Abstract
Cadmium (Cd) is a non‐essential, toxic heavy metal that poses serious threats to both ecosystems and human health. Plants employ various cellular and molecular mechanisms to minimise the impact of Cd toxicity and cell walls function as a defensive barrier during Cd exposure.
In this study, we adopted a quantitative gel‐based proteomic approach (two‐dimensional difference gel electrophoresis) to investigate changes in the abundance of cell wall and soluble proteins in stems of Medicago sativa L. upon long‐term exposure to Cd (10 mg·Cd·kg−1 soil as CdSO 4). Obtained protein data were complemented with targeted gene expression analyses.
Plants were affected by Cd exposure at an early growth stage but seemed to recover at a more mature stage as no difference in biomass was observed. The accumulation of Cd was highest in roots followed by stems and leaves. Quantitative proteomics revealed a changed abundance for 179 cell wall proteins and 30 proteins in the soluble fraction upon long‐term Cd exposure. These proteins are involved in cell wall remodelling, defence response, carbohydrate metabolism and promotion of the lignification process.
The data indicate that Cd exposure alters the cell wall proteome and underline the role of cell wall proteins in defence against Cd stress. The identified proteins are linked to alterations in cell wall structure and lignification process in stems of M. sativa, underpinning the function of the cell wall as an effective barrier against Cd stress.
Keywords: 2‐D DIGE, cadmium, cell wall proteins, gene expression, Medicago sativa, soluble proteins
Introduction
Anthropogenic influence has led to environmental pollution with heavy metals, of which cadmium (Cd) is one of the most common pollutants, released into the environment by mining or smelting activities and through the application of phosphate fertiliser. Its accumulation in crop plants introduces Cd into the human food chain, posing a threat to human health. Although not essential, Cd can enter a plant via the roots and is translocated throughout different tissues by a variety of unspecific transport systems (Clemens & Ma 2016), thereby competing with essential nutrients (Zhang et al. 2014). Cadmium exposure impacts numerous physiological and biochemical processes in plants and leads to limited growth, chlorosis and oxidative stress induced by the production of reactive oxygen species (ROS; Cuypers et al. 2010). Furthermore, plants suffer from water imbalance as an effect of stomatal closure (Perfus‐Barbeoch et al. 2002) and the photosynthetic apparatus is also affected (Sanità Di Toppi & Gabbrielli 1999). Plants have evolved several mechanisms to minimise the induced toxic effects, including detoxification through chelation (Cobbett 2000), compartmentation and sequestration in extracytoplasmatic compartments such as the cell wall (Krzesłowska 2011).
The plant cell wall is a complex structure composed of polysaccharides and proteins, providing mechanical support and rigidity. At the interface between the inside and outside of the cell, it forms a protective barrier, which is important for defence against biotic and abiotic threats to plant cells (Bradley et al. 1992; Brisson et al. 1994; Douchiche et al. 2010a). Cell wall‐localised proteins function in intercellular communication and in the interaction between the cell and its environment. Therefore, the plant cell wall continously undergoes structural modifications in order to adapt to the plant's development stages and environmental conditions (Caffall & Mohnen 2009; Loix et al. 2017). Plants exposed to Cd show concentration‐dependent alterations in cell wall structure. Cell walls become thicker at sites of Cd deposition, with pectin as the main binding site for Cd (Vollenweider et al. 2006). Cadmium accumulation leads to an increased amount of low‐methylesterified pectin in the outer part of the external tangential cell wall (Douchiche et al. 2007), which is mediated by the activity of pectin methylesterase (PME) and increased activity of this enzyme was reported during Cd exposure (Paynel et al. 2009). Low‐methylesterified pectin can bind Cd in competition with calcium (Ca), and Cd‐induced alterations seem to promote the barrier function of the cell wall (Douchiche et al. 2010a; Parrotta et al. 2015). Furthermore, Cd exposure enhances lignification mediated by cell wall‐bound peroxidases (POX), leading to cell wall stiffening and limiting cell growth (Chaoui & El Ferjani 2005).
Multiple molecular techniques have been applied to study abiotic stress responses in plants and the molecular mechanism behind it. Thereby, quantitative protein analyses are an essential tool to identify important players in plant stress responses such as Cd exposure (Villiers et al. 2011; Lopes Júnior et al. 2015) and have been applied in different species and organs (Kieffer et al. 2009; Semane et al. 2010; Hossain et al. 2012). Most studies focus on short‐term Cd exposure in model plant species and not in economically relevant crops (Lee et al. 2010; Dupae et al. 2014). The current study focuses on the non‐model plant Medicago sativa L., which is globally the most important forage legume. High in protein content, M. sativa meets the needs of the feed market. The less digestible stems amount to more than 50% of its biomass, with a high yield in cell wall material. It has a high economic value as the stems can be used for industrial applications such as bioethanol production. Since the composition and structure of cell walls are influenced by altered environmental conditions, this may have an impact on their potential value. Therefore, such alterations to the cell wall are of scientific but also societal and economic interest. Hence M. sativa is often used to study cell wall development and processes (Verdonk et al. 2012; Printz et al. 2016). Recently, an improved protocol to investigate the cell wall proteome was established (Printz et al. 2015). By expanding knowledge about cell wall proteins and their role in plant defence against abiotic stress like Cd exposure will further our understanding of the plant cell wall.
In this study, M. sativa plants were grown on control and Cd‐contaminated soil (10 mg·kg−1 soil) with the aim of identifying effects of this treatment at the proteome level and discover potential Cd‐induced structural effects. Although current literature is dominated by studies on short‐term exposure, long‐term exposure experiments to a realistic Cd concentration, as done in this study, make the data relevant for agricultural practices. Quantification of the stem cell wall and soluble proteome was performed with two‐dimensional difference gel electrophoresis (2‐D DIGE), which enables separation of different protein isoforms and discrimination of modified proteins such as heterogeneous glycosylated cell wall proteins and other processed protein forms. Additionally, targeted gene expression analyses with quantitative real‐time PCR (RT‐qPCR) were used to complement and strengthen the proteomic data. Changes in protein patterns, their influence on cell wall structure and the role of the cell wall as a protective barrier against Cd exposure are discussed.
Material and Methods
Plant material
Medicago sativa L. (cultivar Giulia) seeds were inoculated with Sinorhizobium meliloti. The potting mix was prepared in a single batch using a 2:1 ratio of potting soil to sand. Half of the potting mix was supplemented with 10 mg·Cd·kg−1 soil (added as CdSO4). Sowing was done in May 2015. For the two conditions, one being a control soil without the addition of Cd, and the other Cd‐supplemented (10 mg·Cd·kg−1 soil), 12 × 12 pots were planted. Plants were kept in the greenhouse until the flowering stage was reached (July) and subsequently cut as is agricultural practice. Neither temperature nor photoperiod was controlled during the study. After cutting, plants were kept outside to avoid the observed insect infestation during the first growth cycle. After a re‐growth period until pre‐flowering stage was reached, plants were put back in the greenhouse for one more week before sampling on the 10th of September. No fertiliser was applied. The mineral composition of the two soils was analysed in ten replicates and revealed no significant difference apart from the concentration in Cd (Appendix S1). Stems were separated from leaves and the first two and last two internodes removed to obtain a more homogeneous sample. Five replicates were sampled, with a pool of stem material from 24 pots corresponding to one biological replicate. Samples were ground to a fine powder in liquid nitrogen using a mortar and pestle and kept at −80 °C until further use for quantitative protein analysis and targeted gene expression by qPCR. Additionally, leaf, stem and root samples of each condition were taken to determine their Cd content.
Determination of Cd content by ICP‐MS
Samples of approximately 250 mg fresh weight were oven‐dried for 36 h at 60 °C and mineralized in 7 ml nitric acid and 3 ml hydrogen peroxide using a Multiwave PRO microwave reaction system according to the manufacturer's instructions (Anton Paar, Graz, Austria). Subsequently, the sample volume was adjusted to 25 ml with MilliQ water (Millipore, Darmstadt, Germany) and Cd concentrations in the samples determined by inductively coupled plasma‐mass spectrometry (ICP‐MS). An average concentration was calculated for the two conditions.
Extraction of cell wall proteins
The cell wall proteins were extracted according to (Printz et al. 2015). Four biological replicates were used. Briefly, cell walls were enriched from 7 g ground M. sativa stems using an increasing sucrose gradient (5 mm sodium (Na) acetate pH 4.6, 4 °C supplemented, respectively, with 0.4, 0.6 and 1.0 m sucrose). The final cell wall pellets were washed twice in 5 mm Na acetate (pH 4.6). To extract cell wall proteins, 7.5 ml extraction buffer C (5 mm Na acetate, 200 mm CaCl2, pH 4.6, 4 °C) were added to the cell wall fractions. Samples were placed on a rocking platform (30 min, 4 °C), followed by centrifugation (10,000 × g, 15 min, 4 °C). This step was repeated, and supernatants pooled (stored as CaCl2 fraction, −20 °C). A total of 10 ml extraction buffer E (5 mm Na acetate, 50 mm EGTA, pH 4.6, 4 °C) were added to the pellets, which were shaken vigorously (37 °C, 1 h) and centrifuged (10,000 × g, 15 min, 4 °C). This extraction step was repeated twice, and the supernatants pooled (stored as EGTA fraction, −20 °C). Finally, the pellets were resuspended in 15 ml extraction buffer L (5 mm Na acetate, 3 m LiCl, pH 4.6, 4 °C), placed on a rocking platform overnight (4 °C), centrifuged and supernatants stored as LiCl fraction at −20 °C.
Protein extracts were concentrated with Amicon Ultra‐15 10 K (Millipore) by centrifugation (4700 × g, 4 °C) until a volume of approximately 200 μl was reached. The concentrated extracts were washed and desalted using the ReadyPrep 2‐D Cleanup kit (Bio‐Rad, Hercules, CA, USA) according to the instruction manual. Samples were solubilised in labelling buffer (7 m urea, 2 m thiourea, 2% w/v CHAPS, 30 mm Tris) and protein concentrations determined with the Bradford protein assay (Bio‐Rad).
Extraction of soluble proteins
Extractions were done with trichloroacetic acid (TCA)/phenol‐ sodium dodecyl sulfate (SDS) (Wang et al. 2003). To 300 mg ground plant material 1 ml ice‐cold acetone containing 10% v/v TCA and 0.07% w/v dithiothreitol (DTT) was added, the suspension was briefly vortexed and left at −20 °C overnight. Centrifugation (10,000 × g, 5 min, 4 °C) was followed by two washing steps with ice‐cold acetone (10,000 × g, 3 min, 4 °C), and the final pellet was dried at ambient temperature overnight. Next, 800 μl Tris‐buffered phenol (Invitrogen, Waltham, MA, USA; pH 7.5–7.8) and an equal volume of SDS buffer (30% w/v sucrose, 2% w/v SDS, 0.1 m Tris‐HCL, pH 8.0, 2% 2‐mercaptoethanol) were added to the pellets. After extensive vortexing (10 min) and centrifugation (10,000 × g, 3 min), 300 μl of the upper phenol phase were transferred to a new tube, 1.5 ml pre‐cooled 0.1 m ammonium acetate in methanol added and samples were kept at −20 °C for 2 h. After centrifugation (10,000 × g, 5 min, 4 °C) and two washing steps with pre‐cooled 0.1 m ammonium acetate in methanol (10,000 × g, 3 min, 4 °C), pellets were washed twice with 80% acetone. The obtained protein pellets were dried at ambient temperature and solubilised in labelling buffer (see previous section). Again, the Bradford protein assay (Bio‐Rad) was used to determine the protein concentration in the sample.
The 2‐D DIGE and spot selection
Per condition and fraction (CaCl2, EGTA, LiCl and soluble proteins), 50 μg protein from four replicates were labelled with Cy3 or Cy5. The dyes were swapped between replicates to eliminate the effect of potential preferential labelling on the results. Given that each of the fractions has a highly characteristic protein profile, an internal standard was prepared separately for each of the four fractions. This was composed of 25 μg protein from each of the eight samples (four biological replicates for control and Cd plants) and labelled with Cy2. Samples (one labelled with Cy3, one with Cy5 and the Cy2‐labelled internal standard) were mixed and 9 μl Servalyte pH 3–10 (Serva Electrophoresis, Heidelberg, Germany) and 2.7 μl Destreak Reagent (GE Healthcare, Chicago, IL, USA) were added. The volume was adjusted with lysis buffer (7 m urea, 2 M thiourea, 4% w/v CHAPS) to 450 μl. The labelled samples were loaded onto an Immobiline™ DryStrip NL of 24 cm (GE Healthcare) during overnight rehydration, followed by isoelectric focusing (IEF) in a five‐step programme: (i) constant 100 V·4 h; (ii) linear gradient up to 1000 V·4 h; (iii) constant 1000 V·5 h; (iv) linear gradient up to 10,000 V·6 h; (v) constant 10,000 V until a total of 80,000 V·h was reached. Equilibration of the IE‐strips was done in equilibration buffer (Serva Electrophoresis) supplemented with 6 m urea and 1% w/v DTT for 15 min and subsequently in the same buffer supplemented with 6 m urea and 2.5% w/v iodoacetamide for another 15 min. Strips were applied to 2D HPE™ Large Gel NF‐12.5% (Serva Electrophoresis), 2D electrophoresis was run on a HPE™ tower system according to the manufacturer's instructions and stopped after the front reached the bottom of the gel. Gels were placed into fixation buffer (15% ethanol v/v, 1% w/v citric acid) overnight prior to image acquisition. From each gel three pictures of the different dyes were acquired at different wavelengths [Cy2 488 nm, Cy3 532 nm, Cy5 642 nm (Typhoon FLA 9500; GE Healthcare). Quantitative image analysis was carried out using the SameSpots software (TotalLab). Since the same internal standard is run on each gel of a specific fraction, alignment and normalisation with the image of the internal standard allows comparison of spot volumes across different gels. The software uses an alignment‐based approach and enables automatic statistical analysis for spot detection. When a treatment effect was reported (anova P ≤ 0.05 and detected fold change ≥1.5), the spot was selected for protein identification (Appendices S2, S3 for all spot volumes).
Protein digestion and MS/MS analysis
Selected spots were picked with an Ettan Spot Picker (GE Healthcare). A Freedom EVO II workstation (Tecan, Männedorf, Switzerland) was used for digestion. Briefly, gel plugs were washed with 50 mm ammonium bicarbonate solution in 50% v/v methanol/MilliQ Water (Millipore) for 20 min and dehydrated for 20 min in 75% acetonitrile (ACN). Proteins were digested with 8 μl of a solution containing 5 ng·μl−1 trypsin (Trypsin Gold; Promega, Fitchburg, WI, USA) in 20 mm ammonium bicarbonate (overnight, 37 °C). Digested peptides were extracted from the gel plugs with 50% v/v ACN containing 0.1% v/v trifluoroacetic acid (TFA), dried and resolubilised in 0.7 μl 50% v/v ACN containing 0.1% v/v TFA. Peptides were spotted on a MALDI‐TOF target and 0.7 μl·7 mg·ml−1 α‐cyano‐4‐hydroxycinnamic acid in 50% v/v ACN containing 0.1% v/v TFA was added.
A MALDI mass spectrum was acquired using the Sciex 5800 TOF/TOF (Sciex, Darmstadt, Germany). The ten most abundant peaks, excluding known contaminants, were automatically selected and fragmented. Both MS and MS/MS were submitted to an in‐house MASCOT server (Matrix Science, http://www.matrixscience.com) for database‐dependent identifications against the NCBInr database limited to the taxonomy Viridiplantae (3,334,509 sequences). A second search was performed using the M. sativa sequences downloaded from the Samuel Roberts Noble website (The Alfalfa Gene Index and Expression Atlas Database, AGED, http://plantgrn.noble.org/AGED/index.jsp) (675,756 sequences, 304,231,576 residues). Parameters were a peptide mass tolerance of 100 ppm, a fragment mass tolerance of 0.5 Da, cysteine carbamidomethylation as fixed modification and methionine oxidation, double oxidation of tryptophan, tryptophan to kynurenine as variable modifications. Proteins were considered as identified when at least two peptides passed the MASCOT‐calculated 0.05 threshold score of 40. When high‐quality spectra were not matched to a protein, manual interpretation of the spectra was performed, and/or the search parameters adjusted (semitryptic, single amino acid changes, post‐translational modifications) to increase the sequence coverage of the identified protein. All identifications were manually validated, and their subcellular locations determined using TargetP (Emanuelsson & Nielsen 2000). The standard search parameters were used. In some cases, predictions were corrected based on literature.
Extraction of RNA and cDNA synthesis
The RNA was extracted from 100 mg finely ground stem tissue using the RNAqueouse™ Kit (Life Technologies, Carlsbad, CA, USA) according the manufacturer's instructions. To increase the obtained RNA concentration, all samples were cleaned up by precipitating the RNA with 3 m sodium acetate and 100% isopropanol. The obtained RNA pellet was washed with 70% ethanol and resuspended in RNase‐free water. The RNA concentration and purity were determined using a NanoDrop® ND‐1000 spectrophotometer (Thermo Fischer Scientific, Waltham, MA, USA) (A260/280 and A260/230 ratios between 1.9 and 2.5). Equal amounts (1 μg) of the extracted RNA was DNase‐treated (TURBO DNA‐free™ Kit; Life Technologies) and reverse transcribed following the manufacturer's instructions of the PrimeScript™ RT Reagent Kit (Perfect Real Time; Takara Bio, Shiga, Japan). The cDNA was diluted ten‐fold in 1/10 Tris‐EDTA (TE) buffer (Sigma‐Aldrich, St. Louis, MI, USA) and stored at −20 °C.
Quantitative real‐time PCR
The AGED database was used to obtain the coding sequences for the genes of interest. Specific primer pairs were designed with the Primer3Plus online tool (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and analysed using the OligoAnalyzer 3.1 (https://eu.idtdna.com/calc/analyzer). Primer efficiency was measured using a dilution series of a pooled sample containing an equal amount of all cDNAs in the experiment (six dilution points) (see Appendix S4 Table 1 for primer sequences and efficiencies). Quantitative PCR reactions were performed in a 96 well plate with the 7500 Fast Real Time PCR System (Life Technologies). One amplification reaction contained 2 μl diluted cDNA (or RNase‐free water as “no template” control), forward and reverse primers (100 nm), Fast SYBR® Green Master Mix (Applied Biosystems) and RNase‐free water to a final volume of 10 μl. The PCR conditions were as follows: initial denaturation (20 s at 95 °C), followed by 40 cycles of denaturation (3 s at 95 °C), annealing and elongation (30 s at 60 °C). At the end of the run a melting curve was generated to check the specificity of the amplification reaction. All details of the workflow according to the minimum information for publication of quantitative real time PCR experiments (MIQE) guidelines as described in Bustin et al. (2009) are shown in Appendix S4 Table 2.
Expression values are given as normalised relative expression values. Therefore, the relative expression of each gene was calculated as 2−∆Cq and normalised by the average 2−∆Cq ‐value of the three reference genes cyclophilin, GAPD, PAB4 (Appendix S4 Table 1), which were selected with the GrayNorm algorithm (Remans et al. 2014) out of ten tested reference genes (previously reported for M. sativa; Guerriero et al. 2014). Data from the treated samples were expressed relatively to the data from the untreated samples (set at 1). To determine if the gene expression was significantly affected by Cd, a t‐test was performed (P ≤ 0.05) using Microsoft Excel. Hierarchical clustering of the normalised expression values from all five replicates was done with Clustal 3.0 using an uncentered Pearson correlation and complete linkage clustering (Eisen et al. 1999). The cluster was visualised as a heat map with Java TreeView (Saldanha 2004).
Results
Plant growth
The biomass of the aerial plant parts for each replicate was determined at the end of the experiment. Long‐term Cd exposure did not significantly impact plant growth (110.65 ± 3.78 g for control versus 120.32 ± 4.67 g for Cd‐exposed). Nevertheless, a growth reduction due to Cd exposure was clearly visible at an early growth stage (Fig. 1) but vanished at a more mature plant stage.
Figure 1.
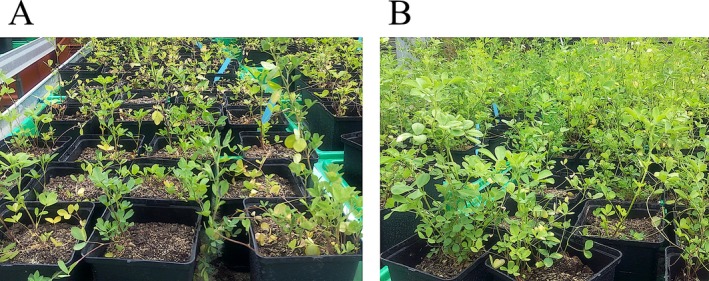
Pictures of M. sativa plants during their first growth period (summer 2015). A: Plants growing on contaminated soil (10 mg·Cd·kg−1 soil). B: Plants growing on uncontaminated soil
Distribution of Cd in M. sativa leaves, stems and roots
The quantity of Cd in M. sativa leaves, stems and roots were assessed via ICP‐MS (Table 1). Cadmium strongly accumulated in the roots and its quantity decreased in aerial plant parts. In stems only 24% of the root Cd quantity was measured, which decreased another 50% in leaves. Changes in the amount of Cd in each tissue between the control and Cd‐exposed plants are significant (P ≤ 0.05).
Table 1.
Cadmium concentration (μg·g−1 DW) in different plant organs of M. sativa given as the mean ± SE of ten biological replicates
| leaves | stems | roots | ||||
|---|---|---|---|---|---|---|
| AVG | SE | AVG | SE | AVG | SE | |
| Ctr | 0.31 | 0.06 | 0.24 | 0.01 | 2.36 | 0.74 |
| Cd | 21.54 | 5.16 | 40.09 | 8.22 | 169.91 | 72.13 |
AVG, average. Significance of difference in Cd quantity: leaves/stems P = 1.03−5, stems/roots P = 2.03−5, roots/leaves P = 4.21−6.
Proteomic analysis
Four different protein fractions were analysed: three different cell wall fractions (CaCl2, EGTA and LiCl) and a fraction containing soluble proteins.
Identification of cell wall proteins
Comparative DIGE analyses revealed a number of spots with a significant change in their abundance: changes in the fluorescence intensity of 107 spots from the CaCl2 fraction, 155 from the EGTA fraction and 87 spots from the LiCl fraction were detected (Appendix S3). After deletion of those spots wherein more than one protein was identified, 228 spots from the different cell wall fractions were considered for biological interpretation (Appendix S5 and S6 Tables 1, 2 and 3). From that dataset, 179 proteins (78.5%) were predicted by TargetP to be secreted proteins targeted to the cell wall. Out of the remaining 49 proteins, 34 are targeted to the chloroplast, three to mitochondria and 12 have no predicted subcellular target site and can be considered as intracellular proteins.
The highest number of proteins was identified in the EGTA fraction (94, of which 76 are secreted). Of the identified proteins, 66 showed an increased abundance when plants were exposed to Cd. In the CaCl2 and LiCl fractions, 73 and 61 proteins were identified, respectively (44 and 59, respectively, are predicted to be secreted). Of those, 45 and 25 proteins, respectively, increased in abundance as a result of Cd exposure. To gather information about the function of the identified proteins, they were clustered according to their predicted biological function using the Blast2Go software (Fig. 2, Table S1). Strikingly, about 50% of the higher‐abundant proteins in each fraction have a designated role in plant defence.
Figure 2.
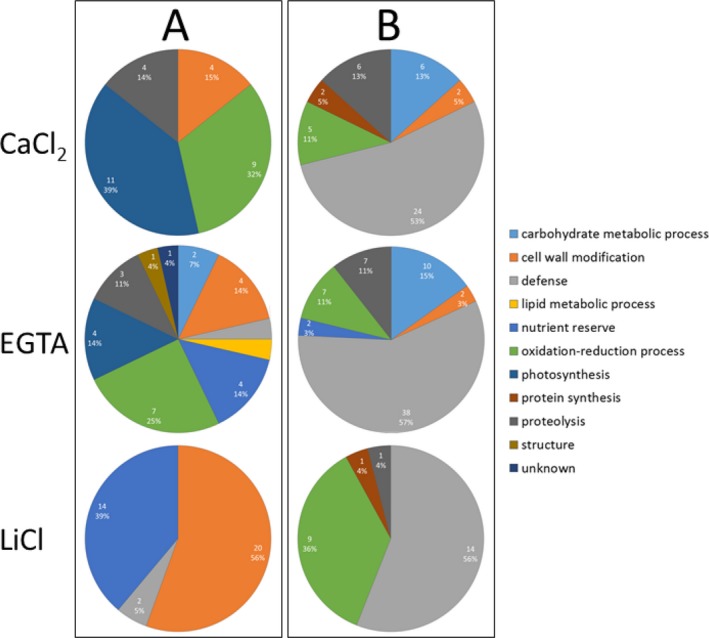
Functional classes of proteins with significant quantitative changes in the cell wall of M. sativa stems in response to Cd. Plants were exposed to 10 mg of Cd·kg−1 soil. Three different cell wall fractions were obtained. Quantitative analyses were performed based on 2‐D DIGE. Clustering into functional classes is based on their predicted biological function using Blast2Go software. A: Functional classes of lower abundant proteins. B: Functional classes of higher abundant proteins.
A large number of identified proteins are involved in carbohydrate metabolic processes, most prevalently glucan endo‐1,3‐β‐glucosidase, but also glycoside hydrolase, family 17, α‐galactosidase‐like protein, which has a higher abundance in Cd‐exposed plants.
For proteins identified in different spots, the intensity of these spots changed in the same direction with a comparable fold change, except for two proteins. In the EGTA fraction different spots containing β‐like galactosidase (spots 1318 and 1433; Appendix S5) and β‐xylosidase/alpha‐L‐arabinofuranosidase (spots 1620 and 1724; Appendix S5) showed an opposite change. The spectra corresponding to spots 1318 and 1433 were clearly different (Appendix S6 Figure 1). An alignment of the identified sequences with the AGED‐database resulted in contig 4097 (spot 1433) and the 40 amino acid longer contig 2997 (spot 1318). Although these contigs showed a different expression profile, no functional distinction between them is known. The identified isoforms of β‐xylosidase/alpha‐L‐arabinofuranosidase had different sequences and matched with two different contigs from M. sativa (contig 15227 for spot 1620, contig 54428 for spot 1724). Both of them have Arabidopsis homologues known to be differently expressed during pathogen infection (Huitema et al. 2003) and development (Hrubá et al. 2005). However, no functional difference between these proteins is known.
Proteins involved in cell wall remodelling are either of lower (fasciclin‐like arabinogalactan protein, polygalacturonase non‐catalytic protein, proline‐rich extensin‐like protein EPR1, dirigent protein 21‐like) or higher (pectinesterase, xyloglucan endotransglucosylase/hydrolase family protein, non‐classical arabinogalactan protein 31‐like, trichome birefringence‐like protein) abundance as a result of Cd exposure. Being involved in oxidation‐reduction processes, a large number of peroxidases were identified with an increased abundance when plants are exposed to Cd. Designated to the same biological process, plastocyanin and plastocyanin‐like domain protein have a lower abundance in stems of Cd‐exposed plants. While the latter are predicted to be secreted, plastocyanin is targeted to the chloroplast. A large number of ferritins were found to be of lower abundance throughout the three cell wall fractions. We identified different ferritin isoforms and distinguished them from their N‐terminal degradation product phytosidirin (Laulhere et al. 1989). The relatively high density of intracellular ferritin multimers might explain the enrichment of these protein complexes in the cell wall‐enriched samples used in this study. Furthermore, proteins with a nutrient reserve function were identified. Auxin‐binding protein ABP 19a and germin‐like protein subfamily 3 member 1 were found to be of lower abundance after Cd exposure, while rhicadhesin receptor protein and bark storage protein increased in abundance. Some secreted proteins with a function in proteolysis were identified. KDEL‐tailed cysteine endopeptidase CEP1 is of lower abundance in Cd‐exposed plants, while the abundance of papain family cysteine protease was higher.
During the MS‐analyses numerous sequence variants, signal sequences, activation/inhibition sequences and some post‐translational modifications were identified (Appendix S5). The latter category includes the confirmation of an α‐β didehydrophenylalanine as a potentially structure‐determining modification in the β‐subunit of polygalacturonase (Sergeant et al. 2017).
Identification of soluble proteins
A significant change in abundance was detected for 52 spots in the soluble fraction (Appendix S3). These spots were picked, leading to identification of 30 proteins. (Appendix S5 and S6 Table 4), which were clustered according to their predicted biological function using the Blast2Go software (Fig. 3, Table 2).
Figure 3.
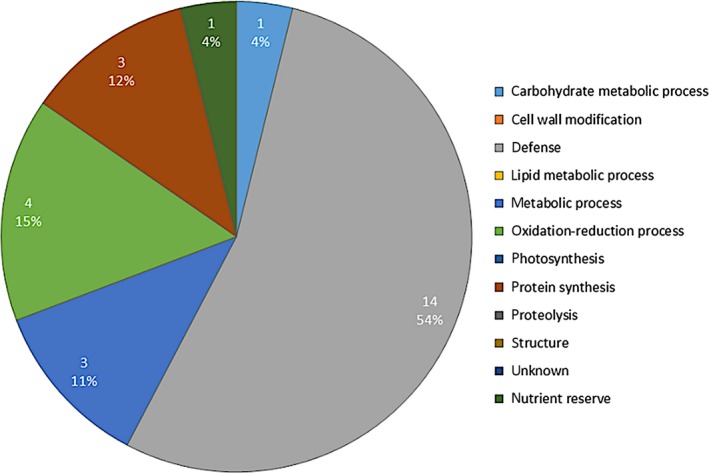
Functional classes of soluble proteins of M. sativa stems with higher abundance in response to Cd. Plants were exposed to 10 mg of Cd·kg−1 soil. Quantitative analyses were performed based on 2‐D DIGE. Clustering into functional classes is based on their predicted biological function using Blast2Go software.
Table 2.
Summary of identified soluble proteins of M. sativa stems that show a Cd‐induced significant abundance change
| annotation | species | regulation | targetP |
|---|---|---|---|
| Carbohydrate metabolic process | |||
| Alpha‐L‐arabinofuranosidase/beta‐D‐xylosidase | M. truncatula | Up | S |
| Defence | |||
| ABA‐responsive protein | M. truncatula | Up | / |
| Epoxide hydrolase‐like protein | M. truncatula | Up | / |
| Polyketide cyclase/dehydrase and lipid transporter | M. truncatula | Up | / |
| Chitinase (Class Ib)/Hevein | M. truncatula | Up | S |
| Chitinase | M. truncatula | Up | S |
| Chitinase class III‐1 | M. sativa | Up | S |
| Class I chitinase | M. sativa | Up | S |
| Plant basic secretory protein (BSP) family protein | M. truncatula | Up | S |
| Pathogenesis‐related protein 1‐like | Brassica rapa | Up | S |
| Metabolic process | |||
| Cobalamin‐independent methionine synthase | M. truncatula | Up | / |
| Sucrose synthase | M. sativa | Up | / |
| Oxidation‐reduction process | |||
| Ferritin | M. truncatula | Down | C |
| Plastocyanin | M. truncatula | Down | C |
| Seed linoleate 9S‐lipoxygenase | M. truncatula | Up | / |
| Protein synthesis | |||
| Glycine‐rich RNA‐binding protein‐like | Arachis duranensis | Up | / |
| Peptidyl‐prolyl cis‐trans isomerase | M. truncatula | Up | C |
| Nutrient reserve | |||
| Rhicadhesin receptor | M. truncatula | Up | S |
C, targeted to the chloroplast; M, targeted to mitochondria; S, secreted; up, increased protein abundance; down, decreased protein abundance. See Appendix S6 Table 4.
Most proteins (26) showed a higher abundance in Cd‐exposed plants and, similar to the cell wall proteins, half of them (54%) are involved in plant defence. Other proteins increasing in abundance are involved in metabolic processes (cobalamin‐independent methionine synthase, sucrose synthase), carbohydrate metabolic processes (β‐xylosidase/α‐L‐arabinofuranosidase), nutrient reserves (rhicadhesin receptor), oxidation‐reduction processes (seed linoleate 9S‐lipoxygenase) or protein synthesis (peptidyl‐prolyl cis‐trans isomerase, glycine‐rich RNA‐binding protein‐like). Of all identified proteins, 43.3% are predicted to be secreted, 16.7% are targeted to the chloroplast, and 40% are without a predicted target site (Appendix S6 Table 4). Proteins identified in one or more of the cell wall fractions and in the soluble protein fraction had abundance changes in the same direction. These include ferritin and plastocyanin, involved in oxido‐reduction processes and of lower abundance in Cd‐exposed plants, as well as defence‐related proteins which were more abundant in all extracted fractions from stems of Cd‐exposed as compared to unexposed plants.
Expression of genes involved in cell wall structure and lignification
Based on our proteomic study, different genes related to cell wall structure and modification were selected for validation via RT‐qPCR. Normalised gene expression values of all five replicates from unexposed and Cd‐exposed plants (Appendix S7) are represented in a heat map (Fig. 4). The expression of analysed genes in M. sativa stems cluster in two main branches. In the upper branch, all genes clearly showed higher expression in Cd‐exposed plants. These include genes encoding different POX isoforms, proteins functioning in carbohydrate metabolic processes (β‐like galactosidase, glycoside hydrolase, β‐xylosidase/α‐L‐arabinofuranosidase, endo‐β‐1,3‐glucanase), pectinesterase/pectinesterase inhibitor and dirigent‐like protein. With the exception of a β‐like galactosidase isoform (contig 2998), all those expression changes were significant (P ≤ 0.05; Table 3).
Figure 4.
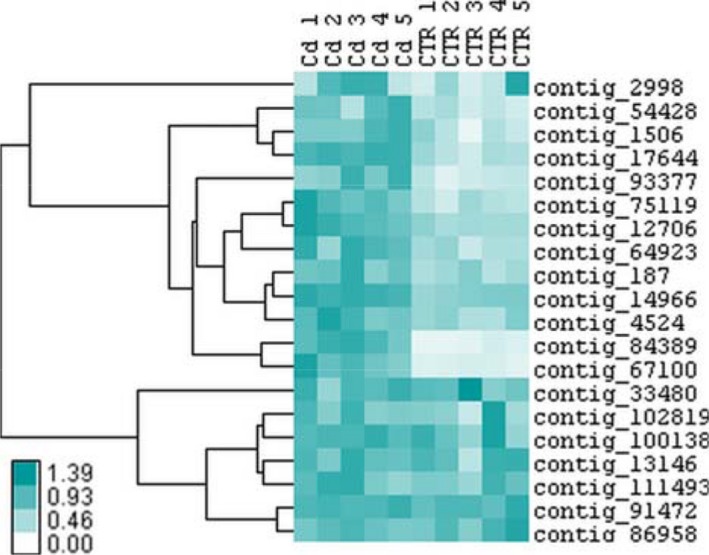
Heat map representation of gene expression data from five biological replicates showing the hierarchical clustering (Pearson uncentred, complete linkage clustering) of cell wall‐related genes in M. sativa stems under long‐term Cd exposure (10 mg·kg−1 soil). Colour intensity is proportional to the actual expression value. Values are provided in Appendix S7.
Table 3.
Relative normalised gene expressions in stems of M. sativa. Plants were exposed to 10 mg·Cd·kg−1 soil. Three reference genes were used for normalisation. Expression levels were calculated relative to the non‐exposed plants
| Gene annotation | Contig_ID M. sativa | Corresponding gene ID M. truncatula | Rel. norm expression ± SE | P‐value |
|---|---|---|---|---|
| Beta‐like galactosidase | 2998 | Medtr1g018200.1 | 1.507 ± 0.325 | 0.323 |
| Beta‐like galactosidase | 1506 | Medtr2g039120.1 | 2.272 ± 0.229 | 0.004 |
| α‐L‐arabinofuranosidase/β‐xylosidase | 187 | Medtr2g008240.1 | 1.656 ± 0.157 | 0.007 |
| α‐L‐arabinofuranosidase/β‐D‐xylosidase | 54428 | Medtr2g034720.1 | 2.299 ± 0.329 | 0.006 |
| Alpha‐galactosidase‐like protein | 33480 | Medtr7g073650.1 | 0.966 ± 0.1 | 0.865 |
| Glucan endo‐1,3‐β‐glucosidase | 75119 | Medtr4g076470.1 | 2.383 ± 0.291 | 0.012 |
| Pectinesterase/pectinesterase inhibitor | 93377 | Medtr7g050950.1 | 2.854 ± 0.364 | 0.001 |
| Non‐classical arabinogalactan protein 31‐like | 91472 | 1.041 ± 0.025 | 0.512 | |
| Trichome birefringence‐like protein | 13146 | Medtr2g015720.1 | 0.895 ± 0.31 | 0.259 |
| Xyloglucan endotransglucosylase/hydrolase family protein | 102819 | Medtr4g126920.1 | 1.065 ± 0.112 | 0.454 |
| Xyloglucanase‐specific endoglucanase inhibitor protein | 100138 | Medtr1g072420.1 | 1.146 ± 0.040 | 0.391 |
| Peroxidase family protein | 12706 | Medtr2g029850.1 | 1.953 ± 0.151 | 0.0003 |
| Class III peroxidase | 17644 | Medtr4g095450.1 | 2.530 ± 0.057 | 2.45·e−6 |
| Lignin biosynthetic peroxidase | 84389 | Medtr2g084020.1 | 5.808 ± 0.522 | 0.002 |
| Peroxidase family protein | 64923 | Medtr6g043240.1 | 2.083 ± 0.235 | 0.005 |
| Class III peroxidase | 14966 | Medtr5g074970.1 | 1.669 ± 0.043 | 5.2·e−6 |
| Glycoside hydrolase, family 17 | 67100 | Medtr2g034440.1 | 4.567 ± 0.494 | 9.584·e−5 |
| Fasciclin‐like arabinogalactan protein | 86958 | Medtr4g059840.1 | 0.920 ± 0.053 | 0.512 |
| Polygalacturonase non‐catalytic protein | 111493 | Medtr8g064530.1 | 1.156 ± 0.143 | 0.405 |
| Dirigent protein 21‐like | 4524 | Medtr1g018200.1 | 1.716 ± 0.170 | 0.006 |
Values are mean ± SE from five biological replicates. P‐values below or equal to 0.05 indicate significant differences.
The second branch is characterised by an unclear distinction between expression levels within stems of Cd‐exposed and unexposed plants, and variable expression between the five replicates of the two conditions was observed, resulting in non‐significant relative expression changes (Fig. 4). Genes in that branch are related to cell wall modification and carbohydrate metabolism.
However, most of the observed significant expression changes were consistent with those observed for protein abundance.
Discussion
Our data show the impact of long‐term Cd exposure on the cell wall and soluble proteome in M. sativa stems. Especially proteins involved in cell wall structure and remodelling are most interesting as they can be connected to structural changes in the cell wall induced by Cd. A brief overview is provided in Fig. 5. Transcription levels from corresponding genes were further investigated to validate our protein data. Several proteins involved in photosynthesis were identified within the cell wall fraction. Although these proteins are not localised at the cell wall, they will be part of the discussion as their abundance is highly affected by Cd exposure, underlining the impairment of photosynthesis due to Cd exposure.
Figure 5.
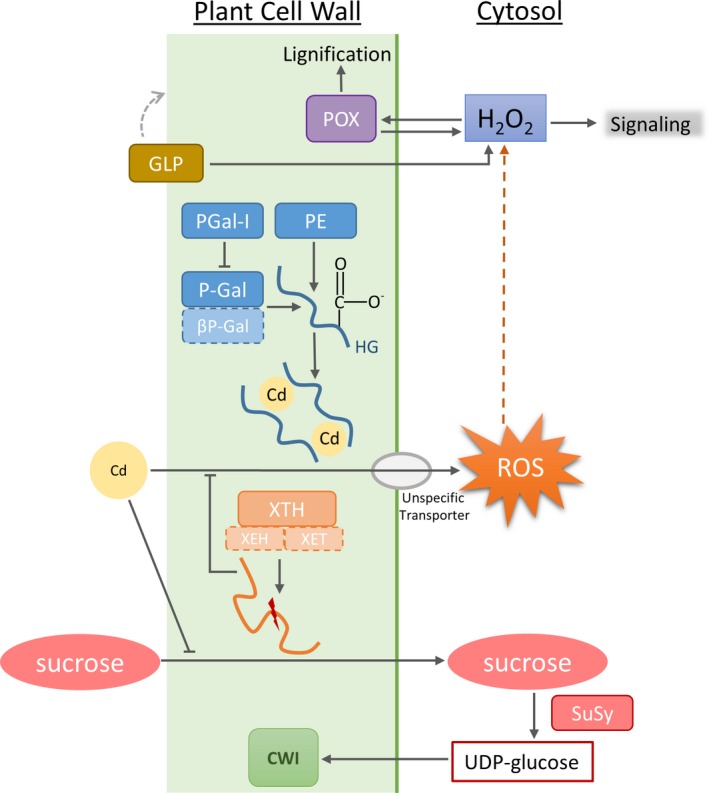
Effects of Cd on cell wall structure. Cd might affect the cell wall structure in different ways. Cd influences lignification by acting on peroxidase (POX) activity. It further changes the activity of pectin methylesterase (PME), which leads to enhanced content of low‐methylesterified pectin and thereby creates binding sites for Cd within the cell wall. This is also supported by the activity of polygalacturonase (P‐Gal) and polygalacturonase β‐subunit (βP‐Gal). The latter protein is negatively influenced by Cd. Cd directly affects the cell wall structure by changing activity of xyloglucan endotransglucosylase/hydrolase (XTH) and sucrose synthase (SuSy). By degrading sucrose, SuSy provides precursor molecules for cell wall synthesis. Intracellular, Cd indirectly triggers production of ROS, which subsequently activates POX activity and also operates as signalling molecules for downstream processes. Germin‐like proteins (GLP) induce ROS production and therefore influence ROS signalling and trigger POX activity.
Plants were grown for 5 months and the secondary shoot was sampled, as the cutting and re‐growing of M. sativa is commonly used in agriculture. Cadmium‐exposed plants exhibited severe effects during and after germination. Seeds grown on Cd‐contaminated soil did not germinate in twice as much pots as those grown on the control soil. Furthermore, Cd‐exposed plants were inhibited in growth during the first weeks after germination (Fig. 1). In a later more mature state, phenotypic differences evened out, and at the end of our experiment (growth of secondary shoots) no significant difference in biomass was observed between control and Cd‐treated M. sativa plants. This observation is in agreement with previous studies showing the strongest impact of heavy metals during germination and the initial stage of growth, while only slight growth inhibition was observed for several plant species in a more mature growth stage after attaining a new homeostasis on mildly polluted soils (Peralta‐Videa et al. 2004; Chaoui & El Ferjani 2005; Wang & Song 2009; Chou et al. 2011). In a previous study undertaken in our laboratory on sunflowers exposed to a polymetallic constraint, this metal sensitivity of the initial growth stage was also observed (Printz et al. 2013). Although stress exposure passes through an alarm phase wherein rapid responses are intiatied to tackle the immediate threat and prevent damage (Lichtenthaler 1996), a recent meta‐study of proteome data indicates that long‐term exposure of plants to low to moderate concentrations of heavy metals results in the establishment of a new homeostatic equilibrium (Dupae et al. 2014).
Cadmium accumulation in roots
In this study, roots had the highest Cd content in Cd‐exposed plants, and we observed a decreasing Cd content from roots > stems > leaves. The assessed Cd content in roots was four and seven times higher than in stems and leaves, respectively (Table 1). Roots are directly exposed to Cd present in the soil. Ion transporters, which are embedded in the plasma membrane of root cells, preferentially take up nutrients from the surrounding soil, but do not discriminate between chemically similar metals and therefore also represent the main entry route for non‐essential metals such as Cd (Clemens 2006; Song et al. 2016). Protective mechanisms occur primarily downstream of the uptake, e.g. by trapping ions inside the cell through selective binding sites with a high affinity. Phytochelatins (PC) are cysteine‐rich peptides synthesised in response to various metal ions, with the ability to bind them. Among metals, Cd is the most potent inducer of PC synthesis (Grill et al. 1989; Clemens et al. 1999). Phytochelatin–Cd complexes are sequestered in the vacuole, supporting the detoxification mechanisms of the cell (Cobbett 2000), and it is assumed that vacuoles of root cells are the major site of metal accumulation. By retaining Cd in the roots and maintaining low root‐to‐shoot translocation, physiological highly important organs such as leaves are protected from Cd‐induced damage, and essential process like photosynthesis can be maintained.
Cadmium affects the accumulation of proteins related to cell wall structure
Effects of Cd exposure are observed in the cell wall proteome and in the TCA/phenol‐extracted soluble proteome. The changes identified in the soluble proteome are consistent with those in the different cell wall fractions. Proteins uniquely identified in the soluble protein fraction include proteins involved in protein biosynthesis. Those proteins are of higher abundance, indicating that the plant is actively synthesising proteins as a protective response to Cd stress. In the soluble protein fraction, a higher abundance was determined for sucrose synthase (SuSy). SuSy breaks down sucrose into fructose and UDP‐glucose (Koch 2004). The latter is used for cellulose biosynthesis, and cell wall‐bound SuSy isoforms might channel UDP‐glucose directly into the cellulose synthase complex (Brill et al. 2011). Cellulose is the main compound of the plant cell wall and during secondary cell wall synthesis a large quantity of UDP‐glucose molecules are required. The needed glucose molecules derive from photosynthesis and carbon fixation through the Calvin Cycle. Throughout all cell wall fractions, a Cd‐induced lower abundance of photosynthetic proteins was observed in this study (Fig. 2, Table S1), which suggests impaired photosynthesis in M. sativa (Al‐Hakimi 2007; Wang et al. 2014). Impaired photosynthesis affects the carbon fixation and reduces the available amount of glucose. The activity of SuSy could compensate for the missing glucose supply from the impaired photosynthesis and through this maintain the integrity of the cell wall during Cd stress.
Generally, proteins of the same functional classification have a change in abundance in the same direction. Even among members of larger protein families (chitinases, germins, POX) a similar directional change or trend was observed. Only in two cases (β‐like galactosidase and β‐xylosidase/α‐L‐arabinofuranosidase), did spots that contained the same protein show significant changes in different directions. Although MS analysis allowed the discrimination of different isoforms in the spots (Appendix S5 and S6 Figure 1), no functional specification of these isoforms was possible.
An important number of proteins with significant abundance changes are involved in cell wall modification and carbohydrate metabolic processes. A variety of these enzymes (including pectinesterase, β‐like galactosidase, α‐galactosidase and polygalacturonase non‐catalytic protein) are responsible for structural modifications, degradation and changes within the pectin network during maturation and as response to stress (Dellapenna et al. 1990; Ali et al. 2004; Chrost et al. 2007; Caffall & Mohnen 2009; Huang et al. 2016). Apart from polygalacturonase non‐catalytic protein, all these proteins were more abundant in Cd‐exposed M. sativa stems (Table S1), which corresponded to increased expression of their encoding genes (Table 3). Cadmium induces changes in the pattern of the pectin homogalacturonane in plant cell walls (Douchiche et al. 2010b) and enhances PME activity (Paynel et al. 2009). The here observed higher abundance of pectinesterase supports elevated levels of low methylesterified homogalacturonane in the cell wall of Cd‐exposed M. sativa stems, creating binding sites for Cd and contributing to Cd deposition in the cell wall (Ramos et al. 2002), which prevents its further entry into the cell and translocation to the leaves. The function of the polygalacturonase non‐catalytic protein remains ambiguous. Data indicate that it limits access of hydrolases by tightly binding to pectin and that it prevents pectin release from the cell wall (Watson et al. 1994; Chun & Huber 1997). Other studies found that overexpression of this protein results in a decreased pectin content and reduced cell adhesion (Liu et al. 2014). In the current study, different isoforms of this protein showed decreased abundance when plants were exposed to Cd, which might support the pectinesterase‐catalysed de‐methylation of pectin.
The backbone of pectin is highly modified with different oligosaccharides and O‐acetyl groups (Mohnen 2008). As a response to Cd, we found β‐xylosidase/α‐L‐arabinofuranosidase either of higher or lower abundance and a higher abundance of trichome birefringence‐like protein (Table S1). The latter is an O‐acetyltransferase, substituting cell wall polysaccharides (hemicellulose, pectin and lignin) with O‐acetyl, which influences their properties and modifies the cell wall (Gille & Pauly 2012). As a bifunctional hydrolase, β‐xylosidase/α‐L‐arabinofuranosidase hydrolyses terminal non‐reduced L‐Ara and D‐Xyl and is involved in reconstruction of the cell wall (Xiong et al. 2007; Chavez Montes et al. 2008). Two spots with a different direction of changes were identified, which can be ascribed to the bifunctional activity or to differences in substrate specificity of the different isoforms.
Furthermore, increased protein abundance of xyloglucan endotransglucosylase/hydrolase (XTH) due to Cd exposure was observed (Table S1). As XTH activity results in the re‐construction of xyloglucan, it promotes cell wall loosening and cell expansion (Osato et al. 2006). Overexpression of this enzyme in Cd‐exposed plants conferred Cd tolerance, correlated with the XTH‐catalysed decline in xyloglucan content in the cell wall (Han et al. 2014). In another group of spots with an increased intensity after Cd stress, a cysteine‐rich secretory protein (CAP) was identified. This small PR‐1 protein has a cd05381 conserved domain with antifungal, as well as cell wall loosening activity. Homologous genes are down‐regulated in salt‐ and Ca2+‐exposed plants (Volkov et al. 2003; Chan et al. 2008) but were highly increased in Crambe abyssinica exposed to arsenate (Paulose et al. 2010). Overexpression of a pepper homologue of this enzyme in tobacco plants resulted in increased resistance to Cd and mercury exposure (Sarowar et al. 2005). Whether this is a result of its direct impact on the cell wall or an indirect impact on ROS generation is not known.
We found lignin biosynthetic peroxidase and other non‐specific cell wall‐linked peroxidases were highly abundant in Cd‐exposed M. sativa stems (Table S1). The corresponding gene transcripts were up‐regulated for all different isoforms and were more than five times higher in Cd‐exposed plants for the lignin biosynthetic peroxidase (Table 3). Lignification results in cell wall stiffening and limits cell growth (Schützendübel et al. 2001; Chaoui et al. 2004). Cadmium is known to induce oxidative stress (Garnier et al. 2006; Cuypers et al. 2011) and H2O2 accumulation (Schützendübel et al. 2001; Rodríguez‐Serrano et al. 2006). As a signalling molecule, H2O2 triggers secondary reactions, such as an increased peroxidase activity, which contributes to increased lignification. Lignification is further supported by dirigent‐like proteins, which bind free radical monolignol species and coordinate their coupling during lignin biosynthesis (Davin & Lewis 2000). A gene encoding a dirigent‐like protein was induced by drought, salt and oxidative stress in Saccharum officinarum, highlighting their important role in plant resistance and defence (Jin‐long et al. 2012), and its involvement in plant responses to metal stress was also observed (Van De Mortel et al. 2006; Chen et al. 2016). Yet, we reported a Cd‐mediated lower abundance of dirigent protein 21‐like (Table S1), although expression of the corresponding gene was significantly induced by Cd (Table 3). Gene expression and protein abundance do not necessarily correlate in a linear manner. Transcription and translation are controlled by different mechanisms which enhance or repress protein synthesis. Thereby, the degree of protein turnover probably has the greatest influence on the transcript‐protein correlation (Maier et al. 2009).
Overall, proteome and transcript data from stems of M. sativa support that Cd alters the cell wall structure and induces lignification as an important mechanism during plant defence. In the case of α‐galactosidase‐like protein, non‐classical arabinogalactan protein 31‐like, trichome birefringence‐like protein, xyloglucan endotransglucosylase/hydrolase family protein, xyloglucanase‐specific endoglucanase inhibitor protein, fasciclin‐like arabinogalactan protein and polygalacturonase non‐catalytic protein, the protein abundance did not correspond with the determined transcript accumulation (Table 3, Fig. 4). For those genes, expression changes after Cd exposure were variable between replicates and therefore no significant changes in mRNA abundance was obtained. However, mRNA transcript levels and protein accumulation do not inevitably correlate due to the complexity of post‐transcriptional mechanisms translating mRNA into proteins, fundamental differences of protein in‐vivo half‐lives as well as experimental limitations and errors. Reports on missing or poor proportional correlation of transcriptome and proteome data can be found in literature (Greenbaum et al. 2003). Therefore, transcriptomic and proteomic data should be seen as independent yet partly correlating information (Vélez‐Bermúdez & Schmidt 2014). Since 2D‐PAGE has the potential to visualize all post‐transcriptional events, although in general without exact identification, correlations between 2D‐based proteome data and mRNA data are generally lower than when using a peptide‐based proteome approach. The change in abundance of a specific proteoform, due to post‐transcriptional events, may therefore be poorly correlated or even reversed to transcriptional regulation. This observation underscores the complementarity of studying biology at the transcriptional and post‐transcriptional level.
Cadmium exposure results in accumulation of stress response proteins
Exposure to Cd induces oxidative stress, which is consistent with an increased abundance of stress‐related proteins in all fractions (Tables S1 and 2, Figs 2 and 3). The majority of these defence proteins are chitinase isoforms. Increased accumulations of chitinases in response to Cd were previously observed in different plants (Békésiová et al. 2008; Mészáros et al. 2014). In general, chitinases are ROS‐regulated and thus the reported increased abundance of chitinases is not specific to Cd and rather reflects the general stress response of M. sativa (Metwally et al. 2003; Hossain & Komatsu 2013; Li et al. 2016).
Germin‐like proteins (GLP) in stem tissue of M. sativa show a decreased abundance when plants were exposed to Cd, even though GLP are involved in the response to abiotic stresses. Formally classified as storage proteins, GLPs are expressed in all plant tissues throughout all development stages and their association to the cell wall has been demonstrated (Vallelian‐Bindschedler et al. 1998; Schweizer & Christoffel 1999; Christensen et al. 2004). Many functions are assigned to GLPs: receptor, structure, enzyme activity (Bernier & Berna 2001), including oxalate oxidase (OxO) and superoxide dismutase (SOD) activity (de los Reyes & McGrath 2003; Gucciardo et al. 2007). Both reactions induce the generation of H2O2, a second messenger molecule during plant stress responses but also a trigger for peroxidase‐induced lignification of the cell wall, as discussed above. A stress‐induced decreased in GLP‐SOD activity in the cell wall was observed previously in cell cultures of Barbula unguiculata (Nakata et al. 2002). Lowering the abundance of GLPs in response to Cd exposure will produce less ROS and prevent additional oxidative stress on top of the already induced oxidative burst by Cd.
Conclusion
Upon long‐term exposure, the roots of M. sativa accumulated Cd, thereby restricting its mobility throughout the plant as further Cd accumulation induces impairment of cellular, biochemical and physiological processes in the aboveground tissues. To test the hypothesis that the cell wall functions as an effective barrier to the entry of Cd, the present study focused on cell wall proteomes of the stems, as stems produce high amounts of cell wall material. Analysis of the cell wall and soluble proteome of the stem revealed a profound impact of Cd. Cadmium influences the abundance of cell wall proteins involved in multiple physiological processes such as plant defence response, oxidation‐reduction processes, carbohydrate metabolism and cell wall remodelling. Transcriptome data of the study overall support these observations.
The identified proteins indicate that long‐term Cd exposure induces alterations in the structure of the cell wall and promotion of the lignification process. Currently, the determination of the cell wall composition and structure and the degree of lignification is ongoing. Although there is a negative impact of Cd stress on the early stages of plant growth, long‐term exposure to Cd does not have an adverse impact on mature plants in terms of biomass production, and the observed transcriptome and proteome changes do not have a significant impact on the produced biomass. This indicates that the mature plants established a new homeostasis and the induced protective mechanisms are more effective in mature than in juvenile plants.
Supporting information
Appendix S1. Nutrient content of the planting soil at the end of the experiment as determined by ICP‐MS. The nutrient concentrations are given as mean ± SE of ten replicates from each condition (μg·g−1 dry weight). Apart from the Cd concentration, no significant changes (P ≤ 0.05) in nutrient composition occurred.
Appendix S2. 2‐D DIGE of the three cell wall fractions and the soluble protein fraction from M. sativa stems. The shown gels were used as master gel for the experiment. Proteins were pre‐labelled with CyDye. Labelled samples were loaded on ImmobilineTM DryStrip NL, 24 cm (GE Healthcare) followed by migration on HPETM Large Gel NF‐12.5% (Serva Electrophoresis). Marked spots were selected for identification using SameSpots software (TatalLab). HMW, high molecular weight; LMW, low molecular weight.
Appendix S3. 2‐D DIGE spot volumes determined with SameSpots software (TotalLab) from spots which show a significant quantitative change after Cd exposure and were chosen for identification. The total and normalised volumes, fold change and P‐value of each spot is given.
Appendix S4. Table 1 provides forward and reverse primer sequences used to determine gene expression levels with RT‐qPCR, including reference genes. Amplicon size and primer efficiency for each gene are indicated. In Table 2 quantitative real‐time PCR parameters according to the Minimum Information for publication of quantitative real‐time PCR experiments (MIQE) guidelines derived from Bustin et al. (2009) are given.
Appendix S5. Complete MASCOT protein identification data of picked spots from the cell wall protein fraction and soluble protein fraction.
Appendix S6. Tables 1–4 contain all identified proteins in each of the four protein fractions and their abundance change after Cd exposure. Fold change and P‐value were obtained using the SameSpots software (TotalLab). Functional classification of each protein was determined using Blast2Go. Subcellular localisation was determined with TargetP. Figure 1 shows MS spectra from spots 1318 and 1433. In both spots the same protein was identified being of contradictory abundance. Spectra are clearly different indicating the presence of different isoforms.
Appendix S7. Normalised expression of genes related to cell wall structure and lignification in stems of M. sativa. Plants were exposed to Cd (10 mg·kg−1 soil) in a long‐term experiment. Data are used for the heat map representation showing the hierarchical clustering of all investigated genes.
Table S1. Summary of identified proteins in the cell wall fractions (CaCl2, EGTA, LiCl) of M. sativa stems that show a Cd‐induced significant abundance change.
Acknowledgements
This study is funded as part of the bilateral project CadWALL (FNR/FWO project INTER/FWO/12/14). JFH, JR, AC, KS and AG conceived the study and designed the experiments. AG prepared the samples and performed most of the experiments and data analyses. JR, GG, KS and EK contributed to analysis of the data. AG and KS drafted the manuscript, which was critically revised and approved by all co‐authors. We especially thank Sébastien Planchon and Laurent Solinhac for technical assistance, Johanna Ziebel for performing all ICP‐MS analyses and Bruno Printz for support. We thank the group of Ann Cuypers at the University of Hasselt for great support and assistance in performing the gene expression analysis.
References
- Al‐Hakimi A.M.A. (2007) Modification of cadmium toxicity in pea seedlings by kinetin. Plant Soil and Environment, 53, 129–135. [Google Scholar]
- Ali Z.M., Chin L.H., Lazan H. (2004) A comparative study on wall degrading enzymes, pectin modifications and softening during ripening of selected tropical fruits. Plant Science, 167, 317–327. [Google Scholar]
- Békésiová B., Hraška Š., Libantová J., Moravčíková J., Matušíková I. (2008) Heavy‐metal stress induced accumulation of chitinase isoforms in plants. Molecular Biology Reports, 35, 579–588. [DOI] [PubMed] [Google Scholar]
- Bernier F., Berna A. (2001) Germins and germin‐like proteins: plant do‐all proteins. But what do they do exactly? Plant Physiology and Biochemistry, 39, 545–554. [Google Scholar]
- Bradley D.J., Kjellbom P., Lamb C.J. (1992) Elicitor‐ and wound‐induced oxidative cross‐linking of a proline‐rich plant cell wall protein: a novel, rapid defense response. The Cell, 70, 21–30. [DOI] [PubMed] [Google Scholar]
- Brill E., van Thournout M., White R.G., Llewellyn D., Campbell P.M., Engelen S., Ruan Y.‐L., Arioli T., Furbank R.T. (2011) A novel isoform of sucrose synthase is targeted to the cell wall during secondary cell wall synthesis in cotton fiber. Plant Physiology, 157, 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson L.F., Tenhaken R., Lamb C. (1994) Function of oxidative cross‐linking of cell wall structural proteins in plant disease resistance. The Plant Cell, 6, 1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. (2009) The MIQE guidelines: minimum information for publication of quantitative real‐time PCR experiments. Clinical Chemistry, 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Caffall K.H., Mohnen D. (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydrate Research, 344, 1879–1900. [DOI] [PubMed] [Google Scholar]
- Chan C.W.M., Wohlbach D.J., Rodesch M.J., Sussman M.R. (2008) Transcriptional changes in response to growth of Arabidopsis in high external calcium. FEBS Letters, 582, 967–976. [DOI] [PubMed] [Google Scholar]
- Chaoui A., El Ferjani E. (2005) Effects of cadmium and copper on antioxidant capacities, lignification and auxin degradation in leaves of pea (Pisum sativum L.) seedlings. Comptes Rendus Biologies, 328, 23–31. [DOI] [PubMed] [Google Scholar]
- Chaoui A., Jarrar B., El Ferjani E. (2004) Effects of cadmium and copper on peroxidase, NADH oxidase and IAA oxidase activities in cell wall, soluble and microsomal membrane fractions of pea roots. Journal of Plant Physiology, 161, 1225–1234. [DOI] [PubMed] [Google Scholar]
- Chavez Montes R.A., Ranocha P., Martinez Y., Minic Z., Jouanin L., Marquis M., Saulnier L., Fulton L.M., Cobbett C.S., Bitton F., Renou J.‐P., Jauneau A., Goffner D. (2008) Cell wall modifications in Arabidopsis plants with altered α‐L‐arabinofuranosidase activity. Plant Physiology, 147, 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Yan W., Sun L., Tian J., Liao H. (2016) Proteomic analysis reveals growth inhibition of soybean roots by manganese toxicity is associated with alteration of cell wall structure and lignification. Journal of Proteomics, 143, 151–160. [DOI] [PubMed] [Google Scholar]
- Chou T.‐S., Chao Y.Y., Huang W.D., Kao C.H. (2011) Effect of magnesium deficiency on antioxidant status and cadmium toxicity in rice seedlings. Journal of Plant Physiology, 168, 1021–1030. [DOI] [PubMed] [Google Scholar]
- Christensen A.B., Thordal‐Christensen H., Zimmermann G., Gjetting T., Lyngkjaer M.F., Dudler R., Schweizer P. (2004) The germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Molecular Plant‐Microbe Interactions, 17, 109–117. [DOI] [PubMed] [Google Scholar]
- Chrost B., Kolukisaoglu U., Schulz B., Krupinska K. (2007) An α‐galactosidase with an essential function during leaf development. Planta, 225, 311–320. [DOI] [PubMed] [Google Scholar]
- Chun J., Huber D.J. (1997) Polygalacturonase isozyme 2 binding and catalysis in cell walls from tomato fruit: pH and β‐subunit effects. Physiologia Plantarum, 101, 283–290. [Google Scholar]
- Clemens S. (2006) Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie, 88, 1707–1719. [DOI] [PubMed] [Google Scholar]
- Clemens S., Ma J.F. (2016) Toxic heavy metal and metalloid accumulation in crop plants and foods. Annual Review of Plant Biology, 67, 489–512. [DOI] [PubMed] [Google Scholar]
- Clemens S., Kim E.J., Neumann D., Schroeder J.I. (1999) Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO Journal, 18, 3325–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbett C.S. (2000) Phytochelatins and their roles in heavy metal detoxification. Plant Physiology, 123, 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuypers A., Plusquin M., Remans T., Jozefczak M., Keunen E., Gielen H., Opdenakker K., Nair A.R., Munters E., Artois T.J., Nawrot T., Vangronsveld J., Smeets K. (2010) Cadmium stress: an oxidative challenge. BioMetals, 23, 927–940. [DOI] [PubMed] [Google Scholar]
- Cuypers A., Smeets K., Ruytinx J., Opdenakker K., Keunen E., Remans T., Horemans N., Vanhoudt N., Van Sanden S., Van Belleghem F., Guisez Y., Colpaert J., Vangronsveld J. (2011) The cellular redox state as a modulator in cadmium and copper responses in Arabidopsis thaliana seedlings. Journal of Plant Physiology, 168, 309–316. [DOI] [PubMed] [Google Scholar]
- Davin L.B., Lewis N.G. (2000) Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiology, 123, 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellapenna D., Lashbrook C.C., Toenjes K., Giovannoni J.J., Fischer R.L., Bennett A.B. (1990) Polygalacturonase isozymes and pectin depolymerization in transgenic rin tomato fruit. Plant Physiology, 94, 1882–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchiche O., Rihouey C., Schaumann A., Driouich A., Morvan C. (2007) Cadmium‐induced alterations of the structural features of pectins in flax hypocotyl. Planta, 225, 1301–1312. [DOI] [PubMed] [Google Scholar]
- Douchiche O., Driouich A., Morvan C. (2010a) Spatial regulation of cell‐wall structure in response to heavy metal stress: cadmium‐induced alteration of the methyl‐esterification pattern of homogalacturonans. Annals of Botany, 105, 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchiche O., Soret‐Morvan O., Chaïbi W., Morvan C., Paynel F. (2010b) Characteristics of cadmium tolerance in “Hermes” flax seedlings: contribution of cell walls. Chemosphere, 81, 1430–1436. [DOI] [PubMed] [Google Scholar]
- Dupae J., Bohler S., Noben J., Carpentier S., Vangronsveld J., Cuypers A. (2014) Problems inherent to a meta‐analysis of proteomics data: a case study on the plants’ response to Cd in different cultivation conditions. Journal of Proteomics, 108, 30–54. [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. (1999) Cluster analysis and display of genome‐wide expression patters. Proceedings of the National Academy of Sciences of the United States of America, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H. (2000) Predicting subcellular localization of proteins based on their N‐terminal amino acid sequence. Journal of Molecular Biology, 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Garnier L., Simon‐Plas F., Thuleau P., Agnel J.P., Blein J.P., Ranjeva R., Montillet J.L. (2006) Cadmium affects tobacco cells by a series of three waves of reactive oxygen species that contribute to cytotoxicity. Plant Cell and Environment, 29, 1956–1969. [DOI] [PubMed] [Google Scholar]
- Gille S., Pauly M. (2012) O‐acetylation of plant cell wall polysaccharides. Frontiers in Plant Science, 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D., Colangelo C., Williams K., Gernstein M. (2003) Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biology, 4, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E., Löffler S., Winnacker E.L., Zenk M.H. (1989) Phytochelatins, the heavy‐metal‐binding peptides of plants, are synthesized from glutathione by a specific gamma‐glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthase). Proceedings of the National Academy of Sciences of the United States of America, 86, 6838–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gucciardo S., Wisniewski J.P., Brewin N.J., Bornemann S. (2007) A germin‐like protein with superoxide dismutase activity in pea nodules with high protein sequence identity to a putative rhicadhesin receptor. Journal of Experimental Botany, 58, 1161–1171. [DOI] [PubMed] [Google Scholar]
- Guerriero G., Legay S., Hausman J.F. (2014) Alfalfa cellulose synthase gene expression under abiotic stress: a hitchhiker's guide to RT‐qPCR normalization. PLoS One, 9, e103808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Sa G., Sun J., Shen Z., Zhao R., Ding M., Deng S., Lu Y., Zhang Y., Shen X., Chen S. (2014) Overexpression of Populus euphratica xyloglucan endotransglucosylase/hydrolase gene confers enhanced cadmium tolerance by the restriction of root cadmium uptake in transgenic tobacco. Environmental and Experimental Botany, 100, 74–83. [Google Scholar]
- Hossain Z., Komatsu S. (2013) Contribution of proteomic studies towards understanding plant heavy metal stress response. Frontiers in Plant Science, 3, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain Z., Hajika M., Komatsu S. (2012) Comparative proteome analysis of high and low cadmium accumulating soybeans under cadmium stress. Amino Acids, 43, 2393–2416. [DOI] [PubMed] [Google Scholar]
- Hrubá P., Honys D., Twell D., Čapková V., Tupý J. (2005) Expression of β‐galactosidase and β‐xylosidase genes during microspore and pollen development. Planta, 220, 931–940. [DOI] [PubMed] [Google Scholar]
- Huang J.H., Kortstee A., Dees D.C.T., Trindade L.M., Schols H.A., Gruppen H. (2016) Modification of potato cell wall pectin by the introduction of rhamnogalacturonan lyase and β‐galactosidase transgenes and their side effects. Carbohydrate Polymers, 144, 9–16. [DOI] [PubMed] [Google Scholar]
- Huitema E., Vleeshouwers V.G.A.A., Francis D.M., Kamoun S. (2003) Active defence responses associated with non‐host resistance of Arabidopsis thaliana to the oomycete pathogen Phytophthora infestans . Molecular Plant Pathology, 4, 487–500. [DOI] [PubMed] [Google Scholar]
- Jin‐long G., Li‐ping X., Jing‐ping F., Ya‐chun S., Hua‐ying F., You‐xiong Q., Jing‐sheng X. (2012) A novel dirigent protein gene with highly stem‐specific expression from sugarcane, response to drought, salt and oxidative stresses. Plant Cell Reports, 31, 1801–1812. [DOI] [PubMed] [Google Scholar]
- Júnior C.A.L., de Sousa Barbosa H., Galazzi R.M., Koolen H.H.F., Gozzo F.C., Arruda M.A.Z. (2015) Evaluation of proteome alterations induced by cadmium stress in sunflower (Helianthus annuus L.) cultures. Ecotoxicology & Environmental Safety, 119, 170–177. [DOI] [PubMed] [Google Scholar]
- Kieffer P., Planchon S., Oufir M., Ziebel J., Dommes J., Hoffmann L., Hausman J.F., Renaut J. (2009) Combining proteomics and metabolite analyses to unravel cadmium stress‐response in poplar leaves. Journal of Proteome Research, 8, 400–417. [DOI] [PubMed] [Google Scholar]
- Koch K. (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Current Opinion in Plant Biology, 7, 235–246. [DOI] [PubMed] [Google Scholar]
- Krzesłowska M. (2011) The cell wall in plant cell response to trace metals: polysaccharide remodeling and its role in defense strategy. Acta Physiologia Plantarum, 33, 35–51. [Google Scholar]
- Laulhere J.P., Laboure A.M., Briat J.F. (1989) Mechanism of the transition from plant ferritin to phytosiderin. Journal of Biological Chemistry, 264, 3629–3635. [PubMed] [Google Scholar]
- Lee K., Bae D.W., Kim S.H., Han H.J., Liu X., Park H.C., Lim C.O., Lee S.Y., Chung W.S. (2010) Comparative proteomic analysis of the short‐term responses of rice roots and leaves to cadmium. Journal of Plant Physiology, 167, 161–168. [DOI] [PubMed] [Google Scholar]
- Li Y., Zhao J., Li Y.‐F., Xu X., Zhang B., Liu Y., Cui L., Li B., Gao Y., Chai Z. (2016) Comparative metalloproteomic approaches for the investigation proteins involved in the toxicity of inorganic and organic forms of mercury in rice (Oryza sativa L.) roots. Metallomics, 8, 663–671. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1996) Vegetation stress: an introduction to the stress concept in plants. Journal of Plant Physiology, 148, 4–14. [Google Scholar]
- Liu H.H.H., Ma Y., Chen N., Guo S., Liu H.H.H., Guo X., Liu H., Guo X., Kang C., Xu Y. (2014) Overexpression of stress‐inducible OsBURP16, the β subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice. Plant Cell and Environment, 37, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loix C., Huybrechts M., Vangronsveld J., Gielen M., Keunen E., Cuypers A. (2017) Reciprocal interactions between cadmium‐induced cell wall responses and oxidative stress in plants. Frontiers in Plant Science, 8, 1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T., Güell M., Serrano L. (2009) Correlation of mRNA and protein in complex biological samples. FEBS Letters, 583, 3966–3973. [DOI] [PubMed] [Google Scholar]
- Mészáros P., Rybanský Ľ., Spieß N., Socha P., Kuna R., Libantová J., Moravčíková J., Piršelová B., Hauptvogel P., Matušíková I. (2014) Plant chitinase responses to different metal‐type stresses reveal specificity. Plant Cell Reports, 33, 1789–1799. [DOI] [PubMed] [Google Scholar]
- Metwally A., Finkemeier I., Georgi M., Dietz K. (2003) Salicylic acid alleviates the cadmium toxicity in barley seedlings. Plant Physiology, 132, 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D. (2008) Pectin structure and biosynthesis. Current Opinion in Plant Biology, 11, 266–277. [DOI] [PubMed] [Google Scholar]
- Nakata M., Shiono T., Watanabe Y., Satoh T. (2002) Salt stress‐induced dissociation from cells of a germin‐like protein with Mn‐SOD activity and an increase in its mRNA in a moss, Barbula unguiculata . Plant and Cell Physiology, 43, 1568–1574. [DOI] [PubMed] [Google Scholar]
- Osato Y., Yokoyama R., Nishitani K. (2006) A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. Journal of Plant Research, 119, 153–162. [DOI] [PubMed] [Google Scholar]
- Parrotta L., Guerriero G., Sergeant K., Cai G., Hausman J.‐F. (2015) Target or barrier? The cell wall of early‐ and later‐diverging plants vs cadmium toxicity: differences in the response mechanisms. Frontiers in Plant Science, 6, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulose B., Kandasamy S., Dhankher O.P. (2010) Expression profiling of Crambe abyssinica under arsenate stress identifies genes and gene networks involved in arsenic metabolism and detoxification. BMC Plant Biology, 10, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paynel F., Schaumann A., Arkoun M., Douchiche O., Morvan C. (2009) Temporal regulation of cell‐wall pectin methylesterase and peroxidase isoforms in cadmium‐treated flax hypocotyl. Annals of Botany, 104, 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta‐Videa J.R., de la Rosa G., Gonzalez J.H., Gardea‐Torresdey J.L. (2004) Effects of the growth stage on the heavy metal tolerance of alfalfa plants. Advances in Environmental Research, 8, 679–685. [Google Scholar]
- Perfus‐Barbeoch L., Leonhardt N., Vavasseur A., Forestier C. (2002) Heavy metal toxicity: cadmium permeates through calcium channels and disturbs the plant water status. The Plant Journal, 32, 539–548. [DOI] [PubMed] [Google Scholar]
- Printz B., Sergeant K., Guignard C., Renaut J., Hausman J.F. (2013) Physiological and proteome study of sunflowers exposed to a polymetallic constraint. Proteomics, 13, 1993–2015. [DOI] [PubMed] [Google Scholar]
- Printz B., Dos Santos Morais R., Wienkoop S., Sergeant K., Lutts S., Hausman J.‐F., Renaut J. (2015) An improved protocol to study the plant cell wall proteome. Frontiers in Plant Science, 6, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Printz B., Guerriero G., Sergeant K., Audinot J.‐N., Guignard C., Renaut J., Lutts S., Hausman J.‐F. (2016) Combining ‐omics to unravel the impact of copper nutrition on alfalfa (Medicago sativa) stem metabolism. Plant and Cell Physiology, 57, 407–422. 10.1093/pcp/pcw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos I., Esteban E., Lucena J.J., Gárate A. (2002) Cadmium uptake and subcellular distribution in plants of Lactuca sp. Cd‐Mn interaction. Plant Science, 162, 761–767. [Google Scholar]
- Remans T., Keunen E., Bex G.J., Smeets K., Vangronsveld J., Cuypers A. (2014) Reliable gene expression analysis by reverse transcription‐quantitative PCR: reporting and minimizing the uncertainty in data accuracy. The Plant Cell, 26, 3829–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Reyes B.G., McGrath J.M. (2003) Cultivar‐specific seedling vigor and expression of a putative oxalate oxidase germin‐like protein in sugar beet (Beta vulgaris L.). Theoretical and Appied Genetics, 107, 54–61. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Serrano M., Romero‐Puertas M.C., Zabalza A., Corpas F.J., Gómez M., Del Río L.A., Sandalio L.M. (2006) Cadmium effect on oxidative metabolism of pea (Pisum sativum L.) roots. Imaging of reactive oxygen species and nitric oxide accumulation in vivo. Plant Cell and Environment, 29, 1532–1544. [DOI] [PubMed] [Google Scholar]
- Saldanha A.J. (2004) Java treeview – extensible visualization of microarray data. Bioinformatics, 20, 3246–3248. [DOI] [PubMed] [Google Scholar]
- Sanità Di Toppi L., Gabbrielli R. (1999) Response to cadmium in higher plants. Environmental and Experimental Botany, 41, 105–130. [Google Scholar]
- Sarowar S., Young J.K., Eui N.K., Ki D.K., Byung K.H., Islam R., Shin J.S. (2005) Overexpression of a pepper basic pathogenesis‐related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant Cell Reports, 24, 216–224. [DOI] [PubMed] [Google Scholar]
- Schützendübel A., Schwanz P., Teichmann T., Gross K., Langenfeld‐Heyser R., Godbold D.L., Polle A. (2001) Cadmium‐induced changes in antioxidative systems, hydrogen peroxide content, and differentiation in Scots pine roots. Plant Physiology, 127, 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer P., Christoffel A. (1999) Transient expression of members of the germin ‐like gene family in epidermal cells of wheat confers disease resistance. The Plant Journal, 20, 541–552. [DOI] [PubMed] [Google Scholar]
- Semane B., Dupae J., Cuypers A., Noben J.P., Tuomainen M., Tervahauta A., Kärenlampi S., Van Belleghem F., Smeets K., Vangronsveld J. (2010) Leaf proteome responses of Arabidopsis thaliana exposed to mild cadmium stress. Journal of Plant Physiology, 167, 247–254. [DOI] [PubMed] [Google Scholar]
- Sergeant K., Printz B., Gutsch A., Behr M., Renaut J., Hausman J.‐F. (2017) Didehydrophenylalanine, an abundant modification in the beta subunit of plant polygalacturonases. PLoS One, 12, e0171990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Jin L., Wang X. (2016) Cadmium absorption and transportation pathways in plants. International Journal of Phytoremediation, 19, 133–141. [DOI] [PubMed] [Google Scholar]
- Vallelian‐Bindschedler L., Mösinger E., Métraux J.P., Schweizer P. (1998) Structure, expression and localization of a germin‐like protein in barley (Hordeum vulgare L.) that is insolubilized in stressed leaves. Plant Molecular Biology, 37, 297–308. [DOI] [PubMed] [Google Scholar]
- Van De Mortel J.E., Villanueva L.A., Schat H., Kwekkeboom J., Coughlan S., Moerland P.D., Loren Ver, van Themaat E., Koornneef M., Aarts M.G.M. (2006) Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens . Plant Physiology, 142, 1127–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vélez‐Bermúdez I.C., Schmidt W. (2014) The conundrum of discordant protein and mRNA expression. Are plants special?. Frontiers in Plant Science, 5, 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdonk J.C., Hatfield R.D., Sullivan M.L. (2012) Proteomic analysis of cell walls of two developmental stages of alfalfa stems. Frontiers in Plant Science, 3, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villiers F., Ducruix C., Hugouvieux V., Jarno N., Ezan E., Garin J., Junot C., Bourguignon J. (2011) Investigating the plant response to cadmium exposure by proteomic and metabolomic approaches. Proteomics, 11, 1650–1663. [DOI] [PubMed] [Google Scholar]
- Volkov V., Wang B., Dominy P.J., Fricke W., Amtmann A. (2003) Thellungiella halophila, a salt‐tolerant relative of Arabidopsis thaliana, possesses effective mechanisms to discriminate between potassium and sodium. Plant Cell and Environment, 27, 1–14. [Google Scholar]
- Vollenweider P., Cosio C., Günthardt‐Goerg M.S., Keller C. (2006) Localization and effects of cadmium in leaves of a cadmium‐tolerant willow (Salix viminalis L.). Environmental and Experimental Botany, 58, 25–40. [Google Scholar]
- Wang C.Q., Song H. (2009) Calcium protects Trifolium repens L. seedlings against cadmium stress. Plant Cell Reports, 28, 1341–1349. [DOI] [PubMed] [Google Scholar]
- Wang W., Scali M., Vignani R., Spadafora A., Sensi E., Mazzuca S., Cresti M. (2003) Protein extraction for two‐dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis, 24, 2369–2375. [DOI] [PubMed] [Google Scholar]
- Wang Y., Jiang X., Li K., Wu M., Zhang R., Zhang L., Chen G. (2014) Photosynthetic responses of Oryza sativa L. seedlings to cadmium stress: physiological, biochemical and ultrastructural analyses. BioMetals, 27, 389–401. [DOI] [PubMed] [Google Scholar]
- Watson C.F., Zheng L., DellaPenna D. (1994) Reduction of tomato polygalacturonase beta subunit expression affects pectin solubilization and degradation during fruit ripening. The Plant Cell, 6, 1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.S., Balland‐Vanney M., Xie Z.P., Schultze M., Kondorosi A., Kondorosi E., Staehelin C. (2007) Molecular cloning of a bifunctional β‐xylosidase/α‐L‐ arabinosidase from alfalfa roots: heterologous expression in Medicago truncatula and substrate specificity of the purified enzyme. Journal of Experimental Botany, 58, 2799–2810. [DOI] [PubMed] [Google Scholar]
- Zhang X., Gao B., Xia H. (2014) Effect of cadmium on growth, photosynthesis, mineral nutrition and metal accumulation of bana grass and vetiver grass. Ecotoxicology and Environmental Safety, 106, 102–108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Nutrient content of the planting soil at the end of the experiment as determined by ICP‐MS. The nutrient concentrations are given as mean ± SE of ten replicates from each condition (μg·g−1 dry weight). Apart from the Cd concentration, no significant changes (P ≤ 0.05) in nutrient composition occurred.
Appendix S2. 2‐D DIGE of the three cell wall fractions and the soluble protein fraction from M. sativa stems. The shown gels were used as master gel for the experiment. Proteins were pre‐labelled with CyDye. Labelled samples were loaded on ImmobilineTM DryStrip NL, 24 cm (GE Healthcare) followed by migration on HPETM Large Gel NF‐12.5% (Serva Electrophoresis). Marked spots were selected for identification using SameSpots software (TatalLab). HMW, high molecular weight; LMW, low molecular weight.
Appendix S3. 2‐D DIGE spot volumes determined with SameSpots software (TotalLab) from spots which show a significant quantitative change after Cd exposure and were chosen for identification. The total and normalised volumes, fold change and P‐value of each spot is given.
Appendix S4. Table 1 provides forward and reverse primer sequences used to determine gene expression levels with RT‐qPCR, including reference genes. Amplicon size and primer efficiency for each gene are indicated. In Table 2 quantitative real‐time PCR parameters according to the Minimum Information for publication of quantitative real‐time PCR experiments (MIQE) guidelines derived from Bustin et al. (2009) are given.
Appendix S5. Complete MASCOT protein identification data of picked spots from the cell wall protein fraction and soluble protein fraction.
Appendix S6. Tables 1–4 contain all identified proteins in each of the four protein fractions and their abundance change after Cd exposure. Fold change and P‐value were obtained using the SameSpots software (TotalLab). Functional classification of each protein was determined using Blast2Go. Subcellular localisation was determined with TargetP. Figure 1 shows MS spectra from spots 1318 and 1433. In both spots the same protein was identified being of contradictory abundance. Spectra are clearly different indicating the presence of different isoforms.
Appendix S7. Normalised expression of genes related to cell wall structure and lignification in stems of M. sativa. Plants were exposed to Cd (10 mg·kg−1 soil) in a long‐term experiment. Data are used for the heat map representation showing the hierarchical clustering of all investigated genes.
Table S1. Summary of identified proteins in the cell wall fractions (CaCl2, EGTA, LiCl) of M. sativa stems that show a Cd‐induced significant abundance change.


