Abstract
Last decade's advancements in optofluidics allowed obtaining an ever increasing integration of different functionalities in lab on chip devices to culture, analyze, and manipulate single cells and entire biological specimens. Despite the importance of optical imaging for biological sample monitoring in microfluidics, imaging is traditionally achieved by placing microfluidics channels in standard bench‐top optical microscopes. Recently, the development of either integrated optical elements or lensless imaging methods allowed optical imaging techniques to be implemented in lab on chip systems, thus increasing their automation, compactness, and portability. In this review, we discuss known solutions to implement microscopes on chip that exploit different optical methods such as bright‐field, phase contrast, holographic, and fluorescence microscopy.
Keywords: optofluidics, optical microscopy, lab on chip, brightfield, fluorescence
Optical microscopy is one of the most widely used imaging tools in material sciences, molecular biology, life sciences, and environmental monitoring. The continuous improvement of spatial and temporal resolution of imaging systems has guided the research and the industrial development of optical microscopy worldwide in the last few centuries. At the same time, new contrast mechanisms have been continuously conceived, implemented, and improved including, among others, brightfield, darkfield, phase contrast, holographic, fluorescence, and Raman microscopies 1.
Several challenges need to be faced for the future development of optical microscopy and its adoption in large scale applications, such as (i) point of care diagnostics in healthcare 2, (ii) environmental and pollution monitoring 3, (iii) field analyses and in situ measurement campaigns 4. A major challenge is the reduction of the production costs, which should be compatible with mass production. The need for low cost microscopes is particularly relevant in all developing countries, considering the lack of analysis laboratories and large imaging infrastructures 5, 6, 7. Then, for a widespread diffusion, the development of portable, automated and easy to use optical microscopes is essential. In addition, future microscopy devices should have a flexible design that allows for customization with various samples. Finally, for the study of large sample populations the microscope must have high throughput capabilities. This is required for the majority of pharmaceutical and drug‐screening studies. Concerning throughput optimization, a possible approach is to enlarge the field of view at given frame rate, so to image a larger amount of samples per time, potentially without affecting the image quality 8. An alternative approach consists in replacing the sample under analysis. This can be done by flowing the sample through a fluidic channel, thus permitting a continuous sample replacement in the detection region 9.
As a matter of fact, a valuable solution, which can fulfill all the previously mentioned requirements is given by the use of Lab On a Chip (LOC) platforms 10. These systems aim to concentrate different instrumentations and functionalities, normally present in biological or chemical laboratories, into a single platform with dimensions that typically range from millimeters to few centimeters. In the field of life sciences, these devices offer the possibility to integrate a broad spectrum of features, including chemical sensing, DNA, protein and single cells analysis, single‐cell sorting and PCR or reactor vessels 11, 12, 13, 14, 15, 16. Optofluidic platforms can be also combined with techniques that allow optimizing the analysis throughput 17, 18. Furthermore, LOC allow one to manipulate cells, treat them mechanically and pharmaceutically 19, create cell aggregates, complex three‐dimensional tissue cultures 20, and even replicate in vivo organ functions 21.
The core of LOC devices is usually a network of micro‐channels that are used to process the sample. The cross‐section of the channels can potentially be smaller than the diameter of a human hair, from tens to hundreds of micrometers, allowing one to manipulate small volumes of liquids (generally between 10−9 and 10−18 l)22. The use of LOC has several benefits: (a) the small size of the device reduces the amount of required reagents, solvents and samples; (b) the integration of processes that would require a whole set of bulk instruments reduces the costs and lowers the risk of sample contamination due to external contact; (c) the use of a microfluidic system allows automatic delivery of samples, fostering high throughput capabilities.
Due to these properties, LOC can be the answer to the limits of standard microscopy, in the form of compact optofluidic platforms where both optical and fluidic components are integrated 23, 24. We will refer at these implementations as Microscopes On Chip (MOC), indicating in particular the devices in which microfluidic channels are integrated with at least one optical component within the lab‐on‐chip.
Optofluidics 25, 26, being the science that originates from the synergy of microfluidics and optics, allows one to combine the advantages of the microfluidic handling with the sensitivity of optical detection 27. Two different approaches can be distinguished: the off‐chip and the on‐chip. The first one envisages the use of microfluidic chips with external bulk optical elements, permitting good sensitivity of the measurements, but with the drawback of a potentially complex alignment of the optics with the fluidic microchannel and a limited portability of the chip. The on‐chip approach instead consists in optical elements fully integrated in the microfluidic platform, guaranteeing the compactness and portability of the devices and, in some cases, allowing a stable alignment of the different components.
In this article, we present a review of different approaches to develop MOCs distinguished on the basis of the implemented optical method. We discuss two main categories, namely transillumination (including bright‐field, phase contrast and holographic) and fluorescence microscopy: each category includes different devices that reinvent the standard approach on a compact platform.
As we will discuss, these configurations not only address and overcome the limitations of standard microscopy (i.e., the throughput, the cost and the portability), but in some cases guarantee diffraction limited resolution (or even super‐resolution), comparable with the one obtained with classical instruments. The samples that can be processed in MOCs range from single cells (10–20 μm) to cellular spheroids (200–300 μm), to worms or even embryos (≈1 mm), underlying the high versatility of these platforms.
An additional feature of microfluidic devices is the possibility to optimize the fluidic circuit 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38 to maximize the device throughput when combined with the proper optical imaging technique, the sample portability, the automation, or to facilitate the measurement protocols to perform imaging under specific sample conditions, such as under different drug exposures.
Due to their relevance in term of portability and low cost, smartphone based MOCs are also described in the last section. In these devices the phone can be used as a valuable tool to provide both the light source and the optical detection, while a microfluidic device is mounted on the phone and used to process the sample 39. The use of the phone, both as source and as detector, directly guarantees the compactness of the system, making it potentially useful as a component of the microscope 40 (Table 1).
Table 1.
MOCs: devices where a lab on chip integrates fluidic and optical components (for illumination or detection). Resolution (column 2) is the highest resolution reported in the corresponding references (Refs). NS indicates Non Specified in the reference. LF indicates that the device is lens free.
| Microscope technique | Spatial resolution | Sensor/ pixel size | Detection objective | Samples | Notes | Refs | |
|---|---|---|---|---|---|---|---|
| TRANSILLUMINATION | Shadow Imaging |
NS | CMOS 10 μm | LF | Caenorhabdtis Elegans | Robust and compact lensless platform. Low spatial resolution, limited by the pixel size. | 43 |
| NS | CCD 10 μm | LF | Blood cells, fibroblasts, stem cells, AML‐12 hepatocytes | 44 | |||
| Nanogrid | 0.5 μm | CCD 20 μm | LF | C. elegans | Subpixel resolution, limited by the aperture size. | 45 | |
| < 1 μm | CMOS 9.9 μm | LF | C. Elegans and Chlamydomonas cells | 46 | |||
| In line DH | 0.6 μm | CMOS 2.2 μm | LF | C. Elegans, Giardia lamblia and Mulberry pollen | Large field of view obtained with subpixel shifted acquisitions. Computationally intensive algorithms are required to reconstruct the images. | 47, 53 | |
| NS | CMOS 5.5 μm | LF | Blood cells | Image acquisition flowing the sample at high flow rates. A pulsed laser is required to obtain neat images of samples at high flow rates. | 54 | ||
| SROFM | 0.75 μm | CMOS 3.2 μm | LF | Euglena gracilis, Entamoeba invadens | No need of mechanical precision translation mechanisms. Combines Nanogrid imaging with sub pixel image shifting. | 55 | |
| Off axis DH | NS | CCD 4.8 μm | 20× 0.5NA Printed lens |
Thalassiosira rotula and C. Elegans | Reduced computational efforts. The optical elements are integrated directly on the microfluidic platform. Coherent source required. | 48, 56 | |
| FLUORESCENCE | Lens Free | NS | CCD 9 μm | LF | Microparticles, White blood cells | Large field of view. Low spatial resolution, limited by the pixel size. | 69, 70, 71 |
| FOFM | 0.6 μm | CMOS 5.2 μm | LF | HeLa cells | Compact lensless platform with resolution improved by a Fresnel zone plate array | 73 | |
| LSFM | NS (transverse) 4.6–7.5 μm(axial) |
EMCCD 16 μm | 40×,0.75NA | Membrane vesicles | Light sheet illumination and sample delivery integrated on the chip | 80 | |
| ≈ 0.8 μm (transverse) ≈ 10 μm (axial) |
CMOS‐CCD 5.6–13 μm | 20×, 0.45NA | Tissue mimics (tumor spheroids) | Light sheet illumination, sample delivery and sample scan integrated on the chip | 81 |
Transillumination MOCs
Transillumination refers to microscopy methods that use light transmitted through a specimen, with the light source (coherent or incoherent) positioned on one side of the sample and the detector on the opposite side. It includes, among others, bright field, phase contrast and holographic microscopy.
Transillumination can be label‐free, allowing in many cases a direct analysis of the sample without the need of pretreatment or staining. In literature there are several examples of devices that use transillumination microscopes integrated on a chip, based on different imaging approaches 23, 24, 41, 42. A very simple but also effective method is the shadow image microscope, which allows lens‐less image acquisition and does not need coherent light sources or complex post image processing. Lange et al. pioneered this field 43, introducing a robust and compact device for the investigation of the effects of spaceflight on Caenorhabdtidis Elegans worms in terms of ageing and longevity. The system includes an LED light source and a microfluidic culture chamber placed above a CMOS camera, so that the specimen casts a shadow on the camera, once illuminated. This permits to monitor the sample behavior even under unconventional settings, such as spaceflight conditions. A device based on a similar principle was presented by Ozcan and Demirci (LUCAS), where shadow images are acquired with a CCD camera placed in proximity to the sample. They demonstrated the possibility to count cells over a very large field of view, two orders of magnitude larger than that of a conventional microscope 44. Despite the great simplicity of this method, one main drawback of the shadow imaging approach is its low resolution. This is limited by the pixel size (usually >3 μm) and it highly affects the quality of the acquired images; smaller pixels may be available, but at the price of a reduced light sensitivity. Indeed, since resolution is an essential aspect in microscopy, several approaches have been adopted to overcome this issue. The Optofluidic Microscope (OFM) 45, 46, allows one to acquire 2D images at a resolution higher than the pixel size without affecting the device simplicity. To increase the resolution with respect to standard shadow imaging, a thin metallic layer with apertures smaller than the pixel size is deposited above the CMOS camera. The subsequent sample translation allows recording the image of the whole sample at high resolution. A simplified version of the device foresees the use of only two aperture grids (1 μm diameter) placed in correspondence to the pixels and tilted with respect to the fluidic channel where the sample is flowing, as shown in Figure 1a. This device was validated on different biological samples (C. elegans, Chlamydomonas cells) demonstrating that it is a valuable tool for cell counting and potentially for screening of diseases. The device resolution is related to the aperture size and decreases by increasing the distance between the specimen and the camera. To reduce possible motion artifacts, the fluid velocity must be maintained constant and sample rotations should be avoided.
Figure 1.
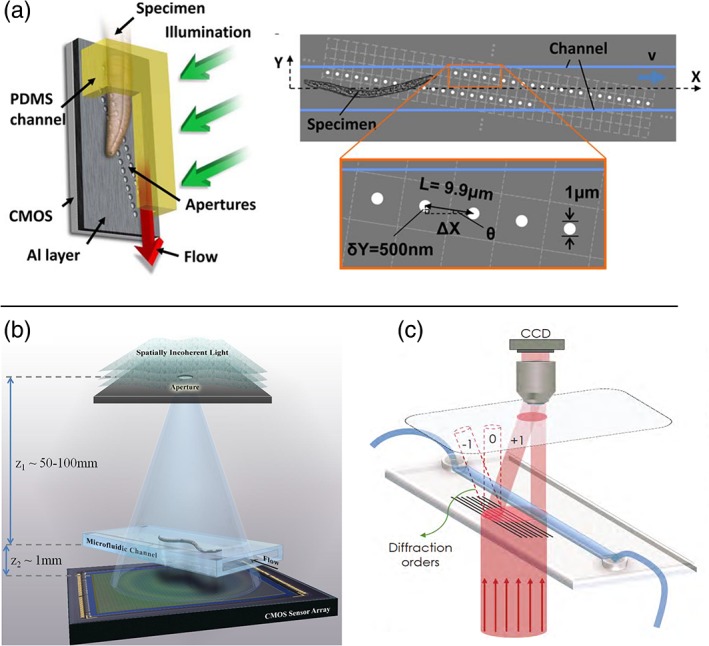
(a) Scheme of the OFM, capable to acquire images at high resolution using a grid of small apertures placed in between the sample and the acquisition camera (Reproduced from Ref. 46, with permission from the National Academy of Sciences). (b) Working principle of the HOM presented by Bishara et al., where a microfluidic channel is placed directly over a CMOS camera and it is illuminated by a partially coherent light source. The device uses digital in line holography and subpixel shift to acquire high resolution images (Reproduced from Ref. 47, with permission from the Optical Society of America). (c) Scheme of the device presented by Bianco et al. 2017, where a diffracting grating is integrated in a commercially available microfluidic chip, which allows off‐axis digital holography by means of a single beam (reproduced from Ref. 48, with permission from Springer Nature). [Color figure can be viewed at http://wieyonlinelibrary.com]
A different approach named Holographic Optofluidic Microscopy (HOM) was presented by Bishara et al., who demonstrated high quality imaging of C. elegans, based on partially coherent in line digital holography (DILH) and multi frame pixel super‐resolution 47. To highlight the advantages of this method, it is useful to first recall the basic principles of in line holography, which is based on the interference of the light scattered by the sample and the unperturbed illumination light. The resulting pattern, which contains information on the phase and the amplitude of the object wave was, in the first implementation of holography, recorded on a photographic target. The reconstruction process mainly consisted in illuminating the target with the same reference beam 48, 49. The original approach has been quickly replaced by digital detection, which is performed by recording the pattern on a sensor and simulating the illumination by the reference beam with a computer, through an iterative wave propagation algorithm 50. With respect to other approaches 51, the configuration introduced by Ozcan's group foresees a large distance between the sample and the light source, with respect to the sample‐camera one, as indicated in Figure 1b. In this way, the fringe magnification is approximately 1 and the field of view equals the sensors active area. Moreover, a source with low coherence can be used, due to the reduced path difference between the two interfering beams. In this specific case an LED source filtered by a large aperture (50–100 μm) placed several centimeter above the sample is used. With this layout, the resolution limit is mainly given by the pixel size, which still may undersample the lens‐free holograms. To overcome this further limitation, it was shown that it is possible to acquire multiple holograms sub‐pixel shifted and to process them through an iterative algorithm capable to reconstruct the image with a resolution higher than the one based on the physical pixel dimension 47. In the HOM device, sub‐pixel shifted frames are acquired while the sample flows through the microfluidic channel placed above the camera. Similar results have been obtained also shifting the light source, which was accomplished in different ways, either scanning the 2D light aperture or using a matrix of fiber‐coupled LED sequentially switched on in a very compact and portable microscope device, and avoiding any mechanical component translation 52, 53. In these devices, using 2.2 μm pixel size a resolution higher than 1 μm was achieved over a 24 mm2 field of view. Another version of DILH on chip, presented by Vercruysse et al. 54 is based on the close proximity of a point source to the microfluidic device; this was obtained using a waveguide, placed just above the microfluidic chip. A nanosecond laser is coupled to the waveguide and the light is scattered whenever a cell flows in the detection region. Scattered and unscattered light interfere on a high‐speed CMOS camera, allowing them to image cells flowing at c.a. 1 mm/s.
Another similar approach is the SROFM device (Sub‐pixel Resolving OFM) presented by Zheng et al. 55 who combined OFM and digital in line holography using a microfluidic channel in direct contact with a CMOS camera, but without the need of a grid of thin aperture, since the resolution in this case is obtained exploiting the sample flow to obtain sub pixel image shifts. The main drawbacks of the methods based on DILH are related to the complexity of the reconstruction algorithm and the fact that a solution of the iterative phase recovery process is not always guaranteed. A different example of MOC based on digital holography was presented by Bianco et al. 48, 56. The authors tried to overcome the limitations of DILH approach presenting a lens based, off‐axis digital holography on chip. Their solution, schematically reported in Figure 1c, foresees the integration on a commercially available microfluidic chip of the microoptical elements necessary for the holographic imaging (i.e., microlenses and diffractive gratings), which are optimized to perform off line holography with a single impinging beam, so as to give rise to a very compact and robust platform for on chip microscopy with no need of any complex external apparatus.
Fluorescence MOCs
Fluorescence microscopy is one of the major tools in biomedicine 57 and plays a relevant role in the observation of cellular and subcellular details in biological specimens. With respect to bright field microscopy, it allows one to investigate a biological system with higher specificity, taking advantage of fluorophores conjugated biomolecules or genetically encoded probes.
Epifluorescence, laser scanning confocal and spinning disk confocal microscopes are the most common equipments in imaging facilities worldwide. Each microscope modality has advantages and limitations compared to the others: epi‐fluorescence microscopes are systems that allow rapid acquisition of fluorescent specimens over a large field of view, but do not provide optical sectioning (the ability to section a cell or a tissue in virtual slices and in 3D). Confocal microscopes have been developed to address this issue, but are strongly affected by photo‐bleaching and photo‐toxicity in live imaging applications.
These fluorescence microscopy configurations have been extensively used in combination with microfluidic devices, primarily for two reasons: i) Microfluidic devices allow growing biological samples directly on a chip, controlling the environment in which cellular systems are growing, injecting nutrients or drugs, and reproducing the dynamic conditions present in a living organ 58. In these systems, microscopy is used as a tool to observe the biological samples within the microfluidic channel, typically in time lapse. These devices have been optimized to study samples in a variety of different spatial scales, from single molecules 59, viruses 60, bacteria 61, single cells 62 to mm‐sized tissues or organisms, including Caenorhabditis elegans 37, 63 and zebrafish embryos 29, 64, 65. ii) Microfluidic devices allow one to flow the sample through a microscope, providing automatic sample scanning and increasing the imaging throughput. Therefore, in the last years, these devices have been optimized for many fluorescence‐based applications: imaging flow cytometers and parallelized imaging devices have been developed 31, 32, 38, 66, 67, 68. In these examples, the microscope is not necessarily integrated on the chip and bulk external instrumentations are still required to image the sample. More compact solutions foresee the integration in a single chip of the fluidic and the optical components for fluorescence excitation and/or detection. A particularly compact MOC is the lens‐less fluorescence microscope 69, 70, 71, 72, which is shown in some of its implementations in Figure 2. In particular, Figure 2a shows how fluorescence is directly detected, integrating an image sensor‐array on the chip. The samples are placed on the sensor (or on a fiber optic faceplate coupled to the sensor) and the excitation light is rejected through total internal reflection that occurs at the bottom facet of the sample substrate. The image is formed by deconvolution of the acquired fluorescence with the point spread function of the lens‐free system, and the resolution is limited to a few microns (< 4 μm) by the pixel size of the sensor (or by the fiber optic diameter in the faceplate). To improve the resolution of lens‐free MOCs, Pang et al. 73 used an integrated Fresnel zone plates array that allowed them to generate an array of focused light spots (Fig. 2b). They collected the fluorescence from samples flowing through the spots and rendered fluorescence microscopy images at a resolution determined by the focused light spot size (0.65 μm).
Figure 2.
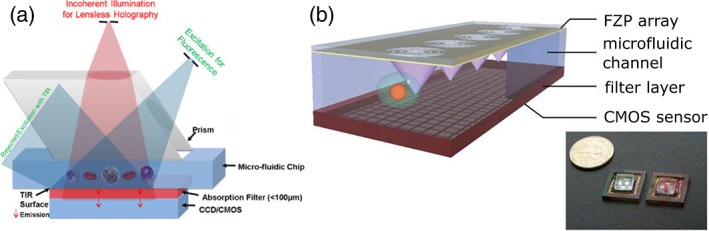
Examples of lens free fluorescence microscopes. (a) Cells are flown in a microfluidic chip that is in attached to a filter coated imaging sensor; fluorescent excitation is achieved using side illumination through a rhomboid prism (reproduced with permission from Ref. 71, with permission from The Royal Society of Chemistry). (b) A Fresnel Zone Plate creates an array of foci inside the channel; the sample flows across the array of focused light spots and the fluorescence emissions are collected by the sensor (reproduced from Ref. 73 with permission from The Royal Society of Chemistry). [Color figure can be viewed at http://wieyonlinelibrary.com]
Within the realm of fluorescence microscopy, a remarkable interest has been dedicated to Light‐Sheet Fluorescence Microscopy (LSFM), also called Selective Plane Illumination Microscopy (SPIM) 74. This technique combines the optical sectioning capability, typical of confocal systems, with the ability to rapidly scan a large field of view, typical of wide‐field epifluorescence microscopy. The major advantages of LSFM include the high acquisition speed over an extended 3D samples, and the reduced photo‐toxicity compared to confocal microscopy. Rapid acquisition is at the basis of high throughput imaging and, for this reason LSFM can deeply benefit from microfluidic integration. As a consequence, several versions of light sheet based MOCs have been presented in literature, each characterized by a different level of integration.
The idea to combine fluidic devices with a light sheet microscope was firstly proposed by Bruns et al. 75. The authors used a glass capillary to hold the sample and delivered a fluorescent marker through the capillary. Wu et al. used a different capillary to deliver a large number of samples to the microscope, demonstrating the first flow cytometer based on LSFM. This system was specifically optimized for phytoplankton analysis 76 and later extended to two‐color imaging 77. Gualda et al. developed a fluidic network to scan large samples (as big as a zebrafish embryo) automatically through a static light sheet 78.
The sample delivery system of the imaging flow cytometer was further integrated in a chip (Fig. 3a) by Regmi et al. 79, 82: a PDMS chip was coupled to a custom light sheet microscope to flow cells at high rate. In parallel, Deschout et al. 80 created the first chip that included an optical component for the generation of the light‐sheet itself (Fig. 3b). Being produced by spin coating deposition, this chip had the advantage to be potentially compatible with mass production, but the light sheet was created by a planar waveguide: the light was, therefore, inherently diverging, and it was difficult to tailor the delivery system to samples that are larger than fluorescent beads or membrane vesicles.
Figure 3.
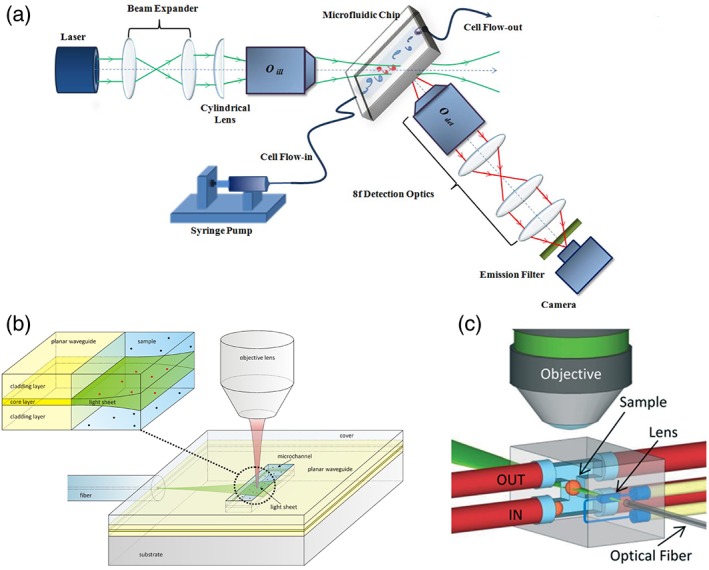
Steps toward Light Fluorescence Microscopy on a chip. (a) A microfluidic chip is integrated in a custom Light sheet Microscope (Reproduced from Ref. 79, with permission from AIP Publishing); used in accordance with the Creative Commons Attribution (CC BY) license). (b) A planar waveguide is integrated on a chip to create a light sheet directly on the sample (Reproduced with permission from Ref. 80, with permission from The Royal Society of Chemistry). (c) An optofluidic lens is integrated on a chip to focus a light sheet in the center of a fluidic channel, where multiple samples are automatically scanned and reconstructed in 3D (reproduced from Ref. 81 with permission from The Royal Society of Chemistry). [Color figure can be viewed at http://wieyonlinelibrary.com]
Both sample delivery and scan through the light‐sheet, were finally integrated by Paiè et al. 81 in a glass chip (Fig. 3c). Here, using femtosecond laser micromachining, an optofluidic device that fully integrates the illumination and sample scanning was realized. The light sheet is formed by an optofluidic cylindrical lens 83 and is focused in a microfluidic channel where the samples flow at a constant velocity through the light sheet, thus allowing automatic imaging of tissue mimics (cellular spheroids).
Future implementations of LSFM on a chip will likely bring to even more compact and portable devices. A possible strategy could come from the combination of lens‐less microscopy with tailored light sheet illumination. Conversely, high‐resolution imaging will be increasingly required for accurate screening of cells and small organisms. So far, some miniaturized chips have fostered high and even super‐resolution light sheet microscopy. In particular, micro‐fabricated reflective components have been developed and tailored for light sheet illumination by Galland et al. and Zagato et al. 84, 85: in these implementations a laser beam is reflected horizontally on the sample by a 45° mirror and forms the light sheet in the image plane of the detection objective of an upright or inverted microscope. The buffer solution can be further exchanged automatically by a microfluidic system coupled with the chip that incorporates the mirrors 86. On the basis of this interest for high resolution devices, we foresee that new solutions that fully integrate super‐resolution light sheet microscopy on a chip will be conceived and implemented in the next future.
Mobile Phone MOCs
In the last decade, there has been an enormous development of portable microscope systems based on mobile phones. Thanks to the high quality electronical and optical components already present in mobile phones, it is possible to implement a compact digital microscope with a reduced number of external components. In the next paragraphs, we will report and discuss the existing attempts to perform digital microscopy with mobile phone based devices, discussing them from an optical point of view. In the most recent examples, a dedicated software application (App) is always developed to control the device electronics and perform data analysis, nevertheless the evaluation of the App performance goes beyond the scope of the present review.
The first attempts to introduce mobile phones into microscopy were focused in using the phone simply as the imaging device (coupling the phone to a traditional microscope) or as a data transmitter 87, 88. It was in 2009 when, for the first time, a research group implemented an entirely integrated mobile phone microscopy system that allowed acquiring bright field and fluorescence images of the specimens under test 7. The device needed external optical components like two lenses (collector and condenser) for the illumination, a microscope objective, an eyepiece for the imaging and filters for excitation and emission. All these components were mounted onto an optical rail system to keep them perfectly aligned with the mobile phone camera. Although the system was smaller than a traditional microscope, it was still cumbersome in comparison with the mobile phone. More recently, Ganguli et al. 89 developed a point‐of‐care platform for the detection of viruses as Zika, Chikungunya, and Dengue from whole blood samples. Here the mobile phone was used to image the fluorescence and to send the data to an external computer for analysis.
Ozcan and coworkers 39, 90 were the first to use a mobile phone microscope to image “moving” specimens. They developed an imaging cytometer that used the phone camera to capture the fluorescence images of a sample flowing inside a microfluidic channel. The excitation was performed through external diodes and, thanks to proper lenses and a filter to remove the excitation residual signal, they acquired fluorescence images that were rapidly analyzed to determine the particle/cell number flowing inside the microfluidic channel. All the optical components and the microfluidic chip were fixed into a mechanical support attached to the mobile phone. A less compact cytometer has been demonstrated by Jagannadh et al., this time exploiting bright field imaging, achieving a counting throughput of 27,000 cells per minute 91.
Over the years the main developments in mobile phone microscopy have been focused in increasing their portability and demonstrating their applicability in several biological and laboratory analysis tests. To increase the portability, it is important to reduce the size and the number of external components. One way consists in avoiding the external device, as demonstrated by Hutchison et al. 92. Their system was extremely compact, they used glass beads as lenses and thanks to a 3D printed holder the lenses and microfluidic chip were fixed to the mobile phone. As proof of concept, their system was used to grow and detect Bacillus anthracis spores. This “miniaturization” concept was also followed by Martínez Vázquez et al. 93 to implement a fluorescence flow cytometer on a mobile phone. Also in this case, all the optical components and the microfluidic chip are fixed to the mobile phone thanks to a dedicated 3D printed holder. Notably, for the first time the illumination in this device is done by exploiting the LED of the mobile phone.
From an analytical point of view, these microscopes have been already used into a broad spectrum of experiments. For example, Cho et al. 94 exploited a smartphone based fluorescence microscope as an in situ monitoring tool for Organ on chips experiments. They developed a dual‐mode assay to monitor the presence and the outflow of γ‐glutamyl transpeptidase (GGT) from the organ on chip by the fluorescence detection of captured and immunoagglutinated anti‐GGT conjugated nanoparticles. Kanakasabapathy et al. have developed an automated CD4 testing (HIV/AIDS) based on a compact and cheap mobile phone cytometer (see Fig. 4). They validated the system using HIV‐infected and uninfected whole blood samples, obtaining a good agreement with traditional laboratory detection systems 95. More recently, Yang et al. 96 have developed a prototype called Mkit, based on a smartphone fluorescence microscope to perform experiments of cell migration and chemotaxis in 3D environments. The platform was validated testing chemotaxis on purified human blood neutrophils that were obtained either directly from cancer cell lines or from blood drops, demonstrating the possibility to use it for on‐site clinical assays.
Figure 4.
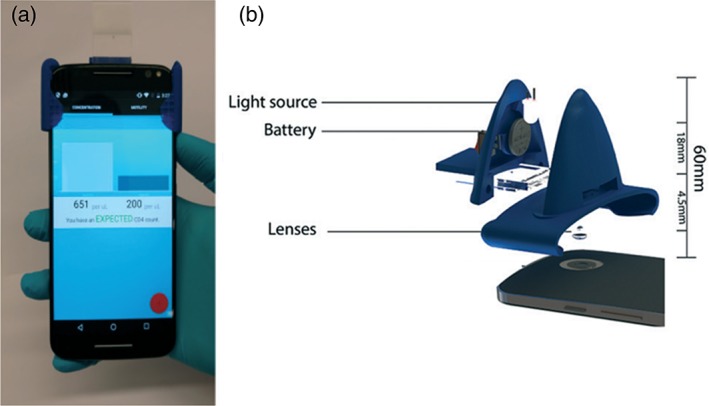
(a) Picture of a smartphone‐based cytometer with the microfluidic chip mounted on it. It is worth noting the compactness of the attached plastic system, (b) schematic view of the external elements needed to perform the bright field microscopy: An external light source and its battery for sample illumination and the optical lens to create the microscope together with the lens already present in the mobile phone (reproduced from Ref. 95 with permission from The Royal Society of Chemistry). [Color figure can be viewed at http://wieyonlinelibrary.com]
Moreover, the versatility of these smartphone microscopes is favoring their use as rapid test readers for laboratory strip tests. For example, in lateral flow immune‐chromatographic assays to detect malaria, HIV or tuberculosis 97 and also in test involving the extraction and identification of Salmonella pathogenic nucleics acids from field samples 98.
Thanks to the cost effective and compactness of the dedicated supports, and the ubiquity of smartphones around the world, we foresee in the next years a fast spread of the mobile phone microscopes, mainly in low resource settings and telemedicine.
Conclusions
MOC are emerging as valuable tools for sample monitoring in applications that include point of care diagnostics and in situ measurements. The essential characteristics of low cost, ease of use, portability, and high throughput can be all satisfied by the development of multifunctional lab‐on‐chips. In this review, we described MOC that integrate optical and fluidic components, because we believe that this integration is the roadmap to the realization of compact and portable systems. We expect that once the microscope is integrated on a chip, low‐consumption power supplies and detectors should be included so as to achieve true portable microscope on chip devices. Here we discussed different solutions reported in literature to implement MOC and we classified them on the basis of the imaging methods, each of them having pros and cons depending on the specific application. The lensless imaging techniques, both in transillumination and in fluorescence microscopy implementations, appear to be particularly appealing for the use of microscopes in areas with limited resources as in developing countries since they allow detection, counting and imaging of cells, with a simple hardware. The integration of optical components, such as the implementation of light‐sheet illumination in microfluidic chips, allows for high‐resolution three‐dimensional imaging with high throughput, which is fundamental for accurate screening of cells. A significant step forward toward the real portability of microscopes has been instead made with the exploitation of smartphones in the microscopes’ realization. We believe that the combination of these compact and low cost implementations will contribute to a wide spread of MOC devices and to their integration in microfluidic platforms for real‐time and in situ analyses.
Literature Cited
- 1. Mertz J. Introduction to optical microscopy. Roberts 2010;138. [Google Scholar]
- 2. Lee SA, Leitao R, Zheng G, Yang S, Rodriguez A, Yang C. Color capable sub‐pixel resolving optofluidic microscope and its application to blood cell imaging for malaria diagnosis. PLoS One 2011;6:e26127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu YC, Shiledar A, Li YC, Wong J, Feng S, Chen X, Chen C, Jin K, Janamian S, Yang Z, et al. Air quality monitoring using mobile microscopy and machine learning. Light Sci Appl 2017;6:e17046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mudanyali O, Oztoprak C, Tseng D, Erlinger A, Ozcan A. Detection of waterborne parasites using field‐portable and cost‐effective lensfree microscopy. Lab Chip 2010;10:2419–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Microfluidic diagnostic technologies for global public health. Nature 2006;442:412–418. [DOI] [PubMed] [Google Scholar]
- 6. Miller AR, Davis GL, Oden ZM, Razavi MR, Fateh A, Ghazanfari M, Abdolrahimi F, Poorazar S, Sakhaie F, et al. Portable, battery‐operated, low‐cost, bright field and fluorescence microscope. PLoS One 2010;5:e11890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Breslauer DN, Maamari RN, Switz NA, Lam WA, Fletcher DA. Mobile phone based clinical microscopy for global health applications. PLoS One 2009;4:e6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Potsaid B, Bellouard Y, Wen JT. Adaptive Scanning Optical Microscope (ASOM): A multidisciplinary optical microscope design for large field of view and high resolution imaging. Opt Express 2005;13:6504–6518. [DOI] [PubMed] [Google Scholar]
- 9. Gualda EJ, Pereira H, Martins GG, Gardner R, Moreno N. Three‐dimensional imaging flow cytometry through light‐sheet fluorescence microscopy. Cytometry Part A 2017;91A:144–151. [DOI] [PubMed] [Google Scholar]
- 10. Figeys D, Pinto D. Lab‐on‐a‐chip: a revolution in biological and medical sciences. Anal Chem 2000;72:330‐A–335‐A. [DOI] [PubMed] [Google Scholar]
- 11. Daridon A, Sequeira M, Pennarun‐Thomas G, Dirac H, Krog JP, Gravesen P, Lichtenberg J, Diamond D, Verpoorte E, de Rooij NF. Chemical sensing using an integrated microfluidic system based on the Berthelot reaction. Sens Actuator B‐Chem 2001;76:235–243. [Google Scholar]
- 12. Khandurina J, McKnight TE, Jacobson SC, Waters LC, Foote RS, Ramsey JM. Integrated system for rapid PCR‐based DNA analysis in microfluidic devices. Anal Chem 2000;72:2995–3000. [DOI] [PubMed] [Google Scholar]
- 13. Andersson H, Van den Berg A. Microfluidic devices for cellomics: a review. Sens Actuator B‐Chem 2003;92:315–325. [Google Scholar]
- 14. Clayton KN, Berglund GD, Linnes JC, Kinzer‐Ursem TL, Wereley ST. DNA microviscosity characterization with particle diffusometry for downstream DNA detection applications. Anal Chem 2017;89:13334–13341. [DOI] [PubMed] [Google Scholar]
- 15. Leary JF. Cytometry of single‐cells for biology and biomedicine In: Tseng FG, Santra T, editors. Essentials of Single‐Cell Analysis. Series in Bioengineering. Berlin, Heidelberg: Springer, 2016; p. 235–255. [Google Scholar]
- 16. Hassan U, Ghonge T, Reddy B, Patel M, Rappleye M, Taneja I, Tanna A, Healey R, Mansury N, Price Z, et al. A point‐of‐care microfluidic biochip for quantification of CD64 expression from whole blood for sepsis stratification. Nat Commun 2017;8:15949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nawaz AA, Chen Y, Nama N, Nissly RH, Ren L, Ozcelik A, Wang L, McCoy JP, Levine SJ, Huang TJ. Acoustofluidic fluorescence activated cell sorter. Anal Chem 2015;87:12051–12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lenshof A, Laurell T. Continuous separation of cells and particles in microfluidic systems. Chem Soc Rev 2010;39:1203–1217. [DOI] [PubMed] [Google Scholar]
- 19. Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, Marquez M, Klibanov AM, Griffiths AD, Weitz DA. Ultrahigh‐throughput screening in drop‐based microfluidics for directed evolution. Proc Natl Acad Sci USA 2010;107:4004–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim T, Doh I, Cho YH. On‐chip three‐dimensional tumor spheroid formation and pump‐less perfusion culture using gravity‐driven cell aggregation and balanced droplet dispensing. Biomicrofluidics 2012;6:34107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bhatia SN, Ingber DE. Microfluidic organs‐on‐chips. Nat Biotechnol 2014;32:760–772. [DOI] [PubMed] [Google Scholar]
- 22. Whitesides GM. The origins and the future of microfluidics. Nature 2006;442:368–373. [DOI] [PubMed] [Google Scholar]
- 23. Wu J, Zheng G, Lee LM. Optical imaging techniques in microfluidics and their applications. Lab Chip 2012;12:3566–3575. [DOI] [PubMed] [Google Scholar]
- 24. Zhao Y, Stratton ZS, Guo F, Lapsley MI, Chan CY, Lin SC, Huang TJ. Optofluidic imaging: Now and beyond. Lab Chip 2013;13:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Psaltis D, Quake SR, Yang C. Developing optofluidic technology through the fusion of microfluidics and optics. Nature 2006;442:381–386. [DOI] [PubMed] [Google Scholar]
- 26. Monat C, Domachuk P, Eggleton BJ. Integrated optofluidics: A new river of light. Nat Photonics 2007;1:106–114. [Google Scholar]
- 27. Fan X, White IM, Shopova SI, Zhu H, Suter JD, Sun Y. Sensitive optical biosensors for unlabeled targets: A review. Anal Chim Acta 2008;620:8–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chung K, Kim Y, Kanodia JS, Gong E, Shvartsman SY, Lu H. A microfluidic array for large‐scale ordering and orientation of embryos. Nat Methods 2011;8:171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin X, Wang S, Yu X, Liu Z, Wang F, Li WT, Cheng SH, Dai Q, Shi P. High‐throughput mapping of brain‐wide activity in awake and drug‐responsive vertebrates. Lab Chip 2015;15:680–689. [DOI] [PubMed] [Google Scholar]
- 30. Goda K, Ayazi A, Gossett DR, Sadasivam J, Lonappan CK, Sollier E, Fard AM, Hur SC, Adam J, Murray C, et al. High‐throughput single‐microparticle imaging flow analyzer. Proc Natl Acad Sci USA 2012;109:11630–11635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Levario TJ, Zhan M, Lim B, Shvartsman SY, Lu H. Microfluidic trap array for massively parallel imaging of Drosophila embryos. Nat Protoc 2013;8:721–736. [DOI] [PubMed] [Google Scholar]
- 32. Chung K, Rivet CA, Kemp ML, Lu H. Imaging single‐cell signaling dynamics with a deterministic high‐density single‐cell trap array. Anal Chem 2011;83:7044–7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paiè P, Bragheri F, Di Carlo D, Osellame R. Particle focusing by 3D inertial microfluidics. Microsyst Nanoeng 2017;3:17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McKenna BK, Evans JG, Cheung MC, Ehrlich DJ. A parallel microfluidic flow cytometer for high‐content screening. Nat Methods 2011;8:401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lau AK, Shum HC, Wong KK, Tsia KK. Optofluidic time‐stretch imaging–an emerging tool for high‐throughput imaging flow cytometry. Lab Chip 2016;16:1743–1756. [DOI] [PubMed] [Google Scholar]
- 36. Rohde CB, Zeng F, Gonzalez‐Rubio R, Angel M, Yanik MF. Microfluidic system for on‐chip high‐throughput whole‐animal sorting and screening at subcellular resolution. Proc Natl Acad Sci USA 2007;104:13891–13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chronis N, Zimmer M, Bargmann CI. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat Methods 2007;4:727–731. [DOI] [PubMed] [Google Scholar]
- 38. Yan W, Wu J, Wong KK, Tsia KK. A high‐throughput all‐optical laser‐scanning imaging flow cytometer with biomolecular specificity and subcellular resolution. J Biophotonics 2018;11:e201700178. [DOI] [PubMed] [Google Scholar]
- 39. Zhu H, Mavandadi S, Coskun AF, Yaglidere O, Ozcan A. Optofluidic fluorescent imaging cytometry on a cell phone. Anal Chem 2011;83:6641–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee SA, Yang C. A smartphone‐based chip‐scale microscope using ambient illumination. Lab Chip 2014;14:3056–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gurkan UA, Moon S, Geckil H, Xu F, Wang S, Lu TJ, Demirci U. Miniaturized lensless imaging systems for cell and microorganism visualization in point‐of‐care testing. J Biotechnol 2011;6:138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Göröcs Z, Ozcan A. On‐chip biomedical imaging. IEEE Rev Biomed Eng 2013;6:29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lange D, Storment CW, Conley CA, Kovacs GT. A microfluidic shadow imaging system for the study of the nematode Caenorhabditis elegans in space. Sens Actuator B‐Chem 2005;107:904–914. [Google Scholar]
- 44. Ozcan A, Demirci U. Ultra wide‐field lens‐free monitoring of cells on‐chip. Lab Chip 2008;8:98–106. [DOI] [PubMed] [Google Scholar]
- 45. Heng X, Erickson D, Baugh LR, Yaqoob Z, Sternberg PW, Psaltis D, Yang C. Optofluidic microscopy—a method for implementing a high resolution optical microscope on a chip. Lab Chip 2006;6:1274–1276. [DOI] [PubMed] [Google Scholar]
- 46. Cui X, Lee LM, Heng X, Zhong W, Sternberg PW, Psaltis D, Yang C. Lensless high‐resolution on‐chip optofluidic microscopes for Caenorhabditis elegans and cell imaging. Proc Natl Acad Sci USA 2008;105:10670–10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bishara W, Zhu H, Ozcan A. Holographic opto‐fluidic microscopy. Opt Express 2010;18:27499–27510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bianco V, Mandracchia B, Marchesano V, Pagliarulo V, Olivieri F, Coppola S, Paturzo M, Ferraro P. Endowing a plain fluidic chip with micro‐optics: a holographic microscope slide. Light Sci Appl 2017;6:e17055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gabor, D. A new microscopic principle. Nature 1948:161:777–778. [DOI] [PubMed] [Google Scholar]
- 50. Garcia‐Sucerquia J, Xu W, Jericho SK, Klages P, Jericho MH, Kreuzer HJ. Digital in‐line holographic microscopy. Appl Opt 2006;45:836–850. [DOI] [PubMed] [Google Scholar]
- 51. Lewis NI, Xu W, Jericho SK, Kreuzer HJ, Jericho MH, Cembella AD. Swimming speed of three species of Alexandrium (Dinophyceae) as determined by digital in‐line holography. Phycologia 2006;45:61–70. [Google Scholar]
- 52. Bishara W, Sikora U, Mudanyali O, Su TW, Yaglidere O, Luckhart S, Ozcan A. Holographic pixel super‐resolution in portable lensless on‐chip microscopy using a fiber‐optic array. Lab Chip 2011;11:1276–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bishara W, Su TW, Coskun AF, Ozcan A. Lensfree on‐chip microscopy over a wide field‐of‐view using pixel super‐resolution. Opt Express 2010;18:11181–11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vercruysse D, Dusa A, Stahl R, Vanmeerbeeck G, de Wijs K, Liu C, Prodanov D, Peumans P, Lagae L. Three‐part differential of unlabeled leukocytes with a compact lens‐free imaging flow cytometer. Lab Chip 2015;15:1123–1132. [DOI] [PubMed] [Google Scholar]
- 55. Zheng G, Lee SA, Yang S, Yang C. Sub‐pixel resolving optofluidic microscope for on‐chip cell imaging. Lab Chip 2010;10:3125–3129. [DOI] [PubMed] [Google Scholar]
- 56. Mandracchia B, Bianco V, Wang Z, Mugnano M, Bramanti A, Paturzo M, Ferraro P. Holographic microscope slide in a spatio‐temporal imaging modality for reliable 3D cell counting. Lab Chip 2017;17:2831–2838. [DOI] [PubMed] [Google Scholar]
- 57. Lichtman JW, Conchello JA. Fluorescence microscopy. Nat Methods 2005;2:910–919. [DOI] [PubMed] [Google Scholar]
- 58. Esch EW, Bahinski A, Huh D. Organs‐on‐chips at the frontiers of drug discovery. Nat Rev Drug Discov 2015;14:248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van Mameren J, Gross P, Farge G, Hooijman P, Modesti M, Falkenberg M, Wuite GJ, Peterman EJ. Unraveling the structure of DNA during overstretching by using multicolor, single‐molecule fluorescence imaging. Proc Natl Acad Sci USA 2009;106:18231–18236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Akin D, Li H, Bashir R. Real‐time virus trapping and fluorescent imaging in microfluidic devices. Nano Lett 2004;4:257–259. [Google Scholar]
- 61. Cattoni DI, Fiche JB, Valeri A, Mignot T, Nöllmann M. Super‐resolution imaging of bacteria in a microfluidics device. PLoS One 2013;8:e76268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tawa K, Yamamura S, Sasakawa C, Shibata I, Kataoka M. Sensitive detection of cell surface membrane proteins in living breast cancer cells using multicolor fluorescence microscopy with a plasmonic chip. ACS Appl Mater Interfaces 2016;8:29893–29898. [DOI] [PubMed] [Google Scholar]
- 63. Larsch J, Ventimiglia D, Bargmann CI, Albrecht DR. High‐throughput imaging of neuronal activity in Caenorhabditis elegans. Proc Natl Acad Sci USA 2013;110:E4266–E4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Candelier R, Murmu MS, Romano SA, Jouary A, Debrégeas G, Sumbre G. A microfluidic device to study neuronal and motor responses to acute chemical stimuli in zebrafish. Sci Rep 2015;5:12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Marquezin CA, Ceffa NG, Cotelli F, Collini M, Sironi L, Chirico G. Image Cross‐Correlation Analysis of time varying flows. Anal Chem 2016. 8;88:7115–7122. [DOI] [PubMed] [Google Scholar]
- 66. Jagannadh VK, Mackenzie MD, Pal P, Kar AK, Gorthi SS. Slanted channel microfluidic chip for 3D fluorescence imaging of cells in flow. Opt Express 2016;24:22144–22158. [DOI] [PubMed] [Google Scholar]
- 67. Balsam J, Bruck HA, Ossandon M, Prickril B, Rasooly A. Streak Imaging Flow Cytometer for Rare Cell Analysis. Biosensors and Biodetection: Methods and Protocols Volume 1: Optical‐Based Detectors. 2017. pp 267–286. [DOI] [PubMed]
- 68. Dashkova V, Malashenkov D, Poulton N, Vorobjev I, Barteneva NS. Imaging flow cytometry for phytoplankton analysis. Methods 2017;112:188–200. [DOI] [PubMed] [Google Scholar]
- 69. Coskun AF, Su TW, Ozcan A. Wide field‐of‐view lens‐free fluorescent imaging on a chip. Lab Chip 2010;10:824–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Coskun AF, Sencan I, Su TW, Ozcan A. Wide‐field lensless fluorescent microscopy using a tapered fiber‐optic faceplate on a chip. Analyst 2011;136:3512–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Coskun AF, Su TW, Sencan I, Ozcan A. Lensless fluorescent microscopy on a chip. J Visualized Exp 2011;54:3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lee SA, Ou X, Lee JE, Yang C. Chip‐scale fluorescence microscope based on a silo‐filter complementary metal‐oxide semiconductor image sensor. Opt Lett 2013;38:1817–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pang S, Han C, Lee LM, Yang C. Fluorescence microscopy imaging with a Fresnel zone plate array based optofluidic microscope. Lab Chip 2011;11:3698–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 2004;305:1007–1009. [DOI] [PubMed] [Google Scholar]
- 75. Bruns T, Schickinger S, Wittig R, Schneckenburger H. Preparation strategy and illumination of three‐dimensional cell cultures in light sheet‐based fluorescence microscopy. J Biomed Opt 2012;17:101518. [DOI] [PubMed] [Google Scholar]
- 76. Wu J, Li J, Chan RK. A light sheet based high throughput 3D‐imaging flow cytometer for phytoplankton analysis. Opt Express 2013;21:14474–14480. [DOI] [PubMed] [Google Scholar]
- 77. Li J, Xu Z. Simultaneous dual‐color light sheet fluorescence imaging flow cytometry for high‐throughput marine phytoplankton analysis. Opt Express. 2017;25:13602–13616. [DOI] [PubMed] [Google Scholar]
- 78. Gualda EJ, Pereira H, Vale T, Estrada MF, Brito C, Moreno N. SPIM‐fluid: open source light‐sheet based platform for high‐throughput imaging. Biomed Opt Express 2015;6:4447–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Regmi R, Mohan K, Mondal PP. High resolution light‐sheet based high‐throughput imaging cytometry system enables visualization of intra‐cellular organelles. AIP Adv 2014;4:097125. [Google Scholar]
- 80. Deschout H, Raemdonck K, Stremersch S, Maoddi P, Mernier G, Renaud P, Jiguet S, Hendrix A, Bracke M, Van den Broecke R, et al. On‐chip light sheet illumination enables diagnostic size and concentration measurements of membrane vesicles in biofluids. Nanoscale 2014;6:1741–1747. [DOI] [PubMed] [Google Scholar]
- 81. Paiè P, Bragheri F, Bassi A, Osellame R. Selective plane illumination microscopy on a chip. Lab Chip 2016;16:1556–1560. [DOI] [PubMed] [Google Scholar]
- 82. Regmi R, Mohan K, Mondal PP. Light sheet based imaging flow cytometry on a microfluidic platform. Microsc Res Tech 2013;76:1101–1107. [DOI] [PubMed] [Google Scholar]
- 83. Paiè P, Bragheri F, Claude T, Osellame R. Optofluidic light modulator integrated in lab‐on‐a‐chip. Opt Express 2017;25:7313–7323. [DOI] [PubMed] [Google Scholar]
- 84. Galland R, Grenci G, Aravind A, Viasnoff V, Studer V, Sibarita JB. 3D high‐and super‐resolution imaging using single‐objective SPIM. Nat Methods 2015;12:641–644. [DOI] [PubMed] [Google Scholar]
- 85. Zagato E, Brans T, Verstuyft S, Van Thourhout D, Missinne J, Van Steenberge G, Demeester J, De Smedt S, Remaut K, Neyts K, et al. Microfabricated devices for single objective single plane illumination microscopy (SoSPIM). Opt Express 2017;25:1732–1745. [DOI] [PubMed] [Google Scholar]
- 86. Meddens MB, Liu S, Finnegan PS, Edwards TL, James CD, Lidke KA. Single objective light‐sheet microscopy for high‐speed whole‐cell 3D super‐resolution. Biomed Opt Express 2016;7:2219–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Frean J. Microscopic images transmitted by mobile cameraphone. Trans R Soc Trop Med Hyg 2007;101:1053. [DOI] [PubMed] [Google Scholar]
- 88. Zimic M, Coronel J, Gilman RH, Luna CG, Curioso WH, Moore DA. Can the power of mobile phones be used to improve tuberculosis diagnosis in developing countries? Trans R Soc Trop Med Hyg 2009;103:638–640. [DOI] [PubMed] [Google Scholar]
- 89. Ganguli A, Ornob A, Yu H, Damhorst GI, Chen W, Sun F, Bhuiya A, Cunningham BT, Bashir R. Hands‐free smartphone‐based diagnostics for simultaneous detection of Zika, Chikungunya, and Dengue at point‐of‐care. Biomed Microdevices 2017;19:73. [DOI] [PubMed] [Google Scholar]
- 90. Zhu H, Sikora U, Ozcan A. Quantum dot enabled detection of Escherichia coli using a cell‐phone. Analyst 2012;137:2541–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jagannadh VK, Adhikari JV, Gorthi SS. Automated cell viability assessment using a microfluidics based portable imaging flow analyzer. Biomicrofluidics 2015;9:024123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hutchison JR, Erikson RL, Sheen AM, Ozanich RM, Kelly RT. Reagent‐free and portable detection of Bacillus anthracis spores using a microfluidic incubator and smartphone microscope. Analyst 2015;140:6269–6276. [DOI] [PubMed] [Google Scholar]
- 93. Martínez Vázquez R, Trotta G, Volpe A, Bernava G, Basile V, Paturzo M, Ferraro P, Ancona A, Fassi I, Osellame R. Rapid prototyping of plastic lab‐on‐a‐chip by femtosecond laser micromachining and removable insert microinjection molding. Micromachines 2017;8:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cho S, Islas‐Robles A, Nicolini AM, Monks TJ, Yoon JY. In situ, dual‐mode monitoring of organ‐on‐a‐chip with smartphone‐based fluorescence microscope. Biosens Bioelectron 2016;86:697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kanakasabapathy MK, Pandya HJ, Draz MS, Chug MK, Sadasivam M, Kumar S, Etemad B, Yogesh V, Safavieh M, Asghar W, et al. Rapid, label‐free CD4 testing using a smartphone compatible device. Lab Chip 2017;17:2910–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yang K, Wu J, Peretz‐Soroka H, Zhu L, Li Z, Sang Y, Hipolito J, Zhang M, Santos S, Hillier C, et al. Mkit: A cell migration assay based on microfluidic device and smartphone. Biosens Bioelectron 2018;99:259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mudanyali O, Dimitrov S, Sikora U, Padmanabhan S, Navruz I, Ozcan A. Integrated rapid‐diagnostic‐test reader platform on a cellphone. Lab Chip 2012;12:2678–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Fronczek CF, San Park T, Harshman DK, Nicolini AM, Yoon JY. Paper microfluidic extraction and direct smartphone‐based identification of pathogenic nucleic acids from field and clinical samples. RSC Adv 2014;4:11103–11110. [Google Scholar]


