Abstract
URI, a member of the prefoldin family of molecular chaperones, functions in the regulation of nutrient-sensitive, mTOR-dependent transcription signaling pathways. Previous studies of several tumor types demonstrated that URI exhibits characteristics similar to those of an oncoprotein. URI has been shown as a mitochondrial substrate of S6 kinase 1 (S6K1), which acts to integrate nutrient and growth factor signals to promote cell growth and survival. Notably, the Akt/mTOR/p70S6K signaling pathway constitutes major negative regulatory mechanism of autophagy. However, the role of URI in autophagy has not been explored. Here, we investigated the involvement of URI in autophagy by manipulating its expression in MGC-803 and HGC-27 cells using siRNA and transfection approaches. GFP-LC3 punctum aggregation was assessed by confocal microscopy, whereas formation of autophagic vesicles was assessed using transmission electron microscopy. NH4Cl was used to inhibit autophagosome-lysosome fusion and to monitor autophagic flux. Expression of LC3-I, LC3-II, beclin1, total and phosphorylated mTOR, and p70S6k was assessed by Western blotting. The results showed that knockdown of URI induced significant autophagic flux in gastric cancer cells. URI regulates the expression of beclin1, which is essential for initiation of conventional autophagy. Levels of p-mTOR (Ser2448) and p-p70S6K (Thr389) increased in URI-overexpressing cells treated with the mTOR inhibitor rapamycin but decreased in URI-silenced cells. The inhibitory effect of URI silencing on mTOR and p70S6K phosphorylation was antagonized by the autophagy inhibitor 3-methyladenine. These results suggest that URI knockdown-induced autophagy is associated with the mTOR/p70S6K signaling pathway, indicating the potential existence of a novel autophagy regulatory mechanism mediated by URI.
Keywords: URI, gastric cancer cells, autophagy, mTOR/p70S6K pathway
Introduction
Autophagy is an intracellular catabolic process in which cytoplasmic macromolecules and organelles (mostly mitochondria) are delivered to lysosomes for subsequent degradation [1,2]. Abnormal impairment or activation of autophagy is closely related to the pathogenesis of cancer [3]. Previous studies have demonstrated that autophagy can be tumor suppressive by eliminating oncogenic or toxic proteins from damaged organelles, or be tumor promoting through intracellular recycling substrates mediated by autophagy [4]. Interestingly, regulators of autophagy, especially those that regulate mTOR (mammalian target of rapamycin) function, can also be either oncogenes or tumor suppressor proteins through a variety of mechanisms [4].
There are multiple signaling molecules or pathways that have been shown to regulate autophagy. As a ser/thr containing protein kinase, mTOR is a key player in mediating mammalian cell growth, proliferation, motility, survival, as well as autophagy [5]. Notably, tumor suppressors (such as PTEN, AMPK etc.) are usually negative regulators of mTOR and stimulate or induce autophagy, while oncogenic proteins (such as PI3K, Ras, and AKT etc.) may activate mTOR and thus, inhibit autophagy. In the meantime, inhibition of autophagy leads to pro-oncogenic events such as oxidative stress and genomic instability, supporting the tumor suppressor mechanism of autophagy, whereas tumors under hypoxia or nutrient starvation, autophagy may promote tumor growth and their resistance to chemotherapy [5].
Recently, increasing evidence has shown that mTOR is a known key player and may also be an important component of complex signaling cascades that control autophagy. Meanwhile, abnormal autophagy has been linked to multiple human diseases, but mostly cancers. Gastric cancer is the second leading cause of cancer-related death worldwide [6].
Overexpression of mTOR and activation of the Akt/mTOR signaling pathway have been observed in gastric cancer, with potential prognostic significance [7,8]. Notably, URI is a transcription factor that belongs to the prefoldin family of molecular chaperones. URI is known to participate in signaling pathway that coordinates nutrients and controls gene expression and is also mTOR-dependent [9,10]. Previous studies have demonstrated that URI amplification and overexpression promotes the survival of S6K1-dependent ovarian cancer cells [11]. We have recently shown that URI promotes cell survival and functions as a chemotherapeutic-resistance factor in gastric cancer cells [12]. As to the relationship between autophagy and gastric cancer, multiple recent studies have suggested that the PI3K/Akt/mTOR pathway is associated with gastric cancer cell autophagy and thus, affecting its progression, chemo-resistance, and its prognosis [13-15].
Based on the association of URI with both gastric cancer and the mTOR/S6K1 signaling pathway, we hypothesized that URI is involved in activation of autophagy in human gastric cancer. Here, we investigated the effect of URI on autophagy in gastric cancer cells and explored potential mechanisms for its effect. Our results suggest that URI participates in the regulation of autophagy via the mTOR/p70S6K signaling pathway.
Materials and methods
Chemicals and antibodies
Opti-MEM and Lipofectamine 2000 transfection reagent were purchased from Invitrogen (Carlsbad, CA). Hiperfect transfection reagent was purchased from QIAGEN (Cat. No. #301705). Rapamycin (RAP) was purchased from Selleck and dissolved in dimethyl sulfoxide (DMSO). 3-Methyladenine (3-MA) was purchased from Sigma-Aldrich Corp. (St. Louis, MO) and diluted to 5 mM before each experiment. Primary antibodies against MAP-LC3 (3868S), RMP (URI) (5844S), S6K1 (2708P), and phosphor-S6K1 (9234P) were purchased from Cell Signaling Technology (Danvers, MA). Polyclonal anti-BECN1 (beclin1) antibody (D160120) was purchased from Sangon Biotech (Shanghai, China). Primary antibodies against mTOR (YT2915) and phosphor-mTOR (YP0176) were obtained from Ruiyingbio. Anti-β-actin (SC-47778) was obtained from Santa Cruz Biotechnology. Secondary antibodies, horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (AB10058) and anti-mouse IgG (D111050) were purchased from Sangon Biotech.
Cell culture
Human gastric carcinoma MGC-803 and HGC-27 cells were gifts from Professor Wei Zhu of Jiangsu University and maintained in Dulbecco’s Modified Eagle Medium (Corning, USA) and RPMI-1640 (Corning) medium, respectively, supplemented with 10% fetal bovine serum (Gibco, New Zealand) and 1% penicillin-streptomycin mixture (Invitrogen). Cells were cultured in a humidified atmosphere of 5% CO2 at 37°C, as described previously [12].
Cell transfection
Results of our previous siRNA gene knockdown experiments demonstrated that of three candidate URI siRNA sequences (siRNA-A, -B, and -C), siRNA-A exhibited the strongest interference of URI expression in MGC-803 and HGC-27 gastric cancer cells [12]. URI siRNA-A and scrambled control sequences, synthesized by Origene Technologies, were as follows: siRNA-A, rArGrArArGrGrUrArGrArUrArArUrGrArCrUrArUrArArUGC; scrambled control, rCrGrUrUrArArUrCrGrCrGrUrArUrArArUrArCrGrCrGrUAT. Transfection was performed using Hiperfect Transfection Reagent and Opti-MEM when cells reached 60-80% confluence. Un-transfected cells served as blank controls. To overexpress URI, the URI expression plasmid pCMV6-URI and its vector control pCMV6-entry (OriGene) were transiently transfected into gastric cancer cells using Lipofectamine 2000 transfection reagent and Opti-MEM. Transfection was performed according to the manufacturer’s protocol and as previously described [12].
Western blotting analysis
Western blotting was performed as described previously [12]. Briefly, cells were washed with ice-cold PBS, lysed in a RIPA buffer (Beyotime Biotechnology, CA, China). Cell lysates were fractionated using SDS-polyacrylamide gel electrophoresis, and proteins were then transferred onto Immobilon-P membranes (Millipore, Billerica, MA). The membranes were probed with specific primary antibodies and then incubated with HRP-conjugated IgG secondary antibodies at 37°C for 1 h. Immunoreactive protein bands were detected using an enhanced chemiluminescence system (Minichemi, China). Expression of β-actin was used as a protein loading control.
GFP-LC3 transient transfection and confocal microscopy
The plasmid pEX-GFP-hLC3WT (#24987) was obtained from Addgene (simply referred to as GFP-LC3). Gastric cells were seeded in 6-well plates and transiently transfected with pEGFP-LC3 using Lipofectamine 2000 transfection reagent and Opti-MEM according to the manufacturer’s protocol. After 48 h of transfection, cells were exposed to 5 mM 3-MA for 12 h. Subsequently, cells were fixed in 4% paraformaldehyde for 15 min. Nuclei were stained with DAPI for 20 min at room temperature in the dark. Treated cells were then examined under a confocal microscope (TCS SP5 II, Leica, Germany), photographed, and counted manually to determine the cellular localization pattern of GFP-LC3 protein. Cells with more than five GFP-LC3 puncta were considered positive. A minimum of 100 random transfected cells were analyzed, and three independent experiments were performed.
Transmission electron microscopy (TEM)
After treatment as described above, cells were fixed in 2.5% glutaraldehyde containing 0.1 M sodium cacodylate and then post-fixed in 2% osmium tetroxide buffer for 1.5 h. The fixed cells were then dehydrated using graded ethanol and routinely embedded in spur resin. Thin sections were cut using an ultramicrotome, stained with 0.3% lead citrate, and examined by TEM at 80 kV (Tecnai12, Philips, Netherlands) at the Testing Center of Yangzhou University. Digital images were obtained using an AMT Imaging System (Advanced Microscopy Techniques Corp., Danvers, MA).
Statistical analyses
All data are expressed as the mean ± SD. One-way analysis of variance followed by Bonferroni’s post-hoc test was performed to test the significance of differences between groups. Statistical analyses were carried out using GraphPad Prism software (v6; GraphPad Software Inc., La Jolla, CA). Statistical significance was set as follows: NS-not significant (P > 0.05); *P ≤ 0.05; **P ≤ 0.01.
Results
URI siRNA-A transfection induces GFP-LC3 punctum aggregation
The autophagosome is an intermediate structure in the dynamic autophagy pathway. Once autophagy is initiated, LC3-I is conjugated to the lipid phosphatidylethanolamine to form LC3-II (membrane-bound form). LC3 tagged at the N-terminus with a fluorescent protein such as GFP (GFP-LC3) has been used to monitor autophagy via fluorescence microscopy, with increases in levels of punctate LC3 or GFP-LC3 reported [16,17]. To evaluate recruitment of LC3-II, gastric cancer cells were transiently transfected with a plasmid encoding GFP-LC3. GFP-LC3-labeled autophagosomes began to appear in the cytoplasm and aggregate into puncta, which could be observed using confocal microscopy. Fluorescence micrographs of representative puncta are shown in Figure 1. Micrographs differed between siRNA-A-transfected and un-transfected cells. In cells transfected with the scrambled control sequence, GFP-LC3 diffused throughout the cytosol. Numerous puncta were observed in siRNA-A-transfected MGC-803 (Figure 1A) and HGC-27 (Figure 1B) cells, but fewer puncta were observed in cells pretreated for 12 h with 3-MA (5 mM), which blocks autophagy at an early stage via inhibition of class III PI3K activity.
Figure 1.
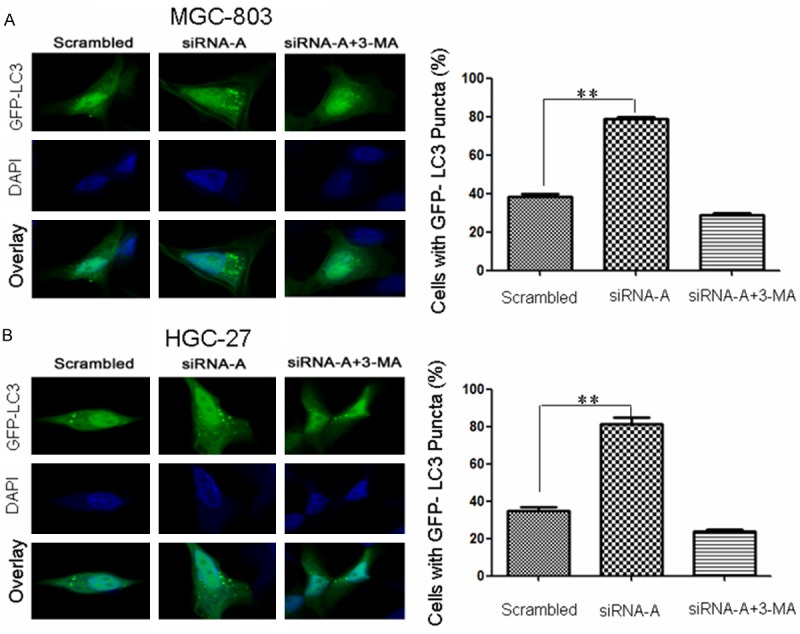
URI siRNA-A transfection enhanced GFP-LC3 puncta aggregation in gastric cancer cells. Cells were transfected with a plasmid encoding EGFP-LC3. Autophagosomes were visualized by the presence of GFP-LC3 puncta (green). DAPI staining (blue) was used to detect nuclei. GFP-LC3 puncta were observed by confocal microscopic analysis of MGC-803 (A) and HGC-27 (B) URI siRNA-A-transfected cells with or without 3-MA treatment. The percentage of cells positive for GFP-LC3 puncta (containing five or more GFP-LC3 punctate dots per cell) is shown (right panel). Data are presented as the mean ± SD of three independent experiments (100 cells were counted for each experiment). **P < 0.01.
URI siRNA-A transfection induces formation of autophagic vesicles
To further confirm the observed induction of autophagy in URI siRNA-A-transfected gastric cancer cells, MGC-803 and HGC-27 cells subjected to URI siRNA interference were analyzed using TEM. As shown in Figure 2, we observed an increase in the number of autophagic vesicles containing cytosolic content and disintegrating material in URI siRNA-A-transfected cells (black arrows, Figure 2B and 2E). Some of these vesicles showed typical double-membrane structure (red arrows). By contrast, such vesicles were not commonly seen in control cells (Figure 2A and 2D) and in cells transfected with siRNA-A and subjected to 3-MA treatment (Figure 2C and 2F).
Figure 2.
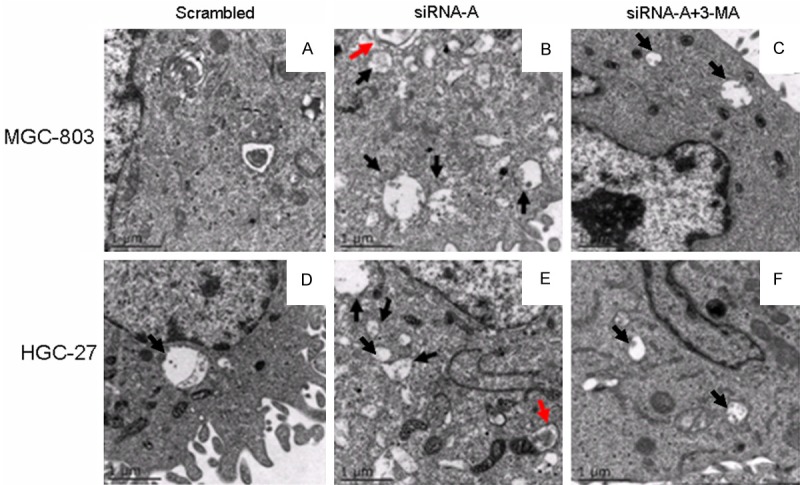
Autophagic vesicles analyzed using TEM. MGC-803 and HGC-27 cells were transfected with URI siRNA-A or scrambled control sequence for 48 h. Ultrastructures of URI siRNA-A-transfected cells subjected to treatment with the autophagy inhibitor 3-MA (5 mM) for 12 h, and those of untreated URI siRNA-A-transfected cells were examined using TEM. Black arrows indicate autophagic vesicles, and red arrows indicate double-membrane structures in particular segments of the autophagic vesicles. URI siRNA-A-transfected cells exhibited more electron-dense mitophagic vesicles (middle panel) compared with cells transfected with scrambled control sequence (left panel) as well as cells transfected with siRNA-A and treated with 3-MA (right panel).
siRNA-mediated silencing of URI induces autophagic flux
Autophagic activity is not always indicated by an increase in the number of autophagic vesicles or the formation of autophagosomes [18]. The term “autophagic flux” is used to describe the dynamic process of autophagosome synthesis, delivery of autophagic substrates to the lysosome, and their subsequent degradation, which is a more reliable indicator of autophagic activity. The accumulation of autophagosomes can represent either increased generation of autophagosomes or a block in autophagosomal maturation and the autophagy pathway. In order to distinguish between these two possibilities, we assessed the autophagic flux using NH4Cl, a commonly used negative regulator of autophagy that inhibits autophagosome-lysosome fusion via acidification within the lysosome [19]. Rap was used in conjunction with URI silencing to induce autophagy. In the case of autophagic flux occurring, LC3-II levels would be higher in the presence of NH4Cl. As shown in Figure 3A and 3B, compared with URI silencing alone and URI silencing plus Rap treatment, the LC3-II/LC3-I ratio in MGC-803 and HGC-27 cells was significantly higher with NH4Cl pretreatment. These results indicate that URI knockdown activates autophagy and induces autophagic flux.
Figure 3.
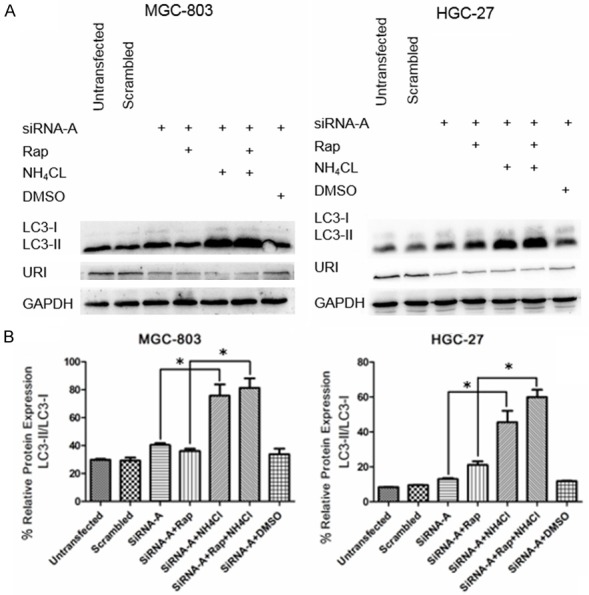
Autophagic flux was activated in URI-silenced MGC-803 and HGC-27 cells. A. MGC-803 and HGC-27 cells were transfected with URI siRNA-A for 24 h and then treated with Rap (5 μM, 12 h), DMSO (0.1% v/v), NH4Cl (10 mM, 4 h) as indicated. Untreated (un-transfected and scrambled control sequence-transfected) samples were used as controls. DMSO was used as a vehicle control for Rap. Levels of LC3-I, LC3-II, and GAPDH in cell lysates were determined by Western blotting. B. Histograms showed comparable trends, in which NH4Cl-treated MGC-803 and HGC-27 cells displayed the highest LC3-II/LC3-I levels, followed by control cells and cells not treated with NH4Cl. Data are presented as the mean ± SD of three independent experiments. *P < 0.05.
URI functions in regulating beclin1 expression
To investigate the role of URI in autophagy in more detail, we examined the expression of beclin1, a core component of the class III phosphatidylinositol 3-kinase (PtdIns3K) complexes that initiate autophagy in mammalian cells [20,21], under conditions of URI overexpression and silencing. As shown in Figure 4A and 4B, immunoblotting analysis indicated that beclin1 expression decreased in URI-overexpressing MGC-803 and HGC-27 cells treated with Rap and increased in URI-silenced MGC-803 and HGC-27 cells. 3-MA inhibited the expression of beclin1 induced by URI silencing. These results suggest that beclin1 plays a role in regulating autophagy mediated by URI.
Figure 4.
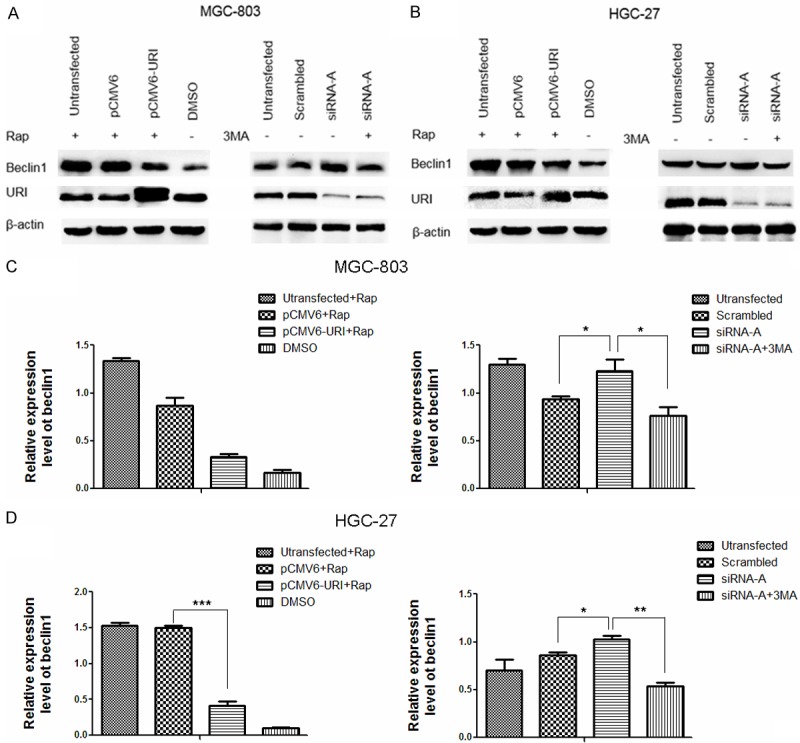
Effect of URI on beclin1 expression. (A and B) Immunoblotting analysis of beclin1 expression in URI-overexpressing MGC-803 and HGC-27 cells treated with Rap (5 μM) for 12 h, with DMSO as a vehicle control (left panel). Beclin1 expression in URI-knockdown MGC-803 and HGC-27 cells in the presence or absence of the autophagy inhibitor 3-MA (5 mM for 12 h) (right panel). (C and D) Bar graph showing the relative quantification of beclin1 expression in immunoblots shown in (A and B). All data are representative of three independent experiments. Data are presented as the mean ± SD and compared between untransfected pCMV6 vs +Rap, scrambled control sequence, or cells transfected with siRNA-A+3MA as indicated. ***P < 0.001, **P < 0.01, *P < 0.05.
URI enhances expression of phosphorylated mTOR and p70S6K
Research has clearly established that the mTOR signaling pathway is the key negative regulator of autophagy [22,23]. We therefore investigated the role of the mTOR pathway in URI silencing-mediated autophagy in MGC-803 and HGC-27 cells. The p70S6 kinase (p70S6K) is a downstream target of mTOR, and its phosphorylation status is as an indicator of mTOR pathway activity [24]. We performed Western blotting to assess levels of p-mTOR and p-p70S6K in pCMV6-URI-transfected cells and URI siRNA-A-transfected cells, with DMSO serving as a vehicle control. As shown in Figure 5A and 5B, levels of p-mTOR (Ser2448) and p-p70S6K (Thr389) were higher in URI-overexpressing cells treated with the mTOR inhibitor Rap. By contrast, URI silencing significantly reduced the levels of phosphorylated mTOR and p70S6K. However, neither URI overexpression nor URI silencing affected the expression of total mTOR and p70S6K. The early stage inhibitor of autophagy, 3-MA, partially reversed the inhibition of mTOR and p70S6K phosphorylation associated with URI silencing.
Figure 5.
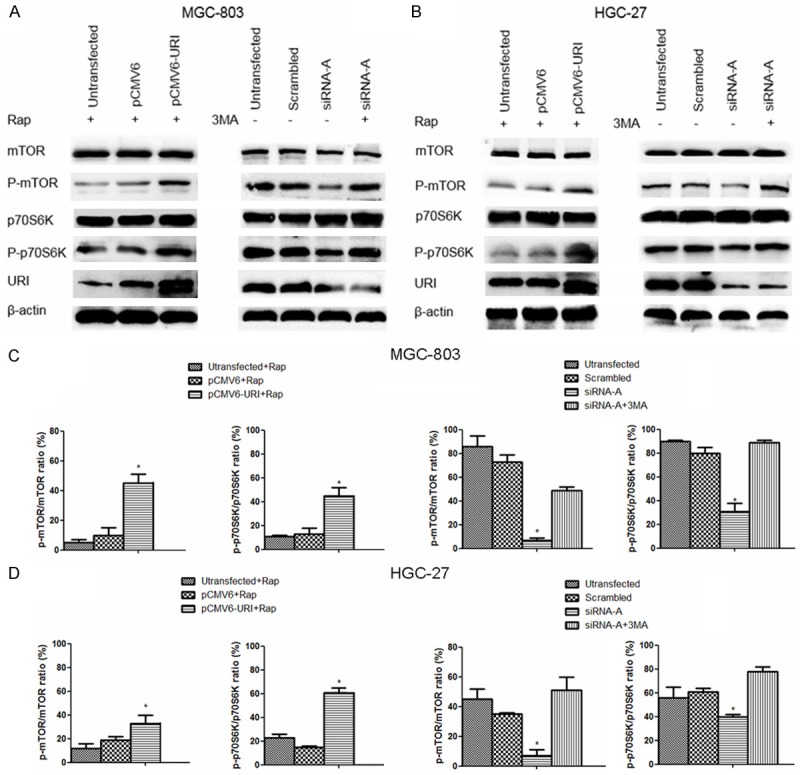
URI overexpression or silencing promotes or inhibits the phosphorylation mTOR and p70S6K, respectively. Levels of total and phosphorylated mTOR and p70S6K were assessed by Western blotting in cells transfected with pCMV6-URI for 48 h with Rap treatment (5 μM) for 12 h (A and B, left panel) and cells transfected with siRNA-A for 48 h with or without 3-MA (5 mM) treatment for 12 h (A and B, right panel). Protein levels were normalized against β-actin. (A and B) Representative immunoblots are shown. (C and D) Relative quantitation of phospho-mTOR (Ser2448)/mTOR and phospho-p70S6K (Thr389)/p70 S6K. All data are representative of three independent experiments. Data are presented as the mean ± SD. *P < 0.05 vs. pCMV6- or scrambled control sequence-transfected cells.
Discussion
URI plays a role in autophagy regulation by affecting autophagic flux
Previous studies demonstrated that URI exhibits properties characteristic of oncogenes [11,25,26]. In the present study, we demonstrated that URI knockdown induces GFP-LC3 punctum aggregation and autophagic vesicle formation in MGC-803 and HGC-27 gastric cancer cells, as evidenced by confocal microscopy and TEM analyses. We also demonstrated that URI knockdown-induced punctum aggregation and vesicle formation are markedly inhibited by the autophagy inhibitor 3-MA, suggesting that URI plays a role in activation of autophagy. To confirm these results, we assessed the autophagic flux in URI-knockdown cells. Cells were treated with NH4Cl to induce impaired fusion of autophagosomes and lysosomes and subsequent accumulation of autophagosomes. Following addition of NH4Cl, we observed a further increase in LC3-II levels in URI-knockdown cells (Figure 1B), demonstrating that URI knockdown induces autophagic flux in MGC-803 and HGC-27 cells.
Beclin1 is a critical component of the class III PtdIns3K complex that initiates autophagy in mammalian cells. However, the role of BECN1 in LC3B lipidation remains controversial [30-32]. A previous study confirmed that although complete loss of BECN1 has little effect on LC3 (MAP1LC3B/LC3B) lipidation, PtdIns3K complex activity and autophagic flux are disrupted in BECN1-/- cells [30]. The effect of URI knockdown on autophagic flux may be related to its regulatory effect on beclin1 expression.
Several recent studies revealed that autophagy induced by different pharmacologic treatments promotes the death of gastric cancer cells [31-35]. We previously reported that URI knockdown promotes apoptosis and attenuates chemoresistance of gastric cancer cells [12]. Collectively, these results suggest that URI knockdown-induced autophagy possibly inhibits cell survival, as previous studies demonstrated that autophagy can be both pro-survival and pro-death in gastric cancer cells [36]. The role of autophagy is assumed to differ in different stages of cancer development [37]. Further investigation is thus needed to obtain a more comprehensive understanding of the complex regulatory mechanisms and roles of autophagy in tumors, including the involvement of URI in autophagy and cancer.
Effect of URI on the autophagy-related mTOR/p70S6K signaling pathway
A variety of protein kinases play integral roles in autophagy. For example, the serine/threonine kinase mTOR negatively regulates autophagy. mTOR/p70S6K signaling is frequently dysregulated in cancer and various metabolic disorders [38,39]. Bahrami et al reported that Monepantel triggers autophagy in human ovarian cancer cells via deactivation of the mTOR/p70S6K signaling pathway [40]. Theurillat et al. demonstrated that URI exhibits characteristics similar to those of oncoproteins. URI amplification affects PI3K-mTOR signaling in ovarian cancer cells downstream, at the level of S6K1. In addition, URI amplification and overexpression in glands of gastric carcinoma in situ correspond with increased S6K1 phosphorylation [11]. In the present study, we demonstrated that phosphorylation of mTOR and p70S6K is induced by URI overexpression in Rap-treated cells but that URI silencing leads to de-phosphorylation of mTOR and p70S6K, which is antagonized by the autophagy inhibitor 3-MA. Taken together, our results suggest that URI knockdown induces autophagy in gastric cancer cells, possibly via the m-TOR/p70S6K signaling pathway, although the exact mechanism and significance of these findings remain to be determined.
Acknowledgements
This study was supported by the innovation program of Jiangsu province (2013, Q.Z.), the leader of the innovation program of Jiangsu province, China (2017, Q.Z.), and the National Natural Science Foundation of China grants, China (Nos. 31271399, 81472047, and 81672229, to Q.Z., J.G., and Y.L).
Disclosure of conflict of interest
None.
References
- 1.Klionsky DJ. Autophagy revisited: a conversation with Christian de Duve. Autophagy. 2008;4:740–743. doi: 10.4161/auto.6398. [DOI] [PubMed] [Google Scholar]
- 2.Jardon MA, Rothe K, Bortnik S, Vezenkov L, Jiang X, Young RN, Lum JJ, Gorski SM. Autophagy: from structure to metabolism to therapeutic regulation. Autophagy. 2013;9:2180–2182. doi: 10.4161/auto.26378. [DOI] [PubMed] [Google Scholar]
- 3.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ávalos Y, Canales J, Bravo-Sagua R, Criollo A, Lavandero S, Quest AF. Tumor suppression and promotion by autophagy. Biomed Res Int. 2014:603980. doi: 10.1155/2014/603980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 7.Tapia O, Riquelme I, Leal P, Sandoval A, Aedo S, Weber H, Letelier P, Bellolio E, Villaseca M, Garcia P, Roa JC. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch. 2014;465:25–33. doi: 10.1007/s00428-014-1588-4. [DOI] [PubMed] [Google Scholar]
- 8.Byeon SJ, Han N, Choi J, Kim MA, Kim WH. Prognostic implication of TSC1 and mTOR expression in gastric carcinoma. J Surg Oncol. 2014;109:812–817. doi: 10.1002/jso.23585. [DOI] [PubMed] [Google Scholar]
- 9.Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, Vigneron M, Peter M, Krek W. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2003;302:1208–1212. doi: 10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- 10.Djouder N, Metzler SC, Schmidt A, Wirbelauer C, Gstaiger M, Aebersold R, Hess D, Krek W. S6K1-Mediated disassembly of mitochondrial URI/PP1g complexes activates a Negative feedback program that counters S6K1 survival signaling. Molecular Cell. 2007;28:28–40. doi: 10.1016/j.molcel.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Theurillat JP, Metzler SC, Henzi N, Djouder N, Helbling M, Zimmermann AK, Jacob F, Sol-termann A, Caduff R, Heinzelmann-Schwarz V, Moch H, Krek W. URI is an oncogene amplified in ovarian cancer cells and is required for their survival. Cancer Cell. 2011;19:317–332. doi: 10.1016/j.ccr.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Hu X, Zhang F, Luo D, Li N, Wang Q, Xu Z, Bian H, Liang Y, Lu Y, Zheng Q, Gu J. URI promotes gastric cancer cell motility, survival, and resistance to adriamycin in vitro. Am J Cancer Res. 2016;6:1420–1430. [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Z, Han F, Yang S, Wu J, Zhan W. Oxamate-mediated inhibition of lactate dehydrogenase induces protective autophagy in gastric cancer cells: involvement of the Akt-mTOR signaling pathway. Cancer Lett. 2015;358:17–26. doi: 10.1016/j.canlet.2014.11.046. [DOI] [PubMed] [Google Scholar]
- 14.Sun Y, Jiang Y, Huang J, Chen H, Liao Y, Yang Z. CISD2 enhances the chemosensitivity of gastric cancer through the enhancement of 5-FU-induced apoptosis and the inhibition of autophagy by AKT/mTOR pathway. Cancer Med. 2017;6:2331–2346. doi: 10.1002/cam4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JF, Wu P, Xia R, Yang J, Huo XY, Gu DY, Tang CJ, De W, Yang F. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer. 2018;17:6. doi: 10.1186/s12943-017-0756-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badr CE, Wurdinger T, Nilsson J, Niers JM, Whalen M, Degterev A, Tannous BA. Lanatoside C sensitizes glioblastoma cells to tumor necrosis factor-related apoptosis-inducing ligand and induces an alternative cell death pathway. Neuro Oncol. 2011;13:1213–1224. doi: 10.1093/neuonc/nor067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 19.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 21.Furuya N, Yu J, Byfi eld M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 22.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu K, Liu P, Wei W. mTOR signaling in tumorigenesis. Biochim Biophys Acta. 2014;1846:638–654. doi: 10.1016/j.bbcan.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, Gu J, Zheng Q, Li M, Lian X, Miao J, Jiang J, Wei W. RPB5-mediating protein is required for the proliferation of hepatocellular carcinoma cells. J Biol Chem. 2011;286:11865–11874. doi: 10.1074/jbc.M110.136929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan JL, Zhang J, Dong LW, Fu WJ, Du J, Shi HG, Jiang H, Ye F, Xi H, Zhang CY, Hou J, Wang HY. URI regulates tumorigenicity and chemotherapeutic resistance of multiple myeloma by modulating IL-6 transcription. Cell Death Dis. 2014;5:e1126. doi: 10.1038/cddis.2014.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng X, Overmeyer JH, Maltese WA. Functional specifi city of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme traffi cking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 28.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 29.Fogel AI, Dlouhy BJ, Wang C, Ryu SW, Neutzner A, Hasson SA, Sideris DP, Abeliovich H, Youle RJ. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol Cell Biol. 2013;33:3675–3688. doi: 10.1128/MCB.00079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He R, Peng J, Yuan P, Xu F, Wei W. Divergent roles of BECN1 in LC3 lipidation and autophagosomal function. Autophagy. 2015;11:740–747. doi: 10.1080/15548627.2015.1034404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu XY, Lv TH, Xie XD, Li J, Su G, Wu H. Antitumour effect of sesquiterpene (+)-chabranol on four human cancer cell lines by inducing apoptosis and autophagy. J Int Med Res. 2012;40:1644–1653. doi: 10.1177/030006051204000503. [DOI] [PubMed] [Google Scholar]
- 32.Rasul A, Yu B, Zhong L, Khan M, Yang H, Ma T. Cytotoxic effect of evodiamine in SGC-7901 human gastric adenocarcinoma cells via simultaneous induction of apoptosis and autophagy. Oncol Rep. 2012;27:1481–1487. doi: 10.3892/or.2012.1694. [DOI] [PubMed] [Google Scholar]
- 33.Lee HW, Jang KS, Choi HJ, Jo A, Cheong JH, Chun KH. Celastrol inhibits gastric cancer growth by induction of apoptosis and autophagy. BMB Rep. 2014;47:697–702. doi: 10.5483/BMBRep.2014.47.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu M, Li CM, Chen ZF, Ji R, Guo QH, Li Q, Zhang HL, Zhou YN. Celecoxib regulates apoptosis and autophagy via the PI3K/Akt signaling pathway in SGC-7901 gastric cancer cells. Int J Mol Med. 2014;33:1451–1458. doi: 10.3892/ijmm.2014.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato Y, Kubo T, Morimoto K, Yanagihara K, Seyama T. High mannose-binding Pseudomonas fluorescens lectin (PFL) downregulates cell surface integrin/EGFR and induces autophagy in gastric cancer cells. BMC Cancer. 2016;16:63. doi: 10.1186/s12885-016-2099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian HR, Yang Y. Functional role of autophagy in gastric cancer. Oncotarget. 2016;7:17641–17651. doi: 10.18632/oncotarget.7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang P, Mizushima N. Autophagy and human diseases. Cell Res. 2014;24:69–79. doi: 10.1038/cr.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 39.Sridharan S, Jain K, Basu A. Regulation of autophagy by kinases. Cancers (Basel) 2011;3:2630–2654. doi: 10.3390/cancers3022630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bahrami F, Pourgholami MH, Mekkawy AH, Rufener L, Morris DL. Monepantel induces autophagy in human ovarian cancer cells through disruption of the mTOR/p70S6K signaling pathway. Am J Cancer Res. 2014;4:558–571. [PMC free article] [PubMed] [Google Scholar]


