Abstract
Dysregulated long non-coding RNAs (lncRNAs) are found in many types of tumors, including esophageal squamous cell carcinoma (ESCC); however, the pattern of expression and function of LINC01296 in esophageal squamous cell carcinoma are unknown. In the current study we showed that LINC01296 expression is significantly higher in ESCC tissues when compared with corresponding adjacent normal tissues. Higher LINC01296 expression was associated with lymph node metastasis, TNM stage, and worse overall survival rate in ESCC patients. Furthermore, functional assays in vitro demonstrated that knockdown of LINC01296 inhibited ESCC cell proliferation, colony formation, migration, and invasiveness. Moreover, RNA immunoprecipitation (RIP) and chromatin immunoprecipitation (ChIP) assays demonstrated that LINC01296 promotes cell proliferation and invasion by epigenetic suppression of KLF2 expression via an interaction with EZH2 in ESCC cells. We also demonstrated that knockdown of LINC01296 inhibited cell growth and up-regulated KLF2 expression in vivo. These results indicate that LINC01296 acts as an oncogene and may serve as a potential target in ESCC treatment.
Keywords: Long non-coding RNA, esophageal squamous cell carcinoma, LINC01296, EZH2, KLF2
Introduction
Esophageal squamous cell carcinoma (ESCC) is a major histopathologic subtype of esophageal cancer [1]. The incidence of ESCC is increasing, and the recurrence rate after surgical resection is extremely high [2]. Despite other advances in chemotherapy, radiation therapy, or molecular targeted therapy for ESCC, the 5-year overall survival rate remains poor [3]. To improve the understanding of the molecular mechanisms underlying ESCC progression is thus warranted.
Long non-coding RNAs (lncRNAs) are a new class of non-coding RNAs without protein-coding capacity [4]. Recently, increasing evidence has revealed that lncRNAs are dysregulated in some cancer types, including ESCC [5]. Several lncRNAs, such as MALAT1 [6], HOTAIR [7], AFAP1-AS1 [8], and BANCR [9], have been reported to have aberrant expression in ESCC, and therefore provide an understanding of lncRNA involvement in the development of ESCC.
The lncRNA, LINC01296, has been reported to function as a tumor-promoting gene in some tumors. For example, LINC01296 is a potential prognostic biomarker for colorectal cancer [10]. Profiling of lncRNAs has identified LINC01296 as a candidate oncogene in bladder cancer patients [11]. Silencing LINC01296 suppresses prostate cancer cell proliferation, migration, and invasion via regulation of the PI3K-Akt-mTOR signaling pathway [12]; however, the pattern of expression and function of LINC01296 in ESCC are unknown.
In the current study we showed that LINC01296 is up-regulated in ESCC. Inhibition of LINC01296 reduced cell proliferation, migration, and invasiveness in ESCC cells. Furthermore, we demonstrated that LINC01296 suppressed KLF2 expression by interacting with EZH2 in vitro and in vivo in ESCC. Therefore, LINC01296 acts as an oncogene and may serve as a potential target in ESCC treatment.
Materials and methods
Patient tissue samples and ESCC cell lines
Seventy-eight fresh ESCC tissues and paired normal tissues were collected in the Department of Thoracic Surgery, The First Affiliated Hospital, College of Medicine, Zhejiang University between 2013 and 2015. All of the patients were diagnosed with ESCC. The clinicopathologic data are shown in Table 1. The patients had no treatment of ESCC, including chemotherapy or radiotherapy, before surgery. The study was approved by the Ethics Committee of The First Affiliated Hospital, College of Medicine, Zhejiang University. All of the patients signed the informed consent. The immortalized human esophageal epithelial cell line, SHEE, and three human ESCC cell lines (EC9706, E106, and TE-1) were purchased from the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum [FBS] (Hyclone, Logan, UT, USA). Cells were maintained at 37°C in a humidified chamber with 5% CO2.
Table 1.
Correlation between LINC01296 expression and clinicopathologic factors
| Clinicopathologic factors | Number of patients | LINC01296 expression | X2 value | p-value | |
|---|---|---|---|---|---|
|
| |||||
| Lower (n=38) | Higher (n=40) | ||||
| Age (years) | 0.641a | 0.423 | |||
| ≤60 | 52 | 27 | 25 | ||
| >60 | 26 | 11 | 15 | ||
| Gender | 0.015a | 0.901 | |||
| Male | 60 | 29 | 31 | ||
| Female | 18 | 9 | 9 | ||
| Tumor size | 3.335a | 0.068 | |||
| <5 cm | 41 | 24 | 17 | ||
| >5 cm | 37 | 14 | 23 | ||
| Infiltration depth | 0.195a | 0.658 | |||
| T1+T2 | 37 | 19 | 18 | ||
| T3+T4 | 41 | 19 | 22 | ||
| Histologic grade | 0.412a | 0.521 | |||
| High and middle | 50 | 23 | 27 | ||
| Low | 28 | 15 | 13 | ||
| Lymph node invasion | 6.022a | 0.014* | |||
| Negative | 51 | 30 | 21 | ||
| Positive | 27 | 8 | 19 | ||
| TNM stage | 6.837a | 0.009* | |||
| I-II | 48 | 29 | 19 | ||
| III-IV | 30 | 9 | 21 | ||
p-value <0.05.
RC table chi-square test.
RNA extraction and qRT-PCR
Total RNA was extracted from tissues and cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The RNA was transcribed into cDNAs using a Prime-Script one-step RT-PCR kit (TaKaRa, Dalian, China). The level of LINC01296 mRNA was detected on a 7500 real-time PCR system (Applied Biosystems, USA). The relative level of mRNA expression was determined using the 2-ΔΔCt method. GAPDH was used as an internal control. The primer sequences were as follows: LINC01296 forward, 5’-AAGTGGCACCAGCCTCACT-3’ and LINC01296 reverse, 5’-CGGCCAAGTTCTTTACCATC-3’; and GAPDH forward, 5’-AGAAGGCTGGGGCT-CATTTG-3’ and GAPDH reverse, 5’-AGGGGCCATCCACAGTCTTC-3’.
Cell Counting Kit-8 assay
Cell viability was analyzed using a Cell Counting Kit-8 kit (CCK8; Dojindo, Tokyo, Japan) according to the manufacturer’s instructions. Briefly, 2000 cells/well were seeded in 96-well plates. Cell viability was tested at 1, 2, 3, 4, and 5 days. The absorbance was read at 450 nm under a microplate reader.
Cell transfection
Two specific siRNA oligonucleotides targeting LINC01296 and negative control siRNA (si-NC) were purchased from GenePharma (Shanghai, China). The target sequences for siRNA-LINC01296 were as follows: siRNA-LINC01296-1 (S1), 5’-GGCUGGAGAAUAUUUCCUATTTT-3’; and siRNA-LINC01296-2 (S2), 5’-CUGAAACAUAUU CCGUGGUTT-3’. The full-length coding sequence for KLF2 was chemical synthesized and sub-cloned into the pcDNA3.1(+) vector (Realgene, Shanghai, China).
Colony formation assay
For the cell colony formation assay, 300 cells/well were plated into 12-well plates and cultured at 37°C in a humidified chamber with 5% CO2. After 14 days, the cell colonies were fixed with methanol and stained with 0.1% crystal violet for 20 min. Then, the cell colonies were counted after twice-washing with H2O.
Transwell migration and invasion assays
Transwell invasion assays were performed using Transwell chambers with (invasion) or without (migration) Matrigel (BD Sciences, Sparks, MD, USA). Briefly, ESCC cells were resuspended and plated in the upper chamber with free FBS medium. Medium supplemented with 10% FBS was in the lower chamber. After 48 h, cells were fixed and stained using 0.1% crystal violet solution for 20 min and counted.
Western blot assay
The protein was extracted from cells and lysed with radioimmunoprecipitation assay buffer (Sigma-Aldrich, USA). The protein extraction was separated using 10% sodium dodecyl sulfate polyacrylamide-gel electrophoresis (SDS-PAGE), then transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were incubated with antibodies to KLF2 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and GAPDH (Santa Cruz Biotechnology, Inc.) at 4°C overnight. Then, the membranes were incubated with HRP-conjugated secondary antibody for 2 h. The blot was detected by enhanced chemiluminescence (ECL) according to the manufacturer’s protocol.
RNA immunoprecipitation (RIP) assay
RIP assay was performed as previously described [13]. A RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) and specific antibody to EZH2 (Millipore) or anti-H3K27me3 (Millipore) were used according to the manufacturer’s instructions. Briefly, cells were lysed and incubated with protein A Sepharose beads and the immunoprecipitated RNA was purified and analyzed by qRT-PCR.
Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation (ChIP) was performed according to the method described in a previous study [14]. The EZ-ChIP Chromatin Immunoprecipitation Kit (Millipore) was used according to the manufacturer’s instructions. Anti-EZH2 (Cell Signaling Technology, USA) or anti-H3K27me3 antibodies (Millipore) were used in the study.
Subcutaneous xenograft in nude mice
EC109 cells (2 × 107/ml) were stably transfected with lentivirus-control or lentivirus-sh LINC01296 plasmid. The cells were injected into either side of the posterior flank of 3-week-old athymic BALB/c mice. The tumor volumes (length × width2 × 0.5) were measured every 7 days in the lv-control or lv-sh LINC01296 groups. Four weeks after injection, the mice were sacrificed and the tumor weights were measured.
Statistical analysis
The data are shown as the mean ± SD from three independent experiments. Differences between groups were analyzed using the two-tailed Student’s t-test. The overall survival rate was analyzed by a Kaplan-Meier curve and the log-rank test method. Pearson correlation analysis was used to assess the correlation between LINC01296 and KLF2 expression. A P< 0.05 was considered statistically significant.
Results
LINC01296 expression is specifically up-regulated in ESCC tissues and cells
Initially, we performed qRT-PCR to analyze the relative expression of LINC01296 in 78 paired samples (ESCC and adjacent normal tissues). As shown in Figure 1A, the results demonstrated that LINC01296 expression is specifically higher in tumor tissues compared with adjacent normal tissues (52/78). Then, we analyzed three ESCC cell lines (EC109, EC9706, and TE-1) and a human esophageal epithelial cell line (SHEE). Compared with normal cells, LINC01296 expression was also significantly up-regulated in ESCC cells (Figure 1B).
Figure 1.
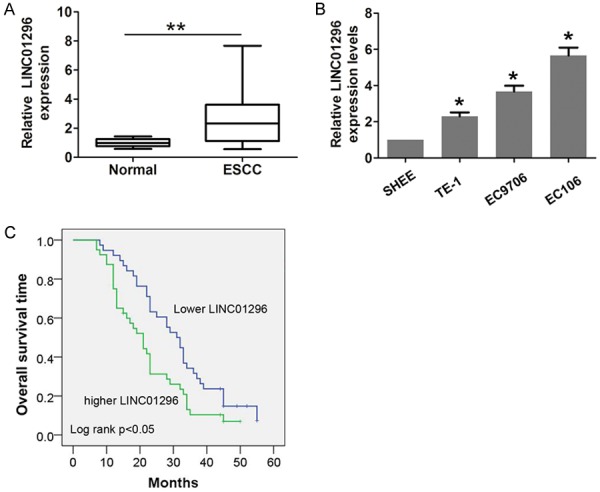
LINC01296 expression was higher in ESCC and associated with poor prognosis. A. The relative expression of LINC01296 in ESCC tissues (n=78) compared with corresponding non-tumor tissues (n=78). LINC01296 expression was examined by qRT-PCR and normalized to GAPDH expression. B. The relative expression of LINC01296 in an immortalized human esophageal epithelial cell line (SHEE) and three human ESCC cell lines (EC9706, EC106, and TE-1) was examined by qRT-PCR and normalized to GAPDH expression. C. ESCC patients with higher LINC01296 expression had a worse overall survival time compared with lower LINC01296 expression. Error bars indicate the mean ± S.D. *P<0.05, **P<0.01.
According to the median expression of LINC01296 (2.65-fold), we classified the patients into the following two groups: higher LINC01296 expression (LINC01296 expression>median expression rate); and lower LINC01296 expression (LINC01296 expression<median expression rate). The association analysis results showed that higher LINC01296 expression was positively correlated with lymph node metastasis (P=0.014, Table 1) and advanced tumor stage (P=0.009, Table 1). To further verify the prognostic value of LINC01296 expression, a Kaplan-Meier curve and the log-rank test method were performed. Indeed, elevated LINC01296 expression predicted a poor outcome compared with lower LINC01296 expression in ESCC patients (Figure 1C).
LINC01296 mediates ESCC cell proliferation, migration, and invasion in vitro
Furthermore, transient transfection was performed using two specific siRNAs targeting LINC01296 to silence LINC01296 expression in EC109 and EC9706 cells (Figure 2A, 2B). As shown in Figure 2C, 2D, cell viability was significantly reduced after LINC01296 was knocked down compared with the si-NC group in EC109 and EC9706 cells. In addition, compared with the si-NC group, cell colonies were dramatically decreased in EC109 and EC9706 cells (Figure 2E, 2F). Next, we assessed the effect of LINC01296 on cell migration and invasion by Transwell assays. The results showed that cell migration and invasion was significantly reduced after LINC01296 knockdown compared with the si-NC group in EC109 and EC9706 cells (Figure 3A-D). These results indicated that LINC01296 promotes cell proliferation, migration, and invasion of ESCC.
Figure 2.
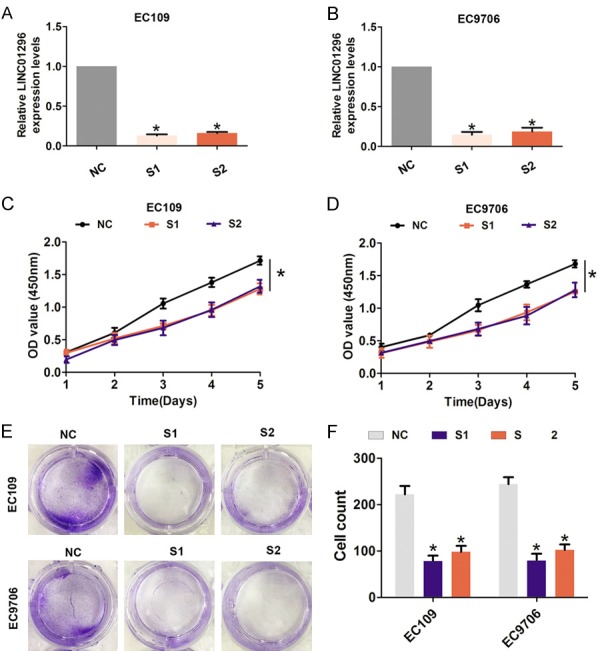
LINC01296 knockdown inhibited ESCC cell proliferation and colony formation ability in vitro. A, B. QRT-PCR analysis of the level of LINC01296 expression following treatment with si-NC, si-LINC01296-1 (S1), or si-LINC01296-1 (S2) in EC106 and EC9706 cells and normalized to GAPDH expression. C, D. CCK8 was performed to assess the cell proliferation rate following treatment with si-NC, si-LINC01296-1 (S1), or si-LINC01296-1 (S2) in EC106 and EC9706 cells. E, F. Cell colony formation assays were performed to assess cell proliferation following treatment with si-LINC01296-1 (S1) or si-LINC01296-1 (S2) in EC106 and EC9706 cells. Error bars indicate the mean ± S.D. *P<0.05.
Figure 3.
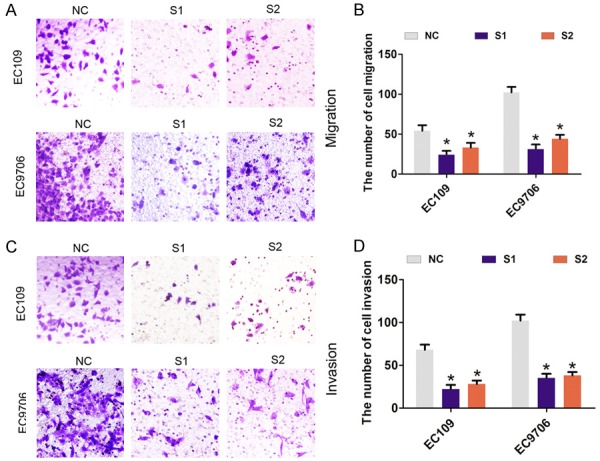
LINC01296 knockdown inhibited ESCC cell migration and invasion in vitro. A, B. Cell migration assays were performed to assess cell migration following treatment with si-NC, si-LINC01296-1 (S1), or si-LINC01296-1 (S2) in EC106 and EC9706 cells. *P<0.05. C, D. Cell invasion assays were performed to assess cell invasion following the treatment with si-NC, si-LINC01296-1 (S1), or si-LINC01296-1 (S2) in EC106 and EC9706 cells. Error bars indicate the mean ± S.D. *P<0.05.
LINC01296 binds to EZH2 and epigenetically silences KLF2 expression in ESCC cells
Recent studies demonstrated that some lncRNAs, such as HOXA-AS2 [15] and AGAP2-AS1 [16], bind to PRC2 enhancing tri-methylation of histone H3 on lysine 27, which causes epigenetic silencing of target genes. To assess whether or not EZH2 is associated with the function of LINC01296 in ESCC cells, we determined the distribution of LINC01296 expression in EC109 and EC9706 cells, and found that LINC01296 was located in the nucleus and cytoplasm (Figure 4A, 4B). According to the knockdown efficiency for LINC01296, we chose siRNA-LINC01296-1 for further silencing experiments. We analyzed several targets of EZH2 (LATS1, P15, P21, KLF2, P57, and E-cadherin) and mRNA level changes after LINC01296 knockdown in EC109 and EC9706 cells. The results showed that KLF2 mRNA expression was significantly up-regulated after LINC01296 silencing in EC109 and EC9706 cells (Figure 4C, 4D). Protein expression was consistently and dramatically increased when LINC01296 was knocked down in EC109 and EC9706 cells (Figure 4E, 4F).
Figure 4.
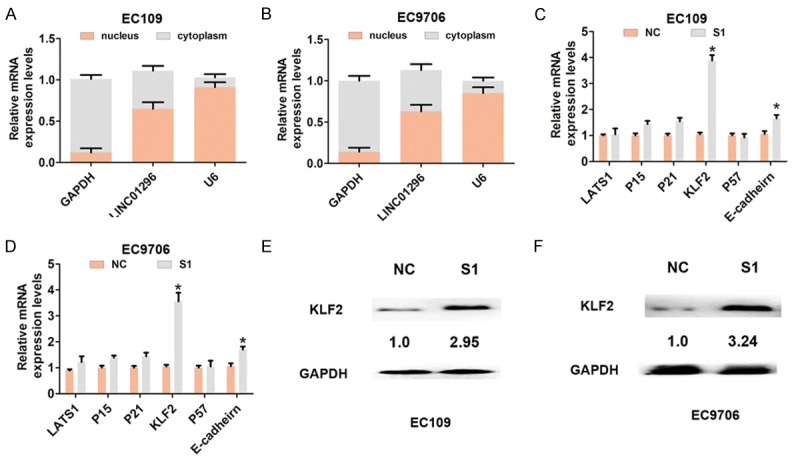
LINC01296 bound to EZH2 and epigenetic silencing KLF2 expression in ESCC cells. A, B. QRT-PCR was used to analyze LINC01296 expression in the nuclei and cytoplasm of EC106 and EC9706 cells. U6 was used as a nucleus marker, and GAPDH was used as a cytosol marker. C, D. QRT-PCR was used to analyze mRNA expression of LATS1, P15, P21, P16, KLF2, P57, and E-cadherin following treatment with si-NC or si-LINC01296-1 (S1) in EC106 and EC9706 cells. E, F. Western blot was used to analyze the expression of KLF2 protein following treatment with si-NC or si-LINC01296-1 (S1) in EC106 and EC9706 cells. Error bars indicate the mean ± S.D. *P<0.05.
The RNA immunoprecipitation assay with EHZ2 or H3K27me3 antibody demonstrated that LINC01296 binds to EZH2 and H3K27me3 in EC109 and EC9706 cells (Figure 5A, 5B). KLF2 was previously reported to be an EZH2 target gene and was epigenetically silenced by H3K27-trimethylation [17]. Furthermore, we assessed the levels of KLF2 mRNA and protein expression when EZH2 was knocked down by qRT-PCR and Western blot analysis. The results demonstrated that the KLF2 mRNA and protein levels were significantly higher compared with the control group (Figure 5C, 5D). To investigate whether or not LINC01296 repressed KLF2 expression by binding to EZH2 and H3K27-trimethylation, we performed the CHIP assay. The analysis showed that EZH2 occupancy and H3K27me3 binding in the promoter regions of KLF2 were significantly reduced when LINC01296 was knocked down in EC109 and EC9706 cells (Figure 5F, 5G). Therefore, the above findings indicated that LINC01296 repressed KLF2 expression by binding to EZH2.
Figure 5.
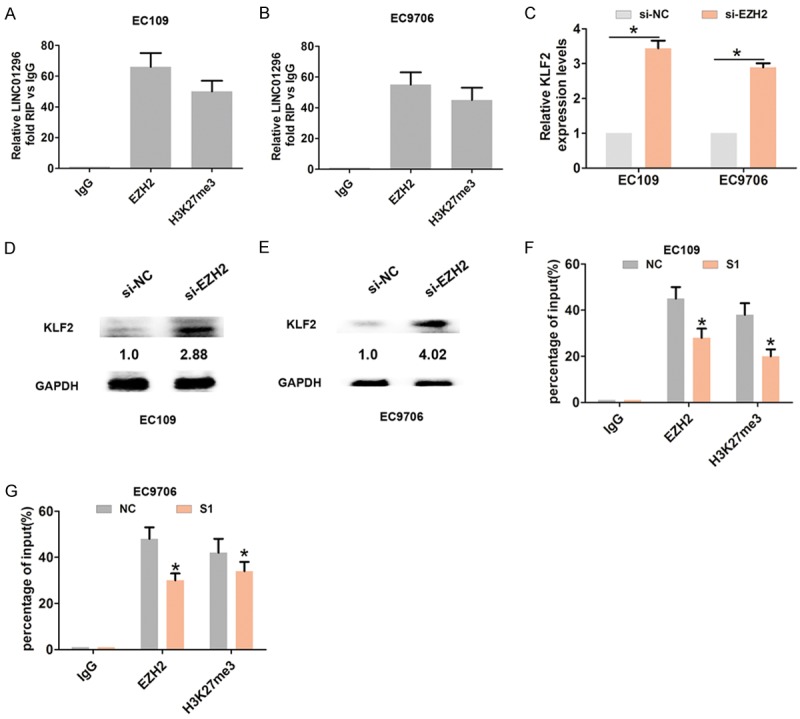
LINC01296 binds to EZH2 and epigenetic silencing of KLF2 expression in ESCC cells. A, B. RIP experiments with anti-EZH2 and anti-H3k27me3 antibodies were performed in EC106 and EC9706 cells. QRT-PCR was used to analyze the expression of LINC01296 in the co-precipitated RNA. The fold enrichment of LINC01296 is relative to the matched IgG control. C. QRT-PCR was used to analyze mRNA expression of KLF2 following treatment with si-NC or si-EZH2 in EC106 and EC9706 cells. D, E. Western blot was used to analyze the expression of KLF2 protein following treatment with si-NC or si-EZH2 in EC106 and EC9706 cells. *P<0.05. F, G. ChIP-qRT-PCR of EZH2 occupancy and H3K27me3 binding in the KLF2 promoters in EC106 and EC9706 cells following treatment with si-NC or si-LINC01296-1 (S1). IgG as a negative control. Error bars indicate the mean ± S.D. *P<0.05.
Overexpression of KLF2 partially mediates the oncogenic function of LINC01296 in ESCC cells
To determine whether or not KLF2 participates in LINC01296-induced promotion of ESCC cell proliferation and invasion, CCK8, colony formation, and cell invasion assays were performed. KLF2 was overexpressed in EC109 cells by transfecting pcDNA3.1-KLF2 (Figure 6A). The CCK8 and colony formation analysis results demonstrated that si-LINC01296 significantly inhibits cell proliferation, while co-transfection with si-LINC01296 and pcDNA3.1-KLF2 partially rescue si-LINC01296-inhibited proliferation in EC109 cells (Figure 6B, 6C). Moreover, the Transwell invasion assay demonstrated that si-LINC01296 significantly inhibits cell invasion, while co-transfection with si-LINC01296 and pcDNA3.1-KLF2 partially rescue si-LINC01296-inhibited invasion in EC109 cells (Figure 6D, 6E). These results suggest that overexpression of KLF2 partially mediate the oncogenic function of LINC01296 in ESCC cells.
Figure 6.
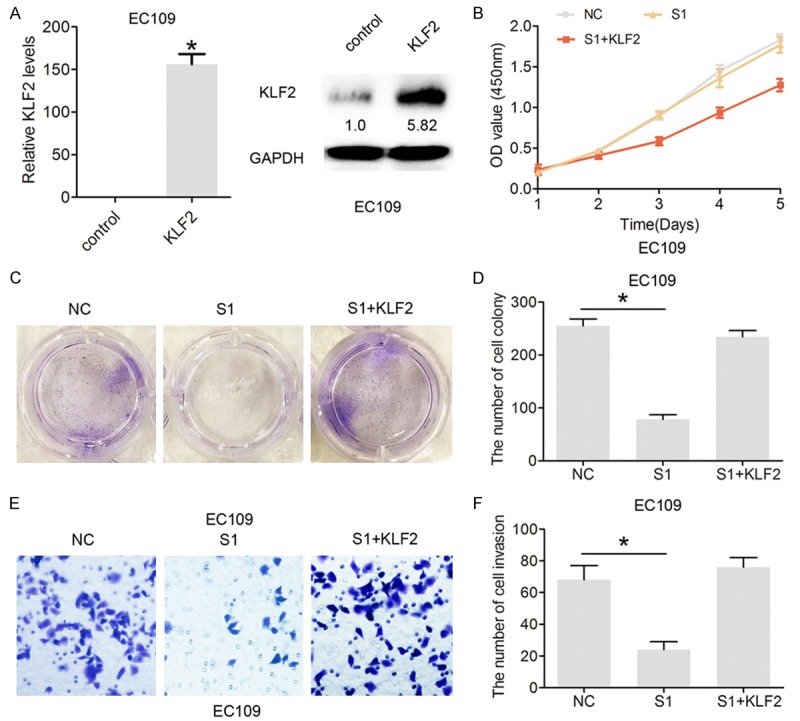
Overexpression of KLF2 partially mediated the oncogenic function of LINC01296 in ESCC cells. A. The relative expression of KLF2 mRNA and protein following treatment with pcDNA3.1 or pcDNA3.1-KLF2 (KLF2) in EC106 cells by qRT-PCR and Western blot analysis. B. CCK8 was performed to assess the cell proliferation rate following treatment with si-NC or si-LINC01296-1 (S1) or pcDNA3.1-KLF2 (KLF2) and si-LINC01296-1 (S1) in EC106 and EC9706 cells. C, D. Cell colony formation was performed to assess cell proliferation following treatment with si-NC or si-LINC01296-1 (S1) or pcDNA3.1-KLF2 (KLF2) and si-LINC01296-1 (S1) in EC106 and EC9706 cells. E, F. Cell invasion assays were performed to assess cell invasion following treatment with si-NC or si-LINC01296-1 (S1) or pcDNA3.1-KLF2 and si-LINC01296-1 (S1) in EC106 and EC9706 cells. Error bars indicate the mean ± S.D. *P<0.05.
Knockdown of LINC01296 inhibits tumor growth and regulates KLF2 expression in vivo
To further detect whether or not LINC01296 expression affects ESCC tumor growth in vivo, lv-sh-LINC01296 or lv-control was inoculated subcutaneously into mice. After 4 weeks, the results demonstrated that the tumor size in the lv-sh-LINC01296 group was dramatically smaller compared with the lv-control group (Figure 7A). The median tumor growth rate and weight in the sh-LINC01296 group was consistently and dramatically reduced compared to the lv-control group (Figure 7B, 7C). Moreover, we showed that KLF2 expression in the sh-LINC01296 group was significantly up-regulated compared to the lv-control group by Western blot analysis (Figure 7D). Thus, these results indicated that LINC01296 regulates KLF2 in vivo.
Figure 7.
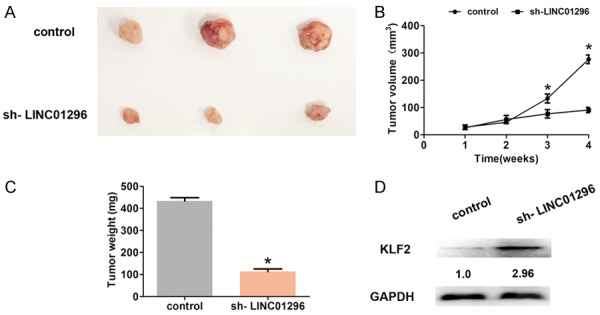
LINC01296 regulated KLF2 expression in vivo. A. The tumor size was shown when tumor was removed from the mice at 4 weeks. B. The tumor volumes were analyzed every 7 days after inoculation. C. The tumor weights were weighted when tumor was removed at 4 weeks. D. Western blot analyses demonstrated that the KLF2 expression was significantly increased after LINC01296 knockdown in vivo. Error bars indicate the mean ± S.D. *P<0.05.
Discussion
Recent studies have shown that lncRNAs function as crucial regulators in several different biologic processes during tumorigenesis, including ESCC. For example, the lncRNA, RP11-766N7.4, functions as a tumor suppressor by regulating the epithelial-mesenchymal transition in ESCC [18]. The lncRNA, UCA1, inhibits ESCC growth by regulating the Wnt signaling pathway [19]. Up-regulation of the lncRNA, SPRY4-IT1, promotes metastasis of ESCC via induction of the epithelial-mesenchymal transition [20]; however, little is known about the expression of LINC01296 associated with ESCC progression. In the current study, we demonstrated that LINC01296 is notably up-regulated in ESCC tissues and cells. Higher LINC01296 expression was shown to be associated with lymph node metastasis, advanced TNM stage, and worse prognosis. Furthermore, knockdown of LINC01296 inhibited cell proliferation, cell colony formation, cell migration, and cell invasion in ESCC cells.
The clinical significance and biologic function of LINC01296 in several tumors has been reported; however, the mechanism underlying LINC01296 in tumors remains unknown. In the current study, we demonstrated that LINC01296 interacts with EZH2. LncRNAs have been shown to bind EZH2, which causes epigenetic silencing of target genes. For example, the lncRNA, BLACAT1, predicts a poor outcome of colorectal cancer and promotes cell proliferation by epigenetic silencing of p15 [21]. TUG1 regulates the expression of LIMK2b, a splice variant of LIM-kinase 2, by binding with enhancer of zeste homolog 2 (EZH2), then promotes cell growth and chemoresistance of NSCLC [22]. The lncRNA, XIST, acts as an oncogene by epigenetic repression of KLF2 in non-small cell lung cancer [23]. Zhang et al. [24] reported that P53-regulates the lncRNA, TUG1, which affects cell proliferation in human non-small cell lung cancer partly through epigenetic regulation of HOXB7 expression. Furthermore, our results demonstrated that KLF2 is a downstream target of LINC01296 and LINC01296 is bound to EZH2 and epigenetic silencing of KLF2 expression in ESCC cells.
KLF2 exhibits tumor suppressor features and down-regulated KLF2 expression promotes cell proliferation and invasion of NSCLC [25]. With respect to ESCC progression, we demonstrated that down-regulation of LINC01296 significantly inhibits cell proliferation and invasion; however, co-transfection with si-LINC01296 and pcDNA3.1-KLF2 partially rescue si-LINC01296 inhibition of proliferation and invasion in ESCC cells. These findings indicated that KLF2 partially mediates the oncogenic function of LINC01296 in ESCC cells.
In conclusion, we demonstrated that LINC01296 is significantly up-regulated in ESCC tissues and cells. Knockdown of LINC01296 inhibited cell proliferation and invasion. Furthermore, LINC01296 interacted with EZH2 and suppressed KLF2 in ESCC cells. These results indicate that LINC01296 acts as an oncogene and may be a potential target of ESCC treatment.
Disclosure of conflict of interest
None.
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 2.Gamliel Z, Krasna MJ. Multimodality treatment of esophageal cancer. Surg Clin North Am. 2005;85:621–630. doi: 10.1016/j.suc.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Han H, Li Y, Zhang Q, Mo K, Chen S. Upregulation of long noncoding RNA HOTTIP promotes metastasis of esophageal squamous cell carcinoma via induction of EMT. Oncotarget. 2016;7:84480–84485. doi: 10.18632/oncotarget.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao X, Zhao R, Chen Q, Zhao Y, Zhang B, Zhang Y, Yu J, Han G, Cao W, Li J, Chen X. MALAT1 might be a predictive marker of poor prognosis in patients who underwent radical resection of middle thoracic esophageal squamous cell carcinoma. Cancer Biomark. 2015;15:717–723. doi: 10.3233/CBM-150513. [DOI] [PubMed] [Google Scholar]
- 7.Ren K, Li Y, Lu H, Li Z, Li Z, Wu K, Li Z, Han X. Long noncoding RNA HOTAIR controls cell cycle by functioning as a competing endogenous RNA in esophageal squamous cell carcinoma. Transl Oncol. 2016;9:489–497. doi: 10.1016/j.tranon.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo HL, Huang MD, Guo JN, Fan RH, Xia XT, He JD, Chen XF. AFAP1-AS1 is upregulated and promotes esophageal squamous cell carcinoma cell proliferation and inhibits cell apoptosis. Cancer Med. 2016;5:2879–2885. doi: 10.1002/cam4.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, Yang T, Xu Z, Cao X. Upregulation of the long non-coding RNA BANCR correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Biomed Pharmacother. 2016;82:406–412. doi: 10.1016/j.biopha.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Qiu JJ, Yan JB. Long non-coding RNA LINC01296 is a potential prognostic biomarker in patients with colorectal cancer. Tumour Biol. 2015;36:7175–7183. doi: 10.1007/s13277-015-3448-5. [DOI] [PubMed] [Google Scholar]
- 11.Seitz AK, Christensen LL, Christensen E, Faarkrog K, Ostenfeld MS, Hedegaard J, Nordentoft I, Nielsen MM, Palmfeldt J, Thomson M, Jensen MT, Nawroth R, Maurer T, Orntoft TF, Jensen JB, Damgaard CK, Dyrskjot L. Profiling of long non-coding RNAs identifies LINC00958 and LINC01296 as candidate oncogenes in bladder cancer. Sci Rep. 2017;7:395. doi: 10.1038/s41598-017-00327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Cheng G, Zhang C, Zheng Y, Xu H, Yang H, Hua L. Long noncoding RNA LINC01296 is associated with poor prognosis in prostate cancer and promotes cancer-cell proliferation and metastasis. Onco Targets Ther. 2017;10:1843–1852. doi: 10.2147/OTT.S129928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol Ther Nucleic Acids. 2016;5:e385. doi: 10.1038/mtna.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Sehgal L, Jain N, Khashab T, Mathur R, Samaniego F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J Transl Med. 2016;14:346. doi: 10.1186/s12967-016-1100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding J, Xie M, Lian Y, Zhu Y, Peng P, Wang J, Wang L, Wang K. Long noncoding RNA HOXA-AS2 represses P21 and KLF2 expression transcription by binding with EZH2, LSD1 in colorectal cancer. Oncogenesis. 2017;6:e288. doi: 10.1038/oncsis.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Sun M, Zang C, Ma P, He J, Zhang M, Huang Z, Ding Y, Shu Y. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 2016;7:e2225. doi: 10.1038/cddis.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taniguchi H, Jacinto FV, Villanueva A, Fernandez AF, Yamamoto H, Carmona FJ, Puertas S, Marquez VE, Shinomura Y, Imai K, Esteller M. Silencing of Kruppel-like factor 2 by the histone methyltransferase EZH2 in human cancer. Oncogene. 2012;31:1988–1994. doi: 10.1038/onc.2011.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao GL, Pan CF, Xu H, Wei K, Liu B, Zhai R, Chen YJ. Long noncoding RNA RP11-766N7.4 functions as a tumor suppressor by regulating epithelial-mesenchymal transition in esophageal squamous cell carcinoma. Biomed Pharmacother. 2017;88:778–785. doi: 10.1016/j.biopha.2017.01.124. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Gao Z, Liao J, Shang M, Li X, Yin L, Pu Y, Liu R. lncRNA UCA1 inhibits esophageal squamous-cell carcinoma growth by regulating the Wnt signaling pathway. J Toxicol Environ Health A. 2016;79:407–418. doi: 10.1080/15287394.2016.1176617. [DOI] [PubMed] [Google Scholar]
- 20.Zhang CY, Li RK, Qi Y, Li XN, Yang Y, Liu DL, Zhao J, Zhu DY, Wu K, Zhou XD, Zhao S. Upregulation of long noncoding RNA SPRY4-IT1 promotes metastasis of esophageal squamous cell carcinoma via induction of epithelial-mesenchymal transition. Cell Biol Toxicol. 2016;32:391–401. doi: 10.1007/s10565-016-9341-1. [DOI] [PubMed] [Google Scholar]
- 21.Su J, Zhang E, Han L, Yin D, Liu Z, He X, Zhang Y, Lin F, Lin Q, Mao P, Mao W, Shen D. Long noncoding RNA BLACAT1 indicates a poor prognosis of colorectal cancer and affects cell proliferation by epigenetically silencing of p15. Cell Death Dis. 2017;8:e2665. doi: 10.1038/cddis.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niu Y, Ma F, Huang W, Fang S, Li M, Wei T, Guo L. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer. 2017;16:5. doi: 10.1186/s12943-016-0575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478:811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 24.Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS, De W, Shu YQ, Wang ZX. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zang C, Nie FQ, Wang Q, Sun M, Li W, He J, Zhang M, Lu KH. Long non-coding RNA LINC01133 represses KLF2, P21 and E-cadherin transcription through binding with EZH2, LSD1 in non small cell lung cancer. Oncotarget. 2016;7:11696–11707. doi: 10.18632/oncotarget.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]


