Abstract
Despite osteoarthritis (OA) and rheumatoid arthritis (RA) being typically age-related, their underlying etiologies are markedly different. We used 1H nuclear magnetic resonance (NMR) spectroscopy to identify differences in metabolite profiles in low volumes of OA and RA synovial fluid (SF). SF was aspirated from knee joints of 10 OA and 14 RA patients. 100 μL SF was analyzed using a 700 MHz Avance IIIHD Bruker NMR spectrometer with a TCI cryoprobe. Spectra were analyzed by Chenomx, Bruker TopSpin and AMIX software. Statistical analysis was undertaken using Metaboanalyst. 50 metabolites were annotated, including amino acids, saccharides, nucleotides and soluble lipids. Discriminant analysis identified group separation between OA and RA cohorts, with 32 metabolites significantly different between OA and RA SF (false discovery rate (FDR) < 0.05). Metabolites of glycolysis and the tricarboxylic acid cycle were lower in RA compared to OA; these results concur with higher levels of inflammation, synovial proliferation and hypoxia found in RA compared to OA. Elevated taurine in OA may indicate increased subchondral bone sclerosis. We demonstrate that quantifiable differences in metabolite abundance can be measured in low volumes of SF by 1H NMR spectroscopy, which may be clinically useful to aid diagnosis and improve understanding of disease pathogenesis.
Keywords: synovial fluid, metabolomics, osteoarthritis, rheumatoid arthritis, nuclear magnetic resonance
Introduction
Osteoarthritis (OA) and rheumatoid arthritis (RA) are the two most common forms of arthritis, which lead to significant disability and substantial reduction in quality of life.1,2 Despite both these chronic conditions being typically age-related with insidious onset, their underlying etiologies are markedly different.3,4 OA is a degenerative joint condition, primarily affecting the hands, hips and knees, with one-third of people in the UK aged over 45 (approximately 8.75 million) requiring treatment.5 This disease is characterized by articular cartilage loss, synovial membrane dysfunction, subchondral bone sclerosis and osteophyte formation with the depletion of matrix proteins driven by proteases including multiple matrix metalloproteinases (MMPs) and a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTSs6−8). We have identified OA synovial fluid (SF) to be enriched in a number of proteins including S100-A10, Collagen type I and CD109.9 RA is a systemic, inflammatory autoimmune disease that primarily affects the synovium of joints,10 presenting as a symmetric polyarthritis with an estimated prevalence of 0.5–1% of the adult population within developed countries.11,12 RA joints are marked by inflammation of the synovium and destruction of articular cartilage and underlying bone. The RA synovium becomes hyperplastic with infiltration by a variety of immunocompetent cells (activated neutrophils in particular13). RA synovial fluid is enriched with cytokines, inflammatory mediators such as leukotrienes, and proteases that degrade the extracellular matrix and hyaluronic acid.13,14
SF is located within the articular joint cavity, providing a pool of nutrients for surrounding tissues but primarily serving as a biological lubricant, containing molecules with low-friction and low-wear properties to articular surfaces.15 As a serum filtrate, not only does SF contain systemic protein/metabolite markers of disease, but also due to its close contact and near proximity to numerous tissues found to be primarily altered during joint pathology (including synovial membrane, cartilage and bone), this biofluid holds significant potential in the discovery of biomarkers whose levels are altered at early stages of disease progression.16
Metabolomics systematically and comprehensively profiles metabolic changes within biological systems, including metabolic pathway analysis and abundance of unique biochemical molecules.17 The small molecules investigated include secondary metabolites, metabolic intermediates and hormones, as well as other molecules involved in cellular signaling.18 Few studies have investigated the whole profile of metabolites within human SF. A recent investigation analyzing 10 mL of each SF sample by liquid chromatography coupled with mass spectrometry (MS), identified 21 differentially expressed metabolites between OA and RA SF.19 These included a generally elevated level of lipid metabolites and phospholipids in RA patients compared to OA patients, although decreased levels of the phospholipid lysophosphatidylcholine was observed in OA. Nuclear magnetic resonance (NMR) spectroscopy is complementary to MS analysis and may offer advantages over MS in the metabolomics analysis of native samples, with a minimal level of sample preparation using a noninvasive and nondestructive method, subsequently producing results which are more reproducible and robust.20 One limiting factor of NMR spectroscopy of SF has previously been the minimum volume required for analysis, with difficulties in analyzing SF from normal human joints in which <200 μL can be aspirated.21 Using the methods described in this study we show that analyzable and reproducible 1H NMR spectra can be produced from just 100 μL of SF, thus increasing the potential applications of this technique when sample volume is limited.
The aim of this study was to apply 1H NMR to study human SF metabolomics, in order to provide novel insights on the underlying pathogenesis of OA and RA, and to optimize protocols for clinically relevant volumes of SF typically available in a clinical setting. In this study we describe protocols for human SF sampling and processing and present a novel approach to inflammation analysis, carrying out full profile metabolite NMR examination on low volumes (100 μL) of SF. This method of biofluid analysis enables further understanding of disease pathogenesis, inflammatory signatures and biomarkers, and may aid in both our comprehension of these conditions and progress toward an earlier diagnosis and/or prognostic indicators to stratify patients to treatment.
Experimental Methods
Sample Cohorts
SF samples from patients with OA and RA were collected in accordance with the declaration of Helsinki. The study that collected OA SF was approved by Maastricht UMC Medical Ethical Committee, approval number 08–4–028, according to Dutch law. The study that collected the RA SF was approved by Sefton Adult Ethics Committee; all patients gave written, informed consent. RA patients (mean age 65.4 years) fulfilled the 1987 American College of Rheumatology (ACR) criteria for RA and were seropositive for rheumatoid factor (RF). OA patients (mean age 67.4 years) were diagnosed by radiographs using Kellgren and Lawrence score, and general orthopedic guidelines carried out by an experienced orthopedic knee surgeon. A diagnosis of RA was excluded for all OA patients. OA and RA groups were sex and age-matched to within 5 years of age. RA patients were receiving multiple therapies including disease-modifying antirheumatic drugs (DMARDs, typically methotrexate), TNF inhibitors and/or prednisolone and had a mean disease duration of 13 years (range 0–35 years). OA patients were typically receiving nonsteroidal anti-inflammatory drugs (NSAIDs) and in end-stage disease.
Sample Collection
OA and RA SF from knee joints was aspirated into heparinized tubes and processed within 1 h. Aliquots of whole SF were centrifuged at >2000g for 5 min and cell-free SF was decanted and frozen at −20 °C.
Sample Preparation for NMR
100 μL of SF was thawed out over ice and diluted to a final volume containing 50% (v/v) SF, 40% (v/v) ddH2O (18.2 MΩ), 10% (v/v) 1 M PO43– pH7.4 buffer (Na2HPO4, VWR, Pennsylvania, US; NaH2PO4, Sigma-Aldrich, Gillingham, UK) in deuterium oxide (2H2O) and 0.0025% (v/v) sodium azide (NaN3, Sigma-Aldrich, Gillingham, UK). Samples were vortexed for 1 min, centrifuged at 13 000g and 4 °C for 2 min and 190 μL transferred into 3 mm outer diameter NMR tubes using a glass pipet.
NMR Setup Acquisition and Processing
SF was analyzed using 1H NMR spectroscopy on a 700 MHz NMR spectrometer Bruker Avance III HD with a TCI cryoprobe and chilled SampleJet autosampler. Software for acquisition and processing was carried out using Topsin 3.1 and IconNMR 4.6.7. 1D 1H standard CPMG-type metabolomics experiments, with optimal water suppression via presaturation, were acquired with the cpmgpr1d filters for small molecules via a Carr–Purcell–Meiboom–Gill (CPMG) sequence. Spectra were acquired at 37 °C with 32 transients a 15 ppm spectral width, 32 K points, 9.6 ms echo time and a 3.1 s interscan delay. Metabolomics data have been deposited to the EMBL-EBI MetaboLights database with the identifier MTBLS564. The complete data set can be accessed at https://www.ebi.ac.uk/metabolights/MTBLS564.
Spectra Processing and Quality Control
Spectra were assessed to conform to minimum reporting standards as outlined by the Metabolomics Society22 to ensure consistent line widths, baseline corrections and water suppression. As commonly used internal standards (TSP and DSS) are known to bind to some proteins including albumins, common in protein-rich fluids such as SF23 we indirectly referenced the spectra via an intrinsic metabolite whose shifts are unaffected by protein presence (glucose). All spectra passing appraisal were then divided into “buckets” that were defined globally for all spectra by the peak limits using Amix software. Spectra were then prepared for statistical analysis by bucketing/binning according to spectral features or peaks; all peaks, both identified and unknown, were included in the bucket table.
Metabolite Annotation and Identification
Metabolites in the SF 1H NMR spectra were initially tentatively annotated using the metabolite discovery software Chenomx (Chenomx, Canada) and, where possible, their identities confirmed using an in-house library of metabolite spectra (further details in Supplementary Table S1).24
Metabolomics Statistical Analysis
Statistical analyses were performed using Metaboanalyst (http://www.metaboanalyst.ca/).25 The bucketed experimental data were normalized by the median and additionally Pareto scaled (for multivariate analysis). Univariate analysis was by t test with application of a false-discovery rate (FDR) adjusted p-value of 0.05. Multivariate analysis was via unsupervised principal component analysis (PCA) followed by partial least squares discriminant analysis (PLS-DA) validated via leave one out cross-validation to automatically determine optimal number of components for the model.
Metabolomics Pathway Analysis
Pathway analysis was carried out using the list of significantly different metabolites between OA and RA SF using Metaboanalyst and with reference to the KEGG (Kyoto Encyclopedia of Genes and Genomes) metabolic pathway database (http://www.kegg.jp/kegg/pathway.html) for human metabolic pathways.25 Pathway enrichment was determined by Hypergeometric test and reported with an FDR-adjusted p-value.
Results
Metabolite Identification
NMR spectra for all cohorts showed a consistent set of metabolite signals present. NMR spectra of extracts were divided into individual spectral bins accounting for one or more multiplets. The number of spectral bins in each extract was 171, of which 126 (73.6%) were assigned to 50 metabolites. Quantile plots identifying the global variation of metabolite abundances within each cohort demonstrated much less variation within the OA cohort than the RA cohort, indicating a more homogeneous OA population (Figure 1). Fifty metabolites were annotated in total, including amino acids, saccharides, nucleotides and soluble lipids (Table 1).
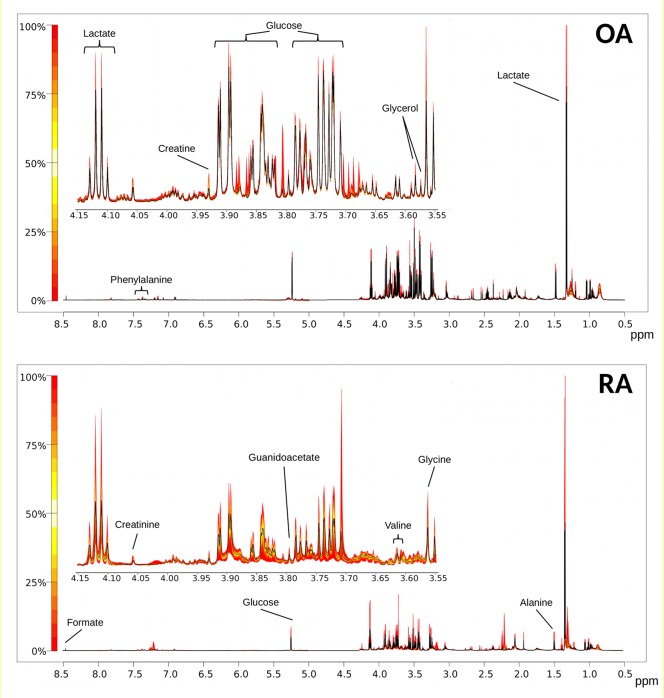
Figure 1.
Quantile Plots of OA and RA spectra depicting the median spectral plot (black line) and variation from the median within each cohort (yellow to red scale) for the full spectral range (8.5–0.5 ppm) and a more detailed region (4.15–3.55 ppm). Peaks of interest are annotated as examples. Note multiple peaks for some metabolites, e.g., glucose.
Table 1. List of 50 Metabolites Detected in OA and RA SF by 1H NMR Spectroscopy.
| Amino Acids |
| 2-Aminobutyrate, 5-Aminolevulinate, Alanine, Asparagine, Creatine, Creatinine, Glutamine, Glycine, Guanidoacetate, Histidine, Isoleucine, Leucine, Lysine, Methionine, n-Acetylamino acid, Ornithine, Phenylalanine, Proline, Sarcosine, Threonine, Tyrosine, Valine |
| Fatty and Organic Acids |
| 2-Hydroxybutyrate, 3-Hydroxybutyrate, 3-Hydroxyisovalerate, Acetate, Acetoacetate, Carnitine, Citrate, Formate, Lactate, Malonate, Pyruvate, Taurine |
| Sugars |
| Acetylated-saccharide, Glucose, Glycerol, Mannose, Myoinositol |
| Other |
| Acetylcholine, Adenosine, Choline, Dimethylamine, Ethanol, Isopropanol, Mobile-lipid, o-Phosphocholine, Oxypurinol, sn-Glycero-3-phosphocholine, Xanthine |
Univariate Comparison of OA and RA SF
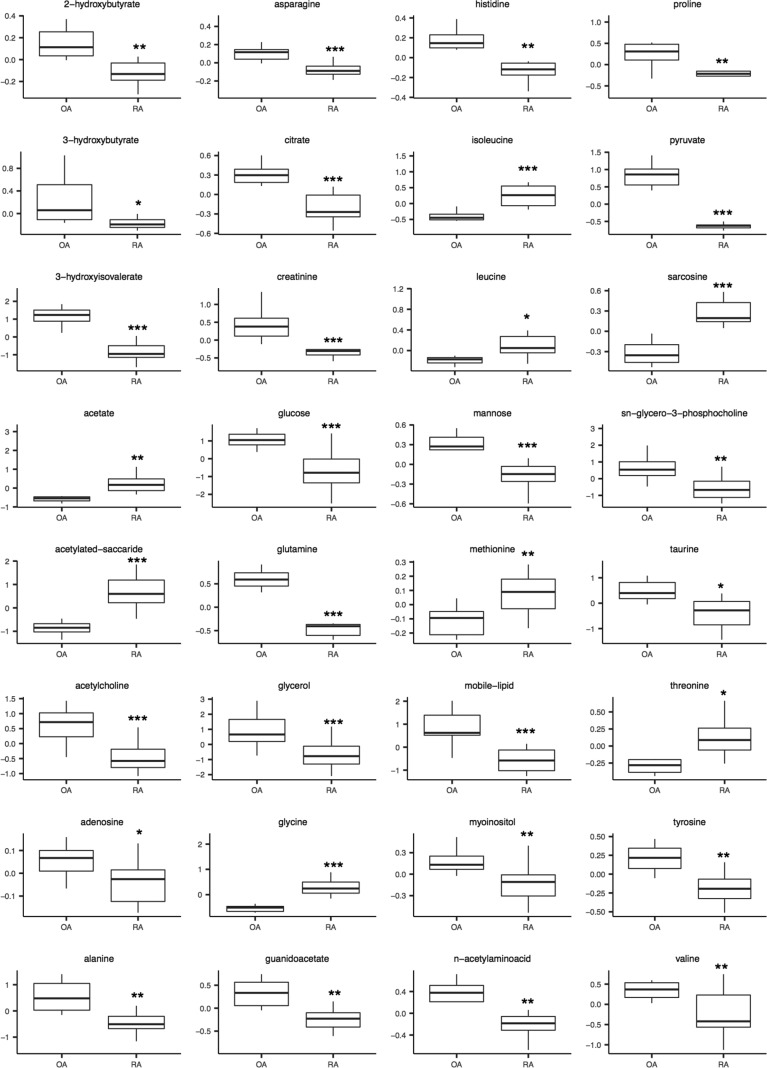
Univariate analysis of the 171 spectral bins identified 91 significantly different bins (including 50 annotated), which corresponded to 32 metabolites which were significantly different (FDR < 0.05) between OA and RA SF. These included citrate, creatinine, glucose, glutamine, glycerol, pyruvate and taurine which were higher in OA (Table 2, Figure 2, Figure 3A). 3-hydroxybutyrate, acetate, isoleucine, leucine, sarcosine and threonine were higher in RA. Acetylated saccharides, an annotation that incorporates commonly acetylated molecules such as heparin and chondroitin sulfate, and monosaccharide subunits N-acetyl glucosamine and N-acetyl galactosamine, were also significantly higher in RA. No significant association was observed between metabolite profile or drug therapy in the RA patients, or with the length of disease activity.
Table 2. List of 32 Significantly Altered Metabolites between OA and RA SF (FDR < 0.05)a.
| metabolite | HMDB ref | higher in OA | higher in RA | fold change OA vs RA | –log10 (p-value) | FDR |
|---|---|---|---|---|---|---|
| 2-Hydroxybutyrate | HMDB00008 | Y | 1.19 | 3.2495 | 1.70 × 10–3 | |
| 3-Hydroxybutyrate | HMDB00357 | Y | 1.59 | 2.3288 | 1.02 × 10–2 | |
| 3-Hydroxyisovalerate | HMDB00754 | Y | 2.22 | 7.6958 | 6.85 × 10–7 | |
| Acetate | HMDB00042 | Y | –1.69 | 2.6401 | 5.56 × 10–3 | |
| Acetylcholine | HMDB00895 | Y | 1.57 | 3.9361 | 5.32 × 10–4 | |
| Acetylated-saccharide | None | Y | –1.49 | 4.5038 | 1.97 × 10–4 | |
| Adenosine | HMDB00050 | Y | 1.94 | 1.6375 | 4.45 × 10–2 | |
| Alanine | HMDB00161 | Y | 1.42 | 2.79 | 4.18 × 10–3 | |
| Asparagine | HMDB00168 | Y | 1.31 | 3.8413 | 5.83 × 10–4 | |
| Citrate | HMDB00094 | Y | 1.67 | 5.972 | 1.65 × 10–5 | |
| Creatinine | HMDB00562 | Y | 1.43 | 4.0608 | 4.48 × 10–4 | |
| Glucose | HMDB00122 | Y | 1.82 | 4.2582 | 3.03 × 10–4 | |
| Glutamine | HMDB00641 | Y | 1.80 | 9.7868 | 9.26 × 10–9 | |
| Glycerol | HMDB00131 | Y | 1.42 | 2.6897 | 5.18 × 10–3 | |
| Glycine | HMDB00123 | Y | –1.64 | 3.9516 | 5.28 × 10–4 | |
| Guanidoacetate | HMDB00128 | Y | 1.19 | 3.283 | 1.61 × 10–3 | |
| Histidine | HMDB00177 | Y | 1.34 | 3.3713 | 1.36 × 10–3 | |
| Isoleucine | HMDB00172 | Y | –1.39 | 4.0006 | 4.88 × 10–4 | |
| Leucine | HMDB00687 | Y | –1.20 | 2.1407 | 1.52 × 10–2 | |
| Mannose | HMDB00169 | Y | 1.73 | 3.5567 | 9.85 × 10–4 | |
| Methionine | HMDB00696 | Y | –1.30 | 2.8572 | 3.69 × 10–3 | |
| Mobile-lipid | None | Y | 1.96 | 4.7317 | 1.42 × 10–4 | |
| Myoinositol | HMDB00211 | Y | 1.24 | 2.48 | 7.61 × 10–3 | |
| n-Acetylamino acid | None | Y | 1.11 | 2.5075 | 7.24 × 10–3 | |
| Proline | HMDB00162 | Y | 1.26 | 3.1978 | 1.85 × 10–3 | |
| Pyruvate | HMDB00243 | Y | 3.44 | 11.994 | 8.61 × 10–11 | |
| Sarcosine | HMDB00271 | Y | –1.56 | 4.6614 | 1.51 × 10–4 | |
| sn-Glycero-3-phosphocholine | HMDB00086 | Y | 1.49 | 3.0967 | 2.27 × 10–3 | |
| Taurine | HMDB00251 | Y | 1.26 | 2.2132 | 1.32 × 10–2 | |
| Threonine | HMDB00167 | Y | –1.22 | 1.6233 | 4.50 × 10–2 | |
| Tyrosine | HMDB00158 | Y | 1.40 | 2.9942 | 2.78 × 10–3 | |
| Valine | HMDB00883 | Y | 1.25 | 2.5638 | 6.45 × 10–3 |
Y = yes.
Figure 2.
Boxplots of representative metabolites identified from univariate analysis as significantly different between OA and RA SF (*p < 0.05, **p < 0.01, ***p < 0.001). Y-axis represents normalized peak intensity following median normalization and Pareto scaling.
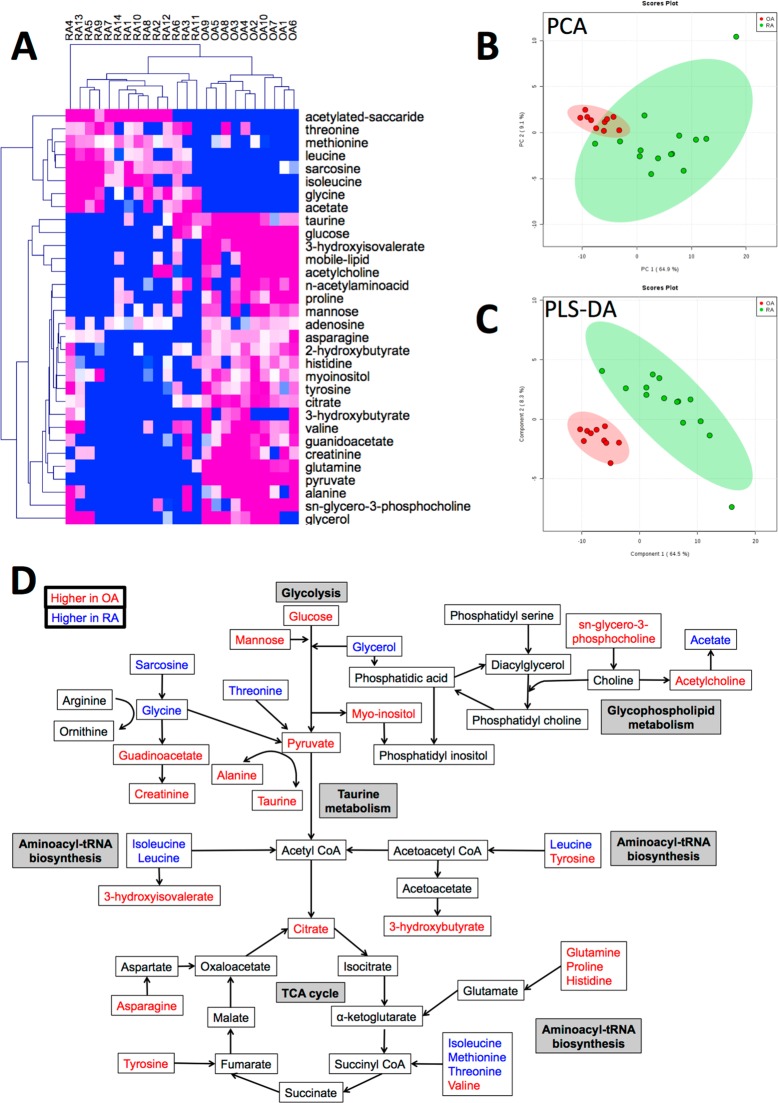
Figure 3.
Comparison of NMR SF metabolome between OA and RA. (A) Heatmap showing metabolites significantly different in OA and RA SF (p < 0.05). Blue = low, white = medium, pink = high concentration. (B) Unsupervised PCA scores plot showing metabolite profile is more variable between RA (green) than OA SF (red). Shading represents 95% confidence region. (C) Supervised multivariate analysis by PLS-DA segregated RA (green) and OA (red) SF samples. Shading represents 95% confidence region. Scores plot is shown for components 1 and 2. (D) Pathway scheme depicting metabolite levels detected in RA and OA SF. Blue = higher in RA SF, Red = higher in OA SF.
Multivariate Analysis Comparison of OA and RA SF
PCA analysis exhibited separation between OA and RA cohorts (Figure 3B). Regression analysis (PLS-DA, partial least-squares discriminant analysis) correctly distinguished between the two groups in 5 components (Figure 3C, R2 0.99448, Q2 0.95496).
Biological Context
In order to determine the metabolic pathways that were differently regulated between OA and RA, the list of 32 significantly different metabolites was uploaded into Metaboanalyst for Pathway Analysis. The results of this analysis are shown in Table 3, and include pathways relating to amino acid synthesis and degradation, taurine and hypotaurine metabolism and glycolysis. With reference to current understanding of metabolic pathways,26 we combined the results of the pathway analysis into a single metabolic flow diagram (Figure 3D) to enable interpretation of the interactions between the metabolites and the different metabolic pathways.
Table 3. Pathway Analysis Using Metaboanalyst and with Reference to the KEGG Database Predicted the Most Enriched Pathways from the List of Metabolites That Were Significantly Different between OA and RA SF (FDR ≤ 0.1)a.
| pathway | total | hits | –log(p) | FDR |
|---|---|---|---|---|
| Aminoacyl-tRNA biosynthesis | 75 | 12 | 24.693 | 1.51 × 10–9 |
| Nitrogen metabolism | 39 | 6 | 12.124 | 2.17 × 10–4 |
| Valine, leucine and isoleucine biosynthesis | 27 | 5 | 11.165 | 3.78 × 10–4 |
| Taurine and hypotaurine metabolism | 20 | 4 | 9.3989 | 1.66 × 10–3 |
| Alanine, aspartate and glutamate metabolism | 24 | 4 | 8.6483 | 2.81 × 10–3 |
| Glycine, serine and threonine metabolism | 48 | 5 | 8.2939 | 3.24 × 10–3 |
| Arginine and proline metabolism | 77 | 6 | 8.1678 | 3.24 × 10–3 |
| Galactose metabolism | 41 | 4 | 6.5415 | 1.44 × 10–2 |
| Glycolysis or Gluconeogenesis | 31 | 3 | 5.0818 | 5.52 × 10–2 |
| Valine, leucine and isoleucine degradation | 40 | 3 | 4.3697 | 1.02 × 10–1 |
Pathway enrichment was determined by Hypergeometric test and reported with an FDR-adjusted p-value. The number of metabolites enriched in our dataset (hits) is shown compared with the total number of metabolites in the KEGG pathway.
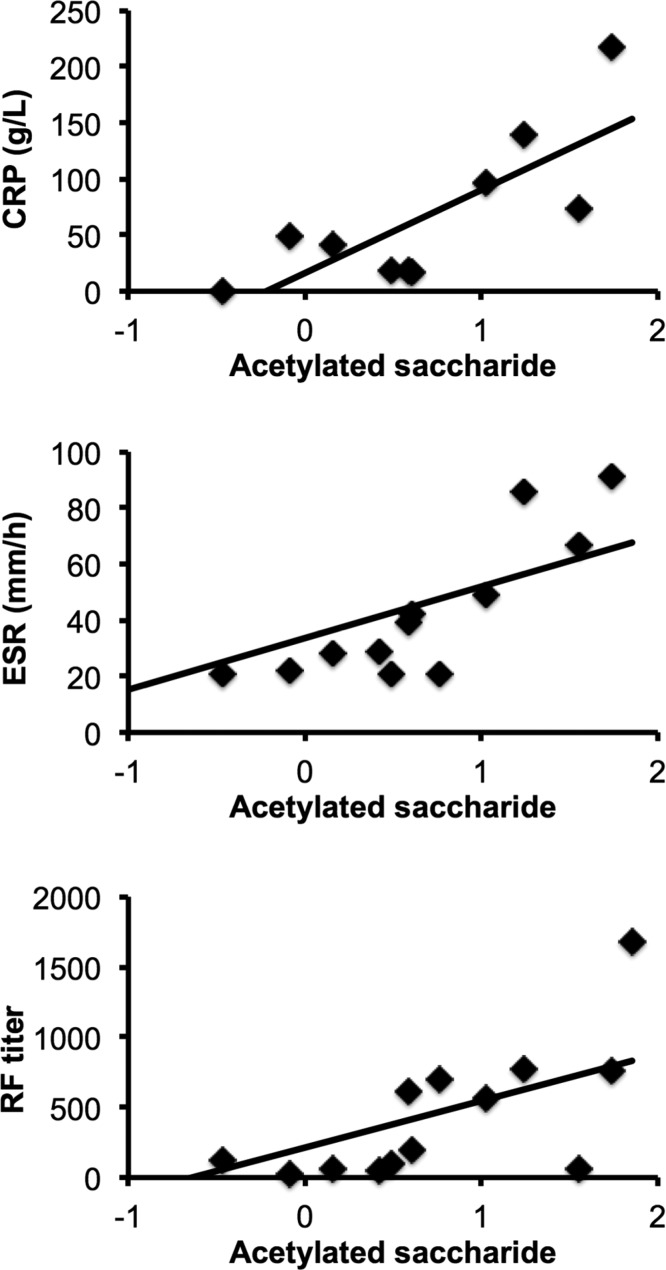
Normalized levels of acetylated saccharide in RA SF correlated positively with serum levels of inflammatory markers (Figure 4), including the acute-phase protein C-related protein (CRP, g/L) (Pearson R2 = 0.78, p = 0.008, n = 10), erythrocyte sedimentation rate (ESR, mm/h) (Pearson R2 = 0.62, p = 0.02, n = 13) and RF titer (Pearson R2 = 0.618, p = 0.018, n = 14).
Figure 4.
Correlation of acetylated saccharide with CRP, ESR and RF titer in RA patients. Levels of acetylated saccharide (median normalized with Pareto scaling) in RA SF correlated positively with serum levels of CRP (Pearson R2 = 0.78, p = 0.008, n = 10), ESR (Pearson R2 = 0.62, p = 0.02, n = 14) and RF titer (Pearson R2 = 0.618, p = 0.018, n = 14).
Discussion
In this study we measured OA and RA SF metabolomes using 1H NMR spectroscopy. This study differs from previous ones in that we have studied the metabolome of SF, the biofluid that is in contact with all the tissues and cells at the site of disease manifestation and joint damage, thereby allowing analysis of the pathophysiology of the whole joint. To date, metabolomic studies in OA and RA have focused on biofluids, including urine, serum and SF using a number of different platforms including gas chromatography–mass spectrometry (GC–MS), liquid chromatography–mass spectrometry (LC–MS) and 1H NMR spectroscopy.19,22,27−37 These studies have provided insight into the metabolites found within human arthritic SF, and the different underlying cellular pathology of OA and RA joints. The RA synovial environment is characterized by proliferation of synovial fibroblasts and angiogenesis. Infiltration of immune cells including neutrophils, macrophages, and T and B cells drive inflammation within the joint, releasing proteases, cytokines, chemokines and lipid mediators that perpetuate inflammation leading to growth of an invasive, inflammatory pannus across the surface of the cartilage. Further dysregulation of neutrophils and osteoclasts within the pannus leads to degradation of the underlying cartilage matrix and bone erosion.13,14 In contrast, while there is some evidence of mild to moderate inflammation and synovial hyperplasia within OA joints, this is markedly less than the levels observed in RA, with the population of infiltrating immune cells consisting mainly of T cells and macrophages.38,39 Synovitis is rare in OA, and the loss of articular cartilage and bone is caused by a dysregulation in proteases including MMPs leading to destruction of the cartilage extracellular matrix.6,7
A common feature of previous studies investigating OA and RA SF metabolites is the activation of metabolic pathways controlling glycolysis and fatty acid metabolism; however most studies compare OA or RA serum metabolites to sera from healthy individuals. Our study presents a direct comparison of OA and RA metabolite profiles in SF using 1H NMR spectroscopy, and provides a novel insight into the differences in metabolic activity within joints of patients with two common forms of arthritis. In addition, we believe this is the first study of this kind to make the raw data fully open access via public repository. In this study we identified 50 metabolites in OA and RA SF: 22 amino acids, 5 sugars, 12 fatty and organic acids, 11 others. Multivariate analysis separated patient SF metabolite profiles based on the levels of 32 metabolites that were significantly different between OA and RA SF. These metabolites were associated with metabolic pathways controlling amino acid synthesis, taurine metabolism, glycophospholipid metabolism, glycolysis and the tricarboxylic acid (TCA) cycle. OA SF had significantly higher levels of substrates for glycolysis and the TCA cycle, including glucose, mannose, pyruvate and citrate. In addition, many amino acids, which feed into glycolysis and the TCA cycle, were higher in OA, including tyrosine, glutamine, proline, histidine, asparagine, taurine and alanine. We have previously identified altered glycolysis pathways in early OA using NMR metabolomics.40 Furthermore, a number of studies have identified altered status of glycolysis and glucose metabolism glycolytic proteins in OA. One study identified these changes in OA compared to normal cartilage.41 In another study of an in vitro model of OA using cartilage explants and label-free mass spectrometry proteomics, 13.8% of up-regulated proteins were related to glycolysis.42
A previous study of urine metabolomics found altered TCA cycle activity in OA.43 The authors hypothesized this was due to changes in cartilage metabolism which resulted in elevations of aconitic acid, isocitric acid and citrate in the urine of OA patients. In addition, a reduction in excretion of glutamine was suggestive of altered energy metabolism in chondrocytes.43 In our study we identified increased citrate and glutamine in OA SF compared to RA, suggestive of altered chondrocyte biology in OA joints. Chondrocytes are extremely dependent on glucose metabolism to drive the extracellular matrix (ECM) biosynthetic machinery, requiring anaerobic glycolysis to generate ATP.44 Indeed, glycolysis constitutes 95% of their energy production.44 Adequate ATP is needed for chondrocytes to respond to stress thus, a high rate of anaerobic glycolysis is essential for ATP generation. It is hypothesized that this increase in glycolysis is an attempt to increase ECM production in the face of MMP-driven degradation.
In this study, we identified lower levels of substrates for glycolysis and the TCA cycle in RA SF, suggesting even higher levels of anaerobic cellular metabolism within RA joints compared to OA. While the levels of lactate did not differ between OA and RA SF in our study, the ratio of lactate to glucose was significantly higher in RA, supporting this theory.13,45 In RA sera, substrates of glycolysis including glucose, valine, alanine, glutamine and tyrosine increase in patients who respond to TNF-inhibitor therapy with etanercept, suggesting the levels of these metabolites increase when inflammation decreases.29 A decrease in 3-hydroxybutyrate levels in RA sera in response to etanercept has also been reported;29 again we observed lower levels of 3-hydroxybutyrate in RA SF compared to OA SF. Lower levels of glucose have been previously reported in RA serum compared to healthy individuals.35 Within the joint, lower glucose levels may be as a consequence of the increase in cellular metabolism in resident SF fibroblasts and infiltrating immune cells, particularly neutrophils,13 as these cells rely solely on glycolysis for energy production.45 Increased hypoxia within RA joints has also been shown to promote glycolysis,46 and is strongly associated with synovial proliferation,47 whereas in OA, no association between synovial proliferation and hypoxia is evident.47 Interestingly taurine was higher in OA versus RA SF, while “taurine and hypotaurine metabolism” was identified in our pathway analysis. Previous metabolomics studies in RA have demonstrated that taurine is increased in RA versus control serum, and in RA SF compared to OA SF.19,36 Taurine is involved in the pathogenesis of subchondral bone sclerosis,48 a feature of both RA and OA.49 It is possible that the higher levels of taurine OA SF identified in our study may be as a consequence of increased subchondral bone sclerosis in OA compared to RA samples, and in addition may be an indicator of the degree of subchondral bone sclerosis along with additional metabolites such as l-carnitine and glycerophospholipids. Patients in our study were in end-stage OA and likely to have high levels of subchondral bone sclerosis associated with advanced disease, whereas in previous studies19 OA SF were collected from patients at an earlier stage of disease progression.
A number of metabolites increased in RA SF indicate activation of ketosis, possibly due to the low glucose levels observed in RA. Metabolites including leucine, isoleucine and threonine were elevated in RA SF. Ketosis occurs via a shift in metabolism during glucose limitation: in the absence of glucose, e.g., via dietary limitation or decreased mobilization of glycogen stores, or during oxygen limitation, metabolism of stored lipids is stimulated to increase the supply of acetyl CoA. 3-Hydroxybutyrate, a key metabolite in ketosis, was previously identified as being elevated in RA serum compared to healthy controls.29 In our study we found levels of this metabolite to be lower in RA compared to OA SF. A number of lipid metabolites have been previously reported to be lower in RA biofluids, with a possible explanation being lipids as a source of energy within the hypoxic joint.36 Alteration of lipid metabolism is associated with changes in membrane composition/permeability, gene expression and protein distribution and function, as well as in cellular functions such as cell growth, proliferation, differentiation, survival, apoptosis, and chemotaxis, implicated in the RA disease process.50
Metabolomic analysis of urine from RA patients prior to commencement of anti-TNF therapy identified higher histamine, glutamine and xanthurenic acid, along with lower levels of ethanolamine as biomarkers of a good response to therapy.30 Serum metabolite biomarkers can also distinguish responders and nonresponders to methotrexate.51 A number of serum metabolites and metabolic pathways have been shown to correlate with markers of inflammation in RA, e.g., CRP, including metabolites associated with arginine metabolism (arginine and ornithine), tryptophan metabolism (serotonin and tryptophan) and branched-chain amino acids (isoleucine, leucine, and valine).31 We did not find correlation of these metabolites with markers of inflammation in RA SF in our study; however, we observed significant correlation of acetylated-saccharide with serum CRP levels, ESR and RF titer. Our assignment of this metabolite as “acetylated saccharide” was based on comparison of 1H 13C 2D spectra to our in-house library of common acetylated molecules including acetylated heparin and acetylated chondroitin sulfate, and to monosaccharide subunits N-acetyl glucosamine and N-acetyl galactosamine (data not shown). While the precise acetylated saccharide cannot be definitively identified from this analysis alone, we are confident in our assignment of the NMR resonances to this class of acetylate-saccharide metabolites. The presence of these metabolites concurs with previous observation of a significant elevation in the activities of exoglycosidases (including β-d-glucuronidase and β-d-N-acetyl-glucosaminidase) in RA compared to OA SF.52 These enzymes are responsible for the degradation of glycosaminoglycans (GAGs) such as heparan sulfate within the cartilage matrix, and of hyaluronic acid within synovial fluid, leading to the reduced viscosity of SF and irreversible damage to cartilage that is a hallmark of RA. Degradation of GAGs by β-d-glucuronidase and β-d-N-acetyl-glucosaminidase liberates both N-acetyl glucosamine monosaccharide and chondroitin sulfate (disaccharide of N-acetyl glucosamine and glucoronic acid). The likely source of exoglycosidases in RA SF is activated neutrophils.13 Neutrophils are also the main source of SF myeloperoxidase, the enzyme responsible for the production of hypochlorous acid, which has also been implicated in the degradation of GAGs and the appearance of acetylated saccharides in SF.53 Interestingly, a strong correlation between β-d-N-acetyl-glucosaminidase activity and RF titer was previously reported.52 Correlation of serum glycoprotein acetyls with CRP and disease activity has also been reported in RA sera.54
One limitation to our study was the heterogeneous nature of the RA patient cohort, which included patients at different stages of disease (from early RA with <1 year diagnosis up to long-standing RA >35 years diagnosis), and on a variety of different treatments including DMARDs such as methotrexate, biologic therapies and glucocorticoids. The heterogeneous nature of RA SFs was evident in the spectral analysis. Despite this, regression analysis (PLS-DA) was able to segregate OA and RA SF samples with a high degree of accuracy. In addition, we were unable to include “normal” SF samples from healthy donors in our study. Definition of normal SF, and in particular the ethical collection of such from living donors, is ambiguous and difficult. Post-mortem SF cannot be defined as having a “normal” metabolome due to the anaerobic changes that take place following death. We believe it would be of great value for a future study of this kind to perform analysis of SF lipids in OA and RA; however, this was outside the scope of our current study. Future studies should also correlate clinical markers of disease activity (e.g., tender and swollen joint counts, disease activity scores), radiographic changes and physical characteristics such as BMI with SF metabolome profiles.
Conclusion
In summary, we have made a direct comparison of SF from RA and OA and found that the metabolic pathways that differ most between the two forms of arthritis are glycolysis, amino acid biosynthesis and taurine and hypotaurine metabolism. In general, metabolites of glycolysis and the TCA cycle were higher in OA compared to RA; these results concur with higher level of inflammation, synovial proliferation and hypoxia found in RA compared to OA. Levels of taurine were also higher in OA indicating increased subchondral bone sclerosis compared to RA. The study has deepened our understanding of the metabolic differences (and hence pathophysiology) of these two forms of arthritis. This study also demonstrates the feasibility of performing 1H NMR metabolomics on small clinical samples (100 μL), hence widening the applicability of the technology within a clinical setting. We speculate that the discriminative power of our approach to analysis of 100 μL SF using 1H NMR spectroscopy may be sensitive enough to identify changes in disease progression that could inform the use of disease modifying therapies or measure the efficacy of future therapies on disease activity/progression in OA and RA.
Acknowledgments
We thank Dr. RC Bucknall for assistance with collection of RA SFs. We thank Dr. PJ Emans and Dr. PZ Feczko for assistance with collection of OA SFs. MJP and JA received funding from The Horse Trust [Grant No G1015], MJP received funding from The Wellcome Trust [Grant No 07471/Z/15/Z], HW received funding from Arthritis Research UK [Grant No 21430] and the University of Liverpool Technology Directorate, LYL received funding for equipment and software from The Medical Research Council [Grant No MR/M009114/1], SC received funding from the 100th Anniversary Chulalongkorn University Fund for Doctoral Scholarship, the Overseas Research Experience Scholarship for Graduate Students, and Chulalongkorn University, TW received funding from the Dutch Arthritis Foundation [grants 15-3-403 and LLP14].
Glossary
Abbreviations
- ACR
American College of Rheumatology
- ADAMTS
A disintegrin and metalloproteinase with thrombospondin motifs
- CRP
C-related protein
- DMARD
Disease-modifying antirheumatic drug
- ECM
Extracellular matrix
- ESR
Erythrocyte sedimentation rate
- FDR
False discovery rate
- GAGs
Glycosaminoglycans
- GC–MS
Gas chromatography–mass spectrometry
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LC–MS
Liquid chromatography–mass spectrometry
- MMP
Matrix metalloproteinases
- MS
Mass spectrometry
- NMR
Nuclear magnetic resonance
- NSAID
Nonsteroidal anti-inflammatory drug
- OA
Osteoarthritis
- PCA
Principal Component Analysis
- PLS-DA
Partial Least Squares Discriminant Analysis
- RA
Rheumatoid arthritis
- SF
Synovial fluid
- TCA
Tricarboxylic acid
- TNF
Tumor necrosis factor
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.8b00455.
Author Contributions
# JRA and SC contributed equally.
The authors declare no competing financial interest.
Notes
Metabolomics data have been deposited to the EMBL-EBI MetaboLights database with the identifier MTBLS564. The complete data set can be accessed at https://www.ebi.ac.uk/metabolights/MTBLS564.
Supplementary Material
References
- Li H.; Hao Z.; Zhao L.; Liu W.; Han Y.; Bai Y.; et al. Comparison of molecular mechanisms of rheumatoid arthritis and osteoarthritis using gene microarrays. Mol. Med. Rep. 2016, 13 (6), 4599–605. 10.3892/mmr.2016.5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprangers M. A.; de Regt E. B.; Andries F.; van Agt H. M.; Bijl R. V.; de Boer J. B.; et al. Which chronic conditions are associated with better or poorer quality of life?. Journal of clinical epidemiology 2000, 53 (9), 895–907. 10.1016/S0895-4356(00)00204-3. [DOI] [PubMed] [Google Scholar]
- Majithia V.; Geraci S. A. Rheumatoid arthritis: diagnosis and management. Am. J. Med. 2007, 120 (11), 936–9. 10.1016/j.amjmed.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Brouwers H.; von Hegedus J.; Toes R.; Kloppenburg M.; Ioan-Facsinay A. Lipid mediators of inflammation in rheumatoid arthritis and osteoarthritis. Best practice & research Clinical rheumatology 2015, 29 (6), 741–55. 10.1016/j.berh.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Reynard L. N.; Loughlin J. Genetics and epigenetics of osteoarthritis. Maturitas 2012, 71 (3), 200–4. 10.1016/j.maturitas.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Struglics A.; Larsson S.; Pratta M. A.; Kumar S.; Lark M. W.; Lohmander L. S. Human osteoarthritis synovial fluid and joint cartilage contain both aggrecanase- and matrix metalloproteinase-generated aggrecan fragments. Osteoarthritis and cartilage 2006, 14 (2), 101–13. 10.1016/j.joca.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Poole A. R. An introduction to the pathophysiology of osteoarthritis. Front. Biosci., Landmark Ed. 1999, 4, D662–70. 10.2741/A463. [DOI] [PubMed] [Google Scholar]
- Truong L. H.; Kuliwaba J. S.; Tsangari H.; Fazzalari N. L. Differential gene expression of bone anabolic factors and trabecular bone architectural changes in the proximal femoral shaft of primary hip osteoarthritis patients. Arthritis research & therapy 2006, 8 (6), R188. 10.1186/ar2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peffers M. J.; McDermott B.; Clegg P. D.; Riggs C. M. Comprehensive protein profiling of synovial fluid in osteoarthritis following protein equalization. Osteoarthritis and cartilage 2015, 23 (7), 1204–13. 10.1016/j.joca.2015.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartok B.; Firestein G. S. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol. Rev. 2010, 233 (1), 233–55. 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. M. 3rd; Matteson E. L. American College of R, European League Against R. My treatment approach to rheumatoid arthritis. Mayo Clin. Proc. 2012, 87 (7), 659–73. 10.1016/j.mayocp.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. L.; Wolfe F.; Huizinga T. W. Rheumatoid arthritis. Lancet 2010, 376 (9746), 1094–108. 10.1016/S0140-6736(10)60826-4. [DOI] [PubMed] [Google Scholar]
- Wright H. L.; Moots R. J.; Edwards S. W. The multifactorial role of neutrophils in rheumatoid arthritis. Nat. Rev. Rheumatol. 2014, 10 (10), 593–601. 10.1038/nrrheum.2014.80. [DOI] [PubMed] [Google Scholar]
- Wright H. L.; Bucknall R. C.; Moots R. J.; Edwards S. W. Analysis of SF and plasma cytokines provides insights into the mechanisms of inflammatory arthritis and may predict response to therapy. Rheumatology (Oxford, U. K.) 2012, 51, 451–9. 10.1093/rheumatology/ker338. [DOI] [PubMed] [Google Scholar]
- Blewis M. E.; Nugent-Derfus G. E.; Schmidt T. A.; Schumacher B. L.; Sah R. L. A model of synovial fluid lubricant composition in normal and injured joints. European cells & materials 2007, 13, 26–39. 10.22203/eCM.v013a03. [DOI] [PubMed] [Google Scholar]
- Ruiz-Romero C.; Blanco F. J. Proteomics role in the search for improved diagnosis, prognosis and treatment of osteoarthritis. Osteoarthritis and cartilage 2010, 18 (4), 500–9. 10.1016/j.joca.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Wang X.; Zhang A.; Han Y.; Wang P.; Sun H.; Song G.; et al. Urine metabolomics analysis for biomarker discovery and detection of jaundice syndrome in patients with liver disease. Mol. Cell. Proteomics 2012, 11 (8), 370–80. 10.1074/mcp.M111.016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukarainen N. Ph.D. Dissertation, NMR Metabolomics Techniques and Mathematical Tools As an Aid in Neurological Disorders; University of Eastern Finland, 2009. http://epublications.uef.fi/pub/urn_isbn_978-951-27-1460-5/urn_isbn_978-951-27-1460-5.pdf. [Google Scholar]
- Kang K. Y.; Lee S. H.; Jung S. M.; Park S. H.; Jung B. H.; Ju J. H. Downregulation of Tryptophan-related Metabolomic Profile in Rheumatoid Arthritis Synovial Fluid. J. Rheumatol. 2015, 42 (11), 2003–11. 10.3899/jrheum.141505. [DOI] [PubMed] [Google Scholar]
- Markley J. L.; Bruschweiler R.; Edison A. S.; Eghbalnia H. R.; Powers R.; Raftery D.; et al. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. 10.1016/j.copbio.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton D.; Whelan M.; Smith E. C.; Williams R.; Blake D. R.; Grootveld M. An investigation of the abnormal metabolic status of synovial fluid from patients with rheumatoid arthritis by high field proton nuclear magnetic resonance spectroscopy. FEBS Lett. 1993, 317 (1–2), 135–8. 10.1016/0014-5793(93)81508-W. [DOI] [PubMed] [Google Scholar]
- Sumner L. W.; Amberg A.; Barrett D.; Beale M. H.; Beger R.; Daykin C. A.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3 (3), 211–21. 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckonert O.; Keun H. C.; Ebbels T. M.; Bundy J.; Holmes E.; Lindon J. C.; et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2 (11), 2692–703. 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- Salek R. M.; Steinbeck C.; Viant M. R.; Goodacre R.; Dunn W. B. The role of reporting standards for metabolite annotation and identification in metabolomic studies. GigaScience 2013, 2 (1), 13. 10.1186/2047-217X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J.; Wishart D. S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Current protocols in bioinformatics 2016, 55, 14.10.1. 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- Berg J. M.; Tymoczko J. L.; Stryer L.. Biochemistry: International Edition, 7th ed.; W. H. Freeman: Basingstoke, 2012. [Google Scholar]
- Kim S.; Hwang J.; Kim J.; Ahn J. K.; Cha H. S.; Kim K. H. Metabolite profiles of synovial fluid change with the radiographic severity of knee osteoarthritis. Jt., Bone, Spine 2017, 84 (5), 605–10. 10.1016/j.jbspin.2016.05.018. [DOI] [PubMed] [Google Scholar]
- Madsen R. K.; Lundstedt T.; Gabrielsson J.; Sennbro C. J.; Alenius G. M.; Moritz T.; et al. Diagnostic properties of metabolic perturbations in rheumatoid arthritis. Arthritis research & therapy 2011, 13 (1), R19. 10.1186/ar3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priori R.; Casadei L.; Valerio M.; Scrivo R.; Valesini G.; Manetti C. (1)H-NMR-Based Metabolomic Study for Identifying Serum Profiles Associated with the Response to Etanercept in Patients with Rheumatoid Arthritis. PLoS One 2015, 10 (11), e0138537. 10.1371/journal.pone.0138537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S. R.; Filer A.; Fitzpatrick M. A.; Fisher B. A.; Taylor P. C.; Buckley C. D.; et al. Metabolic profiling predicts response to anti-tumor necrosis factor alpha therapy in patients with rheumatoid arthritis. Arthritis Rheum. 2013, 65 (6), 1448–56. 10.1002/art.37921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppen B. V.; Fu J.; van Wietmarschen H. A.; Harms A. C.; Koval S.; Marijnissen A. C.; et al. Exploring the Inflammatory Metabolomic Profile to Predict Response to TNF-alpha Inhibitors in Rheumatoid Arthritis. PLoS One 2016, 11 (9), e0163087. 10.1371/journal.pone.0163087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Likhodii S.; Zhang Y.; Aref-Eshghi E.; Harper P. E.; Randell E.; et al. Classification of osteoarthritis phenotypes by metabolomics analysis. BMJ. open 2014, 4 (11), e006286. 10.1136/bmjopen-2014-006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giera M.; Ioan-Facsinay A.; Toes R.; Gao F.; Dalli J.; Deelder A. M.; et al. Lipid and lipid mediator profiling of human synovial fluid in rheumatoid arthritis patients by means of LC–MS/MS. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2012, 1821 (11), 1415–24. 10.1016/j.bbalip.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickiewicz B.; Kelly J. J.; Ludwig T. E.; Weljie A. M.; Wiley J. P.; Schmidt T. A.; et al. Metabolic analysis of knee synovial fluid as a potential diagnostic approach for osteoarthritis. J. Orthop. Res. 2015, 33 (11), 1631–8. 10.1002/jor.22949. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Chen J.; Hu C.; Xie Z.; Li H.; Wei S.; et al. Exploration of the serum metabolite signature in patients with rheumatoid arthritis using gas chromatography-mass spectrometry. J. Pharm. Biomed. Anal. 2016, 127, 60–7. 10.1016/j.jpba.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Young S. P.; Kapoor S. R.; Viant M. R.; Byrne J. J.; Filer A.; Buckley C. D.; et al. The impact of inflammation on metabolomic profiles in patients with arthritis. Arthritis Rheum. 2013, 65 (8), 2015–23. 10.1002/art.38021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugle T.; Kovacs H.; Heijnen I. A.; Daikeler T.; Baisch U.; Hicks J. M.; et al. Synovial fluid metabolomics in different forms of arthritis assessed by nuclear magnetic resonance spectroscopy. Clin. Exp. Rheumatol. 2012, 30 (2), 240–5. [PubMed] [Google Scholar]
- de Lange-Brokaar B. J.; Ioan-Facsinay A.; van Osch G. J.; Zuurmond A. M.; Schoones J.; Toes R. E.; et al. Synovial inflammation, immune cells and their cytokines in osteoarthritis: a review. Osteoarthritis and cartilage 2012, 20 (12), 1484–99. 10.1016/j.joca.2012.08.027. [DOI] [PubMed] [Google Scholar]
- Berenbaum F. Osteoarthritis as an inflammatory disease (osteoarthritis is not osteoarthrosis!). Osteoarthritis and cartilage 2013, 21 (1), 16–21. 10.1016/j.joca.2012.11.012. [DOI] [PubMed] [Google Scholar]
- Peffers M. J.; Riggs C. M.; Phelan M. P.; Clegg P. D. Identification of Disease Specific Metabolic Fingerprints in Early Osteoarthritis. Equine Veterinary Journal 2015, 47 (S48), 13–13. 10.1111/evj.12486_28.26375175 [DOI] [Google Scholar]
- Guo D.; Tan W.; Wang F.; Lv Z.; Hu J.; Lv T.; et al. Proteomic analysis of human articular cartilage: identification of differentially expressed proteins in knee osteoarthritis. Jt., Bone, Spine 2008, 75 (4), 439–44. 10.1016/j.jbspin.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Peffers M. J. Ph.D. Thesis, Proteomic and Transcriptomic Signatures of Cartilage Ageing and Disease; University of Liverpool, 2013. https://livrepository.liverpool.ac.uk/12095/. [Google Scholar]
- Li X.; Yang S.; Qiu Y.; Zhao T.; Chen T.; Su M.; et al. Urinary metabolomics as a potentially novel diagnostic and stratification tool for knee osteoarthritis. Metabolomics 2010, 6 (1), 109–18. 10.1007/s11306-009-0184-0. [DOI] [Google Scholar]
- Heywood H. K.; Knight M. M.; Lee D. A. Both superficial and deep zone articular chondrocyte subpopulations exhibit the Crabtree effect but have different basal oxygen consumption rates. J. Cell. Physiol. 2010, 223 (3), 630–9. 10.1002/jcp.22061. [DOI] [PubMed] [Google Scholar]
- Kramer P. A.; Ravi S.; Chacko B.; Johnson M. S.; Darley-Usmar V. M. A review of the mitochondrial and glycolytic metabolism in human platelets and leukocytes: implications for their use as bioenergetic biomarkers. Redox Biol. 2014, 2, 206–10. 10.1016/j.redox.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biniecka M.; Canavan M.; McGarry T.; Gao W.; McCormick J.; Cregan S.; et al. Dysregulated bioenergetics: a key regulator of joint inflammation. Ann. Rheum. Dis. 2016, 75 (12), 2192–200. 10.1136/annrheumdis-2015-208476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. A.; Kim J. Y.; Hong S. J.; Lee S. H.; Yoo M. C.; Kim K. S.; et al. Synovial proliferation differentially affects hypoxia in the joint cavities of rheumatoid arthritis and osteoarthritis patients. Clin. Rheumatol. 2007, 26 (12), 2023–9. 10.1007/s10067-007-0605-2. [DOI] [PubMed] [Google Scholar]
- Yang G.; Zhang H.; Chen T.; Zhu W.; Ding S.; Xu K.; et al. Metabolic analysis of osteoarthritis subchondral bone based on UPLC/Q-TOF-MS. Anal. Bioanal. Chem. 2016, 408 (16), 4275–86. 10.1007/s00216-016-9524-x. [DOI] [PubMed] [Google Scholar]
- Li G.; Ma Y.; Cheng T. S.; Landao-Bassonga E.; Qin A.; Pavlos N. J.; et al. Identical subchondral bone microarchitecture pattern with increased bone resorption in rheumatoid arthritis as compared to osteoarthritis. Osteoarthritis and cartilage 2014, 22 (12), 2083–92. 10.1016/j.joca.2014.08.015. [DOI] [PubMed] [Google Scholar]
- Guma M.; Tiziani S.; Firestein G. S. Metabolomics in rheumatic diseases: desperately seeking biomarkers. Nat. Rev. Rheumatol. 2016, 12 (5), 269–81. 10.1038/nrrheum.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Chen Z.; Yang S.; Wang Y.; Yu L.; Zhang B.; et al. (1)H NMR-based metabolomic analysis for identifying serum biomarkers to evaluate methotrexate treatment in patients with early rheumatoid arthritis. Exp. Ther. Med. 2012, 4 (1), 165–71. 10.3892/etm.2012.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortutay Z.; Polgar A.; Gomor B.; Geher P.; Lakatos T.; Glant T. T.; et al. Synovial fluid exoglycosidases are predictors of rheumatoid arthritis and are effective in cartilage glycosaminoglycan depletion. Arthritis Rheum. 2003, 48 (8), 2163–72. 10.1002/art.11093. [DOI] [PubMed] [Google Scholar]
- Schiller J.; Arnhold J.; Sonntag K.; Arnold K. NMR studies on human, pathologically changed synovial fluids: role of hypochlorous acid. Magn. Reson. Med. 1996, 35 (6), 848–53. 10.1002/mrm.1910350610. [DOI] [PubMed] [Google Scholar]
- Kononoff A.; Arstila L.; Kautiainen H.; Elfving P.; Savolainen E.; Niinisalo H. Patient reported outcomes correlated with concentrations of glycoprotein acetyls in patients with early untreated rheumatoid arthritis. Ann. Rheum. Dis. 2015, 74 (2), 430.1. 10.1136/annrheumdis-2015-eular.5579.24297378 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.