Abstract
HER2 overexpression is identified on 20–30% breast cancer and other cancers at different levels. Although HER2 targeted monoclonal antibody combined with chemical drugs has shown improved outcomes in HER2 expressing patients, drug resistance and toxicity have limited their efficacy. To overcome drug resistance, cotargeting multiple HER receptors was proven to be effective. EGFR/HER2 dimerization can active PI3K/AKT pathway, and resistance to HER2-targeted drugs is associated with upregulation of EGFR. Here, we developed a novel HER2/EGFR targeted nucleic acid therapeutic to address current drug limits. The new therapeutic is constructed by fusing HER2 aptamer-EGFR siRNA sense strand with HER2 aptamer-EGFR siRNA antisense strand into one molecule: a bivalent HER2 aptamer-EGFR siRNA aptamer chimera (HEH). In breast cancer cell lines, HEH can be selectively taken up into HER2 expressing cells and successfully silence EGFR gene and down regulate HER2 expression. In breast cancer xenograft models, HEH is capable of triggering cell apoptosis, decreasing HER2 and EGFR expression, and suppressing tumor growth. The therapeutic efficacy of HEH is superior to HER2 aptamer only, which suggests that HEH has synergistic effect by targeting HER2 and EGFR. This study demonstrated that HEH has great potential as a new HER2 targeted drug to address toxicity and resistance of current drugs and may provide a cure for many HER2 positive cancers.
Keywords: aptamer, siRNA, HER2, targeted therapeutic, bivalent, siRNA delivery, synergistic treatment
Introduction
Overexpression of HER2 was found in 20–25% of breast tumor patients and associated with increased rate of relapse, poor prognosis, and short overall survival.1,2 HER2 is also overexpressed in other cancers like ovary,3 stomach,4 prostate,5 lung,6 colon,7 and head and neck.8 Although HER2 targeted therapies have significantly improved outcomes of HER2 overexpressing breast cancer, resistance to these therapies remains a clinical challenge. It has been identified that amplification of EGFR correlates with increased metastasis and trastuzumab resistance in breast cancer.9 Dual-targeting approaches by inhibiting EGFR and HER2 have shown remarkably higher antitumor activity than the administration of single agents. For example, trastuzumab in combination with lapatinib (a small molecule dual inhibitor of EGFR and HER2) has significantly improved efficacy in HER2-positive breast cancer,10,11 and synergy has been observed. It is well characterized that the HER family network influences tumor cell growth and response to therapy. Blockade of one HER receptor can often be functionally compensated by another HER family member.12,13 Combinatorial strategies to target multiple HER family members have been increasing interests in cancer treatment.14−16 Many therapies that target HER2 and other HER member receptors are in clinical development for HER2-positive metastatic breast cancer.17 As targeted therapies, monoclonal antibodies and tyrosine kinase inhibitors have significantly improved the life span of HER2-postive breast cancer patients. However, drug resistance and toxicity have hindered the further use these therapies.18−20 To overcome drug resistance and decrease toxicity, there is a compelling need to investigate new types of targeted therapy.
Aptamer-siRNA chimera (AsiC) has emerged as a new type of targeting therapeutics with low immunogenicity, ease of production, and tumor targeting capability.21,22 Aptamers, as nucleic acid antibodies, have shown the efficacy in vivo disease treatment.23,24 By using living cells as targets, cell-specific aptamer can be selected.25 Cell type- and receptor-specific aptamer not only can block cell surface receptors, but also can deliver therapeutic agents into cells.26 AsiCs can be generated by chemically synthesis27,28 or by in vitro transcription29 with low cost and less batch-to-batch variation compared with antibody production.
In this study, we have developed a bivalent HER2 aptamer-EGFR siRNA chimeras that can interfere the functions of HER2 and EGFR receptors and induce HER2 positive breast cancer cell apoptosis. In previous studies, we have developed a platform technology by using bivalent aptamer to deliver two siRNAs into prostate cancer.30 We have proved that bivalent aptamer has antibody-like properties and enables cross-linking cell surface receptors and inducing cell activation, thereby enhancing siRNA internalization. Built on established approach for bivalent aptamer-siRNA chimera construction, in this investigation, we have constructed a bivalent HER2 aptamer-EGFR siRNA chimera. The results demonstrated that new bivalent aptamer chimera is capable of effectively delivering EGFR siRNA into HER2 expressing cells and reducing both HER2 and EGFR protein expression. It is promising that the new chimera alone or by combination with other drugs will provide a new type of tumor targeted treatment for HER2 overexpression cancers.
Materials and Methods
Materials
Antibodies were from Cell Signaling Technology (Danvers, MA). Single stranded DNAs were synthesized by Integrated DNA Technologies (IDT, Coralville, IA). TranscriptAid T7 High Yield Transcription Kits were purchased from Thermo Fisher Scientific. PCR reagents were from Sigma- Aldrich (St Louis, MO). LysoTracker Green DND-26 and Alexa Fluor 488 Annexin V/Dead Cell Apoptosis kits were from life Technologies (Carlsbad, CA). 2′-Fluoro-2′-deoxycytidine-5′-triphosphate, 2′-fluoro-2′-deoxyuridine-5′-triphosphate, and Cy5-labeled 2′-fluoro-labeled aptamers were purchased from TriLink Biotechnologies (San Diego, CA). 2′-Fluoro-modified pyrimidines RNAs were ordered from GE Dharmacon (Chicago, IL).
Cell Culture
BT474, SKBR3, MDA-MB-231, MCF7, and Hs578 T cells were obtained from American Type Culture Collection (Manassas, VA). Cell lines were used within 6 months of receipt from ATCC or resuscitation after cryopreservation in early passages. ATCC uses short tandem repeat (STR) profiling for testing and authentication of cell lines. Cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 units/mL streptomycin and maintained at 37 C in a humidified incubator with 5% CO2.
Mouse
All animal studies were approved by the Institutional Animal Care and Use Committee at Augusta University. Athymic nu/nu mice were purchased from Envigo. All animal procedures and maintenance were conducted in accordance with the institution approved guidelines of Augusta University.
Aptamer-siRNA Chimera Synthesis
The ssDNA templates and primers were synthesized from IDT. For HEH chimera synthesis, two RNAs (RNA1 and RNA2) were generated separately.
RNA1: HER2 aptamer-EGFR sense siRNA.
RNA1 PCR template: 5′-AGCCGCGAGGGGAGGGATAGGGTAGGGCGCGGCTAAAACCTTAGCAGTCTTATCTAATT-3′.
RNA1 5′ primer: 5′-TAATACGACTCACTATAAGCCGCGAGGGGAGGGA-3′. The forward primer contains T7 RNA polymerase promoter site (bolded) (P1).
RNA1 3′primer: 5′-AATTAGATAAGACTGCTAAGGTTTTA-3′. (P2)
RNA2: HER2 aptamer-EGFR antisense siRNA.
RNA2 PCR template: 5′-AGCCGCGAGGGGAGGGATAGGGTAGGGCGCGGCTAAAATTAGATAAGACTGCTAAGGCA-3′.
RNA2 5′-primer: P1.
RNA2 3′-primer: 5′-TGCCTTAGCAGTCTTATCTAATTTTAGCCGCGCCCT-3′ (P3).
RNA1 and RNA2 were generated by in vitro transcription with PCR products as templates. The PCR products were put into T-A cloning pCR2.1 vector (Invitrogen) and sequenced. Transcription was performed with Transcript Aid T7 High Yield Transcription Kits. 2′ F-modified pyrimidines were incorporated into RNAs to replace CTP and UTP. The transcribed RNAs were purified with phenol/chloroform/isoamyl alcohol (25:24:1) (Sigma-Aldrich), precipitated with isopropanol (Sigma-Aldrich) followed by cold 70% ethanol wash. The RNA pellets were dissolved in nuclease free water (IDT). The purification procedures were used for all transcribed RNAs. RNA1 and RNA2 were mixed at a molar ratio of 1:1 and annealed to form one entity by heating at 94 °C for 3 min, followed by slowly cooling to room temperature.
For HER2 aptamer (RNA3) synthesis, RNA1 PCR template and RNA1 5′-primer will be used as the above sequences, and RNA3 3′-primer is 5′-AGCCGCGCCCTACCCTATCCCT-3′ (P4).
For mutant HEH3 synthesis, RNA4 and RNA5 will be separately synthesized and annealed together.
RNA4: mutant HER2 aptamer-EGFR sense siRNA.
RNA4 PCR template: 5′-AGCCAAACGAGGGGGGAGAGGGTGGGGGCGCCTGAAAACCTTAGCAGTCTTATCTAATT-3′.
RNA4 5′ primer: 5′-TAATACGACTCACTATAAGCCAAACGAGGGGGGAGAGGGT-3′ (P5). RNA4 3′ primer: 5′-AATTAGATAAGACTGCTAAGGTTTTCA-3′ (P6).
RNA 5: mutant HER2 aptamer-EGFR antisense siRNA.
RNA5 PCR template: 5′-AGCCAAACGAGGGGGGAGAGGGTGGGGGCGCCTGAAAATTAGATAAGACTGCTAAGGCA-3′.
RNA5 5′-primer: P5.
RNA5 3′-primer: 5′-TGCCTTAGCAGTCTTATCTAATTTTCA-3′ (P7).
For HER2 aptamer-scrambled siRNA synthesis, RNA6 and RNA7 will be separately synthesized and annealed together.
RNA 6: HER2-scrambled sense siRNA.
RNA 6 PCR template: 5′-AGCCAAACGAGGGGGGAGAGGGTGGGGGCGCCTGAAAAAACAGTCGCGTTTGCGACTGG-3′.
RNA 6 5′ primer: P5.
RNA6 3′ primer: 5′-CCAGTCGCAAACGCGACTGTTTTTTCA-3′.
RNA 7: HER2-scrambled antisense siRNA.
RNA 7 PCR template: 5′-AGCCAAACGAGGGGGGAGAGGGTGGGGGCGCCTGAAAACCAGTCGCAAACGCGACTGTT-3′.
RNA7 5′-primer: P5.
RNA7 3′primer: AACAGTCGCGTTTGCGACTGGTTTTCA-3′.
Western Blot Analysis
Whole-cell protein was extracted with RIPA lysis buffer containing 1x Halt Protease Inhibitor Cocktails and quantitated with Bio-Rad Protein Assay. Protein (100 μg per sample) was resolved on 10% SDS-PAGE and transferred to PVDF membrane. After blocking for 2 h at room temperature in 5% milk in TBS/0.1% Tween-20, membrane were incubated overnight at 4 °C with the indicated primary antibodies (HER2, EGFR, Cleaved Caspase-3, GAPDH, 1:1000 dilution, Cell signaling), followed by incubation with horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature. After ECL Western Blotting Substrate (Pierce) was added onto membrane, the signals were captured by the exposure to X-ray film.
Cytotoxicity Assay
Cellular cytotoxicity was quantified by measuring WST-8 formazan using Cell Counting. Cells in RPMI 1640 containing 5% fetal bovine serum were seeded into 96-well plate at a density 5 × 103 per well for 24 h at 37 °C, and then cells were treated with HEH (1 μM), HER2 aptamer (2 μM), EGFR siRNA (1 μM), HScH (1 μM), and muHEH (1 μM) for 72 h at quadruplicate wells without transfection reagents (e.g., Lipofectamine). CCK-8 solution (10 μ l) (Dojindo, Japan) was added to each well and incubated at 37 °C for 4 h. Absorbance at 450 nm was measured using a plate reader.
HEH Stability in Cell Culture Medium
2′ F-modied HEH (0.2 nmoles) were put into 40 μL of RPMI 1640 containing 5% fetal bovine serum, which was the condition for chimera treatment, for different time periods. RNA integrity was detected with 3% agarose gel electrophoresis. HEH intensity was measured with ImageJ.
qRT-PCR Assay
Total RNA from BT474 and MDA-MB-231 cells was extracted with RNAeasy plus kits (Qiagen). The quantity of RNAs was determined by NanoDrop. cDNA was generated with iScript cDNA synthesis kits (Bio-Rad). qRT-PCR analyses were performed using SYBR Green Master Mix (Bio-Rad) and further carried out on a CFX96 Real-Time System (Bio-Rad).
EGFR primers: forward 5′-CCATGCCTTTGAGAACCTAGAA-3′, and reverse 5′-GAGCGTAATCCCAAGGATGTTA-3′.
GAPDH primers: forward 5′-GGTGTGAACCATGAGAAGTATGA-3′, and reverse 5′-GAGTCCTTCCACGATACCAAAG-3′.
Detection of Apoptosis by Flow Cytometry
SKBR3 and BT474 cells were treated with HEH (1 μM) or HER2 apt (2 μM) or muHEH for 48 h and 72 h. The cells were harvested and washed in cold PBS. Cells were stained with Alexa Fluor 488 annexin V–PI solution for 1 h at room temperature. Cells were acquired by BD FACSCalibur and analyzed using BD FACStation software.
Cellular Uptake Assay with Laser Scanning Confocal Microscopy
Cells were seeded into 35 mm glass-bottom Petri dishes for 24 h in RPMI 1640 supplemented with 5% fetal bovine serum. Cy5-labeled HEH (1 μM), HER2 aptamer (2 μM), or EGFR siRNA (1 μM) was added into culture for 12 h at 37 °C. At the same time, LysoTracker Green DND-26 (80 nM) and DAPI (10 μg/mL) were added to the culture medium for imaging. Images were captured using confocal laser scanning microscope (Zeiss 780 inverted). The internalization of treatments was captured by “Z” stacking using an oil-immersion lens (63× magnification). Data were analyzed with Zeiss LSM image Browser.
Cell-type Binding Specificity
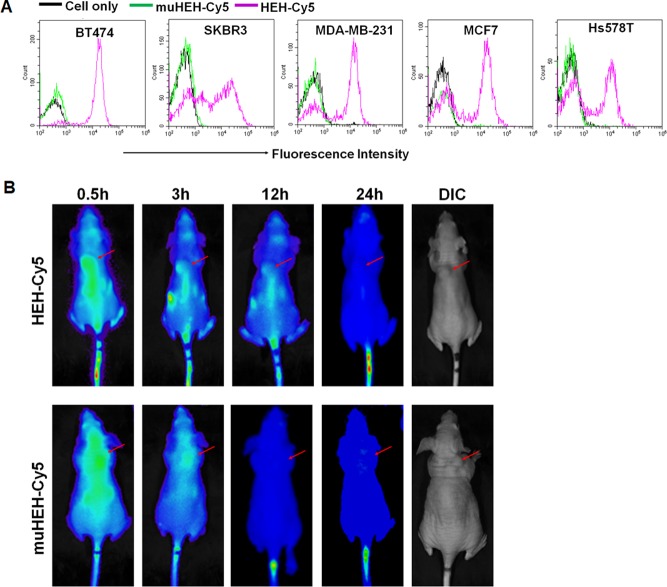
Cells including BT474, SKBR3, MDA-MB-231, MCF7, and Hs578T were grown and harvested. After washing, cells were incubated with Cy5-labeled HEH (2 μM) or Cy3-labeled muHEH (2 μM) in the presence of yeast tRNA (300 μ g/mL) and sperm DNA (500 μg/mL) for 1 h at 37 °C. Cell binding was detected using BD FACSCalibur flow cytometry.
Biodistribution Assay
Athymic nude female mice were implanted with 2 × 106 BT474 cells. After 4 weeks of implantation, tumor-bearing mice (n = 3 per group) were intravenously administered Cy5-labeled HEH (20 nmoles, 200 mL) or an equal mole amount of Cy5-labeled muHEH. The whole-body images were obtained at 0.5 h, 3 h, 12 h, and 24 h using the Xenogen IVIS100 imaging system by setting the wavelength at an excitation of 640 nm and emission at 710 nm.
Mouse Xenograft Models and Drug Administration
Athymic nude female mice (4–6-week old) were obtained from Envigo. BT474 cells (2 × 106) were injected into the flank of the mouse. Once tumor reached 100 mm3, mice were randomly divided into three groups (n = 4). Mice were i.p. injected with PBS (200 μL), HEH (10 nmoles, 200 μL), HER2 aptamer (20 nmoles, 200 μL) three times per week for 4 weeks. Tumor sizes were measured weekly and calculated with the formula V = (L × W2)/2 (W, width; L, length; V, volume). The animals were euthanized 2 days after the last treatment. The tumors and organs were removed.
Histology Assay
Tumor tissue samples and major organs were collected from xenografts and fixed in 10% neutral-buffered formalin and paraffin-embedded. Sections (6 μ m) were cut and mounted on the slides and then deparaffinized in xylene and ethyl alcohol. Each block has a section for HE staining. For immunohistochemistry assay, antigen retrieval was performed in 10 mmol/L citrate buffer (pH6) at 95 °C for 30 min, and then sections were incubated in 3% normal goat serum for 2 h and incubated with primary antibodies: caspase-3 (1:200), HER2 (1:800), and EGFR (1:50). After washing, the sections were incubated with biotinylated secondary antibody (1:200) (Vector Laboratories, Burlingame, CA) for 1 h. Following washing, the sections were incubated with VECTASTAIN ABC reagents for 30 min. The images were captured with a Nuance fluorescence microscope with a bright field imaging system.
Statistical Analysis
GraphPad Prism software was used to perform all statistical analyses. The results were expressed as a mean ± SE. All data were analyzed using two-tailed Student’s t test by comparing with the control group, and P < 0.05 was considered statistically significant.
Results
Synthesis of Bivalent HER2 Aptamer-EGFR siRNA Chimera and Comparison of Cytotoxicity
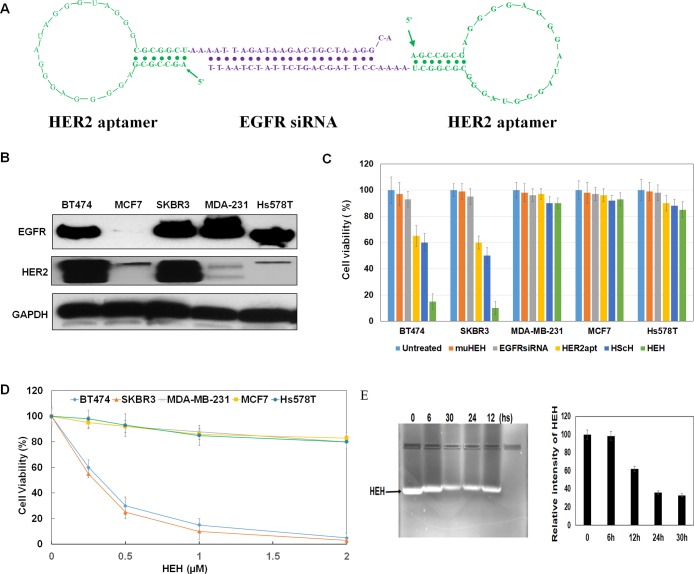
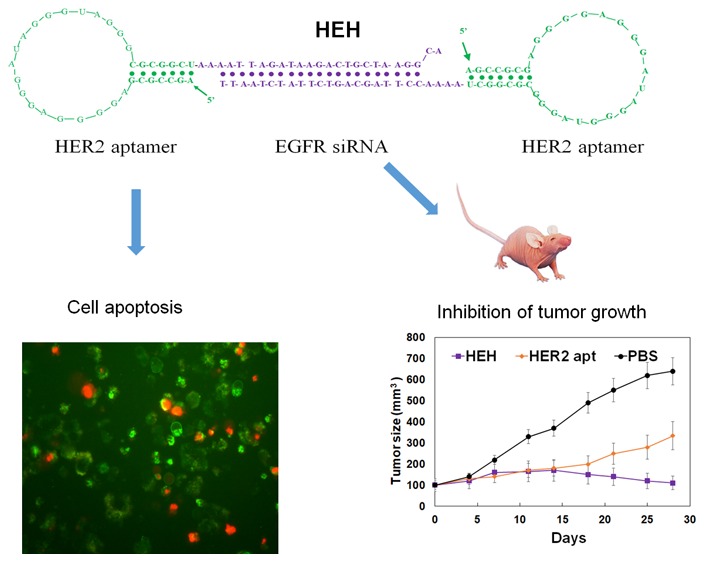
Bivalent aptamer chimera was constructed with our previous established methods.30 HER2 aptamer with 34 nucleic acids was selected by Jeong’s group31 and was used in this investigation. We have proven that this aptamer can specific bind to HER2 expressing tumors in previous studies.32 Briefly, HER2 aptamer containing EGFR sense strand was fused with HER2 aptamer containing EGFR antisense strand to construct a HER2 aptamer-EGFR siRNA-HER2 aptamer chimera (HEH) as shown in Figure 1A. Between aptamer and siRNA, 2–4 unpaired “A”s were added to maintain the flexibility of HER2 aptamer. 2′-Fluoro modified pyrimidines have been incorporated into RNA during transcription to enhance the resistance to nuclease degradation. As shown in Figure 1E, in cell culture medium, HEH has no detectable degradation after 6 h incubation, keeps over 60% integrity after 12 h, and still has over 30% integrity after 30 h incubation.
Figure 1.
Construction and characterization of HEH. (A) Schematic illustration of HER aptamer-EGFR siRNA-HER2 aptamer chimera (HEH). (B) Detection of HER2 and EGFR expression in breast cancer cell lines by Western blot. (C) Evaluation of the cytotoxicity of H2EH3. (D) Dose-dependent cytotoxicity assay of HEH on different breast cancer cell lines. Data are the mean ± SE from three independent experiments. (E) HEH stability in cell culture medium.
Next, the cytotoxicity of HEH was measured comparing with HER2 aptamer only. Prior to cytotoxicity detection, we have evaluated HER2 and EGFR expression among different breast cancer cell lines. As shown in Figure 1B, SKBR3 and BT474 show the high expression of HER2 and EGFR, and cell lines including MDA-MB-231 and Hs578T are HER2 low expression but with high EGFR expression. MCF7 cells are HER2 low and EGFR negative.
Furthermore, cytotoxicity was measured after treated with HEH (1.0 μM), HER2 aptamer (2.0 μM), mutant HEH (muHEH) (1.0 μM), HER2 aptamer-scrambled siRNA-HER2 aptamer (HScH) (1.0 μM), and EGFR siRNA (1.0 μM) in the above five cell lines. Because one HEH has two copies of HER2 aptamer and one copy of EGFR siRNA, we have adjusted aptamer and siRNA with the same amounts (i.e., moles) as HEH. After 72-h treatment, Cell Counting Kit-8 (CCK-8) reagents have been added into the cells. As shown in Figure 1C, HEH shows the significant cytotoxicity to HER2 expressing SKBR3 and BT474 cells, but not to HER2 negative MCF7, MDA-MB-231, and Hs578T, although both of MDA-MB-231 and Hs578T have high EGFR expression. HER2 aptamer only also shows cytotoxicity to SKBR3 and BT474 cells, but much less efficient than HEH. HER2 aptamer can reduce cell viability by 40% in SKBR3 cells and 35% in BT474 cells. However, HEH can reduce cell viability by 90% in SKBR3 cells and 85% in BT474 cells. Notably, EGFR siRNA and muHEH do not show any cytotoxicity to all detected cell lines. That is not surprising since EGFR siRNA without delivery vehicles cannot freely diffuse into cells and will not play gene silencing functionality outside cells. Because of loss of 3-D conformation of HER2 aptamer in muHEH, muHEH is inactive in binding HER2 receptor and thus lost the capabilities of blocking HER2 receptor and inducing EGFR siRNA internalization. Bivalent HER2 aptamer with scrambles siRNA (HScH) has shown the similar treatment effect as HER2 aptamer only since scrambled siRNA does not target any human genes. These results suggest that HEH have cell type- specific cytotoxicity, and bivalent HER2 aptamer-EGFR siRNA has synergistic effect on HER2 positive breast cancer and outperforms single-targeted HER2 aptamer only.
HEH Induced Cell Apoptosis
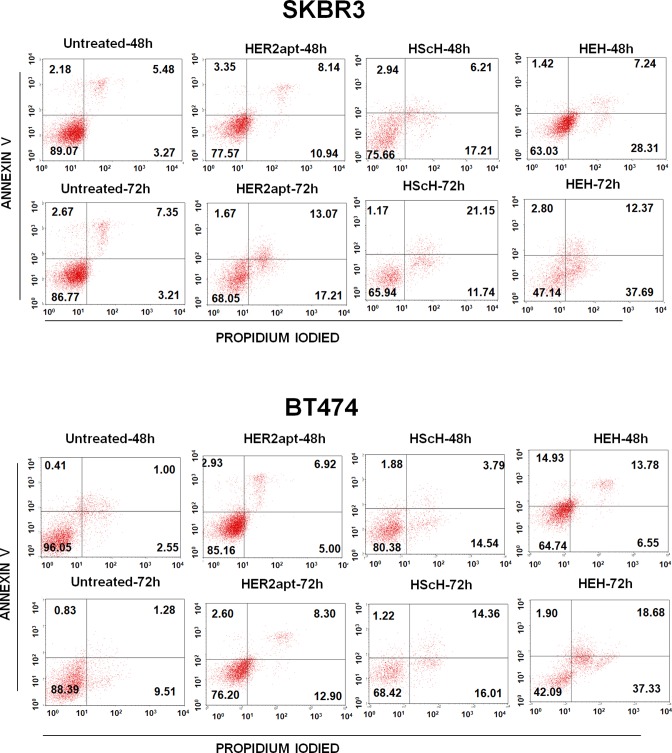
To test if HEH induced cytotoxicity is through triggering apoptosis, HER2 positive SKBR3 and BT474 cells were treated with HER2 aptamer (2 μM), HScH (1 μM), or HEH (1 μM) for 48 h and 72 h. Cell apoptosis and death were measured with Annexin V-Propidium Iodied (PI) staining. As shown in Figure 2, HER2 aptamer, HScH, and HEH can cause time-dependent cell apoptosis and followed by death in SKBR3 and BT474 cells. In SKBR3 cells, compared with untreated cells, upon treatment with HER2 aptamer alone, the rates of apoptotic and necrotic cells increased by 11.5% after 48 h and 16.04% after 72 h incubation, while upon treatment with HEH, apoptotic and necrotic cells increased by 18.72% after 48 h and 39.63% after 72 h incubation. Similar to SKBR3 cells, in BT474 cells, compared with untreated cells, upon treatment with HER2, apoptotic, and necrotic cells increased by 3.91% after 48 h and 24.33% after 72 h, whereas upon treatment with HEH, the rates increase by 12.19% in 48 h and 41.30% in 72 h. HScH showed the similar treatment efficacy as HER2 aptamer only. Overall, HER2 aptamer and HEH can induce cell apoptosis and death; HEH initiates higher levels of cell apoptosis and death than HER2 aptamer only or HScH in both cell lines.
Figure 2.
Detection of cell apoptosis and death by flow cytometry. BT474 and SKBR3 cells were treated with HEH, HScH, and HER2 aptamer for 48 h and 72 h, and then cells were stained with Alexa Fluor 488 Annexin V-Propidium Iodide and analyzed by flow cytometry.
HEH Can Be Internalized into HER2 Expressing Cells and Distribute Cross Cytoplasm
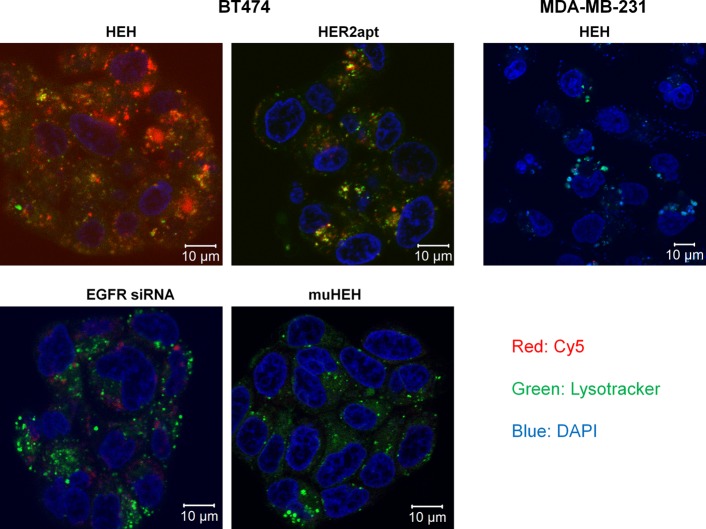
We expect that EGFR siRNA between two HER2 aptamers is able to silence EGFR gene. In previous studies,30 we have demonstrated that bivalent aptamer, like antibody, can cross-link cell surface receptors and active cells and thus induce significantly increased siRNA internalization compared with monovalent aptamer counterparts. To validate if bivalent HER2 aptamer has driven EGFR siRNA internalization and enables endosomal escape, which is the prerequisite of siRNA silencing, Co-focal microscopy was performed to evaluate internalization of Cy5-labled constructs. BT474 cells were treated with Cy5 labeled HEH, HER2 aptamer, muHEH, and EGFR siRNA. Nuclei were stained with DAPI (blue) and endosome/lysosomes were revealed by Lysotracker (green). After 12-h incubation, confocal laser scanning microscopy with Z-stack was performed to evaluate subcellular distribution of treatments. From Z-stack imaging shown in Figure3 and Supporting Information Figure S3, EGFR siRNA treated cells showed marginal amount red Cy5-EGFR siRNA into cells, whereas HEH treated cells showed significantly increased Cy5-HEH that distributes across entire cells. HER2 aptamer only also shows increased Cy5 signals compared with EGFR siRNA. Quantitatively, about 60% HEH (red fluorescence) has escaped from endosome entrapment, while about 40% HEH (yellow dots) is still entrapped in endosomes. Compared with bivalent HEH, HER2 aptamer shows less overall Cy5 signals than HEH but with similar percentage of endosomal escape as HEH. The results suggest that bivalent HEH indeed can induce cargo internalization and enables endosome escape. The z-stack imaging shows very low amount Cy5 signals in EGFR siRNA treated cells because naked EGFR siRNA cannot diffuse freely through cell membrane. In HEH and HER2 aptamer treated cells, Cy5 signals show puncta pattern, which is the feature of internalization through endocytosis. However, Cy5 signals from EGFR siRNA almost evenly distribute in cytoplasm, which means siRNA only has different uptake mechanisms from HER2 receptor mediated endocytosis. SiRNA only uptake has much lower efficiency than endocytosis. muHEH showed similar Cy5 pattern as EGFR siRNA only, which indicates mHEH can not enter to cells through endocytosis. This study also emphasizes that an effective carrier is indeed needed to aid siRNA cellular uptake and suggests that bivalent aptamer is a potent carrier for cell-type specific siRNA delivery.
Figure 3.
Detection of HEH internalization by Z-stack confocal microscopy. Cy5-labeled HEH, muHEH, HER2 aptamer, or EGFR siRNA was added into BT474 cells for 12 h at 37 °C. Lysotracker Green and DAPI were added into cells at the same time as the chimeras. LysoTracker Green was used to show lysosomes and endosomes. DAPI (blue) was used to display nucleus. Confocal laser scanning microscopy with z-stack was performed to show cell binding and internalization.
As a cell control, HER2 negative MDA-MD-231 cells were treated with Cy5-HEH; as shown in Figure 3 and Figure S3, there is very low amount of Cy5-HEH in cytoplasm, which suggests that HEH has a high cell-type specific uptake.
HEH Is Capable of Reducing Expression Levels of EGFR and HER2
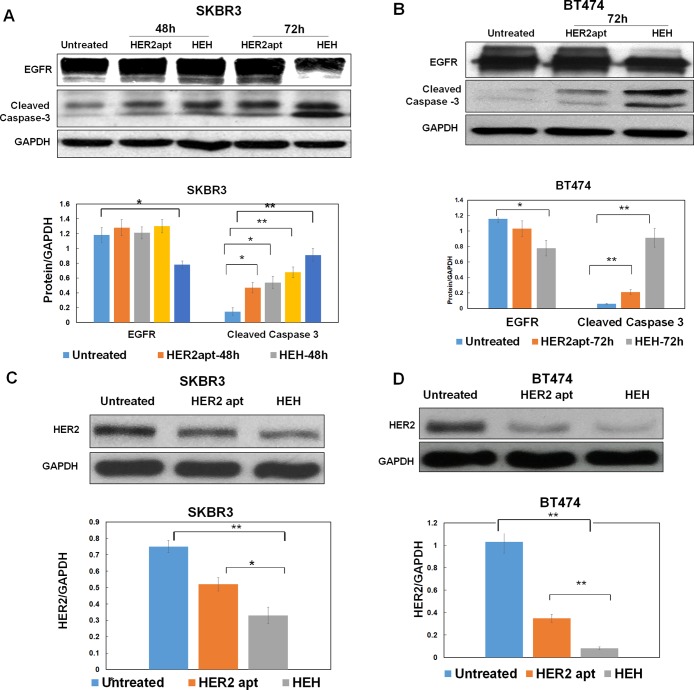
Furthermore, we identify if HEH has induced EGFR silencing and reduced EGFR protein level. Western blot was performed to evaluate EGFR protein expression. As shown in Figure 4A and B, HEH, but not HER2 aptamer, can reduce EGFR expression after 72 h treatment in SKBR3 and BT474 cells. HEH and HER2 aptamer can upregulate Cleaved Caspase-3 in SKBR3 and BT474 cells, while HEH showed higher levels of Cleaved Caspase-3 than HER aptamer only. Next, we tested if HER2 aptamer can behave like antibody. It has been well documented that antibody upon binding to its cell surface receptors can induce receptor-mediated endocytosis and initiate degradation of bound receptors.33,34 As shown in Figure 4C and D, after 72 h treatment, both HER2 aptamer and HEH indeed can significantly reduce HER2 protein expression. The results indicate that co-targeting EGFR and HER2 with HEH enables down regulation of both EGFR and HER2 and effective upregulation of apoptotic executioner Caspase-3, which has translated into inhibit cell growth and induce cell apoptosis on HER2 expressing cancer cell lines.
Figure 4.
Evaluation of protein levels by Western blot. (A) Detection of EGFR and Cleaved Caspase 3 in SKBR3 cells after treatment with HER2 aptamer and HEH for 48 h and 72 h. (B) Detection of EGFR and Cleaved Caspase 3 inBT474 cells after treament with HEH and HER2 aptamer for 72 h. (C) Detection of HER2 in SKBR3 cells after treatment with HER2 aptamer and HEH for 72 h (D) Detection of HER2 in BT474 cells after treatment with HER2 aptamer and HEH for 72 h Quantification of protein levels normalized by GPDH using ImageJ. The results are the mean ± SEM from three independent experiments. ∗P < 0.05, ∗∗P < 0.01.
Through qRT-PCT shown in Figure S2, we also proved that HEH can reduce EGFR mRNA in BT474 cells but not MDA-MB-231 cells. That suggests that HEH indeed can cell-type specific silence EGFR.
HEH Showed Cell Binding Specificity That Is Correlated with HER2 Protein Expression
To explore if HEH has different binding patterns on breast cancer cell lines, breast cancer cells were incubated with Cy5-HEH or Cy5-muHEH and detected with flow cytometry. As shown in Figure 5A, BT474 cells are all positive for HEH; SKBR3 has two populations: one is HEH negative and other one is HEH high expression. Interestingly, MDA-MB-231, MCF7, and Hs578 T also have two cell populations: one is HEH negative and other has certain amount HEH positive. As shown in Figure 1B, indeed, MDA-MB-231, MCF7, and Hs578 T express low amount HER2. Previous studies35 by immunohistochemistry have shown that MDA-MB-231, MCF7, and Hs578 T indeed express low HER2 but not null or absence of HER2. HEH can differentiate SKBR3, MDA-MB-231, MCF7, and Hs578T into two populations that may suggest bivalent aptamer is more sensitive to detect HER2 expressing cells.
Figure 5.
Cell binding specificity and biodistribution of HEH. (A) Evaluation of cell binding specificity by flow cytometry. HER2 positive and HER2 negative breast cancer cell lines were incubated with Cy5 labeled HEH or muHEH for 1 h at 37 °C, and detected with flow cytometry. Black line, cell only; green line, muHEH; light purple, HEH. (B) Biodistribution assay. Athymic female mice were implanted with BT474 cells. After 4 weeks, tumor bearing mice were i.v. injected with Cy5-HEH or Cy5-muHEH. Cy5 fluorescence of whole body was captured at the time points of 0.5 h, 3 h, 12 h, and 24 h using Xenogen IVIS100.
HEH Possesses High Tumor-Targeting Capability in Vivo
To explore tumor-targeting capability and biodistribution of HEH in vivo, tumor-bearing mice (BT474 cell derived xenografts) were i.v. injected with Cy5-HEH or Cy5-mHEH. Biodistribution was monitored with Xenogen IVIS 100 imaging system. As shown in Figure 5B, after 3 h, in HEH treated mice, Cy5 signals can be clearly visualized in tumor sites. The Cy5-HEH signals can last for 12 h in tumor sites. However, in muHEH treated mice, the signals of chimera in tumor site are not clear in 3 h and muHEH chimera has been cleared from the body after 12. The results showed that HEH has tumor targeting capability and its half-life time in vivo can reach 12 h. In HEH- and muHEH-treated groups, after 24-h injection, Cy5 signals disappeared in main bodies except tails. That also indicates that HEH and muHEH will have shorter deposit time in vivo and may cause less side effect on the normal organs than polymer-based siRNA delivery vehicles.
HEH Suppressed HER2 Expressing Breast Tumor Growth
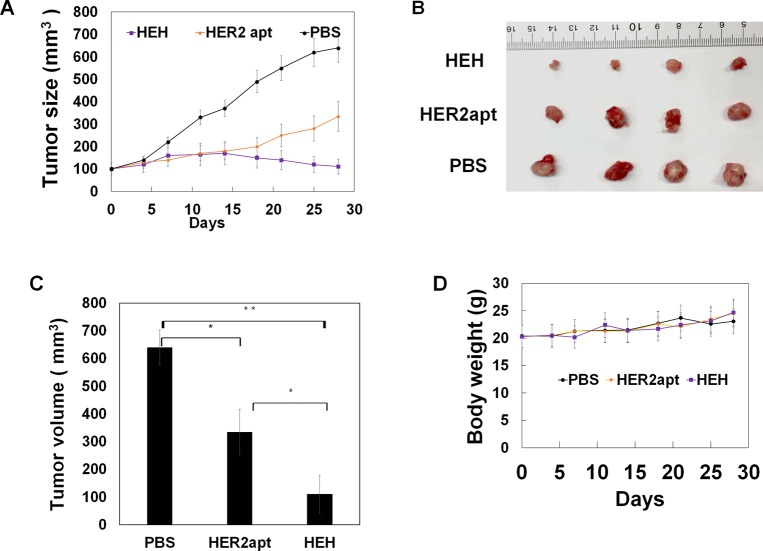
Because HEH can reduce protein expression of HER2 and EGFR and trigger cell apoptosis in vitro, we next evaluated if treatment efficacy can be demonstrated in vivo. BT474 cells were implanted into one flank of athymic nude female mice, after tumor size reach about 100 mm3, HEH (10 nmoles) was intraperitoneally injected into tumor bearing mice three times per week for 4 weeks. Tumor growth was measured weekly with digital caliper meter. As shown in Figure 6A–C, HEH treatment showed pronounced tumor growth inhibition compared with PBS- and HER2 aptamer-treated tumors. HEH treatment has achieved 5–6-fold reduction in tumor sizes compared with the PBS-treated tumors, and two-fold reduction compared with HER2 aptamer treated tumors. HER2 aptamer treatment has one-fold reduction of tumor size compare with PBS controls. These results suggest that cotargeting of HER2 and EGFR has synergistic efficacy in treating HER2 expressing tumors and is superior to HER2 single targeted treatment. Through time-course measurement of body weight (Figure 5D), there is no any changes after HEH and HER aptamer treatments compared with PBS controls.
Figure 6.
HEH inhibits tumor growth in BT474 breast cancer xenografts. Mice with subcutaneous tumors were i.p. injected with HEH and controls (PBS, HER2 apt) for 4 week. (A) Tumor growth curve. Tumor sizes were measured twice a week with digital calipers (n = 4). (B) Dissected tumors after treatment. (C) Quantitation of dissected tumor size from (B) (n = 4). (D) Body weight were measured and averaged (n = 4). ∗p < 0.05; ∗∗p < 0.005. Data represent the mean ± SEM.
HEH Is Capable of Reducing Protein Levels of HER2 and EGFR and Triggering Cell Apoptosis in Vivo
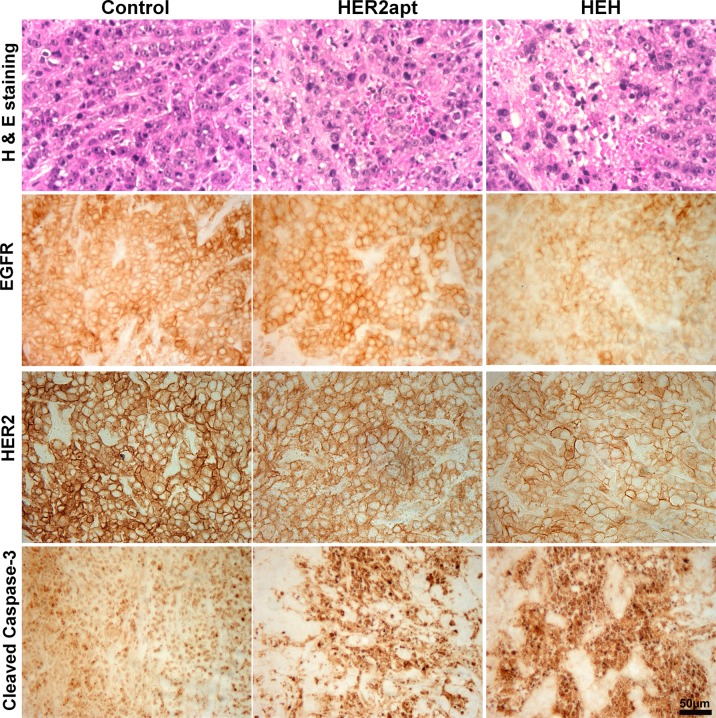
To confirm treatment efficacy, HE staining on excised tumors was performed. As shown in top panel of Figure 7, compared with PBS control, HEH treated tumors were highly vacuolated and contained highly condensed nucleus and cytoplasm. To validate if the observed histological alteration correlated with the occurrence of apoptosis, representative tumor samples were analyzed by IHC to evaluate apoptosis maker, Cleaved Caspase-3.
Figure 7.
Histology analysis of tumor and detection of biomarkers by immunohistochemistry. Formalin-fixed paraffin-embedded sections of xenograft tumors were analyzed with HE staining for detection of morphologic changes and IHC staining for detection of protein levels of EGFR, HER2, and Cleaved Caspase-3. Scale bar, 50 μm.
As shown in Figure 7 (bottom panel), the intensity of cleaved caspase-3 was much increased in HEH-treated tumors compared with PBS-and HER2 aptamer-treated groups. Furthermore, to verify if treatments have reduced HER2 and EGFR in vivo, tumors were examined by IHC staining for detection of HER2 and EGFR expression. As shown in the middle panels of Figure 7, HEH is capable of significantly reducing HER and EGFR in tumor tissue, while HER aptamer alone can reduce HER2 receptor but not EGFR receptor. These findings are consistent with in vitro results in Figure 4. These histology results suggest that HEH enables intervention of EGFR/HER2 concomitantly and inducing apoptosis in vivo, which is translated into significant suppression of tumor growth in xenograft models.
There Is No Detectable Systemic Toxicity after HEH Treatment in Xenografts
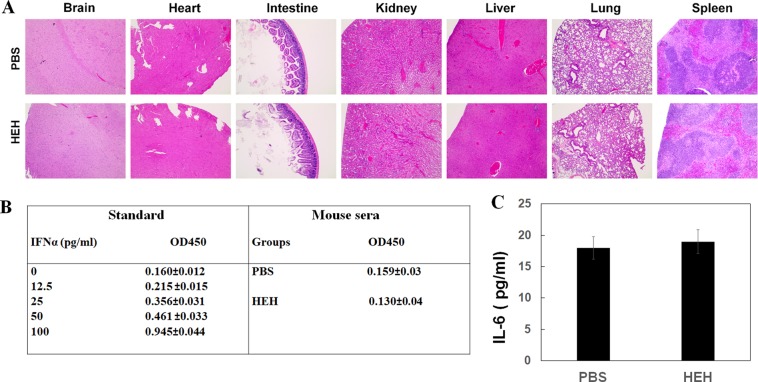
To evaluate potential systemic toxicity, after 4-week treatment, we have performed HE staining on major organs including brain, heart, intestine, kidney, liver, lung, and spleen. There is no obvious histological difference between PBS- and HEH-treated mice (Figure 8A). IFNα and IL-6 in mouse sera have been evaluated with ELISA shown in Figure 8B and C. No statistical difference was identified for IFNα and IL-6 between PBS and HEH groups. That indicates that HEH does not have acute toxicity and not trigger innate immune response. That is consistent with the evaluations from many RNA based chimeras in vivo applications.27,36
Figure 8.
Assessment of systemic toxicity of HEH. (A) Histological examination of organ damage after HEH treatment with HE staining. Detection of mouse serum (B) IFNα and (C) IL-6 with ELISA assay. The results are the mean ± SEM (N = 4).
Discussion
Overexpression of EGFR or HER2 has been found in a wide range of human tumors.37,38 Because of complex mechanisms of HER family signaling pathway activation both inherent and acquired, it is crucial to target multiple HER receptors to achieve synergetic effects compared with targeting a single receptor.15 Bifunctional small molecule tyrosine kinase inhibitor (TKI) Laptinib, which targets both EGFR and HER2, shows the enhanced therapeutic efficacy in HER2 positive breast cancer.39 Anti-HER2 antibody combinations also show the improved treatment efficacy than one type of HER2 antibody.40 It has been revealed that combination of trastuzumab and pertuzumab (both target on HER2) shows significantly increased therapeutic efficacy in trastuzumab-resistant patients with metastatic breast cancer,41 although pertuzumab does not have significant clinical activity as a single agent. To overcome drug resistance and intercept cancer pathway switch, combination treatment is essential and desirable. However, current chemical drugs including TKIs lack tumor targeting specificity and have high toxicity, while antibodies have high immunogenicity and high cost. The need remains for targeted therapeutic agents that are low toxic, low cost, and capable of blocking multiple tumor support pathways.
AsiCs are cell type specific, low immunogenicity, ease of production, thermostable, and less batch-to-batch variation.23,42 Therefore, AsiCs are emerging as a new potent targeted therapeutic. AsiC as a single entity can be easily administrated to patients and is expected to have less cost in passing regulatory approval than multiple-drug combination. In this study, we have used bivalent HER2 aptamer to deliver EGFR siRNA into HER2 positive cells with the approach we have established in previous studies in which bivalent PSMA aptamer has successfully delivered two siRNAs (target on EGFR and surivivn).30 We confirmed that bivalent HER2 aptamer, similar to bivalent PSMA aptamer, can effectively introduce siRNA into cytoplasm and silence target gene. This study shows that HEH has much better therapeutic efficacy than HER2 aptamer. This suggests that targeting of HER2 and EGFR has a synergistic effect on inhibition of HER2 expressing breast cancer growth.
Without delivery aid, free EGFR siRNA does not have gene silencing effect. By insertion into a chimera, an EGFR siRNA was positioned between two HER2 aptamers; in this way, EGFR siRNA can be delivered into cells and play silencing effect. Notably, EGFR siRNA in HEH plays dual functions: as a gene inhibitor and as an adaptor molecule. As an adaptor, EGFR siRNA bridges two single HER2 aptamer together that will increase overall chimera size. HEH molecular weight (MW) is 40.12 KDa, which is much larger than HER2 aptamer only (MW 11.56 kDa); therefore, HEH is supposed to have increased circulation time versus HER2 aptamer only. Within HEH, HER2 aptamer also plays dual functions: an antagonist against HER2 signaling pathway and as a carrier to deliver EGFR siRNA into HER2 expressing cells. In the xenograft treatment, HEH efficacy is superior to HER2 aptamer only, which may attribute longer circulation time of HEH. We will further test this hypothesis in the next experiments.
We did not see any evidence of toxicity or weight loss in HEH treated mice. Many studies have shown that nucleic acid-based AsiCs do not stimulate innate immunity and not elicit antibodies.36,43 Therefore, repeat administration may be allowed for AsiCs. Compared with antibody (150 kDa), HEH is much smaller and thus expected to have higher tissue penetration capability. With chemical modification, RNA serum stability can be easily improved by incorporation of 2′fluoro (F)-pyrimidines into RNAs during transcription. 2′ F-modified RNA can keep stable in circulation for 24–48 h.44 Clinical Phase I and II studies demonstrated that siRNA therapeutics can remain gene knockdown for several weeks in liver and have not shown unacceptable toxicity.45
These studies provide a new tool to target HER2 and EGFR with ease of production and low immunogenicity. By combining with inhibitors to HER346 or IGFR,47 or VEGF,48 HEH is promising to target a wide range of cancers with upregulated HER2 and EGFR. Compared with our previous chimera H2EH3 (HER2 aptamer-EGFR siRNA-HER3 aptamer), which depends on HER2 and HER3 expressing to deliver EGFR siRNA, in HEH, EGFR siRNA delivery will only need HER2 expression that may apply wider cell types than H2EH3. We envision HEH will be a new tool in the cancer-targeted therapeutic toolbox.
By using cell-type specific aptamer, AsiCs can avoid off-target effect and has potential to silence virtually any gene. Two major disadvantages of RNA therapeutics are sensitive to the nuclease degradation and rapid renal clearance due to small size. Fortunately, chemical modification has significantly enhanced the resistance to nuclease and basically solved the problem of serum stability.49 By adding PEG or cholesterol or designing bivalent aptamer, AsiC size can be increased and circulation time can easily reach 24 h to several days.36,50,51 To further improve gene silencing potency of a siRNA in the context of an AsiC, efforts should be put to enhance endosomal escape. With the progress in solving all problems related to aptamer application, AsiC is potential to treat inherently challenging diseases (cancer and HIV) and reduce side-effects inherent to traditional therapies. We envision that RNA-based therapeutics will play dominant roles in future medicine.
Acknowledgments
This work is supported by start-up funding of Augusta University. H.Y.L. thanks the NIH for an F32 fellowship (CA150301).
Glossary
Abbreviations
- HER2
human epidermal growth factor receptor 2
- EGFR
epidermal growth factor receptor
- AsiC
Aptamer-siRNA chimera
- muHEH
mutant HEH
- TKI
tyrosine kinase inhibitor
- IGFR
insulin-like growth factor
- VEGF
vascular endothelial growth factor
- MW
molecular weight
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.molpharmaceut.8b00388.
Annealing efficacy, EGFR mRNA levels, HEH internalization with Z-stack confocal microscopy (PDF)
Author Contributions
⊥ These authors contributed equally. H.Y.L. contributed to experimental design. L.X., X.Y., and H.Y.L. performed all experiments and interpreted data. H.Y.L., N.J.M., and S.C.T. wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Slamon D. J.; Clark G. M.; Wong S. G.; Levin W. J.; Ullrich A.; McGuire W. L. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 235, 177–182. 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Moasser M. M. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 2007, 26, 6469–6487. 10.1038/sj.onc.1210477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon D. J.; Godolphin W.; Jones L. A.; Holt J. A.; Wong S. G.; Keith D. E.; Levin W. J.; Stuart S. G.; Udove J.; Ullrich A. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989, 244, 707–712. 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Abrahao-Machado L. F.; Scapulatempo-Neto C. HER2 testing in gastric cancer: An update. World journal of gastroenterology: WJG 2016, 22, 4619–4625. 10.3748/wjg.v22.i19.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi N.; Salmaninejad A.; Ferdosi S.; Bajestani A. N.; Khaleghiyan M.; Estiar M. A.; Jamali M.; Nowroozi M. R.; Shakoori A. HER2 gene amplification in patients with prostate cancer: Evaluating a CISH-based method. Oncol. Lett. 2016, 12, 4651–4658. 10.3892/ol.2016.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G.; Vyberg M.; Melgaard B.; Askaa J.; Oster A.; O’Byrne K. J. Herceptest: HER2 expression and gene amplification in non-small cell lung cancer. Int. J. Cancer 2001, 92, 480–483. 10.1002/ijc.1214. [DOI] [PubMed] [Google Scholar]
- Kaneko M. K.; Yamada S.; Itai S.; Kato Y. Development of an Anti-HER2Monoclonal Antibody H2Mab-139 Against Colon Cancer. Monoclonal Antibodies Immunodiagn. Immunother. 2018, 37, 59–62. 10.1089/mab.2017.0052. [DOI] [PubMed] [Google Scholar]
- Pollock N. I.; Grandis J. R. HER2 as a therapeutic target in head and neck squamous cell carcinoma. Clin. Cancer Res. 2015, 21, 526–533. 10.1158/1078-0432.CCR-14-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.; He B.; Weber G. F. Growth factor signaling induces metastasis genes in transformed cells: molecular connection between Akt kinase and osteopontin in breast cancer. Mol. Cell. Biol. 2003, 23, 6507–6519. 10.1128/MCB.23.18.6507-6519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris H. A. 3rd; Hurwitz H. I.; Dees E. C.; Dowlati A.; Blackwell K. L.; O’Neil B.; Marcom P. K.; Ellis M. J.; Overmoyer B.; Jones S. F.; Harris J. L.; Smith D. A.; Koch K. M.; Stead A.; Mangum S.; Spector N. L. Phase I safety, pharmacokinetics, and clinical activity study of lapatinib (GW572016), a reversible dual inhibitor of epidermal growth factor receptor tyrosine kinases, in heavily pretreated patients with metastatic carcinomas. J. Clin. Oncol. 2005, 23, 5305–5313. 10.1200/JCO.2005.16.584. [DOI] [PubMed] [Google Scholar]
- Ross J. S.; Slodkowska E. A.; Symmans W. F.; Pusztai L.; Ravdin P. M.; Hortobagyi G. N. The HER-2 receptor and breast cancer: ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist 2009, 14, 320–368. 10.1634/theoncologist.2008-0230. [DOI] [PubMed] [Google Scholar]
- Howe L. R.; Brown P. H. Targeting the HER/EGFR/ErbB family to prevent breast cancer. Cancer Prev. Res. 2011, 4, 1149–1157. 10.1158/1940-6207.CAPR-11-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani E.; Alfieri R.; Giovannetti E.; Cavazzoni A.; La Monica S.; Galetti M.; Fumarola C.; Bonelli M.; Mor M.; Tiseo M.; Peters G. J.; Petronini P. G.; Ardizzoni A. Epidermal growth factor receptor tyrosine kinase inhibitors: current status and future perspectives in the development of novel irreversible inhibitors for the treatment of mutant non-small cell lung cancer. Curr. Pharm. Des. 2013, 19, 818–832. 10.2174/138161213804547222. [DOI] [PubMed] [Google Scholar]
- Shepard H. M.; Brdlik C. M.; Schreiber H. Signal integration: a framework for understanding the efficacy of therapeutics targeting the human EGFR family. J. Clin. Invest. 2008, 118, 3574–3581. 10.1172/JCI36049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A.; Vidal L.; Shaw H.; de Bono J. Dual inhibition of ErbB1 (EGFR/HER1) and ErbB2 (HER2/neu). Eur. J. Cancer 2007, 43, 481–489. 10.1016/j.ejca.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Blumenthal G. M.; Scher N. S.; Cortazar P.; Chattopadhyay S.; Tang S.; Song P.; Liu Q.; Ringgold K.; Pilaro A. M.; Tilley A.; King K. E.; Graham L.; Rellahan B. L.; Weinberg W. C.; Chi B.; Thomas C.; Hughes P.; Ibrahim A.; Justice R.; Pazdur R. First FDA approval of dual anti-HER2 regimen: pertuzumab in combination with trastuzumab and docetaxel for HER2-positive metastatic breast cancer. Clin. Cancer Res. 2013, 19, 4911–4916. 10.1158/1078-0432.CCR-13-1212. [DOI] [PubMed] [Google Scholar]
- Gradishar W. J. Emerging approaches for treating HER2-positive metastatic breast cancer beyond trastuzumab. Ann. Oncol 2013, 24, 2492–2500. 10.1093/annonc/mdt217. [DOI] [PubMed] [Google Scholar]
- Seidman A.; Hudis C.; Pierri M. K.; Shak S.; Paton V.; Ashby M.; Murphy M.; Stewart S. J.; Keefe D. Cardiac dysfunction in the trastuzumab clinical trials experience. J. Clin. Oncol. 2002, 20, 1215–1221. 10.1200/JCO.2002.20.5.1215. [DOI] [PubMed] [Google Scholar]
- Morris P. G.; Hudis C. A. Optimizing dose-dense regimens for early-stage breast cancer. Nat. Rev. Clin. Oncol. 2010, 7, 678–679. 10.1038/nrclinonc.2010.188. [DOI] [PubMed] [Google Scholar]
- Procter M.; Suter T. M.; de Azambuja E.; Dafni U.; van Dooren V.; Muehlbauer S.; Climent M. A.; Rechberger E.; Liu W. T.; Toi M.; Coombes R. C.; Dodwell D.; Pagani O.; Madrid J.; Hall M.; Chen S. C.; Focan C.; Muschol M.; van Veldhuisen D. J.; Piccart-Gebhart M. J. Longer-term assessment of trastuzumab-related cardiac adverse events in the Herceptin Adjuvant (HERA) trial. J. Clin. Oncol. 2010, 28, 3422–3428. 10.1200/JCO.2009.26.0463. [DOI] [PubMed] [Google Scholar]
- Keefe A. D.; Pai S.; Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discovery 2010, 9, 537–550. 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler L. A.; Trifonova R.; Vrbanac V.; Basar E.; McKernan S.; Xu Z.; Seung E.; Deruaz M.; Dudek T.; Einarsson J. I.; Yang L.; Allen T. M.; Luster A. D.; Tager A. M.; Dykxhoorn D. M.; Lieberman J. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J. Clin. Invest. 2011, 121, 2401–2412. 10.1172/JCI45876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. H.; Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discovery 2017, 16, 181–202. 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N.; Adamis A. P. Ten years of anti-vascular endothelial growth factor therapy. Nat. Rev. Drug Discovery 2016, 15, 385–403. 10.1038/nrd.2015.17. [DOI] [PubMed] [Google Scholar]
- Guo K. T.; Paul A.; Schichor C.; Ziemer G.; Wendel H. P. Cell-SELEX: Novel perspectives of aptamer-based therapeutics. Int. J. Mol. Sci. 2008, 9, 668–678. 10.3390/ijms9040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. H.; Rossi J. J. Cell-Specific Aptamer-Mediated Targeted Drug Delivery. Oligonucleotides 2011, 21, 1–10. 10.1089/oli.2010.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa-Geffen A.; Hamar P.; Le M. T.; Wheeler L. A.; Trifonova R.; Petrocca F.; Wittrup A.; Lieberman J. Gene Knockdown by EpCAM Aptamer-siRNA Chimeras Suppresses Epithelial Breast Cancers and Their Tumor-Initiating Cells. Mol. Cancer Ther. 2015, 14, 2279–2291. 10.1158/1535-7163.MCT-15-0201-T. [DOI] [PubMed] [Google Scholar]
- Esposito C. L.; Nuzzo S.; Catuogno S.; Romano S.; de Nigris F.; de Franciscis V. STAT3 Gene Silencing by Aptamer-siRNA Chimera as Selective Therapeutic for Glioblastoma. Mol. Ther.--Nucleic Acids 2018, 10, 398–411. 10.1016/j.omtn.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. H.; Lazar D.; Li H. T.; Xia X.; Satheesan S.; Charlins P.; O’Mealy D.; Akkina R.; Saayman S.; Weinberg M. S.; Rossi J. J.; Morris K. V. Receptor-targeted aptamer-siRNA conjugate-directed transcriptional regulation of HIV-1. Theranostics 2018, 8, 1575–1590. 10.7150/thno.23085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Y.; Yu X.; Liu H.; Wu D.; She J. X. Co-targeting EGFR and survivin with a bivalent aptamer-dual siRNA chimera effectively suppresses prostate cancer. Sci. Rep. 2016, 6, 30346. 10.1038/srep30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. Y.; Jeong S. In vitro selection of RNA aptamer and specific targeting of ErbB2 in breast cancer cells. Nucleic Acid Ther. 2011, 21, 173–178. 10.1089/nat.2011.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.; Ghamande S.; Liu H.; Xue L.; Zhao S.; Tan W.; Zhao L.; Tang S. C.; Wu D.; Korkaya H.; Maihle N. J.; Liu H. Y. Targeting EGFR/HER2/HER3 with a Three-in-One Aptamer-siRNA Chimera Confers Superior Activity against HER2(+) Breast Cancer. Mol. Ther.--Nucleic Acids 2018, 10, 317–330. 10.1016/j.omtn.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahta R. Molecular Mechanisms of Trastuzumab-Based Treatment in HER2-Overexpressing Breast Cancer. ISRN oncology 2012, 2012, 428062. 10.5402/2012/428062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen H. J.; Poulsen T. T.; Dahlman A.; Kjaer I.; Koefoed K.; Sen J. W.; Weilguny D.; Bjerregaard B.; Andersen C. R.; Horak I. D.; Pedersen M. W.; Kragh M.; Lantto J. Pan-HER, an Antibody Mixture Simultaneously Targeting EGFR, HER2, and HER3, Effectively Overcomes Tumor Heterogeneity and Plasticity. Clin. Cancer Res. 2015, 21, 4110–4122. 10.1158/1078-0432.CCR-14-3312. [DOI] [PubMed] [Google Scholar]
- Subik K.; Lee J. F.; Baxter L.; Strzepek T.; Costello D.; Crowley P.; Xing L.; Hung M. C.; Bonfiglio T.; Hicks D. G.; Tang P. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer: Basic Clin. Res. 2010, 4, 35–41. 10.1177/117822341000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassie J. P.; Liu X. Y.; Thomas G. S.; Whitaker R. M.; Thiel K. W.; Stockdale K. R.; Meyerholz D. K.; McCaffrey A. P.; McNamara J. O. 2nd; Giangrande P. H. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009, 27, 839–849. 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwind A.; Fischer O. M.; Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat. Rev. Cancer 2004, 4, 361–370. 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- Chen J. S.; Lan K.; Hung M. C. Strategies to target HER2/neu overexpression for cancer therapy. Drug Resist. Updates 2003, 6, 129–136. 10.1016/S1368-7646(03)00040-2. [DOI] [PubMed] [Google Scholar]
- Ryan Q.; Ibrahim A.; Cohen M. H.; Johnson J.; Ko C. W.; Sridhara R.; Justice R.; Pazdur R. FDA drug approval summary: lapatinib in combination with capecitabine for previously treated metastatic breast cancer that overexpresses HER-2. Oncologist 2008, 13, 1114–1119. 10.1634/theoncologist.2008-0816. [DOI] [PubMed] [Google Scholar]
- Ko B. K.; Lee S. Y.; Lee Y. H.; Hwang I. S.; Persson H.; Rockberg J.; Borrebaeck C.; Park D.; Kim K. T.; Uhlen M.; Lee J. S. Combination of novel HER2-targeting antibody 1E11 with trastuzumab shows synergistic antitumor activity in HER2-positive gastric cancer. Mol. Oncol. 2015, 9, 398–408. 10.1016/j.molonc.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeweiss A.; Chia S.; Hickish T.; Harvey V.; Eniu A.; Hegg R.; Tausch C.; Seo J. H.; Tsai Y. F.; Ratnayake J.; McNally V.; Ross G.; Cortes J. Pertuzumab plus trastuzumab in combination with standard neoadjuvant anthracycline-containing and anthracycline-free chemotherapy regimens in patients with HER2-positive early breast cancer: a randomized phase II cardiac safety study (TRYPHAENA). Ann. Oncol 2013, 24, 2278–2284. 10.1093/annonc/mdt182. [DOI] [PubMed] [Google Scholar]
- Nimjee S. M.; White R. R.; Becker R. C.; Sullenger B. A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. 10.1146/annurev-pharmtox-010716-104558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.; Gantier M. P.; Xiang D.; Bean A. G.; Bruce M.; Zhou S. F.; Khasraw M.; Ward A.; Wang L.; Wei M. Q.; AlShamaileh H.; Chen L.; She X.; Lin J.; Kong L.; Shigdar S.; Duan W. EpCAM Aptamer-mediated Survivin Silencing Sensitized Cancer Stem Cells to Doxorubicin in a Breast Cancer Model. Theranostics 2015, 5, 1456–1472. 10.7150/thno.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.; Zhao S.; Yu X.; Huang S.; Liu H. Y. Simultaneous targeting of CD44 and EpCAM with a bispecific aptamer effectively inhibits intraperitoneal ovarian cancer growth. Theranostics 2017, 7, 1373–1388. 10.7150/thno.17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittrup A.; Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015, 16, 543–552. 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutras A. K.; Fountzilas G.; Kalogeras K. T.; Starakis I.; Iconomou G.; Kalofonos H. P. The upgraded role of HER3 and HER4 receptors in breast cancer. Crit Rev. Oncol Hematol 2010, 74, 73–78. 10.1016/j.critrevonc.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Denduluri S. K.; Idowu O.; Wang Z.; Liao Z.; Yan Z.; Mohammed M. K.; Ye J.; Wei Q.; Wang J.; Zhao L.; Luu H. H. Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance. Genes Dis 2015, 2, 13–25. 10.1016/j.gendis.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpfl M.; Tong D.; Fasching B.; Schuster E.; Obermair A.; Leodolter S.; Zeillinger R. Vascular endothelial growth factor splice variants and their prognostic value in breast and ovarian cancer. Clin. Cancer Res. 2002, 8, 2253–2259. [PubMed] [Google Scholar]
- Lipi F.; Chen S.; Chakravarthy M.; Rakesh S.; Veedu R. N. In vitro evolution of chemically-modified nucleic acid aptamers: Pros and cons, and comprehensive selection strategies. RNA Biol. 2016, 13, 1232–1245. 10.1080/15476286.2016.1236173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomer R. M.; Lewis S. D.; Healy J. M.; Kurz M.; Wilson C.; McCauley T. G. Conjugation to polyethylene glycol polymer promotes aptamer biodistribution to healthy and inflamed tissues. Oligonucleotides 2005, 15, 183–195. 10.1089/oli.2005.15.183. [DOI] [PubMed] [Google Scholar]
- Healy J. M.; Lewis S. D.; Kurz M.; Boomer R. M.; Thompson K. M.; Wilson C.; McCauley T. G. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm. Res. 2004, 21, 2234–2246. 10.1007/s11095-004-7676-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.