Abstract
Primary ovarian sarcomas are extremely rare tumors, and their genomic and transcriptomic alterations remain to be elucidated. We performed whole exome sequencing of primary tumor and matched normal blood samples derived from one patient with ovarian undifferentiated small round cell sarcoma. We identified 8 nonsynonymous somatic mutations, and all mutations were missense or nonsense changes. Next, we performed RNA sequencing of the tumor sample and identified two in‐frame fusion transcripts: MXD4–NUTM1 and ARL6–POT1. Most NUTM1 exons were retained in the MXD4–NUTM1 fusion transcript, and we confirmed an increase in NUTM1 mRNA and protein expression in tumor tissue. Further genomic and transcriptomic analyses might lead to the development of new therapeutic strategies based on the molecular characteristics of ovarian undifferentiated small round cell sarcoma.
Keywords: fusion transcript, MXD4‐NUTM1, NUT rearrangement, ovarian sarcoma, small round cell sarcoma
1. INTRODUCTION
Ovarian sarcomas are rare mesenchymal tumors, and various histological types—such as carcinosarcoma,1 leiomyosarcoma,2 rhabdomyosarcoma,3, 4 endometrial stromal sarcoma5 and fibrosarcoma6—are pathologically defined. Small round cell sarcoma is a heterogeneous group of tumors arising from various sites, and the prognosis of this disease is poor.7, 8 Fusion genes resulting from chromosomal rearrangements have been considered as a significant oncogenic factor in many mesenchymal malignancies,9 and tumor‐specific fusion genes contribute to the classification of small round cell sarcomas into several histologic subtypes,8 that is, EWSR1–FLI1 fusion in Ewing sarcoma10 and PAX3/8 fusions in alveolar rhabdomyosarcoma.11
Recent advances in sequencing technology have enabled the comprehensive detection of fusion genes in the cancer genome and transcriptome. We have used RNA sequencing data to identify numerous fusion genes in various types of tumors12, 13, 14 and developed the fusion gene data portal.
Herein, we report the molecular characteristics of an ovarian undifferentiated small round cell sarcoma based on genomic and transcriptomic analyses.
2. MATERIALS AND METHODS
2.1. Case presentation
We describe a case of 34‐year‐old woman who did not have a significant past medical history or family history. She visited a gynecologist for cancer screening with no symptom. The Pap test was negative, but transvaginal ultrasound examination revealed a large pelvic mass in the pouch of Douglas with ascites. Moreover, transabdominal ultrasound examination also revealed a large mass in the abdominal cavity. Her serum CA125 level was elevated (969 U/mL). MR and CT imaging detected an 8‐cm‐sized tumor that was suspected to be derived from the right ovary in the right‐side pelvic cavity, a 6‐cm‐sized tumor in the lower abdominal cavity, diffuse peritoneal thickening, and ascites. On the other hand, no obvious abnormal findings were noted in the uterus, the left adnexa, and soft tissue (Figure 1A). She was suspected to have cancer of unknown primary origin and was referred to our hospital for further examinations and treatment of this tumor. To confirm a pathological diagnosis, she underwent an exploratory laparotomy. Operative findings revealed ~2350 mL of bloody ascites in the peritoneal cavity, and the ascites cytology was positive for malignant cells. An 8 cm right ovarian solid tumor adhered to the small intestine at multiple sites. Omental cake and diffuse peritoneal dissemination were also observed but their sizes were significantly smaller than the size of the right ovarian solid tumor. A normal‐sized uterus and left adnexa strongly adhered to the right ovarian tumor. We performed partial resection of the right ovarian tumor and biopsy of omental cakes (Figure 1B). Pathological findings demonstrated that tumor tissue consisted of small round cells with abundant eosinophilic cytoplasm and spindle cells, and the cell‐to‐cell junction was unclear in this tumor (Figure 2A). Immunohistochemistry analysis revealed that tumor cells were positive for vimentin, weakly positive for CD99, and negative for pan‐keratin (AE1/AE3), CD10, desmin, MyoD1, myogenin, and SMA (Figure 2B and Supporting Information, Figure 1). Finally, the patient was diagnosed with a stage IIIC ovarian undifferentiated small round cell sarcoma. After the surgery, she was treated with combination chemotherapy, including Ifosfamide and Epirubicin. After 6 courses of the chemotherapy, the tumor size increased, and massive ascites was observed. She did not desire additional therapies. Finally, she died 9 months after the surgery.
Figure 1.
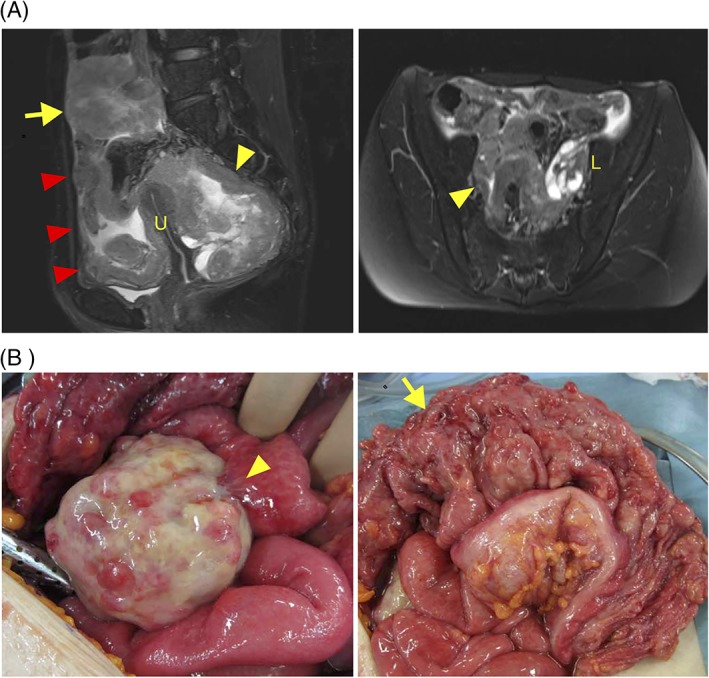
MR imaging and surgical findings. A, T2‐weighted sagittal and axial MRIs are presented. An 8 cm tumor was detected in the right‐side pelvic cavity (yellow arrowhead) and a 6 cm tumor was detected in the lower abdominal cavity (yellow allow). Diffuse peritoneal thickening and ascites were also detected (red arrow). No obvious abnormal findings were noted in the uterus (U) and the left adnexa (L). B, An 8 cm right ovarian solid tumor adhered to small intestine (yellow arrowhead) and Omental cake (yellow arrow) [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 2.
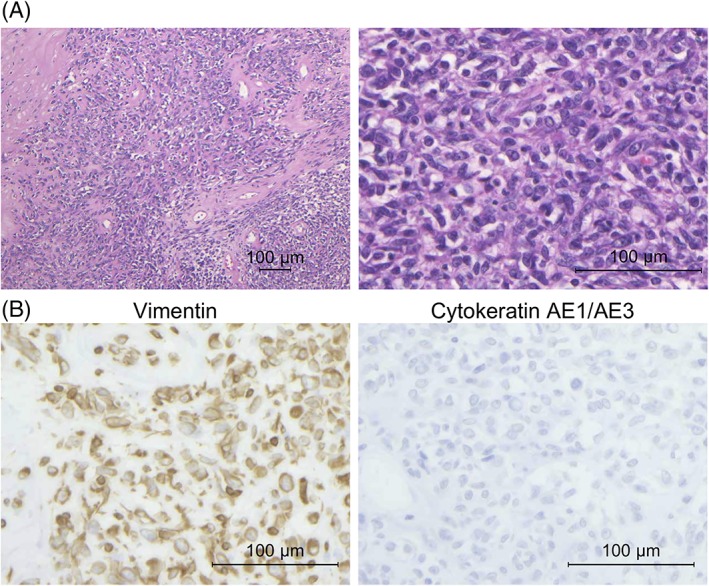
Histological and immunohistochemical findings. A, Representative images of H&E staining. Tumor consisted of small round cells with abundant eosinophilic cytoplasm or spindle cells. Cell‐to‐cell junctions were unclear. (100× and 400× magnification; scale bar, 100 μm). B, Immunohistochemical analysis revealed positive staining for vimentin and negative staining for cytokeratin AE1/AE3 (400× magnification; scale bar, 100 μm) [Color figure can be viewed at http://wileyonlinelibrary.com]
2.2. Clinical samples
The institutional review board approved this study (No. 682 and 837 in Niigata University and No. 29‐16 in National Institute of Genetics). The patient provided written informed consent for the collection of samples and subsequent analysis. Frozen tumor and matched normal blood samples were obtained at the Niigata University Medical and Dental Hospital.
2.3. Whole‐exome sequencing and analysis
Two hundred nanograms of DNA from a patient with ovarian sarcoma and her normal blood sample were used for preparing sequencing libraries using the SureSelect XT Reagent Kit (Agilent Technologies, Santa Clara, CA). Target‐gene enrichment was conducted with SureSelect Human All Exon V5 + lncRNA Kit (Agilent Technologies). The libraries were sequenced with the 2 × 100 bp paired‐end module on the Illumina HiSeq 2500 platform (Illumina).
The Illumina adapter and low‐quality sequences were trimmed using Trimmomatic.15 The paired‐end reads were aligned to the human reference genome (hg19) using Burrows‐Wheeler Aligner (BWA).16 The aligned reads were processed for removal of PCR duplicates using Picard tools (http://broadinstitute.github.io/picard). Local realignments and base‐quality recalibrations were conducted using Genome Analysis Toolkit (GATK).17, 18 Somatic single‐nucleotide variants (SNVs) and short insertions and deletions (indels) were called using Strelka.19 Functional annotation of the identified somatic variants was implemented by ANNOVAR.20
2.4. Detection of putative somatic copy number alterations
We performed copy number analyses using whole‐exome sequencing data with Control‐FREEC.21, 22 The aligned read data from the ovarian sarcoma sample and the matched normal blood sample were used. The sites of the germline variants identified in the matched normal sample were examined for beta allele frequency (BAF) profiles. A threshold of ≥6 copies for gene amplification was used.23 Cancer‐associated genes that were annotated in OncoKB24 were focused in this study.
2.5. RNA sequencing
Total RNA was extracted from frozen a sample using TRIzol (Invitrogen, Carlsbad, CA). The quantity and quality of the extracted RNA were evaluated using the RNA 6000 Nano Assay Kit on the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA integrity value of our sample was 9.8. One microgram of extracted total RNA was used for library preparation, which was conducted using the TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA) according to the manufacturer's protocol. The modal size of the library was ~300 bp. The adapter‐ligated cDNA was amplified with 12 cycles of PCR. The samples were sequenced on the Illumina HiSeq 2500 platform with the 2 × 100 bp paired‐end read module.
We applied the Pipeline for RNA sequencing Data Analysis (PRADA)25 to data from the above RNA sequencing experiments and obtained a list of fusion transcripts and gene expression data for each sample.
2.6. Reverse transcriptase polymerase chain reaction (RT‐PCR) and Sanger sequencing
RT‐PCR and Sanger sequencing were performed as previously reported.13, 14 In brief, total RNA (1 μg) was reverse‐transcribed into cDNA (corresponding to 10 ng of total RNA) using Prime Script II Reverse Transcriptase (Takara Bio, Shiga, Japan). cDNA was subjected to PCR amplification using KAPA Taq DNA polymerase (KAPA Biosystems, Woburn, MA). The reactions were performed in a thermal cycler under the following conditions: 40 cycles at 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute with a final extension at 72°C for 1 minute. PCR products were extracted and purified using NucleoSpin Gel and PCR Clean‐up (Takara Bio) and sequenced on an ABI 3130xl DNA Sequencer (Applied Biosystems, Foster City, CA) using a BigDye Terminator kit (Applied Biosystems). The PCR primers used in this study are presented in Supporting Information, Table 1.
2.7. Quantitative real‐time RT‐PCR
Quantitative real‐time RT‐PCR was performed using the Thermal Cycler Dice Real‐Time System III (Takara Bio). cDNA (corresponding to 10 ng of total RNA) was subjected to real‐time PCR analysis with SYBR Premix Ex Taq II (Takara Bio). The relative quantification method was used to measure the amounts of the respective genes normalized to ACTB. The primers used in this study are presented in Supporting Information, Table 1.
Figure 3.
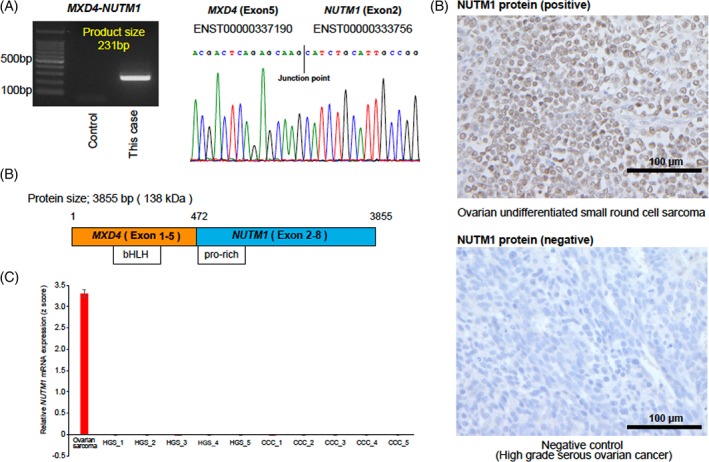
Identification of the MXD4‐NUTM1 fusion gene. A, Identification of the MXD4–NUTM1 fusion transcripts through RT‐PCR and Sanger sequencing. An electropherogram demonstrated the junction point of two genes. B, The schematics of MXD4–NUTM1 fusion transcript. The bHLH domain in MXD4 and pro‐rich domain in NUTM1 were retained in the MXD4–NUTM1 fusion transcript. C, Relative NUTM1 mRNA expression in the undifferentiated ovarian small round cell sarcoma reported in this study compared with epithelial ovarian cancers. Ovarian sarcoma denotes the present case. HGS and CCC denote high‐grade serous carcinoma and clear cell carcinoma, respectively. D, Immunohistochemical staining revealed high expression of NUTM1 with speckled nuclear pattern in the ovarian undifferentiated small round cell sarcoma reported in this study (400× magnification; scale bar, 100 μm) [Color figure can be viewed at http://wileyonlinelibrary.com]
2.8. Immunohistochemical staining
Histological specimens were reviewed by a gynecologic pathologist (TM). In addition, immunohistochemical staining for the NUTM1 protein was performed as previously reported.14, 26 Briefly, after deparaffinization, antigen retrieval was performed with Target Retrieval Solution (10 mM citrate buffer, pH 6.0; Dako) in a microwave for 30 minutes at 96°C. Subsequently, the sections were incubated overnight with primary antibody (C52B1, Cell signaling technology; dilution ratio 1:50) at 4°C. Then, biotinylated anti‐mouse secondary antibodies (Vector Laboratories, Burlingame, CA) were added followed by incubation with ABC reagent (Dako) and 3,3′‐diaminobenzidine (Sigma, St. Louis, MO). Slides were counterstained with hematoxylin. When most of cells were stained, the sample was considered positive. Otherwise, the result was considered negative.
2.9. Western blotting
Western blotting experiments for NUTM1 (C52B1, Cell signaling technology; dilution ratio 1:1000) and anti‐glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) antibody, clone 6C5 (MAB374, Merck Millipore Headquarters; dilution ratio 1:1000) were performed as previously reported.13, 14, 26 Tissues were lysed in RIPA buffer (50 mM Tris at pH 8.0, 150 mM NaCl, 1% Nonidet P‐40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], and 1 mM EDTA) supplemented with protease inhibitors (Roche). The lysates were separated by SDS–polyacrylamide gel electrophoresis on 5%‐20% gradient gels and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). Blots were then incubated with either anti‐mouse or anti‐rabbit horseradish peroxidase‐conjugated secondary antibodies (NA931 and NA934, GE Healthcare; dilution ratio 1:10 000), and visualized by chemiluminescence. Images have been cropped for presentation, and full‐size images are presented in Supporting Information, Figure 3.
3. RESULTS
We performed whole exome sequencing of tumor and matched normal blood samples. We identified 8 nonsynonymous somatic mutations, and mutations were either missense or nonsense changes (Table 1). A stopgain mutation in PRUNE2 (also known as BMCC1) that had an important role in regulating differentiation, survival, and aggressiveness of the tumor cells27 was detected. In addition, genome‐wide copy number analysis revealed that several cancer‐associated genes such as ARID1B, LATS1, PRDM1, CDKN2A, and CDKN2B were harbored heterozygous deletions (Supporting Information, Table 2). However, neither high‐level amplification genes (≥6 copies) nor homozygous deletion genes were detected.
Table 1.
A list of 8 nonsynonymous mutations identified by whole exome sequencing
| Gene | Chr | Pos | Effect | Function in uniprot | Turnor_alt_freq | Accession number |
|---|---|---|---|---|---|---|
| COL9A1 | chr6 | 70935689 | exon31: c.C1798T:p.R600C | Structural component of hyaline cartilage and vitreous of the eye | 0.42 | NM_078485 |
| PRUNE2 | chr9 | 7932190 | exon8:c.G5400A:p.W1800X | Regulating differentiation, survival and aggressiveness of the tumor cells | 0.38 | NM_001308048 |
| C3orf56 | chr3 | 126915725 | exon2:c.G197A:p.G66E | NA | 0.23 | NM_001007534 |
| AGPAT5 | chr8 | 6588234 | exon3:c.G292T:p.D98Y | Converts lysophosphatidic acid (LPA) into phosphatidic acid | 0.21 | NM_018361 |
| ARHGEF17 | chr11 | 73073554 | exon14:c.G4771A:p.A1591T | Acts as guanine nucleotide exchange factor (GEF) for RhoA GTPases | 0.21 | NM_014786 |
| TBC1D10B | chr16 | 30380744 | Exon1: c.C761T:p.P254L | Acts as GTPase‐activating protein for RAB3A, RAB22A, RAB27A, and RAB35 | 0.17 | NM_015527 |
| PER3 | chr1 | 7890053 | Exon19:c.G3046A:p.A1016T | Component of the circadian clock, internal time‐keeping system | 0.04 | NM_001289862 |
| PER3 | chr1 | 7890026 | Exon19:c.A3019G:p.K1007E | Component of the circadian clock, internal time‐keeping system | 0.04 | NM_001289862 |
Next, we performed RNA sequencing of the tumor sample and identified two high‐confidence in‐frame fusion transcripts using the PRADA25 (Table 2). Using RT‐PCR and Sanger sequencing (Figure 2A and Supporting Information, Figure 2), we validated two in‐frame fusion transcripts: MXD4–NUTM1 (exon5–exon2) and ARL6–POT1 (exon2–exon8). Given that the NUTM1 rearrangement was considered as an oncogenic driver event and an attractive therapeutic target in “NUT midline carcinoma (NMC),”28, 29 we focused on the MXD4–NUTM1 fusion transcript to perform subsequent analyses. The transcript allele fraction (TAF) score13 for the NUTM1 was 0.57, suggesting that both MXD4–NUTM1 and wild‐type NUTM1 were expressed in this tumor (Table 1). Next, we compared NUTM1 mRNA expression between this tumor and 10 epithelial ovarian cancer samples using quantitative real‐time PCR. We used the primers targeting common sequences of both MXD4–NUTM1 fusion and wild‐type NUTM1. NUTM1 mRNA expression levels in ovarian undifferentiated small round cell sarcoma were considerably increased compared with epithelial ovarian cancer samples (Figure 2C). Moreover, immunohistochemistry analysis and Western blotting revealed high NUTM1 protein expression in this tumor (Figure 2D and Supporting Information, Figure 3).
Table 2.
A list of 3 novel fusion transcripts identified by RNA sequencing
| 5′ gene | 3′ gene | Locus 5′ gene | Locus 3′ gene | Discordant read (n) | Junction spanning read (n) | maxTAF 5′ gene | maxTAF 3′ gene | Frame |
|---|---|---|---|---|---|---|---|---|
| MXD4 | NUTM1 | 4p16.3 | 15q.14 | 7 | 22 | 0.0594 | 0.5714 | In‐frame |
| ARL6 | POT1 | 3q11.2 | 7q.31.33 | 5 | 6 | 0.25 | 0.13 | In‐frame |
| POT1 | AC110491.1 | 7q.31.33 | 4p16.3 | 2 | 8 | 0.0278 | 0.0081 | NA |
4. DISCUSSION
In this study, we clarified the molecular characteristics of an ovarian undifferentiated small round cell sarcoma using whole exome and RNA sequencing. Although we could not exclude the possibility of metastatic sarcoma in the right ovary completely, we diagnosed an ovarian sarcoma on the basis of clinicopathological findings. We identified a nonsense mutation in PRUNE2 that was reported to be a potentially oncogenic30 but not therapeutically targetable mutated genes at that time. No somatic mutation of cancer‐associated genes frequently mutated in epithelial ovarian cancer—such as TP53, 31 KRAS, 32 PIK3CA,33 and ARID1A 34—was detected.
On the other hand, we identified a novel MXD4–NUTM1 fusion transcript. NUTM1 (NUT midline carcinoma family member 1, also known as C15orf55 or NUT), which maps to chromosome15q14, is typically expressed in a normal testis but not expressed in many other normal and malignant tissues.35, 36 NUTM1 rearrangements causes “NUT midline carcinoma,” which is an aggressive lethal malignancy that occurs in midline anatomical structures, especially the mediastinum, head, and neck.37, 38 Approximately two‐thirds of NMC cases harbor a BRD4–NUTM1 fusion gene, which has been functionally validated as an oncogenic event.39, 40 In the other one‐third of cases, several fusion partners of NUTM1 have been reported in the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman) and recent papers, such as ACIN1, 41, 42, 43 BCOR1, 44 BRD3, 40 BRD9, 41 BPTF, 45 CIC, 46 CUX1, 42 IKZF1, 42, 47 MXD1, 44 NSD3, 48 SLC12A6, 42 ZNF532, 49 and ZNF618. 42 Although MXD1–NUTM1 fusion has been reported in gastric sarcoma, MXD4 has not been previously reported as a fusion partner of NUTM1. Both MXD1 and MXD4 are members of the MAD gene family and regulate MYC, leading to cell growth in differentiating tissues.50 Grayson et al. have identified that MYC is a key transcriptional target of BRD4–NUTM1 fusion and that dysregulation of MYC by BRD4–NUTM1 fusion has a central role in the pathogenesis of in NUT midline carcinoma.51 Our data revealed an increase in both NUTM1 mRNA and protein expression in tumor tissue. Further functional analysis focusing on MYC is needed to prove the significance of NUTM1 fusions whose partners were the MAD gene family such as MXD1 or MXD4 in tumorigenesis, especially sarcomagenesis.
In general, NMC defined by the NUTM1‐fusion gene is histologically epithelial malignancy, especially squamous cell carcinoma that reacts with the antibodies to cytokeratin AE1/AE3 and p63 focally but not myogenin, SMA, chromogranin, synaptophysin, c‐kit, desmin, S‐100, and CD99.28, 52 Most recently, six cases of soft tissue and visceral neoplasms harboring NUTM1 fusion genes have been reported. These tumors occurring outside the anatomic midline were diagnosed with undifferentiated tumors based on histological and immunohistochemical findings, and these tumors are suggested to be “NUT‐associated tumors.”44 Our case harboring the NUTM1 fusion gene would also be classified as a “NUT‐associated tumor,” and undifferentiated mesenchymal, neuroendocrine, and epithelial tumors should be reassessed by immunohistochemical and fusion gene analyses because “NUT‐associated tumors” may be underdiagnosed.44
We also identified an ARL6–POT1 fusion transcript. ARL6 (ADP‐ribosylation factor‐like protein 6),53 which maps to chromosome 3q11, plays essential roles in modulating membrane trafficking and cytoskeletal function. POT1 (protection of telomeres), which maps to chromosome 7q31, has an important role maintaining chromosome stability, and POT1 mutations lead to tumorigenesis in various cancers.54, 55 Although each function of a fused gene may be important in tumorigenesis, we did not focus on the ARL6–POT1 fusion in this study, given that no previous report suggesting oncogenic functions of APL6 fusions or POT1 fusions was available.
Comprehensive genome and transcriptome analysis of one case with ovarian undifferentiated small round cell sarcoma demonstrated unique molecular characteristics, such as a PRUNE2 mutation or MXD4–NUTM1 fusion. More extensive genomic and transcriptomic analyses for a large data set of ovarian undifferentiated small round cell sarcomas would lead to elucidation of the pathogenesis of this rare disease.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Figure S1
Figure S2
Figure S3
Table S1
Table S2
ACKNOWLEDGMENTS
The authors are grateful to Anna Ishida, Junko Kajiwara, Junko Kitayama, and Yumiko Sato for their technical assistance.
Tamura R, Nakaoka H, Yoshihara K, et al. Novel MXD4–NUTM1 fusion transcript identified in primary ovarian undifferentiated small round cell sarcoma. Genes Chromosomes Cancer. 2018;57:557–563. 10.1002/gcc.22668
Funding information JSPS KAKENHI, Grant/Award Numbers: JP16H06279, JP16H06267
REFERENCES
- 1. Bacalbasa N, Balescu I, Dima S, Popescu I. Ovarian sarcoma carries a poorer prognosis than ovarian epithelial cancer throughout all FIGO stages: a single‐center case‐control matched study. Anticancer Res. 2014;34(12):7303‐7308. [PubMed] [Google Scholar]
- 2. Zygouris D, Androutsopoulos G, Grigoriadis C, Arnogiannaki N, Terzakis E. Primary ovarian leiomyosarcoma. Eur J Gynaecol Oncol. 2012;33(3):331‐333. [PubMed] [Google Scholar]
- 3. Nielsen GP, Oliva E, Young RH, Rosenberg AE, Prat J, Scully RE. Primary ovarian rhabdomyosarcoma: a report of 13 cases. Int J Gynecol Pathol. 1998;17(2):113‐119. [DOI] [PubMed] [Google Scholar]
- 4. Qureshi A, Hassan U, Rehman R. Primary ovarian rhabdomosarcoma. BMJ Case Rep. 2011;10:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xie W, Bi X, Cao D, Yang J, Shen K, You Y. Primary endometrioid stromal sarcomas of the ovary: a clinicopathological study of 14 cases with a review of the literature. Oncotarget. 2017;8(38):63345‐63352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ozdemir O, Sari ME, Sen E, Ilgin BU, Guresci S, Atalay CR. Primary ovarian fibrosarcoma: a case report and review of the literature. J Exp Ther Oncol. 2016;11(3):225‐235. [PubMed] [Google Scholar]
- 7. Antonescu C. Round cell sarcomas beyond Ewing: emerging entities. Histopathology. 2014;64(1):26‐37. [DOI] [PubMed] [Google Scholar]
- 8. Watson S, Perrin V, Guillemot D, et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol. 2018;245(1):29‐40. [DOI] [PubMed] [Google Scholar]
- 9. Jain S, Xu R, Prieto VG, Lee P. Molecular classification of soft tissue sarcomas and its clinical applications. Int J Clin Exp Pathol. 2010;3(4):416‐428. [PMC free article] [PubMed] [Google Scholar]
- 10. Delattre O, Zucman J, Melot T, et al. The Ewing family of tumors‐‐a subgroup of small‐round‐cell tumors defined by specific chimeric transcripts. N Engl J Med. 1994;331(5):294‐299. [DOI] [PubMed] [Google Scholar]
- 11. Barr FG. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene. 2001;20(40):5736‐5746. [DOI] [PubMed] [Google Scholar]
- 12. Yoshihara K, Wang Q, Torres‐Garcia W, et al. The landscape and therapeutic relevance of cancer‐associated transcript fusions. Oncogene. 2015;34(37):4845‐4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tamura R, Yoshihara K, Yamawaki K, et al. Novel kinase fusion transcripts found in endometrial cancer. Sci Rep. 2015;5:18657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamura R, Yoshihara K, Saito T, et al. Novel therapeutic strategy for cervical cancer harboring FGFR3‐TACC3 fusions. Oncogene. 2018;7(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114‐2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H, Durbin R. Fast and accurate short read alignment with burrows‐wheeler transform. Bioinformatics. 2009;25(14):1754‐1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Res. 2010;20(9):1297‐1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next‐generation DNA sequencing data. Nat Genet. 2011;43(5):491‐498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small‐variant calling from sequenced tumor‐normal sample pairs. Bioinformatics. 2012;28(14):1811‐1817. [DOI] [PubMed] [Google Scholar]
- 20. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high‐throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boeva V, Zinovyev A, Bleakley K, et al. Control‐free calling of copy number alterations in deep‐sequencing data using GC‐content normalization. Bioinformatics. 2011;27(2):268‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boeva V, Popova T, Bleakley K, et al. Control‐FREEC: a tool for assessing copy number and allelic content using next‐generation sequencing data. Bioinformatics. 2012;28(3):423‐425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023‐1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Torres‐Garcia W, Zheng S, Sivachenko A, et al. PRADA: pipeline for RNA sequencing data analysis. Bioinformatics. 2014;30(15):2224‐2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamawaki K, Ishiguro T, Mori Y, et al. Sox2‐dependent inhibition of p21 is associated with poor prognosis of endometrial cancer. Cancer Sci. 2017;108(4):632‐640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Machida T, Fujita T, Ooo ML, et al. Increased expression of proapoptotic BMCC1, a novel gene with the BNIP2 and Cdc42GAP homology (BCH) domain, is associated with favorable prognosis in human neuroblastomas. Oncogene. 2006;25(13):1931‐1942. [DOI] [PubMed] [Google Scholar]
- 28. Stelow EB. A review of NUT midline carcinoma. Head Neck Pathol. 2011;5(1):31‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stathis A, Zucca E, Bekradda M, et al. Clinical response of carcinomas harboring the BRD4‐NUT oncoprotein to the targeted bromodomain inhibitor OTX015/MK‐8628. Cancer Discov. 2016;6(5):492‐500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu W, McPherson JR, Stevenson M, et al. Whole‐exome sequencing studies of parathyroid carcinomas reveal novel PRUNE2 mutations, distinctive mutational spectra related to APOBEC‐catalyzed DNA mutagenesis and mutational enrichment in kinases associated with cell migration and invasion. J Clin Endocrinol Metab. 2015;100(2):E360‐E364. [DOI] [PubMed] [Google Scholar]
- 31. Cancer Genome Atlas Research . Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609‐615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perren TJ. Mucinous epithelial ovarian carcinoma. Ann Oncol. 2016;27(Suppl 1):i53‐i57. [DOI] [PubMed] [Google Scholar]
- 33. Jones S, Wang TL, Shih Ie M, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330(6001):228‐231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wiegand KC, Shah SP, Al‐Agha OM, et al. ARID1A mutations in endometriosis‐associated ovarian carcinomas. N Engl J Med. 2010;363(16):1532‐1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):eaan2507. [DOI] [PubMed] [Google Scholar]
- 36. Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue‐based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 37. French CA, Kutok JL, Faquin WC, et al. Midline carcinoma of children and young adults with NUT rearrangement. J Clin Oncol. 2004;22(20):4135‐4139. [DOI] [PubMed] [Google Scholar]
- 38. Bauer DE, Mitchell CM, Strait KM, et al. Clinicopathologic features and long‐term outcomes of NUT midline carcinoma. Clin Cancer Res. 2012;18(20):5773‐5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. French CA, Miyoshi I, Kubonishi I, Grier HE, Perez‐Atayde AR, Fletcher JA. BRD4‐NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003;63(2):304‐307. [PubMed] [Google Scholar]
- 40. French CA, Ramirez CL, Kolmakova J, et al. BRD‐NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27(15):2237‐2242. [DOI] [PubMed] [Google Scholar]
- 41. Andersson AK, Ma J, Wang J, et al. The landscape of somatic mutations in infant MLL‐rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47(4):330‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gu Z, Churchman M, Roberts K, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun. 2016;7:13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu YF, Wang BY, Zhang WN, et al. Genomic profiling of adult and pediatric B‐cell acute lymphoblastic leukemia. EBioMedicine. 2016;8:173‐183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dickson BC, Sung YS, Rosenblum MK, et al. NUTM1 gene fusions characterize a subset of undifferentiated soft tissue and visceral tumors. Am J Surg Pathol. 2018;42(5):636‐645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T‐lineage acute lymphoblastic leukemia. Nat Genet. 2017;49(8):1211‐1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schaefer IM, Dal Cin P, Fletcher CDM, Hanna GJ, French CA. CIC‐NUTM1 fusion: a case which expands the Spectrum of NUT‐rearranged Epithelioid malignancies. Genes Chromosomes Cancer. 2018;57(9):446‐451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lilljebjorn H, Henningsson R, Hyrenius‐Wittsten A, et al. Identification of ETV6‐RUNX1‐like and DUX4‐rearranged subtypes in paediatric B‐cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. French CA, Rahman S, Walsh EM, et al. NSD3‐NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism. Cancer Discov. 2014;4(8):928‐941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Alekseyenko AA, Walsh EM, Zee BM, et al. Ectopic protein interactions within BRD4‐chromatin complexes drive oncogenic megadomain formation in NUT midline carcinoma. Proc Natl Acad Sci U S A. 2017;114(21):E4184‐E4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hurlin PJ, Queva C, Koskinen PJ, et al. Mad3 and Mad4: novel max‐interacting transcriptional repressors that suppress c‐myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14(22):5646‐5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grayson AR, Walsh EM, Cameron MJ, et al. MYC, a downstream target of BRD‐NUT, is necessary and sufficient for the blockade of differentiation in NUT midline carcinoma. Oncogene. 2014;33(13):1736‐1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Suzuki S, Kurabe N, Ohnishi I, et al. NSD3‐NUT‐expressing midline carcinoma of the lung: first characterization of primary cancer tissue. Pathol Res Pract. 2015;211(5):404‐408. [DOI] [PubMed] [Google Scholar]
- 53. Wiens CJ, Tong Y, Esmail MA, et al. Bardet‐Biedl syndrome‐associated small GTPase ARL6 (BBS3) functions at or near the ciliary gate and modulates Wnt signaling. J Biol Chem. 2010;285(21):16218‐16230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen C, Gu P, Wu J, et al. Structural insights into POT1‐TPP1 interaction and POT1 C‐terminal mutations in human cancer. Nat Commun. 2017;8:14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gu P, Wang Y, Bisht KK, et al. Pot1 OB‐fold mutations unleash telomere instability to initiate tumorigenesis. Oncogene. 2017;36(14):1939‐1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Table S1
Table S2


