SUMMARY
Cardiac differentiation of human pluripotent stem cells (hPSCs) requires orchestration of dynamic gene regulatory networks during stepwise fate transitions, but often generates immature cell types that do not fully recapitulate properties of their adult counterparts, suggesting incomplete activation of key transcriptional networks. We performed extensive single-cell transcriptomic analyses to map fate choices and gene expression programs during cardiac differentiation of hPSCs, and identified strategies to improve in vitro cardiomyocyte differentiation. Utilizing genetic gain- and loss-of-function approaches, we found that hypertrophic signaling is not effectively activated during monolayer-based cardiac differentiation, thereby preventing expression of HOPX and its activation of downstream genes that govern late stages of cardiomyocyte maturation. This study therefore provides a key transcriptional roadmap of in vitro cardiac differentiation at single-cell resolution, revealing fundamental mechanisms underlying heart development and differentiation of hPSC-derived cardiomyocytes.
Keywords: human pluripotent stem cells, cardiomyocytes, single cell RNA-seq, heart, development, CRISPR, hypertrophy, lineage tracing, HOPX, scdiff
Graphic Abstract
Friedman et al. performed single cell transcriptional analysis over a time-course of in vitro cardiac differentiation from human pluripotent stem cells. They utilized this data to identify the requirement of hypertrophic stimuli for expression of a cardiac regulatory gene, HOPX, to generate cardiomyocytes more accurately reflecting in vivo heart development.
Graphic Abstract

INTRODUCTION
Studies of cardiac development at single cell resolution have provided valuable insights into cell diversity and genetic regulation of cardiovascular differentiation and morphogenesis in vivo (DeLaughter et al., 2016; Li et al., 2016). Human pluripotent stem cells are a key model system to study human cardiovascular developmental biology (Murry and Keller, 2008). However, it is well understood that cardiac differentiation in vitro does not generate cardiomyocytes with the transcriptional profile, cellular diversity, morphometry, or functional maturity of adult in vivo-derived cardiomyocytes (Yang et al., 2014). The fidelity by which cardiac directed differentiation in vitro recapitulates the transcriptional programs governing diverse cell fates generated in vivo has not been well-characterized.
In this study, we report RNA-sequencing data captured from more than forty thousand single cells navigating stage-specific transitions through in vitro cardiac directed differentiation from pluripotency using an established small molecule Wnt modulation protocol (Burridge et al., 2014; Lian et al., 2012). In coordination with a companion paper (Nguyen et al., in review), we utilize the power of this data set to expand our understanding of stem cell directed differentiation as a platform to study cardiovascular development. Since heart development in vivo requires instructive cues from exogenous sources like signaling from endoderm and mechanical forces of heart beat and growth, we aimed to leverage single cell transcriptomic data to identify critical signaling or mechanical strategies for differentiating hPSCs to more accurately understand the identity of and control derivation of cardiac fates. We identify the non-DNA binding homeodomain protein HOPX, a key regulator of heart development (Jain et al., 2015) and hypertrophy (Chen et al., 2002; Kook et al., 2003; Shin et al., 2002) as dysregulated during differentiation and a potential cause for the immature state of hPSC-derived cardiomyocytes in vitro. Taken together, these data provide a unique resource for the field and identify a previously unappreciated strategy for enhancing differentiation of hPSC-derived cardiomyocytes in vitro for applications in cardiovascular biology.
RESULTS
Single Cell RNA-Sequencing Analysis of Cardiac Directed Differentiation
To gain insights into the genetic regulation of cardiovascular development, we performed single cell transcriptional profiling of human iPSCs navigating from pluripotency through stage-specific transitions in cardiac differentiation (Figure 1A). Small molecule Wnt modulation was used as an efficient method to differentiate pluripotent cells toward the cardiac lineage (Burridge et al., 2014; Lian et al., 2012). WTC CRISPRi GCaMP hiPSCs (Mandegar et al., 2016) were chosen as the parental cell line for this study. These cells are genetically engineered with an doxycycline-inducible nuclease-dead Cas9 fused to a KRAB repression domain. The versatility of this line provides a means to use this scRNA-seq data as a reference point for future studies aiming to assess the transcriptional basis of cardiac differentiation at the single cell level. We captured cells at time points corresponding to stage-specific transitions in cell state including pluripotency (day 0), germ layer specification (day 2), and progressing through progenitor (day 5), committed (day 15), and definitive (day 30) cardiac cell states. We harvested a total of 44,020 cells of which 43,168 cells were retained after quality control analysis. In total, we captured expression of 17,718 genes. Dimensionality reduction approaches were used to visualize all 43,168 cells in low-dimensional space (Figure 1B), in which cell’s coordinates were estimated so that they preserve the expression similarity in t-SNE plots (left), and differentiation pseudotime in diffusion plots (right).
Figure 1. Single Cell Analysis of Cardiac Directed Differentiation.
(A) Schematic of protocol for small molecule directed differentiation from pluripotency into the cardiac lineage. hPSC: human pluripotent stem cell; GLS: germ layer specification; PC: progenitor cell: cCD: committed cardiac derivative; dCD: definitive cardiac derivative.
(B) Single cells (n = 43,168 in total) transiting cardiac differentiation beginning at pluripotency (day 0) and transitioning through mesoderm (day 2) into progenitor (day 5) committed (day 15) and definitive (day 30) cardiac derivatives. Data are presented using t-SNE plot, pseudospacing cells by the nonlinear transformation of similarity in gene expression to preserve the local and global distance of cells in multidimensional space when embedded into two dimensional t-SNE space (left), and diffusion plot, pseudospacing cells in a trajectory based on diffusion distance (transition probability) between two cells (right).
(C) Mean gene expression across all cells at individual time points showing proper temporal expression of stage-specific genes governing differentiation into the cardiac lineage. Shown are pluripotency genes (DNMT3B, POU5F1, NANOG), mes-endoderm genes (EOMES, MIXL1, T, MESP1), and genes governing cardiomyocyte differentiation including signaling regulators (TMEM88), transcription factors (ISL1, HAND1, NKX2–5, TBX5, GATA4), calcium handling genes (ATP2A2, PLN) and sarcomere genes (TNNI1, MYH6, MYH7, MYL7). Data are represented as mean ± SEM.
(D) Diffusion plots showing pseudospacing at single cell resolution for gene expression of stage-specific genes during differentiation based on known genetic regulators of cardiac fate specification including POU5F1 (day 0), EOMES (day 2), TMEM88 (day 5), TNNI1 (day 15), and TTN (day 30). Cells are colored in a binary manner. If the cell expresses the gene it is colored according to the day of isolation (0, 2, 5, 15, or 30). Non-expressing cells are shaded gray.
(E) Representation of unsupervised clustering analysis (Nguyen et al., in review) using t-SNE plots to show single cell level expression of stage-specific gene expression at each day of differentiation based on known genetic regulators of cardiac fate specification including POU5F1, EOMES, ISL1, TNNI1, and MYL7. If the cell expresses the gene it is colored according to subpopulation 1–4 in which the cell is associated. Non-expressing cells are shaded gray. Above each t-SNE plot, the percentage of cells expressing the gene in each subpopulation is shown together with the expression histogram and the reference t-SNE plot. UMI: unique molecular identifier.
We generated a time-course gene expression profile using a wide range of known cardiac developmental genes by measuring expression among all cells to reveal the temporally-restricted expression dynamics of stage-specific genes reflecting cardiac fate choices (Figure 1C). To confirm that the differentiation follows known developmental trajectories, we used dimensionality reduction methods (Coifman et al., 2005; Moignard et al., 2015) (Figure 1D) and unsupervised clustering (Clustering at Optimal REsolution (CORE) (Nguyen et al., in review)) (Figure 1E) to analyze the expression of known genes. Overall, these data show that small molecule-mediated cardiac directed differentiation generates distinct populations of cells displaying expected temporal-specific transcriptional profiles. Our parallel computational genomics study (Nguyen et al., in review) presents a web interface (http://computationalgenomics.com.au/shiny/hipsc2cm/), provided as complementary resources for this study.
Phenotypic Diversity and Lineage Heterogeneity During Differentiation
With recent developments in high-resolution transcriptomic mapping of mouse in vivo development of the cardiovascular system (DeLaughter et al., 2016; Li et al., 2016; Peng et al., 2016), we set out to map single cell heterogeneity of human in vitro derived subpopulations against in vivo cell subpopulations. Using previously published approaches, laser microdissection was used to capture spatiotemporal transcriptional data from germ layer cells of mid-gastrula stage (E7.0) embryos (Peng et al., 2016), with an expanded analysis to include early- (E6.5) and late-gastrulation (E7.5) mouse embryos (unpublished data) (Figure S1). To determine phenotypic identities based on gene expression networks governing each human in vitro-derived subpopulation during differentiation, we visualized the spatio-temporal patterns of gene expression in the gastrulating mouse embryo including: EOMES (pan-mesendoderm), MESP1 and MIXL1 (mesoderm), SOX17 and FOXA2 (endoderm), and NKX2–5 (cardiac lineage transcription factor) (Figure 2A, Figure S2).
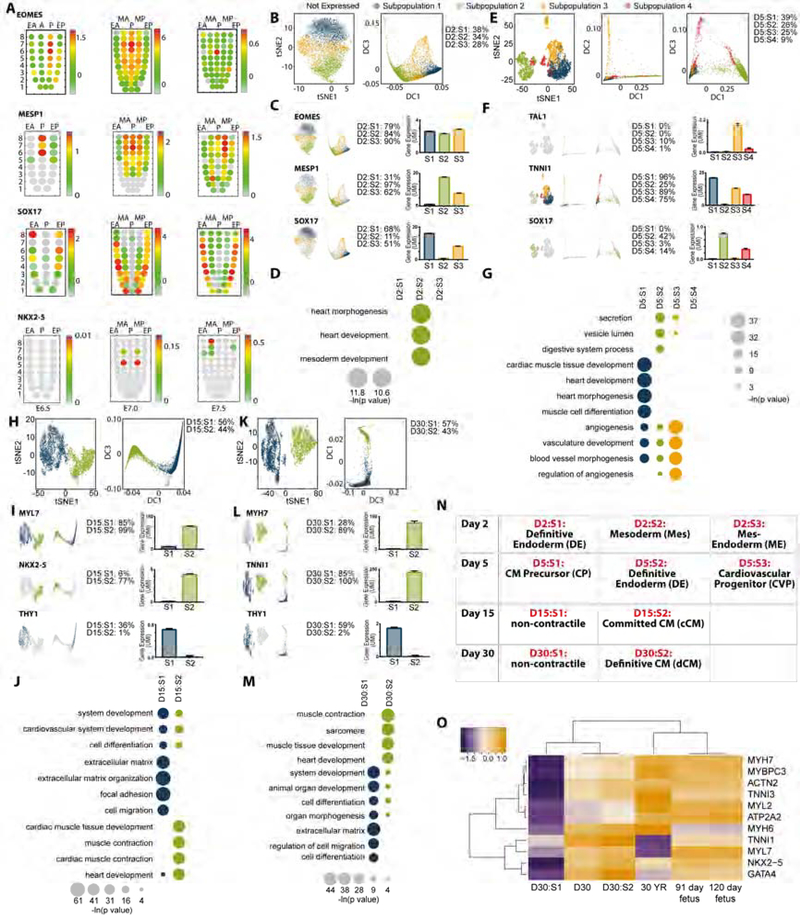
Figure 2. Subpopulation Identification and Characterization.
(A) Corn plots showing spatial domains of EOMES, MESP1, SOX17 and NKX2–5 expression in the mesoderm and endoderm of E6.5, E7.0, and E7.5 mouse embryos during gastrulation (unpublished RNA-seq data for E6.5 and E7.5 mouse embryos and published data for E7.0 mouse embryos (Peng et al., 2016)). Positions of the cell populations (“kernels” in the 2D plot of RNA-Seq data) in the germ layers: the proximal-distal location in descending numerical order (1 = most distal site) and in the transverse plane of the mesoderm and endoderm - Anterior half (EA) and Posterior half (EP) of the endoderm, Anterior half (MA) and Posterior half (MP) of the mesoderm, and Posterior epiblast (P) containing the primitive streak.
(B-M) Below each gene name are shown the following data from left to right: t-SNE plot and diffusion plot of cells expressing each gene, percent of cells expressing gene, expression level of gene in each subpopulation.
(B-D) Analysis of day 2 subpopulations represented by (B) reference t-SNE (left) and diffusion (right) plots and the percent of cells in each subpopulations (D2:S1-S3), (C) analysis of primitive streak genes EOMES (pan-mesendoderm transcription factor), MESP1 (cardiogenic mesoderm transcription factor), and SOX17 (definitive endoderm transcription factor). (D) Gene ontology analysis of differentially expressed genes showing enrichment for networks governing cardiac development enriched in subpopulation 2.
(E-G) Analysis of day 5 progenitor subpopulations represented by (E) reference t-SNE (left) and diffusion (right) plots and the percent of cells in each subpopulations (D5:S1-S4), (F) analysis of progenitor genes TALI (endothelial fate transcription factor), TNNI1 (early development sarcomere isoform of TNNI), and SOX17 (definitive endoderm transcription factor). (G) Gene ontology analysis of differentially expressed genes showing enrichment for networks governing cardiac development (D5:S1), definitive endoderm (D5:S2), and endothelium (D5:S3).
(H-J) Analysis of day 15 subpopulations represented by (H) reference t-SNE (left) and diffusion (right) plots and the percent of cells in each subpopulations (D15:S1-S2), (I) analysis of cardiac genes MYL7 (early development sarcomere isoform of MYL), NKX2–5 (cardiac transcription factor), and THY1 (fibroblast marker). (J) Gene ontology analysis of differentially expressed genes showing enrichment for networks governing extracellular matrix and cell motility (D15:S1) and cardiac development (D15:S2).
(K-M) Analysis of day 30 subpopulations represented by (K) reference t-SNE (left) and diffusion (right) plots and the percent of cells in each subpopulations (D30:S1-S2), (L) analysis of cardiac genes TNNI1 (early development sarcomere isoform of TNNI), MYH7 (mature sarcomere isoform of MYH), and THY1 (fibroblast marker). (M) Gene ontology analysis of differentially expressed genes showing enrichment for networks governing system development and morphogenesis (D30:S1) and cardiac development (D30:S2).
(N) Overall phenotypic determinations of subpopulation identity based on in vivo anchoring genes outlined through stage-specific transitions in differentiation. CM: cardiomyocyte.
(O) Expression of cardiac genes in day 30 hPSC-derived cardiomyocytes relative to expression levels in human foetal and adult heart samples (ENCODE). Gene expression is measured as counts per million mapped reads and each gene is internally normalized to maximum expression. UMI: unique molecular identifier. See also Figure S1–3 and Table S1.
We subsequently dissected the transcriptional phenotype of subpopulations identified during human cardiac directed differentiation. From pluripotency (Figure S3A-B), cells navigate through germ layer specification (day 2), comprising three transcriptionally distinct subpopulations expressing the panmesendoderm gene, EOMES (Figure 2B-C, Figure S2A) with subpopulations expressing genes involved in mesoderm (D2:S2), mesendoderm (D2:S3), and definitive endoderm (D2:S1) (Figure 2B-C and Figure S2D and Figure S3C-D). Gene ontology (GO) analysis of differentially expressed genes between subpopulations indicated that only D2:S2 (34% of cells at day 2) showed significant enrichment for cardiogenic gene networks (Figure 2D, Table S1). At the progenitor stage (day 5), we identified cardiac precursors (D5: S1 and D5:S3) (Figure 2E-G and Figure S3E), a persistent population of definitive endoderm (D5:S2) (Figure 2E-F and Figure S3E), and endothelial cells (D5:S3) (Figure 2E-G). Day 15 and day 30 cells comprised two subpopulations (Figure 2H-M and Figure S3F-G). NKX2–5, MYH6, TTN and other cardiac structural and regulatory genes were identified in S2 (Figure 2H-M and Figure S3F-G). In contrast, S1 was primarily characterized by GO enrichment for genes associated with extracellular matrix deposition, motility, and cell adhesion (Figure 2J and M) which was supported by identification of a significant number of fibroblast-like cells marked by THY1 (CD90) (Figure 2I and L). The co-existence of a non-contractile cell population, which is characterized as non-myocytes, is common in directed cardiac differentiation (Dubois et al., 2011). Taken together, these data show iPSC differentiation into committed (day 15) and definitive (day 30) cardiomyocytes (S2) and non-contractile cells (S1) (Figure 2N). To assess the level of maturity derived from this protocol relative to in vivo human development, we compared day 30 clusters against ENCODE RNA-seq data from foetal and adult hearts (Figure 2O). Using genes that reflect either early foetal (TNNI1, MYH6) vs late stages of heart development (MYH7, TNNI3, MYL2), the most differentiated in vitro derived cardiac population (D30:S2) remains more developmentally immature than even first trimester human hearts.
Lineage Predictions Based on Regulatory Gene Networks Governing Differentiation
We next sought to understand the lineage trajectories and gene regulatory networks governing diversification of cell fates. We implemented a probabilistic method for constructing regulatory networks from single cell time series expression data (scdiff: Cell Differentiation Analysis Using Time-series Single cell RNA-seq Data) (Ding et al., 2018). The algorithm utilizes TF-gene databases to model gene regulation relationships based on the directional changes in expression of TFs and target genes at parental and descendant states.
The algorithm identified three distinct lineages from pluripotency comprising 10 nodes (Table S2 and Figure 3A). Since this algorithm reassigns cells based on regulatory networks, we analyzed the distribution of cell subpopulations based on our CORE cluster classifications as outlined in Figure 2 to establish population identities linking predicted lineages (Figure 3A-B and Figure S4A). The first lineage (N1:N2) diverts from pluripotency into a SOX17/FOXA2/EPCAM+ definitive endoderm population that terminates at day 2 and is comprised almost exclusively of D2:S1 and D2:S3 (Figure 3A-B and Figure S4A). The second lineage, N1:N3:N5, transitions from pluripotency (N1) into node 3 comprised of definitive endoderm (D2:S1) and mesendoderm (D2:S3) and is predicted to terminate at day 5 node 5 comprising FOXA2/EPCAM+ definitive endoderm cells (D5:S2 and D5:S4) (Figure 3A-B and Figure S4A). The third lineage comprises the longest trajectory through differentiation involving transitions in cardiac fate (N 1:N4:N6-N9 and N6-N10). Pluripotent cells (N1) give rise on day 2 to node 4 mesoderm (D2:S2) and mesendoderm (D2:S3) cells with subsequent progression on day 5 into cardiac precursor cells (N6: primarily D5:S1 and D5:S3). From day 5 the algorithm predicts a bifurcation of fate giving rise to THY1+/NKX2–5- non-contractile cardiac derivatives (N8–10: D15:S1 and D30:S1) or NKX2– 5+/MYH6+ committed CM (N7: D15:S2) that progress onto MYH7+/MYL2+ definitive CM (N9: D30:S2) (Figure 3A-B and Figure S4A).
Figure 3. Transcription Factor Regulatory Networks Predict Developmental Fate Choices During Cardiac Differentiation.
(A) Stepwise transitions into cardiac lineages from pluripotency predicted on the basis of gene regulatory networks (GRN) detected between pairwise changes in cell state during differentiation. Circles indicate distinct nodes governed by a common GRN. Since cells can be re-assigned based on the expression of their genes, the re-distribution of subpopulations established by clustering analysis and phenotyping as outlined in Figure 2 are represented as pie charts within each circle indicating the percent of cells from each subpopulation contributing to that node. Each node is numbered N1-N10 for reference.
(B) Phenotypic identity of nodes reflecting stage-specific transitions in cell state through cardiac directed differentiation.
(C) Analysis of transcription factors (TFs) and genes controlling stage-specific regulatory networks underlying cell fate transitions. Mean DE target fold change calculates the fold change for the differentially expressed targets of the TF. DE gene fold change shows up or down-regulated fold change of TF target genes.
(D) Heat map comparing expression across all cells from day 5, 15, and 30 subpopulations for genes involved in progenitor specification, vascular endothelial development, outflow tract development, and primary heart field cardiomyocyte development.
(E) Gene ontology analysis comparing day 30 S1 vs S2 showing gene networks involved in vascular development enriched in S1 vs cardiac muscle development enriched in S2.
(F) t-SNE and diffusion plots for all cells from days 15 and 30 showing expression distribution of the cardiac gene MYH7 (high in S2 at day 15 and 30) relative to outflow tract development genes THY1, PITX2, and BMP4 (high in S1 at day 15 and 30)
(G) The top most differentially expressed genes identified by in vivo single cell analysis comparing outflow tract (OFT) vs ventricular cardiomyocytes (Li et al., 2016) compared against their expression level in D30:S1 vs D30:S2 in vitro derived cardiac derivatives.
(H) Differentially expressed genes between subpopulations D30:S1 and D30:S2 used to assess transcriptional similarity to in vivo cell types (Li et al., 2016; Quaife-Ryan et al., 2017) using Spearman’s correlation analysis. See also Figure S4 and Table S2.
We leveraged the regulatory network predictions to identify key transcription factors and target genes underlying progressive fate changes across all 10 nodes (Figure 3C and Table S2). These data reinforce established mechanisms of cardiac lineage specification. In particular, we found evidence for down-regulation of Wnt/β-catenin signaling (LEF1) between N4-N6 which is required to transition from mesoderm into the cardiac progenitor cell (Palpant et al., 2013; Ueno et al., 2007). From the progenitor node N6 into contractile cardiomyocytes N7:N9, the data show proper down-regulation of progenitor transcription factors such as YY1 and up-regulation of TFs known to control cardiomyocyte differentiation such as NKX2–5 (Figure 3C).
We next sought to understand gene networks underlying specification of non-contractile cardiac derivatives N8:N10, a population currently not well defined although widely used for tissue engineering applications (Thavandiran et al., 2013). The predicted network underlying this transition showed significant down-regulation of cardiac TFs NKX2–5 and MAZ and up-regulation of Pre-B cell leukemia transcription homeobox (PBX1: P = 1.1e−16, mean DE target fold change = 2.72), a transcriptional regulator that activates a network of genes associated with cardiac outflow tract (OFT) morphogenesis (Arrington et al., 2012) (Figure 3C).
We compared expression of a panel of cardiomyocyte, early developmental vascular endothelial, and OFT development genes across all subpopulations comprising transitions from day 5 to 30 (Figure 3D). While early developmental vascular EC differentiation genes (TALI, CDH5) were expressed in D5:S3, these genes were not expressed in D15 or D30. Furthermore, while D15 and D30 S2 cells expressed cardiac sarcomere genes and transcription factors (IRX4 and HCN4), S1 cells uniquely expressed an extensive network of genes associated with OFT development including PITX2, TBX18, HOXA1–3, FGF10, GJA1, and KDR (Figure 3D). To determine the strength of this association, we performed gene ontology analysis of differentially expressed genes between D30:S1 (N10) and D30:S2 (N9) cells. These data show a significant enrichment for gene networks related to vascular development (P = 1.1e−11) and blood vessel morphogenesis (P = 4.7e−9) exclusively within node 10 D30:S1 cells. This finding is supported by single cell visualization showing enrichment of OFT gene expression in S1 vs S2 including THY1 (59% D30:S1 vs 2% D30:S2), BMP4 (70% D30:S1 vs 6% D30:S2), and PITX2 (73% D30:S1 vs 17% D30:S2) (Figure 3E-F).
To link this observation to in vivo cell types, we used single cell RNA-seq data of in vivo heart development (Li et al., 2016) to identify the top most differentially expressed genes between outflow tract and left ventricle (LV). These data show expression of BMP4, RSPO3, TNC, and COL1A2 in D30:S1 and in vivo OFT derivatives and MYL2 and HOPX upregulated in cardiomyocytes (Figure 3G). To assess cell-type specific transcriptional signatures, we identified differentially expressed genes between D30 S1 vs S2 and performed a Spearman rank correlation analysis against expression profiles of in vivo FACS sorted (Quaife-Ryan et al., 2017) or single cell-derived cardiac subtypes (Li et al., 2016). These data show that D30:S1 has a significantly stronger correlation to OFT cells (Spearman’s ρ = 0.442) than to fibroblasts (Spearman’s ρ = 0.219), endothelium (Spearman’s ρ = 0.175), or myocardium (Spearman’s ρ = 0.243) (P < 2.2×10−16 for all pairwise comparisons) (Figure 3H). Collectively, these data indicate that directed differentiation generates definitive cell populations comprising contractile cardiomyocytes and a non-contractile cell type whose transcriptional signature correlates with cardiac outflow tract cells. Due to the complex cellular origins of cardiac outflow tract and the diversity of non-contractile cell types of the heart in vivo, further studies are required to determine the specific identity and biology of these cells and their application in disease modelling and tissue engineering.
HOPX is Dysregulated During in vitro Directed Differentiation from hPSCs
We next aimed to identify dysregulated gene networks with the objective of determining different mechanisms for modelling in vitro differentiation to more accurately reflect in vivo heart development. Focusing on core regulatory genes governing transcriptional networks in heart development, we analyzed 52 transcription factors and epigenetic regulators known to govern diversification of mesoderm and endoderm lineages (Table S3). HOPX, a non-DNA binding homeodomain protein identified in this analysis, has previously been shown to be one of the earliest, specific markers of cardiomyocyte development (Jain et al., 2015), and governs cardiac fate by regulating cardiac gene networks through interactions with transcription factors, epigenetic regulators, and signaling molecules (Chen et al., 2002; Jain et al., 2015). We have also recently shown that HOPX functionally regulates blood formation from hemogenic endothelium (Palpant et al., 2017b). Consistent with mouse heart development, analysis of human foetal development at each trimester indicate a robust activation of HOPX during heart development in vivo (Figure S5A).
Previous studies have shown HOPX is expressed during cardiomyocyte specification at the progenitor stage of mouse development in vivo (Jain et al., 2015) whereas we detected HOPX only in endothelium (D5:C3) and not in cardiac precursor cells (D5:C1) at an equivalent time point (day 5) (Figure 4A-B). Second, in contrast to previous studies in vivo where HOPX lineage traces almost all cardiomyocytes of the heart (Jain et al., 2015), HOPX is detected in only 16% of D30:S2 cardiomyocytes (Figure 4B-D). To rule out stochastic expression in cardiomyocytes due to low sequencing read depth resulting in dropout, we analyzed expression of genes known to regulate cardiac lineage specification and differentiation (Figure 4C-D). While HOPX is rarely detected, its expression level is equivalent to that of other cardiac TFs that are detected in a high percentage of D30:S2 cardiomyocytes (HAND1: 67%, HAND2: 64%, GATA4: 67%, NKX2–5: 86% vs HOPX: 16%) (Figure 4B-D).
Figure 4. HOPX is Rarely Expressed During in vitro Cardiac Directed Differentiation.
(A) Analysis of HOPX expression in eleven subpopulations from day 2 to day 5 of differentiation showing expression as early as day 2 mesoderm and highest expression in day 5 endothelial cells (ECs) and day 30 cardiomyocytes (CMs)
(B) Single cell expression analysis of HOPX at day 2, 5, 15, and 30. Data presented include t-SNE plots indicating distribution and localization of HOPX expressing cells in different subpopulations (bottom), the percentage of HOPX+ cells in each subpopulation (top left), bar graphs showing expression of HOPX in each subpopulation (top middle), and the reference t-SNE plot demarcating subpopulations (top right). Data are represented as mean ± SEM.
(C) Analysis of known genetic regulators of heart development only in subpopulation 2 at day 30 of differentiation.
(D) t-SNE plots of merged data sets from two continuous days for all cells between day 15–30 for each gene showing robust distribution of key cardiac regulatory genes with the exception of HOPX.
(E) Corn plots showing the spatial domains of HOPX expression in the mesoderm and endoderm of E7.0 and E7.5 mouse embryos during gastrulation (unpublished RNA-seq data for E7.5 embryos and published data for E7.0 embryo, (Peng et al., 2016)) (Figure S1).
(F) Single cell expression analysis of E9.5 mouse heart cells (Li et al., 2016) showing HOPX expression relative to markers of cardiomyocytes (MYH7, ACTN2) and endothelial cells (CDH5, PECAM1) (scale bars are Log2(RPM)). Table (right) shows percent of cardiac (MYH7), endothelial (PECAM1), and smooth muscle (TAGLN2) cells co-expressing HOPX in various regions of the developing mouse heart. UMI: unique molecular identifier. See also S4–5 and Table S4.
Assessment of gene expression during mouse gastrulation in vivo shows HOPX expression as early as E6.5 in the proximal portion of the nascent primitive streak (P) (Figure 4E and Figure S5B-C) similar to the expression pattern of MESP1 (Figure S5D-E). From E7.0 to E7.5, HOPX is increasingly expressed throughout the developing endoderm. By E7.5, HOPX displays residual expression in the remaining distal primitive streak, endoderm (EA to EP), and the anterior mesoderm (MA) in coordination with other cardiogenic genes including NKX2–5 and MESP1 (Figure 4E and Figure 2A). We analyzed HOPX expression across diverse cell types contributing to heart development in vivo using single cell transcriptomic analysis of the E9.5 mouse heart (Li et al., 2016). These data indicate that HOPX expression is distributed throughout all chambers and cell types of the heart. While HOPX expression largely coincides with expression of cardiac genes MYH7 and ACTN2, HOPX is also expressed in endothelial cells (CDH5+ and/or PECAM1+), smooth muscle cells (MYH11+ and/or TAGLIN2+), and epicardial cells (WT1+) (Figure 4F and Table S4).
Lineage Trajectory of HOPX-Expressing Cells in vitro
We analyzed the lineage trajectory of HOPX+ cells at single cell resolution during cardiac directed differentiation to determine the core gene networks and transcription factors governing successive fate choices of HOPX expressing cells during cardiac differentiation (Figure S4B-D, and Table S2). At day 2 rare HOPX expressing cells are identified in mes-endoderm (D2:S2 9% and D2:S3 6%) and rarely in definitive endoderm (D2:S1 2%) with the HOPX lineage comprising 2 lineages (N2 and N3) enriched for expression of cardiogenic mesoderm genes such as MESP1 (Figure S3C-D). From day 2 into day 5, HOPX+ cells remain sparse in progenitor cell populations where day 2 N3 splits into two day 5 lineages N4 and N5 (Figure S4B). Based on lineage prediction, an equal proportion of HOPX+ cells give rise to TNNI1+ cardiac precursor cells (N4: 389 cells) or TAL1+ expressing endothelial cells (N5: 381 cells) with both fates governed by established TFs and downstream gene networks required for endothelial (NRP2, KDR) vs cardiac fate specification (TNNI1, TMEM88) (Figure S4C-D and Table S2). Progressing to day 15 of differentiation, HOPX expression remains rare (2–4% of cells) and splits into two separate lineages derived from day 5 cardiac precursor cells (N4). Governed in part by increased NKX2–5 and downregulation of the cardiac progenitor TF YY1, HOPX cardiac cells differentiate into MYL2/IRX4+ cardiomyocytes (N6-N7) while a separate branch governed by TFs such as PBX1 differentiate into noncontractile derivatives (N8-N9) (Figure S4B-D).
Chromatin and Expression Analysis of HOPX in Cardiac Lineage Specification
To determine the epigenetic basis for HOPX dysregulation during in vitro differentiation, we analyzed chromatin and transcriptional regulation at the HOPX locus (Palpant et al., 2017a) (Figure S5F). Chromatin immunoprecipitation data for repressive chromatin (H3K27me3), actively transcribed chromatin (H3K4me3), and gene expression by RNA-seq (Palpant et al., 2017b) show that in the context of cardiac directed differentiation HOPX is epigenetically repressed on the basis of abundant H3K27me3 compared to H3K4me3 in day 5 cardiac precursor cells (Figure S5F). This is consistent with RNA-seq, qRT-PCR, and analysis of HOPX activity in tdTomato knockin HOPX reporter cells showing that HOPX is expressed late during cardiac differentiation, well after sarcomere formation and weeks after the onset of spontaneously beating cells during cardiac directed differentiation (Figures 4B, Figure S5F and S6A-D). The highest level of HOPX expression was observed in cardiomyocyte cultures maintained for 1 year (Figure S5F-G). Collectively, these data show a direct link between chromatin regulation of the HOPX locus and expression of HOPX in cardiac lineage specification in vitro.
HOPX Drives Cardiomyocyte Hypertrophy
To determine the functional role of HOPX, we established conditional HOPX over-expression hPSCs in which a nuclear localized HOPX is targeted to the AAVS1 locus (Figure 5A). Using western blot, qRTPCR, and immunostaining, we show that HOPX transcript and HOPX protein is over-expressed in a doxycycline inducible manner and is nuclear localized (Figure 5A-C). Morphometric analysis of doxtreated cardiomyocytes showed a significant increase in cell area under conditions of HOPX overexpression (Figure 5D) consistent with previous studies showing that forced HOPX expression in vivo causes cardiac hypertrophy (Chen et al., 2002; Kook et al., 2003; Shin et al., 2002). We performed bulk RNA sequencing analysis on control vs HOPX OE cardiomyocytes to determine global transcriptomic changes (Figure 5E-H and Table S5). Analysis of differentially expressed genes showed a significant enrichment of gene ontologies associated with signaling pathways (ERK1–2, IGF-1) and gene networks involved in cell growth and maturation in HOPX OE cardiomyocytes with IGF-1 representing the most highly up-regulated among a panel of known regulators of hypertrophy (Figure 5G-H and Table S5).
Figure 5. HOPX is a Key Regulator of Cardiomyocyte Hypertrophy.
(A) Gene targeting strategy for conditional HOPX over-expression. Schematic shows design of conditionally expressed HOPX-NLS-eGFP construct. Below, western showing doxycycline induction of control (NLS-eGFP) and HOPX OE iPSC lines.
(B) Quantitative PCR analysis of HOPX transcript abundance in control vs HOPX OE iPSCs.
(C) Immunohistochemistry showing nuclear localization of HOPX-GFP in cardiomyocytes.
(D) Cell size analysis showed that HOPX OE treated hiPSC-CMs led to a significant increase in area.
(E-G) Volcano plot (E), quantification of DE genes (F), and gene ontology analysis (G) of significantly differentially expressed genes (−1<log2FC<1; padj > 0.05) identified by RNA-seq of control vs HOPX OE cardiomyocytes.
(H) Genes known to govern hypertrophy showing IGF1 as the most significantly upregulated hypertrophic gene in HOPX OE vs control cells. For heat maps, data are presented as Log10 transformed relative gene expression normalized to HPRT. NLS: nuclear localization signal, eGFP: enhanced green fluorescent protein. Scale bars = 100 μm. * P <0.05 by t test. See also Table S5.
The HOPX Locus is Activated by Hypertrophic Stimulation
Hypertrophic stimulation is modelled poorly in high-density monolayer cardiac-directed differentiation due to the absence of exogenous hypertrophic signals. On the basis that this discrepancy may, at least in part, explain the dysregulation of HOPX in vitro, we tested the reciprocal hypothesis that exogenous hypertrophic stimuli is sufficient to drive HOPX expression in vitro. We implemented an established approach for stimulating hypertrophy (Uesugi et al., 2014) in which high-density monolayer-derived cardiomyocytes are replated at low density at day 10 and analyzed at day 15 (Figure 6A). In keeping with a hypertrophic response, replating cardiomyocytes results in significantly increased cell area, anisotropy (Figure 6B-C) and up-regulation of genes governing cardiac hypertrophic growth including NPPB, MYOCD, EDN1, and IGF-1 (Figure 6D). Importantly, we found that replating cardiomyocytes resulted in a greater than 10 fold increase in HOPX expression (Figure 6E) in coordination with significant increases in expression of myofibrillar isoforms (MYH7, MYL2, TNNI3) and transcription factors (SRF) involved in cardiomyocyte maturation (Figure 6F). To assess HOPX expression at single cell resolution, we utilized HOPX- reporter hESCs in which tdTomato is knocked into the translational start site of HOPX (Palpant et al., 2017b) (Figure S6C). These data show that HOPX is robustly activated uniformly in replated cardiomyocytes (Figure 6G). We further show that treatment with Endothelin-1 (ET-1), a potent stimulus of cardiomyocyte hypertrophy, significantly increases HOPX expression albeit to a much lower level than replated cardiomyocytes (Figure 6H). Taken together, these results indicate that transcriptional activation of the HOPX locus is downstream of hypertrophic signaling.
Figure 6. HOPX Functionally Governs Cardiac Hypertrophy Through the Distal Transcriptional Start Site.
(A) Schematic of in vitro directed differentiation of hPSCs with re-plating at day 10 and analysis at day 15.
(B-C) Representative images (B) and quantification (C) of morphometric changes during replating including cell area and circularity.
(D) Quantitative PCR analysis of a selected panel of hypertrophic genes differentially expressed in the context of replating cardiomyocytes. HD: High density monolayer.
(E-F) Quantitative PCR analysis showing significant increases in HOPX (n = 5–8 biological replicates per condition from 3–4 experiments) (E) among a range of other cardiac transcription factors and myofilament genes (n = 6–8 biological replicates per condition from 3 experiments) (F) involved in cardiomyocyte maturation in replated cardiomyocytes compared to controls.
(G) Immunohistochemistry of HOPX-tdTomato reporter cells showing uniform expression of HOPX in α- actinin+ replated cardiomyocytes.
((H) Treatment with the hypertrophic signaling molecule endothelin-1 (ET1) significantly increases HOPX expression.
(I) UCSC genome browser analysis of transcript variants mapped to the HOPX locus, the position of guide RNAs blocking the proximal (g1) or distal (g4) TSS, and position of qPCR primers amplifying different exons of the HOPX locus.
(J-M) Analysis of gene expression in control high density monolayer cells vs replated cells and HOPX KD replated cells by quantitative PCR for various exons of the HOPX locus as outlined in panel H (J), genes governing cardiomyocyte hypertrophy (K), cardiac myofilament genes (L), and cardiac transcription factors (M).
(N) Morphometric analysis of cell area in control vs HOPX KD cells over 64 hrs of replating.
(O) Schematic lineage tree showing fate choices governed by HOPX during cardiac directed differentiation and a proposed mechanism whereby hypertrophic signaling is identified as a stimulus required for expression of HOPX during in vitro differentiation and showing that HOPX engages with cardiomyocyte hypertrophic growth through its distal transcriptional start site. For heat maps, data are presented as LogJ0 transformed relative gene expression normalized to HPRT. * P < 0.05. See also Figure S6 and Table S6.
Dissecting the Transcriptional Complexity of HOPX Regulation Underlying Cardiomyocyte Hypertrophy
While genetic loss of HOPX did not impact specification of cardiomyocytes (Figure S6E-F), we set out to study the functional requirement and underlying complexity of the HOPX locus in cardiomyocyte hypertrophy. To this end, we utilized CRISPRi loss of function hPSCs to conditionally block HOPX expression at each of its two transcriptional start sites which we term the proximal TSS (g4) and distal TSS (g1) (Figure 6I). Multiple exon-spanning primers were designed to map transcriptional activity across the HOPX locus (Figure 6I). Cells were differentiated into the cardiac lineage +/− dox and analyzed at day 15 of differentiation under standard high density monolayer conditions vs replating. All HOPX transcripts were significantly increased during replating (Figure 6J, Table S6). However, inhibition of the proximal TSS (g4) repressed expression from that locus (HOPX C) with no effect on transcriptional activity from the distal TSS (HOPX A) (Figure 6J). In contrast, inhibition of the distal HOPX TSS (g1) resulted in a global reduction of HOPX expression (HOPX A-E) (Figure 6J). This indicates that HOPX has functionally distinct transcriptional start sites with the distal TSS functioning as the primary target of regulatory factors driving expression of HOPX in the context of hypertrophic stimulation.
To determine the functional requirement of HOPX in hypertrophy, we analyzed hypertrophyrelated genes (Figure 6K and Table S6). Loss of HOPX function from the distal or proximal TSS did not impact expression of any hypertrophic genes tested including IGF-1 (Figure 6K), the most highly upregulated hypertrophy gene in HOPX OE (Figure 5I). We next assessed a panel of cardiac genes associated with maturation. We found that maturation-related myofibrillar genetic isoforms (MYL2, MYH6, MYH7, TNNI3) were significantly depleted when the distal HOPX TSS was inhibited (g1). In contrast, knockdown of the proximal TSS (g4) had small but significant effects on expression of MYL2 and MYH7 but a significant increase in expression of the early fetal MYH6 isoform relative to controls (Figure 6L and Table S6). Expression of selected key cardiac transcription factors were not impacted by HOPX loss of function with the exception of a significant increase in GATA4 with knockdown of the distal TSS (g1) (Figure 6M and Table S6). These data indicate that while genetic networks underlying hypertrophy and early cardiomyocyte myofibrillar development are not dependent on HOPX, we found it plays a key role driving maturation at least in part through regulating expression of late-stage genetic isoforms of myofibrillogenesis with the distal TSS playing a more dominant role compared to the proximal TSS.
We next assessed the impact of HOPX loss of function on morphometric parameters of cardiomyocyte hypertrophy. Cell area was measured at two time points post replating and showed a progressive and significant increase in cell size indicative of cardiomyocyte hypertrophy in control cells and conditions blocking the proximal HOPX TSS (g4) (Figure 6N). However, blocking transcription of the distal HOPX TSS attenuated the hypertrophic growth (Figure 6N). Taken together, we have identified HOPX as a known key epigenetic regulator of cardiovascular development that is dysregulated during in vitro directed differentiation from hPSCs. Using genetic models, we show that HOPX is situated downstream from hypertrophic signaling pathways and is essential for downstream expression of cardiac myofibrillar genetic isoforms involved in cardiomyocyte growth and maturation (Figure 6O).
DISCUSSION
This study provides single cell transcriptional analysis of human cardiac directed in vitro differentiation. Identification and characterization of in vitro derived cell types are supported by spatio-temporal gene expression of the gastrulating mouse embryo and single cell analysis of in vivo heart development. Cardiac directed differentiation protocols using small molecules to modulate Wnt signaling have emerged in recent years as a simple, cost-effective, and reliable method to generate high-purity cardiac derivatives from hPSCs. In light of diverse applications of this protocol in cardiovascular discovery and translational research, the current study provides whole genome-wide analysis of 43,168 transcriptomes undergoing stage-specific changes in gene expression during cardiac differentiation as a resource with which to dissect cell subpopulations at the molecular level.
Analysis of subpopulations during early stages of differentiation indicate a surprising contribution of mesendoderm and definitive endoderm coordinately specified with cardiac fates through the progenitor stage of differentiation. In particular, a minority of cells (34%) comprise MESP1+ cardiogenic mesoderm at day 2 that are predicted to ultimately give rise to all cardiac derivatives at day 30. The interaction between endoderm and mesoderm in governing lineage specification in vivo is well known, and these data suggest that a critical functional role of induction cues provided by directed differentiation protocols is to establish the necessary population stoichiometry of transiently sustained endoderm required to support mesoderm in the derivation of high purity cardiac fates in vitro.
We evaluated lineage trajectories from single cell data by implementing a lineage prediction algorithm, scdiff, specifically designed for learning regulatory networks controlling differentiation from single cell time series data. These data revealed insights into the bifurcation of cardiac precursor cells at day 5 of differentiation into NKX2–5+/MYL2+ ventricular cardiomyocytes and a population of noncontractile cells with transcriptional networks similar to NKX2–5-/PITX2+ cardiac outflow tract (OFT) cells. While previous studies have routinely described a non-contractile THY1+ (CD90+) fibroblast-like cells for tissue engineering applications (Thavandiran et al., 2013), this population remains poorly studied. We provide single cell level transcriptome-wide evidence directly linked to in vivo cell cardiac types that non-contractile THY1+ cells are similar to cardiac OFT derivatives. Of importance, congenital heart disease (CHD) is among the most common forms of congenital defects and OFT anomalies account for roughly 30% of CHD incidences (Thom et al., 2006). Given the complexity of outflow tract differentiation and morphogenesis that involves cell types form diverse origins, future work will require analysis of this population as it pertains to the cellular origins of outflow tract, septum, and other noncontractile cell types of the heart.
It is well-established that in vitro cardiac differentiation does not generate cardiomyocytes with the transcriptional profile, cellular diversity, morphometry, or functional maturity of adult in vivo-derived cardiomyocytes (Yang et al., 2014). Among a panel of candidate regulatory genes we studied, we focused on HOPX as a key developmental regulator of cardiac myoblasts early in heart development in vivo (Jain et al., 2015) that is rarely expressed in in vitro derived cardiomyocytes. We tested the hypothesis that the dysregulation of HOPX was the consequence of deficiencies in directed differentiation accurately mimicking the signaling and mechanical stimuli of the developing heart. To address this, we aimed to understand the basis for activating HOPX and its downstream gene networks in vitro by identifying the upstream cues required for its expression. Utilizing gain and loss of function genetic models, we provide a comprehensive profiling of the complex transcriptional landscape of HOPX as a central regulator of the cardiomyocyte response to hypertrophy. Our data show that the distal TSS is the primary hypertrophy responsive element and regulation of HOPX through this TSS functionally governs gene networks and cellular morphometric growth associated with cardiomyocyte hypertrophy and maturation.
Taken together, this study provide insights into the complexity of cell populations represented in stage-specific transitions from pluripotency and establish a unique reference point for dissecting gene networks involved in human cardiac development and disease. Promoting adult-like phenotypes from in vitro differentiated cell types is essential for the realization of the translational applications of hPSCs in disease modelling and therapy. This study provides evidence that HOPX expression is a key transcriptional regulator near absent during high density cardiac directed differentiation in vitro, requiring hypertrophic stimulation to accurately direct HOPX and its downstream networks underlying the transcriptional and functional maturity of hPSC-derived cardiomyocytes.
STAR ★ METHODS
KEY RESOURCES TABLE
See attached word document.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nathan Palpant (n.palpant@uq.edu.au).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Generation and Maintenance of Human ESC/iPSC Lines
All human pluripotent stem cell studies were carried out in accordance with consent from the University of Queensland’s Institutional Human Research Ethics approval (HREC#: 2015001434). The RUES2-td- Tomato reporter (karyotype: 46, XX; RRID CVCL_VM29) and RUES2 HOPX KO (karyotype; 46, XX; RRID CVCL_VM28) human ES cell lines were generated as previously described (Palpant et al., 2017b). WTC CRISPRi GCaMP hiPSCs (Karyotype: 46, XY; RRID CVCL_VM38), generously provided by M. Mandegar and B. Conklin (UCSF, Gladstone Institute), was generated using a previously described protocol (Mandegar et al., 2016). WTC CRISPRi HOPX g4 (XY; RRID CVCL_VQ28), WTC CRISPRi HOPX g1 (XY; RRID CVCL_VQ27), WTC HOPX-NLS-eGFP (XY; RRID CVCL_VM46), and WTC NLS-eGFP (XY; RRID CVCL_VM47) hiPSCs were generated in this study (see below). All cells were maintained as previously described (Palpant et al., 2017a). Briefly, cells were maintained in mTeSR media with supplement (Stem Cell Technologies, Cat.#05850) at 37° C with 5% CO2. All hESCs, WTC HOPX-NLS-eGFP, and WTC NLS-eGFP lines were maintained on matrigel growth factor reduced basement membrane matrix (Corning, Cat.#356231), while WTC CRISPRi, WTC CRISPRi HOPX g4, and WTC CRISPRi HOPX g4 hiPSC lines were maintained on Vitronectin XF (Stem Cell Technologies, Cat.#07180) coated plates. Despite no overt abnormalities, WTC CRISPRi cells and their derivatives are non-wild type and comprise the sole cell type analysed by scRNA-seq. Further verification of these findings using different cell lines is suggested.
WTC CRISPRi HOPX g4 and g1 hiPSCs:
HOPX-targeted guide RNAs (gRNA) were designed to target sequences near the human HOPX distal and proximal transcription start sites, were cloned into the pQM-u6g-CNKB doxycycline-inducible construct and transfected into WTC CRISPRi GCaMP hiPSCs using GeneJuice Transfection Reagent (Merck, Cat.#70967). Stable clones were selected using successive rounds of re-plating with blasticidine at 10μg/ml (Sigma, Cat.#15205). Populations were tested for knockdown efficiency by qPCR following doxycycline addition at 1 μg/ml (Sigma, Cat.#D9891) continuously from day 0 of cardiac-directed differentiation.
Guide RNAs targeting the HOPX transcription start site.
| gRNA Name | Oligo Sequences 5’ - Forward Primer - 3’ 5’ - Reverse Primer - 3’ |
|---|---|
| gRNA4 | TTGGCCTTCCTTAGAGCCGGAGGT |
| AAACACCTCCGGCTCTAAGGAAGG | |
| gRNA1 | TTGGCTCATTTCAAAGCGTAGATC |
| AAACGATCTACGCTTTGAAATGAG |
WTC HOPX-NLS-eGFP and WTC NLS-eGFP hiPSCs:
We cloned the human HOPX ORF fused to a nuclear localization sequence (CCAAAGAAGAAGCGGAAGGTC) and GFP into the AAVS1 targeting plasmid (pZDonor, Sigma). 1×106 WT WTC hiPSCs were transfected with 0.5μg AAVS1-TALEN, 0.5μg AAVS1-TALEN and 4μg of HOPX-NLS-eGFP or 4μg of NLS-eGFP to generate the HOPX line and the negative control line, respectively, using Amaxa Human stem cell Kit #2 (Lonza, Cat.#VVPH-5022). The cells were then plated with 5mM ROCK inhibitor onto Matrigel-coated plates in mTeSR. Two days following the nucleofection, the cells were selected for puromycin resistance using puromycin at 0.5μg/ml for 48 hours.
METHODS DETAILS
Cell Culture
All human pluripotent stem cell studies were carried out in accordance with consent from the University of Queensland’s Institutional Human Research Ethics approval (HREC#: 2015001434). hESCs and hiPSCs were maintained in mTeSR media (Stem Cell Technologies, Cat.#05850). Unless otherwise specified, cardiomyocyte directed differentiation using a monolayer platform was performed with a modified protocol based on previous reports (Burridge et al., 2014; Lian et al., 2012). On day −1 of differentiation, hPSCs were dissociated using 0.5% EDTA, plated into vitronectin or matrigel coated plates at a density of 1.8 × 105 cells/cm2, and cultured overnight in mTeSR media. Differentiation was induced on day 0 by first washing with PBS, then changing the culture media to RPMI (ThermoFisher, Cat.#11875119) containing 3μΜ CHIR99021 (Stem Cell Technologies, Cat.#72054), 500μg/mL BSA (Sigma Aldrich, Cat.#A9418), and 213μg/mL ascorbic acid (Sigma Aldrich, Cat.#A8960). After 3 days of culture, the media was replaced with RPMI containing 500μg/mL BSA, 213μg/mL ascorbic acid, and 1μΜ Xav-939 (Stem Cell Technologies, Cat.#72674). On day 5, the media was exchanged for RPMI containing 500μg/mL BSA, and 213μg/mL ascorbic acid without supplemental cytokines. From day 7 onwards, the cultures were fed every 2 days with RPMI plus 1x B27 supplement plus insulin (Life Technologies Australia, Cat.#17504001). For endothelin-1 assays, cells were treated with 300nM ET-1 (Sigma-Aldrich, Cat.#E7764) from day 9–15 of directed differentiation. For HOPX over-expression studies (Figure 5), the following protocol was utilized: A monolayer-based directed differentiation protocol was followed to generate hiPSC-CMs, as described previously (Palpant et al., 2017a). On day 15 hiPSC-CMs were enriched by lactate selection (Tohyama et al., 2013).
Quantitative RT-PCR
For quantitative RT-PCR, total RNA was isolated using the RNeasy Mini kit (Qiagen, Cat.#74106). Firststrand cDNA synthesis was generated using the Superscript III First Strand Synthesis System (ThermoFisher, Cat.#18080051). Quantitative RT-PCR was performed using SYBR Green PCR Master Mix (ThermoFisher, Cat.#4312704) on a ViiA 7 Real-Time PCR System (Applied Biosystems). The copy number for each transcript is expressed relative to that of housekeeping gene HPRT1. Quantification of cardiac hypertrophy gene expression was performed using Cardiac Hypertrophy H384 qPCR panels (BioRad, Cat.#10025144) with SYBR Green PCR Master Mix. Samples were run in biological triplicate. The copy number for each transcript is expressed relative to that of housekeeping gene HPRT1. FC was calculated on a gene by gene basis as gene expression divided by control gene expression.
qRT PCR primers
| Gene Name | Forward Primer | Reverse Primer |
|---|---|---|
| HPRT | TGACACTGGCAAAACAATGCA | GGTCCTTTTCACCAGCAAGCT |
| GATA4 | GACCTGGGACTTGGAGGATA | ACAGGAGAGATGCAGTGTGC |
| NKX2–5 | CAAGTGTGCGTCTGCCTTT | CAGCCTTTCTTTTCGGCTCTA |
| MYL4 | TCAAAGAGGCCTTTTCATTG | CGTCTCAAAGTCCAGCATCT |
| MYL2 | TTGGGCGAGTGAACGTGAAAA | CCGAACGTAATCAGCCTTCAG |
| MYH6 | CAAGTTGGAAGACGAGTGCT | ATGGGCCTCTTGTAGAGCTT |
| MYH7 | GGGCAACAGGAAAGTTGGC | ACGGTGGTCTCTCCTTGGG |
| TNNI1 | CCCAGCTCCACGAGGACTGAACA | TTTGCGGGAGGCAGTGATCTTGG |
| TNNI3 | GGAACCTCGCCCTGCACCAG | GCGCGGTAGTTGGAGGAGCG |
| ATP2A2 | TTTCCTACAGTGTAAAGAGGACAACC | TTCCAGGTAGTTGCGGGCCACAAA |
| HOPX A | GCCCAGCTATTTAAGCAGGC | GGGTGCTTGTCGACCTTGTT |
| HOPX B | ATGCTCATTTTCCTGGGCTGT | GGGTGCTTGTCGACCTTGTT |
| HOPX C | CCACCCTCGCGATCTGTCAA | GGGTGCTTGTCGACCTTGTT |
| HOPX D | CAAGGTCGACAAGCACCCGGATTC | GGGCTACTTTCTGGGTGCCA |
| HOPX E | CAAGGTCGACAAGCACCCGGATTC | CATCTCCTTAGTCTGTGACGGA |
| SRF | CGAGATGGAGATCGGTATGGT | GGGTCTTCTTACCCGGCTTG |
Immunofluorescence and Morphometric Analysis
Cells were fixed with 4% paraformaldehyde, permeabilized in PBS containing 0.025% Triton-X, and blocked in PBS containing 1.5% normal goat serum. Cells were stained with alpha-actinin (Clone EA-53; Sigma-Aldrich Cat# A7811, RRID:AB_476766) at 1:800 and dsRed (Clontech Laboratories, Cat# 632496, RRID:AB_10013483) followed by secondary staining with AlexaFluor-594 Donkey Anti-Goat (ThermoFisher, Cat# A-11058, RRID:AB_2534105 lot #1180089, 1:200) or AlexaFluor-594 Goat Anti-Mouse (ThermoFisher, Cat# R37121, RRID:AB_2556549 lot # 1219862, 1:200). Nuclei were counterstained with DAPI. For HOPX over-expression studies (Figure 5), cells were fixed in 4% (vol/vol) paraformaldehyde, blocked for an hour with 5% (vol/vol) normal goat serum (NGS) (Sigma, Cat.#G9023), and incubated overnight with primary antibody in 1% NGS, followed by secondary antibody staining in NGS. Measurements of CM area were performed using Image J software. Analysis was done on a Leica TCS-SPE Confocal microscope using a 40x or 63x objective and Leica Software. Primary antibodies used were: αActinin 1:250 (Clone EA-53; Sigma-Aldrich Cat# A7811, RRID:AB_476766), Titin 1:300 TTN-9 (cTerm) anti-rabbit (MyoMedix, Cat# TTN-9, RRID:AB_2734750), GFP 1:300 anti-rabbit (ThermoFisher, Cat# A-11122, RRID:AB_221569). Secondary antibodies and other reagents used were: DAPI at a concentration of 0.02μg/mL, phalloidin alexa fluor 568 1:250 (ThermoFisher, Cat#A12380), goat anti-mouse alexa fluor 488 (ThermoFisher, Cat# A-11001, RRID:AB_2534069) or goat anti-rabbit alexa fluor 647-conjugated (ThermoFisher, Cat# A-21244, RRID:AB_2535812) secondary antibodies at 1:500.
Flow Cytometry
Cells were fixed with 4% paraformaldehyde (Sigma, Cat.#158127) and permeabilized in 0.75% saponin (Sigma, Cat.#S7900). Cells were labeled for flow cytometry using cardiac troponin T (ThermoFisher, Cat# MA5–12960, RRID:AB_11000742) or APLNR (R&D, Cat# FAB856A, RRID:AB_2044604) and corresponding isotype control. Cells were analyzed using a BD FACSCANTO II (Becton Dickinson, San Jose, CA) with FACSDiva software (BD Biosciences). Data analysis was performed using FlowJo (Tree Star, Ashland, Oregon).
Single Cell Isolation
For each differentiated day 0, 2, 5, 15, and 30 time point, differentiated cells were dissociated with 0.5% EDTA + 0.25% Trypsin (ThermoFisher, Cat.#15400054) and neutralized with foetal bovine serum (GE Healthcare Life Sciences, Cat.#SH30084.03) and DMEM/F12 media (Sigma, Cat.#11320033) (1:1 ratio). For each time point, 2 pooled samples were collected, each pool comprised approximately 12 independent differentiation samples. Cells were centrifuged at 1200 rpm for 4 minutes and resuspended in Dulbecco’s PBS (Gibco; Cat.#14190) with 0.04% bovine serum albumin (Sigma Aldrich, Cat.#B6917) and immediately transported for scRNA-Seq processing. Viable cells were sorted using a Propidium Iodide stain and retained on ice in Dulbecco’s PBS + 0.04 % bovine serum albumin. A Countess automated counter (Invitrogen) was used to check final cell viability using Trypan Blue exclusion.
Single Cell RNA-Sequencing
A Chromium instrument (10X Genomics, Millennium Sciences) was used to partition sorted, viable cell suspensions (8×105-1×106 cells/mL) into single cell droplets using the Single Cell 3’ Library, Gel Bead and Multiplex Kit (version 1, 10X Genomics, Cat.#PN-120233) as per the manufacturer’s protocol. Each time point was run in duplicate, resulting in 10 sample preparations. The samples were loaded into Single Cell 3’ chips (10X Genomics) at a concentration optimized to capture approximately 5,000 cells in individual 1-cell droplets. Single cell libraries were sequenced using an Illumina NextSeq 500 instrument as previously described (Nguyen et al., 2018).
Bioinformatics Processing
Bioinformatics mapping of reads to original transcripts and cells was by cellranger pipeline v1.3.1 by 10X Genomics (http://10xgenomics.com/). We used cellranger mkfastq to prepare demultiplexed raw base call files into library-specific FASTQ files, with the following parameters --use-bases-mask =“Y26n*,I8n*,n*,Y98n*” --ignore-dual-index. The FASTQ files were separately mapped to the GRC38p7 human reference genome using the STAR21 (Dobin et al., 2013) as a part of the cellranger pipeline. Gene expression counts were done using cellranger count based on Gencode v25 annotation and cell identifiers and Unique Molecular Identifiers (UMI) were filtered and corrected with default setting. Raw cellranger count outputs for two biological replicates in each timepoint were aggregated and normalised by a subsampling procedure using cellranger aggr (Zheng et al., 2017). After sample-to- sample normalisation, we filtered outlier genes and cells which were outside the range of 3x median absolute deviation (MAD) of the number of cells with the detected genes, ribosomal reads, mitochondrial reads, and total read mapped to cells to remove noise due to sequencing depth or cell conditions. Further, cells with above 50% ribosomal reads, or above 20% mitochondrial reads, and genes detected in fewer than 0.1% total cells were also removed. Post filtering, cell-to-cell normalisation was done by scran using pooling sizes of 40, 60, 80 and 100 without using a quickClustering option (Lun et al., 2016). Data dimensionality reduction by PCA and t-SNE are previously described in Nguyen et al (Nguyen et al., 2018) and as is implemented in the R ascend package (Senabouth et al., 2017). Briefly, PCA using the R prcomp function was based on the top 1500 most variable genes. We selected the top 10 principal components (PCs) that explained most variance, as confirmed by Scree plot using fa.parallel package. For visualisation, non-linear dimensionality reduction using Rtsne v0.13 package for the top 10 PCs was performed to produce coordinates for cells in 2 dimensional or 3 dimensional tSNE space.
Clustering algorithm was described in Nguyen et al (2017 and 2018) and is implemented in the ascend (Senabouth et al., 2017) as well as the scGPS R packages. Briefly, for each aggregated dataset in each time point, we calculated the top 10 PCs as the input for the CORE clustering algorithm. We first built a high-resolution clustering tree structure based on cell-to-cell Euclidean distance and Ward’s minimum variance with an agglomerative hierarchical clustering (HAC). The branches in the dendrogram tree were dynamically grouped into clusters by a cuttreeDynamic method in the dynamicTreeCut v1.63 package. We performed dynamic clustering 40 times for different height cutoffs spanning 99% of the joining height of the initial dendrogram distance tree. The resulting clusters from the 40 runs were compared by using adjusted Rand indexes (ARI) to find a stable clustering point (Nguyen et al., in review). The optimal clustering point meets two criteria, including robust to changing parameters and less different from a reference with the highest number of clusters. We validated the CORE clustering results using multiple dimensionality reduction and clustering methods, including PCA, tSNE, Multidimentional Scaling (MDS) and Clustering through Imputation and Dimensionality Reduction (CIDR) (Lin et al 2017).
The processed data post filtering, normalisation, and clustering was used as the input for differential expression analysis. We performed DESeq (Anders and Huber, 2010) analysis to find differentially expressed genes between cells in one subpopulation compared to all remaining cells in other subpopulations at a given time-point. We observed that when comparing subpopulations with different number of cells, DESeq was faster and produced more stable performance than DESeq2, consistent to the report by Dal Molin et al (Dal Molin et al., 2017). Briefly, one pseudocount was added to scran cell-to- cell normalised counts, which were then rounded to integer values before estimating dispersion (with the fit Type option set to local) and running negative binomical test function in DESeq. After significant testing, we performed fold change adjustments to subtract mean expression by one, which allowed more accurate estimation of fold changes for lowly expressed genes. Bonferroni correction as applied to account for multiple testing error.
Furthermore, for reproducibility and broader usability of the valuable data resource, we have submitted all data to Array Express and created a web database resource with interactive data mining tools for users to explore the entire dataset without requirement for programming.
Bulk RNA-Sequencing
hiPSC-CMs were harvested for RNA preparation and genome wide RNA-seq (>20 million reads). RNA-seq samples were aligned to hg19 using Tophat, version 2.0.13 (Trapnell et al., 2009). Gene-level read counts were quantified using htseq-count (Anders et al., 2015) using Ensembl GRCh37 gene annotations. Genes with total expression above 1 normalized read count across RNA-seq samples in each binary comparison (e.g., HOPX vs. control) were kept for differential analysis using DESeq (Anders and Huber, 2010). Princomp function from R was used for Principal Component Analysis. TopGO R package (Alexa et al., 2006) was used for Gene Ontology enrichment analysis.
Protein Extraction and Western Blot Analysis
Cells were lysed directly on the plate with a lysis buffer containing 20mM Tris-HCl pH 7.5, 150mM NaCl, 15% Glycerol, 1% Triton X-100, 1M ß-Glycerolphosphate, 0.5M NaF, 0.1M Sodium Pyrophosphate, Orthovanadate, PMSF and 2% SDS (Moody et al., 2017). 25U of Benzonase Nuclease (EMD Chemicals, Gibbstown, NJ) was added to the lysis buffer right before use. Proteins were quantified by Bradford assay (Bio-rad), using BSA (Bovine Serum Albumin) as Standard using the EnWallac Vision. The protein samples were combined with the 4x Laemmli sample buffer, heated (95°C, 5min), and run on SDS-PAGE (protean TGX pre-casted 4%−20% gradient gel, Bio-rad) and transferred to the Nitro-Cellulose membrane (Bio-Rad) by semi-dry transfer (Bio-Rad). Membranes were blocked for 1hr with 5% milk and incubated in the primary antibodies overnight at 4°C. The membranes were then incubated with secondary antibodies (1:10000, goat anti-rabbit [Cat.#1706515; RRID: AB_11125142] or goat anti-mouse [Cat.# 1706516; RRID: AB_11125547] IgG HRP conjugate (Bio-Rad) for 1hr and the detection was performed using the immobilon-luminol reagent assay (EMD Millipore). Primary antibodies are as follows: Alpha tubulin antibody at 1:2000 (Cell Signaling Technology Cat# 2144, RRID:AB_2210548) and anti-GFP anti-rabbit at 1:1000 (Invitrogen, Cat# A-11122, RRID AB_221569).
Genomics Data Sets
Previously published ChIP-seq and gene expression data sets were analyzed for this study. Analysis of cardiac differentiation chromatin dynamics and gene expression by RNA-seq were published previously (Kuppusamy et al., 2015; Palpant et al., 2017b) with data accessed from GEO GSE97080. HOPX gene expression analysis were derived from Stemformatics (Wells et al., 2013) using the following data sets: a dual reporter MESP1-mCherry/NKX2–5 GFP reporter hESC line at day 0 and day 3 of directed differentiation sorted for MESP1 positive vs. negative cells (Stemformatics ID: Hartogh_2015_25187301) (Den Hartogh et al., 2015) and human foetal heart samples isolated at each of three trimesters comparing ventricle and atrial expression (Stemformatics ID: van_den_berg_2015_26209647) (van den Berg et al., 2015). Human fetal heart gene expression data were downloaded from ENCODE (experiment #: ENCSR047LLJ, ENCSR863BUL, ENCSR769LNJ, ENCSR433XCV, and ENCSR675YAS). HOPX expression in engineered tissue, adult heart tissue, and hPSC-CMs were acquired from previous work by Mills et al (Mills et al., 2017).
Gene Ontology Visualization
Gene ontology analysis was performed using DAVID with significance threshold set at FDR < 0.05. The p-values from gene ontology analysis were visualized using the R package corrplot (Wei and Simko, 2016), where the radius of the circle is proportional to the negative natural log of the input p-value.
Spearman Correlation Analysis
We obtained FACS sorted bulk cardiac subtypes (Quaife-Ryan et al., 2017) and single-cell RNA-seq data generated from developing mouse heart (Li et al., 2016). The normalized expression data from these two sources was merged with our scRNA-seq expression data. Mouse Ensembl IDs were converted to human ortholog gene IDs and a new expression matrix was generated using only the 13,490 genes common to all three datasets. Spearman’s rank correlation was used to compare the expression levels of genes between samples, and the significance of the differences between pairs of correlation coefficients were calculated using a Fisher Z-transformation.
iTranscriptome Sample Preparation and Data Analysis
Samples were generated according to the methodology published in (Peng et al., 2016). E6.5, E7.0 and E7.5 embryos (n = 6, 6, and 3 respectively) were cryo-sectioned along the proximal-distal axis. Populations of approximately 20 cells were collected from different regions of the cross-section by laser microdissection and processed for RNA sequencing. Two sets of embryos for each embryonic age were dissected: the first set from the epiblast - E6.5: anterior and posterior; E7.0: anterior, left, right and posterior; E7.5: anterior, left anterior and posterior, right anterior and posterior, posterior. The second set from the three germ layers - E6,5: posterior epiblast and endoderm: anterior and posterior; E7.0: posterior epiblast, mesoderm: anterior and posterior, and endoderm: anterior and posterior; E7.5: posterior epiblast, mesoderm: anterior and posterior, and endoderm: anterior and posterior. Differentially expressed genes (DEGs) were screened first by unsupervised hierarchical clustering method to group samples in the respective regions. Genes with an expression level FPKM>1 and a variance in transcript level across all samples greater than 0.05 were selected. To identify inter-region specific DEGs, each of these selected genes was submitted to a t-test against the level of expression in the other regions. Genes with a p.value< 0.01 and a fold change >2.0 or <0.5 were defined as DEGs. The gene expression pattern (region and level of expression by transcript reads) of the gene of interest was mapped on the corn plots, where each kernel represents the cell population sampled at a defined position in the germ layers, to generate a digital rendition of whole mount in situ hybridization.
Constructing Regulatory Differentiation Networks:
Scdiff
Detailed computational model and derivation for scdiff are provided in (Ding et al., 2018). scdiff software is available on GitHub (https://github.com/phoenixding/scdiff). The method is initialized using Spectral clustering based on cell-to-cell Spearman correlation, followed by an ensemble strategy to determine the optimal K clusters. The model then iteratively connects clusters (representing states in the probabilistic model) between time points using a “Similarity To Ancestor-STA” strategy (with day 0 as the first timepoint), based on expression similarity (Spearman correlation). Cell reassignment is based on a Kalman filter probabilistic model. The initial set of states and their connectivity is iteratively updated by learning trajectory and branching models constrained by transcription factor (TF)-gene interactions via a logistic regression classifier to maximize the ability to predict the expression of target gene based on the interaction data.
Details of the implementation of scdiff are outlined below.
Initial Clustering of Single Cells
scdiff starts by clustering the cells in each of the time points measured. While the original scdiff method used spectral clustering, this method was unable to scale for the large number of cells profiled in this study. We have thus revised scdiff for this study by changing the original clustering methods to a more efficient method. Specifically, we used PCA with 10 dimensions followed by K-means for the initial clustering which led to faster runtime while not greatly affecting performance. To determine the initial number of clusters (k) for each time point we combine 3 widely used clustering quality assessment scores: the Silhouette Score (Rousseeuw, 1987), Davis-Bouldin index (Davies and Bouldin, 1979) and AIC (Akaike, 1998) (Akaike information criterion). We used a bootstrapping strategy to combine these. We first selected a random subset of 90% of the genes. Next, we calculate the Silhouette score, Davis Bouldin score and AIC scores for different k values between 2 and 20 for each time point. We compute a combined score for each of these k values based on the subset of genes selected and repeat this process 100 times (each time with a new random gene subset). We select the optimal k by summing up the scores for each of the possible k values across the 100 repeats.
Initial Model Construction
Initial clustering is based on the time point associated with each cell. However, several recent studies indicate that cells may be unsynchronized with respect to their state even if they are collected at the same time point (Trapnell et al., 2014). Thus, some of the clusters at a specific time point may represent states that are either earlier or later than other clusters in the same time. To address this we next use a correlation-based method to reassign clusters to time points. Once we determined the set of clusters associated with each level (time point), we connect clusters in each level to the most similar cluster (in terms of correlation) at the level directly above it (its parent cluster). This leads to a directed graph with potentially multiple roots (initial set of clusters for the first time points) which structurally represents the initial differentiation model.
Predicting TFs Regulating Differentiation Pathways
An important aspect of scdiff is the ability to both reconstruct and analyze the differentiation pathways based on the set of TFs that regulate various state transitions. We used bulk TF-gene interaction data from (Ernst et al., 2007; Schulz et al., 2012) for this analysis. Following the initial model construction, we first identify a set of differentially expressed (DE) genes for each cluster (state) in our model. Using this set, we identify TFs that are enriched for DE targets based on the hyper-geometric distribution. Next, we check which of the candidate TFs is expressed in the parent node of the state. TFs that are both significantly enriched and expressed are used in the to define a logistic regression function which modifies the likelihood of assigning cells to the different clusters in the model Thus, cell assignment is based on both, expression similarity to other cells in the state and the expression of targets of TFs predicted to regulate this state.
Iterative Assignment of Cells to States
Given initial assignments of cells and TFs to states, we can compute the MLE of the transition and emission noise variance. We next iterate between two steps. The first uses the parameters learned to reassign cells and TFs to states and the second uses the assigned cells and TFs to re-learn model parameters. During the iterative process some states may become empty and if this happens they are removed from the model. The process stops when it converges (no more cells are re-assigned) and the resulting model is returned.
scdiff Parameters Used in This Study
scdiff was run with the following parameters: -k auto -l 1 -s 1 -d 1. For the scdiff input expression matrix, we performed, as described above, a thorough quality check to remove outlier genes and cells (outside 3 × median absolute deviation range) based on mitochondrial, ribosomal genes, library sizes, and number of detected cells. This data processing pipeline was implemented in the ascend pipeline (Senabouth et al., 2017). The preprocessed expression data matrix was then normalized at two levels (as described above): by batches (using the cellranger aggr function), and then by cells (using the deconvolution method in the scran package). The filtered and normalised matrix was then used for scdiff.
QUANTIFICATION AND STATISTICAL ANALYSIS
Unless otherwise noted, all data are represented as mean ± standard error of mean (SEM). Indicated sample sizes (n) represent biological replicates including independent cell culture replicates and individual tissue samples. No methods were used to determine whether data met assumptions of the statistical approach or not. Due to the nature of the experiments, randomization was not performed and the investigators were not blinded. Statistical significance was determined in GraphPad Prism 7 software by using student’s t test (unpaired, two-tailed) or ordinary one-way ANOVA unless otherwise noted. Results were considered to be significant at p < 0.05(*). Statistical parameters are reported in the respective figures and figure legends. All statistical data are represented as mean ± SEM.
Quantification of data used for statistical analysis in this study are described her in detail. For single cell analysis, cell numbers for each time point and subpopulation are as follows: day 0, n = 13,679; day 2, n = 5,905 (n = 2,245 cells in S1, n = 1,994 cells in S2, and n = 1,666 cells in S3); day 5, n = 9,827 (n = 3,850 cells in S1, n = 2,577 cells in S2, n = 2,474 cells in S3, and n = 924 cells in S4); day 15, n = 6,303 (n = 3,520 cells in S1 and n = 2,783 cells in S2); day 30, n = 7,039 (n = 4,038 cells in S1, n = 3,001 cells in S2). For single cell analysis of developing mouse heart (Li et al., 2016) n = 949 cells from two biological replicates.
For iTranscriptome analysis: E6.5 (n = 6) and E7.5 (n = 6) mouse embryos and published data for E7.0 (n = 3) mouse embryos. Color scales represent levels of expression as logi0 of fragments per kilobase million (FPKM + 1).
For Spearman rank correlation analysis (Figure 3H). Values are presented median Spearman’s value p. Significant differences between pairs of correlation coefficients were calculated using a Fisher Z-transformation. P-values for all tests were below the double precision limit of 2.2e-308.
For all results n = 3–16 biological replicates from up to 8 independent experiments.
Immunohistochemistry morphometric analysis: For Figure 6B-C n = 144 cells 16 hours post replating and 77 cells 64 hours post replating. Statistics performed using t test with Welch’s correction, P<0.0001 (****) and P<0.0009 (***). For Figure 6N n = 57–144 biological replicates per condition for g4 cells and n = 52–78 biological replicates per condition for g1 cells.
DATA AND SOFTWARE AVAILABILITY
scRNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-6268.
Supplementary Material
Table S1. Differential Gene Expression and Gene Ontology Analysis of Subpopulations Identified During Cardiac Directed Differentiation at Single Cell Resolution, Related to Figure 2.
Table S2. Computational Analysis of Transcription Factor and Gene Regulatory Networks Underlying Scdiff Prediction of Lineage Trajectories for the Full Single Cell Time Course Data Set, Related to Figure 3 (data demarcated as “ALL”). Computational Analysis of Transcription Factor and Gene Regulatory Networks Underlying Scdiff Prediction of Lineage Trajectories for Only HOPX+ Cells During the Full Time Course Data Set, Related to Figure S4. (Data demarcated as “HOPX”).
Table S5. RNA-Seq Analysis of HOPX Over-Expression Cardiomyocytes vs controls and gene ontology analysis, Related to Figure 5.
Table S6. Statistical Analysis of Gene Expression Shown in Heat Maps in Figure 6J-M.
Highlights.
Single-cell RNAseq during cardiac hPSC differentiation reveals cellular heterogeneity
A key cardiac regulatory gene HOPX is rarely expressed during in vitro differentiation
HOPX is a key in vitro regulator of cardiomyocyte hypertrophy and maturation
ACKNOWLEDGEMENTS
Sequencing was performed by the Institute for Molecular Bioscience Sequencing Facility at the University of Queensland. Assistance with Figure 1A schematic was provided by Suzy Hur. The WTC CRISPRi GCaMP hiPSCs and pQM plasmid backbone were kindly provided by the Conklin lab (UCSF, Gladstone Institute). We thank Prof Richard Harvey (Victor Change Cardiac Research Institute) for assistance reviewing the draft manuscript. Microscopy was performed at the Australian Cancer Research Foundation (ACRF)/Institute for Molecular Bioscience Cancer Biology Imaging Facility. This work was supported by the Australian Research Council (SR1101002) (NJP), the ARC Discovery Early Career Award (DE160100755) (ESW), National Health and Medical Research Council grants 1107599 and 1083405 (JP). ZBJ and JD were supported in part by grant 1R01GM122096 from the National Institute of Health, USA. This work was also supported by a Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01010201 to N.J., XDA01010303 to J.D.J.H), National Key Basic Research and Development Program of China (2014CB964804, 2015CB964500, 2015CB964803), and National Natural Science Foundation of China (91219303, 31430058, 31401261, 91329302, 31210103916, and 91519330). P.P.L.T. is a Senior Principal Research Fellow of the National Health and Medical Research Council of Australia (1110751).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and three tables and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akaike H (1998). Information theory and an extension of the maximum likelihood principle (Springer; ). [Google Scholar]
- Alexa A, Rahnenführer J, and Lengauer T (2006). Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607. [DOI] [PubMed] [Google Scholar]
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrington CB, Dowse BR, Bleyl SB, and Bowles NE (2012). Non-synonymous variants in pre-B cell leukemia homeobox (PBX) genes are associated with congenital heart defects. Eur J Med Genet 55, 235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, et al. (2014). Chemically defined generation of human cardiomyocytes. Nat Methods 11, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Kook H, Milewski R, Gitler AD, Lu MM, Li J, Nazarian R, Schnepp R, Jen K, Biben C, et al. (2002). Hop is an unusual homeobox gene that modulates cardiac development. Cell 110, 713–723. [DOI] [PubMed] [Google Scholar]
- Coifman RR, Lafon S, Lee AB, Maggioni M, Nadler B, Warner F, and Zucker SW (2005). Geometric diffusions as a tool for harmonic analysis and structure definition of data: multiscale methods. Proc Natl Acad Sci U S A 102, 7432–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Molin A, Baruzzo G, and Di Camillo B (2017). Single-Cell RNA-Sequencing: Assessment of Differential Expression Analysis Methods. Front Genet 8, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DL, and Bouldin DW (1979). A cluster separation measure. IEEE transactions on pattern analysis and machine intelligence 2, 224–227. [PubMed] [Google Scholar]
- DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, et al. (2016). Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev Cell 39, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Hartogh SC, Schreurs C, Monshouwer-Kloots JJ, Davis RP, Elliott DA, Mummery CL, and Passier R (2015). Dual reporter MESP1 mCherry/w-NKX2–5 eGFP/w hESCs enable studying early human cardiac differentiation. Stem Cells 33, 56–67. [DOI] [PubMed] [Google Scholar]
- Ding J, Aronow BJ, Kaminski N, Kitzmiller J, Whitsett JA, and Bar-Joseph Z (2018). Reconstructing differentiation networks and their regulation from time series single-cell expression data. Genome Res 28, 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, Gramolini A, and Keller G (2011). SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol 29, 1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Vainas O, Harbison CT, Simon I, and Bar-Joseph Z (2007). Reconstructing dynamic regulatory maps. Mol Syst Biol 3, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Li D, Gupta M, Manderfield LJ, Ifkovits JL, Wang Q, Liu F, Liu Y, Poleshko A, Padmanabhan A, et al. (2015). HEART DEVELOPMENT. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science 348, aaa6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kook H, Lepore JJ, Gitler AD, Lu MM, Wing-Man Yung W, Mackay J, Zhou R, Ferrari V, Gruber P, and Epstein JA (2003). Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J Clin Invest 112, 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy KT, Jones DC, Sperber H, Madan A, Fischer KA, Rodriguez ML, Pabon L, Zhu WZ, Tulloch NL, Yang X, et al. (2015). Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc Natl Acad Sci U S A 112, E2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Xu A, Sim S, Priest JR, Tian X, Khan T, Quertermous T, Zhou B, Tsao PS, Quake SR, et al. (2016). Transcriptomic Profiling Maps Anatomically Patterned Subpopulations among Single Embryonic Cardiac Cells. Dev Cell 39, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, and Palecek SP (2012). Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A 109, E1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A, McCarthy D, and J, M. (2016). A step-by-step workflow for low-level analysis of singlecell RNA-seq data with Bioconductor. F1000Res 5, 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandegar MA, Huebsch N, Frolov EB, Shin E, Truong A, Olvera MP, Chan AH, Miyaoka Y, Holmes K, Spencer CI, et al. (2016). CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell 18, 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RJ, Titmarsh DM, Koenig X, Parker BL, Ryall JG, Quaife-Ryan GA, Voges HK, Hodson MP, Ferguson C, Drowley L, et al. (2017). Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moignard V, Woodhouse S, Haghverdi L, Lilly AJ, Tanaka Y, Wilkinson AC, Buettner F, Macaulay IC, Jawaid W, Diamanti E, et al. (2015). Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat Biotechnol 33, 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody JD, Levy S, Mathieu J, Xing Y, Kim W, Dong C, Tempel W, Robitaille AM, Dang LT, Ferreccio A, et al. (2017). First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry CE, and Keller G (2008). Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661–680. [DOI] [PubMed] [Google Scholar]
- Nguyen Q, Lukowski S, Chiu H, Friedman C, Senabouth A, Bruxner T, Christ A, Palpant N, and Powell J (in review). Determining cell fate specification and genetic contribution to cardiac disease risk in hiPSC-derived cardiomyocytes at single cell resolution. BioRxiv: 229336. [Google Scholar]
- Nguyen QH, Lukowski SW, Chiu HS, Senabouth A, Bruxner TJC, Christ AN, Palpant NJ, and Powell JE (2018). Single-cell RNA-seq of human induced pluripotent stem cells reveals cellular heterogeneity and cell state transitions between subpopulations. Genome Res 28, 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palpant NJ, Pabon L, Friedman CE, Roberts M, Hadland B, Zaunbrecher RJ, Bernstein I, Zheng Y, and Murry CE (2017a). Generating high-purity cardiac and endothelial derivatives from patterned mesoderm using human pluripotent stem cells. Nat Protoc 12, 15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palpant NJ, Pabon L, Rabinowitz JS, Hadland BK, Stoick-Cooper CL, Paige SL, Bernstein ID, Moon RT, and Murry CE (2013). Transmembrane protein 88: A Wnt regulatory protein that specifies cardiomyocyte development. Development (Cambridge) 140, 3799–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palpant NJ, Wang Y, Hadland B, Zaunbrecher RJ, Redd M, Jones D, Pabon L, Jain R, Epstein J, Ruzzo WL, et al. (2017b). Chromatin and Transcriptional Analysis of Mesoderm Progenitor Cells Identifies HOPX as a Regulator of Primitive Hematopoiesis. Cell Rep 20, 1597–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Suo S, Chen J, Chen W, Liu C, Yu F, Wang R, Chen S, Sun N, Cui G, et al. (2016). Spatial Transcriptome for the Molecular Annotation of Lineage Fates and Cell Identity in Mid-gastrula Mouse Embryo. Dev Cell 36, 681–697. [DOI] [PubMed] [Google Scholar]
- Quaife-Ryan GA, Sim CB, Ziemann M, Kaspi A, Rafehi H, Ramialison M, El-Osta A, Hudson JE, and Porrello ER (2017). Multicellular Transcriptional Analysis of Mammalian Heart Regeneration. Circulation 136, 1123–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw PJ (1987). Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. Journal of computational and applied mathematics 20, 53–65. [Google Scholar]
- Schulz MH, Devanny WE, Gitter A, Zhong S, Ernst J, and Bar-Joseph Z (2012). DREM 2.0: Improved reconstruction of dynamic regulatory networks from time-series expression data. BMC Syst Biol 6, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senabouth A, Lukowski S, Alquicira J, Andersen S, Mei X, Nguyen Q, and Powell J (2017). ascend: R package for analysis of single cell RNA-seq data. Biorxiv: 207704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin CH, Liu ZP, Passier R, Zhang CL, Wang DZ, Harris TM, Yamagishi H, Richardson JA, Childs G, and Olson EN (2002). Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell 110, 725–735. [DOI] [PubMed] [Google Scholar]
- Thavandiran N, Dubois N, Mikryukov A, Massé S, Beca B, Simmons CA, Deshpande VS, McGarry JP, Chen CS, Nanthakumar K, et al. (2013). Design and formulation of functional pluripotent stem cell-derived cardiac microtissues. Proc Natl Acad Sci U S A 110, E4698–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom T, Haase N, Rosamond W, Howard VJ, Rumsfeld J, Manolio T, Zheng ZJ, Flegal K, O’Donnell C, Kittner S, et al. (2006). Heart disease and stroke statistics−−2006 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 113, e85–151. [DOI] [PubMed] [Google Scholar]
- Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, et al. (2013). Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 12, 127–137. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, Reinecke H, Moon RT, and Murry CE (2007). Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci U S A 104, 9685–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi M, Ojima A, Taniguchi T, Miyamoto N, and Sawada K (2014). Low-density plating is sufficient to induce cardiac hypertrophy and electrical remodeling in highly purified human iPS cell-derived cardiomyocytes. J Pharmacol Toxicol Methods 69, 177–188. [DOI] [PubMed] [Google Scholar]
- van den Berg CW, Okawa S, Chuva de Sousa Lopes, S.M., van Iperen L, Passier R, Braam SR, Tertoolen LG, del Sol A, Davis RP, and Mummery CL (2015). Transcriptome of human foetal heart compared with cardiomyocytes from pluripotent stem cells. Development 142, 3231–3238. [DOI] [PubMed] [Google Scholar]
- Wei T, and Simko V (2016). corrplot: Visualization of a Correlation Matrix.
- Wells CA, Mosbergen R, Korn O, Choi J, Seidenman N, Matigian NA, Vitale AM, and Shepherd J (2013). Stemformatics: visualisation and sharing of stem cell gene expression. Stem Cell Res 10, 387–395. [DOI] [PubMed] [Google Scholar]
- Yang X, Pabon L, and Murry CE (2014). Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res 114, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Differential Gene Expression and Gene Ontology Analysis of Subpopulations Identified During Cardiac Directed Differentiation at Single Cell Resolution, Related to Figure 2.
Table S2. Computational Analysis of Transcription Factor and Gene Regulatory Networks Underlying Scdiff Prediction of Lineage Trajectories for the Full Single Cell Time Course Data Set, Related to Figure 3 (data demarcated as “ALL”). Computational Analysis of Transcription Factor and Gene Regulatory Networks Underlying Scdiff Prediction of Lineage Trajectories for Only HOPX+ Cells During the Full Time Course Data Set, Related to Figure S4. (Data demarcated as “HOPX”).
Table S5. RNA-Seq Analysis of HOPX Over-Expression Cardiomyocytes vs controls and gene ontology analysis, Related to Figure 5.
Table S6. Statistical Analysis of Gene Expression Shown in Heat Maps in Figure 6J-M.