Abstract
Protein tyrosine phosphatases (PTPs) contribute to a striking variety of human diseases, yet they remain vexingly difficult to inhibit with uncharged, cell-permeable molecules; no inhibitors of PTPs have been approved for clinical use. This study uses a broad set of biophysical analyses to evaluate the use of abietane-type diterpenoids, a biologically active class of phytometabolites with largely nonpolar structures, for the development of pharmaceutically relevant PTP inhibitors. Results of nuclear magnetic resonance analyses, mutational studies, and molecular dynamics simulations indicate that abietic acid can inhibit protein tyrosine phosphatase 1B, a negative regulator of insulin signaling and an elusive drug target, by binding to its active site in a non-substrate-like manner that stabilizes the catalytically essential WPD loop in an inactive conformation; detailed kinetic studies, in turn, show that minor changes in the structures of abietane-type diterpenoids (e.g., the addition of hydrogens) can improve potency (i.e., lower IC50) by 7-fold. These findings elucidate a previously uncharacterized mechanism of diterpenoid-mediated inhibition and suggest, more broadly, that abietane-type diterpenoids are a promising source of structurally diverse—and, intriguingly, microbially synthesizable—molecules on which to base the design of new PTP-inhibiting therapeutics.
Graphical Abstract

Protein tyrosine phosphatases (PTPs), enzymes that catalyze the hydrolytic dephosphorylation of tyrosine residues, regulate a wide range of important physiological processes (e.g., metabolism, appetite, immunity, and memory) and often contribute to diseases that occur when those processes go awry (e.g., diabetes, cancer, autoimmune diseases, and Alzheimer’s disease).1–8 Molecules that inhibit these enzymes are, thus, promising candidates for new therapeutics.9 Unfortunately, to date, the development of potent, biologically active inhibitors of PTPs has been hindered by their positively charged active sites, which bind most tightly to negatively charged molecules with poor membrane permeabilities.10
Protein tyrosine phosphatase 1B (PTP1B) demonstrates, by example, the promise and challenge of developing PTP inhibitors. PTP1B helps regulate insulin, leptin, and epidermal growth factor signaling and exhibits anomalous activity in type 2 diabetes, obesity, and breast cancer;11,12 its inhibition or genetic ablation can restore insulin sensitivity, lower levels of body fat, and slow tumorigenesis in mice.13,14 Efforts to develop potent, reversible inhibitors of PTP1B have generated three mechanistically informative classes of molecules (alternative groupings are also possible): (i) charged substrate analogues that associate closely with the catalytically essential WPD (Trp-Pro-Asp) loop, causing it to adopt a closed conformation,15 (ii) aryl diketoacids, which bind to the active site in a distinct, non-substrate-like manner that stabilizes the WPD loop in an open, inactive conformation,16 and (iii) polycyclic molecules, such as trodusquemine and benzofuran derivatives, which bind to C-terminal allosteric sites that attenuate WPD loop dynamics (Figure 1A).14,17 Inhibitors from the latter two classes tend to exhibit improved membrane permeabilities and molecular selectivities over substrate analogues, but they include a very small number of molecules that have yet to yield clinically approved drugs. The development of new varieties of inhibitors that function through similar or entirely new mechanisms—and, ideally, that possess readily diversifiable structures—could, thus, accelerate the development of new PTP-inhibiting pharmaceuticals.
Figure 1.
Inhibition of PTP1B. (A) Alignments of the backbone of PTP1B in competitively inhibited (yellow and orange, Protein Data Bank entry 2F71) and allosterically inhibited (gray and black, Protein Data Bank entry 1T4J) poses. The binding of substrates and competitive inhibitors to the active site causes the WPD loop to adopt a closed (orange) conformation that stabilizes the C-terminal α7 helix through an allosteric network; this helix is unresolvable in allosterically inhibited, noncompetitively inhibited, and uninhibited structures, which exhibit WPD-open conformations (black). (B) Chemical structure of abietic acid (AA). (C) Initial rates of PTP1B-catalyzed hydrolysis of pNPP in the presence of increasing concentrations of AA. Lines show a fit to a model for mixed inhibition (Table S2A). (D) In this model, the inhibitor (I) binds to the enzyme (E) and the enzyme–substrate complex (ES) with different affinities. Error bars in panel C denote the standard error (n ≥ 3 independent reactions).
This study explores the use of abietane-type diterpenoids, members of the large superfamily (>7000 molecules) of structurally diverse labdane-related diterpenoids,18 as a starting point for the development of pharmaceutically relevant PTP inhibitors. Previous screens of plant extracts have identified variants of these molecules that inhibit PTP1B in a noncompetitive or mixed manner;19,20 the molecular details of that inhibition, however, remain poorly understood. To dissect the non-substrate-like (i.e., not purely competitive) mode of inhibition exhibited by abietane-type diterpenoids, we used a broad set of biophysical analyses to study the inhibition of PTP1B by abietic acid (AA). AA is not an established inhibitor of PTP1B, but it shares a simple, and naturally variable scaffold with several plant-derived inhibitors with more complex structures (Figure 1B);19,20 it, thus, allowed us to examine the influence of minor changes in molecular structure on inhibitor strength and selectivity.
Abietane-type diterpenoids make up a valuable class of molecules for building inhibitors of PTPs for four principal reasons. (i) They are (generally) membrane-permeable and, thus, likely to be bioavailable.21,22 (Abietane-type molecules constitute the main biologically active components of Salvia miltiorrhiza, an herb used in Chinese medicine.23) (ii) The large family of natural products to which they belong includes a diverse set of incrementally varied structures that could facilitate inhibitor optimization.24 (iii) They are readily functionalizable with standard methods of synthetic chemistry and biocatalysis and could, thus, facilitate the preparation of combinatorial compound libraries for drug discovery.25,26 (iv) They can be synthesized in Escherichia coli at high titers (>50 mg/L) and could, thus, lead to the development of pharmaceuticals with low-cost biosynthetic production platforms.27
EXPERIMENTAL SECTION
Materials.
We purchased AA from Arctom Chemicals (Newton, MA), dehydroabietic acid (DeAA) and TCS401 {2-[(carboxycarbonyl)amino]-4,5,6,7-tetrahydro-thieno[2,3-c]-pyridine-3-carboxylic acid hydrochloride} from Santa Cruz Biotechnology (Santa Cruz, CA), isopimaric acid (IA) and dihydroabietic acid (DiAA) from Sigma-Aldrich (St. Louis, MO), continentalic acid (CA) from ChemFaces (Wuhan, China), and BBR {3-(3,5-dibromo-4-hydroxybenzoyl)-2-ethylbenzofuran-6-sulfonic acid-[4-(thiazol-2-ylsulfamyl)phenyl]-amide} from Cayman Chemical (Ann Arbor, MI). The purity and number of batches associated with each compound were as follows: TCS401 (99%, HPLC; two batches), BBR (99.9%, TLC; two batches), AA (>95%, HPLC, 1H NMR; two batches), CA (98%, HPLC; one batch), IA (99%, GC; one batch), and DeAA (96%, 1H NMR; one batch). Multiple batches, when used, had identical reported purities; we verified that they had the same inhibitory effect on PTP1B, in turn, by measuring PTP1B-catalyzed hydrolysis of pNPP in the presence of inhibitor introduced from each batch. The dihydroabietic acid sold by Sigma-Aldrich was impure (relative to the purity of other inhibitors), so we contracted Planta Analytica to isolate it to >97% purity (LC/MS-ELSD) via reversed-phase HPLC. We purchased 15NH4Cl from Cambridge Isotope Laboratories, Inc. (Tewksbury, MA), and additional reagents were from Sigma-Aldrich.
Protein Expression and Purification.
We expressed wild-type and mutant versions of PTP1B by performing the following steps. (i) We transformed BL21(DE3) E. coli cells with a pET21b plasmid harboring PTP1B1–321 fused to a C-terminal six-histidine tag (a kind gift from N. Tonks of Cold Spring Harbor). (ii) We used one colony, thus generated, to transform 20 mL of LB medium (50 mg/L carbenicillin), which we incubated for ~5 h (37 °C, 225 rpm). (iii) We used the 20 mL starter culture to inoculate 1 L of induction medium (20 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl, 4 g/L M9 salts, 4 g/L glucose, and 50 mg/L carbenicillin), which we incubated for 2–3 h (37 °C, 225 rpm). (iv) At an optical density (OD600) of ~0.60, we induced expression of PTP1B with 0.5 mM IPTG and decreased the temperature to 22 °C. We performed induction for 20 h before pelleting and lysing the cells. We expressed TC-PTP1–292 and SHP2237–529 in an analogous fashion. All PTP sequences correspond to the catalytic domain.
We prepared 15N-labeled PTP1B by following a protocol similar to that described above, but we used an induction medium that consisted of the following components (per liter): 100 mL of M9 medium (60 g of Na2HPO4, 30 g of KH2PO4, and 5 g of NaCl in 1 L), 10 mL of trace elements (5 g/L EDTA, 0.83 mg/L FeCl3·6H2O, 13 mg/L CuCl2·2H2O, 10 mg/L CoCl2·6H2O, 10 mg/L H3BO3, and 1.6 mg/L MnCl2·6H2O), 4 g of glucose, 1 mL of 1 M MgSO4, 0.3 mL of 1 M CaCl, 2 g of 15NH4Cl, 300 μL of 0.3 M ZnSO4, 1 mL of biotin (1 mg/mL stock, 1000-fold dilution), 1 mL of thiamin (1 mg/mL), and 50 mg of carbenicillin. After growing the 20 mL starter culture in LB medium for 5 h (step ii above), we pelleted the cells, exchanged the LB medium with 50 mL of induction medium, and incubated the new culture overnight. In the morning, we used the second starter culture to inoculate 1 L of induction medium and completed expression by following step iv from the preceding paragraph.
We purified all PTPs by using fast protein liquid chromatography (FPLC). In brief, we pelleted cells and lysed them with buffer containing the following components for each gram of pellet: 4 mL of B-PER lysis buffer (Thermo Fisher), 1 mg of MgSO4, 2 mg of Nα-p-tosyl-L-arginine methyl ester hydrochloride, 3.5 mg of tris(2-carboxyethyl)phosphine (TCEP), 3.75 μL of phenylmethanesulfonyl fluoride, 0.5 mg of lysozyme, and 10 μL of DNase. After agitating the cell lysate for 1 h, we pelleted the insoluble cell debris (6000 rpm for 20 min) and clarified the resulting supernatant by adding 20% saturated ammonium sulfate, followed by immediate centrifugation. To purify PTPs, we exchanged the lysate into 50 mM Tris-HCl buffer (pH 7.5, 0.5 mM TCEP; HiPrep 26/10 desalting), passed the exchanged solution over a nickel-affinity column (HisTrap HP), eluted the recombinant PTP with 300 mM imidazole, exchanged the eluent into 50 mM HEPES (pH 7.5, 0.5 mM TCEP), passed this solution over an anion exchange column (HiPrep Q HP 16/10 column), and eluted the PTP with a 0 to 1 M NaCl gradient. We stored the final protein (30–50 μM) in 50 mM HEPES buffer (pH 7.5, 0.5 mM TCEP) with 20% glycerol at −80 °C. We purchased all chromatography columns from GE healthcare.
Mutational Analysis.
We generated mutants of PTP1B by using the Quikchange protocol (Stratagene). Briefly, we amplified PTP1B321 with mutagenic primers from Table S1, digested the parent vector with DpnI (60 min, 37 °C), and transformed the mutated plasmid, thus generated, into Turbo Competent E. coli cells (New England Biolabs). We sequenced a subset of plasmids derived from these colonies to confirm the presence of the targeted mutation(s) (QuintaraBio). We evaluated the fractional change in inhibition (F) associated with each mutation as described in SI Note 2. Exact sample sizes are reported in Table S5.
Enzyme Kinetics.
To examine mechanisms of inhibition, we measured PTP-catalyzed hydrolysis of p-nitrophenyl phosphate (pNPP) in the presence of various concentrations of inhibitors. The composition of our reaction mixtures was as follows: PTP (0.05–0.1 μM), pNPP (0.17, 0.33, 0.67, 2, 5, 10, 15, and 20 mM), inhibitors (0, 100, 200, 300, and 400 μM for diterpenoids; 5 μM for BBR; and 100 μM for TCS401), and buffer [50 mM HEPES, 10% DMSO, and 50 μg/mL BSA (pH 7.3)]. We monitored the formation of p-nitrophenol by measuring the absorbance at 405 nm every 5 s on a Spectramax M2 plate reader. For a subset of initial-rate measurements (e.g., those corresponding to high substrate concentrations), we removed late-stage time points that fell outside of the linear regime. We report exact sample sizes (i.e., the number of independently prepared reactions) in Tables S4 and S5.
Statistical Analysis of Kinetic Models.
We evaluated kinetic models in three steps. (i) We used MATLAB’s “nlinfit” and “fminsearch” functions to fit (a) initial-rate measurements collected in the absence of inhibitors to a Michaelis–Menten model and (b) initial-rate measurements collected in the presence and absence of inhibitors to four models of inhibition (Figure S1). The first fit supplied values of kcat and KM, which we used as input parameters for the second. (ii) We used an F-test to compare the fits of (a) a mixed model, which has two parameters, and (b) the nested single-parameter model with the lowest sum of squared errors. We determined p values with MATLAB’s “fcdf” function. (iii) When fits to the mixed model were not superior (p < 0.05) to fits to single-parameter models, we compared single-parameter models to one another by using Akaike’s information criterion (AIC). In brief, we calculated the difference in AIC (Δi) between the best-fit single-parameter model (lowest sum of squared errors), denoted the “reference” model, and each additional single-parameter model, denoted the “test” model. Values of Δi of >5 indicated weak support for the test model and good support for the reference model; values of Δi of >10 indicated very weak support for the test model and strong support for the reference model.28 We tested our analysis on literature-based kinetic data for six well-characterized inhibitors (i.e., inhibitors subjected to extensive biophysical analysis); our results showed good agreement with their reported mechanisms of inhibition (Tables S2P–S2U). Analyses of kinetic data collected in this study are reported in Tables S2A–S2O and discussed in SI Note 4.
Estimation of IC50.
We estimated the half-maximal inhibitory concentration (IC50) of various inhibitors by using kinetic models (parametrized with detailed kinetic assays) to determine the inhibitor concentration required to reduce initial rates of PTP-catalyzed hydrolysis of 20 mM pNPP by 50%. This high substrate concentration minimized the concentration dependence of IC50’s. We used the MATLAB function “nlparci” to determine the confidence intervals of kinetic parameters and propagated those intervals to estimate the corresponding confidence in IC50’s. SI Note 5 examines the relevance of IC50 to binding affinity (i.e., Kd, the dissociation constant of an enzyme–inhibitor complex), and SI Note 7 discusses the influence of DMSO on IC50.
NMR Analysis.
We performed all NMR measurements at 25 °C by using a 750 MHz Bruker spectrometer equipped with a cryogenically cooled CPTXI 5 mm probe and an Avance III console. Our samples contained 75 μM 15N-labeled PTP1B in NMR buffer [50 mM HEPES (pH 6.8), 150 mM NaCl, and 0.5 mM TCEP], 10% d6-DMSO, and 5% D2O. We assigned two-dimensional 1H–15N TROSY-HSQC spectra by using published data deposited in the Biological Magnetic Resonance Bank (entries 26814 and 19223, same buffer). For the AA-bound form of PTP1B, we followed assigned peaks in titration experiments (described below). In total, we assigned 64% of the non-proline residues (PTP1B321); for comparison, BRMB entries 26814 and 19223 include assignments for 80% of nonproline residues. Our lower percentage of assignments was probably caused, in part, by the use of 10% d6-DMSO to solubilize AA; this solvent can cause peaks to shift and/or broaden beyond recognition. Fortunately, assigned residues, which were distributed across the protein and included numerous amino acids near (i.e., <5 Å) known binding sites (Figure 2C), enabled a detailed examination of the response of broadly distributed regions of PTP1B (both buried and surface-exposed) to the binding of AA.
Figure 2.
NMR analysis of the binding of AA to PTP1B. (A) Weighted differences in chemical shifts (Δδ) between 1H–15N HSQC spectra recorded in the absence and presence of AA (10:1 PTP1B:AA). The dashed red line delineates the threshold for values of Δδ larger than two standard deviations (σ) above the mean; gray bars mark residues for which chemical shifts were broadened beyond recognition. (B) Crystal structure of PTP1B (gray, PDB entry 3A5J) highlighting the locations of assigned residues (blue). The bound positions of BBR (green, allosteric site, PDB entry 1T4J) and LZP25 (yellow, active site, PDB entry 3EB1) are overlaid for reference (i.e., we aligned the structures of PDB entries 3A5J, 1T4J, and 3EB1). Residues with significant CSPs (i.e., Δδ > Δδmean + 2σ) are distributed across the protein (red) and, with the exception of two residues in the WPD loop, outside of known binding sites. (C) Detail of the active site (top) and known allosteric site (bottom) with the positions of bound inhibitors overlaid as in panel B.
To analyze the binding of AA, we titrated it into 75 μM PTP1B at molar ratios (AA:PTP1B) of 0:1, 0.75:1, 1.5:1, 3:1, 5:1, and 10:1 and assigned 1H–15N TROSY-HSQC spectra at each point. We calculated chemical shift perturbations (CSPs) by using eq 1.
| (1) |
Figure S5 shows an overlay of 1H–15N TROSY-HSQC spectra of PTP1B and PTP1B-AA (10:1 AA:PTP1B), and Figure 2A shows CSPs calculated from these two spectra.
NMR-Restrained Docking Simulations (i.e., HADDOCK).
To identify residues involved in PTP1B–AA association (i.e., residues that constitute the PTP1B–AA interface), we used HADDOCK version 2.2 (high-ambiguity-driven protein–protein docking).29,30 This platform allows ligands to bind anywhere on a protein surface but enables conformational sampling in only a subset of “active” residues. We used four sets of active residues for our calculations: group 1, residues with large CSPs (i.e., Δδ larger than two standard deviations above the mean); group 2, solvent-exposed residues from group 1 (i.e., those with >0.5 Å2 of solvent-exposed surface area); group 3, solvent-exposed residues within 4 Å of those from group 1; and group 4, solvent-exposed residues within 5 Å of those from group 1. For each set of runs, we used three different crystal structures of PTP1B: ligand-free [Protein Data Bank (PDB) entry 3A5J], allosterically inhibited (PDB entry 1T4J), and competitively inhibited (PDB entry 2F71); in the latter two cases, we removed inhibitors prior to docking analyses. SI Note 1 discusses the results of this analysis.
Molecular Dynamics (MD) Simulations.
We prepared structures of PTP1B and AA for MD simulations by using standard methods. In brief, we used an allosterically inhibited structure of PTP1B (PDB entry 1T4J)17 with the inhibitor removed and protonation state adjusted through the H++ server.31 We note that this structure, which has the WPD loop in the open conformation, is highly similar to the apo structure of PTP1B (the root-mean-square deviations of backbone and heavy atoms between 1T4J and apo structure 3A5J are 0.5 and 1.1 Å, respectively). For AA, we used the LigPrep tool from Schrodinger to predict an initial conformation32 and Epik software to adjust its protonation state (pH 7.0).33 We modeled PTP1B with the Amber/ff14SB force field; for AA, we used the General Amber Force Field (GAFF)34 with the Restrained Electrostatic Potential (RESP)35 model to derive partial charges.
We initialized simulations with an energy-minimized PTP1B (determined with the steepest descent algorithm in a vacuum) positioned in simulation box filled with TIP3P water molecules.36 This box had a minimum distance of 10 Å between protein and box edges. For all simulations, we used Na+ ions to neutralize the net charge and applied a second minimization to relax solvent molecules around the protein. We thermalized and pressurized each system in different steps. Final solvated systems consisted of ~52000 atoms.
We performed MD simulations with an isothermal–isobaric ensemble (p = 1 bar, and T = 300 K) by coupling each simulated system to both (i) a Nose–Hoover thermostat37,38 and (ii) an isotropic Parrinello–Rahman barostat39 with a time constant for coupling of 1 ps. We constrained bonds with LINCS,40 which allowed for an integration time step of 2 fs, and used periodic boundary conditions in all directions. We calculated long-range electrostatic interactions with the particle mesh Ewald (PME)41 method with a real space cutoff of 10 Å and a Fourier spacing of 0.12 Å and estimated van der Waals interactions with a cutoff of 10 Å. We ran all MD simulations with GROMACS-4.642 for >500 ns; analyses reported in this study refer to simulations after 25 ns, a time after which the all-atom RMSDs of PTP1B converge with a standard deviation of 0.1 Å (Figure S8). In our study of bound poses of AA, we defined hydrogen bonds with a maximum donor–acceptor distance of 3.5 Å and a maximum donor–acceptor angle of 30°.
RESULTS AND DISCUSSION
AA Is a Weak Mixed Inhibitor of PTP1B.
We examined the mechanism by which AA inhibits PTP1B by measuring PTP1B-catalyzed hydrolysis of pNPP in the presence of increasing concentrations of AA. Initial rates exhibited saturation behavior indicative of a weak noncompetitive inhibitor [i.e., high concentrations of pNPP did not alleviate inhibition by outcompeting AA (Figure 1C)]. A comparison of four kinetic models of inhibition suggested that measured data fit best to a mixed model (p < 0.001) in which AA engages in a binding mode distinct from the binding mode exhibited by pNPP but exhibits an inhibitory effect sensitive to the presence of bound of pNPP (Figure 1D and Table S2A). This sensitivity could result from either (i) communication (allosteric or steric) between the binding of AA and the binding of pNPP or (ii) the existence of two binding sites for AA (e.g., the active site and a secondary site). Our kinetic analyses could not distinguish between these two possibilities.
AA Binds to the Active Site of PTP1B.
To identify residues involved in the binding of AA to PTP1B, we recorded 1H–15N TROSY-HSQC spectra of PTP1B in the presence and absence of AA (Experimental Section). Weighted differences in chemical shifts (Δδ) between ligand-free and ligand-bound spectra provide a residue-specific metric for binding-induced changes in the local electronic environment; we refer to these differences as “chemical shift perturbations (CSPs)”. Surprisingly, residues that exhibited the largest CSPs (i.e., Δδ more than two standard deviations above the mean) appeared throughout the protein and, with the exception of two residues in the catalytic WPD loop (W179 and V184), outside of known binding sites (Figure 2A–C). This disperse set of residues might suggest that AA binds to multiple sites, but its distribution (five residues appear in flexible loops, and three are buried in regions distal to known binding sites) suggests that changes in protein conformation and/or dynamics are a more likely cause. This effect is consistent with the results of previous NMR analyses, which indicate that the binding of inhibitors to the active site or C-terminal allosteric site (Figure 2C) can trigger CSPs at both sites and within intermediary regions of the protein; PTP1B thus appears to be quite flexible.43,44 The absence of an obvious binding site for AA (i.e., a clustered set of residues with large CSPs) is surprising but could result from multiple bound conformations; this mode of interaction is compatible with the weak inhibitory effect of AA and with the generally low CSPs (i.e., <0.1 ppm) detected in our experiments.45 Our NMR results, taken together, indicate that AA modulates the conformation and/or dynamics of PTP1B by adjusting a broadly distributed set of intraprotein interactions that extends beyond its binding site.
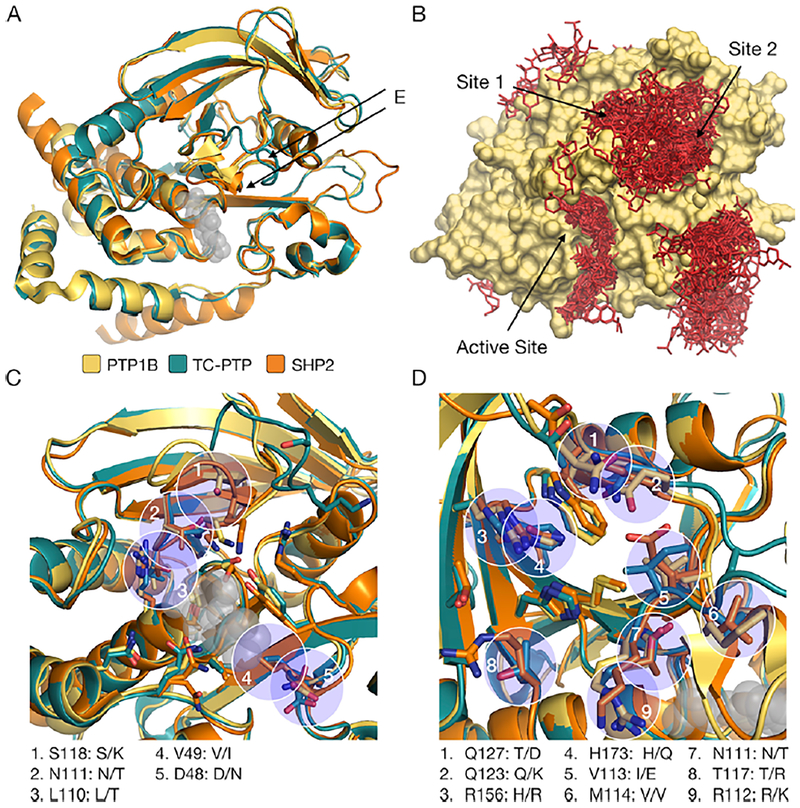
To resolve the binding site of AA more clearly, we used residues with significant CSPs as flexible restraints in HADDOCK (high-ambiguity-driven protein–protein docking), a molecular dynamics platform that combines rigid-body energy minimization with semiflexible and explicit-solvent refinements to predict the structure of biomolecular complexes (Experimental Section).29,30 HADDOCK enables specification of flexible residues but allows ligands to bind anywhere on the surface of a protein. In our analysis, we accounted for the conformational flexibility of PTP1B by using allosterically inhibited, competitively inhibited, and inhibitor-free crystal structures (i.e., structures with slight differences in protein conformation and, thus, surface topography). We explored the contribution of residues proximal (<4–5 Å) to shifted residues, in turn, by including them as flexible restraints in an additional set of HADDOCK analyses. In a total of 12 docking analyses, each of which yielded 1–10 predicted binding pockets, two regions consistently exhibited the highest scores: the active site and a nearby cleft defined by residues W125, Q127, R156, and H175 (Figure 3A and Figure S6A; SI Note 1); we refer to this cleft as “site 1”. Our docking analysis, thus, suggested that AA binds at or near the active site of PTP1B.
Figure 3.
Mutational analysis of the binding site of abietic acid (AA). (A) Crystal structure of PTP1B (gray, PDB entry 3A5J) showing the location of mutations introduced at five sites: the active site (red), the allosteric site (green), site 1 (orange), site 2 (yellow), and the L11 loop (blue). The bound positions of BBR (gray, allosteric site, PDB entry 1T4J) and TCS401 (black, active site, PDB entry 5K9W) are overlaid for reference (i.e., we aligned the structures of PDB entries 3A5J, 1T4J, and 5K9W). (B) Disruptive mutations introduced at each site. Mutations were designed to alter the size and/or polarity of targeted residues. The mutation denoted “YAYA” (Y152A/Y153A), which was identified in a previous study, attenuates allosteric communication between the C-terminus and the WPD loop.43 (C) Fractional change in inhibition (F in eq 1) caused by the mutations from panel B. Five mutations distributed across the protein reduced the level of inhibition by AA and TCS401 but had a negligible effect on inhibition by BBR. The similar effects of most mutations on AA and TCS401 suggest that both inhibitors bind to the active site. Error bars denote the standard error [propagated from n ≥ 9 independent measurements of each initial rate in eq 1 (SI Note 2)].
We probed the binding site of AA further by determining how various disruptive mutations (i.e., mutations that change the size and/or polarity of residues) affect inhibition by AA and two reference inhibitors. Mutations can alter the inhibitory effect of small molecules by modifying the chemical functionality or hydration structure of their binding sites,46 by triggering global changes in protein conformation or dynamics,47 or by stabilizing covalent enzyme–substrate intermediates.43 The sensitivity of an inhibitor to a broadly distributed set of mutations that function through any combination of these mechanisms constitutes a molecular “fingerprint”; when compared between inhibitors, this finger-print can reveal overlapping binding sites. Our references inhibitors—TCS401, which binds to the active site,48 and BBR, which binds to the allosteric site17—enabled such a comparison.
We spread mutations across five sites: the active site, the allosteric site, site 1, site 2 (a cleft that exhibited significant CSPs), and the L11 loop [a loop that facilitates allosteric communication between the α7 helix and the WPD loop43 (Figure 3A,B)]. For each mutant, we evaluated the fractional change in inhibition (F) by using eq 2
| (2) |
where Vo‑mut and Vo‑wt are the uninhibited initial rates of the mutant and wild-type enzyme, respectively, and Vo‑mut(I) and Vo‑wt(I) are the inhibited initial rates (SI Note 2). Figure 3C and Figure S6D show the results of our analysis. AA and TCS401 exhibited similar sensitivities to a range of mutations. Five mutations distributed across the protein weakened the ability of AA and TCS401 to inhibit PTP1B but had a negligible—and, in one case, amplificatory—effect on inhibition by BBR. Two mutations affected only AA or TCS401, and one affected both TCS401 and BBR but not AA. The similar effect of most mutations on AA and TCS401 suggests that both inhibitors bind to the active site. The discrepancy between the effects of a few mutations, in turn, indicates that AA and TCS401 bind to non-overlapping regions of that site, a result consistent with their different structures and inhibitory mechanisms (i.e., mixed vs competitive).
Weak, nonpolar inhibitors such as AA tend to have solubilities that are insufficient for unambiguous identification in X-ray crystal structures; their partial occupancy of a crystal lattice, however, can yield electron density near binding sites.49 Our repeated attempts to collect crystal structures of the PTP1B–AA complex (prepared via co-crystallization and soaking) failed to yield data sets with sufficient density to place AA but, nonetheless, allowed us to detect contiguous regions of positive density in the active site, a site consistent with the results of our kinetic study, NMR-restrained docking simulations (i.e., HADDOCK), and mutational analysis (SI Note 3). Our inability to place a single conformation of AA might suggest that AA adopts multiple conformations (a behavior consistent with the absence of a clustered set of residues with large CSPs in our NMR analysis); the partial positive density, however, was insufficient to place them.
AA Increases Loop Dynamics in PTP1B.
We dissected the mechanism of AA-mediated inhibition in atomic detail by using MD simulations. To begin, we performed a “dynamic docking” analysis.50 We simulated the apo form of PTP1B in the presence of high concentrations of AA and collected snapshots of molecules close (≤2 Å) to its surface every 2.5 ns. This analysis revealed clusters of AA located in the active site and at several nearby regions (Figure S9). Poses in the active site, however, showed less variability in their orientation and position than those located elsewhere on the surface (e.g., they showed only one orientation at slightly different positions along the cleft of the active site); this reduced variability indicates more stable binding.
We examined the influence of AA on protein dynamics, in turn, by comparing simulations of PTP1B in AA-free and AA-bound states (Experimental Section). For this analysis, we chose a stable conformation of AA bound to the active site [i.e., a conformation based on spontaneous docking simulations (Figure S10A)], the site most consistent with the results of our biophysical studies. For a negative control, we selected an alternative low-energy conformation with an opposite orientation (relative to the first); we determined this alternative conformation by using Glide (Shrodinger Suite32) to dock AA to the active site (i.e., it represents one of the predicted poses). Interestingly, in the simulation performed with the second conformation, AA left the binding site and redocked with an orientation that matched that of the first (Figure S10B). We, thus, chose the first conformation of AA to analyze the PTP1B-AA complex.
Our comparison of free and bound states suggests that AA, upon binding, increases the flexibility of the WPD, E, and L10 loops, an effect consistent with the results of our NMR analysis [where the WPD and L10 loops exhibited large CSPs (Figure 4A,B)]. AA exerts this effect in two ways. (i) It sterically occludes F182 from the active site and, thus, prevents the WPD loop from closing, and (ii) it disrupts the formation of hydrogen bonds among R221, E115, and W179, three residues that attenuate loop dynamics by stabilizing the WPD loop in its closed conformation (Figure 4C). The increase in flexibility of the WPD loop propagates to the E and L10 loops through direct noncovalent interactions (for E) or through an increase in backbone dynamics (for L10); all three loops exhibited correlated coaxial motions (Figure 4B). MD simulations, thus, indicate that AA prevents the WPD loop from forming a closed, catalytically competent conformation and enhances the magnitude of conformational dynamics throughout the protein. We did not perform additional MD simulations with poses of AA chosen from clusters located outside of the active site because the position and orientation of AA within those clusters were highly variable (and less stable), and we lacked experimental support for their physical relevance. We discuss the potential importance of alternative clusters below.
Figure 4.
Computational analysis of binding. The results of MD simulations are shown: backbone traces of PTP1B in (A) abietic acid (AA)-free and (B) AA-bound states. The thickness of a trace indicates the amplitude and direction of local motions (Experimental Section). The binding of AA increases the flexibility of the WPD, E, and L10 loops, which exhibit correlated motions along a similar axis. The WPD and L10 loops contain residues with significant CSPs (red), suggesting consistency between the results of MD and NMR analyses. (C) Representative bound conformation of AA (green). Upon binding to the active site, AA (i) forms a hydrogen bond with R221 that weakens a bond between R221 and E115 and (ii) prevents the formation of a hydrogen bond (red) between W179 and R221 that forms when the WPD loop closes. Both effects enhance the conformational dynamics of the WPD loop. (D) The results of docking calculations are consistent with mixed-type inhibition. The binding of AA prevents the WPD loop from closing and disrupts, but does not preclude, the binding of pNPP (spheres).
The mixed mode of inhibition suggested by our kinetic analysis indicates that AA disrupts, but does not preclude, the binding of pNPP. To examine this disruption, and to assess the consistency of our kinetic and MD analyses, we used docking calculations to study the binding of pNPP to the PTP1B–AA complex. Results suggest that pNPP can bind a cleft formed by AA and PTP1B (Figure 4D); this bound conformation, which does not permit closure of the WPD loop, is consistent with the unproductive enzyme–substrate–inhibitor (ESI) complex formed in mixed-type inhibition.
Minor Changes in the Structures of Abietane-Type Diterpenoids Can Yield Large Improvements in Potency.
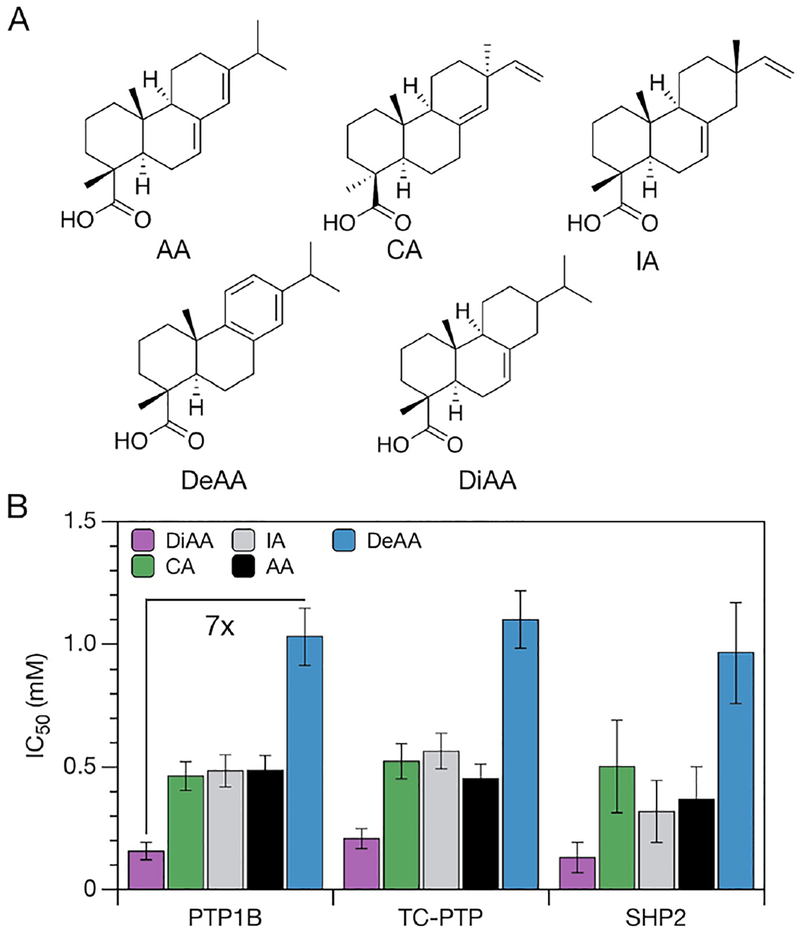
We determined how minor, biologically accessible changes in the structure of AA affect its potency and selectivity by comparing the inhibitory effects of five structurally similar, plant-derived diterpenoids on three sequence-diverse PTPs. The diterpenoids included AA, continentalic acid (CA), isopimaric acid (IA), dehydroabietic acid (DeAA), and dihydroabietic acid (DiAA);21,51–53 these molecules differ in their stereochemistries and/or degrees of saturation (Figure 5A). The PTPs included PTP1B, T-cell protein tyrosine phosphatase (TC-PTP), and protein tyrosine phosphatase nonreceptor type 11 (SHP2); these proteins differ in sequence identity [30–63% (Experimental Section)] and physiological significance. TC-PTP is essential for T-cell function,54 and SHP2 is an immunomodulatory targeted for the treatment of numerous types of cancer.55,56 Results suggested that differences in saturation, but not stereochemistry, had a strong influence on potency [defined in this study as IC50, a metric for the binding affinity of abietane-type diterpenoids (SI Note 5)]: DiAA was 3-fold more potent than AA and 7-fold more potent than DeAA (Figure 5B). Surprisingly, all molecules exhibited similar inhibitory effects on the three PTPs, suggesting that more aggressive structural changes are necessary to adjust selectivity. For reference, aryl diketoacids, noncompetitive inhibitors that also bind to a WPD-open conformation of the active site (Figure S11), exhibit IC50’s on PTP1B and SHP2 that differ by up to 4-fold.16
Figure 5.
Analysis of structurally varied inhibitors. (A) Structural analogues of abietic acid (AA): continentalic acid (CA), isopimaric acid (IA), dehydroabietic acid (DeAA), and dihydroabietic acid (DiAA). (B) Differences in degree of saturation yield pronounced differences in potency (i.e., IC50) but not selectivity. Error bars represent 95% confidence intervals (propagated from the data sets shown in Figures S2–S4).
The identification of inhibitors with a 7-fold difference in potency in a screen of five structurally similar molecules is interesting. For many small-molecule inhibitors, the addition of small structural appendages has surprisingly little influence on binding affinity. An additional hydrogen bond donor or acceptor, for example, might yield favorable changes in enthalpy of binding that are completely compensated by unfavorable changes in entropy of binding;57 this phenomenon, which is termed enthalpy/entropy compensation, represents an important challenge in drug design.58 The affinities of abietane-type diterpenoids for PTPs, by contrast, appear to be sensitive to minor changes in structure; this sensitivity is a desirable attribute for inhibitor optimization.
Alternative Binding Sites Are Unlikely.
The similar inhibitory effect of abietane-type diterpenoids on three sequence-diverse PTPs suggests that these molecules bind to a conserved site. At first glance, a structural alignment indicates that many conserved sites might exist; PTP1B, TC-PTP, and SHP2 exhibit highly similar tertiary structures [i.e., the RMSDs of aligned structures are 0.46 Å for PTP1B and TC-PTP and 0.65 Å for PTP1B and SHP2 (Figure 6A)] and could possess similar topographical features near clusters of AA predicted by dynamic docking (Figure 6B). (Note that structural discrepancies tend to appear at loops; we discuss the E loop, which exhibits a prominent organizational difference between PTP1B and TC-PTP, in SI Note 6.) Closer analysis of the residues that line alternative sites (e.g., site 1), however, indicates that they are poorly conserved relative to the active site (Figure 6C,D). Of the regions where abietane-type diterpenoids could plausibly bind, the active site is, thus, the most consistent with their poor selectivity. Two sets of previous studies support this result. (i) A large-scale structural analysis of PTPs showed that, outside of the active site, their surfaces are highly diverse.59 (ii) Biophysical analyses indicate that most molecules with similar inhibitory effects on different PTPs (in fact, all poorly selective molecules of which we are aware) bind to the active site.60 To summarize, in the absence of co-crystal structures, we cannot provide definitive evidence of a binding site; however, our broad set of analyses (i.e., the results of a kinetic study, an NMR analysis, a mutational analysis, two sets of docking simulations, and an analysis of inhibition on different PTPs) indicates that the active site is the most likely of all possibilities.
Figure 6.
Analysis of alternative binding sites. (A) Alignment of the crystal structures of PTP1B (PDB entry 3A5J), TC-PTP (PDB entry 1L8K), and SHP2 (PDB entry 3B7O) with the bound position of TCS401 (gray spheres, active site, PDB entry 5K9W) overlaid for reference (i.e., we aligned the structures of 3A5J, 1L8K, 3B7O, and 5K9W). Similar tertiary structures (the RMSDs of aligned structures are 0.46 Å for PTP1B and TC-PTP and 0.65 Å for PTP1B and SHP2) suggest that these proteins could exhibit similar topographical features near (B) clusters predicted by dynamic docking. Details of (C) the active site and (D) site 1. Nonconserved residues (blue circles) are labeled for PTP1B and TC-PTP/SHP2. The active site is highly conserved, while site 1 is more variable. This discrepancy, a symptom of the general diversity of PTP surfaces outside of their active sites,59 suggests that, of the sites where abietane-type diterpenoids could plausibly bind, the active site is the most consistent with their similar inhibitory effect on three different PTPs.
CONCLUSIONS AND SPECULATION
Many pharmaceuticals correct anomalous levels of protein phosphorylation by inhibiting protein tyrosine kinases, which catalyze the ATP-dependent phosphorylation of tyrosine residues;61 inhibitors of PTPs, by contrast, have yet to clear clinical trials.10 This study presents evidence that abietane-type diterpenoids provide a promising, and largely untapped, source of readily optimizable PTP-inhibiting leads. Results of detailed kinetic studies, NMR analyses, and MD simulations indicate that AA, a representative abietane-type diterpenoid, can inhibit PTP1B by binding to the active site in a manner that stabilizes the WPD loop in an inactive conformation, and they show that minor changes in the structures of abietane-type diterpenoids (i.e., the addition of hydrogens) can improve potency by 7-fold.
The sensitivity of abietane-mediated inhibition to minor changes in molecular structure suggests an intriguing application for synthetic biology in lead development. The metabolic pathways responsible for synthesizing abietane-type diterpenoids in nature (i.e., pathways comprised of terpene synthases,62 cytochrome P450 monooxygenases,63 and other mutation-sensitive enzymes) could be installed into microbial hosts and combinatorially reconfigured—or, perhaps, evolved—to yield inhibitors with improved potencies and/or selectivities. Recently developed methods for using E. coli for the combinatorial synthesis of structurally varied labdane-related diterpenoids provide a starting point for this approach.25,27,64 We note that the highly nonpolar nature of the diterpenoids examined in this study necessitated the use of high concentrations of co-solvent that reduced their apparent potencies (SI Note 7) and precluded detailed X-ray crystallo-graphic studies; future studies of the binding mode of more soluble and/or potent analogues (e.g., those discovered via synthetic biology) might enable rational, rather than screen-based, approaches to inhibitor optimization.
The results of this study have an interesting implication for previously examined abietane-type diterpenoids with therapeutically relevant activities but poorly understood mechanisms of action. Examples include tanshinone, carnosol, and ferruginol, which exhibit anti-atherogenic, anti-inflammatory, and antibiotic effects, respectively, but lack verified protein targets.65–67 Our findings suggest that these molecules might inhibit one or more PTPs by binding to their highly conserved active sites (PTPs possess regulatory functions consistent with the biological activities of these molecules68,69) and, thus, motivate future analyses of the influence of biologically active abietane-type diterpenoids on PTP-mediated signaling events.
Supplementary Material
ACKNOWLEDGMENTS
The instruments used for NMR spectroscopy are supported by National Institutes of Health (NIH) Grant EB002026. The ALS-ENABLE beamlines, which were used for X-ray crystallography, are supported in part by the NIH (Grant P30 GM124169–01). The Advanced Light Source is a U.S. Department of Energy Office of Science User Facility supported by Contract DE-AC02–05CH11231.
Funding
This work was supported by start-up funds provided by the University of Colorado, Boulder (M.K.H., A.H., A.R., M.Z., E.Y.K., and J.M.F.), a National Science Foundation CAREER Award 1750244 (M.K.H. and J.M.F.), and the Claudia Adams Barr Program for Innovative Cancer Research (H.A.).
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.8b00655.
Supplementary methods, including statistical analyses of kinetic data, crystallization efforts, comparisons of sequence identities and structures, a principal component analysis, docking calculations, and an analysis of binding affinity, and supplementary notes, figures, and tables (PDF)
Kinetic data (XLSX)
Mutational analysis data (XLSX)
The authors declare no competing financial interest.
REFERENCES
- (1).Bence KK, Delibegovic M, Xue B, Gorgun CZ, Hotamisligil GS, Neel BG, and Kahn BB (2006) Neuronal PTP1B regulates body weight, adiposity and leptin action. Nat. Med 12, 917–24. [DOI] [PubMed] [Google Scholar]
- (2).Klaman LD, Boss O, Peroni OD, Kim JK, Martino JL, Zabolotny JM, Moghal N, Lubkin M, Kim Y-B, Sharpe AH, Stricker-Krongrad A, Shulman GI, Neel BG, and Kahn BB (2000) Increased Energy Expenditure, Decreased Adiposity, and Tissue-Specific Insulin Sensitivity in Protein-Tyrosine Phosphatase 1B-Deficient Mice. Mol. Cell. Biol 20, 5479–5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Rhee I, and Veillette a. (2012) Protein tyrosine phosphatases in lymphocyte activation and autoimmunity. Nat. Immunol 13, 439–447. [DOI] [PubMed] [Google Scholar]
- (4).Lee HK, Takamiya K, Han JS, Man H, Kim CH, Rumbaugh G, Yu S, Ding L, He C, Petralia RS, Wenthold RJ, Gallagher M, and Huganir RL (2003) Phosphorylation of the AMPA receptor GluR1 subunit is required for synaptic plasticity and retention of spatial memory. Cell 112, 631–643. [DOI] [PubMed] [Google Scholar]
- (5).Koren S, and Fantus IG (2007) Inhibition of the protein tyrosine phosphatase PTP1B: potential therapy for obesity, insulin resistance and type-2 diabetes mellitus. Best Pract. Res. Clin. Endocrinol. Metab 21, 621–640. [DOI] [PubMed] [Google Scholar]
- (6).Lessard L, Stuible M, and Tremblay ML (2010) The two faces of PTP1B in cancer. Biochim. Biophys. Acta, Proteins Proteomics 1804, 613. [DOI] [PubMed] [Google Scholar]
- (7).Chan G, Kalaitzidis D, and Neel BG (2008) The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev 27, 179. [DOI] [PubMed] [Google Scholar]
- (8).Zhang Y, Kurup P, Xu J, Carty N, Fernandez SM, Nygaard HB, Pittenger C, Greengard P, Strittmatter SM, Nairn AC, and Lombroso PJ (2010) Genetic reduction of striatal-enriched tyrosine phosphatase (STEP) reverses cognitive and cellular deficits in an Alzheimer’s disease mouse model. Proc. Natl. Acad. Sci. U. S. A 107, 19014–19019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Heneberg P (2009) Use of Protein Tyrosine Phosphatase Inhibitors as Promising Targeted Therapeutic Drugs. Curr. Med. Chem 16, 706–733. [DOI] [PubMed] [Google Scholar]
- (10).He R, Yu Z, Zhang R, and Zhang Z (2014) Protein tyrosine phosphatases as potential therapeutic targets. Acta Pharmacol. Sin 35, 1227–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Eden ER, White IJ, Tsapara A, and Futter CE (2010) Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat. Cell Biol. 12, 267–72. [DOI] [PubMed] [Google Scholar]
- (12).Tonks NK, and Muthuswamy SK (2007) A Brake Becomes an Accelerator: PTP1B-A New Therapeutic Target for Breast Cancer. Cancer Cell 11, 214. [DOI] [PubMed] [Google Scholar]
- (13).Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL, Normandin D, Cheng A, Himms-Hagen J, Chan CC, Ramachandran C, Gresser MJ, Tremblay ML, and Kennedy BP (1999) Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283, 1544–1548. [DOI] [PubMed] [Google Scholar]
- (14).Krishnan N, Koveal D, Miller DH, Xue B, Akshinthala SD, Kragelj J, Jensen MR, Gauss C-M, Page R, Blackledge M, Muthuswamy SK, Peti W, and Tonks NK (2014) Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nat. Chem. Biol 10, 558–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Johnson TO, Ermolieff J, and Jirousek MR (2002) Protein tyrosine phosphatase 1B inhibitors for diabetes. Nat. Rev. Drug Discovery 1, 696–709. [DOI] [PubMed] [Google Scholar]
- (16).Liu S, Zeng LF, Wu L, Yu X, Xue T, Gunawan AM, Long YQ, and Zhang ZY (2008) Targeting inactive enzyme conformation: Aryl diketoacid derivatives as a new class of PTP1B inhibitors. J. Am. Chem. Soc 130, 17075–17084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wiesmann C, Barr KJ, Kung J, Zhu J, Erlanson DA, Shen W, Fahr BJ, Zhong M, Taylor L, Randal M, McDowell RS, and Hansen SK (2004) Allosteric inhibition of protein tyrosine phosphatase 1B. Nat. Struct. Mol. Biol 11, 730–737. [DOI] [PubMed] [Google Scholar]
- (18).Peters RJ (2010) Two rings in them all: The labdane-related diterpenoids. Nat. Prod. Rep 27, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Na M, Oh WK, Kim YH, Cai XF, Kim S, Kim BY, and Ahn JS (2006) Inhibition of protein tyrosine phosphatase 1B by diterpenoids isolated from Acanthopanax koreanum. Bioorg. Med. Chem. Lett 16, 3061–3064. [DOI] [PubMed] [Google Scholar]
- (20).Hu C-L, Xiong J, Gao L-X, Li J, Zeng H, Zou Y, and Hu J-F (2016) Diterpenoids from the shed trunk barks of the endangered plant Pinus dabeshanensis and their PTP1B inhibitory effects. RSC Adv 6, 60467–60478. [Google Scholar]
- (21).San Feliciano A, Gordaliza M, Salinero MA, and Miguel Del Corral JM (1993) Abietane acids: Sources, biological activities, and therapeutic uses. Planta Med 59, 485. [DOI] [PubMed] [Google Scholar]
- (22).Aranda FJ, and Villalaín J (1997) The interaction of abietic acid with phospholipid membranes. Biochim. Biophys. Acta, Biomembr 1327, 171–180. [DOI] [PubMed] [Google Scholar]
- (23).Wang X, Morris-Natschke SL, and Lee KH (2007) New developments in the chemistry and biology of the bioactive constituents of Tanshen. Med. Res. Rev 27, 133. [DOI] [PubMed] [Google Scholar]
- (24).Zi J, Mafu S, and Peters RJ (2014) To Gibberellins and Beyond! Surveying the Evolution of (Di)Terpenoid Metabolism. Annu. Rev. Plant Biol 65, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Mafu S, Jia M, Zi J, Morrone D, Wu Y, Xu M, Hillwig ML, and Peters RJ (2016) Probing the promiscuity of ent-kaurene oxidases via combinatorial biosynthesis. Proc. Natl. Acad. Sci. U. S. A 113, 2526–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Rafferty RJ, Hicklin RW, Maloof KA, and Hergenrother PJ (2014) Synthesis of complex and diverse compounds through ring distortion of abietic acid. Angew. Chem., Int. Ed 53, 220. [DOI] [PubMed] [Google Scholar]
- (27).Morrone D, Lowry L, Determan MK, Hershey DM, Xu M, and Peters RJ (2010) Increasing diterpene yield with a modular metabolic engineering system in E. coli: Comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl. Microbiol. Biotechnol 85, 1893–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Burnham KP, and Anderson DR (2002) Model Selection and Multimodel Inference: A Practical Information-theoretic Approach, 2nd ed., Springer-Verlag, New York. [Google Scholar]
- (29).Dominguez C, Boelens R, and Bonvin AMJJ (2003) HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc 125, 1731–1737. [DOI] [PubMed] [Google Scholar]
- (30).Van Zundert GCP, Rodrigues JPGLM, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond ASJ, Van Dijk M, De Vries SJ, and Bonvin AMJJ (2016) The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol 428, 720–725. [DOI] [PubMed] [Google Scholar]
- (31).Anandakrishnan R, Aguilar B, and Onufriev AV (2012) H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res 40, W537–W541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Maestro User Manual 9.7 (2009) Schrödinger Press. [Google Scholar]
- (33).Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, and Uchimaya M (2007) Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules. J. Comput.-Aided Mol. Des 21, 681–91. [DOI] [PubMed] [Google Scholar]
- (34).Wang J, Wolf RM, Caldwell JW, Kollman PA, and Case DA (2004) Development and testing of a general amber force field. J. Comput. Chem 25, 1157–1174. [DOI] [PubMed] [Google Scholar]
- (35).Bayly CI, Cieplak P, Cornell W, and Kollman PA (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem 97, 10269–10280. [Google Scholar]
- (36).Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, and Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 79, 926. [Google Scholar]
- (37).Nosé S (1984) A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys 52, 255–268. [Google Scholar]
- (38).Hoover WG (1985) Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A: At., Mol., Opt. Phys 31, 1695–1697. [DOI] [PubMed] [Google Scholar]
- (39).Parrinello M, and Rahman A (1981) Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys 52, 7182. [Google Scholar]
- (40).Hess B, Bekker H, Berendsen HJC, and Fraaije JGEM (1997) LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem 18, 1463–1472. [Google Scholar]
- (41).Darden T, York D, and Pedersen L (1993) Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys 98, 10089. [Google Scholar]
- (42).Hess B, Kutzner C, van der Spoel D, and Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput 4, 435–447. [DOI] [PubMed] [Google Scholar]
- (43).Choy MS, Li Y, Machado LESF, Kunze MBA, Connors CR, Wei X, Lindorff-Larsen K, Page R, and Peti W (2017) Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery. Mol. Cell 65, 644–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Lipchock JM, Hendrickson HP, Douglas BB, Bird KE, Ginther PS, Rivalta I, Ten NS, Batista VS, and Loria JP (2017) Characterization of Protein Tyrosine Phosphatase 1B Inhibition by Chlorogenic Acid and Cichoric Acid. Biochemistry 56, 96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Williamson MP (2013) Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc 73, 1. [DOI] [PubMed] [Google Scholar]
- (46).Fox JM, Kang KK, Sastry M, Sherman W, Sankaran B, Zwart PH, and Whitesides GM (2017) Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase. Angew. Chem., Int. Ed 56, 3833–3837. [DOI] [PubMed] [Google Scholar]
- (47).Tzeng S-R, and Kalodimos CG (2012) Protein activity regulation by conformational entropy. Nature 488, 236–240. [DOI] [PubMed] [Google Scholar]
- (48).Iversen LF, Andersen HS, Branner S, Mortensen SB, Peters GH, Norris K, Olsen OH, Jeppesen CB, Lundt BF, Ripka W, Møller KB, and Møller NP (2000) Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B. J. Biol. Chem 275, 10300–10307. [DOI] [PubMed] [Google Scholar]
- (49).Lang PT, Holton JM, Fraser JS, and Alber T (2014) Protein structural ensembles are revealed by redefining X-ray electron density noise. Proc. Natl. Acad. Sci. U. S. A 111, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).De Vivo M, and Cavalli A (2017) Recent advances in dynamic docking for drug discovery. Wiley Interdiscip. Rev.: Comput. Mol. Sci 7, e1320. [Google Scholar]
- (51).Fleck EE, and Palkin S (1939) The Presence of Dihydroabietic Acid in Pine Oleoresin and Rosin. J. Am. Chem. Soc 61, 1230–1232. [Google Scholar]
- (52).Jeong SI, Han WS, Yun YH, and Kim KJ (2006) Continentalic acid from Aralia continentalis shows activity against methicillin-resistant Staphylococcus aureus. Phytother. Res 20, 511–514. [DOI] [PubMed] [Google Scholar]
- (53).Smith E, Williamson E, Zloh M, and Gibbons S (2005) Isopimaric acid from Pinus nigra shows activity against multidrug-resistant and EMRSA strains of Staphylococcus aureus. Phytother. Res 19, 538–542. [DOI] [PubMed] [Google Scholar]
- (54).Kleppe M, Lahortiga I, El Chaar T, De Keersmaecker K, Mentens N, Graux C, Van Roosbroeck K, Ferrando AA, Langerak AW, Meijerink JPP, Sigaux F, Haferlach T, Wlodarska I, Vandenberghe P, Soulier J, and Cools J (2010) Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat. Genet 42, 530–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Garcia Fortanet J, Chen CHT, Chen YNP, Chen Z, Deng Z, Firestone B, Fekkes P, Fodor M, Fortin PD, Fridrich C, Grunenfelder D, Ho S, Kang ZB, Karki R, Kato M, Keen N, Labonte LR, Larrow J, Lenoir F, Liu G, Liu S, Lombardo F, Majumdar D, Meyer MJ, Palermo M, Perez L, Pu M, Ramsey T, Sellers WR, Shultz MD, Stams T, Towler C, Wang P, Williams SL, Zhang JH, and Lamarche MJ (2016) Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor. J. Med. Chem 59, 7773–7782. [DOI] [PubMed] [Google Scholar]
- (56).Chen Y-NP, LaMarche MJ, Chan HM, Fekkes P, Garcia-Fortanet J, Acker MG, Antonakos B, Chen CH-T, Chen Z, Cooke VG, Dobson JR, Deng Z, Fei F, Firestone B, Fodor M, Fridrich C, Gao H, Grunenfelder D, Hao H-X, Jacob J, Ho S, Hsiao K, Kang ZB, Karki R, Kato M, Larrow J, La Bonte LR, Lenoir F, Liu G, Liu S, Majumdar D, Meyer MJ, Palermo M, Perez L, Pu M, Price E, Quinn C, Shakya S, Shultz MD, Slisz J, Venkatesan K, Wang P, Warmuth M, Williams S, Yang G, Yuan J, Zhang J-H, Zhu P, Ramsey T, Keen NJ, Sellers WR, Stams T, and Fortin PD (2016) Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 535, 148–52. [DOI] [PubMed] [Google Scholar]
- (57).Lafont V, Armstrong A. a., Ohtaka H, Kiso Y, Mario Amzel L, and Freire E (2007) Compensating enthalpic and entropic changes hinder binding affinity optimization. Chem. Biol. Drug Des 69, 413–422. [DOI] [PubMed] [Google Scholar]
- (58).Fox JM, Zhao M, Fink MJ, Kang K, and Whitesides GM (2018) The Molecular Origin of Enthalpy/Entropy Compensation in Biomolecular Recognition. Annu. Rev. Biophys 47, 223. [DOI] [PubMed] [Google Scholar]
- (59).Barr AJ, Ugochukwu E, Lee WH, King ONF, Filippakopoulos P, Alfano I, Savitsky P, Burgess-Brown NA, Müller S, and Knapp S (2009) Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome. Cell 136, 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Barr AJ (2010) Protein tyrosine phosphatases as drug targets: Strategies and challenges of inhibitor development. Future Med. Chem 2, 1563. [DOI] [PubMed] [Google Scholar]
- (61).Wu P, Nielsen TE, and Clausen MH (2015) FDA-approved small-molecule kinase inhibitors. Trends Pharmacol. Sci 36, 422. [DOI] [PubMed] [Google Scholar]
- (62).Kampranis SC, Ioannidis D, Purvis A, Mahrez W, Ninga E, Katerelos NA, Anssour S, Dunwell JM, Degenhardt J, Makris AM, Goodenough PW, and Johnson CB (2007) Rational conversion of substrate and product specificity in a Salvia monoterpene synthase: structural insights into the evolution of terpene synthase function. Plant Cell 19, 1994–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Nelson DR (2011) Progress in tracing the evolutionary paths of cytochrome P450. Biochim. Biophys. Acta, Proteins Proteomics 1814, 14–18. [DOI] [PubMed] [Google Scholar]
- (64).Jia M, Potter KC, and Peters RJ (2016) Extreme promiscuity of a bacterial and a plant diterpene synthase enables combinatorial biosynthesis. Metab. Eng 37, 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Gao S, Liu Z, Li H, Little PJ, Liu P, and Xu S (2012) Cardiovascular actions and therapeutic potential of tanshinone IIA. Atherosclerosis 221, 604. [DOI] [PubMed] [Google Scholar]
- (66).Johnson JJ (2011) Carnosol: A promising anti-cancer and anti-inflammatory agent. Cancer Lett 305, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Kuźma Ł, Rózalski M, Walencka E, Rózalska B, and Wysokińska H (2007) Antimicrobial activity of diterpenoids from hairy roots of Salvia sclarea L.: Salvipisone as a potential anti-biofilm agent active against antibiotic resistant Staphylococci. Phytomedicine 14, 31–35. [DOI] [PubMed] [Google Scholar]
- (68).Guan KL, and Dixon JE (1990) Protein tyrosine phosphatase activity of an essential virulence determinant in Yersinia. Science 249, 553–556. [DOI] [PubMed] [Google Scholar]
- (69).Andersen JN, and Tonks NK (2004) Protein tyrosine phosphatase-based therapeutics: lessons from PTP1B. Top. Curr. Genet 5, 201–230. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.