Summary
The plant pathogen Pseudomonas syringae pv. phaseolicola, which causes halo blight disease of beans, contains a 106 kb genomic island PPHGI‐1. PPHGI‐1 carries a gene, avrPphB, which encodes an effector protein that triggers a resistance response in certain bean cultivars. Previous studies have shown that when PPHGI‐1 is excised from the bacterial chromosome, avrPphB is downregulated and therefore the pathogen avoids triggering the host’s defence mechanism. Here, we investigate whether the downregulation of avrPphB is caused by the supercoiling of PPHGI‐1. We also investigate the effect of a PPHGI‐1‐encoded type 1A topoisomerase, TopB3, on island stability and bacterial pathogenicity in the plant. Supercoiling inhibitors significantly increased the expression of avrPphB but did not affect the excision of PPHGI‐1. An insertional mutant of topB3 displayed an increase in avrPphB expression and an increase in PPHGI‐1 excision as well as reduced population growth in resistant and susceptible cultivars of bean. These results suggest an important role for topoisomerases in the maintenance and stability of a bacterial‐encoded genomic island and demonstrate that supercoiling is involved in the downregulation of an effector gene once the island has been excised, allowing the pathogen to prevent further activation of the host defence response.
Introduction
Genomic islands are distinct regions of DNA that are present in some strains of bacteria but not others, often containing genes that may be responsible for recombination and mobility such as integrases, and are normally associated with specific integration sites in the genome such as tRNA loci (Hacker and Kaper, 2000; Hacker and Carniel, 2001; van der Meer and Sentchilo, 2003). The plant pathogen Pseudomonas syringae pv. phaseolicola (Pph) strain 1302A (Pph 1302A), which causes halo blight disease of bean, contains a 106 kb genomic island PPHGI‐1, which encodes 100 genes. PPHGI‐1 can exist in one of two forms: (i) it can be integrated within a tRNALYS locus in the chromosome; (ii) it can excise to form a circular molecule. PPHGI‐1 also contains an oriV, which enables it to become a self‐replicating plasmid (Pitman et al., 2005; Neale et al., 2013).
PPHGI‐1 carries an effector gene avrPphB (also called hopAR1) that is specifically upregulated during infection. AvrPphB is then translocated via the type III secretion system from the bacterial cell into the plant cell where its cysteine protease activity is used to target cytoplasmic kinases to disrupt host immunity (Zhu et al., 2004; Zhang et al., 2010); however, in resistant bean plants, avrPphB is recognised by the resistance protein R3 (Jenner et al., 1991; Jackson et al., 2000; Arnold, et al., 2003). When this recognition occurs, the plant cells undergo a hypersensitive response (HR), also known as effector‐triggered immunity, which is a form of programmed cell death (Jones and Dangl, 2006). This leads to an antimicrobial environment at the site of infection, death of the invading bacteria and therefore the plant resists further pathogen infection and spread within the plant (Alfano and Collmer, 2004; Jones and Dangl, 2006). During the HR, it has been observed that an integrase gene (xerC) on PPHGI‐1 is upregulated allowing the island to excise from the chromosome and form a circular structure (Pitman et al., 2005; Neale et al., 2013). PPHGI‐1 can then be lost from the bacterial cell, leading to the loss of the effector gene. Once the effector gene is lost from the bacterial population, the bean plant no longer recognises the invading pathogen, which can rapidly replicate inside the plant and cause disease (Pitman et al., 2005; Lovell et al., 2009). However, an interesting inverse gene expression state exists between the chromosomally located PPHGI‐1 and the excised form: it was observed that when PPHGI‐1 is in its excised circular form, avrPphB is downregulated. This suggests that the circular form of PPHGI‐1 affects gene expression (Godfrey et al., 2011), possibly by supercoiling, and that the reduction in gene expression due to supercoiling may be a way of masking the presence of avrPphB from detection by the host surveillance system.
DNA topoisomerases are enzymes that regulate the coiling and relaxing of the DNA super helix and therefore control the topology of DNA in all cells (Bush et al., 2015). They are a ubiquitous family of enzymes and have important roles to play in many processes including DNA replication, chromosome segregation, recombination and repair (Luttinger, 1995, Vos et al., 2011, Bush et al., 2015). Topoisomerases can also be recruited to active transcriptional units to remove negative supercoils and influence transcription of genes (Peter et al., 2004; Ahmed et al., 2017).
Type I and type II topoisomerases introduce breaks into single‐ and double‐stranded DNA respectively (Terekhova et al., 2013; Bush et al., 2015). Bacterial type I topoisomerases can be further divided into subgroups, for example topoisomerase I and III, both of which are classed as topoisomerase type 1A enzymes (Deweese et al., 2008; Terekhova et al., 2013; Bush et al., 2015). Topoisomerase I removes negative DNA supercoils and works with DNA gyrase (type II topoisomerase) to regulate the level of supercoiling of the chromosomal DNA (Deweese et al., 2008). Topoisomerase III (topo III) enzymes are conserved and ubiquitous and resolve single‐stranded DNA recombination and replication intermediates and in some instances double‐stranded intermediates (Terekhova et al., 2013).
Pitman et al. (2005) used BLASTP analysis to show that the gene PPH.56 from Pph genomic island PPHGI‐1 was related to a topo III topoisomerase enzyme. PPH.56 was therefore designated topB3. The presence of this gene on the island may indicate the topoisomerase specifically influences island topology or expression of island‐based genes. Topoisomerase genes have been associated with other genomic islands. For example, PbTopoIIIβ was described as a topoisomerase III enzyme from Pectobacterium atrosepticum (Pba) strain SCRI1043 that negatively regulates the excision of pathogenicity island HAI2 (Vanga et al., 2012). Inactivation of PbTopoIIIβ on HAI2 caused a 103‐ to 104‐fold increase in excision, led to reduced fitness in vitro and a decrease in the virulence of Pba SCRI1043 in potato. It was therefore suggested that PbTopoIIIβ may be required for stable maintenance of HAI2 in the chromosome of Pba and may control as yet unidentified genes involved in the viability and virulence of Pba SCRI1043 in potato (Vanga et al., 2012). As well as being involved in regulating genomic island topology, topoisomerase III enzymes are also associated with plasmid maintenance, for example the broad host range plasmid RP4 TraE protein exhibits topoisomerase III activity and has been found to have a role in the propagation of plasmids (Li et al., 1997).
We hypothesised that the excised PPHGI‐1 becomes supercoiled, leading to the downregulation of avrPphB and that the island‐encoded topoisomerase topB3 has a role in this process. We investigated the effect of supercoiling inhibitors and a mutation in topB3 on the excision of PPHGI‐1 and the expression of island‐encoded genes. The results show that the supercoiling inhibitors significantly increased the expression of avrPphB but did not affect the expression of the island‐encoded xerC integrase or the excision of PPHGI‐1. However, a mutation in topB3 caused increased avrPphB expression and an increase in xerC expression and PPHGI‐1 excision, as well as reducing the population growth of the pathogen in its host plant. Together, these indicate an intimate relationship of topoisomerase and supercoiling in controlling island excision and gene expression.
Results
Effect of gyrase inhibitors on supercoiling in Pseudomonas syringae pv. phaseolicola
Supercoiling has been implicated in the downregulation of the effector gene avrPphB from Pph 1302A when PPHGI‐1 is excised from the chromosome (Godfrey et al., 2011). Here, supercoiling inhibitors were used to allow the investigation of relaxing of supercoiling on the expression of avrPphB and PPHGI‐1 function. The supercoiling inhibitors chosen were novobiocin, a member of the aminocoumarin group of antibiotics, whose mode of action is to competitively inhibit gyrase ATP binding, and ciprofloxacin, a quinolone antibiotic, which binds to both gyrase enzyme and DNA and stabilises the formation of the gyrase–DNA cleavage complex (Bush et al., 2015).
Firstly, the minimum inhibitory concentrations (MIC) for novobiocin and ciprofloxacin were determined for Pph 1302A. This was done by twofold dilution steps from 512 to 0.0625 µg/ml of each antibiotic in bacterial LB broth, inoculating each broth with 5 × 105 cfu/ml of Pph 1302A and recording CFU/ml of Pph 1302A growth over 18 h (Fig. S1). Moving forward, and for convenience, values of 10 µg/ml for novobiocin and 0.5 µg/ml ciprofloxacin were selected for further work (nearest round numbers at which growth was unaffected), as at these values the growth of Pph 1302A was unaffected.
Next, we tested if the selected concentrations of novobiocin and ciprofloxacin affect supercoiling in Pph. A supercoiling assay using chloroquine gels was performed with a Pph 1302A strain carrying the plasmid pBBR1MCS‐2 (Fig. 1). Topoisomers that are more negatively supercoiled migrate faster in the gel than more relaxed topoisomers (Ó Cróinínet al., 2006). Both novobiocin and ciprofloxacin inhibited supercoiling at the concentrations used, resulting in reduced migration of pBBR1MCS‐2.
Figure 1.
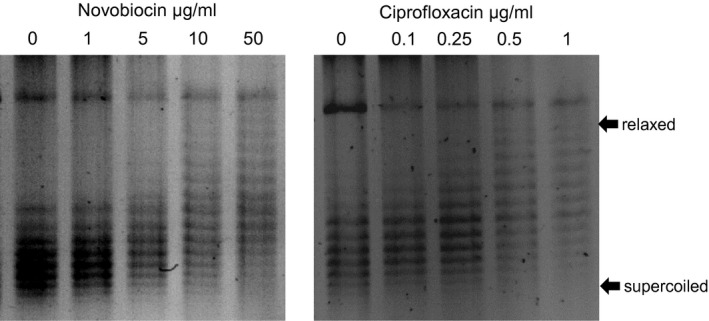
DNA relaxation by supercoiling inhibitors. Plasmid pBBR1MCS‐2, isolated from Pseudomonas syringae pv. phaseolicola 1302A, shows a dose‐dependent relaxation of supercoiling by novobiocin and ciprofloxacin. pBBR1MCS‐2 was isolated following antibiotic treatment and run on a 1% agrose gel + 2.5 µg/ml chloroquine. More negatively supercoiled topoisomers migrate faster in the gel. Direction of migration in agarose gel is from top to bottom.
To provide further support that the selected levels of the supercoiling inhibitors were exerting an effect on supercoiling, the expression levels of gyrase subunit B (gyrB) in Pph 1302A were measured with different treatments. GyrB is one of two subunits found in gyrase (Bush et al., 2015) and inhibition of GyrB by novobiocin has been shown to relax DNA supercoiling (Schröder et al., 2014). Novobiocin and ciprofloxacin both caused a significant (p < 0.05) increase in gene expression of Pph 1302A gyrB when measured in M9 minimal media after five hours using qPCR, whereas there was no change in the expression levels of the control gene acpP (Fig. S2). These results suggest that supercoiling is being affected by the inhibitors as the gyrB gene is being over expressed in an attempt to overcome the inhibitory effects of the antibiotics, as has also been observed in other systems (Sioud et al., 2009; Ferrándiz et al., 2010; Schröder et al., 2014). Overall, we can conclude that the selected concentrations of novobiocin (10 µg/ml) and ciprofloxacin (0.5 µg/ml) do not reduce cell growth but do have an effect of relaxing DNA supercoiling in Pph 1302A. We therefore moved on to look at the effect of relaxing supercoiling on the gene expression of PPHGI‐1‐encoded genes.
The effect of supercoiling inhibitors on genomic island PPHGI‐1 excision and avrPphB gene expression
Godfrey et al. (2011) showed that the expression of the PPHGI‐1‐encoded effector gene avrPphB was downregulated when the genomic island was excised from the chromosome. One reason to explain this downregulation of avrPphB when it is on an excised circular molecule could be due to DNA supercoiling. DNA supercoiling is known to affect gene expression; for example, Ferrándiz et al. (2010) found that > 13% of the genome of Streptococcus pneumoniae exhibited relaxation‐dependent transcription. To test the hypothesis that supercoiling affected avrPphB expression, the effect of novobiocin and ciprofloxacin on PPHGI‐1 excision and gene expression was investigated.
Pph 1302A cells containing chromosomal PPHGI‐1 were treated with the supercoiling inhibitors ciprofloxacin and novobiocin in vitro (Fig. 2) and the levels of island excision and gene expression were quantified. Circular intermediate production measures the amount of a junction fragment of DNA that is produced when PPHGI‐1 is excised from the chromosome and forms a circular molecule (Pitman et al., 2005). XerC is a PPHGI‐1‐encoded integrase protein that is known to be upregulated during PPHGI‐1 excision and topB3 is a PPHGI‐1‐encoded topoisomerase gene (described further below) (Pitman et al., 2005). Both ciprofloxacin and novobiocin treatments resulted in an increased expression of avrPphB (p < 0.05). However, xerC expression and circular intermediate production remained stable. These results suggest that the supercoiling inhibitors do not affect the level of excision of PPHGI‐1 from the bacterial chromosome but that the relaxation of supercoiling allows increased expression of island‐encoded genes, as seen for avrPphB and topB3.
Figure 2.
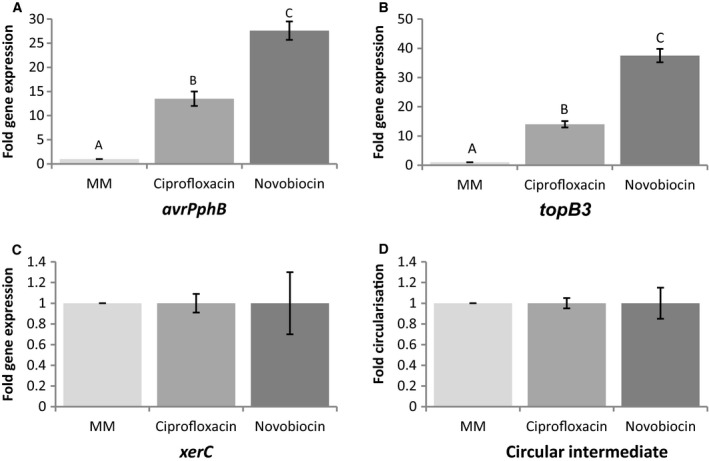
Gene expression and circular intermediate detection in P. syringae pv. phaseolicola strain 1302A after treatment with supercoiling inhibitors. Pph 1302A cells were examined for the effect of supercoiling (SC) inhibitors ciprofloxacin (0.5 µg/ml) and novobiocin (10 µg/ml) on the expression of A. effector gene avrPphB, B. topoisomerase gene topB3, C. integrase gene xerC and D. detection of the circular intermediate. Bacterial cells were incubated in M9 minimal medium (MM) plus SC inhibitors for 5 h before being treated with RNA protect and the RNA extracted (avrPphB, topB3 and xerC) or the cells pelleted and DNA extracted (circular intermediate). Results are displayed as fold expression. avrPphB, xerC and topB3 expressions were standardised by simultaneous qPCR analysis of acpP expression and error bars represent standard error of the mean of three experimental replicates. Letters above bars indicate significant differences at p < 0.05 assessed with Student’s t‐test.
To confirm that the supercoiling inhibitors were affecting avrPphB expression when PPHGI‐1 is in its circular form, an insertional mutant of the xerC integrase gene (Pph 1302A::int, PPH.100) that renders PPHGI‐1 unable to excise from the chromosome (Pitman et al., 2005) was treated with and without 10 µg/ml novobiocin in extracted TG apoplastic fluid. The level of avrPphB expression increased more than 20 times as a result of treatment with novobiocin in wild‐type (WT) Pph 1302A, which can excise PPHGI‐1 from the chromosome (Fig. 3). However, there was no difference in avrPphB expression between Pph 1302A::int with and without novobiocin treatment, suggesting that avrPphB expression is only affected by supercoiling inhibitors when PPHGI‐1 is excised from the chromosome (Fig. 3). This suggests that PPHGI‐1 exists in a supercoiled form when it is excised from the chromosome causing silencing of avrPphB.
Figure 3.
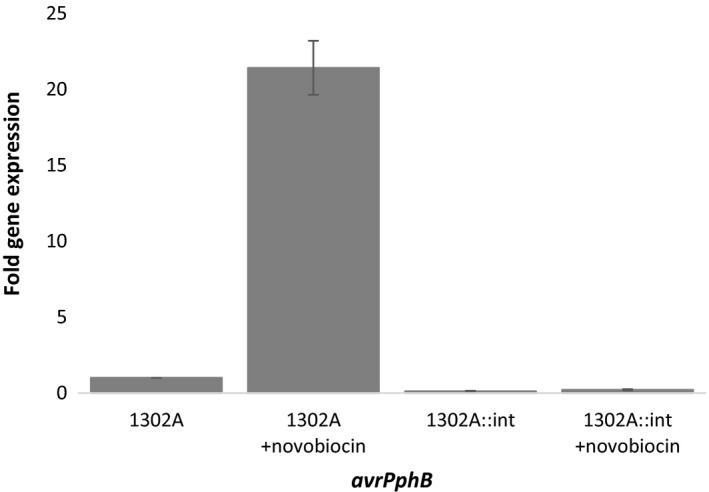
avrPphB expression is only affected by supercoiling inhibitors when PPHGI‐1 is excised from the chromosome. Bacterial cells were incubated in extracted bean cultivar Tendergreen apoplastic fluid for 5 h before the gene expression of avrPphB was measured. avrPphB expression is increased 21 times in wild‐type (WT) Pph 1302A treated with novobiocin, which can excise PPHGI‐1 from the chromosome. However, there is no difference in avrPphB expression between Pph 1302A::int (from which PPHGI‐1 is unable to excise) with and without novobiocin treatment. Results are displayed as fold expression. All data were standardised by simultaneous qPCR analysis of acpP expression and error bars represent standard error of the mean of three experimental replicates.
The effect of topB3 disruption on genomic island PPHGI‐1 excision/loss and avrPphB gene expression
The gene topB3 is a topoIII gene found in the Pph genomic island PPHGI‐1 that is related to PbTopoIIIβ which was previously linked with regulating genomic island excision (Vanga et al., 2012). We hypothesised that this gene may influence the stability of the genomic island within the chromosome and thus the expression of avrPphB. Thus, to further characterise topB3 for its influence on PPHGI‐1 excision, avrPphB expression and fitness of Pph, an insertional knock out of topB3 was created (Pph 1302A::56).
To determine if topB3 has any pleiotropic effects on bacterial growth, growth and competition experiments were carried out. Strain 1302A::56 exhibited no significant difference in in vitro growth in rich medium (LB) over 24h compared to WT Pph 1302A (Fig. S3) when grown individually. Similarly, no difference in relative fitness was observed between Pph 1302AWT and Pph 1302A::56 in in vitro competition where relative fitness was equal to 0.06 (Zhang and Rainey, 2007). However, when the growth of Pph 1302A::56 and Pph 1302A WT was compared in planta, either as individual inoculations or in competition, a small but significant (p < 0.05) reduction in Pph 1302A::56 growth relative to Pph 1302A WT was observed after two and five days, respectively, in both the resistant cultivar TG and the susceptible cultivar Canadian Wonder (CW) (Fig. 4).
Figure 4.
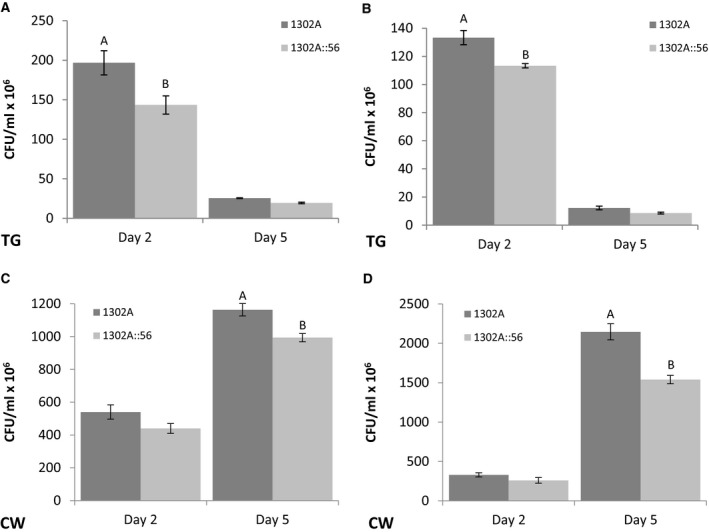
Growth of Pseudomonas syringae pv. phaseolicola 1302A::56 is lower than 1302A wild type in planta. A. Individual growth in resistant host Tendergreen (TG). B. Competitive growth in resistant host TG. C. Individual growth in susceptible host Canadian Wonder (CW). D. Competitive growth in susceptible host CW. Means are of three replicates ±SEM. Letters above bars indicate significant differences at p < 0.05 assessed with Student’s‐t test.
To investigate if topB3 is involved in island excision, bacterial cells were incubated for five hours in an apoplast‐mimicking medium that is known to induce excision and circularisation of PPHGI‐1 (Pitman et al., 2005) and expression of xerC. qPCR was used to compare the expression level of the integrase xerC, which is involved in PPHGI‐1 excision, in the topB3 mutant compared to the WT strain. The level of PPHGI‐1 circular intermediate produced was also examined. Circular intermediate production is used here as a measure of island excision from the chromosome, rather than detecting island loss using our standard bean pod assay (Pitman et al., 2005), because in vitro the island is not lost from the bacteria cell due to the absence of the selection pressure of the resistant plant. xerC showed an increase in the expression of the topB3 mutant compared to the WT strain (Fig. 5A), which is over and above the level seen by the effect of the apoplast alone. This increase in xerC expression also correlated with an increase in circular intermediate formation (Fig. 5B). This supports the hypothesis that topB3 is involved in the regulation of island excision via a direct or indirect effect on xerC. However, given the supercoiling inhibitors had no effect on either xerC expression or circular intermediate formation, this suggests the topB3 mutation must be having an indirect effect on xerC expression. In contrast, avrPphB, which was upregulated in the presence of supercoiling inhibitors, also exhibited an eightfold increase in the expression of topB3 mutant in apoplastic fluid (Fig. 5C) and this observation was confirmed in planta (Fig. 5D). Together, these data suggest that supercoiling appears to control the expression of avrPphB but not xerC, whereas TopB3 negatively regulates the expression of avrPphB and xerC probably independently of the effect of supercoiling.
Figure 5.
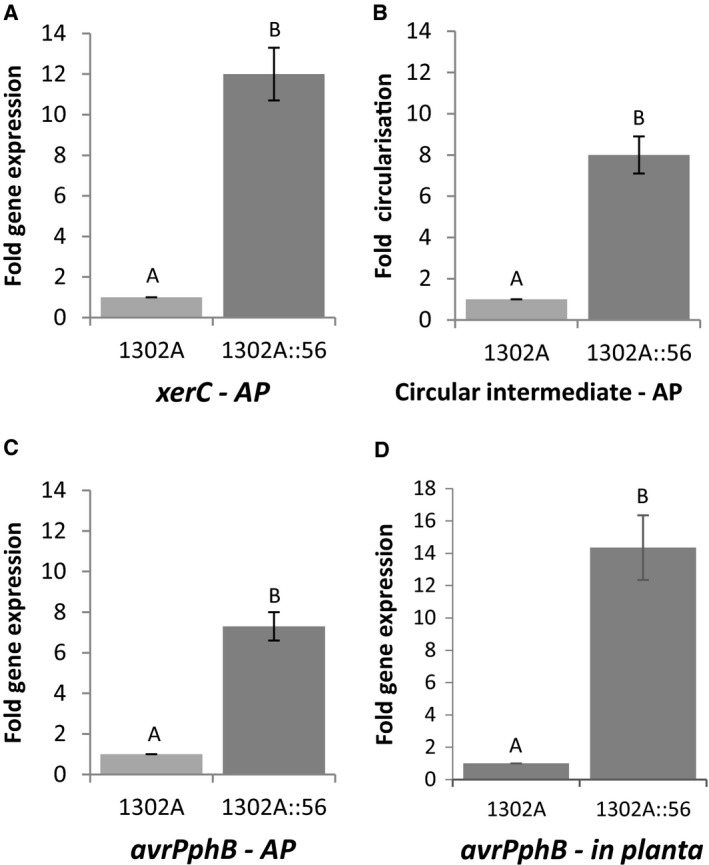
Gene expression and circular intermediate detection in a topoisomerase insertion mutant, strain Pseudomonas syringae pv. phaseolicola 1302A::56. Bacterial cells were inoculated in extracted bean cultivar Tendergreen apoplastic fluid for 5 h before gene expression of A. integrase gene xerC and C. effector gene avrPphB or B. detection of the circular intermediate was measured. Results are displayed as fold expression. D. Bacterial cells were inoculated into bean cultivar Tendergreen leaves and the plants incubated for 5 h before the apoplastic fluid was extracted and gene expression of effector gene avrPphB was measured. avrPphB and xerC expression were standardised by simultaneous qPCR analysis of acpP expression and error bars represent standard error of the mean of three experimental replicates. Letters above bars indicate significant differences at p < 0.05 assessed with Student’s t‐test. AP: extracted TG apoplastic fluid.
To determine if disrupting topB3had any effect on in planta island loss, Pph 1302A::56 was passaged six times through leaves of bean cv. Tendergreen (TG) and 100 colonies were tested for the presence or absence of PPHGI‐1 using TG pods as a screen (Pitman et al., 2005). Strain Pph 1302A::56 showed a high level of island loss (96%), which was very similar to a control insertion mutant in a non‐coding region of PPHGI‐1 (98%) (Fig. S4). It was therefore clear that topB3 is not essential for island loss from the cells.
Discussion
Previous studies have shown that the genomic island PPHGI‐1 can be excised from the bacterial chromosome and that when this happens the expression of avrPphB is downregulated (Godfrey et al., 2011). We have previously speculated that the downregulation of avrPphB during island excision may be due to the supercoiling of the island as it becomes a separate circular molecule from the chromosome, and that this downregulation may effectively ‘hide’ the presence of this avirulence gene from the plant’s surveillance system, thereby reducing the elicitation of the HR response. In order to further examine this, we investigated the effect of supercoiling on PPHGI‐1 structure and avrPphB expression and also investigated an island‐encoded topoisomerase gene, topB3, that had been suggested in other systems to be involved in genomic island stability.
Firstly, we determined the MIC for two supercoiling inhibitors, novobiocin and ciprofloxacin, and found the concentrations of 10 and 0.5 µg/ml, respectively, to have no detectable effect on bacterial growth but to relax the supercoiling of a reporter plasmid pBBR1MCS‐2. These values compared well with other reported levels that were used to investigate supercoiling, for example several studies have shown relaxation of supercoiling with novobiocin at a level of 25 or 50 µg/ml in Salmonella enterica serovar Typhimurium (Ó Cróinín et al., 2006) and 4 and 8 µg/ml in Helicobactor pylori (Ye et al. 2007). Ciprofloxacin has been shown to relax plasmid DNA supercoiling in Escherichia coli K12 (Aleixandre et al., 1991) at a concentration of 0.5–3 µg/ml. Robillard et al. (2012) also demonstrated that ciprofloxacin relaxes supercoiling by 50% in E. coli H560, P. aeruginosa PA02 and Klebsiella pneumoniae MP100 at concentrations of 0.5, 1.0 and 0.5 µg/ml, respectively. When Pph 1302A was treated with novobiocin at 10 µg/ml, the expression of gyrB increased and this was taken as additional evidence that novobiocin affected the supercoiling as an increase in gyrB expression can be seen as an indicator that gyrB is being overexpressed to combat the effect of reduced supercoiling. A similar phenomenon was observed by Sioud et al. (2009) where it was found that during Bacillus subtilis treatment with novobiocin to inhibit gyrase‐maintained supercoiling, both gyrA and gyrB genes were upregulated.
After establishing the levels of supercoiling inhibitors that affect PPHGI‐1, we then investigated their effect on PPHGI‐1 excision from the chromosome and on avrPphB expression. Novobiocin and ciprofloxacin both caused an increase in avrPphB expression but no increase in circularisation of the island suggesting that increased expression is due to a reduction in supercoiling and not an increase in the amount of extra chromosomal island. This supports the theory that supercoiling of the excised PPHGI‐1 inhibits avrPphB expression. It is well established that supercoiling of DNA influences the level of transcription in cells (Pruss and Drlica, 1989; Ma and Wang 2014). For example, Ye et al. (2007) used genome‐wide transcript analysis under conditions of reduced supercoiling and showed both an increase and decrease in the transcription of a number of genes during relaxation of supercoiling by novobiocin. Similarly, Peter et al. (2004) measured the transcriptional response to loss of supercoiling due to topoisomerase gene mutation or addition of topoisomerase inhibitors in E. coli MG1655. They found that the transcription of 306 genes was significantly altered by a change in the level of supercoiling. More recently, Sobetzko (2016) reported that DNA supercoiling is amongst the most influential regulators of gene expression found in bacteria with more than half of all genes sensitive to supercoiling.
An insertional mutant of topB3 (Pph 1302A::56), a PPHGI‐1‐encoded type IA topoisomerase gene, showed increased levels of island circularisation compared to wild type. The Pph 1302A::56 strain also showed increased levels of avrPphB expression. Since topoisomerase inhibitors were found to increase the expression of avrPphB when it was present in the circular intermediate, this suggests that topB3 also contributes to the suppression of avrPphB expression by modulating the supercoiling of the circular intermediate. However, the unexpected observation that avrPphB expression was increased in the topB3 mutant suggests a more complex regulation where TopB3 is actually having a direct effect on gene expression of avrPphB and not through a role in supercoiling. This would be an area for further investigation.
Vanga et al. (2012) showed an increase in the excision of the genomic island HA12 when they inactivated a similar island‐encoded topoisomerase gene PbTopoIIIβ. These authors suggest that PbTopoIIIβ may be responsible for island stability in the chromosome and this may also be the case in the current study. The role of Topo III proteins is not completely understood, but the enzyme is thought to be important for the maintenance of genomic stability (Deweese et al., 2008). Topo III proteins are also known to act as a potent decatenase on DNA rings that have small gaps, but are very poor at relaxing supercoils (Perez‐Cheeks et al., 2012).
Vanga et al. (2012) also observed a reduction in virulence of Pba with PbTopoIIIβ disrupted in potato and we observed a reduction in virulence by the topB3‐disrupted strain on the susceptible bean cultivar CW after five days. We have previously observed that in CW PPHGI‐1 confers a fitness advantage to Pph 1302A (Neale et al., 2016) and it may be that increasing its excision and altering the gene expression of island‐encoded genes has a deleterious effect on the ability of Pph 1302A to cause disease. Here we also observed that the topB3‐disrupted strain caused an increase in the HR response in the early stages of infection as measured by a reduction in bacterial growth in the resistant cultivar (2 days).
Here we have shown that the function of a genomic island, and the expression of a gene carried by it, is affected by supercoiling inhibitors, and that supercoiling may be a way in which, once circularised, the island controls gene expression. We have also shown the importance of an island‐encoded topoisomerase gene for both island stability in the chromosome and island gene expression. However, other factors could be influencing gene expression on PPHGI‐1. Given that the integrase family of tyrosine recombinases can alter the topology of DNA (Grainge and Jayaram, 1999), then it may be possible that the xerC gene on the island could be affecting PPHG‐1 topology and therefore altering gene expression of island‐encoded genes. In addition to plant pathogenic bacteria, pathogens of humans and other animals contain many important GIs including SPI‐1 from Salmonella enterica which also encodes a T3SS (Hensel, 2004), the cag‐pathogenicity island from Helicobactor pylori which encodes a type IV secretion system (Wang et al., 2016) and the locus of enterocyte effacement genomic island in E. coli (Franzin and Sircili, 2015). It would therefore be of interest to see how widespread this phenomenon of regulation of island‐encoded genes being affected by the physical state of the island is and whether island‐encoded topoisomerase genes have a wider role in genomic island stability. Given the well‐established use of supercoiling inhibitors as antimicrobials in a clinical context, it will also be interesting to establish whether they could also potentially be integrated into plant disease control strategies.
Experimental procedures
Bacterial and plant growth conditions
Pph strains (Table 1) were cultured at 25°C for 48 h on King’s B (KB) agar plates (Difco, UK). Overnight cultures were grown in Luria–Bertani media (LB, Difco, UK) at 25°C. Antibiotics and supercoiling inhibitors were used at the following concentrations (μg/ml): kanamycin (Km) 50, novobiocin (1–100) and ciprofloxacin (0.5–5). Bean cultivars Tendergreen (TG) and Canadian Wonder (CW) were grown for 14 days at 23°C, 70% humidity with a 16 h photoperiod.
Table 1.
Bacterial strains
Plant passaging
Overnight bacterial cell suspensions were washed and the pellet resuspended in 10 mM MgCl2 followed by dilution to an optical density of 0.1 (OD600, CFU 8 × 107) before being infiltrated into bean leaves via a syringe and needle. After 7 days, inoculated tissue was dissected and homogenised in 10 mM MgCl2. Bacteria were recovered by brief centrifugation to remove any remaining plant tissue and diluted to their original inoculated optical density, before being re‐infiltrated into fresh leaves. Recovered bacteria were plated onto KB agar at each passage and single colonies screened for their ability to cause disease symptoms or HR by inoculating them into TG bean pods using a sterile cocktail stick.
Growth assays
For in planta growth assays, three replicates of each cell suspension were grown overnight in LB broth and diluted to 8 × 107 CFU/ml (OD600 0.1). Cells were inoculated into 14‐day‐old bean leaves via a syringe and needle either as individual cultures or 50:50 of each strain for competition assays. Plants were incubated for 48 and 120 h. Bacteria were harvested as above and total CFU/ml were counted. Fitness was calculated as per Zhang and Rainey (2007).
For in vitro growth assays, three replicates of each bacterial mutant were grown overnight in LB broth and diluted to 8 × 108 CFU/ml (OD600 1.0). Cells (100 μl) were sub‐cultured into 10 ml fresh LB broth and incubated at 25°C shaking (10 g). At 0, 2, 4, 6, 8, 16 and 24 h, samples were diluted and plated onto KB + Kan and total CFU/ml counted. For competition assays, 50:50 of each strain was used and bacterial cells were diluted and plated onto KB/KB + Kan every hour for 8 h and total CFU/ml counted.
Minimum inhibitory concentrations (MICs)
MICs for novobiocin and ciprofloxacin in Pph were determined using doubling dilution steps from 512 to 0.0625 µg/ml and recording CFU/ml bacterial growth. 10 µg/ml novobiocin and 0.5 µg/ml ciprofloxacin were determined as concentrations that had no effect on bacterial growth.
Separation of plasmid topoisomers by chloroquine gel electrophoresis
The reporter plasmid pBBR1MCS‐2 (Kovach et al., 1994) was electroporated into Pph 1302A following the method of Keen et al. (1992). This strain was then treated with 1, 5, 10, 50 µg/ml novobiocin or 0.1, 0.25, 0.5, 1 µg/ml ciprofloxacin for 4 h. Plasmid DNA was then extracted using the Qiagen mini‐prep plasmid kit and visualised on a 1% agarose gel containing 2.5 µg/ml chloroquine as per Higgins et al. (1988).
PCR and qPCR
For PCR, a standard 25 μl PCR mix was used that consisted of the following: 1 μl bacterial culture, 0.5 μM oligonucleotide primers (Table 2), 12.5 μl mastermix (Taq PCR mastermix, Qiagen) and 9.5 μl sterile deionised water. Standard PCR cycling conditions consisted of 94°C for 10 min, followed by 30 cycles of 94°C for 30 s, X°C for 30 s (dependent on the annealing temperature of primers) and 72°C for 1 min, followed by a final extension step of 72°C for 10 min.
Table 2.
Oligonucleotide primers
| Primer name | Description | Sequence 5′–3′ |
|---|---|---|
| acpP | Housekeeping gene | Forward TTGGCGTCAAATCAGAAGAG |
| Reverse GCTTCTTCGTCAGGGATTTC | ||
| Probe ACCTGGGCGCTGACTCCCTG[Link] | ||
| gyrB | gyrB gene | Forward GATGATGGAATCGGTGTCGAA |
| Reverse TTGGTGAAGCACAACAGGTTCT | ||
| Probe CCCTGCAGTGGAACGACAGCTTCA[Link] | ||
| xerC | PPHGI‐1 encoded xerC intergrase | Forward CGACGATACGGCCTCCAA |
| Reverse AAAGGTGCGGTCGACATCA | ||
| Probe CCCCCTATAGCGGAGCGTCTGGAA[Link] | ||
| QavrPphB | PPHGI‐1 encoded effector gene | Forward CCCATTCCTGGCAATGACA |
| Reverse TTACGCCTGAAGAGGATGCA | ||
| Probe TGGGCGATAAAGGG[Link] | ||
| QCI | Circular intermediate | Forward CATGGGCCTTCCAGATTTTC |
| Reverse CTGCGGTTTGGGATACTGAAC | ||
| Probe CGTAACGCTGAGGCAGGCCCC[Link] | ||
| avrPphB | PPHGI‐1 encoded effector avrPphB | Forward GCGATTGCGTGTCCTTGA |
| Reverse CTGTAAGACCTGAGCCTG |
aProbes labelled with 5′ FAM and 3' TAMRA TaqMan dyes.
After amplification, a 20 μl sample was visualised on a 0.7% agarose gel with 2 μg/ml ethidium bromide in comparison to a DNA molecular marker (Hyperladder, Bioline).
qPCR was used to quantify the expression levels of gyrB, avrPphB, xerC and the amount of circular intermediate produced in vitro. Bacterial cells were incubated in 50:50 1 × M9 Minimal medium with 20% glucose (Sambrook and Russell, 2001) and TG apoplastic fluid. Apoplastic fluid was extracted using the method described by Lovell et al. (2009) where appropriate cultures were spiked with 1–10 µg/ml novobiocin or 0.5–5 µg/ml ciprofloxacin. After 5 h shaking at 25°C, cells were harvested using RNA protect reagent (Qiagen, UK) for RNA extraction or DNA lysis solution (Gentra Systems, UK) for DNA extraction. For quantification of gyrB, avrPphB and xerC expression, RNA was extracted using the RNAeasy kit (Qiagen) followed by a second DNase step of 15 min at 37°C (Promega, UK) (Godfrey et al., 2011). cDNA was synthesised using the TaqMan reverse transcription kit (Qiagen). For circular intermediate quantification, DNA was extracted using the Puregene total DNA extraction kit (Gentra Systems) (Godfrey et al., 2011). For in planta expression, whole TG leaves were inoculated with 0.1 OD600 Pph 1302A and incubated for 5 h before apoplastic fluid was extracted. X‐fold change in gene expression was calculated using WT as the calibrator and acpP as the internal control (Table 2). The gene acpP has been demonstrated to be a stably expressed internal reference gene in Pseudomonas (Lenz et al., 2008; Qi et al., 2014). For the current study, it was tested under a variety of conditions and showed no change in expression (Fig. S5).
Author contributions
The conception or design of the study: HN, DLA.
The acquisition, analysis, or interpretation of the data: HN, DLA, RWJ, GP.
Writing of the manuscript HN, DLA, RWJ, GP.
Supporting information
Acknowledgements
We would like to thank Dr. Tadhg Ó Cróinín (University College Dublin) for his invaluable help with the chloroquine gel supercoiling assays. This work was carried out under the Department for Environment, Food and Rural Affairs’ Plant Health and Seeds Inspectorate license number 51049/202078/1. This research was funded by grants from the Biotechnology and Biological Sciences Research Council, United Kingdom (BB/J016012/1, BB/J014796/1 and BB/J015350/1).
Data underlying this article can be accessed at https://researchdata.uwe.ac.uk/215/ and used under the Creative Commons Attribution licence.
References
- Ahmed, W. , Sala, C. , Hegde, S.R. , Jha, R.K. , Cole, S.T. and Nagaraja, V. (2017) Transcription facilitated genome‐wide recruitment of topoisomerase I and DNA gyrase. PLOS Genetics, 13(5), e1006754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleixandre, V. , Herrera, G. , Urios, A. and Blanco, M. (1991) Effects of ciprofloxacin on plasmid DNA supercoiling of Escherichia coli Topoisomerase I and Gyrase mutants. Antimicrobial Agents and Chemotherapy, 35, 20–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J.R. and Collmer, A. (2004) Type III secretion system effector proteins: double agents in bacteria disease. Annual Review of Phytopathology, 42, 385–414. [DOI] [PubMed] [Google Scholar]
- Arnold, D.L. , Pitman, A. and Jackson, R.W. (2003) Pathogenicity and genomic islands in plant pathogenic bacteria. Molecular Plant Pathology, 4, 407–422. [DOI] [PubMed] [Google Scholar]
- Bush, N. G. , Evans‐Roberts, K. and Maxwell, A. (2015) DNA topoisomerases. Ecosal Plus, 6(2): Available at:doi: 10.1128/ecosalplus. [DOI] [PubMed] [Google Scholar]
- Deweese, J.E. , Osheroff, M.A. and Osheroff, N. (2008) DNA topology and topoisomerases: teaching a "Knotty" subject. Biochemistry and Molecular Biology Education, 37, 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz, M.J. , Martín‐Galiano, A.J. , Schvartzman, J.B. and de la Campa, A.G. (2010) The genome of Streptococcus pneumoniae is organized in topology‐reacting gene clusters. Nucleic Acids Research, 38, 3570–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzin, F.M. and Sircili, M.P. (2015) Locus of enterocyte effacement: a pathogenicity island involved in the virulence of enteropathogenic and enterohemorragic Escherichia coli subjected to a complex network of gene regulation. BioMed Research International 2015: Article ID 534738, 10 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey, S.A.C. , Lovell, H.C. , Mansfield, J.W. , Corry, D.S. , Jackson, R.W. and Arnold, D.L. (2011) The stealth episome: suppression of gene expression on the excised genomic island PPHGI‐1 from Pseudomonas syringae pv. phaseolicola . PLoS Pathogens, 7(3), e1002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainge, I. and Jayaram, M. (1999) The integrase family of recombinase: organization and function of the active site. Molecular Microbiology, 33, 449–456. [DOI] [PubMed] [Google Scholar]
- Hacker, J. and Carniel, E. (2001) Ecological fitness, genomic islands and bacterial pathogenicity. EMBO Reports, 2, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker, J. and Kaper, J.B. (2000) Pathogenicity islands and the evolution of microbes. Annual Review of Microbiology, 54, 641–679. [DOI] [PubMed] [Google Scholar]
- Hensel, M. (2004) Evolution of pathogenicity islands of Salmonella enterica . International Journal of Medical Microbiology, 294, 95–102. [DOI] [PubMed] [Google Scholar]
- Higgins, C.F. , Dorman, C.J. , Stirling, D.A. , Waddell, L. , Booth, I.R. , May, G . et al (1988) A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli . Cell, 52, 569–584. [DOI] [PubMed] [Google Scholar]
- Jackson, R.W. , Mansfield, J.W. , Arnold, D.L. , Sesma, A. , Paynter, C.D. , Murillo, J . et al (2000) Excision from tRNA genes of a large chromosomal region, carrying avrPphB, associated with race change in the bean pathogen, Pseudomonas syringae pv. phaseolicola . Molecular Microbiology, 38, 186–197. [DOI] [PubMed] [Google Scholar]
- Jenner, C. , Hitchin, E. , Mansfield, J. , Walters, K. , Betteridge, P. , Teverson, D . et al (1991) Gene‐for‐gene interactions between Pseudomonas syringae pv. phaseolicola and Phaseolus . Molecular Plant‐Microbe Interactions, 4, 553–562. [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature Reviews, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Keen, N.T. , Shen, H. , Cooksey, D.A. (1992) Introduction of cloned DNA into plant pathogenic bacteria. In: Gurr S.J. (Ed.) Molecular Plant Pathology: A Practical Approach. Oxford: IRL Press, pp. 45–50. [Google Scholar]
- Kovach, M.E. , Phillips, R.W. , Elzer, P.H. , Roop II, R.M. and Peterson, K.M. (1994) pBBR1MCS: a broad‐host‐range cloning vector. BioTechniques, 16, 800–802. [PubMed] [Google Scholar]
- Lenz, A.P. , Williamson, K.S. , Pitts, B. , Stewart, P.S. and Franklin, M.J. (2008) Localized gene expression in Pseudomonas aeruginosa biofilms. Applied and Environmental Microbiology, 74, 4463–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Hiasa, H. , Kumar, U. and DiGate, R.J. (1997) The traE gene of plasmid RP4 encodes a homologue of Escherichia coli DNA topoisomerase III. Journal of Biological Chemistry, 272, 19582–19587. [DOI] [PubMed] [Google Scholar]
- Lovell, H.C. , Mansfield, J.W. , Godfrey, S.A.C. , Jackson, R.W. , Hancock, J.T. and Arnold, D.L. (2009) Bacterial evolution by genomic island transfer occurs via DNA transformation in planta . Current Biology, 19, 1586–1590. [DOI] [PubMed] [Google Scholar]
- Luttinger, A. (1995) The twisted ‘life’ of DNA in the cell: bacterial topoisomerases. Molecular Microbiology, 15, 601–606. [DOI] [PubMed] [Google Scholar]
- Ma, J. and Wang, M. (2014) Interplay between DNA supercoiling and transcription elongation. Transcription, 5, e28636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale, H.C. , Slater, R.T. , Mayne, L.‐M. , Manoharan, B. and Arnold, D.L. (2013) In planta induced changes in the native plasmid profile of Pseudomonas syringae pathovar phaseolicola strain 1302A. Plasmid, 70, 420–424. [DOI] [PubMed] [Google Scholar]
- Neale, H.C. , Laister, R. , Payne, J. , Preston, G. , Jackson, R.W. and Arnold, D.L. (2016) A low frequency persistent reservoir of a genomic island in a pathogen population ensures island survival and improves pathogen fitness in a susceptible host. Environmental Microbiology, 18, 4144–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ó Cróinín, T., Carroll, R.K., Kelly, A. and Dorman, C.J. (2006) Roles for DNA supercoiling and the Fis protein in modulating expression of virulence genes during intracellular growth of Salmonella enterica serovar Typhimurium. Molecular Microbiology, 62, 869–882. [DOI] [PubMed] [Google Scholar]
- Perez‐Cheeks, B.A. , Lee, C. , Hayama, R. and Marians, K.J. (2012) A role for topoisomerase III in Escherichia coli chromosome segregation. Molecular Microbiology, 86, 1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, B.J. , Aesuaga, J. , Breier, A.M. , Khodursky, A.B. , Brown, P.O. and Cozzarelli, N.R. (2004) Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli . Genome Biology, 5, R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman, A. , Jackson, R.W. , Mansfield, J.M. , Kaitell, V. , Thwaites, R. and Arnold, D.L. (2005) Exposure to host resistance mechanisms drives evolution of bacterial virulence in plants. Current Biology, 15, 2230–2235. [DOI] [PubMed] [Google Scholar]
- Pruss, G.J. and Drlica, K. (1989) DNA supercoiling and prokaryotic transcription. Cell, 56, 521–523. [DOI] [PubMed] [Google Scholar]
- Qi, Q. , Preston, G.M. and MacLean, R.C. (2014) Linking system‐wide impacts of RNA polymerase mutations to the fitness cost of rifampin resistance in Pseudomonas aeruginosa . MBio, 5, e01562–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robillard, N.J. , Masecar, B.L. , Brescia, B. and Celesk, R. (2012) Chapter 8. Comparisons of DNA gyrases from different species In: Crumplin G.C. (Ed.) The 4‐Quinolones: Anti Bacterial Agents In Vitro. London: Springer‐Verlag, pp. 103–114. [Google Scholar]
- Sambrook, J. and Russell, D.W. (2001) Molecular Cloning: A Laboratory Manual. vol.3, 3rd ed, Cold Spring Harbor: Cold Spring Harbor Laboratory, ppA2.2. [Google Scholar]
- Schröder, W. , Bernhardt, J. , Marincola, G. , Klein‐Hitpass, L. , Herbig, A. , Krupp, G . et al (2014) Altering gene expression by aminocoumarins: the role of DNA supercoiling in Staphylococcus aureus . BMC Genomics, 16(15), 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud, M. , Boudabous, A. and Cekaite, L. (2009) Transcriptional responses of Bacillus subtillis and thuringiensis to antibiotics and anti‐tumour drugs. International Journal of Molecular Medicine, 23, 33–39. [PubMed] [Google Scholar]
- Sobetzko, P. (2016) Transcription‐coupled DNA supercoiling dictates the chromosomal arrangement of bacterial genes. Nucleic Acids Research, 44, 1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J.D. , Teverson, D.M. , Allen, D.J. and Pastor‐Corrales, M.A. (1996) Identification and origin of races of Pseudomonas syringae pv. phaseolicola from Africa and other bean growing areas. Plant Pathology, 45, 469–478. [Google Scholar]
- Terekhova, K. , Marko, J.F. and Mondragón, A. (2013) Studies of bacterial topoisomerases I and III at the single‐molecule level. Biochemical Society Transactions, 41, 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer, J.R. and Sentchilo, V. (2003) Genomic islands and the evolution of catabolic pathways in bacteria. Current Opinion in Biotechnology, 14, 248–254. [DOI] [PubMed] [Google Scholar]
- Vanga, B.R. , Butler, R.C. , Toth, I.K. , Ronson, C.W. and Pitman, A.R. (2012) Inactivation of PbTopo IIIβ causes hyper‐excision of the pathogenicity island HAI2 resulting in reduced virulence of Pectobacterium atrosepticum . Molecular Microbiology, 84, 648–663. [DOI] [PubMed] [Google Scholar]
- Vos, S.M. , Tretter, E.M. , Schmidt, B.H. and Berger, J.M. (2011) All tangled up: how cells direct, manage and exploit topoisomerase function. Nature Reviews Molecular Cell Biology, 12, 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Ling, F. , Wang, H. , Yu, M. , Zhu, H. , Chen, C . et al (2016) The Helicobacter pylori cag pathogenicity island protein Cag1 is associated with the function of T4SS. Current Microbiology, 73, 22–30. [DOI] [PubMed] [Google Scholar]
- Ye, F. , Brauer, T. , Niehus, E. , Drlica, K. , Josenhans, C. and Suerbaum, S. (2007) Flagellar and global gene regulation in Helicobacter pylori modulated by changes in DNA supercoiling. International Journal of Medical Microbiology, 297, 65–81. [DOI] [PubMed] [Google Scholar]
- Zhang, X.‐X. and Rainey, P.B. (2007) Construction and validation of a neutrally‐marked strain of Pseudomonas fluorescens SBW25. Journal of Microbiological Methods, 71, 78–81. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Li, W. , Xiang, T. , Liu, Z. , Laluk, K. , Ding, X . et al (2010) Receptor‐like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host & Microbe, 7(4), 290–301. [DOI] [PubMed] [Google Scholar]
- Zhu, M. , Shao, F. , Innes, R.W. , Dixon, J.E. and Xu, Z. (2004) The crystal structure of Pseudomonas avirulence protein AvrPphB: a papain‐like fold with a distinct substrate‐binding site. Proceedings of the National Academy of Sciences U S A, 101, 302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


