Abstract
In this paper, three plastomes of Mankyua chejuense, Helminthostachys zeylanica, and Botrychium ternatum in Ophioglossaceae were completely sequenced in order to investigate the plastome evolution and phylogeny of eusporangiate ferns. They were similar to each other in terms of length and the gene orders; however, six unknown open reading frames (ORFs) were found between rps4 and trnL-UAA genes in M. chejuense. Similar sequence regions of six ORFs of M. chejuense were found at the plastomes of Ophioglossum californicum and H. zeylanica, as well as the mitochondrial genome (mitogenome) of H. zeylanica, but not in B. ternatum. Interestingly, the translated amino acid sequences of three ORFs were more similar to the proteins of distantly related taxa such as algae and bacteria than they were to proteins in land plants. It is likely that the six ORFs region arose from endosymbiotic gene transfer (EGT) or horizontal gene transfer (HGT), but further study is needed to verify this. Phylogenetic analyses suggested that Mankyua was resolved as the earliest diverging lineage and that Ophioglossum was subsequently diverged in Ophioglossaceae. This result supports why the plastome of M. chejuense have contained the most ancestral six ORFs in the family.
Introduction
Chloroplast is an apparatus for photosynthesis in plant cells that holds an independent genome compared to nuclear and mitochondrial genomes. The plastomes of land plants are typically 120–160 kb in length and have quadripartite structures1. Because of the strong selective constraint for photosynthesis, land plant plastomes usually contain a set of unique 100–120 photosynthetic and housekeeping genes that originate from cyanobacteria1. Chloroplast gene(s) or duplicated part(s) of plastomes are frequently transferred to the nuclear or mitochondrial genome (mitogenome) through intracellular gene transfer (IGT)s2,3. However, gene transfer to the counter direction is very rare evolutionary events2. Only a few cases of IGT to plastomes have been reported in unrelated plant families, such as Apiaceae4,5, Poaceae6,7, Apocynaceae8, and Anacardiaceae9. In all of these IGT cases, short portions of mitogenome were the donors to plastomes, and there has been no documented case of nuclear genome donor to plastome.
Horizontal plastome capture through hybridization between similar species is one kind of horizontal gene transfer (HGT) that is relatively common in land plants. In addition, HGT has also been documented between far distant organisms such as plant-fungus, plant-bacteria, and plant-virus10. Many land plants live in the symbiotic associations with fungi or bacteria. Therefore, they have relatively high chances of HGT between distant organisms. However, on the plant side, the reported HGTs were engaged in mitogenomes or nuclear genomes, not in plastomes. Land plant plastomes do not normally recombine with other genomes, therefore it is very rare for them to act as a recipient of the HGT in land plant plastomes10. In contrast, in the green alga Euglena myxocylindracea, plastomes show intron gains from bacteria11,12.
So far, more than 2,000 complete plastome sequences are available from public databases, such as NCBI. Plastomes, however, appear to be recalcitrant to the incorporation of foreign DNA by either IGT or HGT9. Only a few families, as mentioned in the previous paragraphs, have been recognized as containing DNA of nonplastome origin. However, most published reports on plastomes have been on those from seed plants. We still have limited complete plastomes for several major fern lineages. Ferns are usually divided into two groups: eusporangiate and leptosporangiate ferns. The eusporangiate ferns form basal paraphyletic assemblages because they include the eusporangiate fern clade. So far, 65 plastomes have been reported in leptosporangiate ferns13–15; in contrast, only nine plastomes in eusporangiate ferns have been sequenced from two species of Marattiales16,17, two species of Psilotales18,19, two species of Equisetales18,20,21, and one species of Ophioglossales18.
The order Ophioglossales of eusporangiate ferns contains a single family Ophioglossaceae, and this family consists of four genera (Ophioglossum, Botrychium, Helmintostachys, and Mankyua)22. Ophioglossum and Botrychium each consist of a number of species and are both relatively common in the northern hemisphere; however, both Helmintostachys and Mankyua are monotypic genera and show restricted distribution patterns in temperate regions of East Asia23,24. Among four genera, Mankyua has recently been described from a volcanic island in the Southern part of Korea as Mankyua chejuense24. It is a rare, endemic, and endangered plant species, and only a couple hundred individuals were reported to live in the specific habitats of small scattered volcanic craters called “Gotjawal” in Jeju island of Korea25.
In the phylogeny of Ophiglossacae, Hauk, et al.26 showed that Ophioglossum was the sister group to the clade of Helminthostachys + Botrychium s.l. using rbcL and trnL-F sequences. However, the phylogenetic studies including Mankyua have shown different phylogenetic relationships among four genera. Sun, et al.27 suggested that M. chejuense was sister to the clade of Botrychium + Helminthostachys and that Ophioglossum was the sister group to the three genera. In contrast, Shinohara, et al.28 suggested that Botrychium was sister to Ophioiglossum + Helminthostachys and that Mankyua was sister to the remaining taxa. In addition to the topological incongruences of the four genera, several nodes in the previous phylogenies of Ophioglossaceae were not strongly supported. As a result, the relationships among four genera in Ophioglossaceae remain unclear.
In this paper, plastomes of M. chejuense, H. zeylanica, and B. ternatum were completely sequenced and compared with previously reported plastomes of O. californiacum in order to investigate the evolution of plastomes in Ophioglossaceae. During this study, we identified approximately 10 kb insertion with six unknown ORFs between rps4 and trnL-UAA genes of M. chejuense plastome. These six ORFs were located in the same direction as those in polycistronic genes. Therefore, we discussed the possible origins of the six ORFs through intensive comparative data analysis. In addition, the phylogeny of the four genera in the family Ophioglossaceae was reconstructed based on coding sequences of the plastome in order to resolve the enigmatic relationships among the four genera in Ophioglossaceae.
Results and Discussion
Genome structure and gene contents of plastomes in Ophioglossaceae
The four completely annotated plastome sequences reported in this study are available from the National Center for Biotechnology Information (NCBI) under the accession numbers of B. ternatum (KM817789), H. zeylanica (KM817788), and M. chejuensis 1,2 (NC017006, KP205433). The row Illumina MiSeq sequence data files also available from the NCBI database (Supplemental Table 1). We sequenced two different accessions of M. chejuensis using different methods: PCR-amplified Sanger sequencing and the MiSeq (Illumina, San Diego) NGS.
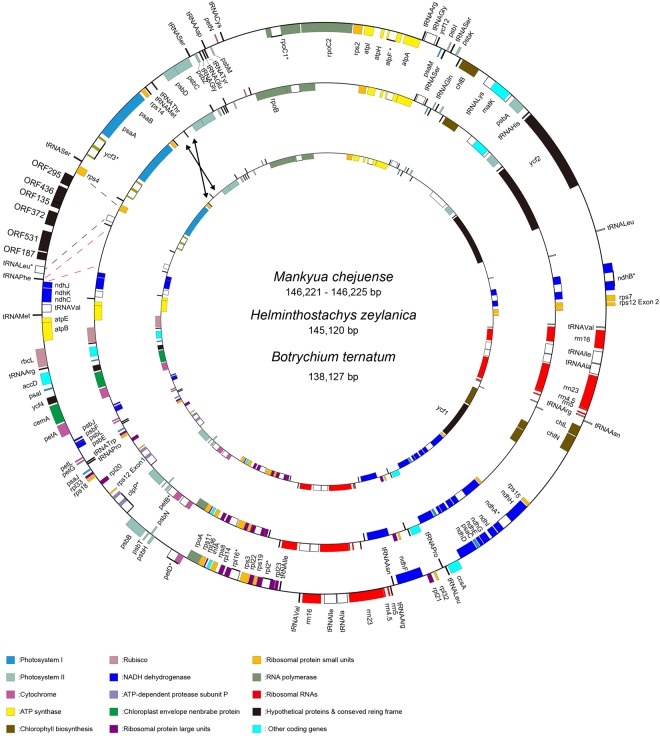
The plastome of M. chejuense sequenced through PCR was 146,221 bp in length with a large single copy (LSC) region of 106,096 bp, a small single copy (SSC) region of 20,613 bp, and two inverted repeat (IR) regions of 9,756 bp each (Fig. 1). It contained 135 genes, including 84 protein coding genes, 8 ribosomal RNAs, 37 transfer RNAs, and six unknown ORFs. Four rRNA and five tRNA genes were duplicated in the IR region (Table 1). Sixteen genes had one intron while the clpP and ycf3 genes each had two introns. The plastome of M. chejuense sequenced through NGS was 146,225 bp. An average coverage depth of the plastome was approximately 400 times (Supplemental Table 1). A total of six poly-T length variations and 45 single nucleotide polymorphisms (SNPs) were found between two plastome sequences of M. chejuense (Supplementary Table S2). Thirty-seven SNPs were found at the coding regions; in particular, SNPs in petB and psbB accounted for 71% of the total SNPs. Interestingly, non-synonymous substitutions were almost three-fold the prevalence of synonymous substitutions in petB and psbB, and petB in plastome of M. chejuense sequenced by PCR method had one premature stop codon caused by substitution (TGG > TGA).
Figure 1.
Maps of three plastomes in Ophioglossaceae. Arrows and dashes refer to inversion and expanded positions, respectively.
Table 1.
Gene list of chloroplast genomes found in four genera in Ophioglossaceae.
| Group of gene | Conserved genes | M. chejuense (PCR) | M. chejuense (NGS) | H. zeylanica | B. ternatum | O. californicum | |
|---|---|---|---|---|---|---|---|
| RNA genes | Ribosomal RNAs | rrn4.5(x2), rrn5(x2), rrn16(x2), rrn23(x2) | |||||
| Transfer RNAs | trnA-UGCa(x2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC, trnH-GUG, trnI-CAU, trnI-GAUa(x2), trnK-UUUa, trnL-CAA, trnL-UAAa, trnL-UAG, trnM-CAU, trnN-GUU(x2), trnP-GGG, trnP-UGG, trnQ-UUG, trnR-ACG(x2), trnR-CCG, trnR-UCU, trnS-CGA, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnV-GAC(x2), trnV-UACa, trnW-CCA, trnY-GUA | φtrnT-UGU | φtrnT-UGU | ||||
| Protein genes | Photosystem I | psaA, psaB, psaC, psaI, psaJ, psaM | |||||
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | ||||||
| Cytochrome | petA, petDa, | φpetB a | petB a | petB a | petB a | petB a | |
| ATP synthase | atpA, atpB, atpE, atpFa, atpH, atpI | ||||||
| Chlorophyll biosynthesis | chlL, chlN | chlB | chlB | chlB | φchlB | chlB | |
| Rubisco | rbcL | ||||||
| NADH dehydrogenease | ndhAa, ndhBa, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | ||||||
| ATP-dependent protease subunit P | clpP a | ||||||
| Chloroplast envelope membrane protein | cemA | ||||||
| Ribosomal proteins | large units | rpl2a, rpl14, rpl16a, rpl20, rpl21, rpl22, rpl23, rpl32, rpl33, rpl36 | |||||
| small units | rps2, rps3, rps7, rps8, rps11, rps12a, rps14, rps15, rps18, rps19 | rps4 | rps4 | rps4 | φrps4 | rps4 | |
| Transcription | RNA polymerase | rpoC1 a | rpoA, φrpoB, rpoC2 | rpoA, φrpoB, rpoC2 | rpoA, rpoB, φrpoC2 | φrpoA, rpoB, φrpoC2 | rpoA, rpoB, rpoC2 |
| Translation | Initiation factor | infA | |||||
| Miscellaneous proteins | accD, ccsA | matK | matK | φmatK | matK | ||
| Hypothetical proteins & Conserved reading frame | ycf2, ycf3a, ycf4, ycf12 | ORF135, ORF187, ORF295, ORF372, ORF436, ORF531, φycf1 | ORF135, ORF187, ORF295, ORF372, ORF436, ORF531, φycf1 | φycf1 | ycf1 | ycf1 | |
(x2): duplicated genes, a: genes having introns φ: pseudogene.
Even though intraspecific variations of plastomes in ferns have been reported21, population studies of M. chejuense have shown extremely low genetic diversity29. Based on our observation of the reproduction of M. chejuense over three years, asexual reproduction by rhizomes was found to be very common. Therefore, these polymorphisms between two plastomes seem to not be genuine. It has been previously shown that after free-living cyanobacteria are engulfed by eukaryotes, numerous genes are translocated from plastids to nucleus30. Nuclear copies of organellar DNAs have frequently been found in land plants31, and they have led to misleading phylogeny results32. Consequently, PCR-amplified sequences of the plastome of M. chejuense might be derived from the nuclear DNA rather than the plastome of M. chejuense, because the nuclear copies of plastid DNA were homologous to their counterparts in the plastome and their primer regions were shared.
The plastome of H. zeylanica was 145,120 bp with an LSC region of 103,088 bp, an SSC of 19,950 bp, and two IR regions of 11,041 bp (Fig. 1). The plastome of B. ternatum was 139,127 bp with an LSC of 99,586 bp, an SSC of 20,569 bp, and two copies of IR with 9,486 bp (Fig. 1). Among four genera in Ophioglossaceae, rps16 and trnT-UGU genes were commonly lost; however, pseudo trnT-UGU remained in the H. zeylanica and B. ternatum plastomes between rps4 and trnL-UAA. The chlB, matK, petB, rpoA, rpoB, rpoC2, rps4, and ycf1 were pseudogenes in at least one plastome, but not in all plastomes. But, the status of pseudogene was not confirmed because we did not study the RNA editing. In addition, there was an inversion of trnT-GGU in the plastome of B. ternatum.
Six ORFs of M. chejuense and similar sequences in Ophioglossaceae
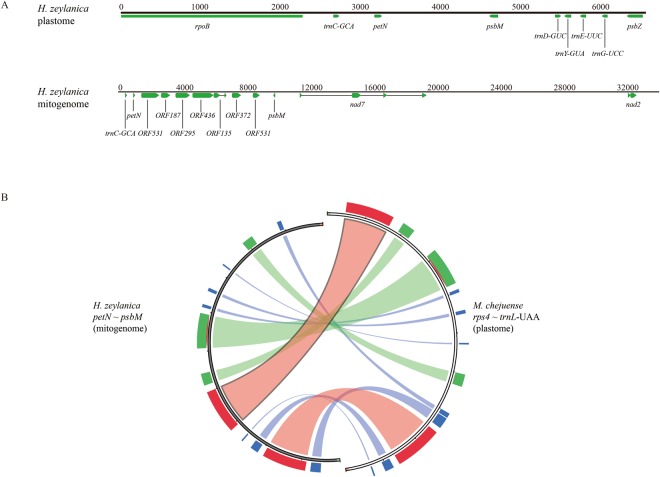
The most distinctive feature among the plastomes of the four genera was the region between trnT-GGU and ndhJ (Fig. 2A). Compared to B. ternatum plastome, the three plastomes of the other genera contained the expanded regions between trnT-GGU and ndhJ, but they were not identical (Fig. 2B). The intergenic spaces (IGSs) of rps4 - trnL (M. chejuense and O. californicum), trnT-GGU - trnfM-CAU (O. californicum and H. zeylanica) and trnF-GAA - ndhJ (H. zeylanica) were 1.5~10 times longer than the IGSs of these regions in B. ternatum. In particular, six unknown ORFs (ORF295, ORF436, ORF135, ORF372, ORF531, and ORF187) were found between rps4 and trnL-UAA genes in M. chejuense, and these six ORFs were located in the same direction as polycistronic genes. The expanded regions in O. californicum and H. zeylanica were partial of six ORFs of M. chejuense with structural mutations (Fig. 2C).
Figure 2.
Alignments of plastomes in Ophioglossaceae. (A) Whole genome alignment. (B) Alignment between trnT-GGU and ndhJ. (C) Visualized alignment by MAUVE.
Contigs generated by the de novo assembly of three sets of NGS data were hit to six ORFs region in the plastome of M. chejuense using blastn in order to investigate translocated ORFs into other genomes such as mitogenome. Two contigs of H. zeylanica with 10-11 coverage depths included sequences similar to six ORFs except for plastome contigs. One contig contained plastome genes of petN and psbM and mitogenome genes of nad7 and nad2 with six ORFs (Fig. 3A). IGS of rps4 - trnL-UAA in the plastome of M. chejuense corresponded highly with petN-psbM in the mitochondrial contig of H. zeylanica, even though there were rearrangements and insertions/deletions (Fig. 3B). Another contig contained a region similar to ORF295 with low similarity. Certain contigs of M. chejuense, H. zeylanica, and B. ternatum had similar sequences of six ORFs; however, they were less than 1,500 bp with less than three coverage depths (almost around 1). These contigs were removed upon further analyses because we could not verify the assembly errors.
Figure 3.
(A) Comparison between plastome and mitogenome of H. zeylanica. (B) Sequence similarity between six ORFs regions of H. zeylanica and M. chejuense. Score/max ratio colouring with blue < = 0.25, green < = 0.5, orange < = 0.75, and red >0.75.
As the total length of NGS data of H. zeylanica was 4.25 Gb, and the lowest 1 C reported in Ophioglossaceae so far was 2.5 Gb33,34, 10-11 coverage depths implied that this contig belonged to mitogenome rather than nuclear genome, even though many plastome and mitogenome sequences were found in nuclear genomes35. The translated amino acid sequences of ORF135, ORF295, and ORF436 between the plastome of M. chejuense and mitogenome of H. zeylanica had over 70% identity while that of ORF372 had 40% identity (Supplementary Fig. 1). The ORF187 and ORF531 of H. zeylanica underwent frame-shift mutations and rearrangement, respectively.
The origin of six ORFs
Based on blastp results (Table 2), the translated amino acid sequence of ORF295 was only similar to the protein of the green alga Roya anglica, which belongs to Streptophyta, and the translated amino acid sequence of ORF436 was similar to the proteins of Chlorophyta, which is a sister group of Streptophyta in Viridiplantae. Interestingly, the translated amino acid sequence of ORF531 was more similar to bacterial proteins than the proteins in Viridiplantae, even though the TrlaMp60 of Treubia lacunosa belonging to Streptophyta was hit to ORF531. Blastn results showed that the plastomes of very few species in ferns contained similar sequences to six ORFs (Supplementary Table S2).
Table 2.
Results of blastp of six ORFs with e-value 10−2.
| Gene | Description | Phylum | Species | Max.score | Total.score | Query.cover | E.value | Ident | Accession |
|---|---|---|---|---|---|---|---|---|---|
| ORF295 | hypothetical protein (chloroplast) | Streptophyta | Roya anglica | 89 | 89 | 65% | 2.00E-17 | 30% | YP_009033761.1 |
| ORF436 | hypothetical protein (chloroplast) | Chlorophyta | Ettlia pseudoalveolaris | 95.5 | 95.5 | 41% | 3.00E-17 | 32% | YP_009105467.1 |
| hypothetical protein (chloroplast) | Chlorophyta | Prasiola crispa | 68.6 | 68.6 | 42% | 4.00E-09 | 27% | AKZ21082.1 | |
| hypothetical protein (chloroplast) | Chlorophyta | Sarcinofilum mucosum | 65.9 | 65.9 | 28% | 5.00E-09 | 35% | YP_009367460.1 | |
| hypothetical protein (chloroplast) | Chlorophyta | Ostreobium sp. HV05007a | 67.4 | 67.4 | 28% | 1.00E-08 | 35% | ARQ82113.1 | |
| hypothetical protein (chloroplast) | Chlorophyta | Gloeotilopsis planctonica | 67.8 | 67.8 | 37% | 2.00E-08 | 29% | AOC61661.1 | |
| ORF531 | hypothetical protein | Cyanobacteria | Pleurocapsa sp. PCC 7319 | 78.6 | 78.6 | 52% | 2.00E-11 | 27% | WP_019503236.1 |
| hypothetical protein | Cyanobacteria | Chondrocystis sp. NIES-4102 | 75.1 | 75.1 | 50% | 2.00E-10 | 27% | WP_096724718.1 | |
| DUF3854 domain-containing protein | Cyanobacteria | Cyanothece sp. CCY0110 | 70.9 | 70.9 | 49% | 4.00E-09 | 28% | WP_008277431.1 | |
| DUF3854 domain-containing protein | Cyanobacteria | Cyanothece sp. CCY0110 | 70.1 | 70.1 | 49% | 7.00E-09 | 29% | WP_008277548.1 | |
| DUF3854 domain-containing protein | Cyanobacteria | Cyanothece sp. PCC 7822 | 69.3 | 69.3 | 21% | 9.00E-09 | 36% | WP_049802779.1 | |
| hypothetical protein | Cyanobacteria | Myxosarcina sp. GI1 | 69.3 | 69.3 | 50% | 1.00E-08 | 25% | WP_052055931.1 | |
| DUF3854 domain-containing protein | Cyanobacteria | Crocosphaera watsonii | 69.3 | 69.3 | 49% | 1.00E-08 | 29% | WP_007304689.1 | |
| DUF3854 domain-containing protein | Cyanobacteria | Tolypothrix bouteillei | 68.9 | 68.9 | 49% | 2.00E-08 | 28% | WP_050044965.1 | |
| hypothetical protein | Cyanobacteria | Pleurocapsa sp. CCALA 161 | 68.2 | 68.2 | 50% | 3.00E-08 | 27% | WP_106238468.1 | |
| DNA primase | Cyanobacteria | Crocosphaera watsonii WH 0402 | 65.5 | 65.5 | 25% | 5.00E-08 | 36% | CCQ65996.1 | |
| ATPase | Cyanobacteria | Aphanothece hegewaldii | 67 | 67 | 48% | 7.00E-08 | 28% | WP_106459345.1 | |
| MULTISPECIES: DUF3854 domain-containing protein | Cyanobacteria | Cyanothece | 66.2 | 66.2 | 36% | 1.00E-07 | 31% | WP_009547941.1 | |
| DUF3854 domain-containing protein | Cyanobacteria | Crocosphaera watsonii | 66.2 | 66.2 | 50% | 1.00E-07 | 26% | WP_007310072.1 | |
| hypothetical protein | Cyanobacteria | Myxosarcina sp. GI1 | 65.9 | 65.9 | 50% | 1.00E-07 | 25% | WP_052055870.1 | |
| DNA primase | Cyanobacteria | Aphanothece hegewaldii | 65.9 | 65.9 | 50% | 2.00E-07 | 24% | WP_106459560.1 | |
| hypothetical protein | Cyanobacteria | Myxosarcina sp. GI1 | 65.5 | 65.5 | 46% | 2.00E-07 | 26% | WP_052056024.1 | |
| DUF3854 domain-containing protein | Cyanobacteria | Cyanothece sp. CCY0110 | 64.7 | 64.7 | 34% | 4.00E-07 | 31% | WP_008278684.1 | |
| hypothetical protein TrlaMp60 | Streptophyta | Treubia lacunosa | 62.4 | 62.4 | 22% | 7.00E-07 | 37% | YP_004927707.1 | |
| hypothetical protein | Firmicutes | Tumebacillus sp. AR23208 | 61.6 | 61.6 | 24% | 1.00E-06 | 27% | WP_087457668.1 | |
| DUF3854 domain-containing protein | Cyanobacteria | Cyanothece sp. CCY0110 | 63.2 | 63.2 | 43% | 1.00E-06 | 27% | WP_008276837.1 | |
| hypothetical protein | Cyanobacteria | Myxosarcina sp. GI1 | 62.4 | 62.4 | 50% | 2.00E-06 | 25% | WP_052056026.1 | |
| hypothetical protein BWK76_02530 | Proteobacteria | Desulfobulbaceae bacterium A2 | 61.6 | 61.6 | 28% | 3.00E-06 | 30% | OQX20052.1 | |
| hypothetical protein | Cyanobacteria | Myxosarcina sp. GI1 | 61.2 | 61.2 | 50% | 4.00E-06 | 24% | WP_052055874.1 | |
| hypothetical protein | Firmicutes | Lachnospiraceae bacterium | 60.8 | 60.8 | 46% | 4.00E-06 | 22% | WP_099450353.1 | |
| hypothetical protein | Cyanobacteria | Myxosarcina sp. GI1 | 59.7 | 59.7 | 50% | 1.00E-05 | 24% | WP_052055951.1 | |
| hypothetical protein C7H79_02365 | Proteobacteria | Nitrosomonas sp. APG5 | 58.5 | 58.5 | 34% | 2.00E-05 | 31% | PSJ18450.1 | |
| hypothetical protein | Actinobacteri | Streptomyces pini | 58.5 | 58.5 | 30% | 3.00E-05 | 25% | WP_093850669.1 | |
| hypothetical protein | Cyanobacteria | Myxosarcina sp. GI1 | 58.2 | 58.2 | 50% | 4.00E-05 | 24% | WP_052056112.1 | |
| hypothetical protein | Actinobacteri | Mycobacterium szulgai | 58.2 | 58.2 | 36% | 4.00E-05 | 24% | WP_082965783.1 | |
| ATP-binding protein | Actinobacteri | Streptomyces coelicolor | 57.8 | 57.8 | 30% | 4.00E-05 | 25% | WP_011030338.1 | |
| hypothetical protein A5657_18130 | Actinobacteri | Mycobacterium szulgai | 57.8 | 57.8 | 36% | 5.00E-05 | 24% | OBK51436.1 | |
| hypothetical protein CBD94_01510 | Proteobacteria | Gammaproteobacteria bacterium TMED234 | 55.5 | 55.5 | 49% | 3.00E-04 | 26% | OUW91419.1 | |
| phage/plasmid primase P4 | Cyanobacteria | Stanieria cyanosphaera | 54.7 | 54.7 | 50% | 4.00E-04 | 25% | WP_015193635.1 | |
| MULTISPECIES: hypothetical protein | Proteobacteria | Alteromonas | 54.7 | 54.7 | 28% | 5.00E-04 | 25% | WP_052010194.1 | |
| phage P4 DNA primase domain-containing protein | Cyanobacteria | Anabaena sp. 90 | 53.9 | 53.9 | 52% | 7.00E-04 | 29% | WP_015081293.1 | |
| hypothetical protein | Cyanobacteria | Nostoc sp. ‘Peltigera malacea cyanobiont’ DB3992 | 53.9 | 53.9 | 30% | 8.00E-04 | 29% | WP_099101112.1 | |
| DNA primase | Firmicutes | Eubacterium aggregans | 53.1 | 53.1 | 34% | 0.001 | 29% | WP_090304657.1 | |
| phage/plasmid primase P4 family C-terminal domain containing protein | Proteobacteria | Desulfovibrio africanus | 53.5 | 53.5 | 20% | 0.001 | 29% | WP_005988925.1 | |
| primase | Firmicutes | Lactobacillus equicursoris | 53.5 | 53.5 | 28% | 0.001 | 23% | WP_008463426.1 | |
| DNA primase | Cyanobacteria | Crocosphaera watsonii WH 0401 | 52.8 | 52.8 | 49% | 0.001 | 22% | CCQ62642.1 | |
| hypothetical protein | Proteobacteria | Thiotrichales bacterium HS_08 | 53.1 | 53.1 | 46% | 0.001 | 24% | WP_103918394.1 | |
| primase | Firmicutes | Lactobacillus delbrueckii | 52.8 | 52.8 | 28% | 0.002 | 23% | WP_003622798.1 | |
| DUF3854 domain-containing protein | Cyanobacteria | Crocosphaera watsonii | 52.4 | 52.4 | 49% | 0.002 | 22% | WP_053074885.1 | |
| MULTISPECIES: hypothetical protein | Proteobacteria | Defluviimonas | 52.4 | 52.4 | 28% | 0.002 | 29% | WP_035839891.1 | |
| phage/plasmid primase P4 | Proteobacteria | Mesorhizobium ciceri | 51.6 | 51.6 | 25% | 0.003 | 25% | WP_013531683.1 | |
| hypothetical protein AMDU1_APLC00062G0028 | Euryarchaeota | Thermoplasmatales archaeon A-plasma | 51.6 | 51.6 | 18% | 0.004 | 34% | EQB70371.1 | |
| hypothetical protein BSZ19_16225 | Proteobacteria | Bradyrhizobium japonicum | 51.2 | 51.2 | 48% | 0.004 | 21% | OSJ33189.1 | |
| hypothetical protein | Cyanobacteria | Aphanizomenon flos-aquae | 51.2 | 51.2 | 52% | 0.004 | 28% | WP_027404306.1 | |
| primase | Proteobacteria | Desulfovibrio vulgaris | 51.6 | 51.6 | 22% | 0.004 | 26% | WP_010939463.1 | |
| primase | Proteobacteria | Desulfovibrio vulgaris | 51.6 | 51.6 | 22% | 0.004 | 26% | WP_011792015.1 | |
| hypothetical protein A5769_14235 | Actinobacteri | Mycobacterium intracellulare | 51.2 | 51.2 | 11% | 0.005 | 38% | OBG17368.1 | |
| hypothetical protein | Actinobacteri | Mycobacterium intracellulare | 51.2 | 51.2 | 11% | 0.005 | 38% | WP_081284074.1 | |
| hypothetical protein | Proteobacteria | Bradyrhizobium japonicum | 50.8 | 50.8 | 48% | 0.006 | 21% | WP_094184029.1 | |
| MULTISPECIES: DNA primase | Firmicutes | Lachnospiraceae | 50.8 | 50.8 | 33% | 0.007 | 27% | WP_066730774.1 | |
| hypothetical protein | Proteobacteria | Sandarakinorhabdus sp. AAP62 | 50.1 | 50.1 | 24% | 0.008 | 22% | WP_017667662.1 | |
| hypothetical protein | Firmicutes | Lachnospiraceae bacterium TWA4 | 50.4 | 50.4 | 34% | 0.009 | 27% | WP_082039423.1 | |
| phage/plasmid primase, P4 family | Firmicutes | Lachnospiraceae bacterium TWA4 | 50.1 | 50.1 | 34% | 0.01 | 27% | KIR03447.1 |
The first question regarding these six ORFs is whether or not they were created through the shuffling process of endogenous sequences in the plastome. The mitogenomes of land plants rapidly evolve structurally36, and direct repeats and inverted repeats have served as good tools for rearrangement37,38. Additionally, double-strand break repairs with non-homologous end-joining affect the dynamic mitogenomic variation39. As a result, novel chimeric ORFs generated by the shuffling process have been reported in the mitogenomes of land plants40,41. However, in contrast with mitogenome, rearrangements of plastome have been generally restricted in land plants1, especially in eusporangiate ferns42. Therefore, we ruled out the possibility of the six ORFs having been generated by the shuffling process of plastome sequences.
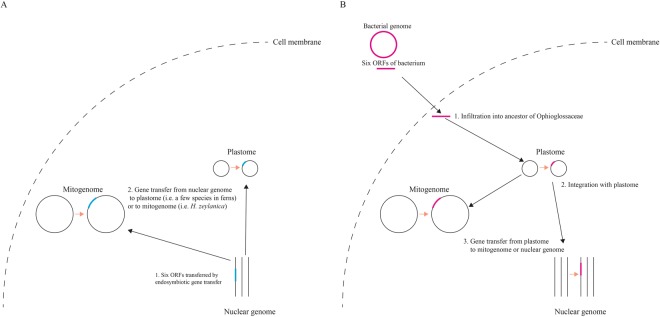
The second question is where the six ORFs region originated from prior to translocation to M. chejuense (or Ophioglossaceae). Gene transfer from other genomes, such as mitogenome or nuclear genome to plastome, were previously thought to occur extremely rarely if at all43; however, recently reported gene transfers from mitogenome to plastome6 have suggested the possibility of gene transfers from nuclear genome to plastome. Knox44 proposed that large ORFs in the plastomes of 51 species belonging to the Campanulaceae sensu lato arose from nuclear genome and Spooner, et al.4 provided the first evidence of a known nuclear sequence transferred into plastome. Martin, et al.45 revealed that massive EGTs have occurred during the evolution from plastid to nucleus in land plants. This implies that plant nuclear genome contains many genes which are orthologous to bacterial genes. Therefore, if the six ORFs region in the plastome of M. chejuense was transferred from the nuclear genome of M. chejuense, they could be similar to the ancient DNAs which remain in green algae or bacteria but not in the plastomes of land plants (Fig. 4A). In addition, as the structures of plastome in land plants have been very conserved throughout evolution with the exception of certain lineages1, it is likely that the translocated six ORFs region in the plastome of M. chejuense keeps its structure.
Figure 4.
Integration models of six ORFs in Ophioglossaceae. (A) Ancient endosymbiotic gene transfer model. (B) Recent horizontal gene transfer model.
Another possible scenario for six ORFs is gene transfer from bacteria to the ancestor of Ophioglossaceae or directly to more ancient clade of ferns (Fig. 4B). Although a few nuclear genome sequences have been reported in land plants and most of them belonged to angiosperms46,47 (https://www.ncbi.nlm.nih.gov/genome), the six ORFs were only similar to the plastomes of very few fern species and the mitogenomes of H. zeylanica. In addition, the amino acid sequences of ORF436 and ORF531 were more similar to the genes of green algae or bacteria, which are distantly related to ferns, than to these of land plants. So far, many HGTs in land plants have been reported, and bacteria, fungi, and viruses have been agents of HGT in certain cases48,49. HGT from bacteria to eukaryotes has been detected in yeast50 along with that from bacteria to organelle51. Therefore, it is conceivable that the six ORFs result from HGT from bacteria to ancestor of Ophioglossaceae or more ancient clade of ferns.
The phylogenetic relationships among genera in eusporangiate ferns
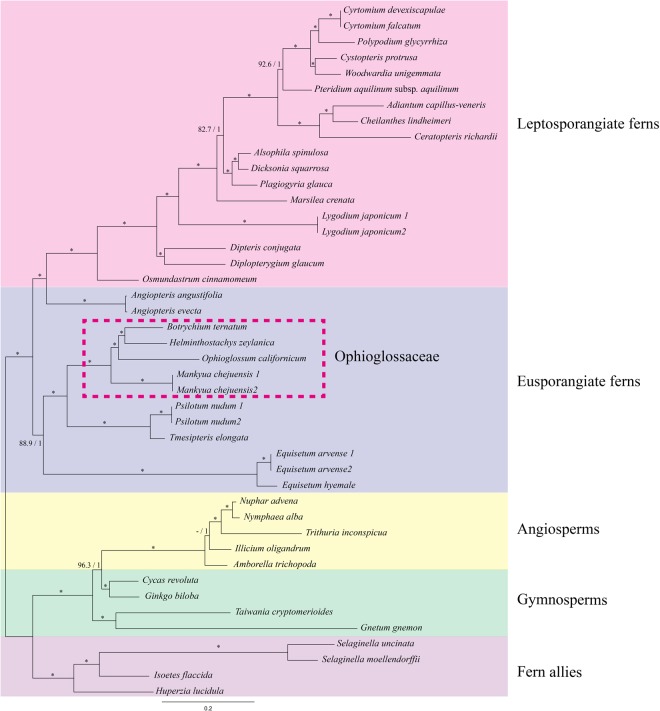
Eusporangiate ferns consist of four major families: Equisetaceae, Ophioglossaceae, Psilotaceae, and Marratiaceae. The generic relationships of eusporangiate ferns have been relatively well resolved by previous studies, except for those of the family Ophioglossaceae. Therefore, our phylogenetic study is focused on the family Ophioglossaceae. Four genera of Ophioglossaceae have different distribution patterns. Both Botrychium and Ophioglossum have cosmopolitan distributions52. H. zeilanica are distributed in Asia from India and Ceyon to South China, Taiwan, and tropical Australia52, but M. chejuense is distributed only in Jeju Island of South Korea24, specifically in twenty areas called “Gotjawal” created by volcanic activity. The trophophore of Mankyua is similar to that of Helminthostachys, but its sporophore is similar to that of Ophioglossum. In addition, Mankyua and Ophioglossum have subterranean vegetative reproduction24. Even though these intermediate features of Mankyua confused its phylogenetic position in Ophioglossaceae, phylogenetic analysis containing Mankyua is rare. Sun, et al.27 presented that Ophioglossum is the sister of Mankyua + Helminthostachys + Botrychium through parsimony analysis using rbcL data, and Shinohara, et al.28 suggested two different phylogenetic position of Mankyua by ML and Bayesian analysis using rbcL and matK. However, the bootstrap values for the clade comprised of more than two genera that were still under 90% according to previous studies.
The phylogenetic relationships among all species used in this paper were almost identical between ML and Bayesian analysis (Fig. 5). Only the topology of ((Amborella, Illicium), (Trithuria (Nuphur, Nymphaea)) was supported as being stronger than that of (Amborella (Illicium (Trithuria (Nuphur, Nymphaea)) in terms of bootstrap value under ML analysis. Eusporangiate ferns, except for the Angiopteris, were monophyly with strong supports, and Ophioglossaceae also formed a clade. The phylogenetic relationships among the four genera in Ophioglossaceae in this study are completely different from those of Sun, et al.27 and Shinohara, et al.28. Mankyua was firstly diverged from a common ancestor of Ophioglossaceae, and then Ophioglossum was subsequently diverged from a common ancestor of Helminthostachys and Botrychium. Finally, Helminthostachys diverged from a sister group with Botrychium. The phylogenetic relationships of Ophioglossum, Helminthostachys, and Botrychium and not for Mankyua correspond with those described in Hauk, et al.26. They described that the ophioglossoid (Ophioglossum s.l.) and botrychioid (Helminthostachys + Botrychium s.l.) diverged relatively early in the evolutional history of the Ophioglossaceae.
Figure 5.
Phylogeny of eusporangiate ferns using 44 complete plastome sequences of ferns and its relatives. Numbers on the branches refer to ML bootstrap/Bayesian posterior probability. Dash and star stand for less than 50/0.5 and 100/1.0, respectively.
Considering molecular phylogenetic analysis and the morphological characters of Ophioglossaceae, it seems that the ancestor of Ophioglossaceae have linear, fleshy spikes and vegetative reproduction. The ophioglossoid derived from a common ancestor have specialized their own trophophore and botrychioid have kept their trophophore and have specialized their own sporophore. In addition, the longitudinally dehiscent of sporangium in H. zeylanica was not plesiomorphic but apomorphic characteristic.
Materials and Methods
Plants materials and DNA extraction
H. zeylanica, M. chejuense, and B. ternatum were sampled at Cambodia and Jeju Island, Korea. The voucher specimens were deposited in the Korea University herbarium (KUS, K.-J. Kim et al., TCA2009-0806; K.-J. Jo et al., 2012–0028; K.-J. Kim et al., 2011–1638; Kim et al., 2012–0053). Total genomic DNA was extracted from fresh leaves using the CTAB method53. The DNA was purified using ultra-centrifugation in a cesium chloride/ethidium bromide gradient, then further purified by dialysis54.
Sequencing of the plastome of M. chejuense by PCR method and assembling
The total genomic DNA of M. chejuense was PCR-amplified in order to construct a plastome map using a series of primer sets designed based on three plastome sequences of Psilotum nudum, Adianthum capillus-veneris, and Angiopteris evecta16,55. Both the long-range PCR method and normal PCR method were employed using overlapping primer sets. The PCR condition for long range amplification was as follows: initial denaturation step for 4 min at 94 °C, then 35 cycle amplifications consisting of 30 sec denaturation at 94 °C, 30 sec annealing at 53~65 °C, and about 1 min/kb extension at 68 °C, followed by an extension period of 7 min at 72 °C. The PCR condition for normal amplification was as follows: initial denaturation step for 4 min at 94 °C, then 35 cycle amplifications consisting of 30 sec denaturation at 94 °C, 30 sec annealing at 47~52 °C, and about 2 min extension at 72 °C, followed by an extension period of 3 min at 72 °C. The PCR products were purified with the MEGAquick-spin kit (iNtRON, Seoul, Korea) and the cleaned products were sequenced in both directions using an ABI 3730XL automatic sequencer. Sequence contigs were assembled using Sequencher 4.7 (Gene Code Corporation, Ann Arbor, MI, USA).
Sequencing of the plastomes of H. zeylanica, M. chejuense, and B. ternatum by NGS and assembling
The genomic DNAs of H. zeylanica, M. chejuense, and B. ternatum were sequenced using MiSeq (Illumina, San Diego, CA, USA) (Supplementary Table S3). The raw reads were trimmed by trimmomatic 0.3656 with LEADING:10 (trimming the leading sequences until quality >10), TRAILING:10 (trimming the trailing sequences until quality >10), SLIDINGWINDOW:4:20 (trimming the window of size four for reads with the average quality less than 20), and MINLEN:50 (removing reads less than 50 bp in length). We followed the assembly method described by Kim, et al.57 using the plastome sequences of O. californicum18 and M. chejuense (NC017006) sequenced through PCR in this paper. Certain regions with low coverages caused by simple sequence repeats were verified using PCR.
Gene annotation
Genes in four plastomes were annotated compared with previously reported plastomes in eusporangiate ferns based on similarity. Coding genes and tRNAs were re-checked by blastp58 and tRNAscan-SE59. ORFs were annotated using with >303 bp in length.
Analyses of six ORFs in Ophioglossaceae
Six ORFs of M. chejuense were searched using blastn with 11 word size and 10−5 e-value and blastp with 3 word size and 10−2 e-value58 in order to investigate the homology with previously reported sequences in GenBank. In order to investigate the translocation of six ORFs into other genomes like nuclear or mitochondrial genome (mitogenome), three NGS raw data were de novo assembled using megahit60 and contigs were hit to six ORFs using blastn58. Mauve61 and Circoletto62,63 were used to visualize sequence similarity between six ORFs contigs in Ophioglossaceae.
Phylogenetic relationships among four genera in Ophioglossaceae
The 44 complete plastome sequences of ferns and their relatives were used to resolve the unclear intergeneric relationships in Ophioglossaceae (Supplementary Table S4). Eighty-four protein coding genes were extracted from each plastome. Each gene was aligned by MAFFT64 and 84 aligned genes were concatenated into a single aligned sequence.
The best-fit nucleotide substitution models for each gene position in a single concatenated sequence were evaluated using Partitionfinder V2.1.165,66. The maximum likelihood (ML) analysis was inferred by RAxML Black Box67 in CIPRES Science Gateway68 and Bayesian inference (BI) analysis was inferred by Mrbayes69 under GTR substitution model with gamma-distributed rate variation and a proportion of invariable sites (ngen = 1,000,000, samplefreq = 200, burninfrac = 0.25).
Electronic supplementary material
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) under grant nos. NRF-2015M3A9B8030588 to KJK and NRF-2018R1D1A3B07048213 to HTK. We thank two anonymous reviewers for helpful comments on the manuscript.
Author Contributions
H.T.K. performed the experiments and data analysis, and wrote a draft of the manuscript, and K.J.K. suggested the idea for the research, guided the experiment and data analysis, and wrote the final version of the manuscript.
Data Availability Statement
The complete sequence data generated during and/or analyzed during the current study are available in the NCBI GenBank repository. All data generated or analyzed during this study are included in this published article and its Supplementary Information files.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34825-6.
References
- 1.Wicke S, Schneeweiss GM, dePamphilis CW, Muller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gandini CL, Sanchez-Puerta MV. Foreign Plastid Sequences in Plant Mitochondria are Frequently Acquired Via Mitochondrion-to-Mitochondrion Horizontal Transfer. Sci. Rep. 2017;7:43402. doi: 10.1038/srep43402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goremykin VV, Salamini F, Velasco R, Viola R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol. Biol. Evol. 2009;26:99–110. doi: 10.1093/molbev/msn226. [DOI] [PubMed] [Google Scholar]
- 4.Spooner DM, Ruess H, Iorizzo M, Senalik D, Simon P. Entire plastid phylogeny of the carrot genus (Daucus, Apiaceae): Concordance with nuclear data and mitochondrial and nuclear DNA insertions to the plastid. Am. J. Bot. 2017;104:296–312. doi: 10.3732/ajb.1600415. [DOI] [PubMed] [Google Scholar]
- 5.Iorizzo M, et al. De novo assembly of the carrot mitochondrial genome using next generation sequencing of whole genomic DNA provides first evidence of DNA transfer into an angiosperm plastid genome. BMC Plant Biol. 2012;12:61. doi: 10.1186/1471-2229-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma PF, Zhang YX, Guo ZH, Li DZ. Evidence for horizontal transfer of mitochondrial DNA to the plastid genome in a bamboo genus. Sci. Rep. 2015;5:11608. doi: 10.1038/srep11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saarela Jeffery M., Wysocki William P., Barrett Craig F., Soreng Robert J., Davis Jerrold I., Clark Lynn G., Kelchner Scot A., Pires J. Chris, Edger Patrick P., Mayfield Dustin R., Duvall Melvin R. Plastid phylogenomics of the cool-season grass subfamily: clarification of relationships among early-diverging tribes. AoB Plants. 2015;7:plv046. doi: 10.1093/aobpla/plv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Straub SC, Cronn RC, Edwards C, Fishbein M, Liston A. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (apocynaceae) Genome Biol. Evol. 2013;5:1872–1885. doi: 10.1093/gbe/evt140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabah Samar O., Lee Chaehee, Hajrah Nahid H., Makki Rania M., Alharby Hesham F., Alhebshi Alawiah M., Sabir Jamal S.M., Jansen Robert K., Ruhlman Tracey A. Plastome Sequencing of Ten Nonmodel Crop Species Uncovers a Large Insertion of Mitochondrial DNA in Cashew. The Plant Genome. 2017;10(3):0. doi: 10.3835/plantgenome2017.03.0020. [DOI] [PubMed] [Google Scholar]
- 10.Bock R. The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci. 2010;15:11–22. doi: 10.1016/j.tplants.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Sheveleva EV, Hallick RB. Recent horizontal intron transfer to a chloroplast genome. Nucleic Acids Res. 2004;32:803–810. doi: 10.1093/nar/gkh225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards TA, et al. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell. 2009;21:1897–1911. doi: 10.1105/tpc.109.065805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei R, et al. Plastid Phylogenomics Resolve Deep Relationships among Eupolypod II Ferns with Rapid Radiation and Rate Heterogeneity. Genome Biol. Evol. 2017;9:1646–1657. doi: 10.1093/gbe/evx107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun MY, Li JR, Li D, Shi L. Complete chloroplast genome sequence of the medical fern Drynaria roosii and its phylogenetic analysis. Mitochondrial DNA B. 2017;2:7–8. doi: 10.1080/23802359.2016.1275835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labiak PH, Karol KG. Plastome sequences of an ancient fern lineage reveal remarkable changes in gene content and architecture. Am. J. Bot. 2017;104:1008–1018. doi: 10.3732/ajb.1700135. [DOI] [PubMed] [Google Scholar]
- 16.Roper, J. M. et al. The complete plastid genome sequence of Angiopteris evecta (G. Forst.) Hoffm. (Marattiaceae). American Fern Journal97, 95–106, doi:10.1640/0002-8444(2007)97[95:Tcpgso]2.0.Co;2 (2007).
- 17.Zhu A, Guo W, Gupta S, Fan W, Mower JP. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016;209:1747–1756. doi: 10.1111/nph.13743. [DOI] [PubMed] [Google Scholar]
- 18.Grewe F, Guo W, Gubbels EA, Hansen AK, Mower JP. Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evol. Biol. 2013;13:8. doi: 10.1186/1471-2148-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong B, Fong R, Collins LJ, McLenachan PA, Penny D. Two new fern chloroplasts and decelerated evolution linked to the long generation time in tree ferns. Genome Biol. Evol. 2014;6:1166–1173. doi: 10.1093/gbe/evu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karol KG, et al. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol. Biol. 2010;10:321. doi: 10.1186/1471-2148-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HT, Kim KJ. Chloroplast genome differences between Asian and American Equisetum arvense (Equisetaceae) and the origin of the hypervariable trnY-trnE intergenic spacer. PLoS One. 2014;9:e103898. doi: 10.1371/journal.pone.0103898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith AR, et al. A classification for extant ferns. Taxon. 2006;55:705–731. doi: 10.2307/25065646. [DOI] [Google Scholar]
- 23.Wagner, W. In Pteridophytes and gymnosperms 193–197 (Springer, 1990).
- 24.Sun BY, Kim MH, Kim CH, Park CW. Mankyua (Ophioglossaceae): a new fern genus from Cheju Island, Korea. Taxon. 2001;50:1019–1024. doi: 10.2307/1224718. [DOI] [Google Scholar]
- 25.Kim CH. Conservation status of the endemic fern Mankyua chejuense (Ophioglossaceae) on Cheju Island, Republic of Korea. Oryx. 2004;38:217–219. doi: 10.1017/S0030605304000377. [DOI] [Google Scholar]
- 26.Hauk WD, Parks CR, Chase MW. Phylogenetic studies of Ophioglossaceae: evidence from rbcL and trnL-F plastid DNA sequences and morphology. Mol. Phylogenet. Evol. 2003;28:131–151. doi: 10.1016/S1055-7903(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 27.Sun B-Y, Baek T-G, Kim Y-D, Kim C-S. Phylogeny of the family Ophioglossaceae with special emphasis on genus Mankyua. Korean Journal of Plant Taxonomy. 2009;39:135–142. doi: 10.11110/kjpt.2009.39.3.135. [DOI] [Google Scholar]
- 28.Shinohara W, et al. The Use of matK in Ophioglossaceae Phylogeny and the Determination of Mankyua Chromosome Number Shed Light on Chromosome Number Evolution in Ophioglossaceae. Systematic Botany. 2013;38:564–570. doi: 10.1600/036364413x670232. [DOI] [Google Scholar]
- 29.Chung MY, et al. Extremely low levels of genetic variation in the critically endangered monotypic fern genus Mankyua chejuense (Ophioglossaceae) from Korea: Implications for conservation. Biochem. Syst. Ecol. 2010;38:888–896. doi: 10.1016/j.bse.2010.09.008. [DOI] [Google Scholar]
- 30.Martin W, Herrmann RG. Gene Transfer from Organelles to the Nucleus: How Much, What Happens, and Why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richly E, Leister D. NUPTs in sequenced eukaryotes and their genomic organization in relation to NUMTs. Mol. Biol. Evol. 2004;21:1972–1980. doi: 10.1093/molbev/msh210. [DOI] [PubMed] [Google Scholar]
- 32.Sorenson MD, Quinn TW. Numts: A challenge for avian systematics and population biology. Auk. 1998;115:214–221. doi: 10.2307/4089130. [DOI] [Google Scholar]
- 33.Bennett, M. & Leitch, I. (Royal Botanic Gardens Kew, 2005).
- 34.Bai C, Alverson WS, Follansbee A, Waller DM. New reports of nuclear DNA content for 407 vascular plant taxa from the United States. Ann Bot. 2012;110:1623–1629. doi: 10.1093/aob/mcs222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleine T, Maier UG, Leister D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 2009;60:115–138. doi: 10.1146/annurev.arplant.043008.092119. [DOI] [PubMed] [Google Scholar]
- 36.Palmer JD, Herbon LA. Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence. J. Mol. Evol. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- 37.Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr. Genet. 2004;46:123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 38.Lonsdale DM, Hodge TP, Fauron CM. The physical map and organisation of the mitochondrial genome from the fertile cytoplasm of maize. Nucleic Acids Res. 1984;12:9249–9261. doi: 10.1093/nar/12.24.9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davila JI, et al. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 2011;9:64. doi: 10.1186/1741-7007-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka Y, Tsuda M, Yasumoto K, Yamagishi H, Terachi T. A complete mitochondrial genome sequence of Ogura-type male-sterile cytoplasm and its comparative analysis with that of normal cytoplasm in radish (Raphanus sativus L.) BMC Genomics. 2012;13:352. doi: 10.1186/1471-2164-13-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jo YD, Choi Y, Kim DH, Kim BD, Kang BC. Extensive structural variations between mitochondrial genomes of CMS and normal peppers (Capsicum annuum L.) revealed by complete nucleotide sequencing. BMC Genomics. 2014;15:561. doi: 10.1186/1471-2164-15-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim HT, Chung MG, Kim KJ. Chloroplast genome evolution in early diverged leptosporangiate ferns. Mol. Cells. 2014;37:372–382. doi: 10.14348/molcells.2014.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leister D. Origin, evolution and genetic effects of nuclear insertions of organelle DNA. Trends Genet. 2005;21:655–663. doi: 10.1016/j.tig.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Knox EB. The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms. Proc. Natl. Acad. Sci. USA. 2014;111:11097–11102. doi: 10.1073/pnas.1403363111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neale, D. B. et al. The Douglas-fir genome sequence reveals specialization of the photosynthetic apparatus in Pinaceae. G3: Genes, Genomes, Genetics, g3. 300078.302017 (2017). [DOI] [PMC free article] [PubMed]
- 47.Li F-W, et al. Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nature plants. 2018;4:460. doi: 10.1038/s41477-018-0188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Won H, Renner SS. Horizontal gene transfer from flowering plants to Gnetum. Proc. Natl. Acad. Sci. USA. 2003;100:10824–10829. doi: 10.1073/pnas.1833775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 50.Hall C, Brachat S, Dietrich FS. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice DW, Palmer JD. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 2006;4:31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clausen RT. A monograph of the Ophioglossaceae. Memoirs of the Torrey Botanical Club. 1938;19:1–177. [Google Scholar]
- 53.Doyle JJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem bull. 1987;19:11–15. [Google Scholar]
- 54.Palmer JD. Isolation and structural analysis of chloroplast DNA. Methods Enzymol. 1986;118:167–186. doi: 10.1016/0076-6879(86)18072-4. [DOI] [Google Scholar]
- 55.Wolf PG, Rowe CA, Sinclair RB, Hasebe M. Complete nucleotide sequence of the chloroplast genome from a leptosporangiate fern, Adiantum capillus-veneris L. DNA Res. 2003;10:59–65. doi: 10.1093/dnares/10.2.59. [DOI] [PubMed] [Google Scholar]
- 56.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim HT, et al. Seven New Complete Plastome Sequences Reveal Rampant Independent Loss of the ndh Gene Family across Orchids and Associated Instability of the Inverted Repeat/Small Single-Copy Region Boundaries. PLoS One. 2015;10:e0142215. doi: 10.1371/journal.pone.0142215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 59.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 61.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darzentas N. Circoletto: visualizing sequence similarity with Circos. Bioinformatics. 2010;26:2620–2621. doi: 10.1093/bioinformatics/btq484. [DOI] [PubMed] [Google Scholar]
- 63.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 66.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 67.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller, M. A., Pfeiffer, W. & Schwartz, T. In Gateway Computing Environments Workshop. 1–8.
- 69.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete sequence data generated during and/or analyzed during the current study are available in the NCBI GenBank repository. All data generated or analyzed during this study are included in this published article and its Supplementary Information files.