SUMMARY
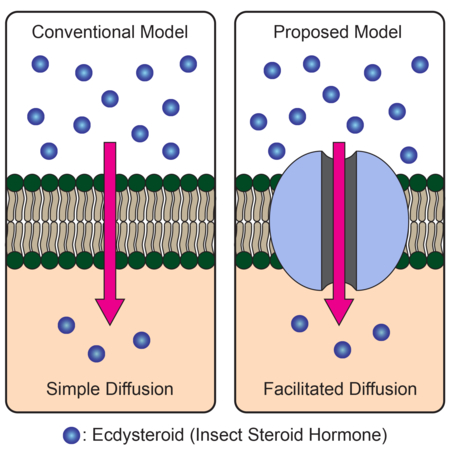
Steroid hormones are a group of lipophilic hormones that are believed to enter cells by simple diffusion to regulate diverse physiological processes through intracellular nuclear receptors. Here, we challenge this model in Drosophila by demonstrating that Ecdysone Importer (EcI), a membrane transporter identified from two independent genetic screens, is involved in cellular uptake of the steroid hormone ecdysone. EcI encodes an organic anion transporting polypeptide of the evolutionary conserved solute carrier organic anion superfamily. In vivo, EcI loss-of-function causes phenotypes indistinguishable from ecdysone-or ecdysone receptor (EcR)-deficient animals, and EcI knockdown inhibits cellular uptake of ecdysone. Furthermore, EcI regulates ecdysone signaling in a cell-autonomous manner and is both necessary and sufficient for inducing ecdysone-dependent gene expression in culture cells expressing EcR. Altogether, our results challenge the simple diffusion model for cellular uptake of ecdysone and may have wide implications for basic and medical aspects of steroid hormone studies.
Graphical Abstract
eTOC Blurb
Steroid hormones are thought to freely enter cells by simple diffusion across lipid bilayers. Okamoto et al. provide evidence that in Drosophila, the insect steroid hormone ecdysone requires a membrane transports that is a part of the evolutionary conserved SLCO family to enter receiving cells from circulation.
INTRODUCTION
Steroid hormones are a group of lipophilic hormones that regulate diverse biological processes. In mammals, glucocorticoids are primary stress hormones that regulate immune response and energy homeostasis (Sapolsky et al., 2000; Rhen and Cidlowski, 2005; Oakley and Cidlowski, 2011), and gonadal steroids control sexual maturation and reproduction (Sisk and Foster, 2004; Wilson and Davies, 2007). They are also involved in various pathological processes, including inflammatory disorders and multiple types of cancer (Clemons and Goss, 2001; Rhen and Cidlowski, 2005; Attard et al., 2009). Genomic functions of steroid hormones are mediated by intracellular nuclear receptors, which regulate the transcription of target genes in the nucleus upon binding of steroid ligands (Mangelsdorf et al., 1995; Nilsson et al., 2001; Mckenna and O’Malley, 2002; King-Jones & Thummel, 2005; Evans and Mangelsdorf, 2014). It is therefore important to understand how intracellular concentration of steroid hormones is regulated at the molecular level, both in physiological and pathological conditions.
Steroid hormones are highly conserved in both animal and plant kingdoms. In insects, the primary steroid hormone ecdysone (more specifically, its active form 20-hydroxyecdysone or 20E and related ecdysteroids) enters its target cells and binds to the ecdysone receptor (EcR), which forms a heterodimer with another nuclear receptor Ultraspiracle (Usp) and activates transcription of multiple genes involved in molting and metamorphosis (Thummel, 1996; Riddiford, 2000; Yamanaka et al., 2013).
It is widely accepted that lipophilic steroid hormones can freely enter and exit cells by simple diffusion across lipid bilayers (Nussey and Whitehead, 2001; Alberts et al, 2015; Urry et al., 2017). This view is supported by previous work on several mammalian steroid hormones (Plagemann and Erbe, 1976; Graff et al., 1977; Giorgi and Stein, 1981). Interestingly, a number of studies also suggested a potential involvement of membrane transporters in cellular uptake of steroid hormones (Milgrom et al., 1973; Rao et al., 1976; Pietras and Szego, 1977). However, a transporter-mediated, facilitated diffusion model of steroid hormone uptake has not been investigated thoroughly.
Recently, we demonstrated in Drosophila that ecdysone is released from an endocrine gland through a vesicle-mediated process, not by simple diffusion (Yamanaka et al., 2015). In the proposed model, ecdysone is loaded into vesicles at high concentrations before it is released into circulation through calcium-regulated exocytosis. As this ecdysone transport happens against the concentration gradient through an ATP-binding cassette (ABC) transporter (Yamanaka et al., 2015), our model does not address the simple diffusion model, which argues that no membrane transporter is required for steroid hormones to pass through lipid bilayers down the concentration gradient. Nonetheless, the model indicated that ecdysone can be contained within vesicles, suggesting that ecdysone cannot freely traverse lipid bilayers in vivo. This led us to hypothesize that cells receiving ecdysone may also require a membrane transporter(s) for its uptake from circulation, which happens down the concentration gradient.
Here, we report the identification and characterization of a Drosophila solute carrier (SLC) transporter, which we named Ecdysone Importer (EcI), involved in cellular uptake of ecdysone. EcI encodes an organic anion transporting polypeptide (OATP), Oatp74D, a member of the solute carrier organic anion (SLCO) superfamily. EcI is highly conserved among insects and other arthropods that utilize ecdysteroids, and its tagged protein localizes to the plasma membrane of the cells in tissues that receive ecdysone. EcI mutants show phenotypes indistinguishable from ecdysone-or EcR-deficient animals, which cannot be rescued by 20E application but can be significantly rescued by a non-steroidal ecdysone agonist. EcI is required for cellular uptake of ecdysteroids from the hemolymph in vivo and regulates ecdysone-dependent gene expression in a cell-autonomous manner. In EcR-expressing cell culture systems, EcI is both necessary and sufficient for the induction of gene expression by 20E added to the media. Collectively, our results challenge the simple diffusion model of ecdysteroid transport across cell membranes, and instead suggest a transporter-mediated, facilitated diffusion mechanism. Evolutionary conservation of SLCO superfamily in metazoans may call for a reconsideration of the simple diffusion model of steroid hormone transport beyond arthropods.
RESULTS
Genetic Screens for Potential Ecdysone Importers
Based on the hypothesis that cells receiving ecdysone from circulation need membrane transporters to facilitate its uptake, we conducted two independent genetic screens in Drosophila. First, we performed an in vivo RNAi screening of putative transporter-encoding genes. Based on the Gene Ontology (GO) terms in FlyBase (Gramates et al., 2017), we selected 537 putative transporter-encoding genes (Figure 1A; Table S1). We specifically knocked down each gene in the salivary glands and examined the effects on ecdysone-dependent expression of a glue gene (Sgs3-GFP) (Biyasheve et al., 2001; Costantino et al., 2008). Knockdown of EcR eliminates Sgs3-GFP expression and reduces the size of salivary gland cells (Figure 1B) (Costantino et al., 2008). From this screen, 51 genes were found to diminish Sgs3-GFP expression when knocked down in the salivary glands. Next, these 51 genes were subsequently tested for their effects on ecdysone-induced fat body remodeling during metamorphosis (Cherbas et al., 2003; Bond et al., 2011). Knockdown of EcR in the fat body blocks ecdysone-dependent cell migration into the pupal head (Figure 1C). From this screen, we identified a single gene, Oatp74D (CG7571), whose knockdown phenocopies EcR knockdown (Figure 1C; Table S1).
Figure 1. Identification of a Putative Ecdysone Importer through Two Independent Genetic Screens.

(A) Scheme of the in vivo RNAi screening to identify ecdysone importers. Candidate genes are listed in Table S1.
(B) Knockdown of EcR or Oatp74D causes the loss of ecdysone-dependent glue-GFP expression in salivary glands in wandering third instar larvae. fkh-Gal4 > UAS-dicer2 was used to induce RNAi in salivary glands. Scale bars, 1 mm (upper panels), 200 µ m (lower panels).
(C) Knockdown of EcR or Oatp74D in the fat body blocks ecdysone-dependent fat body cell migration into the pupal head (arrowheads). Cg-Gal4 > UAS-dicer2, UAS-2xEGFP was used to induce RNAi and label cells in the fat body. hAPF, hours after puparium formation. Scale bar, 1 mm.
(D) Schematic of the in vitro CRISPR screening in S2R+ cells to identify sgRNAs that render resistance to 20E. Cells with mutations on genes essential for 20E-dependent cell cycle arrest (shown in red and blue) are expected to be over-represented in the 20E-treated population.
(E) Gene-level CRISPR score distribution for the in vitro CRISPR screening. Computed CRISPR score (mean Log2[fold-change] of all sgRNAs targeting the same gene) for each gene from 20E-treated versus untreated populations is plotted. Size of bubbles represents number of active sgRNAs for each gene, where active sgRNAs are defined as those conferring 20E context-specific resistance in the top 2% of all sgRNAs in the screen.
Second, as an independent approach, we applied an unbiased in vitro screening strategy to identify Drosophila genes necessary for the ecdysone response. As Drosophila cell-lines are known to arrest the cell cycle in response to ecdysone treatment in an EcR-dependent manner (Koelle et al., 1991), we used a pooled CRISPR knockout screening platform in Drosophila S2R+ cells (Viswanatha et al., 2018) to screen for mutants that allow the cells to grow normally in the presence of 20E (Figure 1D). We screened 25,783 sgRNAs targeting 3,974 genes (Group 1 and Group 2 libraries in Viswanatha et al., 2018) and compared sgRNA abundance after treatment with a partially inhibitory dose of 20E (15 ng/ml; Figure S1A). Next, using next generation sequencing, we identified multiple sgRNAs that were enriched in 20E-treated but not in untreated cells, including those targeting Oapt74D and EcR (Figures 1E, S1B and S1C). Strikingly, Oatp74D was the most enriched gene after 20E treatment, followed by EcR (2nd) and usp (6th) (Figure S1D).
EcI Is an Evolutionarily-Conserved, Ubiquitously-Expressed Membrane Transporter
Identification of the same transporter-encoding gene from the two independent screens strongly suggested its critical role in cellular ecdysone uptake. Based on its putative function, we hereafter call this gene Ecdysone Importer (EcI). EcI is a member of the SLCO gene superfamily that is comprised of membrane-bound transporter proteins called OATPs that are widely present in the animal kingdom (Hagenbuch and Stieger, 2013). Studies on OATPs in fruit flies and other insects are limited, and there are few reports that describe their potential substrates (Torrie et al., 2004; Seabrooke and O’Donnell, 2013; Groen et al., 2017). Phylogenetic analysis indicated that EcI has clear orthologs in insects and other arthropods that utilize ecdysteroids (Table S2; Figures 2A and 2B) (Rewitz and Gilbert, 2008; Niwa and Niwa, 2014; Ogihara et al., 2015).
Figure 2. EcI Is a Conserved Membrane Transporter That Is Expressed Ubiquitously during Development.
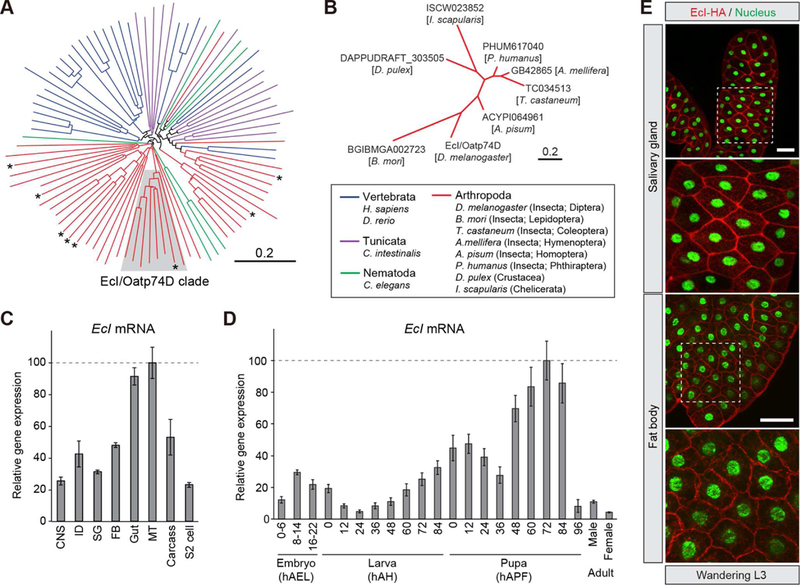
(A, B) EcI/Oatp74D has orthologs in a broad range of insects and other arthropods that utilize ecdysteroids as the molting hormone. (A) Neighbor-joining unrooted phylogenetic tree constructed using entire amino acid sequences of 8 Drosophila melanogaster Oatps (asterisks) and 82 other OATP proteins from vertebrates, a tunicate, arthropods, and a nematode. Protein names and GenBank accession numbers are listed in Table S2. Shaded area indicates the clade containing EcI/Oatp74D. (B) Neighbor-joining unrooted phylogenetic tree constructed using entire amino acid sequences of EcI/Oatp74D clade proteins that are shaded in (A). Scale bars indicate an evolutionary distance of 0.2 amino acid substitutions per position.
(C, D) EcI is expressed ubiquitously during development. (C) Relative expression levels of EcI/Oatp74D in various tissues and S2 cells, as assessed by qRT-PCR. Tissues were dissected from wandering third instar larvae (w1118). CNS, central nervous system; ID, imaginal disc; SG, salivary gland; FB, fat body; MT, Malpighian tubule. (D) Developmental changes in the relative expression level of EcI in the whole body, as assessed by qRT-PCR. Samples were collected from w1118 animals. hAEL, hours after egg laying; hAH, hours after hatching; hAPF, hours after puparium formation. Adult cDNA samples were prepared from flies at 24 hours after eclosion. Values are shown as percentages relative to the maximum level. All values are the means ± SD (n = 3).
(E) HA-tagged EcI is localized at the plasma membrane. fkh-Gal4 or Cg-Gal4 > UAS-EcI-HA wandering third instar larvae were immunostained for HA (red) and nuclei (green). Scale bars, 100 µm.
EcI is expressed ubiquitously with temporal fluctuations during development (Figures 2C and 2D), consistent with the systemic function of ecdysone in controlling stage-specific gene expression (Li and White, 2003; Yamanaka et al., 2013). Compared to the other seven Drosophila OATP-encoding genes, EcI showed dominant expression in most tissues except the Malpighian tubules, in which Oatp58Da, 58Db and 58Dc are dominantly expressed (Figure S2). Silencing the other Drosophila OATP-encoding genes in the salivary glands and fat body showed no discernible defect (Table S3), suggesting a specific role of EcI in ecdysone action. Expression of HA-tagged EcI in the salivary glands and fat body confirmed its primary localization to the plasma membrane (Figure 2E), suggesting that EcI indeed has important functions in transporting ecdysone across cell membranes.
EcI Is Essential for the Larval Developmental Transition
To investigate the loss-of-function phenotype of EcI, we generated two independent EcI null mutants (EcI1 and EcI2) using CRISPR/Cas9-mediated mutagenesis (Figure S3). Although EcI mutants show no discernible defects during embryogenesis and the first 24 hours after hatching (hAH), they show developmental arrest at the end of the first instar larval stage (Figures 3A, 3B, and S4), suggesting a lack of ecdysone signaling required for normal molting. About 5–10% of arrested larvae carry both first and second instar mouth hooks (Figures 3C and S4), a phenotype characteristic of defective ecdysone signaling (Schubiger et al., 1998; Gaziova et al., 2004). Weak ubiquitous expression of EcI using armadillo (arm)-Gal4 is sufficient to rescue the developmental arrest phenotype (Figures 3 and S4), whereas strong ubiquitous overexpression of EcI causes lethality during early development (Table S4), suggesting that appropriate levels of EcI expression are important for normal development.
Figure 3. EcI Is Required for the Larval Developmental Transition.

(A) Developmental changes in body size of control (w1118), EcI mutant (EcI1/EcI2) and EcI mutant rescued by weak ubiquitous expression of EcI (arm-Gal4 > UAS-EcI; EcI1/EcI2). Representative images of animals at different stages were combined into a single panel. hAH, hours after hatching. Scale bar, 1 mm.
(B) Developmental progression and survival rate (%) of control (w1118), EcI mutants (EcI1/EcI1, EcI2/EcI2, and EcI1/EcI2) and EcI transheterozygous mutant rescued by weak ubiquitous expression of EcI (arm-Gal4 > UAS-EcI; EcI1/EcI2). Color bars indicate percentages of first instar larvae (white), second instar larvae (blue), third instar larvae (green), and prepupae/pupae (yellow). Percentages of arrested first instar larvae with double mouth hooks (DM) are shown in light blue. Larval stages were determined by stage specific morphology of larval mouth hooks and posterior spiracles (Figure S4).
(C) Larval mouth hook morphology of control (w1118), EcI transheterozygous mutant (EcI1/EcI2) and EcI transheterozygous mutant rescued by weak ubiquitous expression of EcI (arm-Gal4 > UAS-EcI; EcI1/EcI2) at 96 hAH. Arrows indicate second instar larval mouth hooks observed in the double mouth hooks larva (right lower panel). Scale bars, 25 µm.
In order to further demonstrate the critical role of EcI in ecdysone signaling, we attempted to rescue the larval arrest phenotype of EcI mutants by oral administration of two different compounds (Figure 4A): an endogenous EcR ligand 20E and a non-steroidal ecdysone agonist chromafenozide (CF), which is known to bind directly to Drosophila EcR and mimic 20E effect (Minakuchi et al., 2005). We developed a transient feeding scheme for the administration of these compounds in order to mimic the endogenous ecdysone pulse during the first instar larval stage (Figure 4B). Consistent with our hypothesis that EcI functions as an ecdysone importer in peripheral tissues, feeding EcI mutants with 20E fails to rescue the first instar arrest (Figures 4C and 4D). We also used CF in an alternative rescue experiment, expecting that this non-steroidal, structurally different ecdysone agonist enters peripheral tissues through molecular machinery distinct from EcI. Indeed, CF significantly rescues the developmental arrest phenotype (Figures 4C and 4D), suggesting that EcR function remains intact in EcI mutants.
Figure 4. Developmental Arrest Phenotype of EcI Mutant Can Be Rescued by a Non-Steroidal Ecdysone Agonist.
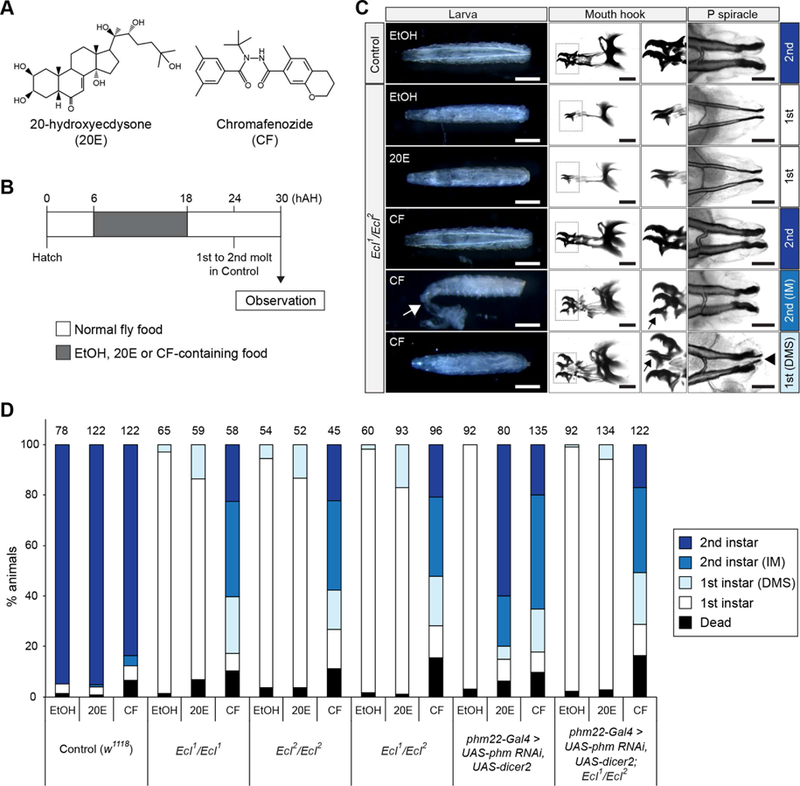
(A) Structures of the endogenous EcR ligand 20-hydroxiecdysone (20E) and a non-steroidal ecdysone agonist chromafenozide (CF).
(B) Transient compound feeding scheme. Larvae were fed on yeast paste with 2% ethanol (EtOH) with or without 1 mM (final concentration) 20E or CF from 6 to 18 hAH. Larvae were then cultured on normal food for an additional 12 hours and larval morphology was observed at 30 hAH. About 80–85% of control larvae complete first-to-second instar molting at 24 hAH.
(C) Representative images of mouth hooks and posterior (P) spiracles of control (w1118) and EcI mutant (EcI1/EcI2) larvae at 30 hAH after 20E or CF feeding. Enlarged mouth hook images correspond to the boxed areas. IM, incomplete molting, where the first instar cuticle remains attached (white arrow). DMS, double mouth hooks and spiracles. Black arrows indicate first instar mouth hooks; arrowhead indicates first instar posterior spiracles. Scale bars, 500 µm (white) and 50 µm (black).
(D) Developmental stages of control (w1118), EcI mutants (EcI1/EcI1, EcI2/EcI2 and EcI1/EcI2), ecdysone-deficient larvae (phm22-Gal4 > UAS-phm RNAi, UAS-dicer2) and ecdysone-deficient larvae in EcI mutant background (phm22-Gal4>UAS-phm RNAi, UAS-dicer2; EcI1/EcI2) after transient feeding of 20E or CF. phm22-Gal4 > UAS-dicer2 was used as a prothoracic gland-specific Gal4 driver. Color bars indicate percentages of first instar larvae (white), first instar larvae with the DMS phenotype (light blue), second instar larvae with IM phenotype (blue), and second instar larvae (dark blue). Numbers of animals observed are shown on top of each bar.
To confirm the step at which EcI is important for ecdysone signaling, we examined the effects of 20E or CF feeding on the EcI mutant with defective ecdysone production. We used one of the “Halloween” genes, which were initially identified as mutants devoid of ecdysone-dependent cuticle deposition during embryogenesis (Gilbert, 2004). The Halloween gene phantom (phm) encodes a cytochrome P450 enzyme essential for ecdysone production in the prothoracic gland, a primary steroidogenic tissue in larval insects (Niwa et al., 2004; Warren et al., 2004). Although the EcI mutant phenotype resembles that of phm RNAi (Danielsen et al., 2016; Ou et al., 2016), the developmental arrest of phm RNAi larvae can be rescued by either 20E or CF application (Figure 4D). In clear contrast, only CF rescues the arrest phenotype of phm RNAi larvae in the EcI mutant background (Figure 4D), demonstrating that EcI functions downstream of ecdysone production but upstream of EcR action.
EcI Functions Cell-Autonomously and Facilitates Cellular Uptake of Ecdysteroids in Vivo
To further examine the functions of EcI in vivo, we analyzed ecdysone-inducible gene expression in the fat body of EcI and EcR knockdown animals. We first confirmed that both EcI and EcR expression levels were significantly reduced in the fat body of EcI and EcR knockdown animals, respectively (Figure S5A). E74A, E75A and E75B are ecdysone-inducible genes whose expression is directly induced by high levels of ecdysone (Karim and Thummel, 1992). When ecdysone is at its basal level (72 hAH), expression levels of these genes are comparable to controls in the fat body of both EcI and EcR knockdown animals (Figure 5A). However, during the white prepupal stage (96 hAH), when ecdysone titer is at its peak, their expression levels are significantly reduced (Figure 5A).
Figure 5. EcI Cell-Autonomously Regulates Ecdysone Signaling and Facilitates Cellular Uptake of Ecdysteroids in Vivo.
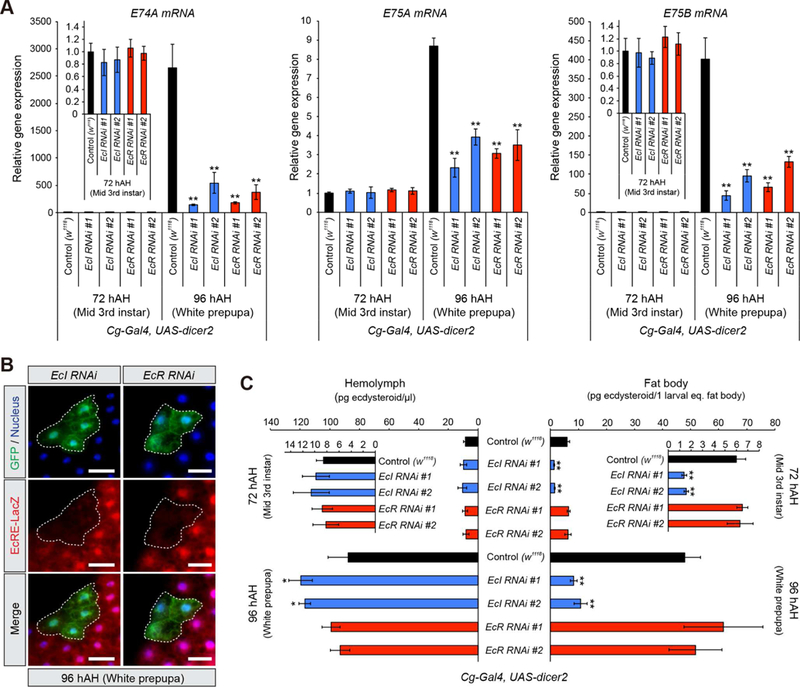
(A) Relative expression levels of ecdysone-inducible genes (E74A, E75A and E75B) in the fat body at 72 and 96 hAH, as assessed by qRT-PCR. Two independent UAS-RNAi lines for EcI and EcR were used. Cg-Gal4 > UAS-dicer2 was used as a fat body-specific Gal4 driver. Values were calculated relative to the expression level of each gene at 72 hAH in control.
(B) Cell-autonomous requirement of EcI and EcR for ecdysone signaling in the fat body. Clones of fat body cells expressing EcI-RNAi #1 or EcR-RNAi #1 with dicer2 were labeled with GFP. EcRE-LacZ reporter gene was used to monitor the activity of ecdysone signaling. hs-flp;; Act>CD2>GAL4, UAS-nlsGFP was used to generate GFP-marked flip-out clones. Flippase activity was induced by 10 min heat shock. The fat body from white prepupae at 96 hAH was immunostained for LacZ (red), GFP (green) and nuclei (blue). Scale bars, 50µm.
(C) Quantification of ecdysteroids in the hemolymph and fat body at 72 and 96 hAH, as assessed by ELISA. UAS-RNAi and Gal4 lines used were the same as in (A).
All values are the means ± SD (n = 3). *p < 0.05, **p < 0.01 from Student’s t test compared to control.
We further examined cell-autonomous effects of EcI knockdown using the FLP-out clonal analysis technique. Consistent with the reduced cell size in salivary gland-specific EcI and EcR knockdown animals (Figure 1B), we found that salivary gland cell sizes are significantly reduced when EcI or EcR levels are reduced by RNAi (Figure S5B). The ecdysone response element (EcRE)-driven LacZ reporter expression is also clearly reduced in both EcI and EcR RNAi clones in the fat body (Figure 5B). These results demonstrate that EcI regulates ecdysone signaling in a cell-autonomous manner. Importantly, neither EcR protein level nor its localization is affected in EcI RNAi clones (Figure S5C).
If EcI is required for cellular uptake of ecdysone from the circulation in vivo, ecdysone levels in target cells would be reduced by downregulation of EcI. To test this, we measured the amount of ecdysteroids in both the hemolymph and fat body of fat body-specific EcI knockdown animals. We found that the amount of ecdysteroids in the fat body is significantly reduced in EcI knockdown animals, but not in EcR knockdown animals (Figure 5C). Ecdysteroid concentration in the hemolymph is not reduced in either EcI or EcR knockdown animals, but rather is slightly increased in EcI knockdown animals (Figure 5C). Taken together, these results indicate that EcI functions as an ecdysone importer in vivo.
EcI Is Both Necessary and Sufficient for EcR-Mediated Ecdysone Action in Culture Cells
In order to further confirm the function of EcI in ecdysone action, a luciferase (Luc) reporter construct with EcRE (EcRE-Luc) and EcR were co-transfected into Drosophila S2 cells, in which EcI is highly expressed (Figure S2). Altering EcI levels in S2 cells by RNAi or overexpression significantly changes dose-and incubation time-dependent induction of Luc activity by 20E (Figures 6A-6C). Although its ligand activity is much lower than that of 20E (Baker et al., 2000), the effect of the 20E precursor ecdysone is also significantly affected (Figures 6A and 6B), suggesting that both 20E and ecdysone are substrates for EcI. Importantly, neither knockdown nor overexpression of EcI in S2 cells affects Luc activity induced by CF (Figures 6A and 6B), consistent with the earlier observation that CF does not require EcI to enter cells (Figure 4).
Figure 6. EcI Is Both Necessary and Sufficient for Ecdysone Action in EcR-Expressing Cells.
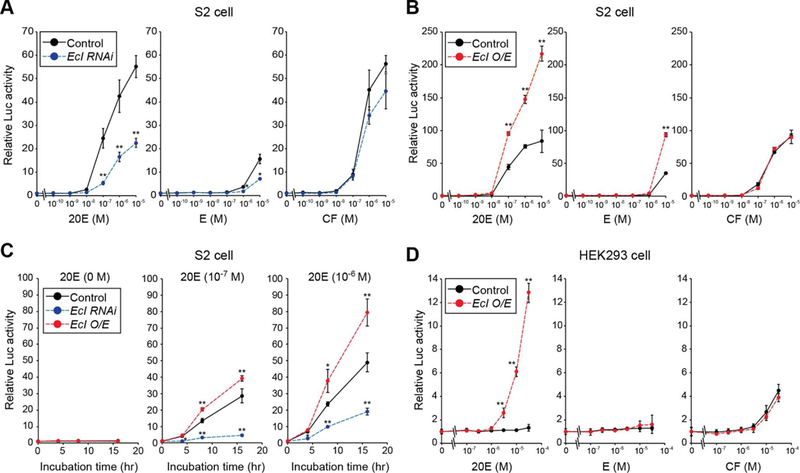
(A, B) Luciferase (Luc) reporter activity was monitored in Drosophila S2 cells treated with EcI double-stranded RNA (A) or overexpressing EcI (B) after 24-hour treatment with 20E, ecdysone (E) or CF. S2 cells were co-transfected with EcREx3-Luc and EcR. usp is endogenously highly-expressed in S2 cells (Baker et al., 2000). O/E, overexpression.
(C) Luc reporter activity in S2 cells treated for 0, 4, 8 or 16 hours with 10−7 M or 10−6 M of 20E.
(D) Luc activity in HEK293 cells treated for 24 hours with 20E, E or CF. EcREx5-Luc was used as a reporter construct. HEK293 cells overexpressing modified EcR (VgEcR) and co-receptor RXR were used for the assay.
Values are relative to the control level (0 M or 0 hour). All values are the means ± SD (n = 3). *p < 0.05, **p < 0.01 from Student’s t test compared to control.
Lastly, we used an ecdysteroid-inducible gene expression system in HEK293 cells to test whether EcI is sufficient for EcR-mediated ecdysone action in a heterologous system (Christopherson et al., 1992; No et al., 1996; Saez et al., 2000). As reported previously (Christopherson et al., 1992; Saez et al., 2000; Dinan and Lafont, 2006), Luc activity is poorly induced by 20E when EcRE-Luc and EcR alone are transfected (Figure 6D). However, when EcI is co-transfected, 20E-inducible Luc activity is dramatically increased, whereas there is no effect on Luc activity induced by CF (Figure 6D). Taken together, our in vitro analyses clearly indicate that EcI is both necessary and sufficient for ecdysone-inducible gene expression in animal cells.
DISCUSSION
Genetic Screens Identify an Evolutionary Conserved Membrane Transporter Necessary for the Ecdysone Response
We conducted two independent in vivo and in vitro genetic screens in Drosophila and identified a single common gene, EcI, as a critical component for ecdysone response. The in vivo screen was focused on putative transporter-encoding genes in the Drosophila genome, whereas the in vitro screen was an unbiased approach to identify genes involved in the ecdysone response. These two complementary approaches provided compelling evidence that EcI is equally indispensable for ecdysone signaling as the nuclear receptors, EcR and Usp. EcI/Oatp74D is a member of the evolutionary conserved SLCO superfamily of SLC transporters, encoding OATPs with 12 predicted transmembrane domains (Hagenbuch and Stieger, 2013). OATPs are membrane influx transporters that mediate ATP-independent cellular uptake of various endogenous substrates, as well as drugs and other xenobiotics. This mode-of-action of OATPs is consistent with our model (Figure 7), whereby EcI on the plasma membrane transports ecdysone into the cytoplasm in a concentration-dependent manner.
Figure 7. Facilitated Diffusion Model of Ecdysteroid Uptake by Target Cells.
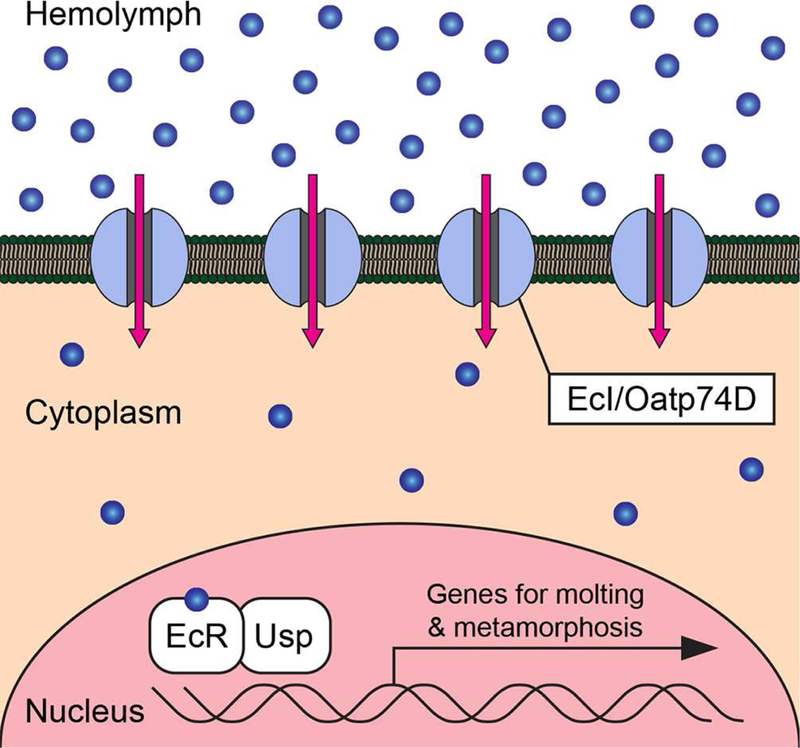
EcI/Oatp74D is required for facilitating cellular uptake of ecdysteroids (blue circles) from the hemolymph into cytoplasm down the concentration gradient. The nuclear receptor complex composed of EcR and Usp regulates the transcription of genes for molting and metamorphosis in the nucleus.
Facilitated Diffusion Is Necessary for Cellular Uptake of Ecdysone
Biology textbooks state that steroid hormones are lipophilic and therefore do not require membrane transporters to traverse phospholipid bilayers (Nussey and Whitehead, 2001; Alberts et al, 2015; Urry et al., 2017). Although the simple diffusion model of steroid hormone transport has been tested for several mammalian steroid hormones in various cell types (Plagemann and Erbe, 1976; Graff et al., 1977; Giorgi and Stein, 1981), the results were often variable and even controversial, leaving the possibility that membrane transporters may be involved in cellular uptake of steroid hormones (Milgrom et al., 1973; Rao et al., 1976; Pietras and Szego, 1977; Yamamoto, 1985). However, despite these studies, no steroid hormone importers have been shown to be important in regulating the physiological functions of steroid hormones in vivo.
It is noteworthy that transporter-mediated steroid hormone trafficking across cell membranes have been demonstrated in many biological systems (Klaassen and Aleksunes, 2010). In most cases, however, transmembrane transport of steroid hormones occurs against their concentration gradient, such as when cells actively eliminate steroid hormones out of the cytoplasm (Kralli et al., 1995; Hock et al., 2000; Karssen et al., 2001; Uhr et al., 2002; Pariante et al., 2004) or load high concentrations of hormones into vesicles (Yamanaka et al., 2015). Such active transport necessarily requires energy, which is often provided by ABC transporters that can couple ATP hydrolysis to substrate transfer (Dean et al., 2001; Rees et al., 2009). These reports do not address the simple diffusion model of steroid hormone transport that involves passive transport of steroid hormones down the concentration gradient across lipid bilayers. In contrast, our current results demonstrate the requirement of EcI for facilitating cellular uptake of ecdysone down the concentration gradient, which clearly challenges the simple diffusion model at least in Drosophila (Figure 7). Given that ecdysteroids are ubiquitously employed as signaling molecules in arthropods and EcI is highly conserved in this phylum, we propose that the transporter-mediated steroid hormone uptake occurs in all arthropod species.
In EcI null mutants, we did not observe any significant embryonic lethality, which is observed in so-called Halloween mutants that cannot produce active ecdysteroids during embryogenesis (Gilbert, 2004; Niwa and Niwa, 2014). Some arrested EcI mutant first instar larvae also showed the double mouth hook phenotype (Figures 3 and S4), indicating defective but low-level ecdysone signaling. Such incomplete block of ecdysone signaling in EcI mutants may either come from maternally-loaded EcI and/or ecdysteroids, or other unknown ecdysone importers that have partially-redundant functions with EcI. Although further studies are warranted to completely eliminate the possibility of simple diffusion of ecdysteroids, our results nonetheless clearly demonstrate that facilitated diffusion mechanism is required for most, if not all, physiological functions of ecdysteroids in vivo.
Although the simple diffusion model has been predominant for all steroid hormones to date, it is highly desirable from a physiological perspective to regulate their cellular uptake, such that cells can adjust the intracellular concentration of steroid hormones. Consistent with this, complete lethality was observed during early development when EcI was overexpressed using strong ubiquitous drivers (Table S4), suggesting that intracellular concentration of ecdysone needs to be regulated at its initial entry into cells. Transgenic rescue of the EcI mutant with a weak ubiquitous driver was significant but incomplete (Figures 3 and S4), further suggesting that the cellular uptake of ecdysone needs to be precisely controlled in different tissues and/or at distinct stages of development. To further investigate this possibility, it will be important to examine the mechanisms through which expression and subcellular localization of EcI is controlled in vivo.
OATPs and Transmembrane Transport of Mammalian Steroid Hormones
Do mammalian steroid hormones also require membrane transporters to diffuse across cell membranes? A key point for consideration is the hydrophobicity of steroid hormones: ecdysone has multiple hydroxyl groups on its carbon backbone, making it relatively more hydrophilic than mammalian steroid hormones. From this perspective, it is conceivable that ecdysone is more similar to other cholesterol-derived substances in mammals, such as bile acids and steroid hormone conjugates. Indeed, mammalian OATPs are known to transport these relatively hydrophilic cholesterol-derivatives across cell membranes (Hagenbuch and Stieger, 2013). Importantly, however, there have been several studies reporting potential functions of OATPs in transmembrane transport of mammalian steroid hormones per se, either in heterologous or pathological conditions (Bossuyt et al., 1996; Hamada et al., 2008; Wright et al., 2011; Obaidat et al., 2012; Green et al., 2017). It is intriguing to hypothesize that these OATPs have redundant functions in mammalian steroid hormone transport, which may explain why their roles in physiological functions of steroid hormones in vivo have been ignored. As our current study clearly indicates the necessity of a membrane transporter in ecdysone signaling, it will be important to investigate the roles of OATPs in regulating the physiological functions of mammalian steroid hormones in vivo.
In conclusion, our results suggest that transporter-mediated, facilitated diffusion is necessary for cellular uptake of steroid hormones in at least one model organism. Our study thus calls for a reconsideration of the simple diffusion model of steroid hormones, which is commonly described in major biology textbooks (Nussey and Whitehead, 2001; Alberts et al, 2015; Urry et al., 2017). It is also noteworthy that OATPs have been recognized as potential drug targets for cancer (Obaidat et al., 2012), and several chemical reagents have already been identified to regulate functions of these membrane transporters (Kalliokoski and Niemi, 2009; El-Gebali et al., 2013). Considering the diverse biological processes regulated by steroid hormones, further investigation of the facilitated diffusion model of steroid hormones may have a tremendous impact on both basic and medical aspects of steroid hormone research.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Flies
All flies were raised at 25°C under 12 hours/12 hours light/dark cycle. The animals were reared on standard fly food containing 6 g Drosophila agar type II (Genesee Scientific, #66–103), 100 g D-(+)-glucose (SIGMA, #G8270–25KG), 50 g inactive dry yeast (Genesee Scientific, #62–106), 70 g yellow corn meal (Genesee Scientific, #62–101), 6 ml propionic acid (SIGMA, #402907–500ML), and 10 ml Tegosept (Genesee Scientific, #20–258) in 1,025 ml of water. All the experiments were conducted under non-crowded conditions.
The control strain was w1118, and transgenic flies are as follows: Act5C-Gal4 (#3954 and #4414), arm-Gal4 (#1560 and #1561), Cg-Gal4 (#7011), da-Gal4 (#55850), TubP-Gal4 (#5138), UAS-EcR RNAi #1 (#9327), UAS-Oatp33Ea RNAi (#50736), UAS-Oatp58Dc RNAi (#44583), UAS-2XEGFP (#6874), EcRE-LacZ (#4516 and #4517) and Sgs3-GFP (#5885) were obtained from the Bloomington Drosophila Stock Center (BDSC); UAS-dicer2 (#60008 and #60009), UAS-EcI (Oatp74D) RNAi #2 (#37295), UAS-EcR RNAi #2 (#37058), UAS-Oatp26F RNAi (#2650 and #109633) , UAS-Oatp30B RNAi (#22983 and #110237), UAS-Oatp33Ea RNAi (#105560), UAS-Oatp33Eb RNAi (#42805 and #100431), UAS-Oatp58Da RNAi (#44122 and #106377), UAS-Oatp58Db RNAi (#100348), UAS-Oatp58Dc RNAi (#39469) and UAS-phm RNAi (#108359) were obtained from the Vienna Drosophila Resource Center (VDRC); UAS-EcI (Oatp74D) RNAi #1 (#7571R-1) and UAS-Oatp58Db RNAi (#HMJ24090) were obtained from the National Institute of Genetics Fly Stock Center. phm22-Gal4 and fkh-Gal4 were obtained from Michael B. O’Connor. hs-flp;; Act>CD2>Gal4, UAS-nlsGFP was obtained from Thomas P. Neufeld. All the RNAi lines used for the genome-wide in vivo RNAi screening are shown in Table S1.
UAS-EcI was generated from cDNA clone GH24467 from the Drosophila Genomics Resource Center (DGRC) and cloned into pUAST vector. UAS-EcI-Flag-HA was generated from cDNA clone UFO0685 from DGRC. EcI1 and EcI2 were generated by the CRISPR/Cas9 system as described in detail below. All new transgenic flies were generated by BestGene Inc.
Cell lines
S2R+ cells containing an attP site and expressing Cas9 used for CRISPR screening (Viswanatha et al., 2018) were maintained in a humidified incubator at 25°C in Schneider’s Drosophila medium (Thermo Fisher Scientific, #21720–024-500ML) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, #10082147) and 1% Penicillin-streptomycin solution (PSS; Thermo Fisher Scientific, #15140–122).
S2 cells used for luciferase reporter assays were obtained from Michael B. O’Connor and maintained in 100 mm x 20 mm Petri dishes (Corning, #430167) in a humidified incubator at 25°C in Shields and Sang M3 (SSM3) insect medium (Sigma-Aldrich, #S3652–500ML) containing 10% Insect Medium Supplement (IMS) (Sigma-Aldrich, #I7267–100ML) and 1% PSS (Thermo Fisher Scientific).
HEK293 cells obtained from Michael E. Adams were maintained in 100 mm x 20 mm Petri dishes (Corning) in a humidified incubator at 37°C and 5% CO2 in Dulbecco’s Modification of Eagle’s Medium (DMEM) with 4.5 mg/ml glucose, L-glutamine, and sodium pyruvate (Corning, #10–013-CV) containing 10% FBS (Corning, #10082–139) and 1% PSS (Thermo Fisher Scientific). Before seeding cells to new plates, they were washed with Dulbecco’s phosphate-buffered saline (Thermo Fisher Scientific, #21–030-CV) and treated with 0.05% Trypsin-EDTA (Thermo Fisher Scientific, #25–052-CI).
METHOD DETAILS
Genome-wide in vivo RNAi screening
Seven virgin females from Gal4 lines (UAS-dicer2; fkh-Gal4, Sgs3-GFP for the first screening, Cg-Gal4, UAS-dicer2, UAS-2xEGFP for the second screening) were crossed with four males from each transgenic UAS-RNAi line (see Table S1 for details), and allowed to lay eggs on standard food in vials for 24–48 hours. For UAS-RNAi lines on the X chromosome, virgin females were collected from RNAi lines and crossed to males from Gal4 lines. Two replicates were collected from the offspring of each cross. Each batch of 50 genetic crosses included a negative and positive control: Gal4 lines crossed to w1118 or UAS-EcR RNAi (BDSC #9327), respectively. In the first screening, phenotypes of F1 offspring were analyzed by observing Sgs3-GFP expression in the salivary glands of wandering third instar larvae. The number of animals (per 25 animals) that showed defects in GFP expression was recorded, and RNAi lines for 51 genes were found to diminish GFP expression in one or more animals. In the second screening, phenotypes of F1 offspring were analyzed by observing defects in GFP-labeled fat body cell migration at 24–36 hours after puparium formation. In this screening, most crosses showed phenotypes other than or in addition to defects in fat body cell migration, such as growth defects and melanization. Since the positive control pupae (Gal4 line crossed to UAS-EcR RNAi) showed a fat body cell migration defect in all animals without any other discernible defect, we used these criteria to evaluate the RNAi lines. This screen identified only one RNAi line, which targets Oatp74D. GFP signals in each larva or pupa on plastic vial walls were observed using a SteREO Discovery.V12 microscope (Zeiss).
In vitro CRISPR screening
CRISPR screens were conducted as described (Viswanatha et al., 2018). S2R+ cells containing an attP site and expressing Cas9 were transfected with the Group 1 and Group 2 attB donor libraries containing 25,783 gene-targeted sgRNAs and 391 control sgRNAs under the Drosophila U6.2 promoter along with an expression vector for the phiC31 integrase (pBS130 from Addgene; Gohl et al., 2011) at a 1:1 molar ratio to mediate recombination. Transfection of S2R+ cells was performed using Effectene transfection reagent (Qiagen) following the manufacturer’s instructions. Approximately 1,500 cells per sgRNA were recombined and >1,500 cells per sgRNA were passaged for 15 days to induce knockout and monitored for percentage of green cells using flow cytometry. Culture was then divided, and half of the cells received 15 ng/ml of 20E (Sigma-Aldrich) while other half was left untreated. Following 30 days, genomic DNA was extracted (Zymo Research) and sgRNA PCR was performed at a sampling of > 1,000 genomes per sgRNA. Next generation sequencing was performed using a NextSeq 500 (Illumina) at the Harvard Biopolymers Facility. Readcounts were demultiplexed and counted using TagDust (Lassmann et al., 2009) and MAGeCK count (Li et al., 2014), respectively. Log2 fold-changes were calculated relative to normalized plasmid readcount values for all CRISPR sgRNAs for which initial readcounts were in the top ninety-fifth percentile (Viswanatha et al., 2018). For 20E resistance screen, two technical replicates were performed. To generate sgRNA-level mean, readcounts from each replicate screen were median-adjusted prior to computing means allowing balanced comparisons. The gene-level CRISPR score is defined as the mean of all log2 fold-changes for all sgRNAs targeting the same gene, as described in Wang et al. 2014.
Cell titer measurements following 20E treatment were conducted using CellTiter-Glo assay (Promega) following the manufacturer’s instructions, and analyzed on a Paradigm luminometer (SpectraMax). Luminescence was normalized to untreated wells.
Phylogenetic tree analysis
We generated unrooted neighbor-joining trees using ClustalW (DNA Data Bank of Japan). We aligned the entire amino acid sequences of 8 Drosophila melanogaster Oatps, including EcI/Oatp74D, and 82 OATP proteins from vertebrates (Human, Homo sapiens; Zebrafish, Danio rerio), tunicate (Sea squirt, Ciona intestinalis), arthropods [holometabolous insects (Silk moth, Bombyx mori; red flour beetle, Tribolium castaneum; western honey bee, Apis mellifera), hemimetabolous insects (pea aphid, Acyrthosiphon pisum; human body louse, Pediculus humanus), crustacean (water flea, Daphnia pulex), chelicerate (black-legged tick, Ixodes scapularis)] and nematode (round worm, Caenorhabditis elegans). The protein names and GenBank accession numbers are listed in Table S2.
Total RNA extraction and quantitative reverse transcription (qRT)-PCR
Animals or dissected tissues were collected in 1.5 ml tubes and immediately flash-frozen in liquid nitrogen. Total RNA from animals or tissues was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Extracted RNA was further purified by RNeasy mini kit (Qiagen) following the manufacturer’s instructions, combined with treatment with RNase-Free DNase Set (Qiagen). cDNA was generated from purified total RNA using PrimeScript RT Master Mix (Takara Bio). qRT-PCR was performed on the CFX connect real-time PCR detection system (Bio-Rad) using SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara Bio). For absolute quantification of mRNAs, serial dilutions of pGEM-T (Promega) plasmids containing coding sequences of the target genes or rp49 were used for standards. After the molar amounts were calculated, transcript levels of the target mRNA were normalized to rp49 levels in the same samples. Three separate samples were collected for each experiment and duplicate measurements were conducted. The primers used are listed in Table S5.
Immunostaining
Tissues were dissected in 1X phosphate-buffered saline (PBS) (Fisher BioReagents), fixed with 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) in PBS (4% PFA/PBS) containing 0.1% TritonX-100 for 20 min at room temperature (RT), and washed multiple times with PBS containing 0.1% TritonX-100 (PBST). Tissues were blocked with 5% normal goat serum (NGS) (Sigma-Aldrich) in PBST for at least 1 hour at RT, incubated overnight at 4°C with the primary antibody mix in PBST containing 5% NGS, washed multiple times with PBST, incubated for 2 hours at RT with the secondary antibody mix in PBST containing 5% NGS, and washed multiple times again with PBST. When needed, DNA was stained with Hoechst 33342 (Life Technologies) at 1:2000 and F-Actin was stained with Alexa Fluor 546 Phalloidin (Life Technologies) at 1:200 for 30 min at RT. After washing, tissues were mounted in Vectashield H-1000 (Vector Laboratories) and observed using a Zeiss Axio Imager M2 equipped with ApoTome.2. Specificity of the signals was established by comparison to appropriate controls.
Primary antibodies used were rat anti-HA (3F10, 1:1000, Roche), mouse anti-beta-galactosidase (40–1a, 1:500, DSHB), mouse anti-EcR (Ag10.2, 1:200, DSHB), and chick anti-GFP (1:500, Abcam). Secondary antibodies used were Alexa Fluor 488 goat anti-chicken (1:500, Life Technologies), Alexa Fluor 546 goat anti-rat (1:500, Life Technologies) and Alexa Fluor 546 goat anti-mouse (1:500, Life Technologies).
Generation of EcI mutants
EcI mutant alleles (EcI1 and EcI2) were generated by the CRISPR/Cas9 system. Pairs of gRNA target sequences (20 bp) were designed near the EcI translational start and stop sites using NIG-FLY Cas9 Target finder (NIG). EcI1 and EcI2 were generated with target pair-I (T1 and T2) and target pair-II (T3 and T4), respectively (Figure S3). Forward and reverse 24-bp oligonucleotides with 20-bp target sequences (Table S5) were annealed to generate a double-strand DNA with 4-bp overhangs on both ends and inserted into BbsI-digested pBFv-U6.2 or pBFv-U6.2B vector provided by the NIG (Kondo and Ueda, 2013). To construct a double-gRNA vector, the first gRNA (T1 or T3) was cloned into pBFv-U6.2 (named pBFv-U6.2-T1 and pBFv-U6.2-T3), whereas the second gRNA (T2 or T4) was cloned into pBFv-U6.2B (named pBFv-U6.2B-T2 and pBFv-U6.2B-T4). A fragment containing the U6 promoter and the first gRNA was cut out from pBFv-U6.2-T1 and pBFv-U6.2-T3, and ligated into pBFv-U6.2B-T2 and pBFv-U6.2B-T4, respectively (named pBFv-U6.2B-T1-T2 and pBFv-U6.2B-T3-T4). These two EcI double-gRNA vectors (pBFv-U6.2B-T1-T2 and pBFv-U6.2B-T3-T4) were independently injected into embryos of yw; nos-cas9 (II-attP40)/CyO flies (BestGene Inc).
For each target pair, surviving G0 males were divided into 10 groups and crossed en masse to TM2/TM6b-Dfd-EGFP (obtained from Takashi Nishimura) virgins. From the progeny of each of these ten crosses, 10 single males were isolated and crossed independently to TM2/TM6b-Dfd-EGFP virgins to establish independent isogenized lines. Among these 100 lines, those showing homozygous lethality (71 and 27 lethal lines from target pair-I and II, respectively) were isolated. To confirm deletions at the EcI locus, we randomly selected 20 lines from each target pair and performed genome DNA extraction and PCR amplification by using primers listed in Table S5. Out of 20 lines, 15 lines from target pair-I and 7 lines from target pair-II possessed deletion mutations in EcI. The PCR products from 5 lines from each target pair were sequenced, and we confirmed complete deletions in all lines. We chose 1 line for each target pair for further analyses and named them EcI1 and EcI2. EcI1 has a 3207-bp deletion including the entire EcI protein coding DNA sequence (CDS), and EcI2 has a 3858-bp deletion including the 5’ untranslated region and almost the entire EcI CDS (Figure S3).
Scoring of developmental progression of EcI mutants
Eggs were laid on grape juice plates with yeast paste at 25°C for 6 hours. After 24 hours, early first instar larvae just after hatching were collected (homozygous mutant larvae were collected by selecting those without the balancer chromosome with GFP), and transferred into vials with standard food (less than 50 animals/vial). Every 24 hours, developmental stages were scored by stage-specific morphology of larval mouth hooks and posterior spiracles.
Pulsed compound feeding assay with 20-hydroxyecdysone and chromafenozide
Eggs were laid on grape juice plates with yeast paste at 25°C for 6 hours. After 24 hours, early first instar larvae just after hatching were collected (homozygous mutant larvae were collected by selecting those without the balancer chromosome with GFP), and transferred into vials with standard food (less than 50 animals/vial). After 6 hours of incubation at 25°C, larvae were transferred and cultured on agar plates with yeast paste containing 2% ethanol (Decon) (for control), 1 mM 20E (Sigma-Aldrich), or 1 mM chromafenozide (Sigma-Aldrich) in 2% ethanol for 12 hours at 25°C. After incubation, larvae were washed and transferred into vials with standard food. After 12 hours of incubation at 25°C, developmental stages were scored by stage-specific morphology of larval mouth hooks and posterior spiracles.
Induction of RNAi clones in the fat body and salivary gland
To induce FLP-out RNAi clones, hs-flp; Act5C>CD2>Gal4, UAS-nlsGFP flies (obtained from Thomas P. Neufeld) were crossed to relevant UAS-RNAi lines, and early first instar larvae were heat shocked at 37°C for either 5 min or 30 min. About 10% of fat body cells and 3–5% of salivary gland cells showed GFP expression after 5-min heat shock, and about 90% of fat body cells and 80% of salivary gland cells showed GFP expression after 30-min heat shock.
Measurement of ecdysteroid concentration
Fat bodies were carefully dissected from 5 mid-third instar larvae (72 hours after hatching, hAH) or white prepupae (96 hAH) in 4% PFA/PBS, briefly rinsed with 4% PFA/PBS, and pooled in 200 µ l of methanol (Fisher chemical, A452–4) on ice. The fat bodies were thoroughly homogenized using pestles with a micro-tube homogenizer. After centrifugation at 4°C for 10 min, the supernatant was pooled on ice, while the pellet was re-extracted by 100 µ l of methanol. The resulting extract was stored at −20°C until use. For hemolymph samples, mid-third instar larvae (72 hAH) or white prepupae (96 hAH) were rinsed in PBS, and dried on tissue paper. The cuticle was carefully torn to release the hemolymph onto a parafilm membrane. 2 µl of hemolymph were collected from 5 mid-third instar larvae or white prepupae and mixed with 200 µ l of methanol on ice. After vortexing, samples were centrifuged at 4°C for 10 min, and the resulting supernatant was stored at −20°C until use.
The sample solutions were dried with a CentriVap concentrator (Labconco) and dissolved in EIA buffer (Cayman Chemical). 20E AChE tracer, 20E EIA antiserum, Precoated (Mouse Anti-Rabbit IgG) ELISA 96-well strip plate, Wash buffer, and Ellman’s reagent were all purchased from Cayman Chemical. The assay was performed according to the manufacturer’s instructions using synthetic 20E (Sigma-Aldrich) as a standard.
Generation of EcI double-strand (ds) RNA
For dsRNA preparation, the 20-bp T7 promoter sequence was attached to the 5’ end of PCR primers for EcI, as listed in Table S5. PCR was carried out using EcI cDNA clone GH24467 (DGRC) as a template. PCR product carrying T7 promoter sequences was used as a template for in vitro transcription using the MEGAscript T7 kit (Ambion) in accordance with the manufacturer’s instructions. The products were incubated at 75°C for 5 min, and then gradually cooled to RT for 2 hours to produce annealed dsRNA. The dsRNA was purified by RNeasy mini kit according to the manufacturer’s instructions, combined with treatment with RNase-Free DNase Set.
Transfection and luciferase reporter assays in S2 cells
S2 cells (obtained from Michael B. O’Connor) at a density of 1 × 106 cells/ml were seeded in 2 ml/well of SSM3 insect medium containing 10% IMS and 1% PSS in a 6-well clear flat bottom multiple well plate (Corning) just before transfection. Transfection of S2 cells was performed using the dimethyldioctadecylammonium bromide (Avanti)-mediated method (Han, 1996). For EcI RNAi experiments, 0.5 µ g/well of pAct5C-EcR plasmid and 0.5 µ g/well of pADH-hspEcRE-LUC firefly luciferase reporter plasmid (both obtained from David J. Mangelsdorf) were transfected, and 10 µ g/well of EcI dsRNA was added at the time of transfection and every 24 hours afterward. For EcI overexpression experiments, 1.0 µg/well of pBRAcPA empty vector (control) or pBRAcPA-EcI plasmids were transfected along with the 0.5 µg/well of pAct5C-EcR and 0.5 µ g/well of pADH-hspEcRE-LUC plasmids. For all experiments, 0.1 µ g/well of pRL-CMV Renilla luciferase reporter plasmid (Promega) was co-transfected and used as a reference. The cells were incubated for 3 days after transfection, after which 100 µ l/well of transfected cells were plated into a 96-well clear flat bottom microplate (Corning). Three hours after incubation, S2 cells were dosed with 25 µl of the indicated compounds in SSM3 insect medium (total 125 µl/well) and harvested after the indicated time of treatment. 50 µ l of the medium in each well was then removed (total 75 µl remaining per well), and 75 µ l/well of Dual-Glo solution (Promega) was added into each well (total 150 µl/well). After incubation at RT in the dark for 30 min, 120 µ l/well of cell lysates were transferred to 96-well solid white flat bottom polystyrene TC-treated plates (Coster). The firefly luciferase activity and co-transfected Renilla luciferase activity were measured subsequently using the Dual-Luciferase Reporter Assay System (Promega) in accordance with the manufacturer’s instructions, and analyzed with GloMax-Multi + Microplate Multimode Reader with Instinct (Promega). For each condition, cells were treated independently in 3 wells of a 96-well plate in each experiment, and the same experiment was conducted multiple times on different days. Results from a representative experiment are shown in Figure 6.
Transfection and luciferase reporter assays in HEK293 cells
HEK cells (obtained from Michael E. Adams) at a density of 4 × 105 cells/ml were seeded in 100 µ l/well of Opti-MEM reduced serum media (Thermo Fisher Scientific) containing 5% FBS and 1% MEM non-essential amino acids (NEAA) solution (Thermo Fisher Scientific) in a 96-well clear flat bottom microplate (Corning). Transfection of HEK293 cells was performed using Attractene transfection reagent (Qiagen) by the fast-forward transfection approach following the manufacturer’s instructions. 50 µl/well of transfection cocktail containing Opti-MEM reduced serum media, Attractene, and DNA plasmids was added to each well, bringing the final volume to 150 µl/well. For all experiments, 0.1 µ g/well of pcDNA3.1 empty vector (control) or pcDNA3.1-EcI were transfected, along with 60 ng/well of pERV3 receptor plasmid (Agilent Technologies) containing a modified ecdysone receptor (VgEcR) and RXR, 36 ng/well of pEGSH-LUC luciferase reporter plasmid (Agilent Technologies), and 0.9 ng/well of pRL-CMV Renilla luciferase reporter plasmid (Promega) as a reference. After 24 hours of incubation at 37°C and 5% CO2, transfection medium was removed and replaced with 150 µl/well of DMEM with 4.5 mg/ml glucose and sodium pyruvate without L-glutamine and phenol red (w-G-SP, wo-G-PR) (Thermo Fisher Scientific) containing 10% FBS, 1% PSS, and 1% MEM NEAA solution. After 48 hours of incubation at 37°C and 5% CO2, medium was removed and replaced with 150 µ l/well of DMEM (w-G-SP, wo-G-PR) containing 1% PSS, 1% MEM NEAA solution, and the indicated compounds at the indicated concentrations. After 24 hours of incubation at 37°C and 5% CO2, 75 µ l of media was removed (total 75 µ l remaining per well) and 75 µl/well of Dual-Glo Solution was added (total 150 µ l/well). After 20 min of incubation at RT in the dark, 120 µl/well of cell lysates were transferred to 96-well solid white flat bottom polystyrene TC-treated plates. The firefly luciferase activity and co-transfected Renilla luciferase activity were measured subsequently using the Dual-Luciferase Reporter Assay System in accordance with the manufacturer’s instructions, and analyzed with GloMax-Multi + Microplate Multimode Reader with Instinct. For each condition, cells were treated independently in 3 wells of a 96-well plate in each experiment, and the same experiment was conducted multiple times on different days. Results from a representative experiment are shown in Figure 6.
Supplementary Material
HIGHLIGHTS.
Insect steroid hormone ecdysone requires a membrane transporter to enter cells
Ecdysone Importer (EcI) is a member of the evolutionary conserved SLCO superfamily
EcI functions cell-autonomously to incorporate ecdysone from circulation
ACKNOWLEDGMENTS
We thank Vienna Drosophila Resource Center, Bloomington Drosophila Stock Center (NIH P40 OD018537), National Institute of Genetics Fly Stock Center, Transgenic RNAi Project at Harvard Medical School (NIH R24 RR032668), T.P. Neufeld, T. Nishimura, and M.B. O’Connor for fly stocks; M.B. O’Connor and M.E. Adams for cell lines; Drosophila Genomics Resource Center (NIH P40 OD010949), National Institute of Genetics Fly Stock Center, and D.J. Mangelsdorf for vectors and cDNA clones; and F.M. Sladek, D.S. Straus, C.G. Pontrello, M.B. O’Connor, and M.E. Adams for comments on the manuscript. The monoclonal antibodies developed by J.R. Sanes, C.S. Thummel and D.S. Hogness were obtained from the Developmental Studies Hybridoma Bank, created by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. This study was supported by a Postdoctoral Fellowship for Research Abroad from the Japan Society for the Promotion of Science to N.O., the Naito Foundation Subsidy for Dispatch of Young Researchers Abroad to N.O., an NIH grant R00 HD073239 from NICHD to N.Y., a research grant from the W.M. Keck Foundation to S.H.Y. and N.Y., and a Pew Biomedical Scholars Award from the Pew Charitable Trusts to N.Y. N.P. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
N.O., R.B., S.H.Y., and N.Y. have a patent pending (U.S. Patent App. No. 15/902,948) relevant to this work.
STAR★METHODS
KEY RESOURCES TABLE
Other sheet
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Naoki Yamanaka (naoki.yamanaka@ucr.edu)
REFERENCES
- Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, and Walter P (2015). Molecular Biology of the Cell, 6th edition (New York: Garland Science; ). [Google Scholar]
- Attard G, Cooper CS, and de Bono JS (2009). Steroid hormone receptors in prostate cancer: a hard habit to break? Cancer Cell 16, 458–462. [DOI] [PubMed] [Google Scholar]
- Baker KD, Warren JT, Thummel CS, Gilbert LI, and Mangelsdorf DJ (2000). Transcriptional activation of the Drosophila ecdysone receptor by insect and plant ecdysteroids. Insect Biochem. Mol. Biol 30, 1037–1043. [DOI] [PubMed] [Google Scholar]
- Biyasheva A, Do TV, Lu Y, Vaskova M, and Andres AJ (2001). Glue secretion in the Drosophila salivary gland: a model for steroid-regulated exocytosis. Dev. Biol 231, 234–251. [DOI] [PubMed] [Google Scholar]
- Bond ND, Nelliot A, Bernardo MK, Ayerh MA, Gorski KA, Hoshizaki DK, and Woodard CT (2011). βFTZ-F1 and Matrix metalloproteinase 2 are required for fat-body remodeling in Drosophila. Dev. Biol 360, 286–296. [DOI] [PubMed] [Google Scholar]
- Bossuyt X, Müller M, Hagenbuch B, and Meier PJ (1996). Polyspecific drug and steroid clearance by an organic anion transporter of mammalian liver. J. Pharmacol. Exp. Ther 276, 891–6. [PubMed] [Google Scholar]
- Cherbas L, Hu X, Zhimulev I, Belyaeva E, and Cherbas P (2003). EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development 130, 271–284. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Mark MR, Bajaj V, and Godowski PJ (1992). Ecdysteroid-dependent regulation of genes in mammalian cells by a Drosophila ecdysone receptor and chimeric transactivators. Proc. Natl. Acad. Sci. USA 89, 6314–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons M, and Goss P (2001). Estrogen and the risk of breast cancer. N. Engl. J. Med 344, 276–22. [DOI] [PubMed] [Google Scholar]
- Costantino BF, Bricker DK, Alexandre K, Shen K, Merriam JR, Antoniewski C, Callender JL, Henrich VC, Presente A, and Andres AJ (2008). A novel ecdysone receptor mediates steroid-regulated developmental events during the mid-third instar of Drosophila. PLoS Genet 4, e1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen ET, Moeller ME, Yamanaka N, Ou Q, Laursen JM, Soenderholm C, Zhuo R, Phelps B, Tang K, Zeng J, et al. (2016). A Drosophila genome-wide screen identifies regulators of steroid hormone production and developmental timing. Dev. Cell 37, 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Hamon Y, and Chimini G (2001). The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid Res 42, 1007–1017. [PubMed] [Google Scholar]
- Dinan L, and Lafont R (2006). Effects and applications of arthropod steroid hormones (ecdysteroids) in mammals. J. Endocrinol 191, 1–8. [DOI] [PubMed] [Google Scholar]
- El-Gebali S, Bentz S, Hediger MA, and Anderle P (2013) Solute carriers (SLCs) in cancer. Mol. 4 Aspects Med 34, 719–734. [DOI] [PubMed] [Google Scholar]
- Evans RM, and Mangelsdorf DJ (2014). Nuclear receptors, RXR, and the Big Bang. Cell 157, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaziova I, Bonnette PC, Henrich VC, and Jindra M (2004). Cell-autonomous roles of the ecdysoneless gene in Drosophila development and oogenesis. Development 131, 2715–2725. [DOI] [PubMed] [Google Scholar]
- Gilbert LI (2004). Halloween genes encode P450 enzymes that mediate steroid hormone biosynthesis in Drosophila melanogaster. Mol. Cell. Endocrinol 215, 1–10. [DOI] [PubMed] [Google Scholar]
- Giorgi EP, and Stein WD (1981) The transport of steroids into animal cells in culture. Endocrinology 108, 688–697. [DOI] [PubMed] [Google Scholar]
- Gohl DM, Silies MA, Gao XJ, Bhalerao S, Luongo FJ, Lin CC, Potter CJ, and Clandinin TR (2011). A versatile in vivo system for directed dissection of gene expression patterns. Nat. Methods 8, 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JC, Wohlhueter RM, and Plagemann PG (1977) Effect of temperature and of cytochalasin B and persantin on the nonmediated permeation of non-electrolytes into cultured Novikoff rat hepatoma cells. J. Biol. Chem 252, 4185–4190. [PubMed] [Google Scholar]
- Gramates LS, Marygold SJ, dos Santos G, Urbano JM, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, et al. (2017). FlyBase at 25: looking to the future. Nucleic Acids Res 45, D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SM, Kaipainen A, Bullock K, Zhang A, Lucas JM, Matson C, Banks WA, and Mostaghel EA (2017). Role of OATP transporters in steroid uptake by prostate cancer cells in vivo. Prostate Cancer Prostatic. Dis 20, 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen SC, LaPlante ER, Alexandre NM, Agrawal AA, Dobler S, and Whiteman NK (2017). Multidrug transporters and organic anion transporting polypeptides protect insects against the toxic effects of cardenolides. Insect Biochem. Mol. Biol 81, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuch B, and Stieger B (2013). The SLCO (former SLC21) superfamily of transporters. Mol. Aspects Med 34, 396–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada A, Sissung T, Price DK, Danesi R, Chau CH, Sharifi N, Venzon D, Maeda K, Nagao K, Sparreboom A, et al. (2008). Effect of SLCO1B3 haplotype on testosterone transport and clinical outcome in caucasian patients with androgen-independent prostatic cancer. Clin. Cancer Res 14, 3312–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K (1996). An efficient DDAB-mediated transfection of Drosophila S2 cells. Nucleic Acid Res 24, 4362–4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock T, Cottrill T, Keegan J, and Garza D (2000). The E23 early gene of Drosophila encodes an ecdysone-inducible ATP-binding cassette transporter capable of repressing ecdysone-mediated gene activation. Proc. Natl. Acad. Sci. USA 97, 9519–9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliokoski A, and Niemi M (2009). Impact of OATP transporters on pharmacokinetics. Br. J. Pharmacol 158, 693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim FD, and Thummel CS (1992). Temporal coordination of regulatory gene expression by the steroid hormone ecdysone. EMBO J 11, 4083–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karssen AM, Meijer OC, Van Der Sandt ICJ, Lucassen PJ, De Lange ECM, De Boer AG, and De Kloet ER (2001). Multidrug resistance P-glycoprotein hampers the access of cortisol but not of corticosterone to mouse and human brain. Endocrinology 142, 2686–2694. [DOI] [PubMed] [Google Scholar]
- King-Jones K, and Thummel CS (2005). Nuclear receptors – a perspective from Drosophila. Nat. Rev. Genet 6, 311–323. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, and Aleksunes LM (2010). Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol. Rev 62, 1–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle MR, Talbot WS, Segraves WA, Bender MT, Cherbas P, and Hogness DS (1991). The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell 67, 59–77. [DOI] [PubMed] [Google Scholar]
- Kondo S, and Ueda R (2013). Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralli A, Bohen SP, and Yamamoto KR (1995). LEM1, an ATP-binding-cassette transporter, selectively modulates the biological potency of steroid hormones. Proc. Natl. Acad. Sci. USA 92, 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Hayashizaki Y, and Daub CO (2009). TagDust—a program to eliminate artifacts from next generation sequencing data. Bioinformatics 25, 2839–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TR, and White KP (2003). Tissue-specific gene expression and ecdysone-regulated genomic networks in Drosophila. Dev. Cell 5, 59–72. [DOI] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, and Liu XS (2014). MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schuütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. (1995). The nuclear receptor superfamily: the second decade. Cell 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NP, and O’Malley BO (2002). Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108, 465–474. [DOI] [PubMed] [Google Scholar]
- Milgrom E, Atger M, and Baulieu EE (1973) Studies on estrogen entry into uterine cells and on estradiol-receptor complex attachment to the nucleus–is the entry of estrogen into uterine cells a protein-mediated process? Biochim. Biophys. Acta 320, 267–283. [DOI] [PubMed] [Google Scholar]
- Minakuchi C, Nakagawa Y, Kamimura M, and Miyagawa H (2005). Measurement of receptor-binding activity of non-steroidal ecdysone agonists using in vitro expressed receptor proteins (EcR/USP complex) of Chilo suppressalis and Drosophila melanogaster. In New Discoveries in Agrochemicals, Clark JM, and Ohkawa H, eds. (Washington DC, USA: American Chemical Society; ), pp. 191–200. [Google Scholar]
- Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, and Gustafsson JA (2001). Mechanisms of estrogen action. Physiol. Rev 81, 1535–1565. [DOI] [PubMed] [Google Scholar]
- Niwa R, Matsuda T, Yoshiyama T, Namiki T, Mita K, Fujimoto Y, and Kataoka H (2004). CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J. Biol. Chem 279, 35942–35949. [DOI] [PubMed] [Google Scholar]
- Niwa R, and Niwa YS (2014). Enzymes for ecdysteroid biosynthesis: their biological functions in insects and beyond. Biosci. Biotech. Biochem 78, 1283–1292. [DOI] [PubMed] [Google Scholar]
- No D, Yao TP, and Evans RM (1996). Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. USA 93, 3346–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussey SS, and Whitehead SA (2001). Endocrinology: An Integrated Approach (London: Taylor & Francis; ). [PubMed] [Google Scholar]
- Oakley RH, and Cidlowski JA (2011). Cellular processing of the glucocorticoid receptor gene and protein: new mechanisms for generating tissue-specific actions of glucocorticoids. J. Biol. Chem 286, 3177–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obaidat A, Roth M, and Hagenbuch B (2012). The expression and function of organic anion transporting polypeptides in normal tissues and in cancer. Annu. Rev. Pharmacol. Toxicol 52, 135–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara MH, Hikiba J, Suzuki Y, Taylor D, and Kataoka H (2015). Ovarian ecdysteroidogenesis in both immature and mature stages of an acari, Ornithodoros moubata. PLoS One 10, e0124953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Q, Zeng J, Yamanaka N, Brakken-Thal C, O’Connor MB, and King-Jones K (2016). The insect prothoracic gland as a model for steroid hormone biosynthesis and regulation. Cell Rep 16, 247–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pariante CM, Thomas SA, Lovestone S, Makoff A, and Kerwin RW (2004). Do antidepressants regulate how cortisol affects the brain? Psychoneuroendocrinology 29, 423–447. [DOI] [PubMed] [Google Scholar]
- Pietras RJ, and Szego CM (1977) Specific binding sites for oestrogen at the outer surfaces of isolated endometrial cells. Nature 265, 69–72. [DOI] [PubMed] [Google Scholar]
- Plagemann PG, and Erbe J (1976). Glucocorticoids–uptake by simple diffusion by cultured Reuber and Novikoff rat hepatoma cells. Biochem. Pharmacol 25, 1489–1494. [DOI] [PubMed] [Google Scholar]
- Rao ML, Rao GS, Höller M, Breuer H, Schattenberg PJ, and Stein WD (1976) Uptake of cortisol by isolated rat liver cells. A phenomenon indicative of carrier-mediation and simple diffusion. Hoppe Seylers Z. Physiol. Chem 357, 573–584. [DOI] [PubMed] [Google Scholar]
- Rees DC, Johnson E, and Lewinson O (2009). ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol 10, 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz KF, and Gilbert LI (2008). Daphnia Halloween genes that encode cytochrome P450s mediating the synthesis of the arthropod molting hormone: evolutionary implications. BMC Evol. Biol 8, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhen T, and Cidlowski JA (2005). Antiinflammatory action of glucocorticoids–new mechanisms for old drugs. N. Engl. J. Med 353, 1711–1723. [DOI] [PubMed] [Google Scholar]
- Riddiford LM, Cherbas P, and Truman JW (2000). Ecdysone receptors and their biological actions. Vitam. Horm 60, 1–73. [DOI] [PubMed] [Google Scholar]
- Saez E, Nelson MC, Eshelman B, Banayo E, Koder A, Cho GJ, and Evans RM (2000). Identification of ligands and coligands for the ecdysone-regulated gene switch. Proc. Natl. Acad. Sci. USA 97, 14512–14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapolsky RM, Romero LM, and Munck AU (2000). How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr. Rev 21, 55–89. [DOI] [PubMed] [Google Scholar]
- Schubiger M, Wade AA, Carney GE, Truman JW, and Bender M (1998). Drosophila EcR-B ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development 125, 2053–2062. [DOI] [PubMed] [Google Scholar]
- Seabrooke S, and O’Donnell MJ (2013). Oatp58Dc contributes to blood-brain barrier function by excluding organic anions from the Drosophila brain. Am. J. Physiol. Cell Physiol 305, C558–C567. [DOI] [PubMed] [Google Scholar]
- Sisk CL, and Foster DL (2004). The neural basis of puberty and adolescence. Nat. Neurosci 7, 1040–1047. [DOI] [PubMed] [Google Scholar]
- Thummel CS (1996). Files on steroids – Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet 12, 306–310. [DOI] [PubMed] [Google Scholar]
- Torrie LS, Radford JC, Southall TD, Kean L, Dinsmore AJ, Davies SA, and Dow JA (2004). Resolution of the insect ouabain paradox. Proc. Natl. Acad. Sci. USA 101, 13689–13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhr M, Holsboer F, and Müller MB (2002). Penetration of endogenous steroid hormones corticosterone, cortisol, aldosterone and progesterone into the brain is enhanced in mice deficient for both mdr1a and mdr1b p-glycoproteins. J. Neuroendocrinol 14, 753–759. [DOI] [PubMed] [Google Scholar]
- Urry LA, Cain ML, Wasserman SA, Minorsky PV, and Reece JB (2017). Campbell Biology, 11th edition (New York: Pearson; ). [Google Scholar]
- Viswanatha R, Li Z, Hu Y, Perrimon N (2018). Pooled genome-wide CRISPR screening for basal and context-specific fitness gene essentiality in Drosophila cells. eLife 7, e36333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, and Lander ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JT, Petryk A, Marqués G, Parvy JP, Shinoda T, Itoyama K, Kobayashi J, Jarcho M, Li Y, O’Connor MB, et al. (2004). Phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: a P450 enzyme critical in ecdysone biosynthesis. Insect Biochem. Mol. Biol 34, 991–1010. [DOI] [PubMed] [Google Scholar]
- Wilson CA, and Davies DC (2007). The control of sexual differentiation of the reproductive system and brain. Reproduction 133, 331–359. [DOI] [PubMed] [Google Scholar]
- Wright JL, Kwon EM, Ostrander EA, Montgomery RB, Lin DW, Vessella R, Stanford JL, and Mostaghel EA (2011). Expression of SLCO transport genes in castration-resistant prostate cancer and impact of genetic variation in SLCO1B3 and SLCO2B1 on prostate cancer outcomes. Cancer Epidemiol. Biomarkers Prev 20, 619–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto KR (1985). Steroid receptor regulated transcription of specific genes and gene networks. Annu. Rev. Genet 19, 209–252. [DOI] [PubMed] [Google Scholar]
- Yamanaka N, Marqués G, and O’Connor MB (2015). Vesicle-mediated steroid hormone secretion in Drosophila melanogaster. Cell 163, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N, Rewitz KF, and O’Connor MB (2013). Ecdysone control of developmental transitions: lessons from Drosophila research. Annu. Rev. Entomol 58, 497–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


