Abstract
Breast cancer is considered to be the most frequently diagnosed malignancy in women worldwide. MicroRNAs (miRNAs) play key roles in the regulation of tumor properties based on their capacity to regulate the expression of tumor-related genes. However, the involvement of miR-196b-5p in breast cancer development is largely unknown. Here, we showed that the expression levels of miR-196b-5p were significantly down-regulated in breast cancer samples and cell lines compared to the matched normal tissues and breast epithelial cell line, respectively. Notably, the expression of miR-196b-5p was negatively associated with lymph node metastasis and the progression of clinical stage in patients with breast cancer. MiR-196b-5p over-expression significantly inhibited the proliferation and migration of MDA-MB-231 and MDA-MB-468 cells in breast cancer. Furthermore, combining bioinformatics prediction and biochemical analyses, we showed that COL1A1 (collagen type I alpha 1 chain) was a direct downstream target gene of miR-196b-5p. Furthermore, overexpression of COL1A1 partly abrogated miR-196b-5p-mediated inhibition of proliferation and migration in MDA-MB-231 and MDA-MB-468 cells. Our data collectively indicate that miR-196b-5p inhibits cell growth and metastasis in breast cancer through down-regulating COL1A1, supporting the targeting of the new miR-196b-5p/COL1A1 axis as a promising effective therapeutic approach for breast cancer.
Keywords: Breast cancer, miR-196b-5p, COL1A1, growth, metastasis
Introduction
Breast cancer is the leading cause of cancer-related death for women worldwide, and metastasis is the most common cause of death in patients with breast cancer [1]. Considerable progress in breast cancer treatment has been made over the past decades; however, due to a lack of specific and effective therapeutic targets, breast cancer treatment remains a challenge [2,3]. Thus, identification of potential molecular mechanism as effective diagnostic and therapeutic targets for this cancer remains an urgent medical need.
MicroRNAs (miRNAs) are a type of endogenous and small non-coding RNAs that are typically 18-24 nucleotides in length [4]. MiRNAs can recognize complementary sequences in the 3’-UTRs of target genes as well as lead to suppress the protein translation of these transcripts and/or degrade target mRNAs [5]. Existing studies have shown that miRNAs are involved in the regulation of multiple pathological processes that contribute to cell growth, differentiation, apoptosis as well as metastasis, playing key roles in the progression of human cancer [6]. Multiple miRNAs, including miR-182, miR-372, and miR-421 are known to play oncogenic roles in the carcinogenesis of breast cancer [7-9], whereas others (miR-485-5p, miR-577, and miR-625) are down-regulated and function as tumor suppressor genes [10-12].
MiR-196b-5p locates on human chromosome 7p15.2. Previous data have revealed that miR-196b-5p is dysregulated and involved in carcinogenesis of several types of human cancer, including colorectal cancer [13], gastric cancer [14,15] and prostate cancer [16]. However, little is known about the role and molecular mechanism of miR-196b-5p in breast cancer. In this study, we investigated the expression patterns of miR-196b-5p in human breast cancer tissues and cell lines, followed by functional analyses in MDA-MB-231 and MDA-MB-468 cells. Our data revealed that miR-196b-5p was down-regulated in breast cancer tissues and cell lines. The expression of miR-196b-5p in breast cancer was inversely correlated with lymph node metastasis and the progression of clinical stage in breast cancer patients. Moreover, our results clearly suggested that miR-196b-5p inhibited cell growth and metastasis in breast cancer via down-regulating COL1A1 (collagen type I alpha 1 chain) expression, further supporting miR-196b-5p as a potential therapeutic target in breast cancer.
Material and methods
Clinical samples and cell lines
Forty breast cancer tissues and matched normal tissues were collected at the Department of General Surgery, Jiangxi Provincial People’s Hospital between January 2009 and October 2015. Samples were frozen immediately at liquid nitrogen and used for extraction of RNA and protein. All patients provided consent for use of samples in study, and this clinical research was also approved by the Ethics Committee of Jiangxi Provincial People’s Hospital (No. JX2016007453). Specimens characteristics and information were described in Table 1. Human breast cancer cell lines (MDA-MB-468 and MDA-MB-231), and the breast epithelial cell line (HBL-100) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China).
Table 1.
Summary of clinicopathological features of patients with breast cancer
| Patient number | Age (years) | Clinical stage | LN metastasis | Perineural invasion | ER status | HER-2 status |
|---|---|---|---|---|---|---|
| 1 | 62 | III-IV | Yes | Yes | Negative | Positive |
| 2 | 64 | III | Yes | No | Positive | Negative |
| 3 | 47 | II | No | No | Positive | Negative |
| 4 | 59 | III-IV | Yes | No | Positive | Negative |
| 5 | 55 | II | No | No | Positive | Negative |
| 6 | 51 | II | No | No | Positive | Negative |
| 7 | 55 | III-IV | Yes | No | Positive | Negative |
| 8 | 63 | I-II | No | No | Positive | Negative |
| 9 | 58 | III-IV | Yes | No | Positive | Positive |
| 10 | 49 | III | Yes | No | Positive | Negative |
| 11 | 56 | II | No | No | Positive | Negative |
| 12 | 59 | III | Yes | No | Positive | Negative |
| 13 | 66 | II | No | No | Positive | Negative |
| 14 | 51 | II | No | No | Positive | Negative |
| 15 | 54 | III-IV | Yes | No | Negative | Positive |
| 16 | 46 | I-II | No | No | Positive | Negative |
| 17 | 58 | IV | Yes | Yes | Negative | Positive |
| 18 | 50 | II | No | No | Positive | Negative |
| 19 | 47 | I-II | No | No | Positive | Negative |
| 20 | 68 | III-IV | Yes | No | Positive | Negative |
| 21 | 63 | III | Yes | No | Positive | Negative |
| 22 | 72 | II | No | No | Positive | Negative |
| 23 | 61 | III | Yes | No | Positive | Negative |
| 24 | 62 | III-IV | Yes | No | Positive | Positive |
| 25 | 41 | II | No | No | Positive | Negative |
| 26 | 65 | III-IV | Yes | No | Positive | Negative |
| 27 | 66 | II | No | No | Positive | Negative |
| 28 | 54 | III | Yes | No | Positive | Negative |
| 29 | 59 | III-IV | Yes | No | Positive | Negative |
| 30 | 60 | IV | Yes | Yes | Negative | Positive |
| 31 | 63 | II | No | No | Positive | Negative |
| 32 | 39 | III | No | No | Positive | Positive |
| 33 | 44 | II | No | No | Positive | Negative |
| 34 | 61 | III | Yes | No | Positive | Negative |
| 35 | 58 | III-IV | Yes | No | Negative | Negative |
| 36 | 58 | II | No | No | Positive | Negative |
| 37 | 45 | IV | Yes | Yes | Positive | Positive |
| 38 | 42 | II | No | No | Positive | Negative |
| 39 | 61 | III-IV | Yes | No | Positive | Negative |
| 40 | 49 | III | No | No | Positive | Negative |
LN: lymph node.
Cell culture and cell transfection
All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA), and incubated at 37°C in a humidified chamber containing 5% CO2. For cell transfection, MDA-MB-468 and MDA-MB-231 cells were seeded into a 6-well plate (6 × 104 cells/well) the day prior to transfection. Then, cells were transiently transfected with 50 nM miR-196b-5p mimics or 50 nM mimics control (RiboBio, Guangzhou, China) by using Lipofectamine 2000 transfection reagent (Invitrogen, CA, USA), according to the manufacturer’s protocols. At 24 h after cell transfection, the cells were collected and used for following analysis.
RNA isolation and real-time quantitative PCR (qRT-PCR)
Total RNA was isolated with the TRIzol® reagent (Invitrogen, CA, USA), according to the manufacturers’ protocols. The RevertAid First Strand cDNA synthesis kit (Thermo Scientific, Massachusetts, USA) was applied for cDNA generation. MiScript SYBR Green PCR Kit (Qiagen, Valencia, CA, USA) was used for PCR reaction. The miR-196b-5p primers were 5’-TAGGTAGTTTCCTGTTGTTGGG-3’ (sense), the antisense primer was provided by the miScript SYBR Green PCR Kit; U6 small nuclear RNA (U6) primers were 5’-CTCGCTTCGGCAGCACA-3’ (sense), 5’-ACGCTTCACGAATTTGCGT-3’ (antisense). The PCR reaction was performed on Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). U6 was used as internal controls. The relative expression levels of miR-196b-5p in breast cancer tissues and cells were quantified by the 2-ΔΔCT method [17].
Dual luciferase reporter assay
The potential miR-196b-5p binding site of COL1A1 was predicted by three computer-aided algorithms including miRanda, TargetScan and PicTar. The mRNA 3’-UTR sequence of COL1A1 was PCR amplified and inserted into the psiCHECK-2 luciferase vector (Promega, Madison, WI, USA) to produce wild type (WT) luciferase reporter plasmid. COL1A1 mRNA 3’-UTR contained mutant (MUT) sequence in the putative binding site of miR-196b-5p was also amplified and inserted into the psiCHECK-2 luciferase vector to produce MUT luciferase reporter plasmid. For dual luciferase reporter assay, MDA-MB-468 and MDA-MB-231 cells were seeded in 24-well plates, co-transfected with the WT or MUT luciferase reporter plasmid and miR-196b-5p mimics or mimics control by using Lipofectamine 2000 transfection reagent, according to the manufacturer’s protocols. After 24 h transfection, the Firefly and Renilla luciferase activities were detected by the GloMax-Multi Jr Single Tube Multimode Reader (Promega, Madison, WI, USA). Firefly luciferase activity was used as an internal control to normalize the transfection efficiency.
Western blot
Breast cancer tissues and cell lines were lysed with RIPA lysis buffer (Invitrogen, CA, USA), according to the manufacturer’s instructions. Equal amount of protein samples (25 µg) were resolved by 10% of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride membranes (PVDF) (Millipore, MA, USA). After blocking with 5% nonfat milk for 2 h, membranes were incubated with the primary antibodies over-night, followed by HRP-linked secondary antibodies (Santa Cruz, USA). GAPDH was used as an internal control. Western blot bands were obtained using Imaging System, and the protein density was quantified with Odyssey v1.2 software (LI-COR Biosciences, NE, USA).
CCK-8 assay
MDA-MB-468 and MDA-MB-231 cells were seeded into a 96-well plate (5× 103 cells/well) the day prior to transfection, and transiently transfected with miR-196b-5p mimics or mimics control. Then, cells were incubated in 10% CCK-8 solution (Dojindo; Kumamoto, Japan) diluted in normal culture media at 37°C, until visible color conversion occurred. Proliferation ratio were determined 0, 24, 48 and 72 h after transfection. The absorbance at 450 nm was measured by amicroplate reader (Bio-Rad, Hercules, CA, USA) after incubation for 2 h at 37°C.
Colony formation assay
After 24 h post-transfection, MDA-MB-468 and MDA-MB-231 cells were seeded into 6-well plates (~3 × 103/well) and were cultured for 10 days at 37°C in an atmosphere containing 5% CO2. Subsequently, the cells were fixed with methanol (Sigma, USA) and stained with 0.1% crystal violet (Sigma, USA). Image was then captured and the number of colony was counted under a light microscope (Eclipse TS100; Nikon Corporation, Tokyo, Japan).
Wound scratch assay
Wound scratch assay was used to evaluate the migratory ability of breast cancer cells. MDA-MB-468 and MDA-MB-231 cells were seeded at density of approximately 6 × 104 cells/well in a 6-well plate and transfected with miR-196b-5p mimics or mimics control as above. Wounds were made with 10-µl pipette tips after cells confluence reached 75% at 6 h post-transfection. Then, cells were then washed with PBS to remove residual free-floating cells and debris. Finally, cells were continued to incubate at a humidified chamber. Different stages of wound healing were observed along the scrape line using a standard caliper and representative mages were photographed.
Transwell migration assay
Before the cells were seeded, Corning Costar Transwell 24-well plates (Corning, Corning, NY, USA) were placed in DMEM medium for 1 h at 37°C. A total of 2 × 104 cells were seeded in the wells after 24 h post-transfection, and were cultured in 500 µl DMEM medium without FBS. Normal growth medium was placed in the bottom wells. After incubation for 24 h at 37°C, the cells that did not migration through the pores were carefully removed by cotton swabs. Then, the migratory cells were fixed with methanol (Sigma, USA) and stained with 0.1% crystal violet (Sigma, USA). Image was captured and number of migratory cells was counted under a light microscope.
Statistical analysis
Statistical analyses were performed using SPSS 21.0 (International Business Machines, Armonk, NY, USA). Values are expressed as the mean ± standard deviation of results from three independent experiments. The differences were assessed using a two-tailed Student’s t-test, while the MTT data was examined by one-way ANOVA. P<0.05 was considered to indicate a statistically significant difference between values.
Results
Expression levels of miR-196b-5p in human breast cancer tissues and cell lines
To investigate miR-196b-5p expression in breast cancer, we initially examined its expression in two breast cancer cell lines (MDA-MB-468 and MDA-MB-231) and a breast epithelial cell line (HBL-100). We found that miR-196b-5p levels was lower in MDA-MB-468 and MDA-MB-231 cells, compared with HBL-100 (Figure 1A, P<0.05). We further investigated miR-196b-5p expression in 40 pairs of breast cancer tissues and matched normal tissues. Consistently, miR-196b-5p was down-regulated to a significant extent in tumor samples, relative to matched normal tissues (Figure 1B, P<0.05).
Figure 1.
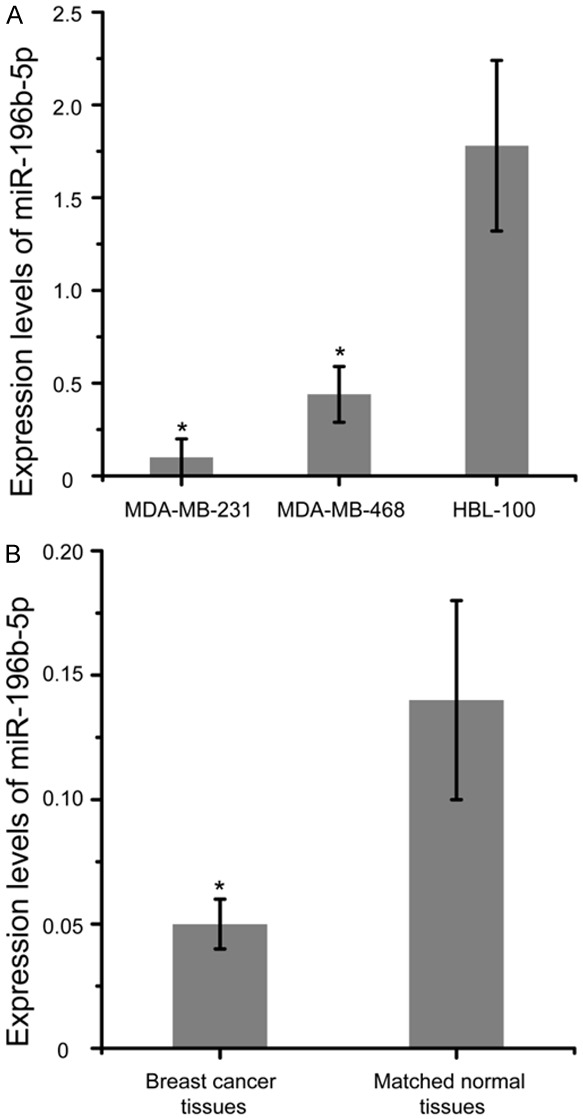
miR-196b-5p was down-regulated in breast cancer tissues and cell lines. A. Relative expression of miR-196b-5p in two breast cancer cell lines (MDA-MB-468 and MDA-MB-231) and a breast epithelial cell line (HBL-100). B. qRT-PCR was applied to detect miR-196b-5p levels in 40 pairs of breast cancer tissues and matched normal tissues. *P<0.05. qRT-PCR, real-time Quantitative PCR.
Analysis of COL1A1 expression in human breast cancer tissues and cell lines
Then, we examined the relative expression of COL1A1 in MDA-MB-468, MDA-MB-231 and HBL-100 by Western blot analysis. In the two breast cancer cell lines, the levels of COL1A1 was found higher than in the HBL-100 cells (Figure 2A, P<0.05). We further found that the expression of COL1A1 were much higher in the breast cancer tissues than in matched normal tissues (Figure 2B, P<0.05).
Figure 2.
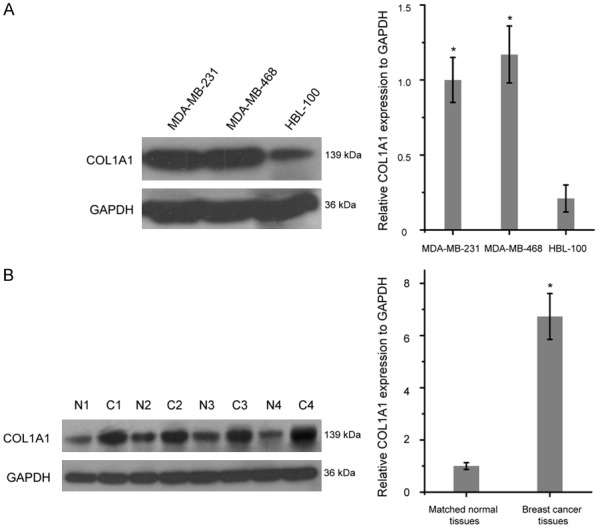
COL1A1 was high expressed in breast cancer tissues and cell lines. A. Western blot analysis of COL1A1 expression in MDA-MB-468, MDA-MB-231 and HBL-100. B. COL1A1 was up-regulated in breast cancer tissues (C) compared with matched normal tissues (N). *P<0.05. COL1A1, collagen type I alpha 1 chain.
miR-196b-5p expression is associated with lymph node metastasis and the progression of clinical stage in patients with breast cancer
Based on the miR-196b-5p expression measured by qRT-PCR, the 40 cases of breast cancer patients were divided into high or low miR-196b-5p expression group using the median expression level as the cut-off point (0.052; range from 0.016 to 0.279). The associations between miR-196b-5p expression levels and clinical characteristics were evaluated by chi-square test. As shown in Table 2, the results revealed that the expression of miR-196b-5p was negatively associated with lymph node metastasis and the progression of clinical stage in breast cancer patients (P<0.05).
Table 2.
Association between miR-196b-5p expression and clinical characteristics in patients with breast cancer
| Clinicopathologic feature | Patients | miR-196b-5p | P-value | |
|---|---|---|---|---|
|
| ||||
| High | Low | |||
| Age (years) | 1.0 | |||
| ≥50 | 30 | 12 | 18 | |
| <50 | 10 | 4 | 6 | |
| ER status | 1.0 | |||
| Positive | 35 | 14 | 21 | |
| Negative | 5 | 2 | 3 | |
| HER-2 status | 0.15 | |||
| Positive | 8 | 5 | 3 | |
| Negative | 32 | 11 | 21 | |
| LN metastasis | 0.0* | |||
| Present | 21 | 4 | 17 | |
| Not identified | 19 | 12 | 7 | |
| Clinical stage | 0.0* | |||
| I, II | 17 | 15 | 2 | |
| III, IV | 23 | 1 | 22 | |
| Perineural invasion | 0.67 | |||
| Present | 4 | 2 | 2 | |
| Not identified | 36 | 14 | 22 | |
P<0.05.
miR-196b-5p over-expression inhibited the proliferation of breast cancer cells in vitro
Next, after transfecting MDA-MB-468 and MDA-MB-231 cells with miR-196b-5p mimics or mimics control for 48 h, the miR-196b-5p expression was explored by qRT-PCR. As a result, transfection of miR-196b-5p mimics led to a significant increase in its expression compared to cells treated with mimics control (Figure 3A and 3C, P<0.05). To explore the function of miR-196b-5p on proliferation of breast cancer cells, CCK-8 assay was performed in MDA-MB-468 and MDA-MB-231 cells. After transfection with miR-196b-5p mimics, the proliferation of MDA-MB-468 and MDA-MB-231 cells was relatively inhibited compared with the mimic control groups, respectively (Figure 3B and 3D, P<0.05).
Figure 3.
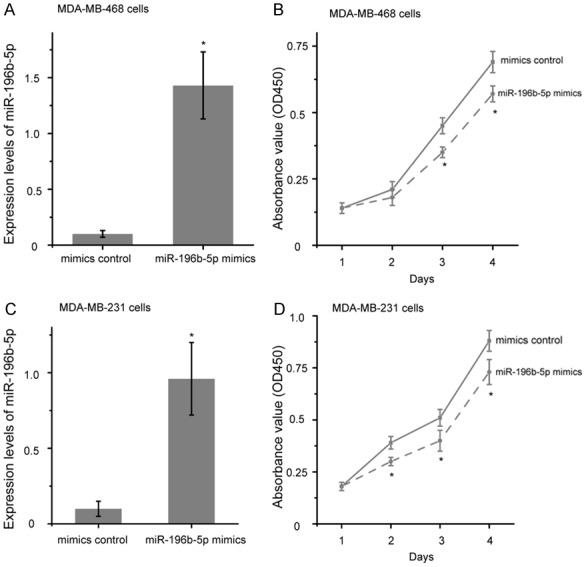
Over-expression of miR-196b-5p suppressed the proliferation of breast cancer cells. Relative expression levels of miR-196b-5p were detected in MDA-MB-468 (A) and MDA-MB-231 (C) cells by qRT-PCR after transfection with miR-196b-5p mimics or mimics control. (B) The cell proliferation of MDA-MB-468 (B) and MDA-MB-231 (D) cells was determined by CCK8 assay after transfection with miR-196b-5p mimics or mimics control. *P<0.05.
Restoration of miR-196b-5p suppressed the migration of breast cancer cells in vitro
To investigate the effect of miR-196b-5p on breast cancer cell metastasis, a wound healing assay was used. Images of the scratches were captured at 0 and 24 h after transfection, the results showed that over-expression of miR-196b-5p repressed cell migration in MDA-MB-468 and MDA-MB-231 cells compared with the mimic control groups, respectively (Figure 4, P<0.05).
Figure 4.
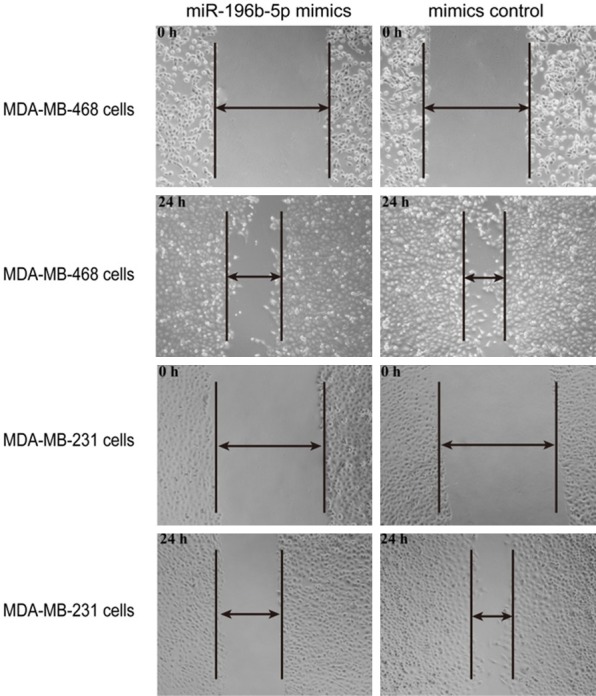
Over-expression of miR-196b-5p inhibited the migration of breast cancer cells. Cell migration ability was determined at 0 h and 24 h in MDA-MB-468 and MDA-MB-231 cells by wound healing assay after treatment with miR-196b-5p mimics or mimics control. *P<0.05.
COL1A1 was a direct target of miR-196b-5p
Bioinformatics analysis was used to identify potential targets of miR-196b-5p, and from these results, COL1A1 was selected as a potential target for further study. Seven nucleotides in miR-196b-5p were shown to have complementary to the 3’-UTR of COL1A1 mRNA (Figure 5A). Spearman’s correlation analysis showed a significant inverse correlation between COL1A1 mRNA and miR-196b-5p expression in breast cancer (Figure 5B, P<0.05). To further confirm our finding that miR-196b-5p could directly targeting COL1A1 expression, dual luciferase reporter assays were performed. As shown in Figure 5C, co-transfection of MDA-MB-468 and MDA-MB-231 cells with WT luciferase reporter plasmid and miR-196b-5p mimics caused a significant decrease in the luciferase activity compared with that of the mimics control groups (P<0.05). By contrast, co-transfection of MUT luciferase reporter plasmid and miR-196b-5p mimics in MDA-MB-468 and MDA-MB-231 cells showed no obvious change in fluorescence compared with that of the mimics control groups (Figure 5D). These results indicate that miR-196b-5p exerts an inhibitory effect on COL1A1 expression via binding to the 3’-UTR of COL1A1 mRNA, and that COL1A1 is therefore a direct target of miR-196b-5p in breast cancer.
Figure 5.
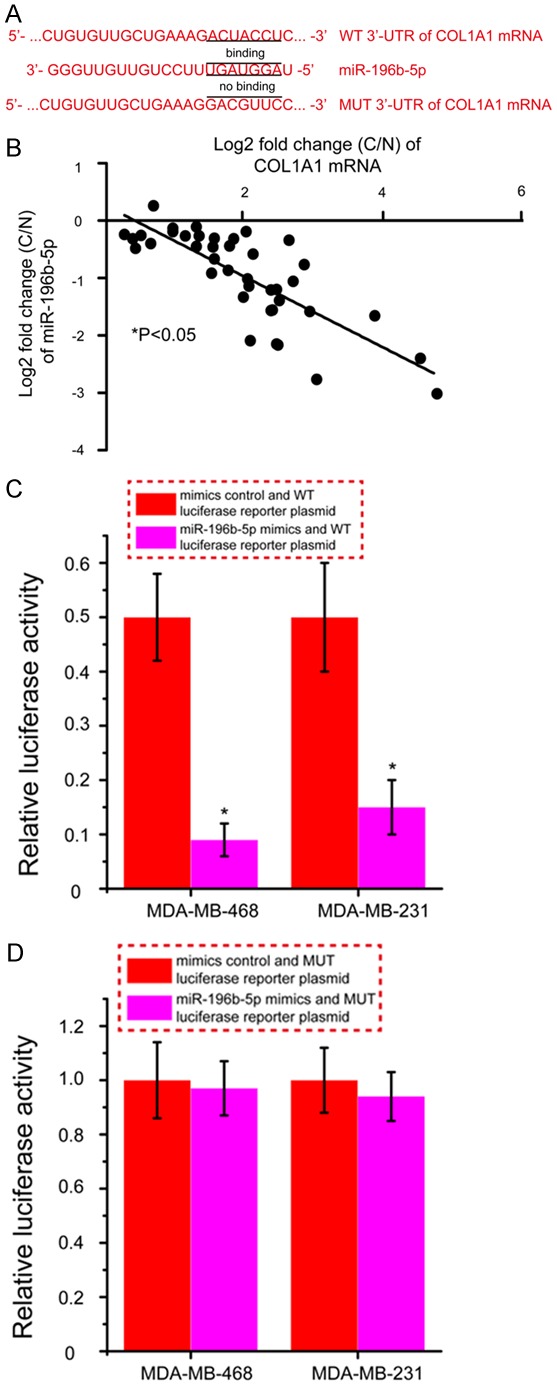
COL1A1 is a direct target gene of miR-196b-5p. A. Bioinformatics analysis of the interaction of miR-196b-5p and its binding site within the 3’-UTR of COL1A1 mRNA. B. miR-196b-5p levels was inversely correlated with COL1A1 mRNA expression in breast cancer. C. Relative luciferase activity in MDA-MB-468 and MDA-MB-231 cells after co-transfected WT luciferase reporter plasmid and miR-196b-5p mimics or mimics control. D. Relative luciferase activity in MDA-MB-468 and MDA-MB-231 cells after co-transfected MUT luciferase reporter plasmid and miR-196b-5p mimics or mimics control. *P<0.05. C, breast cancer tissues; N, matched normal tissues; WT, wild type; MUT, mutant.
Over-expression of COL1A1 partly abrogated miR-196b-5p-induced inhibitory effects on breast cancer cells
COL1A1 was identified as a direct target of miR-196b-5p in breast cancer, as demonstrated by dual luciferase reporter assays. The role of COL1A1 in miR-196b-5p -induced inhibition on breast cancer cells remained unknown. As presented in Figure 6, colony formation assay showed that the number of colony of MDA-MB-468 and MDA-MB-231 cells was significantly increased following co-transfection with the COL1A1 overexpressed vector and miR-196b-5p mimics as compared with cells treated with control vector and miR-196b-5p mimics (P<0.05). Additionally, transwell migration assay revealed that over-expression of COL1A1 and up-regulation of miR-196b-5p could promote the migration of MDA-MB-468 and MDA-MB-231 cells as compared with cells treated with control vector and miR-196b-5p mimics (Figure 7, P<0.05). These data strongly indicate that over-expression of COL1A1 in breast cancer cells partially reverses the inhibitory effects of miR-196b-5p on cell proliferation and migration.
Figure 6.
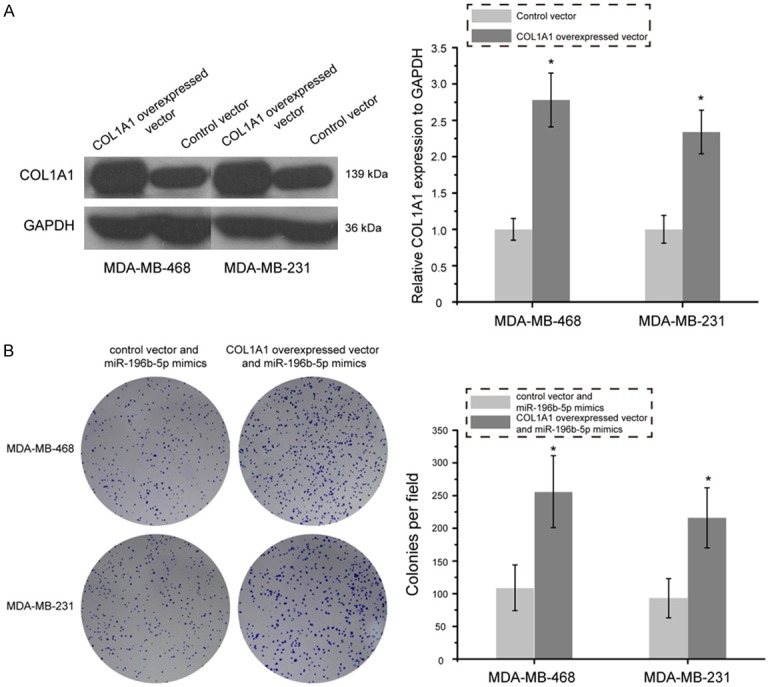
Restoration of COL1A1 partly abrogated miR-196b-5p-induced inhibitory effect on proliferation of breast cancer cells. A. Western blot analysis of COL1A1 expression in MDA-MB-468 and MDA-MB-231 cells after treatment with COL1A1 overexpressed vector and control vector. B. Cell proliferation was measured by colony formation assay in MDA-MB-468 and MDA-MB-231 cells transfected with COL1A1 overexpressed vector and miR-196b-5p mimics or control vector and miR-196b-5p mimics. *P<0.05.
Figure 7.
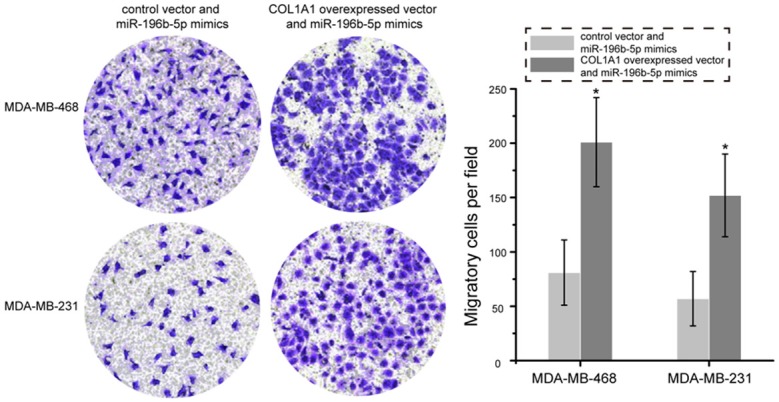
Overexpression of miR-196b-5p partly abrogated miR-196b-5p-mediated inhibition of MDA-MB-468 and MDA-MB-231 cells migration. Images of migration in the MDA-MB-468 and MDA-MB-231 cells transfected with COL1A1 overexpressed vector and miR-196b-5p mimics or control vector and miR-196b-5p mimics. *P<0.05.
Discussion
Emerging evidence has shown that miRNAs are dysregulated in various human cancers and are defined as tumor suppressors or oncogenes [18]. Moreover, several studies have demonstrated a crucial role for miRNAs in the diagnosis and therapy of breast cancer [19]. MiR-196b-5p has been reported to be associated with metastases and poor outcomes in two independent colorectal cancer patient cohorts, and miR-196b-5p inhibition led to significantly increased colorectal cancer cell migration/invasion and metastases formation [13]. However, there are no literature reports investigating the function, and potential mechanisms of miR-196b-5p in breast cancer.
In this study, qRT-PCR showed that the levels of miR-196b-5p was downregulated in breast cancer tissues in comparison with matched normal tissues. In addition, we also observed the expression of miR-196b-5p was markedly lower in the MDA-MB-468 and MDA-MB-231 cells than that in the HBL-100 cells. Interesting, the expression of miR-196b-5p was negatively associated with lymph node metastasis and the progression of clinical stage in breast cancer patients. The differential expression of miR-196b-5p was indicative of the fact that miR-196b-5p might be involved in the development of breast cancer. This paper highlighted the utilization of the gain of function techniques to explore the function of miR-196b-5p on cell growth and metastasis in MDA-MB-468 and MDA-MB-231 cells. In our experiments, over-expression of miR-196b-5p inhibited cell proliferation and migration in MDA-MB-468 and MDA-MB-231 cells in vitro, leading to the conclusion that miR-196b-5p exerts tumor suppressor activity and impedes breast cancer growth and metastasis.
To explore the potential mechanism by which miR-196b-5p suppresses growth and metastasis of breast cancer, we identified COL1A1 as a directly target gene of miR-196b-5p. First, COL1A1 was remarkably up-regulated in breast cancer samples and cell lines compared to the matched normal tissues and breast epithelial cell line. Second, COL1A1 mRNA expression was significant inverse correlated with miR-196b-5p levels in breast cancer. Third, a complimentary binding region for miR-196b-5p was found in the 3’-UTR of COL1A1 mRNA. Fourth, over-expression of miR-196b-5p led to significant suppression of the activity of WT luciferase reporter plasmid, while mutation of the “seed region” abolished this effect.
Collagen type I alpha 1 (COL1A1) has been proved to be closely related to tumors, it could regulate tumors as an oncogene [20-22]. For example, Mori et al. [23] have found that COL1A1 contributed to the differentiation and metastasis abilities of human bladder cancer. He et al. [24] revealed that COL1A1 upregulation promoted proliferation, invasion, and mitosis of oral squamous cell carcinoma. Liu et al. [25] have confirmed that COL1A1 could induce breast cancer metastasis, and serve as a new prognostic biomarker and a potential therapeutic target for breast cancer. COL1A1 has been reported to be a target gene of miR-129-5p in gastric cancer [26], and miR-133a-3p in oral squamous cell carcinoma [24]. Here, we found that COL1A1 was a target gene of miR-196b-5p, and over-expression of COL1A1 in breast cancer cells partially reverses the inhibitory effects of miR-196b-5p on cell proliferation and migration. These data strong demonstrate that miR-196b-5p inhibits cell growth and metastasis in breast cancer via regulation of COL1A1 expression.
Conclusion
Taken together, the present study identified that miR-196b-5p expression was significantly down-regulated in breast cancer. Notably, over-expression of miR-196b-5p represses tumor cell growth and metastasis via regulation of COL1A1. Therefore, miR-196b-5p/COL1A1 axis may serve as a novel therapeutic target in breast cancer.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 3.Ha R, Chow D, Wynn R. Global trend in breast cancer imaging research 1992-2012: bibliometric study. AJR Am J Roentgenol. 2014;202:696–697. doi: 10.2214/AJR.13.11993. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Wang L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell Biosci. 2011;1:31. doi: 10.1186/2045-3701-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang CH, Chu PY, Hou MF, Hung WC. MiR-182 promotes proliferation and invasion and elevates the HIF-1alpha-VEGF-A axis in breast cancer cells by targeting FBXW7. Am J Cancer Res. 2016;6:1785–1798. [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng X, Chen J, Huang Z. miR-372 promotes breast cancer cell proliferation by directly targeting LATS2. Exp Ther Med. 2018;15:2812–2817. doi: 10.3892/etm.2018.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu TB, Chen HS, Cao MQ, Guo FD, Cheng XY, Han ZB, Li MQ. MicroRNA-421 inhibits caspase-10 expression and promotes breast cancer progression. Neoplasma. 2018;65:49–54. doi: 10.4149/neo_2018_170306N159. [DOI] [PubMed] [Google Scholar]
- 10.Wang M, Cai WR, Meng R, Chi JR, Li YR, Chen AX, Yu Y, Cao XC. miR-485-5p suppresses breast cancer progression and chemosensitivity by targeting survivin. Biochem Biophys Res Commun. 2018;501:48–54. doi: 10.1016/j.bbrc.2018.04.129. [DOI] [PubMed] [Google Scholar]
- 11.Yin C, Mou Q, Pan X, Zhang G, Li H, Sun Y. MiR-577 suppresses epithelial-mesenchymal transition and metastasis of breast cancer by targeting Rab25. Thorac Cancer. 2018;9:472–479. doi: 10.1111/1759-7714.12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou WB, Zhong CN, Luo XP, Zhang YY, Zhang GY, Zhou DX, Liu LP. miR-625 suppresses cell proliferation and migration by targeting HMGA1 in breast cancer. Biochem Biophys Res Commun. 2016;470:838–844. doi: 10.1016/j.bbrc.2016.01.122. [DOI] [PubMed] [Google Scholar]
- 13.Stiegelbauer V, Vychytilova-Faltejskova P, Karbiener M, Pehserl AM, Reicher A, Resel M, Heitzer E, Ivan C, Bullock M, Ling H, Deutsch A, Wulf-Goldenberg A, Adiprasito JB, Stoeger H, Haybaeck J, Svoboda M, Stotz M, Hoefler G, Slaby O, Calin GA, Gerger A, Pichler M. miR-196b-5p regulates colorectal cancer cell migration and metastases through interaction with HOXB7 and GALNT5. Clin Cancer Res. 2017;23:5255–5266. doi: 10.1158/1078-0432.CCR-17-0023. [DOI] [PubMed] [Google Scholar]
- 14.Lee SW, Park KC, Kim JG, Moon SJ, Kang SB, Lee DS, Sul HJ, Ji JS, Jeong HY. Dysregulation of MicroRNA-196b-5p and MicroRNA-375 in gastric cancer. J Gastric Cancer. 2016;16:221–229. doi: 10.5230/jgc.2016.16.4.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao L, Chen Z, Peng D, Soutto M, Zhu S, Bates A, Zhang S, El-Rifai W. Methylation of the HOXA10 promoter directs miR-196b-5pdependent cell proliferation and invasion of gastric cancer cells. Mol Cancer Res. 2018;16:696–706. doi: 10.1158/1541-7786.MCR-17-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porretti J, Dalton GN, Massillo C, Scalise GD, Farre PL, Elble R, Gerez EN, Accialini P, Cabanillas AM, Gardner K, De Luca P, De Siervi A. CLCA2 epigenetic regulation by CTBP1, HDACs, ZEB1, EP300 and miR-196b-5p impacts prostate cancer cell adhesion and EMT in metabolic syndrome disease. Int J Cancer. 2018;143:897–906. doi: 10.1002/ijc.31379. [DOI] [PubMed] [Google Scholar]
- 17.Schmittgen TD, Livak KJ. Analyzing realtime PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 18.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 19.Nana-Sinkam SP, Croce CM. MicroRNAs as therapeutic targets in cancer. Transl Res. 2011;157:216–225. doi: 10.1016/j.trsl.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Oleksiewicz U, Liloglou T, Tasopoulou KM, Daskoulidou N, Gosney JR, Field JK, Xinarianos G. COL1A1, PRPF40A, and UCP2 correlate with hypoxia markers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2017;143:1133–1141. doi: 10.1007/s00432-017-2381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Ding Y, Li A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World J Surg Oncol. 2016;14:297. doi: 10.1186/s12957-016-1056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, Wang Y, Zhang J, Zhong J, Yang R. COL1A1 promotes metastasis in colorectal cancer by regulating the WNT/PCP pathway. Mol Med Rep. 2018;17:5037–5042. doi: 10.3892/mmr.2018.8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mori K, Enokida H, Kagara I, Kawakami K, Chiyomaru T, Tatarano S, Kawahara K, Nishiyama K, Seki N, Nakagawa M. CpG hypermethylation of collagen type I alpha 2 contributes to proliferation and migration activity of human bladder cancer. Int J Oncol. 2009;34:1593–1602. doi: 10.3892/ijo_00000289. [DOI] [PubMed] [Google Scholar]
- 24.He B, Lin X, Tian F, Yu W, Qiao B. MiR-133a-3p inhibits oral squamous cell carcinoma (OSCC) proliferation and invasion by suppressing COL1A1. J Cell Biochem. 2018;119:338–346. doi: 10.1002/jcb.26182. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Shen JX, Wu HT, Li XL, Wen XF, Du CW, Zhang GJ. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov Med. 2018;25:211–223. [PubMed] [Google Scholar]
- 26.Wang Q, Yu J. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochem Cell Biol. 2018;96:19–25. doi: 10.1139/bcb-2016-0254. [DOI] [PubMed] [Google Scholar]


