 Both Strychnos nux-vomica Linn. (SNV) and Tripterygium wilfordii Hook F (TwHF) have received extensive attention due to their excellent clinical efficacies.
Both Strychnos nux-vomica Linn. (SNV) and Tripterygium wilfordii Hook F (TwHF) have received extensive attention due to their excellent clinical efficacies.
Abstract
Both Strychnos nux-vomica Linn. (SNV) and Tripterygium wilfordii Hook F (TwHF) have received extensive attention due to their excellent clinical efficacies. However, clinical applications of SNV and TwHF have been limited by their narrow therapeutic windows and severe kidney toxicities. In this paper, based on ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC/Q-TOF-MS), endogenous metabolites after administration of SNV and TwHF extracts were detected, and biomarkers were screened successfully. Additionally, the levels of Cr and BUN in serum and pathological findings of kidneys were detected and observed. Finally, both biochemical and pathological tests of the SNV group and TwHF group indicated that kidney damage had occurred. After comparison with the normal saline group, 15 nephrotoxic biomarkers were selected from the SNV group, and 17 nephrotoxic biomarkers were selected from the TwHF group. The experimental results showed that there are some differences in the mechanisms of nephrotoxicity induced by SNV and TwHF, which are significant for revealing the mechanisms of renal injury of different medicines.
1. Introduction
Strychnos nux-vomica Linn. (SNV) is bitter, cold, and has high toxicity. It is known for dredging collaterals, relieving pain, dispersing swelling, cooling blood and dissipating heat. It is commonly used in the treatment of rheumatoid arthritis, chronic bronchitis, malignant tumors and other diseases.1 Tripterygium wilfordii Hook F (TwHF) is widely used in the treatment of rheumatoid arthritis, systemic lupus erythematosus, psoriasis, and other diseases, and it is known for activating blood and removing stasis, clearing away heat and detoxifying, and eliminating swelling and dispelling knots.2 Due to the unique curative effects of SNV and TwHF, almost no similar Chinese medicine can completely replace them. SNV and TwHF are commonly used traditional Chinese medicines in clinical practice, and both have very good research values and development prospects. However, both SNV and TwHF have high toxicities, and the therapeutic dose is close to the toxic dose,3,4 which easily leads to human kidney toxicity and even acute renal failure.5 Renal toxicity occurs frequently, and the poisoning consequences are serious, resulting in restrictions on the use and promotion of SNV and TwHF. In recent years, the toxicity of Chinese herbal medicines, especially kidney toxicity, has gradually attracted the attention of scholars both in China and abroad. However, there is lack of systematic and in-depth research on the mechanism of toxicity of traditional Chinese medicine for kidney damage.6 Therefore, in this study, the mechanism of nephrotoxicity was studied using commonly used SNV and TwHF for clinical experiments.
Metabonomics is the study of the types, quantities and changes of endogenous metabolites in biological systems. It is a novel drug toxicity evaluation technique that combines experiments and information calculations.7 The development of metabolomics has brought new opportunities and challenges for the evaluation of toxicity and the study on mechanisms of toxicity of Chinese medicine.8 Metabonomics can evaluate toxic changes produced by organisms from an overall perspective, predict toxicity, and find toxicity-related biomarkers, thus elucidating the toxicity mechanism.9 The aim of this technique is the end product of the whole metabolic network of the body, which reflects the true state of the body and represents the whole function of the body, coinciding with Chinese medicine's emphasis on integrity and dialectics while treating diseases.10 The use of metabolomics analysis technology to study traditional Chinese medicine has important scientific significance and economic value for understanding the pharmacological effects and material basis for producing toxic and side effects as well as the proper use of Chinese medicine in the treatment of diseases.
Compared with other detection techniques used in metabolomics, such as gas chromatography-mass spectrometry (GC-MS) and nuclear magnet resonance (NMR), ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC/Q-TOF-MS) with better throughput, better detection limits, increased chromatographic resolution, and improved mass spectrographic data quality has been extensively applied in many metabonomic studies.11,12 For example, L. Zhu et al.11 used UPLC/Q-TOF-MS to explore the molecular mechanism of T-2 toxin-induced cartilage destruction in Wistar rats; the endogenous metabolite profile of serum was determined by UPLC/Q-TOF in the model group. It was found that 8 metabolites exhibited significant changes in these rats with articular cartilage destruction. Xinwen Dong et al.13 evaluated the urinary metabolomics of dietary di(2-ethylhexyl) phthalate (DEHP) in rats using UPLC/Q-TOF-MS and studied the toxicity mechanism of DEHP. The UPLC-MS-based metabolomic method was extensively applied for the renal toxicity study of traditional Chinese medicines. UPLC-MS is considered suitable for large-scale untargeted metabolic profiling in complex biological samples, and it provides important technical conditions for studying the mechanism of nephrotoxicity.14,15 For example, UPLC-MS metabolomics was used to study the nephrotoxicity of R. alismatis in rats, and it was also employed to investigate urinary metabolites from melamine-exposed children.16 Ying-Yong Zhao et al. used UPLC-QTOF/HDMS to study the nephrotoxicity mechanism of aristolochic acid and explain the mechanism of renal toxicity from the disorder of metabolic pathways and changes in biomarkers.17–19 Lipids with small molecular weights such as glycerolipids, sphingolipids, glycerophospholipids (GPs), and fatty acids have diverse and complex functions with respect to health and diseases. Moreover, they also play an important role in the regulation of normal renal function and pathogenesis of kidney illness. UPLC-MS can also be used in lipid metabolomics to detect these compounds.20
In our study, ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC/Q-TOF-MS) was used to detect the endogenous metabolic fingerprints of renal toxicity induced by SNV and TwHF. By using principal component analysis (PCA) and partial least squares-discriminant analysis (PLS-DA), the independent sample t-test was used for variables of variable importance projection (VIP) value >1 extracted from the PLS-DA model. The substances with p < 0.05 were considered statistically significant. Subsequently, their respective potential biomarkers are screened out and looking for their common and differential plasma markers. Combined with the database of metabolic pathways, possible common and differential mechanisms of nephrotoxicity of the two medicines were explored, which can provide support for the early detection and safety evaluation of the kidney toxicity in traditional Chinese medicine. It is also significant for the rational clinical use of the Chinese medicines SNV and TwHF and for early warning of their toxicity.
2. Materials and methods
2.1. Materials and reagents
TwHF (Hebei Qiyi Tang Pharmaceutical Co., Ltd, China, batch number 1404266), SNV (Anguo Shengshan Pharmaceutical Co., Ltd China, batch number 141018): medicinal materials were identified by Associate Professor Hu Jing of Tianjin University of Traditional Chinese Medicine. Acetonitrile (HPLC-grade, Oceanpak, Sweden), formic acid (HPLC-grade, ROE, USA), ethanol (HPLC-grade, Oceanpak, Sweden), distilled water (Hangzhou WaHaha, China), normal saline (Shandong Qi Pharmaceutical Co., Ltd, China), automatic biochemical analyzer 7020 (Hitachi, Japan), ALLLEGRATM-64R high speed centrifuge (Beckman, United States), KQ-300DV numerical control ultrasonic cleaner (Jiangsu Kunshan Ultrasonic Instrument Co., Ltd, China), XK96-A Fast Mixer (Jiangyan Xinkang Medical Instruments Co., Ltd, China), Waters Acquity UPLC liquid chromatograph (Waters, USA), Waters Xevo G2 Q-TOF-MS mass spectrometer (Waters, USA), UPLC HSS C18 column (2.1 mm × 100 mm, 1.7 μm) (Waters, USA), and R-220SE rotary evaporator (BUCHI, Germany) were used in this study.
2.2. Extraction of traditional Chinese medicine
SNV and TwHF were tested in accordance with the Chinese Pharmacopoeia 2015 Edition and content identification under SNV and TwHF. The above-mentioned two medicinal materials were crushed and weighed separately, and 10 times the amount of water was added. The water bath was heated and refluxed for 1 hour. Then, 8 times the amount of water was added for the second time; the water bath was heated and refluxed for 1 hour. The two filtrates were combined and recovered under reduced pressure and constant volume.
2.3. Animal treatment
Thirty male Wistar rats weighing 200 ± 10 g were provided by Sibei Fu (Beijing, China) Experimental Animal Science and Technology Co., Ltd [SCXK (Jing) 2012-0001] and housed at the Experimental Animal Center of Tianjin Institute of Radiology animal laboratory in SPF conditions. The rats were raised in controlled environmental conditions with room temperature of 23 ± 2 °C, relative humidity of 35 ± 5%, and a light–dark cycle of 12 h. After a week of adaptation, the rats were randomly divided into three groups: normal saline group (NS), TwHF group (TwHF) and SNV group (SNV). The rats in each group were given adequate water and food. According to 10–25 times of the clinical dosage (based on the crude medicines), the dosage of each group of rats was obtained by calculating body surface area. The normal saline group was given 1 ml of normal saline. The doses and sample collection for each group are shown in Table 1. This study was approved by the Animal Ethics Committee of Tianjin University of Traditional Chinese Medicine under permit number TCM-2012-078F01. All experimental procedures were conducted in accordance with Chinese national legislation and local guidelines.
Table 1. The dose and sample collection of each group.
| Group | Medicine | Quantity | Dose | Mode of administration | Modeling |
| NS | Normal saline | 10 | 1 mL | Gavage, continuous administration | 7 days |
| TwHF | TwHF extracted solution | 10 | 60 g kg–1 d–1 | Gavage, continuous administration | 7 days |
| SNV | SNV extracted solution | 10 | 0.1 g kg–1 d–1 | Gavage, continuous administration | 7 days |
2.4. Pathological and biochemical examination
Rats in each group were sacrificed after taking blood samples. The same portion of the kidney was taken from each group, and blood stains were washed with normal saline. The kidney tissues were then immersed in a 10% formaldehyde solution and stored. The kidney tissues were stained with hematoxylin and eosin (H&E), and pathological changes were observed under an optical microscope. The paraffin section was dewaxed and hydrated with dimethylbenzene, stained with hematoxylin for 10 minutes, differentiated, stained with eosin, and dehydrated. Dimethylbenzene was transparent, and neutral resin was used to seal the pieces. The pathological features of kidneys were observed under 100× optical microscope, and optical microscopy and histological evaluation were employed to observe the damage to the kidney tissue. Cr and BUN contents in serum were determined by an automatic biochemistry analyzer.
2.5. Sample collection and preparation
Before samples were collected, all animals were subjected to fasting for 12 hours. Blood samples (10 mL) from rats in each group were collected via abdominal aorta blood draws 7 days after administration. Blood was centrifuged at 3500 rpm at 4 °C for 15 min to isolate the supernatant. The supernatant was centrifuged at 3500 rpm at 4 °C for 8 min and then, it was extracted again and stored at –80 °C in a freezer for subsequent biochemical analyses. The other 5 mL of whole blood was centrifuged at 3500 rpm and 4 °C for 15 min, and the supernatant was taken as serum and stored in a refrigerator at –80 °C for biochemical indicator detection.21
Plasma samples were stored at –80 °C for metabonomic analysis. Before testing, the plasma samples were re-dissolved at room temperature. Acetonitrile (300 μL) was added to each 100 μL of plasma for protein precipitation. The mixture was vortexed and mixed for 1 min, sonicated in an ice water bath for 10 min, and centrifuged at 13 000 rpm at 4 °C for 15 min.22 The supernatant was removed and analyzed by UPLC/Q-TOF-MS. Quality control (QC) samples were used for methodological studies. The same amount of plasma was obtained from each sample and mixed as QC samples.23 QC samples were prepared using the same sample preparation procedure. The specific preparation process for QC is as follows: the plasma samples frozen in the refrigerator at –80 °C were thawed at room temperature. Each sample was pipetted with 20 μL of plasma sample in the same centrifuge tube, and the mixture was vortexed for 1 min. Then, 300 μL of acetonitrile was added to 100 μL of plasma, vortexed for 1 min, ultrasonicated for 10 min and centrifuged at 13 000 rpm for 15 min at 4 °C. Then, 200 μl supernatant was removed into a vial for UPLC/Q-TOF-MS analysis.
2.6. Chromatographic and mass spectrometric conditions
Data acquisition was performed on the UPLC/Q-TOF-MS system (Waters, USA). Five μL aliquot of the supernatant was injected into an ACQUITY UPLC HSS C18 column (2.1 × 100 mm, 1.7 μm, Waters). The column temperature was set to 40 °C, and the flow rate was 0.3 mL min–1. A binary solvent system of mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile) was used for the UPLC separation system. Gradient elution was used in this experiment, and the gradient program was designed as follows: 0–0.5 min, A: 99–99%; 0.5–2 min, A: 99–50%; 2–9 min, A: 50–1%; 9–10 min, A: 1–1%; 10–10.5 min, A: 1–99%; and 10.5–12 min, A: 99–99%. The UPLC system was coupled to Q-TOF-MS equipped with a positive ion mode electrospray ionization device. The MS parameters were as follows: drying gas flow, 10 mL min–1; drying gas temperature, 350 °C; capillary voltage, 310 kPa; desolvation gas flow, 600 L h–1; counter-blowing nitrogen, 50 L h–1; capillary ionization voltage, 2.1 kV; evaporative and auxiliary gases, high-purity nitrogen. The range of data acquisition was 50–1000 Da, and all samples were injected randomly.
2.7. Method validation
Before sample analysis, QC samples were injected six times to monitor instrument stability. The six QC samples were then parallel processed and injected to assess the repeatability of the method. Moreover, a QC sample was injected once every six hours. QC samples were stored at 4 °C and analyzed within 24 hours to ensure sample and instrument stability. The entire system was determined to be under stable conditions before injecting the samples.
2.8. Data processing
In this study, the UPLC/Q-TOF-MS technique was used to perform metabolomic analysis of plasma of the rat model for toxicity induced by SNV and TwHF. Through multivariate statistical analysis and data integration analysis, toxicity-related biomarkers of TwHF and SNV were searched, and their biological mechanisms were explained. The specific data processing procedure was as follows: raw data was collected by the Masslynx V4.1 software (Waters Corp., Manchester, UK) in the workstation, and multivariate statistical analysis was performed using the SIMCA-P + 11.5 statistical software (Umetrics AB, Umea, Sweden). First, unsupervised discriminant analysis was performed using PCA to eliminate outlier samples. Next, supervised partial least squares-discriminant analysis (PLS-DA) was performed, and compounds that significantly contributed to the classification (VIP > 1) were selected as candidate markers. Subsequently, SPSS version 17.0 (SPSS Inc., Chicago, IL) was used to perform an independent sample T-test to determine whether the metabolite exhibited significant change from the statistical point of view. Finally, the substance exhibiting p-value < 0.05 was used as a marker for significant differences. At the same time, potential biomarkers were identified by using the mass number (m/z value) of markers in the HMDB (; http://www.hmdb.ca/) database and the KEGG (; http://www.genome.jp/kegg/) database as well as in literature.24
3. Results and discussion
3.1. Histopathological assessment and biochemical analysis
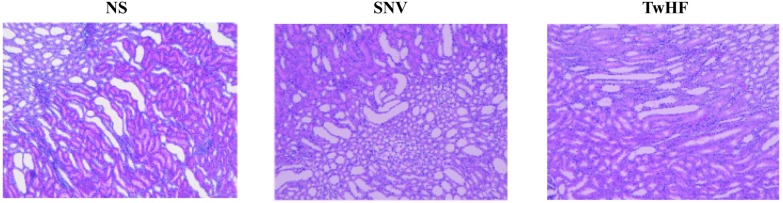
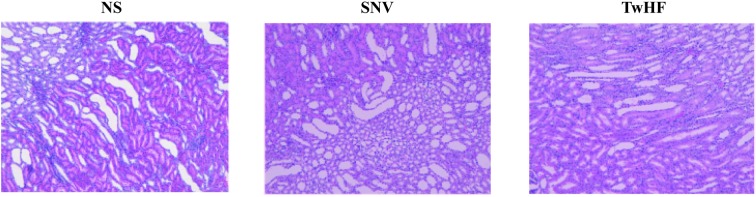
Histopathological sections can intuitively reflect the degree of organ damage.25 After 7 days of administration, histopathological results (Fig. 1) showed that the kidneys in the normal saline group did not exhibit significant injuries. The kidneys of the administration group compared with those of the normal saline group exhibited varying degrees of interstitial loose (the interstitial of the renal cells are in a loose state) and inflammatory cell infiltration; the proximal and distal tubules showed atrophy and dilation, and there was slight expansion of the renal pelvis. The results demonstrated that TwHF and SNV caused pathological changes in the kidneys under the conditions of this experiment, which resulted in damage to the kidneys, thus revealing nephrotoxicity.
Fig. 1. Histopathological sections of the kidney (100× magnification).
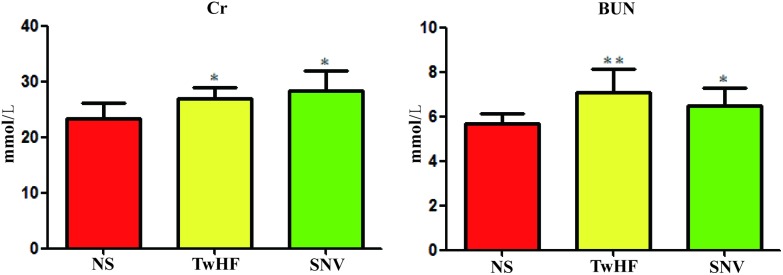
The detection and analysis of biochemical zymogram in serum have important implications in understanding the degree of organ damage caused by toxic effects.26 The change in serum biochemical indicators Cr and BUN reflects the damage cause by medicines to kidneys to a certain extent. In this study, we compared the Cr and BUN T-test results of the normal saline group and drug delivery group and evaluated whether kidney damage was caused by SNV or TwHF. After 7 days of administration, the levels of Cr and BUN in SNV and TwHF groups were significantly different (p-value < 0.05) compared to those of the normal saline group. The two indicators increased, as shown in Fig. 2. Biochemical results showed that both SNV and TwHF lead to kidney damage.
Fig. 2. Content changes of Cr and BUN in normal saline group, TwHF group and SNV group (compared with normal saline group: *P < 0.05, **P < 0.01).
3.2. Method validation
The method validation results are shown in Table 2. In method validation, the peak area must have an RSD value of less than 15%, and the retention time must have an RSD value of less than 5% to meet metabolomics requirements. As can be seen from the results shown below, instrument precision, method repeatability, and sample stability all met the requirements of metabolomics.
Table 2. The results of the experimental methodology.
| Experiment name | RSD (peak area) | RSD (Rt) |
| Instrument precision | <10.6% | <0.6% |
| Method repeatability | <10.8% | <0.2% |
| Sample stability | <12.0% | <0.2% |
3.3. Metabolic profiling and data processing
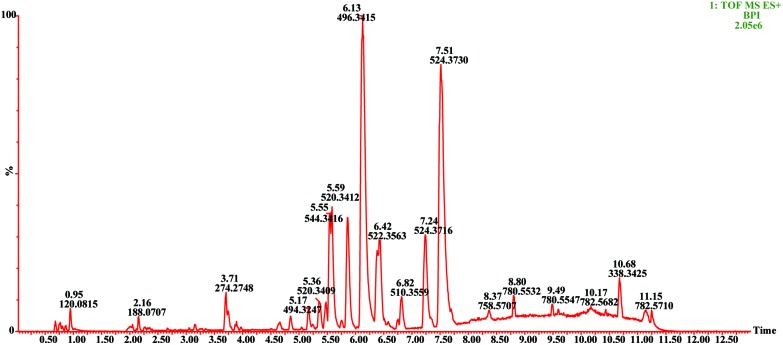
UPLC/Q-TOF-MS analysis was applied to plasma metabolites of different groups of rats. The BPI map of plasma QC samples under positive ion mode is shown in Fig. 3.
Fig. 3. BPI diagram of plasma QC sample in positive ion mode.
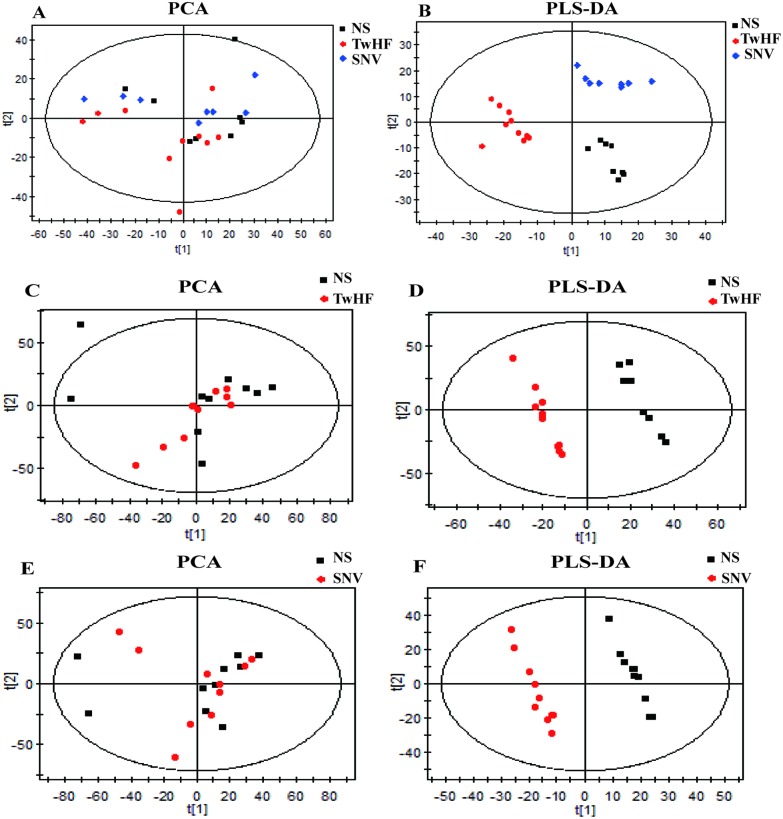
In this experiment, plasma sample data obtained by UPLC/Q-TOF-MS were subjected to a metabolomic study using the SIMCA-P + 11.5 multivariate statistical analysis software. First, unsupervised data analysis was performed using PCA; it can be observed from Fig. 4(A) that there was no good distinction between the three groups, i.e., the SNV, the TwHF and the normal saline groups. Therefore, we inferred that there is a need for further supervised metabolomic analysis using PLS-DA. Fig. 4(B) shows that the three groups exhibited significant classification and aggregation, indicating that both SNV and TwHF caused significant changes in the body's endogenous metabolites and that the two groups were distinctly differentiated at the metabolic level.
Fig. 4. Results of multivariate statistical analysis. (A) PCA score plots of TwHF and SNV compared with those of NS groups; (B) PLS-DA score plots of TwHF and SNV compared with those of NS groups; (C–F) PCA score plots and PLS-DA score plots of TwHF and SNV compared with those of NS groups, respectively.
In this experiment, first, PCA was used to eliminate outlier samples and then, PLS-DA was used to determine different groups of differential metabolites. In Fig. 4, the PCA scores of the normal saline group and TwHF group (Fig. 4(C)) and those of the normal saline group and SNV group (Fig. 4(E)) are shown. The samples outside the confidence interval are outlier samples, which should be excluded. Second, based on the PLS-DA model, as shown in Fig. 4(D) and (F), we further obtained potential information and screened the potential biomarkers of toxicity between the normal saline group and the drug groups. The values of R2 and Q2 in the PLS-DA model established in this experiment were high, indicating that the multivariate statistical model is good, which ensures the accuracy of the analysis results. These markers were then subjected to a student T-test to obtain a significant change (p < 0.05) of nephrotoxicity biomarkers. The m/z values of the markers were used to find possible substances in the HMDB (; http://www.hmdb.ca/) database and the Chemspider (; http://www.chemspider.com/) database, and candidate biomarkers were identified using MS/MS analysis, metabolite database information, and literature.
Finally, in this experiment, through comparison with the saline group, 15 toxic biomarkers of SNV and 17 toxic biomarkers of TwHF were screened. There were differences in the types of markers for the two drugs. Specific ion information of the markers is shown in Table 3, which mainly includes LPC (16:1), LPC (20:5), LPC (22:5), thymidine, LPC (18:4) and other nephrotoxic biomarkers. The main metabolic pathways involved are phenylalanine, tyrosine and tryptophan biosynthetic pathways, phenylalanine metabolic pathways, and tryptophan metabolic pathways. The results showed that both SNV and TwHF caused significant metabolic profile changes in rats.
Table 3. Identified potential biomarkers in the SNV group and TwHF group.
| No. | TR (min) | m/z obsd | m/z calcd | ppm | Metabolite | Formula | Fragment | P value | AUC | Tendency | Medicine |
| 1 | 6.74 | 548.3717 | 548.3716 | 0.18 | LPC(20:2) | C28H54NO7P | 570.3 [M + Na]+ | 0.033 | 0.825 | ↓ | TwHF and SNV |
| 548.3 [M + H]+ | |||||||||||
| 184.0 [M + H – C23H40O3]+ | |||||||||||
| 104.1 [M + H – C23H41O6P]+ | |||||||||||
| 2 | 3.03 | 347.221 | 347.2222 | –3.46 | 19-Hydroxydeoxycorticosterone | C21H30O4 | 385.1 [M + K]+ | 0.040 | 0.797 | ↑ | SNV |
| 369.2 [M + Na]+ | |||||||||||
| 347.2 [M + H]+ | |||||||||||
| 329.2 [M + H – H2O]+ | |||||||||||
| 109.1 [M + H – C15H22O3]+ | |||||||||||
| 97.1 [M + H – C15H22O3]+ | |||||||||||
| 3 | 0.77 | 162.1124 | 162.1125 | –0.62 | l-Carnitine | C7H15NO3 | 162.1 [M + H]+ | 0.027 | 0.859 | ↓ | SNV |
| 103.0 [M + H – C3H9N]+ | |||||||||||
| 4 | 4.84 | 490.2911 | 490.291 | 0.20 | LPC(14:0) | C22H46NO7P | 490.2 [M + Na]+ | 0.035 | 0.775 | ↓ | TwHF and SNV |
| 468.3 [M + H]+ | |||||||||||
| 450.3 [M + H – H2O]+ | |||||||||||
| 184.0 [M + H – C17H32O3]+ | |||||||||||
| 104.1 [M + H – C17H33O6P]+ | |||||||||||
| 5 | 2.99 | 190.0867 | 190.0862 | 2.63 | 3-Indolepropionic acid | C11H11NO2 | 190.1 [M + H]+ | 0.037 | 0.775 | ↓ | TwHF |
| 144.1 [M + H – CH2O2]+ | |||||||||||
| 6 | 6.02 | 546.3556 | 546.3554 | 0.37 | LPC (20:3) | C28H52NO7P | 546.4 [M + H]+ | 0.026 | 0.938 | ↓ | TwHF and SNV |
| 528.4 [M + H – H2O]+ | |||||||||||
| 184.1 [M + H – C23H38O3]+ 104.1 [M + H – C23H39O6P]+ | |||||||||||
| 7 | 5.51 | 478.2932 | 478.2928 | 0.84 | LPE (18:2) | C23H44NO7P | 478.3 [M + H]+ | 0.028 | 0.797 | ↑ | SNV |
| 337.3 [M + H – C21H37O3]+ | |||||||||||
| 8 | 2.14 | 205.0969 | 205.0972 | –1.46 | Tryptophan | C11H12N2O2 | 205.1 [M + H]+ | 0.029 | 0.875 | ↓ | TwHF and SNV |
| 188.1 [M + H – NH3]+ | |||||||||||
| 9 | 3.72 | 431.2764 | 431.2768 | –0.93 | Cholic acid | C24H40O5 | 431.3 [M + Na]+ | 0.042 | 0.750 | ↓ | TwHF and SNV |
| 391.2 [M + H – H2O]+ | |||||||||||
| 10 | 2.13 | 232.1526 | 232.1543 | –7.32 | Isobutyryl-l-carnitine | C11H21NO4 | 232.2 [M + H]+ | 0.041 | 0.797 | ↓ | TwHF and SNV |
| 173.0 [M + H – C3H9N]+ | |||||||||||
| 85.0 [M + H – C7H18NO2]+ | |||||||||||
| 11 | 0.8 | 90.0549 | 90.0549 | 0.00 | Alanine | C3H7NO2 | 90.0 [M + H]+ | 0.015 | 0.828 | ↓ | TwHF and SNV |
| 72.0 [M + H – H2O]+ | |||||||||||
| 12 | 5.78 | 508.3397 | 508.3398 | –0.20 | LPE (20:1) | C25H50NO7P | 508.3 [M + H]+ | 0.006 | 0.875 | ↓ | TwHF |
| 367.4 [M + H – C21H37O3]+ | |||||||||||
| 13 | 2.02 | 166.0865 | 166.0863 | 1.20 | Phenylalanine | C9H11NO2 | 166.1 [M + H]+ | 0.018 | 0.788 | ↓ | TwHF and SNV |
| 120.1 [M + H – CH2O2]+ | |||||||||||
| 14 | 5.57 | 520.3406 | 520.3398 | 1.54 | LPC (18:2) | C26H50NO7P | 520.3 [M + H]+ | 0.020 | 0.862 | ↑ | TwHF |
| 502.3 [M + H – H2O]+ | |||||||||||
| 184.1 [M + H – C21H36O3]+ 104.1 [M + H – C21H37O6P]+ | |||||||||||
| 15 | 6.19 | 400.3423 | 400.3421 | 0.50 | Palmitoylcarnitine | C23H45NO4 | 400.3 [M + H]+ | 0.010 | 0.888 | ↓ | TwHF and SNV |
| 341.2 [M + H – C3H9N]+ 144.1 [M + H – C16H32O2]+ 85.0 [M + H – C19H41NO2]+ | |||||||||||
| 16 | 5.18 | 494.3244 | 494.3241 | 0.61 | LPC (16:1) | C24H48NO7P | 494.3 [M + H]+ | 0.048 | 0.763 | ↓ | TwHF and SNV |
| 476.3 [M + H – H2O]+ | |||||||||||
| 184.1 [M + H – C19H34O3]+ 104.0 [M + H – C19H35O6P]+ | |||||||||||
| 17 | 5.59 | 542.3222 | 542.3241 | –3.50 | LPC (20:5) | C28H48NO7P | 542.3 [M + H]+ | 0.007 | 0.875 | ↓ | TwHF |
| 524.3 [M + H – H2O]+ | |||||||||||
| 259.1 [M + H – C17H33NO2]+ | |||||||||||
| 184.0 [M + H – C23H34O3]+ | |||||||||||
| 125.0 [M + H – C26H43NO3]+ | |||||||||||
| 104.1 [M + H – C23H35O6P]+ | |||||||||||
| 18 | 6.17 | 570.3545 | 570.3554 | –1.58 | LPC (22:5) | C30H52NO7P | 570.4 [M + H]+ | 0.022 | 0.862 | ↓ | TwHF |
| 552.3 [M + H – H2O]+ | |||||||||||
| 184.0 [M + H – C25H38O3]+ | |||||||||||
| 125.0 [M + H – C28H47NO3]+ | |||||||||||
| 104.1 [M + H – C25H38O6P]+ | |||||||||||
| 19 | 2.52 | 243.1007 | 243.0975 | 13.16 | Thymidine | C10H14N2O5 | 243.1 [M + H]+ | 0.029 | 0.800 | ↓ | TwHF and SNV |
| 127.1 [M + H – C5H8O3]+ | |||||||||||
| 20 | 5.18 | 516.3059 | 516.3085 | –5.04 | LPC (18:4) | C26H46NO7P | 516.3 [M + H]+ | 0.022 | 0.813 | ↓ | TwHF and SNV |
| 498.3 [M + H – H2O]+ | |||||||||||
| 184.0 [M + H – C21H32O3]+ | |||||||||||
| 104.1 [M + H – C21H33O6P]+ |
3.4. Optimization by ROC curve
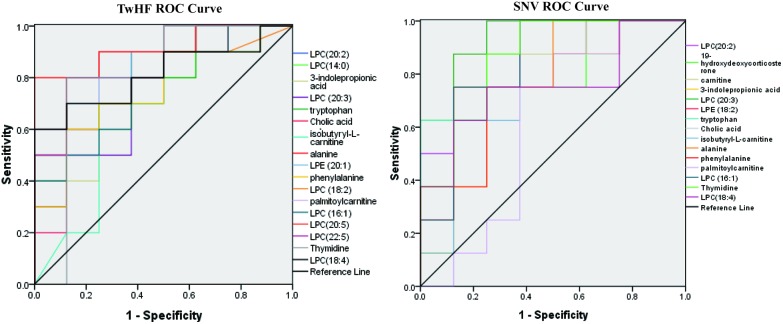
Through the above-mentioned experiments, we have found the toxic biomarkers of TwHF and SNV. To further explore whether these markers are meaningful, we use ROC curves to verify them. The ROC curve is widely used to evaluate sensitivity and specificity of markers. Diagnostic efficiency is evaluated by the area under the curve (AUC). If AUC is greater than 0.7, the biomarker is relatively unique and can be used as a diagnostic biomarker.27 Combined with the information of the screened toxicological markers, ROS curves and binary logistic regression model are established using the SPSS 17.0 software (SPSS Inc., Chicago, IL) to evaluate sensitivity and specificity of the biomarkers and screen more diagnostic biomarkers.21 Through the ROC curve, as shown in Fig. 5, we obtain AUCs for SNV and TwHF markers. It is found that AUCs of all substances are greater than 0.7, and their distribution is in the confidence interval of 95%, indicating that these biomarkers have good diagnostic capabilities for toxicities of the two medicines.
Fig. 5. Roc curves of biomarkers in TwHF and SNV groups.
3.5. Metabolomic pathway analysis (MetPA)
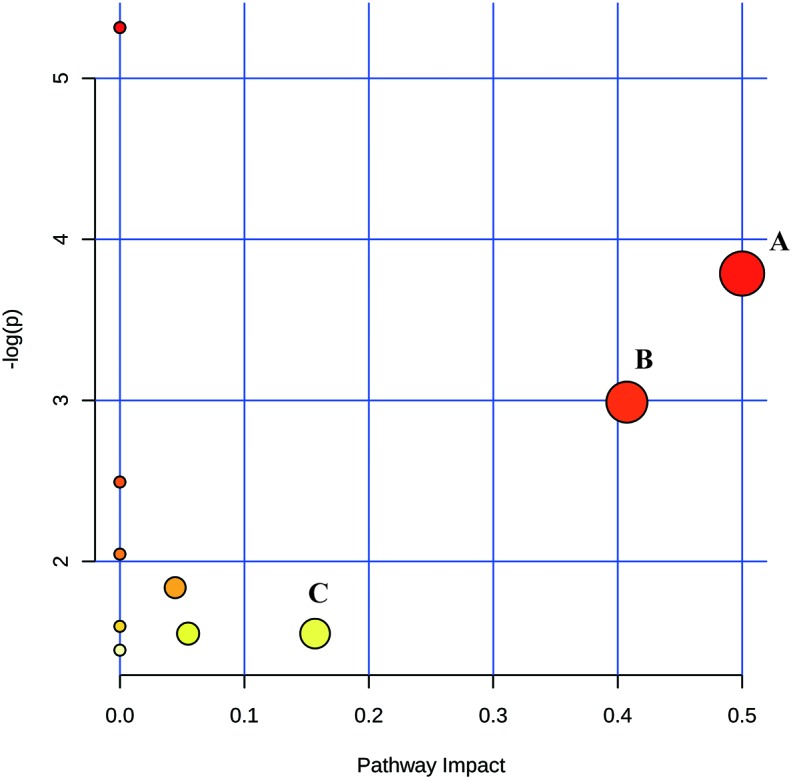
MetPA uses the KEGG metabolic pathway as the back-end knowledge base and expresses the analysis results in an intuitive way; thus, it is widely applied in metabolomics research.28 We analyzed the metabolic pathways of selected toxic biomarkers of SNV and TwHF. The results are shown in Fig. 6. In MetPA analysis, the metabolic pathways with a value greater than 0.1 were considered to be associated with kidney toxicities of SNV and TwHF. It could be concluded from Fig. 6 that TwHF and SNV affected the body's nephrotoxicity mainly by affecting phenylalanine, tyrosine and tryptophan biosynthetic pathways, phenylalanine metabolic pathway and tryptophan metabolic pathway. In addition, the metabolic pathways affecting nephrotoxicity of SNV and TwHF were consistent, and the pathways affected by the two drugs were not different. There were differences in the types of toxic biomarkers of the two drugs, and these differences did not affect the main metabolic pathways.
Fig. 6. Disturbed pathways (A: phenylalanine, tyrosine and tryptophan biosynthetic pathways; B: phenylalanine metabolic pathway; C: tryptophan metabolic pathway).
3.6. Biological significance of biomarkers for nephrotoxicity
3.6.1. Phospholipid metabolism
LPC is produced in vivo by PC under the action of phospholipase A2 or by lysophospholipase I during the lipid oxidation process.29 LPCs are a class of endogenous phospholipids and a type of inflammatory factor. LPCs in millimolar concentrations have the effects of aggregating monocytes and promoting the production of inflammatory cytokines in macrophages.30,31 After rats were given extracts of TwHF and SNV, most LPC substances in plasma were reduced, indicating that the rats under the action of medicines were in a state of severe immunosuppression. It has been reported that changes in the levels of LPCs in plasma may be related to drug nephrotoxicity.32,33 When the kidney is damaged, the body may consume a large amount of PCs to remove peroxide and achieve a normal oxidative stress level, which can affect the metabolism of glycerol phospholipid, resulting in decrease in the content of LPCs. This agrees with our experimental results that nephrotoxicity causes decrease in the content of LPC. Among the biomarkers screened, we found that LPC (20:2), LPC (14:0), LPC (20:3), LPE (18:2), LPE (20:1), LPC (18:2), LPC (16:1), LPC (20:5), LPC (22:5), and LPC (18:4) participated in the metabolism of phospholipids. LPE (18:2) was found only in the biomarkers of the SNV group. LPE (20:1), LPC (18:2), LPC (20:5), and LPC (22:5) were found only in the TwHF group. The other markers were found in both groups. It could be seen that the extent of phospholipid changes induced by the two drugs was different, leading to differences in these markers. The significant changes in plasma phospholipids in the two groups also indicated that SNV and TwHF could inhibit or interrupt the metabolism of phospholipids. The metabolism of lipids in the two groups was disordered, but the markers that caused the disorder were slightly different. In this experiment, many phospholipids were significantly changed by TwHF. Therefore, we can conclude that abnormal plasma phospholipid metabolism is closely related to kidney injury induced by traditional Chinese medicines.
3.6.2. Amino acid metabolism
Amino acids are important carbon and nitrogen sources in organisms as well as precursors of proteins and nucleotides. They are involved in the synthesis of proteins and nucleotides in the body.34 Renal damage is often accompanied by metabolic disorders of various amino acids including a decrease in the proportion of essential amino acids and non-essential amino acids and a decrease in the level of serum branched-chain amino acids, which is associated with an increase in the consumption of protein and amino acids in patients with renal injury.35 In this experiment, alanine, phenylalanine, 3-indolepropionic acid, and tryptophan were selected as markers associated with amino acid metabolism, and it was found that the levels of all four amino acids significantly reduced. Phenylalanine is an essential amino acid in the human body and is used for protein synthesis in vivo. Phenylalanine can produce ketones and carbohydrates. After kidney injury, it is necessary to accelerate the repair of the tissue, and consumption of phenylalanine can provide the energy required for this process.36 Tryptophan is an important precursor metabolite of the neurotransmitter 5-hydroxytryptamine in the body. Tryptophan is one of the 8 essential amino acids in the human body and is used in most of the body for protein synthesis.37 In this experiment, the decrease in tryptophan in the plasma of the two groups showed that kidney function was abnormal due to SNV and TwHF, which seriously affected the metabolism of tryptophan.
3-Indolepropionic acid is also a substance in the tryptophan metabolism pathway, and its level was also significantly reduced compared to that of the normal saline group. In the administration groups, only the TwHF group showed significant change in the content of 3-indolepropionic acid. Therefore, when TwHF caused damage to the kidney, it interfered with tryptophan metabolism, resulting in decrease in the levels of phenylalanine, tryptophan and 3-indolepropionic acid. At the same time, 3-indolepropionic acid can inhibit oxidative stress reaction, remove free radicals, prevent free radicals from attacking biological macromolecules and causing tissue cell damage, and delay aging of the body and other nervous system diseases.38 The decrease of 3-indolepropionic acid in the plasma of TwHF group may be related to renal tissue damage caused by TwHF, which indicated the difference in nephrotoxicities caused by TwHF and SNV.
In this study, significant reductions in amino acids in the SNV group and TwHF group suggest disorder of amino acid metabolism. This indicates that abnormal metabolism of amino acids is induced by drugs in rats.
3.6.3. Energy metabolism
l-Carnitine, palmitoylcarnitine and isobutyryl-l-carnitine are markers of energy metabolism. After the body is damaged, the level of carnitine in plasma also changes. Carnitine compounds have been considered as reliable biomarkers for determining whether energy metabolism is abnormal.39 Carnitine compounds are related to metabolism and transport of fatty acids in animals. Their main function is to transport long chain fatty acids from outside the mitochondrial membrane to the membrane to promote beta oxidation of fatty acids and conversion of fat metabolism to energy.40 Palmitoylcarnitine is an acylcarnitine that transfers lipids to mitochondria, providing energy necessary for cell growth and survival. Fatty acids can be transported to mitochondria by binding to l-carnitine under the action of carnitine palmitoyltransferase-1. In this experiment, three carnitine markers were significantly reduced in the administration groups, indicating that both SNV and TwHF caused disorder of energy metabolism, which resulted in slow energy supply and a series of toxic reactions in the body.
l-Carnitine was found only in the TwHF group. It is a type of special amino acid that exists widely in the body tissue, and it is necessary for energy metabolism of animals. l-Carnitine plays an important role in fat metabolism and has anti-inflammatory, anti-oxidative and anti-apoptotic effects.41l-Carnitine can effectively reduce inflammatory factors in serum, thereby improving the inflammatory state of heart, lung, kidney and other important organs in failure.42 Significant decrease in l-carnitine in the TwHF group indicated that TwHF affected the content of l-carnitine and then affected the metabolism of fatty acids and the inflammatory state of the body. There are subtle differences in the energy metabolism abnormalities that result from nephrotoxicity caused by SNV.
3.6.4. Pyrimidine metabolism
Thymidine is a pyrimidine deoxynucleoside. Thymidine in the SNV group and TwHF group was significantly reduced, indicating disorder of pyrimidine metabolism in rats with renal injury.
3.6.5. Bile acid metabolism
Cholic acid is a major bile acid. The kidney plays a significant role in the process of bile acid recirculation.43 Kidney injury leads to disorder of bile acid metabolism, which causes decrease in the content of cholic acid in plasma of SNV and TwHF groups. Abnormal bile acid metabolism is also one of the important mechanisms of drug-induced kidney damage.
4. Conclusion
In recent years, the safety of traditional Chinese medicine has become a social concern. Toxicity and adverse reactions caused by the use of traditional Chinese medicine have become areas of extreme interest for the academic community. Because the chemical compositions of traditional Chinese medicines are complex and the extraction and separation of most components are difficult, there are limitations in evaluating the toxicity of traditional Chinese medicines using in vivo and in vitro toxicology experimental methods. The toxicity of traditional Chinese medicine can be studied rapidly and effectively using small-molecule metabolic markers. Using metabonomics, we can also systematically study the toxicities of traditional Chinese medicine and its compounds. At the same time, it can also rapidly give an early warning of toxicity before the tissues and organs are pathologically damaged. It can also be used in the pharmaceutical industry and the national drug testing department. The significance of finding biomarkers is early prediction of toxicity of traditional Chinese medicine. The toxic biomarkers of SNV and TwHF selected in this experiment are advantageous in the evaluation of medicine safety. In this experiment, we established a metabolomics analysis method for medicine toxicity, which can reflect the changes in plasma small-molecule metabolites in vivo and provide toxicity assessment of renal and other tissues at the stage of biochemical detection while the tissue is pathologically undamaged. It can identify toxic substances and toxicity routes faster and more accurately than traditional methods, providing a basis for medicine safety evaluation and secondary development. At the same time, the biological significance of the nephrotoxicity of SNV and TwHF interference with normal metabolism in vivo is explained, which provides a reference for comprehensive and clear explanation of toxicity mechanisms. By comparing rat nephrotoxicities of SNV and TwHF, we inferred that both SNV and TwHF could cause experimental renal functional changes in rats with different properties and in varying degrees. This study provides a meaningful reference for the study of nephrotoxicity mechanism of traditional Chinese medicine.
Conflicts of interest
There are no conflicts of interest to declare.
Acknowledgments
This project was supported by the National Natural Science Foundation of China (No. 81573825).
References
- Patel K., Laloo D., Singh G., Gadewar M., Patel D. Chin. J. Integr. Med. 2017:1–13. doi: 10.1007/s11655-016-2514-1. [DOI] [PubMed] [Google Scholar]
- Ma B., Hu T., Li P., Yuan Q., Lin Z., Tu Y., Li J., Zhang X., Wu X., Wang X., Huang L., Gao W. Ecology and evolution. 2017;7:8612–8623. doi: 10.1002/ece3.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. X., Du F. Y., Liu H. X., Ji J. B., Xing J. J. Ethnopharmacol. 2015;162:238–243. doi: 10.1016/j.jep.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Prat S., Hoizey G., Lefrancq T., Saint-Martin P. J. Forensic Sci. 2015;60:816–817. doi: 10.1111/1556-4029.12706. [DOI] [PubMed] [Google Scholar]
- Brown A. C. Food Chem. Toxicol. 2017;107:502–519. doi: 10.1016/j.fct.2016.07.024. [DOI] [PubMed] [Google Scholar]
- Xu X. L., Yang L. J., Jiang J. G. Expert Opin. Drug Metab. Toxicol. 2016;12:149–159. doi: 10.1517/17425255.2016.1132306. [DOI] [PubMed] [Google Scholar]
- Coen M. Drug Metab. Rev. 2015;47:29–44. doi: 10.3109/03602532.2014.982865. [DOI] [PubMed] [Google Scholar]
- Cao H., Zhang A., Zhang H., Sun H., Wang X. Phytother. Res. 2015;29:159–166. doi: 10.1002/ptr.5240. [DOI] [PubMed] [Google Scholar]
- Zong L., Xing J., Liu S., Liu Z., Song F. Ecotoxicol. Environ. Saf. 2018;147:26–33. doi: 10.1016/j.ecoenv.2017.08.028. [DOI] [PubMed] [Google Scholar]
- Shi J., Cao B., Wang X.-W., Aa J.-Y., Duan J.-A., Zhu X.-X., Wang G.-J., Liu C.-X. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2016;1026:204–216. doi: 10.1016/j.jchromb.2015.10.014. [DOI] [PubMed] [Google Scholar]
- Zhu L., Zhao Z. J., Ren X. B., Li Q., Ding H., Sun Z., Kao Q. J., Wang L. H. Biomed. Environ. Sci. 2018;31:76–80. doi: 10.3967/bes2018.009. [DOI] [PubMed] [Google Scholar]
- Wang C., Feng R., Li Y., Zhang Y., Kang Z., Zhang W., Sun D. J. Toxicol. Lett. 2014;229:474–481. doi: 10.1016/j.toxlet.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Dong X., Zhang Y., Dong J., Zhao Y., Guo J., Wang Z., Liu M., Na X., Wang C. Environ. Sci. Pollut. Res. Int. 2017;24:16659–16672. doi: 10.1007/s11356-017-9091-5. [DOI] [PubMed] [Google Scholar]
- Chen D. Q., Chen H., Chen L., Tang D. D., Miao H., Zhao Y. Y. Chem.-Biol. Interact. 2016;252:114–130. doi: 10.1016/j.cbi.2016.03.028. [DOI] [PubMed] [Google Scholar]
- Zhao Y. Y. Clin. Chim. Acta. 2013;422:59–69. doi: 10.1016/j.cca.2013.03.033. [DOI] [PubMed] [Google Scholar]
- Zhao Y.-Y., Lin R.-C. Adv. Clin. Chem. 2014;65:69–89. [PubMed] [Google Scholar]
- Zhao Y.-Y., Tang D. D., Chen H., Mao J. R., Bai X., Cheng X. H., Xiao X. Y. Bioanalysis. 2015;7:685–700. doi: 10.4155/bio.14.309. [DOI] [PubMed] [Google Scholar]
- Zhao Y. Y., Wang H. L., Cheng X. L., Wei F., Bai X., Lin R. C., Vaziri N. D. Sci. Rep. 2015;5:1–13. doi: 10.1038/srep12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Cao G., Chen D. Q., Wang M., Vaziri N. D., Zhang Z. H., Mao J. R., Bai X., Zhao Y. Y. Redox Biol. 2016;10:168–178. doi: 10.1016/j.redox.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Y., Vaziri N. D., Lin R. C. Adv. Clin. Chem. 2015;68:153–175. doi: 10.1016/bs.acc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Yin J., Xie J., Guo X., Ju L., Li Y., Zhang Y. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2016;1033:428–435. doi: 10.1016/j.jchromb.2016.08.042. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang L., Ju L., Deng H., Zhang Z., Hou Z., Xie J., Wang Y., Zhang Y. Toxicol. Sci. 2016;150:390–399. doi: 10.1093/toxsci/kfw001. [DOI] [PubMed] [Google Scholar]
- Xie G., Lu L., Qiu Y., Ni Q., Zhang W., Gao Y. T., Risch H. A., Yu H., Jia W. J. Proteome Res. 2015;14:1195–1202. doi: 10.1021/pr501135f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Dong W., Liu R., Wang Y., Li Y. Nanotoxicology. 2018;12:18–31. doi: 10.1080/17435390.2017.1415389. [DOI] [PubMed] [Google Scholar]
- Guo X., GU C., Xu Y., Li Y., Zhang Y. Drug Eval. Res. 2017;40:472–478. [Google Scholar]
- Khalid M., Ashraf M. Circ. Res. 1993;72:725–736. doi: 10.1161/01.res.72.4.725. [DOI] [PubMed] [Google Scholar]
- Zhang J., Huang Z., Chen M., Xia Y., Martin F. L., Hang W., Shen H. Fertil. Steril. 2014;102:44–53e12. doi: 10.1016/j.fertnstert.2014.03.033. [DOI] [PubMed] [Google Scholar]
- Mika A., Wojtowicz W., Zabek A., Mlynarz P., Chmielewski M., Sledzinski T., Stepnowski P. J. Pharm. Biomed. Anal. 2018;149:1–8. doi: 10.1016/j.jpba.2017.10.037. [DOI] [PubMed] [Google Scholar]
- Tsutsumi T., Yamakawa S., Ishihara A., Yamamoto A., Tanaka T., Tokumura A. Toxicol. Rep. 2015;2:121–129. doi: 10.1016/j.toxrep.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson K. E., Andersson L., Nilsson J., Bjorkbacka H. Biochem. Biophys. Res. Commun. 2008;370:348–352. doi: 10.1016/j.bbrc.2008.03.087. [DOI] [PubMed] [Google Scholar]
- Radu C. G., Yang L. V., Riedinger M., Au M., Witte O. N. Proc. Natl. Acad. Sci. U. S. A. 2004;101:245–250. doi: 10.1073/pnas.2536801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang X., Zhou H., Fan S., Wang Y., Zhang L., Ju L., Wu X., Wu H., Zhang Y. RSC Adv. 2014;4:8260–8270. [Google Scholar]
- Zhang X., Li Y., Zhou H., Fan S., Zhang Z., Wang L., Zhang Y. J. Pharm. Biomed. Anal. 2014;97:151–156. doi: 10.1016/j.jpba.2014.04.036. [DOI] [PubMed] [Google Scholar]
- Halsey C. R., Lei S., Wax J. K., Lehman M. K., Nuxoll A. S., Steinke L., Sadykov M., Powers R., Fey P. D. mBio. 2017;8:1–19. doi: 10.1128/mBio.01434-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylis C. Am. J. Physiol.: Renal. Physiol. 2008;294:F1–F9. doi: 10.1152/ajprenal.00424.2007. [DOI] [PubMed] [Google Scholar]
- Li M.-H., Ruan L.-Y., Liu Y., Xu H.-D., Chen T., Fu Y.-H., Jiang L., Wang J.-S. Toxicol. Res. 2015;4:1374–1388. [Google Scholar]
- Winn S. R., Scherer T., Thony B., Ying M., Martinez A., Weber S., Raber J., Harding C. O. Mol. Genet. Metab. 2018;123:6–20. doi: 10.1016/j.ymgme.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyan Y. J., Poeggeler B., Omar R. A., Chaint D., Frangione B., Ghiso J., Pappolla M. J. Biol. Chem. 1999;274:21937–21942. doi: 10.1074/jbc.274.31.21937. [DOI] [PubMed] [Google Scholar]
- Fujieda Y., Manno A., Hayashi Y., Rhodes N., Guo L., Arita M., Bamba T., Fukusaki E. PLoS One. 2013;8:e66270. doi: 10.1371/journal.pone.0066270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghonimy A., Zhang D. M., Farouk M. H., Wang Q. Int. J. Mol. Sci. 2018;19:1–11. doi: 10.3390/ijms19041008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier C., Lindquist C., Bjørndal B., Rossmann C. R., Svardal A., Hallström S., Berge R. K. PLoS One. 2018;13:e0194978. doi: 10.1371/journal.pone.0194978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek Mahdavi A., Mahdavi R., Kolahi S. J. Am. Coll. Nutr. 2016;35:597–603. doi: 10.1080/07315724.2015.1068139. [DOI] [PubMed] [Google Scholar]
- Kurz M., Brachvogel V., Matter H., Stengelin S., Thüring H., Kramer W. Proteins: Struct., Funct., Bioinf. 2003;50:312–328. doi: 10.1002/prot.10289. [DOI] [PubMed] [Google Scholar]