Abstract
We examined the signaling pathways and cell type-specific responses of interferon regulatory factor (IRF)5, an immune-regulatory transcription factor. We show that the protein kinases IKKα, IKKβ, IKKε and TBK1 each confer IRF5 phosphorylation/dimerization, thus extending the family of IRF5 activator kinases. Among primary human immune cell subsets, we found that IRF5 is most abundant in plasmacytoid dendritic cells (pDCs). Flow cytometric cell imaging revealed that IRF5 is specifically activated by endosomal Toll-like receptor (TLR) signaling. Comparative analyses revealed that IRF3 is activated in pDCs uniquely through RIG-I-like receptor (RLR) signaling. Transcriptomic analyses of pDCs show that the partitioning of TLR7/IRF5 and RLR/IRF3 pathways confers differential gene expression and immune cytokine production in pDCs, linking IRF5 with immune regulatory and proinflammatory gene expression. Thus, TLR7/IRF5 and RLR-IRF3 partitioning serve to polarize pDC response outcome. Strategies to differentially engage IRF signaling pathways should be considered in the design of immunotherapeutic approaches to modulate or polarize the immune response for specific outcome.
Introduction
Interferon Regulatory Factors (IRFs) are transcription factors that regulate the intricate gene networks essential for coordinating an appropriate and effective immune response(1, 2). In particular, IRF3 and IRF7 have been extensively studied and shown to regulate the induction of type I interferons (IFNs) and other cytokines in response to pattern recognition receptor (PRR) recognition of pathogen associated molecular patterns (PAMPs) during virus infection(3, 4). During RNA virus infection, viral PAMP RNA motifs are recognized by RIG-I-like receptors (RLRs), leading to RLR signaling activation and interaction with the adaptor MAVS(5). MAVS recruits TANK-binding kinase 1 (TBK1), which phosphorylates IRF3 and IRF7, leading to the homodimerization of these IRFs and translocation into the nucleus to induce gene expression(6). Stimulation of some Toll-like receptors (TLRs) also activates IRF3 and IRF7 to induce type I interferons (IFNs)(7).
In contrast to IRF3 and IRF7, IRF5 regulation and function are less well-characterized. Mouse studies revealed essential roles of IRF5 in the production of IFNβ and proinflammatory mediators including interleukin (IL)-6, IL-12, and tumor necrosis factor α (TNFα)(8–10) (11) (12). In humans, carriers of autoimmune risk haplotypes at the IRF5 locus exhibit elevated levels of IFNα(13–16), and dendritic cells (DCs) from these carriers produced elevated TNFα and IL-12 upon TLR stimulation(17, 18). In HEK293 cells overexpressing TLR7, the TLR7/8 agonist R848 induced activation of an IRF5 reporter, accompanied by the translocation of IRF5-GFP into the nucleus(19). TBK1 was reported to phosphorylate IRF5(19, 20), and a kinase-dead mutant of TBK1 or the related IKKε inhibited the TLR7-dependent activation of a Gal4-IRF5 reporter(19). These results suggest that TBK1 and IKKε activate both IRF5 and IRF3. Subsequent reports identified IKKβ as the activating kinase of IRF5(21, 22). MAVS overexpression also was shown to induce IRF5 dimerization in HEK293T cells(21, 22), and RIG-I and IRF5 co-expression rescued cytokine production defects in Irf3/5/7-deficient mouse cells(23), suggesting that both TLR and RLR stimulation may activate IRF5. Of note is that these studies have typically relied on overexpression approaches often in epithelial cell lines to evaluate IRF5 functions. In human plasmacytoid DC (pDC) cell lines, RNAi-mediated IRF5 knockdown experiments have established IRF5 as a crucial mediator type I IFN induction(21, 24). However, the relevant regulatory pathways, endogenous steps of IRF5 activation, and IRF5 transcriptional signatures have not been defined nor directly compared to signaling from other IRFs.
Here we evaluated the IRF5 activation process and defined the IRF5 transcriptome. Our study shows that each IKK kinase can direct IRF5 phosphorylation for dimerization/activation and that IRF5 is variably present in different primary immune cell subsets wherein it is highly abundant in pDCs. Using robust assays to assess and compare endogenous IRF5 and IRF3 activation in pDCs, we show that IRF5 is activated upon endosomal TLR7/8 stimulation, whereas RLR stimulation leads to IRF3 activation. High throughput transcriptomic analyses reveal that triggering pDCs through TLR7/IRF5 and RLR/IRF3 pathways leads to a response that induces specific gene expression profiles and the production of distinct sets of cytokines for differential immune activation. This unique partitioning of TLR7/IRF5 and RLR/IRF3 signaling serves to drive expanded proinflammatory, immune regulatory, and antiviral actions of pDCs.
Materials and Methods
Cells
CAL-1 was gifted by Dr. Takahiro Maeda (Nagasaki University). PMDC05 cells were from Miwako Narita (Niigata University). BJAB and Ramos were gifted by Dr. Ed Clark (University of Washington). MUTZ-3 was purchased from DSMZ. Jurkat, THP-1 and HEK293T were purchased from ATCC. HEK293T was cultured in DMEM supplemented with 5% FBS, L-glutamine, and sodium pyruvate. MUTZ-3 was cultured in α-MEM supplemented with 20% FBS and 20% conditioned medium from 5637 cells (purchased from DSMZ). All other cell lines used in this study were cultured in RPMI 1640 supplemented with 10% FBS, L-glutamine, sodium pyruvate, HEPES, and β-mercaptoethanol. THP-1 cells were differentiated in complete RPMI supplemented with 40nM PMA (Sigma) for 18–24 hours. MUTZ-3 cells were differentiated in complete conditioned α-MEM supplemented with 20ng/ml recombinant human IL-4 (Peprotech) and 50ng/ml recombinant human GM-CSF (Peprotech) for 5 days, and then 12ng/ml recombinant human TNFα (Peprotech) for an additional 2 days. All cells were grown at 37°C in 5% CO2.
Whole blood was obtained from healthy individuals under IRB approval. LRS chambers containing white blood cell concentrate were obtained from Bloodworks NW. Peripheral blood mononuclear cells (PBMCs) were purified using Ficoll-Paque Plus (GE Healthcare). To obtain pDCs, purified PBMCs were negatively selected by an EasySep Human Plasmacytoid DC Enrichment Kit (STEMCELL Technologies). Autologous PBMCs were added back to the purified pDCs such that final pDCs consisted of 5–10% of population to allow for sufficient cell number for subsequent analysis.
Chemicals
LTA-SA, FSL-1, FLA-ST, poly(I:C), LPS, R837, R848, CpG-A (ODN 2216) and CpG-B (ODN 2006) were purchased from Invivogen.
Viruses
Sendai virus (SeV; Cantell strain) was purchased from Charles River and cells were infected at 20–200 HAU/ml.
Transfection
CAL-1 and PMDC05 cells were transfected using the Amaxa 96-well shuttle nucleofector system (Lonza) according to manufacturer’s protocol. 1ug of RNA was transfected into 1 × 106 cells/well in 16-well strips using buffer SF and program CM130.
Immunoblot
Cells were lysed in NP-40 buffer and then clarified. Protein was quantified with Bradford Protein Assay (Bio-Rad). For SDS-PAGE, 10–50ug of lysate was separated by 7.5% Tris-glycine gels with SDS. For Mn2+-Phos-Tag SDS-PAGE, 20–40ug of lysate was separated by 8% gel containing 50uM Phos-Tag reagent (Wako). For IRF5 Native-PAGE, 10ug of lysate was separated by 3–12% Bis-Tris NativePAGE gels (Invitrogen). For IRF3 Native-PAGE, 10ug of lysate was separated by 7.5% Tris-Glysine gels without SDS. Native gels were soaked in running buffer with SDS for 30 minutes at room temperature prior to transfer. All gels were transferred to PVDF-FL (Millipore) membranes, blocked in PBS or TBS blocking buffer (Li-COR Biosciences), and labeled with primary and secondary antibodies according to the manufacturer’s instructions. Blots were analyzed with the Odyssey Infrared Imaging System (LI-COR Biosciences). The following primary antibodies were used in this study: anti-FLAG (M2; Sigma), anti-phospho-IRF5 (a kind gift from Dr. Phil Cohen; University of Dundee), anti-IRF5 (A303–385 or A303–386; Bethyl), anti-LSD1 (C69G12; Cell Signaling), anti-HA (Y11; Santa Cruz Biotechnology), anti-Actin (C4; Santa Cruz Biotechnology), anti-IRF3 (dimer) (AR-1; previously described(25)), anti-IRF3 (D83B9; Cell Signaling). AF680- or AF790-conjugated secondary antibodies were purchased from Jackson Immunoresearch.
ImageStream imaging flow cytometry
PBMCs or enriched pDCs were stained with the following cell surface markers: CD3-FITC (UCHT1, BD Biosciences), CD14-AF488 (M5E2, BD Biosciences), CD19-FITC (HIB19, BD Biosciences), CD56-AF488 (B159, BD Biosciences), CD11c-FITC, HLA-DR-PE (L243, Biolegend), BCDA2-PE (AC144, Miltenyi), FITC-conjugated lineage cocktail panel 1 (BD Biosciences). Cells were fixed and permeabilized using the Cytofix/perm kit (BD), and stained with IRF5-AF647 antibody (EPR6094, Abcam) and DAPI (Life Technologies). Stained cells were acquired on an ImageStreamX Mark II imaging flow cytometer at 60x magnification (Amnis/EMD Millipore). At least 10,000 cells per condition were analyzed. Data were compensated and analyzed using the IDEAS software. Following standard analysis procedures(26), single focused cells were analyzed using Nuclear Localization Wizard to calculate similarity score (SS) between IRF5 and DAPI, which quantifies the correlation of pixel values of the two stains on a per cell basis. Representative images of cells and/or SS plotted on histograms are displayed.
Luminex
Forty five cytokines/chemokines were measured using a Cytokine/Chemokine/Growth Factor 45-Plex Human ProcartaPlex Panel 1 immunoassay kit (eBioscience): BDNF; Eotaxin/CCL11; EGF; FGF-2; GM-CSF; GROα/CXCL1; HGF; NGFβ; LIF; IFNα; IFNγ; IL-1β; IL-1α; IL-1Rα; IL-2; IL-4; IL-5; IL-6; IL-7; IL-8/CXCL8; IL-9; IL-10; IL-12p70; IL-13; IL-15; IL-17α; IL-18; IL-21; IL-22; IL-23; IL-27; IL-31; IP-10/CXCL10; MCP-1/CCL2; MIP-1α/CCL3; MIP-1β/CCL4; RANTES/CCL5; SDF-1α/CXCL12; TNFα; TNFβ/LTA; PDGF-BB; PLGF; SCF; VEGF-A; VEGF-D. Biological triplicates were used for each treatment condition, and each sample was prepared and run in technical duplicate according to manufacturer’s protocol. Standards provided by manufacturer were run on each plate to create a standard curve for each analyte. Data were acquired using a Luminex 200 system and analyzed using Bio-Plex Manager software (Bio-Rad). Concentration of each analyte was determined by standard curve method. Levels beyond standard curve were assigned the highest standard concentration. Undetected values were assigned the limit of detection to allow for calculation of fold change over baseline.
RNA sequencing and transcriptomic analysis
Total RNA was purified from cells using an RNeasy kit in RLT buffer (QIAGEN). RNA with a Bioanalyzer (Agilent, USA) RNA integrity number (RIN) of ≥8 was sent to Seattle Genomics (University of Washington) for cDNA library construction and next-generation sequencing on the Illumina Nextseq 500 instrument. Raw FASTQ files were demultiplexed and quality checked through FastQC. Adapters and rRNA sequences that remained were digitally removed using Cutadapt (version 1.8.3) and Bowtie2 (version 2.2.5). The remaining reads (~20 million reads per sample) were mapped to the human genome (build NCBI_build 37.2) using the STAR aligner (version 2.4.2), and alignments were further converted into gene counts using HTSeq (version 0.6.0).
Statistical analysis of RNA-seq data was performed using the R statistical software (version 3.4.0). Gene counts were filtered by sum counts >75 across all samples to remove unexpressed and lowly-expressed genes. Samples were voom normalized using the limma package (version 3.32.2), and differential expression was calculated using linear regression against a defined contrast matrix followed by eBayes testing (all within the limma package). All samples were contrasted against appropriate negative controls. Genes designated as significantly differentially expressed (DE) had a fold change over control >2 (above or below), with a Benjamini-Hochberg adjusted p-value <0.05. Heat maps of DE genes were created using a modified version of the heatmap2 function within the gplots package. Red = increased expression and blue = reduced expression. Sequence data have been deposited into GEO with accession number GSE108526 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108526).
Expression plasmids
IRF5 was gifted by Dr. Savan Ram. IKKα was gifted by Dr. Kim Orth (UT Southwestern). IKKβ was purchased from Dharmacon. IKKε and TBK1 were gifted by Dr. Tom Maniatis (Columbia University). All open reading frames were confirmed by DNA sequence analysis.
Results
Activation of endogenous IRF5 by IKK family kinases and RLR signaling
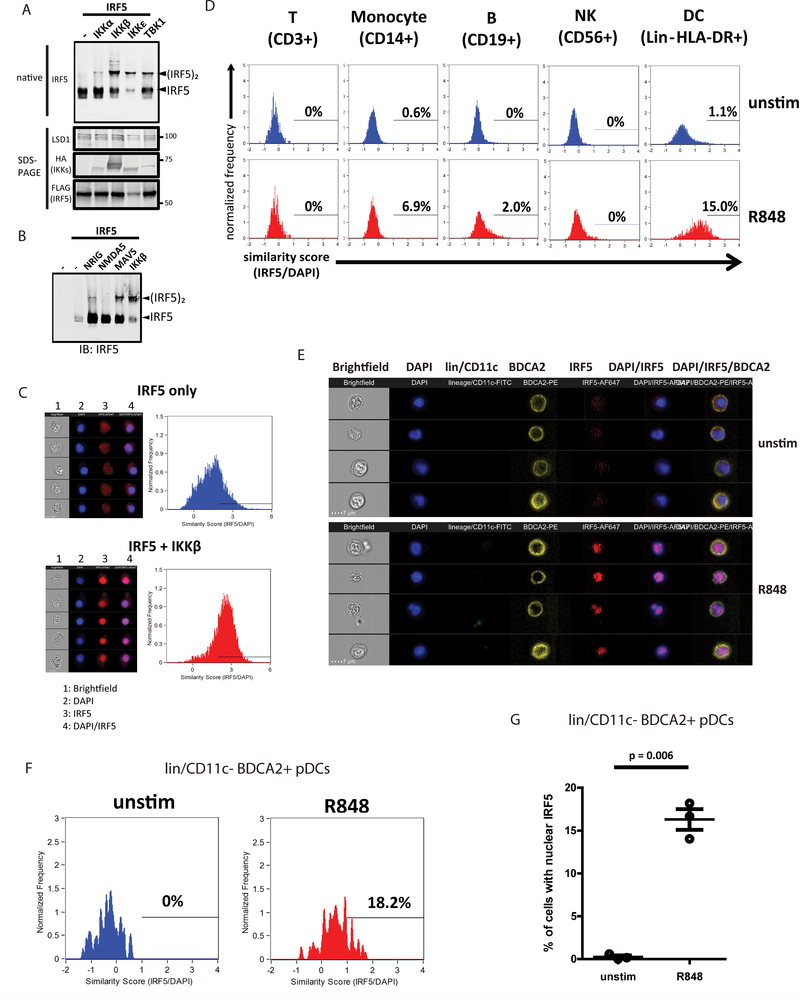
To evaluate IRF5 function, we first sought to develop a sensitive IRF5 dimerization assay to measure the abundance of monomeric/resting and active/dimeric IRF5 in response to PRR signaling. This task was important because to date, most IRF5 activation assays have been performed with overexpressing IRF5 and detecting with antibodies against the epitope tags instead of IRF5 itself(19, 20, 27, 28). Moreover, though multiple IRF5 antibodies are available, a recent report highlighted the issues misleading the understanding of IRF5 activation processes due to use of non-specific/unreliable antibodies(29). IKKβ has been shown to activate IRF5 by phosphorylating serine 462 (S462) that facilitates subsequent dimerization(21, 22, 30). We found that the Bis-Tris NativePage system consistently allowed separation and detection of IRF5 dimers when epitope tagged-IRF5 was co-expressed with IKKβ in HEK293T cells (Fig. S1A). Using a phospho-S462 specific IRF5 antibody, we confirmed that dimeric but not monomeric IRF5 was indeed phosphorylated at S462 (Fig S1A). We also tested several IRF5 antibodies and identified one (A303–385/A303–386; Bethyl) that detects both monomeric and dimeric forms of IRF5. When increasing amount of IKKβ expression plasmid was co-transfected with fixed amount of IRF5 into HEK293T cells, a corresponding increase of IRF5 dimer abundance was detected (Fig. S1B). To determine the ability of IKK family kinases (IKKα, IKKβ, IKKε and TBK1)(19, 21, 22, 31) to induce IRF5 dimerization, we co-expressed each with IRF5, finding that all four kinases variably induced IRF5 activation (Fig. 1A). Previous studies implicated the RLR pathway in IRF5 activation(23). Overexpression of the constitutively active form of RLRs (N-RIG and N-MDA5) or MAVS(32, 33) led to moderate level of IRF5 dimerization (Fig. 1B). Thus, different IKK family kinases and RLR pathway signaling have the potential to induce IRF5 activation.
Figure 1. IRF5 is activated by TLR7/8 stimulus in primary human pDCs.
(a) Native PAGE immunoblot analysis of IRF5 (top) and SDS-PAGE (bottom) immunoblot analysis of LSD1 (loading control), HA (IKK kinases), and FLAG (IRF5) in HEK293T cells overexpressing IRF5 and indicated IKK family kinases. (b) Native PAGE immunoblot analysis of IRF5 in HEK293T cells overexpressing IRF5 and indicated RLR pathway components or IKKβ (positive control). (c) ImageStream analysis of HEK293T cells overexpressing IRF5 alone or IRF5+IKKβ. Brightfield, DAPI (blue), IRF5-AF647 (red) and merged DAPI/IRF5 channels are shown for 5 representative images. Histograms showing similarity score of IRF5-AF647 and DAPI are shown for IRF5 only (blue) and IRF5+IKKβ (red) expressing cells. (d) ImageStream analysis of PBMCs unstimulated or treated with 1ug/ml R848 for 1.5 hours. Cells were labeled with CD3-FITC, CD56-AF488, CD19-FITC, CD14-AF488, or FITC-conjugated lineage panel together with HLA-DR-PE, and analyses were performed on cells gated on the indicated cell surface markers. Similarity scores were calculated for IRF5-AF647 and DAPI and displayed in histograms for unstimulated (top panel) or R848-treated (bottom panel) cells. (e) ImageStream analysis of purified primary human pDCs unstimulated or treated with 1ug/ml R848 for 1.5 hours. The analysis was gated on lineage/CD11c- and BDCA2+ cells. Brightfield, DAPI (blue), lineage/CD11c-FITC (green), BDCA2-PE (yellow), IRF5-AF647 (red) and merged DAPI/IRF5 and DAPI/IRF5/BDCA2 channels are shown for 5 representative images of unstimulated (top panel) or R848-treated (bottom panel) pDCs. (f) ImageStream analysis of lineage/CD11c- and BDCA2+ pDCs as described in (e). Similarity scores were calculated for IRF5-AF647 and DAPI and displayed in histograms for unstimulated (blue) or R848-treated (red) cells. (g) ImageStream analysis of lineage/CD11c- and BDCA2+ pDCs as described in (e). The percentage of cells with similarity score above 1 was calculated. T-test was performed and p-value is displayed between unstimulated and R848-treated pDCs across three independent experiments. Immunoblot experiments in (a-c) were performed at least 3 times. ImageStream experiments in (d-g) were performed once with 3 donors.
Single cell analysis of IRF5 activation
To validate dimerization as a marker for IRF5 activation, we used ImageStream imaging flow cytometry to measure IRF5 nuclear translocation under similar conditions that confer IRF5 dimerization. We first tested antibody specificity by transfecting HEK293T cells with constructs encoding empty vector (EV) or IRF5. EV-transfected cells showed no IRF5 signal, while IRF5-transfected cells showed specific IRF5 staining (Fig. S1C). To assess subcellular localization of IRF5, we used the IDEAS software to calculate a similarity score based on the pixel overlap between nucleus (DAPI) and IRF5 staining. A high similarity score represents high degree of overlap, thus indicating nuclear localization. We analyzed HEK293T cells transfected with IRF5 alone or co-transfected with IRF5 and IKKβ. We observed marked shift toward higher similarity score when IRF5 was co-transfected with IKKβ, corresponding to increased number of cells with nuclear IRF5 (Fig. 1C). These results indicate that IKKβ induced IRF5 dimerization as well as IRF5 translocation into the nucleus. These two orthogonal assays thus allow examination of the activation process of endogenous IRF5.
TLR7/8 signaling drives variable IRF5 activation in PBMC subsets
Since IRF5 is critical for modulating the immune response(8, 34–37), we evaluated IRF5 activation in primary peripheral blood mononuclear cells (PBMCs). We isolated and stimulated PBMCs with R848, a TLR7/8 agonist previously shown to activate IRF5 (19, 38, 39). Cells were stained with surface markers corresponding to T cells (CD3+), monocytes (CD14+), B cells (CD19+), natural killer (NK) cells (CD56+), or DCs (lineage lacking CD3, CD14, CD16, CD19, CD20, and CD56, but HLA-DR+) and then were fixed, permeabilized and stained with anti-IRF5 antibody and DAPI. Cells were then analyzed by ImageStream to assess the percentage of cells within each subset having nuclear/activate IRF5 upon R848 treatment. T cells (CD3+) and NK cells (CD56+) exhibited little to no IRF5 expression (Fig. S1D) nor did we detect nuclear IRF5 translocation in these cells upon R848 treatment (Fig. 1D). B cells (CD19+) had detectable level of IRF5 and a low frequency of cells responded to R848 treatment (Figs. S1D and 1D). Monocytes (CD14+) displayed varying levels of IRF5 expression, likely representing the heterogeneous nature of this cell subset(40) (Fig. S1D) wherein a more prominent shift of CD14+ cells with nuclear IRF5 was detected in response to R848 treatment (Fig. 1D). DCs (lineage- HLA-DR+), which included conventional DCs (cDCs) and pDCs within our staining scheme, expressed IRF5 in a range of levels (Fig. S1D) and exhibited the most robust IRF5 nuclear accumulation in response to R848 among the PBMC cell populations (see Fig. 1D).
To further subset the DC population, we concurrently stained PBMCs with FITC-antibodies specific to lineage markers and CD11c, a marker of cDCs(41), together with PE-antibody to BDCA2, a pDC-specific marker. We defined pDCs as lineage/CD11c-FITC- and BDCA2-PE+ (Fig. 1E), and observed consistent IRF5 accumulation in the nucleus in response to TLR7/8 stimulation (Figs. 1F and 1G). pDCs thus represent a major IRF5-repsonsive cell type.
IRF5 is activated by endosomal TLR stimulation in pDCs
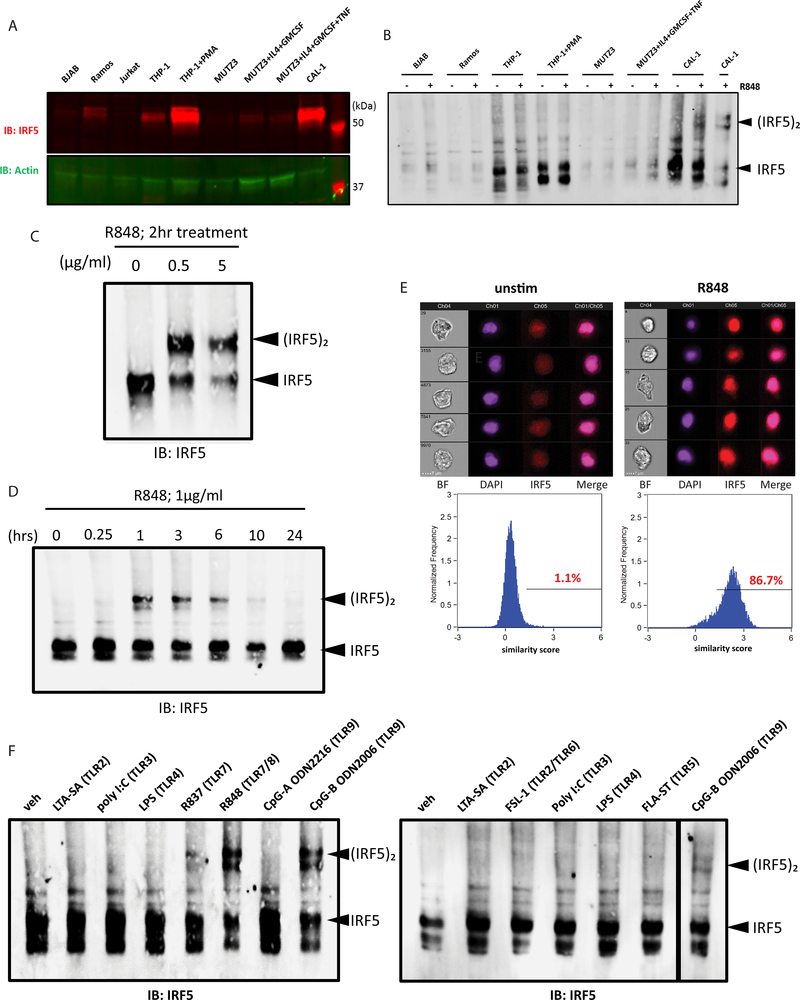
Because we observed modest IRF5 response in B cells and monocytes upon R848 treatment in primary PBMCs, we tested IRF5 activation in human cell line models corresponding to each cell type. We first measured IRF5 protein level in a panel of cell lines (Fig. 2A), and found that Jurkat T cells(42) had nearly undetectable level of IRF5, followed by B cell lines (BJAB and Ramos(43, 44)) that had low IRF5 expression, consistent with our PBMC datasets (Fig. S1D). THP-1 monocytic cell line(45) had moderate level of IRF5 expression, which significantly increased upon differentiation into macrophage-like cells by PMA(46). MUTZ3 DC precursor cell line(47) expressed little to undetectable level of IRF5, but induced IRF5 expression upon differentiation into immature DCs by IL-4 and GM-CSF or into mature DCs by additional TNFα treatment(48). CAL-1 pDC cell line(49) had the highest level of IRF5 expression amongst the panel of cell lines tested. We then stimulated BJAB, Ramos, THP-1 (undifferentiated and PMA-differentiated), MUTZ3 (undifferentiated or IL-4/GM-CSF/TNFα differentiated), and CAL-1 with R848. Only CAL-1 cells robustly activated IRF5 upon R848 treatment (Fig. 2B). These results indicate that IRF5 is differentially regulated among the different cell types/lines and PBMC subsets.
Figure 2. IRF5 is activated by TLR7/8 stimulation specifically in pDCs.
(a) Immunoblot analysis of IRF5 expression in a panel of cell lines. Actin was used as a loading control. (b) Native PAGE immunoblot analysis of IRF5 in a panel of cell lines treated with 1ug/ml R848 for 2 hours. (c) Native PAGE immunoblot analysis of IRF5 in CAL-1 cells treated with increasing dose of R848 for 2 hours (d) Native PAGE immunoblot analysis of IRF5 in CAL-1 cells treated with 1ug/ml R848 over a time course of 0–24 hours. (e) ImageStream analysis of CAL-1 cells unstimulated (left) and treated with 1ug/ml R848 (right) for 1.5 hours. Brightfield, DAPI (purple), IRF5-AF647 (red) and merged DAPI/IRF5 channels are shown for 5 representative images (top panel). Similarity score were calculated for IRF5-AF647 and DAPI and displayed in histograms (bottom panel). Percentage of cells with similarity score above 1 is indicated. (f) Native PAGE immunoblot analyses of IRF5 in CAL-1 cells treated with a panel of TLR agonists at 5ug/ml for 4 hours. All data are representative of at least three independent experiments.
Due to extremely low frequency of pDCs in PBMCs, we utilized CAL-1 cells to study IRF5 regulation as they have been shown to recapitulate multiple aspects of human pDC physiology with an ability to mirror the response of primary human pDCs to stimulation(24, 49–51). Consistent with primary pDCs, we observed rapid and robust IRF5 activation upon R848 treatment of CAL-1 pDCs (Figs. 2C and 2D). Using ImageStream analysis, we confirmed that the percentage of cells with nuclear IRF5 markedly increased upon R848 stimulation as compared to untreated cells (Fig. 2E), in agreement with results from the dimerization assay. We did not detect IRF7 dimer formation upon TLR7/8 stimulation in R848-treated CAL-1 cells (Fig. S1E).
We focused on CAL-1 cells to identify the PRR signaling pathways that activate IRF5. We stimulated CAL-1 pDCs with a panel of agonists: LTA-SA (TLR2), FSL-1 (TLR2/6), poly(I:C) (TLR3), LPS (TLR4), FLA-ST (TLR5), R837 (TLR7), R848 (TLR7/8), CpG-A ODN2216 or CpG-B ODN2006 (TLR9) to assess whether IRF5 is activated by other TLRs. We observed IRF5 dimerization when CAL-1 cells were stimulated with R837 and R848. We also observed IRF5 dimerization upon stimulation with CpG-B, but not CpG-A, corroborating with previous findings that TLR9 stimulation activates IRF5 in primary human pDCs(24). These data indicate that endogenous IRF5 is activated downstream of endosomal TLRs, including TLR7, 8 and 9, in pDCs (Fig. 2F).
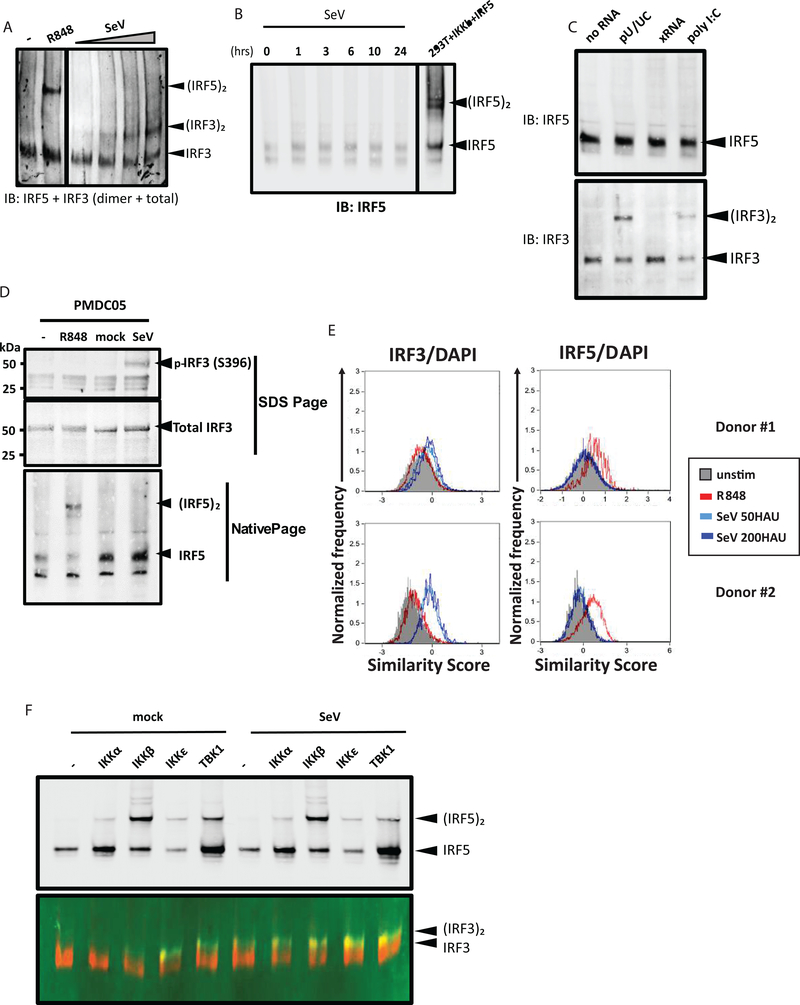
PRR-specific activation of IRF3 and IRF5
Previous studies showed that RLR signaling in pDCs can drive IFN production independently of TLR signaling(52). To determine whether CAL-1 cells had a functional RLR pathway, we infected cells with increasing dosage of Sendai virus (SeV) and assayed for the activation of IRF3, which is activated by RLR signaling(53). Upon SeV infection, we detected robust IRF3 activation/dimerization in CAL-1 cells, but IRF5 remained latent/monomeric (Fig. 3A and 3B). We also evaluated RLR signaling by transfection of the poly-U/UC motif from the Hepatitis C virus (HCV) genome, a known RIG-I-specific PAMP(54–56), or by high molecular weight poly(I:C) that activates MDA5(57). The non-stimulatory × region of the HCV genome (xRNA) was used as a negative control RNA. We simultaneously assessed IRF3 and IRF5 activation from the same cell extracts. We found that while IRF3 activation/dimerization occurred in cells transfected with poly-U/UC or poly(I:C) but not with xRNA, IRF5 remained in its latent/monomeric form upon each treatment (Fig. 3C). These results show that in CAL-1 pDCs, RLR signaling via RIG-I and MDA5 is fully intact to activate IRF3, whereas TLR7/8 signaling activates IRF5. To confirm these observations, we employed a Phos-Tag western blot assay to detect phosphorylated/activated forms of IRF3 and IRF5. The Phos-Tag reagent binds to phosphate groups of phosphorylated proteins to mediate mass shift for identification of phosphorylated isoforms by immunoblot(58). We identified phosphorylated/Phos-Tag bound IRF3 but not IRF5 in cells infected with SeV, while phosphorylated/Phos-Tag-bound IRF5 but not IRF3 was found in cells treated with R848 (Fig. S1F).
Figure 3. PRR-specific activation of IRF3 and IRF5 in pDCs.
(a) Native PAGE immunoblot analysis of IRF5, total IRF3 and IRF3 dimers in CAL-1 cells untreated and treated with 1ug/ml R848 for 2 hours, or infected with SeV at 25–200 HAU/ml for 16 hours. (b) Native PAGE immunoblot analysis of IRF5 in CAL-1 cells infected with SeV over a time course of 0–24 hours. Lysate from HEK293T cells overexpressing IRF5 and IKKβ was included in the same gel as positive control. (c) Native PAGE immunoblot analysis of IRF5 (top) or total IRF3 and IRF3 dimers (bottom) in CAL-1 cells transfected with no RNA, HCV polyU/UC PAMP RNA (pU/UC), HCV xRNA negative control (xRNA), or poly(I:C). Cells were harvested at 6 hours post transfection. (d) Immunoblot analysis of phosphor-IRF3 (S396) and total IRF3 (top panel), and native PAGE immunoblot analysis of IRF5 (bottom panel) in PMDC05 cells untreated and treated with 1ug/ml R848 for 2 hours, or mock- and SeV-infected for 14 hours. (e) ImageStream analysis of primary pDCs unstimulated (gray shaded), treated with 1ug/ml R848 (red) for 2 hours, or infected with SeV with (light blue and dark blue) for 6 hours. Results from two individual donors are shown. Similarity score were calculated for IRF3-AF488 and DAPI (left panel) and IRF5-AF647 and DAPI (right panel). (f) Native PAGE immunoblot analysis of IRF5 (top), or total IRF3 and IRF3 dimers (bottom) in HEK293T cells transfected with IRF5 alone (−) or IRF5 along with indicated IKK kinases. Cells were allowed to recover for 24 hours, then mock- or SeV-infected at 100 HAU/ml for 16 hours. Immunoblot experiments in (a-d, f) were performed at least 3 times. ImageStream experiments in (e) was performed 4 times, each time with 1 donor.
To assure that differential IRF3 and IRF5 activation in pDCs are not a spurious feature of CAL-1 pDCs, we utilized an additional human pDC cell line, PMDC05, which displays many hallmarks of pDCs and has been used for studying pDC biology(59, 60). We observed similar partitioning of IRF3 and IRF5 signaling in these cells, where SeV infection activated IRF3 but not IRF5, and R848 treatment activated IRF5 but not IRF3 (Fig. 3D). We then obtained primary human pDCs from healthy donors and tested IRF3 and IRF5 activation upon R848 treatment or SeV infection using ImageStream. Similar to our results in CAL-1 and PMDC05 cells, we observed IRF3 translocation into the nucleus upon SeV infection but not R848 treatment, whereas R848 treatment resulted in IRF5 nuclear translocation while IRF3 remained cytoplasmic in the primary human pDCs from each donor (Fig. 3E). These results show that activation of IRF3 and IRF5 are regulated by distinct PRR pathways in human pDCs.
IRF3 and IRF5 are both activated by IKK family kinases(21, 22, 28, 61). To test which activates the respective IRF, we ectopically expressed the four IKK kinases in HEK293T cells and assayed for IRF3 and IRF5 dimerization. While all IKKs induced IRF5 dimerization to various extents, only IKKε and TBK1 induced IRF3 dimerization (Fig. 3F). We infected the cells with SeV to test whether overexpression of IKKs sensitized the cells to RLR pathway activation upon virus infection. SeV infection alone induced IRF3 dimerization but not IRF5, and infection did not enhance IRF5 dimerization upon IKK overexpression (Fig. 3F). Together our datasets show that IRF3 and IRF5 activation are partitioned in pDCs wherein endosomal TLR signaling activates IRF5 but not IRF3, while IRF3 activation links specifically with RLR signaling.
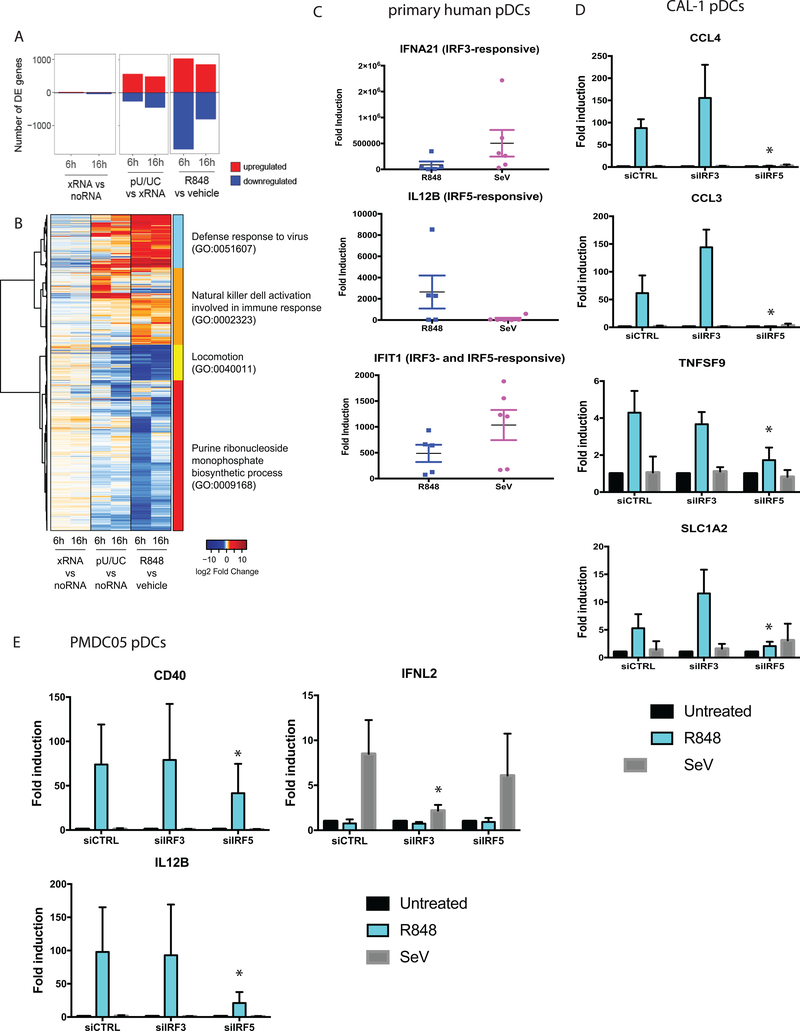
TLR7/8 and RLR signaling direct differential and overlapping IRF3 and IRF5 transcriptomes
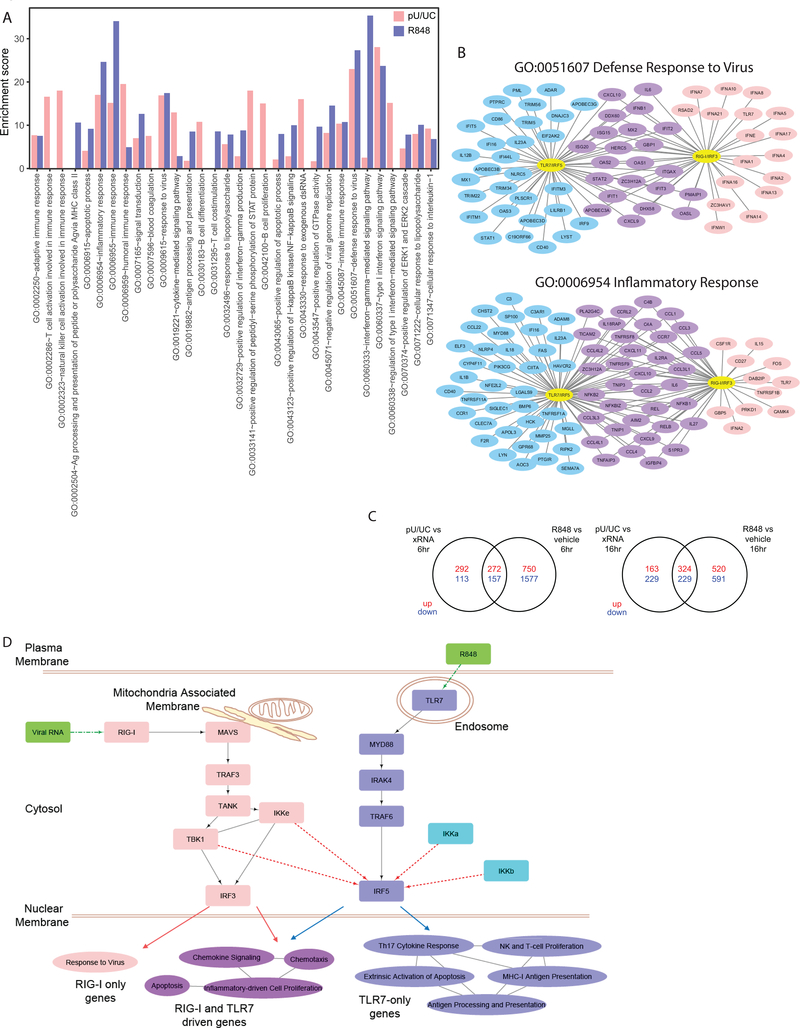
To determine how the partitioning of PRR signaling through TLR7/IRF5 and RLR/IRF3 directs the innate immune response in pDCs, we performed transcriptional profiling of CAL-1 cells by RNA-seq analysis. We focused on identifying IRF5- and IRF3-responsive genes respectively regulated by R848 treatment and poly-U/UC transfection. Control treatments included vehicle and transfection with non-signaling X-RNA or no RNA. X-RNA had little effect on gene expression when compared to no RNA transfection. Because X-RNA transfection resulted in mild changes in expression of some genes as compared to vehicle control (data not shown), indicating that the transfection process may trigger gene expression changes, we opted to use X-RNA as the proper control for poly-U/UC transfection. We identified differentially expressed (DE) genes (fold change>2; p<0.05) in R848-treated cells as compared to vehicle-treated cells, and in poly-U/UC-transfected cells as compared to X-RNA-transfected cells (Fig. 4A). Overall, the number of DE genes in R848 treatment exceeded that in poly-U/UC transfection, especially at the earlier time point (Fig. 4A). Analysis of gene function within the global DE gene set revealed distinct expression signatures of TLR7/IRF5 and RLR/IRF3 signaling (Fig. 4B). We examined the immune related gene modules and observed remarkable differences in R848- and pU/UC-specific programs that included differential innate immune response, inflammatory response, and immune activation driven by each stimulus representing distinct RLR/IRF3 and TLR7/IRF5 axes (Fig. S2).
Figure 4. RLR/IRF3 and TLR7/IRF5 partitioning regulate different sets of genes in pDCs.
(a) RNAseq analysis of CAL-1 cells treated with vehicle or 1ug/ml R848, or transfected with xRNA (control) or poly-U/UC RNA. Number of differentially expressed (DE) genes (Fold change > 2, p-value < 0.05) upregulated (red) or downregulated (blue) at 6 and 16 hour time points compared to control samples are shown. (b) Heatmap showing gene expression profiles of all genes that are DE in at least one comparison (R848/vehicle-6hrs, R848/vehicle-16hrs, pU/UC/xRNA-6hrs, and pU/UC/xRNA-16hrs), visualized by log2 fold change over control sample. (c) Quantitative real-time PCR of indicated genes in primary human donor pDCs treated with 1ug/ml R848 for 6 hours, or infected with SeV (100HAU/ml) for 9 hours. Data represent results from 6 individual donors. (d) Quantitative real-time PCR of indicated genes in CAL-1 pDCs transfected with siRNAs against IRF3 or IRF5, followed by treatment with 1ug/ml R848 for 6 hours, or infection with SeV (100HAU/ml) for 9 hours. Data represent results from biological triplicates. (e) Quantitative real-time PCR of indicated genes in PMDC05 pDCs transfected with siRNAs against IRF3 or IRF5, followed by treatment with 1ug/ml R848 for 6 hours, or infection with SeV (100HAU/ml) for 9 hours. RNAseq experiment was performed once with biological triplicates. Experiments with primary pDCs and pDC cell lines (c-e) were performed 4 times; quantitative real-time PCR data represent results from 4 biological replicates. *p<0.05.
To ensure that the RLR/IRF3 and TLR7/IRF5 signaling bifurcation is a physiologically relevant process in pDCs, we evaluated the expression of IRF3- and IRF5-responsive gene targets in R848-treated or SeV-infected primary human pDCs freshly isolated from healthy donors. We chose to assess the expression of individual genes rather than entire gene modules as biomarkers of IRF3 or IRF5 actions in order to validate each pathway marked by its responding gene as guided by the respective transcriptomics signature. Corroborating with our RNA-seq results, we observed induction of IFNA21, a known IRF3-responsive gene, upon SeV infection but occurring to a much lesser extent in R848 treatment. We assessed expression level of IL12B, an IRF5-responsive gene, and found that it was largely induced in R848 treatment but not upon SeV infection. IFIT1, a gene responsive to both IRF3 and IRF5, was induced both upon R848 treatment and SeV infection (Fig. 4C). We note that other factors, such as IRF7, likely play a role in regulating these genes in primary human pDCs, such that response variation is expected in primary human pDCs compared to a homogenous cell line. Importantly, given the expected donor-to-donor variability in responses, the major trends in IRF3- and IRF5-regulated gene expression were conserved in primary pDCs and pDC cell lines.
To definitively show the unique dependence of IRF3 and IRF5 in RLR and TLR7 signaling respectively, we first performed knockdown studies using siRNA and evaluated the expression of IRF3- and IRF5-repsonsive genes in CAL-1 pDCs upon SeV infection or R848 treatment. We observed a dependence of IRF5 but not IRF3 in the induction of CCL4, CCL3, TNFSF9, and SLC1A2, which are all R848- but not SeV-induced genes via IRF5 and IRF3, respectively (Fig.4D). We further validated these results by knocking down IRF3 or IRF5 in PMDC05 pDCs, another human pDC cell line (59). We observed similar dependence of IRF5 in the induction of R848-induced genes (IL12B and CD40) (Fig. 4E). Further, we observed dependence on IRF3, but not IRF5, in IFNλ2 expression, an SeV-induced gene (Fig. 4E). Together these results indicate that the observed partitioning of RLR/IRF3 and TLR7/IRF5 signaling is an intrinsic feature of pDCs.
To reveal the biological pathways regulated by the two axes, we performed functional analysis on all the DE genes from each treatment group to compare the enriched pathways. R848 and poly-U/UC stimulation resulted in a shared and distinct set of pathway enrichment (Fig. S3). Fig. 4C shows the top 20 most enriched gene ontology (GO) terms for DE genes regulated by RLR/IRF3 and TLR7/IRF5 activation. The top most enriched GO categories unique in R848-treated cells included antigen processing and presentation (GO:0002504) and T cell costimulation (GO:0031295), pointing to a bias towards activating adaptive immunity in response to IRF5 activation. On the other hand, GO terms unique in poly-U/UC-transfected cells included immune cell activation (GO:0002323 and GO:0002286), STAT protein activation (GO:0033141), and response to RNA (GO:0043330), showing a polarization towards cell-intrinsic innate immune activation driven by RLR/IRF3 signaling.
We observed that GO terms enriched by both stimuli are often driven by distinct genes (Fig. 5A and Fig. S3). For example, within the antiviral response (GO:0051607) and inflammatory response (GO:0006954) modules, poly-U/UC stimulation specifically induced the expression of various type I IFN subtypes, whereas R848 treatment signaled the induction of inflammatory cytokine genes, genes of immune activation, and molecules involved in pathogen sensing. The NF-kB pathway for inflammatory response featured prominently amongst DE genes shared by both poly-U/UC and R848 stimulations in these GO categories, suggesting a common immune activation pathway by both stimuli (Fig. 5B). Additional GO categories displayed a similar expression pattern, where TLR7/IRF5 and RLR/IRF3 signaling resulted in overlapping and distinct sets of DE genes (Fig. S4).
Figure 5. TLR7/IRF5 and RLR/IRF3 regulate genes with distinct functions.
(a) Bar graphs showing the enrichment score of the top 20 most enriched gene ontology (GO) terms in DE genes from R848/vehicle and poly-U/UC/xRNA comparisons. poly-U/UC stimulation is shown in pink, R848 treatment is shown in purple. No bar is shown for categories that are not enriched in the specific comparison. (b) Networks showing genes within GO categories “defense response to virus” (GO:0051607) and “inflammatory response” (GO:0006954) that are DE in R848 treatment (blue), poly-U/UC stimulation (pink), or both (purple). (c) Venn diagrams of upregulated (red) or downregulated (blue) genes in R848/vehicle and pU/UC/xRNA samples at 6 (top) and 16 (bottom) hour time points. (d) Diagram showing RLR and TLR pathway components leading to IRF3 and IRF5 activation using ImmuneDB analysis. Solid lines denote known pathway components. Dotted lines denote potential regulatory relationships demonstrated in our study. Pathway categories enriched in DE gene sets only in R848/vehicle, poly-U/UC/xRNA, or both in RNAseq analysis are displayed on the bottom. Lines between categories denote presence of shared genes in the categories. GO term enrichment induced by poly-U/UC is in pink, those induced by R848 are in blue, those shared by poly-U/UC and R848 stimulation are in purple. RNAseq experiment was performed once with biological triplicates.
We then examined DE genes in each stimulus to distinguish them into three groups: 1) those only induced by R848 treatment, 2) those only induced by poly-U/UC stimulation, and 3) those induced by both. We identified shared DE genes between R848 and poly-U/UC stimulations, but the majority of DE genes were respectively segregated into TLR7/IRF5 response and RLR/IRF3 response sets especially at the earlier time point (Fig. 5C). We performed DAVID functional analysis(62) on each group of DE genes to respectively reveal the pathways of TLR7/IRF5 and RLR/IRF3 activation in pDCs. We found that the pathways shared by both stimuli included chemotaxis, chemokine production, inflammatory response genes, and apoptosis (Fig. 5D). DE genes uniquely responsive to TLR7/IRF5 signaling by R848 were enriched for NK and T cell proliferation, Th17 response, and antigen processing/presentation. In contrast, poly-U/UC stimulation of RLR/IRF3 signaling enriched for genes that respond to virus infection (Fig. 5D). These results indicate that while IRF5 and IRF3 both serve to program pDCs for immune response activation, the TLR7/IRF5 and RLR/IRF3 pathways mediate differential immune polarization by pDCs to support an inflammatory response favoring T cell activation (TLR7/IRF5) or antiviral response (RLR/IRF3).
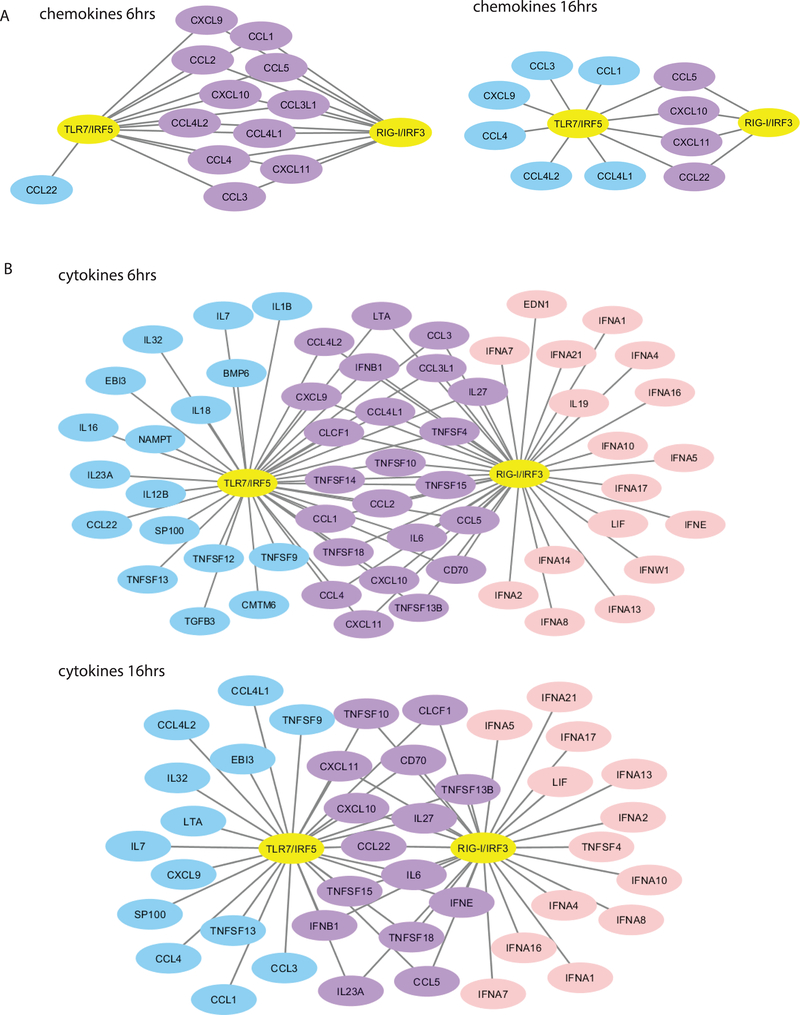
Because RLR/IRF3 and TLR7/IRF5 activation regulated genes that polarize toward distinct immune response programs, we examined the expression of chemokine genes. Chemokines play a major role in immune polarization(63, 64), and we found that most DE chemokine genes are regulated by both RLR/IRF3 and TLR7/IRF5 at the early time point (Fig. 6A). At the later time point, both RLR/IRF3 and TLR7/IRF5 regulated CCL5, CXCL10, CXCL11 and CCL22, but TLR7/IRF5 stimulation uniquely induced other chemokines. These results indicate that both pathways stimulated an initial burst of chemokines, but bifurcate in their chemokine profiles later that likely polarize the immune response by recruiting specific subsets of immune cells through chemokine actions.
Figure 6. IRF3 and IRF5 differentially regulate chemokine and cytokine gene expression in pDCs.
(a) Networks showing chemokine genes that are DE at 6 and 16 hrs post stimulation with R848 and poly-U/UC. (b) Networks showing cytokine genes that are DE at 6 and 16 hrs post stimulation with R848 and poly-U/UC. DE genes are shown for R848 treatment (blue), poly-U/UC stimulation (pink), or both (purple). RNAseq experiment was performed once with biological triplicates.
We also assessed DE cytokine gene expression (Fig. 6B). At 6 hours, poly-U/UC stimulation induced the expression of 12 different IFNA subtypes as well as IFNE and IFNW1, none of which was induced by R848 treatment. At 16 hours, 11 IFNA subtypes continued to be uniquely induced by poly-U/UC stimulation, while both poly-U/UC and R848 treatment induced IFNE. This expression profile reveals a dynamic regulation of type I IFN subtype expression signaled by both the RLR/IRF3 and TLR7/IRF5 pathways. On the other hand, R848 treatment induced proinflammatory cytokines as well as TGFB, which respectively are crucial for initiating the inflammatory response and supporting T cell differentiation(63). Other cytokines, such as the TNF super family cytokines, also displayed overlapping and distinct expression patterns differentially induced by RLR/IRF3 and TLR7/IRF5 signaling.
Taken together, these data show that TLR7/8 and RLR preferentially utilize IRF5 and IRF3 respectively to shape the immune response that is relevant to the danger at hand.
Differential cytokine and chemokine expression profiles regulated by IRF3 and IRF5 in pDCs
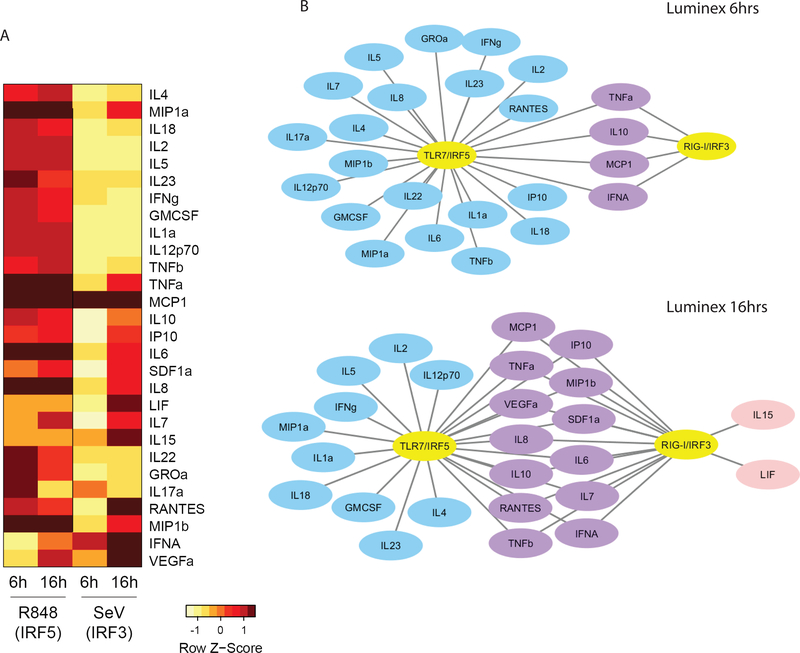
To further evaluate and validate the differential profiles of cytokines/chemokines produced in response to TLR7/IRF5 and RLR/IRF3 signaling, we analyzed cell supernatants for a panel of 45 secreted cytokines/chemokines using a Luminex multiplex assay. We found 28 analytes to be elevated by more than 2-fold after R848 treatment or SeV infection of CAL-1 cells in at least 1 time point (Fig. 7A). R848 treatment resulted in a more rapid and robust response overall than SeV infection in CAL-1 pDCs, likely due to the temporal nature of direct TLR7/8 stimulation compared to the processes of virus replication and PAMP signaling through RLRs(65). Of the panel of factors evaluated, some were highly induced by R848 but exhibited much lower induction in cells infected with SeV. These included major inflammatory cytokines such as IL-18, IL-1, IL-4, TNF, IL-6, and IL-8/CXCL8, as well as cytokines involved in immune activation and polarization including IL-2, IL-5, IL-23, IL-10, IL-22, IL-12, IFNγ, GM-CSF, GROα/CXCL1, MIP-1α/CCL3, and MIP-1β/CCL4. Some factors were produced robustly upon RLR/IRF3 signaling (LIF, IL-15, RANTES/CCL5, and IFNα), while the rest were induced in response to both TLR7/IRF5 and RLR/IRF3 signaling (Fig. 7B). While there are slight differences between the transcriptomic and proteomic datasets, likely indicating additional levels of regulation in cytokine production beyond transcription, these datasets are in overall agreement that TLR7/IRF5 signaling resulted in the preferred induction of proinflammatory cytokines in general, whereas RLR/IRF3 strongly favors type I IFN production and an antiviral response.
Figure 7. Differential cytokine and chemokine production by IRF3 and IRF5.
(a) Luminex analysis of CAL-1 cells treated with 1ug/ml R848 or infected with 100HAU/ml SeV for 6 or 16 hours. Fold change over untreated/mock-infected cells were calculated for each analyte, and those with fold change above 2 are displayed in the heap map. Log2 of fold change (log2(treatment) -log2 (baseline)) for each analyte was calculated and values were row-normalized and displayed as z-scores. (b) Luminex analysis in (a) displayed as networks showing 6 and 16 hour time points. Blue denotes factors only induced with R848 treatment, pink denotes factors only induced with SeV infection, purple denotes factors induced by both treatments. Luminex experiment was performed once with biological triplicates analyzed in technical duplicates.
Discussion
In this study we developed and utilized robust assays to detect endogenous IRF5 activation in primary human cells and cell lines. We revealed that IRF5 activation occurs in response to endosomal TLR signaling in a cell type-dependent manner. Examination of PBMC subsets showed that pDCs exhibit a robust response through the TLR7/IRF5 pathway. The high responsiveness of IRF5 activation in response to R848 treatment in pDCs compared to other cell subsets was recapitulated in both PMDC05 and CAL-1 pDC cell lines. Moreover, we demonstrate that RLR signaling in pDCs causes activation of IRF3 but not IRF5. Secreted cytokine/chemokine profiling and transcriptomic analyses showed distinct gene regulation profiles between IRF5 and IRF3. These results reveal a partitioning of TLR and RLR signaling that leads to activation of differential downstream transcription factors to direct appropriately tuned immune programs in pDCs.
While IRFs are well-studied and recognized as crucial factors in mediating PRR signaling, our study clarified several important aspects of IRF biology. We note that the regulation of IRF5 activity in vivo has remained elusive despite extensive studies. Important insights were provided by knockout mouse studies(8, 66), but key differences between human and mice necessitate careful examination, especially since earlier mouse studies harbored an additional confounding mutation(11, 12). In humans, much of our knowledge in IRF5 regulation has been gleaned from overexpression studies often in irrelevant cell types, sometimes using unreliable reagents(19–22, 29). Recently, several studies mainly using RNAi-knockdown approaches to examine endogenous IRF5 regulation support the notion that much remains to be learned about the multifaceted functions IRF5(21, 24, 67). A first major feature of our study is that we have now identified a relevant human cell type that provides an appropriate context to study IRF5 functions, and we have developed tools to interrogate endogenous IRF5 regulation. Second, RLR signaling is sometimes considered dispensable in pDCs(68), even though others have provided evidence for its utilization in specific contexts(52, 69). In our study and others’, RLR signaling has been shown to be capable in regulating IRF5 activation(22, 23). Our observations clearly show that the RLR pathway is intact and utilized in pDCs for pathogen sensing, but importantly its stimulation activates IRF3 and not IRF5. This outcome is clinically important as expression of both IRF3 and IRF5 in pDCs has been shown to play a role in systemic lupus erythematosus (SLE) pathology(70–74). Our study implies that different stimuli and cellular pathways can signal IRF3 and IRF5 activation underlying disease in SLE patients. Of note, our observations also highlighted the cell type differences in IRF5 regulation, raising the possibility that RLRs may regulate IRF5 activity in other cell types. Third, despite the lack of TLR7/IRF7 signaling in CAL-1 cells(24), we demonstrate that these cells are a useful model to study pDC biology(75, 76). In this case the lack of IRF7 activation serendipitously allowed us to define the relative contribution of IRF3 and IRF5 in pDCs, which otherwise could be masked by dominant effects of IRF7. Indeed, others have used CAL-1 cells to define processes of TLR9 signaling to distinct IRF family members(77). That we observed similar activation of IRF5 in response to TLR7/8 stimulation in primary pDCs, our results reflect relevant pDC physiology. In this regard, we note that besides IRF3 and IRF5, IRF7 plays an important role in PRR signaling in pDCs as demonstrated by many studies (78–80). In most cell types, IRF7 is IFN-inducible but IRF3 and IRF5 are constitutively expressed (81–84). Constitutive expression of IRF7 is a unique feature of pDCs, in which IRF7 has been shown to mediate late IFN production (24). It is possible that IRF3 and IRF5 mediate the acute response of pDC to specific TLR or RLR stimuli, while IRF7 serves an important role to amplify these responses and drive the high level production of IFN. However, we refrain from drawing any conclusion about IRF7 in this study, which focuses on the regulation and function of IRF3 and IRF5 in pDCs. Finally, IRFs share a common DNA binding domain (DBD) and are often hypothesized to have similar gene targets. Our study shows that despite sharing a conserved DBD, IRF members regulate disparate set of genes. We note that NF-kB activation is a shared component response of both TLR7 and RLR signaling(5) and likely contributes to the co-regulation of gene expression shared by TLR7/IRF5 and RLR/IRF3 pathways in the pDCs, as suggested by our transcriptomic and GO analyses.
The partitioning of IRF5 and IRF3 activation in pDCs presents mechanistic insights into how pDCs can be activated in different ways to exert various immune effector functions and immune polarization. Our observations are supported by recent studies that pDCs have diverse functions in immune regulation going beyond IFN production, to produce proinflammatory cytokines, and to mediate antigen presentation and T cell co-stimulation(64, 85–88). We expanded the repertoire of cytokines and chemokines that are produced by pDCs in response to differential stimulation of PRRs, and demonstrated the multifaceted functions of pDCs in response to TLR7/IRF5 and RLR/IRF3 pathway signaling. Our observations overall support a role for pDCs as more than professional type I IFN producing cells, but as a regulator of immune polarization and response. Thus, differential TLR versus RLR signaling outcome should be considered for targeting pDCs in immunotherapy and vaccine strategies aimed at modulating and enhancing specific components of the innate and adaptive immune response through differential usage of IRFs.
Supplementary Material
Acknowledgements
We thank all colleagues who generously shared reagents. We thank the Cell Analysis Facility (University of Washington) for help with ImageStream analysis, and Seattle Genomics (University of Washington) for providing RNA sequencing service. We thank all members of the Gale lab for experimental input and constructive criticisms.
This work was supported by Croucher Foundation Postdoctoral Fellowship and NIH NRSA F32 Postdoctoral Fellowship to KTC (AI115935), and NIH grants AI104002, AI100625, and AI083019.
Footnotes
Disclosure of Conflicts of Interest
The authors declare no competing financial interests.
Supplemental Materials
Please refer to Supplemental Materials for Supplemental Figures.
References
- 1.Barnes B, Lubyova B, and Pitha PM. 2002. On the role of IRF in host defense. J Interferon Cytokine Res 22: 59–71. [DOI] [PubMed] [Google Scholar]
- 2.Honda K, and Taniguchi T. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 6: 644–658. [DOI] [PubMed] [Google Scholar]
- 3.Ning S, Pagano JS, and Barber GN. 2011. IRF7: activation, regulation, modification and function. Genes Immun 12: 399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiscott J 2007. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem 282: 15325–15329. [DOI] [PubMed] [Google Scholar]
- 5.Loo YM, and Gale M Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity 34: 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kell AM, and Gale M Jr. 2015. RIG-I in RNA virus recognition. Virology 479-480: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton GM, and Kagan JC. 2009. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol 9: 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, and Taniguchi T. 2005. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 434: 243–249. [DOI] [PubMed] [Google Scholar]
- 9.Paun A, Reinert JT, Jiang Z, Medin C, Balkhi MY, Fitzgerald KA, and Pitha PM. 2008. Functional characterization of murine interferon regulatory factor 5 (IRF-5) and its role in the innate antiviral response. J Biol Chem 283: 14295–14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yasuda K, Richez C, Maciaszek JW, Agrawal N, Akira S, Marshak-Rothstein A, and Rifkin IR. 2007. Murine dendritic cell type I IFN production induced by human IgG-RNA immune complexes is IFN regulatory factor (IRF)5 and IRF7 dependent and is required for IL-6 production. J Immunol 178: 6876–6885. [DOI] [PubMed] [Google Scholar]
- 11.Purtha WE, Swiecki M, Colonna M, Diamond MS, and Bhattacharya D. 2012. Spontaneous mutation of the Dock2 gene in Irf5−/− mice complicates interpretation of type I interferon production and antibody responses. Proc Natl Acad Sci U S A 109: E898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasuda K, Nundel K, Watkins AA, Dhawan T, Bonegio RG, Ubellacker JM, Marshak-Rothstein A, and Rifkin IR. 2013. Phenotype and function of B cells and dendritic cells from interferon regulatory factor 5-deficient mice with and without a mutation in DOCK2. Int Immunol 25: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, and Crow MK. 2008. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis Rheum 58: 2481–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niewold TB, Kelly JA, Kariuki SN, Franek BS, Kumar AA, Kaufman KM, Thomas K, Walker D, Kamp S, Frost JM, Wong AK, Merrill JT, Alarcon-Riquelme ME, Tikly M, Ramsey-Goldman R, Reveille JD, Petri MA, Edberg JC, Kimberly RP, Alarcon GS, Kamen DL, Gilkeson GS, Vyse TJ, James JA, Gaffney PM, Moser KL, Crow MK, and Harley JB. 2012. IRF5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus. Ann Rheum Dis 71: 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rullo OJ, Woo JM, Wu H, Hoftman AD, Maranian P, Brahn BA, McCurdy D, Cantor RM, and Tsao BP. 2010. Association of IRF5 polymorphisms with activation of the interferon alpha pathway. Ann Rheum Dis 69: 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berggren O, Alexsson A, Morris DL, Tandre K, Weber G, Vyse TJ, Syvanen AC, Ronnblom L, and Eloranta ML. 2015. IFN-alpha production by plasmacytoid dendritic cell associations with polymorphisms in gene loci related to autoimmune and inflammatory diseases. Hum Mol Genet 24: 3571–3581. [DOI] [PubMed] [Google Scholar]
- 17.Hedl M, and Abraham C. 2012. IRF5 risk polymorphisms contribute to interindividual variance in pattern recognition receptor-mediated cytokine secretion in human monocyte-derived cells. J Immunol 188: 5348–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, Plenge RM, Koeuth T, Ortmann WA, Hom G, Bauer JW, Gillett C, Burtt N, Cunninghame Graham DS, Onofrio R, Petri M, Gunnarsson I, Svenungsson E, Ronnblom L, Nordmark G, Gregersen PK, Moser K, Gaffney PM, Criswell LA, Vyse TJ, Syvanen AC, Bohjanen PR, Daly MJ, Behrens TW, and Altshuler D. 2007. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci U S A 104: 6758–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, Fitzgerald KA, and Golenbock DT. 2005. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J Biol Chem 280: 17005–17012. [DOI] [PubMed] [Google Scholar]
- 20.Cheng TF, Brzostek S, Ando O, Van Scoy S, Kumar KP, and Reich NC. 2006. Differential activation of IFN regulatory factor (IRF)-3 and IRF-5 transcription factors during viral infection. J Immunol 176: 7462–7470. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Pelaez M, Lamont DJ, Peggie M, Shpiro N, Gray NS, and Cohen P. 2014. Protein kinase IKKbeta-catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proc Natl Acad Sci U S A 111: 17432–17437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren J, Chen X, and Chen ZJ. 2014. IKKbeta is an IRF5 kinase that instigates inflammation. Proc Natl Acad Sci U S A 111: 17438–17443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, Vick SC, Clepper L, Thackray L, Brassil MM, Virgin HW, Nikolich-Zugich J, Moses AV, Gale M Jr., Fruh K, and Diamond MS. 2013. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinhagen F, McFarland AP, Rodriguez LG, Tewary P, Jarret A, Savan R, and Klinman DM. 2013. IRF-5 and NF-kappaB p50 co-regulate IFN-beta and IL-6 expression in TLR9-stimulated human plasmacytoid dendritic cells. Eur J Immunol 43: 1896–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rustagi A, Doehle BP, McElrath MJ, and Gale M Jr. 2013. Two new monoclonal antibodies for biochemical and flow cytometric analyses of human interferon regulatory factor-3 activation, turnover, and depletion. Methods 59: 225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.George TC, Fanning SL, Fitzgerald-Bocarsly P, Medeiros RB, Highfill S, Shimizu Y, Hall BE, Frost K, Basiji D, Ortyn WE, Morrissey PJ, and Lynch DH. 2006. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J Immunol Methods 311: 117–129. [DOI] [PubMed] [Google Scholar]
- 27.Balkhi MY, Fitzgerald KA, and Pitha PM. 2008. Functional regulation of MyD88-activated interferon regulatory factor 5 by K63-linked polyubiquitination. Mol Cell Biol 28: 7296–7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang Foreman HC, Van Scoy S, Cheng TF, and Reich NC. 2012. Activation of interferon regulatory factor 5 by site specific phosphorylation. PLoS One 7: e33098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D, De S, Li D, Song S, Matta B, and Barnes BJ. 2016. Specific detection of interferon regulatory factor 5 (IRF5): A case of antibody inequality. Sci Rep 6: 31002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W, Lam SS, Srinath H, Jiang Z, Correia JJ, Schiffer CA, Fitzgerald KA, Lin K, and Royer WE Jr. 2008. Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5. Nat Struct Mol Biol 15: 1213–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balkhi MY, Fitzgerald KA, and Pitha PM. 2010. IKKalpha negatively regulates IRF-5 function in a MyD88-TRAF6 pathway. Cell Signal 22: 117–127. [DOI] [PubMed] [Google Scholar]
- 32.Sumpter R Jr., Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, and Gale M Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79: 2689–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DT, Weinman SA, Lemon SM, and Gale M Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A 103: 6001–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thackray LB, Shrestha B, Richner JM, Miner JJ, Pinto AK, Lazear HM, Gale M Jr., and Diamond MS. 2014. Interferon regulatory factor 5-dependent immune responses in the draining lymph node protect against West Nile virus infection. J Virol 88: 11007–11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paun A, Bankoti R, Joshi T, Pitha PM, and Stager S. 2011. Critical role of IRF-5 in the development of T helper 1 responses to Leishmania donovani infection. PLoS Pathog 7: e1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang CM, Roy S, Nielsen E, Paul M, Maul R, Paun A, Koentgen F, Raval FM, Szomolanyi-Tsuda E, and Pitha PM. 2012. Unique contribution of IRF-5-Ikaros axis to the B-cell IgG2a response. Genes Immun 13: 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krausgruber T, Saliba D, Ryzhakov G, Lanfrancotti A, Blazek K, and Udalova IA. 2010. IRF5 is required for late-phase TNF secretion by human dendritic cells. Blood 115: 4421–4430. [DOI] [PubMed] [Google Scholar]
- 38.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, and Akira S. 2002. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol 3: 196–200. [DOI] [PubMed] [Google Scholar]
- 39.Jurk M, Heil F, Vollmer J, Schetter C, Krieg AM, Wagner H, Lipford G, and Bauer S. 2002. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol 3: 499. [DOI] [PubMed] [Google Scholar]
- 40.Sprangers S, de Vries TJ, and Everts V. 2016. Monocyte Heterogeneity: Consequences for Monocyte-Derived Immune Cells. J Immunol Res 2016: 1475435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collin M, McGovern N, and Haniffa M. 2013. Human dendritic cell subsets. Immunology 140: 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider U, Schwenk HU, and Bornkamm G. 1977. Characterization of EBV-genome negative “null” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int J Cancer 19: 621–626. [DOI] [PubMed] [Google Scholar]
- 43.Menezes J, Leibold W, Klein G, and Clements G. 1975. Establishment and characterization of an Epstein-Barr virus (EBC)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt’s lymphoma. Biomedicine 22: 276–284. [PubMed] [Google Scholar]
- 44.Klein G, Giovanella B, Westman A, Stehlin JS, and Mumford D. 1975. An EBV-genome-negative cell line established from an American Burkitt lymphoma; receptor characteristics. EBV infectibility and permanent conversion into EBV-positive sublines by in vitro infection. Intervirology 5: 319–334. [DOI] [PubMed] [Google Scholar]
- 45.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, and Tada K. 1980. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int J Cancer 26: 171–176. [DOI] [PubMed] [Google Scholar]
- 46.Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, and Tada K. 1982. Induction of maturation in cultured human monocytic leukemia cells by a phorbol diester. Cancer Res 42: 1530–1536. [PubMed] [Google Scholar]
- 47.Hu ZB, Ma W, Zaborski M, MacLeod R, Quentmeier H, and Drexler HG. 1996. Establishment and characterization of two novel cytokine-responsive acute myeloid and monocytic leukemia cell lines, MUTZ-2 and MUTZ-3. Leukemia 10: 1025–1040. [PubMed] [Google Scholar]
- 48.Masterson AJ, Sombroek CC, De Gruijl TD, Graus YM, van der Vliet HJ, Lougheed SM, van den Eertwegh AJ, Pinedo HM, and Scheper RJ. 2002. MUTZ-3, a human cell line model for the cytokine-induced differentiation of dendritic cells from CD34+ precursors. Blood 100: 701–703. [DOI] [PubMed] [Google Scholar]
- 49.Maeda T, Murata K, Fukushima T, Sugahara K, Tsuruda K, Anami M, Onimaru Y, Tsukasaki K, Tomonaga M, Moriuchi R, Hasegawa H, Yamada Y, and Kamihira S. 2005. A novel plasmacytoid dendritic cell line, CAL-1, established from a patient with blastic natural killer cell lymphoma. Int J Hematol 81: 148–154. [DOI] [PubMed] [Google Scholar]
- 50.Steinhagen F, Meyer C, Tross D, Gursel M, Maeda T, Klaschik S, and Klinman DM. 2012. Activation of type I interferon-dependent genes characterizes the “core response” induced by CpG DNA. J Leukoc Biol 92: 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pelka K, and Latz E. 2013. IRF5, IRF8, and IRF7 in human pDCs - the good, the bad, and the insignificant? Eur J Immunol 43: 1693–1697. [DOI] [PubMed] [Google Scholar]
- 52.Stone AE, Giugliano S, Schnell G, Cheng L, Leahy KF, Golden-Mason L, Gale M Jr., and Rosen HR. 2013. Hepatitis C virus pathogen associated molecular pattern (PAMP) triggers production of lambda-interferons by human plasmacytoid dendritic cells. PLoS Pathog 9: e1003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, and Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5: 730–737. [DOI] [PubMed] [Google Scholar]
- 54.Schnell G, Loo YM, Marcotrigiano J, and Gale M Jr. 2012. Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I. PLoS Pathog 8: e1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saito T, Owen DM, Jiang F, Marcotrigiano J, and Gale M Jr. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454: 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uzri D, and Gehrke L. 2009. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol 83: 4174–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saito T, and Gale M Jr. 2008. Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J Exp Med 205: 1523–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, and Koike T. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5: 749–757. [DOI] [PubMed] [Google Scholar]
- 59.Narita M, Watanabe N, Yamahira A, Hashimoto S, Tochiki N, Saitoh A, Kaji M, Nakamura T, Furukawa T, Toba K, Fuse I, Aizawa Y, and Takahashi M. 2009. A leukemic plasmacytoid dendritic cell line, PMDC05, with the ability to secrete IFN-alpha by stimulation via Toll-like receptors and present antigens to naive T cells. Leuk Res 33: 1224–1232. [DOI] [PubMed] [Google Scholar]
- 60.Yamahira A, Narita M, Iwabuchi M, Uchiyama T, Iwaya S, Ohiwa R, Nishizawa Y, Suzuki T, Yokoyama Y, Hashimoto S, Takizawa J, Sone H, and Takahashi M. 2014. Activation of the leukemia plasmacytoid dendritic cell line PMDC05 by Toho-1, a novel IDO inhibitor. Anticancer Res 34: 4021–4028. [PubMed] [Google Scholar]
- 61.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, and Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4: 491–496. [DOI] [PubMed] [Google Scholar]
- 62.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, and Lempicki RA. 2007. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8: R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turner MD, Nedjai B, Hurst T, and Pennington DJ. 2014. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta 1843: 2563–2582. [DOI] [PubMed] [Google Scholar]
- 64.Swiecki M, and Colonna M. 2015. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol 15: 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Errett JS, Suthar MS, McMillan A, Diamond MS, and Gale M Jr. 2013. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol 87: 11416–11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dai P, Cao H, Merghoub T, Avogadri F, Wang W, Parikh T, Fang CM, Pitha PM, Fitzgerald KA, Rahman MM, McFadden G, Hu X, Houghton AN, Shuman S, and Deng L. 2011. Myxoma virus induces type I interferon production in murine plasmacytoid dendritic cells via a TLR9/MyD88-, IRF5/IRF7-, and IFNAR-dependent pathway. J Virol 85: 10814–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stone RC, Feng D, Deng J, Singh S, Yang L, Fitzgerald-Bocarsly P, Eloranta ML, Ronnblom L, and Barnes BJ. 2012. Interferon regulatory factor 5 activation in monocytes of systemic lupus erythematosus patients is triggered by circulating autoantigens independent of type I interferons. Arthritis Rheum 64: 788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoneyama M, and Fujita T. 2007. Function of RIG-I-like receptors in antiviral innate immunity. J Biol Chem 282: 15315–15318. [DOI] [PubMed] [Google Scholar]
- 69.Szabo A, Magyarics Z, Pazmandi K, Gopcsa L, Rajnavolgyi E, and Bacsi A. 2014. TLR ligands upregulate RIG-I expression in human plasmacytoid dendritic cells in a type I IFN-independent manner. Immunol Cell Biol 92: 671–678. [DOI] [PubMed] [Google Scholar]
- 70.Santana-de Anda K, Gomez-Martin D, Monsivais-Urenda AE, Salgado-Bustamante M, Gonzalez-Amaro R, and Alcocer-Varela J. 2014. Interferon regulatory factor 3 as key element of the interferon signature in plasmacytoid dendritic cells from systemic lupus erythematosus patients: novel genetic associations in the Mexican mestizo population. Clin Exp Immunol 178: 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akahoshi M, Nakashima H, Sadanaga A, Miyake K, Obara K, Tamari M, Hirota T, Matsuda A, and Shirakawa T. 2008. Promoter polymorphisms in the IRF3 gene confer protection against systemic lupus erythematosus. Lupus 17: 568–574. [DOI] [PubMed] [Google Scholar]
- 72.Lee YH, and Song GG. 2009. Association between the rs2004640 functional polymorphism of interferon regulatory factor 5 and systemic lupus erythematosus: a meta-analysis. Rheumatol Int 29: 1137–1142. [DOI] [PubMed] [Google Scholar]
- 73.Kelly JA, Kelley JM, Kaufman KM, Kilpatrick J, Bruner GR, Merrill JT, James JA, Frank SG, Reams E, Brown EE, Gibson AW, Marion MC, Langefeld CD, Li QZ, Karp DR, Wakeland EK, Petri M, Ramsey-Goldman R, Reveille JD, Vila LM, Alarcon GS, Kimberly RP, Harley JB, and Edberg JC. 2008. Interferon regulatory factor-5 is genetically associated with systemic lupus erythematosus in African Americans. Genes Immun 9: 187–194. [DOI] [PubMed] [Google Scholar]
- 74.Reddy MV, Velazquez-Cruz R, Baca V, Lima G, Granados J, Orozco L, and Alarcon-Riquelme ME. 2007. Genetic association of IRF5 with SLE in Mexicans: higher frequency of the risk haplotype and its homozygozity than Europeans. Hum Genet 121: 721–727. [DOI] [PubMed] [Google Scholar]
- 75.Karrich JJ, Balzarolo M, Schmidlin H, Libouban M, Nagasawa M, Gentek R, Kamihira S, Maeda T, Amsen D, Wolkers MC, and Blom B. 2012. The transcription factor Spi-B regulates human plasmacytoid dendritic cell survival through direct induction of the antiapoptotic gene BCL2-A1. Blood 119: 5191–5200. [DOI] [PubMed] [Google Scholar]
- 76.Karrich JJ, Jachimowski LC, Libouban M, Iyer A, Brandwijk K, Taanman-Kueter EW, Nagasawa M, de Jong EC, Uittenbogaart CH, and Blom B. 2013. MicroRNA-146a regulates survival and maturation of human plasmacytoid dendritic cells. Blood 122: 3001–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steinhagen F, Rodriguez LG, Tross D, Tewary P, Bode C, and Klinman DM. 2016. IRF5 and IRF8 modulate the CAL-1 human plasmacytoid dendritic cell line response following TLR9 ligation. Eur J Immunol 46: 647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Izaguirre A, Barnes BJ, Amrute S, Yeow WS, Megjugorac N, Dai J, Feng D, Chung E, Pitha PM, and Fitzgerald-Bocarsly P. 2003. Comparative analysis of IRF and IFN-alpha expression in human plasmacytoid and monocyte-derived dendritic cells. J Leukoc Biol 74: 1125–1138. [DOI] [PubMed] [Google Scholar]
- 79.Kerkmann M, Rothenfusser S, Hornung V, Towarowski A, Wagner M, Sarris A, Giese T, Endres S, and Hartmann G. 2003. Activation with CpG-A and CpG-B oligonucleotides reveals two distinct regulatory pathways of type I IFN synthesis in human plasmacytoid dendritic cells. J Immunol 170: 4465–4474. [DOI] [PubMed] [Google Scholar]
- 80.Dai J, Megjugorac NJ, Amrute SB, and Fitzgerald-Bocarsly P. 2004. Regulation of IFN regulatory factor-7 and IFN-alpha production by enveloped virus and lipopolysaccharide in human plasmacytoid dendritic cells. J Immunol 173: 1535–1548. [DOI] [PubMed] [Google Scholar]
- 81.Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant MJ, LePage C, DeLuca C, Kwon H, Lin R, and Hiscott J. 1999. Interferon regulatory factors: the next generation. Gene 237: 1–14. [DOI] [PubMed] [Google Scholar]
- 82.Zhang L, and Pagano JS. 2002. Structure and function of IRF-7. J Interferon Cytokine Res 22: 95–101. [DOI] [PubMed] [Google Scholar]
- 83.Taniguchi T, Ogasawara K, Takaoka A, and Tanaka N. 2001. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol 19: 623–655. [DOI] [PubMed] [Google Scholar]
- 84.Xu WD, Pan HF, Xu Y, and Ye DQ. 2013. Interferon regulatory factor 5 and autoimmune lupus. Expert Rev Mol Med 15: e6. [DOI] [PubMed] [Google Scholar]
- 85.Kamogawa-Schifter Y, Ohkawa J, Namiki S, Arai N, Arai K, and Liu Y. 2005. Ly49Q defines 2 pDC subsets in mice. Blood 105: 2787–2792. [DOI] [PubMed] [Google Scholar]
- 86.Guiducci C, Ott G, Chan JH, Damon E, Calacsan C, Matray T, Lee KD, Coffman RL, and Barrat FJ. 2006. Properties regulating the nature of the plasmacytoid dendritic cell response to Toll-like receptor 9 activation. J Exp Med 203: 1999–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ogata M, Ito T, Shimamoto K, Nakanishi T, Satsutani N, Miyamoto R, and Nomura S. 2013. Plasmacytoid dendritic cells have a cytokine-producing capacity to enhance ICOS ligand-mediated IL-10 production during T-cell priming. Int Immunol 25: 171–182. [DOI] [PubMed] [Google Scholar]
- 88.Osawa Y, Iho S, Takauji R, Takatsuka H, Yamamoto S, Takahashi T, Horiguchi S, Urasaki Y, Matsuki T, and Fujieda S. 2006. Collaborative action of NF-kappaB and p38 MAPK is involved in CpG DNA-induced IFN-alpha and chemokine production in human plasmacytoid dendritic cells. J Immunol 177: 4841–4852. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.