB.0000000000001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallographica. Section A, Foundations of Crystallography. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Singh V, Yeoh BS, Chassaing B, Zhang B, Saha P, Xiao X. Microbiota-Inducible innate immune siderophore binding protein lipocalin 2 is critical for intestinal homeostasis. C cell mol gastroenterol hepatol. Elsevier Inc. 2016;2:482–498. doi: 10.1016/j.jcmgh.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisinni L, Cendron L, Favaro G, Zanotti G. Helicobacter pylori acidic stress response factor HP1286 is a YceI homolog with new binding specificity. FEBS Journal. 2010;277:1896–1905. doi: 10.1111/j.1742-4658.2010.07612.x. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut Microbiota analysis of crohn disease patients. PNAS. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens WZ, Burns AR, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJ. The composition of the zebrafish intestinal microbial community varies across development. The ISME journal. 2016;10 doi: 10.1038/ismej.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taormina MJ, Jemielita M, Stephens WZ, Burns AR, Troll JV, Parthasarathy R, Guillemin K. Investigating bacterial-animal symbioses with light sheet microscopy. The Biological Bulletin. 2012;223:7–20. doi: 10.1086/BBLv223n1p7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares R, Pathak SK. Helicobacter pylori Secreted Protein HP1286 Triggers Apoptosis in Macrophages via TNF-Independent and ERK MAPK-Dependent Pathways. Frontiers in Cellular and Infection Microbiology. 2017;7:1–16. doi: 10.3389/fcimb.2017.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyonaga T, Matsuura M, Mori K, Honzawa Y, Minami N, Yamada S, Kobayashi T, Hibi T, Nakase H. Lipocalin 2 prevents intestinal inflammation by enhancing phagocytic bacterial clearance in macrophages. Scientific Reports. 2016;6:35014. doi: 10.1038/srep35014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Intestinal growth and differentiation in zebrafish. Mechanisms of Development. 2005;122:157–173. doi: 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. A Guide for the Laboratory Use of Zebrafish (Danio Rerio) 4th ed. Eugene: University of Oregon Press; 2000. The Zebrafish Book. [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. PNAS. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles TJ, Jemielita M, Baker RP, Schlomann BH, Logan SL, Ganz J, Melancon E, Eisen JS, Guillemin K, Parthasarathy R. Host gut motility promotes competitive exclusion within a model intestinal Microbiota. PLOS Biology. 2016;14:e1002517. doi: 10.1371/journal.pbio.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles TJ, Wall ES, Schlomann BH, Hay EA, Parthasarathy R, Guillemin K. Modernized tools for streamlined genetic manipulation and comparative study of wild and diverse proteobacterial lineages. mBio. 2018;9:e01877-18. doi: 10.1128/mBio.01877-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Yeoh BS, Vijay-Kumar M. Lipocalin 2: an emerging player in iron homeostasis and inflammation. Annual Review of Nutrition. 2017;37:103–130. doi: 10.1146/annurev-nutr-071816-064559. [DOI] [PubMed] [Google Scholar]
- Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132:562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Cao H, Cover TL, Washington MK, Shi Y, Liu L, Chaturvedi R, Peek RM, Wilson KT, Polk DB. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. Journal of Clinical Investigation. 2011;121:2242–2253. doi: 10.1172/JCI44031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]