Abstract
MicroRNA (miRNA) are well known to target 3’ untranslated regions (3’UTR) in mRNAs to silence gene expression at post-transcriptional levels. Multiple reports have also indicated the capability of miRNAs to target protein-coding sequences (CDS); however, miRNAs have been generally believed to function in a similar mechanism(s) regardless of the location of their action sites. We herein report a class of miRNA recognition elements (MREs) that exclusively function in CDS regions in humans. Through functional and mechanistic characterization of these “unusual” MREs, we demonstrate that CDS-targeted miRNAs require extensive base pairings in the 3’ side rather than the 5’ seed; cause gene silencing in an Argonaute-dependent, but GW182-independent manner; and repress translation by inducing transient ribosome stalling instead of mRNA destabilization. These findings reveal distinct mechanisms and functional consequences for miRNAs to target CDS versus 3’UTR and suggest that CDS-targeted miRNAs may enlist a translational quality control (QC)-related mechanism to regulate translation in mammalian cells.
Introduction
MicroRNAs (miRNAs) are small noncoding RNAs (ncRNAs) of 21–22nt in length that function in post-transcriptional regulation of gene expression1,2. These small ncRNAs bind their target sequences in mRNAs, known as miRNA responsive elements (MREs), through imperfect base-pairing interactions. It has been well established that base-pairings via their the 5’ portion of sequences from 2–7nt, the “seed”, are particularly important for miRNA recognition of target mRNAs3. These base-pairing interactions take place in the RNA-Induced Silencing Complex (RISC) in which the 5’ portion of miRNA is exposed for initial base-pairings followed by a conformational change that allows additional base-pairings in the 3’ portion4,5. Thus, while nonessential, 3’ base-pairings may strength miRNA targeting. This so-called seed base-pairing rule has been the basis for development of various computational algorithms for predicting miRNA target sites in mRNAs6. However, there are numerous non-canonical MREs that have been described in literature7,8, indicating a degree of flexibility for miRNA-mRNA interactions, which may be influenced by a variety of parameters, including RNA secondary structures and RNA binding proteins that may enhance or repress miRNA targeting in the cell9.
It has been a general consensus in the field that miRNAs selectively target 3’UTRs of mRNA, as a functional MRE in 3’UTR generally loses its ability to mediate miRNA action when moved to the protein-coding sequence (CDS)10. This selectivity likely results from the action of elongating ribosome, which may strip off the RISC in CDS, but not in 3’UTR. However, various reports have characterized a number of functional MREs in CDS11–13. Because a main protein component of the RISC is an Argonaute protein (mammalian cells encode 4 such Argonaute paralogs, Ago1–4), genome-wide RISC targeting sites have been surveyed by UV crosslinking Immunoprecipitation sequencing (CLIP), which has revealed roughly equal distributions of Ago peaks in CDS and 3’UTR in both mammalian cells14,15. This suggests that the RISC may target the CDS regions more extensively than we thought, but such potential CDS MREs have not been widely appreciated nor characterized in any detail. It has thus remained largely unclear whether those Ago CLIP peaks represent some transient sampling events of the RISC in CDS or at least a fraction of those peaks reflect functional miRNA actions.
The functional consequence of miRNA action has been a major mystery in the field16, and little progress has been made since such review 10 years ago. Initially, using reporter-based assays, miRNA was found to induce translation repression as well as mRNA decay17–19. However, later genomic scale analysis of mRNA levels (by RNA-seq) and translation efficiency (by ribosome profiling normalized against transcript levels) suggests that miRNA-induced mRNA decay is the dominant functional consequence in mammalian cells20. However, careful analysis of miRNA-induced responses on endogenous gene products suggests that translation repression appears to occur ahead of mRNA decay21. The mechanism for miRNA-mediated mRNA decay has been well elucidated: The RISC initially recruits GW182 (mammalian cells encode 3 GW182 paralogs), which in turn attracts both decapping and deadenylase complexes to de-protect mRNA ends, leading mRNA decay by both 5’−3’ and 3’−5’ RNA exonucleases16,22. In contrast, many different mechanisms have been proposed to explain miRNA-mediated translational repression, ranging from interference of translation initiation23,24, to blockage of ribosome elongation25,26, to ribosome drop-off accompanying with degradation of nascent peptide27. To date, none of these mechanisms has been substantiated. Via whatever mechanism, it has been generally accepted that the RISC is part of the polysome28, suggesting that the RISC is associated with translating mRNA to modulate translation and/or degradation29.
In the current study, we pursued a MRE initially identified in the CDS region of the p53-activating kinase DAPK3 (ref30). Interestingly, we found that this MRE is representative of a class of previously unrecognized MREs that exclusively function in CDS. Extensively mutational analysis revealed that this class of MREs requires extensive 3’ base-pairings with a minimal requirement for 5’ base-pairings in the seed. These CDS MREs effectively repress translation, but do not seem to affect mRNA stability; require an Ago, but not GW182. Ribosome profiling experiments showed that these MREs induce a measurable degree of ribosome stalling, but without evident ribosome queuing, suggesting that CDS RISC may have both local and global impacts on translation, and we provide initial evidence that such regulation may be related to a mechanism involved in translation quality control.
Results
A MRE exclusively functions in CDS to repress translation
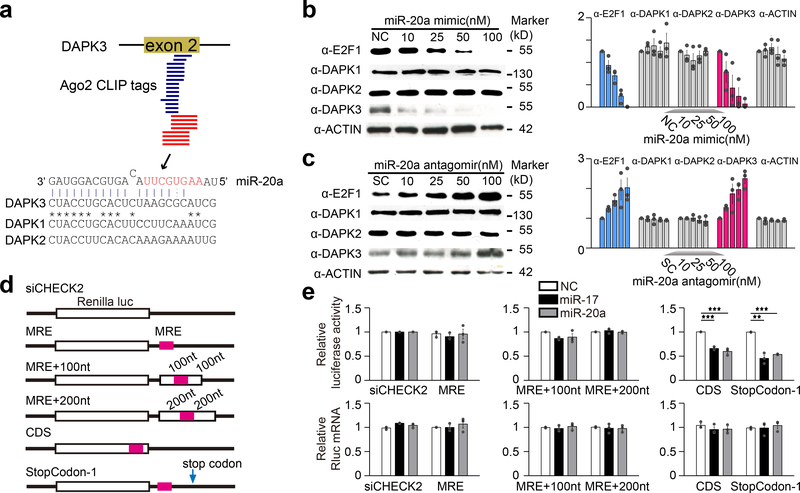
We previously identified and characterized a target site for both miR-17 and miR-20a in the second coding exon of the p53-activating kinase DAPK3 (ref30). These two miRNAs differ by a single nucleotide in the 5’ end, which does not affect their base-pairing interactions. Since most of the following studies employed miR-20a, we thus use miR-20a throughout the report to represent both miR-17 and miR-20a. Several approaches confirmed that this site is an authentic MRE: The site was identified in two separate Ago2 mapping experiments14,15 as well as by CLASH mapping of miRNA:mRNA interactions in HEK293T cells8 (Fig.1a). Importantly, DAPK3 protein levels were specifically repressed by a miR-20a mimic and elevated by a miR-20a antagomir, similar to the expected effects on the E2F1 protein via the characterized miR-20a MRE in its 3’UTR31 (Fig.1b,c; Supplementary Fig.1a-d). However, we detected no change in endogenous DAPK3 mRNA levels (Supplementary Fig.1e,f), suggesting that this CDS MRE mediates translational repression, rather than mRNA decay.
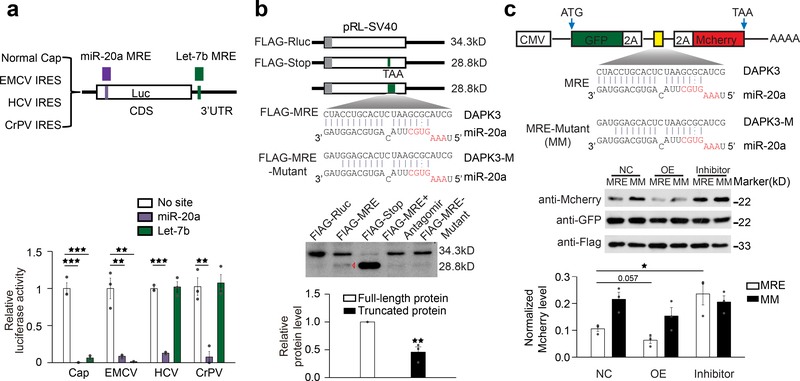
Fig. 1. miR-20a target site in DAPK3 functions exclusively in CDS.
a, Location of a miR-20a-responsive MRE in the second coding exon of the DAPK3 gene. Blue lines: Ago2 HITS-CLIP tags identified by Darnell and Colleagues15; red lines: Ago2 CLIP-seq identified in our previous study14. b,c, Western blotting analysis of miR-20a targets in response to the miR-20a mimic (b) or antagomir (c) in transfected HeLa cells. Data were quantified by ImageJ and presented on the right. d, Luciferase reporter constructs containing the miR-20a MRE from DAPK3with various lengths of flanking sequences (MRE+100nt or MRE+200nt) in Renilla 3’ UTR, or in a CDS region or 1nt deletion in the stop codon to allow the use of a downstream TAA stop codon. e, Results of the luciferase reporter assays (upper panels) and quantification of luciferase mRNA (lower panels). Quantitative results were based on 3 independent experiments and expressed as mean+/−SEM. **p<0.01; ***p<0.001 (Student’s t-test). See also Supplementary Fig.1 for dosage-dependent effects of the miR-20a mimic and antagomir.
This MRE is atypical of “standard” MREs in that it lacks typical seed base-pairing with its cognate miRNA. To characterize this unusual MRE, we inserted it into a standard dual luciferase reporter where the MRE is present in the 3’UTR of the Renilla luciferase. Unexpectedly, the MRE was non-functional in this context (Fig.1d,e). To ensure that the loss of function was not caused by sequence effects, we inserted varying amounts of “flanking’ sequences derived from exon 2 of DAPK3, but still failed to record any effect. In contrast, when we placed the MRE in CDS 6nt upstream of the stop codon or mutated the stop codon to allow the use of a downstream stop codon, thereby switching the MRE from 3’UTR to CDS, both reporters responded to transfected miR-20a mimic (Fig.1d,e). Again, we observed no change at the mRNA levels under all of these conditions (Fig.1e). Therefore, this MRE appears to function only in CDS, unlike all previously reported CDS MREs that were also functional when placed in 3’UTR11,12,32–34, and in contrast to typical MREs that lost functionality when moved to CDS10.
CDS MRE requires extensive 3’ base-pairings for functioning
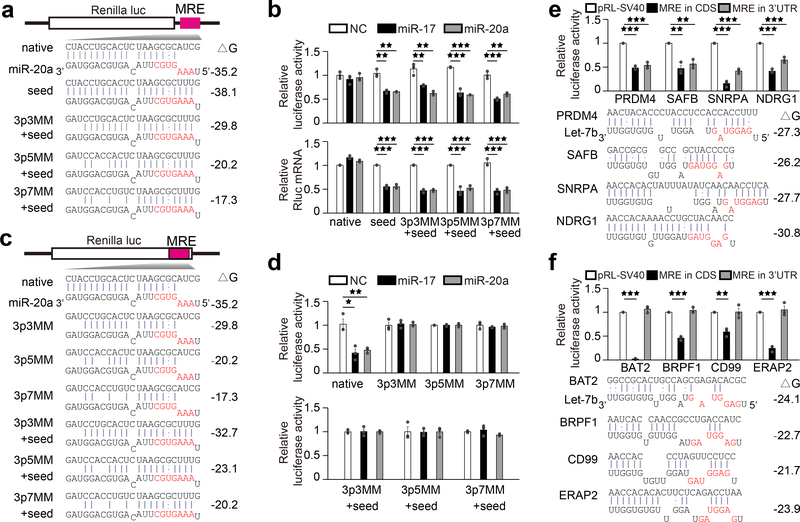
We noted that while this atypical MRE lacked conventional 5’ seed base-pairings with target mRNA, it had extensive base-pairing potential at 3’ end, consistent with the directly mapped miRNA targeting sites in CDS in C. elegans35. When mutations were introduced into the MRE to re-establish seed base-pairing (Fig.2a), the MRE placed in 3’UTR became responsive to transfected miR-20a mimic (Fig.2b) and 3’ base-pairing became nonessential (Fig.2a,b). To determine if 3’ base-pairing was vital for the MRE to function in CDS, we introduced mutations into the MRE and found that 3’ base-pairing was indeed critical for the MRE to function, even after restoring seed base-pairing (Fig.2c,d). We also tested the MRE in 5’UTR and found that, similar to its behavior in 3’UTR, it largely lacked the effect, but restoration of seed base-pairing regained repression of the luciferase reporter activity, and with restored seed, 3’ base-pairing was not critically required (Supplementary Fig.1g-i).
Fig. 2. CDS and 3’UTR targeted miRNA requires distinct base pairings at the 3’ end and 5’ seed.
a, Luciferase reporter constructs containing the miR-20a MRE in DAPK3(native) in Renilla 3’ UTR or mutated to restore base-pairing in 5’seed coupled with progressive mismatches (3MM, 5MM and 7MM) in 3’ end. Seed sequences are highlighted in red and Gibbs free energies (ΔG) of base-pairing potentials are shown in right. b, The luciferase assay results (upper panels) and relative luciferase mRNA levels (lower panels). c, Luciferase constructs containing the miR-20a MRE in the Renilla CDS region with increase in mismatches (3MM, 5MM and 7MM) in the 3’ end, without or with restored base pairing in the 5’ seed (+seed). Calculated base-pairing potentials (ΔG) are shown in right. d, The luciferase assay results. e,f, Let-7b target sites identified in CDS of the genes indicated were inserted in CDS (black bar) or 3’UTR (gray bar) of the Renilla reporter. Shown are deduced base-pairing in each case and the luciferase results of the Let-7b sites that function in both CDS and 3’UTR (e) or only in CDS (f). Quantitative data in b,d,e,f were based on 3 independent experiments and expressed as mean+/−SEM. *p<0.05; **p<0.01; ***p<0.001 (Student’s t-test). See also Supplementary Fig.1–3 for additional mutational analysis results.
We next sought to determine if this miR-20a MRE was unique. Mining of the publicly available CLASH data8, we identified two classes of putative MREs. The first class (i.e. those in PRDM4, SAFB, SNRPA, and NDRG1) showed both 5’ and 3’ base-pairing potential with Let-7b, and when inserted into the luciferase reporter, all repressed luciferase activity regardless in 3’UTR or CDS (Fig.2e). In contrast, the second class (i.e. those in BAT2, BRPF1, CD99, and ERAP2) exhibited pronounced similarity to the miR-20a MRE, with substantial 3’ pairing potential with Let-7b, but lacking typical base-pairing at the 5’ seed, and when placed into the luciferase reporter, all exclusively functioned in CDS context (Fig.2f). Like the miR-20a MRE in DAPK3, none of these MREs that functioned only in CDS caused mRNA decay, in contrast to the four MREs in the first class, all causing mRNA reduction (Supplementary Fig.1j,k). We further showed that all of the effects on the luciferase activity and mRNA levels were Let-7 dependent (Supplementary Fig.1l,m). Western blotting with available antibodies further confirmed the expected effects of transfected Let-7b mimic or antagomir on endogenous CD99, NDRG1, and SAFB proteins (Supplementary Fig.1n). As with the miR-20a MRE in DAPK3, extensive mutational analysis on BAT2, BRPF1, CD99, and ERAP2 demonstrated the central requirement of 3’ base-pairings for each of these MREs to function in CDS, with or without restored 5’ seed (Supplementary Fig.2), and with full 3’ base-pairings, a minimal of 3 base-pairings in the 5’ seed was found to be critical, as shown on the ERAP2 MRE (Supplementary Fig.3). Collectively, these results define a novel class of MREs wherein substantive 3’ base-pairings with minimal base-pairings at the 5’ seed confer their repressive activities exclusively in CDS.
The function of CDS targeted miRNA depends on Ago, but GW182
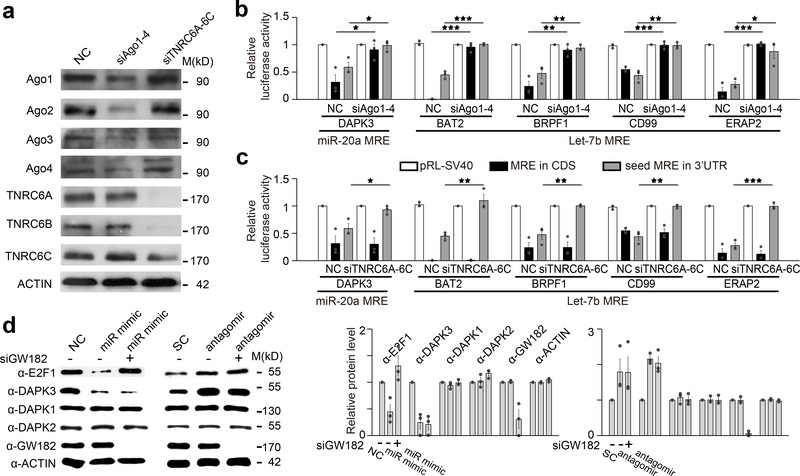
It has been well established that most, if not all, of miRNA-mediated repression by “standard” MREs in 3’UTR result from targeted mRNA destabilization20,29,36, which fully agrees with our current data (Fig.2b; Supplementary Fig.1j; Supplementary Fig.3). By contrast, with all atypical MREs that function in CDS, we failed to detect mRNA reduction that could account for reduced luciferase activity (Fig.1e; Supplementary Fig.1e,f; Supplementary Fig.1k). Interestingly, even with those MREs that function in both CDS and 3’UTR, we did not detect mRNA reduction in the context of CDS (see Supplementary Fig.1j), suggesting that CDS targeted miRNAs may function via a mechanism different from that used by 3’UTR targeted miRNAs. This prompted us to ask if the same protein cofactors that mediate standard MRE function are also required for CDS MRE to function. An Ago protein and GW182 (a.k.a. TNRC6A) are both required for standard MRE function in 3’UTR17,37–40. To test whether Ago is required, we analyzed multiple atypical MREs in Ago2 null mouse embryonic fibroblast (MEFs)39 as well as in HeLa cells treated with a pool of siRNAs against Ago1–4. The function of these atypical MREs was partially compromised in Ago2 null MEFs and completely abolished in Ago1–4 depleted HeLa cells, indicating the requirement of an Ago protein for CDS MRE to function (Fig.3a,b; Supplementary Fig.4a). By contrast, knockdown of all three TNRC6 family members, a treatment that prevented standard MRE to function in 3’UTR, did not affect the activity of CDS MRE either based on reporter assays (Fig.3c; Supplementary Fig.4b) or on the response of endogenous DAPK3 (Fig.3d; Supplementary Fig.4c).
Fig. 3. CDS-targeted miRNAs function in an Ago-dependent but GW182-independent manner.
a, Confirmation of siRNA knockdown of Ago1–4 and GW182 family member (TNRC6A-C) by western blotting. b,c, Luciferase reporter assays on five MREs from the indicated genes inserted into the CDS region (black bars) or the MREs each with restored 5’ seed in 3’UTR (gray bars) of the Rellina luciferase reporter in transfected HeLa cells, which were treated with either non-specific control (NC) or a pool of siRNAs against Ago1–4 (b)or three GW182 family members (c). *p<0.05; **p<0.01; ***p<0.001 (Student’s t-test). d, Western blotting analysis of E2F1, DAPK1, DAPK2 and DAPK3 in HeLa cells transfected with miR-20a mimic or antagomir, with or without siGW182. Quantitative data (by ImageJ) on the right were based on 3 independent experiments and expressed as mean+/−SEM. See Supplementary Fig.4 for additional data upon single knockdown of Ago2 or GW182 and the two repeated western blotting results.
Lack of mRNA decay due to inefficient recruitment of GW182
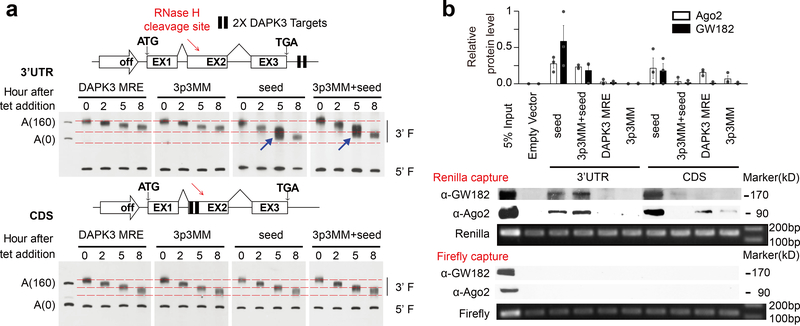
As Ago-associated TNRC6 proteins are required for bridging mRNA decay machineries to induce deadenylation followed by decapping17,37, the TNRC6-independent function of atypical MREs might thus explain unchanged mRNA levels. To provide further evidence for this, we inserted the miR-20a MRE and variants with different base-pairing potentials in CDS or 3’UTR into an inducible β-globin gene and performed a pulse-chase experiment to detect accelerated deadenylation (ADA) by Northern blotting (Fig.4a), as described earlier41. We found that, with the MRE inserted in 3’UTR, extensive base-pairing in the 5’ seed, but not 3’ end, was required for ADA (Fig.4a, arrows in upper panel). By contrast, we detected little effect with the same MRE inserted in CDS despite the increased base-pairing potential at the 3’ end or the 5’ seed (Fig.4a, lower panel). These findings suggest that sufficient base-pairing in the 5’ seed is central for the recruitment of mRNA decay machineries in 3’UTR to trigger mRNA decay, but such recruitment may be prevented by elongating ribosome in CDS.
Fig. 4. Inefficient recruitment of GW182 to CDS RISC.
a, Time-course deadenylation assay by Northern blotting. Two copies of the miR-20a native MRE from DAPK3 or its derivatives were inserted into a β-globin minigene (TBG) expressed from a tet-inducible promoter either in the 3’UTR (upper left panel) or CDS (lower left panel). 3p3MM: 3 mismatches in the 3’ end; seed: restored seed in the 5’ end; and combination of both mutations. The two 3’UTR reporters with restored seed regardless of 3’ mismatches showed accelerated deadenylation (ADA), as highlighted by the two blue arrows in the upper right panel, while the 3’UTR constructs carrying the native or mutant MRE from DAPK3 or all CDS reporters exhibited identical, unaltered deadenylation kinetics (lower right panel). b, Western blot analysis of Ago2 and GW182 associated with the Renilla luciferase reporters carrying perfect seed base-pairing (“seed”) or imperfect seed with or without additional 3nt mismatches in the 3’ end (“3p3MM+seed” and “3p3MM”) in CDS or 3’UTR. Equal amounts of captured reporter mRNAs were detected by RT-PCR. Quantified data from 3 independent capture experiments are shown on the top. See Supplementary Fig.4d for the other two repeated capture results.
To directly evaluate RISC recruitment, we engineered a Renilla reporter to carry the miR-20a MRE or variations in CDS or 3’UTR. Using the Firefly reporter as a negative control, we performed mRNA capture with biotinylated antisense oligonucleotides followed by Western blotting for Ago2 and GW182 (Fig.4b). We found that seed base-pairing alone was required for both Ago2 and GW182 to bind the Renilla mRNA that contains the MRE in 3’UTR. By contrast, extensive base-pairing interactions at both the 5’ seed and the 3’ end were required for stable binding of Ago2 and GW182 to the reporter mRNA carrying the MRE in CDS, implying that under certain conditions, some very stable RISC in CDS might also cause a measurable degree of mRNA decay. Interestingly, in CDS, the miR-20a MRE in DAPK3 with strong base-pairing interactions at the 3’ end, but minimal base-pairings in the 5’ seed still allowed a degree of Ago2 recruitment, but not GW182. Quantification of three independent capture experiments confirmed these results (Fig.4b, top panel; Supplementary Fig.4d). These data suggest that translating ribosome may prevent CDS RISC from stably recruiting additional co-factors to mediate mRNA decay.
Evidence for aborted translation induced by CDS-targeted miRNA
To investigate how miRNA might repress translation when acting in CDS, we took advantage of a report showing that the effect of 3’UTR-targeted Let-7b could be progressively bypassed by virus-derived internal ribosome entry sites (IRESs)42. Using this set of these reporters, we confirmed the inhibitory effect of Let-7b on the cap-dependent reporter as well as on the reporter containing the encephalomyocarditis virus (EMCV) IRES that bypasses the requirement for eIF4E (Fig.5a). As demonstrated earlier42, the effect of Let-7b was greatly diminished on the reporter carrying the hepatitis C virus (HCV) IRES, which bypasses the requirement for initiation factor-dependent recruitment of 40S, but not 60S ribosome, and on the reporter containing the cricket paralysis virus (CrPV) IRES, which requires no initiation factors to promote translational elongation (Fig.5a). Under the same conditions, the CDS MRE from DAPK3 mediated translational repression on all of these reporters (Fig.5a).
Fig. 5. CDS targeted miRNA induces aborted translation.
a, Luciferase assays of the reporters carrying normal cap or IRES from EMCV, HCV or CrPV, each of which also contains the miR-20a MRE in the CDS or the Let-7bMRE in 3’UTR. b, Upper panel: Diagram of FLAG-tagged luciferase constructs containing a stop codon or carrying a wild-type or mutant miR-20a MRE from DAPK3 in CDS. The expected protein sizes are indicated on the right. Lower panel: Western blotting of the luciferase reporter proteins expressed in transfected HeLa cells. The second to last lane was from cells co-transfected with the reporter containing the miR-20a MRE and the miR-20a antagomir. The data are representative of three independent experiments and quantified at bottom with the other two repeats shown in Supplementary Fig.4e. c, Upper panel: Diagram of theGFP-2A-MRE-2A-RFP constructs containing either wild-type or mutant miR-20a MRE. Lower panel: Western blotting of the reporter proteins expressed in transfected HeLa cells. A FLAG-tag Renilla was co-transfected to provide a loading control. The data are representative of three independent experiments and quantified at bottom with the other two repeats shown in Supplementary Fig.4f. Data in a,b,c were expressed as mean+/−SEM. *p<0.05; **p<0.01; ***p<0.001 (Student’s t-test).
To further probe for the mechanism underlying translational repression, we inserted the MRE from DAPK3 into the luciferase reporter at the location expected to truncate the full-length 34kD FLAG-luciferase protein to ~28kD in transfected HeLa cells, and for comparison, we also engineered a stop codon or a non-functional MRE with 3 base mismatches at the 3’ end in the same location (Fig.5b, upper panel). Under these reporter-based, overexpression conditions, while the wild-type luciferase gave rise to the expected 34kD band, we indeed detected a 28kD band with the reporter containing the functional MRE in CDS, although the level of such band was much lower that induced by the stop codon (note a minor degree of translational read-through with the stop codon-containing reporter) (Fig.5b). Importantly, such truncated product depended on the functional MRE, as such band was significantly attenuated with the mutant MRE inserted in the same location as well as when the cells were co-transfected with a miR-20a antagomir. Quantification of three independent experiments confirmed these results (Fig.5b, lower panel; Supplementary Fig.4e).
To further substantiate such aborted translation, we took a “2A” strategy, in which a P2A self-cleavage peptide sequence was inserted between GFP and MCherry to allow translation of the two reporters from a single mRNA43,44. This enabled us to detect MCherry reduction relative to GFP in response to a functional MRE in CDS. As expected, based on three independent experiments, we found that MCherry was reduced in response to wild-type miR-20a MRE from DAPK3 in comparison with a mutant MRE. miR-20a overexpression caused a further effect (note a reduction of MCherry even with the mutant MRE, likely resulting from miR-20a overexpression) and the miR-20a antagomir diminished such MRE-dependent effect (Fig.5c; Supplementary Fig.4f). Together, these data demonstrated aborted translation by a CDS-targeted miRNA.
CDS-targeted miRNA induces a degree of ribosome stalling
A RISC in the middle of CDS might cause local ribosome stalling followed by drop-off as a mechanism to induce translation abortion. To test this possibility, we performed ribosome profiling to characterize ribosome binding behavior relative to miRNA actions on endogenous targets20. For this purpose, we had to first localize RISC complexes in both CDS and 3’UTR for comparison and in response to overexpression of a specific miRNA. From two independent Ago2 eCLIP experiments that demonstrated the reproducibility of our libraries (Supplementary Fig.5a), we identified specific Ago2 peaks before and after miR-20a transfection (Fig.6a). We found enriched seed sequences in 3’UTR based on predicted seeds underlying mapped Ago2 peaks for top 20 expressed miRNAs in HeLa cells (Supplementary Fig.5b, left) and highly enriched seed sequences for miR-20a underlying miR-20a overexpression-induced Ago2 peaks in 3’UTR (Supplementary Fig.5b, right; Supplementary Fig.5c). Importantly, relative to unaltered Ago2 peaks or random locations in mRNAs, we found elevated base-pairing potential with miR-20a among induced Ago2 peaks in both CDS and 3’UTR, which is evident from using combined Ago2 eCLIP data (Fig.6b) or based on independent repeats of the Ago2 eCLIP experiment (Supplementary Fig.5d). Together, these data strongly support that the identified Ago2 peaks reflect the miRNA action sites on endogenous mRNAs.
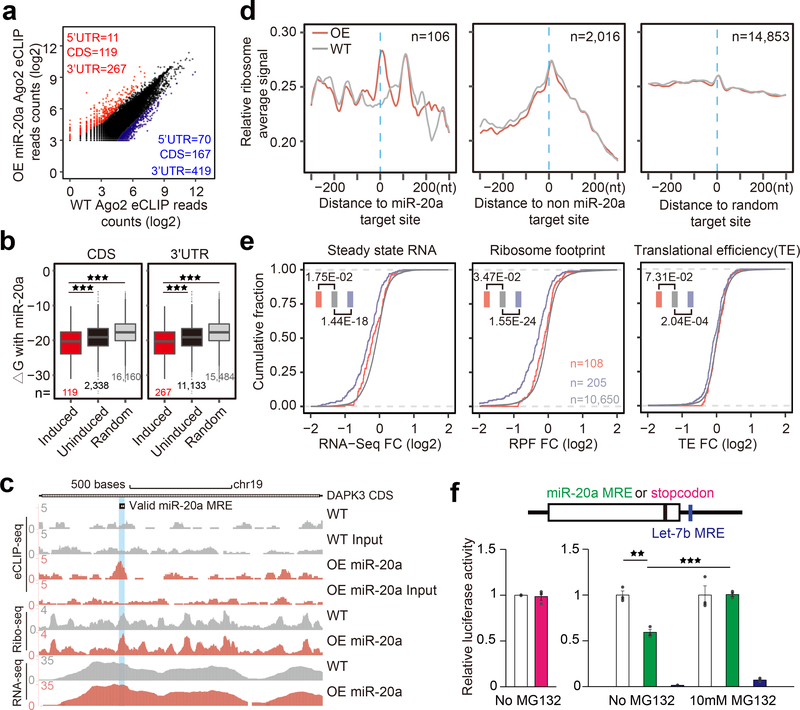
Fig. 6. Impact of CDS-targeted miRNAs on ribosome binding.
a, Scatter plot of combined Ago2 eCLIP data before and after miR-20a overexpression. Red and blue: Induced and decreased Ago2 peaks, separately counted in 5’UTR, CDS, and 3’UTR. b, Boxplots of base-pairing potential (based on ΔG) between miR-20a and sequences underlying miR-20a overexpression-induced Ago2 peaks in CDS (left) or 3’UTR (right) relative to uninduced Ago2 peaks or random sites. The median value for each group is shown with a horizontal gray line. Filled boxes extend from the first to the third quartile. The upper/lower whisker extends from the hinge to the highest/lowest value that is within 1.5 * IQR of the hinge, where IQR is the inter-quartile range. Data beyond the end of the whiskers are outliers and plotted as points. *p<0.05; **p<0.01; ***p<0.001 (Student’s t-test). c, UCSC genome browser view of Ago2 eCLIP-seq, Ribo-seq, and RNA-seq tracks on the DAPK3 transcript before and after miR-20a overexpression, all based on combined reads from independent replicates of each experiment. d, Meta-gene analysis of RFPs on miR-20a overexpression-induced Ago2 peaks (left), all other Ago2 peaks (middle) or on random sites (right). Gray and red tracks: RPFs before and after miR-20a overexpression, respectively. e, Accumulative distribution of RNA-seq, Ribo-seq, and translation efficiency fold-change (FC) in response to miR-20a overexpression. Red and blue: Transcripts containing miR-20a induced Ago2 peaks in CDS or 3’UTR (note that transcripts containing Ago2 in both CDS and 3’UTR were not included in the analysis); Gray: Transcripts without significant Ago2 binding peak. See Supplementary Fig.5 for additional Ago2 eCLIP and ribosome profiling related data. f, Evidence for CDS-targeted miRNA-induced degradation of nascent polypeptide. Left, luciferase assay of the reporters containing a normal stop codon or the stop codon moved to 6nt upstream. Right, luciferase assay of the reporters carrying the miR-20a target site in the CDS region near the 3’end with or without MG132 treatment. Quantitative results were based on 3 independent experiments and expressed as mean+/−SEM. **p<0.01; ***p<0.001 (Student’s t-test).
We next performed parallel ribosome profiling and RNA-seq under the same conditions and with independent replicates to show the reproducibility of our libraries (Supplementary Fig.5e). As expected, we observed typical length distribution of ribosome-protected fragments (RPFs), and more importantly, the 3nt periodicity (Supplementary Fig.5f,g). Although the ribosome profiling data on individual transcripts tend to be noisy, we nevertheless inspected the data on the characterized DAPK3 transcript, and by using combined mapped reads from repeated experiments, we observed a clear induction of Ago2 and a modest increase in RFP on the characterized miR-20a MRE in response to miR-20a overexpression, while RNA-seq profiles remained largely the same (Fig.6c).
We then performed meta-gene analysis based on the Ago2 CLIP-seq data in Fig.6a, as well as on the miR-20a-induced peaks in CDS. For this meta-gene analysis, we omitted transcripts with Ago2 peaks too close to the translation start or stop site to avoid natural ribosome pausing events. By aligning RPFs relative to the center of miR-20a induced Ago2 peaks, we detected elevated RPFs on the peak center (Fig.6d, left). We also observed increased RPFs on the center of Ago2 peaks not induced by miR-20a, and as expected, these accumulated RPFs were not affected by miR-20a overexpression (Fig.6d, middle). No RFP accumulation was detected on randomly selected sites (Fig.6d, right). We reached the same conclusion based on Ago2 peaks identified from independent Ago2 eCLIP experiments (Supplementary Fig.5h,i).
Translation repression without evident reduction of ribosome occupancy
Given induced ribosome stalling by CDS targeted miRNAs, we next sought evidence for potentially compromised translation efficiency based on RPFs normalized against transcript levels. To this end, we plotted fold-changes in RNA-seq, RPFs, and translation efficiency (RPF density/RNA-seq density), all in an accumulative fashion, but separately for transcripts without Ago2 peak (gray) or with Ago peaks in 3’UTR (blue) or CDS (red) (Fig.6e). We found RNA decay with transcripts containing miR-20a induced Ago2 peaks in 3’UTR as observed earlier20. This validated our approach based on mapped, rather than predicted, miRNA action sites in 3’UTR. We observed a minor transcript reduction with Ago2 peaks in CDS, indicating that some stable RISC in CDS might also cause a degree of mRNA destabilization. Surprisingly, however, we saw little difference in translation efficiency between transcripts with or without Ago2 binding in CDS (Fig.6e).
We further validated this finding using a panel of HeLa cell lines stably expressing a luciferase reporter without a MRE or with a functional miR-20a MRE from DAPK3 either in CDS or 3’UTR. As we observed in transfected cells, the MRE repressed translation without reducing mRNA decay only in CDS (note that such atypical MRE is not functional in 3’UTR) (Supplementary Fig.6a,b). We next performed polysome profiling on these cells. Based on the average of three independent polysome experiments, we found that all reporter transcripts with or without a MRE showed similar polysome profiles to that of endogenous Actin mRNA (Supplementary Fig.6b,c,d). These data are consistent with our ribosome profiling data, together suggesting that CDS-targeted miRNAs do not appear to alter the overall ribosome occupancy on their mRNA targets, despite a measurable degree of ribosome stalling that can be correlated to translation repression.
CDS-targeted miRNA may employ a related mechanism for translation QC
We suspected that CDS-targeted miRNAs might repress translation via a related mechanism for translation quality control (QC) due to defective tRNA loading45. In this published study, the defect induced much stronger ribosome stalling than what we observed with CDS MREs, but did not alter overall polysome profile. Such translation QC mechanism has been extensively characterized in yeast, which is also known to operate in mammalian cells, and a key functional consequence is the induced degradation of nascent polypeptide by the proteasome system46. To provide initial evidence for this mechanism, we engineered a miR-20a MRE near the 3’ end of the luciferase reporter but still within the CDS region. Placing a stop codon in the same location did not affect the luciferase activity, indicating that the very last C-terminal sequence is not required for luciferase activity (Fig.6f, bottom left). In contrast, the presence of the miR-20a MRE reduced the luciferase activity by endogenous miR-20a, and importantly, the effect could be “rescued” with the proteasome inhibitor MG132 (Fig.6f, bottom right). These observations indicate that aborted translation is coupled with degradation of nascent polypeptide by the proteasome system, suggesting that CDS-targeted miRNA may exploit a related translational QC mechanism.
Discussion
In this study, we provide multiple lines of evidence for a novel class of MREs that exclusively function in CDS to repress translation without inducing mRNA decay. We show that the function of these MREs requires minimal 5’ base-pairings in the seed and extensive 3’ base-pairings with their target miRNAs. Our results extend the already extensive range of miRNA actions in mammalian cells, consistent with numerous Ago binding events detected in CDS by CLIP14,15,47 and miRNA-mRNA interactions captured by RNA ligation8,35,48. Although several reports have documented such miRNA action in CDS11,12,49, it has been generally assumed that those CDS targeted miRNAs employ the same mechanism as those acting in 3’UTR. Our data reveal that this is not the case as the function of CDS-targeted miRNA depends on an Ago, but not GW182 or its paralogs, thus providing a distinct mechanism to account for their predominant effect on translation, but not mRNA decay, opposite to the functional consequence induced by miRNA acting in 3’UTR.
Such atypical CDS MREs do not seem to trigger nonsense-mediated RNA decay (NMD), likely because those CDS-targeted miRNAs function after the “pioneer translation” in the cytoplasm50. It has been reported that a strong RNA secondary structure can induce so-called No-Go Decay (NGD), but such mechanism has only been documented with a reporter-based assay in yeast51, not on any endogenous transcript either in yeast or mammalian cells and a unknown endonuclease involved has remained unidentified. Instead, we provide initial evidence that CDS-targeted miRNA might use a mechanism related to that used in translation QC, which has been mainly characterized in the Non-Stop Decay (NSD) pathway52. Interestingly, NGD and NSD pathways appear to utilize the same machinery consisting of a heterodimer Hbs1 and Dom34 (Pelota in mammals), which are structurally related to the translation termination factor eRF1 and eRF3, respectively. The NSD pathway is normally activated on mRNA that lacks a natural stop codon, which catalyzes ribosome split, tRNA release, ubiquitination of nascent peptide, and mRNA decay46. This machinery appears to also function in the middle of CDS in response to a tRNA loading defect, which causes strong ribosome stalling, but not mRNA decay45. However, it has been reported that suboptimal codons are extensively correlated to mRNA instability53, although it remains unclear whether suboptimal codons trigger mRNA decay. We provide initial evidence that a CDS-targeted miRNA induces degradation of nascent peptide, suggesting a potential mechanistic link to the NSD pathway, thus pointing to a new direction in understanding how CDS-targeted miRNA may repress translation in future studies.
During this investigation, we initially thought that CDS-targeted miRNA might cause translation abortion by inducing ribosome stalling followed by ribosome drop-off. We provide evidence for such aborted translation on two separate reporters. Interestingly, we note that most miRNAs target mRNAs in CDS regions in plant, which has been demonstrated to induce translation abortion at the miRNA target site54. Unexpectedly, however, our ribosome profiling data provide no evidence for the anticipated ribosome queuing (ribosome build-up in the front of a stalling site and drop after such site). As a matter of fact, such ribosome queuing is rarely seen on most transcripts exhibiting strong ribosome stalling, even in bacteria depleted of a key elongation factor55. This raises a formal possibility that internal ribosome stalling may ultimately transmit a signal(s) that affects the overall competence of affected mRNA in translation. One possibility is the sequestration of such mRNA along with bound ribosome in P-bodies56,57, although this has now been a subject of active debate58,59. Another possibility is the activation of an eIF2α kinase (GCN2) in response to stalled ribosome, thereby linking translation elongation to initiation60, but such activated translational regulator would be expected to exert global effects on translation, rather than on affected mRNAs. By whatever mechanism, translational repression due to internal ribosome stalling does not seem to trigger extensive ribosome queuing in general nor be reflected at the calculated translation efficiency, which clearly requires further investigation in future studies.
Methods
Cell culture and antibodies
HeLa and 293Tcells were cultured in MEDM plus 10% FBS. Both cells were determined to be free from mycoplasma contamination. The antibodies used were from different vendors: Anti-DAPK1 (Abcam, ab109382; ABclonal, A5741), Anti-DAPK2 (Abgent, AP7033A), Anti-DAPK3 (Abgent, AJ1236b; Thermal scientific, PA5–27700), Anti-ACTIN (PTGCN, 60008-I-Ig), Anti-E2F1 (Abcam, ab179445), Anti-GW182 (Bethyl, A302–329A), Anti-TNRC6B (Abnova), Anti-TNRC6C (Bethyl, A303–969A), Anti-Ago1 (Abcam, ab105104), Anti-Ago2 (Abnova, H00027161-M01), Anti-Ago3 (Proteintech, 19692-I-AP), Anti-Ago4 (CST, 6913S), Anti-CD99 (Proteintech, 60354-I-Ig; ABclonal, A2028), Anti-NDRG1 (Abcam, ab124698), Anti-SAFB (Abnova, H00006294-M04), Anti-FLAG (ABclonal, AE005), Anti-GFP (Proteintech, 66002-I-Ig), anti-Mcherry (ABclonal, AE002). Western blotting results were quantified by the ImageJ software.
RT-PCR and Real-time PCR
For PCR analysis, total RNA was extracted from cells with Trizol (Life Technology) and ~1ug RNA was treated with RQI DNase followed by random priming with N9 (Takara) by M-MLV reverse transcriptase. Real-time PCR was performed with Master SYBR Green kit (TIANGEN) on the Rotor Gene 6000 Real-time Analyzer. All primers and miRNA sequences used in this study are listed in Supplementary Table 1.
Luciferase reporter constructs and dual luciferase assays
The siCHECK2 reporter vector was digested with XhoI and NotI. The miR-20a MRE from DAPK3 was inserted into the vector after annealing oligonucleotides consisting of MRE F (5’-TCGAGCTACCTGCACTCTAAGCGCATCGGC-3’) and MRE R (5’-GGCCGCGATGCGCTTAGAGTGCAGGTAGC-3’). Specific mutations on the target site were introduced by PCR-based site-specific mutagenesis.
The luciferase reporters containing EMCV, CrPV and HCV IRES cloned in the pRL-SV40 reporter were gifts of Dr. Martin Bushell. NdeI and SacI sites were inserted in the 5’ and 3’ end of the Renilla luciferase reporter followed by inserting the annealed oligonucleotides containing the miR-20a target site from DAPK3.
HeLa cells were seeded at ~45% confluency in 24-well plates and transfected the next day with 10ng siCHECK2 alone or in combination with 100ng pGL3 and 15ng of pRL plasmids. The luciferase activity was measured 48hrs after transfection using the Promega dual luciferase kit.
Deadenylation assay
The TBG-3UTR-DAPK3–2T plasmid and variants were constructed by inserting two copies of the miR-20a target sequence form DAPK3 into the 3’UTR of a rabbit β-globin gene between the NheI and XbaI sites. The TBG-ORF-DAPK3–2T and variants were similarly constructed at the NcoI site within the second exon of the β-globin gene in the reporter. All β-globin reporter genes were expressed from a tet-off promoter as described41. HeLa-tTA cells (Clontech) were seeded in 35-mm plates in DMEM containing 20ng/mL tet one day before transfection and then transfected with 1μg of the TBG reporter plasmid together with 50 pmols of the miR-20a mimic. Other steps of the deadenylation assay were performed as previously described41.
Detection of translation termination by CDS-targeted miRNA
The Renilla luciferase was FLAG-tagged and the expression plasmid was further modified by inserting a TAA stop codon at the position nt1479 (from ATG) or a WT or mutant miR-20a MRE from DAPK3. HeLa cells were seeded at ~30% confluency in 24-well plates and transfected with either non-specific RNA oligonucleotide control (NC) or the miR-20a antagomir. After RNA transfection for 12 hrs, cells were fed with fresh media and then transfected with individual reporter plasmids. Cells were harvested for western blotting 48 hrs after transfection.
We followed the previously established “2A” strategy to construct the GFP-2A-MRE-2A-RFP series of reporters43,44, FLAG-tagged Renilla was co-transfeced to provide loading control. Western blotting was performed 48hrs after transfecting non-specific RNA oligonucleotide control (NC) or specific miR-20a mimic or antagomir.
Polysome profiling
Stable transfected HeLa cells were plated on 10 cm dishes. Cells were collected and lyzed with the lysis buffer (10mM Tris-HCl, pH 7.4, 10mM NaCl, 3mM MgCl2, 1% Triton X-100, 2% Tween 20, 1% DOC) for 15 min on ice. After centrifugation at 12000g at 4oC for 15 min, the supernatant was collected, layered onto a 10–50% sucrose gradient and centrifuged at 4°C in the Beckman SW40 rotor at 36,000 rpm for 4 hrs. A total of 26 fractions were collected from each gradient with continual OD260 recording for RNA analysis by real-time RT-PCR.
Ribosome profiling and associated experiments
To link miRNA binding to translational behavior of ribosomes, we performed Ago2 CLIP-seq, RNA-seq, and ribosome profiling on before and after miR-20a overexpression. For Ago2 CLIP-seq, we followed the eCLIP procedure as described61. HeLa cells grown on 15 cm plates were first UV irradiated at 400 mj and collected by scraping the cells. After IP, the 3’ RNA adapter was ligated to RNA fragments on beads, which was followed by reverse transcription and ligation of a 3’ single-stranded DNA adapter to the cDNA. Libraries were sequenced by Hi-Seq 2500.
Ribosome profiling (Ribo-seq) and RNA-seq were performed as previously described with modifications20. For Ribo-seq, transfected cells were collected and resuspended with ice-cold lysis buffer (10 mM Tris-Cl pH7.4, 5mM MgCl2, 100 mM KCl, 1% Triton X-100) and placed on ice for 20 min. The cell lysates were centrifuged at 12000g for 20 min, and then the supernatant was loaded on a 10%−50% linear sucrose gradient and centrifuged at 36400 rpm for 3 hrs in SW41Ti rotor (Beckman). After centrifugation, ribosome and polyribosome fractions were collected and monitored by UV254. The collected fractions were pelleted by ultracentrifuge at 50,000 rpm in SW60 rotor for 6 hrs and ribosomal bound RNAs were digested with RNase I (Ambion). The RNA fragments were size selected (~30 nt) and rRNA depleted by antisense oligo and RNase H strategy62. Ribosome protected fragments (RPFs) were ligated with the 5’ and 3’ adaptor and sequenced on Hi-Seq 2500.
A fraction of total RNA from the same cells was each isolated using TRIZOL Reagent (Ambion) and fragmented using reverse transcriptase buffer (Invitrogen) at 94oC for 10 min. RNA fragments were size selected (180–220 nt) and rRNA depleted by antisense oligo and RNase H strategy, and then ligated with 5′ and 3′ adaptor. The libraries were sequenced on Hi-Seq X10.
Sequencing data analysis
Adaptors in single-end sequencing reads were trimmed by using the cutadapt program with parameters ‘-m16 -e 0.2’ plus ‘-a AGATCGGAAGAGCACACGTCTGAACTC’ for eCLIP-seq and ‘-a ATCTCGTATGCCGTCTTCTGCTTGAAA’ for Ribo-seq data, respectively. Gene annotation from human refGene (hg19) was downloaded from UCSC genome browser for mapping. The ensemble of mRNA sequences named as hmg was constructed by joining mRNA sequences (with 1kb N) of coding genes, and the longest isoform was chosen as the representative isoform for each transcript. The first random 6nt of eCLIP-seq reads were used to remove PCR products, and only those equal to or longer than 16 nt were mapped to hmg by using Bowtie2 (ref63) in the end-to-end model. The first 4 nt barcode sequence from Ribo-seq and RNA-seq reads was removed before mapping to hmg by using STAR64. For each group of mapped reads with same genomic position and same barcode, only one is retained to avoid PCR products.
For each replicate group of eCLIP-seq libraries, peak calling was performed by using CLIPper61 on combined reads from WT and OE miR-20a samples. The Ago2 peaks were filtered on the relative enrichment of Ago2 CLIP reads counts over those in input control samples65. The reads counts of peaks were computed by using featureCount program66 and the enrichment was calculated with edgeR package67. The peaks corresponding to miR-20a were computed in a similar manner by comparing eCLIP under miR-20a OE over WT. The miR-20a targets under Ago2 peaks were predicted by using RNAhybrid68 under -b mode. Random sites were generated by shuffling eCLIP peaks with BEDTools program69.
Two replicates of Ribo-seq and RNA-seq were all normalized to 1M total reads. Ribo-seq signals were normalized to RNA-seq signals before meta-gene analysis of RPFs on the center miR-20a peaks. For comparison of the ribosome profile between WT and miR-20a OE, the −300 nt to 300 nt intervals centered at miR-20a target sites were used, and only CDS region 60 nt away from start and stop codon were included for meta-gene analysis to avoid potential bias at translation initiation or termination sites. The signals of the interval centered at each site were normalized by its maximum value to remove potential site bias. The signals for all target sites in WT and OE were averaged by position in the meta-profiles, and normalized by mean-value for comparison of the relative differences between WT and OE conditions around target sites.
Statistical analysis
Data shown for each experiment was based on 3 or 4 technical replicates, as indicated in individual figure legends. Data are presented as means ± s.e.m, and P values were determined by two-tailed Student’s t test. All experiments were further confirmed by biological repeats. A Life Science Reporting Summary for this paper is available.
Data Availability Statement
All deep sequencing data from this study have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE115146. Source data for the graphs in all figures are available online. Other data are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
This work was supported by grants from the Ministry of Science and Technology of China (2017YFA0506400, YZ and XDF), the National Natural Science Foundation of China (31670827 and 91640115, YZ), the Chinese 111 program grant (B06018, XDF), Chinese Academy of Science foundation grant (22KJZD-EW-L12, XZ) and NIH grants (HG004659, GM049369 and GM052872, XDF). pRL-SV40 plasmids carrying IRESs from EMCV, HCV and CrPV are generous gifts of M. Bushell (Medical Research Council Toxicology Unit, UK).
Footnotes
Competing Interests Statement
The authors declare no competing financial interests.
References
- 1.Pasquinelli AE MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13, 271–82 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Jonas S & Izaurralde E Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16, 421–33 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP MicroRNAs: target recognition and regulatory functions. Cell 136, 215–33 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y et al. A new transmyocardial degradable stent combined with growth factor, heparin, and stem cells in acute myocardial infarction. Cardiovasc Res 84, 461–9 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Schirle NT & MacRae IJ The crystal structure of human Argonaute2. Science 336, 1037–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajewsky N microRNA target predictions in animals. Nat Genet 38 Suppl, S8–13 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Lal A et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol Cell 35, 610–25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helwak A, Kudla G, Dudnakova T & Tollervey D Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153, 654–65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Kouwenhove M, Kedde M & Agami R MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 11, 644–56 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Gu S, Jin L, Zhang F, Sarnow P & Kay MA Biological basis for restriction of microRNA targets to the 3’ untranslated region in mammalian mRNAs. Nat Struct Mol Biol 16, 144–50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duursma AM, Kedde M, Schrier M, le Sage C & Agami R miR-148 targets human DNMT3b protein coding region. RNA 14, 872–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tay Y, Zhang J, Thomson AM, Lim B & Rigoutsos I MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455, 1124–8 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Hausser J, Syed AP, Bilen B & Zavolan M Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res 23, 604–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue Y et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 152, 82–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi SW, Zang JB, Mele A & Darnell RB Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–86 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filipowicz W, Bhattacharyya SN & Sonenberg N Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9, 102–14 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Wu L, Fan J & Belasco JG MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A 103, 4034–9 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagga S et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Giraldez AJ et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Guo H, Ingolia NT, Weissman JS & Bartel DP Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Djuranovic S, Nahvi A & Green R miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huntzinger E & Izaurralde E Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12, 99–110 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Pillai RS et al. Inhibition of translational initiation by Let-7 microRNA in human cells. Science 309, 1573–1576 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Humphreys DT, Westman BJ, Martin DIK & Preiss T MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proceedings of the National Academy of Sciences of the United States of America 102, 16961–16966 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seggerson K, Tang LJ & Moss EG Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Developmental Biology 243, 215–225 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Petersen CP, Bordeleau ME, Pelletier J & Sharp PA Short RNAs repress translation after initiation in mammalian cells. Molecular Cell 21, 533–542 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Nottrott S, Simard MJ & Richter JD Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nature Structural & Molecular Biology 13, 1108–1114 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Maroney PA, Yu Y & Nilsen TW MicroRNAs, mRNAs, and translation. Cold Spring Harb Symp Quant Biol 71, 531–5 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Tat TT, Maroney PA, Chamnongpol S, Coller J & Nilsen TW Cotranslational microRNA mediated messenger RNA destabilization. Elife 5, e12880 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai Z et al. Oncogenic miR-17/20a Forms a Positive Feed-forward Loop with the p53 Kinase DAPK3 to Promote Tumorigenesis. J Biol Chem 290, 19967–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV & Mendell JT c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–43 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Forman JJ, Legesse-Miller A & Coller HA A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A 105, 14879–84 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elcheva I, Goswami S, Noubissi FK & Spiegelman VS CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol Cell 35, 240–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takagi S et al. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem 285, 4415–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broughton JP, Lovci MT, Huang JL, Yeo GW & Pasquinelli AE Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol Cell 64, 320–333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eichhorn SW et al. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell 56, 104–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behm-Ansmant I et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20, 1885–98 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakymiw A et al. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol 7, 1267–74 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Liu J et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–41 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Liu J et al. A role for the P-body component GW182 in microRNA function. Nat Cell Biol 7, 1261–6 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Y et al. MicroRNA-mediated repression of nonsense mRNAs. Elife 3, e03032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meijer HA et al. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 340, 82–5 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Donnelly ML et al. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J Gen Virol 82, 1027–41 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Kim JH et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6, e18556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishimura R et al. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buskirk AR & Green R Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos Trans R Soc Lond B Biol Sci 372(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hafner M et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grosswendt S et al. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol Cell 54, 1042–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nottrott S, Simard MJ & Richter JD Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol 13, 1108–14 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Ishigaki Y, Li X, Serin G & Maquat LE Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106, 607–17 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Doma MK & Parker R Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–4 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shoemaker CJ, Eyler DE & Green R Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330, 369–72 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Presnyak V et al. Codon optimality is a major determinant of mRNA stability. Cell 160, 1111–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwakawa HO & Tomari Y Molecular insights into microRNA-mediated translational repression in plants. Mol Cell 52, 591–601 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Woolstenhulme CJ, Guydosh NR, Green R & Buskirk AR High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep 11, 13–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhattacharyya SN, Habermacher R, Martine U, Closs EI & Filipowicz W Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–24 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Parker R & Sheth U P bodies and the control of mRNA translation and degradation. Mol Cell 25, 635–46 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Decker CJ & Parker R P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4, a012286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aizer A et al. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J Cell Sci 127, 4443–56 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Ishimura R, Nagy G, Dotu I, Chuang JH & Ackerman SL Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Nostrand EL et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13, 508–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adiconis X et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat Methods 10, 623–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loeb GB et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell 48, 760–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–30 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rehmsmeier M, Steffen P, Hochsmann M & Giegerich R Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–17 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All deep sequencing data from this study have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE115146. Source data for the graphs in all figures are available online. Other data are available from the corresponding author upon reasonable request.